Abstract
Diet during pregnancy influences the future health of a woman's offspring, with outcomes differing depending on the child's sex. Because the placenta buffers the fetus from the mother, we examined the impact of diet and fetal sex on placental gene expression in mice fed either a very-high-fat, low-fat, chow diet of intermediate caloric density. At day 12.5 of pregnancy, placental RNA was extracted and analyzed by microarray. The expression of 1,972 genes was changed more than 2-fold (P < 0.05) in comparisons across diet in at least one of the three groups. Female placentae demonstrated more striking alterations in gene expression in response to maternal diet than male placentae. Notably, each diet provided a distinctive signature of sexually dimorphic genes, with expression generally higher in genes (651 out of 700) from female placentae than those from male placentae. Several genes normally considered as characteristic of kidney function were affected by diet, including genes regulating ion balance and chemoreception. The placenta also expressed most of the known olfactory receptor genes (Olfr), which may allow the placenta to sense odorant molecules and other minor dietary components, with transcript levels of many of these genes influenced by diet and fetal sex. In conclusion, gene expression in the murine placenta is adaptive and shaped by maternal diet. It also exhibits pronounced sexual dimorphism, with placentae of females more sensitive to nutritional perturbations than placentae of males.
Keywords: nutrition, obesity, olfaction, kidney, sexual dimorphism
More than 50% of all pregnant American women are considered to be either overweight, with a body mass index (BMI) of 25–29.9 kg/m2, or obese, with a BMI of ≥30 kg/m2 (1). Compared with normal-weight women, these women tend to consume more calories, especially from fat (2), and to have a higher rate of adverse pregnancy outcomes (3). It is also clear that marked sex differences in perinatal outcome and infant mortality exist in human populations, with sons at increased risk for preterm and postterm mortality and birth defects (4, 5). Moreover, poor maternal diet during pregnancy might predispose offspring to so-called “adult-onset” diseases, including stroke, coronary heart disease, type 2 diabetes mellitus, and hypertension (6). This in utero programming leading to disease later in life is now generally referred to as either the “developmental origins of health and disease” or the “fetal origin of adult disease” (6).
Maternal diet also may influence the sex of offspring born in certain mammalian species, including mice and humans (7–11). High-calorie diets generally favor birth of males over females, whereas low-calorie diets tend to favor females over males (7, 9–11). In humans and mice, food restriction and a suboptimal diet during the period around conception and early pregnancy also lead to a surfeit of daughters, most probably due to selective loss of male fetuses, the most vulnerable sex in utero (11–13).
Sons and daughters also are at differential risk for various late-onset diseases, apparently related to either the mother's diet or body condition while pregnant (14–17). For instance, sons of obese mothers are more likely than daughters to become obese and insulin-resistant as they mature, even though no differences in birth weight may be evident (18). In rats, male offspring exposed to maternal undernutrition during the preimplantation period (16) or to moderately increased sodium intake exhibit elevated blood pressure (19). Because predisposition to adult diseases such as diabetes, hypertension, and cardiovascular disease may originate in utero and be shaped by what the mother consumes during her pregnancy, a molecular-level investigation of whether male and female conceptuses respond differently to the same maternal diet is of interest.
Scant information exists on how maternal diet influences global gene expression in the conceptus. Because the placenta is the primary communication and nutrient acquisition organ for the fetus (20) and presumably acts to maintain fetal homeostasis, it is an appropriate organ to use to examine how differences in maternal food consumption are sensed by the developing offspring. However, to date, only one study has explored the impact of altering maternal diet on global placental gene response (21). The investigators found that maintaining murine dams on a 50%-reduced protein content but otherwise isocaloric diet for 10.5–17.5 days postcoitus led to deleterious consequences in placental gene expression, with up-regulation of genes governing the p53 pathway, apoptosis, growth inhibition, and epigenetic processes but simultaneous decreased expression of genes mediating nucleotide metabolism (21). The investigators did not consider the possibility that male and female conceptuses might exhibit different responses to the imposed diet, however.
Only a handful of studies have reported differing expression of individual gene products in male and female human placentae (22–24), and only one study has studied the phenomenon globally through the use of microarrays (25). Examining different regions of human term placentae, Sood et al. (25) found a handful of autosome-linked genes that exhibit sexually dimorphic expression patterns. In general, these genes are up-regulated in females compared with males (25). As far as we are aware, no study, excluding the aforementioned study on acute protein reduction (21), has addressed the impact of differences in quality of maternal diet on placental gene expression in any species, nor has there been a systematic investigation of the relationship between diet and expression of sexually dimorphic genes that might begin to explain the different sensitivities of male and female fetuses to what the mother eats. Accordingly, we sought to examine how maternal diet might influence the full range of placental gene expression in male and female conceptuses at around midpregnancy, when the morphological development of the placenta is complete (26), yet the gonads are just beginning to form.
Results
Effects of Maternal Diet and Uterine Position on Placental Weight.
The low-fat (LF) (high-carbohydrate) diet, very-high-fat (VHF) diet, and chow (C; Purina 5015 complete life-cycle diet, used routinely for pregnant and lactating mice) used here were nutritionally complete. Consistent with previous studies (7, 27), dams on the VHF diet were heavier (51.2 ± 2.2 g; P < 0.01) at the time of conceptus collection compared with those on the LF and C diets (37.7 ± 1.7 g and 42.8 ± 1.9 g, respectively). Consistent with our previous studies, there was a tendency toward proportionately more female conceptuses in the LF group (Fig. S1); however, only 7 dams with 37 viable fetuses were recovered from the VHF group, an insufficient number for a statistical assessment of sex ratio. An analysis of the sex of additional conceptuses collected more recently from comparably aged dams on the VHF diet (n = 10) at day 12.5 provided a sex ratio of 36 males to 27 females (57.1%; P = 0.08), suggesting that the dietary effects on offspring sex ratio are already established by this stage of pregnancy. Unexpectedly, placentae from conceptuses implanted in the left uterine horn were on average heavier than those in the right horn in the C and VHF groups (27.8 ± 1.0 mg vs. 22.3 ± 1.1 mg; P < 0.01) (Table S1). There was a modest effect of fetal sex on placental weight, with female placentae on average weighing more than male placentae (26.9 ± 1.0 mg vs. 23.8 ± 1.1 mg; P < 0.05). The combined impact of all three factors—uterine position, maternal diet, and fetal sex—on placental weights is depicted in Table S1 and Fig. S1.
Effects of Maternal Diet on Global Placental Gene Expression Patterns.
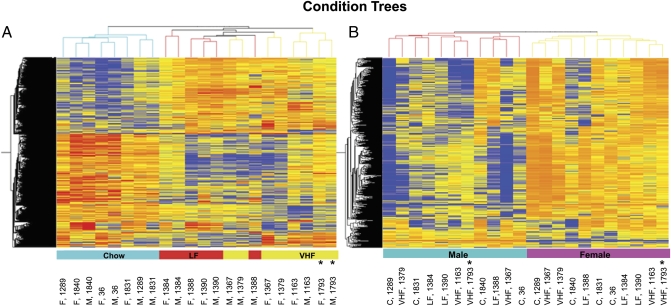
As illustrated on the heat map in Fig. 1A, which was based on hierarchical clustering of 22 samples and 1,972 genes (∼5% of the total) whose expression was changed by >2-fold with P < 0.05 in comparisons in at least one of the three dietary groups, the analysis sorted the placental RNA samples according to maternal diet. Placentae from the dams on the C diet formed a distinct cluster that distinguished them from the placentae of conceptuses of dams on the LF and VHF diets. On the other hand, while the placental samples from LF and VHF females clustered separately, there was some overlap of samples from LF and VHF males. Thus, despite the differences in caloric density between LF and VHF diets, the gene regulation patterns in the placentae of LF and VHF conceptuses were more like each other than like those in placentae of the C conceptuses. The overall conclusion based on these results is that the diet consumed by the dam has a wide-ranging influence on gene expression in the placenta at day 12.5 of pregnancy, and different diets can be distinguished according to the pattern of gene expression they evoke in the placenta.
Fig. 1.
(A) Heat map based on maternal diet effects on placental gene expression. Placentae from dams on the C diet are well separated from the placentae of conceptuses from dams on the LF and VHF diets. Despite the differences in caloric density between two defined diets, the gene regulation patterns in placentae of LF and VHF conceptuses were more like each other than like those in placentae of C conceptuses. Gene tree clustering on 1,972 genes, whose expression was changed by >2-fold with P < 0.05, is able to distinguish placentae from dams on the LF and VHF diets. (B) Heat map based on fetal sex effects on placental gene expression. Placentae gene expression patterns of male conceptuses clearly clustered separately from the placentae of females, when data on the total regulated genes (with 2-fold differences) across all dietary groups are compared (P < 0.05). Samples marked with an asterisk are those from the left as opposed to the right uterine horn. The close grouping of these with the other samples suggests that uterine implantation site has no affect on placental gene expression patterns.
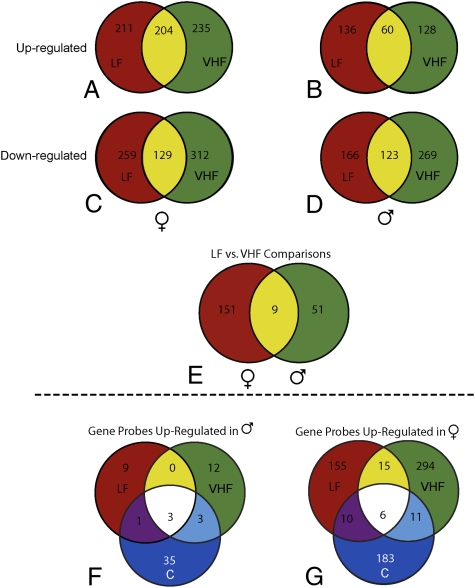
The Venn diagrams shown in Fig. 2 A–E analyze the gene expression data derived from the VHF and LF diets relative to the C diet and confirm distinct gene expression patterns for each diet (Dataset S1). For example, in females, only 204 out of 650 up-regulated genes (relative to the C group) and 129 of 700 down-regulated genes (also relative to the C group) were common to the LF and VHF placental groups (Fig. 2 A and C). Somewhat similar data were obtained for males (Fig. 2 B and D). Of particular note is the observation that when placentae from the LF and VHF groups were compared with each other rather than with the C group, 211 genes showed at least a 2-fold difference in expression (Fig. 2E and Dataset S1), but only 9 of these regulated genes were common to both male and female placentae, and females accounted for almost two-thirds of the differentially expressed genes.
Fig. 2.
Venn diagrams of placental gene expression patterns influenced by maternal diet and fetal sex. (A–D) Diagrams comparing the placental genes that were up-regulated (A and B) and down-regulated (C and D) in female and male conceptuses from dams on the LF and VHF diet after subtracting from the C diet group. (E) Diagram comparing the number of differentially expressed genes in female and male placentae in the LF group versus the VHF group (≥2-fold; P < 0.05, for a total of 200 autosomal and 11 sex chromosomal genes). (F and G) Sexually dimorphically expressed genes in at least one of the three diet groups (≥2-fold; P < 0.05, for a total of 736 autosomal and sex chromosome genes). The up-regulated genes that overlap in the male placentae of all diet groups were Ddx3y (represented by two probes) and Eif2s3y. The down-regulated genes that overlap in the male placentae of all diet groups were Xist (represented by three probes), Klk7, Crabp1, and Rimklb (4933426K21Rik).
Effects of Fetal Sex on Global Placental Gene Expression Patterns.
The initial assignment of fetal sex for each placenta was based on X/Y FISH analysis of cells removed from the fetus. The presence of Y-specific transcripts (Ddx3y, represented by 2 probes, and Eif2s3yb, represented by a single probe) only in the male tissue and of a X-inactive–specific transcript (Xist, represented by three probes) in the female tissue confirmed the accuracy of the FISH sex determination (Dataset S2, Fig. 2 F and G, and Table S2).
In the subsequent heat map clustering analysis (Fig. 1B), the effects of fetal sex were assessed by combining both autosomal and sex chromosomal genes. Considerable sexual dimorphism was clearly evident in the placentae from all three groups. A total of 700 autosomally located genes demonstrated sexually dimorphic expression within at least one of the three dietary groups (Dataset S2 and Table S2). In almost all of these cases, (651 out of 700), the genes were up-regulated in the female tissue but not in the male tissue (Fig. 2 F and G and Table S2). Very few of these genes differentially expressed between males and females from dams on the same diet were common to more than one of the diets (Fig. 2 F and G); in other words, each of the diets elicited its own specific pattern of sexually dimorphic gene expression, generally involving up-regulation of genes in the female placentae but not in the male placentae. Three autosomal genes (Klk7, Crabp1, and Rimklb/4933426K21Rik) displayed sexually dimorphic differences across the three maternal diet groups (Fig. 2G), with each of the genes up-regulated at least 2-fold in the female placentae compared with the male placentae.
Quantitative RT-PCR Analysis.
To confirm the microarray results, we selected 16 genes (listed in the legend to Fig. S2 and in Table S3) identified by the microarray data as being expressed in a sexually dimorphic manner in at least one diet group and considered to have some potential relevance to normal placental physiology. To assess the correlation between the microarray and quantitative real-time data for all 16 genes in the 11 RNA samples, female-to-male expression ratios obtained by quantitative RT-PCR (the y-axis in Fig. S2) were plotted against the equivalent values obtained from the microarray analysis (the x-axis). In general, the quantitative RT-PCR and microarray data agreed quite well for most samples, with an overall correlation coefficient of 0.87, emphasizing the robustness of the microarray data.
Effects of Diet and Sex on Endocrine Pathways.
The placenta is an endocrine organ that produces hormones linking the needs of the conceptus to maternal physiology. We examined whether diet influenced expression of a number of genes with known roles in the production of placental hormones and whether any of these genes were sexually dimorphic. In rodents, placental expression of prolactin-related proteins, including those previously referred to as placental lactogens (now termed Prl3b and Prl3d, respectively), are considered essential to maintenance of pregnancy (28, 29). As in previous microarray studies (30), here placental lactogens were among the most highly expressed genes in the murine placenta at day 12.5 of pregnancy (Table S4). Although some genes in the extensive Prl family (i.e., Prl2c3, Prl3b1, Prl3d2, Prl5a1, and Prl7c1) were modestly up-regulated in male placentae versus female placentae in the C group (P < 0.05), only 2 of these genes were up-regulated in male placentae in the VHF and LF groups (Prl5a1 in the VHF group and Prl7c1 in the LF group) (P < 0.05) (Table S4). The gene for growth hormone (somatotropin 1), Gh1, was down-regulated in male placentae in the VHF group (P < 0.05). Whereas Lha (luteinizing hormone α subunit gene) was unaffected by both maternal diet and fetal sex, Lhb was up-regulated in male placentae from LF dams and down-regulated in male placentae from VHF dams (P < 0.05) (Table S4).
The expression of two steroid nuclear receptor genes also was found to be influenced by fetal sex and maternal diet. The gene encoding estrogen receptor (ER) α, Esr1, was up-regulated in female placentae compared with male placentae in the C group (P < 0.05), but was not differentially expressed in the LF and VHF groups (Table S3 and Dataset S2). The ERβ gene, Esr2, demonstrated no significant changes in expression. On the other hand, the gene encoding the androgen receptor (Ar), which is located on the X-chromosome, was up-regulated in females compared with males in the VHF group (P < 0.05) (Dataset S2).
Only a few changes were noted in genes implicated in either steroidogenesis or steroid catabolism, with females less affected than males. In male placentae in the LF group, Hsd3b5, which catalyzes the conversion of pregnenolone to progesterone and of 21-hydroxypregnenolone to 11-deoxycorticosterone, demonstrated a >2-fold increase in expression relative to adjacently placed female placentae in the same dietary group (P < 0.05) (Dataset S2), whereas the related genes Hsd3b2 and Hsd3b6 were up-regulated in LF males compared with C males (P < 0.05) (Dataset S1 and Fig. S3). Males from VHF dams expressed less Prmt3 (whose product converts 2-hydroxy-17β-estradiol to 2-methoxy-17β-estradiol) than males from C dams (Dataset S1). In contrast, males from LF dams had greater expression of several genes encoding enzymes involved in androgen metabolism, but less expression of Sts (steroid sulfatase gene), compared with males from C dams (Dataset S1 and Fig. S4).
Our findings also suggest dietary regulation of the two genes encoding the rate-limiting enzymes of prostaglandin and leukotriene synthesis, Ptgs1 (Cox1) and Ptgs2 (Cox2). Ptgs1 transcript concentration was increased by 2.8-fold (P < 0.04) and Ptgs2 transcript concentration was increased by 1.8-fold (P < 0.20) in female placentae from VHF dams compared with female placentae from C dams (Dataset S1). In addition, the primary prostaglandin-metabolizing enzyme, Hpgd, was up-regulated in female placentae from VHF dams compared to those of C dams (Dataset S1). Comparing female placentae from LF and C dams showed that in the former, Ptgs2 was up-regulated by 2.0-fold (P < 0.03), but Ptgs1 was up-regulated by only 1.4-fold (P < 0.4) (Dataset S1).
Gene Differences Between Male and Female Placentae from LF Dams and VHF Dams.
Because our original goal was to determine whether some of the diet-induced changes in gene expression might account for the contrasting offspring sex ratios observed for dams fed the VHF and LF diets, we further scrutinized the 211 genes that exhibited <2-fold differences between the male and female placentae in LF and VHF dams (Dataset S1). No one gene or set of genes stood out as a potential candidate, although male placentae of VHF dams displayed an ∼3-fold greater Esr1 expression and reduced Ghr expression compared with male placentae from LF dams, indicating a diet effect. Other differentially expressed genes included Bhmt, Gpc6, Vipr1, Runx1, Agtrap, several peptidase inhibitors involved in the complement/coagulation cascade, and serine peptidases, including Htra1.
Discussion
In this paper we make two important observations regarding gene expression in day-12.5 murine placenta and the effects of maternal diet on these expression patterns. The first observation is that diet has a marked influence on global gene expression, with each diet providing a specific signature in terms of regulated genes. The second observation is that female fetuses respond more robustly to dietary influences than males, and, perhaps most surprisingly, each diet endows the placenta with a distinct pattern of sexually dimorphic gene expression.
The finding that placental gene expression responds to diet is not entirely unexpected, given that the placenta provides a metabolic and physical interface between the maternal blood supply and the fetus and thus would be expected to be influenced by variations in the quality and quantity of circulating nutrients available to it (20). The food fed to the dams in our studies was in the form of two defined diets, one in high in carbohydrates but low in fats (LF) and the other low in carbohydrates but high in fats (VHF), as well as a commercial diet based largely on soybean meal but, like the other two, also nutritionally complete. Because these three diets differed considerably in caloric density (calories from fat: LF, 10%; VHF, 60%; C, 26%), it was not surprising that the dams on the VHF diet were significantly heavier than those on the other two diets. On the other hand, the LF and VHF diets differed only in their relative content of fats and carbohydrates and were otherwise identical, possibly explaining why the dietary responses to these two diets differed from one another to a lesser degree than from the C diet (Fig. 1A). Although the VHF dams tended to be obese and to have higher circulating concentrations of free fatty acids than the more normally proportioned dams on the LF diet (31), placental weight was not increased (Table S1). We also found no apparent bias toward regulation of genes associated with particular metabolic pathways, including lipid and energy metabolism, in either group. Thus, despite the fact that our data do not provide specific insight into how the placenta responds to increased availability of fatty acids or other compounds derived from dietary intake, a critical function may be to buffer the fetus from maternal surfeit and shortage and from potentially harmful dietary components. The marked deviation of the patterns of regulated gene expression in the C diet relative to the LF and VHF diets is perhaps a reflection of the distinct composition (including protein and other natural products from soybean meal) in the former versus the refined nature of the latter diets. Importantly, chow-based diets, although economical, are rarely uniform in composition, and thus each such diet is likely to provide its own microarray signature. Soy-based chow diets also vary in terms of content of estrogenic compounds (32). Accordingly, the up-regulation of the gene encoding ERα (Esr1) in female tissue from dams on the C diet and the androgen receptor gene (Ar) in female tissue from dams on the VHF diet (Table S3 and Dataset S2) could make the placentae in these groups more susceptible to estrogens and androgens, as well as to endocrine disruptors, such as diethylstilbestrol, bisphenol A, vinclozolin, and dichlorodiphenyltrichloroethane (33, 34). Even though the placenta is the guardian of the fetus, some of its responses to maternal diet could possibly have deleterious consequences to the fetus.
One group of genes whose expression in the placenta was unexpected was the large Olfr family, which encodes olfactory receptors. Although normalized intensity values (those in the upper 25th percentile of genes providing a signal on the arrays) were generally low, each of 931 Olfr gene probes gave a positive “call” in at least one placental sample. Two of these genes (Olfr571 and Olfr1009) exhibited a mean >2-fold increase in normalized expression level across all dietary groups, and 52 of the genes (including Olfr571) demonstrated a >2-fold change in expression (P < 0.05) in response to at least one of the diets (Table S3). Eight genes (Olfr91, Olfr433, Olfr520, Olfr786, Olfr1381, Olfr1383, Olfr1394, and Olfr1395) were significantly up-regulated in female placentae from dams on the C or VHF diets, whereas Olfr 154 was up-regulated in male placentae from dams on the C diet (Table S3). Accordingly, the presence of a functional olfaction system could allow the placenta to “sense” changes in levels of odorant molecules to control uptake or excretion of such compounds. Such a role has recently been proposed for olfactory receptors in the murine kidney (35). Indeed, several other genes characteristic of the kidney, especially genes involved in ion and fluid movement, including Agtrap, Aqp9, Anxa6, and Ccr3, were expressed in a dietary and sexually dimorphic manner in day-12.5 murine placentae (Table S3 and Dataset S1 and Dataset S2). Taken together, these data emphasize the role of the placenta in chemoreception and control of fluid and ion balance, functions clearly in common with the kidney.
As mentioned earlier, two of the most surprising outcomes of our experiments were (i) the large number of genes that were regulated in a sexually dimorphic manner and (ii) each maternal diet gave rise to a distinct pattern of sexually dimorphic gene expression in the placentae. Based on these findings, any study that seeks to examine genes that are differentially expressed between sons and daughters within the placenta must take into account maternal diet as an important variable. In particular, placentae from female conceptuses were more sensitive to diet changes than placentae from male conceptuses, and this phenomenon was generally manifested as increased gene expression in females (Fig. 2 A and G), with the effect distributed across both autosomal and sex chromosomes (Table S2). Such a tendency toward up-regulation of genes in female placentae relative to adjacent male placentae appears to account for the great majority of the sexual dimorphism in gene expression observed in our study (Fig. 2 F and G). This increased sensitivity of the female placentae to changes in the maternal environment might serve as a buffer to protect the female against in utero perturbations, thus possibly accounting for the decreased risk for adult diseases in daughters versus sons born to women who were eating a high-fat diet and/or were obese at time of pregnancy (14–17), although this hypothesis remains to be tested. It would be of interest to explore whether or not the increased expression of a particular autosomal gene in female placentae relative to male placentae is biallelic or manifested through just one of the parental alleles, and, if the latter, whether a particular pattern of allelic bias in expression persists across the entire group of sexually dimorphic genes. Allelic exclusion is not an uncommon phenomenon (36); it has been reported for Olfr genes in olfactory sensory neurons (37). The X chromosome is a special case, of course, because paternal X is selectively subjected to silencing in the extraembryonic lineages of the mouse (38). Thus, with the exception of Xist, the sexually dimorphic expression of X-linked genes, such as Ar and Ogt, which were up-regulated in female placentae (Dataset S2), is presumably due entirely to the maternally inherited allele.
The underlying cause of the sexual dimorphism reported here is unclear. A connection to sex hormone differences between male and female conceptuses is unlikely. Although day 12.5 approximates the earliest morphological stage of testis differentiation and the appearance of androgen-producing Leydig cells (39), intratesticular testosterone concentrations are negligible at this point, and masculinization of the male fetus is not yet evident (40). In any case, sexually dimorphic differences have been observed as early as the blastocyst stage in several mammalian species, when hormonal influence on either development rates (41) or gene expression (42) can be ruled out. Like those described here, the differences in gene expression patterns between males and female noted in mouse blastocysts by Kobayashi et al. (42) were generally modest in magnitude and involved higher transcript concentrations in females than in males. One possible explanation for both sets of observations is that the male and female conceptuses might have been out of phase with one another. To minimize this possibility, we took particular care to select adjacent placentae from the same uterine horn, because position can affect fetal growth (43), and, with one exception, made our selection from the right uterine horn, because left/right inequalities in uterine function have been reported (44). Nonetheless, the greater average weight of the female placentae (Table S1) might reflect subtle developmental differences between the sexes.
The reason why a maternal high-fat (low-carbohydrate) diet favors survival of sons and a maternal low-fat (high-carbohydrate) diet results in more daughters continues to elude us (7, 9–11). One possibility is that the selective events occur through pressures imparted by the uterine environment and its interaction with trophoblast earlier than day 12.5 of pregnancy. However, our studies provide several candidate genes (including those mediating uptake of nutrients and ions, genes in the Olfr family, steroidogenic enzymes, and steroid receptor genes) expressed by the placenta that might be important for conceptus survival and are markedly influenced by the interaction of maternal diet and fetal sex.
Materials and Methods
Animals.
All animal experiments were approved by the University of Missouri's Animal Care and Use Committee and performed in accordance with National Institutes of Health (NIH) Animal Care and Use Guidelines. NIH Swiss female mice were placed on one of three diets beginning at 5 weeks of age: C (Purina 5015 chow; 4.35 kcal/g), LF (3.8 kcal/g; Research Diets), and VHF (5.2 kcal/g, 54% from lard; Research Diets). Mice were bred at 35–40 weeks of age and euthanized at 12.5 days postcoitus. Fetal and whole placental samples were selectively collected from viable fetuses. The position of each conceptus within the uterus was documented, and each placenta was weighed. For placental weight comparisons, the placentae from 12 C dams, 14 LF dams, and 7 VHF dams were analyzed.
X/Y FISH Analysis to Determine Conceptus Sex.
The sex of each fetus was determined by X/Y FISH, as described previously (45).
RNA Isolation and Microarray Analysis of Placentae.
Based on the X/Y FISH analysis results, one female placenta and one male placenta were selected from the middle uterine region of each dam for evaluation of placental gene expression by microarray analysis. For almost all of the samples chosen for microarray analysis, placentae from adjacent male and female conceptuses on the right side of the uterine horn were analyzed. However, one VHF dam (no. 1793) did not have an adjacent pair of female and male conceptuses in the right uterine horn; for this dam, we analyzed the placentae of female and male conceptuses implanted in the left miduterine horn region. Placental RNA was isolated by TriReagent (Sigma-Aldrich). After first-strand and second-strand cDNA preparation, cRNA target material was prepared and hybridized to Agilent Whole Murine Genome 4 × 44K arrays. Slides were scanned with an Agilent G2565 Microarray Scanner, and data were analyzed with Agilent Feature Extraction and GeneSpring GX v7.3.1 software. Comparisons were made between diet groups within the same sex and between male and female conceptuses from four C dams, three LF dams, and four VHF dams. Each placental sample was processed and analyzed independently; that is, no samples from any of the diets or fetal sexes were pooled.
Quantitative RT-PCR.
Expression of select placental genes was verified by quantitative RT-PCR, as described by Davis et al. (46). The same placental RNA samples used for the microarray analysis were used for the RT-PCR studies, except for one VHF female–male placental pair in which there was insufficient RNA.
Statistical Analysis.
The placental weight from each fetus was analyzed using the SAS general linear model (GLM) procedure (SAS Institute), with maternal diet, position in uterus (left horn vs. right horn), and fetal sex as main factors. Differences in placental weight among groups were evaluared by Fisher's least-significant difference, with P < 0.05 considered significant. The raw microarray data were analyzed with GeneSpring GX v7.3 software (Agilent Technologies) and normalized to the 75th percentile of each array (i.e., 25% provided normalized values >1) to compare individual expression values across arrays for diet comparisons. For fetal sex comparisons, genes were further normalized to the median expression within each dam pair. Only genes with values exceeding background intensity in at least three samples of either condition for each comparison were used for further analysis. Volcano plots were used to filter for genes differentially expressed by ≥2-fold and with P ≤ 0.05. P values for diet and fetal sex comparisons were calculated using a two-sample t test assuming unequal variance.
For quantitative RT-PCR, the relative expression levels in placental tissue for the selected genes were normalized to endogenous reference 18S by using the formula 2-Cttarget /2-Ct18s, where Ct is the threshold cycle. The female-to-male gene expression ratio was then determined and analyzed by using SAS software, with diet as the main effect. All data are expressed as mean ± SEM. Differences are considered significant at P < 0.05.
Supplementary Material
Acknowledgments
We thank Denise Warzak, Kara Stowers, Benjamin Bockting, Amy Desaulniers, and Jackman Eschenroeder for their assistance with mouse husbandry and Donald L. Connor and Howard A. Wilson for their assistance with figure preparation. This work was supported by National Institutes of Health Grant HD 44042.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/cgi/content/full/1000440107/DCSupplemental.
References
- 1.Ogden CL, et al. Prevalence of overweight and obesity in the United States, 1999–2004. JAMA. 2006;295:1549–1555. doi: 10.1001/jama.295.13.1549. [DOI] [PubMed] [Google Scholar]
- 2.Ehrenberg HM, Mercer BM, Catalano PM. The influence of obesity and diabetes on the prevalence of macrosomia. Am J Obstet Gynecol. 2004;191:964–968. doi: 10.1016/j.ajog.2004.05.052. [DOI] [PubMed] [Google Scholar]
- 3.Smith GCS, Shah I, Pell JP, Crossley JA, Dobbie R. Maternal obesity in early pregnancy and risk of spontaneous and elective preterm deliveries: A retrospective cohort study. Am J Public Health. 2007;97:157–162. doi: 10.2105/AJPH.2005.074294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drevenstedt GL, Crimmins EM, Vasunilashorn S, Finch CE. The rise and fall of excess male infant mortality. Proc Natl Acad Sci USA. 2008;105:5016–5021. doi: 10.1073/pnas.0800221105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cui W, et al. Sex differences in birth defects: A study of opposite-sex twins. Birth Defects Res A Clin Mol Teratol. 2005;73:876–880. doi: 10.1002/bdra.20196. [DOI] [PubMed] [Google Scholar]
- 6.Barker DJ. Maternal nutrition, fetal nutrition, and disease in later life. Nutrition. 1997;13:807–813. doi: 10.1016/s0899-9007(97)00193-7. [DOI] [PubMed] [Google Scholar]
- 7.Rosenfeld CS, et al. Striking variation in the sex ratio of pups born to mice according to whether maternal diet is high in fat or carbohydrate. Proc Natl Acad Sci USA. 2003;100:4628–4632. doi: 10.1073/pnas.0330808100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stolkowski J, Choukroun J. Preconception selection of sex in man. Isr J Med Sci. 1981;17:1061–1067. [PubMed] [Google Scholar]
- 9.Fountain ED, et al. Effects of diets enriched in omega-3 and omega-6 polyunsaturated fatty acids on offspring sex ratio and maternal behavior in mice. Biol Reprod. 2008;78:211–217. doi: 10.1095/biolreprod.107.065003. [DOI] [PubMed] [Google Scholar]
- 10.Mathews F, Johnson PJ, Neil A. You are what your mother eats: Evidence for maternal preconception diet influencing fetal sex in humans. Proc Biol Sci. 2008;275:1661–1668. doi: 10.1098/rspb.2008.0105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meikle DB, Drickamer LC. Food availability and secondary sex ratio variation in wild and laboratory house mice (Mus musculus) J Reprod Fertil. 1986;78:587–591. doi: 10.1530/jrf.0.0780587. [DOI] [PubMed] [Google Scholar]
- 12.Meikle DB, Thornton MW. Premating and gestational effects of maternal nutrition on secondary sex ratio in house mice. J Reprod Fertil. 1995;105:193–196. doi: 10.1530/jrf.0.1050193. [DOI] [PubMed] [Google Scholar]
- 13.Rivers JP, Crawford MA. Maternal nutrition and the sex ratio at birth. Nature. 1974;252:297–298. doi: 10.1038/252297a0. [DOI] [PubMed] [Google Scholar]
- 14.Grigore D, Ojeda NB, Alexander BT. Sex differences in the fetal programming of hypertension. Gend Med. 2008;5(Suppl A):S121–S132. doi: 10.1016/j.genm.2008.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ozaki T, Nishina H, Hanson MA, Poston L. Dietary restriction in pregnant rats causes gender-related hypertension and vascular dysfunction in offspring. J Physiol. 2001;530:141–152. doi: 10.1111/j.1469-7793.2001.0141m.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kwong WY, Wild AE, Roberts P, Willis AC, Fleming TP. Maternal undernutrition during the preimplantation period of rat development causes blastocyst abnormalities and programming of postnatal hypertension. Development. 2000;127:4195–4202. doi: 10.1242/dev.127.19.4195. [DOI] [PubMed] [Google Scholar]
- 17.Woods LL, Weeks DA, Rasch R. Programming of adult blood pressure by maternal protein restriction: Role of nephrogenesis. Kidney Int. 2004;65:1339–1348. doi: 10.1111/j.1523-1755.2004.00511.x. [DOI] [PubMed] [Google Scholar]
- 18.Mingrone G, et al. Influence of maternal obesity on insulin sensitivity and secretion in offspring. Diabetes Care. 2008;31:1872–1876. doi: 10.2337/dc08-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Langley-Evans SC, Jackson AA. Captopril normalizes systolic blood pressure in rats with hypertension induced by fetal exposure to maternal low protein diets. Comp Biochem Physiol Physiol. 1995;110:223–228. doi: 10.1016/0300-9629(94)00177-u. [DOI] [PubMed] [Google Scholar]
- 20.Jansson T, Powell TL. IFPA 2005 Award in Placentology Lecture. Human placental transport in altered fetal growth: Does the placenta function as a nutrient sensor? A review. Placenta. 2006;27(Suppl A):S91–S97. doi: 10.1016/j.placenta.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 21.Gheorghe CP, Goyal R, Holweger JD, Longo LD. Placental gene expression responses to maternal protein restriction in the mouse. Placenta. 2009;30:411–417. doi: 10.1016/j.placenta.2009.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lehavi O, Aizenstein O, Evans MI, Yaron Y. 2nd-trimester maternal serum human chorionic gonadotropin and alpha-fetoprotein levels in male and female fetuses with Down syndrome. Fetal Diagn Ther. 2005;20:235–238. doi: 10.1159/000083911. [DOI] [PubMed] [Google Scholar]
- 23.Steier JA, Bergsjø PB, Thorsen T, Myking OL. Human chorionic gonadotropin in maternal serum in relation to fetal gender and utero-placental blood flow. Acta Obstet Gynecol Scand. 2004;83:170–174. doi: 10.1111/j.0001-6349.2004.00407.x. [DOI] [PubMed] [Google Scholar]
- 24.Brown MJ, Cook CL, Henry JL, Schultz GS. Levels of epidermal growth factor binding in third-trimester and term human placentas: Elevated binding in term placentas of male fetuses. Am J Obstet Gynecol. 1987;156:716–720. doi: 10.1016/0002-9378(87)90085-8. [DOI] [PubMed] [Google Scholar]
- 25.Sood R, Zehnder JL, Druzin ML, Brown PO. Gene expression patterns in human placenta. Proc Natl Acad Sci USA. 2006;103:5478–5483. doi: 10.1073/pnas.0508035103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mueller BR, Bale TL. Sex-specific programming of offspring emotionality after stress early in pregnancy. J Neurosci. 2008;28:9055–9065. doi: 10.1523/JNEUROSCI.1424-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alexenko AP, et al. The contrasting effects of ad libitum and restricted reeding of a diet very high in saturated fats on sex ratio and metabolic hormones in mice. Biol Reprod. 2007;77:599–604. doi: 10.1095/biolreprod.107.062174. [DOI] [PubMed] [Google Scholar]
- 28.Simmons DG, Rawn S, Davies A, Hughes M, Cross JC. Spatial and temporal expression of the 23 murine prolactin/placental lactogen–related genes is not associated with their position in the locus. BMC Genomics. 2008;9:352. doi: 10.1186/1471-2164-9-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Soares MJ. The prolactin and growth hormone families: Pregnancy-specific hormones/cytokines at the maternal-fetal interface. Reprod Biol Endocrinol. 2004;2:51. doi: 10.1186/1477-7827-2-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gheorghe C, Mohan S, Longo LD. Gene expression patterns in the developing murine placenta. J Soc Gynecol Investig. 2006;13:256–262. doi: 10.1016/j.jsgi.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 31.Whyte JJ, et al. Maternal diet composition alters serum steroid and free fatty acid concentrations and vaginal pH in mice. J Endocrinol. 2007;192:75–81. doi: 10.1677/JOE-06-0095. [DOI] [PubMed] [Google Scholar]
- 32.Heindel JJ, vom Saal FS. Meeting report. Batch-to-batch variability in estrogenic activity in commercial animal diets: importance and approaches for laboratory animal research. Environ Health Perspect. 2008;116:389–393. doi: 10.1289/ehp.10524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kelce WR, Gray LE, Wilson EM. Antiandrogens as environmental endocrine disruptors. Reprod Fertil Dev. 1998;10:105–111. doi: 10.1071/r98051. [DOI] [PubMed] [Google Scholar]
- 34.Vandenberg LN, Maffini MV, Sonnenschein C, Rubin BS, Soto AM. Bisphenol-A and the great divide: A review of controversies in the field of endocrine disruption. Endocr Rev. 2009;30:75–95. doi: 10.1210/er.2008-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pluznick JL, et al. Functional expression of the olfactory signaling system in the kidney. Proc Natl Acad Sci USA. 2009;106:2059–2064. doi: 10.1073/pnas.0812859106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ohlsson R, Tycko B, Sapienza C. Monoallelic expression: “There can only be one.”. Trends Genet. 1998;14:435–438. doi: 10.1016/s0168-9525(98)01583-2. [DOI] [PubMed] [Google Scholar]
- 37.Chess A, Simon I, Cedar H, Axel R. Allelic inactivation regulates olfactory receptor gene expression. Cell. 1994;78:823–834. doi: 10.1016/s0092-8674(94)90562-2. [DOI] [PubMed] [Google Scholar]
- 38.Latham KE. X chromosome imprinting and inactivation in the early mammalian embryo. Trends Genet. 1996;12:134–138. doi: 10.1016/0168-9525(96)10017-2. [DOI] [PubMed] [Google Scholar]
- 39.Brennan J, Capel B. One tissue, two fates: Molecular genetic events that underlie testis versus ovary development. Nat Rev Genet. 2004;5:509–521. doi: 10.1038/nrg1381. [DOI] [PubMed] [Google Scholar]
- 40.O'Shaughnessy PJ, et al. Fetal development of Leydig cell activity in the mouse is independent of pituitary gonadotroph function. Endocrinology. 1998;139:1141–1146. doi: 10.1210/endo.139.3.5788. [DOI] [PubMed] [Google Scholar]
- 41.Larson MA, Kimura K, Kubisch HM, Roberts RM. Sexual dimorphism among bovine embryos in their ability to make the transition to expanded blastocyst and in the expression of the signaling molecule IFN-tau. Proc Natl Acad Sci USA. 2001;98:9677–9682. doi: 10.1073/pnas.171305398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kobayashi S, et al. Comparison of gene expression in male and female mouse blastocysts revealed imprinting of the X-linked gene, Rhox5/Pem, at preimplantation stages. Curr Biol. 2006;16:166–172. doi: 10.1016/j.cub.2005.11.071. [DOI] [PubMed] [Google Scholar]
- 43.Barr M, Jr, Jensh RP, Brent RL. Fetal weight and intrauterine position in rats. Teratology. 1969;2:241–246. doi: 10.1002/tera.1420020308. [DOI] [PubMed] [Google Scholar]
- 44.Wiebold JL, Becker WC. Inequality in function of the right and left ovaries and uterine horns of the mouse. J Reprod Fertil. 1987;79:125–134. doi: 10.1530/jrf.0.0790125. [DOI] [PubMed] [Google Scholar]
- 45.Whyte JJ, Roberts RM, Rosenfeld CS. Fluorescent in situ hybridization for sex chromosome determination before and after fertilization in mice. Theriogenology. 2007;67:1022–1031. doi: 10.1016/j.theriogenology.2006.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Davis AM, Mao J, Naz B, Kohl JA, Rosenfeld CS. Comparative effects of estradiol, methyl-piperidino-pyrazole, raloxifene, and ICI 182 780 on gene expression in the murine uterus. J Mol Endocrinol. 2008;41:205–217. doi: 10.1677/JME-08-0029. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.