Abstract
Numerous human genes display dual coding within alternatively spliced regions, which give rise to distinct protein products that include segments translated in more than one reading frame. To resolve the ensuing protein structural puzzle, we identified 67 human genes with alternative splice variants comprising a dual-coding region at least 75 nucleotides in length and analyzed the structural status of the protein segments they encode. The inspection of their amino acid composition and predictions by the IUPred and PONDR® VSL2 algorithms suggest a high propensity for structural disorder in dual-coding regions. In the case of +1 frameshifts, the average level of disorder in the two frames is similarly high (47.2% in the ancestral frame, 58.2% in the derived frame, with the average level of disorder in human proteins being approximately 30%), whereas in the case of -1 frameshifts, there is a significant tendency to become more disordered upon shifting the frame (16.7% in the ancestral frame, 56.3% in the derived frame). The regions encoded by the derived frame are mostly disordered (disorder percentage > 50%) in 39 out of 62 cases, which strongly suggests that structural disorder enables these protein products to exist and function without the need of a highly evolved 3D fold. The potential advantages are also demonstrated by the appearance of novel functions and the high incidence of transcripts escaping nonsense-mediated decay. By discussing several examples, we demonstrate that dual coding may be an effective mechanism for the evolutionary appearance of novel intrinsically disordered regions with new functions.
Keywords: alternative splicing, nonsense-mediated decay, unstructured protein
The process of alternative splicing (AS), in which different combinations of exons are joined together in mRNA maturation, enables several protein isoforms to be encoded by a single gene (1, 2). It is estimated that more than 75% of mammalian genes are alternatively spliced (1, 3) and in about 50% of all AS events the reading frame is altered (4), i.e., a certain stretch of DNA has the potential to be translated in different reading frames. The use of such alternative reading frames (ARFs), however, is often suppressed by a premature termination codon (PTC) that results in nonsense-mediated decay (NMD) of the mRNA product (5, 6). In mammals, a stop codon followed by an exon–exon junction more than 50–55 nucleotides downstream is recognized as a PTC (7) that regulates gene expression and/or acts as a surveillance mechanism against potentially harmful protein products.
A major concern with dual-coding in ARFs is that it gives rise to two intertwined polypeptide sequences which are highly unlikely to both result in two properly folded functional proteins. Thus, dual-coding has long been thought to be prevalent only in viruses and prokaryotes that are under pressure to maintain a compact genome (8, 9). Only relatively recently, results on functional pairs of proteins derived from ARFs (10–16) and bioinformatic studies of conserved overlapping open reading frames (ORFs) (16–19) have pointed to the likely importance of the use of ARFs in eukaryotes.
An enigmatic issue largely overlooked thus far is the protein structural impact of this phenomenon. Because folding of a polypeptide chain to a unique 3D state is a highly evolved feature, the chance is very low that dual coding would result in two sequences that are both capable of folding into well-defined, functional, 3D structures. It is thought that coevolution of overlapping viral reading frames has been made possible by the presence of structural disorder (8, 9). In the much more relaxed eukaryotic genomes, the structural conundrum delayed the recognition of now accepted cases of such functional protein pairs translated in two ARFs, such as XLαs/ALEX (14), p16INK1α/p19ARF (15), 4E-BP3/MASK (20), and pXBP1(U)/pXBP1(S) (21). These observations and further structural considerations raise the idea that the structural puzzle of dual coding can be solved by assuming the structural disorder of the proteins involved, at least in one of the ARFs. If any of the alternative polypeptides lack a well-structured 3D fold (i.e., intrinsically disordered), both variants could exist and function without violating our basic notions of gene organization and protein structure.
This idea draws from the recent realization that intrinsically disordered proteins or regions of proteins (IDPs/IDRs) exist and function without a well-defined 3D structure, yet they are surprisingly common in proteomes (22–25). Their functions either directly stem from their ability to fluctuate over an ensemble of structural states (entropic chain functions) or from molecular recognition, in which the disordered segment undergoes local folding induced by the partner (26, 27). Structural disorder correlates with AS (28) and it often provides functional advantages, such as the increased speed of binding, extended binding surfaces, and the ability to adapt to the structure of different partners (29). IDPs are enriched in disorder-promoting polar and charged amino acids and depleted in order-promoting bulky hydrophobic amino acids (30), and structural disorder is predictable by a variety of bioinformatic algorithms.
In accord, we carried out a thorough bioinformatic analysis of the dual-coding regions of human alternative splice variants. For both their original and derived reading frames, we analyzed their amino acid composition and predicted their disorder by two algorithms, IUPred (31, 32) and PONDR® VSL2 (33). By all measures, we found a positive correlation between structural disorder and dual coding in ARFs. We discuss in detail the structural rationale of this finding and also extend our observation to several confirmed and probable functions related to the derived frames of the dual-coding regions.
Results
Amino Acid Composition of Dual-Coding Regions.
Ninety-seven dual-coding human genes encoding for at least 25 residues in both alternative frames have been selected by BLAST homology searches in the Reference Sequence Database (RefSeq), as described in Methods (with an average length of 49 and a median of 38 residues; Table S1). Although the RefSeq database is one of the most reliable sources of mRNA data, and we only used the transcripts with a “reviewed” status, this dataset still could contain annotation errors and mRNA sequences without evidence at protein level. As a further verification step, we excluded cases where we did not find evidence for both isoforms at the protein level, which resulted in 67 genes in our final dataset. We could confidently identify the original (ancient) and the (novel) derived state of the region in 62 cases.
Because amino acid composition is the primary determinant of protein disorder (34), we started our study with analyzing the amino acid frequencies of these dual-coding regions of human alternative splice variants. First, to investigate the effect of a frameshift on an arbitrary sequence, we compared a sample RefSeq dataset to its +1 and -1 frameshifted versions and to IDPs/IDRs in the DisProt database (25) (Table 1). Our results show that the majority of order-promoting residues are underrepresented in the frameshifted RefSeq dataset, whereas several disorder-promoting residues, such as the structure-breaker proline (35), are overrepresented. Almost all the observed differences caused by shifting the frame are statistically significant (Chi-square test with Bonferroni correction, P < 0.0025).
Table 1.
Changes in amino acid frequencies due to frameshift in an average nucleotide sequence
| W | F | Y | I | M | L | V | N | C | T | A | G | R | D | H | Q | K | S | E | P | X | |
| RefSeq +1 | +3 | -2 | -2 | -2 | -1 | 0 | -3 | -2 | +2 | +3 | -1 | -2 | +5 | -4 | -1 | -1 | -1 | +4 | -5 | +3 | +5 |
| RefSeq -1 | +2 | -2 | -2 | -3 | -2 | -2 | -3 | -2 | +3 | -2 | 0 | +4 | +4 | -2 | +3 | +1 | -3 | 0 | -3 | +3 | +5 |
| DisProt | -1 | -2 | -1 | -2 | -1 | -4 | -1 | 0 | -2 | 0 | +1 | 0 | -1 | +2 | -1 | +1 | +3 | +1 | +3 | +2 |
Frameshifted human RefSeq datasets and IDPs/IDRs in DisProt were compared to the original human RefSeq dataset. Nonsignificant differences are marked with zero. The numbers represent the difference in the percentage of the appropriate residue (0–1%: +1, 1–2%: +2, etc.). Amino acid frequencies were compared by Chi-square test with the Bonferroni correction (p < 0.0025).
Next, we compared the amino acid composition of dual-coding regions to these controls, i.e., ancestral frames to the RefSeq database and derived frames to the frameshifted RefSeq datasets. Intriguingly, -1 and +1 frameshifted dual-coding regions show different patterns of change. In case of the +1 frameshifted splice variants, the ancestral and derived frames differ similarly from their controls, i.e., they show a further decrease in certain order-promoting amino acids (Ile, Met, Asn) and an increase in disorder-promoting residues (Ala, Pro). The most remarkable change is a further increase in the frequency of Pro (3–5% compared to the frameshifted RefSeq sequence). In case of the -1 frameshifted variants, the ancestral frame contains less Lys, Ser, and Glu and more Leu, whereas the derived frame resembles the amino acid composition of the +1 shifts. The characteristic pattern in the ancestral frame of the -1 splice variants could be explained by the appearance of stop codons from the codons of Lys, Ser, and Glu upon shifting the frame.
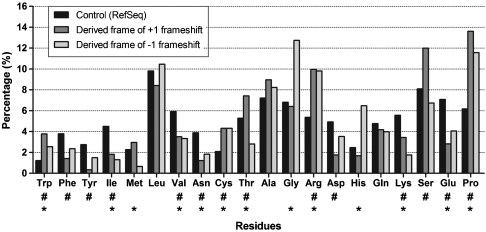
The amino acid composition of derived frames is compared to that of an average protein in Fig. 1. Rather consistently, they have a lower level of hydrophobic residues (Phe, Tyr, Ile, Met, Val) compensated by small and/or hydrophilic amino acids (Cys, Gly, Arg, His, Ser), with a conspicuous increase in proline to about 12–14% in both types of frameshifts. The significance of this value is underscored by the average proline content of proteins in UniProt/Swiss-Prot (< 5%), and even of disordered proteins in DisProt (< 8%). Overall, the direction of these changes in amino acid composition upon frameshift suggests an increase in the level of structural disorder.
Fig. 1.
Amino acid frequencies of regions encoded by derived alternative reading frames. The amino acid composition of regions encoded by the derived frames shifted either +1 or -1 relative to their ancestral frame were compared to the composition of the translated human RefSeq dataset as a control. Amino acids are shown in the order of their increasing disorder-promoting potential, as defined by the TOP-IDP scale (30). Significant differences are indicated by # (+1 shifted derived frames) and * (-1 shifted derived frames), calculated by Chi-square test with the Bonferroni correction (p < 0.0025).
High Predicted Disorder in Dual-Coding Regions.
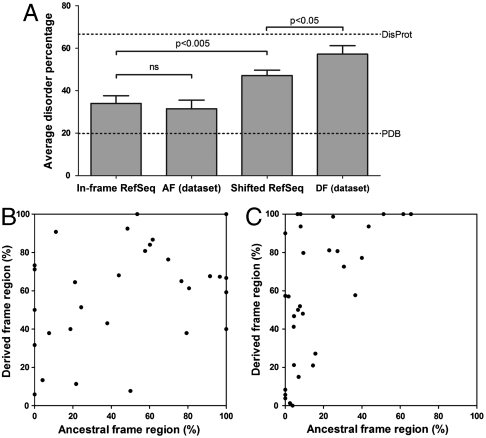
Because amino acid composition is only a rough descriptor of structural disorder, we predicted protein disorder in the alternative frames of the dual-coding regions by sequence-based bioinformatic predictors IUPred (31, 32) and PONDR® VSL2 (33), and calculated the percentage of disordered residues (for cutoff values; see Methods). With the COILS software, we could not detect any predicted coiled-coil structure that would be misinterpreted as a disordered region. The calculated percentages of disordered residues are given in Table S1. The overall high propensity for disorder is shown in Fig. 2A: Predicted disorder of regions encoded by the derived frame is significantly higher (Mann–Whitney test, p < 0.05) than that of random sequences in the frameshifted RefSeq dataset and approaches that of fully disordered proteins in DisProt.
Fig. 2.
Disorder percentage of the ancestral and the derived frames of dual-coding regions. (A) Average predicted disorder ± SEM of random in-frame and frameshifted regions of RefSeq, ancestral (AF) and derived (DF) frames of dual-coding human genes (datasets were compared by the Mann–Whitney test). Average disorder content of Protein Data Bank and DisProt are indicated by dotted lines. (B) The change in disorder caused by a +1 frameshift, as estimated by the average of IUPred and PONDR® VSL2 disorder percentages. (C) The change in disorder caused by a -1 frameshift, as estimated by the average of IUPred and PONDR® VSL2 disorder percentages.
When individual pairs are considered, in most cases at least one of the ARFs encodes for a protein region with high predicted disorder, only very rarely show both the ancestral and derived frames a pattern of order (geneIDs 1185, 4170, 122769). Upon shifting the frame, structural disorder tends to increase overall, as demonstrated by the averages of disorder percentages in ancestral frames (19.6% by IUPred or 43.3% by PONDR® VSL2) vs. derived frames (41.2% by IUPred or 73.2% by PONDR® VSL2).
The two possible directions of the shift, however, differ in this regard: In the case of +1 frameshift, there is no general direction in the change of disorder, either the ancestral or the derived frame can be disordered (Fig. 2B). In the case of the -1 frameshift, the region encoded by the ancestral frame tends to be more ordered and the derived frame tends to be more disordered (Fig. 2C). On the whole, mostly disordered regions are prevalent in our dataset, they are found in at least one frame 17/30 (IUPred) or 27/30 (PONDR® VSL2) of the +1 frameshifted cases, and 13/32 (IUPred) or 22/32 (PONDR® VSL2) of the -1 frameshifted cases.
Region Encoded by the Derived Frame Often Enables the Transcript to Escape NMD.
NMD is the first line of defense against truncated proteins that would appear in the cell due to the generation of a PTC (6) by a shift in the reading frame. We can infer functional selection if the alternative (derived) frame is long enough to reach the end of the NMD zone (past 50 base pairs upstream of the last exon–exon junction).
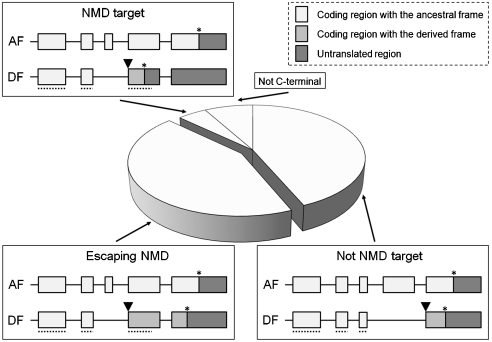
To this end, we analyzed our dataset for the NMD status of the splice variants encoded by the derived frame (Fig. 3). Only a few cases were found where the dual-coding region is not at the C terminus (5/62) or the short derived frames potentially lead to NMD degradation (3/62). Almost half of the dual-coding regions (27/62) begin in the last exon (or in a very short exon before the last), i.e., they are not targets of NMD. Interestingly, the same number of proteins with derived frames (27/62) escape NMD because of their sufficiently long coding regions.
Fig. 3.
The relation of derived frames to NMD. The pie chart shows the ratio of distinct types of NMD-related behavior (nontarget, target, escaping, and not relevant) of derived ARFs of dual-coding regions. Of 62 cases when the original and derived frames could be assigned, in not-NMD targets (Bottom Right, n = 27) the dual-coding region starts (▾) in the last exon, so the PTC (*) does not elicit NMD. If the dual-coding region starts before the last exon, and the PTC appears more than 50 base pairs upstream of the last exon–exon junction, the transcript will be degraded by NMD (Top, n = 3). If the novel coding region is long enough to pass the end of the NMD zone (⋯), the PTC will not induce NMD, and the transcript survives (Bottom Left, n = 27).
The mRNAs of these altered proteins (carrying the truncated common sequence and a different C-terminal segment) are not degraded, and the functional significance of the loss of their original C terminus and/or the function of the novel C terminus can manifest itself. Because we observed high predicted disorder at the protein level in 22 out of 27 cases, we can assume an evolutionary advantage of structural disorder in NMD-escaping variants.
Select Examples of Dual-Coding Splice Variants Show Well-Defined Functions for Both Isoforms.
The frequent escape from NMD and a high level of structural disorder suggest that dual coding may represent an effective genetic strategy of generating functional variation at the protein level. A search in the literature unveils several cases of such functional pairs demonstrating exciting biology.
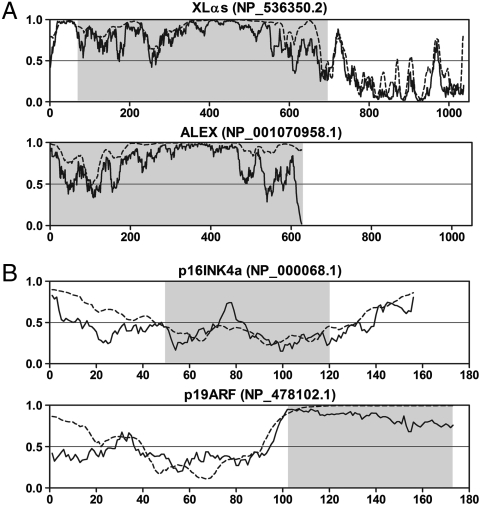
The most extraordinary finding is the Guanine nucleotide-binding protein G(s) (GNAS) complex locus (geneID 2778), which encodes for several isoforms of the stimulatory G-protein alpha subunit (14). The transcript that encodes XLαs, the longest G(s)α subunit variant, includes a second overlapping ORF within its N-terminal XL domain, which encodes the structurally unrelated protein ALEX (14). The XL domain of XLαs and ALEX are completely disordered (Fig. 4A) and function by binding to each other, in which ALEX regulates XLαs activity (14). This isoform pair has the longest known dual-coding region in the human proteome, spanning more than 1,800 base pairs.
Fig. 4.
Disorder prediction for the XLαs/ALEX and p16INK4a/p19ARF pairs. (A) Two isoforms expressed from the GNAS complex locus (GeneID: 2778) with an extreme long dual-coding region. (B) CDKN2A gene (GeneID: 1029) products with different first exons, and a typically highly disordered pattern in the derived frame. Lines represent the disorder prediction for the whole protein (─, IUPred; ⋯, PONDR® VSL2). Gray shaped regions cover the fragments corresponding to the dual-coding region. Residues above the threshold of 0.5 are predicted to be disordered.
Another interesting pair is the INK4a/ARF tumor suppressor locus (geneID 1029). ARFs of this locus generate two distinct products: the p16INK4a protein, a cyclin-dependent kinase inhibitor that functions upstream of the retinoblastoma protein, and the p19ARF protein, which blocks MDM2 inhibition of p53 activity (15). Remarkably, both these unrelated proteins are capable of inducing cell cycle arrest, although through completely different mechanisms (36). Probably encoded by the original frame, p16INK4a adopts a well-defined α-helical structure (37). On the other hand, p19ARF has an N-terminal domain, whereas the second half of the protein is comprised of an entirely disordered segment coded by the derived frame (Fig. 4B). The asymmetric evolution of the two coding frames reported by Szklarczyk et al. (16) can be also explained by this finding, because the fully disordered region encoded by the derived frame could tolerate more nonsynonymous mutations without the evolutionary constraints dictated by a folded structure. In fact, anomalously high mutation rates were reported in IDPs/IDRs by recent comparative studies (38, 39).
Effective Generation of Functional Variants by the Elimination of a Functional Domain by Structural Disorder in the Alternative Frame.
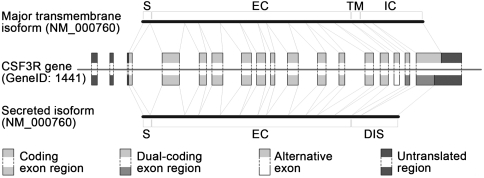
Several isoform pairs revealed that dual coding combined with structural disorder can generate functional variants of a protein by replacing a domain with a disordered segment. The resulting protein, which is viable due to structural disorder and has an altered function and/or cellular localization due to the loss of a key element, can act as an inhibitor of the original protein. The immediate evolutionary potential of this mechanism is most apparent in the case of transmembrane (TM) proteins. Our dual-coding dataset contains several TM proteins, and in 12 cases (GeneIDs 177, 858, 958, 960, 1438, 1441, 2204, 4179, 8784, 79412, 80739, 80835) the splice event not only shifts the reading frame but also results in the loss of the TM region (e.g., CSF3R; see Fig. 5). These single-pass type I membrane proteins have receptor or cell-adhesion functions, and deletion of their TM region leads to a change in localization, i.e., secretion to the extracellular space because they lack other transmembrane regions in either frame, as suggested by both HMMTOP (40) and TMHMM (41) predictions.
Fig. 5.
An example for a TM localization/functional switch. A TM protein (CSF3R) in which exon skipping and frameshift results in the loss of its membrane-spanning region and subsequent secretion into the extracellular space. The upper transcript represents the major TM isoform containing the intracellular region, which is encoded by the ancestral frame (light gray). As a result of AS, the exon containing the TM region is excluded from the transcript in the bottom, resulting in a secreted isoform that has a novel disordered C-terminal segment. S, signal sequence; EC, extracellular part; TM, transmembrane region; IC, intracellular part; DIS, disordered region.
Some isoform pairs in this TM subgroup are well characterized as receptor–inhibitor pairs (42–44), either because the secreted form can deplete the ligand of the receptor from the extracellular space or it can form a dimer with the receptor without generating a signal in the cell (see Table 2). The protein regions encoded by the derived frames are mostly disordered in 10 out of 12 cases (Table S1), which contributes to function by allowing the transformation of a TM protein to a soluble one.
Table 2.
Sudden functional change may be caused by replacing a functional region with a disordered segment
| Gene (GeneID) | Original function (with AF) | Novel function (with DF) | Disorder status | Ref. |
| AGER (177) | Receptor for advanced glycosylation end products | Soluble inhibitor (ligand depletion) | FD | (38) |
| CD40 (958) | Member of the tumor necrosis factor receptor superfamily | Soluble inhibitor (interference with the TM form) | MD | (39) |
| CSF2RA (1438) | Colony stimulating factor 2 receptor | Soluble inhibitor (ligand depletion) | MD | (40) |
| GNRHR (2798) | Gonadotropine-releasing hormone receptor | Results in impaired insertion of the original receptor into the membrane | FD | (10) |
| HSF4 (3299)* | Transcriptional inhibitor of heat shock genes | Transcriptional activator of heat shock genes | MD | (11) |
| MCL1 (4170) | Inhibits apoptosis with a complete Bcl-2 domain | Promotes apoptosis with a severely truncated (> 80%) Bcl-2 domain | ND | (9) |
| XBP1 (7494)* | Transcriptional regulator of MHC class II genes | Negative regulator of the other isoform | MD | (12) |
The case of three TM proteins and four other examples show that AS may elicit dual coding that leads to the replacement of a functional domain or region with a viable disordered segment. Because part of the original function is retained, such a change has the potential to generate an inhibitor of the ancestral protein at the expense of a minimal evolutionary investment. MHC, major histocompatibility complex.
*Cases where the ancestral and derived frame could not be determined.
There are also other genes where the change in the reading frame eliminates part of the protein that is functionally important, thus the frameshifted variant can function as an inhibitor. Such protein/inhibitor pairs are thought to exist in the case of the XBP1 transcription factor [geneID 7494; (13)], the gonadotropin-releasing hormone receptor [geneID 2798; (11)], the heat shock transcription factor 4 [geneID 3299; (12)], and MCL1, an apoptosis regulator protein that belongs to the Bcl-2 family [geneID 4170; (10)]. With the exception of the apoptosis regulator protein, protein disorder in the alternative frames prevails in these variants (see Table 2 and Table S1).
Discussion
A unique consequence of AS is the generation of distinct mRNAs, which can be translated in alternative reading frames. The ensuing dual coding raises serious concerns due to the codon codependency of the overlapping frames and the structural integrity of the encoded proteins. Due to these concerns, dual coding is generally thought to prevail in pathogenic organisms under pressure to maintain a compact genome (8, 9) and only recent results pointed to its possible importance in mammals in general and in the human genome in particular. It occurred to us that folding of two sequentially intertwined, yet unrelated proteins is very unlikely to occur, and this structural and functional conundrum can only be resolved by assuming structural disorder of the protein product generated in at least one of the frames as suggested for products of overlapping viral genes (8, 9). To address this issue, we approached the structural status of proteins encoded by human genes that contain a dual-coding segment at least 75 base pairs in length. The amino acid composition of protein sequences and disorder prediction by IUPred and PONDR® VSL2 suggest that in the case of a -1 frameshift, the ancestral frame tends to be more ordered and the derived frame more disordered, whereas in the case of a +1 frameshift, both frames have a similar high level of disorder. By both predictors, the majority of protein segments involved are mostly disordered. Because AS is known to correlate with structural disorder (28), this correlation might be a consequence of the involvement of AS. This, however, is limited, because we only analyzed the dual-coding region and not the AS region that precedes it. Thus, the upstream AS event does not strongly discriminate between ordered and disordered regions that lie downstream. Furthermore, in certain cases, the dual-coding region even precedes the AS region.
Thus, the observed strong correlation suggests an intimate association between dual coding and structural disorder, which probably reflects selection for such segments at various levels, such as (i) evasion of cellular surveillance mechanisms eliminating misfolded proteins, (ii) prevention of protein aggregation of potentially misfolded proteins, and (iii) rapid selection of novel functions enabled by the functional plasticity and permissivity of IDPs. We will discuss these features in detail.
Protein folding is a highly evolved functional feature, and effective cellular surveillance mechanisms, such as the ubiquitin/proteasome system (45), have evolved to eliminate misfolded proteins that lose function and are prone to aggregate. Although the polypeptide chain of IDPs is exposed all the time, due to their lack of hydrophobic residues (23, 34) they are resistant to spontaneous elimination by such mechanisms. In fact, many IDPs are long-lived proteins and structural disorder does not cause rapid clearance of proteins from the cell (46), whereas a newly emerging random sequence with the amino acid composition of ordered proteins would most probably expose its hydrophobic residues leading to its rapid elimination. In contrast, a random hydrophilic IDP sequence generated by dual coding is unlikely to be immediately disposed of by the cell.
A novel, random sequence also faces the risk of aggregation initiated by its exposed hydrophobic patches (47). Whereas IDPs are known to form pathological aggregates (48), often they are resistant to denaturing conditions, such as high temperature (23) and structural disorder is anticorrelated with aggregation propensity (49). Because globular proteins are highly amyloidogenic, their primary defense against aggregation is fast and effective folding (50), of which a random sequence generated by frameshift is unlikely to be capable. The mark of selection against aggregation is probably also evident in the high frequency of Pro and Gly, which can inhibit the formation of aggregates (51), and also in high charge, i.e., extreme pI (34), inhibitory to aggregation (52).
Probably the most exciting feature of structural disorder in the derived sequences is their enhanced capacity to generate novel function. IDPs often function by short linear motifs, which very effectively arise in evolution by random mutations (53), their functions in general rely on very limited sequence information (26) that persist in the face of rapid evolutionary changes (39). In line with these features, ancestral reading frames are usually under stronger selection (16, 18), and alternatively spliced exons that alter the reading frame have a higher mutation rate than those which preserve the reading frame (4). In our dataset, there are several isoform pairs where both the ancestral and the derived variants have well-described functions (e.g., ALEX/XLαs and INK4a/ARF) or the novel function of the derived isoform results from replacing a functionally important segment with a disordered region, such as in the case of the secreted soluble forms of TM proteins. AS with frameshift could be especially important in these cases, because the simple exclusion of the TM region by AS without a frameshift would result in a secreted protein with the original intracellular region, inducing unwanted processes in the extracellular space. Structural disorder in this case enables the generation of a truncated, yet viable protein product that lacks the intracellular segment and is functionally related to the original protein.
In summary, we demonstrated that protein regions encoded by alternative reading frames have a high propensity for disorder due to selective advantages in protein viability and function. Whereas the small effective population size of humans might have enabled to fix stochastic effects in AS leading to dual coding in ARFs, the statistical overrepresentation of structural disorder in the derived frame—even compared to the random shifted sequences—and the functional association of several well-studied examples suggest that selection for the lack of structure and associated functions has occurred. Structural disorder and the underlying biased amino acid composition in the derived frame can help escape NMD, cause a functional switch, or simply help evade clearance at the protein level as a functionally neutral soluble “junk sequence.” On a longer evolutionary timescale, random mutations compatible with the disordered state can drive functional adaptation, hence, dual coding in ARFs is also likely to have contributed to the sudden evolutionary rise of IDPs in eukaryotes. Although dual coding in prokaryotes evolved to compact their genome, the described phenomenon in eukaryotes participates in the extension of complexity and plasticity.
Methods
Generation of the Dataset of Dual-Coding Genes.
As a source of sequence data and AS annotation, we used the human entries from the National Center for Biotechnology Information RefSeq (54). We only selected mRNA sequences marked as reviewed, which are highly reliable and valid transcripts. To refine the homologous regions between splice variants, a BLAST (55) search was run on the database of +1 and -1 frameshifted translations of RefSeq, with the original translated sequence as the query. Only dual-coding regions of more than 25 residues were analyzed, which is about the practical minimum required to carry an individual function by a disordered region (56). Finally, we determined the protein evidence level of the transcripts (transcript, protein, and functional protein levels), and filtered out genes, where at least one of the isoforms have evidence only at transcript level. Thus, keeping protein pairs with the highest reliability only, our final dataset contains 67 isoform pairs. Our dataset is most similar to that of Liang and Landweber (18), which is based on almost the same RefSeq database, and shows somewhat less similarity to other related datasets generated by de novo bioinformatic methods (16, 17, 19).
To determine the ancestral reading frame, we searched for homologues of the dual-coding regions and compared the location of stop codons in both frames in other species (18). We also considered the presence of other splice variants and the organization of exons of the gene, as well as the presence of conserved domains at the protein level. This procedure enabled the determination of the ancestral and the derived frames in 62 cases. Finally, we divided the final dataset into two groups according to the position of the derived frame relative to the ancestral frame, +1 splice variants (frameshifted in the 3′ direction), and -1 splice variants (frameshifted in the 5′ direction). For a flowchart on the procedure applied for selecting data and obtaining results see Fig. S1.
Disorder Prediction.
Protein disorder was predicted by two algorithms, IUPred (31, 32) and PONDR® VSL2 (33), which are based on different principles and apply different approaches for disorder prediction. These predictors assign a disorder tendency score (between zero and one) for each amino acid and, as a measure of protein disorder, we calculated the percentage of residues above a cutoff value. In the case of PONDR® VSL2 we used the default cutoff value of 0.5, whereas a cutoff value of 0.4 was used for IUPred because this is the average score for disordered regions in the DisProt database (25) predicted by IUPred (57). To avoid mispredicting coiled-coil regions as disordered, we used the COILS software (58) with > 50% probability within a 21-residue window as an ordered segment. For the generation of the random shifted control dataset see Fig. S2.
Detection of Possible Degradation by NMD.
Because the alternative reading frame may contain a novel PTC, we also analyzed the transcripts for their NMD status. To this end, we applied the widely accepted criterion that a transcript is destined for NMD if its coding sequence ends > 50 bp upstream of a splice site. We classified the NMD status of proteins coded by the derived frame into four groups: (i) dual-coding region is not in the C terminus, (ii) possible NMD target because of the presence of a PTC, (iii) dual-coding region is in the last exon, or starts near the last exon–exon junction, and (iv) it escapes NMD due to the length of the derived frame. Proteins were only placed into the third group if the length of their derived frame was the cause of NMD escape, as it was significantly longer in the NMD zone (region where a stop codon results in NMD) than a random frameshifted sequence, assuming an average of one stop codon per 20 residues.
Functional Characterization.
Functional information on dual-coding genes was gathered from different sources: (i) annotations about function and localization in the UniProt database, (ii) UniProt keywords and Gene Ontology cross references (59, 60), (iii) domain information in the Pfam database (61), and (iv) extensive search in the literature for functional information. As a well-defined structural and functional attribute, we also mapped the TM regions of annotated TM proteins using the Topology Data Bank of Transmembrane Proteins and UniProt databases (40, 60). If the splice variant of a TM protein lost the TM region, the new sequence was analyzed for the appearance of any novel TM region using the HMMTOP and the TMHMM software (40, 41).
Statistical Analysis and Programming.
Chi-square analysis with a 95% confidence interval and the Bonferroni correction (with p-value threshold reduced to 0.0025) were used to compare the amino acid content of the different datasets. Differences in predicted disorder were analyzed by the Mann–Whitney test. Perl scripts (SI Appendix) and other compiled software (e.g., IUPred, HMMTOP, etc.) were executed locally.
Supplementary Material
Acknowledgments.
The authors thank Dr. Zsuzsanna Dosztanyi for helpful discussions. This research was supported by Grants OTKA K60694 and NK71582 from the Hungarian Scientific Research Fund, and ETT 245/2006 from the Hungarian Ministry of Health.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/cgi/content/full/0907841107/DCSupplemental.
References
- 1.Blencowe BJ. Alternative splicing: New insights from global analyses. Cell. 2006;126:37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 2.Kim E, Goren A, Ast G. Alternative splicing: Current perspectives. Bioessays. 2008;30:38–47. doi: 10.1002/bies.20692. [DOI] [PubMed] [Google Scholar]
- 3.Johnson JM, et al. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- 4.Zhang C, Krainer AR, Zhang MQ. Evolutionary impact of limited splicing fidelity in mammalian genes. Trends Genet. 2007;23:484–488. doi: 10.1016/j.tig.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 5.Lewis BP, Green RE, Brenner SE. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc Natl Acad Sci USA. 2003;100:189–192. doi: 10.1073/pnas.0136770100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McGlincy NJ, Smith CW. Alternative splicing resulting in nonsense-mediated mRNA decay: What is the meaning of nonsense? Trends Biochem Sci. 2008;33:385–393. doi: 10.1016/j.tibs.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 7.Nagy E, Maquat LE. A rule for termination-codon position within intron-containing genes: when nonsense affects RNA abundance. Trends Biochem Sci. 1998;23:198–199. doi: 10.1016/s0968-0004(98)01208-0. [DOI] [PubMed] [Google Scholar]
- 8.Karlin D, Ferron F, Canard B, Longhi S. Structural disorder and modular organization in Paramyxovirinae N and P. J Gen Virol. 2003;84:3239–3252. doi: 10.1099/vir.0.19451-0. [DOI] [PubMed] [Google Scholar]
- 9.Rancurel C, Khosravi M, Dunker AK, Romero PR, Karlin D. Overlapping genes produce proteins with unusual sequence properties and offer insight into de novo protein creation. J Virol. 2009;83:10719–10736. doi: 10.1128/JVI.00595-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bingle CD, et al. Exon skipping in Mcl-1 results in a bcl-2 homology domain 3 only gene product that promotes cell death. J Biol Chem. 2000;275:22136–22146. doi: 10.1074/jbc.M909572199. [DOI] [PubMed] [Google Scholar]
- 11.Grosse R, Schoneberg T, Schultz G, Gudermann T. Inhibition of gonadotropin-releasing hormone receptor signaling by expression of a splice variant of the human receptor. Mol Endocrinol. 1997;11:1305–1318. doi: 10.1210/mend.11.9.9966. [DOI] [PubMed] [Google Scholar]
- 12.Tanabe M, et al. The mammalian HSF4 gene generates both an activator and a repressor of heat shock genes by alternative splicing. J Biol Chem. 1999;274:27845–27856. doi: 10.1074/jbc.274.39.27845. [DOI] [PubMed] [Google Scholar]
- 13.Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 14.Klemke M, Kehlenbach RH, Huttner WB. Two overlapping reading frames in a single exon encode interacting proteins—A novel way of gene usage. EMBO J. 2001;20:3849–3860. doi: 10.1093/emboj/20.14.3849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sharpless NE. INK4a/ARF: A multifunctional tumor suppressor locus. Mutat Res. 2005;576:22–38. doi: 10.1016/j.mrfmmm.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 16.Szklarczyk R, Heringa J, Pond SK, Nekrutenko A. Rapid asymmetric evolution of a dual-coding tumor suppressor INK4a/ARF locus contradicts its function. Proc Natl Acad Sci USA. 2007;104:12807–12812. doi: 10.1073/pnas.0703238104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chung WY, Wadhawan S, Szklarczyk R, Pond SK, Nekrutenko A. A first look at ARFome: dual-coding genes in mammalian genomes. PLoS Comput Biol. 2007;3:e91. doi: 10.1371/journal.pcbi.0030091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liang H, Landweber LF. A genome-wide study of dual coding regions in human alternatively spliced genes. Genome Res. 2005;16:190–196. doi: 10.1101/gr.4246506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ribrioux S, Brungger A, Baumgarten B, Seuwen K, John MR. Bioinformatics prediction of overlapping frameshifted translation products in mammalian transcripts. BMC Genomics. 2008;9:122. doi: 10.1186/1471-2164-9-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Poulin F, Brueschke A, Sonenberg N. Gene fusion and overlapping reading frames in the mammalian genes for 4E-BP3 and MASK. J Biol Chem. 2003;278:52290–52297. doi: 10.1074/jbc.M310761200. [DOI] [PubMed] [Google Scholar]
- 21.Yoshida H, Oku M, Suzuki M, Mori K. pXBP1(U) encoded in XBP1 pre-mRNA negatively regulates unfolded protein response activator pXBP1(S) in mammalian ER stress response. J Cell Biol. 2006;172:565–575. doi: 10.1083/jcb.200508145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 23.Tompa P. Intrinsically unstructured proteins. Trends Biochem Sci. 2002;27:527–533. doi: 10.1016/s0968-0004(02)02169-2. [DOI] [PubMed] [Google Scholar]
- 24.Tompa P. The interplay between structure and function in intrinsically unstructured proteins. FEBS Lett. 2005;579:3346–3354. doi: 10.1016/j.febslet.2005.03.072. [DOI] [PubMed] [Google Scholar]
- 25.Sickmeier M, et al. DisProt: The database of disordered proteins. Nucleic Acids Res. 2007;35:D786–793. doi: 10.1093/nar/gkl893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tompa P, Fuxreiter M. Fuzzy complexes: Polymorphism and structural disorder in protein–protein interactions. Trends Biochem Sci. 2008;33:2–8. doi: 10.1016/j.tibs.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 27.Wright PE, Dyson HJ. Linking folding and binding. Curr Opin Struct Biol. 2009;19:1–8. doi: 10.1016/j.sbi.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Romero PR, et al. Alternative splicing in concert with protein intrinsic disorder enables increased functional diversity in multicellular organisms. Proc Natl Acad Sci USA. 2006;103:8390–8395. doi: 10.1073/pnas.0507916103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tompa P, Szasz C, Buday L. Structural disorder throws new light on moonlighting. Trends Biochem Sci. 2005;30:484–489. doi: 10.1016/j.tibs.2005.07.008. [DOI] [PubMed] [Google Scholar]
- 30.Campen A, et al. TOP-IDP-scale: A new amino acid scale measuring propensity for intrinsic disorder. Protein Peptide Lett. 2008;15:956–963. doi: 10.2174/092986608785849164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dosztanyi Z, Csizmok V, Tompa P, Simon I. IUPred: Web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 2005;21:3433–3434. doi: 10.1093/bioinformatics/bti541. [DOI] [PubMed] [Google Scholar]
- 32.Dosztányi Z, Csizmok V, Tompa P, Simon I. The pairwise energy content estimated from amino acid composition discriminates between folded and instrinsically unstructured proteins. J Mol Biol. 2005;347:827–839. doi: 10.1016/j.jmb.2005.01.071. [DOI] [PubMed] [Google Scholar]
- 33.Peng K, Radivojac P, Vucetic S, Dunker AK, Obradovic Z. Length-dependent prediction of protein intrinsic disorder. BMC Bioinformatics. 2006;7:208. doi: 10.1186/1471-2105-7-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Uversky VN, Gillespie JR, Fink AL. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins. 2000;41:415–427. doi: 10.1002/1097-0134(20001115)41:3<415::aid-prot130>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 35.Williamson MP. The structure and function of proline-rich regions in proteins. Biochem J . 1994;297:249–260. doi: 10.1042/bj2970249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Quelle DE, Zindy F, Ashmun RA, Sherr CJ. Alternative reading frames of the INK4a tumor suppressor gene encode two unrelated proteins capable of inducing cell cycle arrest. Cell. 1995;83:993–1000. doi: 10.1016/0092-8674(95)90214-7. [DOI] [PubMed] [Google Scholar]
- 37.Byeon IJ, et al. Tumor suppressor p16INK4A: Determination of solution structure and analyses of its interaction with cyclin-dependent kinase 4. Mol Cell. 1998;1:421–431. doi: 10.1016/s1097-2765(00)80042-8. [DOI] [PubMed] [Google Scholar]
- 38.Brown CJ, et al. Evolutionary rate heterogeneity in proteins with long disordered regions. J Mol Evol. 2002;55:104–110. doi: 10.1007/s00239-001-2309-6. [DOI] [PubMed] [Google Scholar]
- 39.Daughdrill GW, Narayanaswami P, Gilmore SH, Belczyk A, Brown CJ. Dynamic behavior of an intrinsically unstructured linker domain is conserved in the face of negligible amino acid sequence conservation. J Mol Evol. 2007;65:277–288. doi: 10.1007/s00239-007-9011-2. [DOI] [PubMed] [Google Scholar]
- 40.Tusnady GE, Simon I. The HMMTOP transmembrane topology prediction server. Bioinformatics. 2001;17:849–850. doi: 10.1093/bioinformatics/17.9.849. [DOI] [PubMed] [Google Scholar]
- 41.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 42.Brown CB, Beaudry P, Laing TD, Shoemaker S, Kaushansky K. In vitro characterization of the human recombinant soluble granulocyte-macrophage colony-stimulating factor receptor. Blood. 1995;85:1488–1495. [PubMed] [Google Scholar]
- 43.Tone M, Tone Y, Fairchild PJ, Wykes M, Waldmann H. Regulation of CD40 function by its isoforms generated through alternative splicing. Proc Natl Acad Sci USA. 2001;98:1751–1756. doi: 10.1073/pnas.98.4.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yonekura H, et al. Novel splice variants of the receptor for advanced glycation end-products expressed in human vascular endothelial cells and pericytes, and their putative roles in diabetes-induced vascular injury. Biochem J . 2003;370:1097–1109. doi: 10.1042/BJ20021371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 46.Tompa P, Prilusky J, Silman I, Sussman JL. Structural disorder serves as a weak signal for intracellular protein degradation. Proteins. 2008;71:903–909. doi: 10.1002/prot.21773. [DOI] [PubMed] [Google Scholar]
- 47.Dobson CM. Getting out of shape. Nature. 2002;418:729–730. doi: 10.1038/418729a. [DOI] [PubMed] [Google Scholar]
- 48.Uversky VN, Fink AL. Conformational constraints for amyloid fibrillation: The importance of being unfolded. Biochim Biophys Acta. 2004;1698:131–153. doi: 10.1016/j.bbapap.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 49.Linding R, Schymkowitz J, Rousseau F, Diella F, Serrano L. A comparative study of the relationship between protein structure and beta-aggregation in globular and intrinsically disordered proteins. J Mol Biol. 2004;342:345–353. doi: 10.1016/j.jmb.2004.06.088. [DOI] [PubMed] [Google Scholar]
- 50.Monsellier E, Chiti F. Prevention of amyloid-like aggregation as a driving force of protein evolution. EMBO Rep. 2007;8:737–742. doi: 10.1038/sj.embor.7401034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rauscher S, Baud S, Miao M, Keeley FW, Pomes R. Proline and glycine control protein self-organization into elastomeric or amyloid fibrils. Structure. 2006;14:1667–1676. doi: 10.1016/j.str.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 52.Chiti F, Stefani M, Taddei N, Ramponi G, Dobson CM. Rationalization of the effects of mutations on peptide and protein aggregation rates. Nature. 2003;424:805–808. doi: 10.1038/nature01891. [DOI] [PubMed] [Google Scholar]
- 53.Neduva V, Russell RB. Linear motifs: Evolutionary interaction switches. FEBS Lett. 2005;579:3342–3345. doi: 10.1016/j.febslet.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 54.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): A curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 56.Dunker AK, et al. Intrinsically disordered protein. J Mol Graph Model. 2001;19:26–59. doi: 10.1016/s1093-3263(00)00138-8. [DOI] [PubMed] [Google Scholar]
- 57.Fuxreiter M, Tompa P, Simon I. Local structural disorder imparts plasticity on linear motifs. Bioinformatics. 2007;23:950–956. doi: 10.1093/bioinformatics/btm035. [DOI] [PubMed] [Google Scholar]
- 58.Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 59.Ashburner M, et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jain E, et al. Infrastructure for the life sciences: Design and implementation of the UniProt website. BMC Bioinformatics. 2009;10:136. doi: 10.1186/1471-2105-10-136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Finn RD, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.