Abstract
Paramutation is the ability of specific DNA sequences to communicate in trans to establish meiotically heritable expression states. Paramutation at the maize b1 locus is mediated by seven unique noncoding transcribed tandem repeats of 853 bp that are required to establish and maintain the meiotically heritable expression and distinct chromatin states associated with b1 paramutation. In this study, we report the identification of a CXC-domain protein CBBP (CXC domain b1-repeat binding protein) that binds to a defined region within the b1 tandem repeat sequence in vivo and in vitro. When CBBP is expressed from a transgene in maize, it can induce a silent state at the b1 locus that is heritable in progeny no longer containing the transgene, and the silent epiallele is capable of silencing an active epiallele, characteristic of paramutation. Accumulation of the CBBP protein correlates with b1 silencing in transgenic and nontransgenic plants. The ability of CBBP to form multimers and to bind to the b1 tandem repeats suggests a model for counting the number of b1 repeats. In contrast to previously identified proteins involved in paramutation, CBBP does not share similarity to the known components of the Arabidopsis RNAi heterochromatin silencing pathway. Thus, this study defines another class of protein that is involved in heritable gene silencing.
Keywords: chromatin, gene silencing
Paramutation was initially described at the maize r1 locus (1) as an interaction between specific alleles that leads to heritable changes in expression. Since that time, several other examples of paramutation were identified in maize and in other species, including other plants, fungi, and animals (see refs. 2–5 for review). Recent findings demonstrated an essential role for RNAi in paramutation, as four cloned genes involved in paramutation encode proteins closely related to RNAi pathway components (6–9).
Herein we focus on paramutation at b1, a locus that encodes a transcription factor required to activate the purple anthocyanin pigment biosynthetic pathway. Two epialleles that participate in paramutation, B-I and B′, have the same DNA sequences (10) but distinct epigenetic states (11). B-I has a high transcription rate, resulting in homozygous dark purple plants, whereas B′ plants are lightly pigmented because of a reduced transcription rate. In heterozygous plants, B-I is always changed (paramutated) to B′. The new B′ epiallele is fully capable of paramutating naïve B-I epialleles in subsequent generations (11). The B′ epiallele is extremely stable; changes from B′ to B-I have never been observed in wild-type genetic backgrounds. In contrast, B-I is unstable and can spontaneously change to B′ at variable frequencies of 0.1–10% (11, 12).
The key sequences required for b1 paramutation are seven unique tandem repeats of an 853-bp noncoding DNA located ∼100 kb upstream of the b1 transcription start site (10). Although this sequence and the number of repeats are identical in B-I and B′ epialleles, the DNA methylation pattern and chromatin structure differ (10). The b1 alleles that have only one copy of the sequence that is repeated in B-I and B′ do not participate in paramutation (referred to as neutral alleles). Studies of an allelic series that differed only in the number of repeats (1, 3, 5, and 7 repeats) revealed that paramutation requires repeats and that the strength of paramutation correlates with the number of repeats, suggesting a mechanism exists that is able to count the number of repeats (10).
The repeats are transcribed on both strands (7), and siRNAs are produced (9). Given that an RNA-dependent silencing mechanism is required for paramutation, it is intriguing that repeats are transcribed at similar levels from both B′ and B-I epialleles and, even from the neutral b1-K55 allele with a single repeat unit (7). These data suggest mechanisms and tandem repeat RNA levels may be involved in repeat counting. Important questions that need to be addressed are why multiple repeats are required for paramutation and what is the nature of the repeat counting mechanism. One approach toward addressing these questions is to identify proteins binding to the repeat sequence. Herein we report the identification of CBBP, a DNA binding protein that is associated with the repeats in vivo. We also demonstrate that when expressed from a transgene, CBBP can trigger heritable silencing of B-I reminiscent of, but distinct from, B′.
Results
Identification of CBBP, a CXC Domain Protein, Which Interacts with the b1 Repeat Sequence Required for Paramutation.
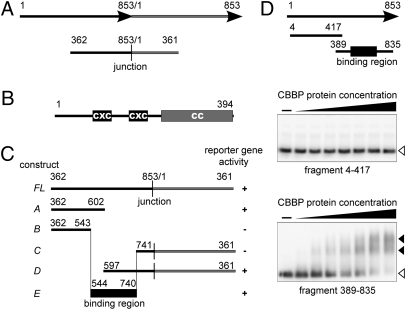
To identify proteins capable of binding to the b1 repeats in a sequence-specific manner, we used the yeast one-hybrid technology. The bait DNA fragment consisted of the 853-bp fragment, encompassing base pairs 362–853 of one repeat unit and base pairs 1–361 of the following repeat unit (Fig. 1A). This approach could potentially identify proteins that are able to discriminate between the multiple and single repeat unit alleles by recognition of the junction unique to multiple repeat alleles.
Fig. 1.
CBBP interacts with b1 repeats. (A) A schematic representation of the b1 repeat fragment used in the yeast one-hybrid library screen shown relative to the native arrangement. (Upper) Two repeat units are shown as black and gray arrows. (Lower) Structure of the DNA fragment used in the screen. (B) A schematic representation of the CBBP domain structure. CXC domains are shown as black boxes, and the conserved CC motif in the C terminus is shown as a gray box. (C) Deletion fragments used in mapping the CBBP binding region within the repeat junction fragment. The sequences remaining in the deletion fragments are shown relative to the full length (FL) fragment shown in A. Reporter gene activity in yeast one-hybrid assays is indicated to the right as + for active and − for inactive. (D) Recombinant CBBP binds to the b1 repeat unit in vitro. (Top) Schematic representation of a single b1 repeat unit and the two fragments used in electrophoretic mobility shift assays. (Middle and Bottom) Recombinant CBBP (0.5–5 picomoles) was incubated with radioactively labeled DNA fragments (10 femtomoles) and analyzed by electrophoresis. The position of free probe DNA is indicated to the right as open triangles, shifted bands are indicated as filled triangles.
While screening 4 × 106 clones, we identified eight cDNA clones from the same gene we refer to as cbbp (CXC domain b1-repeat binding protein), because of the results described below. The longest recovered cbbp cDNA is likely to contain the full-length coding sequence as the corresponding genomic sequence (accession number AC211184_3) revealed an in-frame STOP codon located 339 base pairs upstream of the first Met codon, without a potential initiation codon in between (Fig. S1). The predicted 394-aa protein contains two CXC domains encompassing amino acids 84–125 and 160–202 (Fig. 1B). The region containing the CXC domains in the soybean CCP1 protein has been shown to interact with DNA in a sequence-specific manner (13). The similarity between CBBP and CCP1 is restricted to the CXC domains. CCP1 was shown to repress leghemoglobin gene expression; however, there is no report for its involvement in heritable silencing. The Drosophila CXC domain proteins Tombola and Mip120 have been found in chromatin-associated regulatory complexes (14–16).
Five maize sequences potentially encoding CXC domain proteins are found in the current version of the NCBI database, including the putative full-length cbbp cDNA (EU967670). The plant CXC domain protein family can be divided into several subclasses based on phylogenetic reconstruction (17). The similarity between subclasses does not extend beyond the CXC domains. The subclass containing CBBP appears unique to monocots, with rice and maize each having two proteins with high sequence similarity throughout their entire coding region. The C-terminal part of CBBP contains a previously unidentified motif encompassing amino acids 325–392 that is highly conserved among CBBP, some of the CACTA En/Spm-like transposases, and many other uncharacterized plant proteins (Fig. 1B). We termed this motif CC (CACTA/CBBP). This motif was not detected in proteins outside of the plant kingdom. CBBP is the only CC motif protein that also has CXC domains, except for the closely related protein mentioned above.
CBBP Interacts with a Defined Region in the Repeats.
To further map the CBBP binding region in the b1 repeats, we constructed a series of deletions within the repeat sequence and tested them in the yeast one-hybrid assay (Fig. 1C). In one deletion series, the shortest fragment that showed reporter gene activity was fragment A. The next construct in the series, fragment B, did not show any detectable signal, suggesting that a binding site was located between positions 544 and 602. An additional binding region was detected by examining the series deleting from the other direction. A robust signal was observed with the reporter construct D, but not construct C, the next in the series. These data suggest multiple binding sites are located in the region between 544 and 740. Construct E, containing only the binding region defined by the deletion series analyses, produced strong reporter gene activity, verifying the above results.
CBBP Interacts with b1 Repeats in Vitro.
To determine whether CBBP can bind the b1 repeat unit sequence in vitro, recombinant CBBP purified from Escherichia coli was used in electrophoretic mobility shift assays (EMSA) with the probe fragments containing base pairs 4–417 or 389–835 (Fig. 1D). We did not observe any band retardation in the assay performed with fragment 4–417 (Fig. 1D). In contrast, fragment 389–835, which included the binding region defined in the yeast one-hybrid assays, produced a shifted band (Fig. 1D). Increasing the CBBP concentration resulted in additional shifted bands, suggesting formation of higher order complexes, which could result from multiple binding sites, CBBP protein multimerization (see below), or both. These results demonstrate that CBBP can bind to the b1 sequence in vitro.
CXC Domains Are Required for Interaction with DNA.
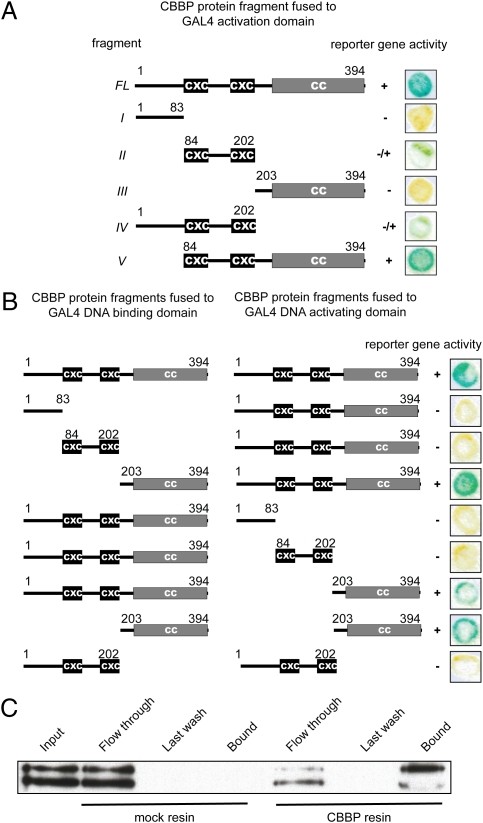
To identify the region mediating DNA binding in CBBP, we prepared a series of deletion proteins and tested them in the yeast one-hybrid assay (Fig. 2A). Only the fragment that contained the CXC domains (fragment II) showed detectible expression of the reporter gene. We therefore assayed two additional constructs containing CXC domains with either the N-terminal (fragment IV) or the C-terminal part with CC motif (fragment V). Only fragment V showed robust expression of the reporter gene. These data indicate that the CXC domain region is capable of mediating DNA binding and that the presence of the C terminus increased the assay signal significantly. Two possible roles for the C terminus could be stabilization of the protein structure or multimerization.
Fig. 2.
Deletion analysis of the CBBP protein. (A) CXC domain region of CBBP mediates DNA binding. Schematic representation of CBBP deletion fragments tested in yeast one-hybrid assays. Reporter gene activity in yeast one-hybrid assays is indicated to the right as + for active, +/− for weakly active and − for inactive. Respective yeast colony phenotypes are shown to the right. Western blots using the GAL4 activation domain indicated all proteins tested were equivalently expressed. (B) C-terminal region of CBBP mediates multimerization. Schematic representation of CBBP fragments used in yeast two-hybrid assays. Reporter gene expression for each combination of fusion proteins is indicated to the right as + for active or − for inactive, with representative yeast colony phenotypes shown. (C) CBBP is capable of multimerization in vitro. Recombinant CBBP protein was incubated with intein-CBP (mock resin) and CBBP-intein-CBP fusion proteins (CBBP resin) bound to the chitin beads. Ten percent of each: input, flow through, last wash, and bound fractions were separated by SDS/PAGE, and the CBBP protein was detected by Western blot. The two bands detected by αCBBP antibody are likely to represent some type of modification or slight proteolytic degradation. BSA used to block the beads is comigrating with the doublet in the bound fraction (last lane) and likely is influencing the signal of the larger band.
CBBP Can Form Multimers.
The presence of multiple bands in the gel-shift assay suggested that CBBP might be capable of multimerization. To test this hypothesis, we used the yeast two-hybrid assay (Fig. 2B). The experiment with the full-length protein fused to both the GAL4 DNA binding and GAL4 activation domain resulted in a positive signal, indicating multimer formation. For simplicity, we use the term multimers to mean at least dimers are formed. To identify the protein fragment mediating multimer formation, we used the deletion fragments indicated in Fig. 2A in combination with the full-length protein. Only the C-terminal fragment showed a positive signal indicating multimerization. To verify these results, we used recombinant CBBP protein in a pull-down assay with CBBP-Intein-Chitin Binding Domain (CBD) fusion protein bound to chitin beads (Fig. 2C). Intein-CBD protein bound to the beads was used as a negative control. In this assay, CBBP was able to form multimers, verifying the data obtained with the yeast two-hybrid assay and demonstrating CBBP multimers can form in the absence of DNA.
Transgenic Expression of cbbp Induces B-I Silencing.
To investigate the potential role of CBBP in b1 regulation and paramutation and to determine whether CBBP is bound to the b1 tandem repeats in vivo, we generated transgenic plants expressing FLAG-cbbp under control of the maize ubiquitin promoter. The original FLAG-cbbp transgenic lines were generated in a genetic background with b1 alleles that are not expressed, do not participate in paramutation, and contain a single copy of the repeat unit. For simplicity, these alleles are referred to as b. To investigate potential binding to the b1 tandem repeats and potential effects of CBBP on B-I expression, we crossed the FLAG-cbbp transgenic plants with B-I/B-Peru plants. The B-Peru allele contains one copy of the repeat unit, does not participate in paramutation, and does not accumulate significant anthocyanin in the plant body. B-I is dominant with respect to plant body pigmentation and results in dark purple plants because of a high transcription rate. At a low frequency, the B-I allele can spontaneously change to B′, the silent paramutagenic epiallelle. As stated previously, the B-I allele contains seven copies of the repeat unit.
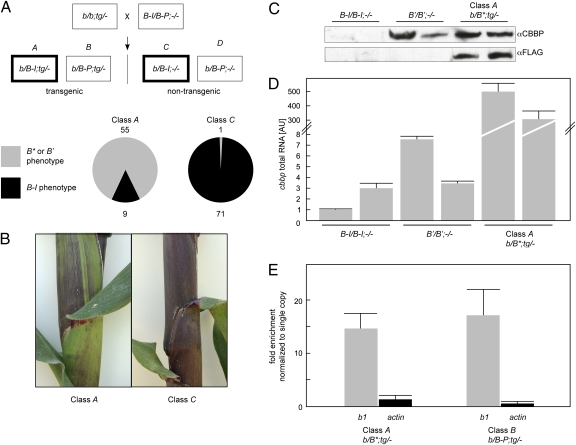
The cross produced four classes of progeny (Fig. 3A). To investigate the potential role of CBBP on B-I expression, we analyzed plant pigmentation in transgenic b/B-I progeny (class A). In these genetic backgrounds, the pigmentation phenotype is directly proportional to B-I transcription and reduced pigment is a phenotypic manifestation of B-I silencing. We analyzed 64 class A plants resulting from nine independent transgene integration events. Of these 64 plants, 55 showed reduced levels of anthocyanin (Fig. 3 A and B and Table S1). The high frequency of silencing observed was unlikely to be caused by spontaneous paramutation of B-I to B′, suggesting the presence of the FLAG-cbbp transgene-induced B-I silencing. To estimate spontaneous paramutation, we scored the pigment phenotype of the b/B-I;-/- nontransgenic siblings (class C). In those plants, the B-I allele was never exposed to the FLAG-cbbp transgene; therefore, silencing is because of spontaneous paramutation. Among the 72 plants analyzed, we found only one plant with reduced pigmentation (Fig. 3A and Table S1). These data suggested that the B-I silencing in the class A plants was occurring primarily because of the transgene. We termed the FLAG-cbbp-induced silent state B*.
Fig. 3.
Transgenic expression of FLAG-cbbp silences B-I. (A Top Upper) Crossing scheme. The parental genotypes are indicated in the crossing scheme with all expected progeny classes shown below. The progeny classes used in the experiment are indicated with a thicker box. Presence of the transgene is indicated by the abbreviation tg. Class A progeny revealed the FLAG-CBBP effect on B-I silencing, class C was a control for the frequency of spontaneous changes of B-I to B′. (A Bottom Lower) Summary of the pigment phenotypes in Class A and C progeny. B* is used to indicate a former B-I epiallele that is now silent. Total numbers of B* and B′ individuals (light plants) is indicated above the pie charts, and the total numbers of B-I individuals (dark plants) are indicated below the pie charts. (B) Typical pigmentation phenotypes of class A and class C plants. (C) Western blot analysis of total CBBP protein levels in two of the same individuals shown in D. (D) Total cbbp RNA in samples from class A and nontransgenic B-I and B′ seedlings was analyzed by quantitative RT-PCR, normalized to actin and the results are shown in arbitrary units (AU). Mean values from three independent experiments on individual seedlings from the same genotype or the same transgenic event are shown with error bars indicating SD. (E) ChIP assays were performed on sheath tissue from class A and class B plants by using αFLAG antibody. Quantitative PCR was used to measure FLAG-CBBP binding to the b1 repeats (gray bars) and actin (black bars). Fold enrichment is adjusted to the number of b1 repeat units in the respective genotypes to allow direct comparison between b1 genotypes and actin. Data are expressed as fold enrichment as compared with the no-antibody control. Mean values from two independent experiments are shown with error bars indicating SD. Respective genotypes are shown below.
To determine whether the observed phenotypic changes correlated with decreased B-I mRNA levels, we analyzed steady-state b1 transcript levels in selected class A individuals and in a control nontransgenic class C sibling by quantitative RT-PCR (Fig. S2A). A reduction in b1 mRNA levels (10- to 50-fold decrease) was observed in all three transgenic plants tested, indicating that the phenotypic changes in pigment levels were due to reduced b1 transcript levels. The same transgenic plants showed 100- to 600-fold increase in total cbbp mRNA levels as measured by qRT-PCR (Fig. S2B).
To verify that the transgene was translated, we performed Western blot analysis, which confirmed the presence of FLAG-tagged protein in the extracts (Fig. 3C). To estimate the relative CBBP protein levels in B-I, B′, and in transgenic class A plants, we performed a Western blot with αCBBP antibody (Fig. 3C), using seedling tissue that gave a relatively strong signal. The results indicated similar CBBP levels in B′ and transgenic plants despite much higher cbbp transcript levels in the same transgenic plants (Fig. 3D). B-I and B′ showed a similar range of cbbp mRNA levels (Fig. 3D), yet CBBP protein was undetectable in B-I plants (Fig. 3C), suggesting regulatory mechanisms operating at the protein synthesis or degradation levels. These data indicate that B-I silencing in B* plants correlates with production of CBBP protein from the transgene.
CBBP Interacts with the b1 Repeats in Vivo.
To determine whether FLAG-CBBP was bound to the b1 repeats in vivo, we used the chromatin immunoprecipitation (ChIP) assay with αFLAG antibody (Fig. 3E). Immunoprecipitated DNA was analyzed by quantitative PCR with primers specific to the previously defined CBBP binding region. Actin, which was not predicted to interact with CBBP, was used as an internal reference sequence to estimate the nonspecific background signal. To allow direct comparison with actin, which is a single copy gene, PCR signals obtained with primers specific to the CBBP binding region in class A plants were divided by four to account for the eight copies of the b1 repeat unit present in the diploid genome of this genotype (seven repeats in B* and one in b), relative to class B plants with two copies (one in B-Peru and one in b). A 15-fold enrichment was observed in the class A genotype relative to the no-antibody control reactions, indicating that FLAG-CBBP was associated with the b1 repeats in transgenic plants. Similar levels of CBBP binding per repeat unit in class A and class B indicated FLAG-CBBP can associate with a single repeat unit and does not require repeats to bind. This result indicates the junction fragment unique to repeats is not necessary to binding. In addition, because the overall binding was higher with more repeats, this result suggests that multiple CBBP proteins are associated with the multiple repeat alleles. Actin signal was very weak, indicating that the CBBP interactions with the b1 repeat unit were specific. We could not directly test whether CBBP is bound to the b1 repeats in nontransgenic B-I or B′ plants because we have been unable to obtain CBBP antibodies that are productive for ChIP assays.
FLAG-CBBP–Induced Silent State Is Heritable and Paramutagenic.
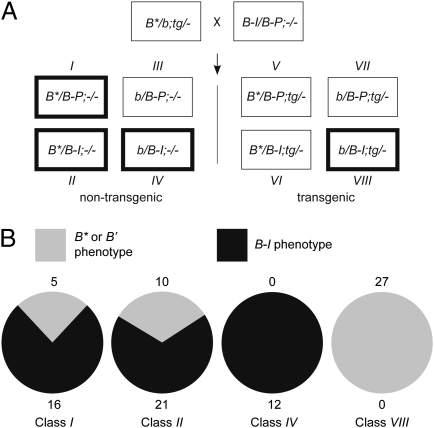
Paramutation establishes an extremely stably silenced B′ epiallele, as changes from B′ to B-I have never been observed in wild-type plants. To test whether the B* silent state is also heritable and paramutagenic, we crossed B*/b; tg/- (class A in previous experiment) plants with B-I/B-Peru;-/- plants (Fig. 4A). If the newly established silencing was not heritable in the absence of the FLAG-cbbp transgene, we would predict that the B* state would revert to a higher expressing B-I state, resulting in darkly pigmented plants in Class I progeny. If instead silencing established a heritably silenced epiallele, we would predict progeny plants with low b1 expression and light pigmentation in class I. Both light (B*) and dark (B-I) plants were observed in class I (Fig. 4B and Table S2), indicating that the silent B* state was heritable, but B* was not as stably silenced as B′.
Fig. 4.
Heritability and paramutagenicity of B* epiallele. (A) Parental genotypes are shown in the crossing scheme with all eight progeny classes shown below. The progeny classes used in the experiment are indicated by thick boxes. Class numbers are indicated above or below the boxes. Class I progeny was used to assay B* heritability. Class II was used to assay B* heritability and paramutagenicity. Class IV was used to monitor spontaneous paramutation frequency. Class VIII was used to assay for the continued ability of the transgene to induce silencing. (B) Phenotype frequencies in each of the progeny classes. Total numbers of B* and B′ individuals (lightly pigmented) is indicated above the pie charts and the total numbers of B-I individuals (darkly pigmented) are indicated below the pie charts.
Another distinctive attribute of the B′ epiallele is its ability to paramutate B-I. If the silenced B* epiallele, established in the FLAG-cbbp transgenic plants, was paramutagenic, lightly colored plants in the B*/BI;-/- (class II) progeny would be predicted from B* silencing the naïve B-I allele. In contrast, if B* was not paramutagenic, dark plants would be expected in B*/B-I;-/- class II progeny, even if B* remained stably silenced. We observed a significant number of light plants in Class II progeny, which could be due to B* being paramutagenic, B-I spontaneously changing to B′, or both. Class IV progeny served as a control to monitor the spontaneous paramutation frequency and none was observed in those families. These results demonstrate that transgenic FLAG-cbbp expression changed B-I to a silent epiallele B*, which functionally resembles paramutagenic B′, although silencing of B* is not as penetrant and B* is not as stably silenced as B′.
The b1 tandem repeats are differently methylated and show differential sensitivity to DNaseI in B′ relative to B-I, indicating different chromatin structures (10). To assess whether B* chromatin resembled B′, we analyzed DNA methylation levels at Sau96I sites within the b1 tandem repeats. In B′, the Sau96I sites are fully methylated, whereas in B-I a single Sau96I site is not methylated. In the three transgenic B* individuals tested, the DNA methylation levels at those sites were the same as B-I not B′ (Fig. S3), suggesting that B* is a distinct epiallele relative to B′.
Discussion
Yeast one-hybrid screening identified a CXC domain protein, CBBP, which was shown to interact with the b1 tandem repeats in vitro and in vivo. In addition to CBBP, we identified two homeodomain proteins that also bound to the b1 repeats in vitro. However, because these two proteins have not yet been further characterized, their functional significance for b1 paramutation is unknown. Transgenic expression of FLAG-cbbp can lead to reduced B-I transcript levels and conversion of B-I to a heritably silent and paramutagenic B* epiallele. These results provide a unique function for a CXC domain protein in heritable gene repression and paramutation. Below, we discuss models for the role of CBBP in b1 paramutation.
Potential Role for CBBP Multimer Formation in B-I Silencing.
Two interesting aspects of CBBP are its ability to bind to the B* tandem repeats, which have at least two binding sites per repeat unit, and its ability to form multimers. These properties might provide a repeat counting mechanism. The domain mediating multimer formation is the C-terminal region that contains the conserved CC motif, which is shared with some transposases. In these transposases, dimer formation is hypothesized to play a crucial role in transposase function (18) by bringing the inverted repeats to which the transposase binds in close proximity. By analogy, interactions of CBBP molecules bound to more than one repeat unit might lead to a chromatin structure that prevents access of transcription activators resulting in transcriptional repression. There is a quantitative response to the number of repeats (10) and more CBBP is bound to alleles with multiple repeats, thus one speculation is that CBBP binding to multiple repeats and formation of higher order multimers could provide a mechanism for counting the number of repeats. It is also possible that CBBP multimer formation could play a role in trans interactions between repeats on each chromosome, similar to that observed for Zeste in Drosophila (19, 20). With such models, the stability of the repressive chromatin might depend on the number of interacting CBBP molecules.
Model for CBBP-Mediated Repression.
Expression of FLAG-cbbp in transgenic plants frequently induced silencing of B-I. One possibility is that when CBBP accumulates to a certain level, as it does in B′ and in transgenic FLAG-cbbp plants, it can bind the b1 repeats and induce repressive chromatin states. CBBP might be a subunit of a chromatin modification complex or it could provide an interaction platform for a chromatin modification complex, a role hypothesized for two Drosophila CXC domain proteins (21, 22). Another possibility is that expression of CBBP from the ubiquitin promoter results in expression in distinct tissues or developmental times relative to the endogenous cbbp promoter, and this altered expression leads to silencing. The cbbp mRNA is expressed in numerous tissues (Fig. S4), but the observation that cbbp mRNA levels do not always predict protein levels will require numerous experiments to fully explore this possibility.
Model for Differential Regulation of CBBP.
Our results demonstrate that in seedlings, the CBBP protein is not detectable in B-I, yet it is present in B′, even though the cbbp mRNA levels are equivalent. This result suggests differential posttranscriptional control of CBBP in B-I relative to B′, in spite of B′ and B-I being epialleles in very similar genetic backgrounds. The observation that there is a similar amount of CBBP protein in the transgenic lines and in B′, yet transgenic lines have much higher mRNA levels, further suggests posttranscriptional regulation of CBBP is occurring, either at the protein synthesis or degradation levels. One possibility is that in B-I and B′, CBBP forms distinct complexes and it is more susceptible to degradation in the B-I versus B′ complexes. How this might occur is a mystery because B-I and B′ are essentially identical except for the chromatin and transcription differences at b1.
Our data show that the B* epiallele is paramutagenic in the absence of FLAG-cbbp, indicating that FLAG-CBBP is not required to maintain the B* state. One possibility could be because once the heritable B* chromatin state is established in the transgenic line, it can be maintained independent of CBBP. Consistent with this idea, CBBP protein was not detectable in Class I (B*/b nontransgenic lines). To date, CBBP protein has only been detected in B′ nontransgenic lines and in all of the transgenic FLAG-CBBP genotypes tested, including FLAG-CBBP transgenic stocks carrying the B-Peru/b alleles with no tandem repeats. In the transgenic lines, the FLAG tag could confer stability on CBBP, leading to a gain of function phenotype. It is also possible the increased amounts of transgene mRNA levels results in more of FLAG-CBBP protein than the putative regulatory pathway operating in B-I can process and, therefore, more CBBP protein accumulates. Additional experiments will be required to explore the role of CBBP in B-I silencing and to investigate the relation between CBBP and the RNAi components mediating paramutation.
Materials and Methods
Yeast One-Hybrid Library Screening.
The fragment from the B-I, B′ repeats containing 362–853 bp from one repeat unit and 1–361 bp from the next repeat unit was inserted into pHISi1 and pLacZi reporter plasmids (Clontech). A GAL4-AD fusion cDNA library was prepared from 5-day-old B′ seedling mRNA by Invitrogen. Individual clones (4 × 106) were screened according to the Clontech yeast one-hybrid system manual. Three different proteins were identified CBBP and two homeodomain proteins.
ExoIII/S1 Mapping of the CBBP Binding Region.
Deletions were introduced to pLacZi-based reporter plasmid with the repeat fragment (Fig. 1A) by using the ExoIII/S1 exonuclease kit (Fermentas). This procedure resulted in two series of constructs with deletions beginning from either end. Deletions were tested in the yeast one-hybrid assay for β-galactosidase by using protocols in the Clontech yeast one-hybrid system manual.
Recombinant CBBP Production and Purification.
To remove multiple rare codons present in the cbbp cDNA, the sequence was redesigned and synthetic DNA was ordered from Genescript. The nucleotide sequence is available upon request. cDNA was inserted into pTYB-2 (New England Biolabs) vector. Recombinant protein was expressed in ER322 E. coli cells and purified according to the manufacturer instructions. The protein identity was verified by mass spectrometry. Recombinant protein was ≈90% pure as assayed by SDS/PAGE.
Electrophoretic Mobility Shift Assays.
To generate 32P-labeled repeat DNA fragments, PCR was performed in the presence of 32P dATP by using P1 and P2 primers for the 1–417 b1 repeat fragment and P3 and P4 primers for the b1 repeat 389–835 fragment (Table S3). DNA (10 femtomoles) was incubated with recombinant CBBP (0.5–5 picomoles) in HEMG [20 mM Hepes-KOH (pH 7.8), 1 mM MgCl2, 1 mM DTT, 100 μg/mL BSA, 10% glycerol] supplemented with 100 KCl on ice for 20 min. A nonspecific competing DNA, Xenopus laevis AT-rich oocyte-type 5S rRNA gene cloned in pBR322 plasmid, was used at the concentration of 300 ng/μl. The reaction products were separated by electrophoresis in a 1.5% agarose 0.5× TBE gel and visualized with the phosphorimager.
Mapping of the CBBP Domains.
The cbbp cDNA fragments shown in Fig. 2 were amplified by PCR using P5-P6, P7-P8, P9-P10, P5-P8 and P7-P10 primer pairs, respectively (Table S3). PCR products were inserted into pDEST22 to generate the GAL4 activation domain fusions and pDEST32 to generate the GAL4 DNA binding domain fusions (Invitrogen). Yeast one- and two hybrid assays were performed according to the manufacturer's instructions.
Generation of Transgenic Plants.
The putative full-length cbbp cDNA was inserted into the pEARLEY M402 vector (K. Earley and C. Pikaard, unpublished data) that includes a N-terminal FLAG tag. HiII embryos were transformed using Agrobacterium tumefaciens at the Iowa State Transformation Facility (Ames, IA), and callus clones expressing FLAG-cbbp were regenerated. Transgene presence and b1 allele identity were determined by PCR using P11-P12, P13-14 or P15-16 primers, respectively (Table S3).
RNA Analysis.
cDNA first strand was synthesized by using ProtoScript II RT-PCR kit (New England Biolabs) according to manufacturer's instructions. Q-PCR was performed with the BioRad MyIQ Single-Color Real-Time PCR Detection System. Primers used were cbbp cDNA, primers P17-P18; actin cDNA, primers P19-P20; b1 cDNA, primers P21-P22 (Table S3). Experiments were done in triplicate and normalized to actin.
Immunoassays.
To generate αCBBP antibodies, two rabbits were immunized with VAPRESKKAAEVRG peptide. Antibody was produced at Pacific Immunology. Western blots were performed by using seedling tissue and ECL+ (GE Healthcare) according to the manufacturer's instructions.
ChIP was performed with sheath tissue as described (23) by using either M2 αFLAG or αCBBP antibodies; only M2 αFLAG antibodies yielded positive results. Chromatin was precipitated by using M2 αFLAG affinity gel (Sigma). IgG-agarose resin (Sigma) was used in the respective no-antibody control reactions. Precipitated DNA was analyzed by Q-PCR as described above. Primers P23-P24 and P19-20 were used to amplify CBBP binding region and actin, respectively, and Q-PCR signals were normalized to input DNA. To calculate the fold enrichment, Q-PCR background signals obtained in no-antibody control ChIP reactions were subtracted from respective reactions signals. Data were collected from two independent experiments.
For the pull-down assay, purified recombinant CBBP was incubated with either CBBP-Intein-Chitin Binding Domain fusion or Intein-Chitin Binding domain (mock control) bound to chitin beads (New England Biolabs) for 20 min in HEMG supplemented with 150 mM KCl without DTT. After washing, bound proteins were analyzed by Western blot with αCBBP antibody.
Supplementary Material
Acknowledgments
We thank Lyudmila Sidorenko for guidance on the maize field experiments, Rosa Jaime-Frias for providing maize stocks, Jack Gardiner for providing B73 tissues, and Mario Arteaga-Vazquez for permission to cite his unpublished data. This work was supported by National Institutes of Health Grant DPIOD575 (to V.L.C.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/1001576107/DCSupplemental.
References
- 1.Brink RA. A genetic change associated with the R Locus in maize which is directed and potentially reversible. Genetics. 1956;41:872–889. doi: 10.1093/genetics/41.6.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chandler VL. Paramutation: From maize to mice. Cell. 2007;128:641–645. doi: 10.1016/j.cell.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 3.Chandler VL, Stam M. Chromatin conversations: mechanisms and implications of paramutation. Nat Rev Genet. 2004;5:532–544. doi: 10.1038/nrg1378. [DOI] [PubMed] [Google Scholar]
- 4.Stam M, Mittelsten Scheid O. Paramutation: An encounter leaving a lasting impression. Trends Plant Sci. 2005;10:283–290. doi: 10.1016/j.tplants.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 5.Suter CM, Martin DI. Paramutation: The tip of an epigenetic iceberg? Trends Genet. November 27, 2009 doi: 10.1016/j.tig.2009.11.003. 10.1016/j.tig.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hale CJ, Stonaker JL, Gross SM, Hollick JB. A novel Snf2 protein maintains trans-generational regulatory states established by paramutation in maize. PLoS Biol. 2007;5:e275. doi: 10.1371/journal.pbio.0050275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alleman M, et al. An RNA-dependent RNA polymerase is required for paramutation in maize. Nature. 2006;442:295–298. doi: 10.1038/nature04884. [DOI] [PubMed] [Google Scholar]
- 8.Erhard KF, Jr, et al. RNA polymerase IV functions in paramutation in Zea mays. Science. 2009;323:1201–1205. doi: 10.1126/science.1164508. [DOI] [PubMed] [Google Scholar]
- 9.Sidorenko L, et al. A dominant mutation in mediator of paramutation2, one of three second-largest subunits of a plant-specific RNA polymerase, disrupts multiple siRNA silencing processes. PLoS Genet. 2009;5:e1000725. doi: 10.1371/journal.pgen.1000725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stam M, Belele C, Dorweiler JE, Chandler VL. Differential chromatin structure within a tandem array 100 kb upstream of the maize b1 locus is associated with paramutation. Genes Dev. 2002;16:1906–1918. doi: 10.1101/gad.1006702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Patterson GI, Thorpe CJ, Chandler VL. Paramutation, an allelic interaction, is associated with a stable and heritable reduction of transcription of the maize b regulatory gene. Genetics. 1993;135:881–894. doi: 10.1093/genetics/135.3.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coe EH. The properties, origin, and mechanism of conversion-type inheritance at the B locus in maize. Genetics. 1966;53:1035–1063. doi: 10.1093/genetics/53.6.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cvitanich C, et al. CPP1, a DNA-binding protein involved in the expression of a soybean leghemoglobin c3 gene. Proc Natl Acad Sci USA. 2000;97:8163–8168. doi: 10.1073/pnas.090468497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lewis PW, et al. Identification of a Drosophila Myb-E2F2/RBF transcriptional repressor complex. Genes Dev. 2004;18:2929–2940. doi: 10.1101/gad.1255204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Korenjak M, et al. Native E2F/RBF complexes contain Myb-interacting proteins and repress transcription of developmentally controlled E2F target genes. Cell. 2004;119:181–193. doi: 10.1016/j.cell.2004.09.034. [DOI] [PubMed] [Google Scholar]
- 16.Beall EL, et al. Discovery of tMAC: A Drosophila testis-specific meiotic arrest complex paralogous to Myb-Muv B. Genes Dev. 2007;21:904–919. doi: 10.1101/gad.1516607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang Z, et al. Molecular evolution of the CPP-like gene family in plants: insights from comparative genomics of Arabidopsis and rice. J Mol Evol. 2008;67:266–277. doi: 10.1007/s00239-008-9143-z. [DOI] [PubMed] [Google Scholar]
- 18.Trentmann SM, Saedler H, Gierl A. The transposable element En/Spm-encoded TNPA protein contains a DNA binding and a dimerization domain. Mol Gen Genet. 1993;238:201–208. doi: 10.1007/BF00279548. [DOI] [PubMed] [Google Scholar]
- 19.Duncan IW. Transvection effects in Drosophila. Annu Rev Genet. 2002;36:521–556. doi: 10.1146/annurev.genet.36.060402.100441. [DOI] [PubMed] [Google Scholar]
- 20.Bickel S, Pirrotta V. Self-association of the Drosophila zeste protein is responsible for transvection effects. EMBO J. 1999;9:2959–2967. doi: 10.1002/j.1460-2075.1990.tb07488.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang J, Benson E, Bausek N, Doggett K, White-Cooper H. Tombola, a tesmin/TSO1-family protein, regulates transcriptional activation in the Drosophila male germline and physically interacts with always early. Development. 2007;134:1549–1559. doi: 10.1242/dev.000521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Beall EL, et al. Role for a Drosophila Myb-containing protein complex in site-specific DNA replication. Nature. 2002;420:833–837. doi: 10.1038/nature01228. [DOI] [PubMed] [Google Scholar]
- 23.Haring M, et al. Chromatin immunoprecipitation: Optimization, quantitative analysis and data normalization. Plant Methods. 2007;3:11. doi: 10.1186/1746-4811-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.