Abstract
Studies here respond to two long-standing questions: Are human “pre/pro-B” CD34+CD10−CD19+ and “common lymphoid progenitor (CLP)/early-B” CD34+CD10+CD19− alternate precursors to “pro-B” CD34+CD19+CD10+ cells, and do the pro-B cells that arise from these progenitors belong to the same or distinct B-cell development pathways? Using flow cytometry, gene expression profiling, and Ig VH-D-JH sequencing, we monitor the initial 10 generations of development of sorted cord blood CD34highLineage− pluripotential progenitors growing in bone marrow S17 stroma cocultures. We show that (i) multipotent progenitors (CD34+CD45RA+CD10−CD19−) directly generate an initial wave of Pax5+TdT− “unilineage” pre/pro-B cells and a later wave of “multilineage” CLP/early-B cells and (ii) the cells generated in these successive stages act as precursors for distinct pro-B cells through two independent layered pathways. Studies by others have tracked the origin of B-lineage leukemias in elderly mice to the mouse B-1a pre/pro-B lineage, which lacks the TdT activity that diversifies the VH-D-JH Ig heavy chain joints found in the early-B or B-2 lineage. Here, we show a similar divergence in human B-cell development pathways between the Pax5+TdT− pre/pro-B differentiation pathway that gives rise to infant B-lineage leukemias and the early-B pathway.
Keywords: B1 and B2 cells; CD19, a Pax5 reporter stepwise acquisition; layered B-lymphocyte lineages; single-cell gene and protein expression profiles; umbilical cord blood
Whereas models of human B-lineage development propose a single hierarchical development pathway from CD34+ stem cells (SCs) to B cells (1–6), studies to track the order of emergence of B-cell differentiation stages in the mouse have documented distinct B-cell lineages, termed B-1 and B-2 (7–9).
Ordered stages in the current human model are termed the “common lymphoid progenitor” (CLP) and the “early-B,” “progenitor-B,” and “precursor-B” subsets that follow it (i.e., SC → CLP → early-B → pro-B → pre-B → B) (5). In this pathway, CLP and early-B stages, which both express the CD34+CD45RA+CD10+CD19− surface phenotype, are actually “multilineage” progenitors of B, T, dendritic (DC), and natural killer (NK) cells (5, 10–12).
Pro-B cells, which retain CD34 and CD10 expression but also express CD19 (CD34+CD10+CD19+), constitute the first Pax-5+ committed B-lineage stage, which is followed by pre-B (CD34−CD10+CD19+) cells that have ceased expression of CD34 and now express the surface μH-VpreB-λ5/CD79 surrogate Ig receptor complex (1–5, 13–15). Development of the latter CD34− pre-B cells and their B-cell and Ig-secreting plasma cell progeny can be reconstituted after coculture of purified CD34+Lineage− (Lin−) progenitors with bone marrow (BM) stroma lines (e.g., S17) (15–18).
Before the CLP subset and the above developmental pathway were recognized, a subset called “pre/pro-B,” which expresses the CD34+CD10−CD19+ surface phenotype, was commonly considered to be the first human B-lineage stage (19). It occurs in fetal liver, fetal bone marrow (FBM), and umbilical cord blood (CB) (19–21). An FBM pre/pro-B cell stage was shown to distinctively lack TdT and to bear CD7 unlike concurrent conventional CD10+CD19− B-cell progenitors, leading to the idea that these cells might belong to a distinct B lineage (20). This lack of TdT-mediated “N-nucleotide additions” between D and JH segments is further associated with differential V-D-J gene segment usage, enabling the creation of unique Ig repertoire specificities that develop before 2 months of age (22). Schroeder and coworkers (22) raised but did not pursue the possibility that the IgH rearrangements conducted in the absence of TdT may occur in B-cell progenitors distinct from CLP/early-B cells because the latter are TdT+/high from the beginning of lymphopoiesis onward (1–3, 12). The possibility that CLP/early-B and pre/pro-B cells are alternative pro-B-cell precursors in two distinct pathways has lain fallow, although, as we show here, this is indeed the case at least in CB CD34+ SC development.
Results
CD19 Expression Precedes CD10 Acquisition in the First Wave of B-Lineage Development from CB CD34highCD10−CD19− Progenitors in S17 BM Stroma Cocultures.
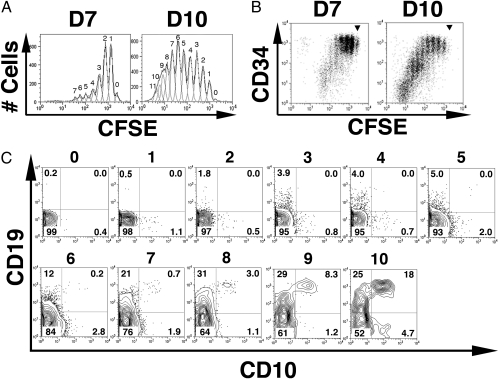
Carboxyfluorescein succinimidylester (CFSE) analysis was used to examine the order of CD10 and CD19 acquisition by CFSEhighCD34highCD10−CD19− “double-negative” progenitors purified from CB mononuclear cells as a function of cell division (23). Their growth and differentiation were promoted on progenitor coculture with S17 BM stroma (24), without added recombinant cytokines as described before (15). The emerging B-lineage stages were defined by CD10 and CD19 analyses gated in CD34 and CFSE expression levels, following conventions outlined in the introductory section of this study.
The CD34high double-negative cell progeny divided actively up to 7 or 11 times after 7 or 10 days, respectively (Fig. 1A), and the CD34+ cell divisions were developmentally asymmetrical (Fig. 1 B and C). Most cells in the first to fourth generations retained the original very bright CD34 levels typically found in human SCs, consistent with self-renewal of CD34highLin− cells (4, 16, 23). A few CD34 cells with intermediate expression levels (CD34int) and CD34high cells continued to divide but maintained their CD34 levels in the fifth to 11th generations. However, from the fourth generation onward, most CD34high cells started a stepwise differentiation wave toward CD34int, which was continued by most of the cells in the fifth to seventh generations and completed in the eighth generation.
Fig. 1.
In the first wave of development, CD34highCD10−CD19− precursors asymmetrically self-renew and differentiate to acquire CD19 before CD10 in S17 cocultures. CFSEhighCD34highCD10−CD19− precursors were purified by FACS (99.8 ± 0.1% pure, n = 8) from CB mononuclear cells loaded with CFSE; labeled with CD34-, CD10-, and CD19-specific antibodies; and cultured on S17 stroma for 7 days (D7) or 10 days (D10). The CFSEhighCD34highCD10−CD19− progeny was stained with CD34-, CD10-, and CD19-specific antibodies and submitted to flow cytometry analyses to quantify CFSE content (numbers on top of each peak designate the number of progeny divisions, with 0 representing no division) (A) and surface CD34 levels in each generation dot-plot analysis of CD34 expression vs. CFSE content (B), using the same samples as in A [position of undivided cells is shown (▾)]. (C) Day 10 expression of CD10 and CD19 in gated CD34+ cells ordered by their progeny generation, as defined by CFSE levels and indicated by numbers on top of the plot. Numbers in quadrants represent the frequency of positive cells. The input cell number was 104 cells per well, and average yields were 2.76 ± 0.9 × 104 cells per well at D7 and 15.04 ± 3.5 × 104 cells per well at D10. Cells in parallel wells were pooled, as indicated before for FACS analyses (18). Data are representative of the results from eight independent experiments.
Remarkably, that CD34int cell progeny started to acquire surface expression of either CD10 or CD19 and at different generations during the initial 10 divisions (Fig. 1C), with pre/pro-B cells developing before CLP/early-B cells. CD34 down-modulation in the fifth to seventh generations was paralleled by a wave of pre/pro-B cell genesis with characteristic stepwise acquisition of surface CD19 without CD10 expression (CD34intCD10−CD19low→int→high phenotypes in generations 6, 7, and 8, respectively, when these pre/pro-B cells accounted for approximately one-third of CD34+ cells). In contrast, CLP/early-B cells remained at low frequencies from the fifth to ninth generations and expanded from the 10th generation, even after a large number of pro-B (CD34+CD10+CD19+) cells had already emerged in the seventh to 10th generations and accounted for approximately one-fifth of the CD34+ cell progeny.
The time kinetic was optimized here to enable visualization of pre/pro-B, CLP/early-B, and pro-B cells, the CD34+ progenitors that were undetectable in prior reports (15–18). Although, many CD34int cells lose CD34 expression between the ninth and 11th generations, this CD34− progeny subset was only approximately one-fifth of cells in 10-day cultures and it included an ~25% pre-B-cell fraction (Fig. S1). The latter pre-B stage predominates after 20–30 days, however, when the pathway for cellular generation from CLPs is also evident (Fig. S2A). This is in agreement with the kinetics for the burst of conventional B-lineage development from fresh CB SCs or CLPs in other systems (17, 18). Indeed, fresh CB CLPs (CD34+CD10intCD19−) developed into an early-B-committed intermediate (CD10highCD19−) (11) and 45 ± 7% pro-B (CD10highCD19+) cells after 21 days (yield of 146.3 ± 15-fold initial seeded cells) (Fig. S2B).
CD34highLin− progeny proliferation was assayed in 20-day cultures by multiplex detection of CD10/CD19-staining and active DNA synthesis inspected 4 h after an EdU (a thymidine analogue) pulse in a FACS. Cultures proliferated actively, albeit at different rates by phenotype: pre/pro-B stage > pro-B > CLP (CD10−CD19high 41 ± 5.2% > CD10+CD19high 20 ± 3.5% > CD10intCD19− 7.44 ± 2.1%). DNA synthesis rates are not reported for pre/pro-B cells, but data may be compared with an average of 20–40% BM pre-B cells in S/G2 + M ex vivo phases or 25% in the fraction of large dividing CD34−CD19+ pre-B cells in vitro (1, 15). Collectively, it indicates a comparatively efficient B-lineage development from CD34+ CB progenitors in the S17 system (15, 24) that may involve S17-derived cytokines (16) and includes pre/pro-B-cell and CLP/early-B-cell differentiation with different onset kinetics.
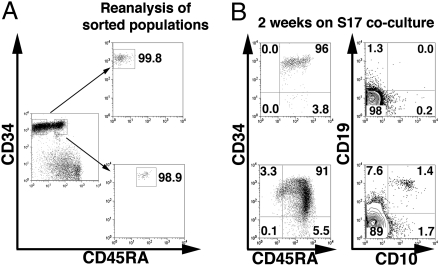
The above wave of pre/pro-B development before CLP/early-B-cell differentiation challenges the paradigm that the CLP is the sole pro-B-cell precursor (5). We next asked whether the proposed CLP or pre/pro-B stage could originate alternatively from the pluripotent CB precursors that further lack the CD45RA CLP marker (10). CD34highCD45RA−CD10−CD19− and CD34high-CD45RAlow/intCD10−CD19− cells (Fig. 2A, Left) were purified by FACS (Fig. 2A, Right), and their fate was assessed after 12–15 days in S17 cocultures (Fig. 2B). CD34highCD45RA−CD10−CD19− cells differentiated into CD34highCD45RAint and then CD34highCD45RAhigh intermediate populations (Fig. 2B, Upper Left), within which cells started to express either CD10 or CD19 in an alternative manner. Whereas pre/pro-B and CLP/early-B cells were scarce but readily evident, pro-B cells represented only 6 × 10−3 total CD34+CD45RA+ cells (Fig. 2B, Upper Right). Notably, when more differentiated CD34highCD45RAlow/intCD10−CD19− precursors were used (Fig. 2A, Bottom Right), they developed further into a CD34int/lowCD45RAhigh stage that showed markedly higher frequencies of pre/pro-B and CLP/early-B-cell progeny and also generated larger pro-B cell numbers (Fig. 2B, Lower). Collectively, pre/pro-B and CLP/early-B cells are in alternate B-lineage pathways branching after CD45RA acquisition, as defined by the order of CD10 and CD19 acquisition by individual cells from precursor–product assays.
Fig. 2.
CD34highCD45RA−Lin− pluripotent precursors develop into a CD34highCD45RAlow/intLin− intermediate stage that acquires either CD10 or CD19 in a mutually exclusive manner. (A) Magnetically enriched CB CD34high cells were labeled with antibodies against CD34, CD45RA, CD10, and CD19 antigens, and CD34highCD45RA−CD10−CD19− or CD34highCD45RAlow/intCD10−CD19− populations gated as indicated (Left) were sorted. (Right) Reanalysis and purity of sorted cells (99.9 ± 0.1% and 99.0 ± 0.2%, respectively; n = 4). (B) CD34, CD45RA, CD10, and CD19 expression patterns in CD34highCD45RA−Lin− (Upper) and CD34highCD45RAlow/intLin− (Lower) progenies, respectively, after 2 weeks of S17 cocultures. The input cell number was 103 cells per well, and the 2-week yield was 50.78 ± 6.9 × 103 or 304.38 ± 45 × 103 cells per well for CD34highCD45RA−CD10−CD19− or CD34highCD45RAlow/intCD10−CD19− progeny, respectively. Data are representative of four independent experiments.
Single-Cell Gene Expression Profiles and Precursor–Product Differentiation Assays Show That Unilineage Pre/Pro-B Cells Are in a Distinct Developmental Pathway Alternative to Multilineage CLPs.
The gene expression profiles and precursor potential of human pre/pro-B cells generated in vitro have not been examined. First, multiplex gene expression profiles of B-lineage commitment and specification genes were comparatively analyzed in single pre/pro-B, CLP/early-B, pro-B cells, and the CD34+ double negative precursors (Table 1) following methods reported for fresh CB and BM B-cell progenitors (2, 21). Day 13–16 S17 cocultures (from three distinct donors reported in Fig. 3) were chosen because all the subpopulations were represented in sufficient numbers at this culture time to be sorted with high purity into the PCR tubes.
Table 1.
Expression profiles of B-lineage commitment and specification genes in individual CD34+ sorted cells from CD34high/S17 cocultures during the initial wave of B-cell development (percentages of individual positive cells)
| CD34+10−19− | CD34+10+19− | CD34+10−19+ | CD34+10+19+ | |
| Pax5 | 4.10 | 6.67 | 89.47 | 88.23 |
| TdT | <1 | <1 | <1 | <1 |
| Rag-1 | <1 | 6.67 | 70.58 | 40.00 |
| VpreB | <1 | 6.67 | 76.47 | 66.67 |
| CD79a | 4.10 | 6.67 | 76.47 | 86.67 |
| VH | <1 | <1 | <1 | <1 |
Multiplex RT-PCR analyses of gene profiles of individual cells from the indicated populations were carried out in single cells sorted from 1- to 2-week CD34highLin− pluripotent precursor/S17 cocultures according to established protocols (18–36 cells per subpopulation from three independent donors, total of 75 cells) (2, 21). Results are expressed as the percentage of individual cells showing mRNA expression of the indicated gene among the total number of single cells analyzed in each population. Only samples showing GAPDH amplification were considered (21).
Fig. 3.
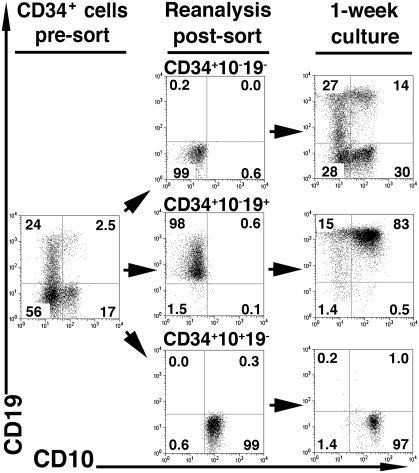
Precursor–product relations among CD34highLin− progeny developed in vitro delineate two distinct B lineage development pathways. Highly pure CB CD34highCD10−CD19− precursors were cocultured on S17 stroma for 13–16 days; labeled with antibodies to CD34, CD10, and CD19 to sort CD34+CD10−CD19−, CD34+CD10−CD19+, and CD34+CD10+CD19− progenitors (>99%, >99%, and 98.5 ± 0.3% pure on reanalysis, respectively); and replated on fresh S17 for an additional 1 week. (Middle) Purity of sorted populations. (Right) CD10 and CD19 expression in each sorted subset differentiated progeny. The input cell number was 103 cells per well, and 13-day yields were 289 ± 48 × 103, 224 ± 59 × 103, and 72 ± 14 × 103 cells per well for CD34+CD10−CD19−, CD34+CD10−CD19+, and CD34+CD10+CD19− progenitors, respectively. Data are representative of four independent experiments.
Results show that 9 of 10 single pre/pro-B and pro-B cells expressed Pax5 message and a high proportion of them also coexpressed Rag-1, VpreB, and CD79a mRNA but did not express recombined V@DJH message (VHDJH−, μH−) (Table 1) consistent with bona fide pre/pro-B and pro-B stages, respectively. The latter gene expression profiles are coincident with the mRNA expression patterns from fresh CB cells of the same phenotype (21). In contrast to pre/pro-B-cell and pro-B-cell homogeneous B-lineage profiles, only ~1/20 of the individual CLP/early-B cells analyzed in parallel expressed Pax5, Rag-1, VpreB, or CD79a, and many did it in stochastic combinations of some of those genes. These findings, together with CD3ε or pre-Tα mRNA expression patterns in this population (8.3% and 12.3%, respectively), define CD34+CD10+CD19− by phenotype and genotype as a CLP with a minor early-B-cell fraction (11, 21). This is again similar to the findings reported for fresh CB counterparts (21).
Aliquots of the purified CD34+ double-negative CLPs and pre/pro-B subpopulations (Fig. 3, Middle) were cultured to compare in parallel their gene expression profiles and their developmental potential in distinct differentiation conditions. In S17 cocultures, pre/pro-B cells showed increased CD19 surface expression and developed mainly into pro-B cells (Fig. 3, Right Middle). CLPs developed after 1 week of S17 reculture (Fig. 3, Right Bottom) into phenotypically homogeneous progeny of early-B cells (11). Finally, we show that the more primitive double-negative precursor population indeed contained progenitors that entered into the two alternate differentiation pathways proposed here: They generated either pre/pro-B cells that generate pro-B cells or alternatively developed into CLPs that differentiate after 1 week of culture in early-B cells that raises pro-B cells at 3 weeks (Fig. S2).
Recently, fresh CLPs (defined by a CD34+CD45RA+CD10low-CD19− phenotype) have been shown to have a multilineage potential with a low B-lineage-committed precursor frequency (i.e., ~1/87) (11). Accordingly, the CLPs generated here (also a CD34+CD45RA+CD10lowCD19− phenotype) are multipotent and give rise to NK cells or DCs after 6 days in IL-15 or GM-CSF + IL-4 conditions (10, 11, 17), respectively. Remarkably, the Pax-5+ pre/pro-B cells entered a cell death program under the same NK cell or DC standard development conditions (Figs. S3 and S4). Collectively, Pax-5+ pre/pro-B cell “unilineage” commitment reinforces the notion that they are in an alternate pathway to the CLP multilineage pathway that includes T-cell, NK cell, and DC precursors as well as some Pax-5+ early-B cells (10, 11, 21).
We also examined the patterns of CD7 expression in the two B lineages shown in the S17 system, because a fraction of FBM pre/pro-B cells defined by three-color analyses were reported to express a CD7+TdT− phenotype not found in the “conventional” B-lineage precursors (CD7−CD10+TdT+) from the same FBM samples (20). We found that during the initial wave of development in our cultures, CD7high expression profiles distinguished most of the pre/pro-B cells generated from the CLP/early-B cells (Fig. S5). Ordering of the CD34high → pre/pro-B → pro-B → pre-B stages after five-color analyses of live cultured cells revealed a developmental modulation of CD7 levels that coordinated with key CD34, CD19, and CD10 expression changes among the cells differentiated in vitro but could not be directly compared with the reported findings in FBMs (20) (Fig. S6). These findings led us to assess TdT expression and function directly in the proposed alternate pathway.
Regulation of TdT Expression in the Pre/Pro-B Pathway: Low Level of TdT-Mediated N-Nucleotide Insertion Associated with Differential V-D-JH Gene Usage.
The pre/pro-B and pro-B cells generated in 10-day S17 cultures did not express TdT mRNA (Table 1); this genotype differs in part from the fresh CB counterpart population, in which ~55–91% of cells bear TdT (21). The pre-B-cell progeny (CD34−CD10+CD19+) was 100% Pax5+ and expressed TdT in one-third of individual cells in 3-week cultures, and TdT levels increased steadily with culture age, as reported in gene expression studies and single-cell TdT staining analyses (15, 21).
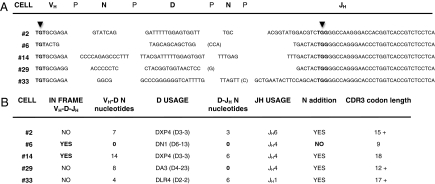
We previously sequenced VHDJH transcripts from single pre/pro-B cells circulating in CB, which included ~5% cμH+L− and 9–45% TdT− subsets (21). The IgH chain third complementary region (HCDR3), which includes the V@DJH and D@JH coding joints where TdT inserts “N-nucleotides,” had neither the typical fetal nor adult prototype HCDR3 patterns proposed for conventional CLP/early-B cells (3, 12, 21, 22). Here, we tested whether the HCDR3 in fresh CB pre/pro-B-cell progeny harbors VH@DJH rearrangements without N-additions, as occurs before 2 months of age; this process cannot be explained by the precursor cells in conventional CLP/early-B-cell pathways, as proposed by Schroeder and coworkers (22). We show additional HCDR3 sequences from single-cell progenitors from a distinct CB donor (Fig. 4). Collectively, approximately one-third of the individual pre/pro-B-lineage cells examined had VHDJH transcripts with no N-addition at the V@DJH or D@JH joints; this is typical of third-month fetal HCDR3 sequences (25). The average N-addition length between D and JH was approximately three nucleotides, similar to that reported for total fetal type HCDR3 with DJH usage associated with low N-addition numbers (22). This differs significantly from typical postnatal HCDR3, which shows extensive N-additions independent of D and JH usage (22, 25–27). As predicted, some joints without N-addition had “P-insertions” (short germline sequences palindromic to the coding joint ends of D-JH genes), because P-insertions occur when TdT is absent (28). Finally, the low TdT expression levels and functional activity in the CB pre/pro-B lineage was associated with frequent use of distal 5′-upstream DH segments and distant 3′-downstream JH elements. In addition, it did not include the JH proximal D7–27 (DQ52) gene, which is used preferentially by the fetal CLP/early-B-cell lineage (22). All these characteristics define D-JH usage strikingly similar to that of fetal μH transcripts with low N-insertion numbers (22, 26).
Fig. 4.
CB alternate B-cell progenitor HCDR3. (A) VH-D-JH sequences from individual cells of CD34+CD19+IgM− phenotype, with each segment contribution. Thirty-six cells were sorted and analyzed; all were GAPDH+Pax5+, but only 5 of 36 expressed mRNA transcripts for VH-D-JH rearrangements, a frequency of pre-B-I cells in this CB B-cell progenitor compartment (~14%) that is consistent with our prior report (21). Consensus HCDR3 definition is the interval between the marked (▾) TGT (92 cysteine) and TGG (103 tryptophan) codons (22). (B) Defined contribution to the sequence of VH segments, P junctions, TdT N-region insertions, D genes, and JH sequences to the HCDR3 length and diversity is depicted for every single cell. Sequences were analyzed using the ImMunoGeneTics software tools, which are freely available at http://www.imgt.org. That approximately three of five rearrangements were out of frame is a feature expected in sμH− B-cell precursors and confirms the progenitor and unselected nature of these cell repertoires (22).
Discussion
We show that pre/pro-B cells and CLP/early-B cells are alternate pro-B-cell precursors in two distinct B lineages using multiplex analyses of cell division, surface phenotypes, gene expression profiles, and HCDR3 sequences to visualize their precursor–product relations at the single-cell level. The first pathway is initiated from unilineage CD34+ pre/pro-B cells that acquire CD19 in a stepwise fashion before CD10 and contains cells that lack TdT in the initial wave of in vivo and in vitro B-cell development. This population accordingly harbors HCDR3 repertoires that include Igs with no N-addition in their D-JH joints and frequently uses distal 5′-upstream DH segments and distant 3′-downstream JH elements; these HCDR3 sequences are found neither in conventional late CLP/early-B-lineage cells nor in total repertoires generated after 2 months of age. The second pathway originates from multilineage CD34+ CLP/early-B cells that, in turn, are known to acquire CD10 before CD19 and express TdT from the beginning of CLP ontogeny onward.
Here, we used CB CD34+ SC and B-cell precursors, because the number and generative capacity of human CD34+ SC and B-cell progenitors in vitro and in vivo are greater in CB than in adult or pediatric BM; this has led to their extensive application in reparative medicine (16, 29). The primitive CD34+ SCs originate in subaortic patches in the aorta-gonad-mesonephros region, enter the blood circulation, colonize several embryo organs, and promote transient multifocal B-lymphopoiesis that features frequent TdT− pre-B cells and B-1a/CD5+ B cells; both in mice and humans, all these processes take place before the organogenesis of the BM cavities (6–8, 30–33).
The origin of CB B-cell progenitors is, however, unknown in both species (21, 34), but pre/pro-B cells and B-1a progenitors are rare or undetectable in adult BM (8, 35). In adult BM, human pre/pro-B-cell frequencies are well below those detected in CB (21, 35), and mouse B-1 cells belong to the B-1b lineage that bears TdT activity and are, by that criterion, like the B-2 lineage (8, 36). The mouse B-1a- and B-2-lineage biology is markedly different (7–9) and may resemble aspects of the human pre/pro-B and CLP/early-B-cell pathways, at least in CB and S17 in vitro systems analyzed, respectively.
Classical schemes of blood cell formation are described in terms of discrete developmental points, with a single route for each major cell type. Recent reports nonetheless show that the B-cell development process is much more plastic than originally thought and generates multiple B lineages with distinct biological features that are medically relevant (9, 37–39). Mouse B-1a cells have unilineage precursors that acquire CD19 before the conventional CLP marker appears days before B2-lineage onset in the fetus and show the absence of TdT-mediated N-nucleotide addition at their HCDR3 (7, 8, 36). This HCDR3 repertoire is essential for generating perinatal antibodies with nonrandom “germline-encoded” specificities (“canonical” Igs); in mice, these antibodies are needed for acquisition of complete protective immune defense from lethal bacterial pathogens and production of natural autoantibodies (37, 28). These findings suggest the need for studies to determine whether the pre/pro-B population has similar functions in humans.
HCDR3 inspection in adult B cells allowed mapping the origin of B-cell chronic leukemia to the fetal emergence of B-1a progenitors (39). The origin of human infant acute B-lymphoblastic leukemia (B-ALL) has been backtracked by others to an in utero transformation event following the detection of transformed preleukemic cells in CB years before clinical onset of disease (40). Recent results show that the actual frequency of Pax-5+ early-B-committed cells in the CLP population is lower than generally expected (11, 21); this, together with their T-cell potential, might explain the large proportion of DQ52-bearing rearrangements in acute T-lymphoblastic leukemia and their very low frequency in B-ALL, without requiring that DQ52 expression protect against B-ALL disease (22, 41). Although B-ALL is the most frequent pediatric cancer, the known phenotype and Ig repertoire of infant B-ALL could not be accommodated by any cell target in the CLP/early-B lineage model of human development (27). We propose that pre/pro-B cells are the normal infant B-ALL counterparts: Both are phenotypically CD34+CD10−CD19+ and harbor typical N-insertion-lacking HCDR3 sequences that we map to the initial pre/pro-B lineage wave.
Materials and Methods
Human Cells: Phenotypic Characterization and Sorting.
CB cells from term healthy donors were processed as before (21) after obtaining the mother's informed consent and the approval of the Alcala University Ethics Committee. CD34high cells were enriched by means of the CD34 MicroBead kit and AutoMACS (Miltenyi Biotec) following the manufacturer's instructions (96–98% purity). Antibodies were CD34-APC (clone 581), CD34-peridinin chlorophyll protein (PerCP)-Cy5.5 (8G12), CD19-phycoerythrin (PE) and CD19-allophycocyanin (APC) (HIB19), anti-HLA-DR-PerCP (L243), CD1a-APC (HI149), CD80-PE (L307.4), CD83-APC (HB15e), and CD56-PE (NCAM16.2) from Becton Dickinson; CD10-FITC and CD10-PE (MEM-78), CD45RA-FITC and CD45RA-PE-Cy5.5 (MEM-56), CD16-FITC (3G8), CD3-APC (54.1), and CD7-PE (clone 6B7; Invitrogen); biotin aμH (SA-DA4) and streptavidin-APC from Southern Biotech; and CD86-FITC (BU63) from Serotec. Isotype-matched antibodies (Becton Dickinson) were the background controls. Dead cells were excluded from analyses with 7-AAD or LIVE/DEAD-fixable-violet (Invitrogen). Multicolor analyses and sortings were done with a FACSCalibur or FACSAria (Becton Dickinson). FACS data were analyzed using FlowJo software (Tree Star) with “logicle” compensation and fluorescence-minus-one background controls (42).
CFSE and Click-It labeling.
CB mononuclear cells were resuspended in 37 °C prewarmed PBS at 107 cells/mL loaded with 1 μM CFSE (Vybrant CFDA SE cell tracer kit; Invitrogen) by incubation for 10 min at 37 °C protected from light. The reaction was stopped by adding cold 10% FBS. Ten CD34high/S17 cocultures of progenitors not labeled with CFSE were run in parallel to discard the dye's interference in reported progenitor development assays. Cell division generation numbers were defined with the FlowJo Proliferation Platform Tool (Tree Star). The Click-It/EdU proliferation assay (Invitrogen) was run as instructed by the manufacturer.
Progenitor Cell Cultures.
The S17 line (24) was maintained in complete medium: RPMI 1640 with glutamax (Lonza) and 5 × 10−5 M 2-mercaptoethanol (Fluka), plus 10% (vol/vol) FBS, at 37 °C in an 8% (vol/vol) CO2 atmosphere. B-cell differentiation conditions in progenitor/S17 cocultures were as reported (21) in complete medium plus 3% (vol/vol) FBS in 24-well plates (Falcon 353047; Becton Dickinson).
Single-Cell Multiplex RT-PCR.
Multiplex RT-PCR analysis was performed according to free full-text article protocols (2, 21). Sets of 75 individual cells from three independent CD34high/S17 cocultures were analyzed and always gave GAPDH+ amplification.
Supplementary Material
Acknowledgments
We thank Dr. Zapico and the staff of the Alcala University Hospital Department of Gynecology and Obstetrics for CB, Dr. Dorshkind (University of California at Los Angeles) for his generous gift of the S17 stroma cells, Dr. Rodriguez-Marcos (Centro de Biología Molecular) and Dr. Sanz (Rochester University) for their critical reading of the manuscript, and Dr. Mark (Centro de Biotecnología) for editorial assistance. This work was supported by the Ministries of Health (Grants FIS04/1889 and FIS/CIBEREHD), Research and Innovation (Grants GEN-2001-4856-C13-13 and SAF2004-8138), and Madrid State (Grant MITIC-CM-BIO-0189/2006). N.M.-A. was a Madrid State Research Fellow.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/cgi/content/full/0907942107/DCSupplemental.
References
- 1.Nishimoto N, et al. Normal pre-B cells express a receptor complex of μ heavy chains and surrogate light-chain proteins. Proc Natl Acad Sci USA. 1991;88:6284–6288. doi: 10.1073/pnas.88.14.6284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ghia P, et al. Ordering of human bone marrow B lymphocyte precursors by single-cell polymerase chain reaction analyses of the rearrangement status of the immunoglobulin H and L chain gene loci. J Exp Med. 1996;184:2217–2229. doi: 10.1084/jem.184.6.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Davi F, et al. Early onset of immunoglobulin heavy chain gene rearrangements in normal human bone marrow CD34+ cells. Blood. 1997;90:4014–4021. [PubMed] [Google Scholar]
- 4.Berenson RJ, et al. Antigen CD34+ marrow cells engraft lethally irradiated baboons. J Clin Invest. 1988;81:951–955. doi: 10.1172/JCI113409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.LeBien TW. Fates of human B-cell precursors. Blood. 2000;96:9–23. [PubMed] [Google Scholar]
- 6.Nuñez C, et al. B cells are generated throughout life in humans. J Immunol. 1996;156:866–872. [PubMed] [Google Scholar]
- 7.de Andrés B, et al. The first 3 days of B-cell development in the mouse embryo. Blood. 2002;100:4074–4081. doi: 10.1182/blood-2002-03-0809. [DOI] [PubMed] [Google Scholar]
- 8.Montecino-Rodriguez E, Leathers H, Dorshkind K. Identification of a B-1 B cell-specified progenitor. Nat Immunol. 2006;7:293–301. doi: 10.1038/ni1301. [DOI] [PubMed] [Google Scholar]
- 9.Herzenberg LA, Tung JW. B cell lineages: Documented at last! Nat Immunol. 2006;7:225–226. doi: 10.1038/ni0306-225. [DOI] [PubMed] [Google Scholar]
- 10.Galy A, Travis M, Cen D, Chen B. Human T, B, natural killer, and dendritic cells arise from a common bone marrow progenitor cell subset. Immunity. 1995;3:459–473. doi: 10.1016/1074-7613(95)90175-2. [DOI] [PubMed] [Google Scholar]
- 11.Six EM, et al. A human postnatal lymphoid progenitor capable of circulating and seeding the thymus. J Exp Med. 2007;204:3085–3093. doi: 10.1084/jem.20071003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bertrand FE, 3rd, et al. Ig D(H) gene segment transcription and rearrangement before surface expression of the pan-B-cell marker CD19 in normal human bone marrow. Blood. 1997;90:736–744. [PubMed] [Google Scholar]
- 13.Kozmik Z, Wang S, Dörfler P, Adams B, Busslinger M. The promoter of the CD19 gene is a target for the B-cell-specific transcription factor BSAP. Mol Cell Biol. 1992;12:2662–2672. doi: 10.1128/mcb.12.6.2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cobaleda C, Schebesta A, Delogu A, Busslinger M. Pax5: The guardian of B cell identity and function. Nat Immunol. 2007;8:463–470. doi: 10.1038/ni1454. [DOI] [PubMed] [Google Scholar]
- 15.Fluckiger AC, et al. In vitro reconstitution of human B-cell ontogeny: From CD34(+) multipotent progenitors to Ig-secreting cells. Blood. 1998;92:4509–4520. [PubMed] [Google Scholar]
- 16.Arakawa-Hoyt J, et al. The number and generative capacity of human B lymphocyte progenitors, measured in vitro and in vivo, is higher in umbilical cord blood than in adult or pediatric bone marrow. Bone Marrow Transplant. 1999;24:1167–1176. doi: 10.1038/sj.bmt.1702048. [DOI] [PubMed] [Google Scholar]
- 17.Rossi MI, et al. B lymphopoiesis is active throughout human life, but there are developmental age-related changes. Blood. 2003;101:576–584. doi: 10.1182/blood-2002-03-0896. [DOI] [PubMed] [Google Scholar]
- 18.Johnson SE, Shah N, Panoskaltsis-Mortari A, LeBien TW. Murine and human IL-7 activate STAT5 and induce proliferation of normal human pro-B cells. J Immunol. 2005;175:7325–7331. doi: 10.4049/jimmunol.175.11.7325. [DOI] [PubMed] [Google Scholar]
- 19.Nadler LM, et al. B cell origin of non-T cell acute lymphoblastic leukemia. A model for discrete stages of neoplastic and normal pre-B cell differentiation. J Clin Invest. 1984;74:332–340. doi: 10.1172/JCI111428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grümayer ER, Griesinger F, Hummell DS, Brunning RD, Kersey JH. Identification of novel B-lineage cells in human fetal bone marrow that coexpress CD7. Blood. 1991;77:64–68. [PubMed] [Google Scholar]
- 21.Sanz E, Alvarez-Mon M, Martínez-A C, de la Hera A. Human cord blood CD34+Pax-5+ B-cell progenitors: Single-cell analyses of their gene expression profiles. Blood. 2003;101:3424–3430. doi: 10.1182/blood-2002-07-2244. [DOI] [PubMed] [Google Scholar]
- 22.Shiokawa S, et al. IgM heavy chain complementarity-determining region 3 diversity is constrained by genetic and somatic mechanisms until two months after birth. J Immunol. 1999;162:6060–6070. [PubMed] [Google Scholar]
- 23.Oostendorp RA, Audet J, Miller C, Eaves CJ. Cell division tracking and expansion of hematopoietic long-term repopulating cells. Leukemia. 1999;13:499–501. doi: 10.1038/sj.leu.2401373. [DOI] [PubMed] [Google Scholar]
- 24.Collins LS, Dorshkind K. A stromal cell line from myeloid long-term bone marrow cultures can support myelopoiesis and B lymphopoiesis. J Immunol. 1987;138:1082–1087. [PubMed] [Google Scholar]
- 25.Sanz I. Multiple mechanisms participate in the generation of diversity of human H chain CDR3 regions. J Immunol. 1991;147:1720–1729. [PubMed] [Google Scholar]
- 26.Schroeder HW, Jr, Wang JY. Preferential utilization of conserved immunoglo-bulin heavy chain variable gene segments during human fetal life. Proc Natl Acad Sci USA. 1990;87:6146–6150. doi: 10.1073/pnas.87.16.6146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wasserman R, et al. Predominance of fetal type DJH joining in young children with B precursor lymphoblastic leukemia as evidence for an in utero transforming event. J Exp Med. 1992;176:1577–1581. doi: 10.1084/jem.176.6.1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tuaillon N, Capra JD. Evidence that terminal deoxynucleotidyltransferase expression plays a role in Ig heavy chain gene segment utilization. J Immunol. 2000;164:6387–6397. doi: 10.4049/jimmunol.164.12.6387. [DOI] [PubMed] [Google Scholar]
- 29.Broxmeyer HE, et al. High-efficiency recovery of functional hematopoietic progenitor and stem cells from human cord blood cryopreserved for 15 years. Proc Natl Acad Sci USA. 2003;100:645–650. doi: 10.1073/pnas.0237086100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tavian M, Hallais MF, Péault B. Emergence of intraembryonic hematopoietic precursors in the pre-liver human embryo. Development. 1999;126:793–803. doi: 10.1242/dev.126.4.793. [DOI] [PubMed] [Google Scholar]
- 31.Coulomb-L'Hermin A, et al. Stromal cell-derived factor 1 (SDF-1) and antenatal human B cell lymphopoiesis: Expression of SDF-1 by mesothelial cells and biliary ductal plate epithelial cells. Proc Natl Acad Sci USA. 1999;96:8585–8590. doi: 10.1073/pnas.96.15.8585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Solvason N, Kearney JF. The human fetal omentum: A site of B cell generation. J Exp Med. 1992;175:397–404. doi: 10.1084/jem.175.2.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bofill M, et al. Human B cell development. II. Subpopulations in the human fetus. J Immunol. 1985;134:1531–1538. [PubMed] [Google Scholar]
- 34.Rolink A, Haasner D, Nishikawa S, Melchers F. Changes in frequencies of clonable pre B cells during life in different lymphoid organs of mice. Blood. 1993;81:2290–2300. [PubMed] [Google Scholar]
- 35.Ciudad J, et al. Immunophenotypic analysis of CD19+ precursors in normal human adult bone marrow: Implications for minimal residual disease detection. Haematologica. 1998;83:1069–1075. [PubMed] [Google Scholar]
- 36.Holodick NE, Repetny K, Zhong X, Rothstein TL. Adult BM generates CD5+ B1 cells containing abundant N-region additions. Eur J Immunol. 2009;39:2383–2394. doi: 10.1002/eji.200838920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Benedict CL, Kearney JF. Increased junctional diversity in fetal B cells results in a loss of protective anti-phosphorylcholine antibodies in adult mice. Immunity. 1999;10:607–617. doi: 10.1016/s1074-7613(00)80060-6. [DOI] [PubMed] [Google Scholar]
- 38.Coutinho A, Kazatchkine MD, Avrameas S. Natural autoantibodies. Curr Opin Immunol. 1995;7:812–818. doi: 10.1016/0952-7915(95)80053-0. [DOI] [PubMed] [Google Scholar]
- 39.Gu H, Förster I, Rajewsky K. Sequence homologies, N sequence insertion and JH gene utilization in VHDJH joining: Implications for the joining mechanism and the ontogenetic timing of Ly1 B cell and B-CLL progenitor generation. EMBO J. 1990;9:2133–2140. doi: 10.1002/j.1460-2075.1990.tb07382.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Greaves M. Infection, immune responses and the aetiology of childhood leukaemia. Nat Rev Cancer. 2006;6:193–203. doi: 10.1038/nrc1816. [DOI] [PubMed] [Google Scholar]
- 41.Mizutani S, et al. Rearrangement of immunoglobulin heavy chain genes in human T leukaemic cells shows preferential utilization of the D segment (DQ52) nearest to the J region. EMBO J. 1986;5:3467–3473. doi: 10.1002/j.1460-2075.1986.tb04671.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Herzenberg LA, Tung J, Moore WA, Herzenberg LA, Parks DR. Interpreting flow cytometry data: A guide for the perplexed. Nat Immunol. 2006;7:681–685. doi: 10.1038/ni0706-681. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.