Abstract
The cytosolic domain of Notch is a membrane-tethered transcription factor. Ligand binding ultimately leads to γ-secretase cleavage within the transmembrane domain, allowing the intracellular domain to translocate to the nucleus and activate target gene transcription. Constitutive Notch signaling has been associated with human cancers such as T cell acute lymphoblastic leukemia (T-ALL). As tetraspanins have been implicated in many different signaling processes, we assessed their potential contribution to Notch signaling. We used a genetic assay in Caenorhabditis elegans to identify TSP-12 as a positive factor for Notch activity in several cellular contexts. Then, using a cell culture system, we showed that two human TSP-12 orthologs, TSPAN33 and TSPAN5, promote Notch activity and are likely to act at the γ-secretase cleavage step. We also acquired evidence for functional redundancy among tetraspanins in both C. elegans and human cells. Selective inhibition of tetraspanins may constitute an anti-NOTCH therapeutic approach to reduce γ-secretase activity.
Keywords: cancer, development, γ-secretase
The receptor Notch mediates cell–cell interactions that specify cell fate during animal development. The intracellular domain of Notch is a transcriptional coactivator that is tethered to the membrane until it is released by ligand-induced proteolytic processing events (reviewed in refs. 1, 2). Notch is present at the cell surface as a heterodimer between N- and C-terminal fragments formed by proteolytic cleavage at “Site 1” during transit of the receptor to the cell surface. When a ligand of the Delta/Serrate/LAG-2 family binds to the extracellular domain of Notch, the juxtamembrane “Site 2” region is exposed to a disintegrin and metalloprotease (ADAM) protease activity. ADAM cleavage and ectodomain shedding creates a substrate for the presenilin-containing γ-secretase complex, which subsequently cleaves the truncated Notch at “Site 3” within the transmembrane domain and releases the intracellular domain from its membrane tether. The intracellular domain then travels to the nucleus and forms a complex with the sequence-specific DNA binding protein CBF1/Suppressor of Hairless/LAG-1, Mastermind/SEL-8, and other factors, activating target gene transcription.
In Caenorhabditis elegans, missense mutations in the extracellular domain that cause constitutive or elevated Notch signaling result in distinctive developmental abnormalities and overt mutant phenotypes (3, 4). Genetic screens in C. elegans, conducted by identifying mutagen-induced suppressors of these mutant phenotypes, have yielded many core components and modulators of Notch signaling (reviewed in ref. 1). Characterization of such suppressors has illuminated the mechanism of Notch signal transduction and modes of regulation of Notch activity.
In mammals, constitutive or elevated Notch signaling is associated with cancers such as T cell acute lymphoblastic leukemia (T-ALL). Constitutive Notch activity in such cancers can result from missense mutations in the extracellular domain that, like the hyperactive C. elegans mutants mentioned above, promote Site 2 cleavage (5). Alternatively, constitutive Notch activity can result from the expression of truncated forms that mimic the Site 3 cleavage product (6). The identification of endogenous components that can be inhibited to reduce constitutive Notch activity is of interest due to the potential of such factors to serve as targets for anticancer therapies.
Here, we have taken a genetic suppressor approach in C. elegans to explore the potential contribution of tetraspanins to Notch signaling. Tetraspanins are defined by a characteristic protein topology, consisting of four transmembrane domains as well as a number of conserved amino acid residues (7). The tetraspanin superfamily is large: the C. elegans genome encodes 21 tetraspanins and the human genome encodes 33 tetraspanins. In animals, the tetraspanin proteins can be divided into four main subfamilies, each comprising several different groups of orthologs (8). Functional analysis has been relatively uninformative as to biological roles of tetraspanins because, with rare exceptions (9), removal of individual tetraspanins, or even multiple tetraspanins, has had undetectable or minimal effects in C. elegans (9), Drosophila (10), or mammalian immune cells (11, 12), possibly as a consequence of functional redundancy (e.g., ref. 10).
When we began our study, tetraspanins were attractive candidates for potential Notch modulators because they had been found to associate physically with several signaling proteins, including ADAM10 (13), and had been implicated in organizing signaling microdomains at the cell surface (reviewed in ref. 7). More recently, the tetraspanins CD81 and CD9 were found to associate in a complex with γ-secretase and promote cleavage of β-amyloid precursor protein (β-APP) (14), and various tetraspanins were found to associate with ADAM10 and also to promote cleavage of β-APP (15). However, until now, there has been no biochemical or functional evidence implicating tetraspanins in Notch signaling.
To assess tetraspanins for a potential role in Notch signaling, we used RNAi in C. elegans to deplete individual tetraspanins and found that depletion of tsp-12 alone suppressed the effects of a constitutively active form of Notch. We then performed further genetic characterization in both C. elegans and a human cell culture system. Our results implicate members of the “RDS” subfamily of tetraspanins as playing a conserved role in facilitating Notch signaling by promoting the γ-secretase cleavage step and provide evidence for significant functional redundancy among related tetraspanin proteins.
Results
Identification of tsp-12 as a Positive Factor for Notch Activity in the C. elegans Germ Line.
There are two C. elegans Notch proteins, LIN-12 and GLP-1. Missense mutations that alter the extracellular domain of either protein cause constitutive or elevated activity and distinctive mutant phenotypes (3, 16, 17). Because RNAi depletion of known components of the Notch signaling system robustly suppresses the sterility of the missense allele, glp-1(ar202), we used suppression of this allele to assess the effect of tetraspanin gene activity depletion on Notch activity.
glp-1(ar202) is temperature-sensitive; at 25 °C, nearly all worms are sterile with germline tumors (17). When “feeding RNAi” of each of the 21 individual tetraspanin genes was performed, only tsp-12(RNAi) suppressed this sterility. Suppression of glp-1(ar202) by loss of tsp-12 was confirmed by using tsp-12(ok239), a null allele of tsp-12 (Materials and Methods) (Table 1A). In addition, tsp-12(ok239) synergizes with the glp-1(bn18ts) hypomorphic allele in the reproductive system: at 20 °C, glp-1(bn18ts) is completely fertile, yet the glp-1(bn18); tsp-12(ok239) strain exhibits highly penetrant sterility (Table 1B). These results indicate that tsp-12 promotes Notch activity in the germ line.
Table 1.
tsp-12 and tsp-14 are positive factors for LIN-12/Notch and GLP-1/Notch activity in C. elegans
| A. Suppress glp-1(ar202) sterility | |
| Genotype | Fertile (suppressed) |
| glp-1(ar202) | 0/46 (0%) |
| glp-1(ar202); tsp-12(0) | 10/41 (24%) |
| B. Enhance glp-1(bn18) sterility | |
| Genotype | Sterile (enhanced) |
| Wild type | 0/29 (0%) |
| tsp-12(0) | 0/26 (0%) |
| glp-1(bn18) | 1/30 (3%) |
| glp-1(bn18); tsp-12(0) | 23/29 (79%) |
| C. Suppress lin-12(d) | |
| Genotype | Egl+ (suppressed)* |
| lin-12(n302) | 0/148 (0%) |
| lin-12(n302); tsp-12(0) | 14/126 (11%) |
| lin-12(n676) | 0/227 (0%) |
| lin-12(n676); tsp-12(0) | 5/253 (2%) |
| D. Enhance glp-1(bn18) and rescue with tsp-12(+) | |
| Genotype | Emb (enhanced) |
| Wild type | 1/752 (0%) |
| tsp-12(0) | 14/600 (2%) |
| glp-1(bn18) | 3/384 (1%) |
| glp-1(bn18); tsp-12(0) | 367/399 (92%) |
| glp-1(bn18); tsp-12(0); arIs119[tsp-12(+)] | 100/506 (20%) |
| glp-1(bn18); tsp-12(0); arIs120[tsp-12(+)] | 37/285 (13%) |
| glp-1(bn18); tsp-12(0); arIs135[lacks tsp-12(+)] | 395/428 (92%) |
| glp-1(bn18); tsp-12(0); arIs136[lacks tsp-12(+)] | 344/415 (83%) |
| E. Enhance glp-1(e2142) lethality | |
| Genotype | Emb (enhanced) |
| Wild type | 3/1190 (0%) |
| tsp-12(0) | 7/719 (1%) |
| glp-1(e2142) | 80/1091 (7%) |
| glp-1(e2142); tsp-12(0) | 311/695 (45%) |
| F. Synthetic lethal with sup-17 | |
| Genotype of segregants from sup-17; tsp-12(0)/dpy-20 | Viable and fertile |
| sup-17(n1258); dpy-20 | 35/66 (53%) |
| sup-17(n1258); tsp-12(0)/dpy-20 | 31/66 (47%)† |
| sup-17(n1258); tsp-12(0) | 0 (0%)† |
| G. Synthetic lethal with adm-4 | |
| Genotype | Emb (synthetic lethal) |
| adm-4(ok265) | 1/368 (0%) |
| tsp-12(0); adm-4(ok265) | 45/275 (16%) |
| H. Synthetic lethal with tsp-14 | |
| Genotype of segregants from tsp-12; tsp-14/dpy-8 | Obtained |
| tsp-12(0); tsp-14(0) | 0 (0%) |
| tsp-12(0); tsp-14(0)/dpy-8(e130) | 88 (64%) |
| tsp-12(0); dpy-8(e130) | 49 (36%) |
| I. tsp-14 suppresses glp-1(ar202) sterility | |
| Genotype | Fertile (suppressed) |
| glp-1(ar202) | 0/65 (0%) |
| glp-1(ar202); tsp-14(0) | 27/69 (39%) |
| glp-1(ar202); tsp-12(0) | 16/72 (22%) |
Eggs from bleached adults were placed on plates without food for 1 day at 20 °C. (A) Synchronized L1 worms were then cloned and placed at 24 °C for 3 days and plates scored for progeny. (B) Newly hatched worms were shifted from 15 °C to 20 °C. Adults were assayed for a Glp- phenotype after day 5. (C) Strains were maintained at 20 °C and scored in parallel. A two-tailed Fisher's Exact Test comparing lin-12(n302) to lin-12(n302); tsp-12(ok239) indicated that the difference is significant (P < 0.0001). However, in comparison of the stronger lin-12(n676) allele and lin-12(n676); tsp-12(ok239), the difference falls slightly below statistical significance (P = 0.063). (D) L4 worms were shifted from 15 °C to 20 °C, and progeny produced during two consecutive days were assessed. (E) L4 worms were kept at 15 °C, and progeny laid on four consecutive days were assessed. (F) Progeny laid by sup-17(n1258); tsp-12(ok239)/dpy-20(e1282) at 20 °C were cloned as L1 worms and kept at 20 °C. The dpy-20 mutation balances tsp-12(ok239). (G) Strains were maintained at 20 °C. (H) tsp-12(ok239); tsp-14(niDf7)/dpy-8(e130) adults laid eggs at 20 °C and progeny were assayed as in F. The dpy-8 mutation balances tsp-14(niDf7). (I) Experiment performed as in A.
*Sterile worms were excluded from scoring.
†21 worms arrested as larvae, died as adults, or laid a handful of dead progeny; these were presumed to be homozygous for both sup-17 and tsp-12(ok239). Moreover, the underrepresentation of heterozygotes suggests that loss of tsp-12 may be semidominant for inducing synthetic lethality.
tsp-12 Is a Positive Factor for Notch Activity in Other Cellular Contexts.
If tsp-12 plays a general role in Notch signaling, then tsp-12 deletion might be expected to reduce Notch activity in contexts other than the germ line. These roles might be revealed by genetic interactions with mutant alleles of glp-1 and lin-12, or with alleles of other genes in the Notch signal transduction pathway.
We examined the effect of tsp-12(ok239) on lin-12 activity in the somatic gonad, where LIN-12-mediated signaling between two defined somatic gonadal cells causes one cell to become a ventral uterine precursor cell (VU) and the other to become an anchor cell (AC) (1). In “lin-12(d)” mutants, which carry missense mutations that constitutively activate LIN-12, both cells become VUs and no AC is formed. Because the AC is required to induce vulval development, lin-12(d) mutants cannot lay eggs. We found that deletion of tsp-12 restored the ability of lin-12(d) mutants to lay eggs (Table 1C), indicating that an AC was made and constitutive LIN-12 activity was reduced in this somatic gonadal context. As lin-12(d) alleles result in ligand-independent activity in the AC/VU decision (3), these results also suggest that tsp-12 acts in the signal-receiving cell to promote Notch pathway activity (see below).
GLP-1 controls several cell fate decisions during embryonic development, so reducing glp-1 activity results in embryonic lethality (18). We found that tsp-12(ok239) synergizes with glp-1 partial loss-of-function alleles to cause embryonic lethality. glp-1(bn18) and tsp-12(ok239) single mutants exhibit a negligible embryonic lethality (Emb) phenotype at 20 °C, but glp-1(bn18); tsp-12(ok239) double mutants have a highly penetrant Emb phenotype (Table 1D). Similarly, at 15 °C, glp-1(e2142) is 7% Emb, but deletion of tsp-12 in a glp-1(e2142) background increases Emb penetrance to 45% (Table 1E).
Synergistic interactions between tsp-12(ok239) and mutations in other core components of the Notch signaling pathway are consistent with the interpretation that Notch activity is lowered in different contexts by tsp-12 deletion. Phenotypic enhancement was observed when tsp-12(ok239) was combined with a partial loss-of-function allele of sup-17, the ADAM10 ortholog and major Site 2 protease in C. elegans (Table 1F) and with a null allele of adm-4, which encodes the ortholog of mammalian ADAM17/TACE and appears to be partially redundant with sup-17 in some contexts (19) (Table 1G). Nearly complete lethality was also observed with a partial loss-of-function allele of sel-8, which encodes an essential nuclear factor that appears to play the role of mammalian Mastermind (20, 21).
Human TSPAN33, an Ortholog of C. elegans TSP-12, Is a Positive Factor for NOTCH1 Activity.
Reciprocal BLAST searches suggested that there are six likely human TSP-12 orthologs, listed here in decreasing order of relatedness: TSPAN5, TSPAN17, TSPAN14, TSPAN33, TSPAN15, and TSPAN10. The main regions of conservation between TSP-12 and these apparent human orthologs are in the intracellular loop and in the second extracellular loop, the location of many protein–protein interaction sites (7). This assessment of orthology is in agreement with a more extensive phylogenetic analysis, which places this set of proteins (as well as C. elegans TSP-12) into the retinal degeneration slow (RDS) subfamily of tetraspanins (8).
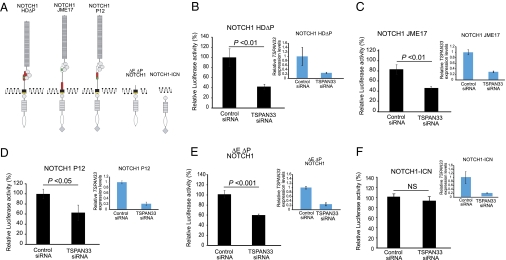
To assess whether TSP-12 orthologs play a conserved role in promoting Notch signaling, we tested the effect of TSPAN depletion on the activity of constitutively active NOTCH1 in human cells. We selected three different NOTCH1 oncogenic mutations found in T-ALL tumors that, like the C. elegans glp-1(ar202) and lin-12(d) mutations, have alterations in the heterodimerization (HD) domain causing ligand hypersensitive or ligand-independent activity (22, 23). NOTCH1 L1601P-ΔPEST combines a missense mutation in the HD domain with a truncating mutation in the intracellular PEST domain, and NOTCH1 Jurkat JME17 and NOTCH1 P12 ICHIKAWA each contain an in-frame insertion in the vicinity of the HD domain (Fig. 1A).
Fig. 1.
Inhibition of human TSPAN33 down-regulates NOTCH1 signaling (A) Structure of constitutively active NOTCH1 mutant proteins. The location of the NOTCH1 HD mutations is indicated in red; the PEST domain is represented by a diamond. (B–E) NOTCH/RBPJ luciferase reporter assays in HeLa cells expressing NOTCH1 L1601P-ΔPEST (NOTCH1 HD-ΔP) (B), NOTCH1 Jurkat JME17 (NOTCH1 JME17) (C), NOTCH1 P12 ICHIKAWA (NOTCH1 P12) (D), ΔEΔPEST-NOTCH1 (E), or the intracellular form of NOTCH1 (NOTCH1-ICN) (F) in the presence of a siRNA targeting TSPAN33 or a control inactive siRNA (control). NOTCH/RBPJ reporter activity after normalization with respect to the Renilla luciferase transfection control is as shown as relative levels compared with those in present in control siRNA transfected cells. Quantitative RT-PCR analysis of TSPAN33 demonstrating reduced levels of expression upon siRNA knockdown compared to siRNA control is shown in adjacent bar graphs on the right. Here and in Fig. 2, the error bars show SD; experiments with the T-ALL mutants were performed four times with single knock-down and two times with combined knock-down of TSPAN5 and TSPAN33; experiments with ΔEΔPEST-NOTCH1 and NOTCH1-ICN were performed respectively three and four times with both single and combined knock-down of TSPAN5 and TSPAN33. In addition, we performed statistical analysis on the measured reporter activity in control and RNAi cell populations using the Student's t test; values were considered statistically significant at P < 0.05. We note that comparable levels of TSPAN33 transcript reduction were achieved in each of the RNAi experiments shown. In addition, we note that for all experiments, the luciferase activity was in the linear range of detection; for the data presented here, the absolute value of luciferase activity in the control was 0.43 for NOTCH1-ICN and 0.87 for ΔE-NOTCH1, indicating that under these conditions, they were of comparable allelic strength.
Transfection of siRNAs targeting each of the four human tetraspanins most closely related to TSP-12 demonstrated adequate reduction in mRNA levels for only TSPAN5 and TSPAN33 in HeLa cells, so we focused on these tetraspanins (Figs. 1 and 2). We found that depletion of TSPAN33 caused a strong and reproducible reduction in the activity of all three NOTCH1 T-ALL mutant alleles (Fig. 1 B–D). These results suggest that TSPAN33 acts as positive factor promoting constitutive NOTCH1 activity. Single knockdown of TSPAN5 did not reproducibly reduce Notch pathway activity. However, experiments described below indicate that TSPAN5 plays a positive role in promoting Notch signaling that can be revealed when TSPAN33 is also depleted.
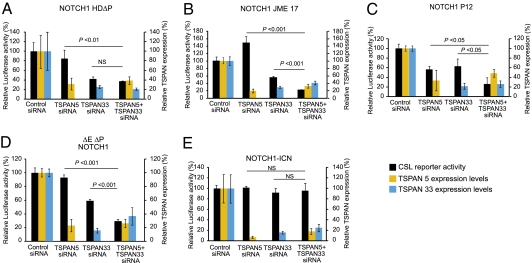
Fig. 2.
TSPAN33 and TSPAN5 redundantly down-regulate NOTCH1 activation. (A–E) NOTCH/RBPJ luciferase reporter assays in HeLa cells expressing NOTCH1 L1601P-ΔPEST (NOTCH1 HD-ΔP) (A), NOTCH1 Jurkat JME17 (NOTCH1 JME17) (B), NOTCH1 P12 ICHIKAWA (NOTCH1 P12) (C), ΔEΔPEST-NOTCH1 (D), or the intracellular form of NOTCH1 (NOTCH1-ICN) (E) in the presence of a siRNAs targeting TSPAN33, TSPAN5, or a control inactive siRNA (control). NOTCH/RBPJ reporter activity after normalization with respect to the Renilla luciferase transfection control is as shown as relative levels compared to those present in control siRNA transfected cells. Quantitative RT-PCR analysis of TSPAN5 (orange bars) and TSPAN33 (blue bars) demonstrating reduced levels of expression upon siRNA knockdown compared with siRNA control and similar knockdown levels in single SiRNA and double siRNA transfected cells is shown. The changes in NOTCH/RBPJ reporter activity in HeLa cells expressing the different NOTCH1 alleles in the presence of TSPAN5 knock-down did not reach statistical significance except for the P12 ICHIKAWA mutant.
TSPAN33 Facilitates the γ-Secretase Cleavage Step of Notch Activation.
Next, we assessed which step of constitutive Notch signal transduction was affected by TSPAN33 depletion by employing a functional assay previously used to demonstrate that the transmembrane cleavage of Notch depends on presenilin and nicastrin (24–27). This assay utilizes constitutively active truncated forms of Notch that mimic cleavage products produced during signal transduction (Fig. 1A). ΔEΔP-NOTCH1 contains a short extracellular stub and transmembrane tether for the intracellular domain, and therefore mimics the Site 2 cleavage product produced by the ADAM proteases; this form depends on γ-secretase, but not ADAM protease cleavage for activity. NOTCH-ICN1 consists of the intracellular domains of the receptor and is similar in form to the Site 3 γ-secretase-mediated cleavage product. This form bypasses the need for both the ADAM and the γ-secretase cleavage events. ΔEΔP-NOTCH1 also lacks the PEST domain and this additional stabilization makes ΔEΔP-NOTCH1 similar to NOTCH-ICN1 in inducing high levels of reporter activation in luciferase assays (Fig. 1).
We found that depletion of TSPAN33 by RNAi significantly reduced the activity of the γ-secretase substrate ΔEΔP-NOTCH1 but not the activity of the untethered NOTCH-ICN1 form (Fig. 1 E and F). The experimental conditions were such that the two Notch forms had comparable activity in the control siRNA cultures and similar reductions in TSPAN33 levels in the experimental siRNA cultures (Fig. 1). These results, together with our data showing that T-ALL NOTCH1 oncogenic mutants require TSPAN33 activity, implicate TSPAN33 as contributing to the γ-secretase cleavage of NOTCH1.
Evidence for Functional Redundancy Within the RDS Subfamily of Tetraspanins.
In C. elegans, the null allele tsp-12(ok239) does not have a discernable phenotype on its own. The lack of an overt phenotype may reflect functional redundancy with other tetraspanins and/or a modulatory (as opposed to a required) role for tsp-12 for Notch activity.
To explore possible functional redundancy between tsp-12 and other tetraspanins in C. elegans, we looked for synthetic phenotypes that appeared when individual tetraspanins were depleted by RNAi in the tsp-12(ok239) background. Preliminary results using RNAi suggested that tsp-14 may be functionally redundant with tsp-12, and this inference was confirmed using the available tsp-14(niDf7) null allele: we found that, while neither tsp-12(ok239) nor tsp-14(niDf7) has an overt phenotype, the double mutant displays a strong synthetic embryonic lethality (Table 1H), although we do not know whether this phenotype reflects a reduction in Notch activity per se. However, although tsp-14(RNAi) did not suppress glp-1(ar202) in our initial survey, the tsp-14(niDf7) null allele suppresses glp-1(ar202) (Table 1I), suggesting that tsp-14 promotes glp-1 activity in C. elegans germ line development.
We also explored potential functional redundancy between human TSPAN5 and TSPAN33. As described above, depletion of TSPAN33 reduces the activity of constitutively active forms of NOTCH1. We found that, although depletion of TSPAN5 alone had weak and/or variable effects on the activity of constitutively active Notch proteins across different experiments, depletion of TSPAN5 along with TSPAN33 often appeared to synergize to reduce the activity of membrane-bound, constitutively active forms (Fig. 2 A–D). Consistent with the interpretation that tetraspanin facilitates γ-secretase cleavage of Notch, combined TSPAN33 and TSPAN5 RNAi had no effect on the activity of the soluble NOTCH-ICN protein (Fig. 2E). Our results suggest that members of the RDS subfamily of tetraspanins play partially redundant roles in NOTCH1 γ-secretase processing and activation in humans.
Reduction of CD9 or CD81 Does Not Reduce the Activity of Leukemic Forms of NOTCH1.
Wakabayashi et al. (14) reported that two tetraspanins of the CD subfamily, CD9 and CD81, physically associate with γ-secretase and also showed that CD81 RNAi reduces γ-secretase processing of β-APP. We assessed the effect of depleting the activity of CD9 or CD81 individually in HeLa cells by RNAi and measured the effect on the activity of different leukemic mutant forms of NOTCH1 as above in two separate experiments. Overall, we observed no effect on NOTCH1 activity (Fig. S1) except for a small and variable redu-ction in the case of the NOTCH JME17 allele. These results suggest that, unlike TSPAN33, CD9 and CD81 are not likely to play individually significant roles in γ-secretase processing of NOTCH1.
Discussion
Our analysis indicates that C. elegans TSP-12 and its human ortholog TSPAN33 facilitate Notch signaling: reduction or loss of these tetraspanins suppresses the effects of constitutively active mutant Notch proteins, and loss of TSP-12 enhances the effects of hypomorphic mutations in several components of the Notch pathway. Moreover, under comparable conditions, loss of TSPAN33 suppresses signaling by a transmembrane form of activated Notch that requires γ-secretase activity but not the activity of a cytosolic form that bypasses the need for γ-secretase. Although tsp-12 appears to be expressed in somatic cells in addition to the germ line, all of the genetic interactions we have described are consistent with cell autonomous function of tetraspanins in Notch signal transduction in the receiving cell: in both systems, loss of the relevant tetraspanin suppresses the effects of constitutively active forms that do not require external ligand, as is particularly evident in the restoration of an anchor cell in lin-12(d) mutant backgrounds and in the reduction of constitutive Notch activity of ΔEΔP-NOTCH1 in the cell culture experiments. Our results taken together suggest that members of this tetraspanin subfamily facilitate γ-secretase cleavage of Notch.
We also provide evidence that members of the RDS subfamily of tetraspanins display functional redundancy. In C. elegans, loss of TSP-14, like loss of TSP-12, suppresses the sterility associated with abnormal proliferation of the germ line in hermaphrodites with elevated GLP-1/Notch activity, and concomitant loss of both TSP-12 and TSP-14 results in synthetic embryonic lethality. In human cells, concomitant reduction of TSPAN33 and TSPAN5 appears to suppress activated Notch synergistically. The redundancy that we infer from our experiments, which may be more extensive than assessed in this study, complicates a full evaluation of whether tetraspanins are core or modulatory components of the Notch signaling pathway.
Before considering possible molecular mechanisms by which these tetraspanins facilitate Notch activity, we note that our genetic results support the interpretation that these tetraspanins promote γ-secretase processing of Notch per se rather than promoting other signaling events occurring downstream of γ-secretase cleavage because the ΔEΔP-NOTCH1 transmembrane form (Site 2 cleavage mimic) and the ICN-NOTCH1 cytosolic form (Site 3 cleavage mimic) are affected differentially by TSPAN33 depletion. If tetraspanins were acting by promoting the activity of an intersecting signaling pathway that augments the ability of nuclear Notch to activate its targets, we would instead expect that depletion of tetraspanin should reduce the activity of both of these forms of Notch. Thus, although it is possible that TSPAN33 and TSPAN5 also promote the activity of other signaling pathways, we believe that such roles are not likely to be the cause of the effects on Notch activity that we have measured here.
Wakabayashi et al. (14) identified the tetraspanins CD9 and CD81 as proteins that physically associate with γ-secretase and promote γ-secretase processing of β-APP. Using the same conditions as we used for TSPAN33, we performed an initial assessment of the potential contribution of CD9 or CD81 to constitutive Notch activity and did not observe strong or consistent reduction on reporter activation by leukemic forms of NOTCH1. Although the limitations of RNAi and potentially extensive functional redundancy within and among tetraspanin subfamilies does not allow for a definitive conclusion, this observation suggests the intriguing possibility that different tetraspanin-mediated microdomains may favor γ-secretase cleavage of different substrates.
In view of the proposed role of tetraspanins in establishing or maintaining membrane microdomains, Wakabayashi et al. (14) suggested that CD9 and CD81 modulate the proteolytic activity of γ-secretase or its interaction with β-APP. We favor a similar model in which members of the RDS subfamily of tetraspanins promote γ-secretase activity through promoting physical association between γ-secretase and Notch in membrane microdomains. However, our genetic data do not rule out the possibility that tetraspanin affects one or more aspects of protein trafficking, so that only transmembrane forms of activated Notch would be vulnerable to TSPAN depletion; but even a more general effect on trafficking could ultimately modulate the accessibility of activated Notch to γ-secretase.
Our results implicating individual tetraspanins of the RDS subfamily in promoting constitutive Notch signaling are interesting from a potential therapeutic perspective, because targeting individual tetraspanins might partially reduce constitutive Notch activity without causing the major side effects associated with strongly and globally reducing Notch or presenilin activity using γ-secretase inhibitors. Monoclonal antibodies and RNAi have been proposed as possible therapeutic approaches to modulating signaling processes involving tetraspanins (28). Depending on the specific interactions in the putative complex that includes γ-secretase, specific tetraspanin(s) and Notch, it may also be possible to design specific small molecule or peptide inhibitors that block this interaction. Furthermore, if different tetraspanins do promote γ-secretase cleavage of different substrates selectively, it may be possible to develop γ-secretase inhibitors that are disease specific. For example, such inhibitors could distinctly target Alzheimer's disease or specifically treat Notch-driven tumors.
Materials and Methods
C. elegans Genetics.
The known Notch pathway alleles used in our studies are described in SI Materials and Methods. Experiments were performed at 20 °C unless otherwise indicated. Strains carrying glp-1(bn18) or glp-1(ar202) alleles were always maintained at 15 °C because these double mutants are particularly sensitive to temperatures above 15 °C, even for short periods of time and therefore were handled and transferred at this temperature to prevent sterility.
tsp-12(ok239) is an internal deletion of much of the coding region and part of the 5′ flanking region (www.wormbase.org). We confirmed that genetic interactions resulted from the lack of tsp-12 activity in this allele, and not from background mutations or polar effects on downstream genes in the predicted tsp-12 operon, by showing that the synthetic Emb phenotype of glp-1(bn18); tsp-12(ok239) is rescued by wild-type sequences provided by plasmid p809 present in transgenes arIs119 and arIs120 created by bombardment (see Supplemental Material). These tsp-12(+) arrays efficiently rescued the synthetic Emb phenotype of glp-1(bn18); tsp-12(ok239) (Table 1D). As a negative control for potential transgene marker effects, we used arrays arIs135 and arIs136; these arrays, which are similar to ar119 and arIs120, yet lack the tsp-12(+) coding region, did not rescue the glp-1(bn18); tsp-12(ok239) Emb phenotype (Table 1D).
tsp-14(niDf7) is a deletion/insertion allele (www.wormbase.org) resulting in the removal of at least three transmembrane domains from all isoforms and therefore is likely to be a null mutation. The synthetic lethality of tsp-12(ok239); tsp-14(niDf7) appeared to be rescued by arEx1243, a transgene that includes a fosmid carrying tsp-12(+).
Cells and Cell Culture.
Growth conditions and methods for RNAi and qPCR are given in SI Materials and Methods.
Notch1 Plasmid Constructs.
The pcDNA3 NOTCH1 L1601P-ΔPEST encodes a double mutant form of NOTCH1 with a missense mutation in the heterodimerization domain (HD) (substitution of L to P at position 1601) plus ΔPEST (truncation at position 2472) tagged with a FLAG tag epitope in the C terminus. The pcDNA3 NOTCH1 L1601P-ΔPEST construct was a gift from Dr. I. Aifantis at New York University, New York, NY (29). The pcDNA3 NOTCH1 Jurkat JME17 mutant was generated by cloning a partial NOTCH1 transcript (exons 19–29) amplified by PCR from Jurkat cells, which contains an internal tandem duplication of 51 bases within exon 28 of the NOTCH1 gene, into the unique Bam1H and NotI restriction sites of pcDNA3 NOTCH1. The pcDNA3 NOTCH1 P12 ICHIKAWA mutant was generated by cloning a partial NOTCH1 transcript (exons 19–29) amplified by PCR from P12-ICHIKAWA cells, which harbor an internal tandem duplication of 42 bases within exon 27 of the NOTCH1 gene, into the unique Bam1H and NotI restriction sites of pcDNA3 NOTCH1. Wild-type ICN1 (amino acids 1752–2545) was amplified from full-length human NOTCH1 by PCR and cloned in the unique EcoR1 and XhoI sites of pcDNA3. ΔEΔP-NOTCH1 was generated as previously described (30).
Luciferase Reporter Assays.
NOTCH1 expression plasmids (pcDNA3), control siRNA, TSPAN5, TSPAN33, CD9, and CD81 siRNA were transiently transfected using Lipofectamine 2000 (Invitrogen) transfection reagent together with the pGaLUC artificial luciferase reporter construct (31) (a gift from Dr. Honjo at Kyoto University, Kyoto, Japan), which contains six tandem CSL binding sites. pRL, a vector that expresses the Renilla luciferase gene under the control of the CMV promoter, was used as internal control. Total DNA was kept constant by adding empty vector as needed. All transfections were carried out in triplicate. Cell lysates were harvested 48 h posttransfection and luciferase assays were carried out using the Dual Luciferase Assay System (Promega) on a LUMAT LB 9507 luminometer (Berthold Technologies).
Supplementary Material
Acknowledgments
We thank Paul Fox and Tim Schedl for help and advice; Mark Edgley for his helpful analysis of tsp-14(niDf7), and the C. elegans Gene Knockout Consortium for tsp-12(ok239). Some of the strains used in this study were provided by the Caenorhabditis Genetics Center (CGC), which is supported by the National Institutes of Health-National Center for Research Resources. We gratefully acknowledge the expert technical assistance of Michael Hadler, Xinlan Zhou, and Richard Ruiz. We are indebted to Gary Struhl, Dan Shaye, Rafi Kopan, Tim Schedl, and David Greenstein for helpful comments on the manuscript. This work was supported by the National Institutes of Health (R01CA095389 to I.G. and R01CA120196 to A.F.) and the Leukemia and Lymphoma Society (Grants 1287-08 and 6237-08 to A.F.). C.D.D. was a postdoctoral associate and I.G. is an investigator of the Howard Hughes Medical Institute. A.F. is a Leukemia & Lymphoma Society Scholar.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/1001647107/DCSupplemental.
References
- 1.Greenwald I. LIN-12/Notch signaling in C. elegans. WormBook. 2005;8:1–16. doi: 10.1895/wormbook.1.10.1. Aug. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kopan R, Ilagan MX. The canonical Notch signaling pathway: Unfolding the activation mechanism. Cell. 2009;137:216–233. doi: 10.1016/j.cell.2009.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Greenwald I, Seydoux G. Analysis of gain-of-function mutations of the lin-12 gene of Caenorhabditis elegans. Nature. 1990;346:197–199. doi: 10.1038/346197a0. [DOI] [PubMed] [Google Scholar]
- 4.Greenwald IS, Sternberg PW, Horvitz HR. The lin-12 locus specifies cell fates in Caenorhabditis elegans. Cell. 1983;34:435–444. doi: 10.1016/0092-8674(83)90377-x. [DOI] [PubMed] [Google Scholar]
- 5.Weng AP, et al. Activating mutations of NOTCH1 in human T cell acute lymphoblastic leukemia. Science. 2004;306:269–271. doi: 10.1126/science.1102160. [DOI] [PubMed] [Google Scholar]
- 6.Ellisen LW, et al. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 1991;66:649–661. doi: 10.1016/0092-8674(91)90111-b. [DOI] [PubMed] [Google Scholar]
- 7.Hemler ME. Tetraspanin functions and associated microdomains. Nat Rev Mol Cell Biol. 2005;6:801–811. doi: 10.1038/nrm1736. [DOI] [PubMed] [Google Scholar]
- 8.Garcia-España A, et al. Origin of the tetraspanin uroplakins and their co-evolution with associated proteins: Implications for uroplakin structure and function. Mol Phylogenet Evol. 2006;41:355–367. doi: 10.1016/j.ympev.2006.04.023. [DOI] [PubMed] [Google Scholar]
- 9.Moribe H, et al. Tetraspanin protein (TSP-15) is required for epidermal integrity in Caenorhabditis elegans. J Cell Sci. 2004;117:5209–5220. doi: 10.1242/jcs.01403. [DOI] [PubMed] [Google Scholar]
- 10.Fradkin LG, Kamphorst JT, DiAntonio A, Goodman CS, Noordermeer JN. Genomewide analysis of the Drosophila tetraspanins reveals a subset with similar function in the formation of the embryonic synapse. Proc Natl Acad Sci USA. 2002;99:13663–13668. doi: 10.1073/pnas.212511099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Spriel AB, et al. A regulatory role for CD37 in T cell proliferation. J Immunol. 2004;172:2953–2961. doi: 10.4049/jimmunol.172.5.2953. [DOI] [PubMed] [Google Scholar]
- 12.Knobeloch KP, et al. Targeted inactivation of the tetraspanin CD37 impairs T-cell-dependent B-cell response under suboptimal costimulatory conditions. Mol Cell Biol. 2000;20:5363–5369. doi: 10.1128/mcb.20.15.5363-5369.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yan Y, Shirakabe K, Werb Z. The metalloprotease Kuzbanian (ADAM10) mediates the transactivation of EGF receptor by G protein-coupled receptors. J Cell Biol. 2002;158:221–226. doi: 10.1083/jcb.200112026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wakabayashi T, et al. Analysis of the gamma-secretase interactome and validation of its association with tetraspanin-enriched microdomains. Nat Cell Biol. 2009;11:1340–1346. doi: 10.1038/ncb1978. [DOI] [PubMed] [Google Scholar]
- 15.Xu D, Sharma C, Hemler ME. Tetraspanin12 regulates ADAM10-dependent cleavage of amyloid precursor protein. FASEB J. 2009;23:3674–3681. doi: 10.1096/fj.09-133462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berry LW, Westlund B, Schedl T. Germ-line tumor formation caused by activation of glp-1, a Caenorhabditis elegans member of the Notch family of receptors. Develop-ment. 1997;124:925–936. doi: 10.1242/dev.124.4.925. [DOI] [PubMed] [Google Scholar]
- 17.Pepper AS, Killian DJ, Hubbard EJ. Genetic analysis of Caenorhabditis elegans glp-1 mutants suggests receptor interaction or competition. Genetics. 2003;163:115–132. doi: 10.1093/genetics/163.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Priess JR. Notch signaling in the C. elegans embryo. WormBook. 2005;June 25:1–16. doi: 10.1895/wormbook.1.4.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jarriault S, Greenwald I. Evidence for functional redundancy between C. elegans ADAM proteins SUP-17/Kuzbanian and ADM-4/TACE. Dev Biol. 2005;287:1–10. doi: 10.1016/j.ydbio.2005.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doyle TG, Wen C, Greenwald I. SEL-8, a nuclear protein required for LIN-12 and GLP-1 signaling in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2000;97:7877–7881. doi: 10.1073/pnas.97.14.7877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Petcherski AG, Kimble J. LAG-3 is a putative transcriptional activator in the C. elegans Notch pathway. Nature. 2000;405:364–368. doi: 10.1038/35012645. [DOI] [PubMed] [Google Scholar]
- 22.Malecki MJ, et al. Leukemia-associated mutations within the NOTCH1 heterodimerization domain fall into at least two distinct mechanistic classes. Mol Cell Biol. 2006;26:4642–4651. doi: 10.1128/MCB.01655-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sulis ML, et al. NOTCH1 extracellular juxtamembrane expansion mutations in T-ALL. Blood. 2008;112:733–740. doi: 10.1182/blood-2007-12-130096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Strooper B, et al. A presenilin-1-dependent gamma-secretase-like protease mediates release of Notch intracellular domain. Nature. 1999;398:518–522. doi: 10.1038/19083. [DOI] [PubMed] [Google Scholar]
- 25.Struhl G, Greenwald I. Presenilin is required for activity and nuclear access of Notch in Drosophila. Nature. 1999;398:522–525. doi: 10.1038/19091. [DOI] [PubMed] [Google Scholar]
- 26.Struhl G, Greenwald I. Presenilin-mediated transmembrane cleavage is required for Notch signal transduction in Drosophila. Proc Natl Acad Sci USA. 2001;98:229–234. doi: 10.1073/pnas.011530298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chung HM, Struhl G. Nicastrin is required for Presenilin-mediated transmem-brane cleavage in Drosophila. Nat Cell Biol. 2001;3:1129–1132. doi: 10.1038/ncb1201-1129. [DOI] [PubMed] [Google Scholar]
- 28.Hemler ME. Targeting of tetraspanin proteins–potential benefits and strategies. Nat Rev Drug Discov. 2008;7:747–758. doi: 10.1038/nrd2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thompson BJ, et al. The SCFFBW7 ubiquitin ligase complex as a tumor suppressor in T cell leukemia. J Exp Med. 2007;204:1825–1835. doi: 10.1084/jem.20070872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kopan R, Schroeter EH, Weintraub H, Nye JS. Signal transduction by activated mNotch: Importance of proteolytic processing and its regulation by the extracellular domain. Proc Natl Acad Sci USA. 1996;93:1683–1688. doi: 10.1073/pnas.93.4.1683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sakai T, et al. Functional replacement of the intracellular region of the Notch1 receptor by Epstein-Barr virus nuclear antigen 2. J Virol. 1998;72:6034–6039. doi: 10.1128/jvi.72.7.6034-6039.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.