Abstract
Regulatory T (Treg) cells that express the Foxp3 transcription factor are essential for lymphoid homeostasis and immune tolerance to self. Other nonimmunological functions of Treg cells, such as controlling metabolic function in adipose tissue, are also emerging. Treg cells originate primarily in the thymus, but can also be elicited from conventional T cells by in vivo exposure to low-dose antigen or homeostatic expansion or by activation in the presence of TGFβ in vitro. Treg cells are characterized by a distinct transcriptional signature controlled in part, but not solely, by Foxp3. For a better perspective on transcriptional control in Treg cells, we compared gene expression profiles of a broad panel of Treg cells from various origins or anatomical locations. Treg cells generated by different means form different subphenotypes and were identifiable by particular combinations of transcripts, none of which fully encompassed the entire Treg signature. Molecules involved in Treg cell effector function, chemokine receptors, and the transcription factors that control them were differentially represented in these subphenotypes. Treg cells from the gut proved dissimilar to cells elicited by exposure to TGFβ in vitro, but instead they resembled a CD103+Klrg1+ subphenotype preferentially generated in response to lymphopenia.
Keywords: Foxp3, microarray
Regulatory T (Treg) cells characterized by stable expression of the Foxp3 transcription factor are involved in the maintenance of lymphoid homeostasis in a number of immunological contexts: They maintain tolerance to self and control autoimmune deviation, help regulate responses to pathogens or allergens, and help maintain a balance with commensal microbial flora (1–4). Although other T cell lineages may also partake in such regulatory functions (5), the central role played by Foxp3+ Treg cells is highlighted by the devastating multiorgan autoimmune disease that appears in Foxp3-deficient scurfy mice or human patients (6).
In keeping with their multiple impacts, several molecular mediators of Treg cell activities have been described, although the actual in vivo relevance and relative importance of these mechanisms have yet to be clearly demarcated (7, 8). More recently, targeted gene ablation in Treg cells demonstrated that the control of particular effector functions in conventional T (Tconv) cells requires distinct programs in Treg cells (9–11). Interestingly, these programs appear to involve the same controlling factors in Treg cells and in the Tconv cells they regulate (e.g., Irf4 in Th2 cells and in the Treg cells that control them) (9).
Two distinct origins for Foxp3+ cells have been reported. First, Foxp3+ cells are generated in the thymus as an alternative lineage at the time of positive selection into the conventional CD4+ T cell lineage (12, 13). These thymic Foxp3+ cells have a distinctive T cell receptor (TCR) repertoire that distinguishes them from Tconv cells, and these TCRs track to Treg cell pools in peripheral lymphoid organs, where they constitute the majority, if not all, of the Treg pool (14–16). Second, mature CD4+ T cells from peripheral lymphoid organs can be converted experimentally to Foxp3 positivity under a variety of conditions in vivo: chronic suboptimal stimulation by agonist peptide (17–19), exposure to orally administered agonist (20–22), or during lymphopenia-driven homeostatic expansion (23–25). These in vivo-converted cells are functionally effective in several suppression assays (17, 22, 23, 25). In addition, naive CD4+ Tconv cells activated in vitro in the presence of IL-2 and TGFβ induce Foxp3 and acquire some characteristics of Treg cells, including some suppressive properties (26–28); on the other hand, the phenotype of TGFβ-induced Treg cells is unstable (29), these cells are not suppressive in all assays, and converted cells acquire only a segment of the Treg transcriptional signature (30).
That naive Tconv cells can induce Foxp3 de novo led to the suggestion that such conversion might be an important element in dampening immune responses to self or to foreign antigens, the generation of new regulatory cells acting as an immediate negative feedback on an inflammatory response. On the other hand, the true contribution of such converted cells to the composition and function of Treg cell pools in peripheral lymphoid organs or in inflamed tissues remains unclear. Recent evidence suggests that deletion of conserved noncoding DNA elements within the Foxp3 promoter can be used to track some of these converted populations, whose presence may be limited to specific anatomic locations such as the gut-associated lymphoid tissues (31). To more precisely delineate the types of Foxp3+ cells elicited by conversion in different circumstances, we performed a broad gene-expression profiling study of Foxp3+ cells. We aimed to determine whether in vivo conversion of CD4+ Tconv cells could fully reproduce the transcriptional signature of normal Treg cells isolated from unmanipulated tissues and to assess the genomic heterogeneity of normal Treg cell pools. The composite data, which can be browsed in extenso via a unique web display (http://cbdm.hms.harvard.edu/TregSubphenotypes/heatmap.html), argue for a marked heterogeneity between different populations. The subphenotypes could also be distinguished among Treg cells of secondary lymphoid organs and gut tissue, albeit not in the expected manner.
Results
Heterogeneity in Expression of Treg Cell Signature Genes in Foxp3+ T Cells.
CD4+Foxp3+ Treg cells isolated from lymph nodes (LNs) and spleen have a characteristic and reproducible gene-expression profile when compared to Foxp3−CD4+ Tconv cells (12, 30). We and others have previously shown that Foxp3 alone is insufficient to engender the whole Treg signature (30). Thus, it was of interest to know how the Treg signature is reproduced among Foxp3+ cells generated by conversion in response to different experimental manipulations.
We chose two models of in vivo conversion. In the first, Foxp3+ cells were induced by administration of antigen at low dose, delivered to steady-state dendritic cells (DCs) by recombinant antiDEC-205 antibodies fused to the influenza hemagglutinin (HA) peptide 107–119 (17, 32). As previously reported, adoptive transfer of congenically marked CD25–CD4+ Tconv cells from HA-reactive transgenic mice into immunocompetent recipient animals, followed by a single injection of 50 ng antiDEC205-(HA107–119) fusion protein led to conversion of >30% of the donor Tconv cells to a Foxp3+ phenotype by 3 weeks after administration (over a background of <0.7% in control-treated animals). CD25high (hereafter referred to as “DEC-pept converted”) and nonconverted CD25− cells were sorted at this time for gene-expression profiling on Affymetrix M430v2 arrays. In the second model, conversion of naive CD4+ T cells was induced by transferring them into the lymphopenic environment of RAG-deficient mice (25). Introduction of 105 purified GFP−CD4+ cells from BDC2.5/NOD.Thy1.1.Foxp3gfp reporter mice typically induced Foxp3 expression in 5–10% of donor Tconv cells after 10–14 days. Two weeks after transfer, donor cells were identified by the Thy1.1 congenic marker, and Foxp3-GFP+ (hereafter “homeostatically converted”) or nonconverted Foxp3-GFP− CD4+ T cells were sorted.
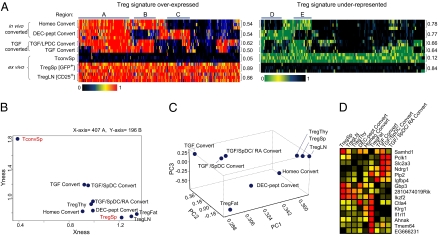
Fig. 1A compares the expression profiles of 603 transcripts of the canonical Treg signature (30) in these in vivo-converted Foxp3+ cells with those from standard ex vivo Treg cells (from spleens or lymph nodes of 6- to 8-week-old C57BL/6 mice, sorted as CD4+GFP+ or CD4+CD25high, respectively) and with profiles previously obtained from in vitro-converted Foxp3+ T cells (cells grown with antiCD3/28 +IL2+TGFβ or with small intestine lamina propria (LP) DCs as a source of retinoic acid to enhance conversion) (33). All datasets analyzed in this study are listed in Table S1. This “SignatureMatch” representation normalizes the level of transcripts from a particular signature between two reference populations. Here the ex vivo Tconv cells and Treg cells are taken as minimum and maximum, respectively. Treg cells isolated using either CD25hi or Foxp3-GFP+ were very similar, with this mode of analysis (Fig. 1A Bottom). In contrast, Foxp3+ T cells converted in vitro in the presence of TGFβ expressed only a fraction of the Treg signature, consistent with previous reports. This was also true of in vivo-converted Foxp3+ T cells, which showed a mosaic pattern of signature transcripts. All converted cells expressed those genes clustered in “region A”, the vast majority of which are related to T cell proliferation/activation (Fig. S1). Regions B and E, however, correspond to signature genes that were characteristic of cells converted both in vitro by TGFβ and in vivo by homeostatic cues but not by DEC-pept, whereas regions C and D showed the opposite pattern. Overall, however, these converted cells were more similar to normal Treg than to Tconv cells, as evidenced by the population plot in Fig. 1B, where populations are ordered according to their distance, integrated over all transcripts of the Treg signature, to splenic Treg and Tconv cells taken as a reference.
Fig. 1.
Distinct genomic profiles in Foxp3+ cells are elicited by different means. (A) “SignatureMatch” analysis showing the normalized expression of Treg signature probes (30) across different Treg cell populations. Raw expression values were normalized to 1 or 0 for spleen Treg and spleen Tconv, respectively, and displayed as a heat map where red represents the expression value of a gene at the same or a greater level than what was found for spleen Treg, whereas black represents the expression value of a gene that is at the same or a lower level than that of spleen Tconv (Upper). (Right) The underexpressed Treg signature, green representing the expression value of a gene at the same or a greater level than what was found in spleen Tconv and black representing the expression value of a gene that is at the same or a lower level than that of spleen Treg. Highlighted gene regions are discussed in the text (and in Fig. S1). Additional description of cell types can be found in Table S1. (B) “2D reference plots” with Treg overexpressed and Treg underrepresented genes on the x and the y axis, respectively. The relative coordinates for each population are calculated for these two traits and plotted on the 2D panel (47). (C) Principal component analysis. Three components are displayed and the different T cell groups are plotted relative to the component distribution. (D) Most influential genes that discriminate between the first three principal components are shown as a heat map.
To better depict the global relationships between these Foxp3+ cells, we used principal component analysis, a mathematical procedure that extracts the principal elements of variance from multidimensional data (Fig. 1C). In this analysis, based on normalized expression values of Treg signature transcripts, cells converted in vitro in the presence of TGFβ clustered together in one corner, and homeostatically converted cells were the closest to unmanipulated Treg cells; and ex vivo fat Treg cells segregated to a different corner, in keeping with their distinct profiles (34).
The transcripts having the largest influence on these three principal components are listed in Fig. 1D, highlighting the heterogeneity between Treg cell populations. These transcripts encode functionally important surface molecules such as Ctla4 or transcription factors such as Ikzf2 (a.k.a. Helios), a member of the Ikaros family already reported to be expressed at higher levels in ex vivo Treg cells relative to converted Treg cells (25).
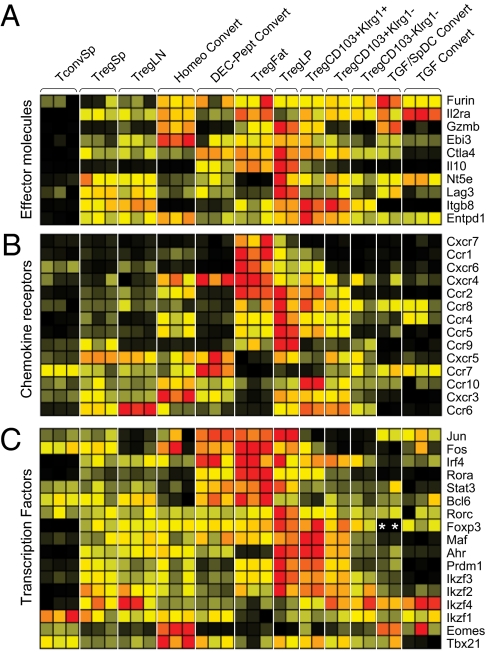
A more systematic analysis of transcripts encoding the major functional molecules reported to be involved in Treg cell inhibitory function is depicted in Fig. 2. Ctla4 transcripts were indeed overrepresented in DEC-pept converted cells (and in fat Tregs), whereas Entpd1 (CD39) and Ebi3 (a component of IL35) were preferentially expressed in homeostatically converted cells. Because Treg cell localization and trafficking can play a critical role in their functional abilities, we also analyzed chemokine receptor expression in these Foxp3+ populations. Here again, heterogeneous patterns were found for many of these genes (Fig. 2B), with Ccr6 transcripts predominantly found in LN Treg cells whereas Ccr10 and Cxcr3 dominated in homeostatically converted cells. Finally, heterogeneous patterns were observed for transcription factors (TFs) that belong to the Treg signature or are known to be necessary for particular facets of Treg function (9–11) (Fig. 2C). Whereas Eomes and Tbx21 (Tbet) transcripts were very prominent in homeostatically converted cells, Jun, Fos, and Irf4 were strongly expressed in DEC-pept converted Treg cells (as well as in fat Tregs). A complete compendium of gene-expression values within these Treg cell populations can be viewed and searched online (http://cbdm.hms.harvard.edu/TregSubphenotypes/heatmap.html).
Fig. 2.
Effector, homing, and transcription factor molecules in Treg phenotypes. Comparison is shown of normalized expression values of candidate effector molecules (A), chemokine receptors (B), and transcription factors (C). Data are row normalized and presented as a heat map where red and black represent the highest- and lowest-expressed genes, respectively. *, inactive probe due to genetic knock-in.
These data indicate that the heterogeneity of gene expression within Foxp3+ T cells does encompass elements critical to their anatomical localization, effector functions, and transcriptional programs.
Identifying Foxp3+ Treg Cell Subphenotypes from Secondary Lymphoid Tissues.
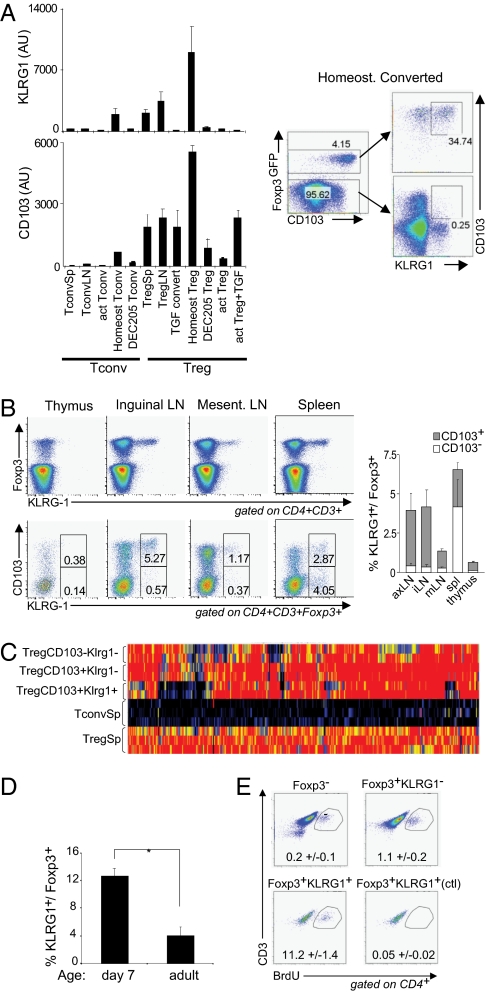
As the Treg cell subphenotypes revealed in the above studies were elicited under experimental—sometimes rather contrived—conditions, it was important to determine whether these phenotypes are indeed represented in normal lymphoid organs of unmanipulated mice. We first examined expression datasets from converted Foxp3+ cells for transcripts encoding cell surface markers that might be used to uniquely identify analogous populations in normal tissues. For homeostatically converted cells, promising candidates were Itgae, which encodes the adhesion molecule αEβ7 (CD103), and Klrg1, a member of the killer cell lectin-like receptor family: This combination of transcripts seemed much reduced on Foxp3+ cells elicited by TGFβ or antigen (Fig. 3A). Indeed, cell-surface staining showed that homeostatically converted Foxp3+ cells were uniformly CD103+, and roughly half expressed Klrg1 (Fig. 3A Right). In CD4+ cells of normal B6 mice, Klrg1 expression was almost exclusively restricted to Foxp3+ cells and among those was predominantly found in CD103+Klrg1+ “double-positive” cells (Fig. 3B). These cells accounted for ∼5% of Treg cells isolated from s.c. LNs, but were less frequent in the mesenteric LNs or spleen. To compare their contribution to the overall Treg signature, we generated expression profiles from Klrg1/CD103 single and double positive cells from pooled LNs. As illustrated in Fig. 3C (see Table S2 for a listing), each subset appeared distinct, with different but complementary “holes” in the overall Treg signature.
Fig. 3.
Normal equivalents of homeostatically converted Treg cells. (A) (Left) RNA expression of Klrg1 and CD103 in different Treg and Tconv populations determined by microarray (values are shown in arbitrary units). Protein expression of Klrg1 and CD103 in homeostatically converted and nonconverted CD4 T cells was determined by FACS (Right). (B) Expression pattern of Foxp3, Klrg1, and CD103 in CD4 T cells isolated from different organs from C57BL/6 mice. Representative dot plots are shown (Left) and summarized as means ± SD (Right). (C) Gene expression prolife of CD103−KLRG1−, CD103+KLRG1−, and CD103+KLRG1+ Foxp3+Treg cells isolated from peripheral LN. Data are represented as “Signature Match” analysis of the Treg overexpressed signature (30). (D) Comparison of the fraction of KLRG1-expressing cells among Foxp3+ Treg cells in 7-day-old perinatal and 6-week-old adult mice. Means ± SD of three mice per group are shown. * indicates significant difference by t test (P < 0.05). (E) Comparison of BrdU incorporation of KLRG1-positive and -negative Treg cells. Representative dot plots with the means ± SD of the gated population are shown for three independent experiments.
Klrg1+ Treg cells also resemble homeostatically converted Treg cells in that they were enriched, relative to adults, in peripheral LNs of 7-day-old mice (Fig. 3D). Lymphopenia-driven homeostatic proliferation is physiological in the first week of age in mice and in part drives the expansion of T cell pools (35). In addition, Foxp3+Klrg1+ cells cycle very extensively, a 10-fold greater proportion of cells incorporating BrdU during a 2-h pulse label, relative to Foxp3+Klrg1− counterparts (Fig. 3E). This activated status is reflected by the fact that Klrg1+ Treg cells express globally higher levels of an activation signature (Fig. S2). Klrg1 expression on Treg cells is not merely a marker of proliferation, however, because proliferation in vitro induced by antiCD3/CD28 and IL-2 does not induce Klrg1 (Fig. 3A).
Can Conversion Mediated by TGFβ Be Tracked in Vivo?
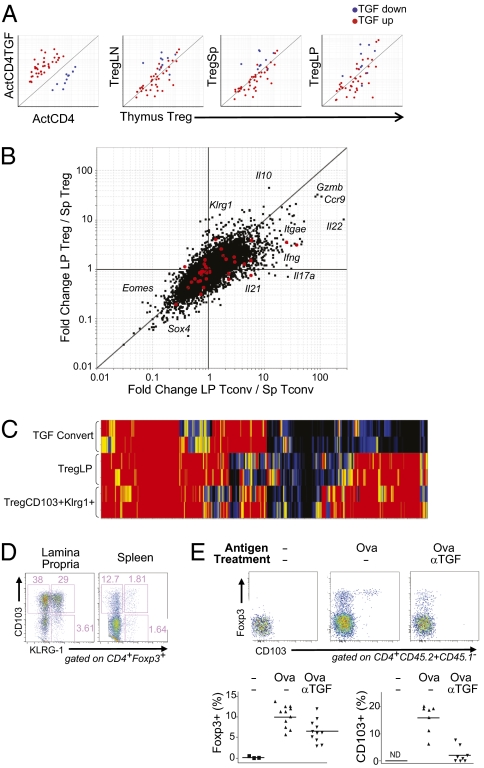
In parallel, we asked whether hallmarks of TGFβ-converted Treg cells could be identified in vivo. Several authors have suggested that such cells may contribute significantly to the Treg pools, particularly in the gut-associated lymphoid tissue (GALT), where TGFβ and retinoic acid are most abundant and where subsets of DCs are particularly efficacious at supporting conversion to Foxp3 positivity (4). Unfortunately, no unique cell-surface marker that would allow unequivocal identification of a TGFβ-converted cell could be deduced from the microarray data. Instead, we generated a “TGF signature” by selecting a set of transcripts affected by TGFβ treatment of Tconv and Treg cells (from TGFβ-treated natural Treg cells and from TGFβ-converted cells), but independent of the Treg signature. We then assessed the relative level of transcripts from this gene set in expression profiles of Treg cells from different organs of standard mice. Should secondary conversion induced by TGFβ be a numerically important contributor, one might expect to detect the TGF signature in Treg cells of peripheral organs, relative to the thymus where it is generally accepted to be uninvolved (at least in the adult) (36). This TGF signature was clearly biased in TGFβ-treated cells taken as a positive control (Fig. 4A Left). On the other hand, no bias in the TGF signature transcripts was observed for Treg cells from various peripheral organs, even for Treg cells derived from the LP of B6 mice (Fig. 4A). In addition, this signature showed no positive bias in homeostatically converted Foxp3+ cells, consistent with the fact that administration of blocking anti-TGFβ had no effect on homeostatic conversion, further establishing that this phenomenon is independent of TGFβ (Fig. S3).
Fig. 4.
In vivo equivalents of TGFβ-converted Tregs? (A) TGFβ signature probes were plotted on several groups for comparison, from left to right, of in vitro TGFβ converted T cells vs. activated T cells and lymph node (LN)-, spleen (Sp)-, or lamina propria (LP)-derived Treg cells compared to thymus Tregs. (B) Fold-change–fold-change plots showing the influence of the lamina propria environment on gene expression in Treg and Tconv cells. The y axis compares the expression profiles between LP and Sp Treg cells, whereas the x axis compares the expression profiles of LP and Sp Tconv cells. Highlighted in red are the TGFβ signature genes. (C) SignatureMatch profile of the Treg Up signature plotted using Sp Treg cells as the full signature and Sp Tconv cells as the baseline. Groups shown are TGFβ-converted T cells compared to LP or CD103+Klrg1+ Treg cells. (D) The Foxp3+CD103+Klrg1+ phenotype is found at the highest frequency in the LP. Cells were gated on being positive for both CD4 and Foxp3. (E) OT-II conversion mediated by oral ovalbumin is enhanced by the presence of TGFβ. Congenically marked (CD45.2+) naive OT-II CD4+ T cells (CD25−CD44lo) were identified in the mesenteric lymph nodes of CD45.1+ recipient mice 7 days after i.v. transfer. Recipient mice received either normal drinking water (no Ova) or water supplemented with 1.5% ovalbumin (PBS and anti-TGF). Mice were also injected with either anti-TGF antibody (1D11, 1 mg/mouse × 3 over 7 days) or PBS. FACS plots show the expression of Foxp3 and CD103 on OT-II donor cells. Graphs to the right summarize the data for individual mice from three independent experiments. P values were determined by t test.
As another means of identifying TGFβ-converted T cells in vivo, we analyzed more extensively the gene-expression profiles from LP Foxp3+ and Foxp3− CD4+ T cells in the small intestine (Fig. 4B). Here again, the TGFβ signature (highlighted in red) was not skewed as a whole, as would have been expected if there were a strong influence of TGFβ. We noted, however, striking differences in the LP profiles vis-a-vis their lymphoid tissue counterparts. First, there was a general effect of tissue localization, the diagonal disposition of transcripts on the plot indicating that many changes affected both Treg and Tconv cells in the GALT environment (e.g., a marked induction of Ccr9 and Gzmb and underexpression of Sox4 and Eomes). Second, Foxp3−CD4+ Tconv cells showed marked induction of many transcripts encoding effector cytokines (Ifng, Il17a, Il21, and Il22, but not IL4). Third, Foxp3+ Treg cells also showed several particularities: Il10 mRNA was present at a very high level, and there was a strong induction of Klrg1 and Itgae (CD103), which suggested a relationship with the Klrg1+ Treg cell population described above. This relationship was also observed when taking into account all of the Treg signature transcripts: LP Treg cells were clearly more similar to the CD103+Klrg1+ subset than to TFGβ-elicited Treg cells (Fig. 4C). The relationship was further confirmed by flow cytometry (Fig. 4D): Most Foxp3+ Treg cells in the LP expressed CD103 and many of those were KLRG1+. Thus, the genomic profile of LP Treg cells is most similar to that found in homeostatically proliferating or converted Treg cells, rather than those induced by TGFβ.
Finally, we searched for an impact of TGFβ on cells converted to Foxp3+ by exposure to oral antigen, taking as a model Foxp3-negative OT-II T cells transferred into hosts also fed with oral ovalbumin (Ova). Conversion in this model is restricted by antigen exposure and confined to the GALT (37) and had been shown to be influenced by TGFβ (20). Seven days after oral exposure to Ova, Foxp3+ cells were detectable among mesenteric lymph node T cells (Fig. 4E). A minority of these cells expressed CD103, but all of them were Klrg1 negative, consistent with the absence of this marker on DEC-pept converted Foxp3+ cells, as noted above. Recipient mice were treated with a TGFβ depleting monoclonal antibody during the Ova exposure period, which resulted in a modest but significant reduction in the number of converted cells (Fig. 4E Right); the efficacy of TGFβ inhibition was confirmed by the strong down-regulation of CD103 on both the transferred (Fig. 4E) and host T cells (Fig. S4). Together, these observations suggest that TGFβ might partake in Ag-induced conversion in the GALT, but that it does not leave a profound imprint under steady-state conditions.
Discussion
The field of immunology has a long history of defining lymphocyte lineages and sublineages, mainly driven by the availability of monoclonal antibodies that distinguish different populations. Here, primarily using genomic tools rather than cell-surface markers, we demonstrated that cells globally termed Tregs are a heterogeneous population. There appeared to be a mosaic of genes differentially expressed in discrete populations of thymus-derived or converted ex vivo Foxp3+ Treg cells. The expression profiles of the Treg cell populations presented complementary holes in the canonical Treg signature. Thus, the canonical signature of bulk Treg cells, defined by us and others, is really a composite derived from cells of diverse subphenotypes. Certainly, there is precedent for the subphenotypes observed here, as variability in the surface expression of CD103, Klrg1, or Cxcr3 and corresponding functional diversity have been described previously (38–41).
One caveat is that, whereas the comparisons always included matched Treg and Tconv sets, they were performed on variable genetic backgrounds and/or with Treg cells sorted on the basis of either CD25 or GFP reporter expression. These variations are unlikely to make much contribution to the holes observed here: The Treg signature in NOD and B6 mice is very similar (42), and the comparison of CD25+ or GFP+ sorted Treg cells showed very few differences on this scale.
The microarray data revealed variations in specific sets of transcripts, but also fluctuations in “bedrock genes” of the Treg signature, found in Treg cells everywhere (such as Il2ra, Ctla4, or Foxp3). Some of these fluctuations impact functionally relevant molecules, either potential effector molecules or chemokine receptors guiding the homing of these various Treg cell populations to different anatomical compartments. In this light, it may be more correct to consider these subphenotypes not as distinct and invariant sublineages (Treg1, Treg2, etc.), but rather as overlapping states that can be adopted in response to distinct differentiation cues or anatomic locations.
Different modes of conversion resulted in different subphenotypes. When driven by homeostatic forces, conversion (and the following proliferative expansion) resulted primarily in a CD103+Klrg1+ phenotype, whereas the more “subliminal” drive of antigen delivered to nonactivated DCs led to a different, perhaps more quiescent, phenotype with high levels of Ctla4 and Il10.
Recent results have argued that particular transcriptional modules in Treg cells are required to regulate different facets of effector T cell activity and that this matching is achieved by involving transcriptional control elements characteristic of the very cells being regulated. The prototypical Th1 transcription factor T-bet (Tbx21) is required to control Th1-type functions, the Th2/17 related transcription factor Irf4 to control Th2-like helper activities, and Stat3 to control Th17 functions (9–11). These and other TFs showed a very heterogeneous distribution across the different Treg subphenotypes. In this context, the patterns displayed in Fig. 2 lead one to speculate that the suppressive functions dependent on Irf4 would be more effectively performed by Ag-converted Treg cells, those dependent on Tbet optimally promoted by homeostatically converted Treg cells.
TGFβ and Treg cells have often been associated, in part because of the similarity of phenotypes elicited by Treg and TGFβ deficiencies (43) and in part because of the ease with which Foxp3+ cells can be elicited in TGFβ-supplemented cultures (26). We searched for an imprint of the influence of TGFβ in vivo, particularly in the LP, which has been described as a major site of TGFβ action and as a tolerogenic environment. We did not find an overrepresentation of TGFβ-influenced genes in LP Treg cells; of course, a signature derived in vitro in the presence of a single cytokine might be masked in vivo by other influences or because of differences in dose and timing. Instead, however, LP Tregs had a transcriptional profile very similar to that of CD103+Klrg1+ homeostatically converted Treg cells. This similarity is likely connected to the highly activated state of Tconv cells in that compartment, with strong transcriptional activity at several proinflammatory loci such as Ifng, Il17a, Il22, and Il21. By these criteria, the LP appears not to be a quiescent and tolerogenic environment.
There are therapeutic implications to this heterogeneity of Treg subphenotypes. Exploratory trials to harness Treg cell activities to control autoimmune diseases are underway. The general strategy entails amplification and transfer of Treg cell populations or combination therapies that would attempt to convert/expand Ag-specific Treg cells. Yet, precisely what Tregs cells would patients be receiving? And would they be effective, or even deleterious, for that particular context?
Materials and Methods
Mice.
NOD/LtJ, C57BL/6, BALB/c, B6.CD45.1, OT-II/B6, BDC2.5/NOD.Thy1.1.Foxp3-GFP (12), NOD.Rag1−, and HA6.5/BALB/c.Thy1.2.Rag2− (44) mice were bred under specific pathogen-free conditions, under Institutional Animal Care and Use Committee protocol JDC99-20.
In Vivo Conversion.
For homeostatic conversion, Foxp3-GFP negative T cells (CD4+CD8a−B220−CD11b/c−) were sorted from BDC2.5/NOD.Thy1.1.Foxp3-GFP mice and transferred i.v. (1 × 105 cells/mouse) to NOD.RAG10/0 recipients. After 14 days, Thy1.1+ CD4+ Foxp3-GFP+ or Foxp3-GFP− T cells were sorted from spleen and peripheral lymph nodes (cervical, axillary, and inguinal) of three to five NOD.RAG10/0 recipient mice. For anti-TGFβ blocking experiments, mice received injections of 1 mg anti-TGFβ (1D11) antibody every second day until day 12. For Ag-driven conversion, CD4+ T cells (CD4+ CD25− 6.5+) were sorted from HA6.5/BALB/c.Thy1.2.Rag2− and transferred i.v. (2 × 106/mouse) to BALB/c recipients, which received a single-dose injection of 50 ng of DEC-HA as described previously (17). After 3 weeks, B220−CD8α−CD11b/c−CD4+thy1.2+ CD25+ and CD25− cells were isolated by flow cytometry. For conversion by oral antigen, CD4+CD25−CD44lo T cells (B220−CD8a−CD11b/c−) were sorted from the spleens of OT-II/B6 TCR transgenic mice and injected i.v. (106 cells/mouse) into B6.CD45.1 recipients. Mice were then fed ovalbumin in drinking water (1.5% wt/vol) for 7 days as described previously (20, 37). For TGFβ blockade, mice were injected i.p. with anti-TGF antibody (1D11, 1 mg/mouse per injection) at days 0, 3, and 5 relative to cell transfer. For BrdU labeling, 6-week-old B6 mice were injected with 1 mg of BrdU (Sigma); s.c. LN cells were stained 2 h later for BrdU incorporation.
Lamina Propria Analysis.
Lamina propria T cells from 6- to 8-week-old C57BL/6 Foxp3-GFP mice were isolated by enzymatic digestion of the small intestine after removal of Peyer's patches and epithelial layers as described previously (37). CD4+ T cells were enriched by MACS and sorted on the basis of Foxp3-GFP+ (Treg cell) or Foxp3-GFP− (Tconv cell), CD3+CD4+B220−CD8a− and CD11b/c−. Additional experiments assessed CD103 and Klrg1 expression in lamina propria CD4+Foxp3+ T cells by flow cytometry.
Microarray Analysis.
For microarray profiling, cells were enriched by MACS then flow sorted into TRIzol reagent as described (45). RNA was amplified for two rounds (MessageAmp aRNA; Ambion), biotin labeled (BioArray High Yield RNA Transcription Labeling; Enzo), and purified using the RNeasy Mini Kit (Qiagen). The resulting cRNAs were hybridized to M430 2.0 chips (Affymetrix). All cell populations analyzed were generated in duplicate or triplicate. Datasets are available at NCBI under accession no. GSE20366.
Raw data were normalized using the RMA algorithm implemented in the “Expression File Creator” module from the GenePattern suite (46). Data were visualized using the “Multiplot” and SignatureMatch modules. SignatureMatch tests how well a signature is achieved in expression profiles. It uses normalized expression values, which are standardized relative to two reference populations that define the expression minima and maxima for each transcript of the signature (here, transcripts of the Treg signature, with Treg and Tconv cells as the max and min references). “PopulationPlots” positions cell populations in a two-dimensional frame of reference, created using the expression values of sets of genes that most distinguish two reference populations (x and y axes being defined by the values for the genes overexpressed in one reference population relative to the other); expression values of these gene sets were normalized relative to the reference populations (defined as 0 and 1), and the x and y coordinates of test populations were then calculated by averaging the values for each gene set. Principal components analysis was performed in S-Plus; the coefficients for the first three components were used as coordinates of each population.
Supplementary Material
Acknowledgments
We thank J. Pagan and K. Hattori for assistance with mice, J. LaVecchio and G. Buruzala for help with cytometry, J. Perez and K. Leatherbee for microarrays, and S. Davis for computational analysis. This work was supported by grants from the Juvenile Diabetes Research Foundation (4-2007-1057) and the National Institutes of Health (7R01AI51530), by Joslin Diabetes Center's National Institute of Diabetes and Digestive and Kidney Diseases-funded Diabetes Endocrinology Research Care core facilities (D.M. and C.B.), and by National Institutes of Health Grant R37 AI053102 (to H.v.B.). M.F. was supported by postdoctoral fellowships from the German Research Foundation (Emmy-Noether Fellowship, FE 801/1-1) and the Charles A. King Trust Postdoctoral Fellowship. J.A.H. was supported by a postdoctoral fellowship from the Canadian Institutes of Health Research and the Canadian Diabetes Association.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/1002006107/DCSupplemental.
References
- 1.Zheng Y, Rudensky AY. Foxp3 in control of the regulatory T cell lineage. Nat Immunol. 2007;8:457–462. doi: 10.1038/ni1455. [DOI] [PubMed] [Google Scholar]
- 2.Sakaguchi S, Yamaguchi T, Nomura T, Ono M. Regulatory T cells and immune tolerance. Cell. 2008;133:775–787. doi: 10.1016/j.cell.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 3.Sakaguchi S, et al. Foxp3+ CD25+ CD4+ natural regulatory T cells in dominant self-tolerance and autoimmune disease. Immunol Rev. 2006;212:8–27. doi: 10.1111/j.0105-2896.2006.00427.x. [DOI] [PubMed] [Google Scholar]
- 4.Belkaid Y, Tarbell K. Regulatory T cells in the control of host-microorganism interactions (*) Annu Rev Immunol. 2009;27:551–589. doi: 10.1146/annurev.immunol.021908.132723. [DOI] [PubMed] [Google Scholar]
- 5.O'Garra A, Vieira P. T(H)1 cells control themselves by producing interleukin-10. Nat Rev Immunol. 2007;7:425–428. doi: 10.1038/nri2097. [DOI] [PubMed] [Google Scholar]
- 6.Ziegler SF. FOXP3: Of mice and men. Annu Rev Immunol. 2006;24:209–226. doi: 10.1146/annurev.immunol.24.021605.090547. [DOI] [PubMed] [Google Scholar]
- 7.Vignali DA, Collison LW, Workman CJ. How regulatory T cells work. Nat Rev Immunol. 2008;8:523–532. doi: 10.1038/nri2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shevach EM. Mechanisms of foxp3+ T regulatory cell-mediated suppression. Immunity. 2009;30:636–645. doi: 10.1016/j.immuni.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 9.Zheng Y, et al. Regulatory T-cell suppressor program co-opts transcription factor IRF4 to control T(H)2 responses. Nature. 2009;458:351–356. doi: 10.1038/nature07674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koch MA, et al. The transcription factor T-bet controls regulatory T cell homeostasis and function during type 1 inflammation. Nat Immunol. 2009;10:595–602. doi: 10.1038/ni.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chaudhry A, et al. CD4+ regulatory T cells control TH17 responses in a Stat3-dependent manner. Science. 2009;326:986–991. doi: 10.1126/science.1172702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fontenot JD, et al. Regulatory T cell lineage specification by the forkhead transcription factor foxp3. Immunity. 2005;22:329–341. doi: 10.1016/j.immuni.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 13.Lio CW, Hsieh CS. A two-step process for thymic regulatory T cell development. Immunity. 2008;28:100–111. doi: 10.1016/j.immuni.2007.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsieh CS, Zheng Y, Liang Y, Fontenot JD, Rudensky AY. An intersection between the self-reactive regulatory and nonregulatory T cell receptor repertoires. Nat Immunol. 2006;7:401–410. doi: 10.1038/ni1318. [DOI] [PubMed] [Google Scholar]
- 15.Pacholczyk R, Ignatowicz H, Kraj P, Ignatowicz L. Origin and T cell receptor diversity of Foxp3+CD4+CD25+ T cells. Immunity. 2006;25:249–259. doi: 10.1016/j.immuni.2006.05.016. [DOI] [PubMed] [Google Scholar]
- 16.Wong J, et al. Adaptation of TCR repertoires to self-peptides in regulatory and nonregulatory CD4+ T cells. J Immunol. 2007;178:7032–7041. doi: 10.4049/jimmunol.178.11.7032. [DOI] [PubMed] [Google Scholar]
- 17.Kretschmer K, et al. Inducing and expanding regulatory T cell populations by foreign antigen. Nat Immunol. 2005;6:1219–1227. doi: 10.1038/ni1265. [DOI] [PubMed] [Google Scholar]
- 18.Apostolou I, von Boehmer H. In vivo instruction of suppressor commitment in naive T cells. J Exp Med. 2004;199:1401–1408. doi: 10.1084/jem.20040249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Verginis P, McLaughlin KA, Wucherpfennig KW, von Boehmer H, Apostolou I. Induction of antigen-specific regulatory T cells in wild-type mice: Visualization and targets of suppression. Proc Natl Acad Sci USA. 2008;105:3479–3484. doi: 10.1073/pnas.0800149105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mucida D, et al. Oral tolerance in the absence of naturally occurring Tregs. J Clin Invest. 2005;115:1923–1933. doi: 10.1172/JCI24487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Coombes JL, et al. A functionally specialized population of mucosal CD103+ DCs induces Foxp3+ regulatory T cells via a TGF-beta and retinoic acid-dependent mechanism. J Exp Med. 2007;204:1757–1764. doi: 10.1084/jem.20070590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Curotto de Lafaille MA, et al. Adaptive Foxp3+ regulatory T cell-dependent and -independent control of allergic inflammation. Immunity. 2008;29:114–126. doi: 10.1016/j.immuni.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 23.Knoechel B, Lohr J, Kahn E, Bluestone JA, Abbas AK. Sequential development of interleukin 2-dependent effector and regulatory T cells in response to endogenous systemic antigen. J Exp Med. 2005;202:1375–1386. doi: 10.1084/jem.20050855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Curotto de Lafaille MA, Lino AC, Kutchukhidze N, Lafaille JJ. CD25− T cells generate CD25+Foxp3+ regulatory T cells by peripheral expansion. J Immunol. 2004;173:7259–7268. doi: 10.4049/jimmunol.173.12.7259. [DOI] [PubMed] [Google Scholar]
- 25.Haribhai D, et al. A central role for induced regulatory T cells in tolerance induction in experimental colitis. J Immunol. 2009;182:3461–3468. doi: 10.4049/jimmunol.0802535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen W, et al. Conversion of peripheral CD4+CD25− naive T cells to CD4+CD25+ regulatory T cells by TGF-beta induction of transcription factor Foxp3. J Exp Med. 2003;198:1875–1886. doi: 10.1084/jem.20030152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fantini MC, et al. Cutting edge: TGF-beta induces a regulatory phenotype in CD4+CD25− T cells through Foxp3 induction and down-regulation of Smad7. J Immunol. 2004;172:5149–5153. doi: 10.4049/jimmunol.172.9.5149. [DOI] [PubMed] [Google Scholar]
- 28.Wan YY, Flavell RA. Identifying Foxp3-expressing suppressor T cells with a bicistronic reporter. Proc Natl Acad Sci USA. 2005;102:5126–5131. doi: 10.1073/pnas.0501701102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Floess S, et al. Epigenetic control of the foxp3 locus in regulatory T cells. PLoS Biol. 2007;5:e38. doi: 10.1371/journal.pbio.0050038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hill JA, et al. Foxp3 transcription-factor-dependent and -independent regula-tion of the regulatory T cell transcriptional signature. Immunity. 2007;27:786–800. doi: 10.1016/j.immuni.2007.09.010. [DOI] [PubMed] [Google Scholar]
- 31.Zheng Y, et al. Role of conserved non-coding DNA elements in the Foxp3 gene in regulatory T-cell fate. Nature. 2010;463:808–812. doi: 10.1038/nature08750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kretschmer K, Heng TS, von Boehmer H. De novo production of antigen-specific suppressor cells in vivo. Nat Protoc. 2006;1:653–661. doi: 10.1038/nprot.2006.105. [DOI] [PubMed] [Google Scholar]
- 33.Hill JA, et al. Retinoic acid enhances Foxp3 induction indirectly by relieving inhibition from CD4+CD44hi cells. Immunity. 2008;29:758–770. doi: 10.1016/j.immuni.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feuerer M, et al. Lean, but not obese, fat is enriched for a unique population of regulatory T cells that affect metabolic parameters. Nat Med. 2009;15:930–939. doi: 10.1038/nm.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Min B, et al. Neonates support lymphopenia-induced proliferation. Immunity. 2003;18:131–140. doi: 10.1016/s1074-7613(02)00508-3. [DOI] [PubMed] [Google Scholar]
- 36.Feuerer M, Hill JA, Mathis D, Benoist C. Foxp3+ regulatory T cells: Differentia-tion, specification, subphenotypes. Nat Immunol. 2009;10:689–695. doi: 10.1038/ni.1760. [DOI] [PubMed] [Google Scholar]
- 37.Sun CM, et al. Small intestine lamina propria dendritic cells promote de novo generation of Foxp3 T reg cells via retinoic acid. J Exp Med. 2007;204:1775–1785. doi: 10.1084/jem.20070602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huehn J, et al. Developmental stage, phenotype, and migration distinguish naive- and effector/memory-like CD4+ regulatory T cells. J Exp Med. 2004;199:303–313. doi: 10.1084/jem.20031562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suffia I, Reckling SK, Salay G, Belkaid Y. A role for CD103 in the retention of CD4+CD25+ Treg and control of Leishmania major infection. J Immunol. 2005;174:5444–5455. doi: 10.4049/jimmunol.174.9.5444. [DOI] [PubMed] [Google Scholar]
- 40.Stephens GL, Andersson J, Shevach EM. Distinct subsets of FoxP3+ regulatory T cells participate in the control of immune responses. J Immunol. 2007;178:6901–6911. doi: 10.4049/jimmunol.178.11.6901. [DOI] [PubMed] [Google Scholar]
- 41.Beyersdorf N, Ding X, Tietze JK, Hanke T. Characterization of mouse CD4 T cell subsets defined by expression of KLRG1. Eur J Immunol. 2007;37:3445–3454. doi: 10.1002/eji.200737126. [DOI] [PubMed] [Google Scholar]
- 42.D'Alise AM, et al. The defect in T-cell regulation in NOD mice is an effect on the T-cell effectors. Proc Natl Acad Sci USA. 2008;105:19857–19862. doi: 10.1073/pnas.0810713105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wahl SM, Wen J, Moutsopoulos N. TGF-beta: A mobile purveyor of immune privilege. Immunol Rev. 2006;213:213–227. doi: 10.1111/j.1600-065X.2006.00437.x. [DOI] [PubMed] [Google Scholar]
- 44.Klein L, Khazaie K, von Boehmer H. In vivo dynamics of antigen-specific regulatory T cells not predicted from behavior in vitro. Proc Natl Acad Sci USA. 2003;100:8886–8891. doi: 10.1073/pnas.1533365100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yamagata T, Mathis D, Benoist C. Self-reactivity in thymic double-positive cells commits cells to a CD8 alpha alpha lineage with characteristics of innate immune cells. Nat Immunol. 2004;5:597–605. doi: 10.1038/ni1070. [DOI] [PubMed] [Google Scholar]
- 46.Reich M, et al. GenePattern 2.0. Nat Genet. 2006;38:500–501. doi: 10.1038/ng0506-500. [DOI] [PubMed] [Google Scholar]
- 47.Yamagata T, Benoist C, Mathis D. A shared gene-expression signature in innate-like lymphocytes. Immunol Rev. 2006;210:52–66. doi: 10.1111/j.0105-2896.2006.00371.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.