Abstract
Seed dormancy provides a strategy for flowering plants to survive adverse natural conditions. It is also an important agronomic trait affecting grain yield, quality, and processing performance. We cloned a rice quantitative trait locus, Sdr4, which contributes substantially to differences in seed dormancy between japonica (Nipponbare) and indica (Kasalath) cultivars. Sdr4 expression is positively regulated by OsVP1, a global regulator of seed maturation, and in turn positively regulates potential regulators of seed dormancy and represses the expression of postgerminative genes, suggesting that Sdr4 acts as an intermediate regulator of dormancy in the seed maturation program. Japonica cultivars have only the Nipponbare allele (Sdr4-n), which endows reduced dormancy, whereas both the Kasalath allele (Srd4-k) and Sdr4-n are widely distributed in the indica group, indicating prevalent introgression. Srd4-k also is found in the wild ancestor Oryza rufipogon, whereas Sdr4-n appears to have been produced through at least two mutation events from the closest O. rufipogon allele among the accessions examined. These results are discussed with respect to possible selection of the allele during the domestication process.
Keywords: domestication, preharvest sprouting, quantitative trait locus
Plant seeds are major sources of human nutrition, either directly or indirectly, and are the major means of crop propagation. Consequently, various seed traits, including dormancy, have been selected through crop domestication (1, 2). Seed dormancy has both advantages and disadvantages for plants—especially crops—in terms of cultivation and utilization, because weak dormancy leads to uniform germination, whereas deep dormancy prevents preharvest sprouting but inhibits germination. Preharvest sprouting often occurs under favorable temperature and humidity at maturity, resulting in reduced grain quality and germinability. Thus, controlling seed dormancy is a very important goal in the breeding of rice and other cereals.
Seed dormancy is genetically controlled by the genotypes of both the mother plant and the embryo. The former affects the nature of the tissues surrounding the embryo, such as the seed coat (testa). These tissues impose dormancy and act as physical barriers to radicle growth on imbibition. Whereas this “coat-imposed” dormancy depends on the anatomy of the seed, which varies among species, embryonic dormancy is controlled more finely by the developmental program. Embryonic dormancy is acquired during seed maturation, in which the plant hormone abscisic acid (ABA) plays a fundamental role (3). Studies in Arabidopsis have revealed the framework of the regulatory network for seed maturation, which is controlled by several master transcription factors, including ABI3, which is orthologous to maize VP1 and rice OsVP1 (4–6). Mutations in these genes profoundly affect the acquisition of seed dormancy. In addition to these maturation-related regulatory genes, mutants have been isolated by direct screening for reduced-dormancy phenotypes in Arabidopsis. Among these, REDUCED DORMANCY4 (RDO4), renamed HISTONE MONOUBIQUITINATION1 (HUB1), has been molecularly identified, and the results of this identification point to the importance of chromatin modification in seed dormancy (7). Studies of natural variations in seed dormancy in Arabidopsis have led to the cloning of a quantitative tract locus (QTL), DELAY OF GERMINATION 1 (DOG1), which affects embryonic dormancy (8). DOG1 encodes a member of a plant-specific protein family with a domain shared by the D class bZIP DNA-binding proteins.
Despite such remarkable findings, our knowledge of the molecular mechanisms underlying seed dormancy is far from complete. Information about the molecular identities of the genes controlling dormancy is even more limited in crops than in the model plant Arabidopsis. Particularly in cereals, there is a need to identify the genes contributing to natural variations in dormancy, to improve crops and better understand the history of domestication. Many studies have been performed to detect QTLs for seed dormancy in cereals, but thus far no genes have been molecularly identified (9–12).
In previous work, we detected five QTLs for dormancy in backcross inbred lines (BILs) derived from crosses between the japonica cultivar Nipponbare and the indica cultivar Kasalath (9). We have continued to perform map-based cloning of these QTLs. Here we report the molecular cloning of one of these QTLs, Seed dormancy 4 (Sdr4). Sdr4 encodes a novel protein with an amino acid sequence that has no similarity to proteins with known functions; it may act as a seed dormancy-specific regulator that is under the control of the seed global regulator OsVP1. Furthermore, haplotype analysis of the Sdr4 region has revealed that Sdr4 acts as an important determinant of seed dormancy in rice cultivars and might have been involved in rice domestication.
Results
Isolation and Characterization of the Kasalath Allele of Sdr4, Which Confers Preharvest Sprouting Resistance and Deeper Dormancy.
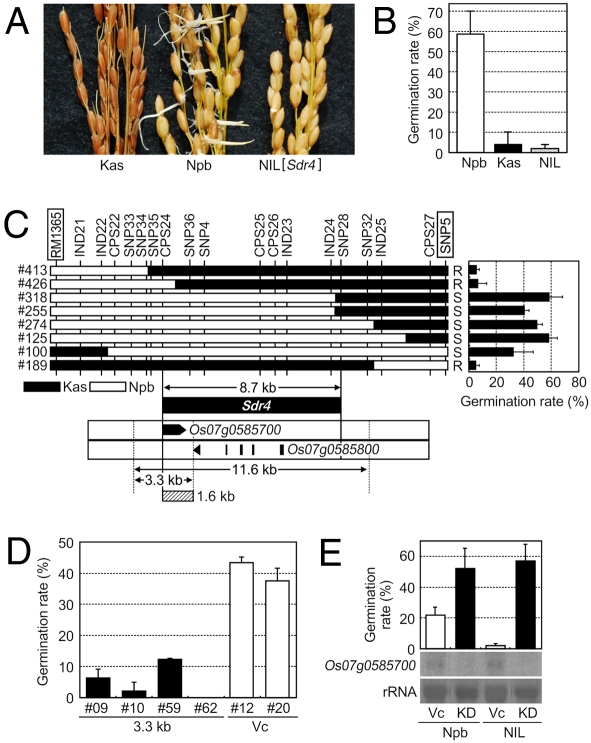
We developed a nearly isogenic line of Sdr4 (NIL[Sdr4]) with a 7.5-Mb Kasalath segment in a Nipponbare background (Fig. S1A). The germination rate of NIL seeds collected 6 weeks after heading (WAH) was very low (2%), even lower than that of Kasalath (4%), whereas Nipponbare had a high germination rate (58%), indicating that the Kasalath allele of Sdr4 (Sdr4-k) had a strong positive effect on seed dormancy (Fig. 1 A and B). From a population of 2,515 plants, we identified 8 plants with recombination in the vicinity of Sdr4 (Fig. S1 B–D). Progeny testing delimited the candidate region to <8.7 kb (Fig. 1C). By transforming Nipponbare with subfragments of this region, we found that a 3.3-kb Kasalath fragment, which contained a single gene, Os07g0585700, substantially decreased germination compared with the control (Fig. 1D and Fig. S2 A and B). Furthermore, RNAi knockdown of Os07g0585700 in both Nipponbare and NIL[Sdr4] increased germination rates to above that of the control Nipponbare (Fig. 1E). In addition, seeds of two independent mutants (M25 and M100) carrying the same seven-aa deletion in the Os07g0585700-encoded protein were less dormant than those of wild-type Nipponbare seeds (see below). Thus, we conclude that Os07g0585700 is Sdr4. The results also indicate that Sdr4-n remained functional, although it was not as effective as Sdr4-k in conferring dormancy.
Fig. 1.
Genetic effects, delimitation of candidate genomic region, and genetic complementation of Sdr4. (A) Germination of Kasalath (Kas), Nipponbare (Npb), and NIL[Sdr4] (NIL). (B) Germination rates of seeds from panicles sampled 6 weeks after heading. (C) Delimitation of Sdr4. Graphical genotypes of the Sdr4 region in eight plants in which recombination occurred between RM1365 and SNP5 (Fig. S1) are shown at the left. Black and white regions represent chromosomal segments homozygous for the Kas and Npb alleles, respectively. The bar graph at the right shows the germination rates of recombinants. Preharvest sprouting resistance is denoted by “R,” and sensitive is denoted by “S.” Genes predicted by RAP-DB (http://rapdb.dna.affrc.go.jp/) (52) in the Npb genomic sequence around the Sdr4 candidate region are indicated by closed arrows and closed boxes. Two genes were annotated in the region. The 8.7-kb Sdr4 candidate region was defined by linkage analysis. The 3.3-kb fragment was used for complementation analysis. The 1.6-kb region with FNP is indicated by a hatched box. (D) Bar graph showing germination rates of T3 seeds harboring the Kas fragment (3.3 kb) or empty vector (Vc). (E) RNA gel blot analysis of Os07g0585700 gene expression in knockdown (KD) and Vc lines. Knockdown transgenic lines were generated from Npb and NIL. rRNA stained with methylene blue shows equal loading of RNAs (rRNA). Germination rates 7 days after imbibition are shown as averages of three biological repeats with standard deviation.
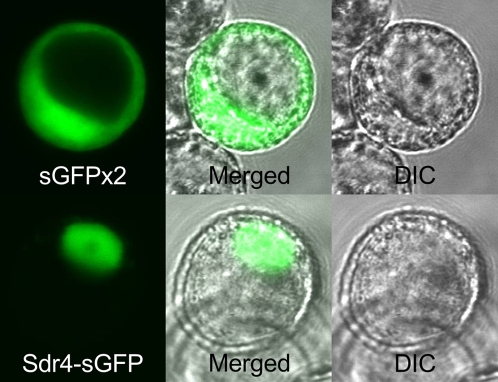
Sdr4 contained no intron, and the deduced amino acid sequence of the encoded protein was not homologous with those sequences of proteins with known functions (Fig. S3A). A putative bipartite-type nuclear localization signal (13) was found in the N-terminal region. Nuclear localization of Sdr4 was confirmed by expression of Sdr4-k–GFP (green fluorescent protein) fusion protein in cultured rice cells (Fig. 2).
Fig. 2.
Subcellular localization of Sdr4. The 35S promoter–driven dimer of sGFP and the Sdr4-k–sGFP fusion were transiently expressed in oc cells, and the localizations were investigated under a confocal laser scanning microscope (Left). Differential interference contrast images (Right) and merged images (Middle) are shown.
Prediction of Functional Nucleotide Polymorphisms of Sdr4-k.
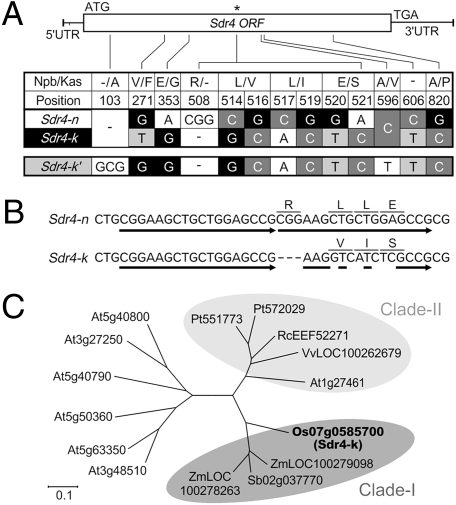
From the results of complementation analysis and genetic mapping, we expected to find functional nucleotide polymoprphisms (FNPs) to explain the functional difference between Sdr4-n and Sdr4-k located within a 1.6-kb region (Fig. 1C) corresponding to the protein-coding region and 3′-UTR. In addition to a SNP in the 3′-UTR, the coding region contained clustered sequence changes corresponding to one InDel and three amino acid substitutions, along with four simple SNPs, three of which resulted in amino acid changes (Fig. 3A). The presence of an 18-bp direct repeat in Sdr4-n, which could have been created by double-strand cleavage and repair in the corresponding region of Sdr4-k, as is seen in maize waxy mutants (14), suggests that the clustered sequence changes resulted from a single event (Fig. 3B). Such polymorphisms were associated with a change in the amino acid sequence from −(K)VIS270–274 (Sdr4-k) to R(K)LLE270–275 (Sdr4-n). The dipeptide sequence Val-Ile (VI) in Sdr4-k was well preserved in a conserved sequence block among related proteins across species, including dicots, whereas other amino acids associated with simple SNPs were not well conserved (Fig. 3C and Fig. S4); thus, the polymorphisms most likely were FNPs.
Fig. 3.
InDels and SNPs between Sdr4-n and Sdr4-k, and amino acid sequence comparison among Sdr4 and its homologs. (A) Haplotypes derived from 11 SNPs and two InDels of the Sdr4 coding region. (B) Sequences of two alleles around the SNP cluster, indicated by the asterisk in A. The 18-nt direct repeats are indicated by arrows. (C) Sdr4 homologs were searched by BLAST. Aligned sequences by Genetyx version 9 (Genetyx) are shown by unrooted phylogenetic tree using MEGA4 (53).
Expression and Localization of Sdr4.
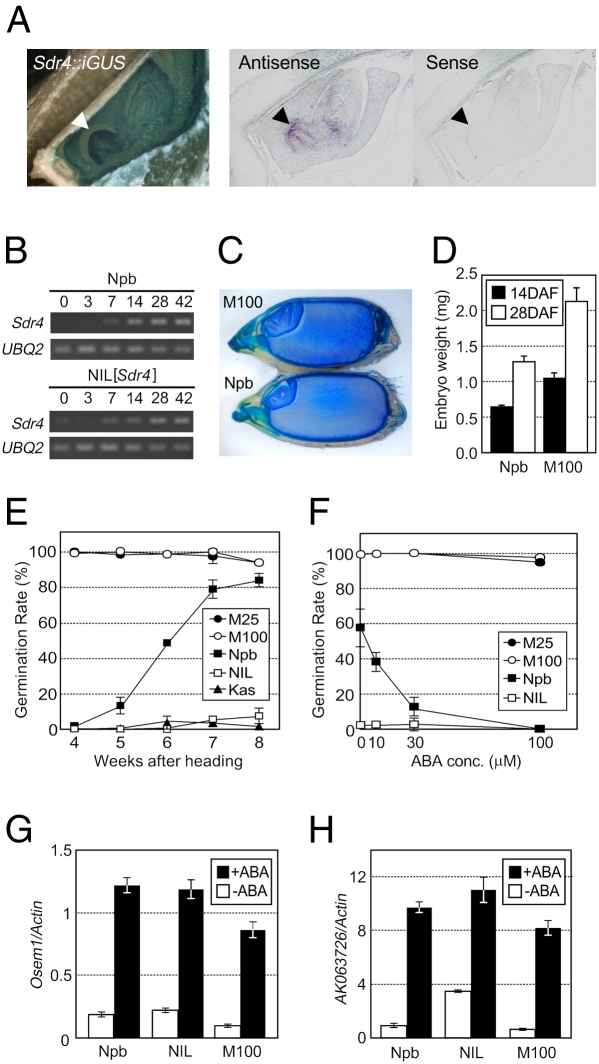
Sdr4 mRNA was detected preferentially in the seed tissues 14 days after flowering (DAF) (Fig. S5). Promoter–GUS transgenics and in situ hybridization suggest that Sdr4 mRNA was expressed throughout the embryo but preferentially in the radicle; the in situ hybridization signals suggest that it also was expressed in the shoot (Fig. 4A). Sdr4 mRNA began to accumulate in the seed at 7 DAF, and its level increased as the seed matured (Fig. 4B). These expression patterns during the course of seed maturation were in accordance with the set of cis-elements predicted in the Sdr4 promoter; there were seven RY repeats (CATGCA), which are important for seed-specific gene expression and are the target of the VP1/ABI3 subfamily of B3 domain transcription factors (15), along with an ABA response element [ABRE; ACGTGG/T(C)] and an ABRE-related coupling element (CE) (Fig. S3B) (16–18). One RY repeat is closely linked to an ABRE, and another is closely linked to an ABRE-CE and an ABRE. The combination of these elements and the close linkages are frequently seen in seed maturation–related genes (19).
Fig. 4.
Morphological and physiological analysis of sdr4 mutant. (A) GUS staining of sliced seed of Nipponbare (Npb) with Sdr4-n:GUS. The radicle is indicated by an arrowhead. For tissue-specific expression of Sdr4 mRNA, in situ hybridization used Sdr4-specific antisense and sense probes on sections of Npb seed. The radicle is indicated by an arrowhead. (B) Temporal changes in level of Sdr4 mRNA during the ripening period, based on semiquantitative RT-PCR, at 0, 3, 7, 14, 28, and 42 days after heading. The UBQ2 was used as a control to show equal loading. (C) Longitudinal slices of the seeds of Npb and the sdr4 mutant were stained by toluidine blue. (D) Embryos were harvested (n = 50) and weighed, with three repeats. (E) Germination rates of Npb, NIL[Sdr4] (NIL), two sdr4 mutants, and Kasalath (Kas) at various time points after heading were determined. (F) Germination rates just after harvest (6 WAH) of fresh seeds treated with different concentrations of ABA. (G) Embryoless half-seeds were treated with 30 μM ABA or left untreated for 24 h; expression levels of Osem1 were determined by real-time PCR. (H) AK063726 expression levels in the embryoless half-seeds were measured.
Characterization of the Loss-of-Function Mutant of Sdr4.
The seeds of two independent sdr4 mutant lines, M25 and M100, contained embryos larger than those of the wild type (Fig. 4 C and D) and were completely nondormant (nearly 100% germination at 4 WAH; Fig. 4E). The loss of dormancy was associated with severely reduced ABA sensitivity. Germination of sdr4 seeds at 6 WAH was not inhibited by ABA at 100 μM, whereas that of Nipponbare seeds was completely inhibited (Fig. 4F); however, no significant differences in the ABA content of seeds sampled at 6 WAH were seen (83 ng/g fresh weight for Nipponbare, 87 ng/g for NIL(Sdr4), 83 ng/g for M25, and 78 ng/g for M100). Despite the ABA insensitivity of seed germination, the primary ABA-signaling mechanism, which leads to ABA-induced gene expression, was not affected by the sdr4 mutation. Expression of two genes for the ABA-inducible late-embryogenesis–abundant proteins (LEA) Osem1 and AK063726, having ABREs and a CE, was normally induced by 30 μM ABA in embryoless half-seeds of the M100 mtant (Fig. 4 G and H) (20, 21).
Analysis of Upstream (Sdr4 Regulator) and Downstream Genes.
The presence of ABREs and RY repeats in the Sdr4 promoter, together with an expression pattern typical of that of maturation-related genes, prompted us to investigate whether Sdr4 expression is regulated by OsVP1, a global regulator of seed maturation (6). Sdr4 expression in embryos at 28 DAF were substantially reduced in Osvp1 mutant embryos (Fig. 5B and Fig. S6B). This finding suggests that the regulation exerted by Sdr4 was, at least in part, integrated into the global seed maturation program directed by OsVP1.
Fig. 5.
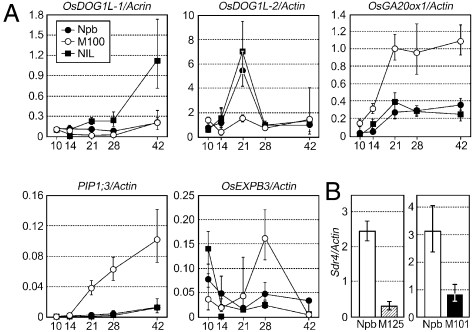
Expression analysis of dormancy and germination related genes in sdr4 and Osvp1 mutants. (A) Temporal changes (DAF) in mRNA levels of OsDOG1-like genes, gibberellin biosynthesis gene (OsGA20ox-1), aquaporin gene (PIP1;3), and expansin gene (OsEXPB3) in embryos were monitored by real-time PCR in Nipponbare (Npb), the sdr4 mutant (M100), and NIL[Sdr4] (NIL), with three biological repeats. Expression levels are shown as ratios to Actin-1 gene expression. (B) Sdr4 gene expression in wild-type (Npb), Osvp1-1 (M125), and Osvp1-2 (M101) at 28 DAF was determined.
To relate Sdr4 more closely to known seed dormancy and germination mechanisms, we examined the effects of sdr4 mutation on the expression of several genes potentially related to dormancy and germination. Although little is known about regulators of embryonic dormancy other than those of seed maturation and ABA signaling in cereals, we examined the expression of the three closest rice homologs of Arabidopsis DOG1(8) (a QTL identified as a positive regulator of seed dormancy) as potential positive regulators of seed dormancy. the levels of expression of two of the three OsDOG1-like genes were lower in sdr4 mutant embryos than in wild-type Nipponbare or NIL[Sdr4] (Fig. 5A and Fig. S7 A and B). In addition, OsDOG1-like-1 (OsDOG1L-1) expression was significantly higher in NIL[Sdr4] than in Nipponbare. It was difficult to judge this gene to be orthologous to Arabidopsis DOG1 from the phylogenetic relationships, due to the presence of three highly related DOG1-like genes in Arabidopsis, some of whose mutations were reported to not affect dormancy. However, the observed down-regulation in the mutant and up-regulation in NIL[Sdr4] suggest that OsDOG1L-1 is a positive regulator of dormancy in rice, and that Sdr4, at least in part, controls seed dormancy via the regulation of these OsDOG1-like genes.
In Arabidopsis nondormant mutants with mutations in seed maturation regulators such as abi3, germinative and postgerminative programs operate prematurely in the developing seed (22). Consequently, we examined the expression of several germination-related genes: a gibberellin biosynthesis gene (OsGA20ox-1) (23), aquaporin genes (PIP1;3 and PIP2;2) (24), and an expansin gene (OsEXPB3) (25) (Fig. 5A and Fig. S7C). Expression of these genes is induced in nondormant seeds on imbibition (26). Expression levels of all of these genes were significantly higher in the sdr4 mutant than in the wild-type Nipponbare or NIL[Sdr4], consistent with the positive regulation of Sdr4 by OsVP1.
The foregoing expression analysis results suggest that Sdr4 plays a regulatory, rather than a structural or metabolic, role in the promotion of dormancy and inhibition of germination, as also suggested by the nuclear localization of the gene product.
Natural Variations in Sdr4 in Cultivars and Wild Rice.
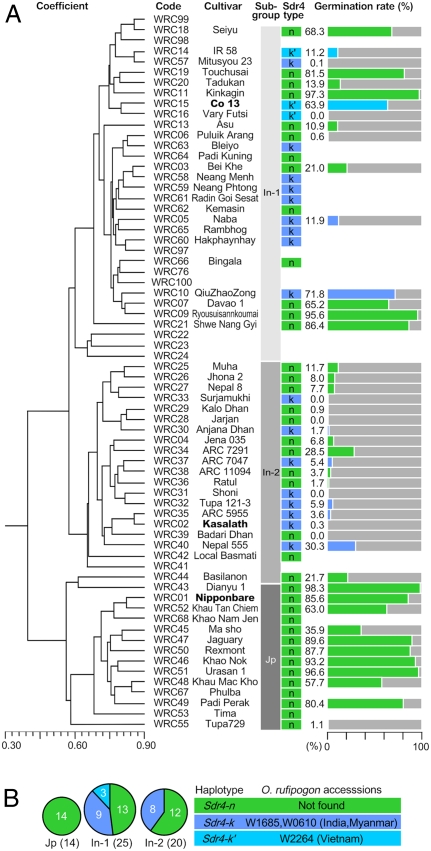
Sequence analysis of 59 cultivars from the world rice core collection revealed the existence of only three haplotypes, Sdr4-n, Sdr4-k, and Sdr4-k′, in the coding sequences (Figs. 3A and 6A) (27). Interestingly, all japonica cultivars had only Sdr4-n, whereas both Sdr4-n and Sdr4-k were found in indica In-2, and all three haplotypes were found in indica In-1 (Fig. 6B). SNP analysis of the chromosomal regions containing Sdr4-n in indica cultivars revealed that in most cases, the japonica haplotypes with Sdr4-n were flanked by indica haplotypes, although some complex haplotypes were found as well (Fig. S8). Therefore, we considered the Sdr4-n in indica cultivars to be the result of introgression from japonica sources. The seeds of cultivars with Sdr4-n had lower dormancy than those of cultivars with Sdr4-k (P = 0.012 by the F test). These results strongly suggest that the functional differences in Sdr4 contribute substantially to the variations in dormancy of Asian rice accessions, although the involvement of other loci is suggested by the presence of some indica cultivars with Sdr4-k(k′) showing high germination rates and some indica cultivars with Sdr4-n showing low germination rates (Fig. 6A).
Fig. 6.
Natural variations in the Sdr4 coding region. (A) SNPs and germination rates at 6 WAH in cultivars in the world rice core collection. Cultivars, ordered according to the genetic distance based on RFLPs, are shown. Germination rates at 6 WAH are indicated in mauve-blue or sky-blue bars (cultivars with Sdr4-k or Sdr4-k′, respectively) or green bars (cultivars with Sdr4-n); the 100% scale is indicated by the gray bars. (B) The frequency of each haplotype in japonica and indica (In-1 and In-2) lines is summarized, and wild relatives showing the same haplotypes as the cultivars are shown.
We also identified the sequences of Sdr4 in 46 accessions of Oryza rufipogon, the wild ancestor of O. sativa (28, 29) (Table S1). Our analysis revealed 23 variations, mostly with sequences closer to the sequence of Sdr4-k than to the sequence of Sdr4-n. In particular, two of these variations (W1685 and W0610) were identical to Sdr4-k, and one variation (W2264) was identical to Sdr4-k′. Thus, Sdr4-k and Sdr4-k′ in indica were inherited from these subgroups of the wild ancestor. In contrast, the Sdr4-n sequence was not found in any of the 46 accessions.
Discussion
We isolated and characterized the preharvest-sprouting–resistance gene Sdr4, which encodes a protein with no similarity to proteins of known function. However, in light of the findings that Sdr4 was located in the nucleus, was transcriptionally regulated by OsVP1 (the global regulator of seed maturation), and was involved in the regulation of gene expression related to seed dormancy and germination, Sdr4 might be involved in the gene expression machinery as, for example, a transcription factor. The sdr4 mutant had a nondormant phenotype, as well as a larger embryo. In addition, the sdr4 embryo precociously expressed germinative or postgerminative genes. Precocious expression of these genes in the mutant resembled the mutant expression of seed maturation regulators, including ABI3/VP1 (30, 31), and was consistent with the result that expression of Sdr4 was under the control of OsVP1. The ABA insensitivity of sdr4 seeds with respect to germination also was in accordance with OsVP1 control of Sdr4 expression. In addition, ABA levels and basic ABA signaling as monitored by LEA gene induction in response to this hormone were not significantly affected in the mutant. This suggests that Sdr4 is a specialized regulator in the seed maturation program responsible for the dormancy pathway. Furthermore, the Sdr4-dependent expression of OsDOG1L-1, a rice homolog of Arabidopsis DOG1, is in line with this suggestion, although the role of OsDOG1L-1 in dormancy remains to be determined.
In addition to the seed coat, such tissues as the endosperm and coleorhiza surrounding the radicle are known to play important roles in the imposition of seed dormancy. These tissues act as barriers and need to be broken for the radicle to emerge—that is, to germinate. It has been recently shown that in barley, the coleorhiza undergoes a reduction in ABA metabolism and content during after-ripening; these changes ultimately allow the imbibed radicle to grow (32). It remains to be elucidated whether Sdr4 plays a role in the establishment of such a function of the coleorhiza. The preferential expression of Sdr4 in the radicle and shoot tissues suggests that this gene plays a role in regulating the growth potential of the radicle and shoot, which counteracts the imposed barriers. In light of the aforementioned functions of the coleorhiza in dormancy in rice, it should be noted that a recent study found that qLTG3-1, which limits germination at low temperatures, is preferentially expressed in the coleorhiza and epiblast (33).
In crop plants, such as rice, wheat, and barley, seed dormancy must be finely controlled, because a excessively high or low level of dormancy can cause severe problems with seed germination and preharvest sprouting, respectively (34). Thus, cereals require appropriate levels of dormancy to maintain quality and yield. Genetic approaches have been used in wheat, barley, and several cereals, and numerous QTLs have been reported (35, 36). Rice chromosome 7 demonstrates synteny with wheat chromosome 2D (37). Four dormancy QTLs (including Sdr4) have been identified on rice chromosome 7, (9, 10). Because QTLs related to seed dormancy have been reported on wheat chromosome 2DS (35, 38), determining the locus of the wheat ortholog of Sdr4 is of interest.
Most of the genes involved in seed dormancy that have been isolated so far are involved in ABA synthesis and ABA signal transduction (34, 39, 40). Thus, modulation of these genes is predicted to have pleiotropic effects on seed maturation events, such as desiccation tolerance and storage material accumulation (41). In contrast, we can expect modification of Sdr4 to not be accompanied by effects other than those on seed dormancy, because Sdr4 is a seed dormancy–specific regulator of the seed maturation program. Cloning of Sdr4 in rice will provide a unique opportunity to explore the genetic control and modification of seed dormancy in these crops.
The recent identification of FNPs in several cloned genes for agronomically important traits, including those for grain shattering (Sh4, qSH1), pericarp color (Rc), seed width (qSW5), and amylose content (Waxy), has allowed us to speculate on the rice domestication process (42–45). One current view of the process of domestication of rice from O. rufipogon proposes that the japonica and indica subspecies arose from independent subgroups of wild ancestors (1). It has been suggested that W1943, an O. rufipogon accession, is a close relative of the subpopulation from which the japonica group originated; this suggestion is based on the patterns of short interspersed nuclear transposable elements (46) and cDNA sequences (47). In fact, three accessions from China and India, including W1943, have Sdr4 sequences closer than those of other accessions to Sdr4-n. The differences between Sdr4-n and Sdr4-W1943 appear to have been generated by two events, a single nucleotide substitution and clustered substitutions of the potential FNP caused by a double-strand break and repair (see above). Because dormancy is considered a vital domestication trait in rice, we can reasonably expect that these mutations were selected during the domestication of japonica. We can speculate that, in their wisdom, ancestral farmers would not have failed to use the rare mutation events that produced a useful degree of reduction in function of Sdr4, rather than loss of function, which would have led to a problem of vivipary. The prevalent introgression of Sdr4-n in indica cultivars supports the profitability of this allele in cultivation, similar to that of the rc allele and wx (44, 48). Obviously, the Sdr4 allele is not the only dormancy-related domestication gene. A recent transcriptomic study in Arabidopsis indicates that the mechanisms for establishing dormancy and controlling after-ripening potential can be differentiated (22). Our results suggest that Sdr4 is involved in the former mechanism, although the possibility of participation in both mechanisms has not been excluded. Reduced seed dormancy during domestication could have been achieved by the selection of alleles of genes related to either type of mechanism. Presumably, selection was made based on both germination synchronization and the duration of the after-ripening period. Therefore, genes related to after-ripening potential may well be identified as domestication genes in the future.
Our molecular identification of Sdr4 should not only provide clues to rice domestication, but also shed light on the molecular mechanisms of seed dormancy. More specifically, our results placing Sdr4 in the central regulatory network of seed maturation provide opportunities for a more comprehensive understanding of seed development, which will be further enhanced by the cloning of other identified dormancy QTLs.
Methods
Plant Materials for Mapping.
We crossed Nipponbare (a japonica cultivar) and Kasalath (an indica cultivar) and then performed repeated backcrossing with Nipponbare as the recurrent parent. BC4F2 plants in which the Sdr4 region was heterozygous were selected to produce a mapping population. We used 100 and 2,515 self-pollinated progeny plants (00F2#53) for coarse-resolution and high-resolution mapping of Sdr4, respectively. With this population, Sdr4 was mapped in the interval between markers SNP1 and SNP8 (Fig. S1C). Twenty-eight BC4F2 plants in which recombination occurred in the interval between IND3 and IND8 were selected (Fig. S1D). The self-pollinated progeny of these plants (BC4F3) were used for progeny testing. The germination rates of three plants fixed for each allele were determined. The molecular markers used for high-resolution mapping are listed in Table S2A.
Germination Testing.
Three panicles sampled at 4, 5, 6, 7, or 8 WAH were wrapped with paper towels and dipped in water. After the water was briefly drained off, the panicles were incubated in the dark at 30 °C for 1 week, and seed germination was scored.
Isolation of sdr4 and Osvp1 Mutants.
The sequences of Sdr4 and Osvp1 were determined in 16 mutant lines exhibiting vivipary. The plants were selected from a mutant panel (49), a library of rice mutants created by tissue culture of Nipponbare. The same deletion of 21 nucleotides causing a 7-aa deletion in the C-terminal conserved–sequence block of Sdr4 was found in three lines—M25, M26, and M100—derived from independent mutagenesis. This phenotype of the M100 line was complemented using a 3.3-kb fragment (Fig. 1C and Fig. S6A). Mutations in Osvp1 were found in two lines, one line with a 32-nt deletion resulting in a frame shift generating a truncation before the B3 domain (M125; Osvp1-1), and the other line with an amino acid substitution in the B3 domain (M101; Osvp1-2) (Fig. S6B). Genetic analysis found that the phenotype and the Osvp1-2 mutation were colocalized within a 131-kb region. This result supports the hypothesis that the viviparous phenotype was due to this substitution (Fig. S6C). Mutant lines M25, M26, M100, M101, and M125 corresponded to ND2054, ND2126, NE2331, NE2465, and NE5113, respectively, in the Tos17 mutant panel database (http://tos.nias.affrc.go.jp/).
Transgenic Complementation and Knockdown.
A BAC library of Kasalath genomic DNA was screened by PCR for a clone with the Sdr4 region, from which the NruI–NruI 11.6-kb fragment was subcloned into the EcoRV site of pBluescript II SK(+) (Agilent Technologies), resulting in pBS-Sdr4KN. The 11.6-kb KpnI–SmaI fragment, the 11.2-kb KpnI–SmaI fragment with internal deletion of a 0.4-kb BamHI–BamHI fragment, or the 3.3-kb BamHI–ApaLI fragment of pBS-Sdr4KN was inserted into pPZP2Hlac, yielding pPZP-Sdr4KN, pPZP-Sdr4KNd, or pPZP-Sdr4KNa, respectively. To produce a construct for knockdown, a fragment containing the 3′-UTR (nt 1,104–1,294 of cDNA) and part of the GUS gene (345 nt) as a spacer were amplified using primers with the created restriction sites, digested with the appropriate enzymes, and then inserted immediately into pPZP-Ha3(+), resulting in pPZP-35S-Sdr4i. These plasmids were introduced into Agrobacterium (EHA101) and transformed into Nipponbare, M100 mutant, or NIL[Sdr4] by an Agrobacterium-mediated rapid method (50).
RNA Preparation and Real-Time PCR.
RNAs were purified using the RNeasy Plant Kit with on-column DNaseI treatment (Qiagen). cDNA obtained by reverse-transcription reaction using a ReverTra Ace alpha kit (Toyobo) was amplified by conventional PCR or subjected to real-time PCR using an SYBR Green–based kit (QPK-212; Toyobo) and an ABI 7900HT system (Applied Biosystems). Primer sequences for the amplifications are listed in Table S2B.
Measurement of ABA.
Seeds (500 mg) harvested at 6 WAH were extracted in aqueous methanol and purified by HPLC. Their ABA content was quantified by GC-MS as described by Iuchi et al. (51).
Subcellular Localization of Sdr4.
The Sdr4 coding sequence was inserted in front of the 5′ end of the GFP (sGFP) in pCaMV35S-sGFP(S65T)-nos3′, resulting in p35S-Sdr4n-GFP. This plasmid was introduced into the protoplasts of cultured rice cells (oc cells) by electroporation, and localization of the GFP fluorescence was observed under a confocal laser scanning microscope (FluoView; Olympus).
Sequencing of Sdr4 in Rice Accessions.
The genomic sequences around the Sdr4 ORF were amplified by PCR with the primer sets 87535F (5′-ccg ccc acg cct tct taa cc-3′) and 88688R (5′-aaa gtt tgc tcc ggc ttg atg c-3′). The PCR products were purified with a MagExtractor PCR and Gel Extraction Kit (Toyobo) and used directly for sequence reaction.
Statistical Methods.
Association of the Sdr4 allele with germination rate was tested using a general linear model. The significance of associations between the allele and traits was based on the F test. All statistical analyses were done with SPSS version 15 J for Windows (SPSS Inc.).
Supplementary Material
Acknowledgments
We thank Dr. Y. Niwa (University of Shizuoka) for kindly providing the sGFP(T65S) gene. We also acknowledge the help of T. Ando of the STAFF Institute and S. Toki, Y. Eguchi, and T. Tokunaga of the National Institute of Agrobiological Science. The wild rice accessions used in this study were distributed by the National Institute of Genetics with the support of the National Bioresource Project of the Ministry of Education, Culture, Sports, Science and Technology of Japan. This work was supported by grants from the Ministry of Agriculture, Forestry and Fisheries of Japan (Integrated Research Project for Plant, Insect, and Animal using Genome Technology Grant GD-3003 and Genomics for Agricultural Innovation Grant IPG-0010).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. AB506455 (Sdr4-k) and AB510199 (Sdr4-n)].
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/cgi/content/full/0911965107/DCSupplemental.
References
- 1.Kovach MJ, Sweeney MT, McCouch SR. New insights into the history of rice domestication. Trends Genet. 2007;23:578–587. doi: 10.1016/j.tig.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 2.Izawa T, Konishi S, Shomura A, Yano M. DNA changes tell us about rice domestication. Curr Opin Plant Biol. 2009;12:185–192. doi: 10.1016/j.pbi.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 3.Hilhorst HWM. In: Seed Development, Dormancy and Germination. Bradford KJ, Nonogaki H, editors. Sheffield, UK: Blackwell; 2007. pp. 50–71. [Google Scholar]
- 4.Koornneef M, Hanhart CJ, Hilhorst HW, Karssen CM. In vivo inhibition of seed development and reserve protein accumulation in recombinants of abscisic acid biosynthesis and responsiveness mutants in Arabidopsis thaliana. Plant Physiol. 1989;90:463–469. doi: 10.1104/pp.90.2.463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McCarty DR, et al. The Viviparous-1 developmental gene of maize encodes a novel transcriptional activator. Cell. 1991;66:895–905. doi: 10.1016/0092-8674(91)90436-3. [DOI] [PubMed] [Google Scholar]
- 6.Hattori T, Terada T, Hamasuna ST. Sequence and functional analyses of the rice gene homologous to the maize Vp1. Plant Mol Biol. 1994;24:805–810. doi: 10.1007/BF00029862. [DOI] [PubMed] [Google Scholar]
- 7.Liu Y, Koornneef M, Soppe WJ. The absence of histone H2B monoubiquitination in the Arabidopsis hub1 (rdo4) mutant reveals a role for chromatin remodeling in seed dormancy. Plant Cell. 2007;19:433–444. doi: 10.1105/tpc.106.049221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bentsink L, Jowett J, Hanhart CJ, Koornneef M. Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis. Proc Natl Acad Sci USA. 2006;103:17042–17047. doi: 10.1073/pnas.0607877103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lin SY, Sasaki T, Yano M. Mapping quantitative trait loci controlling seed dormancy and heading date in rice, Oryza sativa L., using backcross inbred lines. Theor Appl Genet. 1998;96:997–1003. [Google Scholar]
- 10.Gu XY, Kianian SF, Foley ME. Multiple loci and epistases control genetic variation for seed dormancy in weedy rice (Oryza sativa) Genetics. 2004;166:1503–1516. doi: 10.1534/genetics.166.3.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guo L, et al. QTL analysis of seed dormancy in rice (Oryza sativa L.) Euphytica. 2004;140:155–162. [Google Scholar]
- 12.Gu XY, Liu T, Feng J, Suttle JC, Gibbons J. The qSD12 underlying gene promotes abscisic acid accumulation in early developing seeds to induce primary dormancy in rice. Plant Mol Biol. 2009 doi: 10.1007/s11103-009-9555-1. in press. [DOI] [PubMed] [Google Scholar]
- 13.Robbins J, Dilworth SM, Laskey RA, Dingwall C. Two interdependent basic domains in nucleoplasmin nuclear targeting sequence: Identification of a class of bipartite nuclear targeting sequence. Cell. 1991;64:615–623. doi: 10.1016/0092-8674(91)90245-t. [DOI] [PubMed] [Google Scholar]
- 14.Wessler S, Tarpley A, Purugganan M, Spell M, Okagaki R. Filler DNA is associated with spontaneous deletions in maize. Proc Natl Acad Sci USA. 1990;87:8731–8735. doi: 10.1073/pnas.87.22.8731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Baumlein H, Nagy I, Villarroel R, Inze D, Wobus U. Cis-analysis of a seed protein gene promoter: The conservative RY repeat CATGCATG within the legumin box is essential for tissue-specific expression of a legumin gene. Plant J. 1992;2:233–239. [PubMed] [Google Scholar]
- 16.Shen Q, Zhang P, Ho TH. Modular nature of abscisic acid (ABA) response complexes: Composite promoter units that are necessary and sufficient for ABA induction of gene expression in barley. Plant Cell. 1996;8:1107–1119. doi: 10.1105/tpc.8.7.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hobo T, Asada M, Kowyama Y, Hattori T. ACGT-containing abscisic acid response element (ABRE) and coupling element 3 (CE3) are functionally equivalent. Plant J. 1999;19:679–689. doi: 10.1046/j.1365-313x.1999.00565.x. [DOI] [PubMed] [Google Scholar]
- 18.Hattori T, Totsuka M, Hobo T, Kagaya Y, Yamamoto-Toyoda A. Experimentally determined sequence requirement of ACGT-containing abscisic acid response element. Plant Cell Physiol. 2002;43:136–140. doi: 10.1093/pcp/pcf014. [DOI] [PubMed] [Google Scholar]
- 19.Suzuki M, Ketterling MG, McCarty DR. Quantitative statistical analysis of cis-regulatory sequences in ABA/VP1- and CBF/DREB1-regulated genes of Arabidopsis. Plant Physiol. 2005;139:437–447. doi: 10.1104/pp.104.058412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hattori T, Terada T, Hamasuna S. Regulation of the Osem gene by abscisic acid and the transcriptional activator VP1: Analysis of cis-acting promoter elements required for regulation by abscisic acid and VP1. Plant J. 1995;7:913–925. doi: 10.1046/j.1365-313x.1995.07060913.x. [DOI] [PubMed] [Google Scholar]
- 21.Ross C, Shen QJ. Computational prediction and experimental verification of HVA1-like abscisic acid responsive promoters in rice (Oryza sativa) Plant Mol Biol. 2006;62:233–246. doi: 10.1007/s11103-006-9017-y. [DOI] [PubMed] [Google Scholar]
- 22.Carrera E, et al. Seed after-ripening is a discrete developmental pathway associated with specific gene networks in Arabidopsis. Plant J. 2008;53:214–224. doi: 10.1111/j.1365-313X.2007.03331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oikawa T, Koshioka M, Kojima K, Yoshida H, Kawata M. A role of OsGA20ox1, encoding an isoform of gibberellin 20-oxidase, for regulation of plant stature in rice. Plant Mol Biol. 2004;55:687–700. doi: 10.1007/s11103-004-1692-y. [DOI] [PubMed] [Google Scholar]
- 24.Liu HY, et al. The role of water channel proteins and nitric oxide signaling in rice seed germination. Cell Res. 2007;17:638–649. doi: 10.1038/cr.2007.34. [DOI] [PubMed] [Google Scholar]
- 25.Lee Y, Kende H. Expression of beta-expansins is correlated with internodal elongation in deepwater rice. Plant Physiol. 2001;127:645–654. [PMC free article] [PubMed] [Google Scholar]
- 26.Howell KA, et al. Mapping metabolic and transcript temporal switches during germination in rice highlights specific transcription factors and the role of RNA instability in the germination process. Plant Physiol. 2009;149:961–980. doi: 10.1104/pp.108.129874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kojima Y, Ebana K, Fukuoka S, Nagamine T, Kawase M. Development of an RFLP-based rice diversity research set of germplasm. Breed Sci. 2005;55:431–440. [Google Scholar]
- 28.Second G. Oribin of the genetic diversity of cultvated rice (Oryza spp.): Study of the polymorphism scored at 40 isozyme loci. Jpn J Genet. 1982;57:25–57. [Google Scholar]
- 29.Garris AJ, Tai TH, Coburn J, Kresovich S, McCouch S. Genetic structure and diversity in Oryza sativa L. Genetics. 2005;169:1631–1638. doi: 10.1534/genetics.104.035642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nambara E, Keith K, McCourt P, Naito S. Isolation of an internal deletion mutant of the Arabidopsis thaliana ABI3 gene. Plant Cell Physiol. 1994;35:509–513. [PubMed] [Google Scholar]
- 31.Hoecker U, Vasil IK, McCarty DR. Signaling from the embryo conditions Vp1-mediated repression of alpha-amylase genes in the aleurone of developing maize seeds. Plant J. 1999;19:371–377. doi: 10.1046/j.1365-313x.1999.00521.x. [DOI] [PubMed] [Google Scholar]
- 32.Barrero JM, Talbot MJ, White RG, Jacobsen JV, Gubler F. Anatomical and transcriptomic studies of the coleorhiza reveal the importance of this tissue in regulating dormancy in barley. Plant Physiol. 2009;150:1006–1021. doi: 10.1104/pp.109.137901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fujino K, et al. Molecular identification of a major quantitative trait locus, qLTG3-1, controlling low-temperature germinability in rice. Proc Natl Acad Sci USA. 2008;105:12623–12628. doi: 10.1073/pnas.0805303105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bentsink L, Soppe W, Koornneef M. In: Seed Development, Dormancy and Germination. Bradford KJ, Nonogaki H, editors. Oxford, UK: Blackwell; 2007. pp. 113–132. [Google Scholar]
- 35.Anderson JA, Sorrells ME, Tanksley SD. RFLP analysis of genomic regions associated with resistance of preharvest sprouting in wheat. Crop Sci. 1993;33:453–459. [Google Scholar]
- 36.Han F, et al. Verification of barley seed dormancy loci via linked molecular markers. Theor Appl Genet. 1996;92:87–91. doi: 10.1007/BF00222956. [DOI] [PubMed] [Google Scholar]
- 37.Luo MC, et al. Genome comparisons reveal a dominant mechanism of chromosome number reduction in grasses and accelerated genome evolution in Triticeae. Proc Natl Acad Sci USA. 2009;106:15780–15785. doi: 10.1073/pnas.0908195106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ren XB, Lan XJ, Liu DC, Wang JL, Zheng YL. Mapping QTLs for pre-harvest sprouting tolerance on chromosome 2D in a synthetic hexaploid wheat x common wheat cross. J Appl Genet. 2008;49:333–341. doi: 10.1007/BF03195631. [DOI] [PubMed] [Google Scholar]
- 39.Fujii H, et al. In vitro reconstitution of an abscisic acid signaling pathway. Nature. 2009;462:660–664. doi: 10.1038/nature08599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Umezawa T, et al. Type 2C protein phosphatases directly regulate abscisic acid–activated protein kinases in Arabidopsis. Proc Natl Acad Sci USA. 2009;106:17588–17593. doi: 10.1073/pnas.0907095106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Finkelstein RR, Gampala SS, Rock CD. Abscisic acid signaling in seeds and seedlings. Plant Cell. 2002;14(Suppl):S15–S45. doi: 10.1105/tpc.010441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li C, Zhou A, Sang T. Rice domestication by reducing shattering. Science. 2006;311:1936–1939. doi: 10.1126/science.1123604. [DOI] [PubMed] [Google Scholar]
- 43.Konishi S, et al. An SNP caused loss of seed shattering during rice domestication. Science. 2006;312:1392–1396. doi: 10.1126/science.1126410. [DOI] [PubMed] [Google Scholar]
- 44.Sweeney MT, et al. Global dissemination of a single mutation conferring white pericarp in rice. PLoS Genet. 2007;3:e133. doi: 10.1371/journal.pgen.0030133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shomura A, et al. Deletion in a gene associated with grain size increased yields during rice domestication. Nat Genet. 2008;40:1023–1028. doi: 10.1038/ng.169. [DOI] [PubMed] [Google Scholar]
- 46.Cheng C, et al. Polyphyletic origin of cultivated rice: Based on the interspersion pattern of SINEs. Mol Biol Evol. 2003;20:67–75. doi: 10.1093/molbev/msg004. [DOI] [PubMed] [Google Scholar]
- 47.Lu T, et al. Collection and comparative analysis of 1888 full-length cDNAs from wild rice Oryza rufipogon Griff. W1943. DNA Res. 2008;15:285–295. doi: 10.1093/dnares/dsn018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamanaka S, Nakamura I, Watanabe KN, Sato Y. Identification of SNPs in the waxy gene among glutinous rice cultivars and their evolutionary significance during the domestication process of rice. Theor Appl Genet. 2004;108:1200–1204. doi: 10.1007/s00122-003-1564-x. [DOI] [PubMed] [Google Scholar]
- 49.Miyao A, et al. A large-scale collection of phenotypic data describing an insertional mutant population to facilitate functional analysis of rice genes. Plant Mol Biol. 2007;63:625–635. doi: 10.1007/s11103-006-9118-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Toki S, et al. Early infection of scutellum tissue with Agrobacterium allows high-speed transformation of rice. Plant J. 2006;47:969–976. doi: 10.1111/j.1365-313X.2006.02836.x. [DOI] [PubMed] [Google Scholar]
- 51.Iuchi S, Kobayashi M, Yamaguchi-Shinozaki K, Shinozaki K. A stress-inducible gene for 9-cis-epoxycarotenoid dioxygenase involved in abscisic acid biosynthesis under water stress in drought-tolerant cowpea. Plant Physiol. 2000;123:553–562. doi: 10.1104/pp.123.2.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tanaka T, et al. The Rice Annotation Project Database (RAP-DB): 2008 update. Nucleic Acids Res. 2008;36:D1028–D1033. doi: 10.1093/nar/gkm978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.