Abstract
Differentiation-inducing factor 1 (DIF-1) is a polyketide-derived morphogen which drives stalk cell formation in the developmental cycle of Dictyostelium discoideum. Previous experiments demonstrated that the biosynthetic pathway proceeds via dichlorination of the precursor molecule THPH, but the enzyme responsible for this transformation has eluded characterization. Our recent studies on prokaryotic flavin-dependent halogenases and insights from the sequenced Dd genome led us to a candidate gene for this transformation. In this work, we present in vivo and in vitro evidence that chlA from Dd encodes a flavin-dependent halogenase capable of catalyzing both chlorinations in the biosynthesis of DIF-1. The results provide in vitro characterization of a eukaryotic oxygen-dependent halogenase and demonstrate a broad reach in biology for this molecular tailoring strategy, notably its involvement in the differentiation program of a social amoeba.
Keywords: Dictyostelium development, enzymatic halogenation, polyketide, natural products
Secondary metabolites are widely produced by microorganisms, ostensibly as defensive molecules to inhibit the growth of competitors in the natural environment. Though antibiosis is a widely appreciated effect of this secondary metabolism, there is a developing understanding that such molecules can also modulate more complex inter- and intraspecies interactions (1–3). For instance, quorum sensing molecules secreted by various bacterial species serve as indicators of local population density and modulate coordinated responses (e.g., luminescence, virulence) in appropriate environmental conditions (4). Bacillus subtilus has also been shown recently to generate a paracrine signal, using the small molecule surfactin to induce differentiation of a subset of cells during biofilm formation (5).
Social amoebas, represented by the model species Dictyostelium discoideum, also use a sophisticated system of chemical messengers in the regulation of their unique life cycle (6, 7). In nutrient-rich conditions, Dictyostelium exists as single-cell amoeba which grow and reproduce by binary fission. Starvation of the solitary amoeba induces a cAMP-mediated aggregation response to initiate a multicellular stage of development. Aggregated cells first form an undifferentiated mound comprised of up to 105 cells. This grouping of cells then reorganizes into a motile slug form by which point cells show clear evidence of differentiation towards stalk or spore formation. After migrating along chemical, thermal, and light gradients to the soil surface, the organism completes its development into a fruiting body in which the sorus composed of reproductive spores is lifted above the substrate by a cellulose-rich stalk structure.
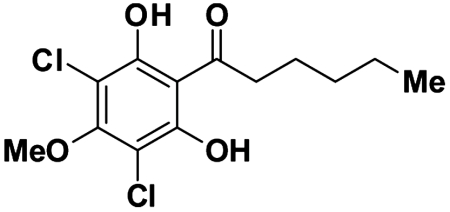
Small molecule signals have been shown to mediate multiple steps in this developmental process. Differentiation-inducing factor 1 (DIF-1, Fig. 1) was one of the first morphogens to be identified in Dictyostelium (8, 9). It rapidly induces prestalk-specific gene transcription in responsive cells and is the primary signal directing the differentiation of cells into the prestalk-O (pstO) lineage (10, 11). The stalk cells are not competent for reproduction, so the individual cells responding to the DIF-1 signal are sacrificed for the reproductive benefit of the overall community, representing an interesting case of evolved altruism. DIF-1 biosynthesis and activity has been a long-standing topic of interest due to its essential role in the proper developmental program of Dictyostelium. DIF-1 has also garnered interest as a lead compound with various biological activities in mammalian cells (12–14).
Fig. 1.
The chemical structure of DIF-1.
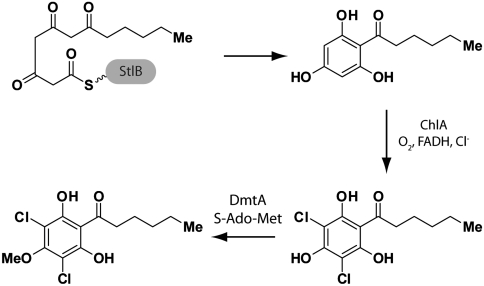
Early studies in Dictyostelium helped to define a three-step biosynthetic pathway for production of DIF-1 (15). Formation of the core polyketide (2,4,6-trihydroxyphenyl)-1-hexan-1-one (THPH) is followed in order by dichlorination of the aromatic ring and O-methylation. The first and last steps of this pathway have been well characterized. In 2006, the Kay and Noel laboratories reported a unique Type I-Type III fused polyketide synthase, StlB, which is responsible for production of THPH (16). In vitro, the purified Type III domain was observed to catalyze formation of THPH from hexanoyl-CoA and three molecules of malonyl-CoA. Previously, the Kay laboratory identified dmtA as the gene encoding the O-methyltransferase (10). Purified DmtA protein was shown to methylate both monochloro- and dichloro-THPH. Both stlB- and dmtA- strains form irregular slug and fruiting body structures, defining a “DIF-less” phenotype (10, 16).
Chlorination is an essential step in the maturation of DIF-1. Not only is chlorination of THPH a prerequisite for methylation by DmtA (15), but DIF-1 is 25 times more potent than its monochloro analogue (the known metabolite DIF-3) in inducing stalk cell differentiation (17). The chlorination of THPH has only been observed as an enzymatic activity in crude lysates of Dictyostelium (15). Chlorinating activity is low in vegetative cells but rises substantially at the end of aggregation, mimicking the rise in enzymatic activity seen for the methyltransferase. The chlorinase activity requires components from both the cytosolic and membrane/organelle fractions of the preparation, with the soluble component likely being a small molecule (R.R.K., unpublished). The enzymatic activity was stimulated by very high levels (50 mM) of hydrogen peroxide, leading to an initial hypothesis that a chloroperoxidase enzyme was responsible for this transformation. Since these initial results, the sequencing of the Dictyostelium genome (18) and advances in the understanding of enzymatic halogenation have allowed new insight into this critical step in DIF-1 biosynthesis.
Studies of halogenated natural products from microbial sources have recently defined a new family of O2-and flavin-dependent halogenases which are distinct from the peroxide-dependent haloperoxidases characterized many years ago in fungi (19). The mechanism of halogenation is initiated by reaction of reduced flavin cofactor with molecular oxygen in one enzyme active site. The bridgehead peroxyflavin thus generated reacts with chloride ion to produce hypochlorous acid which diffuses through a tunnel approximately 10 Å in length to a second active site (20). In this site, hypochlorous reacts first with a conserved lysine residue to generate a metastable lysine-chloramine; it is this chloramine species which serves as the proximal donor of Cl+ to an electron rich substrate (21). Genes for flavin-dependent halogenases are found in a number of prokaryotic and fungal biosynthetic gene clusters for secondary metabolites. These compounds include a wide variety of antibacterials (e.g. chlortetracycline), antifungals (e.g., kutznerides), and anticancer agents (e.g., rebeccamycin) (22–24). In vitro biochemical characterization has been performed on prokaryote-derived enzymes which act on amino acids (either free in solution or bound to carrier proteins), though genetic and bioinformatic approaches have implicated these halogenases in the tailoring of diverse polyketide and eukaryote-derived natural products as well (25, 26).
Sequencing of the Dictyostelium genome allowed identification of a putative flavin-dependent halogenase which we have termed chlA (27). In consideration of the two-component nature of the crude enzymatic activity, the genomic clustering of chlA with stlB (see below), and the sequence homology of ChlA to known halogenases, we hypothesized that ChlA is responsible for the dichlorination of THPH in the biosynthesis of DIF-1. Through a combination of in vitro and in vivo experiments, this study shows that ChlA is the essential halogenase in DIF-1 biosynthesis and that it employs the same mechanistic logic used by prokaryotic halogenases. In doing so, we demonstrate that this enzymatic machinery plays an essential role in the biosynthesis of signaling molecules to direct the developmental program of a eukaryotic organism.
Results and Discussion
Genetic Context and Transcription of chlA.
The chlA gene (DDB_G0290825 at dictyBase, http://dictybase.org/) is located on chromosome 5 adjacent to stlB, and divergently transcribed from it, with 2.7 kb of intervening noncoding sequence. There are no introns present in chlA and the GC content is 31%, consistent with overall gene GC content of 27% in Dictyostelium. A highly similar gene (79% amino acid identity) is present in the genome of Dictyostelium purpureum, a dictyostelid which may also use DIF-1 as a morphogen.
A restriction-enzyme-mediated insertional mutant at the chlA locus (V31853) was previously identified in a genetic screen for developmental phenotypes and was reported to show aberrant aggregation, cAMP wave formation, slug motility, and culmination (28). As part of a detailed phylogenetic analysis of halogenases involved in cyanopeptolin and aeruginosin biosynthesis, Cadel-Six et al. have previously described the close sequence relationship of ChlA to known and predicted prokaryotic halogenases (29).
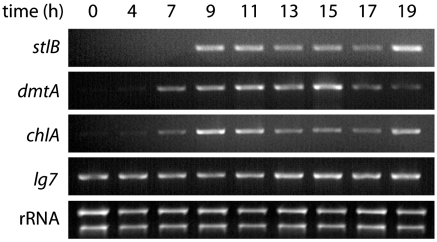
Up-regulation of stlB and dmtA transcription has been observed during development, coinciding with the first finger-slug developmental stages and production of DIF-1 (10, 16). Enzymatic chlorinating activity has also been shown to reach a maximum at this time. To assess if the activation of chlA transcription coincides with these events, we collected RNA from developing cells at consecutive points in the developmental process. RT-PCR was used to detect the presence of transcripts corresponding to chlA as well as the known genes stlB and dmtA (Fig. 2). Transcription of chlA is up-regulated during early development in synchrony with stlB and dmtA. Low levels of chlA transcription were also observed at 0 and 4 h consistent with a previous observation that lysates of undeveloped cells exhibit very weak in vitro chlorinating activity (15).
Fig. 2.
RT-PCR analysis shows transcription of chlA is up-regulated along with stlB and dmtA at 7–9 h of development. lg7 (mitochondrial large rRNA) and total rRNA serve as PCR and loading controls, respectively.
Purification and Biochemical Analysis of ChlA.
In order to characterize the enzymatic activity of ChlA, the protein was targeted for overproduction and purification. Because of initial solubility problems in Escherichia coli, we pursued purification by homologous overexpression in Dictyostelium. The native gene was cloned from genomic DNA and inserted into the pTX-FLAG expression vector (30) with N-terminal FLAG and C-terminal c-myc tags for affinity purification. Expression in this vector is under the control of the constitutive actin15 promoter. Electroporation and G418 selection provided a polyclonal population of transformants expressing soluble ChlA during axenic growth as evidenced by Western blot (Fig. S1A). The observed solubility of the protein here contrasts with the previous studies which suggested the protein component of the chlorinase activity was located in the insoluble/organelle fraction of the crude lysate. It is possible that the native protein expressed during development is localized to a specific organelle or otherwise processed in such a way as to associate with insoluble membranes. Overproduction and/or epitope tagging of the protein could be affecting any such process. Large-scale growth followed by lysis and immunoaffinity purification with an anti-FLAG resin provided partially purified ChlA for biochemical analysis (Fig. S1B). The ChlA K86A mutant, deficient in the conserved catalytic lysine residue, was generated by site-directed mutagenesis and purified by the identical procedure.
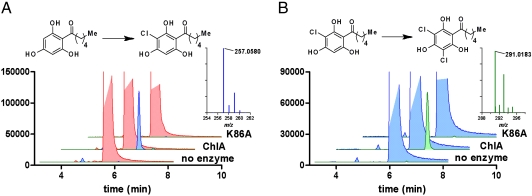
Studies of prokaryotic halogenases have established the requirement for FADH, O2, and chloride ion for catalysis. The halogenase activity of ChlA was successfully reconstituted in an in vitro assay comprising these reagents. Reduced flavin cofactor was provided by in situ reduction of FAD by the NAD(P)H-dependent flavin reductase SsuE from E. coli (31), although halogenase activity was also observed in the absence of SsuE, presumably stimulated by an unknown copurifying flavin reductase. Treatment of THPH with ChlA for 2 h at room temperature afforded a substoichiometric amount of a new compound whose HPLC retention time and UV spectrum (λmax = 286 nm) match that of synthetic (3-chloro-2,4,6-trihydroxyphenyl)-1-hexan-1-one (Cl-THPH, Fig. S2). Further analysis of the reaction mixture by high resolution liquid chromatography-mass spectrometry (LCMS) confirmed the identity of Cl-THPH based on its retention time, molecular weight ([M-H]- = 257.0580, 0.39 ppm error), and isotopic distribution (Fig. 3A). A trace amount of (3,5-dichloro-2,4,6-trihydroxyphenyl)-1-hexan-1-one (Cl2-THPH, [M-H]- = 291.0183, 2.7 ppm error) was also detected, though the intensity of higher molecular weight isotopomers was insufficient to establish the distribution ratio. We reasoned that the trace formation of Cl2-THPH resulted from the low concentration of the Cl-THPH intermediate, likely far below its Km. To verify that ChlA is indeed capable of catalyzing the second halogenation event, we separately incubated purified enzyme with high levels (500 μM) of Cl-THPH. In this case Cl2-THPH was readily detected by LCMS (Fig. 3B) and matched the spectral data for the synthetic standard. When ChlA K86A was tested there was no increase in product formation relative to no-enzyme control reactions, consistent with the proposed role of Lys-86 chloramine formation (Fig. 3). Additional control experiments were run in the absence of exogenous NADH and FAD and showed no formation of chlorinated products confirming the requirement for these redox cofactors.* Insolubility of the THPH and Cl-THPH substrates above 500 μM prevented accurate measurement of Km values, but we estimate them to be in the 200–400 μM range.
Fig. 3.
Extracted ion chromatograms for in vitro activity of ChlA. (A) Conversion of THPH (red, truncated) to Cl-THPH (blue) is catalyzed by ChlA, but not the ChlA K86A mutant. Inset MS shows observed spectrum for Cl-THPH ([M-H]-: predicted = 257.0581, observed = 257.0580, 0.39 ppm error). (B) Conversion of Cl-THPH (blue, truncated) to Cl2-THPH (green). Inset MS shows observed spectrum for Cl2-THPH ([M-H]-: predicted = 291.0191, observed = 257.0183, 2.7 ppm error).
Because of the slow conversion of THPH into either Cl-THPH or Cl2-THPH, we sought further evidence to verify that product formation is enzyme dependent. In a time course experiment, yield was dependent on length of incubation in a roughly linear fashion consistent with steady state enzymatic behavior (Fig. S3). The reason for the low enzymatic activity is not clear at this point in our studies. It is possible that the coupling efficiency between the heterologous flavin reductase and halogenase is not optimal in these in vitro reactions; in vivo, the unidentified flavin reductase endogenous to Dictyostelium might be more effective in channeling the reduced cofactor into the halogenase active site. Alternatively, it is possible that only a fraction of the purified enzyme is catalytically active. Nonetheless, the data support our initial hypothesis that ChlA is capable of catalyzing both chlorination events in the biosynthesis of DIF-1.
In Vivo Analysis of DIF-1 Production.
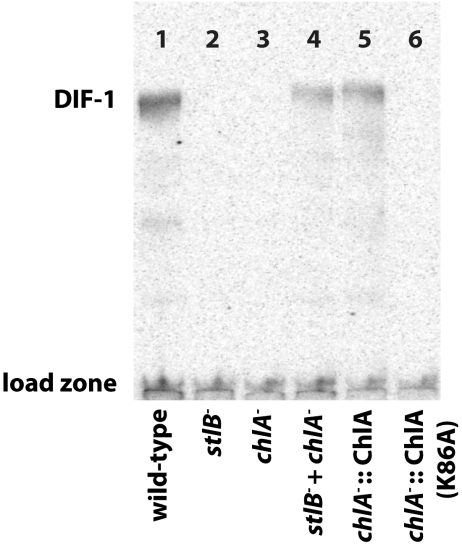
To directly assess the role of ChlA in production of DIF metabolites in Dictyostelium, the gene was targeted by homologous recombination to produce a large internal deletion, spanning amino acids 18-273, and thus including the essential residue K86. Two individual blasticidin resistant clones of the desired genomic structure were verified by PCR analysis. Developing cells were labeled with 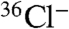 to permit identification of chlorinated metabolites by autoradiography. Organic-soluble metabolites were extracted after 16–17 h of development, at the slug/early culminate stage, and analyzed by radio-TLC. In each of three separate trials, the parental Ax2 strain produces a detectable amount of the DIF-1 compound whereas the mutant strains are completely deficient in chlorinated compounds (Fig. 4, lanes 1 and 3).
to permit identification of chlorinated metabolites by autoradiography. Organic-soluble metabolites were extracted after 16–17 h of development, at the slug/early culminate stage, and analyzed by radio-TLC. In each of three separate trials, the parental Ax2 strain produces a detectable amount of the DIF-1 compound whereas the mutant strains are completely deficient in chlorinated compounds (Fig. 4, lanes 1 and 3).
Fig. 4.
Radio-TLC analysis of DIF-1 production in vivo. The knockout strain chlA- (HM1522) is unable to produce detectable DIF-1 during development, unlike its wild-type parent, Ax2. Complementation can be achieved by codeveloping with a stlB- strain (HM1154) or by expression of ChlA, but not by expression of ChlA K86A.
A possible explanation for the failure of the chlA- mutant to make DIF is that the adjacent stlB gene, or its promoter, is inadvertently disrupted, thus blocking THPH production. To rule out this scenario, chlA- and stlB- (HM1154) strains were plated as single and mixed populations and labeled with 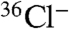 . Individually, the chlA- and stlB- strains are deficient in production of DIF-1, but codevelopment of the two strains restores production of DIF-1 (Fig. 4, lanes 2–4). Because of the cell-permeable nature of these metabolites, we can conclude that the chlA- strain is capable of producing THPH while the stlB- strain is capable of using this substrate to synthesize Cl2-THPH. In this way, the two strains are able to complement each other and reconstitute a fully functional DIF-1 biosynthetic pathway. Additionally, inclusion of THPH in the agarose substratum restores DIF-1 production in stlB- but not chlA- strains (Fig. S4).
. Individually, the chlA- and stlB- strains are deficient in production of DIF-1, but codevelopment of the two strains restores production of DIF-1 (Fig. 4, lanes 2–4). Because of the cell-permeable nature of these metabolites, we can conclude that the chlA- strain is capable of producing THPH while the stlB- strain is capable of using this substrate to synthesize Cl2-THPH. In this way, the two strains are able to complement each other and reconstitute a fully functional DIF-1 biosynthetic pathway. Additionally, inclusion of THPH in the agarose substratum restores DIF-1 production in stlB- but not chlA- strains (Fig. S4).
To demonstrate the in vivo activity of cloned ChlA, we transformed the chlA- strain with the expression plasmids used for protein purification. Expression of wild-type ChlA restored the production of DIF-1 as evidenced by 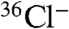 radio-TLC, whereas expression of the ChlA K86A mutant failed to restore DIF-1 biosynthesis (Fig. 4, lanes 5–6). These results provide additional confirmation that a deficiency in ChlA disrupts DIF-1 biosynthesis and that K86 is an essential residue for catalysis.
radio-TLC, whereas expression of the ChlA K86A mutant failed to restore DIF-1 biosynthesis (Fig. 4, lanes 5–6). These results provide additional confirmation that a deficiency in ChlA disrupts DIF-1 biosynthesis and that K86 is an essential residue for catalysis.
Developmental Phenotype of chlA-.
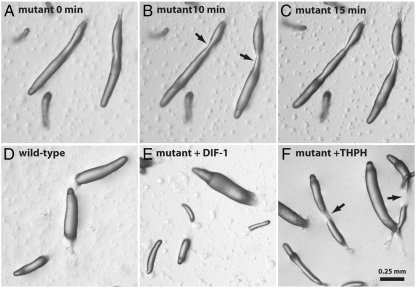
Analyses of stlB- and dmtA- strains have previously defined a “DIF-less” phenotype associated with disruption of the DIF biosynthetic pathway (10, 16, 32, 33). The chlA- strain exhibits the same deficiencies in slug structure and fruiting body formation seen in these other strains. Most notably, mutant cells reproducibly form elongated slugs that often break apart shortly after touching down on the substrate (Fig. 5 A–C). In addition, spore cells are unable to properly ascend the weakened stalk and the collapsed fruiting body structures lead to a disheveled appearance at the end of development.
Fig. 5.
The developmental phenotype of a chlA- mutant. Slugs formed by the mutant (HM1522) are elongated and fragile, breaking apart shortly after touching down on the agar surface (A–C). Wild-type Ax2 slug morphology (D) is recovered in the mutants by supplementing the agar with 100 nM DIF-1 (E), but not with the halogenase substrate THPH (F).
Development of the mutant strains on agar containing 100 nM DIF-1 restores wild-type slug morphology to the strains (Fig. 5 D and E), suggesting that a deficiency in this metabolite is responsible for the observed phenotype. In addition, development in the presence of Cl2-THPH also rescues wild-type development, demonstrating that this intermediate lies downstream of the blocked step in DIF-1 biosynthesis (Fig. S5). When developed in the presence of 100 nM THPH, the mutant cells retain the fragile slug phenotype (Fig. 5F), again indicating that disruption of the adjacent stlB gene is not responsible for the DIF-less phenotype seen in HM1522.
As a final test of our hypothesis, we evaluated the developmental phenotype of the chlA- strain transformed with expression vectors containing the epitope-tagged chlA gene or the gene encoding for the K86A mutant (Fig. S5). Expression of the wild-type gene restored proper slug development and fruiting body formation, whereas the K86A mutant retained the DIF-less phenotype. In addition to verifying that the chlA gene alone is sufficient to restore normal development, these data suggest that the tagged construct achieves sufficient activity in vivo to allow critical flux through the DIF pathway.
Conclusions
The biochemical basis for halogenation of DIF-1 has remained poorly defined since the first enzymatic activity was detected in crude lysates (15). The combination of in vitro and in vivo experiments in this study conclusively demonstrate that both chlorination events in this pathway are carried out by the flavin-dependent halogenase ChlA. The chlA gene is clustered with stlB, the polyketide synthase involved in THPH formation, the first step in DIF-1 biosynthesis. Increased transcription of chlA coincides with rises in stlB and dmtA transcription at ∼9–11 h of development. The chlorination of THPH and Cl-THPH can be reconstituted in vitro using purified ChlA protein. Lysine-86 has been identified as an essential catalytic residue, consistent with the lysine-chloramine model of halogenation proposed for this family.
In vivo, disruption of chlA leads to the characteristic DIF-less phenotype of fragile slug formation and collapsed fruiting bodies. More directly, the synthesis of DIF-1 and related metabolites is also inhibited by this mutation. ChlA deficiency can be complemented by codevelopment with a stlB- mutant, by development on agar containing DIF-1 or Cl2-THPH, and by transformation with an expression plasmid for ChlA. These experiments therefore draw direct links between the biochemistry of ChlA seen in vitro, the in vivo production of DIF-1, and the physiological process of Dictyostelium development.
This study also serves to complete the molecular characterization of the DIF-1 biosynthetic pathway (Fig. 6). The three-step process first proposed is now shown to be composed of THPH formation by an unusual Type I-Type III polyketide synthase StlB, dichlorination by the flavin-dependent halogenase ChlA, and O-methylation by DmtA. Biochemically, the characterization of ChlA provides a foundation for the further analysis of polyketide halogenases. Upon optimization of expression and reconstitution methods we can further explore kinetic parameters and substrate specificity, with an eye towards manipulating specificity determinants for the generation of novel analogues.
Fig. 6.
The complete molecular-level description of the DIF-1 biosynthetic pathway, consisting of StlB-mediated polyketide formation, ChlA-mediated dichlorination, and DmtA mediated O-methylation.
Finally, our results illustrate the broad reach of halogenation chemistry in biological systems by showing that a flavin-dependent halogenase plays an essential role in the maturation of a chemical signal for cell differentiation in a eukaryotic organism. It serves as a reminder that, beyond defensive roles in competitive environments, small “secondary” metabolites can serve as vital signals for essential biological processes in higher organisms.
Materials and Methods
Dictyostelium Growth and Manipulation.
Dictyostelium strains Ax2 (Kay laboratory, Dictyostelium Stock Center ID DBS0235521) and Ax4 were propagated on lawns of Klebsiella aerogenes or axenically in HL5 medium according to standard published protocols (34). Genomic DNA was isolated by phenol:chloroform extraction (35). Cloning vectors were propagated in E. coli Top-10 cells (Invitrogen). PCR reactions were performed using Phusion High Fidelity polymerase in 50 ul reaction volumes. The chlA gene was amplified using primers ATTATTCGAGCTCATGGATACAAATATTATTAATC (forward) and ATTATTCGAGCTCATACTTCCAATCAACAATATTATT (reverse), digested with SacI (sites underlined), and ligated into SacI-digested pTX-FLAG vector (30) to provide N-terminal FLAG and C-terminal c-myc tags. The expression vector for ChlA K86A was constructed by PCR-mediated site-directed mutagenesis of the wild-type expression vector with primers ATAGAGAATCATCCACCAGCATTTTCATTACAATTTCAT and ATGAAATTGTAATGAAAATGCTGGTGGATGATTCTCTAT (Ala codons underlined). Plasmids were transformed into Dictyostelium by electroporation in H50 buffer (36). Development of Dictyostelium strains was induced by plating ∼4 × 107 cells on KK2 (20 mM K1K2PO4, pH 6.2) plates supplemented with 0.1 mM CaCl2 and 2 mM MgSO4. Developmental structures were visualized and photographed on an Olympus SZX12 stereomicroscope equipped with a Olympus HC-300z/OL digital camera. RNA was isolated using the RNeasy mini kit (Qiagen). RT-PCR was carried out with the Thermoscript reverse transcriptase and Platinum Taq polymerase (Invitrogen). Primers for stlB, dmtA, and lg7 were previously reported (16); primers for chlA were GTCAAGATGTGACATGCTCTTGGAC (forward) and TGACCACCTCTATGATTATCCATGAATCC (reverse). The chlA knockout strain was generated by homologous recombination using plasmid pRRK2, which is based on plasmid pLPBLP (37) and has a blasticidin resistance cassette flanked by approximately 1.6 kb on either side of genomic sequence at the locus (CCCCAAATTTTATCT to GTTGTATTTTCAATT in the genome) and produces a deletion of amino acids 18-273. Two resistant clones (HM1522, HM1523) were verified to be chlA- by multiple PCR analyses and showed identical developmental and labeling phenotypes. HM1522 was used for the experiments presented herein.
Overproduction and Purification of ChlA.
Expression constructs in pTX-FLAG for ChlA and ChlA K86A were transformed into Dictyostelium Ax4 and maintained under constant selection with 20 μg/mL G418. Expression of protein was confirmed by Western blotting with anti-FLAG and anti-c-myc antibodies conjugated to horseradish peroxidase. For purification, cells were grown in 2L portions to 2–3 × 106 cells/ml and harvested by centrifugation at 500 g. Cells were resuspended in 4 ml/g (wet weight) of lysis buffer (20 mM HEPES, 300 mM NaCl, pH 7.5). A protease inhibitor cocktail (Roche) was added to 1X according to manufacturer instructions. Cells were lysed by two passes through a cell disruptor at 10,000–15,000 psi and the lysate cleared by centrifugation at 35,000 rpm for 35 min. The lysate was incubated with anti-FLAG M2-agarose conjugate (Sigma) for 2 h at 4 °C. The resin was washed with lysis buffer until the A280 of eluent was below 0.03. Bound protein was eluted with 200 ng/μl 3X FLAG peptide (Sigma). The resin was washed with 0.1 M glycine pH 3.5 and reequilibrated with lysis buffer. The flow through from the first round of purification was reused for a second round and the two batches combined. The eluted protein was washed with ∼3 volumes of lysis buffer and passed through a 30 kD molecular weight cutoff filter to reduce the concentration of 3X FLAG peptide. Concentrated protein was then flash frozen and stored at -80 °C until use. Protein yields were approximately 0.7 mg per liter of culture; total protein concentration was estimated by A280 measurement.
In Vitro Assay of ChlA Activity.
THPH was synthesized according to the method of Masento et al. (38); Cl-THPH, and Cl2-THPH were synthesized according to the method of Gokan et al. (39). A typical assay was run in 20 mM HEPES, 150 mM NaCl, pH 7.5 with 67 μM ChlA, 4.5 μM SsuE, 10 mM NADH, 100 μM FAD, and 500 μM substrate (either THPH or Cl-THPH). ChlA concentration is reported as the upper limit, calculated by assuming 100% purity. Final reaction volumes were 10–20 μl. Reactions were quenched by dilution with 300 μl buffer followed by immediate extraction with three portions of 300 μl EtOAc. The organic extract was dried and redissolved in 75 μl 5% DMSO. HPLC analysis was carried out on a Beckman instrument equipped with a Phenomenex Luna C18 column and diode array detector using a gradient of acetonitrile in water with 0.1% TFA as modifier. LCMS analysis was carried out on an Agilent 1200 series HPLC interfaced to an Agilent 6250 Q-TOF. Chromatography was run on a Phenomenex Gemini-NX C18 column using a gradient of acetonitrile in water with 0.1% formic acid modifier and mass spectra were collected in the negative mode. LCMS data was analyzed with MassHunter Qualitative Analysis software (Agilent).
Radio-TLC Analysis of Chlorinated Metabolites.
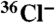 radio-TLC experiments were carried out as previously described (15). Briefly, cells were allowed to develop on an agar surface (1.8% electrophoresis-grade agarose) containing 10% DIFlab (100% is 12 mM KH2PO4, 8 mM Na2HPO4), 0.1 mM MgSO4 at pH 6.7, with 0.05 or 0.1 μCi ml-1
radio-TLC experiments were carried out as previously described (15). Briefly, cells were allowed to develop on an agar surface (1.8% electrophoresis-grade agarose) containing 10% DIFlab (100% is 12 mM KH2PO4, 8 mM Na2HPO4), 0.1 mM MgSO4 at pH 6.7, with 0.05 or 0.1 μCi ml-1 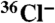 . After 16–17 h of starvation, cells were harvested and organic components extracted by the method of Bligh and Dyer (26). TLC was performed on activated Whatman LK6D silica plates developed with 60/40/2 ethyl acetate/hexane/acetic acid. Radio-labeled compounds were detected by exposure to phosphor imager plates in a lead box to reduce background.
. After 16–17 h of starvation, cells were harvested and organic components extracted by the method of Bligh and Dyer (26). TLC was performed on activated Whatman LK6D silica plates developed with 60/40/2 ethyl acetate/hexane/acetic acid. Radio-labeled compounds were detected by exposure to phosphor imager plates in a lead box to reduce background.
Supplementary Material
ACKNOWLEDGMENTS.
We thank Prof. Ralph Isberg and Ms. Tamara O’Conner (Tufts University) for providing strain Ax4 and technical assistance to C.S.N., and Dr. Michael Myre (Massachusetts General Hospital) for plasmid pTX-FLAG. This work was supported by National Institutes of Health Grant GM20011 (to C.T.W.) and core funding from the MRC (R.R.K.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/1001681107/DCSupplemental.
*We attempted to stimulate the in vitro reaction by addition of hydrogen peroxide as had been done in the original experiments with crude lysates but observed a decrease in ChlA activity. The reason for this discrepancy is a subject of ongoing experimentation.
References
- 1.Camilli A, Bassler BL. Bacterial small-molecule signaling pathways. Science. 2006;311:1113–1116. doi: 10.1126/science.1121357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shank EA, Kolter R. New developments in microbial interspecies signaling. Curr Opin Micrbiol. 2009;12:205–214. doi: 10.1016/j.mib.2009.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yim G, Huimi Wang H, Davies Frs J. Antibiotics as signalling molecules. Philos T Roy Soc B. 2007;362:1195–1200. doi: 10.1098/rstb.2007.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Antunes LCM, Ferreira RBR. Intercellular communication in bacteria. Crit Rev Microbiol. 2009;35:69–80. doi: 10.1080/10408410902733946. [DOI] [PubMed] [Google Scholar]
- 5.López D, Vlamakis H, Losick R, Kolter R. Paracrine signaling in a bacterium. Genes Dev. 2009;23:1631–1638. doi: 10.1101/gad.1813709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chisholm RL, Firtel RA. Insights into morphogenesis from a simple developmental system. Nat Rev Mol Cell Biol. 2004;5:531–541. doi: 10.1038/nrm1427. [DOI] [PubMed] [Google Scholar]
- 7.Kessin RH. Dictyostelium: Evolution, Cell Biology, and the Development of Multicellularity. Cambridge, UK: Cambridge Univ Press; 2001. [Google Scholar]
- 8.Morris HR, Taylor GW, Masento MS, Jermyn KA, Kay RR. Chemical structure of the morphogen differentiation inducing factor from Dictyostelium discoideum. Nature. 1987;328:811–814. doi: 10.1038/328811a0. [DOI] [PubMed] [Google Scholar]
- 9.Town CD, Gross JD, Kay RR. Cell differentiation without morphogenesis in Dictyostelium discoideum. Nature. 1976;262:717–719. doi: 10.1038/262717a0. [DOI] [PubMed] [Google Scholar]
- 10.Thompson CR, Kay RR. The role of DIF-1 signaling in Dictyostelium development. Mol Cell. 2000;6:1509–1514. doi: 10.1016/s1097-2765(00)00147-7. [DOI] [PubMed] [Google Scholar]
- 11.Williams JG, et al. Direct induction of Dictyostelium prestalk gene expression by DIF provides evidence that DIF is a morphogen. Cell. 1987;49:185–192. doi: 10.1016/0092-8674(87)90559-9. [DOI] [PubMed] [Google Scholar]
- 12.Kubohara Y. Effects of differentiation-inducing factors of Dictyostelium discoideum on human leukemia K562 cells: DIF-3 is the most potent anti-leukemic agent. Eur J Pharmacol. 1999;381:57–62. doi: 10.1016/s0014-2999(99)00548-8. [DOI] [PubMed] [Google Scholar]
- 13.Myre MA, et al. Reduced amyloidogenic processing of the amyloid beta-protein precursor by the small-molecule differentiation inducing factor-1. Cell Signal. 2009;21:567–576. doi: 10.1016/j.cellsig.2008.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Omata W, et al. Dictyostelium differentiation-inducing factor-1 induces glucose transporter 1 translocation and promotes glucose uptake in mammalian cells. FEBS J. 2007;274:3392–3404. doi: 10.1111/j.1742-4658.2007.05872.x. [DOI] [PubMed] [Google Scholar]
- 15.Kay RR. The biosynthesis of differentiation-inducing factor, a chlorinated signal molecule regulating Dictyostelium development. J Biol Chem. 1998;273:2669–2675. doi: 10.1074/jbc.273.5.2669. [DOI] [PubMed] [Google Scholar]
- 16.Austin MB, et al. Biosynthesis of Dictyostelium discoideum differentiation-inducing factor by a hybrid type I fatty acid-type III polyketide synthase. Nat Chem Biol. 2006;2:494–502. doi: 10.1038/nchembio811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Insall R, Kay RR. A specific DIF binding protein in Dictyostelium. EMBO J. 1990;9:3323–3328. doi: 10.1002/j.1460-2075.1990.tb07532.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eichinger L, et al. The genome of the social amoeba Dictyostelium discoideum. Nature. 2005;435:43–57. doi: 10.1038/nature03481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Neumann CS, Fujimori DG, Walsh CT. Halogenation strategies in natural product biosynthesis. Chem Biol. 2008;15:99–109. doi: 10.1016/j.chembiol.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 20.Dong C, et al. Tryptophan 7-halogenase (PrnA) structure suggests a mechanism for regioselective chlorination. Science. 2005;309:2216–2219. doi: 10.1126/science.1116510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yeh E, Blasiak LC, Koglin A, Drennan CL, Walsh CT. Chlorination by a long-lived intermediate in the mechanism of flavin-dependent halogenases. Biochemistry. 2007;46:1284–1292. doi: 10.1021/bi0621213. [DOI] [PubMed] [Google Scholar]
- 22.Dairi T, Nakano T, Aisaka K, Katsumata R, Hasegawa M. Cloning and nucleotide sequence of the gene responsible for chlorination of tetracycline. Biosci Biotechnol Biochem. 1995;59:1099–1106. doi: 10.1271/bbb.59.1099. [DOI] [PubMed] [Google Scholar]
- 23.Heemstra JR, Walsh CT. Tandem action of the O2- and FADH2-dependent halogenases KtzQ and KtzR produce 6,7-Dichlorotryptophan for kutzneride assembly. J Am Chem Soc. 2008;130:14024–14025. doi: 10.1021/ja806467a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yeh E, Garneau S, Walsh CT. Robust in vitro activity of RebF and RebH, a two-component reductase/halogenase, generating 7-chlorotryptophan during rebeccamycin biosynthesis. Proc Natl Acad Sci USA. 2005;102:3960–3965. doi: 10.1073/pnas.0500755102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang S, et al. Functional characterization of the biosynthesis of radicicol, an Hsp90 inhibitor resorcylic acid lactone from Chaetomium chiversii. Chem Biol. 2008;15:1328–1338. doi: 10.1016/j.chembiol.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 26.Winter JM, et al. Molecular basis for chloronium-mediated meroterpene cyclization: Cloning, sequencing, and heterologous expression of the napyradiomycin biosynthetic gene cluster. J Biol Chem. 2007;282:16362–16368. doi: 10.1074/jbc.M611046200. [DOI] [PubMed] [Google Scholar]
- 27.Vaillancourt FH, Yeh E, Vosburg DA, Garneau-Tsodikova S, Walsh CT. Nature’s inventory of halogenation catalysts: Oxidative strategies predominate. Chem Rev. 2006;106:3364–3378. doi: 10.1021/cr050313i. [DOI] [PubMed] [Google Scholar]
- 28.Sawai S, Guan X-J, Kuspa A, Cox E. High-throughput analysis of spatio-temporal dynamics in Dictyostelium. Genome Biol. 2007;8:R144. doi: 10.1186/gb-2007-8-7-r144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cadel-Six S, et al. Halogenase genes in nonribosomal peptide synthetase gene clusters of Microcystis (Cyanobacteria): Sporadic distribution and evolution. Mol Biol Evol. 2008;25:2031–2041. doi: 10.1093/molbev/msn150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Levi S, Polyakov M, Egelhoff TT. Green fluorescent protein and epitope tag fusion vectors for Dictyostelium discoideum. Plasmid. 2000;44:231–238. doi: 10.1006/plas.2000.1487. [DOI] [PubMed] [Google Scholar]
- 31.Eichhorn E, van der Ploeg JR, Leisinger T. Characterization of a two-component alkanesulfonate monooxygenase from Escherichia coli. J Biol Chem. 1999;274:26639–26646. doi: 10.1074/jbc.274.38.26639. [DOI] [PubMed] [Google Scholar]
- 32.Rieu JP, Saito T, Delanoe-Ayari H, Sawada Y, Kay RR. Migration of Dictyostelium slugs: Anterior-like cells may provide the motive force for the prespore zone. Cell Motil Cytoskeleton. 2009;66:1073–1086. doi: 10.1002/cm.20411. [DOI] [PubMed] [Google Scholar]
- 33.Saito T, Kato A, Kay RR. DIF-1 induces the basal disc of the Dictyostelium fruiting body. Dev Biol. 2008;317:444–453. doi: 10.1016/j.ydbio.2008.02.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fey P, Kowal AS, Gaudet P, Pilcher KE, Chisholm RL. Protocols for growth and development of Dictyostelium discoideum. Nat Protoc. 2007;2:1307–1316. doi: 10.1038/nprot.2007.178. [DOI] [PubMed] [Google Scholar]
- 35.Pilcher KE, Fey P, Gaudet P, Kowal AS, Chisholm RL. A reliable general purpose method for extracting genomic DNA from Dictyostelium cells. Nat Protoc. 2007;2:1325–1328. doi: 10.1038/nprot.2007.180. [DOI] [PubMed] [Google Scholar]
- 36.Gaudet P, Pilcher KE, Fey P, Chisholm RL. Transformation of Dictyostelium discoideum with plasmid DNA. Nat Protoc. 2007;2:1317–1324. doi: 10.1038/nprot.2007.179. [DOI] [PubMed] [Google Scholar]
- 37.Faix J, Kreppel L, Shaulsky G, Schleicher M, Kimmel AR. A rapid and efficient method to generate multiple gene disruptions in Dictyostelium discoideum using a single selectable marker and the Cre-loxP system. Nucleic Acids Res. 2004;32:e143. doi: 10.1093/nar/gnh136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Masento MS, et al. Differentiation-inducing factor from the slime mould Dictyostelium discoideum and its analogues. Synthesis, structure and biological activity. Biochem J. 1988;256:23–28. doi: 10.1042/bj2560023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gokan N, et al. Structural requirements of Dictyostelium differentiation-inducing factors for their stalk-cell-inducing activity in Dictyostelium cells and anti-proliferative activity in K562 human leukemic cells. Biochem Pharmacol. 2005;70:676–685. doi: 10.1016/j.bcp.2005.06.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.