Abstract
Biologists have long used model organisms to study human diseases, particularly when the model bears a close resemblance to the disease. We present a method that quantitatively and systematically identifies nonobvious equivalences between mutant phenotypes in different species, based on overlapping sets of orthologous genes from human, mouse, yeast, worm, and plant (212,542 gene-phenotype associations). These orthologous phenotypes, or phenologs, predict unique genes associated with diseases. Our method suggests a yeast model for angiogenesis defects, a worm model for breast cancer, mouse models of autism, and a plant model for the neural crest defects associated with Waardenburg syndrome, among others. Using these models, we show that SOX13 regulates angiogenesis, and that SEC23IP is a likely Waardenburg gene. Phenologs reveal functionally coherent, evolutionarily conserved gene networks—many predating the plant-animal divergence—capable of identifying candidate disease genes.
Keywords: angiogenesis, bioinformatics, evolution, gene-phenotype associations, homology
Biochemical and molecular functions of a given protein are generally conserved between organisms; this observation is fundamental to biological research. For example, in x-ray crystallography studies, one can often choose the organism from which the protein is most easily crystallized to facilitate the study of the protein's biochemical function. On the other hand, even with a conserved gene, disruption of function may give rise to radically different phenotypic outcomes in different species. For example, mutating the human RB1 gene leads to retinoblastoma, a cancer of the retina, yet disrupting the nematode ortholog contributes to ectopic vulvae (1, 2). Thus, although a gene's “molecular” functions are conserved, the “organism-level” functions need not be. When a conserved gene is mutated, the resulting organism-level phenotype is an emergent property of the system. This bedrock principle underlying the use of model organisms not only allows us to study important aspects of human biology using mice or frogs, but also permits exploration of inherently multicellular processes, such as cancer, using unicellular organisms like yeast.
Within this paradigm, once a molecular function has been discovered in one organism, it should be predictable in other organisms: GSK3 homologs in yeast are kinases, and such GSK3 homologs in every other organism will generally be kinases. In contrast, the emergent organism-level phenotypes are far less predictable between organisms, in part because relationships between genes and phenotypes are many-to-many. Manipulation of GSK3 perturbs nutrient and stress signaling in yeast, anteroposterior patterning and segmentation in insects, dorsoventral patterning in frogs, and craniofacial morphogenesis in mice (3–5). Recognizing functionally equivalent organism-level phenotypes between model organisms can therefore be nonobvious, especially across large evolutionary distances.
However, the ability to recognize equivalent phenotypes between different model organisms is important for the study of human diseases. Given the success of studies in model systems (genes and phenotypes have been associated in model organisms at a far higher rate than for humans) (Fig. 1A), it seems likely that useful and tractable models for human disease await discovery, currently hidden by differences in the emergent appearance of phenotype in diverse model organisms. Although a framework exists for discussing complex gene-phenotype relationships across evolution, we lack simple—and importantly, quantifiable—methods for discovering new gene-phenotype relationships from existing data.
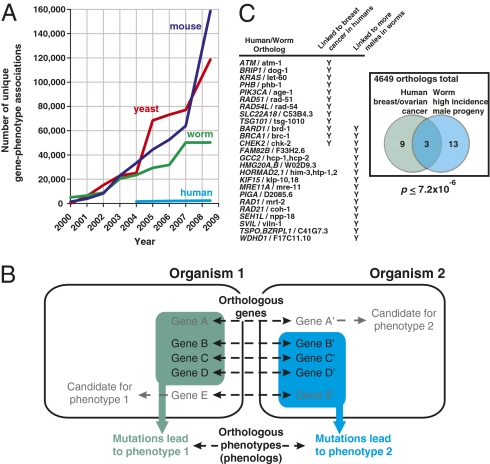
Fig. 1.
Number of unique gene-phenotype associations, identification of phenologs, and the example of a worm model of breast cancer. (A) The rate of associating genes to organism-level phenotypes in model organisms greatly exceeds that in humans (data from refs. 8–11, 14). Thus, appropriate mapping of model organism phenotypes to human diseases could significantly accelerate discovery of human disease gene associations. Orthologous phenotypes (phenologs) offer one such approach. (B) Phenologs can be identified based on significantly overlapping sets of orthologous genes (gene A is orthologous to A', B to B', etc.), such that each gene in a given set (green box or cyan box) gives rise to the same phenotype in that organism. The phenotypes may differ in appearance between organisms because of differing organismal contexts. As gene-phenotype associations are often incompletely mapped, genes currently linked to only one of the orthologous phenotypes become candidate genes for the other phenotype; that is, the gene A' is a new candidate for phenotype 2. (C) An example of a phenolog mapping high incidence of male C. elegans progeny to human breast/ovarian cancers (details in text).
As a foundation for a quantifiable approach to identifying equivalent phenotypes, we introduce the notion of orthologous phenotypes (phenologs), defined as phenotypes related by the orthology of the associated genes in two organisms. Phenologs are the phenotype-level equivalent of gene orthologs. Two phenotypes are thus said to be orthologous if they share a significantly larger set of common orthologous genes than would be expected at random (i.e., are enriched for the same orthologous genes) (Fig. 1B), even if the phenotypes may appear dissimilar.
Phenologs, therefore, are evolutionarily conserved outputs arising from disruption of any of a set of conserved genes (Fig. 1B, green and blue). These outputs manifest as different traits or defects in different organisms because of the organism-specific roles played by each set of genes. One example, noted above, is the human retinoblastoma eye cancer and the Caenorhabditis elegans ectopic vulvae. These phenotypes are orthologous, as failure of equivalent genes (the Rb pathway)—performing conserved molecular functions but in different contexts—leads to different phenotypes in the different organisms (1, 2).
By quantifying the equivalence of mutational phenotypes between different organisms, we demonstrate that orthologous phenotypes may be found objectively, and that these phenologs suggest nonobvious models for human disease. We demonstrate the power of this approach by defining a unique yeast model that effectively predicts vertebrate angiogenesis genes and a plant model that predicts genes involved in vertebrate craniofacial defects that are associated with human congenital malformations.
Results and Discussion
Phenologs are identified by assembling known gene-phenotype associations for two organisms—considering only genes that are orthologous between the two organisms—and searching for interorganism phenotype pairs with significantly overlapping sets of genes. Significance is derived from three observations: (i) the total number of orthologs in organism 1 that give rise to phenotype 1; (ii) the total number of orthologs in organism 2 that give rise to phenotype 2; and (iii) the number of orthologs shared between these two sets. Formally, significance of a phenolog is calculated from the hypergeometric probability of observing at least that many shared orthologs by chance (SI Materials and Methods).
Fig. 1C shows an example of these observations: the set of human-worm genes associated with X-linked breast/ovarian cancer in human significantly overlaps the set of genes whose mutations lead to a high frequency of male progeny in C. elegans. Male C. elegans are determined by a single X chromosome, hermaphrodites by two copies; thus, X chromosome nondisjunction leads to higher frequencies of males (6). Human breast/ovarian cancers may derive from X chromosome abnormalities (7), supporting the notion that this phenolog is identifying a useful disease model. Human orthologs of the 13 additional genes associated with this worm trait are thus reasonable candidate genes for involvement in breast/ovarian cancers. Nine of these genes were not yet linked to breast cancer in the databases we employed, but could be confirmed as such in the primary literature (Table S1). The remaining four genes (GCC2, PIGA, WDHD1, SEH1L) are thus implicated as breast cancer candidate genes. This rate of literature confirmation is 268-fold higher than that expected based on the current annotation rate in the Online Mendelian Inheritance in Man database (8), providing significant support for the utility of the worm phenotype to predict and suggest additional genes relevant to human breast cancer. (Note that the estimated fold-improvements we present here depend upon consistent literature curation and, thus, should be interpreted cautiously.)
Systematic Discovery of Phenologs.
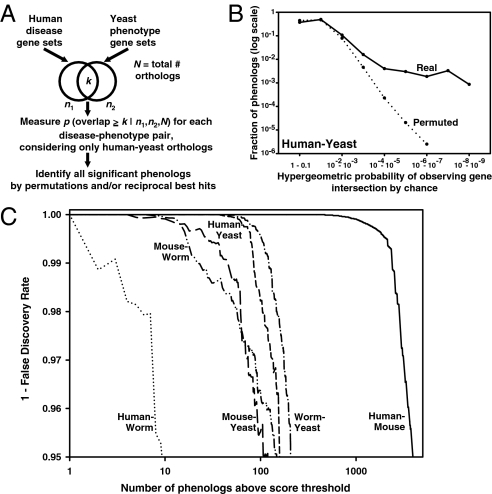
To systematically discover phenolog relationships, we collected from the literature a set of 1,923 human disease-gene associations (8), 74,250 mouse gene-phenotype associations (9), 27,065 C. elegans gene-phenotype associations (10), and 86,383 yeast gene-phenotype associations (11–14). The dataset spans ∼300 human diseases and > 6,000 model organism phenotypes. With these data and the sets of orthologous gene relationships between each pair of organisms (15), we quantitatively examined the overlap of each interorganism phenotype pair, measuring their significance (Fig. 2A). To correct for testing multiple hypotheses, we repeated all analyses 1,000 times with randomly permuted gene-phenotype associations, then calculated a false-discovery rate (FDR) based upon the observed null distribution of scores (Fig. 2B and Fig. S1). We observed 154 significant phenologs (5% FDR) between human diseases and yeast mutational phenotypes, 3,755 between human and mouse, 147 between mouse and worm, 119 between mouse and yeast, 206 between yeast and worm, and 9 between human and worm (the low number stems from limited mutational data in both species) (Fig. 2C).
Fig. 2.
Systematic identification of phenologs. (A) For a pair of organisms, sets of genes known to be associated with mutational phenotypes are assembled, considering only orthologous genes between the two organisms. Pairs of mutational phenotypes—one phenotype from each organism, each associated with a set of genes—are then compared to determine the extent of overlap of the associated gene sets, calculating the significance of overlap by the hypergeo-metric probability. Comparison of the distribution of observed probabilities with those derived from the same analysis following permutation of gene-phenotype associations reveals that many more orthologous phenotypes are observed than expected by random chance, as shown in B for the case of the human-yeast comparison (also Fig. S1), and summarized for each organism pair in C.
Many specific, intuitively obvious, phenologs were revealed by this analysis, especially for the comparison of mouse and human phenotypes. Our analysis recapitulates many known mouse models of disease, providing an important positive control for our approach; Table 1 lists other specific examples of both known and previously undescribed equivalences. For example, one of the most significant phenologs identified between human disease and mouse mutational phenotypes is that linking Bardet-Biedl syndrome with four mouse traits, each of which relates to the disruption of ciliary function (abnormal brain ventricle/choroid plexus morphology, small hippocampus, enlarged third ventricle, absent sperm flagella; all P ≤ 10−11), consistent with the apparent molecular defects in Bardet-Biedl syndrome (16). These phenologs thus suggest mouse ciliary defects provide a powerful model for studying human Bardet-Biedl syndrome, consistent with its recognized utility in this regard. Similarly, human cataracts are observed to be phenologous to mouse cataracts (P ≤ 10−24), human obesity is phenologous to mouse obesity (P ≤ 10−14), human deafness to mouse deafness (P ≤ 10−29), human retinitis to mouse retinal degeneration (P ≤ 10−26), and human goiter to mouse enlarged thyroid glands (P ≤ 10−8). Thus, the calculation of phenologs correctly identifies many known mouse models of human diseases and therefore has the potential to identify new models. More generally, cross-validated tests of phenologs confirm their strong predictability for genes associated with a substantial portion of human diseases (Fig. S2).
Table 1.
Examples from the >6,200 significant phenologs detected among human diseases and mouse, yeast, worm, and Arabidopsis mutant phenotypes
| Phenotype1 | Phenotype2 | n1 | n2 | k | P value | PPV | ||
| Hs | Cataracts | Mm | Cataracts | 19 | 47 | 11 | 6 × 10−24 | 1.00 |
| Hs | X-linked conductive deafness | Mm | Circling | 47 | 50 | 12 | 2 × 10−20 | 1.00 |
| Hs | Bardet-Biedl syndrome | Mm | Absent sperm flagella | 11 | 5 | 4 | 8 × 10−13 | 1.00 |
| Mm | Lymphoma | Sc | CANR mutator high | 14 | 11 | 6 | 1 × 10−11 | 1.00 |
| Hs | Zellweger syndrome | Sc | Reduced number of peroxisomes | 8 | 6 | 4 | 1 × 10−9 | 1.00 |
| Hs | Susceptible to autism | Mm | Abnormal social investigation | 5 | 16 | 3 | 1 × 10−8 | 1.00 |
| Mm | Abnormal heart development | At | Defective response to red light | 25 | 9 | 4 | 3 × 10−7 | 1.00 |
| Hs | Refsum disease | At | Defective protein import into peroxisomal matrix | 4 | 5 | 2 | 1 × 10−5 | 1.00 |
| Mm | Absent posterior semicircular canal | At | Shade avoidance defect | 2 | 4 | 2 | 1 × 10−6 | 0.99 |
| Mm | Spleen hypoplasia | Sc | Uge (enlarged cells) | 5 | 16 | 3 | 3 × 10−6 | 0.99 |
| Mm | Gastrointestinal hemorrhage | Ce | Abnormal body wall muscle cell polarization | 6 | 3 | 2 | 4 × 10−6 | 0.98 |
| Hs | Mental retardation | At | Cotyledon development defects | 13 | 5 | 2 | 1 × 10−4 | 0.98 |
| Hs | Congenital disorder of glycosylation | Sc | CID 604586 sensitive | 10 | 25 | 3 | 2 × 10−4 | 0.98 |
| Hs | Hemolytic anemia | Sc | Hydroxyurea sensitive | 11 | 23 | 3 | 2 × 10−4 | 0.98 |
| Hs | Amyotrophic lateral sclerosis | Sc | Increased resistance to wortmannin | 2 | 34 | 2 | 2 × 10−4 | 0.97 |
n1 indicates the number of orthologs in organism 1 with phenotype1, n2 the number in organism 2 with phenotype2, and k the number in both sets. The significance of each phenolog is assessed by the hypergeometric probability (P value), the positive predictive value (PPV) when considering multiple testing (1 – FDR), and the reciprocal best-hit criterion (bold text). At, Arabidopsis; Ce, worm; Hs, human; Mm, mouse; Sc, yeast.
Much of the powerful conceptual framework established for determining homology and orthology for gene sequences may also be applicable to phenologs. For example, many of the algorithmic approaches used to identify orthologous genes might also be applied to the identification of phenologs. We explored this notion for one effective and easily automated approach to identify orthologous sequences, the reciprocal best hit (RBH) strategy. The RBH criterion holds that genes X and Y are orthologs if genes X and Y are the most similar to each other (reflexively) when searched genome-wide. We adapted the RBH criterion to the identification of phenologs to identify the most equivalent phenotypes between two organisms from among those assayed, by asking if the phenotypes have the most significant (lowest P value) gene overlaps with each other when searched against all phenotypes in their respective organisms. Such analysis gives a second criterion for identifying phenologs, useful for legitimate phenologs with poor P values because of limited phenotypic data sets. Examples of such RBH phenologs are indicated in Table 1.
Mouse and Yeast Phenologs Predict Unique Angiogenesis Genes.
The power of the phenolog framework lies in discovery of nonobvious disease models. We identified just such a phenolog between abnormal angiogenesis in mutant mice and reduced growth rate of yeast deletion strains when grown in the hypercholesterolemia drug lovastatin (8 mouse, 67 yeast, 5 shared genes, P ≤ 10−6) (Fig. 3A). This observation suggests that budding yeast, which obviously lack blood vessels, could potentially model the genetics of mammalian vasculature formation and could be used to iden-tify previously unrecognized genes affecting this process.
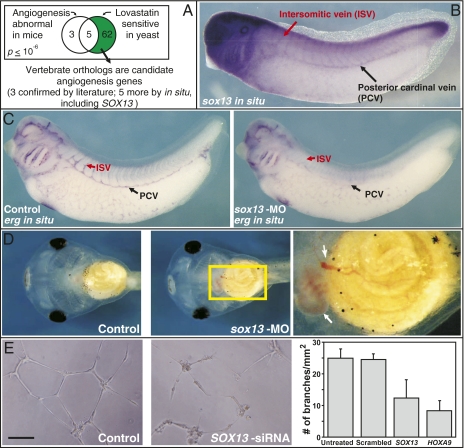
Fig. 3.
Example of a nonobvious disease model revealed by phenologs: a yeast model of angiogenesis. (A) The sets of 8 genes (considering only mouse/yeast orthologs) associated with mouse angiogenesis defects and 67 genes associated with yeast hypersensitivity to the hypercholesterolemia drug lovastatin significantly overlap, suggesting that the yeast gene set may predict angiogenesis genes. This prediction was verified in Xenopus embryos for eight genes (three from literature support and five based upon vascular expression patterns) (Fig. S3) and studied in detail for the case of the transcription factor sox13. (B) sox13 is expressed in developing Xenopus vasculature, as measured by in situ hybridization (also Fig. S4). (C) Morpholino (MO) knockdown of sox13 induces defects in vasculature, measured using in situ hybridization versus the vasculature markers erg (defects observed in 31 of 49 animals tested) or agtrl1 (12 of 19 animals tested) (Fig S5). Such defects are rare in untreated control animals and five base pair mismatch morpholino (MM) knockdowns (0 of 22 control animals tested with agtrl1, 2 of 46 tested with erg; 5 of 28 MM animals tested with erg). (D) Hemorrhaging (white arrows) is apparent in stage 45 Xenopus embryos because of dysfunctional vasculature following sox13 morpholino knockdown (12 of 50 animals tested; two also showed unusually small hearts with defective morphology; Right: magnification of yellow boxed region in Middle), but is rare in control animals (1 of 45 tested untreated animals, 1 of 22 sox13-MM knockdown animals tested). All phenotypes in Figs. 3 and 4 are significantly different from controls by χ2 tests (P < 0.001). (E) In an in vitro human umbilical vein endothelial cell model of angiogenesis, knockdown of human SOX13 by siRNA disrupts tube formation (an in vitro model for capillary formation) to an extent comparable to knockdown of a known effector of angiogenesis (HOXA9) and significantly more than untreated cells or cells transfected with an off-target (scrambled) negative control siRNA. (Scale bar, 100 μm.)
We identified five shared genes between these processes. In yeast, they are the MAP kinases SLT2, PBS2, and HOG1, the calcineurin B gene CNB1, and the uncharacterized gene VPS70. Strikingly, mutations of their mouse orthologs (MAPK7, MAP2K1, MAPK14, PPP3R1, and the prostate-specific membrane antigen PSMA, respectively) all confer strong angiogenesis defects (e.g., MAPK7 deletion causes defective blood vessel and cardiac development) (17), and ablation in adult mice leads to leaky blood vessels (18). Similarly, PSMA regulates angiogenesis by modulating integrin signal transduction (19). Thus, this conserved set of genes was alternately repurposed to regulate cell wall stress and biogenesis in yeast cells (20) or to regulate proper formation and maintenance of blood vessels in mice.
Phenologs are most important for their ability to map gene-phenotype associations from one organism to another. In other words, phenologs suggest unobserved associations in one organism from observed associations in another (Fig. 1B, gene E and gene A'). Thus, examination of phenologs should substantially improve the rate of discovery of new genes over the rate expected by chance (here ∼1 in 267, as estimated from the frequency of known angiogenesis genes). We might a priori expect a discovery rate comparable to the fraction of genes already verified by the phenolog; for the yeast angiogenesis model, a likely discovery rate of ∼1 in 13, for a ∼21-fold higher discovery rate than random expectation. Our model therefore asserts that some of the 62 additional genes associated with lovastatin sensitivity in yeast would be predicted to be involved in angiogenesis. In fact, literature confirms three of the corresponding mouse genes to function in angiogenesis: the known target of lovastatin, HMG-CoA reductase, whose role in angiogenesis has been previously observed (21), the sirtuin SIRT1, whose disruption in zebrafish and mice caused defective blood vessel formation and blunted ischemia-induced neovascularization (22), and the casein kinase CSNK2A1, inhibitors of which inhibit mouse retinal neovascularization (23).
For phenologs to be useful, they must be able to predict entirely new gene-phenotype associations. To this end, we examined the 59 remaining genes not already associated with angiogenesis for conserved function using the frog Xenopus. Using whole-mount in situ hybridization, we examined mRNA expression of the Xenopus orthologs of these genes. Consistent with our hypothesis, we found that five of the genes (orthologs of SOX13, RAB11B, HMHA1, TCEA1/TCEA3, and TBL1XR1) were robustly and predominantly expressed in the developing vasculature (Fig. 3B and Fig. S3). These expression data suggest an overall discovery rate (8 of 62) of this phenolog 34 times higher than expected given the current annotation rate.
Finally, we directly assayed the role of one of these genes, SOX13, in angiogenesis. SOX13 is a transcription factor that is known to regulate T lymphocyte differentiation (24). The gene is expressed in mouse arterial walls (25), although it is also expressed in 30 of 45 assayed tissues in the National Center for Biotechnol-ogy Information Unigene Expressed Sequence Tag database. (The Xenopus ortholog of SOX13 was previously referred to as Xenopus xSOX12, but in accordance with recent Xenopus gene nomenclature guidelines, we refer to this gene now as sox13, Gene ID 397727.) We found this gene to be prominently expressed in the posterior cardinal veins, intersomitic veins, and developing heart, consistent with a role affecting developing vasculature (Fig. 3B and Fig. S4). We knocked down sox13 using microinjection of morpholino antisense oligonucleotides (MO) and assayed for vasculature defects by in situ hybridization to the vasculature reporter genes erg and agtrl1 (previously called X-msr, Gene ID 399306). Knockdown of sox13 resulted in severe defects in vascular development, with morphant animals largely lacking intersomitic and posterior cardinal veins (Fig. 3C and Fig. S5). By later stages, hemorrhaging was apparent in morphants because of the defective vasculature (Fig. 3D).
This in vivo requirement for sox13 in Xenopus was then confirmed in humans using siRNA-induced knockdown of SOX13 in an in vitro human umbilical vein endothelial cell angiogenesis assay (Fig. 3E). Thus, SOX13 is a unique regulator of angiogenesis, discovered in the absence of any previous functional data linking it to angiogenesis, on the basis of orthology between mouse angiogenesis defects and yeast lovastatin sensitivity. Notably, these data also demonstrate that differentiation both of blood cells (24) and blood vessels are controlled by the same transcription factor.
Human/Arabidopsis Phenologs Predict Vertebrate Regulators of Craniofacial Development.
Phenologs provide a quantitative framework for identifying cases of extremely distant homology [“deep homology” (26)] of functionally coherent gene systems. This creates an opportunity to use very distantly related species as human disease models. We tested this approach by systematically searching for plant models of human disease. We collected 22,921 gene-phenotype associations—spanning 1,711 unique phenotypes—for the mustard plant Arabidopsis thaliana and analyzed these for phenologs with fungal and animal phenotypes. Hundreds of orth-ologous phenotypes were evident (Fig. 4A and Fig. S6), including 897, 733, 172, and 48 between Arabidopsis and yeast, mice, worms, and humans, respectively (5% FDR). The human-plant phenologs suggest mappings between specific plant mutational phenotypes and diverse human diseases, including cancers, peroxisomal disorders, such as Refsum disease and Zellweger syndrome, and a variety of birth defects (Table 1).
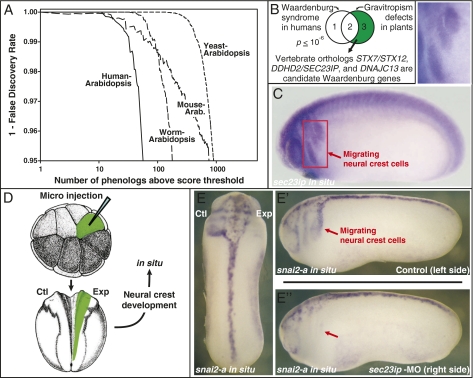
Fig. 4.
Phenologs reveal plant models of human disease, including a model of Waardenburg syndrome (WS) neural crest defects. (A) Many orthologous phenotypes are observed between Arabidopsis and worms, yeast, mouse, and humans, with hundreds more than expected by chance. Many mammalian/plant phenologs relate to vertebrate developmental defects, including models for WS and other birth defects. (B) Considering only human/Arabidopsis orthologs, the three known WS genes significantly overlap the five genes associated with negative gravitropism defects in Arabidopsis. The plant gene set suggests unique candidate WS genes. (C) In situ hybridization versus candidate sec23ip in developing Xenopus embyros confirms neural crest cell expression. (D) Unilateral morpholino knockdown of sec23ip induces (E) defects in neural crest cell migration on the side with the knockdown (E'') but not the control side (E'), measured using in situ hybridization versus two independent markers of neural crest cells, snai2-a (defects observed in 23 of 35 animals tested) and twist (8 of 14 animals tested) (Fig S7). Such defects are rare in untreated control animals and off-target morpholino (OM) knockdowns (0 of 21 control animals tested with snai2-a; 1 of 14 OM animals tested with snai2-a; 0 of 14 OM animals tested with twist).
We observed a striking plant-human phenolog relating negative gravitropism defects to Waardenburg syndrome (Fig. 4B). This congenital syndrome stems from defects in the developing neural crest and is characterized by craniofacial dysmorphology, abnormal pigmentation, and hearing loss [in fact, it accounts for 2–5% of cases of human deafness (27)]. In particular, this phenolog suggested that a set of three vesicle trafficking genes involved in directing plant growth in response to gravitational cues might also serve to direct neural crest cell migration and differentiation in developing animal embryos.
Encouragingly, one of the identified proteins (STX12) is known to interact with the protein encoded by the pallid gene in mice (28), whose mutational phenotypes, including pigmentation and ear defects, are consistent with Waardenberg syndrome (29). The remaining two proteins had no support in the literature, and we therefore evaluated the three mammalian orthologs of these genes by whole mount in situ hybridization in developing Xenopus embryos. Strikingly, we found that sec23ip was prominently expressed in migrating neural crest cells (Fig. 4C). We used targeted microinjection of sec23ip morpholinos to knock down this gene specifically in the neural crest. Unilateral targeting of sec23ip MOs (Fig. 4D) resulted in marked defects in neural crest cell migration patterns specifically on the injected side (Fig. 4E and Fig. S7), thus confirming a role for this gene in neural crest cell development. Notably, SEC23IP physically associates with SEC23, a component of the COPII complex that controls ER-to-Golgi trafficking, and mutations in SEC23 underlie Cranio-lenticular-sutural dysplasia, another congenital human disease related to neural crest development (30). Thus, SEC23IP, identified here on the basis of orthology to plant gravitropism defects, is both a promising candidate gene for Waardenburg syndrome and also provides insights into the emerging link between COPII function and craniofacial development in vertebrates (31). Our success rate of one in two for finding Waardenburg-relevant genes represents a 550-fold improvement over the current annotation rate of ∼1 in 1,100 genes. Notably, in spite of the extremely dissimilar associated phenotypes, phenologs can identify functionally coherent gene sets that predate the divergence of plants and animals.
Phenologs Reveal Deeply Homologous Modular Subnetworks.
Phenologs imply that although phenotypes diverge, the orthology of the underlying gene networks—and the networks’ immediate functional output—is conserved. We might therefore expect genes involved in a given phenolog to represent a coherent biological module, and thus to be highly interconnected in gene networks. Moreover, we might expect that the genes already confirmed to show the signature phenotypes in both organisms (e.g., the intersection labeled by k in Fig. 2A) would be even more highly interconnected than the genes associated with the signature phenotype in only one organism; these latter genes might or might not belong to this subnetwork, as multiple mechanisms might give rise to the phenotype. Evidence in current gene networks of more linkages among the genes in each such intersection would support this notion of phenologs recapitulating modular subnetworks. We therefore systematically tested all significant phenologs involving yeast and worm genes for the genes’ connectivity in available functional networks (32). We find the network connectivity of genes in phenolog intersections to be significantly higher (P < 0.0001; Wilcoxon signed-rank) than the phenolog genes outside of the intersections, which nonetheless show significantly higher network connectivity than random size-matched gene sets (P < 0.0001) (Fig. S8). Additional tests confirm that genes in phenolog intersections are no more enriched for homologous genes than are phenolog genes outside of the intersections (Fig. S9), ruling out trivial discovery of “deep paralogs.” These observations indicate that phenologs identify evolutionarily conserved subnetworks of genes relevant to particular phenotypes or diseases, yet still predicting as yet undiscovered candidate genes significantly better than random expectation. Phenologs may inherently identify systems such as those found by aligning protein interaction networks across species (33). Indeed, direct searches for evolutionarily conserved subnetworks composed largely of genes with similar phenotypes might provide an alternate strategy for phenolo discovery.
Conclusions
Phenologs reflect the innate modularity of gene systems and identify adaptive reuse of those systems, creating a rich framework for comparing mutational phenotypes with potential for finding nonobvious models of human disease. Cross-validated tests indicate phenologs show utility for roughly one-third to one-half of tested human genetic diseases (Fig. S2). Given a phenolog for a human disease, any approach for associating more genes with the model organism trait (e.g., a genetic screen) will suggest additional new human disease gene candidates. In addition to associating unique genes with modeled diseases, such models can provide mechanistic understanding in simplified model organisms for understanding aspects of more complex human diseases.
Phenologs thus bridge the molecular definitions of homologous and orthologous genes (34) with classic definitions of homologous structures from Owen (35) and Darwin (36), deriving from considerations both of gene heredity and of the traits/structures affected by perturbing the genes, concepts falling within the general field of evolutionary developmental biology (evo-devo) (37). The conserved gene systems revealed by the plant-vertebrate phenologs illustrate a more ancient homology than the “deep homology” of metazoans that is currently a focus of evolutionary developmental biology (26). These phenologs should bring attention to the potentially extensive molecular toolkit within the last common eukaryotic ancestor, which facilitated the parallel evolution of complex multicellular organisms. This comparative approach provides a simultaneously deeper and wider view of the evolution of life and points the way to a greater synthesis of evolutionary developmental biology and modern medicine.
Materials and Methods
Detailed information regarding the collection of phenotypes, identification of nonredundant phenotype sets, calculation of orthologs, calculation of phenologs, and tests of subnetwork modularity can be found in the SI Materials and Methods. Animal care met the principles and guidelines of the Institute for Laboratory Animal Research “Guide for Care and Use of Laboratory Animals” and the University of Texas at Austin Institutional Animal Care and Use Committee. Details of Xenopus laevis embryo manipulations and tube formation assays can be found in the SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Greg Weiss for Fig. 1A, and Andrew Fraser, Andrew Ellington, and Jim Bull for critical discussion. This work was supported by grants from the Texas Advanced Research Program, the National Science Foundation, the National Institutes of Health, and the Welch Foundation (F-1515), and a Packard Fellowship (to E.M.M.), a National Science Foundation graduate fellowship (to J.O.W.), and grants from the National Institute of General Medical Sciences and The March of Dimes (to J.B.W.). J.B.W. is an Early Career Scientist of the Howard Hughes Medical Institute. T.J.P. is supported by a Postdoctoral Research Initiative Seed Grant from the Texas Institute for Drug and Diagnostic Development.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/cgi/content/full/0910200107/DCSupplemental.
References
- 1.Dryja TP, et al. Homozygosity of chromosome 13 in retinoblastoma. N Engl J Med. 1984;310:550–553. doi: 10.1056/NEJM198403013100902. [DOI] [PubMed] [Google Scholar]
- 2.Lu X, Horvitz HR. lin-35 and lin-53, two genes that antagonize a C. elegans Ras pathway, encode proteins similar to Rb and its binding protein RbAp48. Cell. 1998;95:981–991. doi: 10.1016/s0092-8674(00)81722-5. [DOI] [PubMed] [Google Scholar]
- 3.Kassir Y, Rubin-Bejerano I, Mandel-Gutfreund Y. The Saccharomyces cerevisiae GSK-3 beta homologs. Curr Drug Targets. 2006;7:1455–1465. doi: 10.2174/1389450110607011455. [DOI] [PubMed] [Google Scholar]
- 4.Kim L, Kimmel AR. GSK3 at the edge: regulation of developmental specification and cell polarization. Curr Drug Targets. 2006;7:1411–1419. doi: 10.2174/1389450110607011411. [DOI] [PubMed] [Google Scholar]
- 5.Liu KJ, Arron JR, Stankunas K, Crabtree GR, Longaker MT. Chemical rescue of cleft palate and midline defects in conditional GSK-3beta mice. Nature. 2007;446:79–82. doi: 10.1038/nature05557. [DOI] [PubMed] [Google Scholar]
- 6.Hodgkin J, Horvitz HR, Brenner S. Nondisjunction mutants of the nematode Caenorhabditis elegans. Genetics. 1979;91:67–94. doi: 10.1093/genetics/91.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Richardson AL, et al. X chromosomal abnormalities in basal-like human breast cancer. Cancer Cell. 2006;9:121–132. doi: 10.1016/j.ccr.2006.01.013. [DOI] [PubMed] [Google Scholar]
- 8.Amberger J, Bocchini CA, Scott AF, Hamosh A. McKusick's Online Mendelian Inheritance in Man (OMIM) Nucleic Acids Res. 2008;37(Database issue):D793–D796. doi: 10.1093/nar/gkn665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eppig JT, Blake JA, Bult CJ, Kadin JA, Richardson JE, Mouse Genome Database Group The mouse genome database (MGD): new features facilitating a model system. Nucleic Acids Res. 2007;35(Database issue):D630–D637. doi: 10.1093/nar/gkl940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen N, et al. WormBase: a comprehensive data resource for Caenorhabditis biology and genomics. Nucleic Acids Res. 2005;33(Database issue):D383–D389. doi: 10.1093/nar/gki066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dwight SS, et al. Saccharomyces Genome Database (SGD) provides secondary gene annotation using the Gene Ontology (GO) Nucleic Acids Res. 2002;30:69–72. doi: 10.1093/nar/30.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hillenmeyer ME, et al. The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science. 2008;320:362–365. doi: 10.1126/science.1150021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McGary KL, Lee I, Marcotte EM. Broad network-based predictability of Saccharomyces cerevisiae gene loss-of-function phenotypes. Genome Biol. 2007;8:R258. doi: 10.1186/gb-2007-8-12-r258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saito TL, et al. SCMD: Saccharomyces cerevisiae Morphological Database. Nucleic Acids Res. 2004;32(Database issue):D319–S322. doi: 10.1093/nar/gkh113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Remm M, Storm CE, Sonnhammer EL. Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J Mol Biol. 2001;314:1041–1052. doi: 10.1006/jmbi.2000.5197. [DOI] [PubMed] [Google Scholar]
- 16.Ross AJ, et al. Disruption of Bardet-Biedl syndrome ciliary proteins perturbs planar cell polarity in vertebrates. Nat Genet. 2005;37:1135–1140. doi: 10.1038/ng1644. [DOI] [PubMed] [Google Scholar]
- 17.Regan CP, et al. Erk5 null mice display multiple extraembryonic vascular and embryonic cardiovascular defects. Proc Natl Acad Sci USA. 2002;99:9248–9253. doi: 10.1073/pnas.142293999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hayashi M, et al. Targeted deletion of BMK1/ERK5 in adult mice perturbs vascular integrity and leads to endothelial failure. J Clin Invest. 2004;113:1138–1148. doi: 10.1172/JCI19890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Conway RE, et al. Prostate-specific membrane antigen regulates angiogenesis by modulating integrin signal transduction. Mol Cell Biol. 2006;26:5310–5324. doi: 10.1128/MCB.00084-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bermejo C, et al. The sequential activation of the yeast HOG and SLT2 pathways is required for cell survival to cell wall stress. Mol Biol Cell. 2008;19:1113–1124. doi: 10.1091/mbc.E07-08-0742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Demierre MF, Higgins PD, Gruber SB, Hawk E, Lippman SM. Statins and cancer prevention. Nat Rev Cancer. 2005;5:930–942. doi: 10.1038/nrc1751. [DOI] [PubMed] [Google Scholar]
- 22.Potente M, et al. SIRT1 controls endothelial angiogenic functions during vascular growth. Genes Dev. 2007;21:2644–2658. doi: 10.1101/gad.435107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ljubimov AV, et al. Involvement of protein kinase CK2 in angiogenesis and retinal neovascularization. Invest Ophthalmol Vis Sci. 2004;45:4583–4591. doi: 10.1167/iovs.04-0686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Melichar HJ, et al. Regulation of gammadelta versus alphabeta T lymphocyte differentiation by the transcription factor SOX13. Science. 2007;315:230–233. doi: 10.1126/science.1135344. [DOI] [PubMed] [Google Scholar]
- 25.Roose J, et al. High expression of the HMG box factor sox-13 in arterial walls during embryonic development. Nucleic Acids Res. 1998;26:469–476. doi: 10.1093/nar/26.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shubin N, Tabin C, Carroll S. Deep homology and the origins of evolutionary novelty. Nature. 2009;457:818–823. doi: 10.1038/nature07891. [DOI] [PubMed] [Google Scholar]
- 27.Nayak CS, Isaacson G. Worldwide distribution of Waardenburg syndrome. Ann Otol Rhinol Laryngol. 2003;112:817–820. doi: 10.1177/000348940311200913. [DOI] [PubMed] [Google Scholar]
- 28.Huang L, Kuo YM, Gitschier J. The pallid gene encodes a novel, syntaxin 13-interacting protein involved in platelet storage pool deficiency. Nat Genet. 1999;23:329–332. doi: 10.1038/15507. [DOI] [PubMed] [Google Scholar]
- 29.Theriault LL, Hurley LS. Ultrastructure of developing melanosomes in C57 black and pallid mice. Dev Biol. 1970;23:261–275. doi: 10.1016/0012-1606(70)90098-9. [DOI] [PubMed] [Google Scholar]
- 30.Boyadjiev SA, et al. Cranio-lenticulo-sutural dysplasia is caused by a SEC23A mutation leading to abnormal endoplasmic-reticulum-to-Golgi trafficking. Nat Genet . 2006;38:1192–1197. doi: 10.1038/ng1876. [DOI] [PubMed] [Google Scholar]
- 31.Fromme JC, et al. The genetic basis of a craniofacial disease provides insight into COPII coat assembly. Dev Cell. 2007;13:623–634. doi: 10.1016/j.devcel.2007.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee I, Li Z, Marcotte EM. An improved, bias-reduced probabilistic functional gene network of baker's yeast, Saccharomyces cerevisiae. PLoS One. 2007;2:e988. doi: 10.1371/journal.pone.0000988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kelley BP, et al. Conserved pathways within bacteria and yeast as revealed by global protein network alignment. Proc Natl Acad Sci USA. 2003;100:11394–11399. doi: 10.1073/pnas.1534710100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fitch WM. Distinguishing homologous from analogous proteins. Syst Zool. 1970;19:99–113. [PubMed] [Google Scholar]
- 35.Owen R. Lectures on Comparative Anatomy and Physiology of the Invertebrate Animals. London: Longmans, Brown, Green and Longmans; 1843. [Google Scholar]
- 36.Darwin C. On the Origin of Species by Means of Natural Selection, or the Preservation of Favoured Races in the Struggle for Life. London: John Murray; 1859. [PMC free article] [PubMed] [Google Scholar]
- 37.Arthur W. The emerging conceptual framework of evolutionary developmental biology. Nature. 2002;415:757–764. doi: 10.1038/415757a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.