Abstract
Sibling Paenibacillus dendritiformis bacterial colonies grown on low-nutrient agar medium mutually inhibit growth through secretion of a lethal factor. Analysis of secretions reveals the presence of subtilisin (a protease) and a 12 kDa protein, termed sibling lethal factor (Slf). Purified subtilisin promotes the growth and expansion of P. dendritiformis colonies, whereas Slf is lethal and lyses P. dendritiformis cells in culture. Slf is encoded by a gene belonging to a large family of bacterial genes of unknown function, and the gene is predicted to encode a protein of approximately 20 kDa, termed dendritiformis sibling bacteriocin. The 20 kDa recombinant protein was produced and found to be inactive, but exposure to subtilisin resulted in cleavage to the active, 12 kDa form. The experimental results, combined with mathematical modeling, show that subtilisin serves to regulate growth of the colony. Below a threshold concentration, subtilisin promotes colony growth and expansion. However, once it exceeds a threshold, as occurs at the interface between competing colonies, Slf is then secreted into the medium to rapidly reduce cell density by lysis of the bacterial cells. The presence of genes encoding homologs of dendritiformis sibling bacteriocin in other bacterial species suggests that this mechanism for self-regulation of colony growth might not be limited to P. dendritiformis.
Keywords: bacterial competition, bacterial growth inhibition, growth regulation, Paenibacillus dendritiformis, subtilisin
In adverse conditions, Gram-positive bacteria have the ability to transition into a dormant, endospore phase (1) to ensure survival. Research on Bacillus subtilis has revealed that during starvation, competing bacteria within the same colony can lyse their siblings and use them as a nutrient source to delay sporulation (2, 3). In this complex process known as bacterial cannibalism, some bacterial prespores secrete a molecule that kills their neighbors, while inducing an immunization system that ensures their own survival. The evolutionary advantage is clear: Although some cells die, the strain survives. Another process that leads to bacterial death is known as fratricide or siblicide (4–6). For example, in Streptococcus pneumoniae, some cells in a population are killed by their siblings or close relatives in a process linked to induction of competence (5). The release of DNA from lysed cells (allolysis) may increase genetic diversity when there is variation in the population. Additionally, the increased release of virulence factors from lysed cells during host infection would be advantageous to virulent streptococci (5). The antimicrobial compounds secreted during cannibalism or fratricide (2–6) are called bacteriocins, and usually have a narrow spectrum of activity as they kill only closely related bacteria, which compete for the same resources.
Competition among bacteria is not limited to members of single colonies. Recently we found (7) that Paenibacillus dendritiformis (T morphotype) secretes inhibiting agents that are lethal above a threshold. When two neighboring sibling colonies (colonies initiated from the same bacterial culture) grew from droplet inoculation, all the bacteria at the interface (approximately 250 μm) of the inhibited regions died. Although P. dendritiformis are able to produce spores, death rather than sporulation occurred under these conditions. The inhibition was greater for lower initial nutrients levels (7) but persisted even for relatively high nutrient levels, which excludes bacterial cannibalism (killing due to lack of nutrients).
Here we analyze the proteins secreted by competing sibling P. dendritiformis colonies and show that they include subtilisin, which stimulates cell growth and reproduction, and a lethal protein. A mathematical model shows how colonies maintain their growth by self-regulating the secretion of subtilisin. If regulation is interrupted by the presence of another colony, the number of bacteria increases locally; this triggers the secretion of the lethal protein to rapidly reduce bacterial density.
Results
Analysis of Inhibiting Material.
Two sibling colonies of P. dendritiformis (T morphotype) were inoculated simultaneously on an agar plate at equal distances from the plate’s center. After a few days of growth (depending on the initial separation between colonies), the speed of each colony’s front facing its neighbor starts to decrease and the growth front finally stops, leaving a gap between the colonies (Fig. 1A) (7). Our previous studies showed that bacteria at the inhibited interfaces were dead, and material extracted from the agar gel between two colonies was found to kill single growing colonies (7). To identify the lethal factor, we have now extracted proteins from the agar gel between two colonies and analyzed them by SDS-polyacrylamide gel.
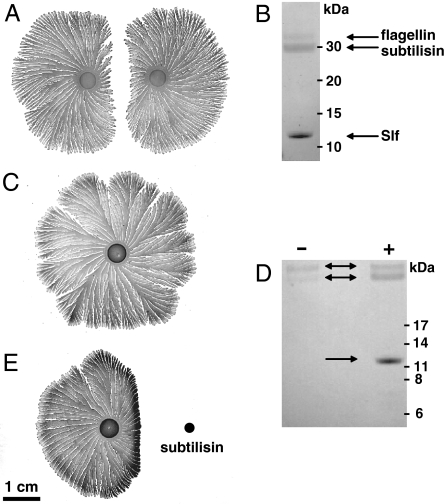
Fig. 1.
Colonies of P. dendritiformis (T morphotype) grown for one week on 1.5% agar with 2 g/L peptone, and analysis of proteins released into the medium. (A) Two sibling colonies grown after simultaneous inoculation of an agar plate at the same distance from the plate’s center. (B) Proteins extracted from the agar between two colonies, analyzed by SDS–PAGE gel electrophoresis and identified by peptide sequencing as flagellin at 32 kDa, subtilisin at 30 kDa, and Slf at 12 kDa (Arrows). (C) A single colony of P. dendritiformis grown on an agar plate. (D) SDS–PAGE electrophoresis of proteins extracted from agar surrounding a single colony, without (indicated by - or with +) added subtilisin, as shown in panels C and E. (E) A single colony with subtilisin (0.1 mg dissolved in 5 μL) added 1 d after inoculation (Black Dot).
Protein bands were detected at 32, 30, and 12 kDa (Fig. 1B). In contrast, extracts from the agar surrounding single growing colonies (Fig. 1C) showed only two protein bands, 32 and 30 kDa (Fig. 1D, Left Lane). The protein bands were analyzed by Edman degradation sequencing, and the genes encoding each of these proteins were identified using Blast analysis against the assembled genome of P. dendritiformis (8). The Blast analysis resulted in one perfect match between each of the partial protein sequences and a corresponding gene in the P. dendritiformis genome. The predicted P. dendritiformis amino acid sequences for each of these proteins are given in Fig. 2 and in SI Text. Based on BLASTP analysis against the nonredundant protein sequences database at the National Center for Biotechnology Information (http://blast.ncbi.nlm.nih.gov/Blast.cgi), the genes encoding the 32 and 30 kDa proteins have been annotated in the P. dendritiformis genome as flagellin (GenBank accession no. GQ891985) and subtilisin Carlsberg (GenBank accession no. GQ891986), respectively. The 12 kDa protein corresponded to a gene predicted to encode a larger, 173 amino acid, protein. This protein belongs to the DUF1706 family of conserved hypothetical proteins. The highest identity (57%) was found with a protein of unknown function of the Geobacillus sp. Y412MC10 (9). There was also identity (53%) with an uncharacterized protein IRC4 (increased recombination center) associated with recombination in yeast (10), but no other insights into function were obtained from analysis of the protein sequence. The P. dendritiformis gene was assigned GenBank accession no. GQ891987 and was named dfsB (dendritiformis sibling bacteriocin).
Fig. 2.
The list of the 173 amino acids of the DfsB protein corresponding to the dfsB gene. Large letters indicate the segment in position 5-169 amino acids, which is associated with a conserved Pfam family domain. Bold letters indicate the segment of the isolated protein Slf. The detected peptide (Edman sequencing) is underlined.
To determine whether one of these proteins might cause cell death, flagellin and subtilsin purified from the agar medium and commercial subtilisin (Carlsberg, P8038 Sigma) were each added at relatively high levels (0.1 mg for commercial subtilisin) near a growing single colony. Flagellin had no effect on colony growth or morphology, but subtilisin (both the P. dendritiformis protein and the commercial one) inhibited growth, and only dead cells were found in the inhibited area (Fig. 1E). Material extracted from agar near subtilisin-inhibited regions was found to contain a protein of the same size (12 kDa) detected in the region between two colonies (Fig. 1D Right Lane); the identity was confirmed by amino acid sequence analysis. This suggests that high subtilisin levels trigger secretion of this third protein, which is involved in the inhibition process and cell death.
Identification of the Killing Factor.
To further characterize the effect of the proteins on P. dendritiformis colony growth, various concentrations of subtilisin were placed near growing colonies (Fig. 3A). Low subtilisin levels (Fig. 3A, Upper Line) promoted bacterial reproduction and colony expansion, whereas higher subtilisin levels (Fig. 3A, Middle Line) initially promoted expansion but at later times inhibited expansion. Very high subtilisin levels (Fig. 3A, Lower Line) resulted in cell death at the inhibited interface. In low-nutrient liquid medium, subtilisin stimulated growth, even at the maximum concentration (20 mg in 3 mL of culture) tested (Fig. 3B). Cell shape and motility were unaffected in liquid culture, as determined by optical microscopy. A major difference between agar plates and liquid media is that the number of bacteria per unit volume is 1,000 times greater on agar plates; hence, it is suggested that high subtilisin levels will initially promote reproduction even on agar plates if the colony can expand fast enough. Indeed, when two colonies are competing, there is faster expansion at the facing fronts at the early stages of growth (SI Text), likely because of the added low level of subtilisin from the neighboring colony. A colony’s expansion is limited by surface tension (11); it cannot expand fast enough to create new space for the reproducing bacteria. Thus, an increase in subtilisin results in an increase of bacterial density and consequently nutrient stress.
Fig. 3.
Effect of subtilisin and Slf on colony growth. (A) Subtilisin was added (at the positions indicated by arrows) to single colonies growing on an agar gel, 1 d after inoculation; the images give the time elapsed (in hours) after subtilisin was added. The amount of subtilisin dissolved in a 5 μL water inoculation droplet was: Upper row, 0.001 mg; only attraction of growing colonies was observed. Middle row, 0.01 mg; attraction occurred only initially. Lower row, 0.1 mg; only inhibition was observed (see Fig. 1E). (B) Subtilisin (Carlsberg, P8038 Sigma) was added in different amounts (20, 10, 5, 2.5, 0.5, 0.25, 0.05, 0.025, 0.005, 0.0025, 0.0005, 0.00025, 0.00005 mg) to low-nutrient liquid media at 30 °C, after each tube was inoculated with 1 × 106 bacteria taken from a rich liquid media culture. The optical density of each tube was measured 24 h after inoculation (OD = 1 at 650 nm corresponds to approximately 108 bacteria/mL, verified by counting colonies on LB plates after appropriate dilution). The arrow marks the amount of subtilisin (0.1 mg) added to low-nutrient media agar plates near a growing colony to show strong inhibition and cell death. Error bars stem from 5 independent experiments. (C) Slf was introduced at the black dot near a single growing colony 4 days after inoculation. The faint region (shown at higher magnification in the inset) corresponds to lysed cells; growing branches were absent. Only a single band appeared in the gel electrophoresis results for the isolated Slf (Insert, Lower Right). In both A and C the growth conditions were 1.5% agar with 2 g/L peptone nutrient.
To further test the hypothesis that subtilisin is a growth promoter, two experiments were conducted: (i) Bacteria were inoculated on a hard agar gel (1.7%), where almost no growth occurs even for very high peptone levels (40 g/L) [the cutoff for growth of P. dendritiformis is 1.75% agar (7)]. However, no growth was detected even at very low subtilisin level on the 1.7% agar gel. (ii) At a lower agar concentration (1.5%), high subtilisin levels were found to inhibit growth, trigger secretion of the third protein and cause cell death (Fig. 1E). However, if surfactant (0.0006% Brij 30) was initially added to the medium (11), the colony expanded; high subtilisin levels were found to promote colony growth, and the third protein was not detected. These experiments show that even on an agar plate, subtilisin promotes reproduction, independently of how much is added. Consequently, bacteria become overpopulated, which likely triggers the production of the 12 kDa protein, reducing the number of bacteria.
To determine whether the 12 kDa protein, designated sibling lethal factor (Slf), is the killing factor, it was extracted from the agar and separated from the other proteins by dialysis with a 12–14 kDa cutoff. HPLC indicated that no other inhibiting compounds, including proteins or small peptides, were present in the dialyzed extract (SI Text). The extracted Slf placed near a single growing colony inhibited growth (Fig. 3C). This protein was found to be inhibitory only; no growth stimulation was found even at low levels of the protein (SI Text). In addition, when Slf was added to liquid cultures before the liquid was inoculated, no growth was detected. For grown liquid cultures, high levels of Slf lysed bacterial cells. More importantly, evidence of cell lysis was visible at the edge of inhibited colonies (see faint region in the magnified image in Fig. 3C). Colony branches exposed to the lethal protein were destroyed within a few hours (SI Text). These data indicate that Slf is the killing factor.
Activation of Lethality.
Slf secreted into the medium has an approximate molecular weight of 12 kDa based on its migration in polyacrylamide gels, but the predicted protein sequence of the gene is 173 amino acids, or 20 kDa (Fig. 2). The segment in position 5–169 amino acids (larger letters) is associated with a conserved Pfam family domain in many bacteria. For reasons specified below, we associate the bold segment with the isolated protein. This segment starts with the sequenced part of the detected peptide (underlined) and continues downstream to the end of the protein. The smaller size of the purified protein (12 kDa) indicates that the protein is processed or cleaved during secretion.
To determine whether the lethality was a property of the 20 kDa protein or if the protein required cleavage to be active, the DNA encoding the 20 kDa DfsB protein was cloned into an expression vector, and the protein was synthesized and purified as described in Materials and Methods. After purification, the 20 kDa protein was treated with commercial subtilisin (Carlsberg, P8038 Sigma), and the products were examined by SDS–PAGE gel electrophoresis (SI Text). The subtilisin treatment resulted in conversion of the 20 kDa protein to a 12 kDa protein that comigrated with the 12 kDa protein isolated from the agar medium. Colonies were exposed to both the uncleaved 20 kDa species and the 12 kDa processed protein. The P. dendritiformis bacteria were unaffected by the 20 kDa protein, but the 12 kDa fragment lysed growing colonies, as observed for the 12 kDa protein that was extracted from the area of inhibition between colonies (Fig. 3C).
Model.
The above results leave several unanswered key questions. In particular, why is Slf produced by neighboring colonies (or when subtilisin is added near single colonies), but not by a single colony? By answering this question we could potentially show the evolutionary advantage of bacteria dying in the inhibited region in the presence of another colony, rather than simply sporulating (P. dendritiformis spores are shown in SI Text).
In this paper we suggest an approach in which bacteria, nutrients, prespores, subtilisin, and Slf are modeled as continuous fields, and the outer effective envelope of the colony is given by a smooth, time-dependent curve (which is in accord with the experimental observation that the bacteria swim in a lubrication layer (12) that has a sharp, well-defined edge). The advantage of this approach is that the system is described as a free boundary problem, and the time evolution of both the continuous fields and that of the envelope can be modeled and simulated in a consistent and efficient way. This is suited to the problem at hand where we are more interested in describing a coarse-grained behavior of the colony rather than the exact shape of the edge of the lubrication layer. Details of the model are given in SI Text. The envelope of growing neighboring colonies is depicted in Fig. 4B.
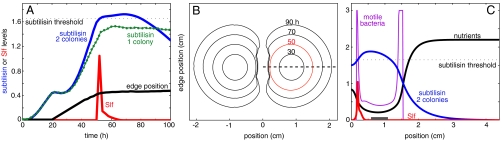
Fig. 4.
Numerical simulation of colony competition. (A) The subtilisin levels (in arbitrary units) at the moving edge of a colony as a function of time, both for a single colony (Green Dots) and for the inhibited interface in the case of two colonies (Blue Line). After 40 h, a competing colony senses its neighbor, and after 50 h the subtilisin level crosses a prefixed threshold (Horizontal Dashed Black Line). The red curve shows the level of Slf for the inhibited colony; no Slf is secreted in the single colony case. The black line shows the position of the edge of a colony growing toward a neighbor. The colony’s edge starts to move after a lag time of 20 h, and then begins slowing down just before Slf is secreted. (B) The positions of the edges of competing colonies at 30, 50, 70, and 90 h. (C) Levels of motile bacteria (Purple), subtilisin (Blue), nutrients (Black), and Slf (Red), measured along the horizontal dashed line in B, 50 h after inoculation. The gray rectangle represents the initial inoculation droplet. For each panel the model parameters and the initial conditions were the same; the initial distance between the colonies was 1.2 cm and the diameter of the dish was 8.8 cm, as in the experiments.
The main result from the model is the identification of a negative feedback loop that regulates subtilisin concentration at the front of a growing colony. Fig. 4C shows the profile of the bacteria, nutrient, subtilisin, and Slf concentrations immediately after Slf was produced. One of the key features of the profile is that inside the colony, subtilisin concentration exceeds a critical threshold. This does not trigger production of Slf because the motile bacterial concentration in this region is low. Assuming that subtilisin increases bacterial reproduction, we find that high subtilisin levels at the front increase bacterial density. This, in turn, increases the expansion rate of the colony, and the bacteria at the front move farther away from the location of the subtilisin maximum. Subsequently, subtilisin levels at the front decrease. An opposite chain of events happens at low subtilisin concentrations. Combining the two effects we find that subtilisin levels for a single colony reach a steady state in which the concentration of bacteria is close to maximal. Thus, subtilisin concentration at the front of the colony is regulated. This also accounts for the constant expansion rate of the colony (7).
An external source of subtilisin, such as a neighboring colony, can disrupt the above regulatory mechanism (Fig. 4A). Simulations show that disruption can happen in two ways: (i) due to added subtilisin from the neighboring colony, and (ii) due to nutrient depletion in the inhibited region between colonies, which increases the sporulation rate. The additional prespores increase the subtilisin level further (13). Another effect of nutrient depletion is the slowing down of the colony’s front. As explained previously, the bacteria at the front get closer to the location of the subtilisin maximum, and as a result, the reproduction rate becomes faster than the ability of the colony to expand (11). The bacterial stress cannot be resolved through sporulation, as (i) it requires additional nutrients (1–3), which may not be present, and (ii) sporulating bacteria are assumed to secrete high levels of subtilisin (13), which would make the situation even worse. It is suggested that to ensure survival the bacteria must quickly reduce the population level. The model predicts that secretion of Slf quickly reduces the bacterial population, which is consistent with the laboratory observations (Fig. 4B).
Testing Slf on Other Bacteria.
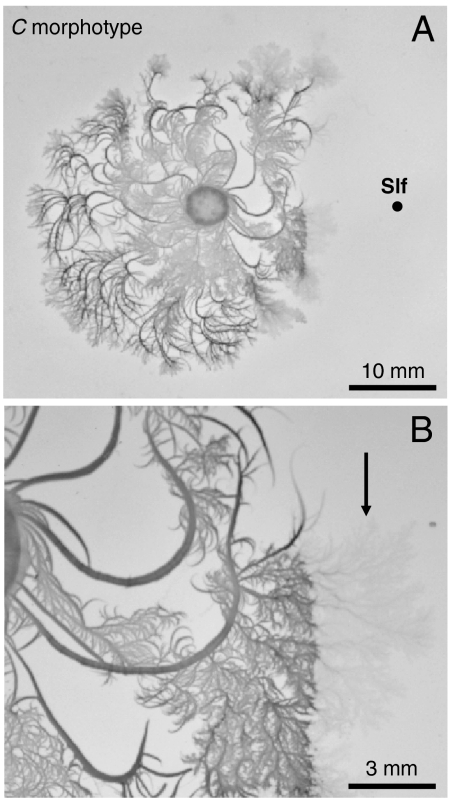
The toxic protein Slf was tested on P. dendritiformis C morphotype (14), and found to be lethal, lysing the cells (Fig. 5). However, Slf had no effect on the closely related species Bacillus subtilis. This suggests that Slf has a narrow spectrum of activity, similar to other bacteriocins (15, 16).
Fig. 5.
Slf (Black Point) placed near a single growing P. dendritiformis C morphotype colony (0.7% agar gel with 2 g/L peptone nutrient). (A) Inhibition of the colony. (B) Magnified image, which has a faint region (marked with an arrow) where bacteria are lysed. The concentration of Slf is same as in Fig. 3C.
Discussion
Bacteria in the natural environment must cope with constant changes in nutrient availability, physical conditions, available space, and competitors. Long-term survival favors those organisms that can adapt to environmental changes and regulate growth and development for optimal efficiency. P. dendritiformis is an excellent model system for studying bacterial competition and regulation of population size (7, 17). These motile bacteria grow as complex branching colonies on low-nutrient surfaces (12, 18, 19), and exhibit inhibition of growth in the presence of encroaching siblings (7).
An approach to assessing the role of Slf and subtilisin could be the construction of knockout mutations and the corresponding complemented strains in each gene of interest. However, suitable genetic systems are not yet available for P. dendritiformis, and we have used biochemical analysis as an alternative and equally valid approach to determining the mechanism of competition and killing. Using these methods, we have shown that inhibition is due to secretion of a lethal protein, Slf. This protein has no homology to any known bacteriocin or toxin and may represent a new class of antibacterial factors. Slf is secreted into the environment and can be isolated from the agar between competing colonies. N-terminal sequencing of the purified protein and comparison of the amino acid sequence with the genome identified the dfsB gene. The gene is annotated as a conserved hypothetical, predicted to encode a protein of 20.4 kDa. The protein is apparently processed and cleaved between lys-71 and pro-72, and the secreted form is the 12 kDa carboxy terminus of the protein. This secreted form of the protein is biologically active and causes lysis of P. dendritiformis on surfaces or in liquid medium.
Our previous studies showed that the killing of neighboring colonies occurred when a secreted factor reached a threshold concentration. The present analysis and testing of factors secreted by P. dendritiformis indicates that it is a second factor, subtilisin, which triggers the threshold response. Subtilisin, a serine protease, is secreted by P. dendritiformis and can be detected around single colonies. The concentration at the front of converging colonies is higher, and this appears to lead to the secretion of Slf. This behavior can be modeled mathematically, and as predicted by the model, exposure of a single colony of P. dendritiformis to a high concentration of subtilisin results in Slf secretion and death. Taken together, the data suggest that subtilisin and Slf are part of a complex system to regulate population spread and density in response to environmental conditions. On the surface of relatively low-nutrient agar, the bacteria are actively motile and spread outward, forming highly branched colonies. Subtilisin is secreted into the environment and its proteolytic activity may break down proteins to provide more easily used nutrient sources to the bacteria. The concentration of subtilisin is proportional to the cell density and may serve as a quorum sensor to control the growth of cells at the edge of the colony. When the local nutrient supply is exhausted, a proportion of the bacteria cease growing and initiate sporulation, allowing the colony to survive unfavorable conditions. However, when the bacteria are faced with invasion by a competing colony, a more drastic response occurs.
As the edges of neighboring colonies approach each other, the local concentration of subtilisin increases sharply, and exceeds the threshold for regulated colony growth (compare to Fig. 4A). This results in secretion of the active form of Slf (Fig. 4A, Red Curve), which lyses cells in the zone between the two colonies and prevents either colony from invading the other’s territory.
Bacteria are known to secrete a wide variety of bacteriocins and other antimicrobials for competing with other bacterial species or strains in the same ecological niche. For instance, the bacterium Proteus mirabilis (20) is capable of movement on solid surfaces by swarming motility. Boundaries form between swarming colonies of different P. mirabilis strains but not between colonies of a single strain (sibling). A fundamental requirement for boundary formation is the ability to discriminate between self and nonself. In our case, Slf was found to be lethal to P. dendritiformis but not to other bacteria, even closely related ones. These observations support the idea that the Slf acts as a sibling bacteriocin that is secreted for killing bacteria of the same strain and not for general killing of other competing species. Recently, much attention has been given to the special class of toxins that are secreted by stressed bacteria (either nutrient or antibiotic stress) to kill sibling cells within the same colony by cannibalism or fratricide (2–6). Both Slf and these toxins kill sibling cells, pointing to a possible similar mechanism. However, we did not find evidence that Slf is produced in an isolated, nutrient-starved colony; thus, we suggest that the Slf represents a new class of toxins that are most effective for regulating intercolony competition.
It is likely that this phenomenon is not restricted to P. dendritiformis. Bioinformatics analyses of the full-length, 20 kDa, DfsB protein revealed that it is a member of a large family of conserved hypothetical genes found in a wide range of Gram-positive bacteria such as Geobacillus sp. Y412MC10 (9). A close homolog (designated IRC4) is found in the yeast Saccharomyces cerevisiae, where it has been implicated in recombination (10). However, the gene is not found in other yeasts or in any of the other sequenced eukaryotic genomes. The position of the gene, at the end of a S. cerevisiae chromosome in a region that is not syntenic with other yeast, suggests that it may have been acquired from a bacterial source at some point in the evolution of S. cerevisiae.
The wide distribution and conservation of the family domain that encodes the Slf proteins suggests the existence of a new class of Slf-like proteins. Slf is the first member of this family to be isolated and characterized. It will be of interest to determine whether other members of this class serve the same function in promoting survival of bacteria in the natural environment by regulating the competition between sibling colonies growing in the same niches.
Materials and Methods
Strain, Growth Media, and Growth Pattern Experiments.
Bacterial strains used were P. dendritiformis (T morphotype) (21), P. dendritiformis (C morphotype) (14) and B. subtilis 3610. The low-nutrient medium contained NaCl (5 g/L), K2HPO4 (5 g/L) and Bacto Peptone (2 g/L). Difco Agar (Becton Dickinson) was added for solid media at concentration 1.5–1.75% for the T morphotype and 0.7% for the C morphotype. Twelve mL of molten agar was poured into 8.8 cm diameter Petri dishes, which were dried for 4 d at 25 °C and 50% humidity until the weight decreased by 1 g. Where indicated, a nonionic commercial surfactant, Brij 35 (Sigma), was added at a final concentration of 0.0006%) to the agar medium prior to autoclaving (11). The bacteria are maintained at -80 °C in Luria Broth (Sigma) with 25% glycerol. Luria Broth was inoculated with the frozen stock and grown for 24 h at 30 °C with shaking; it reached an OD650 of 1.0, corresponding to approximately 1 × 108 bacteria/mL.
For recording the growth of the colony over time, the plates were mounted on a rotating stage inside a 1 m3 chamber maintained at 30.0 ± 0.5 °C and 90 ± 2% humidity, as described previously (7).
Isolation and Identification of Secreted Proteins.
Secreted proteins were extracted from the agar between competing colonies (7) and separated by polyacrylamide gel electrophoresis on 1 mm NuPAGE 4–12% Bis-Tris gels (Invitrogen). Standard markers used were Invitrogen Novex sharp PreStained Protein Standards LC 5801 (for high molecular weight) and Molecular Weight Marker for Peptides (MW-SDS-17S, Sigma) (for low molecular weight). Following electrophoresis, protein bands were transferred to a membrane using a mini electro-wet blotter (Invitrogen), operated at 100 mA for 1 h. The Blotting buffer contains 20% methanol and dichlorodiphenyltrichloroethane. The N-terminal amino acid sequence of each band was determined by Edman sequencing (Pulse liquid ABI Procise 462).
The 12 kDa protein was separated by dialysis from the larger proteins in the material extracted f. The precipitated proteins were resuspended in 1 mL water and placed in dialysis tubing with a 12–14 kDa cutoff (SERVA, Servapor, dialysis tubing 44144). The material was dialyzed against distilled water and the dialysate collected. SDS–PAGE electrophoresis indicated a single protein of 12 kDa (Fig. 3C, Right Lower Inset).
High-Performance Liquid Chromatography.
The 12 kDa protein was analyzed and further purified by reverse Phase HPLC (Gold Gradient 126B, Beckman) and Karat software. Column: 0.8 × 250 mm C18 column. Buffer A: 0.1% TFA in water (LC-grade). Buffer B: 100% acetonnitrile (LC-grade), ca. 0.8% TFA. Flow rate 200 μL/ min. Absorption was measured for 2 wavelengths: 214 nm (absorption of peptide bonds) and 280 nm (for detecting aromatic amino acids residues). The 32 kDa protein (flagellin) and the 30 kDa protein (subtilisin) were also purified by reverse Phase HPLC.
Isolation of Slf.
The dfsB gene was amplified from P. dendritiformis using primers:
dfsBF 5′-CGCGGATCCGCAAGCTACGAATACACCTCCAAA-3′ and
dfsBR 5′-CCCAAGCTTTTACGCTTGTCTCTGATGTTTTTT-3′
BamHI and HindIII restriction sites (underlined) were incorporated into the forward (BamH1) and reverse (HindIII) primers. The PCR produced the expected 522 bp fragment. The PCR product was gel purified and ligated into the multiple cloning site of pGEM-T Easy (Promega), and transformed into JM109 Escherichia coli competent cells. The dfsB gene was excised from pGEM-T Easy plasmid using BamHI and HindIII, gel purified and ligated to BamHI and HindIII digested pQE30 His-tag expression plasmid (Qiagen). The orientation and correct DNA sequence of the inserted fragment was verified by DNA sequencing using ABI PRISM 3100 Genetic Analyzer.
Purification of the dfsB Protein.
The dfsB plasmid was transferred to E. coli strain BL-21 and overexpression of the amino terminal His-tagged DfsB was induced by the addition of 0.1 mM isopropyl β-D-1-thiogalactopyranoside to mid log phase cells (OD650 = 0.6). Following induction for 2 h at 37 °C, cells were harvested and lysed by sonication in buffer containing 20 mM sodium phosphate, 500 mM NaCl, 30 mM imidazole, 1 mM PMSF and 450 mg/L lysozyme. The crude lysate was incubated with Nickel Sepharose beads (Ni Sepharose 6 Fast Flow—GE Healthcare) to bind the His-tagged protein. The resin was then washed with phosphate buffer to remove proteins that did not specifically bind to the nickel column. The His-tag protein was eluted from the nickel column using 500 mM Imidazole. Expression and production of DfsB was monitored by gel electrohphoresis and Western blot analysis using antibodies against the Histidine residues.
Supplementary Material
Acknowledgments.
We thank I. Brainis (Tel Aviv University) for providing the bacterial strain and the growth protocol; S.A. Craig and E.R. Murphy (University of Texas, Austin) for helping us with electrophoresis gel-runs and protein precipitation; K.D. Linse (University of Texas, Austin) for advice and for help with Edman protein sequence and HPLC; and B. Engquist and R. Tsai (University of Texas, Austin) for valuable suggestions on modeling and numerical algorithms. The sequencing effort was supported by the Tauber Family Foundation and was conducted under the Tauber Initiative (Tel Aviv University) in collaboration with GeneBee group at Moscow State University and the Genome center at the Weizmann Institute and DYN-GS Israel. This work was supported by R. A. Welch Foundation Grant F-1573 (E.L.F.), National Institutes of Health Grant AI50669 (S.M.P.), National Science Foundation-sponsored Center for Theoretical Biological Physics Grants PHY-0216576 and 0225630 (E.B.J.), the University of California at San Diego (E.B.J.), and the Sid W. Richardson Foundation (H.L.S.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/1001062107/DCSupplemental.
References
- 1.Errington J. Regulation of endospore formation in Bacillus subtilis. Nat Rev Microbiol. 2003;1:117–126. doi: 10.1038/nrmicro750. [DOI] [PubMed] [Google Scholar]
- 2.Gonzalez-Pastor JE, Hobbs EC, Losick R. Cannibalism by sporulating bacteria. Science. 2003;301:510–513. doi: 10.1126/science.1086462. [DOI] [PubMed] [Google Scholar]
- 3.Ellermeier CD, Hobbs EC, Gonzalez-Pastor JE, Losick R. A three-protein signaling pathway governing immunity to a bacterial cannibalism toxin. Cell. 2006;124:549–559. doi: 10.1016/j.cell.2005.11.041. [DOI] [PubMed] [Google Scholar]
- 4.Claverys JP, Havarstein LS. Cannibalism and fratricide: mechanisms and raisons d’etre. Nature. 2007;5:219–229. doi: 10.1038/nrmicro1613. [DOI] [PubMed] [Google Scholar]
- 5.Guiral S, Mitchell TJ, Martin B, Claverys JP. Competence-programmed predation of noncompetent cells in the human pathogen Streptococcus pneumoniae: Genetic requirements. Proc Natl Acad Sci USA. 2005;102:8710–8715. doi: 10.1073/pnas.0500879102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Havarstein LS, Martin B, Johnsborg O, Granadel C, Claverys JP. New insights into pneumococcal fratricide: Relationship to clumping and identification of a novel immunity factor. Mol Microbiol. 2006;59:1297–1307. doi: 10.1111/j.1365-2958.2005.05021.x. [DOI] [PubMed] [Google Scholar]
- 7.Be’er A, et al. Deadly competition between sibling bacterial colonies. Proc Natl Acad Sci USA. 2009;106:428–433. doi: 10.1073/pnas.0811816106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. The assembly of the de novo P. dendritiformis genome was obtained using the 454 Life Sciences deep-sequencing technologies with 19X coverage and 100 bp reads long. The reads were assembled using the Newbler assembler. The assembled contigs were run through JCVI’s prokaryotic annotation pipeline.
- 9.Finn RD, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alvaro D, Lisby M, Rothstein R. Genome-Wide Analysis of Rad52 Foci Reveals Diverse Mechanisms Impacting Recombination. Plos Genet. 2007;3:2439–2449. doi: 10.1371/journal.pgen.0030228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Be’er A, et al. Paenibacillus dendritiformis bacterial colony growth depends on surfactant but not on bacterial motion. J Bacteriol. 2009;191:5758–5764. doi: 10.1128/JB.00660-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ben-Jacob E, Cohen I, Levine H. Cooperative Self-Organization of Microorganism. Adv Phys. 2000;49:395–554. [Google Scholar]
- 13.Kim JH, Kim BG. Construction of spore mutants of Bacillus subtilis for the development as a host for foreign protein production. Biotechnol Lett. 2001;23:999–1004. [Google Scholar]
- 14.Ben-Jacob E, et al. Cooperative Formation of Chiral Patterns during Growth of Bacterial Colonies. Phys Rev Lett. 1995;75:2899–2902. doi: 10.1103/PhysRevLett.75.2899. [DOI] [PubMed] [Google Scholar]
- 15.Swe PM, Cook GM, Tagg JR, Jack RW. Mode of action of dysgalacticin: A large heat-labile bacteriocin. J Antimicrob Chemother. 2009;63:679–686. doi: 10.1093/jac/dkn552. [DOI] [PubMed] [Google Scholar]
- 16.Heng NCK, et al. The large antimicrobial proteins (bacteriocins) of streptococci. Int Congr Ser. 2006;1289:351–354. [Google Scholar]
- 17.Ben-Jacob E, et al. Bacterial cooperative organization under antibiotic stress. Physica A. 2000;282:247–282. [Google Scholar]
- 18.Ben-Jacob E. Bacterial self-organization: Co-enhancement of complexification and adaptability in a dynamic environment. Philos T R Soc S-A. 2003;361:1283–1312. doi: 10.1098/rsta.2003.1199. [DOI] [PubMed] [Google Scholar]
- 19.Ben-Jacob E, Cohen I. Cooperative formation of bacterial patterns. In: Shapiro JA, Dworkin M, editors. Bacteria as Multicellular Organisms. London: Oxford Univ Press; 1997. pp. 394–416. [Google Scholar]
- 20.Gibbs KA, Urbanowski ML, Greenberg EP. Genetic determinants of self identity and social recognition in bacteria. Science. 2008;321:256–259. doi: 10.1126/science.1160033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ben-Jacob E, Cohen I, Gutnick DL. Cooperative organization of bacterial colonies: From genotype to morphotype. Annu Rev Microbiology. 1998;52:779–806. doi: 10.1146/annurev.micro.52.1.779. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.