Abstract
A century after the identification of a coenzymatic activity for NAD+, NAD+ metabolism has come into the spotlight again due to the potential therapeutic relevance of a set of enzymes whose activity is tightly regulated by the balance between the oxidized and reduced forms of this metabolite. In fact, the actions of NAD+ have been extended from being an oxidoreductase cofactor for single enzymatic activities to acting as substrate for a wide range of proteins. These include NAD+-dependent protein deacetylases, poly(ADP-ribose) polymerases, and transcription factors that affect a large array of cellular functions. Through these effects, NAD+ provides a direct link between the cellular redox status and the control of signaling and transcriptional events. Of particular interest within the metabolic/endocrine arena are the recent results, which indicate that the regulation of these NAD+-dependent pathways may have a major contribution to oxidative metabolism and life span extension. In this review, we will provide an integrated view on: 1) the pathways that control NAD+ production and cycling, as well as its cellular compartmentalization; 2) the signaling and transcriptional pathways controlled by NAD+; and 3) novel data that show how modulation of NAD+-producing and -consuming pathways have a major physiological impact and hold promise for the prevention and treatment of metabolic disease.
Nicotinamide adenine dinucleotide (NAD+) is long known as one of the main cofactors of single oxidoreductase reactions, but in recent years NAD+ is also emerging as a signaling molecule. In this review, we discuss the biosynthesis of NAD+ as well as its consumption by sirtuins, PARPs and cADP-ribose synthases. We also discuss the therapeutical potential of manipulating NAD+ levels as a treatment for metabolic and neurodegenerative diseases, as well as longevity.
I. Introduction
- II. NAD+ Biosynthesis and Salvage Pathways
- A. NAD+ biosynthesis
- B. NAD+ salvage pathways
- C. Substrate preference for NAD+ biosynthesis
- III. Role of NAD+ in Cellular Redox State
- A. Cellular NAD(H) and NADP(H): binding to proteins, and the metabolite indicator method
- B. The mitochondrial energy-linked transhydrogenase and the regulation of NAD+/NADH- and NADP+/NADPH-redox states
- C. Regulation of the mitochondrial and cytosolic NAD(H)-redox state
IV. NAD+ as a Signaling Molecule
- V. NAD+-Consuming Enzymes in Mammals (I)—Sirtuins
- A. Sirtuins as NAD+ sensors
- B. Biological consequences of NAD+ signaling through sirtuins
- VI. NAD+-Consuming Enzymes in Mammals (II)—PARPs
- A. PARPs as modulators of intracellular NAD+ levels
- VII. NAD+-Consuming Enzymes in Mammals (III)—cADP-Ribose Synthases
- A. CD38 as a regulator of NAD+ availability
VIII. Subcellular NAD+ Homeostasis
- IX. Therapeutical Potential of NAD+ Metabolism
- A. Regulation of NAD+ biosynthesis and salvage
- B. Therapeutic compounds
- C. Endocrine regulation and NAD+
- D. Type 2 diabetes (T2DM)
- E. Neurodegenerative disease
- F. Other pathophysiological states
- G. Longevity
X. Conclusions and Future Perspectives
I. Introduction
Since its initial discovery more than a century ago as “cozymase,” a cofactor in fermentation, NAD+ has received abundant attention in research, among others from four Nobel prize laureates (1). The breakthrough finding of the function of NAD+ by Otto Warburg in the 1930s significantly improved our knowledge of the chemistry of enzymatic reactions and the role of NAD+ therein. In several cellular subcompartments, either oxidized or reduced NAD serves in transhydrogenase reactions catalyzed by various oxidoreductase enzymes. It was only in the last decade, however, that the full extent of the function of NAD+ began to emerge with the identification of NAD+-consuming proteins, such as the sirtuins, that in turn function as metabolic regulators. By their NAD+ dependence, these proteins serve as metabolic sensors in the cell that can regulate downstream metabolic pathways. The relevance of these findings is illustrated by studies that imply these metabolic regulators in aging-related diseases such as type 2 diabetes mellitus (T2DM) and cancer. Therapeutic approaches targeting these pathways are currently being tested in clinical trials.
In this review, we will give an overview of the current state of knowledge of NAD+ metabolism. We will discuss its biosynthesis, both the primary biosynthesis from tryptophan and the NAD+ salvage pathways from the niacins, i.e., nicotinic acid (NA), nicotinamide (NAM), and NAM riboside (NR). We will touch upon its long-known function as a cofactor in oxidoreductase reactions and its role in maintaining the cellular redox state. Its recently attributed function as a substrate for metabolic regulatory proteins will be extensively reviewed, as well as its potential as a therapeutic target for the treatment of diseases that involve an important metabolic component.
II. NAD+ Biosynthesis and Salvage Pathways
A. NAD+ biosynthesis
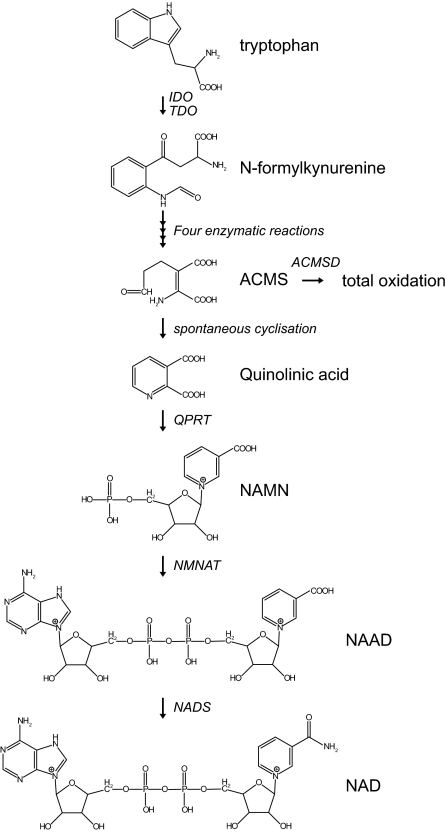
The primary biosynthesis of NAD+ starts with the essential amino acid L-tryptophan, which is taken up from the diet (Fig. 1) (2). The importance of dietary tryptophan is stressed by the human disease pellagra, which is caused by insufficiency of NAM, one of the intermediates of the NAD+ biosynthesis from tryptophan. As a result, patients suffer from diarrhea, dermatitis, and dementia and ultimately can die (3), but the disease can easily be treated by dietary supplementation of tryptophan or niacin (vitamin B3; indicating NA, NAM, and the recently discovered NR) (4). The first, rate-limiting step in the biosynthesis of NAD+ is the conversion of tryptophan to N-formylkynurenine by either indoleamine 2,3-dioxygenase (IDO) or tryptophan 2,3-dioxygenase (TDO), both requiring molecular oxygen. In mammals, TDO is the major enzyme contributing to NAD+ biosynthesis in the liver. In extrahepatic tissues the cytosolic IDO plays an important role, with highest activity reported in lung, spleen, and small intestine (5,6). Four subsequent enzymatic conversions result in the formation of the unstable α-amino-β-carboxymuconate-ε-semialdehyde (ACMS), which is a branchpoint in the tryptophan catabolic pathway (2). ACMS can be enzymatically converted to α-amino-β-muconate-ε-semialdehyde by ACMS decarboxylase directing the pathway to complete oxidation to CO2 and water. Alternatively, ACMS can undergo spontaneous cyclization forming quinolinic acid, which subsequently serves as a precursor for NAD+ (2). This latter nonenzymatic possibility seems to be only relevant in case the supply of ACMS is such that the enzymatic capacity is limiting, making this step another control point of NAD+ biosynthesis. In the dedicated NAD+ biosynthesis from tryptophan, quinolinic acid is condensed with 5-phospho-α-d-ribose 1-diphosphate, forming NA mononucleotide (NAMN), a reaction catalyzed by the enzyme quinolinate phosphoribosyltransferase (QPRT). This enzyme represents a second rate-limiting step in the biosynthesis of NAD+. QPRT is mainly expressed in liver and kidney (7), but it was also purified from brain (8). In brain, a reduction of QPRT activity was shown to be associated with epilepsy (9) and Huntington disease (10), possibly because accumulation of quinolinic acid results in activation of N-methyl-d-aspartate receptors (9,10,11,12,13,14). NAMN is converted to NA adenine dinucleotide (NAAD) using AMP by the enzyme NAM mononucleotide (NMN) adenylyltransferase (NMNAT), of which three isoforms have been described in humans (15,16,17). Human NMNAT1 is a nuclear enzyme that is ubiquitously expressed, with highest expression in heart and skeletal muscle and relatively low expression in brain (18,19). In contrast, NMNAT2 is highly expressed in brain (16,20) and is reported to be localized in the Golgi and cytosol (16,21). Its localization in the Golgi might coincide with the expression of tankyrases, members of the poly(ADP-ribose) polymerase (PARP) family. Upon NAD+ consumption by tankyrases, NMNAT2 could be involved in the repletion of the NAD+ pool in the Golgi. The best characterized targets of tankyrases reside, however, in the nucleus, leaving the question open whether tankyrase-mediated NAD+ consumption is indeed linked to NAD+ repletion in the Golgi (21). NMNAT3 is localized in both mitochondria and the cytosol and is mainly expressed in lung and spleen, tissues in which the other two isoforms are hardly expressed (17). Although the diverse subcellular localization suggests different pools of NAD+, which are synthesized separately, it is difficult to comprehend this process, e.g., why mitochondrial NAD+ biosynthesis is high in lung. Clarification of the subcellular localization of all the NAD+ biosynthetic enzymes could shed further light on this matter. The final step in the primary biosynthesis of NAD+ includes the ATP-dependent amidation of NAAD by NAD+ synthase using glutamine as a donor (22). Although in prokaryotes ammonia also serves as an amide donor, it remains controversial whether this activity is also relevant in humans (23). The human glutamine-dependent NAD+ synthase (NADsyn1) is mainly expressed in small intestine, liver, kidney, and testis, but other tissues showed expression as well (22). The whole body homeostasis of NAD+ biosynthesis, including potential fluxes of metabolites between organs, will be discussed in Section II.C.
Figure 1.
De novo NAD+ biosynthesis from tryptophan. The de novo biosynthesis of NAD+ starts with the conversion of tryptophan to N-formylkynurenine catalyzed by either IDO or TDO. N-Formylkynurenine is subsequently converted in four individual steps to the unstable ACMS, which can undergo either enzymatic conversion directed to total oxidation or nonenzymatic cyclization to quinolinic acid. The final step of the dedicated de novo biosynthesis of NAD+ is comprised of the QPRT-catalyzed formation of NAMN. NAMN is subsequently converted to NAAD by one of the NMNAT enzymes. The final step in the biosynthesis of NAD+ is the amidation of NAAD by the NAD synthase enzyme.
B. NAD+ salvage pathways
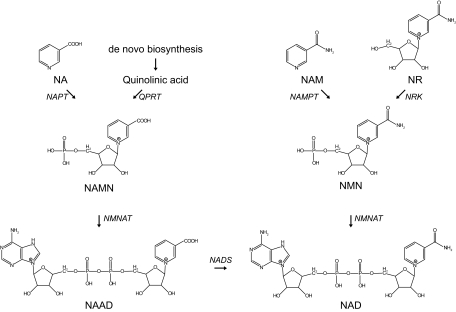
Although NAD+ can be synthesized de novo from tryptophan, it is assumed that the main source of NAD+ is from salvage pathways, which require the uptake of other NAD+ precursors from the diet. As mentioned in Section II.A, the dietary deficiency of tryptophan results in pellagra, which can be overcome by supplementation of the vitamin niacin. The dietary niacin, consisting of NA, NAM, and NR, can serve as an NAD+ precursor by means of the salvage pathways (Fig. 2). In mammals, NAM is thought to be the main niacin-derived NAD+ precursor (24,25), but the pathway for NA is also conserved (7). NA is converted to NAMN as a starting point of the so-called Preiss-Handler pathway (26). This reaction uses 5-phospho-α-d-ribose 1-diphosphate as a substrate and is catalyzed by NA phosphoribosyltransferase (NAPT), which in rats and mice is highly expressed in liver and kidney (27). After the initial conversion of NA, the Preiss-Handler pathway converges with the aforementioned primary NAD+ biosynthesis from tryptophan (Figs. 1 and 2).
Figure 2.
Mammalian NAD+ salvage pathway. NAD+ is synthesized in the NAD+ salvage pathway from its precursors NA, NAM, or NR. The initial step in NAD+ synthesis from NA, the so-called Preiss-Handler pathway, is catalyzed by NAPT and results in the formation of NAMN, which can also be derived from de novo NAD+ biosynthesis. In an identical reaction, but catalyzed by a different enzyme, NAM is converted by NAMPT forming NMN, which is also the product of phosphorylation of NR by NRK. The subsequent conversion of both NAMN and NMN is catalyzed by the same enzyme, i.e., NMNAT. In the case of NAMN, this reaction is followed by amidation, finally producing NAD+. NADS, NAD synthase.
Two pathways exist to convert the other niacin-derived molecule NAM to NAD+, each of which is exclusive in specific organisms. In lower organisms, such as bacteria and yeast, NAM is converted to NA by nicotinamidase, followed by integration in the Preiss-Handler pathway (28,29,30). In contrast, mammals lack the nicotinamidase activity but instead convert NAM to NAM mononucleotide (NMN) by one of the NAM phosphoribosyltransferase (NAMPT) enzymes (31), an enzymatic activity that has not been described for lower organisms (7). The enzyme has been reported to reside both intracellularly (iNAMPT) and extracellularly (eNAMPT), raising important questions regarding transport fluxes of NAD+ precursors, topics that will be discussed in Section II.C. Interestingly but confusingly, the eNAMPT isoform has also been described as a cytokine (pre-B cell colony-enhancing factor) involved in early B cell formation (32,33). eNAMPT, however, does not act as an insulin-like hormone, as described earlier (34,35). NAMPT, which is the rate-limiting step of this part of the pathway, is highly expressed in brown adipose tissue and liver and is undetectable in brain and pancreas (36). By virtue of its function in the conversion of NAM to NAD+, hence lowering NAM levels and increasing NAD+, NAMPT is considered an important regulatory enzyme with respect to the NAD+ consumers, notably the aging-associated histone deacetylase SIRT1. Several studies have shown the impact of NAMPT on cellular function. For instance, in human vascular smooth muscle cells, reduced NAMPT expression resulted in premature senescence, whereas a significant delay in senescence was observed upon overexpression of NAMPT (37). Furthermore, two very recent studies showed that NAMPT expression is regulated in a circadian fashion (38,39). The core clock components of the circadian machinery regulate the recruitment of SIRT1 to the NAMPT promoter to increase NAMPT expression. This is followed by NAD+ biosynthesis, which in turn will activate sirtuins as well as other NAD+-dependent enzymes. In a negative feedback loop, SIRT1 will repress the clock components and thereby NAMPT expression (38,39). It is suggested that through this mechanism, NAMPT (38,39) and SIRT1 (40,41) may play a crucial role in the circadian regulation of metabolism. After the NAMPT reaction, NMN is converted to NAD+ by NMNAT, which is able to condense the adenylyl moiety to both NAMN and NMN (16,17,18,42).
A third NAD+ salvage pathway, which was long known as the only NAD+ biosynthesis pathway in certain bacteria, is comprised of the phosphorylation of NR to NMN by NR kinase (NRK), after which one of the NMNAT enzymes catalyzes the formation of NAD+. Recently, this pathway was shown to be highly conserved and also active in yeast and humans, for which two NRK enzymes have been described (43). The NRK1 isoform is ubiquitously expressed, whereas NRK2 localization seems to be restricted to heart, brain, and muscle (44). The discovery of NR as a nutrient in cow milk (43) poses an interesting opportunity for therapeutic intervention in NAD+-dependent metabolism, as will be discussed in Section IX.
C. Substrate preference for NAD+ biosynthesis
The existence of four pathways for NAD+ biosynthesis originating from four independent precursors raises questions regarding the fluxes of metabolites. As mentioned in Sections II.A and II.B, some of the enzymes involved in NAD+ biosynthesis are strictly localized to a limited number of organs. Also, some enzymes, for instance NMNAT1-3, clearly show different subcellular localization. To date there is no consensus with respect to the question of the preferred substrate of NAD+. Feeding of rats with tryptophan, NA, or NAM showed that the first resulted in the highest NAD+ concentration in liver, suggesting that this is the preferred substrate at least in this organ (45). These results were confirmed in isolated hepatocytes, showing that NA and NAM are normally taken up from the medium but are not used for NAD+ biosynthesis (46). In tissues that lack the complete de novo NAD+ biosynthesis pathway, NAM is thought to be preferred over NA as the main precursor for NAD+ biosynthesis. This is illustrated by the fact that mice receiving an ip injection of NA only have a transient increase in NAD+ concentration accompanied by a high excretion rate of nicotinuric acid in urine, accounting for 36% of the administered NA. In contrast, NAM injection results in a more stable increase in NAD+ levels, with very limited nicotinuric acid excretion (25). Whether or not NAM is directly transported into the cell or is metabolized extracellularly remains unknown. Because both intra- and extracellular NAMPT have been reported, as well as circulating NMN (36), it seems plausible that both pathways contribute to NAD+ biosynthesis. In addition, the lack of NAMPT in some tissues suggests that these either use other substrates for NAD+ biosynthesis or convert NAM in the circulation. Recently, an NR transporter (Nrt1) has been described in yeast (47). Nrt1 belongs to the family of sodium-coupled transporters and was essential for optimal NAD+ synthesis and for growth of yeast on NR. NAD+ synthesis from NA and NAM was not affected by deletion of Nrt1 (47). The human ortholog of Nrt1 is yet unknown, although some transporters have been identified that are capable of transporting the NR analogs benzamide riboside and tiazofurin (48). Despite the significant advances in the field of NAD+ metabolism, especially since sirtuins have come to center stage, it remains to be established how the various pathways interact and which precursors are preferentially used in vivo. The elucidation of the transport pathways could advance this field of research and clarify the relevant pathways for NAD+ synthesis.
III. Role of NAD+ in Cellular Redox State
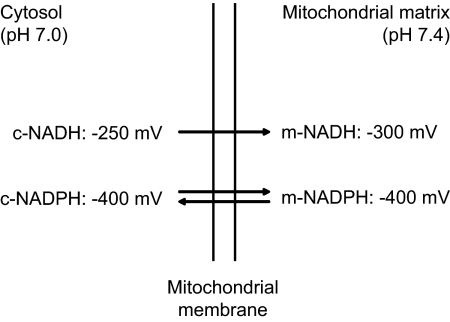
Pioneering work by Warburg and co-workers in the 1930s has led to the identification of the NAM dinucleotides NAD(H) and NADP(H) (49,50). Indeed, NADP+ was discovered as the coenzyme of the glucose-6-phosphate dehydrogenase reaction, whereas NAD+ turned out to be the obligatory cofactor of fermentation. Later studies revealed that NAD and NADP play an indispensable role in cellular oxidation/reduction reactions, with the NAD+/NADH couple primarily driving oxidation reactions and the NADP+/NADPH couple driving reductive reactions. The underlying basis for this remarkable difference between the NAD+/NADH- and NADP+/NADPH-redox couples has to do with the fact that the redox potentials of the two redox couples are widely different, with the NADP(H)-redox couple being much more reduced than the NAD(H)-redox couple (Fig. 3). The mitochondrial enzyme energy-linked transhydrogenase is responsible for this phenomenon as will be discussed in Section III.B.
Figure 3.
Redox potentials of the mitochondrial and cytosolic NAD(H) and NADP(H) systems in the liver. c, Cytosolic; m, mitochondrial.
The contents of the NAM nucleotides in distinct cell types and tissues differ markedly, especially if NADP plus NADPH is concerned. For instance, in rat liver total NAD, i.e., NAD+ plus NADH, amounts to some 800 nmoles/g wet weight, whereas total NADP amounts to about 420 nmoles/g wet weight. For skeletal muscle, however, these values are 590 and 30 nmoles/g wet weight, respectively. Apart from the differences in absolute levels, there is also the fact that NAD and NADP are heavily compartmentalized, with mitochondria containing high levels of NAD(H) and NADP(H). This explains why the percentage of NAD and NADP present in mitochondria, compared with the total amount of tissue, is higher in heart tissue than in liver tissue, especially if NAD is concerned (35 vs. 20%; Ref. 51), considering that mitochondria are more abundant in heart compared with liver.
A. Cellular NAD(H) and NADP(H): binding to proteins, and the metabolite indicator method
It has long been established that NAD(H) and NADP(H) are predominantly bound to intracellular proteins and that the free concentrations of NAD+, NADH, NADP+, and NADPH are much lower than the total concentrations as determined in protein-free tissue extracts obtained by acidification (NAD+ and NADP+) and alkalinization (NADH and NADPH). The existence of these binding sites, which bind NADH and NADPH more avidly than NAD+ (and NADP+) complicates the determination of the true redox state of the NAD(H)- and NADP(H)-redox couples within any tissue or cell.
To circumvent this problem, the so-called metabolite indicator method has been conceived (51). This elegant method is based on the notion that a particular NAD+/NADH-ratio may be calculated from the concentrations of a certain oxidized substrate and reduced substrate, participating in a NAD(H)-linked dehydrogenase reaction, provided that the selected dehydrogenase catalyzes a near-equilibrium reaction and is localized in one particular subcellular compartment only. If these conditions are fulfilled, the redox state of NAD(H) and NADP(H) can be calculated from the concentrations of the metabolites as measured in tissue homogenates using the following equation: [NAD+]/[NADH] = 1/K × [oxidized substrate]/ [reduced substrate]. It soon became clear that it would even be technically more convenient if the oxidized and reduced substrates could actually be measured in the extracellular space, which implies that both of the metabolites would need to permeate the plasma membrane rapidly.
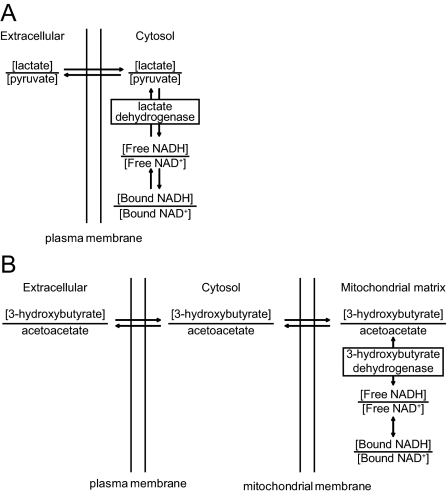
For the cytosolic and mitochondrial NAD-redox states, excellent metabolite indicator methods have been developed, which include the lactate dehydrogenase reaction for the cytosolic NAD(H)-redox state (Fig. 4A) and the 3-hydroxybutyrate dehydrogenase reaction for the mitochondrial NAD(H)-redox state (Fig. 4B). On the basis of lactate dehydrogenase and 3-hydroxybutyrate dehydrogenase as specific indicators of the cytosolic and mitochondrial NAD-redox states, it has long been recognized that the mitochondrial NAD-redox state is more reduced than that of the cytosol (some 50 mV), corresponding to a 40-fold difference in the NAD+/NADH ratio in the two compartments (Fig. 3).
Figure 4.
Metabolite indicators for NAD+/NADH. A, Lactate dehydrogenase as specific metabolite indicator reaction for cytosolic NAD+/NADH. B, Mitochondrial 3-hydroxybutyrate dehydrogenase as a specific metabolite indicator reaction for mitochondrial NAD+/NADH.
Such ideal metabolite indicator systems are not available for the cytosolic and mitochondrial NADP(H)-redox couples. Using the isocitrate dehydrogenase, malic enzyme, and the pentose phosphate dehydrogenase systems, the cytosolic NADP(H)-redox state has been found to be much more reduced than the NAD(H)-redox state (redox potential of around −400 mV) (Fig. 3). Subsequent studies, notably from Zuurendonk et al. (52) using the digitonin-fractionation technique, have clearly shown that the mitochondrial NADP(H)-redox state is approximately the same as the cytosolic one.
B. The mitochondrial energy-linked transhydrogenase and the regulation of the NAD+/NADH- and NADP+/NADPH-redox states
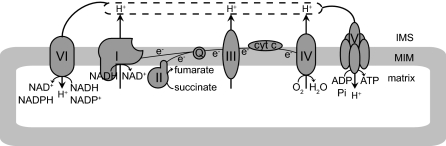
The energy-linked NAM nucleotide transhydrogenase catalyzes the following reaction: NADH + NADP+ ↔ NAD+ + NADPH (53,54) (Fig. 5).
Figure 5.
The role of the energy-linked transhydrogenase for the redox state. Schematic representation of the mitochondrial respiratory chain system, including complex VI, which is the energy-linked transhydrogenase catalyzing the reaction: NADH + NADP+ ↔ NAD+ + NADPH. The fact that complex VI is driven by the proton gradient across the mitochondrial membrane (just like the F1F0-ATP-ase) drives the transhydrogenase reaction far to the site of NADP reduction so that the transhydrogenase reaction is virtually unidirectional (NADH + NADP+ → NAD+ + NADPH). MIM, Mitochondrial inner membrane; cytc, cytochrome c; IMS, intermembrane space.
Because the midpoint potentials of the NAD- and NADP-redox couples are virtually identical, the equilibrium constant of the transhydrogenase reaction would normally be around 1, which would imply very similar redox potentials for the NAD- and NADP-redox couples resulting in similar NAD+/NADH and NADP+/NADPH ratios. Interestingly, however, the transhydrogenase catalyzing this reaction is an integral mitochondrial membrane protein, acting as a proton pump. For this reason, the energy-linked transhydrogenase is also referred to as complex VI of the respiratory chain (Fig. 5). Just like the mitochondrial F0F1-ATP-ase (complex V), the transhydrogenase is driven by the proton gradient across the mitochondrial membrane, which drives the equilibrium of the NADH + NADP+ ↔ NAD+ + NADPH far to the right. The property of the transhydrogenase being a proton pump explains why the mitochondrial NADP(H)-redox state is so much more reduced than the mitochondrial NAD(H)-redox state (Fig. 3).
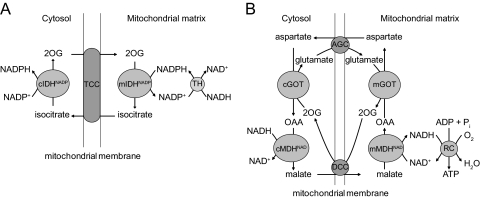
The mitochondrial NADP(H)-redox state dictates the cytosolic NADP(H)-redox state by virtue of the fact that the NADP(H)-redox states in the two compartments are in direct contact with one another via the 2-oxoglutarate/isocitrate NADP(H)-redox shuttle, made up of the mitochondrial and cytosolic NADP-linked isocitrate dehydrogenases and the 2-ketoglutarate/(iso)citrate carrier (Fig. 6A). The mitochondrial and cytosolic NAD(H)-redox states are also in direct contact with one another, again via specific redox shuttles, of which the malate-aspartate shuttle is probably the most important next to the α-glycerophosphate shuttle (Fig. 6B). These redox shuttles allow the transfer of reducing equivalents across membranes, which in general are impermeable toward the various NAM nucleotides.
Figure 6.
Redox shuttles. A, Schematic description of the NADP(H) redox shuttle: the NADPH as produced in the transhydrogenase (TH) reaction is transduced to the cytosol via the 2-oxoglutarate (2OG)–isocitrate redox shuttle in which mitochondrial NADPH first reacts with 2-oxoglutarate and bicarbonate as catalyzed by the mitochondrial NADP-linked isocitrate dehydrogenase (mIDHNADP) to produce isocitrate and NADP+, followed by the export of isocitrate to the cytosolic in exchange for 2-oxoglutarate. The isocitrate now present in the cytosol space is then converted back into 2-oxoglutarate and bicarbonate via the cytosolic NADPH-linked isocitrate dehydrogenase (cIDHNADP), followed by the uptake of 2-oxoglutarate into the mitochondrial space in exchange for isocitrate. TCC, Tricarboxylate. B, Schematic description of the malate/aspartate NAD(H)-redox shuttle. The NADH that is produced in the cytosol, for instance during glycolysis, is first converted into malate via cytosolic NAD+-linked malate dehydrogenase (mMDHNAD), after which the malate is transported into the mitochondria in exchange for 2-oxoglutarate (20G). Intramitochondrial malate is then converted into oxaloacetate (OAA), during which NADH is generated, which can now be used in the respiratory chain (RC) with formation of ATP. The oxaloacetate is not very permeable to mitochondrial membranes and is therefore first converted into aspartate via the glutamate oxaloacetate transaminase (GOT; cGOT, cytosolic GOT; mGOT, mitochondrial GOT), followed by the export of aspartate in exchange for glutamate. Cytosolic aspartate is subsequently converted back into cytosolic oxaloacetate, thus completing the cycle. Note that cytosolic NADH can also be reoxidized via other redox shuttles including the α-glycerol-3-phosphate shuttle in which the NADH first reacts with dihydroxyacetonephosphate to generate glycerol-3-phosphate plus NAD+, after which glycerol-3-phosphate directly transfers its electrons to the respiratory chain at the level of ubiquinone via the membrane-bound enzyme α-glycerol-3-phosphate dehydrogenase, thus completing the cycle. DCC, Dicarboxylate carrier; AGC, aspartate-glutamate carrier.
C. Regulation of the mitochondrial and cytosolic NAD(H)-redox state
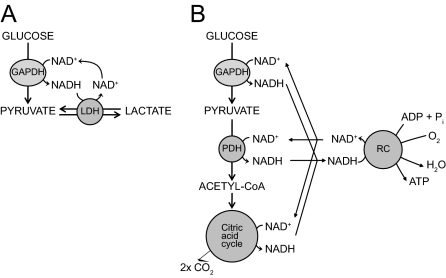
Under any particular condition, the NAD(H)-redox state is determined by the rate at which NADH is generated and the rate at which NADH is reconverted back into NAD+. In an intact cell that oxidizes glucose, NADH generated in the glyceraldehyde 3-phosphate dehydrogenase reaction can be reoxidized to NAD+ either via the lactate dehydrogenase reaction, as is the case in erythrocytes (Fig. 7A), or via pyruvate dehydrogenase, the citric acid cycle, and the mitochondrial respiratory chain, as in most other eukaryotic cells (Fig. 7B). In the latter case, mitochondrial and cytosolic NAD(H)-redox states will be the complicated end result of all the steps involved in the generation of NAD, including: 1) glycolytic enzymes, catalyzing the formation of pyruvate from glucose; 2) the mitochondrial pyruvate transporter and the pyruvate dehydrogenase complex; 3) the citric acid cycle; and 4) the respiratory chain. In the respiratory chain, NAD(H)-reoxidation is determined by the rate at which ATP is consumed in conjunction with the rate at which uncoupled respiration takes place, for instance under conditions of high uncoupling protein expression as in brown adipose tissue or in other tissues under certain conditions. Although the initial work on the redox biochemistry already started early in the previous century, the full extent of its implications is still poorly understood. Clearly, the many interactions between these pathways complicate this field of research, which definitely warrants further investigation.
Figure 7.
Regulation of mitochondrial and cytosolic redox state. A, Anaerobic metabolism of glucose into lactate with NADH produced in the glyceraldehyde-3-phosphate dehydrogenase (GADPH) reaction and with lactate dehydrogenase (LDH) reconverting NADH back into NAD+. B, Schematic representation of the aerobic oxidation of glucose with particular attention for the sites at which NADH is formed including: 1) the GADPH reaction during glycolysis; 2) the pyruvate dehydrogenase (PDH) complex; and 3) the citric acid cycle, followed by the reoxidation of NADH back to NAD+ by the respiratory chain (RC) with the concomitant formation of ATP from ADP + phosphate.
IV. NAD+ as a Signaling Molecule
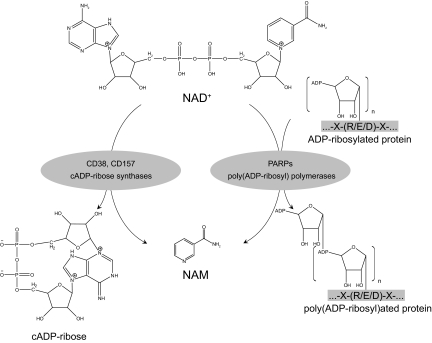
The coenzymatic activity of NAD+, together with the tight regulation of its biosynthesis and bioavailability, meets all the preliminary requirements to act as a metabolic monitoring system. This notion roots in the central role that NAD+ and NADH have as hydride-accepting and hydride-donating coenzymes in the reactions catalyzed by key enzymes of the glycolytic pathway, the respiratory chain, and in the redistribution of the electron equivalents generated from these catabolic pathways into de novo biosynthesis of macromolecules. During the last decade, however, it has become clear that NAD+ not only acts as a coenzyme for oxidoreductases, but also as a substrate that is consumed in certain reactions. In these reactions, NAD+ contributes as a donator of ADP-ribose. Three major families of enzymes can cleave NAD+ in mammals: sirtuins, ADP-ribose transferases, including PARPs, and cyclic ADP (cADP)-ribose synthases. These NAD+-cleaving enzymes are furthermore of medical relevance because they not only are indirectly affecting NAD+ bioavailability but also have a major impact on energy metabolism, cell survival, and aging. The blending of these two facts has led to the hypothesis that NAD+-consuming activities, mostly sirtuins, could act as energy sensors through NAD+ and, consequently, trigger appropriate adaptive responses.
To be true metabolic sensors, the activity of NAD+-consuming proteins must respond to physiological changes in NAD+ levels. Such a consideration is far from obvious because the previously mentioned NAD(H) compartmentalization and transport mechanisms add several layers of complexity. In addition, it should be noted that many of these mechanisms show a clear tissue specificity. For example, in heart and myocytes up to 75% of total intracellular NAD+ was found in mitochondria (55), whereas hepatocytes contain most NAD+ in the cytosol (56). The intracellular NAD+ concentration that has been described (0.4–0.7 mm) (57) might therefore not represent the actual in situ concentration when specific localization is taken into account, also because present techniques cannot differentiate between free and protein-bound NAD(H). For this reason, it is currently impossible to precisely couple physiological NAD+ metabolism and the activation of NAD+-consuming enzymes. Current data, albeit mostly correlative, clearly indicate that this coupling exists, but further technical leeway will be necessary to transform this concept into a factual reality.
V. NAD+-Consuming Enzymes in Mammals (I)—Sirtuins
Sirtuins are a family of NAD+-dependent protein deacetylases with similarity to the yeast silent information regulator 2 (Sir2). In general, sirtuins reverse acetyl modifications of lysine residues on histones and other proteins in a reaction that, unlike other previously characterized histone deacetylases, consumes NAD+, releasing NAM, O-acetyl ADP ribose, and the deacetylated substrate (58). The yeast Sir2 gained major scientific interest when it was linked to transcriptional silencing, within the context of yeast aging and senescence. In a seminal paper, it was shown that extra copies of Sir2 increased life span by 30%, whereas ablation of the Sir2 gene had the opposite effects, reducing life span by 50% (59). The NAD+-dependence of its deacetylase activity was fertile ground to sow the hypothesis that Sir2 could act as a metabolic sensor, capable of modulating gene expression according to the metabolic state of the cell (60). Supporting this hypothesis, several studies indicated that Sir2 could be a critical mediator of the beneficial effects of calorie restriction (CR) on yeast life span (61,62). The findings in yeast were soon transposed to metazoans such as Caenorhabditis elegans (63) and Drosophila melanogaster (64), which also lived longer with extra copies of Sir2 homologs.
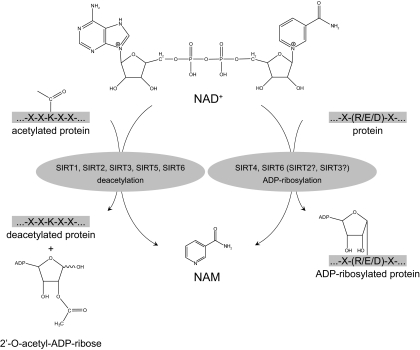
In mammals, seven homologs of Sir2 have been described, namely SIRT1–7 (Fig. 8), which are ubiquitously expressed and share a conserved catalytic core comprising 275 amino acids (see Refs. 65 and 66 for review). The multiplicity of sirtuin genes in higher eukaryotes has been associated with a divergence of the subcellular localization of the proteins that they encode to fulfill specialized functions. Consistent with a strong role of yeast Sir2 in the regulation of chromatin structure and gene expression, SIRT1, SIRT 6, and SIRT 7 are nuclear proteins, which are enriched in the nucleoplasm, in heterochromatin, and in nucleoli, respectively (67). In contrast, SIRT2 is mainly localized in the cytoplasm, although it can also regulate gene expression by deacetylating transcription factors that shuttle from the cytoplasm to the nucleus (68), and it contributes to chromatin compaction upon disassembly of the cell nucleus during mitosis (69). The remaining members of the sirtuin family (SIRT 3, SIRT 4, and SIRT 5) are predominantly mitochondrial proteins (67,70,71). In addition, the compartmentalization of the sirtuins might not be static, but rather displays dynamic characteristics, with shuttling between cellular compartments being induced at different stages during development or in response to cellular stress. This has been illustrated by recent reports demonstrating that SIRT1 can shuttle between nuclear and cytoplasmic compartments (72) and that the SIRT1 substrate realm also comprises cytosolic proteins, such as acetyl-coenzyme A (CoA) synthetase 1 (73). Similarly, SIRT3 has also been reported to translocate between the nucleus and mitochondria in response to cellular stress (74), although this is still a matter of debate (75). In addition to their different subcellular localization, all mammalian sirtuins do not have similar enzymatic activities. Indeed, SIRT1 and SIRT5 seem to act exclusively as deacetylases (76,77), whereas SIRT4 and SIRT6 act as mono-ADP-ribosyl transferases (78,79), and SIRT2 and SIRT3 display both activities (76,80). In the case of SIRT7, no clear activity has been reported as of yet, although it has been proposed to act as a deacetylase (81).
Figure 8.
Sirtuin enzymatic activities. Sirtuins display at least two different NAD+-consuming activities, both of which render NAM as a product. In mammals, SIRT1, SIRT2, SIRT3, SIRT5, and SIRT7 act as deacetylase enzymes, using NAD+ to cleave acetyl groups from ε-acetyl lysine residues of target proteins in a reaction that, in addition to NAM, generates 2′-O-acetyl-ADP-ribose. SIRT4 and SIRT6, rather, act as mono-ADP-ribosyl transferases, in a reaction where the ADP-ribosyl moiety of NAD+ is transferred to a substrate protein. Despite their predominant deacetylase activity, SIRT2 and SIRT3 also display ADP-ribosyl transferase activity.
A. Sirtuins as NAD+ sensors
Diverse approaches aimed at altering intracellular NAD+ levels and energy metabolism have consistently been shown to impact on sirtuin activity, although NAD+ is an abundant metabolite whose physiological concentrations rarely fluctuate more than 2-fold (82,83,84,85). Kinetic studies have determined that the Km values of sirtuins for NAD+ are in the range of 100–300 mm (58,86), which is surprising considering that the intracellular amounts of this metabolite clearly exceed the Km (Table 1). As mentioned before, however, (sub)cellular differences as well as the variability between the different NAD+ analytical methods could cause this discrepancy. Concentrations of freely available NAD+ might therefore be lower and thereby function as a metabolic sensor. Finally, distinct sirtuins may have different Km values for NAD+, further complicating the picture.
Table 1.
NAD+ concentrations
| Type of sample | NAD+ amount/concentration |
|---|---|
| Plasma (human) | 50–100 nm (239) |
| Muscle | 200–500 μmol/kg protein (83,84) |
| White adipose tissue | 10 μmol/kg protein (83) |
| Liver | 400–800 μmol/kg protein (82,83) |
| Mammalian cultured cells | 200–500 μmol/kg protein (83,85); 300–500 μm (95,240) |
| Mitochondria | 0.5–4 mmol/kg protein (95,123); 250 μm (95) |
| Cytosol | Unknown |
| Nucleus | 70 μm–estimate (241) |
| Enzymes | Km for NAD+ |
| Sir2 | ∼100 μm (242) |
| SIRT1 | ∼150–200 μm (243) |
| SIRT2 | ∼100 μm (243) |
| SIRT3 | ∼280 μm (244) |
| SIRT4 | Unknown |
| SIRT5 | Unknown |
| SIRT6 | Unknown |
| SIRT7 | Unknown |
| PARP1 | 20–60 μm (245,246) |
| PARP2 | ∼130 μm (246) |
| CD38 | ∼15–25 μm (85,153,247) |
NAD+ metabolism might also impact on sirtuins in other ways. It has been proposed that NADH competes with NAD+ for binding to sirtuins and inhibits the catalytic activity of sirtuins (87). Importantly, to do so, NADH must reach concentrations around the millimolar range, which are unlikely to be reached physiologically in cells (88). It remains plausible, however, that NADH concentrations might impact on sirtuin activity in the absence of competitive binding, but by affecting NAD+ levels because the metabolism of both molecules is interconnected (89). A second, but more likely, metabolite that could critically modulate sirtuin activity is NAM, an end-product of the sirtuin reaction (58). NAM can noncompetitively bind to the sirtuins and acts as a very potent inhibitor of sirtuin activity (90,91). Kinetic studies show that NAM acts at a Km between 30 and 200 μm, depending on the sirtuin (91). The in vivo relevance of this inhibition needs to be explored further. Although not much is known about the concentration of NAM in mammalian tissues and cells, the limited information [concentration in fasted human plasma = 0.3 μm (24)] points toward a role in NAD+ biosynthesis [Km= 0.92 μm (57)] rather than sirtuin inhibition. Localized regulation of NAM levels, however, might influence its fate. In this way, low levels of NAM could stimulate NAD+ biosynthesis, whereas localized higher concentrations of NAM could inhibit sirtuin activity in situ.
Although the information on localized (subcellular) NAM concentrations is lacking, some studies indicate that the NAM concentrations are indeed compatible with a role as a physiological inhibitor of certain sirtuins (89). One further difficulty is that, unlike NAD+, NAM diffuses through membranes (92), making it extremely challenging to determine whether there is compartmentalization in its abundance.
All the above data indicate that the mechanisms regulating sirtuin activity are not yet completely solved and that it is not clear whether the link between intracellular NAD+ levels and sirtuin activity is correlative or causal. This is, to a great extent, due to the fact that obtaining precise measurements of NAD+ and derived metabolites is technically demanding. Another shortcoming comes from the challenge of manipulating biological systems in such a way to specifically affect NAD+ without inflicting major metabolic perturbations. As discussed in Section IV, NAD+ levels are continuously modulated by a complex network of reactions (93), and determining when, how, and where NAD+ levels change is an extremely ambitious goal. For example, recent evidence suggests that NAD+/NADH mitochondrial shuttles are crucial for proper maintenance of the nuclear and cytosolic NAD+ homeostasis upon energy stress (94). In relation to sirtuins, it seems unlikely also that NAD+ or NAM could act as the sole switch controlling sirtuin activity because that would imply that, at the same time, several sirtuins would be activated and deacetylating a large number substrates. A recent report from our lab provides evidence that the increase in NAD+ must be accompanied by a targeting signal on the deacetylation substrate, such as a phosphorylation (or perhaps other) posttranslational mark, which would confer specificity to the sirtuin reaction (83). Additionally, specific sirtuin activation and function might be achieved by differential organelle-specific variations in NAD+ metabolism (95).
B. Biological consequences of NAD+ signaling through sirtuins
1. SIRT1
Among all mammalian sirtuins, SIRT1 is by far the best characterized. In general, SIRT1 is activated in situations of energy stress, such as fasting (82,96), exercise (83), or low glucose availability (85), which also increase NAD+ intracellular levels. Although our understanding of mammalian SIRT1 biology is still surprisingly weak, it seems clear that SIRT1 actions are strongly linked with metabolic homeostasis. Importantly, SIRT1 critically regulates the activity of a number of transcription factors and cofactors by modulating their acetylation status (see Ref. 97 for review). This list of transcriptional regulators includes the peroxisome proliferator-activated receptor γ (PPARγ), PPARγ coactivator-1α (PGC-1α), p53, and the FOXO family of transcription factors, all of which are key metabolic regulators. Although deacetylation has normally been associated with transcriptional repression, this is not always the case for SIRT1 targets. Deacetylation of the FOXO transcription factors, for example, seems to confer target gene specificity (98,99). In other cases, such as for PGC-1α, deacetylation is tightly linked with enhanced transcriptional activation (82,100). In support of this, mutation of PGC-1α acetylation sites to arginine, which mimics the deacetylated state, markedly increases basal PGC-1α transcriptional activity (82). Finally, the spectrum of SIRT1 substrates expands beyond transcriptional regulators, as has been demonstrated by the work showing that SIRT1 can directly regulate the activity of acetyl-CoA synthetase through deacetylation (73).
The relevance of SIRT1 in the control of whole body glucose homeostasis is the result of the combined action on different tissues (101,102), and it has a critical role during fasting (103). In the fasted liver, for example, NAD+ levels are 50% higher than in control conditions (82). This increase in NAD+ content correlates with increased SIRT1-mediated deacetylation and activation of the coactivator PGC-1α (82), which then promotes gluconeogenesis by coactivating a set of transcription factors, including hepatocyte nuclear factor-4 (104), glucocorticoid receptor (105), and FOXOs (106). Similarly, NAD+ content increases by more than 100% in cultured myotubes upon glucose deprivation (85,96), and around 45% in fasted muscle (C. Cantó and J. Auwerx, unpublished observations), both situations leading to increased deacetylation of SIRT1 targets, such as PGC-1α and FOXOs (96,107).
SIRT1 might also contribute to whole body metabolic homeostasis by acting on other tissues. In adipocytes, activation of SIRT1 by resveratrol decreases fat accumulation, whereas inhibition of SIRT1 results in triglyceride accumulation (108). This can be explained because deacetylation of PPARγ by SIRT1 promotes the docking of PPARγ with the corepressors NCoR (nuclear receptor corepressor) and SMRT (silencing mediator of retinoid and thyroid hormone receptors) (108). In the pancreas, SIRT1 regulates insulin secretion by decreasing uncoupling protein-2 expression and mitochondrial uncoupling, allowing a more efficient ATP synthesis (109,110). Additionally, SIRT1 action prevents oxidative damage in the pancreas, not only by decreasing the production of reactive oxygen species in the mitochondria, but also by enhancing FOXO and PGC-1α-mediated transcription of detoxification genes (111,112). Not surprisingly, the effects of SIRT1 on pancreatic function go hand in hand with those of NAD+ metabolism (36,57), suggesting again a causal relationship where SIRT1 translates alterations of NAD+ levels into transcriptional events.
2. Other sirtuins
Although less is known about the cellular roles of the other sirtuins as well as their interplay with NAD+ levels, some interesting functions have already been attributed to other sirtuins that we will discuss here. SIRT2 is mostly known as a tubulin deacetylase (76) that is down-regulated in human gliomas (113), which are among the most frequent malignant brain tumors. This observation, together with the fact that ectopic expression of SIRT2 in glioma cells lines is enough to decrease colony formation (113), suggested that SIRT2 could have a tumor suppressor role. It is interesting to note that gliomas, as several other tumors, are characterized by dramatically enhanced glycolytic rates without NADH reoxidation in the mitochondria and by the activation of the NAD+ consuming poly(ADP-ribosyl)ation. This would deplete NAD+ levels, which would keep sirtuin activity low. Further evidence supporting this hypothesis comes from experiments showing that SIRT2 controls the mitotic exit in the cell cycle and delays cell cycle progression through mitosis (114). Furthermore, SIRT2 acts as a mitotic checkpoint that prevents chromosomal instability and the formation of hyperploid cells in the early metaphase (115). Recently, SIRT2 in oligodendrocytes has been suggested to contribute to myelinization processes (116). Manipulation of NAD+ metabolism has not been properly studied in this context, and, therefore it will be necessary to relate the dynamics of SIRT2 activation to changes in NAD+ levels in future studies. Another recent interesting report indicates that SIRT2 might inhibit adipogenesis by deacetylating FOXO1 and promoting its binding to, and inactivation of, the proadipogenic nuclear receptor, PPARγ (68). The dynamics of NAD+ metabolism during adipogenesis have not been clearly established. Preliminary evidence in C2C12 myocytes indicates that the NAD+/NADH equilibrium is shifted to the reduced form during differentiation (117), which could be consistent with an inhibitory role of SIRT2 or other sirtuins on cellular differentiation. In this sense, adipocyte differentiation has been linked to both SIRT1 and SIRT2, casting some doubt on whether this is due to a specific effect on a given sirtuin or due to the global inactivation of sirtuins due to alterations in NAD+ levels.
SIRT3 action has been related to adaptive thermogenesis (80), mitochondrial function (80), energy homeostasis (118), and cellular survival during genotoxic stress (95). Intracellular ATP levels are lower in mouse embryonic fibroblast cells from SIRT3 knockout mice (118). This defect may possibly be explained by different reasons. First, SIRT3 seems to interact with and deacetylate at least one of the subunits of complex I of the mitochondrial respiratory chain (118). Because this interaction seems to be necessary for normal complex I function, it could contribute to the defects in ATP generation. An alternative, or complementary, explanation is that SIRT3 regulates the acetylation status of additional mitochondrial proteins, among which are glutamate dehydrogenase (GDH) (119) and acetyl-CoA synthetase 2 (120), which could also affect mitochondrial and global energy metabolism. Despite the major impact that such a function should have on global metabolism, no robust phenotype was observed in SIRT3−/− mice (119). Interestingly, even upon massive cytosolic NAD+ depletion, mitochondrial NAD+ levels are reported to be maintained (95). Although still a matter of debate (121), it was suggested that this was due to the fact that mitochondria are equipped with all the necessary enzymatic activities required for NAD+ salvage, including NAMPT and NMNAT3, enabling them to maintain mitochondrial NAD+ levels and NAD+-dependent activities. This latter observation further underscores that the transfer of NAD equivalents between the cytosol and mitochondria is tightly regulated during stress. Interestingly, mitochondrial NAD+ levels are increased upon fasting (95), suggesting that SIRT3 is more active and increases energy production in such conditions. If this is the case, it may not be the only mechanism capable of doing so because SIRT3 knockout mice can normally adapt to fasting (119).
Despite the presence of a conserved sirtuin domain, SIRT4 does not seem to possess significant deacetylase activity in vitro (78). Instead, SIRT4 modulates target protein activities through ADP-ribosylation. GDH, for example, can be ADP-ribosylated and inhibited by SIRT4 in pancreatic β-cells (78). GDH controls amino acid-stimulated insulin secretion by regulating glutamine and glutamate oxidative metabolism (78). Consequently, inhibition of GDH activity by SIRT4 decreases insulin secretion in mouse pancreatic β-cells in response to amino acids (78,122). Interestingly, two different sirtuins, i.e., SIRT1 and SIRT4 seem have contrary actions on insulin secretion (110). This observation suggests that NAD+ production might be confined to specific subcellular compartments to achieve a desired specific activation of sirtuins and, hence, trigger the appropriate physiological response.
Like SIRT3 and SIRT4, SIRT5 is a mitochondrial sirtuin and has deacetylase activity. The identification of carbamoyl phosphate synthetase 1 (CPS-1) as one of its deacetylation targets in liver (123) has provided one of the very first clues on the possible biological functions of SIRT5. CPS-1 is the committed and principal regulated enzyme in the urea cycle. During fasting situations, where NAD+ increases in the mitochondria, SIRT5 would deacetylate and activate CPS-1, which would then help detoxify the excess ammonia produced when amino acids are used as an energy source. A striking contradiction in this hypothesis is that mitochondrial NAD+ levels are approximately 250 mm under basal conditions (95), a concentration at which CPS-1 is already almost maximally activated by SIRT5 (123), hence leaving a very narrow window for further CPS-1 activation. This caveat might be explained by tissue-specific differences in mitochondrial NAD+, the fact that total NAD+ definitely does not mirror unbound NAD+ as available for sirtuin utilization or, alternatively, the existence of NAD+-independent mechanisms involved in the regulation of SIRT5.
SIRT6 was initially suggested to possess exclusively ADP-ribosylation activity (79), but recent evidence indicates that SIRT6 can also deacetylate histones and DNA polymerase β (124), a DNA repair enzyme. Coherent with this latter role, several lines of evidence suggest that SIRT6 controls genomic DNA stability and DNA repair. SIRT6−/− mice die prematurely, displaying severe defects, such as lymphopenia, loss of sc fat, decreased bone mineral density, a profound imbalance in glucose homeostasis, and decreased IGF-I levels (124). This phenotype has been suggested to mimic multiple pathologies found in elderly humans. An effect on DNA repair was hence proposed to explain the phenotype in SIRT6−/− mice, raising the hypothesis that SIRT6 could play an essential role in maintaining organ integrity as animals age.
SIRT7 is localized in the nucleolus as was initially described as a component of the RNA polymerase I (Pol I) transcriptional machinery (125). SIRT7 positively regulates transcription of ribosomal DNA (rDNA) during elongation, which, depending on the cell type, can account for up to 60% of total transcription in metabolically active states (125,126). SIRT7 overexpression increases Pol I-mediated transcription in a NAD+-dependent manner, whereas SIRT7 knockdown or inhibition of its catalytic activity decreases the Pol I-mediated transcription and its association with rDNA (125). As a consequence, the depletion of SIRT7 stopped cell proliferation and triggered apoptosis (125). The antiapoptotic effects of SIRT7 have also been observed in cardiomyocytes (81), where it was suggested that SIRT7 could directly control p53 acetylation. SIRT7 knockout mice display reduced mean and maximum life span, linked to heart hypertrophy, inflammatory cardiomyopathy, and hyperacetylation of p53 (81). Considering SIRT7 as a positive regulator of rDNA, it is striking that presumably SIRT7 may also have an antiproliferative role, as evidenced in loss- and gain-of-function cell-based models (127). In addition, the tumorigenic potential of several cell lines inversely correlates with SIRT7 expression (127). Upcoming research will bring some light in our understanding of SIRT7 biology.
VI. NAD+-Consuming Enzymes in Mammals (II)—PARPs
More than 45 yr ago, Chambon et al. (128) described how the addition of NAD to liver nuclear extracts stimulated the synthesis of poly(ADP-ribose). This finding led to the description of ADP-ribosylation activities, where NAD+ is used as substrate for a cellular process in which the ADP-ribose moiety is not transferred to an acetyl group, as happens with most sirtuins, but instead, it is linked to an amino-acid acceptor (129) (Fig. 9). This reaction is referred as mono- or poly(ADP-ribosyl)ation, and it constitutes a highly dynamic process because the half-life of ADP polymers is very short (130). Mono(ADP-ribosyl)ation reactions are catalyzed by ADP-ribosyl transferases, and this is a common reaction in most higher eukaryotes (1). Most of the time, however, attached ADP-ribosyl groups are built as polymers, a reaction that is catalyzed by PARPs. PARPs, the most abundant ADP-ribosyl transferases, constitute a superfamily of up to 17 members sharing a conserved catalytic domain containing the highly conserved PARP motif, which forms the active site (131). Among all these members PARP1 and PARP2 have been most widely studied so far because they account for virtually all PARP activity in the cell (132). PARP1 and PARP2 are ubiquitous nuclear proteins, and their targets are involved in the maintenance of chromatin structure and DNA metabolism. PARP2 is the least active one of the two, accounting for only 5–10% of total PARP activity in response to DNA damage (133). For detailed reviews on PARP functions, the reader is referred to some recent excellent reviews in Refs. 129 and 134.
Figure 9.
PARPs and cADP-ribose synthases as NAD+-consuming enzymes. Intracellular NAD+ content largely depends on the activities of two different families of enzymes: PARPs and cADP-ribose synthases (CD38 and CD157), both of which render NAM as an end-product. PARPs catalyze a reaction in which the ADP-ribose moiety is transferred to a substrate protein. PARPs can transfer multiple ADP-ribosyl groups and form long chains and branches of ADP-ribosyl polymers. cADP-ribose synthases use NAD+ to generate cADP-ribose, which acts as an intracellular secondary messenger.
A. PARPs as modulators of intracellular NAD+ levels
In contrast to sirtuins, activation of PARPs may not depend so much on NAD+, since the Km is situated in the lower micromolar range (135). A second piece of evidence supporting this concept comes from experiments that have shown that the catalytic activity of PARPs seems to be regulated by binding to DNA breaks through their DNA-binding domains. This physical interaction stimulates PARP catalytic activity by more than 500-fold (136). Despite these observations, some studies have pointed out that NAD+ availability can influence the length of poly(ADP-ribosyl) polymers synthesized by PARP1 (137). Despite this nonlimiting role of NAD+ on PARP activity, PARP action has a major impact on global NAD+ metabolism, which in turn, and as discussed below in this section, may affect sirtuin function.
Several reports suggest that PARP activity constitutes the main NAD+ catabolic activity in the cell (138,139,140). Consequently, PARPs critically affect the intracellular NAD+ levels, forcing the cell to continuously synthesize NAD+ from de novo or salvage pathways to maintain cellular viability. This fast recycling is consistent with the short half-life of NAD+, which is estimated around 1 to 2 h (141,142,143). The detrimental action of PARPs on NAD+ pools becomes evident in experiments where cells are treated with genotoxic agents to promote DNA damage. These agents lead to a sustained activation of PARP activity and a concomitant decrease in NAD+ levels to 10–20% of their normal levels within 5–15 min (138,139). This can have further consequences because NAD+ depletion lowers ATP generation rates. Considering that ATP is required for the resynthesis of NAD+ (144), a feedforward loop culminating in a major NAD+ depletion would be expected under severe genotoxic conditions. Given the nuclear nature of the PARPs, the quest for a fast nuclear regenerator of NAD+ levels has been intensive. Initially, NAD+ was thought to freely interchange between the nucleus and the cytosol through specialized pore complexes in the nuclear membrane (21). For this reason, it was shocking that the predominant form of NMNAT in mammals, NMNAT1, is a nuclear protein (21), suggesting that nuclear NAD+ production might be required to compensate for the high rates of NAD+ breakdown caused by PARP activation. Very preliminary studies indicating that PARP1 and NMNAT1 interact and modulate each other’s enzymatic activities seem to support this hypothesis (42,145).
The fact that PARP activities consume such a high amount of NAD+ raised the hypothesis that PARPs and sirtuins could compete for the same limiting intracellular NAD+ pool. Some reports suggest that this may actually be the case. Experiments in myocytes, where DNA damage was induced with H2O2, demonstrated that increased PARP activity depleted cellular NAD+ levels and reduced SIRT1 deacetylase activity (146). Repletion of cellular NAD+ levels either by adding NAD+ directly to the culture medium or by overexpressing NAD+ biosynthetic enzymes was enough to recover cell viability, but only in the presence of SIRT1 (146). Higher acetylation levels of p53 are also observed after H2O2 treatment, probably reflecting a decrease in SIRT1 activity (146). PARP activity, therefore, critically adjusts NAD+ levels according to cell damage, and the fluctuations in NAD+ set by PARP might act as a permissive or repressive determinant for sirtuin activity. Interestingly, this relationship may also work in the reverse direction because activation of SIRT1 with resveratrol reduces PARP1 activity (147). Conversely, there is a sharp increase in PARP activity in cells where SIRT1 has been genetically ablated (147). This interesting interplay of NAD+-dependent enzymes opens the window for new therapeutic strategies for the activation or inhibition of their respective activity.
VII. NAD+-Consuming Enzymes in Mammals (III)—cADP-Ribose Synthases
cADP-ribose synthases are a pair of ectoenzymes, also known as the lymphocyte antigens CD38 and CD157. Both enzymes arose from a gene duplication event and are highly conserved in most eukaryotes all along the evolutionary scale (148). CD38 is a type II glycosylated protein with a single transmembrane domain near its N terminus (149). It is approximately 25% identical to CD157 (149). Both share 10 cysteine residues, making them very similar at the structural level, and they are ubiquitously distributed (149) (Fig. 9). CD38 and CD157 are multifunctional enzymes that use NAD+ as a substrate to generate second messengers, such as cADP-ribose, which contributes to calcium mobilization (149). The enzymatic activity of CD38 is highly dependent on the pH (149), suggesting that the in vivo activity of this promiscuous enzyme may change according to its environment. For further information on the basics of CD38 and CD157, we refer to recent reviews in Refs. 150,151,152.
A. CD38 as a regulator of NAD+ availability
As happened with the PARPs, CD38 displays a Km for NAD+ in the low micromolar range (153), indicating that, a priori, intracellular NAD+ levels may not be limiting its catalytic activity. The stoichiometry of the reaction catalyzed by CD38, however, involves a massive amount of NAD+, around 100 molecules, to yield a single cADP-ribose (154). This led to the hypothesis that CD38, even if it is not highly expressed, could act as a major intracellular regulator of NAD+ levels. Experiments devoted to answering this question identified CD38 as one of the main cellular NADases in mammalian tissues and confirmed CD38 as a critical regulator of cellular NAD+ (155). CD38−/− mice displayed up to a 30-fold increase in intracellular NAD+ levels (155). It must be noted that physiological increases in NAD+ levels upon fasting or CR generally do not exceed 2-fold changes (82,84). Presumably, such an increase should drive toward enhanced sirtuin activity. Supporting this hypothesis, CD38 depletion was sufficient to increase SIRT1 activity via the up-regulation of intracellular NAD+ levels (156). Consequently, CD38−/− mice phenocopy the situation of SIRT1 activation, with decreased p53 acetylation levels, enhanced energy expenditure, and resistance to high-fat diet obesity (157). Preliminary experiments in which CD38−/− mice were fed with the putative sirtuin inhibitor, sirtinol, showed that the protection against diet-induced obesity was lost (157). Whether this phenotype is exclusively due to sirtuin activation, however, is still unclear. Nevertheless, these findings seem to attribute a role as a regulator of cellular NAD levels to CD38 and indicate that CD38 may serve as a pharmacological target for boosting sirtuin activity.
VIII. Subcellular NAD+ Homeostasis
As pointed out in the previous sections, the biosynthesis and consumption of NAD+ involves many proteins that are localized to various subcellular compartments and are active in different circumstances. The subcellular localization of NAD+ and its related proteins is especially the subject of recent research because this has important implications for intervention in these pathways. As indicated in Table 2, all components for the de novo biosynthesis of NAD+ from tryptophan are localized in the cytosol. Although NAD+ could be transported to other subcellular compartments, it seems plausible that these compartments also have autonomous NAD+ biosynthesis, especially considering the high NAD+ consumption rates that require a compensatory repletion. Indeed, the nucleus and mitochondria, both of which require NAD+ repletion due to the sirtuin- and/or PARP-mediated consumption, are capable of (re)synthesizing NAD+ from NAM. The tight interplay between NAD+ synthesis and consumption is also nicely illustrated by a recent study showing that NMNAT1 is recruited to SIRT1 target gene promoters, where it regulates its deacetylase activity (158). Whether or not this is due to the localized increase in NAD+ biosynthesis is unknown, but this could pose an attractive mechanism of regulation.
Table 2.
Localization and disease relevance of NAD+ synthesizing and consuming enzymes
| Enzyme | Tissue specificity | Subcellular localizationa | Disease relevance | Ref. |
|---|---|---|---|---|
| NAD biosynthesis and salvage enzymes
|
||||
| IDO | Ubiquitous | Cytosol | 5,6 | |
| TDO | Liver | Cytosol | 7,8 | |
| QPRT | Liver, kidney (brain) | Cytosol | Epilepsy, Huntington disease | 1,7,8,9,10 |
| NMNAT1 | Ubiquitous, high in muscle, heart | Nucleus | Wallerian degeneration | 18,19,213,214 |
| NMNAT2 | Brain | Golgi, cytosol | 8,20,21 | |
| NMNAT3 | Ubiquitous; high in lung, spleen; low in brain | Mitochondria, cytosol | 17 | |
| NADsyn1 | Ubiquitous | Cytosol | 1,22 | |
| NAPT | High in liver and kidney | Cytosol | 27 | |
| iNAMPT | High in brown adipose tissue, liver; low in brain, pancreas | Cytosol, nucleus, mitochondria | Cancer, T2DM, neutrophil survival | 8,36,95,184,230,231 |
| eNAMPT | Plasma | Ectoenzyme | T2DM | 32,33,36 |
| NRK1 | Ubiquitous | Unknown | 43,44 | |
| NRK2 | Heart, brain, muscle | Unknown | 43,44 | |
| NAD-consuming enzymes | ||||
| SIRT1 | Ubiquitous | Nucleus (cytoplasm) | Longevity, obesity, T2DM, cancer, cardiovascular and neurodegenerative disease | 61,62,63,64,65,67,101,102,206,207,223,224,232,234 |
| SIRT2 | Ubiquitous | Cytoplasm (nucleus) | 65,67 | |
| SIRT3 | Ubiquitous | Mitochondria (nucleus) | 65,67 | |
| SIRT4 | Ubiquitous | Mitochondria | Longevity | 65,67,236 |
| SIRT5 | Ubiquitous | Mitochondria | 65,67 | |
| SIRT6 | Ubiquitous | Nucleus | Longevity | 65,67,124,235 |
| SIRT7 | Ubiquitous | Nucleolus | Longevity | 65,67,81,127 |
| PARP1 | Ubiquitous | Nucleus | 128,248 | |
| PARP2 | Ubiquitous | Nucleus | 246,248 | |
| CD38 | Ubiquitous, mainly immune cells | Ectoenzyme (nucleus, cytoplasm) | 152 |
The main subcellular localization is indicated. For some enzymes secondary localization is shown in parentheses.
Although the examination of the compartmentalization of NAD+ homeostasis is so far limited by technical difficulties, new techniques have been developed recently that will allow better analysis. Several probes are available that can be specifically targeted to a single subcellular compartment and could thereby be suitable for the thorough assessment of the redox state of organelles (159,160). These probes, based on either the catalytic domain of PARP or a reduction-oxidation-sensitive GFP, will allow better defining of the subcellular redox conditions.
Another technique that has been developed to analyze the cellular redox state is two-photon microscopy of NAD(P)H (161). This technique is based on the measurement of autofluorescence of NADH and NADPH and allows analysis in living cells. Although a global view of the cellular or even subcellular redox state can be obtained, the lack of specificity of the measurement may preclude its use for further analysis of NAD+ subcellular compartmentalization.
IX. Therapeutical Potential of NAD+ Metabolism
The therapeutic relevance of NAD+ has long been recognized. Although it was not yet known that NAD+ deficiency was the underlying cause of the disease, pellagra was known as a deficiency of niacin (4). Dogs suffering from “canine black tongue” regained appetite only 2 h after niacin administration, reverted their retarded growth, and appeared completely normal after 3 d of treatment (4,162). The current incidence of pellagra is low due to improved nutritional status, with a notable exception of incidental cases of pellagra in patients with alcoholism or malnutrition disorders (163). More recently, NAD+ as a therapeutic target has gained interest mainly because of the discovery of sirtuins, which can seriously impact on mitochondrial function and therefore be beneficial for a large array of disorders.
In this section, we will touch upon the regulation of NAD+ biosynthesis and salvage, which is requisite for its potential use as a therapeutic target. We will also refer to therapeutic options to intervene in NAD+ metabolism as a potential treatment of NAD+-related diseases. We will give an in-depth view of pathophysiological states that are associated with NAD+ metabolism, such as type 2 diabetes and neurodegenerative disease, and discuss the involvement of NAD+ in longevity.
A. Regulation of NAD+ biosynthesis and salvage
Several of the NAD+ biosynthetic steps were shown to be subject to regulation, induced by either pharmacological or (patho-)physiological means. The initial rate-limiting step catalyzed by either TDO or IDO is regulated. The fact that TDO has a short half-life of approximately 2 h makes it a useful target for regulation (2). TDO is induced by tryptophan, thereby increasing tryptophan catabolism and potentially enhancing NAD+ biosynthesis. Additionally, TDO expression and activity are regulated by glucocorticoids, for which TDO contains a glucocorticoid response element (164). The increased TDO activity limits tryptophan availability for protein synthesis, and consequently, other available amino acids will be used for gluconeogenesis (2). In contrast to TDO, IDO is not induced by its substrate tryptophan but rather by inflammatory stimuli, such as viral, bacterial, and parasitic infections (165,166,167,168). The induction is mainly mediated by interferon-γ by virtue of an interferon-stimulated response element (169). Although the mechanisms are not well understood, IDO induction is thought to result in local tryptophan depletion in combination with increased tryptophan metabolite concentrations, thereby modulating the T cell response by selective proliferation and apoptosis regulation (170,171,172). Whether or not NAD+ levels are also affected by the increased IDO activity remains an open question, but it is debatable because most tissues and/or cell types lack the QPRT enzyme. It is therefore more likely that the increased IDO activity results in increased tryptophan oxidation. Along with the increased IDO activity, kynurenine 3-hydroxylase and kynureninase, intermediary enzymes in the pathway from tryptophan to ACMS, are also induced on exposure to lipopolysaccharide and proinflammatory cytokines (8).
The QPRT enzyme activity was markedly increased in liver of rats that were fed with various peroxisome proliferators, in combination with an inhibitory effect on ACMS decarboxylase, together leading to a significant increase in NAD+ concentrations (173). ACMS decarboxylase was also reported to be decreased by T4 and high-fat diet, whereas increased activity was observed in diabetic rats, glucocorticoid treatment, and high-protein diet (173,174). In the NAD+ salvage pathway, regulation has been described for eNAMPT. In lymphocytes, eNAMPT transcription is increased by exposure to pokeweed mitogen (32). Whether this increase results in higher NAD+ biosynthesis activity or in enhanced cytokine-like signaling activity of this two-faced protein is not known. In rats in which the sciatic nerve was transected, all tested mRNAs encoding NAD+ biosynthetic enzymes, but most strikingly the mRNA of NRK2, were increased, providing a potential mechanism for preventing neuronal degeneration (175). As will be discussed later, the increase in NAD+ metabolism might be beneficial for protection against neuronal degeneration.
B. Therapeutic compounds
1. Increasing NAD+
To benefit from increased metabolic activity associated with sirtuin activation, strategies could be employed to increase the concentration of NAD+. Most obvious would be treatment with one of the NAD+ precursors (Fig. 10A). Along this line, it was shown that incubation of human kidney cells with NA resulted in a substantial increase in NAD+ content. Interestingly, this increase was not inhibited by NAD+ (27). Also, high doses of NA given to rats robustly increased NAD+ levels in both liver and blood (176). Such high doses of NA, as used for the treatment for hypercholesterolemia (see Section IX.F), are, however, not always well tolerated. Because it is unlikely that the effects of high amounts of NA on hypercholesterolemia are sirtuin-dependent, one might speculate that lower doses of NA, not causing adverse effects, may well be beneficial for increasing NAD+ levels and activating sirtuins.
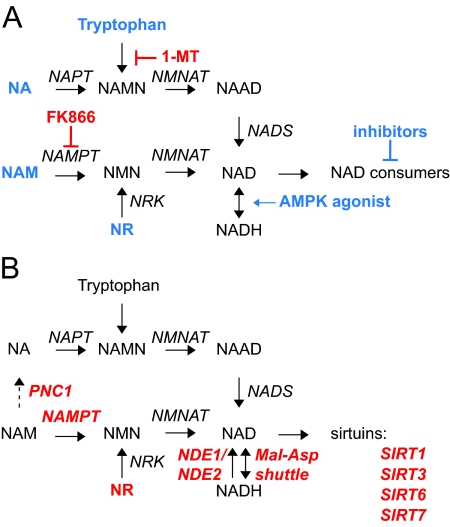
Figure 10.
Therapeutical strategies associated with NAD+ metabolism. A, Several therapeutical strategies have been described to either increase (blue) or decrease (red) NAD+ content. B, Various enzymes or metabolites linked to NAD+ metabolism were implicated in longevity and/or its related health issues establishing the link between NAD+ and healthspan. The dashed arrow indicates a metabolic conversion that is not present in mammals. 1-MT, 1-Methyltryptophan.
NAM has been evaluated for its use as a T1DM treatment by means of its protective effect on pancreatic β-cells. As will be discussed later, however, no positive effect was observed from long-term NAM treatment (177). Adverse effects of NAM have hardly been reported, except for hepatic toxicity at high doses (≥3 g/d) (178). The fact that the Km of NAMPT is quite low suggests that additional NAM might not have the desired result because the enzyme will already be operating at its maximal activity with endogenous NAM levels. Worse even, administered NAM might decrease sirtuin activity because NAM has been reported to act as an inhibitor (90,91).
Since its discovery as an NAD+ precursor in humans, NR has not yet been used in a clinical study. It could potentially, however, be a very useful compound to increase NAD+ levels and enhance its downstream effects. As mentioned before, both NA and NAM have relevant downsides, either the NA-induced flushing or the NAM-mediated inhibition of sirtuins. Because the NA receptor GPR109B, which is in part responsible for the NA-induced adverse effects, is specific for NA, one could expect that NR will not cause flushing. The inhibitory effect of NAM on sirtuin activity could also be circumvented by using NR. More research is, however, needed to experimentally demonstrate that NR indeed has the desired effect without the NA- and/or NAM-associated side effects.
Recently, we have shown that treatment of myotubes as well as whole muscle with 5-aminoimidazole-4-carboxamide-1-β-D-riboside, an activator of AMP-activated protein kinase (AMPK), resulted in an increase of NAD+ concentrations. This AMPK-induced increase in NAD+ was associated with an increase in SIRT1-mediated PGC-1α deacetylation and subsequent activation of oxidative mitochondrial activity (83). In line with these data, administration of 5-aminoimidazole-4-carboxamide-1-β-D-riboside, which works as an exercise mimetic, markedly improved endurance, making this pathway an attractive target for the treatment of metabolic disease (179). Further research should elucidate the respective contributions of PPARβ/δ and/or PGC-1α in this process.
Treatment of cells with 2-deoxyglucose has been shown to result in a decrease in the lactate:pyruvate ratio, indicative for an increased NAD+/NADH ratio (180). This increase was associated with SIRT1 activation, whereas other SIRTs were unaffected (180). In rats, treatment with low-dose 2-deoxyglucose resulted in physiological effects that mimic CR, although the effects seem less pronounced (181).
Finally, NAD+ levels could be elevated by selective inhibition of NAD+ consumers. For example, inhibition of the PARPs or CD38 could increase the NAD+ pool leading to the activation of other NAD+ consumers, such as the sirtuins. The competition for the same limiting pool of NAD+ might hence confer a novel mechanism to influence the activity of NAD+ consumers. The fact that one pathway of NAD+ consuming enzymes is blocked or inhibited could, however, also cause potential side effects, and therefore further studies are required to evaluate whether such a strategy may have a therapeutic value.
2. Decreasing NAD+
Selective lowering of NAD+ levels could be an efficient approach for the growth inhibition or elimination of undesired cells. For instance, inhibition of NAD+ biosynthesis could be applied in insecticides, microbial infections, and for cancer cells. The efficacy of inhibition of primary biosynthesis of NAD+ from tryptophan in insects has already been demonstrated before, although its use as an insecticide should be carefully evaluated because the inhibitors have a profound teratogenic effect in chick embryos (182) and could therefore severely impact on the environment. Interfering with NAD+ metabolism of infectious microorganisms has recently been reviewed (183). By determining the enzyme characteristics of the NAD+ biosynthesis enzymes in both the host and the microorganism, specific inhibitors could be developed that selectively target the pathogen. Decreasing NAD+ biosynthesis as a treatment for cancer has also already shown promising results (31). The small molecule FK866, which is a potent inhibitor of NAMPT, is already being tested in clinical cancer trials. FK866 was shown to inhibit NAMPT at low concentrations and cause cell death in cells that rely on high NAM to NAD+ turnover, as is the case for cancer cells (184). In several cell models, FK866 showed great promise by depleting NAD+ levels and thereby influencing not only tumorigenesis, but also metastasis and angiogenesis (31,184). The effects of NAMPT expression on oxidative stress response and PARP-mediated cell death could also seriously contribute to the beneficial effects of FK866 in anticancer treatment (37,146). Inhibition of NAD+ biosynthesis from tryptophan has also been implicated in cancer because inhibition of IDO by 1-methyltryptophan is suggested to prevent the escape of cancer cells from immunological clearance. This matter is still debated and is the subject of an excellent review (185).
Altogether, lowering NAD+ levels seems to be a relevant therapeutic approach to hamper metabolism of potentially harmful cells, such as the case for infections or cancer. Careful consideration, however, has to be made when it comes to such therapies because the inhibition might also affect the healthy tissues and/or cells, thereby causing severe adverse effects. Also, the fact that lowering NAD+ (as seen with FK866) as well as increasing NAD+ and subsequent SIRT1 activation could have antitumor effects clearly demonstrates that the mechanisms are complicated and that more research is warranted to clarify whether NAD+ metabolism is indeed an attractive target for drug design.
C. Endocrine regulation and NAD+
Physiological responses are modulated by multiple endocrine signaling pathways. As NAD+, endocrine pathways are affected by environmental cues and nutritional factors, suggesting that NAD+ might eventually mediate some endocrine responses and/or vice versa. The possible regulation of longevity by SIRT1 has spurred research into possible interactions of NAD+ signaling and endocrine pathways. Findings in several species have linked endocrine signaling to life span. For example, mutations in many genes of the insulin/IGF-I signaling pathway affect life span in lower eukaryotic organisms (186). Similarly, decreases in IGF-I and insulin signaling prolong life span in mice (187). Interestingly, NAD+-dependent enzymes such as sirtuins are known to influence insulin and IGF-I signaling. The activity of SIRT1 orthologs in different species influences the insulin/IGF signaling pathway at diverse steps (63,188). In addition, SIRT1 might participate in the regulation of insulin and IGF release (36,110). Of note, SIRT1 knockout mice are rarely born, but when they are, they overexpress IGF-binding protein-1, which decreases the free levels of IGF-I, rendering a dwarf phenotype. Remarkably, another sirtuin, SIRT6, may also determine serum IGF-I levels (124), although the molecular link between these observations is not clear. It must be mentioned that SIRT1 and SIRT6 knockout mice present multiple abnormalities, which can explain their reduced life span and why reduced IGF-I levels do not promote longevity in this model. In general, the effects of SIRT1 activation have a tendency to oppose those of IGF on longevity, although in the long term or in insulin-resistant states, they might both favor insulin sensitivity through repression of PTP1b (189). It is particularly relevant that sirtuin activity influences FOXO transcriptional activity, which is another evolutionary conserved player in the regulation of life span (190). Insulin/IGF-I negatively regulate FOXO activity through Akt-mediated phosphorylation. Deacetylation of FOXO by sirtuins seems to contribute to target-gene specification and may inhibit insulin-stimulated FOXO1 phosphorylation. Interestingly, insulin/IGF-I action generally increases glycolysis in cells, which should theoretically decrease the NAD+/NADH ratio and, therefore, shut down sirtuin activity. Consequently, it seems likely that SIRT1 and insulin/IGF-I actions negatively regulate each other. Other NAD+-consuming enzymes, such as those of the PARP family, have also been linked to insulin action and secretion. PARP1 activation in the pancreas leads to NAD+ depletion, which compromises ATP levels and insulin secretion (191). Remarkably, PARP1-deficient mice are resistant to streptozotocin-induced β-cell death and diabetes (192,193). Conversely, in peripheral tissues, insulin (194) and IGF-I (195) action inhibit PARP activity, probably as a way to preserve NAD+ levels. NAD+-consuming enzymes might also regulate the expression and release of other families of growth factors. Specific SIRT1 activating molecules promote a substantial increase in fibroblast growth factor (FGF) 21 expression in the liver (102), probably as a consequence of increased PPARα action (102,196). This observation was consolidated recently in a mouse model of mild SIRT1 overexpression (197). FGF21 regulation could as such contribute to the positive metabolic effects of SIRT1 activation (198). Of note, FGF21 and FGF23 action have already been linked to aging phenomena because the Klotho family of transmembrane “anti-aging” proteins was demonstrated to work as a crucial coreceptor for FGF-R signaling (199,200).
Recent findings evidence that endocrine communication and NAD+-consuming enzymes clearly expand beyond insulin/IGF-I and FGF signaling. For example, SIRT1 directly represses DNA binding and transactivation activity of the estrogen and androgen receptors through NAD+-dependent deacetylation (201,202), clearly suggesting that metabolism may directly modulate ligand-induced hormone signaling. Similarly, PARP1 can bind at the promoter of SMARCB1, a member of the SWI/SNF complex that modulates steroid sensitivity (203). In addition, PARP1 seems to participate in the regulation of estradiol target genes by forming part of a transcriptional “activation” complex together with TopoIIβ (204). Intracellular lipid signaling also seems to be closely related to NAD+ consuming proteins, through the modulation of PPARγ activity. SIRT1 negatively regulates PPARγ by docking with its cofactors NCoR and SMRT (108). Interestingly, another NAD+-consuming protein, PARP2, positively regulates PPARγ activity (205). These observations suggest a model where NAD+ consumption through competing enzymes (SIRT1 and PARP2) may provide opposite outcomes on commonly regulated targets. Together, these findings imply that we may be at the tip of the iceberg in our understanding of possible cross-regulation between NAD+-consuming enzymes and hormone action, providing potential molecular mechanisms on how metabolism or stress situations can modulate hormone sensitivity.
D. Type 2 diabetes (T2DM)
The increasing prevalence of obesity in Western societies is a major health problem, especially because of its association with T2DM and cardiovascular disease. An important role has emerged for the sirtuins in metabolic homeostasis. As discussed, especially the NAD+-consuming transcriptional cofactor SIRT1 has received major attention for its role in increasing mitochondrial activity, boosting exercise performance, and improving thermogenic activity, ultimately preventing obesity-induced diabetes. Increasing SIRT1 activity, by using pharmacological levels of resveratrol, prevented the development of diet-induced obesity and the concurrent metabolic disease in mice (101,206). Although these results look highly promising, the specificity of the compound as well as the required dose (400 mg/kg/d, i.e., 30 g/d for an average person of 75 kg) is precluding its widespread therapeutic use. Compounds such as SRT1720, which more specifically target SIRT1 and have the similar effect on T2DM at lower doses, might facilitate clinical development (102,207).
A role for NAMPT has recently been elucidated in the pancreatic β-cell (36). Because homozygosity of NAMPT mutations results in embryonic lethality, mice were studied that are heterozygous for NAMPT mutations (NAMPT+/−). Intracellular NAMPT protein levels were reduced in both female and male mice, whereas only females also displayed reduced eNAMPT content. This reduction in both NAMPT protein variants resulted in decreased NMN levels in plasma of female NAMPT+/− mice only, which in turn impaired glucose-stimulated insulin secretion. Importantly, administration of NMN rescued these defects (36), suggesting that the maintenance of NAD+ levels is crucial for pancreatic function. It will be interesting to clearly determine whether the rescue by NMN requires SIRT1. An independent line of research that indicated the importance of NAD+ metabolism in β-cells was based on the use of mouse genetic reference populations, where it was established that the NAM nucleotide transhydrogenase (Nnt) increases the susceptibility to T2DM by its role in insulin secretion in the β-cell (208,209,210). Whether polymorphisms in genes involved in NAD+ synthesis or salvage pathways that associate with T2DM exist in humans is yet unknown, but is worth studying.
Recently, the results were reported of the European Nicotinamide Diabetes Intervention Trial evaluating the long-term effects of NAM treatment on the development of type 1 diabetes mellitus (T1DM) (177). Based on the rationale that NAM can prevent β-cell damage and thereby T1DM, a randomized double-blind placebo-controlled trial was performed testing NAM treatment before the onset of the T1DM. Healthy volunteers that had a first-degree relative suffering from T1DM were included in the study, but unfortunately no difference was observed between the NAM-treated and placebo-treated groups in the development of T1DM after 5 yr of treatment (177).
E. Neurodegenerative disease
As early as 1850, Augustus Waller described the process of lesion-induced neuronal degeneration, known today as Wallerian degeneration. Wallerian degeneration typically occurs within 2 d after lesioning and involves highly programmed microtubular rearrangement, blebbing and fragmentation of the distal axon, followed by phagocytosis (211). Although not fully appreciated, Wallerian degeneration is thought to resemble neurodegenerative diseases and therefore is a relevant model for diseases, such as Alzheimer, Parkinson disease, and multiple sclerosis (212). Interestingly, a dominant C57BL/6 mouse mutant, termed WldS for Wallerian degeneration slow, has been described that shows delayed (2 wk) Wallerian degeneration. The protective gene in this mouse was found to consist of the N-terminal 70 amino acids of a ubiquitination assembly factor (Ube4B), the full coding sequence of NMNAT1, and a small linking region (213,214). Crossing the WldS mouse with mouse models for demyelinating motoneuron or Charcot-Marie-Tooth disease resulted in improved pathology of these mice (215,216). Also, in the central nervous system the WldS mouse was protected from pathological states, such as Parkinson disease (212,217). It is still not fully understood which part of the chimeric WldS protein conveys the protective effect to the phenotype of the WldS mouse. It has been hypothesized that increasing NAD+ concentration via NMNAT1 and the consequent SIRT1 activation might play a role (212,218,219). Although there is still some doubt about whether the positive effect on the Wallerian degeneration is mediated by NMNAT1 (220) or NAD+ levels (221), protection against axonopathy by expression of NMNAT1, NMNAT3, or NAMPT (in combination with its substrate NAM), or by the addition of the NAD+, NMN, NAMN, or NR in dorsal root ganglia undergoing transection (175,218) argue in support of the involvement of NAD. Finally, the aforementioned increase in expression of NAD+ biosynthesis genes, notably the 20-fold induction of Nrk2, suggests a neuroprotective role for NAD+ metabolism (175).
Another link between neurodegeneration and NAD+ metabolism lies in the fact that many neurodegenerative disorders, including Parkinson’s, Alzheimer’s, and Huntington’s disease, have been tightly linked to mitochondrial dysfunction, notably the generation of reactive oxygen species (222). Activation of SIRT1 in models for these diseases was shown to significantly improve the pathology (223,224). The effects are likely to be mediated by PGC-1α because PGC-1α-deficient mice suffer from neurodegenerative lesions, and PGC-1α was previously shown to be an important regulator of oxidative stress defense proteins (112,225). Although ultimate causal evidence that the protective effects of SIRT1 and PGC-1α are a direct consequence of NAD+ biosynthesis is lacking, all the evidence to date points toward the fact that increases in NAD+ and SIRT1 activity are correlative events unlikely to act independently. Further clarification of the involvement of the NAD+/SIRT1/PGC-1α pathway in neurodegeneration could pave the way for future therapeutic approaches to treat these devastating conditions.
F. Other pathophysiological states
Besides T2DM and neurodegenerative diseases, several other pathological states have been linked to NAD+ metabolism. Deficiency of the niacins has been shown to result in increased sensitivity to the mutagen ethylnitrosourea, which is used as a model for secondary effects of chemotherapy. Rats treated with ethylnitrosourea when fed with a niacin-deficient diet developed almost four times more malignancies, mostly leukemias, than rats fed a niacin-sufficient diet (226). This protective effect of niacin might be interesting for the prevention of secondary leukemia in patients receiving chemotherapy for another (primary) cancer.
Recently, several papers emerged on the role of SIRT1 in cancer. Although SIRT1 is involved in the deacetylation and activation of the proapoptotic p53 and FOXO (see Section V.B), some contradicting results have been published that implicate SIRT1 in a tumorigenic role, especially in a p53-deficient background, as is the case for most tumors (for excellent review specifically on this topic, we recommend Ref. 227).
Already in the 1950s, high doses of NA were given to lower cholesterol and triglyceride levels and to improve the high-density lipoprotein/low-density lipoprotein ratio (228). It has been demonstrated that the NA receptor GPR109A is essential for some of these beneficial effects of NA (229). It was recently hypothesized that NA treatment could at least in part improve lipid levels through the increase in NAD+ levels and concurrent SIRT1 activation (44). Although this could be an attractive model, no data are available to confirm/contradict this idea, such as studies with specific SIRT1 activators in a hypercholesterolemia background. Also, the dose required to achieve optimal results is quite high (>1 g/d), making it less likely that all hypolipidemic effects can be attributed to SIRT1 activation.
Recently, NAMPT has been linked to neutrophilic granulocyte survival and myeloid differentiation. During sepsis, NAMPT levels were shown to rise in combination with a delay in caspase-8- and caspase-3-mediated neutrophil apoptosis (230). Besides granulocyte survival, granulocyte differentiation is also influenced by NAMPT. The main mediator of myeloid differentiation is the cytokine granulocyte colony-stimulating factor (G-CSF), which is widely applied as a therapy in patients suffering from neutropenia. It was shown that expression of G-CSF and the G-CSF receptor was dependent on NAMPT in a SIRT1-dependent manner. Interestingly, the authors used NAM treatment (10–20 mg/kg/d for 1 wk) in control individuals and showed that granulocyte differentiation was increased and involved the NAD+/SIRT1/G-CSF pathway (231). Because no adverse effects were observed, although the treatment was short, NAM could be considered as a potential treatment for those suffering from neutropenia.
G. Longevity
As aging-associated health improves, it is not surprising that the NAD+ metabolic pathways also have been suggested to play an important role in longevity. Several enzymes involved in either NAD+ biosynthesis or consumption pathways have been associated with aging and associated health issues (Fig. 10B). Although some effects are until now only established in lower organisms, such as yeast, research is ongoing to extend this phenomenon to mammals because improving “healthspan” is an attractive target for pharmacological intervention, considering the aging population and the increase of aging-associated mortality and morbidity. It has been well established that one of the best strategies to increase life span in a range of species is CR (232). The concept that enhanced mitochondrial function upon CR is contributing to its beneficial effects on healthspan has recently been extended to humans, in which a general improvement in metabolism occurred during CR (233).
The beneficial effect of CR is in part mediated via SIRT1/Sir2. In organisms spanning from yeast over D. melanogaster, C. elegans, to mice, CR impacts on health in a SIRT1/Sir2-dependent fashion (61,62,63,64,232). Recently, it was also shown that activation of SIRT1 using resveratrol in mice fed a high-calorie diet improved metabolism (101,206) and significantly extended life span (234). Other sirtuins that have been reported to affect life span in mouse are SIRT6 (124,235) and SIRT7 (81,127). Studies on sirtuins and human health- and/or life span are limited. Nevertheless, SIRT3 gene polymorphisms were associated with increased life span in a small study in humans (236), whereas SIRT1 gene polymorphisms could affect healthspan through a stimulating effect on energy expenditure (101). Further human studies will certainly clarify the role of the sirtuins on human healthspan and life span in the not too distant future.
For other NAD+ consumers such as PARP or CD38, evidence pointing to a role in longevity is lacking. In yeast, however, two other NAD-related processes were implicated in life span extension. Overexpression of two mitochondrial NADH dehydrogenases, Nde1 and Nde2, resulted in an increased NAD+/NADH ratio, thereby activating Sir2 and in turn increasing life span (87). In a similar mechanism, overexpression of the malate-aspartate shuttle components resulted in a Sir2-mediated extension of life span, whereas deletion of multiple shuttle components resulted in a deficiency of CR-mediated life span extension (94).
Other factors that could have a major contribution to healthspan include NAD+ biosynthesis enzymes. As can be anticipated, SIRT1 can only exert its beneficial function in the presence of sufficient amounts of NAD+. This has been illustrated by the fact that the Npt1 yeast mutant, lacking NAPT enzymatic function and hence the most important part of total NAD+ biosynthesis, does not extend life span by CR (237). Within the NAD+ biosynthesis pathway NAMPT has been described to affect the aging process. Reduced NAMPT expression, as was also observed in aging cells, resulted in premature senescence of human smooth muscle cells, whereas overexpression of NAMPT delayed senescence in combination with enhanced oxidative stress defense (37). Although the mechanism is different, yeast mutant lacking the nicotinamidase Pnc1, which is essential for NAD+ biosynthesis from NAM in bacteria and yeast, also showed a reduced life span, most likely due to sirtuin inhibition by NAM, because artificial depletion of NAM in the Pnc1 mutant restored life span extension by CR (90). Finally, it was very recently demonstrated that the NR pathway is indispensable for the life span extension effect of CR in yeast (238).
Altogether, it can be concluded that, although not so much clear-cut evidence is known to date on the exact mechanism of life span extension in mammals, the NAD+ metabolic pathway seems likely involved. Further research in this direction should clarify the role of NAD+ and define whether it will be possible to increase life span and healthspan by targeting NAD+.
X. Conclusions and Future Perspectives
During the last few years, it has become clear that the cellular role of NAD+ expands far beyond its classic participation as a cofactor in reductive biosynthesis and oxidative breakdown. NAD+ has emerged as a signaling molecule whose levels critically depend on the metabolic status and cellular stress derived from toxicity. In this review we have summarized several interesting molecules that, through NAD+ breakdown, can heavily impact on metabolism, cellular viability, and signaling pathways. Interestingly, only one family of them, sirtuins, requires elevated NAD+ levels to enhance their activity, making them suitable to act as metabolic NAD+ sensors. Other families of NAD+ consumers show higher Km for NAD+, and therefore, their activity may not be limited in the standard situations, when NAD+ is abundant in the cell, approximately 10-fold higher than the Km (Table 1). As discussed in Section VI.A, however, the action of these other NAD+ consumers may critically impact on NAD+ levels, therefore determining whether they reach a permissive threshold for sirtuin activation. It is interesting to note that sirtuins have virtually opposed functions compared with other NAD+ consumers. For example, PARP activity has been linked to chromatin relaxation to facilitate DNA repair and gene expression, whereas sirtuin action on histones is normally associated to gene silencing. The massive NAD+ consumption by members of the PARP or CD38 family explains why a reduction in their activities can potentially be linked to sirtuin activation as a consequence of increased intracellular NAD+ content. The variety and different localization of sirtuins, however, should throw some caution on generalizations about their functions. As discussed in Section V.A, it seems very unlikely that the fine regulation of cellular and physiological adaptations could rely on a system where all sirtuins are activated simultaneously. In fact, some sirtuins seem to have opposing action. Therefore, additional regulatory mechanisms might be affecting sirtuin activity. Similarly, it has to be pointed out that our current knowledge on the compartmentalization of NAD+ and derived metabolites is still very poor, and recent technical advances may help to bring light to our understanding on how the intracellular specifics of NAD+ homeostasis could lead to differential and specific sirtuin activation. Because NAD+ metabolism holds such an important position in total cellular metabolism, both as a cofactor for individual enzymes and as a substrate for NAD+ consuming proteins, such as sirtuins, it could pose an attractive target for the treatment of various pathologies. Especially in the field of sirtuin biology, linked to the prevention of aging and its related diseases like obesity, T2DM, and cancer, NAD+ is being studied as a modulator of whole metabolic networks, which in turn could greatly influence the metabolic state of the cell. The recent development of compounds targeted on the biosynthesis and salvage of NAD+, for instance FK866, which is already being tested in clinical trials, holds promise for treating these diseases. More research, however, will be needed to understand the full extent of the consequences, also on the long term, of interfering in these crucial NAD+ metabolic pathways and to comprehend the possible beneficial effects of this on healthspan.
Footnotes
R.H.H. is supported by a Rubicon fellowship of the Netherlands Organization for Scientific Research. C.C. is supported by an European Molecular Biology Organization long-term fellowship. The work in the laboratory of the authors is supported by grants of the Ecole Polytechnique Fédérale de Lausanne, National Institutes of Health (Grant DK59820), the EU Ideas programme (Sirtuins; ERC-2008-AdG-23118), and the Swiss National Science Foundation (Grant SNF 31003A-124713).
Disclosure Summary: The authors have nothing to declare.
First Published Online December 9, 2009
Abbreviations: ACMS, α-Amino-β-carboxymuconate-ε-semialdehyde; AMPK, AMP-activated protein kinase; cADP, cyclic ADP; CoA, coenzyme A; CPS-1, carbamoyl phosphate synthetase 1; CR, calorie restriction; eNAMPT, extracellular NAMPT; FGF, fibroblast growth factor; G-CSF, granulocyte colony-stimulating factor; GDH, glutamate dehydrogenase; IDO, indoleamine 2,3-dioxygenase; iNAMPT, intracellular NAMPT; NA, nicotinic acid; NAAD, NA adenine dinucleotide; NAD, NAM adenine dinucleotide; NADP, NAD phosphate; NAM, nicotinamide; NAMN, NA mononucleotide; NAMPT, NAM phosphoribosyltransferase; NAPT, NA phosphoribosyltransferase; NMN, NAM mononucleotide; NMNAT, NMN adenylyltransferase; NR, NAM riboside; NRK, NR kinase; Nrt1, NR transporter; PARP, poly(ADP-ribose) polymerase; PGC, PPAR γ coactivator; PPAR, peroxisome proliferator-activated receptor; QPRT, quinolinate phosphoribosyltransferase; rDNA, ribosomal DNA; Sir2, silent information regulator 2; SIRT, sirtuin; T1DM, type 1 diabetes mellitus; T2DM, type 2 diabetes mellitus; TDO, tryptophan 2,3-dioxygenase.
References
- Berger F, Ramírez-Hernández MH, Ziegler M 2004 The new life of a centenarian: signalling functions of NAD(P). Trends Biochem Sci 29:111–118 [DOI] [PubMed] [Google Scholar]
- Bender DA 1983 Biochemistry of tryptophan in health and disease. Mol Aspects Med 6:101–197 [DOI] [PubMed] [Google Scholar]
- Hegyi J, Schwartz RA, Hegyi V 2004 Pellagra: dermatitis, dementia, and diarrhea. Int J Dermatol 43:1–5 [DOI] [PubMed] [Google Scholar]
- Elvehjem C, Madden R, Strong F, Woolley D 1937 Relation of nicotinic acid and nicotinic acid amide to canine black tongue. J Am Chem Soc 59:1767–1768 [Google Scholar]
- Yamazaki F, Kuroiwa T, Takikawa O, Kido R 1985 Human indolylamine 2,3-dioxygenase. Its tissue distribution, and characterization of the placental enzyme. Biochem J 230:635–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo Y, Boyd CA 2000 Human placental indoleamine 2,3-dioxygenase: cellular localization and characterization of an enzyme preventing fetal rejection. Biochim Biophys Acta 1500:119–124 [DOI] [PubMed] [Google Scholar]
- Rongvaux A, Andris F, Van Gool F, Leo O 2003 Reconstructing eukaryotic NAD metabolism. Bioessays 25:683–690 [DOI] [PubMed] [Google Scholar]
- Magni G, Amici A, Emanuelli M, Orsomando G, Raffaelli N, Ruggieri S 2004 Enzymology of NAD+ homeostasis in man. Cell Mol Life Sci 61:19–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldblum S, Rougier A, Loiseau H, Loiseau P, Cohadon F, Morselli PL, Lloyd KG 1988 Quinolinic-phosphoribosyl transferase activity is decreased in epileptic human brain tissue. Epilepsia 29:523–529 [DOI] [PubMed] [Google Scholar]
- Foster AC, Whetsell Jr WO, Bird ED, Schwarcz R 1985 Quinolinic acid phosphoribosyl transferase in human and rat brain: activity in Huntington’s disease and in quinolinate-lesioned rat striatum. Brain Res 336:207–214 [DOI] [PubMed] [Google Scholar]
- Du F, Okuno E, Whetsell Jr WO, Köhler C, Schwarcz R 1990 Distribution of quinolinic acid phosphoribosyltransferase in the human hippocampal formation and parahippocampal gyrus. J Comp Neurol 295:71–82 [DOI] [PubMed] [Google Scholar]
- Heyes MP, Saito K, Crowley JS, Davis LE, Demitrack MA, Der M, Dilling LA, Elia J, Kruesi MJ, Lackner A, Larsen SA, Lee K, Leonard HL, Markey SP, Martin A, Milstein S, Mouradian MM, Pranzatelli MR, Quearry BJ, Salazar A, Smith M, Strauss SE, Sunderland T, Swedo SW, Tourtellotte WW 1992 Quinolinic acid and kynurenine pathway metabolism in inflammatory and non-inflammatory neurological disease. Brain 115:1249–1273 [DOI] [PubMed] [Google Scholar]
- Okuno E, White RJ, Schwarcz R 1988 Quinolinic acid phosphoribosyltransferase: purification and partial characterization from human liver and brain. J Biochem 103:1054–1059 [DOI] [PubMed] [Google Scholar]
- Stone TW, Perkins MN 1981 Quinolinic acid: a potent endogenous excitant at amino acid receptors in CNS. Eur J Pharmacol 72:411–412 [DOI] [PubMed] [Google Scholar]
- Kornberg A 1948 The participation of inorganic pyrophosphate in the reversible enzymatic synthesis of diphosphopyridine nucleotide. J Biol Chem 176:1475 [PubMed] [Google Scholar]
- Raffaelli N, Sorci L, Amici A, Emanuelli M, Mazzola F, Magni G 2002 Identification of a novel human nicotinamide mononucleotide adenylyltransferase. Biochem Biophys Res Commun 297:835–840 [DOI] [PubMed] [Google Scholar]
- Zhang X, Kurnasov OV, Karthikeyan S, Grishin NV, Osterman AL, Zhang H 2003 Structural characterization of a human cytosolic NMN/NaMN adenylyltransferase and implication in human NAD biosynthesis. J Biol Chem 278:13503–13511 [DOI] [PubMed] [Google Scholar]
- Emanuelli M, Carnevali F, Saccucci F, Pierella F, Amici A, Raffaelli N, Magni G 2001 Molecular cloning, chromosomal localization, tissue mRNA levels, bacterial expression, and enzymatic properties of human NMN adenylyltransferase. J Biol Chem 276:406–412 [DOI] [PubMed] [Google Scholar]
- Fernando FS, Conforti L, Tosi S, Smith AD, Coleman MP 2002 Human homologue of a gene mutated in the slow Wallerian degeneration (C57BL/Wld(s)) mouse. Gene 284:23–29 [DOI] [PubMed] [Google Scholar]
- Sood R, Bonner TI, Makalowska I, Stephan DA, Robbins CM, Connors TD, Morgenbesser SD, Su K, Faruque MU, Pinkett H, Graham C, Baxevanis AD, Klinger KW, Landes GM, Trent JM, Carpten JD 2001 Cloning and characterization of 13 novel transcripts and the human RGS8 gene from the 1q25 region encompassing the hereditary prostate cancer (HPC1) locus. Genomics 73:211–222 [DOI] [PubMed] [Google Scholar]
- Berger F, Lau C, Dahlmann M, Ziegler M 2005 Subcellular compartmentation and differential catalytic properties of the three human nicotinamide mononucleotide adenylyltransferase isoforms. J Biol Chem 280:36334–36341 [DOI] [PubMed] [Google Scholar]
- Hara N, Yamada K, Terashima M, Osago H, Shimoyama M, Tsuchiya M 2003 Molecular identification of human glutamine- and ammonia-dependent NAD synthetases. Carbon-nitrogen hydrolase domain confers glutamine dependency. J Biol Chem 278:10914–10921 [DOI] [PubMed] [Google Scholar]
- Bieganowski P, Brenner C 2003 The reported human NADsyn2 is ammonia-dependent NAD synthetase from a pseudomonad. J Biol Chem 278:33056–33059 [DOI] [PubMed] [Google Scholar]
- Jacobson EL, Dame AJ, Pyrek JS, Jacobson MK 1995 Evaluating the role of niacin in human carcinogenesis. Biochimie 77:394–398 [DOI] [PubMed] [Google Scholar]
- Collins PB, Chaykin S 1972 The management of nicotinamide and nicotinic acid in the mouse. J Biol Chem 247:778–783 [PubMed] [Google Scholar]
- Preiss J, Handler P 1958 Biosynthesis of diphosphopyridine nucleotide. I. Identification of intermediates. J Biol Chem 233:488–492 [PubMed] [Google Scholar]
- Hara N, Yamada K, Shibata T, Osago H, Hashimoto T, Tsuchiya M 2007 Elevation of cellular NAD levels by nicotinic acid and involvement of nicotinic acid phosphoribosyltransferase in human cells. J Biol Chem 282:24574–24582 [DOI] [PubMed] [Google Scholar]
- Frothingham R, Meeker-O'Connell WA, Talbot EA, George JW, Kreuzer KN 1996 Identification, cloning, and expression of the Escherichia coli pyrazinamidase and nicotinamidase gene, pncA. Antimicrob Agents Chemother 40:1426–1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghislain M, Talla E, François JM 2002 Identification and functional analysis of the Saccharomyces cerevisiae nicotinamidase gene, PNC1. Yeast 19:215–224 [DOI] [PubMed] [Google Scholar]
- Joshi JG, Handler P 1962 Purification and properties of nicotinamidase from Torula cremoris. J Biol Chem 237:929–935 [PubMed] [Google Scholar]
- Garten A, Petzold S, Körner A, Imai S, Kiess W 2009 Nampt: linking NAD biology, metabolism and cancer. Trends Endocrinol Metab 20:130–138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samal B, Sun Y, Stearns G, Xie C, Suggs S, McNiece I 1994 Cloning and characterization of the cDNA encoding a novel human pre-B-cell colony-enhancing factor. Mol Cell Biol 14:1431–1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Zhang Y, Dorweiler B, Cui D, Wang T, Woo CW, Brunkan CS, Wolberger C, Imai S, Tabas I 2008 Extracellular Nampt promotes macrophage survival via a nonenzymatic interleukin-6/STAT3 signaling mechanism. J Biol Chem 283:34833–34843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuhara A, Matsuda M, Nishizawa M, Segawa K, Tanaka M, Kishimoto K, Matsuki Y, Murakami M, Ichisaka T, Murakami H, Watanabe E, Takagi T, Akiyoshi M, Ohtsubo T, Kihara S, Yamashita S, Makishima M, Funahashi T, Yamanaka S, Hiramatsu R, Matsuzawa Y, Shimomura I 2005 Visfatin: a protein secreted by visceral fat that mimics the effects of insulin. Science 307:426–430 [DOI] [PubMed] [Google Scholar]
- Fukuhara A, Matsuda M, Nishizawa M, Segawa K, Tanaka M, Kishimoto K, Matsuki Y, Murakami M, Ichisaka T, Murakami H, Watanabe E, Takagi T, Akiyoshi M, Ohtsubo T, Kihara S, Yamashita S, Makishima M, Funahashi T, Yamanaka S, Hiramatsu R, Matsuzawa Y, Shimomura I 2007 Retraction. Science 318:565 [DOI] [PubMed] [Google Scholar]
- Revollo JR, Körner A, Mills KF, Satoh A, Wang T, Garten A, Dasgupta B, Sasaki Y, Wolberger C, Townsend RR, Milbrandt J, Kiess W, Imai S 2007 Nampt/PBEF/Visfatin regulates insulin secretion in β cells as a systemic NAD biosynthetic enzyme. Cell Metab 6:363–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Veer E, Ho C, O'Neil C, Barbosa N, Scott R, Cregan SP, Pickering JG 2007 Extension of human cell lifespan by nicotinamide phosphoribosyltransferase. J Biol Chem 282:10841–10845 [DOI] [PubMed] [Google Scholar]
- Nakahata Y, Sahar S, Astarita G, Kaluzova M, Sassone-Corsi P 2009 Circadian control of the NAD+ salvage pathway by CLOCK-SIRT1. Science 324:654–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey KM, Yoshino J, Brace CS, Abrassart D, Kobayashi Y, Marcheva B, Hong HK, Chong JL, Buhr ED, Lee C, Takahashi JS, Imai S, Bass J 2009 Circadian clock feedback cycle through NAMPT-mediated NAD+ biosynthesis. Science 324:651–654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, Guarente LP, Sassone-Corsi P 2008 The NAD+-dependent deacetylase SIRT1 modulates CLOCK-mediated chromatin remodeling and circadian control. Cell 134:329–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asher G, Gatfield D, Stratmann M, Reinke H, Dibner C, Kreppel F, Mostoslavsky R, Alt FW, Schibler U 2008 SIRT1 regulates circadian clock gene expression through PER2 deacetylation. Cell 134:317–328 [DOI] [PubMed] [Google Scholar]
- Schweiger M, Hennig K, Lerner F, Niere M, Hirsch- Kauffmann M, Specht T, Weise C, Oei SL, Ziegler M 2001 Characterization of recombinant human nicotinamide mononucleotide adenylyl transferase (NMNAT), a nuclear enzyme essential for NAD synthesis. FEBS Lett 492:95–100 [DOI] [PubMed] [Google Scholar]
- Bieganowski P, Brenner C 2004 Discoveries of nicotinamide riboside as a nutrient and conserved NRK genes establish a Preiss-Handler independent route to NAD+ in fungi and humans. Cell 117:495–502 [DOI] [PubMed] [Google Scholar]
- Bogan KL, Brenner C 2008 Nicotinic acid, nicotinamide, and nicotinamide riboside: a molecular evaluation of NAD+ precursor vitamins in human nutrition. Annu Rev Nutr 28:115–130 [DOI] [PubMed] [Google Scholar]
- Bender DA, Magboul BI, Wynick D 1982 Probable mechanisms of regulation of the utilization of dietary tryptophan, nicotinamide and nicotinic acid as precursors of nicotinamide nucleotides in the rat. Br J Nutr 48:119–127 [DOI] [PubMed] [Google Scholar]
- Bender DA, Olufunwa R 1988 Utilization of tryptophan, nicotinamide and nicotinic acid as precursors for nicotinamide nucleotide synthesis in isolated rat liver cells. Br J Nutr 59:279–287 [DOI] [PubMed] [Google Scholar]
- Belenky PA, Moga TG, Brenner C 2008 Saccharomyces cerevisiae YOR071C encodes the high affinity nicotinamide riboside transporter Nrt1. J Biol Chem 283:8075–8079 [DOI] [PubMed] [Google Scholar]
- Damaraju VL, Visser F, Zhang J, Mowles D, Ng AM, Young JD, Jayaram HN, Cass CE 2005 Role of human nucleoside transporters in the cellular uptake of two inhibitors of IMP dehydrogenase, tiazofurin and benzamide riboside. Mol Pharmacol 67:273–279 [DOI] [PubMed] [Google Scholar]
- Warburg O, Christian W 1936 Pyridin, der wasserstoffübertragende Bestandteil von Gärungsfermenten. Helvetica Chimica Acta 19:E79–E88 [Google Scholar]
- Warburg O, Christian W, Griese A 1935 Wasserstoffübertragendes Co-Ferment, seine Zusammensetzung und Wirkungsweise. Biochem Z 282:157–165 [Google Scholar]
- Klingenberg M, Buecher T 1960 Biological oxidations. Annu Rev Biochem 29:669–708 [DOI] [PubMed] [Google Scholar]
- Zuurendonk PF, Akerboom TPM, Tager JM 1976 Metabolite distribution in isolated rat-liver cells and equilibrium relationships of mitochondrial and cytosolic dehydrogenases. In: Tager JM, Soling HD, Williamson JR, eds. Use of isolated liver cells and kidney tubulus in metabolic studies. New York: American Elsevier Publishing Company; 17–27 [Google Scholar]
- Hoek JB, Rydström J 1988 Physiological roles of nicotinamide nucleotide transhydrogenase. Biochem J 254:1–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rydström J 2006 Mitochondrial NADPH, transhydrogenase and disease. Biochim Biophys Acta 1757:721–726 [DOI] [PubMed] [Google Scholar]
- Di Lisa F, Ziegler M 2001 Pathophysiological relevance of mitochondria in NAD(+) metabolism. FEBS Lett 492:4–8 [DOI] [PubMed] [Google Scholar]
- Tischler ME, Friedrichs D, Coll K, Williamson JR 1977 Pyridine nucleotide distributions and enzyme mass action ratios in hepatocytes from fed and starved rats. Arch Biochem Biophys 184:222–236 [DOI] [PubMed] [Google Scholar]
- Revollo JR, Grimm AA, Imai S 2004 The NAD biosynthesis pathway mediated by nicotinamide phosphoribosyltransferase regulates Sir2 activity in mammalian cells. J Biol Chem 279:50754–50763 [DOI] [PubMed] [Google Scholar]
- Sauve AA, Wolberger C, Schramm VL, Boeke JD 2006 The biochemistry of sirtuins. Annu Rev Biochem 75:435–465 [DOI] [PubMed] [Google Scholar]
- Kaeberlein M, McVey M, Guarente L 1999 The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev 13:2570–2580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L 2000 Sir2 links chromatin silencing, metabolism, and aging. Genes Dev 14:1021–1026 [PubMed] [Google Scholar]
- Howitz KT, Bitterman KJ, Cohen HY, Lamming DW, Lavu S, Wood JG, Zipkin RE, Chung P, Kisielewski A, Zhang LL, Scherer B, Sinclair DA 2003 Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature 425:191–196 [DOI] [PubMed] [Google Scholar]
- Lin SJ, Kaeberlein M, Andalis AA, Sturtz LA, Defossez PA, Culotta VC, Fink GR, Guarente L 2002 Calorie restriction extends Saccharomyces cerevisiae lifespan by increasing respiration. Nature 418:344–348 [DOI] [PubMed] [Google Scholar]
- Tissenbaum HA, Guarente L 2001 Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature 410:227–230 [DOI] [PubMed] [Google Scholar]
- Rogina B, Helfand SL 2004 Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc Natl Acad Sci USA 101:15998–16003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dali-Youcef N, Lagouge M, Froelich S, Koehl C, Schoonjans K, Auwerx J 2007 Sirtuins: the ‘magnificent seven’, function, metabolism and longevity. Ann Med 39:335–345 [DOI] [PubMed] [Google Scholar]
- Michan S, Sinclair D 2007 Sirtuins in mammals: insights into their biological function. Biochem J 404:1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michishita E, Park JY, Burneskis JM, Barrett JC, Horikawa I 2005 Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol Biol Cell 16:4623–4635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jing E, Gesta S, Kahn CR 2007 SIRT2 regulates adipocyte differentiation through FoxO1 acetylation/deacetylation. Cell Metab 6:105–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquero A, Scher MB, Lee DH, Sutton A, Cheng HL, Alt FW, Serrano L, Sternglanz R, Reinberg D 2006 SirT2 is a histone deacetylase with preference for histone H4 Lys 16 during mitosis. Genes Dev 20:1256–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onyango P, Celic I, McCaffery JM, Boeke JD, Feinberg AP 2002 SIRT3, a human SIR2 homologue, is an NAD-dependent deacetylase localized to mitochondria. Proc Natl Acad Sci USA 99:13653–13658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B, North BJ, Frye RA, Ott M, Verdin E 2002 The human silent information regulator (Sir)2 homologue hSIRT3 is a mitochondrial nicotinamide adenine dinucleotide-dependent deacetylase. J Cell Biol 158:647–657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanno M, Sakamoto J, Miura T, Shimamoto K, Horio Y 2007 Nucleocytoplasmic shuttling of the NAD+-dependent histone deacetylase SIRT1. J Biol Chem 282:6823–6832 [DOI] [PubMed] [Google Scholar]
- Hallows WC, Lee S, Denu JM 2006 Sirtuins deacetylate and activate mammalian acetyl-CoA synthetases. Proc Natl Acad Sci USA 103:10230–10235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scher MB, Vaquero A, Reinberg D 2007 SirT3 is a nuclear NAD+-dependent histone deacetylase that translocates to the mitochondria upon cellular stress. Genes Dev 21:920–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallows WC, Albaugh BN, Denu JM 2008 Where in the cell is SIRT3?—functional localization of an NAD+-dependent protein deacetylase. Biochem J 411:e11–e13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- North BJ, Marshall BL, Borra MT, Denu JM, Verdin E 2003 The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol Cell 11:437–444 [DOI] [PubMed] [Google Scholar]
- Vaziri H, Dessain SK, Ng Eaton E, Imai SI, Frye RA, Pandita TK, Guarente L, Weinberg RA 2001 hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell 107:149–159 [DOI] [PubMed] [Google Scholar]
- Haigis MC, Mostoslavsky R, Haigis KM, Fahie K, Christodoulou DC, Murphy AJ, Valenzuela DM, Yancopoulos GD, Karow M, Blander G, Wolberger C, Prolla TA, Weindruch R, Alt FW, Guarente L 2006 SIRT4 inhibits glutamate dehydrogenase and opposes the effects of calorie restriction in pancreatic β cells. Cell 126:941–954 [DOI] [PubMed] [Google Scholar]
- Liszt G, Ford E, Kurtev M, Guarente L 2005 Mouse Sir2 homolog SIRT6 is a nuclear ADP-ribosyltransferase. J Biol Chem 280:21313–21320 [DOI] [PubMed] [Google Scholar]
- Shi T, Wang F, Stieren E, Tong Q 2005 SIRT3, a mitochondrial sirtuin deacetylase, regulates mitochondrial function and thermogenesis in brown adipocytes. J Biol Chem 280:13560–13567 [DOI] [PubMed] [Google Scholar]
- Vakhrusheva O, Smolka C, Gajawada P, Kostin S, Boettger T, Kubin T, Braun T, Bober E 2008 Sirt7 increases stress resistance of cardiomyocytes and prevents apoptosis and inflammatory cardiomyopathy in mice. Circ Res 102:703–710 [DOI] [PubMed] [Google Scholar]
- Rodgers JT, Lerin C, Haas W, Gygi SP, Spiegelman BM, Puigserver P 2005 Nutrient control of glucose homeostasis through a complex of PGC-1α and SIRT1. Nature 434:113–118 [DOI] [PubMed] [Google Scholar]
- Cantó C, Gerhart-Hines Z, Feige JN, Lagouge M, Noriega L, Milne JC, Elliott PJ, Puigserver P, Auwerx J 2009 AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature 458:1056–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Bruno J, Easlon E, Lin SJ, Cheng HL, Alt FW, Guarente L 2008 Tissue-specific regulation of SIRT1 by calorie restriction. Genes Dev 22:1753–1757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulco M, Cen Y, Zhao P, Hoffman EP, McBurney MW, Sauve AA, Sartorelli V 2008 Glucose restriction inhibits skeletal myoblast differentiation by activating SIRT1 through AMPK-mediated regulation of Nampt. Dev Cell 14:661–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood JG, Rogina B, Lavu S, Howitz K, Helfand SL, Tatar M, Sinclair D 2004 Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature 430:686–689 [DOI] [PubMed] [Google Scholar]
- Lin SJ, Ford E, Haigis M, Liszt G, Guarente L 2004 Calorie restriction extends yeast life span by lowering the level of NADH. Genes Dev 18:12–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt MT, Smith BC, Jackson MD, Denu JM 2004 Coenzyme specificity of Sir2 protein deacetylases: implications for physiological regulation. J Biol Chem 279:40122–40129 [DOI] [PubMed] [Google Scholar]
- Yang T, Sauve AA 2006 NAD metabolism and sirtuins: metabolic regulation of protein deacetylation in stress and toxicity. AAPS J 8:E632–E643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson RM, Bitterman KJ, Wood JG, Medvedik O, Sinclair DA 2003 Nicotinamide and PNC1 govern lifespan extension by calorie restriction in Saccharomyces cerevisiae. Nature 423:181–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitterman KJ, Anderson RM, Cohen HY, Latorre-Esteves M, Sinclair DA 2002 Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast sir2 and human SIRT1. J Biol Chem 277:45099–45107 [DOI] [PubMed] [Google Scholar]
- van Roermund CW, Elgersma Y, Singh N, Wanders RJ, Tabak HF 1995 The membrane of peroxisomes in Saccharomyces cerevisiae is impermeable to NAD(H) and acetyl-CoA under in vivo conditions. EMBO J 14:3480–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker BM, Overkamp KM, van Maris AJ, Kötter P, Luttik MA, van Dijken JP, Pronk JT 2001 Stoichiometry and compartmentation of NADH metabolism in Saccharomyces cerevisiae. FEMS Microbiol Rev 25:15–37 [DOI] [PubMed] [Google Scholar]
- Easlon E, Tsang F, Skinner C, Wang C, Lin SJ 2008 The malate-aspartate NADH shuttle components are novel metabolic longevity regulators required for calorie restriction-mediated life span extension in yeast. Genes Dev 22:931–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Yang T, Baur JA, Perez E, Matsui T, Carmona JJ, Lamming DW, Souza-Pinto NC, Bohr VA, Rosenzweig A, de Cabo R, Sauve AA, Sinclair DA 2007 Nutrient-sensitive mitochondrial NAD+ levels dictate cell survival. Cell 130:1095–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerhart-Hines Z, Rodgers JT, Bare O, Lerin C, Kim SH, Mostoslavsky R, Alt FW, Wu Z, Puigserver P 2007 Metabolic control of muscle mitochondrial function and fatty acid oxidation through SIRT1/PGC-1α. EMBO J 26:1913–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feige JN, Auwerx J 2008 Transcriptional targets of sirtuins in the coordination of mammalian physiology. Curr Opin Cell Biol 20:303–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, Hu LS, Cheng HL, Jedrychowski MP, Gygi SP, Sinclair DA, Alt FW, Greenberg ME 2004 Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science 303:2011–2015 [DOI] [PubMed] [Google Scholar]
- Kobayashi Y, Furukawa-Hibi Y, Chen C, Horio Y, Isobe K, Ikeda K, Motoyama N 2005 SIRT1 is critical regulator of FOXO-mediated transcription in response to oxidative stress. Int J Mol Med 16:237–243 [PubMed] [Google Scholar]
- Lerin C, Rodgers JT, Kalume DE, Kim SH, Pandey A, Puigserver P 2006 GCN5 acetyltransferase complex controls glucose metabolism through transcriptional repression of PGC-1α. Cell Metab 3:429–438 [DOI] [PubMed] [Google Scholar]
- Lagouge M, Argmann C, Gerhart-Hines Z, Meziane H, Lerin C, Daussin F, Messadeq N, Milne J, Lambert P, Elliott P, Geny B, Laakso M, Puigserver P, Auwerx J 2006 Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1α. Cell 127:1109–1122 [DOI] [PubMed] [Google Scholar]
- Feige JN, Lagouge M, Canto C, Strehle A, Houten SM, Milne JC, Lambert PD, Mataki C, Elliott PJ, Auwerx J 2008 Specific SIRT1 activation mimics low energy levels and protects against diet-induced metabolic disorders by enhancing fat oxidation. Cell Metab 8:347–358 [DOI] [PubMed] [Google Scholar]
- Rodgers JT, Puigserver P 2007 Fasting-dependent glucose and lipid metabolic response through hepatic sirtuin 1. Proc Natl Acad Sci USA 104:12861–12866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon JC, Puigserver P, Chen G, Donovan J, Wu Z, Rhee J, Adelmant G, Stafford J, Kahn CR, Granner DK, Newgard CB, Spiegelman BM 2001 Control of hepatic gluconeogenesis through the transcriptional coactivator PGC-1. Nature 413:131–138 [DOI] [PubMed] [Google Scholar]
- Herzig S, Long F, Jhala US, Hedrick S, Quinn R, Bauer A, Rudolph D, Schutz G, Yoon C, Puigserver P, Spiegelman B, Montminy M 2001 CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature 413:179–183 [DOI] [PubMed] [Google Scholar]
- Puigserver P, Rhee J, Donovan J, Walkey CJ, Yoon JC, Oriente F, Kitamura Y, Altomonte J, Dong H, Accili D, Spiegelman BM 2003 Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1α interaction. Nature 423:550–555 [DOI] [PubMed] [Google Scholar]
- Suchankova G, Nelson LE, Gerhart-Hines Z, Kelly M, Gauthier MS, Saha AK, Ido Y, Puigserver P, Ruderman NB 2009 Concurrent regulation of AMP-activated protein kinase and SIRT1 in mammalian cells. Biochem Biophys Res Commun 378:836–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picard F, Kurtev M, Chung N, Topark-Ngarm A, Senawong T, Machado De Oliveira R, Leid M, McBurney MW, Guarente L 2004 Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-γ. Nature 429:771–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moynihan KA, Grimm AA, Plueger MM, Bernal-Mizrachi E, Ford E, Cras-Méneur C, Permutt MA, Imai S 2005 Increased dosage of mammalian Sir2 in pancreatic β cells enhances glucose-stimulated insulin secretion in mice. Cell Metab 2:105–117 [DOI] [PubMed] [Google Scholar]
- Bordone L, Motta MC, Picard F, Robinson A, Jhala US, Apfeld J, McDonagh T, Lemieux M, McBurney M, Szilvasi A, Easlon EJ, Lin SJ, Guarente L 2006 Sirt1 regulates insulin secretion by repressing UCP2 in pancreatic β cells. PLoS Biol 4:e31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura YI, Kitamura T, Kruse JP, Raum JC, Stein R, Gu W, Accili D 2005 FoxO1 protects against pancreatic β cell failure through NeuroD and MafA induction. Cell Metab 2:153–163 [DOI] [PubMed] [Google Scholar]
- St-Pierre J, Drori S, Uldry M, Silvaggi JM, Rhee J, Jäger S, Handschin C, Zheng K, Lin J, Yang W, Simon DK, Bachoo R, Spiegelman BM 2006 Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell 127:397–408 [DOI] [PubMed] [Google Scholar]
- Hiratsuka M, Inoue T, Toda T, Kimura N, Shirayoshi Y, Kamitani H, Watanabe T, Ohama E, Tahimic CG, Kurimasa A, Oshimura M 2003 Proteomics-based identification of differentially expressed genes in human gliomas: down-regulation of SIRT2 gene. Biochem Biophys Res Commun 309:558–566 [DOI] [PubMed] [Google Scholar]
- Dryden SC, Nahhas FA, Nowak JE, Goustin AS, Tainsky MA 2003 Role for human SIRT2 NAD-dependent deacetylase activity in control of mitotic exit in the cell cycle. Mol Cell Biol 23:3173–3185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T, Hiratsuka M, Osaki M, Yamada H, Kishimoto I, Yamaguchi S, Nakano S, Katoh M, Ito H, Oshimura M 2007 SIRT2, a tubulin deacetylase, acts to block the entry to chromosome condensation in response to mitotic stress. Oncogene 26:945–957 [DOI] [PubMed] [Google Scholar]
- Li W, Zhang B, Tang J, Cao Q, Wu Y, Wu C, Guo J, Ling EA, Liang F 2007 Sirtuin 2, a mammalian homolog of yeast silent information regulator-2 longevity regulator, is an oligodendroglial protein that decelerates cell differentiation through deacetylating α-tubulin. J Neurosci 27:2606–2616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulco M, Schiltz RL, Iezzi S, King MT, Zhao P, Kashiwaya Y, Hoffman E, Veech RL, Sartorelli V 2003 Sir2 regulates skeletal muscle differentiation as a potential sensor of the redox state. Mol Cell 12:51–62 [DOI] [PubMed] [Google Scholar]
- Ahn BH, Kim HS, Song S, Lee IH, Liu J, Vassilopoulos A, Deng CX, Finkel T 2008 A role for the mitochondrial deacetylase Sirt3 in regulating energy homeostasis. Proc Natl Acad Sci USA 105:14447–14452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombard DB, Alt FW, Cheng HL, Bunkenborg J, Streeper RS, Mostoslavsky R, Kim J, Yancopoulos G, Valenzuela D, Murphy A, Yang Y, Chen Y, Hirschey MD, Bronson RT, Haigis M, Guarente LP, Farese Jr RV, Weissman S, Verdin E, Schwer B 2007 Mammalian Sir2 homolog SIRT3 regulates global mitochondrial lysine acetylation. Mol Cell Biol 27:8807–8814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwer B, Bunkenborg J, Verdin RO, Andersen JS, Verdin E 2006 Reversible lysine acetylation controls the activity of the mitochondrial enzyme acetyl-CoA synthetase 2. Proc Natl Acad Sci USA 103:10224–10229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitani T, Okuno S, Fujisawa H 2003 Growth phase-dependent changes in the subcellular localization of pre-B-cell colony-enhancing factor. FEBS Lett 544:74–78 [DOI] [PubMed] [Google Scholar]
- Ahuja N, Schwer B, Carobbio S, Waltregny D, North BJ, Castronovo V, Maechler P, Verdin E 2007 Regulation of insulin secretion by SIRT4, a mitochondrial ADP-ribosyltransferase. J Biol Chem 282:33583–33592 [DOI] [PubMed] [Google Scholar]
- Nakagawa T, Lomb DJ, Haigis MC, Guarente L 2009 SIRT5 Deacetylates carbamoyl phosphate synthetase 1 and regulates the urea cycle. Cell 137:560–570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostoslavsky R, Chua KF, Lombard DB, Pang WW, Fischer MR, Gellon L, Liu P, Mostoslavsky G, Franco S, Murphy MM, Mills KD, Patel P, Hsu JT, Hong AL, Ford E, Cheng HL, Kennedy C, Nunez N, Bronson R, Frendewey D, Auerbach W, Valenzuela D, Karow M, Hottiger MO, Hursting S, Barrett JC, Guarente L, Mulligan R, Demple B, Yancopoulos GD, Alt FW 2006 Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell 124:315–329 [DOI] [PubMed] [Google Scholar]
- Ford E, Voit R, Liszt G, Magin C, Grummt I, Guarente L 2006 Mammalian Sir2 homolog SIRT7 is an activator of RNA polymerase I transcription. Genes Dev 20:1075–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grummt I, Pikaard CS 2003 Epigenetic silencing of RNA polymerase I transcription. Nat Rev Mol Cell Biol 4:641–649 [DOI] [PubMed] [Google Scholar]
- Vakhrusheva O, Braeuer D, Liu Z, Braun T, Bober E 2008 Sirt7-dependent inhibition of cell growth and proliferation might be instrumental to mediate tissue integrity during aging. J Physiol Pharmacol 59(Suppl 9):201–212 [PubMed] [Google Scholar]
- Chambon P, Weill JD, Mandel P 1963 Nicotinamide mononucleotide activation of new DNA-dependent polyadenylic acid synthesizing nuclear enzyme. Biochem Biophys Res Commun 11:39–43 [DOI] [PubMed] [Google Scholar]
- Hassa PO, Haenni SS, Elser M, Hottiger MO 2006 Nuclear ADP-ribosylation reactions in mammalian cells: where are we today and where are we going? Microbiol Mol Biol Rev 70:789–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wielckens K, Schmidt A, George E, Bredehorst R, Hilz H 1982 DNA fragmentation and NAD depletion. Their relation to the turnover of endogenous mono(ADP-ribosyl) and poly(ADP-ribosyl) proteins. J Biol Chem 257:12872–12877 [PubMed] [Google Scholar]
- Bürkle A 2005 Poly(ADP-ribose). The most elaborate metabolite of NAD+. FEBS J 272:4576–4589 [DOI] [PubMed] [Google Scholar]
- Shieh WM, Amé JC, Wilson MV, Wang ZQ, Koh DW, Jacobson MK, Jacobson EL 1998 Poly(ADP-ribose) polymerase null mouse cells synthesize ADP-ribose polymers. J Biol Chem 273:30069–30072 [DOI] [PubMed] [Google Scholar]
- Schreiber V, Amé JC, Dollé P, Schultz I, Rinaldi B, Fraulob V, Ménissier-de Murcia J, de Murcia G 2002 Poly(ADP-ribose) polymerase-2 (PARP-2) is required for efficient base excision DNA repair in association with PARP-1 and XRCC1. J Biol Chem 277:23028–23036 [DOI] [PubMed] [Google Scholar]
- Schreiber V, Dantzer F, Ame JC, de Murcia G 2006 Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol 7:517–528 [DOI] [PubMed] [Google Scholar]
- Ueda K, Ogata N, Kawaichi M, Inada S, Hayaishi O 1982 ADP-ribosylation reactions. Curr Top Cell Regul 21:175–187 [DOI] [PubMed] [Google Scholar]
- D'Amours D, Desnoyers S, D'Silva I, Poirier GG 1999 Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem J 342:249–268 [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Gonzalez R, Mendoza-Alvarez H 1995 Dissection of ADP-ribose polymer synthesis into individual steps of initiation, elongation, and branching. Biochimie 77:403–407 [DOI] [PubMed] [Google Scholar]
- Goodwin PM, Lewis PJ, Davies MI, Skidmore CJ, Shall S 1978 The effect of γ radiation and neocarzinostatin on NAD and ATP levels in mouse leukaemia cells. Biochim Biophys Acta 543:576–582 [DOI] [PubMed] [Google Scholar]
- Skidmore CJ, Davies MI, Goodwin PM, Halldorsson H, Lewis PJ, Shall S, Zia'ee AA 1979 The involvement of poly(ADP-ribose) polymerase in the degradation of NAD caused by γ-radiation and N-methyl-N-nitrosourea. Eur J Biochem 101:135–142 [DOI] [PubMed] [Google Scholar]
- Berger NA 1985 Poly(ADP-ribose) in the cellular response to DNA damage. Radiat Res 101:4–15 [PubMed] [Google Scholar]
- Williams GT, Lau KM, Coote JM, Johnstone AP 1985 NAD metabolism and mitogen stimulation of human lymphocytes. Exp Cell Res 160:419–426 [DOI] [PubMed] [Google Scholar]
- Rechsteiner M, Hillyard D, Olivera BM 1976 Turnover at nicotinamide adenine dinucleotide in cultures of human cells. J Cell Physiol 88:207–217 [DOI] [PubMed] [Google Scholar]
- Elliott G, Rechsteiner M 1975 Pyridine nucleotide metabolism in mitotic cells. J Cell Physiol 86(Suppl 2):641–651 [DOI] [PubMed] [Google Scholar]
- Chappie JS, Cànaves JM, Han GW, Rife CL, Xu Q, Stevens RC 2005 The structure of a eukaryotic nicotinic acid phosphoribosyltransferase reveals structural heterogeneity among type II PRTases. Structure 13:1385–1396 [DOI] [PubMed] [Google Scholar]
- Ruggieri S, Gregori L, Natalini P, Vita A, Emanuelli M, Raffaelli N, Magni G 1990 Evidence for an inhibitory effect exerted by yeast NMN adenylyltransferase on poly(ADP-ribose) polymerase activity. Biochemistry 29:2501–2506 [DOI] [PubMed] [Google Scholar]
- Pillai JB, Isbatan A, Imai S, Gupta MP 2005 Poly(ADP-ribose) polymerase-1-dependent cardiac myocyte cell death during heart failure is mediated by NAD+ depletion and reduced Sir2α deacetylase activity. J Biol Chem 280:43121–43130 [DOI] [PubMed] [Google Scholar]
- Kolthur-Seetharam U, Dantzer F, McBurney MW, de Murcia G, Sassone-Corsi P 2006 Control of AIF-mediated cell death by the functional interplay of SIRT1 and PARP-1 in response to DNA damage. Cell Cycle 5:873–877 [DOI] [PubMed] [Google Scholar]
- Malavasi F, Deaglio S, Ferrero E, Funaro A, Sancho J, Ausiello CM, Ortolan E, Vaisitti T, Zubiaur M, Fedele G, Aydin S, Tibaldi EV, Durelli I, Lusso R, Cozno F, Horenstein AL 2006 CD38 and CD157 as receptors of the immune system: a bridge between innate and adaptive immunity. Mol Med 12:334–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HC 2006 Structure and enzymatic functions of human CD38. Mol Med 12:317–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortolan E, Vacca P, Capobianco A, Armando E, Crivellin F, Horenstein A, Malavasi F 2002 CD157, the Janus of CD38 but with a unique personality. Cell Biochem Funct 20:309–322 [DOI] [PubMed] [Google Scholar]
- Partida-Sánchez S, Randall TD, Lund FE 2003 Innate immunity is regulated by CD38, an ecto-enzyme with ADP-ribosyl cyclase activity. Microbes Infect 5:49–58 [DOI] [PubMed] [Google Scholar]
- Malavasi F, Deaglio S, Funaro A, Ferrero E, Horenstein AL, Ortolan E, Vaisitti T, Aydin S 2008 Evolution and function of the ADP ribosyl cyclase/CD38 gene family in physiology and pathology. Physiol Rev 88:841–886 [DOI] [PubMed] [Google Scholar]
- Cakir-Kiefer C, Muller-Steffner H, Oppenheimer N, Schuber F 2001 Kinetic competence of the cADP-ribose-CD38 complex as an intermediate in the CD38/NAD+ glycohydrolase-catalysed reactions: implication for CD38 signalling. Biochem J 358:399–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dousa TP, Chini EN, Beers KW 1996 Adenine nucleotide diphosphates: emerging second messengers acting via intracellular Ca2+ release. Am J Physiol 271:C1007–C1024 [DOI] [PubMed] [Google Scholar]
- Aksoy P, White TA, Thompson M, Chini EN 2006 Regulation of intracellular levels of NAD: a novel role for CD38. Biochem Biophys Res Commun 345:1386–1392 [DOI] [PubMed] [Google Scholar]
- Aksoy P, Escande C, White TA, Thompson M, Soares S, Benech JC, Chini EN 2006 Regulation of SIRT 1 mediated NAD dependent deacetylation: a novel role for the multifunctional enzyme CD38. Biochem Biophys Res Commun 349:353–359 [DOI] [PubMed] [Google Scholar]
- Barbosa MT, Soares SM, Novak CM, Sinclair D, Levine JA, Aksoy P, Chini EN 2007 The enzyme CD38 (a NAD glycohydrolase, EC 3.2.2.5) is necessary for the development of diet-induced obesity. FASEB J 21:3629–3639 [DOI] [PubMed] [Google Scholar]
- Zhang T, Berrocal JG, Frizzell KM, Gamble MJ, DuMond ME, Krishnakumar R, Yang T, Sauve AA, Kraus WL 2009 Enzymes in the NAD+ salvage pathway regulate SIRT1 activity at target gene promoters. J Biol Chem 284:20408–20417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niere M, Kernstock S, Koch-Nolte F, Ziegler M 2008 Functional localization of two poly(ADP-ribose)-degrading enzymes to the mitochondrial matrix. Mol Cell Biol 28:814–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzländer M, Fricker MD, Müller C, Marty L, Brach T, Novak J, Sweetlove LJ, Hell R, Meyer AJ 2008 Confocal imaging of glutathione redox potential in living plant cells. J Microsc 231:299–316 [DOI] [PubMed] [Google Scholar]
- Xu C, Zipfel W, Shear JB, Williams RM, Webb WW 1996 Multiphoton fluorescence excitation: new spectral windows for biological nonlinear microscopy. Proc Natl Acad Sci USA 93:10763–10768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehn CJ, Elvehjem CA 1937 Further studies on the concentration of the antipellagra factor. J Biol Chem 118:693–699 [Google Scholar]
- Sauve AA 2008 NAD+ and vitamin B3: from metabolism to therapies. J Pharmacol Exp Ther 324:883–893 [DOI] [PubMed] [Google Scholar]
- Comings DE, Muhleman D, Dietz G, Sherman M, Forest GL 1995 Sequence of human tryptophan 2,3-dioxygenase (TDO2): presence of a glucocorticoid response-like element composed of a GTT repeat and an intronic CCCCT repeat. Genomics 29:390–396 [DOI] [PubMed] [Google Scholar]
- Heyes MP, Saito K, Jacobowitz D, Markey SP, Takikawa O, Vickers JH 1992 Poliovirus induces indoleamine-2,3-dioxygenase and quinolinic acid synthesis in macaque brain. FASEB J 6:2977–2989 [DOI] [PubMed] [Google Scholar]
- Sanni LA, Thomas SR, Tattam BN, Moore DE, Chaudhri G, Stocker R, Hunt NH 1998 Dramatic changes in oxidative tryptophan metabolism along the kynurenine pathway in experimental cerebral and noncerebral malaria. Am J Pathol 152:611–619 [PMC free article] [PubMed] [Google Scholar]
- Yoshida R, Hayaishi O 1978 Induction of pulmonary indoleamine 2,3-dioxygenase by intraperitoneal injection of bacterial lipopolysaccharide. Proc Natl Acad Sci USA 75:3998–4000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida R, Urade Y, Tokuda M, Hayaishi O 1979 Induction of indoleamine 2,3-dioxygenase in mouse lung during virus infection. Proc Natl Acad Sci USA 76:4084–4086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konan KV, Taylor MW 1996 Importance of the two interferon-stimulated response element (ISRE) sequences in the regulation of the human indoleamine 2,3-dioxygenase gene. J Biol Chem 271:19140–19145 [DOI] [PubMed] [Google Scholar]
- Fallarino F, Grohmann U, Vacca C, Bianchi R, Orabona C, Spreca A, Fioretti MC, Puccetti P 2002 T cell apoptosis by tryptophan catabolism. Cell Death Differ 9:1069–1077 [DOI] [PubMed] [Google Scholar]
- Hwu P, Du MX, Lapointe R, Do M, Taylor MW, Young HA 2000 Indoleamine 2,3-dioxygenase production by human dendritic cells results in the inhibition of T cell proliferation. J Immunol 164:3596–3599 [DOI] [PubMed] [Google Scholar]
- Lee GK, Park HJ, Macleod M, Chandler P, Munn DH, Mellor AL 2002 Tryptophan deprivation sensitizes activated T cells to apoptosis prior to cell division. Immunology 107:452–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin M, Ohnishi M, Iguchi S, Sano K, Umezawa C 1999 Peroxisome-proliferator regulates key enzymes of the tryptophan-NAD+ pathway. Toxicol Appl Pharmacol 158:71–80 [DOI] [PubMed] [Google Scholar]
- Ikeda M, Tsuji H, Nakamura S, Ichiyama A, Nishizuka Y, Hayaishi O 1965 Studies on the biosynthesis of nicotinamide adenine dinucleotide. II. A role of picolinic carboxylase in the biosynthesis of nicotinamide adenine dinucleotide from tryptophan in mammals. J Biol Chem 240:1395–1401 [PubMed] [Google Scholar]
- Sasaki Y, Araki T, Milbrandt J 2006 Stimulation of nicotinamide adenine dinucleotide biosynthetic pathways delays axonal degeneration after axotomy. J Neurosci 26:8484–8491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson TM, Rawling JM, Roebuck BD, Kirkland JB 1995 Large supplements of nicotinic acid and nicotinamide increase tissue NAD+ and poly(ADP-ribose) levels but do not affect diethylnitrosamine-induced altered hepatic foci in Fischer-344 rats. J Nutr 125:1455–1461 [DOI] [PubMed] [Google Scholar]
- Gale EA, Bingley PJ, Emmett CL, Collier T 2004 European Nicotinamide Diabetes Intervention Trial (ENDIT): a randomised controlled trial of intervention before the onset of type 1 diabetes. Lancet 363:925–931 [DOI] [PubMed] [Google Scholar]
- Knip M, Douek IF, Moore WP, Gillmor HA, McLean AE, Bingley PJ, Gale EA 2000 Safety of high-dose nicotinamide: a review. Diabetologia 43:1337–1345 [DOI] [PubMed] [Google Scholar]
- Narkar VA, Downes M, Yu RT, Embler E, Wang YX, Banayo E, Mihaylova MM, Nelson MC, Zou Y, Juguilon H, Kang H, Shaw RJ, Evans RM 2008 AMPK and PPARδ agonists are exercise mimetics. Cell 134:405–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Wang SY, Fleuriel C, Leprince D, Rocheleau JV, Piston DW, Goodman RH 2007 Metabolic regulation of SIRT1 transcription via a HIC1:CtBP corepressor complex. Proc Natl Acad Sci USA 104:829:833 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Lane MA, Ingram DK, Roth GS 1998 2-Deoxy-D-glucose feeding in rats mimics physiologic effects of calorie restriction. J Anti Aging Med 1:327–337 [Google Scholar]
- Moscioni AD, Engel JL, Casida JE 1977 Kynurenine formamidase inhibition as a possible mechanism for certain teratogenic effects of organophosphorus and methylcarbamate insecticides in chicken embryos. Biochem Pharmacol 26:2251–2258 [DOI] [PubMed] [Google Scholar]
- Magni G, Di Stefano M, Orsomando G, Raffaelli N, Ruggieri S 2009 NAD(P) biosynthesis enzymes as potential targets for selective drug design. Curr Med Chem 16:1372–1390 [DOI] [PubMed] [Google Scholar]
- Hasmann M, Schemainda I 2003 FK866, a highly specific noncompetitive inhibitor of nicotinamide phosphoribosyltransferase, represents a novel mechanism for induction of tumor cell apoptosis. Cancer Res 63:7436–7442 [PubMed] [Google Scholar]
- Löb S, Königsrainer A, Rammensee HG, Opelz G, Terness P 2009 Inhibitors of indoleamine-2,3-dioxygenase for cancer therapy: can we see the wood for the trees? Nat Rev Cancer 9:445–452 [DOI] [PubMed] [Google Scholar]
- Kenyon C 2005 The plasticity of aging: insights from long-lived mutants. Cell 120:449–460 [DOI] [PubMed] [Google Scholar]
- Liang H, Masoro EJ, Nelson JF, Strong R, McMahan CA, Richardson A 2003 Genetic mouse models of extended lifespan. Exp Gerontol 38:1353–1364 [DOI] [PubMed] [Google Scholar]
- Yang T, Fu M, Pestell R, Sauve AA 2006 SIRT1 and endocrine signaling. Trends Endocrinol Metab 17:186–191 [DOI] [PubMed] [Google Scholar]
- Sun C, Zhang F, Ge X, Yan T, Chen X, Shi X, Zhai Q 2007 SIRT1 improves insulin sensitivity under insulin-resistant conditions by repressing PTP1B. Cell Metab 6:307–319 [DOI] [PubMed] [Google Scholar]
- Russell SJ, Kahn CR 2007 Endocrine regulation of ageing. Nat Rev Mol Cell Biol 8:681–691 [DOI] [PubMed] [Google Scholar]
- Yamamoto H, Uchigata Y, Okamoto H 1981 Streptozotocin and alloxan induce DNA strand breaks and poly(ADP-ribose) synthetase in pancreatic islets. Nature 294:284–286 [DOI] [PubMed] [Google Scholar]
- Burkart V, Wang ZQ, Radons J, Heller B, Herceg Z, Stingl L, Wagner EF, Kolb H 1999 Mice lacking the poly(ADP-ribose) polymerase gene are resistant to pancreatic β-cell destruction and diabetes development induced by streptozocin. Nat Med 5:314–319 [DOI] [PubMed] [Google Scholar]
- Pieper AA, Brat DJ, Krug DK, Watkins CC, Gupta A, Blackshaw S, Verma A, Wang ZQ, Snyder SH 1999 Poly(ADP-ribose) polymerase-deficient mice are protected from streptozotocin-induced diabetes. Proc Natl Acad Sci USA 96:3059–3064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horváth EM, Benko R, Gero D, Kiss L, Szabó C 2008 Treatment with insulin inhibits poly(ADP-ribose)polymerase activation in a rat model of endotoxemia. Life Sci 82:205–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckert S, Farrahi F, Perveen Ghani Q, Aslam R, Scheuenstuhl H, Coerper S, Königsrainer A, Hunt TK, Hussain MZ 2006 IGF-I-induced VEGF expression in HUVEC involves phosphorylation and inhibition of poly(ADP-ribose)polymerase. Biochem Biophys Res Commun 341:67–72 [DOI] [PubMed] [Google Scholar]
- Badman MK, Pissios P, Kennedy AR, Koukos G, Flier JS, Maratos-Flier E 2007 Hepatic fibroblast growth factor 21 is regulated by PPARα and is a key mediator of hepatic lipid metabolism in ketotic states. Cell Metab 5:426–437 [DOI] [PubMed] [Google Scholar]
- Banks AS, Kon N, Knight C, Matsumoto M, Gutiérrez-Juárez R, Rossetti L, Gu W, Accili D 2008 SirT1 gain of function increases energy efficiency and prevents diabetes in mice. Cell Metab 8:333–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharitonenkov A, Shiyanova TL, Koester A, Ford AM, Micanovic R, Galbreath EJ, Sandusky GE, Hammond LJ, Moyers JS, Owens RA, Gromada J, Brozinick JT, Hawkins ED, Wroblewski VJ, Li DS, Mehrbod F, Jaskunas SR, Shanafelt AB 2005 FGF-21 as a novel metabolic regulator. J Clin Invest 115:1627–1635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurosu H, Ogawa Y, Miyoshi M, Yamamoto M, Nandi A, Rosenblatt KP, Baum MG, Schiavi S, Hu MC, Moe OW, Kuro-o M 2006 Regulation of fibroblast growth factor-23 signaling by Klotho. J Biol Chem 281:6120–6123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Uehara Y, Motomura-Matsuzaka K, Oki J, Koyama Y, Kimura M, Asada M, Komi-Kuramochi A, Oka S, Imamura T 2008 β-Klotho is required for fibroblast growth factor (FGF) 21 signaling through FGF receptor (FGFR) 1c and FGFR3c. Mol Endocrinol 22:1006–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MY, Woo EM, Chong YT, Homenko DR, Kraus WL 2006 Acetylation of estrogen receptor α by p300 at lysines 266 and 268 enhances the deoxyribonucleic acid binding and transactivation activities of the receptor. Mol Endocrinol 20:1479–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu M, Liu M, Sauve AA, Jiao X, Zhang X, Wu X, Powell MJ, Yang T, Gu W, Avantaggiati ML, Pattabiraman N, Pestell TG, Wang F, Quong AA, Wang C, Pestell RG 2006 Hormonal control of androgen receptor function through SIRT1. Mol Cell Biol 26:8122–8135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pottier N, Cheok MH, Yang W, Assem M, Tracey L, Obenauer JC, Panetta JC, Relling MV, Evans WE 2007 Expression of SMARCB1 modulates steroid sensitivity in human lymphoblastoid cells: identification of a promoter SNP that alters PARP1 binding and SMARCB1 expression. Hum Mol Genet 16:2261–2271 [DOI] [PubMed] [Google Scholar]
- Ju BG, Lunyak VV, Perissi V, Garcia-Bassets I, Rose DW, Glass CK, Rosenfeld MG 2006 A topoisomerase IIβ-mediated dsDNA break required for regulated transcription. Science 312:1798–1802 [DOI] [PubMed] [Google Scholar]
- Bai P, Houten SM, Huber A, Schreiber V, Watanabe M, Kiss B, de Murcia G, Auwerx J, Ménissier-de Murcia J 2007 Poly(ADP-ribose) polymerase-2 [corrected] controls adipocyte differentiation and adipose tissue function through the regulation of the activity of the retinoid X receptor/peroxisome proliferator-activated receptor-γ [corrected] heterodimer. J Biol Chem [Erratum (2008) 283:5972] 282:37738–37746 [DOI] [PubMed] [Google Scholar]
- Baur JA, Pearson KJ, Price NL, Jamieson HA, Lerin C, Kalra A, Prabhu VV, Allard JS, Lopez-Lluch G, Lewis K, Pistell PJ, Poosala S, Becker KG, Boss O, Gwinn D, Wang M, Ramaswamy S, Fishbein KW, Spencer RG, Lakatta EG, Le Couteur D, Shaw RJ, Navas P, Puigserver P, Ingram DK, de Cabo R, Sinclair DA 2006 Resveratrol improves health and survival of mice on a high-calorie diet. Nature 444:337–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne JC, Lambert PD, Schenk S, Carney DP, Smith JJ, Gagne DJ, Jin L, Boss O, Perni RB, Vu CB, Bemis JE, Xie R, Disch JS, Ng PY, Nunes JJ, Lynch AV, Yang H, Galonek H, Israelian K, Choy W, Iffland A, Lavu S, Medvedik O, Sinclair DA, Olefsky JM, Jirousek MR, Elliott PJ, Westphal CH 2007 Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature 450:712–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman HC, Hugill A, Dear NT, Ashcroft FM, Cox RD 2006 Deletion of nicotinamide nucleotide transhydrogenase: a new quantitive trait locus accounting for glucose intolerance in C57BL/6J mice. Diabetes 55:2153–2156 [DOI] [PubMed] [Google Scholar]
- Aston-Mourney K, Wong N, Kebede M, Zraika S, Balmer L, McMahon JM, Fam BC, Favaloro J, Proietto J, Morahan G, Andrikopoulos S 2007 Increased nicotinamide nucleotide transhydrogenase levels predispose to insulin hypersecretion in a mouse strain susceptible to diabetes. Diabetologia 50:2476–2485 [DOI] [PubMed] [Google Scholar]
- Freeman H, Shimomura K, Horner E, Cox RD, Ashcroft FM 2006 Nicotinamide nucleotide transhydrogenase: a key role in insulin secretion. Cell Metab 3:35–45 [DOI] [PubMed] [Google Scholar]
- Saxena S, Caroni P 2007 Mechanisms of axon degeneration: from development to disease. Prog Neurobiol 83:174–191 [DOI] [PubMed] [Google Scholar]
- Coleman M 2005 Axon degeneration mechanisms: commonality amid diversity. Nat Rev Neurosci 6:889–898 [DOI] [PubMed] [Google Scholar]
- Conforti L, Tarlton A, Mack TG, Mi W, Buckmaster EA, Wagner D, Perry VH, Coleman MP 2000 A Ufd2/D4Cole1e chimeric protein and overexpression of Rbp7 in the slow Wallerian degeneration (WldS) mouse. Proc Natl Acad Sci USA 97:11377–11382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mack TG, Reiner M, Beirowski B, Mi W, Emanuelli M, Wagner D, Thomson D, Gillingwater T, Court F, Conforti L, Fernando FS, Tarlton A, Andressen C, Addicks K, Magni G, Ribchester RR, Perry VH, Coleman MP 2001 Wallerian degeneration of injured axons and synapses is delayed by a Ube4b/Nmnat chimeric gene. Nat Neurosci 4:1199–1206 [DOI] [PubMed] [Google Scholar]
- Ferri A, Sanes JR, Coleman MP, Cunningham JM, Kato AC 2003 Inhibiting axon degeneration and synapse loss attenuates apoptosis and disease progression in a mouse model of motoneuron disease. Curr Biol 13:669–673 [DOI] [PubMed] [Google Scholar]
- Samsam M, Mi W, Wessig C, Zielasek J, Toyka KV, Coleman MP, Martini R 2003 The Wlds mutation delays robust loss of motor and sensory axons in a genetic model for myelin-related axonopathy. J Neurosci 23:2833–2839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sajadi A, Schneider BL, Aebischer P 2004 Wlds-mediated protection of dopaminergic fibers in an animal model of Parkinson disease. Curr Biol 14:326–330 [DOI] [PubMed] [Google Scholar]
- Araki T, Sasaki Y, Milbrandt J 2004 Increased nuclear NAD biosynthesis and SIRT1 activation prevent axonal degeneration. Science 305:1010–1013 [DOI] [PubMed] [Google Scholar]
- Press C, Milbrandt J 2008 NMNAT delays axonal degeneration caused by mitochondrial and oxidative stress. J Neurosci 28:4861–4871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conforti L, Fang G, Beirowski B, Wang MS, Sorci L, Asress S, Adalbert R, Silva A, Bridge K, Huang XP, Magni G, Glass JD, Coleman MP 2007 NAD(+) and axon degeneration revisited: Nmnat1 cannot substitute for Wld(S) to delay Wallerian degeneration. Cell Death Differ 14:116–127 [DOI] [PubMed] [Google Scholar]
- Sasaki Y, Vohra BP, Lund FE, Milbrandt J 2009 Nicotinamide mononucleotide adenylyl transferase-mediated axonal protection requires enzymatic activity but not increased levels of neuronal nicotinamide adenine dinucleotide. J Neurosci 29:5525–5535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MT, Beal MF 2006 Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature 443:787–795 [DOI] [PubMed] [Google Scholar]
- Parker JA, Arango M, Abderrahmane S, Lambert E, Tourette C, Catoire H, Néri C 2005 Resveratrol rescues mutant polyglutamine cytotoxicity in nematode and mammalian neurons. Nat Genet 37:349–350 [DOI] [PubMed] [Google Scholar]
- Qin W, Yang T, Ho L, Zhao Z, Wang J, Chen L, Zhao W, Thiyagarajan M, MacGrogan D, Rodgers JT, Puigserver P, Sadoshima J, Deng H, Pedrini S, Gandy S, Sauve AA, Pasinetti GM 2006 Neuronal SIRT1 activation as a novel mechanism underlying the prevention of Alzheimer disease amyloid neuropathology by calorie restriction. J Biol Chem 281:21745–21754 [DOI] [PubMed] [Google Scholar]
- Lin J, Wu PH, Tarr PT, Lindenberg KS, St-Pierre J, Zhang CY, Mootha VK, Jäger S, Vianna CR, Reznick RM, Cui L, Manieri M, Donovan MX, Wu Z, Cooper MP, Fan MC, Rohas LM, Zavacki AM, Cinti S, Shulman GI, Lowell BB, Krainc D, Spiegelman BM 2004 Defects in adaptive energy metabolism with CNS-linked hyperactivity in PGC-1α null mice. Cell 119:121–135 [DOI] [PubMed] [Google Scholar]
- Boyonoski AC, Spronck JC, Gallacher LM, Jacobs RM, Shah GM, Poirier GG, Kirkland JB 2002 Niacin deficiency decreases bone marrow poly(ADP-ribose) and the latency of ethylnitrosourea-induced carcinogenesis in rats. J Nutr 132:108–114 [DOI] [PubMed] [Google Scholar]
- Brooks CL, Gu W 2009 How does SIRT1 affect metabolism, senescence and cancer? Nat Rev Cancer 9:123–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul R, Hoffer A, Stephen JD 1955 Influence of nicotinic acid on serum cholesterol in man. Arch Biochem 54:558–559 [DOI] [PubMed] [Google Scholar]
- Tunaru S, Kero J, Schaub A, Wufka C, Blaukat A, Pfeffer K, Offermanns S 2003 PUMA-G and HM74 are receptors for nicotinic acid and mediate its anti-lipolytic effect. Nat Med 9:352–355 [DOI] [PubMed] [Google Scholar]
- Jia SH, Li Y, Parodo J, Kapus A, Fan L, Rotstein OD, Marshall JC 2004 Pre-B cell colony-enhancing factor inhibits neutrophil apoptosis in experimental inflammation and clinical sepsis. J Clin Invest 113:1318–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skokowa J, Lan D, Thakur BK, Wang F, Gupta K, Cario G, Brechlin AM, Schambach A, Hinrichsen L, Meyer G, Gaestel M, Stanulla M, Tong Q, Welte K 2009 NAMPT is essential for the G-CSF-induced myeloid differentiation via a NAD(+)-sirtuin-1-dependent pathway. Nat Med 15:151–158 [DOI] [PubMed] [Google Scholar]
- Bordone L, Guarente L 2005 Calorie restriction, SIRT1 and metabolism: understanding longevity. Nat Rev Mol Cell Biol 6:298–305 [DOI] [PubMed] [Google Scholar]
- Civitarese AE, Carling S, Heilbronn LK, Hulver MH, Ukropcova B, Deutsch WA, Smith SR, Ravussin E 2007 Calorie restriction increases muscle mitochondrial biogenesis in healthy humans. PLoS Med 4:e76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson KJ, Baur JA, Lewis KN, Peshkin L, Price NL, Labinskyy N, Swindell WR, Kamara D, Minor RK, Perez E, Jamieson HA, Zhang Y, Dunn SR, Sharma K, Pleshko N, Woollett LA, Csiszar A, Ikeno Y, Le Couteur D, Elliott PJ, Becker KG, Navas P, Ingram DK, Wolf NS, Ungvari Z, Sinclair DA, de Cabo R 2008 Resveratrol delays age-related deterioration and mimics transcriptional aspects of dietary restriction without extending life span. Cell Metab 8:157–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michishita E, McCord RA, Berber E, Kioi M, Padilla-Nash H, Damian M, Cheung P, Kusumoto R, Kawahara TL, Barrett JC, Chang HY, Bohr VA, Ried T, Gozani O, Chua KF 2008 SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature 452:492–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellizzi D, Rose G, Cavalcante P, Covello G, Dato S, De Rango F, Greco V, Maggiolini M, Feraco E, Mari V, Franceschi C, Passarino G, De Benedictis G 2005 A novel VNTR enhancer within the SIRT3 gene, a human homologue of SIR2, is associated with survival at oldest ages. Genomics 85:258–263 [DOI] [PubMed] [Google Scholar]
- Lin SJ, Defossez PA, Guarente L 2000 Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science 289:2126–2128 [DOI] [PubMed] [Google Scholar]
- Lu SP, Kato M, Lin SJ 2009 Assimilation of endogenous nicotinamide riboside is essential for calorie restriction-mediated life span extension in Saccharomyces cerevisiae. J Biol Chem 284:17110–17119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Reilly T, Niven DF 2003 Levels of nicotinamide adenine dinucleotide in extracellular body fluids of pigs may be growth-limiting for Actinobacillus pleuropneumoniae and Haemophilus parasuis. Can J Vet Res 67:229–231 [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Hara N, Shibata T, Osago H, Tsuchiya M 2006 The simultaneous measurement of nicotinamide adenine dinucleotide and related compounds by liquid chromatography/electrospray ionization tandem mass spectrometry. Anal Biochem 352:282–285 [DOI] [PubMed] [Google Scholar]
- Fjeld CC, Birdsong WT, Goodman RH 2003 Differential binding of NAD+ and NADH allows the transcriptional corepressor carboxyl-terminal binding protein to serve as a metabolic sensor. Proc Natl Acad Sci USA 100:9202–9207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai S, Armstrong CM, Kaeberlein M, Guarente L 2000 Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 403:795–800 [DOI] [PubMed] [Google Scholar]
- Smith BC, Hallows WC, Denu JM 2009 A continuous microplate assay for sirtuins and nicotinamide-producing enzymes. Anal Biochem 394:101–109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin L, Galonek H, Israelian K, Choy W, Morrison M, Xia Y, Wang X, Xu Y, Yang Y, Smith JJ, Hoffmann E, Carney DP, Perni RB, Jirousek MR, Bemis JE, Milne JC, Sinclair DA, Westphal CH 2009 Biochemical characterization, localization, and tissue distribution of the longer form of mouse SIRT3. Protein Sci 18:514–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza-Alvarez H, Alvarez-Gonzalez R 1993 Poly(ADP-ribose) polymerase is a catalytic dimer and the automodification reaction is intermolecular. J Biol Chem 268:22575–22580 [PubMed] [Google Scholar]
- Amé JC, Rolli V, Schreiber V, Niedergang C, Apiou F, Decker P, Muller S, Höger T, Ménissier-de Murcia J, de Murcia G 1999 PARP-2, a novel mammalian DNA damage-dependent poly(ADP-ribose) polymerase. J Biol Chem 274:17860–17868 [DOI] [PubMed] [Google Scholar]
- Sauve AA, Munshi C, Lee HC, Schramm VL 1998 The reaction mechanism for CD38. A single intermediate is responsible for cyclization, hydrolysis, and base-exchange chemistries. Biochemistry 37:13239–13249 [DOI] [PubMed] [Google Scholar]
- Johansson M 1999 A human poly(ADP-ribose) polymerase gene family (ADPRTL): cDNA cloning of two novel poly(ADP-ribose) polymerase homologs. Genomics 57:442–445 [DOI] [PubMed] [Google Scholar]