Abstract
The nuclear receptor and bona fide oncogene, steroid receptor coactivator-3 (SRC-3, AIB1), acts as a master transcriptional regulator of breast cancer by transducing growth signals via the estrogen receptor α (ER). In this resource paper, we present the genome-wide localization analysis of SRC-3 chromatin affinity sites in MCF-7 human breast cancer chromatin and compare the cis binding sites to global cartographies for ER and FoxA1. By correlating their gene proximal binding sites to integrated gene expression signatures, and in combination with gene ontology analyses, we provide a functional classification of estradiol-induced gene regulation that further highlights an intricate transcriptional control of interdependent cellular pathways by SRC-3. Furthermore, by presenting proteomics analyses of in vivo SRC-3- and ER-associated proteins, we give strong evidence to support the idea that the interpretative power of SRC-3 in estrogen signaling is mediated through the formation of distinct, cell state-dependent protein complexes. Altogether, we present the first approach in complementary comparative analyses that converges results obtained by three discovery-driven methods (cistromics, transcriptomics, and proteomics) into testable hypotheses, thus providing a valuable resource for follow-up studies that further our understanding of estrogen signaling in human diseases in general and breast cancer in particular.
This manuscript describes SRC-3 cistrome and holocomplexes in MCF-7 cells, and integrates public resources for a broad characterization and functional classification of estradiol-induced gene regulation.
The identification of target genes is of great interest when multifaceted genomic interdependencies have to be solved before providing translational projects with frameworks that allow for studying diseases and testing interventions. This is particularly true for alignments for which susceptibility loci show strong associations for disease markers, such as for the sex hormone nuclear receptors (NRs), which play pivotal roles in reproduction, development, cell homeostasis, and in the pathobiology of various cancers. NRs are transcription factors (TFs) that control target gene expression by responding to their cognate ligands and recruiting a heterogeneous and functionally distinct group of molecules, the coregulators, that, together, sense, refine, and integrate cellular signals by tethering various enzymatic activities to specific transcription sites. These coregulators modify chromatin for TF binding to DNA and mediate and regulate proximal and distal components of gene expression that include transcription initiation, elongation, and splicing (1,2). More than 300 coregulators have been described for NRs alone (www.nursa.org) (3).
Steroid receptor coactivator-3 (SRC-3), also known as amplified in breast cancer-1 (AIB1), ACTR, NCOA3, p/CIP, RAC3, and TRAM-1, is a tumorigenic NR coactivator (4) and a common target for cellular growth programs. SRC-3 was classified as an oncogene based on its genetic amplification (4), cell growth and tumorigenesis studies in mice (5,6), and overall high tumor incidence in clinical reports (7,8). Conversely, by occasionally functioning as a tumor suppressor (9,10), SRC-3 demonstrates divergent effects on cell proliferation. Functional diversity can be afforded by associated proteins, which are selected distinctly under control of extensive posttranslational coding of the SRC-3 protein (11,12,13). Vice versa, varied posttranslational modifications imply SRC-3 as a recipient of multiple cell-signal pathways (8,14,15,16,17) and as an integrator in elucidating processes of cell regulation by converging signals to the transcription unit for modulation of target gene expression.
SRC-3 is required for maximal activity of estrogen receptor α (ER) and is a rate-limiting factor for estrogen-dependent growth of ER-positive human MCF-7 breast cancer cells (18,19). Upon estrogen binding, ER can both stimulate and repress target gene transcription by directly binding to cognate DNA sequences or by recruitment to DNA through tethering to other TFs such as AP-1 or Sp1. Potential high-affinity ER binding sites, along with adjacent binding elements for auxiliary TFs, recently have been identified in MCF-7 cells using chromatin immunoprecipitation (ChIP)-based methods (20,21,22,23,24,25,26,27,28,29).
A better picture of the phenotypic features that certain binding sites impart on target gene expression is obtained when genome-wide location analyses for non-DNA-binding coregulators are included in TF cartographies. Due to their intimate functional relationships, this approach is particularly true for SRC-3 and ER. We thus mapped genome-wide SRC-3 chromatin affinity sites in MCF-7 breast cancer cells by chromatin cross-linking and compared the location data with existing global cartographies for ER and FoxA1. Next, we correlated their gene proximal binding sites to integrated RNA expression signatures obtained from a collection of MCF-7 microarray datasets. Finally, we identified potential auxiliary factors for SRC-3 coactivator function using mass spectrometry (MS)-based proteomics analyses of in vivo nuclear SRC-3 and ER protein complexes. These proteomics findings support the idea that the interpretive power of SRC-3 in cell signaling is mediated through the formation of distinct protein complexes. Altogether, we provide an example of successful bridging of high-throughput technologies for the global characterization of the transcriptional impact of the SRC-3 coregulator.
Results
Identification of SRC-3 binding sites in MCF-7 breast cancer chromatin
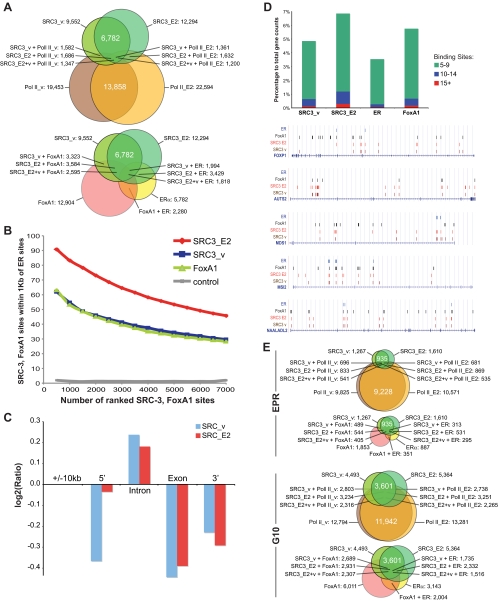
SRC-3 and RNA polymerase (Pol) II ChIP DNA from ethanol (vehicle) and 17β-estradiol (E2)-treated MCF-7 cells were amplified for Illumina sequencing. About 9 million tags of each sample mapped to unique locations in the human genome. We used the model-based analysis of ChIP-sequencing approach to normalize ChIP samples against MCF-7 input controls. A P value cutoff of 1e−10 (corresponding to <1% false discovery rate) produced 9552 high-confidence SRC-3 sites for vehicle-treated MCF-7 cells (SRC3_v) and 12,294 SRC-3 binding sites for ligand-treated cells (SRC3_E2); location data are provided in BED format in Supplemental Material 1 (SM1) published on The Endocrine Society’s Journals Online web site at http://mend.endojournals.org; note that the contents of Supplemental Materials SM1–SM9 are described on the Supplemental Index). The two datasets shared 6782 peaks for which the sequence centers were within 1 kb of each other. Similar assay and analysis conditions yielded 22,594 RNA Pol II sites in E2-treated cells (Tx_E2) and 19,453 RNA Pol II sites in vehicle-control cells (Tx_v) (Fig. 1A). About 40% (8736 sites) of the detected Pol II activity occurred de novo upon E2 treatment, and only a relatively small portion (13–16%) of the SRC-3 peaks showed colocalization with RNA Pol II sites.
Figure 1.
Genome-wide MCF-7 cartographies of SRC-3, RNA Pol II, and comparative location analyses with recalculated global maps of ER and FoxA1 binding sites. High-confidence ChIP sites for SRC-3 and RNA Pol II in vehicle (SRC3_v, Tx_v) and 1 h E2-treated MCF-7 cells (SRC3_E2, Tx_E2), ER, and HNF3A (FoxA1). BED files containing chromosome annotation and start-stop positions of the ChIP sites are provided as Supplemental Material SM1. A, ChIP binding site counts and proximity relationships. Venn illustrations of binding sites with sequence centers within 1 kb of each other for SRC-3 and RNA Pol II (top) and for SRC-3, ER, and FoxA1 (bottom). B, Proximity distribution of SRC-3, FoxA1, and ER binding sites. The x-axis represents numbers of SRC-3 and FoxA1 binding sites ranked by P values and binned at 500, and the y-axis represents percentage of SRC-3 and FoxA1 sites that are within 1 kb of ER binding sites. Random selected 7000 regions on the arrays were used as control. Approximately 10% of SRC-3 ChIP-Seq platform-specific sequences have been removed to allow for a fair cross-platform comparison. A reciprocal proximity distribution of ER binding sites compared with SRC-3 sites is provided in Supplemental Material SM2. C, Intragenic SRC-3 sites are overrepresented in intronic sequences. Log2 values are shown for ratios of expected numbers of SRC-3 sites over actual counts for the 10-kb gene-flanking regions (5′, 3′) and for intron and exon sequences. Binding site counts were normalized for the average length of total exon and intron sequences (8 × 316 bp and 7 × 5747 bp, respectively) according to the RefSeq model gene. D, Genome-wide intragenic SRC-3, FoxA1, and ER binding site frequency distribution; top, stacked columns representation of gene counts that have five to nine, 10–14, and 15 or more respective binding sites in the gene-transcribed region; bottom, UCSC Genome Browser illustrations of select genes showing array-type distributions of SRC-3 and FoxA1 binding sites. Sequence positions and other generic UCSC annotations were removed for clarity. E, Gene counts. Venn illustrations for genes with SRC-3 and RNA Pol II, and with SRC-3, ER, and FoxA1 binding sites in the EPR (−7.5 to +2.5 kb of TSS), and in the gene-transcribed regions ±10 kb (G10). The diagrams in A and E were drawn proportionally.
All genomic mappings of ER binding sites published to date in human cells used MCF-7 and similar E2 treatment conditions (30). To allow for comparative analyses between SRC-3 and available ER data, we reanalyzed the published global maps of ER and FoxA1 binding sites in MCF-7 cells (21,31), which produced 5782 high-confidence ER and 12,904 FoxA1 sites.
A binding site proximity calculation for SRC-3, ER, and FoxA1 showed that many more SRC-3 sites have their sequence centers within 1 kb of ER (SRC3_v 21%, SRC3_E2 28%) and FoxA1 (SRC3_v 35%, SRC3_E2 29%) binding sites than with Pol II sites (SRC3_v 14%, SRC3_E2 13% for Tx_E2) (Fig. 1A). Being a coactivator for ER, a significant overlap of SRC-3 with ER binding sites was expected in E2-treated cells. The intersections of SRC-3 with FoxA1 binding sites, however, is remarkable when considering that SRC-3 has not yet been functionally liked to FoxA1 biology.
Carroll and Lupien and colleagues analyzed the ChIP DNA by hybridization to the Affymetrix human tiling 1.0 microarrays representing the nonrepetitive human genome sequence (21,31), whereas we used the Illumina sequencing approach. To allow for a fair peak comparison between ChIP-chip and ChIP-Seq assay types, we removed about 10% of the ChIP-Seq platform-specific SRC-3 peaks residing in sequences that are not present on tiled arrays and again determined the numbers of proximal binding sites, which we ranked by their respective P values. We found 91% of the top 500, or 54% of the top 5000, scoring SRC3_E2 sites within 1 kb from reported ER binding sites, whereas only 62% (34%) of the top 500 (5000) scoring SRC3_v sequences were proximal to ER sites (Fig. 1B). The distribution of SRC3_v sites is similar to the spreading of FoxA1 sites relative to ER positions. Being a rather promiscuous transcriptional coactivator interacting with many TFs other than ER, we expected to find significant SRC3_v binding sites before estrogen signaling, whereas a significant sequence overlap with ER binding sites was anticipated for E2-stimulated cells.
Genome-wide location analysis of SRC-3 sites
We then mapped the high-scoring ChIP-Seq binding sites to reference genes of the NCBI human genome database (Hg18). Recent genomic analyses have indicated that for many E2-regulated genes, the ER binding sites are located far distal from promoters of their target genes, whereas for other target genes, the receptor binding sites reside within their promoter regions. We therefore determined the number of genes that have high-confidence binding sites for SRC-3, Pol II, ER, and FoxA1 within different ranges of RefSeq gene annotations (Table 1). Like ER, SRC-3 does not show an overall preference for gene-proximal locations; less than 3% of all SRC-3 and about 4% of ER binding sites are in classical promoter regions (±500 bp from transcription start site, TSS), and only 13–15% map to an extended promoter region (EPR; 7.5 kb upstream to 2.5 kb downstream of TSS) often covered by commercial promoter arrays. About 61–65% of SRC-3, ER, and FoxA1 sites, but 95% of Pol II sites, were found within ±10 kb of gene-transcribed regions (Table 1), suggesting that about 40% of SRC-3, ER, and FoxA1 ChIP binding sites map to remote DNA locations and intergenic regions.
Table 1.
Tabular comparison of genome-wide location analyses for SRC-3, RNA Pol II, ER, and FoxA1 binding sites
| ChIP assay | SRC3_v
|
SRC3_E2
|
Tx_v
|
Tx_E2
|
ER
|
FoxA1
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. | % | No. | % | No. | % | No. | % | No. | % | No. | % | |
| 100 | ||||||||||||
| Total BS | 9,552 | 100 | 12,294 | 100 | 19,453 | 100 | 22,594 | 100 | 5,782 | 100 | 12,904 | 100 |
| BS | 7,395 | 77.40 | 9,815 | 79.80 | 19,022 | 97.80 | 22,199 | 98.30 | 4,682 | 81.00 | 9,716 | 75.30 |
| Gene hits | 24,711 | 33,852 | 118,048 | 138,929 | 16,422 | 30,556 | ||||||
| Nrd genes | 13,126 | 40.10 | 14,944 | 45.70 | 20,589 | 62.90 | 20,796 | 63.50 | 9,184 | 28.10 | 15,879 | 48.50 |
| G10 BS | 5,825 | 61.00 | 7,651 | 62.20 | 18,314 | 94.10 | 21,469 | 95.00 | 3,733 | 64.60 | 8,170 | 63.30 |
| Gene hits | 7,078 | 9,369 | 33,915 | 39,731 | 4,674 | 9,920 | ||||||
| Nrd genes | 4,493 | 13.70 | 5,364 | 16.40 | 12,794 | 39.10 | 13,281 | 40.60 | 3,143 | 9.60 | 6,011 | 18.40 |
| EPR BS | 1,198 | 12.50 | 1,617 | 13.20 | 10,889 | 56.00 | 12,695 | 56.20 | 870 | 15.00 | 1,693 | 29.30 |
| Gene hits | 1,376 | 1,848 | 14,599 | 17,035 | 980 | 1,997 | ||||||
| Nrd genes | 1,267 | 3.90 | 1,610 | 4.90 | 9,825 | 30.00 | 10,571 | 32.30 | 887 | 2.70 | 1,853 | 5.70 |
| P BS | 228 | 2.40 | 331 | 2.70 | 7,509 | 38.60 | 8,601 | 38.10 | 244 | 4.20 | 521 | 9.00 |
| Gene hits | 247 | 353 | 8,681 | 9,956 | 256 | 582 | ||||||
| Nrd genes | 246 | 0.80 | 348 | 1.10 | 8,029 | 24.50 | 8,781 | 26.80 | 251 | 0.80 | 578 | 1.80 |
| Total | 32,735 | 100 | ||||||||||
| RefSeq genes | ||||||||||||
Total number of high-confidence ChIP sites (Total BS) and counts for binding sites (BS), genes (Gene Hits), and nonredundant genes (Nrd genes) are given for different gene regions: TSS ±100 kb (100); the gene-transcribed region ±10 kb (G10); EPR, TSS −7.5 kb, +2.5 kb (EPR); and TSS ±0.5 kb (P). Percentage calculations for genes (italic) are relative to 32,735 nonredundant NCBI RefSeq genes.
Within the genomic organization of genes, we found a remarkably high number of SRC-3 sites in introns. When normalized for the average length of exon and intron sequences (316 and 5747 bp, respectively, according to the NCBI reference assembly for the human gene model), SRC3_E2 as well as SRC3_v binding sites were found to be enriched in introns and underrepresented in exons and in the gene 3′-flanking 10-kb regions (Fig. 1C). The positioning of SRC-3 sites within intronic sequences appeared random. Although the counts for SRC3_E2 sites in the gene 5′ 10-kb region was as expected for a sequence of that length, SRC3_v sites were found underrepresented in this region, indicating that the number of SRC-3 binding sites in MCF-7 DNA promoter sequences is dependent on estrogen signaling in the cell.
A visual interrogation using the University of California Santa Cruz (UCSC) Genome Browser illustrated that many SRC-3 sites coalign with ER and FoxA1 binding sites also within the gene-transcribed regions; this observation is in agreement with our genome-wide binding sites proximity calculations. We found a considerable number of SRC-3 sites mapping to FoxA1 sites that have no partnering ER binding site, hinting at a hitherto undocumented possible shared biology between SRC-3 and FoxA1. Furthermore, we noticed uneven distributions of the binding sites; although many genes not unexpectedly have few sites for ER, FoxA1, and SRC-3, we found a respectable number of genes that have multiple SRC-3 and FoxA1 binding sites spread over their entire transcribed region. A genome-wide binding site frequency distribution calculation in fact identified many more genes with entire arrays of high-confidence SRC-3 or FoxA1 binding sites than genes with comparable numbers of ER sites (Fig. 1D). Such a clustering of 10 or more DNA affinity sites for SRC-3 and FoxA1, many of which coalign, is different from the more anchorite DNA binding of ER and implies a SRC-3 biology that is distinct from just serving as a transcriptional coactivator for ER.
Redundant binding sites, however, may not necessarily convert into a different, or a more pronounced, use of a target gene than a single high-affinity site can impart on gene expression. We thus determined the number of genes that have any number of ChIP binding sites for SRC-3, Pol II, ER, and FoxA1 in MCF-7 cells. Because we already established a preference of SRC-3 for intron sequences, in addition to analyzing the EPR normally selected for target gene evaluations for TFs, we also interrogated the entire gene-transcribed region including the gene 10-kb flanking sequences. Venn representations of the binary relations and intersections of gene counts with respective ChIP binding sites strikingly illustrate an intragenic preference of SRC-3 binding as well as a predilection of SRC-3 to associate with genes that also bind ER and FoxA1 (Fig. 1E), hinting at a shared biology of SRC-3 with both TFs.
Integrated expression analysis
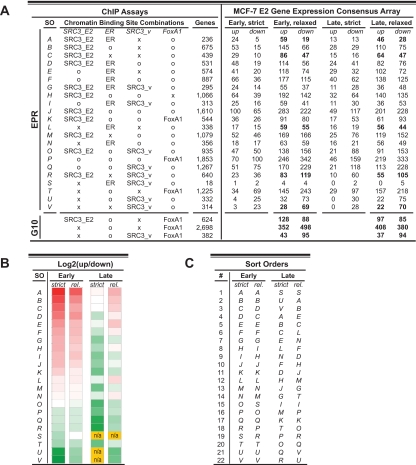
To identify possible target genes for SRC-3, and to better understand the global features of SRC-3-mediated ER transcription, we next correlated ChIP binding with E2-regulated gene expression. A large number of independent genome-wide RNA expression profiles have been made available for MCF-7 cells, allowing us to exploit the abundance of data and to work with consensus expression signatures that resulted from common array features across 10 independent microarray sets (32). This MCF-7 E2 gene expression consensus array has 13,182 genes, which we analyzed for SRC-3, ER, and FoxA1 ChIP binding sites in the EPR for the effects of E2 at an early (3–4 h) or late (24 h) time point. A high-stringency evaluation (strict), which required a gene to have the same direction of regulation in all microarray datasets, allowed 1446 genes at the early time point and 1649 genes late, whereas a more relaxed cutoff offered 7589 genes early and 6778 genes late for our analyses. The consensus array has about as many genes up-regulated as it has down-regulated at the early time point (713 [3609] up-regulated, 733 [3980] down-regulated), results from the relaxed analysis shown in closed brackets, whereas there are more E2 down-regulated expression signatures for the late time point (Supplemental Material SM4).
Figure 2 summarizes our analyses by listing gene expression signatures for targets on the consensus array that have different ChIP binding site combinations in the EPR (Fig. 2A) and by heat-map representations of the fold differences between the numbers of up- and down-regulated genes at early and late time points (Fig. 2B). Overall, our analyses suggest that E2-induced binding of SRC-3 to promoter regions supports the up-regulation of early target gene expression. For instance, potential target genes that have SRC3_E2 and FoxA1 binding sites in the EPR show more early up-regulated expression signatures than genes that have FoxA1 but don’t have SRC-3 sites (Fig. 2A, rows K and T, respectively). Genes that have both, ER and SRC3_E2, but don’t have SRC3_v binding sites in the EPR, constitute the gene group with the highest percentage of early E2 up-regulated genes (83% [76%], row A). Genes with SRC3_v binding sites in the EPR, on the other hand, show overall more down-regulated expression signatures; down-regulation is especially striking for genes that do not have additional binding sites for SRC3_E2 (row U) or ER (row V). The presence of SRC-3 in promoters of target genes that are down-regulated by E2 upon dissociation of the coregulator from the DNA implies that SRC-3 in the absence of E2 imparts a different biology on E2 target genes than it does during estrogen signaling.
Figure 2.
Integrated MCF-7 E2 gene expression analysis. Consensus gene expression signatures from multiple independent MCF-7 RNA microarray experiments were analyzed for direction of E2-induced expression and promoter occupancy for SRC-3, ER, and FoxA1. Overall, E2-induced SRC-3 binding to promoters correlates with early up-regulated expression profiles. A, Gene counts for potential targets that have different combinations of SRC3_v, SRC3_E2, ER, and FoxA1 binding sites (left) in the EPR or in gene-transcribed regions ±10 kb (G10); counts are for genes on the NCBI reference genome assembly and for genes on the MCF-7 E2 consensus array that show agreement in gene expression signatures at 3–4 h (early) and at 24 h (late) of E2 exposure. A stringent (strict) analysis required consensus with regard to the direction of expression in all arrays (five arrays for the early, seven arrays for the late time point), whereas a different cutoff (relaxed) demanded agreement in the direction of expression outside the log2(fold change) greater than or equal to ±0.15 in four arrays for the early and in five arrays for the late time point. O, Optional (binding site may or may not be present); SO, sort order (sorted for a gradual showing of the early-strict gene counts); X, absence of respective binding site. Additional information on the MCF-7 E2 gene expression consensus array is provided in Supplemental Material SM4. Numbers in bold indicate gene sets used for the functional classification of E2 gene regulation using the DAVID public resource (provided in Supplemental Materials SM5 and SM6). B, Heat-map representations of the fold differences between numbers of up- and down-regulated genes [log2 (up/down)] at early and late time points and for both stringencies of analysis: strict and relaxed (rel.). The log2 values range from 2.26 (red) to −2.94 (green). n/a, Not applicable due to zero gene counts for dividend or divisor. Sort order (SO) is the same as in A. The heat-maps illustrate that multiple binding site combinations result in E2 up-regulated expression signatures at the early, and overall down-regulated E2-signatures at the late time point. C, Different sort orders of the fold differences for both stringencies and both time points of the gene counts shown in A. Pair-wise comparisons of equal letters between strict and relaxed analysis show close positional neighborhoods at different sort orders, indicating similar overall expression profiles for both stringencies of analysis at the early time point.
The fold differences between up- and down-regulated gene expression signatures for most binding site combinations turned out highly similar for both stringencies of evaluation (Fig. 2C), which encouraged us to use the larger gene sets obtained from the relaxed analyses for ontology studies to possibly correlate functional gene regulation with promoter occupancy.
Functional classification of gene regulations
We next interrogated groups of prospective SRC-3, ER, and FoxA1 target genes that share ChIP binding signatures and that have common expression profiles on the MCF-7 E2 gene expression arrays for enrichments of biological themes such as gene ontology terms, function-predicting protein domains, and enzymatic activities and pathways. We used the web-based DAVID Bioinformatics Resources database for finding clusters of biological themes in the related gene groups; a synopsis of the analysis and lists of enriched annotation terms for the gene groups analyzed are provided as Supplemental Material SM5.
Pair-wise comparisons of enriched terms between groups of genes that only have SRC3_v or SRC3_E2 or ER binding sites in the EPR produced no more than empty intersections when a threshold cutoff of P < 0.1 was used on the annotations. Likewise, not a single enriched ontology was found shared between sets of genes that either have SRC3_E2 and ER ChIP binding sites, or only have ER or SRC3_E2 binding sites, respectively (Supplemental Material SM5), indicating that gene expression controlled by SRC-3 and ER put forth different biogenic responses than target genes that have binding sites for either factor alone. Albeit showing a more disbanded biology affecting many cellular systems, such distinct E2-dependent SRC-3 biology also was indicated for genes that have SRC-3 and FoxA1 binding sites in the gene-transcribed region, because none of the enriched functional annotations were shared between FoxA1 targets that also have SRC3_E2, or also have SRC3_v sites, respectively (Supplemental Material SM5).
Overall, the functional classification of gene expression as investigated in this study produced distinct classes of enriched biological themes that showed rather low functional redundancies, suggesting a quite subtle transcriptional control of cellular pathways by SRC-3 in response to estrogen and in combination with ER and FoxA1.
Cooperating transcription factors
The intricate transcriptional control of interdependent cellular pathways and interpretation of estrogen signaling by SRC-3 raises the question of how a single transcription coregulator can impart such levels of signaling interpretation power. The association of SRC-3 with other distinct cooperating TFs would constitute protein complexes that are formed differently depending on promoter or distal cis-regulatory enhancer sequences and hence are likely to regulate receptor activity and coregulator recruitment distinctly.
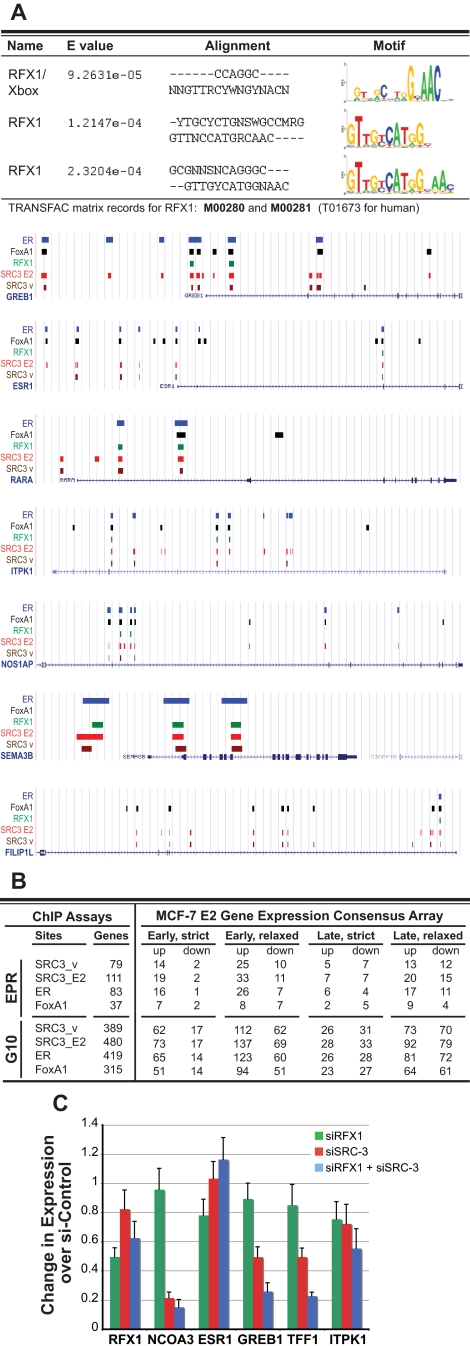
A search for enriched motifs in the SRC3_E2 ChIP dataset revealed the anticipated ER response elements and FoxA1/HNF3α binding sites as well as 24 of the 58 motifs identified for the ER cistrome (21), including motifs for AP-1, BACH2, and many different GATA and forkhead TFs (Supplemental Material SM6). We also found ER cooperating factors such as c-MYC, SP1, and different PAX binding motifs previously reported by other groups (22,28) as well as potential binding sites for NRs (ER, ERR, GR, HNF4α, PPARα, PPARγ, PXR, RORα, RORβ, RXRα, and VDR), for the zinc finger proteins ZNF219 and ZNF423, and DNA binding motifs for other TFs such as NF-κB, p53, Nkx2-5, and SMAD4 as well as XBP1 and regulatory factor X1 (RFX1) (Supplemental Material SM6).
The RFX1 DNA binding motif (Fig. 3A) was unexpectedly found in 695 SRC3_E2 ChIP binding sites, which mapped to 480 genes within the 10-kb gene margin and to 111 genes within the EPR. We focused our attention to RFX1 as a predictive test of the combinatorial power of our cistromic, transcriptomic, and proteomic approaches. Potential RFX1/SRC-3 target genes are up-regulated early by E2 in MCF-7 cells (Fig. 3B); this expression signature is 94% [79%] up if the genes also have ER binding sites in the promoter. Notable potential RFX1/SRC-3 target genes by motif sequence association are the known ER targets pS2 (TFF1), growth regulation by estrogen in breast cancer 1 (GREB1), IGFBP4, WISP2, RARα, EEIG1 (FAM102A), PTPγ (PTPRG), and cyclin D1 (CCND1), all of which are early up-regulated by E2 in MCF-7, and the NRs ER and RXRα, which show down-regulated E2 signatures on the consensus array.
Figure 3.
RFX1 likely is a cooperating TF for SRC-3/ER-E2 transcription. Sequence motif analysis as well as RNA expression and biochemical interaction data together suggest RFX1 as an auxiliary factor for a subset of SRC-3/ER-target genes. A, Top, RFX1/X-box DNA binding motifs as analyzed using the STAMP resource were enriched in SRC3_E2 ChIP sequences; below, UCSC Genome Browser illustrations of the genomic locations of RFX1 motif sequences (green) and ChIP sequences for SRC3_v, SRC3_E2, FoxA1, and ER for select target genes. Sequence positions and other generic UCSC annotations were removed for clarity. B, Tabular representation of gene counts for potential RFX1 target genes with additional respective ChIP binding sites for ER, FoxA1, and SRC-3 in the EPR or in the gene-transcribed regions ±10 kb (G10) and for genes on the MCF-7 E2 gene expression consensus array. Genes with RFX1 sequence motifs show overall E2-up-regulated expression signatures. C, RNAi experiments show that RFX1 and SRC-3 cooperate in controlling GREB1 and pS2 (TFF1) gene expression in untreated, cycling MCF-7 cells transfected with nontargeting siRNA controls or siRNA pools directed to RFX1 or to SRC-3, as described in Materials and Methods. mRNA levels for RFX1, SRC-3 (NCOA3), and ER (ESR1) as well as for the predicted targets GREB1, pS2 (TFF1), and ITPK1 were determined by qPCR. Results shown are the average of three individual RNAi experiments with three quantifications for each target gene. Error bars show sd. Expression levels were normalized to nontargeting control siRNA.
A functional classification of MCF-7 E2-up-regulated genes with binding sites for all three factors, RFX1, SRC-3, and ER, produced enriched ontologies such as developmental processes, anatomical structure development/morphogenesis, and disease mutation. Gene list comparisons with external databases showed that 80 (17%) of the potential RFX1/SRC-3 target genes have human disease associations in Morbid Map or are listed (with a 2.7-fold overrepresentation) in cancer candidate gene databases (Supplemental Material SM7).
RNA interference directed against RFX1 and SRC-3 showed that both factors cooperate in controlling transcription of at least a subset of putative target genes. Although different small interfering RNA (siRNA) pools against RFX1 showed only limited interference in our assays, the reduction of RFX1 expression was sufficient to significantly lower expression of the GREB1 and the pS2 (TFF1) genes when endogenous SRC-3 levels also were limited in MCF-7 cells (Fig. 3C).
A physical association in vivo between SRC-3, ER, and RFX1 is supported by mutual affinity coprecipitation experiments we carried out in MCF-7 cells. We detected SRC-3 and ER protein in immunopurified RFX1 complexes by Western blotting (data not shown), and reciprocally, we also identified ER and RFX1 protein in SRC-3 precipitates by MS (Fig. 4). Together with the genomic colocalization of independent DNA binding sites and the resulting collective consensus expression patterns of potential target genes, these biochemical data imply a previously unknown protein complex that brings together, among other proteins, ligand-activated ER, coactivator SRC-3, and transcriptional activator RFX1 for early up-regulation of E2-responsive target genes. Although the RFX1 TF has not yet been functionally linked to ER-controlled gene expression, our results demonstrate the power of a combined genome-wide location analysis, microarray expression, and proteomic analysis and strongly suggest RFX1 to be a hitherto uncharacterized cooperating factor for ligand-activated, SRC-3-coactivated ER transcription.
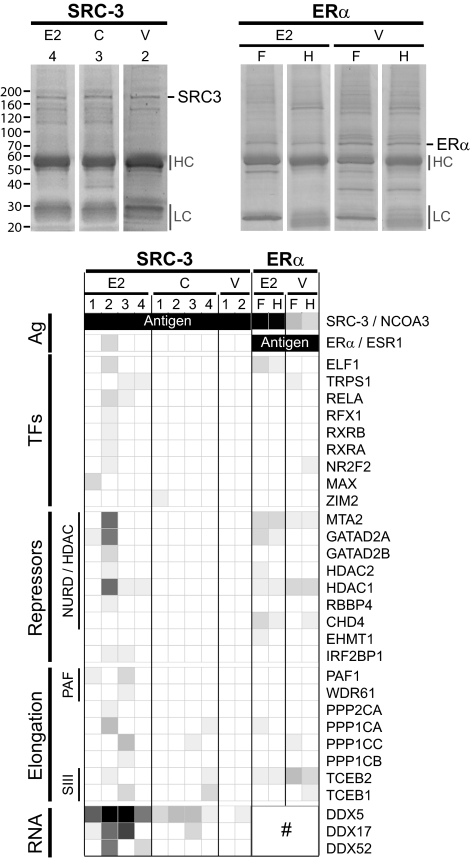
Figure 4.
SRC-3 and E2 define proteomics context for ER transcription. IP/MS identified treatment-dependent SRC-3 and ER interactomes from nuclear extracts of E2-induced and untreated MFC-7 breast cancer cells. Top, Photographs of Coomassie Blue-stained SDS-PAGE of SRC-3 (left) and ERα (right) IPs from nuclear extracts of E2-treated or control cells (C, untreated cycling cells; V, hormone-starved, ethanol vehicle-treated cells). Gel annotations are for the antigens (SRC-3, ERα) and select experiments [4, 3, and 2 for SRC-3 IPs with a mixture of A300-347A and A300-348A antibodies; F and H for ER IPs using F-10 (sc-8002) and HC-20 (sc-543) antibodies], for Ig (HC, high chain; LC, low chain), and molecular size markers (left). Bottom, Grid-map representation of relative protein abundances of SRC-3 and ER-associated proteins in complexes identified by IP/MS experiments. This compilation is a highly restricted list; more detailed analyses are provided in Supplemental Material SM8. Enriched proteins that show discernable differences in their associations in different cell treatment conditions are arranged in groups as annotated to the left: top two rows of the graph show the antigens (Ag), followed by transcription factors (TF), repressor proteins (Repressors) including components of the NURD/HDAC complex, proteins that function in transcription elongation (Elongation) such as subunits of the PAF-1 protein complex and of the transcription factor B (SIII) complex, and components of the RNA processing machinery (RNA). Gene symbols of the identified proteins are shown to the right. Changes in DDX5, DDX17, and DDX52 associations with ER cannot be evaluated because both antibodies used for ER IPs show cross-reaction with these DDX proteins (marked with #).
SRC-3 and estrogen define proteomics context for ER transcription
The functionally distinct gene regulation by SRC-3 and its DNA colocalization with different cooperating TFs imply different protein complexes that likely are formed depending upon particular cell signals that elicit specific functions in transcription. We thus further studied ER- and SRC-3-associated proteins by co-immunoprecipitation (co-IP) of in vivo steady-state protein complexes in MCF-7 cells followed by identification of complex components by MS.
As predictable for a ligand-activated NR, we found different proteins associated with ER depending on whether or not E2 was at the receptor’s disposal in the cell. In the nuclear fraction, we isolated apo-ER associated with proteins that are involved in chromosomal fiber formation, DNA repair/fidelity, and ubiquitin-dependent protein catabolic processes. These proteins were either not present or identified at largely diminished quantities in the complex formed by ligand-activated ER, which we isolated together with SRC-3, different TFs (e.g. SP1), and many components of the nucleosome remodeling and histone deacetylation complex (NURD/HDAC) repressor complex (Fig. 4 and Supplemental Material SM8). SRC-3 always was the highest enriched protein when E2-induced complexes were compared with ER precipitates from vehicle-treated cells. We also copurified a total of 26 known NR coregulators along with nuclear ER (Supplemental Material SM8), including the NR interacting protein 1, ER-associated protein 140 (NCOA7), and the estrogen-responsive finger protein TRIM25. The erythroblast transformation specific domain E74-like protein ELF1 was the highest enriched TF to be associated with liganded ER, whereas the E2-responsive GREB1 protein, thought to be critically involved in hormone-induced growth of breast and prostate cancers (33,34), was only found together with apo-ER (Supplemental Material SM8). IPs using different ER antibodies yielded a high degree of similarity in the identified proteins, suggesting a rather robust and stable nature of ER complexes.
The situation was different for SRC-3, which protein complexes turned out to be rather dynamic and cell-state dependent. We identified quite different sets of proteins associated with SRC-3 in untreated (cycling) and E2-treated MCF-7 cells, whereas in vehicle-treated cells, distinctiveness in the proteomics context of SRC-3 was made apparent by the conspicuous overall absence of enriched associated proteins (Fig. 4 and Supplemental Material SM8), likely reflecting a cell state for which a response was not evolved. Many of the identified SRC-3-interacting proteins participate in transcription and in signal transduction, supporting the role of SRC-3 as integrator of intracellular signaling into transcriptional control. The preference for such interaction partners in cycling cells suggests E2-independent pathways for SRC-3 in converging cell signals to transcription units for cell homeostasis and cell fate determination.
In the nuclear fraction of E2-treated MCF-7 cells, in addition to ER and RFX1, we identified many TFs that previously have been functionally liked to ER biology, such as the NRs RXRα, RXRβ, and COUP-TFII (NR2F2), the MYC-associated factor X (MAX), and the NF-κB p65 subunit RELA as well as enhanced at puberty 1 (EAP1/C14orf4), which is a transcriptional regulator of the female neuroendocrine reproductive axis (35), and deleted in breast-cancer 1 (DBC-1/KIAA1967), which is a principal determinant of ER expression and survival function in human breast cancer cells (36). We also identified protein-modifying enzymes functionally implemented in various signaling axes and in transcription elongation and RNA biology, such as subunits of the TF B (SIII) complex, catalytic subunits of protein phosphatase 1 and RNA Pol II-associated factor PAF1 as well as DEAD box proteins and RNA binding motif proteins (Fig. 4 and Supplemental Material SM8). These findings suggest that in E2-treated MCF-7 cells, SRC-3 congregates cell signals triggered by estrogenic responses not only by direct modulation of ER transactivation but also for accenting distal components of transcription, including elongation and splicing (see Discussion).
Quite remarkable when considering the overall up-regulation of SRC-3 target genes by E2 is the collection of corepressor proteins associated with SRC-3 in E2-stimulated cells. With HDAC1 and HDAC2, the HDAC-interacting metastasis-associated protein 2 (MTA2), the GATA zinc finger domain containing 2A and 2B (GATAD2A/2B), and retinoblastoma binding protein 4 (RBBP4), we identified the major components of the NURD repressor complex (37) as SRC3-E2-associated proteins. Because all these corepressor proteins showed relative enrichments for SRC-3 association if compared with respective proteins identified in ER-E2 extracts (Fig. 4), it is possible that, in addition to each factor having is own set of chromatin targets, it is SRC-3 that also brings NURD and other transcriptional corepressors into E2-responsive transcription units.
Overall, the number and diversity of enriched proteins identified in complexes in extracts from different cell treatment conditions hint at the key role SRC-3 is thought to play in responding to cellular signals. By assembling distinct protein complexes, SRC-3 conducts different signals to the transcription unit and also uses distinct sets of DNA binding sites to direct target gene expression.
Discussion
In this study, we report the genome-wide identification of SRC-3 chromatin affinity sites in MCF-7 breast cancer cells, which we then mapped to the ER and FoxA1 cistromes and to consensus MCF-7 microarray E2 expression signatures. We used enriched functional annotations from target gene sets with common E2 expression and promoter binding site patterns to further delineate SRC-3 functions in transcription and used biochemical characterizations of ER- and SRC-3-associated proteins to provide a molecular framework for SRC-3 in interpreting estrogen signaling.
SRC-3, ER, and FoxA1 chromatin binding sites
As expected based on their concerted action in transcription, we found a significant positional overlap between SRC3_E2 and ER ChIP binding sites, including on promoters for known SRC-3 target genes such as TFF1, XBP1, DSCAM, NRIP1, and SOD1 (20) but also on promoters for previously unrecognized ER targets, such as ERBB2, which only recently was shown to be under the control of ER and SRC-3 (29). The fact that we found more gene proximal SRC3_E2 sites than ER binding sites that have been reported previously, we could attribute to SRC-3 also binding to ERβ, although ostentatious SRC-3 and undocumented ER binding sites due to aggressive or restrained peak calling are also possible. Nonetheless, because evolutionarily conserved near-consensus ER binding sites occur much more frequently than indicated by available ChIP data (38,39), the SRC3_E2 chromatin affinity sites aid our identification of hitherto unrecognized ER target genes.
A startling finding was the striking number of DNA coalignments between SRC-3 and FoxA1 binding sites. Both factors were reported to be indispensable for maximal E2 response in MCF-7 cells: SRC-3 by virtue of its transcriptional coactivation function (18,19) and FoxA1 by defining a cis-regulatory unit for ER transactivation (20,27). Coalignments of SRC-3 and FoxA1 sites were mostly observed in the gene-transcribed regions, and often in the absence of ER binding sites. Yet, we could not detect a possible physical association between SRC-3 and FoxA1 in reciprocal IPs followed by Western or MS identification (data not shown). Additional experiments, which are beyond the scope of this paper, are required to examine the suspected shared spatiotemporal biology of this observation.
Chromatin expression domains and fuel stations
The particular array-type distribution of intragenic SRC-3 and FoxA1 binding sites beg for interpretations. First, like FoxA1, SRC-3 by its own right can play the role of a pioneer factor by anchoring enzymatic activities (e.g. histone acetyltransferase) to nucleosomal DNA for successive chromatin unfolding and maintenance of open chromatin. With many transcript lengths exceeding 10 kb, it is natural to think that chromatin-unfolding mechanisms may exist along the entire gene-transcribed region to permit uninterrupted DNA Pol II activity and transcript extension.
Second, SRC-3-containing protein complexes strategically positioned along a gene could serve as refueling stations for the transcription machinery. The fuel ought to be the intrinsic transcription coactivation function of SRC-3 itself and/or enzymatic activities from SRC-3-interacting proteins brought to the intragenic chromatin for use by RNA polymerase. To this end, we have identified two components of the PAF1 protein complex that were associated with SRC-3 in nuclear extracts of E2-treated MCF-7 cells (PAF1 and WDR61, Fig. 4). The PAF1 complex was reported to bind throughout the coding regions of genes and to be responsible for Ser2 phosphorylation of the heptapeptide repeats in the carboxyl-terminal domain of the POLR2A subunit of RNA Pol II (40,41). PAF1 also is required for methylation of histone H3 on lysine 4 (H3K4me) (42,43). Hypermethylated H3K4 is the epigenetic signature that defines lineage-specific FoxA1 recruitment sites in chromatin (31), therefore linking transcriptional elongation to chromatin methylation and providing an interpretation for shared SRC-3 and FoxA1 DNA binding sites.
Third, we also identified DEAD-box (DDX) and other RNA-binding proteins in SRC-3 IPs. DEAD-box RNA helicases function in all aspects of RNA biology (44). Some, like DDX17 (p72) and DDX5 (p68), associate with and coregulate NRs, thereby also playing roles in the regulation of alternative splicing of genes driven by steroid-sensitive promoters (45,46,47). DDX17 and DDX5, for examples, were shown to act as ER subtype-selective coactivators by bridging activation domains of ER with a hetero-nucleoprotein coactivator complex that also contains steroid receptor RNA activator (SRA) (48) and SRC family proteins (49). Altogether, and in concert with the genome-wide location analysis, our proteomics results provide further support and biochemically link SRC-3 to transcriptional regulation beyond the initiation steps.
Corepressor complexes
The strong identification of components of the NURD corepressor module in E2-induced SRC-3 and ER complexes raises the questions of whether corepressors are required for ER/SRC-3-controlled transcription. Although repressors associated with E2-activated ER can account for possible repressive activities on a subset of E2 target genes (50,51), this repression is likely to occur independently, because genes that share E2-induced binding of SRC-3 and ER are preferentially up-regulated in MCF-7 E2 cells (Fig. 2). Similarly, also RFX1 was reported to interact with HDAC1 to repress target gene expression (52,53), yet we find RFX1 in both ER and SRC-3 precipitates and by ChIP sequence alignments on promoters of ER target genes that form the tightest clusters of E2 up-regulated genes on the consensus expression array. At the same time, because SRC-3 and ER show enrichment in HDAC1 and other transcriptional corepressors, it is equally possible that corepressor complexes are recruited by the binary ER/SRC-3 complex to E2 target genes. This would indicate an extraordinary role of HDACs in transcriptional activation and also suggest a new role for SRC-3 in coordinating transcription by possibly neutralizing forceful deacetylase activities. Such a swing function in transcription control has recently been reported also for the NURD-corepressor complex via the previously unrecognized function of MTA1 in stimulating BCAS3 or PAX5 expression (54,55). Finally, it is possible also that a gene must be repeatedly reset by activators and repressors to maintain efficient repetitive transcription. To this end, corepressors have been reported to enhance transcription of select target genes in cells (56,57,58). On the other hand, deacetylase activity brought into the gene-transcribed region may render the chromosomal structure unfavorable for repetitive transcription, marking the gene as transcribed, which would attribute a memory function to SRC-3. More detailed studies are required to distinguish between these possibilities.
In summary, by bridging genome-wide location and expression analyses with targeted proteomics studies of multiprotein complexes, we present an approach in complementary comparative analyses that converges results obtained by discovery-driven methods into testable hypotheses. Such observations provide valuable resources for follow-up studies by other investigators that will further our understanding of E2 signaling and extend the value of SRC-3 in conveying key physiological functions by congregating and interpreting cellular signals to transcription units for controlled target gene expression.
Materials and Methods
Cell culture and ChIP
We adhered to previously described ChIP protocols to allow our results to be compared with other global cartographies of ER and FoxA1 binding sites (see introductory section for references). MCF-7 cells were maintained in DMEM supplemented with 10% fetal bovine serum (FBS), nonessential amino acids, and antibiotic/antimycotic. For stimulation with E2, MCF-7 cells were cultured in phenol red-free DMEM containing 5% charcoal/dextran-treated FBS for 72 h, followed by the addition of 10 nm E2 (or ethanol vehicle). After 1 h, cells were fixed with 1% formaldehyde for 30 min and quenched with 0.125 m glycine. Preparation of chromatin and ChIP was carried out essentially as described by Soutoglou and Talianidis (59). Anti-SRC-3 antibody was sc-7216 from Santa Cruz Biotechnology (Santa Cruz, CA). ChIP enrichment of known SRC-3 target sites were assessed by quantitative real-time PCR (qPCR).
ChIP sequencing
ChIP and input DNA samples were amplified using the Illumina ChIP-Seq DNA sample prep kit (Illumina, Inc., San Diego, CA). Briefly, the ends of DNA fragments were first converted into phosphorylated blunt ends using T4 DNA polymerase, Klenow enzyme, and T4 polynucleotide kinase. The blunt-ended fragments were prepared for ligation by adding an adenine to the 3′-ends using Klenow fragment (3′–5′ exo minus) and dATP. Illumina adaptors were added using DNA ligase. The library was size selected to a length of 175–225 bp on a 2% agarose gel and then amplified for 18 cycles with Phusion polymerase (Finnzymes, Espoo, Finland). The resulting DNA libraries were tested by qPCR at the same specific genomic regions as the original ChIP DNA to assess the quality of the amplification reactions. DNA libraries were sent to Illumina Sequencing Services for sequencing on a Genome Analyzer II. Sequences (35 bases) were aligned to the human genome using the Eland software.
Sequence analysis and data handling
The model-based analysis of ChIP-sequencing (MACS) software (60) was used to normalize ChIP sample against MCF-7 input control. A P value cutoff of 1e−10 produced the ChIP sequences presented in this work. The ER and FoxA1 ChIP-chip raw data from Affymetrix tiling arrays (21,31) were reanalyzed using the model-based analysis of tiling-arrays (MAT) algorithm (61) and updated it to the most recent human genome sequence (Hg18) using the xMAN program (62). ChIP binding sites were mapped to genes as curated by the NCBI reference genome assembly (Build 36.3, March 2008), which annotations were downloaded from the NCBI Map Viewer FTP site and integrated into a database-driven network of relational tables designed using guidelines based on the normalization theory and the concept of constraints to enforce data integrity (63). All data handling (including establishing gene associations, filtering of microarray expression signatures and analyzing clusters of functional annotations for pulled gene lists with common expression and binding site signatures as well as the filtering of proteomics IP/MS data) were carried out using custom-made software running on FileMaker/Apple MacIntosh platforms. A local copy of MDscan (64) was used for motif finding, and DNA-binding motif similarities were analyzed using the STAMP web server (65). For gene-functional classifications, we used the public Database for Annotation, Visualization, and Integrated Discovery (DAVID, http://david.abcc.ncifcrf.gov/) (66) running default standard conditions; batch queries produced clusters of annotations that we further analyzed for common terms between different query sets using a P value cutoff of 0.1 on the annotations. JColorGrid software (67) was used for transforming numerical data into color grids for Figs. 2 and 4 and Supplemental Material SM8.
MCF-7 E2 gene expression consensus array
The consensus array is based on a weighted metaanalysis across 10 independent published datasets that address the effect of E2 in MCF-7 cells at early (3–4 h) and late (24 h) time points (32) and made available through the NURSA web site (www. nursa.org/gems). Gene expression signatures were queried with two different stringencies: a strict analysis required consensus with regard to the direction of E2-regulated gene expression (all up or all down) as well as combined q-value (combined measure of significance corrected for multiple testing by using a false discovery rate method) of less than 0.05 in all individual arrays (five arrays for the early, seven arrays for the late time points), whereas the relaxed analysis looked for consensus in the direction of expression outside the log2(fold change) greater than or equal to ±0.15 in four arrays for the early and in five arrays for the late time point. Supplemental Material SM4 has additional information on the MCF-7 E2 consensus array.
RNA interference (RNAi), RNA isolation, and qPCR
ON-TARGET plus siRNA duplex SMARTpools against RFX1 and SRC3 as well as nontargeting control were purchased from Dharmacon Inc. (Lafayette, CO). siRNA was delivered to MCF-7 cells using Lipofectamine RNAiMAX reagent (Invitrogen, Carlsbad, CA) according to manufacturer’s instructions. After 48–60 h RNAi, MCF-7 cells were collected and total RNA was isolated using QIAGEN (Valencia, CA) RNeasy mini kit. Approximately 500 ng RNA was used for cDNA generation using Superscript III reverse transcriptase (Invitrogen). Real-time SYBR Green qPCR measurements were done on an ABI 7700 thermocycler (Applied Biosystems, Foster City, CA) with primers specific for the target genes (see Supplemental Material SM9).
Nuclear extract preparation
Nuclear extracts from cycling (DMEM with phenol red; 5% FBS), vehicle-treated, or E2-treated (10−8 m for 2 h in DMEM without phenol red, 5% charcoal-stripped FBS) MCF-7 cells were prepared using modified Dignam’s method essentially as described (68). All buffers in this procedure were supplemented freshly with 10 mm β-mercaptoethanol and protease inhibitor cocktail (phenylmethylsulfonyl fluoride, pepstatin A, leupeptin, and benzamidine). Briefly, cells were trypsinized, collected by centrifugation, and washed in PBS buffer. Cells were then swollen in 10 vol hypotonic buffer (10 mm Tris-HCl, pH 7.5; 1.5 mm MgCl2; 10 mm KCl) for 10 min on ice, collected by centrifugation at 1000 × g, and homogenized using loose pestle B in a glass Dounce homogenizer. Nuclei were pelleted by centrifugation at 3700 × g for 15 min and then resuspended in 0.5 vol low-salt buffer (20 mm Tris-HCl, pH 7.5; 1.5 mm MgCl2; 300 mm KCl; 0.2 mm EDTA; 25% glycerol). Nuclear proteins were extracted by drop-wise addition of high-salt buffer (same as low-salt buffer except 900 mm KCl was used), and nuclear debris were pelleted by centrifugation at 25,000 × g for 20 min at 4 C. Nuclear extracts were then dialyzed in BC-150 buffer (20 mm Tris-HCl, pH 7.5; 0.2 mm EDTA; 150 mm KCl; 20% glycerol) until extract KCl concentration reached 160 mm. Nuclear extracts were then cleared from precipitate by another 20-min round of centrifugation at 25,000 × g, aliquoted, and snap-frozen in liquid nitrogen.
IP and MS identification of SRC-3 and ERα complexes
For each IP experiment, 1 ml (approximately 10 mg total protein) of nuclear extract was used per treatment condition. Extracts were precleared by centrifugation at 100,000 × g for 20 min at 25 C before antibody addition. Nine micrograms of anti-SRC-3 antibody (mixture of A300-347A and A300-348A; Bethyl Laboratories Inc., Montgomery, TX) or either one of the three anti-ERα antibodies from Santa Cruz Biotechnology [D-12 (sc-8005), F-10 (sc-8002), or HC-20 (sc-543)] were incubated with extracts for 4 h at 4 C. Extracts were then cleared again by ultracentrifugation, and the supernatants were incubated with 35 μl 50% protein A-Sepharose CL4B beads slurry (GE Healthcare, Piscataway, NJ) for 1 h at 4 C. Beads-bound immunocomplexes were briefly washed in NETN buffer (20 mm Tris-HCl, pH 7.5; 1 mm EDTA; 150 mm NaCl; 0.5% Nonidet P-40) and resuspended in 40 μl 1× Laemmli buffer. IPs were resolved on 4–20% Tris-glycine NOVEX SDS-PAGE (Invitrogen) and protein banding visualized with Coomassie Blue. Gel processing, in-gel trypsin digestion, and MS identification were done as described (69). Proteins were identified using SeQuest software (Thermo Scientific, Pittsburgh, PA); protein identifications were manually verified, and putative nonspecific interactions were filtered against an in-house database of common nonspecific identifications. Relative protein abundances of associated proteins (spectral counts divided by the molecular weight and normalized to the protein abundance of the antigen) in SRC-3 and ER complexes were compared between treatments to compile a restricted list of interacting proteins that display discernable differences between treatments in their associations with their respective antigens.
Western blot analysis
RFX1 complexes were immunoprecipitated essentially as described for IP/MS, using 300 μl nuclear extract, 5 μg RFX1 (H-230, catalog item sc-48809; Santa Cruz), and 20 μl 50% protein A slurry. IP proteins were analyzed by Western blotting using anti-SRC-3 (Bethyl) and anti-ERα (Santa Cruz; F-10) or antibodies, followed by horseradish peroxidase-conjugated secondary antibody and visualization with SuperSignal West Pico chemiluminescent reagent (Thermo Scientific) using Fluorochem FC2 multi-imager with DE-500FC (AlfaInnotech Inc., San Leandro, CA).
Supplementary Material
Footnotes
This work was generously supported by Genpathway Inc. and by National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) Grant U19 DK062434 (NURSA), National Institutes of Health Grant 5 R01 HD08188, NIDDK Grant 5 P01 DK059820, and National Cancer Institute Grant P30CA125123 (Baylor College of Medicine Cancer Center Core).
Disclosure Summary: R.B.L., Y.B., A.M., L.W., W.L., J.Q., and B.W.O. have nothing to declare. P.L. is employed by Genpathway Inc. M.H. was previously employed by Genpathway Inc.
First Published Online February 24, 2010
Abbreviations: ChIP, Chromatin immunoprecipitation; E2, 17β-estradiol; EPR, extended promoter region; ER, estrogen receptor α; FBS, fetal bovine serum; GREB1, gene regulated breast cancer 1; IP, immunoprecipitation; MS, mass spectrometry; NR, nuclear receptor; NURD/HDAC, nucleosome remodeling and histone deacetylation complex; Pol, polymerase; qPCR, real-time quantitative PCR; RFX1, regulatory factor X1; RNAi, RNA interference; siRNA, small interfering RNA; SRC-3, steroid receptor coactivator-3; TF, transcription factor; TSS, transcription start site.
References
- McKenna NJ, Lanz RB, O'Malley BW 1999 Nuclear receptor coregulators: cellular and molecular biology. Endocr Rev 20:321–344 [DOI] [PubMed] [Google Scholar]
- Rosenfeld MG, Lunyak VV, Glass CK 2006 Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev 20:1405–1428 [DOI] [PubMed] [Google Scholar]
- Margolis RN, Evans RM, O'Malley BW 2005 The nuclear receptor signaling atlas: development of a functional atlas of nuclear receptors. Mol Endocrinol 19:2433–2436 [DOI] [PubMed] [Google Scholar]
- Anzick SL, Kononen J, Walker RL, Azorsa DO, Tanner MM, Guan XY, Sauter G, Kallioniemi OP, Trent JM, Meltzer PS 1997 AIB1, a steroid receptor coactivator amplified in breast and ovarian cancer. Science 277:965–968 [DOI] [PubMed] [Google Scholar]
- Kuang SQ, Liao L, Zhang H, Lee AV, O'Malley BW, Xu J 2004 AIB1/SRC-3 deficiency affects insulin-like growth factor I signaling pathway and suppresses v-Ha-ras-induced breast cancer initiation and progression in mice. Cancer Res 64:1875–1885 [DOI] [PubMed] [Google Scholar]
- Torres-Arzayus MI, Font de Mora J, Yuan J, Vazquez F, Bronson R, Rue M, Sellers WR, Brown M 2004 High tumor incidence and activation of the PI3K/AKT pathway in transgenic mice define AIB1 as an oncogene. Cancer Cell 6:263–274 [DOI] [PubMed] [Google Scholar]
- Lanz RB, Lonard DM, O'Malley BW 2008 Nuclear receptor coregulators in human diseases. In: Kumar R, O'Malley BW, eds. NR coregulators and human diseases. Hackensack, NJ: World Scientific Publishing Co. [Google Scholar]
- Yan J, Tsai SY, Tsai MJ 2006 SRC-3/AIB1: transcriptional coactivator in oncogenesis. Acta Pharmacol Sin 27:387–394 [DOI] [PubMed] [Google Scholar]
- Zhang H, Kuang SQ, Liao L, Zhou S, Xu J 2004 Haploid inactivation of the amplified-in-breast cancer 3 coactivator reduces the inhibitory effect of peroxisome proliferator-activated receptor γ and retinoid X receptor on cell proliferation and accelerates polyoma middle-T antigen-induced mammary tumorigenesis in mice. Cancer Res 64:7169–7177 [DOI] [PubMed] [Google Scholar]
- Coste A, Antal MC, Chan S, Kastner P, Mark M, O'Malley BW, Auwerx J 2006 Absence of the steroid receptor coactivator-3 induces B-cell lymphoma. EMBO J 25:2453–2464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han SJ, Lonard DM, O'Malley BW 2009 Multi-modulation of nuclear receptor coactivators through posttranslational modifications. Trends Endocrinol Metab 20:8–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Liang YY, Feng XH, Tsai SY, Tsai MJ, O'Malley BW 2008 Essential phosphatases and a phospho-degron are critical for regulation of SRC-3/AIB1 coactivator function and turnover. Mol Cell 31:835–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Malley BW, Qin J, Lanz RB 2008 Cracking the coregulator codes. Curr Opin Cell Biol 20:310–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Font de Mora J, Brown M 2000 AIB1 is a conduit for kinase-mediated growth factor signaling to the estrogen receptor. Mol Cell Biol 20:5041–5047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannì M, Parrella E, Raska Jr I, Gaillard E, Nigro EA, Gaudon C, Garattini E, Rochette-Egly C 2006 P38MAPK-dependent phosphorylation and degradation of SRC-3/AIB1 and RARα-mediated transcription. EMBO J 25:739–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu RC, Qin J, Yi P, Wong J, Tsai SY, Tsai MJ, O'Malley BW 2004 Selective phosphorylations of the SRC-3/AIB1 coactivator integrate genomic responses to multiple cellular signaling pathways. Mol Cell 15:937–949 [DOI] [PubMed] [Google Scholar]
- Yi P, Feng Q, Amazit L, Lonard DM, Tsai SY, Tsai MJ, O'Malley BW 2008 Atypical protein kinase C regulates dual pathways for degradation of the oncogenic coactivator SRC-3/AIB1. Mol Cell 29:465–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azorsa DO, Cunliffe HE, Meltzer PS 2001 Association of steroid receptor coactivator AIB1 with estrogen receptor-α in breast cancer cells. Breast Cancer Res Treat 70:89–101 [DOI] [PubMed] [Google Scholar]
- List HJ, Lauritsen KJ, Reiter R, Powers C, Wellstein A, Riegel AT 2001 Ribozyme targeting demonstrates that the nuclear receptor coactivator AIB1 is a rate-limiting factor for estrogen-dependent growth of human MCF-7 breast cancer cells. J Biol Chem 276:23763–23768 [DOI] [PubMed] [Google Scholar]
- Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, Fox EA, Silver PA, Brown M 2005 Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 122:33–43 [DOI] [PubMed] [Google Scholar]
- Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, Wang Q, Bekiranov S, Sementchenko V, Fox EA, Silver PA, Gingeras TR, Liu XS, Brown M 2006 Genome-wide analysis of estrogen receptor binding sites. Nat Genet 38:1289–1297 [DOI] [PubMed] [Google Scholar]
- Cheng AS, Jin VX, Fan M, Smith LT, Liyanarachchi S, Yan PS, Leu YW, Chan MW, Plass C, Nephew KP, Davuluri RV, Huang TH 2006 Combinatorial analysis of transcription factor partners reveals recruitment of c-MYC to estrogen receptor-α responsive promoters. Mol Cell 21:393–404 [DOI] [PubMed] [Google Scholar]
- Jin VX, Leu YW, Liyanarachchi S, Sun H, Fan M, Nephew KP, Huang TH, Davuluri RV 2004 Identifying estrogen receptor α target genes using integrated computational genomics and chromatin immunoprecipitation microarray. Nucleic Acids Res 32:6627–6635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kininis M, Chen BS, Diehl AG, Isaacs GD, Zhang T, Siepel AC, Clark AG, Kraus WL 2007 Genomic analyses of transcription factor binding, histone acetylation, and gene expression reveal mechanistically distinct classes of estrogen-regulated promoters. Mol Cell Biol 27:5090–5104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon YS, Garcia-Bassets I, Hutt KR, Cheng CS, Jin M, Liu D, Benner C, Wang D, Ye Z, Bibikova M, Fan JB, Duan L, Glass CK, Rosenfeld MG, Fu XD 2007 Sensitive ChIP-DSL technology reveals an extensive estrogen receptor α-binding program on human gene promoters. Proc Natl Acad Sci USA 104:4852–4857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laganière J, Deblois G, Giguère V 2005 Functional genomics identifies a mechanism for estrogen activation of the retinoic acid receptor α1 gene in breast cancer cells. Mol Endocrinol 19:1584–1592 [DOI] [PubMed] [Google Scholar]
- Laganière J, Deblois G, Lefebvre C, Bataille AR, Robert F, Giguère V 2005 Location analysis of estrogen receptor α target promoters reveals that FOXA1 defines a domain of the estrogen response. Proc Natl Acad Sci USA 102:11651–11656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CY, Vega VB, Thomsen JS, Zhang T, Kong SL, Xie M, Chiu KP, Lipovich L, Barnett DH, Stossi F, Yeo A, George J, Kuznetsov VA, Lee YK, Charn TH, Palanisamy N, Miller LD, Cheung E, Katzenellenbogen BS, Ruan Y, Bourque G, Wei CL, Liu ET 2007 Whole-genome cartography of estrogen receptor α binding sites. PLoS Genet 3:e87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurtado A, Holmes KA, Geistlinger TR, Hutcheson IR, Nicholson RI, Brown M, Jiang J, Howat WJ, Ali S, Carroll JS 2008 Regulation of ERBB2 by oestrogen receptor-PAX2 determines response to tamoxifen. Nature 456:663–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kininis M, Kraus WL 2008 A global view of transcriptional regulation by nuclear receptors: gene expression, factor localization, and DNA sequence analysis. Nucl Recept Signal 6:e005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupien M, Eeckhoute J, Meyer CA, Wang Q, Zhang Y, Li W, Carroll JS, Liu XS, Brown M 2008 FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell 132:958–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochsner SA, Steffen DL, Hilsenbeck SG, Chen ES, Watkins C, McKenna NJ 2009 GEMS (Gene Expression MetaSignatures), a Web resource for querying meta-analysis of expression microarray datasets: 17β-estradiol in MCF-7 cells. Cancer Res 69:23–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rae JM, Johnson MD, Cordero KE, Scheys JO, Larios JM, Gottardis MM, Pienta KJ, Lippman ME 2006 GREB1 is a novel androgen-regulated gene required for prostate cancer growth. Prostate 66:886–894 [DOI] [PubMed] [Google Scholar]
- Rae JM, Johnson MD, Scheys JO, Cordero KE, Larios JM, Lippman ME 2005 GREB 1 is a critical regulator of hormone dependent breast cancer growth. Breast Cancer Res Treat 92:141–149 [DOI] [PubMed] [Google Scholar]
- Heger S, Mastronardi C, Dissen GA, Lomniczi A, Cabrera R, Roth CL, Jung H, Galimi F, Sippell W, Ojeda SR 2007 Enhanced at puberty 1 (EAP1) is a new transcriptional regulator of the female neuroendocrine reproductive axis. J Clin Invest 117:2145–2154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trauernicht AM, Kim SJ, Kim NH, Boyer TG 2007 Modulation of estrogen receptor α protein level and survival function by DBC-1. Mol Endocrinol 21:1526–1536 [DOI] [PubMed] [Google Scholar]
- Xue Y, Wong J, Moreno GT, Young MK, Côté J, Wang W 1998 NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol Cell 2:851–861 [DOI] [PubMed] [Google Scholar]
- Bourdeau V, Deschênes J, Métivier R, Nagai Y, Nguyen D, Bretschneider N, Gannon F, White JH, Mader S 2004 Genome-wide identification of high-affinity estrogen response elements in human and mouse. Mol Endocrinol 18:1411–1427 [DOI] [PubMed] [Google Scholar]
- Krum SA, Miranda-Carboni GA, Lupien M, Eeckhoute J, Carroll JS, Brown M 2008 Unique ERα cistromes control cell type-specific gene regulation. Mol Endocrinol 22:2393–2406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller CL, Porter SE, Hoffman MG, Jaehning JA 2004 The Paf1 complex has functions independent of actively transcribing RNA polymerase II. Mol Cell 14:447–456 [DOI] [PubMed] [Google Scholar]
- Pokholok DK, Hannett NM, Young RA 2002 Exchange of RNA polymerase II initiation and elongation factors during gene expression in vivo. Mol Cell 9:799–809 [DOI] [PubMed] [Google Scholar]
- Krogan NJ, Dover J, Wood A, Schneider J, Heidt J, Boateng MA, Dean K, Ryan OW, Golshani A, Johnston M, Greenblatt JF, Shilatifard A 2003 The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol Cell 11:721–729 [DOI] [PubMed] [Google Scholar]
- Ng HH, Robert F, Young RA, Struhl K 2003 Targeted recruitment of Set1 histone methylase by elongating Pol II provides a localized mark and memory of recent transcriptional activity. Mol Cell 11:709–719 [DOI] [PubMed] [Google Scholar]
- Fuller-Pace FV 2006 DExD/H box RNA helicases: multifunctional proteins with important roles in transcriptional regulation. Nucleic Acids Res 34:4206–4215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auboeuf D, Hönig A, Berget SM, O'Malley BW 2002 Coordinate regulation of transcription and splicing by steroid receptor coregulators. Science 298:416–419 [DOI] [PubMed] [Google Scholar]
- Clark EL, Coulson A, Dalgliesh C, Rajan P, Nicol SM, Fleming S, Heer R, Gaughan L, Leung HY, Elliott DJ, Fuller-Pace FV, Robson CN 2008 The RNA helicase p68 is a novel androgen receptor coactivator involved in splicing and is overexpressed in prostate cancer. Cancer Res 68:7938–7946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hönig A, Auboeuf D, Parker MM, O'Malley BW, Berget SM 2002 Regulation of alternative splicing by the ATP-dependent DEAD-box RNA helicase p72. Mol Cell Biol 22:5698–5707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanz RB, McKenna NJ, Onate SA, Albrecht U, Wong J, Tsai SY, Tsai MJ, O'Malley BW 1999 A steroid receptor coactivator, SRA, functions as an RNA and is present in an SRC-1 complex. Cell 97:17–27 [DOI] [PubMed] [Google Scholar]
- Watanabe M, Yanagisawa J, Kitagawa H, Takeyama K, Ogawa S, Arao Y, Suzawa M, Kobayashi Y, Yano T, Yoshikawa H, Masuhiro Y, Kato S 2001 A subfamily of RNA-binding DEAD-box proteins acts as an estrogen receptor α coactivator through the N-terminal activation domain (AF-1) with an RNA coactivator, SRA. EMBO J 20:1341–1352 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Stossi F, Likhite VS, Katzenellenbogen JA, Katzenellenbogen BS 2006 Estrogen-occupied estrogen receptor represses cyclin G2 gene expression and recruits a repressor complex at the cyclin G2 promoter. J Biol Chem 281:16272–16278 [DOI] [PubMed] [Google Scholar]
- Stossi F, Madak-Erdogan Z, Katzenellenbogen BS 2009 Estrogen receptor α represses transcription of early target genes via p300 and CtBP1. Mol Cell Biol 29:1749–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang KR, Nemoto T, Yokota Y 2007 RFX1 mediates the serum-induced immediate early response of Id2 gene expression. J Biol Chem 282:26167–26177 [DOI] [PubMed] [Google Scholar]
- Xu Y, Sengupta PK, Seto E, Smith BD 2006 Regulatory factor for X-box family proteins differentially interact with histone deacetylases to repress collagen α2(I) gene (COL1A2) expression. J Biol Chem 281:9260–9270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gururaj AE, Singh RR, Rayala SK, Holm C, den Hollander P, Zhang H, Balasenthil S, Talukder AH, Landberg G, Kumar R 2006 MTA1, a transcriptional activator of breast cancer amplified sequence 3. Proc Natl Acad Sci USA 103:6670–6675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasenthil S, Gururaj AE, Talukder AH, Bagheri-Yarmand R, Arrington T, Haas BJ, Braisted JC, Kim I, Lee NH, Kumar R 2007 Identification of Pax5 as a target of MTA1 in B-cell lymphomas. Cancer Res 67:7132–7138 [DOI] [PubMed] [Google Scholar]
- Peterson TJ, Karmakar S, Pace MC, Gao T, Smith CL 2007 The silencing mediator of retinoic acid and thyroid hormone receptor (SMRT) corepressor is required for full estrogen receptor α transcriptional activity. Mol Cell Biol 27:5933–5948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perissi V, Aggarwal A, Glass CK, Rose DW, Rosenfeld MG 2004 A corepressor/coactivator exchange complex required for transcriptional activation by nuclear receptors and other regulated transcription factors. Cell 116:511–526 [DOI] [PubMed] [Google Scholar]
- Huang HJ, Norris JD, McDonnell DP 2002 Identification of a negative regulatory surface within estrogen receptor α provides evidence in support of a role for corepressors in regulating cellular responses to agonists and antagonists. Mol Endocrinol 16:1778–1792 [DOI] [PubMed] [Google Scholar]
- Soutoglou E, Talianidis I 2002 Coordination of PIC assembly and chromatin remodeling during differentiation-induced gene activation. Science 295:1901–1904 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nussbaum C, Myers RM, Brown M, Li W, Liu XS 2008 Model-based analysis of ChIP-Seq (MACS). Genome Biol 9:R137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson WE, Li W, Meyer CA, Gottardo R, Carroll JS, Brown M, Liu XS 2006 Model-based analysis of tiling-arrays for ChIP-chip. Proc Natl Acad Sci USA 103:12457–12462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Carroll JS, Brown M, Liu S 2008 xMAN: extreme MApping of OligoNucleotides. BMC Genomics 9(Suppl 1):S20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codd EF 1998 A relational model of data for large shared data banks. 1970. MD Comput 15:162–166 [PubMed] [Google Scholar]
- Liu XS, Brutlag DL, Liu JS 2002 An algorithm for finding protein-DNA binding sites with applications to chromatin-immunoprecipitation microarray experiments. Nat Biotechnol 20:835–839 [DOI] [PubMed] [Google Scholar]
- Mahony S, Benos PV 2007 STAMP: a web tool for exploring DNA-binding motif similarities. Nucleic Acids Res 35:W253–W258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA 2009 Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44–57 [DOI] [PubMed] [Google Scholar]
- Joachimiak MP, Weisman JL, May BCh 2006 JColorGrid: software for the visualization of biological measurements. BMC Bioinformatics 7:225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung SY, Malovannaya A, Wei J, O'Malley BW, Qin J 2005 Proteomic analysis of steady-state nuclear hormone receptor coactivator complexes. Mol Endocrinol 19:2451–2465 [DOI] [PubMed] [Google Scholar]
- Luo H, Li Y, Mu JJ, Zhang J, Tonaka T, Hamamori Y, Jung SY, Wang Y, Qin J 2008 Regulation of intra-S phase checkpoint by ionizing radiation (IR)-dependent and IR-independent phosphorylation of SMC3. J Biol Chem 283:19176–19183 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.