Summary
Acute respiratory infections (ARI) are a common reason for seeking medical attention and the threat of pandemic influenza will likely add to these numbers. Using human viral challenge studies with live rhinovirus, respiratory syncytial virus, and influenza A, we developed peripheral blood gene expression signatures that distinguish individuals with symptomatic ARI from uninfected individuals with > 95% accuracy. We validated this “acute respiratory viral” signature - encompassing genes with a known role in host defense against viral infections - across each viral challenge. We also validated the signature in an independently acquired dataset for influenza A and classified infected individuals from healthy controls with 100% accuracy. In the same dataset, we could also distinguish viral from bacterial ARIs (93% accuracy). These results demonstrate that ARIs induce changes in human peripheral blood gene expression that can be used to diagnose a viral etiology of respiratory infection and triage symptomatic individuals.
Introduction
Acute respiratory infections (ARI) are among the most common reasons for seeking medical attention in the United States (Hong et al., 2004; Johnstone et al., 2008). Rhinovirus (HRV), influenza, and respiratory syncytial virus (RSV) are recognized as leading etiologies of ARI in adults (Peltola et al., 2008). Viral ARIs are generally self-limited, but can lead to disease exacerbation among individuals with prior pulmonary disease (Johnston, 1995; Rakes et al., 1999). Most adults experience at least one HRV infection per year (Arruda et al., 1997); (Schaller et al., 2006). Adult RSV infections may be self-limited or lead to airways obstruction and morbidity (Falsey et al., 2005). Influenza infection remains common, with associated significant health-care and societal costs (Gums et al., 2008). Early detection of influenza A can facilitate individual treatment decisions, as well as provide early data to forecast an epidemic/pandemic (Memoli et al., 2008).
HRV, RSV, and influenza are all spread by droplet inhalation, and upon contact with the respiratory epithelium, these viruses initiate a cytokine and chemokine response that orchestrates proliferation, chemotaxis and amplification of inflammatory cells (Bhoj et al., 2008; Kirchberger et al., 2007). Nasal epithelial inflammation produced on contact with virus triggers a coordinated host response that may result from infection limited to the upper respiratory tract or spread to the lower respiratory tract with bronchiolitis and pneumonia. Understanding the host responses to these common infections will allow for better understanding of disease pathobiology and provide a basis for development of novel diagnostic methodologies for distinguishing viral respiratory infection from respiratory disease caused by other common pathogens.
Peripheral blood leukocytes are a reservoir and migration point for cells representing all aspects of the host immune response. Gene expression patterns obtained from these cells can discriminate between complex physiologic states (Aziz et al., 2007), exposures to pathogens (Ramilo et al., 2007; Simmons et al., 2007), immune modifiers (e.g., LPS) (Boldrick et al., 2002; Kobayashi et al., 2003), and environmental exposures (Dressman et al., 2007; Meadows et al., 2008; Wang et al., 2005). While current infectious disease diagnostics rely on pathogen-based detection (Chiarini et al., 2008; Lambert et al., 2008; Robinson et al., 2008), the development of reproducible means for extracting RNA from whole blood, coupled with advanced statistical methods for analysis of complex datasets, now allows the possibility of classifying infections based on host gene expression profiling that reveal pathogen specific signatures of disease.
To realize the potential of genome-scale information requires a paradigm shift in the way complex, large-scale data are viewed, analyzed and utilized. The biology of infection, the host response and the ensuing disease process are highly complex. Our previous work in defining the complexity of the cancer phenotype using gene expression analysis has defined approaches involving successive sub-categorization of patients according to combinations of both clinical and genomic risk factors, highlighting the predictive value of multiple genomic patterns (Acharya et al., 2008; Garman et al., 2008; Xu et al., 2008). The role of formal statistical models to incorporate, evaluate, and weigh multiple gene expression patterns is fundamental to this methodology. We have shown that specific classes of statistical tree models are capable of such synthesis and can improve prediction and classification for individual patients. One core methodology that underlies our comprehensive models uses statistical prediction tree models, and the expression data enters into these models signatures (estimated “factors”) that are candidate predictive factors in statistical tree models. This approach to molecular characterization and candidate gene identification has provided significant value in recent work (Acharya et al., 2008; Garman et al., 2008; Lucas et al., 2006; Meadows et al., 2008; Seo et al., 2006), uncovering patterns of non-linear associations between gene expression and phenotypic outcomes(Brieman, 2001; Kooperberg et al., 2001; Ruczinski, 2003).
Using three human viral challenge cohorts for HRV, RSV, and influenza A, we developed a robust blood mRNA expression signature that classifies symptomatic human respiratory viral infection. Factor analysis (Carvalho et al., 2008) of mRNA expression data revealed a pattern of gene expression common across symptomatic individuals from all viral challenges. This was termed the “acute respiratory viral” bio-signature of disease, that encompassed transcripts of genes known to be related to viral infection and the overall immune response. Further, this signature could accurately classify influenza A infection in an independent community-based cohort. For this signature to serve as an important diagnostic indicator of viral respiratory infection, and for the purpose of clinical triage and treatment decisions, it should be distinct from the overall response to bacterial respiratory tract infections. An analysis of publically available peripheral blood-based gene expression data from patients with bacterial infection indicated that the acute respiratory viral signature was viral infection-specific and could distinguish patients with viral and bacterial infections as well as healthy controls. Moreover, bacterial and viral respiratory infections could be accurately classified using this gene expression signature. This work emphasizes the important concept that capturing the human host response to pathogen exposure may serve as the basis for both diagnostic testing as well as a window into the fundamental biology of infection.
Results
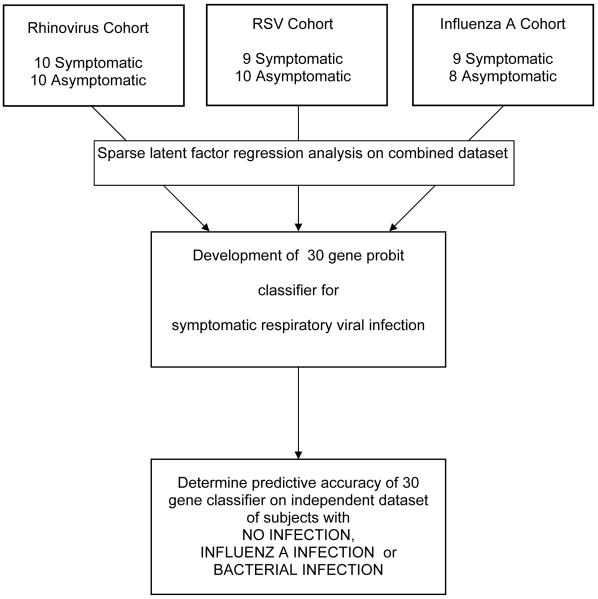
Organization and data flow are shown in Figure 1. Exposures were performed on independent cohorts and datasets combined for analysis.
Figure 1.
Consort diagram of study organization. Three unique cohorts of healthy volunteers were infected with one of three respiratory viruses (HRV, RSV or influenza A). Combined data was analyzed using sparse latent factor regression with leave-one-out cross validation. Subsequent validation occurred using a dataset available from the public domain.
Viral challenge
HRV
The attack rate was 50%, as ten of the 20 inoculated subjects developed ARI-like symptoms and had confirmed viral shedding (Table 1; Supplemental Figures 1 and 2). Peak symptoms occurred at 48 hours (n=2), 72 hours (n=4) or 96 hours (n=4) post inoculation (median 72 hours).
Table 1.
Description of experimental cohorts
| Cohort | Number Exposed |
Number Symptomatic |
Median Time “T”: Time to Peak Symptoms |
Corresponding Time Used for Asymptomatic Subjects |
|---|---|---|---|---|
| Rhinovirus | 20 | 10 | 72 hours | 72 hours |
| RSV | 20 | 9 | 141.5 hours | 141.5 hours |
| Influenza | 17 | 9 | 80 hours | 86 hours |
RSV
The attack rate was 45%, as nine of the 20 inoculated subjects developed ARI-like symptoms and had confirmed viral shedding (Table 1, Supplemental Figures 1 and 2). One subject (RSV020) had late symptoms and uninterpretable culture data and was excluded. Peak symptoms occurred at 93.5 hours (n=1), 117.5 hours (n=1), 141.5 hours (n=5) and 165.5 hours (n=1) post inoculation (median 141.5 hours).
Influenza
The attack rate was 53%, as nine of the 17 inoculated subjects developed ARI-like symptoms and had confirmed viral shedding (Table 1, Supplemental Figures 1 and 2). Peak symptoms occurred at 50 hours (n=1), 62 hours (n=2), 74 hours (n=2), 86 hours (n=2), 98 (n=1) and 110 hours (n=1) post inoculation (median 80 hours).
A common blood RNA based viral response signature differentiates adults with symptomatic HRV, RSV, or influenza A infection from uninfected individuals
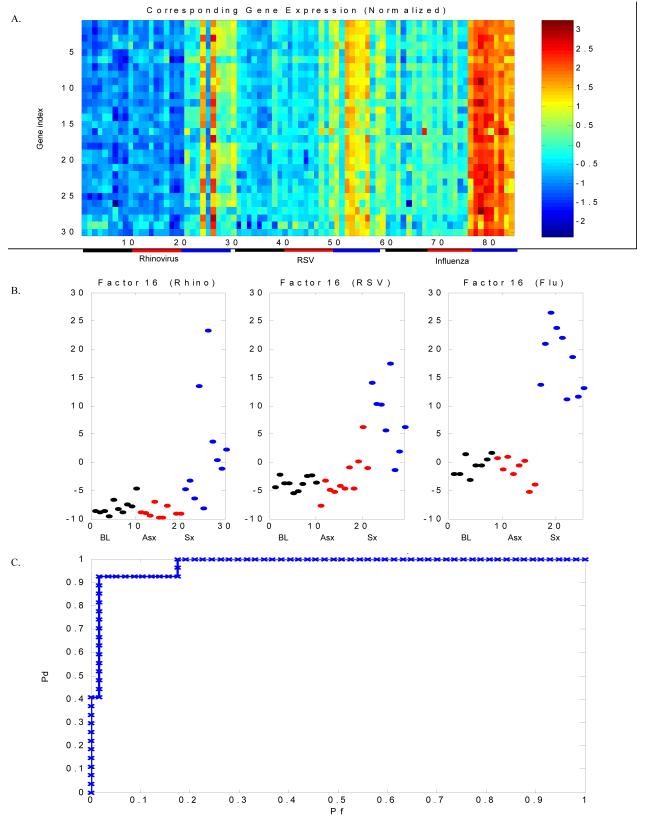
We first combined data from each challenge and analyzed it as a single dataset. Eighty-four timepoints were included in the analysis (HRV: 10 baseline, 10 symptomatic, 10 matched timepoint asymptomatic; RSV: 10 baseline, 9 symptomatic, 10 matched timepoint asymptomatic; influenza: 8 baseline, 9 symptomatic, 8 matched timepoint asymptomatic). Twenty factors were developed using all available probes and a single factor (Factor 16) could best discriminate symptomatic (infected) subjects (HRV, RSV or influenza A) from asymptomatic (uninfected) individuals. Baseline (pre-inoculation) gene expression was indistinguishable from the matched timepoint of asymptomatic subjects (Figure 2). Baseline gene expression in subjects who became symptomatic was indistinguishable from those who remained asymptomatic (data not shown). The top 30 predictive genes contained in Factor 16 are known to characterize host response to viral infection (Supplemental Table 1). These 30 genes were used as features for the sparse probit regression model to perform leave-one-out cross validation and generate an ROC curve (Figure 2) to estimate performance of the model. Leave-one-out cross validation correctly identified 96.5% of infected subjects (misclassification rate 3.5%, 3/84). These data - from three distinct viral challenge experiments – demonstrate a clear acute respiratory viral response factor as a common feature of peak infection.
Figure 2. An acute respiratory viral gene expression signature characterizes symptomatic respiratory viral infection.
Experimentally infected adult subjects with symptomatic HRV, RSV or influenza A infection can be distinguished from uninfected individuals by a distinct group of genes (“factor”) demonstrating differential expression among symptomatic individuals as compared to asymptomatic individuals. For each viral challenge, peripheral blood was drawn for whole blood gene expression analysis at scheduled time points post intranasal inoculation of virus. Whole blood gene expression was determined pre-inoculation (baseline), at time of peak symptoms for each symptomatic individual and a matched timepoint for each asymptomatic individual. A) Heat map representing gene expression for genes contained in Factor 16. Columns represent subjects and correspond to points in Figure 1B, with the first 10 columns representing baseline gene expression of asymptomatic individuals in the HRV challenge, the next 10 columns representing timepoints matched to peak symptoms for the asymptomatic subjects in the HRV cohort and the following 10 columns representing time of peak symptoms for the 10 subjects who developed symptomatic HRV infection. A similar layout continues for the RSV and influenza cohorts. Blue and red represent extremes of gene expression, with visually apparent differences between baseline and matched timepoints in the asymptomatic individuals versus time of peak symptoms in symptomatic individuals. The initial models were built without label information for each subject (asymptomatic versus symptomatic, baseline timepoint versus infected/matched timepoint). This design allowed for the model to cluster individuals based on expression patterns alone, thus minimizing bias in factor organization. Bars underneath represent individual groups (black = baseline, red = asymptomatic, blue = symptomatic). P-value (ANOVA) for the difference in factor scores between symptomatic and asymptomatic subjects at time T for the combined dataset is < 1×10−16; for rhinovirus 2.5 × 10−5, for RSV is 2.3 × 10−7 and for influenza is 5.0 × 10−13). B) Factor plots representing categorization of asymptomatic and symptomatic subjects at baseline (black), matched timepoint to peak symptoms (asymptomatic, red) and peak symptoms (symptomatic, blue). C) Leave-one-out cross validation correctly identifies 97% of individuals with viral infection versus no infection (3/84 misclassified). Pd = probability of detection; Pf = probability of false discovery.
To further validate the robust acute respiratory viral response signature, we next analyzed each dataset (HRV, RSV and influenza A) separately to identify a factor that characterized symptomatic viral infection for each individual dataset (Supplemental Figure 3). We performed sparse probit regression on the 30 genes with the highest factor loading values in each factor, and this data was used for leave-one-out cross validation and generation of an ROC curve to estimate factor performance. Notably, the p-values associated with each factor (i.e. the likelihood that this group of genes would not be selected randomly) were 2.33 ×10−5 (HRV), 2.29 × 10−7 (RSV), and 4.95 × 10−13 (influenza) (www.gather.duke.edu). The individual challenge-specific factors were used as a de facto “training set” to classify subjects from the other challenges. As shown in Supplemental Figure 4 and Table 2, when the model was trained on any individual dataset, prediction of symptomatic versus asymptomatic was >96%. This supports the conclusion that, at peak viral respiratory infection symptoms, the host response converges to encompass a gene expression program highly characteristic of response to viral infection. Supplemental Figure 5 shows overlap between genes represented in the factors predictive for the individual viruses. Most genes contained in the individual virus factors were present in the acute respiratory viral factor. Genes unique to an individual virus factor include the following: SOCS1 (HRV) and FCGR1A, GBP1, LAP3, ETV7 and FCGR1B (RSV). Complete gene lists for the individual virus factors and the acute respiratory viral factor are listed in Supplemental Table 1. Genes represented in these factors were highly representative of host response to viral infection, including RSAD2, interferon response elements and the OAS gene family.
Table 2.
Intra-dataset probit classification cross-validation results. The error rate is shown based on the top gene (noted in parentheses) selected from the training set probit classifier. For this model, the top 40 genes from the training set discriminative factor were used to build the probit classifier for testing in the validation dataset
| Test | Rhinovirus | RSV | Influenza |
|---|---|---|---|
| Train | |||
| Rhinovirus | 1/30 (RSAD2) |
2/29 (RTP4) |
0/25 (ISG15) |
| RSV | 1/30 (RSAD2) |
2/29 (RTP4) |
0/25 (ISG15) |
| Influenza | 1/30 (RSAD2) |
2/29 (RTP4) |
0/25 (ISG15) |
Validation of an acute respiratory viral peripheral blood gene expression signature for experimentally induced symptomatic HRV, RSV, or influenza using data from an independent set of individuals with symptomatic community acquired influenza A infection
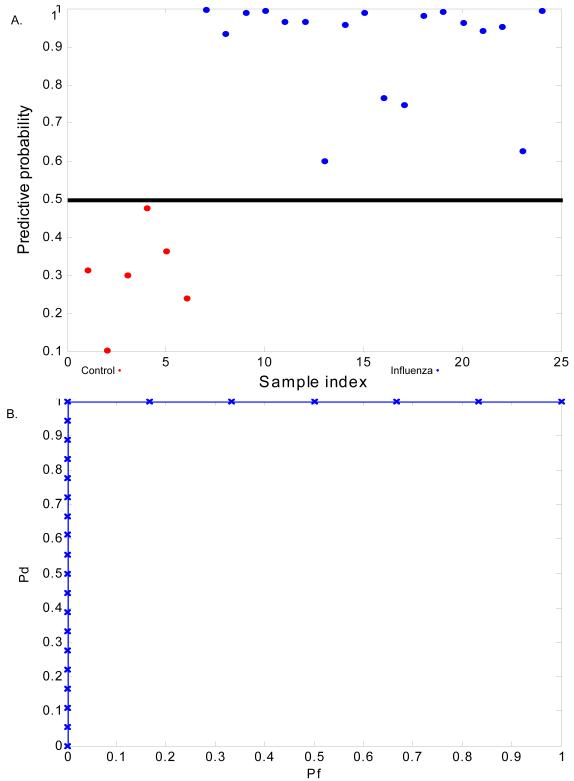
Given the strong viral response signature that distinguished symptomatic HRV, RSV, and influenza infection from uninfected subjects, we sought to confirm the specificity of this response to viral infection diagnosed in a community setting. We utilized two methods to validate our acute respiratory viral signature using microarray datasets derived from PBMC mRNA from a published study (Ramilo et al., 2007) of viral respiratory infection ascertained a from cohort of pediatric patients with microbiologically proven influenza A infection with linked gene expression data. First, we used the acute respiratory viral classifier built on the combined three challenge datasets to predict disease state (uninfected vs influenza A infection) in the literature cohort (Figure 3). Despite differences in subject ascertainment in the experimental cohort and the literature cohort [as well as other potential confounders (such as age and demographics)], we were able to accurately classify subjects as influenza A infected versus no infection in the literature cohort. This classification of subjects in this cohort was highly accurate [100% (23/23) for influenza infected versus no infection] (Figure 3b). Prediction of viral infection in a pre-existing dataset using genes identified as discriminative in an experimental dataset reinforces the robust nature of both the methodology and the classifier.
Figure 3.
Acute respiratory viral factor derived from the three experimental cohorts (HRV, RSV, and influenza) predicts subjects with culture-proven influenza infection from an independent dataset with a high degree of accuracy. The acute respiratory viral classifier built on the combined three challenge datasets was used to predict disease state (uninfected versus influenza A infection) in the literature cohort. A) Predictive capability of the acute respiratory viral factor to classify subjects with no infection (red) versus influenza A infection (blue). X-axis represents the individual subjects and y-axis represents the decision threshold. 0.5 is chosen as the threshold for generation of the subsequent ROC curves. B) Prediction of influenza A infected versus healthy hospitalized control subjects using the acute respiratory viral classifier. Classification of subjects in the literature cohort was highly accurate [100% (23/23) for influenza infected versus no infection].
In the second approach, we re-analyzed the raw gene expression data from the literature data set [14] using the same methods that were utilized to generate the HRV, RSV, and influenza expression signatures. Similar to our analysis of the HRV-, RSV-, and influenza-infected cohorts, twenty factors were built using the entire gene set from all persons in the literature cohort (Supplemental Figure 6). These factors were used to build a classifier that distinguished persons with influenza A (n = 18) from healthy controls (n = 6 pediatric subjects hospitalized for elective surgery). The top 30 genes in this factor were used as features for the sparse probit regression model to perform leave-one-out cross validation and generate ROC curves to estimate performance of the algorithm. Leave-one-out cross validation correctly identified 100% of the 24 individuals in this dataset. Of the 27 unique genes represented in the literature cohort factor, 20 were also present in the acute respiratory viral factor derived from the experimental cohorts. Of the 28 unique genes represented in the acute respiratory viral factor derived from our experimental cohorts, 20 were also present in the literature cohort factor. The probit function was also used to discriminate between influenza A infection and bacterial infection, with cross-validation correctly classifying 90/97 subjects (misclassification rate 7%). This finding further supports the acute respiratory viral factor derived above is a robust disease signature at time of peak symptoms. Predictive performance of each gene contained in the probit function generated from the acute respiratory viral factor to predict pathogen class in the independent dataset is shown in Supplemental Figure 7.
Peripheral blood gene expression signatures can discriminate between individuals with symptomatic HRV, RSV or influenza A virus and bacterial infection
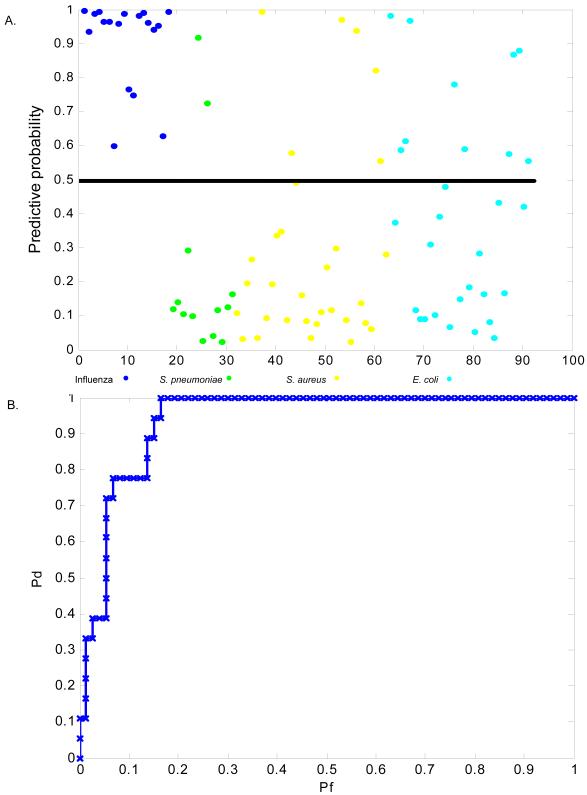
We next sought to further show that our acute respiratory viral gene expression factor was specific for viral infections. We used microarray datasets available in the literature [14] derived from PBMC mRNA from a cohort of pediatric patients with microbiologically proven S. pneumoniae, S. aureus, or E. coli infections [(S. pneumoniae (n=13), S. aureus (n=31), or E. coli (n=29)]. We used the acute respiratory viral classifier built on the three combined challenge datasets to predict disease state (influenza A infection versus bacterial infection) in the literature cohort (Figure 4). Classification of subjects in the literature cohort was highly accurate: 80% (73/91) for influenza infected versus any bacterial infection (Figure 4) and 93% (31/33) for influenza infected versus pneumococcal infection (data not shown). This analysis confirms specificity of the viral infection signature to discriminate not only between subjects with acute respiratory viral infection and uninfected subjects, but also from subjects with acute bacterial infections, including bacterial respiratory infection. Ultimately, the differentiation that is most valuable clinically may be discriminative between host response to viral respiratory tract infection and bacterial pneumonia (i.e. S. pneumoniae infection). Thus, despite inherent differences in sample acquisition and study design between the experimental HRV, RSV, and influenza cohorts and the literature cohort, these analyses confirm the robust nature of gene expression signatures that differentiate subjects with respiratory viral infection from subjects with bacterial infections, including pneumococcal infection, and from healthy subjects.
Figure 4.
Acute respiratory viral factor derived from the three experimental cohorts (HRV, RSV, and influenza) distinguishes subjects from an independent dataset with culture-proven influenza infection versus bacterial infection (blue = influenza A; green = S. pneumoniae; yellow = S. aureus; turquoise = E. coli) with a high degree of accuracy. B) Prediction of bacterial infection (any) versus influenza A infection using the pan-respiratory viral classifier. Classification is accurate [80%, (73/91)] for influenza A infection versus any bacterial infection.
Discussion
We performed three independent human viral challenge studies (HRV, RSV, and influenza) to define host-based peripheral blood gene expression patterns characteristic of response to viral respiratory infection. The results provide clear evidence that a unique biologically relevant peripheral blood gene expression signature classifies respiratory viral infection with a remarkable degree of accuracy. These findings underscore the conserved nature of the host response to viral infection, which is also evident in the cross-validation between experimental cohorts. The “acute respiratory viral” gene expression signature derived from these cohorts was validated in an independently derived external dataset, and, importantly, can distinguish respiratory viral infection from bacterial infection. These findings provide compelling evidence that peripheral blood gene expression can function as a biomarker for specific classes of infectious pathogens and may potentially serve as a useful diagnostic for triaging treatment decisions for ARI.
Discrimination between infectious causes of illness is a critical component of acute care of the medical patient as such distinctions facilitate both triage and treatment decisions. While traditional culture, antigen-based, and PCR based diagnostics are useful in pathogen classification, these assays are not without limitations(Bryant et al., 2004; Campbell and Ghazal, 2004). Current rapid diagnostic methods are lacking in sensitivity, with influenza and RSV tests (e.g. BinaxNOW antigen testing) reporting sensitivities of 53-80% (Jonathan, 2006; Landry et al., 2008; Rahman et al., 2008) or are labor-intensive, such as direct-fluorescent antibody (DFA) testing. Categorizing infection based on host response is an emerging hypothesis that not only enhances our diagnostic capabilities, but may provide additional insight into the pathobiology of infection. We have identified gene expression patterns that characterize host response to viral infection and that identify infected individuals with a high degree of accuracy. Several lines of evidence validate our findings, including the internal cross validation between exposure cohorts as well as validation with the free-living influenza A and bacterial infection pediatric cohort (Ramilo et al., 2007). Other investigators have identified host gene expression patterns – in nasal epithelium – that are associated with viral infection. Differentially expressed genes in nasal epithelium exposed to HRV 16 (in vitro and from experimentally infected subjects) were similar to those found in the current study in peripheral blood (Proud et al., 2008). In particular, RSAD2 (viperin), a potential antiviral molecule (Chin and Cresswell, 2001; Jiang et al., 2008; Wang et al., 2007b), was the most highly differentially expressed gene in nasal epithelium between infected and uninfected individuals at 48 hours post inoculation. Our HRV (HRV-16) predictive factor included RSAD2 (viperin) and the probit regression model selected it as the key differentially expressed gene in blood for determining infected state in the HRV cohort. Whole blood gene expression studies looking at RSV infection in hospitalized infants shared differentially expressed genes with the RSV factor found in our study, with a predominance of interferon-response elements, FCγ1AR, and OAS3 (Fjaerli et al., 2006). Finally, data from the naturally-occurring influenza A/bacterial infection study (Ramilo et al., 2007) confirmed a distinct host response signature to viral infection occurring both in this cohort and our experimentally infected cohorts. Taken together, this provides strong evidence for highly accurate in vivo detection of human viral respiratory infection through analysis of peripheral blood gene expression. Notably, different peripheral blood immune cell types induce varying gene expression programs in response to pathogen exposure. Thus, the peripheral blood gene expression signatures derived and validated in these cohorts may only be applicable to individuals without underlying immune deficiencies. Additional studies in immune deficient populations will be needed to generalize the current findings to these rare but clinically important patient subsets.
Evident from the genes in each factor, signatures that discriminate subjects with symptomatic respiratory viral infections from healthy subjects and subjects with bacterial infection contain biologically plausible gene networks involved in host viral response. The acute respiratory viral factor was most heavily represented by genes in the interferon signaling canonical pathway (p = 9.75 × 10−9) and the pattern recognition pathway for bacteria and viruses (p = 5.67 × 10−5). This over-representation of interferon response elements remained when individual viral challenges were analyzed as separate entities (HRV p = 1.38 × 10−10, RSV p = 2.25 × 10−9, influenza p = 1.25 × 10−7). (www.ingenuity.com). Overlap between the genes defining each factor (discriminating symptomatic individuals versus asymptomatic individuals OR discriminating viral respiratory infection from bacterial infection) was strong. Baseline gene expression among all challenge subjects was similar and indistinguishable from the later timepoints for asymptomatic subjects and classification of subjects from one cohort based on the other cohorts was remarkably accurate. Discovery of discriminant factors for disease states such as this one is inherently blind to biology, as the model is not aware of data labels. Despite differences in study design, commonalities between experimentally infected adults with HRV, RSV, or influenza A and community infected children with influenza A predominated over virus-specific aspects of each signature. However, when selecting the gene or genes with greatest discriminating power for leave-one-out cross validation, the model chose different genes for each viral illness (HRV: RSAD2; RSV: RTP4; influenza A: ISG15; viral vs. bacterial: IFI27, RSAD2, IFI6, CXCL10, FLJ20035, GBP1 and SIGLEC1 and viral vs. S. pneumoniae: RSAD2). Thus, with careful exploration of disease biology or with additional cohorts for validation, disease specific markers of infection may arise, adding parity to the diagnostic signatures. Overlap is minimal with differentially expressed genes from other studies of peripheral blood response to environmental stress found in a study of humans exposed to ionizing radiation, and the genotoxic stress of chemotherapy and LPS (Dressman et al., 2007; Meadows et al., 2008), decreasing the likelihood that these genes are part of a generalized response program inherent to immune effector cells.
Despite data acquisition and processing differences, gene expression patterns derived from publically available microarray data for individuals with influenza A infection were similar to those with experimentally acquired symptomatic HRV, RSV, or influenza A infection. Genes found to characterize the response to respiratory viral infection in our cohorts overlap with genes found in many gene expression studies of host response to viral infections, both in vivo (Bhoj et al., 2008; Proud et al., 2008; Ramilo et al., 2007) and in vitro (Jenner and Young, 2005). This generalizability of the respiratory viral response signature finding illustrates that the host response to respiratory viral infections is robust and conserved such that it can be discerned in divergent patient populations (healthy adult volunteers experimentally infected with HRV or RSV and children hospitalized with influenza A). Second, this finding illustrates the dominance of a pathogen specific response at time of peak symptoms over a generalized “infection” response, as discrimination between viral and bacterial infection is possible. The ability of these signatures to differentiate between pathogen classes (viral versus bacterial) provides a marked distinction between these findings and current methods of infectious or inflammatory illness classification (e.g. peripheral white blood cell count or measurement of inflammatory markers such as C-reactive protein). The sensitivity and specificity of these markers in both our experimental setting and when applied to a cohort from the literature data represent an improvement on the performance of current rapid (e.g. rapid antigen testing) diagnostics as well as current culture-based diagnostics. A combination of these tests may ultimately prove to offer the best sensitivity and specificity for disease diagnosis. These data provide an important backbone to the concept that host peripheral blood gene expression may be a valuable tool alone or in conjunction with standard microbiologic testing for infectious diseases. Validation in an additional community based cohort, as well as developing signatures to diagnose pre-symptomatic viral respiratory infections is desirable.
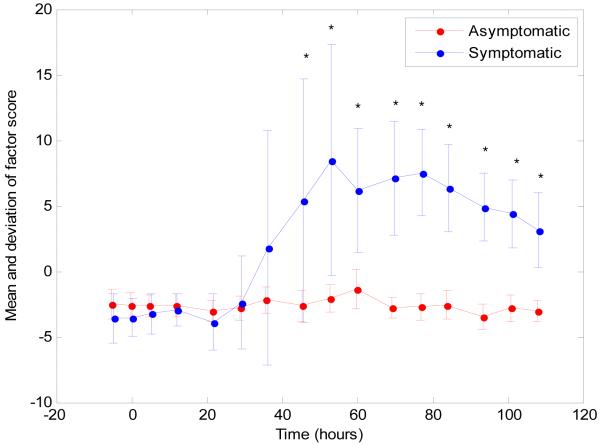
An important question that arises is whether the changes in host gene expression described here occur before peak symptoms? While still preliminary, we have time course data on subsets of these cohorts. The factor analysis was applied using the RSV, HRV and influenza data from all samples at all times, from which the factor discussed above [Factor 16] was constituted. In Figure 5 we plot the factor score (strength) of the discriminative factor, as a function of time. Two curves are depicted, representing the average factor scores, averaged separately for those that would eventually be symptomatic, and those that would not. The differences in f scores between individuals who remain asymptomatic and those who become symptomatic reach statistical significance (p = 0.028) at 45.5 hours following inoculation. This factor was found to be detectable prior to development of peak symptoms among symptomatic individuals. Thus, using host response as the diagnostic paradigm, presymptomatic diagnosis may be possible.
Figure 5.
Detection of the acute respiratory viral factor occurs earlier than time of peak symptoms. Factor trajectory for the acute respiratory viral factor described in Figure 2 is shown for the symptomatic (blue) and asymptomatic (red) subjects from the influenza challenge study. Notably, factor 16 is detectable prior to the timing of peak symptoms. Each point represents the average factor score for the samples that fall into that group, with error bars representing the standard deviation. For example, the blue dot at Time 0 represents all samples from subjects immediately post inoculation who will subsequently become symptomatic (9 subjects). A t-test was performed at teach timepoint for difference in factor score from those who will become symptomatic from those who will remain asymptomatic. The difference between factor scores for symptomatic and asymptomatic became significant at P < 0.03 at 45.5 hours and continued through the end of the measurements. * = p <0.03.
Signature validation across experimentally infected cohorts illustrates the robust nature of the host response to viral infection. Additional validation of the gene expression signatures in other community-based cohorts would elevate these findings to a true diagnostic test that could enhance or supersede traditional microbiologic based diagnostics. Additionally, such data would be extremely valuable if it could be used to either diagnose infection class prior to standard microbiologic studies (i.e. in the early phases of disease) or indicate prognosis following disease acquisition or therapeutic intervention. In our study, we were able to utilize an easily obtained sample (peripheral blood) to characterize response to a respiratory infection. While development of a diagnostic test that utilizes host gene expression to characterize or predict infectious diseases is not yet possible from the data generated in this study, it represents an important advance showing that peripheral blood gene expression can be used to characterize host response to infection.
Experimental Procedures
All exposures were approved by the relevant institutional review boards (IRBs) and conducted according to the Declaration of Helsinki. Funding for this study was provided by the US Defense Advanced Research Projects Agency (DARPA) through contract N66001-07-C-2024.
Human viral challenges
HRV Cohort (n=20)
We recruited healthy volunteers via advertisement to participate in the HRV challenge study through an active screening protocol at the University of Virginia (Charlottesville, VA). Subjects who met inclusion criteria underwent informed consent and pre-screening for serotype-specific anti-HRV approximately two weeks prior to study start date. On the day prior to inoculation, subjects underwent repeat HRV antibody testing as well as baseline laboratory studies, including complete blood count, serum chemistries, and hepatic enzymes. On day of inoculation, 106 TCID50 GMP HRV serotype 39 (Charles River Laboratories, Malvern PA) was inoculated intranasally according to published methods (Drake et al., 2000; Gwaltney et al., 1992; Turner, 2001). Subjects were admitted to the quarantine facility for 48 hours following HRV inoculation and remained for 48 hours following inoculation. Blood was sampled into RNA PAXGene™ collection tubes (PreAnalytix; Franklin Lakes, NJ) at pre-determined intervals post inoculation. Nasopharyngeal (NP) lavage samples were obtained from each subject daily for HRV titers to accurately gauge the success and timing of the HRV inoculation. Following the 48th hour post inoculation, subjects were released from quarantine and returned for three consecutive mornings for sample acquisition and symptom score ascertainment.
RSV Cohort (n=20)
A healthy volunteer intranasal challenge with RSV A was performed in a manner similar to the HRV challenge. The RSV challenge was performed by Retroscreen Virology, Ltd (London, UK) in 20 pre-screened volunteers who provided informed consent. On day of inoculation, a dose of 104 TCID50 RSV (serotype A) manufactured and processed under current good manufacturing practices (cGMP) by Meridian Life Sciences, Inc. (Memphis, TN USA) was inoculated intranasally per standard methods. Blood and NP lavage collection methods were similar to the HRV cohort, but continued throughout the quarantine. Due to the incubation period of RSV A, subjects were not released from quarantine until after the 288th hour AND were negative by rapid RSV antigen detection (BinaxNow Rapid RSV Antigen; Inverness Medical Innovations, Inc).
Influenza Cohort (n=17)
A healthy volunteer intranasal challenge with influenza A A/Wisconsin/67/2005 (H3N2) was performed at Retroscreen Virology, Ltd (Brentwood, UK) in 17 pre-screened volunteers who provided informed consent. On day of inoculation, a dose of 106 TCID50 Influenza A manufactured and processed under current good manufacturing practices (cGMP) by Baxter BioScience, (Vienna, Austria) was diluted and inoculated intranasally per standard methods at a varying dose (1:10, 1:100, 1:1000, 1:10000) with four to five subjects receiving each dose. Due to the incubation period, subjects were not released from quarantine until after the 168th hour. Blood and NP lavage collection continued throughout the duration of the quarantine. All subjects received oral oseltamivir (Roche Pharmaceuticals) 75 mg by mouth twice daily at day 6 following inoculation and were negative by rapid antigen detection (BinaxNow Rapid Influenza Antigen; Inverness Medical Innovations, Inc) at time of discharge.
Case Definitions
Symptoms were recorded twice daily using standardized symptom scoring(Jackson et al., 1958). The modified Jackson Score requires subjects to rank symptoms of upper respiratory infection (stuffy nose, scratchy throat, headache, cough, etc) on a scale of 0-3 of “no symptoms”, “just noticeable”, “bothersome but can still do activities” and “bothersome and cannot do daily activities”. Modified Jackson scores were tabulated to determine if subjects became symptomatic from the respiratory viral challenge. A modified Jackson score of >= 6 over the quarantine period was the primary indicator of successful viral infection(Turner, 2001) and subjects with this score were denoted as “symptomatic, infected” Viral titers from daily nasopharyngeal washes were used as corroborative evidence of successful infection using quantitative culture (Barrett et al., 2006; Jackson et al., 1958; Turner, 2001).
Subjects were classified as “asymptomatic, not infected” if the Jackson score was less than 6 over the five days of observation and viral shedding was not documented after the first 24 hours subsequent to inoculation. Standardized symptom scores tabulated at the end of each study to determine attack rate and time of maximal symptoms (time “T”).
Sample Collections
Subjects had the following samples taken 24 hours prior to inoculation with virus (baseline), immediately prior to inoculation (pre-challenge) and at set intervals following challenge: peripheral blood for serum and plasma, peripheral blood for RNA PAXgene™, NP wash for viral culture/PCR, urine, and exhaled breath condensate (EBC). For the HRV challenge, peripheral blood was taken at baseline, then at 4 hour intervals for the first 24 hours, then 6 hour intervals for the next 24 hours, then 8 hour intervals for the next 24 hours and then 24 hour intervals for the remaining 3 days of the study. For the RSV and influenza challenges, peripheral blood was taken at baseline, then at 8 hour intervals for the initial 120 hours and then 24 hours for two further days. For all cohorts, NP washes, urine and EBCs were taken at baseline and every 24 hours. Samples were aliquoted and frozen at −80°C immediately. This study is focused on comparison of baseline samples with RNA PAXgene™ samples taken at time of peak symptoms. Paxgene™ RNA from the timepoint of maximal symptoms was chosen for hybridization to Affymetrix U133a human microarrays for further analysis. For all results reported, gene expression signatures were evaluated at the time of maximal symptoms following viral inoculation for symptomatic subjects and a matched timepoint for asymptomatic subjects. Baseline (pre-inoculation) samples were also analyzed.
Community influenza and bacterial infection (“literature”) cohort
Raw data from Ramilo, et al, (Ramilo et al., 2007)was obtained from the public domain database GEO (www.ncbi.nlm.nih.gov/geo/projectIDGSE6269) and were analyzed independently using methods described below.
RNA purification/microarray analysis
RNA was extracted at Expression Analysis (Durham, NC) from whole blood using the PAXgene™ 96 Blood RNA Kit (PreAnalytiX, Valencia, CA) employing the manufacturer’s recommended protocol. Complete methodology can be viewed in the Supplementary Methods. Hybridization and microarray data collection was performed at Expression Analysis (Durham, NC) using the GeneChip® Human Genome U133A 2.0 Array (Affymetrix, Santa Clara, CA).
Statistical Analysis
Using just the data from the influenza challenge, we tested (Kruskal-Wallis) each probe for differential expression between subjects who were sick vs healthy at Time T. Due to the small sample size, there were no probes showing significant association after correction for multiple hypotheses (Bonferroni). We then analyzed jointly the results from all three trials in an ANOVA framework. In addition to the intercept term, we included in the design matrix indicators of sick versus healthy, t0 versus tmax, and indicator for each of rhinovirus and RSV, and interaction terms for rhinovirus – sick and RSV – sick (Supplemental Analysis). Following RMA normalization of raw probe data, sparse latent factor regression analysis was applied to each dataset (Aziz et al., 2007; Carvalho et al., 2008; Lucas et al., 2006; Wang et al., 2007a). This reduces the dimensionality of the complex gene expression array dataset assuming that many of the probe sets on the expression array chip are highly interrelated (targeting the same genes or genes in the same pathways). Dimension reduction is performed by constructing factors (groups of genes with related expression values). These are used in a sparse linear regression framework to explain the variation seen in all of the probe sets. By default, most of the coefficients in this linear regression are zero. Thus, a small number (e.g. 20) of factors explain variation seen in any single dataset. Factor loadings are defined as the coefficients of the factor regression, and, to explore the biological relevance any particular factor, we examine the genes that are “in” that factor -- the genes that show significantly non-zero factor loadings. “Factor scores” are defined as the vector that best describes the co-expression of the genes in a particular factor. Both factor loadings and factor scores are fit to the data concurrently. While 20 factors were used for the results reported here, we also considered 30 and 40, with minimal effect on the significant factor loadings. The initial models were derived using an unsupervised process (Acharya et al., 2008) (i.e. the model classified subjects based on gene expression pattern alone, without a priori knowledge of infection status). The top 30 genes in each factor were used as features for the sparse probit regression model to perform leave-one-out cross validation and generate ROC curves to estimate performance of the algorithm. The probit regression model selects the “top” predictive gene from the gene set for sample classification and generation of an ROC curve. Validation of the factor most discriminative between the asymptomatic and symptomatic state was performed using labeled data. Validation between datasets (HRV, RSV, and influenza A) was performed by training the regression model on one set of data (i.e. one viral exposure) and using this model to predict health or disease in a different data set (i.e. a different viral exposure). Validation of the model using the publically available dataset was performed by utilizing the joint factor analysis on the viral exposure dataset (HRV, RSV, and influenza), building a probit classifier using the top 30 genes from the most predictive factor and applying this classifier to the publically available dataset to estimate the predictive performance of the acute respiratory viral classifier.
Supplementary Material
Acknowledgements
The authors wish to acknowledge Daphne Jones, Stephanie Dobos and Kyle Breitschwerdt for data management; L. Brett Caram, MD for protocol design; and Anil Potti, MD for critical review of the manuscript. This work was supported by funding from the Defense Advanced Projects Research Agency (DARPA) lN66001-07-C-0092 (G.S.G.)
Contributor Information
Aimee K. Zaas, Division of Infectious Diseases and International Health; Department of Medicine; Institute for Genome Sciences and Policy; Duke University School of Medicine; Durham, NC.
Minhua Chen, Department of Electrical and Computer Engineering; Duke University, Durham, NC.
Jay Varkey, Division of Infectious Diseases and International Health; Department of Medicine; Institute for Genome Sciences and Policy; Duke University School of Medicine; Durham, NC.
Timothy Veldman, Institute for Genome Sciences and Policy; Duke University; Durham, NC.
Alfred O. Hero, III, Department of Electrical Engineering and Computer Science; University of Michigan, Ann Arbor, MI.
Joseph Lucas, Institute for Genome Sciences and Policy; Duke University; Durham, NC.
Yongsheng Huang, Department of Electrical Engineering and Computer Science; University of Michigan, Ann Arbor, MI.
Ronald Turner, University of Virginia School of Medicine; Charlottesville, VA.
Anthony Gilbert, Retroscreen Virology; Brentwood, UK.
Robert Lambkin-Williams, Retroscreen Virology; Brentwood, UK.
N. Christine Øien, Institute for Genome Sciences and Policy; Duke University; Durham, NC.
Bradly Nicholson, Division of Infectious Diseases; Durham Veteran’s Affairs Medical Center; Durham, NC.
Stephen Kingsmore, National Center for Genome Research; Santa Fe, NM.
Lawrence Carin, Department of Electrical and Computer Engineering; Duke University, Durham, NC.
Christopher W. Woods, Division of Infectious Diseases and International Health; Department of Medicine; Institute for Genome Sciences and Policy; Duke University School of Medicine; Durham, NC.
Geoffrey S. Ginsburg, Institute for Genome Sciences and Policy; Duke University; Durham, NC.
References
- Acharya CR, Hsu DS, Anders CK, Anguiano A, Salter KH, Walters KS, Redman RC, Tuchman SA, Moylan CA, Mukherjee S, et al. Gene expression signatures, clinicopathological features, and individualized therapy in breast cancer. Jama. 2008;299:1574–1587. doi: 10.1001/jama.299.13.1574. [DOI] [PubMed] [Google Scholar]
- Arruda E, Pitkaranta A, Witek TJ, Jr., Doyle CA, Hayden FG. Frequency and natural history of rhinovirus infections in adults during autumn. J Clin Microbiol. 1997;35:2864–2868. doi: 10.1128/jcm.35.11.2864-2868.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz H, Zaas A, Ginsburg GS. Peripheral blood gene expression profiling for cardiovascular disease assessment. Genomic Med. 2007;1:105–112. doi: 10.1007/s11568-008-9017-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett B, Brown R, Voland R, Maberry R, Turner R. Relations among questionnaire and laboratory measures of rhinovirus infection. Eur Respir J. 2006;28:358–363. doi: 10.1183/09031936.06.00002606. [DOI] [PubMed] [Google Scholar]
- Bhoj VG, Sun Q, Bhoj EJ, Somers C, Chen X, Torres JP, Mejias A, Gomez AM, Jafri H, Ramilo O, Chen ZJ. MAVS and MyD88 are essential for innate immunity but not cytotoxic T lymphocyte response against respiratory syncytial virus. Proc Natl Acad Sci U S A. 2008;105:14046–14051. doi: 10.1073/pnas.0804717105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boldrick JC, Alizadeh AA, Diehn M, Dudoit S, Liu CL, Belcher CE, Botstein D, Staudt LM, Brown PO, Relman DA. Stereotyped and specific gene expression programs in human innate immune responses to bacteria. Proc Natl Acad Sci U S A. 2002;99:972–977. doi: 10.1073/pnas.231625398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brieman L. Statistical modeling: the two cultures. Statistical Science. 2001;16:199–215. [Google Scholar]
- Bryant PA, Venter D, Robins-Browne R, Curtis N. Chips with everything: DNA microarrays in infectious diseases. Lancet Infect Dis. 2004;4:100–111. doi: 10.1016/S1473-3099(04)00930-2. [DOI] [PubMed] [Google Scholar]
- Campbell CJ, Ghazal P. Molecular signatures for diagnosis of infection: application of microarray technology. J Appl Microbiol. 2004;96:18–23. doi: 10.1046/j.1365-2672.2003.02112.x. [DOI] [PubMed] [Google Scholar]
- Carvalho C, Lucas J, Wang Q, Chang J, Nevins JR, West M. High dimensional sparse factor modelling: applications in gene expression genomics. Journal of American Statistical Association. 2008 doi: 10.1198/016214508000000869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiarini A, Palmeri A, Amato T, Immordino R, Distefano S, Giammanco A. Detection of bacteria and yeast species by the BACTEC 9120 automated system with the routine use of aerobic, anaerobic, and fungal media. J Clin Microbiol. 2008 doi: 10.1128/JCM.01014-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin KC, Cresswell P. Viperin (cig5), an IFN-inducible antiviral protein directly induced by human cytomegalovirus. Proc Natl Acad Sci U S A. 2001;98:15125–15130. doi: 10.1073/pnas.011593298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drake CL, Roehrs TA, Royer H, Koshorek G, Turner RB, Roth T. Effects of an experimentally induced rhinovirus cold on sleep, performance, and daytime alertness. Physiol Behav. 2000;71:75–81. doi: 10.1016/S0031-9384(00)00322-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dressman HK, Muramoto GG, Chao NJ, Meadows S, Marshall D, Ginsburg GS, Nevins JR, Chute JP. Gene expression signatures that predict radiation exposure in mice and humans. PLoS Med. 2007;4:e106. doi: 10.1371/journal.pmed.0040106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falsey AR, Hennessey PA, Formica MA, Cox C, Walsh EE. Respiratory syncytial virus infection in elderly and high-risk adults. N Engl J Med. 2005;352:1749–1759. doi: 10.1056/NEJMoa043951. [DOI] [PubMed] [Google Scholar]
- Fjaerli HO, Bukholm G, Krog A, Skjaeret C, Holden M, Nakstad B. Whole blood gene expression in infants with respiratory syncytial virus bronchiolitis. BMC Infect Dis. 2006;6:175. doi: 10.1186/1471-2334-6-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garman KS, Acharya CR, Edelman E, Grade M, Gaedcke J, Sud S, Barry W, Diehl AM, Provenzale D, Ginsburg GS, et al. A genomic approach to colon cancer risk stratification yields biologic insights into therapeutic opportunities. Proc Natl Acad Sci U S A. 2008;105:19432–19437. doi: 10.1073/pnas.0806674105. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Gums JG, Pelletier EM, Blumentals WA. Oseltamivir and influenza-related complications, hospitalization and healthcare expenditure in healthy adults and children. Expert Opin Pharmacother. 2008;9:151–161. doi: 10.1517/14656566.9.2.151. [DOI] [PubMed] [Google Scholar]
- Gwaltney JM, Jr., Hendley O, Hayden FG, McIntosh K, Hollinger FB, Melnick JL, Turner RB. Updated recommendations for safety-testing of viral inocula used in volunteer experiments on rhinovirus colds. Prog Med Virol. 1992;39:256–263. [PubMed] [Google Scholar]
- Hong CY, Lin RT, Tan ES, Chong PN, Tan YS, Lew YJ, Loo LH. Acute respiratory symptoms in adults in general practice. Fam Pract. 2004;21:317–323. doi: 10.1093/fampra/cmh319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson GG, Dowling HF, Spiesman IG, Boand AV. Transmission of the common cold to volunteers under controlled conditions. I. The common cold as a clinical entity. AMA Arch Intern Med. 1958;101:267–278. doi: 10.1001/archinte.1958.00260140099015. [DOI] [PubMed] [Google Scholar]
- Jenner RG, Young RA. Insights into host responses against pathogens from transcriptional profiling. Nat Rev Microbiol. 2005;3:281–294. doi: 10.1038/nrmicro1126. [DOI] [PubMed] [Google Scholar]
- Jiang D, Guo H, Xu C, Chang J, Gu B, Wang L, Block TM, Guo JT. Identification of three interferon-inducible cellular enzymes that inhibit the replication of hepatitis C virus. J Virol. 2008;82:1665–1678. doi: 10.1128/JVI.02113-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston SL. Natural and experimental rhinovirus infections of the lower respiratory tract. Am J Respir Crit Care Med. 1995;152:S46–52. doi: 10.1164/ajrccm/152.4_Pt_2.S46. [DOI] [PubMed] [Google Scholar]
- Johnstone J, Majumdar SR, Fox JD, Marrie TJ. Viral Infection in Adults Hospitalized with Community Acquired Pneumonia: Prevalence, Pathogens and Presentation. Chest. 2008 doi: 10.1378/chest.08-0888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonathan N. Diagnostic utility of BINAX NOW RSV--an evaluation of the diagnostic performance of BINAX NOW RSV in comparison with cell culture and direct immunofluorescence. Ann Clin Microbiol Antimicrob. 2006;5:13. doi: 10.1186/1476-0711-5-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchberger S, Majdic O, Stockl J. Modulation of the immune system by human rhinoviruses. Int Arch Allergy Immunol. 2007;142:1–10. doi: 10.1159/000095993. [DOI] [PubMed] [Google Scholar]
- Kobayashi SD, Braughton KR, Whitney AR, Voyich JM, Schwan TG, Musser JM, DeLeo FR. Bacterial pathogens modulate an apoptosis differentiation program in human neutrophils. Proc Natl Acad Sci U S A. 2003;100:10948–10953. doi: 10.1073/pnas.1833375100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooperberg C, Ruczinski I, LeBlanc ML, Hsu L. Sequence analysis using logic regression. Genet Epidemiol. 2001;21(Suppl 1):S626–631. doi: 10.1002/gepi.2001.21.s1.s626. [DOI] [PubMed] [Google Scholar]
- Lambert SB, Whiley DM, O’Neill NT, Andrews EC, Canavan FM, Bletchly C, Siebert DJ, Sloots TP, Nissen MD. Comparing nose-throat swabs and nasopharyngeal aspirates collected from children with symptoms for respiratory virus identification using real-time polymerase chain reaction. Pediatrics. 2008;122:e615–620. doi: 10.1542/peds.2008-0691. [DOI] [PubMed] [Google Scholar]
- Landry ML, Cohen S, Ferguson D. Real-time PCR compared to Binax NOW and cytospin-immunofluorescence for detection of influenza in hospitalized patients. J Clin Virol. 2008;43:148–151. doi: 10.1016/j.jcv.2008.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas JE, Carvalho CM, Wang Q, Bild A, Nevins JR, West M. Bayesian Inference for Gene Expression and Proteomics. Cambridge University Press; 2006. Sparse statistical modelling in gene expression genomics; pp. 155–176. [Google Scholar]
- Meadows SK, Dressman HK, Muramoto GG, Himburg H, Salter A, Wei Z, Ginsburg G, Chao NJ, Nevins JR, Chute JP. Gene expression signatures of radiation response are specific, durable and accurate in mice and humans. PLoS ONE. 2008;3:e1912. doi: 10.1371/journal.pone.0001912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Memoli MJ, Morens DM, Taubenberger JK. Pandemic and seasonal influenza: therapeutic challenges. Drug Discov Today. 2008;13:590–595. doi: 10.1016/j.drudis.2008.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltola V, Waris M, Osterback R, Susi P, Ruuskanen O, Hyypia T. Rhinovirus transmission within families with children: incidence of symptomatic and asymptomatic infections. J Infect Dis. 2008;197:382–389. doi: 10.1086/525542. [DOI] [PubMed] [Google Scholar]
- Proud D, Turner RB, Winther B, Wiehler S, Tiesman JP, Reichling TD, Juhlin KD, Fulmer AW, Ho BY, Walanski AA, et al. Gene Expression Profiles During In Vivo Human Rhinovirus Infection: Insights into the Host Response. Am J Respir Crit Care Med. 2008 doi: 10.1164/rccm.200805-670OC. [DOI] [PubMed] [Google Scholar]
- Rahman M, Vandermause MF, Kieke BA, Belongia EA. Performance of Binax NOW Flu A and B and direct fluorescent assay in comparison with a composite of viral culture or reverse transcription polymerase chain reaction for detection of influenza infection during the 2006 to 2007 season. Diagn Microbiol Infect Dis. 2008;62:162–166. doi: 10.1016/j.diagmicrobio.2007.10.012. [DOI] [PubMed] [Google Scholar]
- Rakes GP, Arruda E, Ingram JM, Hoover GE, Zambrano JC, Hayden FG, Platts-Mills TA, Heymann PW. Rhinovirus and respiratory syncytial virus in wheezing children requiring emergency care. IgE and eosinophil analyses. Am J Respir Crit Care Med. 1999;159:785–790. doi: 10.1164/ajrccm.159.3.9801052. [DOI] [PubMed] [Google Scholar]
- Ramilo O, Allman W, Chung W, Mejias A, Ardura M, Glaser C, Wittkowski KM, Piqueras B, Banchereau J, Palucka AK, Chaussabel D. Gene expression patterns in blood leukocytes discriminate patients with acute infections. Blood. 2007;109:2066–2077. doi: 10.1182/blood-2006-02-002477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JL, Lee BE, Kothapalli S, Craig WR, Fox JD. Use of throat swab or saliva specimens for detection of respiratory viruses in children. Clin Infect Dis. 2008;46:e61–64. doi: 10.1086/529386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruczinski I, Kooperberg C, LeBlanc ML. Logic Regression. J Computational and Graphical Statistics. 2003;12:475–511. [Google Scholar]
- Schaller M, Hogaboam CM, Lukacs N, Kunkel SL. Respiratory viral infections drive chemokine expression and exacerbate the asthmatic response. J Allergy Clin Immunol. 2006;118:295–302. doi: 10.1016/j.jaci.2006.05.025. quiz 303-294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo D, Ginsburg GS, Goldschmidt-Clermont PJ. Gene expression analysis of cardiovascular diseases: novel insights into biology and clinical applications. J Am Coll Cardiol. 2006;48:227–235. doi: 10.1016/j.jacc.2006.02.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmons CP, Popper S, Dolocek C, Chau TN, Griffiths M, Dung NT, Long TH, Hoang DM, Chau NV, Thao le TT, et al. Patterns of host genome-wide gene transcript abundance in the peripheral blood of patients with acute dengue hemorrhagic fever. J Infect Dis. 2007;195:1097–1107. doi: 10.1086/512162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner RB. Ineffectiveness of intranasal zinc gluconate for prevention of experimental rhinovirus colds. Clin Infect Dis. 2001;33:1865–1870. doi: 10.1086/324347. [DOI] [PubMed] [Google Scholar]
- Wang Q, Carvalho CM, Lucas JE, West M. BFRM: Bayesian factor regression modeling. Bulletin of the International Society of Bayesian Analysis. 2007a;14:4–5. [Google Scholar]
- Wang X, Hinson ER, Cresswell P. The interferon-inducible protein viperin inhibits influenza virus release by perturbing lipid rafts. Cell Host Microbe. 2007b;2:96–105. doi: 10.1016/j.chom.2007.06.009. [DOI] [PubMed] [Google Scholar]
- Wang Z, Neuburg D, Li C, Su L, Kim JY, Chen JC, Christiani DC. Global gene expression profiling in whole-blood samples from individuals exposed to metal fumes. Environ Health Perspect. 2005;113:233–241. doi: 10.1289/txg.7273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M, Kao MC, Nunez-Iglesias J, Nevins JR, West M, Zhou XJ. An integrative approach to characterize disease-specific pathways and their coordination: a case study in cancer. BMC Genomics. 2008;9(Suppl 1):S12. doi: 10.1186/1471-2164-9-S1-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.