Abstract
Urine has become one of the most attractive biofluids in clinical proteomics, for its procurement is easy and noninvasive and it contains sufficient proteins and peptides. Urinary proteomics has thus rapidly developed and has been extensively applied to biomarker discovery in clinical diseases, especially kidney diseases. In this review, we discuss two important aspects of urinary proteomics in detail, namely, sample preparation and proteomic technologies. In addition, data mining in urinary proteomics is also briefly introduced. At last, we present several successful examples on the application of urinary proteomics for biomarker discovery in kidney diseases, including diabetic nephropathy, IgA nephropathy, lupus nephritis, renal Fanconi syndrome, acute kidney injury, and renal allograft rejection.
Keywords: Clinical proteomics, Urinary proteomics, Biomarker, Kidney diseases
1. Introduction
With the completion of the human genome project in 2001, approximately 25 000 human genes have been identified by genomics technology and then placed into databases. However, it is impossible to get enough information on diseases solely by studying the genome, for proteins are responsible for the phenotypes of cells. Thus, more and more researchers have focused on the proteome studies in diseases.
Concerning kidney diseases, proteomics is a promising approach for the detection of novel biomarkers in biological fluids such as urine, plasma, and serum. Among these, urine is regarded as the most attractive proteomic sample due to several advantages: (1) it is noninvasive and easy to be obtained in large amounts, (2) proteins and peptides in urine are quite stable and less complex, and (3) the amount and composition of urinary proteome directly reflect changes in functions of the kidney and the urogenital tract. Although, on the other hand, urine is also a difficult proteomic sample due to its wide variability in protein concentrations, this can be compensated by standardization based on urinary creatinine (Vestergaard and Leverett, 1958) or urinary housekeeping peptides, which are present almost ubiquitously in human urine independent of age, sex, health, and drug (Theodorescu et al., 2005). Recently, urinary proteomics has become an important and efficient approach for biomarker discovery in kidney diseases. In this review, we shall discuss preparation of urine samples and main technological approaches as crucial steps for urinary proteomics. Additionally, we want to describe the data mining of proteome analysis shortly and present several examples on the successful application of urinary proteomics for biomarker discovery in kidney diseases.
2. Urine sample preparation
2.1. Sample collection: first-void or midstream urine
Collection of midstream urine is always considered as the standard for almost all urine analysis. While no significant differences were observed between first-void and midstream urine in males, there are significant variations in female first-void urine compared with midstream urine, probably due to bacterial contamination (Schaub et al., 2004). Other studies showed that approximately 40% of bacterial contamination in female urine samples derived from the skin contamination (Finkel, 2006; Lifshitz and Kramer, 2000). However, by using a prostatic fluid indicative panel, Theodorescu et al. (2008) discovered a panel of 12 novel prostate cancer (PCa) biomarkers in the first-void urine, in addition to collection of sufficient amounts of prostatic fluid. Thus, first-void urine may be recommended in prostate diseases. But sometimes it is really difficult to differentiate first-void urine and midstream urine; for example, urine samples of newborns are usually collected with a urine collection bag. The differentiation of first-void and midstream urine, therefore, needs extensive investigations and further elucidations.
2.2. Sample handling
Lee et al. (2008) used standard Bradford assay to examine four common methods of protein extraction: ethanol precipitation, vacuum centrifugation, microconcentration, and reverse phase trapping column. They found, vacuum centrifugation method yielded the highest protein concentration, probably due to its inability in salt removal. However, when comparing the numbers of proteins and peptides identified and cumulative spectral count by liquid chromatography coupled to mass spectrometry (LC-MS), no significant differences were observed among the four methods. Therefore, when choosing a suitable approach for protein extraction, a number of other factors, other than a method, such as the technical cost, speed, and compatibility with downstream protocols need to be considered.
The effect of multiple freeze-thaw cycles in urine sample handling on the urinary proteome is another concern. By use of surface-enhanced laser desorption/ionization time-of-flight mass spectrometry (SELDI-TOF-MS), Schaub et al. (2004) showed that there were no significant changes of urine protein profiles before freezing and after 1 to 4 freeze-thaw cycles, but intensities at some weak peaks became undetectable after the fifth freeze-thaw cycle. Fiedler et al. (2007), also using SELDI-TOF-MS, found that some relative intensities of mass signals were altered in once-frozen samples, whereas in recurrent freeze-thaw cycles (3×), no further significant variation was detected. In addition, it is also reported that the levels of some urinary proteins were changed in 4–7 freeze-thaw cycles (Thongboonkerd, 2007). The discrepancy of these findings may be explained by the lack of technical and biological replicates. In general, it is safe to avoid multiple freeze-thaw cycles in clinical urine sample handling in order to minimize the errors in experiment process.
2.3. Sample storage
Weissinger et al. (2007) reported that no significant alterations of urine proteome were observed in urine samples stored at −20 °C for several years. Klasen et al. (1999) reported that the contents of urine sample including albumin, transferrin, IgG, α1-microglobulin, and β2-microglobulin were more stable when stored at −70 °C than at −20 °C. Zerefos and Vlahou (2008) reported that occasional changes of protein profile were observed for a 24-h storage at 4 °C; thus, shorter storage time (up to 6 h) at 4 °C may be a good choice. Therefore, according to these findings, it can be concluded that proteins and polypeptides are quite stable in urine samples. The stability might be due to the fact that the urine storage time in the bladder before voiding is sufficient to complete proteolytic degradation by endogenous proteases. However, exosomes in urine represent a less stability. Zhou et al. (2006a) reported that storage at −20 °C resulted in a major loss (72.6%) of urinary exosome-associated proteins, whereas storage at −80 °C resulted only in a mild loss (14%). Thus, for a limited time, the urine can be stored at 4 °C, and for a relatively long time, it is best to freeze at −80 °C before analysis.
3. Technical aspects for urinary proteomics
3.1. Mass spectrometry-based proteomic technologies
Mass spectrometry (MS) is widely used as a method for protein identification. In this review, we shall introduce it briefly [For more detailed information, see (Feng et al., 2008; Palmblad et al., 2009)]. There are two main components of a mass spectrometer, namely, the ionization technique and the mass analyzer. Matrix-assisted laser desorption/ionization (MALDI) and electrospray ionization (ESI) are the most commonly-used ionization technique to volatilize and ionize proteins and peptides for mass analysis with high sensitivity and accuracy. There are four basic different types of mass analyzers: Time-of-flight (TOF), quadrupole (Q), ion trap (IT), and Fourier transform ion cyclotron resonance (FTICR). For MALDI, the sample is mixed with a matrix, followed by spotting onto a target for co-crystallization. At last, proteins in the sample are ionized via laser pulses. ESI ionizes proteins out of a solution. This method generates charged droplets in a high-voltage field; solvent from the charged droplets evaporates, and then multiply charged ions are generated. For a detailed overview on the advantages and disadvantages of MALDI and ESI, please refer to the review by Mischak et al. (2007).
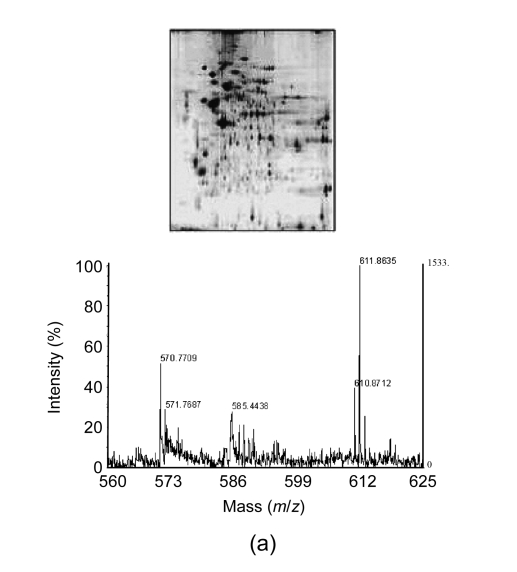
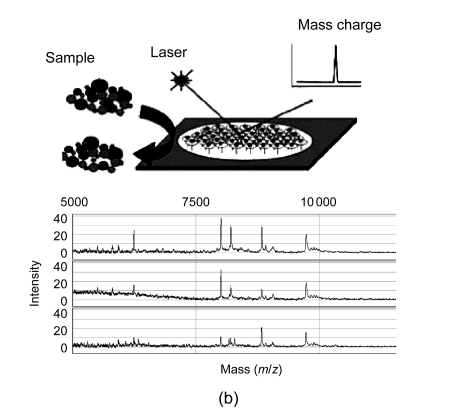
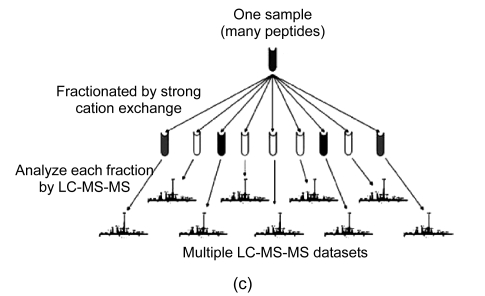
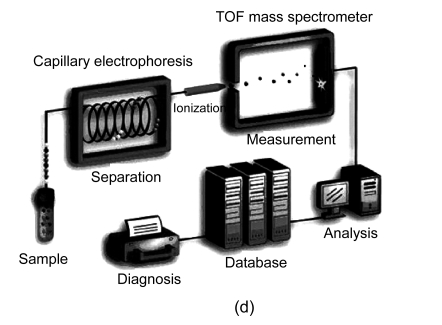
In general, four different types of MS-based proteomic technologies are used in proteomics, namely, two-dimensional gel electrophoresis coupled to mass spectrometry (2DE-MS), surface-enhanced laser desorption/ionization coupled to mass spectrometry (SELDI-MS), liquid chromatography coupled to mass spectrometry (LC-MS), and capillary electrophoresis coupled to mass spectrometry (CE-MS) (Fig. 1). Their advantages and disadvantages are summarized in Table 1. In the following section, we shall discuss the four proteomic platforms in turn.
Fig. 1.
Four different types of MS-based proteomic technologies
(a) 2DE-MS; (b) SELDI-MS; (c) LC-MS; (d) CE-MS
Table 1.
Summary of advantages and disadvantages of different MS-based proteomic technologies
| Technology | Advantages | Disadvantages |
| 2DE-MS | Widely available, applicable to large molecules | Time-consuming, technically demanding, difficult analysis for smaller protein (<10 kDa) and highly hydrophobic proteins |
| SELDI-MS | Ease of operation, automation, high throughput capability, low sample volume required | Low reproducibility, lack of comparability, low-resolution MS |
| LC-MS | High sensitivity, automation, multidimensionality | Time-consuming, sensitive to interfering compounds |
| CE-MS | High resolution, high selectivity, fast separation, automation, low cost | Not well suited for the analysis of high-molecular-weight proteins (>20 kDa) |
3.1.1. Two-dimensional gel electrophoresis coupled to mass spectrometry (2DE-MS)
2DE was first reported for protein separation by O′Farrell (1975) and is still widely used today. In 2DE, proteins are separated in the first dimension by isoelectric focusing (IEF) (proteins migrate to their isoelectric point in a pH-gradient), and then in the second dimension by sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) (proteins migrate on the basis of their molecular masses). Once the separation is finished, the proteins are stained, analyzed by computer-assisted programs, and identified by MS [for detailed experiment procedures, see review by López (2007) and Penque (2009)]. 2DE is time-consuming, technically demanding, and has low reproducibility. In addition, it is difficult to analyze smaller proteins (<10 kDa) and highly hydrophobic proteins by 2DE. Recently, the concept of two-dimensional difference gel electrophoresis (2D-DIGE) was introduced to reduce gel-to-gel variability. In 2D-DIGE, two samples are differentially labeled with fluorescence dyes (Cy3 and Cy5) and mixed together and then separated within the same 2DE gel (Timms and Cramer, 2008). An internal standard labeled with a third dye (i.e., Cy2) can be also incorporated, thereby allowing more accurate quantitative analysis. Although it is satisfactory to compare two samples with 2D-DIGE, the comparison of several different experiments remains challenging.
3.1.2. Surface-enhanced laser desorption/ionization coupled to mass spectrometry (SELDI-MS)
SELDI reduces the complexity of a biological sample by selective interactions of polypeptides with different active surfaces (hydrophilic materials, reversed-phase materials, lectin or antibody affinity reagents). After the interaction phase, only a small fraction of polypeptides can bind to the surface of the SELDI chip, depending on the concentration, pH, salt content, presence of interfering compounds like lipids, etc., while the unbound samples are washed away. A matrix is added to the sample surface to absorb energy and to allow vaporization and ionization by laser for further MS detection. SELDI-MS is widely used to detect biomarkers in a variety of diseases due to its ease of operation and its high throughput (Huang et al., 2009). However, there are several limitations for SELDI-MS approach including low reproducibility and comparability of datasets due to different chip surfaces and conditions, as well as low resolution of the mass spectrometer. Recently, material-enhanced laser desorption/ionization (MELDI) with enlarged active binding surfaces has been introduced to improve the low reproducibility of binding to SELDI surfaces (Najam-ul-Haq et al., 2007). In addition, more appropriate mass spectrometers, such as MALDI-TOF/TOF instruments, are described to solve the low resolution of the mass spectrometer (Orvisky et al., 2006).
3.1.3. Liquid chromatography coupled to mass spectrometry (LC-MS)
LC provides a powerful fractionation method that is compatible with virtually any type of mass spectrometers. This method separates large amounts of analytes on an LC-column with high sensitivity. The column contains the sorbent materials with various physical, chemical, and immunological properties. When the sample dissolves in a solvent and subsequently moves through the column, the peptides in the sample can be separated by elution at different time points depending on their separation characteristics. A sequential separation using different matrices in two independent steps provides a multidimensional fractionation that can generate large amount of information. Multidimensional protein identification technology (MudPIT) or a 2D liquid-phase fractionation approach is well-suited for in-depth analysis of body fluids such as urine. However, LC-MS is time-consuming, and sensitive to interfering compounds and precipitation of analytes on LC-column materials. Therefore, application of LC-MS is not suitable to routine clinical analysis. Recently, monoliths as stationary phase have been used in nano-LC and micro-LC separations due to their fritless design, ease of preparation, and high permeability (Liu et al., 2009; Wu et al., 2008). The application of monoliths can improve the separation efficiency and analytical throughput.
3.1.4. Capillary electrophoresis coupled to mass spectrometry (CE-MS)
CE-MS is a widely-used MS-based approach for the proteomic analysis of body fluids such as urine. This approach is based on CE as a front-end fractionation device coupled to a mass spectrometer. CE separates proteins with high resolution based on their migration through a buffer-filled capillary column in an electrical field (300 to 500 V/cm). Several advantages of CE-MS are listed as follows: (1) it provides fast and robust separation using an inexpensive capillaries, (2) it is compatible with most buffers and analytes, (3) it provides a stable constant flow, which may avoid interfering subsequent MS detection by buffer gradients, (4) CE can be interfaced with almost any mass spectrometer, and (5) it has the ability to recondition fast with NaOH after each run. A limitation of CE-MS is that it is not well suited for the analysis of high-molecular-weight proteins (>20 kDa) as they tend to precipitate in the acidic buffers that are generally used for CE-MS analysis. However, it is not a considerable drawback for CE-MS, because large proteins can be removed by ultrafiltration, and urinary proteome contains a great number of various peptides and low-molecular weight proteins. Another limitation of CE-MS is that only small sample volume can be loaded onto the capillary, leading to a lower selectivity compared with LC. Improved methods of ionization by micro- and nano-ion sprays, as well as improvements of the detection limits of mass spectrometers, can resolve the problem to a large extent. Advantages and disadvantages of CE-MS in regard to biomarker discovery and clinical applications have been described in a recent review (Mischak et al., 2009).
3.2. Protein microarrays
As a non-MS-based approach, protein microarrays can be used to discover proteomic biomarkers in biofluid samples, including serum, plasma, and urine. More detailed information can be referred to a recent review (Caiazzo et al., 2009). Protein microarrays are designed to print specific antibodies or antigens onto a support surface, generally a slide or membrane. A single sample is hybridized to the array. The captured antigens or antibodies are subsequently detected. This approach allows for immune detection of multiple proteins. However, the wide applications of protein microarrays are restricted for several limitations, including requirement for highly specific proteins or antibodies and conservation of protein functionality during immobilization, as well as the provision of the required sensitivity.
4. Data mining
Once data have been generated, they are submitted to data mining algorithms. Generally, there are two categories of data mining approaches: the unsupervised and the supervised. The unsupervised learning approaches, such as K-means clustering, principal component analysis (PCA), and hierarchal clustering, are analogous to clustering, which can identify natural clusters on the data. While the supervised learning approaches, such as classification and regression trees, Bayesian classification, neural networks, genetic algorithms, and support vector machine, are analogous to classification, they need to divide samples into a validation set and a training set (if the training set is too large, it can be subdivided into a training set and a test set). Feature selection and model-building are performed on the training set and their performances are assessed in the test set in order to obtain better accuracy (Ahmed, 2009). In addition, cross-validation techniques such as k-fold cross-validation and leave-one-out cross-validation can be used to maximize the training set size and avoid falsely low error estimates (Dakna et al., 2009). A detailed discussion of the advantages and disadvantages of certain algorithms is beyond the scope of this review [for more detailed information, see (Fung et al., 2005)]. Generally, each data mining algorithm must be matched to the specific statistical problems to be addressed.
5. Clinical applications of urinary proteomics in kidney diseases
5.1. Diabetic nephropathy (DN)
DN is a serious complication of diabetes. According to World Health Organization, approximately 10%–20% of people with diabetes die of kidney failure. However, regarding to diagnosis of DN, routine renal biopsy is invasive and the microalbuminuria level has inadequate sensitivity and specificity. Thus, noninvasive and earlier biomarkers are urgently warranted to better identify and treat individuals at high risk for DN.
Bellei et al. (2008) applied 2DE and ESI-Q-TOF MS/MS to identify urine biomarkers for the early renal alterations in type 2 diabetes patients. Urine samples were obtained from type 2 diabetic patients with normoalbuminuria (n=10), type 2 diabetic nephropathy patients (n=12), and health subjects (n=12). Comparing the two patients groups with health subjects, 11 differentially-expressed proteins were identified. Of these, 3 proteins (the prostatic acid phosphatase precursor, the ribonuclease, and the kallikrein-3) decreased and 8 proteins progressively increased, including transthyretin precursor, immunoglobulin kappa (Igκ) chain C region, Igκ chain V-II region Cum, Igκ-chain V-III region SIE, carbonic anhydrase 1, plasma retinol-binding protein, β2-microglobulin precursor, and β2-glycoprotein 1.
Lapolla et al. (2009) used MALDI-MS to discover protein biomarkers in urine samples from diabetic, nephropathic, and diabetic-nephropathic patients, as well as healthy subjects. Proteins with masses of 1219, 1912, and 2049 Da were found to be suitable to distinguish among the four groups and they were identified as originating forms of uromodulin, the collagen α-1 (I) chain precursor, and the collagen α-5 (IV) chain precursor by MALDI-TOF/TOF.
Recently, several groups have reported the biomarkers associated with DN in Chinese population. In our own laboratory, we have used SELDI-TOF-MS to discover urinary protein biomarkers for detection of the early stage DN. Urine samples were obtained from 106 diabetic patients with normo-, micro-, or macroalbuminuria and 50 healthy subjects. Four proteins of mass 4239.0, 4453.5, 5281.1, and 5898.5 Da were detected as the potential biomarkers for DN with a specificity of 80% and a sensitivity of 75%. The work is now in progress on identification and validation of these proteins involved in early prediction of DN (Gu et al., 2008).
Jiang et al. (2009a) used 2DE and MALDI-TOF/MS to analyze urine samples from type 2 diabetes patients with normoalbuminuria (DM group), microalbuminuria (DN1 group), macroalbuminuria (DN2 group), and control group. A total of 12 proteins were identified as candidate biomarkers. Among them, E-cadherin was increased by 1.3-, 5.2-, and 8.5-fold in DM, DN1, and DN2 groups, respectively, compared with control group. By using Western blot analysis, they verified that the expression of E-cadherin increased in urine samples from DM, DN1, and DN2 groups. Meanwhile, by using enzyme-linked immunosorbent assay (ELISA) analysis, urinary sE-cadherin-to-creatinine ratio was found significantly increased in DN1 and DN2 groups compared with DM and control groups. Furthermore, the decreased expression of E-cadherin in renal tubular epithelial cells of DN was determined by immunohistochemical staining. Thus, E-cadherin may be regarded as a novel DN-related biomarker. In another study, they also used 2DE and MALDI-TOF-MS to discover protein biomarkers for monitoring the development and progression of DN. Urine samples were obtained from 12 DN patients (6 type 1 and 6 type 2 diabetic patients) and 6 healthy controls. A protein was identified as orosomucoid and was further detected by immunoturbidimetry assay in urine samples from 90 type 1 and type 2 diabetic patients with normo-, micro- and macroalbuminuria and 30 healthy controls. The results show that urinary orosomucoid excretion rate (UOER) was progressively up-regulated with increasing albuminuria. Thus, urinary orosomucoid may be regarded as a biomarker for DN patients (Jiang et al., 2009b).
5.2. Immunoglobulin A (IgA) nephropathy
IgA nephropathy (IgAN) is the most common type of primary glomerulonephritis clinically. It is characterized by the presence of IgA predominant or codominant immunoglobulin deposits in the glomerular mesangium. The diagnosis of IgAN can be made by renal biopsy, which is effective but associated with significant risks. Thus, noninvasive biomarkers would be desirable for predicting the diagnosis and prognosis of this disease prior to renal biopsy.
Haubitz et al. (2005) used CE-MS to find the IgAN-specific urinary polypeptide pattern that can distinguish IgAN (n=45) from normal controls (n=57) with a sensitivity of 100% and a specificity of 90%, and from membranous nephropathy (n=13) with a sensitivity of 77% and a specificity of 100%. Three of the polypeptides with masses of 2752.4, 2427.1, and 2057.3 Da have been sequenced and identified as the fragments of serum albumin. In addition, compared with the patterns established earlier in minimal change disease (n=16), focal segmental glomerulosclerosis (FSGS) (n=10), and diabetic nephropathy (n=23), the pattern of IgAN had both sensitivity and specificity of 100%.
Rocchetti et al. (2008) employed 2DE to obtain urine protein spots from controls (n=20), IgA patients responsive to angiotension converting enzyme inhibitors (ACEI) therapy (IgAN R) (n=9), and IgA patients unresponsive to ACEI therapy (IgAN NR) (n=9). By using nano-high performance liquid chromatography coupled with electrospray ionization-tandem MS (nano-HPLC-ESI-MS/MS), three of the spots were identified as kininogen, inter-α-trypsin inhibitor heavy chain H4 precursor (ITIH4), and transthyretin. Moreover, the levels of the three urinary proteins were significantly different between IgAN R and IgAN NR patients. By using immunoblotting, the authors confirmed that the very low levels of urinary kininogen could be a useful marker for prediction of the poor response to the ACEI therapy of IgAN patients.
5.3. Lupus nephritis
Lupus nephritis is a common and serious complication of systemic lupus erythematosus (SLE). Traditionally available serological biomarkers are not sensitive or specific to detect early active SLE nephritis, such as the third and fourth components of complement (C3, C4), and anti-double stranded DNA antibodies (ADNA).
Recently, the application of urinary proteomic profiling has been described using SELDI-TOF-MS in the noninvasive diagnosis of lupus nephritis. Mosley et al. (2006) reported that two protein ions at m/z 3340 and 3980 Da have been detected, which can distinguish between active and inactive lupus nephritis with 92% sensitivity and specificity each. The two biomarkers can predict early relapse and remission of lupus nephritis, but the peptides were not further isolated and identified. In a pilot study involving only children and young adults with SLE or juvenile idiopathic arthritis (JIA), a panel of eight novel biomarkers with peaks at m/z of 2.7, 22, 23, 44, 56, 79, 100, and 133 kDa for SLE nephritis was found. Suzuki et al. (2009) then identified the 23-kDa band as lipocalin-type prostaglandin-D synthetase (L-PGDS), the 56-kDa band as α1-acid-glycoprotein (AGP) or orosomucoid, the 79-kDa band as transferring (Tf), the 133-kDa band as ceruloplasmin (Cp), and the remaining four bands as albumin or albumin fragments, respectively. By using immunonephelometry and ELISA, they further indicated that these proteins may be useful to predict the future course of LN. Among the selected 27 differentially-expressed protein ions in different phases of a lupus nephritis flare, Zhang et al. (2008) identified three potential low-molecular-weight (LMW) biomarkers, namely, the 20 and 25 amino-acid isoforms of hepcidin, fragments of α1-antitrypsin and albumin. Of those, hepcidin appears to have a significant relationship with proinflammatory cytokines in lupus kidneys. Now further evaluation of the hepcidin is undergoing.
5.4. Renal Fanconi syndrome (FS)
Cutillas et al. (2003) used nanoflow LC/ESI-MS/MS to discover more than 100 polypeptides in urine from three patients with Dent’s disease (a form of FS). The smallest peptide with the mass of 1.4 kD was identified as bradykinin. In larger polypeptides, the authors also found the complement components and albumin. More recently, Cutillas et al. (2004) applied three different methods (μLC, 2DE, and multidimensional LC of isotopically labeled proteins) to assess the qualitation and quantitation of urinary proteome in Dent’s disease. They found that several vitamin and prosthetic group carrier proteins, apolipoproteins, complement components, and potentially bioactive peptides were excreted at higher levels in patients with Dent’s disease than in normal controls. Recently, Drube et al. (2009) used CE-MS to detect urinary LMW proteome profiling of the study group (7 paediatric patients with cystinosis and 6 patients with ifosfamide-induced FS) compared with control subjects (54 healthy volunteers and 45 patients suffering from other renal diseases). The results showed that the urinary proteome derived from osteopontin, uromodulin, and collagen alpha-1. The decreased levels of osteopontin and uromodulin indicated the impaired function of tubular excretion in FS patients. While collagen alpha-1 was either increased or decreased, this indicates that early starting tubular fibrosis may be the reason for the development of renal insufficiency in patients with FS.
5.5. Acute kidney injury (AKI)
AKI, previously referred as acute renal failure (ARF), is a common clinical problem with in-hospital incidence of 5% to 30%–50% in intensive care units. Despite the current improvements in diagnostics and therapeutics, the mortality and morbidity associated with AKI remain high.
Nguyen et al. (2005) used SELDI-TOF-MS to identify urinary biomarker patterns to predict AKI in children undergoing cardiopulmonary bypass (CPB). SELDI-TOF-MS analysis of AKI patients at baseline (t=0) versus at 2 and 6 h post-CPB revealed that proteins of 6.4, 28.5, 43 and 66 kDa were significantly enhanced as urine biomarkers. The sensitivity and specificity of the 28.5-, 43- and 66-kDa biomarkers for the prediction of AKI at 2 h post-CPB were 100%. The area under the receiver-operating characteristic (ROC) curve for the three biomarkers is 0.98. Furthermore, the 6.4-kDa urinary biomarker displayed the highest increase in peak intensity at 2 h post-CPB. A further study using tandem mass spectrometry (MS/MS) has identified the protein of 6.4 kDa as aprotinin (Nguyen et al., 2008). The authors subsequently employed SELDI-TOF-MS and functional assays to quantify urinary aprotinin levels in pediatric patients undergoing CPB. The result showed that urinary aprotinin levels at 2 h after initiation of CPB were extremely powerful biomarkers for the development of AKI and its adverse effects. In the subjects receiving aprotinin therapy, urinary aprotinin levels may also be regarded as a predictor of acute kidney injury. Ho et al. (2009) also used SELDI-TOF-MS to identify urinary biomarker patterns from 22 patients with AKI and 22 patients without AKI before, during, and after CPB surgery. Two novel peaks with masses of 2.43 and 2.78 kDa were detected to be more prominent in postoperative non-AKI urine samples. The 2.78-kDa protein was finally identified as hepcidin-25, an important regulator in iron homeostasis. The increase of hepcidin-25 in non-AKI patients may suggest a specific role for iron sequestration in modulating AKI.
Urinary exosomes containing both membrane proteins and cytosolic proteins are normally secreted into the urine from all nephron segments. Recently, the exosomes are regarded as a source of urinary biomarkers (Hoorn et al., 2005). Zhou et al. (2006b) used two-dimensional difference in gel electrophoresis, MALDI-TOF/TOF, and LC-MS/MS to identify urinary protein biomarkers in exosomes from rats subjected to kidney injury by cisplatin injection. Urinary exosomal fetuin-A was validated by Western blotting to be increased by 31.6-fold in the early phase of ischemia/reperfusion and increased in three patients on intensive care units with AKI compared with those without AKI. Thus, urinary exosomal fetuin-A may be a potential biomarker of AKI.
5.6. Renal allograft rejection
Renal transplantation has emerged as the best renal replacement therapy for many patients with end-stage renal failure. Acute rejection (AR) is the major impediment for the long-term allograft survival. At present, the diagnosis of renal allograft rejection is made by renal biopsy, which is inconvenient and costly, and is associated with significant risks. Therefore, there is a high need to develop noninvasive and reliable methods for detecting biomarkers of rejection.
Schaub et al. (2005) used SELDI-TOF-MS to discover three prominent peak clusters, which were found in 17 of 18 (94%) patients with AR episodes but only in 4 of 22 (18%) patients without clinical and histologic evidence for rejection and in 0 of 28 normal controls. A follow-up study revealed that these proteins in the mass ranges of 5270–5550 Da and 10 530–11 000 Da derived from non-tryptic cleaved forms of β2-microglobulin. In vitro experiments demonstrated that the cleavage of intact β2-microglobulin requires a urine pH <6 and the presence of aspartic proteases. Compared with stable transplanted patients and healthy individuals, patients with acute tubulointerstitial rejections had lower urine pH and higher amounts of aspartic proteases. These factors ultimately lead to increased amounts of cleaved urinary β2-microglobulin, which may be regarded as a useful biomarker of acute tubular injury. In another study, two proteins (β-defensin-1 and α1-antichymotrypsin) were identified by O′Riordan et al. (2007) as potentially useful candidate biomarkers of acute rejection.
Quintana et al. (2009) used mass spectrometry to obtain polypeptide patterns from urine from two groups, including 32 with chronic allograft dysfunction (14 with pure interstitial fibrosis and tubular atrophy and 18 with chronic active antibody-mediated rejection) and 18 control subjects (8 stable recipients and 10 healthy control subjects). In their study, they found 14 protein ions best discriminated between the two groups. These protein ions could identify patients with chronic allograft dysfunction with a sensitivity of 100%.
6. Conclusion
Urinary proteomics has great potential in biomarker discovery of kidney diseases and non-kidney diseases such as coronary artery atherosclerosis (von Zur Muhlen et al., 2009), prostate cancer (Theodorescu et al., 2008), and urinary bladder cancer (Saito et al., 2005). To date, more and more laboratories have engaged in urinary proteomic research and a great number of satisfying advances have been achieved, among which biomarker discovery in kidney diseases discussed above is one of the most exciting and significant ones. In addition, urinary proteome analysis can provide information to better understand the renal (patho)physiology.
At present, in order to minimize the errors, standardizations of urinary proteome analysis are urgently needed. The recommended steps of biomarker discovery can include (1) a clear clinical question, (2) protocols for sampling and sample preparation to minimize the inevitable biological variability in urine, (3) analysis by technological platforms with high throughput and high reproducibility, (4) analysis by appropriate statistical methods combined with the clear clinical question, (5) validation of the potential biomarkers by analyzing larger patient groups and running blinded samples, and (6) sequencing of the potential biomarkers.
Footnotes
Project supported by the National Natural Science Foundation of China (No. 30500598) and the Medical Health Science Research Foundation of Zhejiang Province, China (No. 2007B102)
References
- 1.Ahmed FE. Utility of mass spectrometry for proteome analysis: Part II. Ion-activation methods, statistics, bioinformatics and annotation. Expert Rev Proteomics. 2009;6(2):171–197. doi: 10.1586/epr.09.4. [DOI] [PubMed] [Google Scholar]
- 2.Bellei E, Rossi E, Lucchi L, Uggeri S, Albertazzi A, Tomasi A, Iannone A. Proteomic analysis of early urinary biomarkers of renal changes in type 2 diabetic patients. Proteomics Clin Appl. 2008;2(4):478–491. doi: 10.1002/prca.200780109. [DOI] [PubMed] [Google Scholar]
- 3.Caiazzo RJJr, Maher AJ, Drummond MP, Lander CI, Tassinari OW, Nelson BP, Liu BCS. Protein microarrays as an application for disease biomarkers. Proteomics Clin Appl. 2009;3(2):138–147. doi: 10.1002/prca.200800149. [DOI] [PubMed] [Google Scholar]
- 4.Cutillas PR, Norden AG, Cramer R, Burlingame AL, Unwin RJ. Detection and analysis of urinary peptides by on-line liquid chromatography and mass spectrometry: application to patients with renal Fanconi syndrome. Clin Sci (Lond) 2003;104(5):483–490. doi: 10.1042/CS20020342. [DOI] [PubMed] [Google Scholar]
- 5.Dakna M, He Z, Yu WC, Mischak H, Kolch W. Technical, bioinformatical and statistical aspects of liquid chromatography-mass spectrometry (LC-MS) and capillary electrophoresis-mass spectrometry (CE-MS) based clinical proteomics: a critical assessment. J Chromatogr B Analyt Technol Biomed Life Sci. 2009;877(13):1250–1258. doi: 10.1016/j.jchromb.2008.10.048. [DOI] [PubMed] [Google Scholar]
- 6.Drube J, Schiffer E, Mischak H, Kemper MJ, Neuhaus T, Pape L, Lichtinghagen R, Ehrich JH. Urinary proteome pattern in children with renal Fanconi syndrome. Nephrol Dial Transplant. 2009;24(7):2161–2169. doi: 10.1093/ndt/gfp063. [DOI] [PubMed] [Google Scholar]
- 7.Feng X, Liu X, Luo Q, Liu BF. Mass spectrometry in systems biology: an overview. Mass Spectrom Rev. 2008;27(6):635–660. doi: 10.1002/mas.20182. [DOI] [PubMed] [Google Scholar]
- 8.Fiedler GM, Baumann S, Leichtle A, Oltmann A, Kase J, Thiery J, Ceglarek U. Standardized peptidome profiling of human urine by magnetic bead separation and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Clin Chem. 2007;53(3):421–428. doi: 10.1373/clinchem.2006.077834. [DOI] [PubMed] [Google Scholar]
- 9.Finkel E. Cloudy or clear? Best forecast for urine cultures. CAP Today. 2006;20(11):86–90. [Google Scholar]
- 10.Fung ET, Weinberger SR, Gavin E, Zhang F. Bioinformatics approaches in clinical proteomics. Expert Rev Proteomics. 2005;2(6):847–862. doi: 10.1586/14789450.2.6.847. [DOI] [PubMed] [Google Scholar]
- 11.Gu W, Zou LX, Shan PF, Chen YD. Analysis of urinary proteomic patterns for diabetic nephropathy by ProteinChip. Proteomics Clin Appl. 2008;2(5):744–750. doi: 10.1002/prca.200780083. [DOI] [PubMed] [Google Scholar]
- 12.Haubitz M, Wittke S, Weissinger EM, Walden M, Rupprecht HD, Floege J, Haller H, Mischak H. Urine protein patterns can serve as diagnostic tools in patients with IgA nephropathy. Kidney Int. 2005;67(6):2313–2320. doi: 10.1111/j.1523-1755.2005.00335.x. [DOI] [PubMed] [Google Scholar]
- 13.Ho J, Lucy M, Krokhin O, Hayglass K, Pascoe E, Darroch G, Rush D, Nickerson P, Rigatto C, Reslerova M. Mass spectrometry-based proteomic analysis of urine in acute kidney injury following cardiopulmonary bypass: a nested case-control study. Am J Kidney Dis. 2009;53(4):584–595. doi: 10.1053/j.ajkd.2008.10.037. [DOI] [PubMed] [Google Scholar]
- 14.Hoorn EJ, Pistkun T, Zietse R, Gross P, Frokiaer J, Wang NS, Gonzales PA, Star RA, Knepper MA. Prospects for urinary proteomics: exosomes as a source of urinary biomarkers. Nephrology (Carlton) 2005;10(3):283–290. doi: 10.1111/j.1440-1797.2005.00387.x. [DOI] [PubMed] [Google Scholar]
- 15.Huang F, Clifton J, Yang X, Rosenquist T, Hixson D, Kovac S, Josic D. SELDI-TOF as a method for biomarker discovery in the urine of aristolochic-acid-treated mice. Electrophoresis. 2009;30(7):1168–1174. doi: 10.1002/elps.200800548. [DOI] [PubMed] [Google Scholar]
- 16.Jiang H, Guan G, Zhang R, Liu G, Cheng J, Hou X, Cui Y. Identification of urinary soluble E-cadherin as a novel biomarker for diabetic nephropathy. Diabetes Metab Res Rev. 2009;25(3):232–241. doi: 10.1002/dmrr.940. [DOI] [PubMed] [Google Scholar]
- 17.Jiang H, Guan G, Zhang R, Liu G, Liu H, Hou X, Cheng J. Increased urinary excretion of orosomucoid is a risk predictor of diabetic nephropathy. Nephrology (Carlton) 2009;14(3):332–337. doi: 10.1111/j.1440-1797.2008.01053.x. [DOI] [PubMed] [Google Scholar]
- 18.Klasen IS, Reichert LJ, de Kat Angelino CM, Wetzels JF. Quantitative determination of low and high molecular weight proteins in human urine: influence of temperature and storage time. Clin Chem. 1999;45(3):430–432. [PubMed] [Google Scholar]
- 19.Lapolla A, Seraglia R, Molin L, Williams K, Cosma C, Reitano R, Sechi A, Ragazzie E, Traldi P. Low molecular weight proteins in urines from healthy subjects as well as diabetic, nephropathic and diabetic-nephropathic patients: a MALDI study. J Mass Spectrom. 2009;44(3):419–425. doi: 10.1002/jms.1520. [DOI] [PubMed] [Google Scholar]
- 20.Lee RS, Monigatti F, Briscoe AC, Waldon Z, Freeman MR, Steen H. Optimizing sample handling for urinary proteomics. J Proteome Res. 2008;7(9):4022–4030. doi: 10.1021/pr800301h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lifshitz E, Kramer L. Outpatient urine culture: dose collection technique matter. Arch Intern Med. 2000;160(16):2537–3540. doi: 10.1001/archinte.160.16.2537. [DOI] [PubMed] [Google Scholar]
- 22.Liu J, Chen CF, Tsao CW, Chang CC, Chu CC, Devoe DL. Polymer microchips jntegrating solid-phase extraction and high-performance liquid chromatography using reversed-phase polymethacrylate monoliths. Anal Chem. 2009;81(7):2545–2554. doi: 10.1021/ac802359e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.López JL. Two-dimensional electrophoresis in proteome expression analysis. J Chromatogr B Analyt Technol Biomed Life Sci. 2007;849(1-2):190–202. doi: 10.1016/j.jchromb.2006.11.049. [DOI] [PubMed] [Google Scholar]
- 24.Mischak H, Julian BA, Novak J. High-resolution proteome/peptidome analysis of peptides and low-molecular-weight proteins in urine. Proteomics Clin Appl. 2007;1(8):792–804. doi: 10.1002/prca.200700043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mischak H, Coon JJ, Novak J, Weissinger EM, Schanstra JP, Dominiczak AF. Capillary electrophoresis-mass spectrometry as a powerful tool in biomarker discovery and clinical diagnosis: an update of recent developments. Mass Spectrom Rev. 2009;28(5):703–724. doi: 10.1002/mas.20205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mosley K, Tam FW, Edwards RJ, Crozier J, Pusey CD, Lightstone L. Urinary proteomic profiles distinguish between active and inactive lupus nephritis. Rheumatology (Oxford) 2006;45(12):1497–1504. doi: 10.1093/rheumatology/kel351. [DOI] [PubMed] [Google Scholar]
- 27.Najam-ul-Haq M, Rainer M, Trojer L, Feuerstein I, Vallant RM, Huck CW, Bakry R, Bonn GK. Alternative profiling platform based on MELDI and its applicability in clinical proteomics. Expert Rev Proteomics. 2007;4(4):447–452. doi: 10.1586/14789450.4.4.447. [DOI] [PubMed] [Google Scholar]
- 28.Nguyen MT, Ross GF, Dent CL, Devarajan P. Early prediction of acute renal injury using urinary proteomics. Am J Nephrol. 2005;25(4):318–326. doi: 10.1159/000086476. [DOI] [PubMed] [Google Scholar]
- 29.Nguyen MT, Dent CL, Ross GF, Harris N, Manning PB, Mitsnefes MM, Devarajan P. Urinary aprotinin as a predictor of acute kidney injury after cardiac surgery in children receiving aprotinin therapy. Pediatr Nephrol. 2008;23(8):1317–1326. doi: 10.1007/s00467-008-0827-9. [DOI] [PubMed] [Google Scholar]
- 30.O′Farrell PH. High resolution two-dimensional electrophoresis of proteins. J Biol Chem. 1975;250(10):4007–4021. [PMC free article] [PubMed] [Google Scholar]
- 31.O′Riordan E, Orlova TN, Podust VN, Chander PN, Yanagi S, Nakazato M, Hu R, Butt K, Delaney V, Goligorsky MS. Characterization of urinary peptide biomarkers of acute rejection in renal allografts. Am J Transplant. 2007;7(4):930–940. doi: 10.1111/j.1600-6143.2007.01733.x. [DOI] [PubMed] [Google Scholar]
- 32.Orvisky E, Drake SK, Martin BM, Abdel-Hamid M, Ressom HW, Varghese RS, An Y, Saha D, Hortin GL, Loffredo CA, et al. Enrichment of low molecular weight fraction of serum for MS analysis of peptides associated with hepatocellular carcinoma. Proteomics. 2006;6(9):2895–2902. doi: 10.1002/pmic.200500443. [DOI] [PubMed] [Google Scholar]
- 33.Palmblad M, Tiss A, Cramer R. Mass spectrometry in clinical proteomics–from the present to the future. Proteomics Clin Appl. 2009;3(1):6–17. doi: 10.1002/prca.200800090. [DOI] [PubMed] [Google Scholar]
- 34.Penque D. Two-dimensional gel electrophoresis and mass spectrometry for biomarker discovery. Proteomics Clin Appl. 2009;3(2):155–172. doi: 10.1002/prca.200800025. [DOI] [PubMed] [Google Scholar]
- 35.Quintana LF, Solé-Gonzalez A, Kalko SG, Bañon-Maneus E, Solé M, Diekmann F, Gutierrez-Dalmau A, Abian J, Campistol JM. Urine proteomics to detect biomarkers for chronic allograft dysfunction. J Am Soc Nephrol. 2009;20(2):428–435. doi: 10.1681/ASN.2007101137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rocchetti MT, Centra M, Papale M, Bortone G, Palermo C, Centonze D, Ranieri E, Paolo SD, Gesualdo L. Urine protein profile of IgA nephropathy patients may predict the response to ACE-inhibitor therapy. Proteomics. 2008;8(1):206–216. doi: 10.1002/pmic.200700492. [DOI] [PubMed] [Google Scholar]
- 37.Saito M, Kimoto M, Araki T, Shimada Y, Fujii R, Oofusa K, Hide M, Usui T, Yoshizato K. Proteome analysis of gelatin-bound urinary proteins from patients with bladder cancers. Eur Urol. 2005;48(5):865–871. doi: 10.1016/j.eururo.2005.04.028. [DOI] [PubMed] [Google Scholar]
- 38.Schaub S, Wilkins J, Weiler T, Sangster K, Rush D, Nickerson P. Urine protein profiling with surface-enhanced laser-desorption/ionization time-of-flight mass spectrometry. Kidney Int. 2004;65(1):323–332. doi: 10.1111/j.1523-1755.2004.00352.x. [DOI] [PubMed] [Google Scholar]
- 39.Schaub S, Wilkins JA, Antonovici M, Krokhin O, Weiler T, Rush D, Nickerson P. Proteomic-based identification of cleaved urinary β2-microglobulin as a potential marker of acute tubular injury in renal allograft. Am J Transplant. 2005;5(4 Pt 1):729–738. doi: 10.1111/j.1600-6143.2005.00766.x. [DOI] [PubMed] [Google Scholar]
- 40.Suzuki M, Wiers K, Brooks EB, Greis KD, Haines K, Klein-Gitelman MS, Olson J, Onel K, O′Neil KM, Silverman ED, et al. Initial validation of a novel protein biomarker panel for active pediatric lupus nephritis. Pediatr Res. 2009;65(5):530–536. doi: 10.1203/PDR.0b013e31819e4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Theodorescu D, Fliser D, Wittke S, Mischak H, Krebs R, Walden M, Ross M, Eltze E, Bettendorf O, Wulfing C, et al. Pilot study of capillary electrophoresis coupled to mass spectrometry as a tool to define potential prostate cancer biomarkers in urine. Electrophoresis. 2005;26(14):2797–2808. doi: 10.1002/elps.200400208. [DOI] [PubMed] [Google Scholar]
- 42.Theodorescu D, Schiffer E, Bauer HW, Douwes F, Eichhorn F, Polley R, Schmidt T, Schöfer W, Zürbig P, Good DM, et al. Discovery and validation of urinary biomarkers for prostate cancer. Proteomics Clin Appl. 2008;2(4):556–570. doi: 10.1002/prca.200780082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thongboonkerd V. Practical points in urinary proteomics. J Proteome Res. 2007;6(10):3881–3890. doi: 10.1021/pr070328s. [DOI] [PubMed] [Google Scholar]
- 44.Timms JF, Cramer R. Difference gel electrophoresis. Proteomics. 2008;8(23-24):4886–4897. doi: 10.1002/pmic.200800298. [DOI] [PubMed] [Google Scholar]
- 45.Vestergaard P, Leverett R. Constancy of urinary creatinine excretion. J Lab Clin Med. 1958;51(2):211–218. [PubMed] [Google Scholar]
- 46.von Zur Muhlen C, Schiffer E, Zuerbig P, Kellmann M, Brasse M, Meert N, Vanholder RC, Dominiczak AF, Chen YC, Mischak H, et al. Evaluation of urine proteome pattern analysis for its potential to reflect coronary artery atherosclerosis in symptomatic patients. J Proteome Res. 2009;8(1):335–345. doi: 10.1021/pr800615t. [DOI] [PubMed] [Google Scholar]
- 47.Weissinger EM, Schiffer E, Hertenstein B, Ferrara JL, Holler E, Stadler M, Kolb HJ, Zander A, Zurbig P, Kellmann M, et al. Proteomic patterns predict acute graft-versus-host disease after allogeneic hematopoietic stem cell transplantation. Blood. 2007;109(12):5511–5519. doi: 10.1182/blood-2007-01-069757. [DOI] [PubMed] [Google Scholar]
- 48.Wu R, Hu L, Wang F, Ye M, Zou H. Recent development of monolithic stationary phases with emphasis on microscale chromatographic separation. J Chromatogr A. 2008;1184(1-2):369–392. doi: 10.1016/j.chroma.2007.09.022. [DOI] [PubMed] [Google Scholar]
- 49.Zerefos PG, Vlahou A. Urine sample preparation and protein profiling by two-dimensional electrophoresis and matrix-assisted laser desorption ionization time of flight mass spectroscopy. Methods Mol Biol. 2008;428:141–157. doi: 10.1007/978-1-59745-117-8_8. [DOI] [PubMed] [Google Scholar]
- 50.Zhang X, Jin M, Wu H, Nadasdy T, Nadasdy G, Harris N, Green-Church K, Nagaraja H, Birmingham DJ, Yu CY, et al. Biomarkers of lupus nephritis determined by serial urine proteomics. Kidney Int. 2008;74(6):799–807. doi: 10.1038/ki.2008.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhou H, Yuen PS, Pisitkun T, Gonzales PA, Yasuda H, Dear JW, Gross P, Knepper MA, Star RA. Collection, storage, preservation, and normalization of human urinary exosomes for biomarker discovery. Kidney Int. 2006;69(8):1471–1476. doi: 10.1038/sj.ki.5000273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhou H, Pisitkun T, Aponte A, Yuen PS, Hoffert JD, Yasuda H, Hu X, Chawla L, Shen RF, Knepper MA, et al. Exosomal Fetuin-A identified by proteomics: a novel urinary biomarker for detecting acute kidney injury. Kidney Int. 2006;70(10):1847–1857. doi: 10.1038/sj.ki.5001874. [DOI] [PMC free article] [PubMed] [Google Scholar]