Abstract
Myotonic dystrophy (DM1) is associated with expression of expanded CTG DNA repeats as RNA (CUGexp RNA). To test whether CUGexp RNA creates a global splicing defect, we compared skeletal muscle of two mouse DM1 models, one expressing a CTGexp transgene, and another homozygous for a defective Mbnl1 gene. Strong correlation in splicing changes for ~100 new Mbnl1-regulated exons indicates loss of Mbnl1 explains >80% of the splicing pathology due to CUGexp RNA. In contrast, only about half of mRNA level changes can be attributed to loss of Mbnl1, indicating CUGexp RNA has Mbnl1-independent effects, particularly on mRNAs for extracellular matrix (ECM) proteins. We propose that CUGexp RNA causes two separate effects: loss of Mbnl1 function, disrupting splicing, and loss of another function that disrupts ECM mRNA regulation, possibly mediated by MBNL2. These findings reveal unanticipated similarities between DM1 and other muscular dystrophies.
Myotonic dystrophy (DM) is a triplet repeat expansion disease, one of a group that includes Huntington disease, Fragile X syndrome, and Friedreich’s ataxia1,2. The common form, DM1, is caused by a CTG-repeat expansion (CTGexp) in the 3′-untranslated region of the DMPK gene, leading to myotonia, muscle degeneration, reduced heart function, ocular cataracts, and nervous system dysfunction3. RNA containing CUG repeats (CUGexp RNA) accumulates in nuclear foci4,5. Based on its autosomal dominant inheritance, a leading hypothesis for DM1 is that CUGexp RNA is toxic. Mice engineered to express CUGexp RNA show many symptoms of myotonic dystrophy6,7. In addition to RNA splicing (see below), CUGexp RNA has been proposed to disrupt a wide variety of cellular processes through several mechanisms, including “leaching” of transcription factors8, processing into small RNAs that trigger inappropriate gene silencing9, or through PKC-dependent signaling pathways10. Of these, the contribution of splicing perturbations is most clear given the physiological relevance of several splicing alterations that occur when CUGexp RNA is expressed5,7,11. For example, aberrant splicing of transcripts from the chloride channel gene CLCN1 is responsible for the myotonia in skeletal muscle12–15, and aberrant splicing of insulin receptor transcripts is associated with insulin resistance16.
Among proteins that bind CUGexp RNA are homologs of Drosophila muscleblind (muscleblind-like proteins, Mbnl1 through 3)17,18. Mbnl proteins contain 4 CCCH-type zinc fingers that recognize a YGCY motif repeated within the CUG repeat (CUGCUG), and can function in splicing regulation19–21. CUG repeats can form long dsRNA structures in vitro22, however crystal structures of Mbnl1 zinc fingers complexed with RNA indicates that the Watson-Crick faces of the binding site 5′-GC-3′ dinucleotide residues are buried in the protein and unavailable for duplex formation20. The colocalization of CUGexp RNA and Mbnl1 in nuclear foci suggests a place where sequestration occurs4,11,17.
Besides Mbnl1, the altered function of splicing factors CUGBP1 and hnRNP H have been proposed to play a role in DM1 pathogenesis23–26. To test of the role of Mbnl1, a homozygous Mbnl1 mutant mouse (Mbnl1ΔE3/ΔE3) was created27. Like mice expressing CUGexp RNA, mice deficient in Mbnl1 show characteristics of myotonic dystrophy including aberrant splicing11,19,27. Some but not all of the DM-like symptoms and aberrant splicing of six exons are rescued by AAV-mediated expression of Mbnl1 in CUGexp RNA expressing mice28, leaving open the possibility that CUGexp RNA has other mechanisms of action. In addition, the broader impact on splicing of the loss or sequestration of Mbnl1 beyond the few genes tested so far, is unknown.
To determine the extent to which the loss of Mbnl1 explains the splicing and gene expression defects caused by CUGexp RNA, we compared mRNA in skeletal muscle of HSALR mice7 expressing CUGexp RNA, to those in Mbnl1ΔE3/ΔE3 mice27 using splicing-sensitive microarrays29,30. We find that global splicing perturbations are remarkably congruent in these mice, and identify more than 200 splicing events altered upon loss of Mbnl1. Testing human exons orthologous to these new mouse Mbnl1-dependent exons suggests that human DM1 patients suffer many of the same mis-splicing events, identifying new potential splicing markers for the human disease. Mbnl1 RNA binding sequences were greatly enriched near the affected exons suggesting that CUGexp RNA affects splicing primarily through Mbnl1. In contrast to splicing, many changes in mRNA level are found in CUGexp RNA expressing muscle but not in the Mbnl1 mutant, indicating a distinct second defect caused by CTGexp DNA. This defect appears to be unusually focused on genes expressing extracellular matrix (ECM) components and their regulators, some of which are known to play roles in other forms of muscular dystrophy and connective tissue diseases.
Results
Splicing perturbations in muscle of HSALR and Mbnl1ΔE3/ΔE3 mice
If Mbnl1 sequestration is the main cause of gene expression perturbation in DM1, then transcripts from muscle genetically lacking Mbnl1 should appear broadly similar to those expressing CUGexp RNA. The extent of this similarity should provide a strong quantitative estimate of the extent to which the Mbnl1 sequestration hypothesis can explain the disease. To test this directly, RNA was extracted from quadriceps muscle of age-matched males carrying either the HSALR transgene expressing CUGexp RNA (HSALR)7 or homozygous for the Mbnl1 knockout allele (Mbnl1ΔE3/ΔE3)27, or homozygous wild type Mbnl1 in the same (FBV) background (wt), and analyzed on splicing-sensitive microarrays as described previously29,30. Experience with this method indicates that differences in the log2 of the skip/include ratio between two samples (Fig. 1a, Sepscore, see Methods) with absolute value > 0.3 can be validated by RT-PCR about 85% of the time29,30. We observe 246 events in Mbnl1ΔE3/ΔE3 mice and 221 events in HSALR mice that exceed this score, distributed among different splicing modes (alternative cassette exons, alternative 5′ or 3′ splice sites, and mutually exclusive cassette exons, Fig. 1b and Supplementary Table 1). Even at this relatively crude level of analysis, nearly 80% of splicing events altered in HSALR mice are also altered in Mbnl1ΔE3/ΔE3 mice. About half show increased exon skipping after loss of Mbnl1, and half show decreased skipping, indicating formally that Mbnl1 contributes to both activation and repression of splicing.
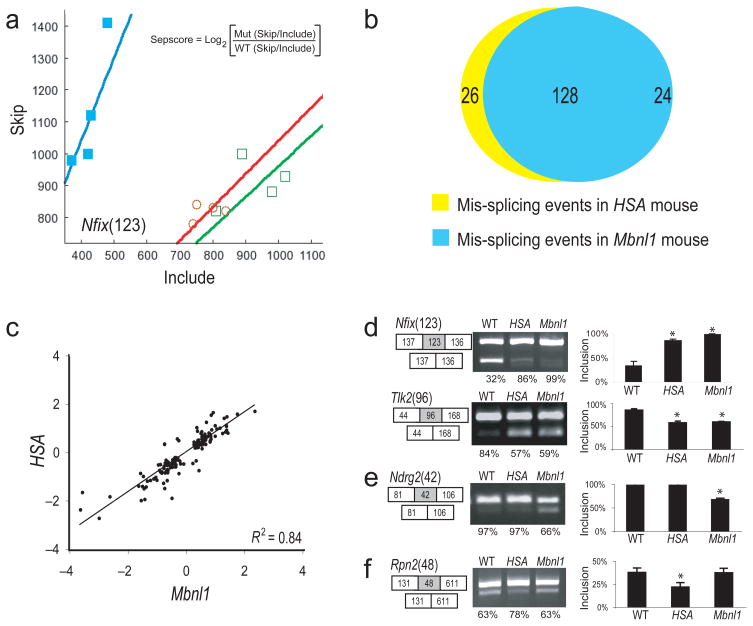
Figure 1.
Comparison of splicing perturbation in the two mouse models. (a) Plot of skip/include ratio array data for an exon from Nfix in wt (filled squares), HSALR (open circles) and Mbnl1ΔE3/ΔE3 (open squares) mice (n = 4). Separation score is the log2 ratio of skipping probe set intensity versus include probe set intensity in a mutant relative to wild-type (see inset, also see Methods). (b) Diagram showing numbers of aberrant splicing events in HSALR mice and Mbnl1ΔE3/ΔE3 mice with |sepscore| ≥ 0.3. (c) Correlation of sepscore values for aberrant splicing in HSALR compared to Mbnl1ΔE3/ΔE3 mice (Pearson R2 = 0.84). (d–f) Validation of members from different classes of events by RT-PCR. (d) Exons either up (Nfix) or down (Tlk) regulated in both HSALR and Mbnl1ΔE3/ΔE3 mice, (e) an exon only affected in Mbnl1ΔE3/ΔE3 mice, or (f) only affected in HSALR mice. The numbers in parentheses represent the length in nt of the affected exon (gray box), shown with flanking exons (white boxes). The RT-PCR products from three individual mice were quantified using the Bioanalyzer, and inclusion rates were calculated. Samples were judged as different from wild type if a t-test indicated that the sample was unlikely to be from the wild type distribution with p < 0.05 (asterisks).
To evaluate whether the two disease models share quantitatively similar changes in splicing, we compared the Sepscores for each splicing event altered (with |Sepscore| ≥ 0.3) in both models. The R2 value for this comparison is 0.84, suggesting that Mbnl1 loss of function explains more than 80% of the splicing phenotype caused by CUGexp RNA expression (Fig. 1c). We used RT-PCR to validate a subset of the alternative cassette exons, as shown for a 123 nt cassette exon repressed by Mbnl1 in the Nfix gene (encoding NF-I/X CAAT box-binding transcription factor, Fig. 1d, see Methods). The 93 nt exon of the Tlk gene represents Mbnl1 contribution to splicing activation, as skipping is increased in the mutants (Fig. 1d). Of 33 other events tested, all but one behaved as expected from the arrays (FDR = 0.03, Supplementary Fig. 1 and Supplementary Table 1). Of these, 28 are affected in both HSALR and Mbnl1ΔE3/ΔE3 mice (Fig. 1d; Supplementary Fig. 1a); 4 appear affected only in Mbnl1ΔE3/ΔE3 (e. g. Ndrg2, Fig. 1e; Supplementary Fig. 1b); and 1 appears affected only in HSALR (Fig. 1f; Supplementary Fig. 1c). The concordance observed by arrays (Fig. 1c) was confirmed by RT-PCR reactions quantified on an Agilent Bioanalyzer (R2 = 0.88, Supplementary Fig. 1d).
We found four splicing events affected in Mbnl1ΔE3/ΔE3 but not in HSALR mice (Fig. 1e, Supplementary Fig. 1b). These might compete effectively for Mbnl1 against CUGexp RNA, and thus may not be inhibited except at levels higher than those achieved by this transgene. We found only one event affected specifically in HSALR mice (Fig. 1f), suggesting that CUGexp RNA transgene expression can cause splicing defects by mechanisms other than simple loss of Mbnl1. Possibly the regulation of this exon is the responsibility of other factors that function in the Mbnl1 knockout mouse but not in HSALR, such as Mbnl2, which is also sequestered by CUGexp RNA31,32, or CUGBP1 whose activity is altered in CUGexp RNA expressing cells24, or by some other indirect or combinatorial mechanism (see below). Given that Mbnl2 is sequestered by CUGexp RNA31,32, our results also mean that Mbnl2 is largely unable to compensate for the loss of Mbnl1 in muscle, otherwise we would have found many more splicing events perturbed in the HSALR but not in the Mbnl1ΔE3/ΔE3 mouse. We conclude that the vast majority of splicing dysregulation in the CUGexp RNA-expressing muscle is due to catastrophic loss of Mbnl1 splicing factor activity.
Sequence signatures of Mbnl1-regulated exons in muscle
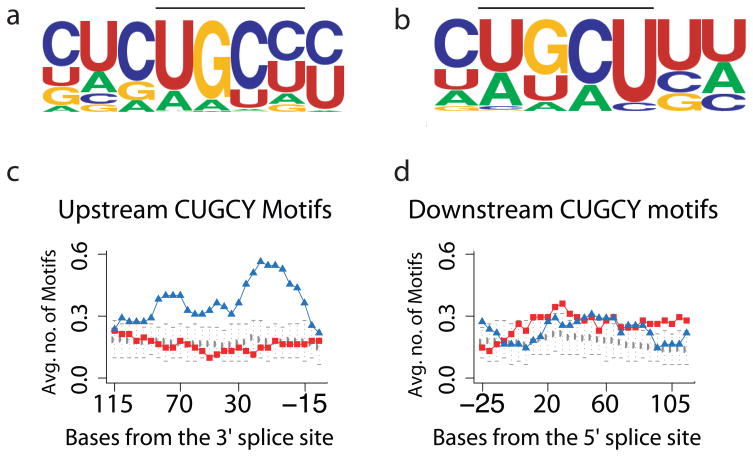
To identify motifs associated with Mbnl1-regulated exons, we used Improbizer, which identifies sequence motifs present in a set of sequences as compared to a background sequence set (See Methods). We contrasted the introns upstream and downstream of three groups of exons (Supplementary Table 2): Mbnl1-activated exons (more skipping in Mbnl1ΔE3/ΔE3 than wild type); Mbnl1-repressed exons, (more inclusion in Mbnl1ΔE3/ΔE3); and background exons (detected in our experiments but showing no significant splicing change). Improbizer strongly recognizes enrichment of a YGCY-containing motif (CUGCY) in the Mbnl1-repressed and Mbnl1-activated exons (Fig. 2a, b), consistent with interpretation of mutagenesis experiments on several splicing substrates18,33,34, and with crystal complexes of Mbnl1 zinc fingers with RNA20. We mapped the distribution of positions of CUGCY motifs to upstream and downstream intron regions (Fig. 2c, d). Introns upstream of Mbnl1-repressed exons, as well as part of the exon itself, are significantly enriched for CUGCY motifs, which often occur multiple times in individual pre-mRNAs. In contrast the corresponding region of Mbnl1-activated exons is not enriched (Fig. 2c). In the intron downstream of affected exons, both Mbnl1-repressed and Mbnl1-activated exons show slight but significant enrichment in CUGCY elements above the background exons (Fig. 2d). Enrichment for CUGBP1 or hnRNP H motifs is not observed (Supplementary Fig. 2). This pattern argues that the splicing defects are primarily due to loss of Mbnl1, and suggests that regulation by Mbnl1 often involves its direct binding to regions near the exons it regulates.
Figure 2.
RNA motifs found by bioinformatics analysis near exons altered in the mouse models. (a–b) Improbizer identifies a motif containing CUGCY upstream of Mbnl1-repressed exons (a) and downstream of Mbnl1-activated exons (b). (c–d) Mapping CUGCY elements upstream (c) and downstream (d) of the Mbnl1-repressed and Mbnl1-activated exons. Each point represents the average frequency of CUGCY element for the 55 Mbnl1-repressed exons (blue triangles), or 66 Mbnl1-activated exons (red squares) or 790 expressed alternative cassette exons that did not change in the experiment (grey circles). Error bars indicate plus and minus two standard deviations of the mean frequency distribution for this population of background exons.
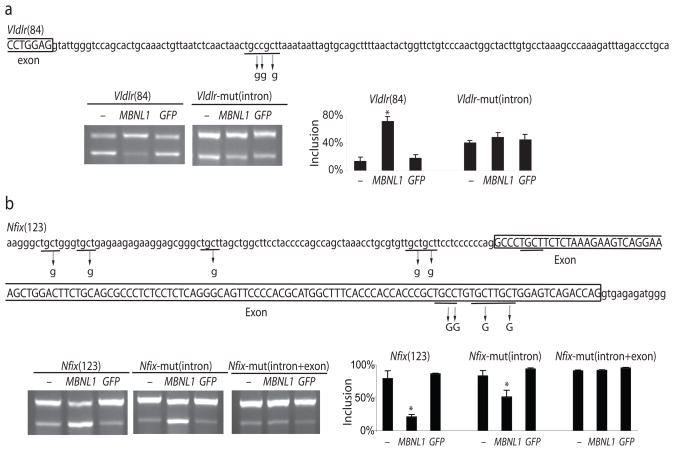
To confirm the function of YGCY motifs in newly identified exons, we cloned the Mbnl1-activated exon from the Vldlr gene and the Mbnl1-repressed exon from the Nfix gene, each with their native flanking intron sequences, into a splicing reporter plasmid. We then mutated copies of the motif in each and tested them in mouse embryo fibroblasts lacking Mbnl1 (derived from the Mbnl1ΔE3/ΔE3 mouse), along with a construct expressing an Mbnl1-GFP fusion protein or GFP alone (Fig. 3a, b). Splicing activation is promoted by the Mbnl1-GFP fusion protein for the wild type, but not the mutant Vldlr splicing reporter, indicating that the motif downstream of the exon is important for Mbnl1-mediated splicing activation (Fig. 3a). Splicing repression is promoted by the Mbnl1-GFP fusion protein for the wild type Nfix reporter, and is only partly compromised by alteration of four copies of the motif in the intron upstream of the exon. Complete loss of repression is achieved once three additional motif copies in the exon itself are altered (Fig. 3b). These results confirm by reconstruction that newly identified exons depend on Mbnl1 for their correct splicing, and furthermore show that regulation is mediated by sequence motifs enriched in Mbnl1-regulated exons.
Figure 3.
YGCY motifs mediate Mbnl1-dependent splicing repression and activation. Intron (lower case) and exon sequences (upper case and boxed) for the (a) Vldlr(84) and (b) Nfix(123) exons are shown, and mutations that disrupt YGCY motifs are labeled with arrows. Mouse embryo fibroblasts lacking endogenous Mbnl1 (derived from the Mbnl1ΔE3/ΔE3 mouse) were transfected with the splicing reporter and either no expression plasmid (−) or a plasmid producing Mbnl1-GFP protein (MBNL1) or just GFP protein (GFP) and exon inclusion was measured by RT-PCR using the bioanalyzer. Significant splicing changes (t-test derived p < 0.05) are labeled with an asterisk.
Mis-splicing events discovered in mouse are observed in human DM1
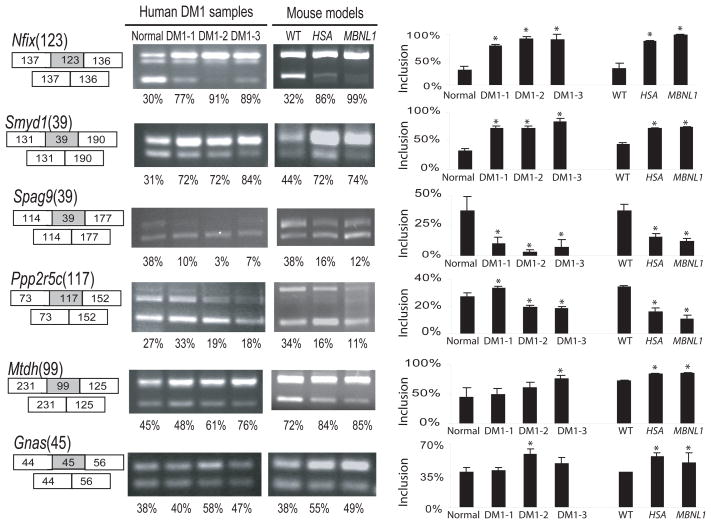
A handful of splicing defects are known in human DM1 patients11. To determine whether human DM1 patients share newly identified splicing changes with the mouse DM1 models, we aligned affected mouse exons to the human genome and found 49 that are conserved and orthologous to Mbnl1-dependent mouse exons, 39 of which show significant evidence of alternative splicing in humans, according to the Alt Events track on the UCSC Genome Browser35. We tested six for perturbation of splicing in human DM1 patients. Three (in Nfix, Smyd1, and Spag9) are perturbed in all three DM1 patients tested, as compared to a normal human control, whereas three others (Gnas, Mtdh and Ppp2r5c) are clearly affected only in either patients 2 or 3 or both (Fig. 4). To rigorously determine the value of such changes in monitoring human DM1, more DM1 patients and unaffected people would need to be tested. Nonetheless, these and other11 splicing changes indicate that Mbnl1-dependent events observed in the mouse are altered in human DM1. Thus exons affected in the mouse models represent a rich source of potential markers for the detection and evaluation of the human disease, as well as for testing the efficacy of treatments.
Figure 4.
Test of human DM1 patients for splicing perturbations newly predicted from mouse model microarray data. Skeletal muscle RNA from 3 individual DM1 patients and one normal human control were compared to those from HSALR and Mbnl1ΔE3/ΔE3 mice by RT-PCR. Inclusion rates were determined on a Bioanalyzer. Statistically significant differences from the normal individual human are indicated by an asterisk, however this captures only technical variation in the RT-PCR protocol, as a single original RNA donation was processed in triplicate. For mouse samples, three individuals of each genetic type were compared, so that statistical differences here arise from both biological and technical variation. Note that extent of genetic variation between the humans is unknown (except at the CTGexp locus), but is likely much greater than the differences between the mouse groups.
Many changes in HSALR mice are not explained by loss of Mbnl1
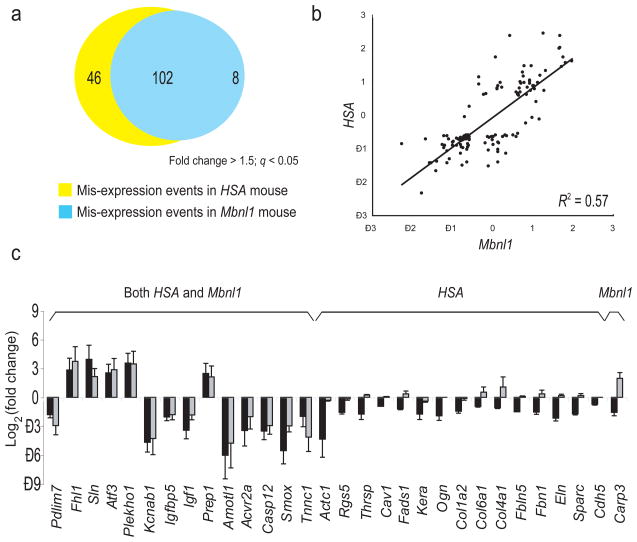
Besides impacting splicing, Mbnl1 loss could lead to transcript-level changes by other direct or indirect mechanisms, including nonsense mediated decay caused by aberrant splicing (AS-NMD)36, altered mRNA stability, or indirectly through mis-splicing of transcription factor or splicing factor mRNAs, all of which could combine to alter the transcriptome37. If Mbnl1 sequestration is the only mechanism by which CTGexp DNA alters gene expression, these myriad effects should be highly similar in both mouse models. To test this, we compared gene expression changes in HSALR mice to Mbnl1ΔE3/ΔE3 mice using the array probe intensities for all the constitutive (always included) exons in each gene (see Methods). After filtering out genes with probes affected by cross-hybridization to the massive amounts of CUGexp RNA in the HSALR mouse samples (see Methods), we identified 148 genes whose muscle transcript levels were significantly altered in HSALR mice, and 110 in the Mbnl1ΔE3/ΔE3 mice (≥ 1.5 fold change; q < 0.05 cutoff, Fig. 5a). Of the 148 changes observed in HSALR mice, 102 (69%) also appear in Mbnl1ΔE3/ΔE3 mice, suggesting that, as for splicing, the underlying reason for those expression changes is loss of Mbnl1, either directly or indirectly through other factors. The 46 genes (31%) that change only in HSALR mice indicate that CTGexp DNA imposes a second layer of dysregulation not shared by mice lacking Mbnl1. Overall, Mbnl1 loss explains a little more than half of the transcript level variance between wild type and HSALR mice (Fig. 5b; R2 = 0.57). We named the larger class of genes whose mRNA levels are altered in both models primarily due to loss of Mbnl1 “class I” genes, and those altered only in the HSALR but not Mbnl1ΔE3/ΔE3 mice “class II” genes. In a more complex study using a very different array platform37, similar gene classes were identified that contained different genes (see Discussion).
Figure 5.
Comparison of altered mRNA levels in the two mouse models. (a) Numbers of genes with altered mRNA levels in HSALR and Mbnl1ΔE3/ΔE3 mice (cutoff: Fold change ≥ 1.5; q ≤ 0.05). (b) Comparison of magnitudes of the log2(fold change) in mRNA levels in the two mouse models (Spearman R2 = 0.57). (c) Validation of altered mRNA levels found on arrays by quantitative RT-PCR. Quantification of triplicate data with error bars (± SD) is shown, where black bars represent change in HSALR mice, and grey bars represent change in Mbnl1ΔE3/ΔE3 mice.
To validate the transcript level changes, we used quantitative RT-PCR (qPCR, Fig. 5c and Supplementary Table 3). Of changes observed only in the HSALR mice, more genes are down-regulated than up-regulated (Fig. 5c). There is good agreement with the microarray data, however the magnitude of the change detected is larger by qPCR, likely because of the smaller dynamic range of arrays. Because the class II changes do not occur in the Mbnl1ΔE3/ΔE3 mouse (at least by 12–14 weeks of age), they are unlikely to be a simple consequence of a loss of Mbnl1. However, the HSALR mouse also suffers a loss of Mbnl1 function28, and it remains possible that Mbnl1 loss is necessary but not sufficient to produce the class II changes. We conclude that a substantial number of transcript level changes occur in the HSALR mouse that do not occur in the Mbnl1ΔE3/ΔE3 mouse.
To capture class II gene functions, we relaxed the array-measured fold change cut off from 1.5 to 1.3 fold, since qPCR indicated that fold change is underestimated in this data. Because the filtering of cross hybridizing CUGexp RNA in the samples is incomplete, some genes will artificially appear to be up-regulated in the HSALR mouse but not in the Mbnl1ΔE3/ΔE3 mouse. We avoided such genes by only considering those significantly down regulated in the HSALR mouse relative to wild type. To ensure that the genes on the list were specific to CTGexp effects, we required that their q values (FDRs) in both the HSALR vs wild type and the HSALR vs Mbnl1ΔE3/ΔE3 mouse comparisons were < 0.05, and that their q values in the Mbnl1 vs wild type comparisons were > 0.05. This resulted in a set of 93 genes down regulated in the HSALR mouse but not changed in the Mbnl1ΔE3/ΔE3 mouse (Supplementary Table 4).
Distinct impacts of CUGexp imposed through different sets of genes
Using the splicing changes, and the class I and class II gene expression change lists, we searched for functionally related gene classes that might provide insight into the disease. We performed a Gene Ontology (GO) analysis38, using the background set of all genes expressed in the experiment (~6000 genes of the ~10,000 on the array). Splicing changes were not significantly enriched in any GO category (not shown), suggesting that Mbnl1-regulated exons are distributed in several broad functional classes of genes. Class I genes, whose expression changes presumably arise as an indirect consequence of the loss of Mbnl1 in splicing, are enriched insulin and insulin-like growth factor signaling and glucose metabolism (Supplementary Table 5), suggesting that loss of Mbnl1 has strong but specific effect on glucose metabolism in muscle cells. Other muscle cell terms such as contractile fiber and muscle development are also significantly associated with this gene set.
Class II genes are very significantly enriched for those encoding or regulating components of the extracellular matrix (ECM), with one fifth of the genes (18/93) carrying this functional label (Supplementary Table 6). Fifteen are orthologous to human genes involved in other forms of muscular dystrophy, myopathies, and several connective tissue diseases, or their deletion in mouse creates a similar defect (Table 1, Supplementary Table 7). We conclude that the coordinate dysregulation of a large set of genes associated with ECM function is caused by a second effect of CTGexp DNA that is not explained by simple loss of Mbnl1.
Table 1.
| Gene | Protein Name | Function in ECM and cell adhesion | Human Disease or mouse knockout phenotype |
|---|---|---|---|
| COL15A1 | procollagen, type XV | Adhesion function in skeletal and cardiac muscle | Col15a1-deficient mice have skeletal myopathy and cardiovascular defects |
| COL1A1 | procollagen, type I, alpha 1 | Abundant, ECM organization | osteogenesis imperfecta Type I; Ehlers-Danlos syndrom (EDS) |
| COL1A2 | procollagen, type I, alpha 2 | Abundant, ECM organization | osteogenesis imperfecta Type I |
| COL4A1 | procollagen, type IV, alpha 1 | ECM organization | Familial Porencephaly; Alport syndrome |
| COL6A1 | procollagen, type VI, alpha 1 | Anchors the muscle basement to the ECM | Ullrich congenital muscular dystrophy; Bethlem myopathy |
| ELN | elastin | ECM organization | Supravalvular Aortic Stenosis; Williams-Beuren Syndrome |
| ENG | endoglin(TGF- β type III receptor) | Both the extracellular and intracellular domains of endoglin interact with TβR-II and ALK-5 and its cytoplasmic domain, which is phosphorylated by ALK-5 and TβR-II | Hereditary Hemorrhagic Telangiectasia of Rendu, Osler, and Weber |
| FBLN5 | fibulin 5 | Elastin fiber formation | Autosomal Recessive Cutis Laxa, Type I; age- related muscular degeneration |
| FBN1 | fibrilin-1 | Elastin fiber assembly and formation | Marfan Syndrome |
| FMOD | fibromodulin | inhibits type I collagen fibrillogenesis | Ehlers-Danlos Syndrome, Type I |
| KERA | keratocan | keratan sulfate proteoglycan of the extracellular matrix. Based on studies in the mouse, keratocan expression is more limited to the cornea in the adult organism. | Cornea Plana 2 |
| LOX | lysyl oxidase | Formation and repair of the extracellular matrix (ECM) by oxidizing lysine residues in elastin and collagen | Lox knockout mice have cardiovascular dysfunction |
| PLOD1 | procollagen- lysine, 2- oxoglutarate 5- dioxygenase 1 | lysyl hydroxylase that controls cross- links | Ehlers-Danlos syndrome type VIA, Nevo Syndrome |
| POSTN | periostin, osteoblast specific factor | Regulation of collagen fiber diameter and crosslinking | Postn knockout mice have cardiac valve disease |
| TNC | tenascin C | Regulation of muscle strength | Tnc-deficient mice have weak muscles |
We analyzed the sequences associated with the 5′ and 3′ untranslated regions of the down regulated class II genes. Using the frequency of 7-mer sequence words, we find that sequences containing the Mbnl1 motif (YGCY) are enriched in the untranslated regions of the 93 class II mRNAs. There is a significant enrichment of GGUGCUA in the 3′UTRs as well as enrichment of several related words in the 5′ UTRs (UGCCUGC, CCUGCCU, UGUGCCU; Supplementary Table 8). Due to the extreme similarity between the RNA binding domains of Mbnl1 and Mbnl2, it seems likely that the two proteins bind highly related RNA sequences21,39. Since Mbnl2 is sequestered by CUGexp RNA31,32, these results lead to the strong hypothesis that expression of class II genes might be altered through loss of Mbnl2 function mediated through binding to the untranslated regions of the mRNAs. The enrichment of Mbnl binding sites observed in the 93 class II mRNAs is also observed in the subgroup of class II genes that encode ECM components (Supplementary Table 8). Mbnl2 colocalizes with integrin α3 mRNA in the cytoplasm and promotes its expression at the cell surface40. Since integrins are ECM components, it is tempting to propose that Mbnl2 plays a general role in promoting correct localization, stability, and translation of mRNAs encoding ECM proteins and their regulators.
Discussion
Loss of Mbnl1 is the major cause of CUGexp RNA splicing toxicity
Our genome-wide test of the Mbnl1 sequestration hypothesis identified a large set of mouse muscle splicing events that depend on Mbnl1, and compared them to splicing changes caused by toxic CUGexp RNA. Strikingly, genetic loss of Mbnl1 accounts for about 80–90% of the phenotype in the CUGexp RNA expressing mice (Fig. 1, Supplementary Fig. 1). The extreme similarity of global splicing defects supports a sequestration model in which CUGexp RNA creates a catastrophic loss of Mbnl1 splicing function in DM1. This is confirmed by the presence of Mbnl1 binding motifs in and near exons whose splicing is compromised by loss of Mbnl1 (Figs. 2, 3). Our analysis was unable to identify enrichment of motifs for other splicing factors implicated in DM1 pathogenesis, such as CUGBP1 (refs. 23, 25) or hnRNP H23 and neither we (this study) nor Kalsotra et al.25, who studied the developing heart, found strong co-enrichment of CUGBP1 motifs with Mbnl1-dependent exons. We also found rare splicing changes specific to each model (Fig. 1). Thus, although Mbnl1 loss is the major contributor to splicing defects in the HSALR mouse, other factors also contribute in more subtle ways.
A cascade of other perturbations due to Mbnl1 loss also occurs. These encompass changes in transcript level (Fig 5, see also ref. 37). Fully 69% of the transcript level changes observed in the Mbnl1ΔE3/ΔE3 mouse are also observed in the HSALR mouse (class I genes). Many of these show no altered splicing, but they could depend directly on Mbnl1 (if it has a role in mRNA stabilization), or could be altered through indirect effects consequent to the loss of splicing regulation for transcription or other RNA stability factors. The broad impact of Mbnl1 loss makes following the threads of each perturbation to the mechanistic source of each disease symptom a daunting prospect. Whether direct or indirect, perturbations that arise from loss of Mbnl1 are pervasive and complex, and may explain the medical complexities of the human disease. The mouse models reveal defects conserved in humans DM1 patients (Fig. 4), suggesting that diagnostic tests for a wide spectrum of genes that depend on Mbnl1 function may presage DM1 onset, or allow sensitive evaluation of therapies.
What causes perturbations not attributable to Mbnl1 loss?
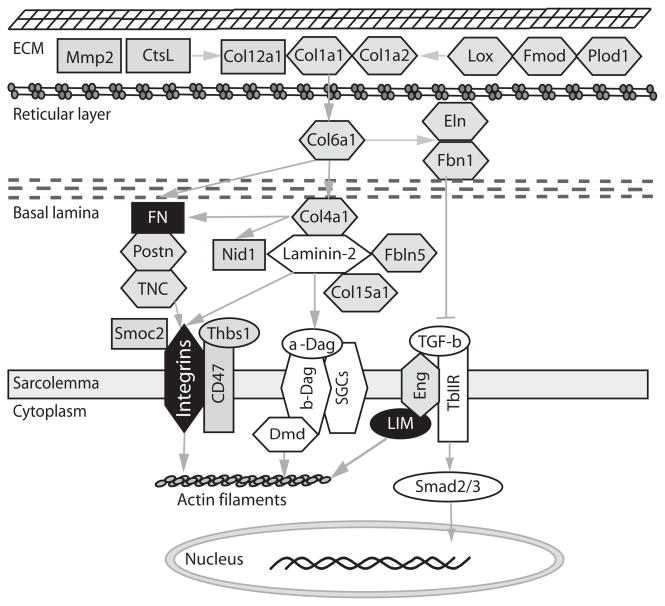
A second class of gene expression perturbations occurs in CUGexp RNA-expressing mice, but not in mice lacking Mbnl1 (class II genes). A surprising fraction is associated with ECM function (Fig. 6, Supplementary Tables 6 and 7). We propose that their misregulation is due to loss of function of a second factor. The leading candidate for such a factor is Mbnl2, which like Mbnl1, is sequestered by CUGexp RNA5,31,32,41. The RNA binding domains of Mbnl1 and Mbnl2 are nearly identical, and untranslated regions of class II genes are enriched in Mbnl RNA motifs (Supplementary Table 8), supporting this hypothesis. A mouse Mbnl2 mutant has a relatively mild phenotype that includes muscle fibrosis, however expression of residual Mbnl2 has not been strictly ruled out in this system42. The enrichment of ECM genes among the class II set (Fig. 6, Supplementary Tables 6 and 7) combined with the finding that Mbnl2 colocalizes with integrin α3 mRNA and promotes localization of the integrin protein at focal adhesions40 makes it reasonable to postulate that Mbnl2 plays a general role in promoting correct expression of ECM molecules.
Figure 6.
ECM proteins whose mRNA levels are altered by CUGexp RNA expression. The proteins whose mRNAs are misregulated in HSALR but not Mbnl1ΔE3/ΔE3 mice (class II genes) are shown in grey, and other proteins in the same network are shown in white. Proteins in black are those whose mRNAs have splicing perturbations in both HSALR and Mbnl1ΔE3/ΔE3 mice. Proteins in hexagons are known to be associated with other muscular dystrophies or connective tissue diseases (see text), and grey arrows denote regulatory interactions.
Tracing CUGexp RNA toxicity from gene to disease phenotype
Loss of Mbnl1 through sequestration by CUGexp RNA contributes to changes in muscle morphology and excitability as indicated by studies with Mbnl1-deficient mice27. These include aberrant splicing of CLCN1 causing myotonia14,15,43, mis-splicing of Camk2g causing interference in excitation–contraction coupling44, or alteration of Acvr2a expression disturbing regulation of muscle growth45. A large number of new candidates associated with ion channels, cytoskeleton and myotubule composition, glucose metabolism and signal transduction may lead to additional defects (Supplementary Tables 1 and 3).
Despite this, loss of Mbnl1 does not produce the progressive muscle wasting observed in DM1. Orengo et al. reported that transgenic mice with 960 CTG repeats in DMPK 3′UTR display muscle loss not observed in Mbnl1ΔE3/ΔE3 mice6,27. We found that CUGexp RNA, but not genetic loss of Mbnl1, causes alteration of a broad set of ECM mRNAs (Fig. 6) that could worsen with age or increased repeat expansion, as observed in the human disease. Consistent with this, mutations in some of these same ECM genes cause muscular dystrophies or connective tissue diseases (Table 1, refs. 46–48). For example, defects in collagen I cause osteogenesis imperfecta with muscle weakness49, whereas mutations in Col6a1 are responsible for Ullrich congenital muscular dystrophy50. Col15a1 is involved in maintaining ECM structural integrity, and Col15a1-deficient mice show progressive muscle fiber degeneration51. Defects in elastin assembly also affect muscle strength and regeneration. Fibrillin-1 (FBN1) mutation causes Marfan syndrome with muscle involvement52,53. Loss of Fibulin-5 contributes to age-related muscular degeneration54,55. Tenascin C interacts with integrin (integrin α7 mutations cause congenital muscular dystrophy), and TNC-deficient mice show reduced muscle strength56. Unlike EpA960 transgenic mice6, HSALR mice do not develop severe muscle weakness and wasting, so more extreme alterations in ECM gene expression may promote a more severe dystrophic phenotype. Together these data overwhelmingly point to the idea that a loss of regulation of ECM function and the consequent effects on cell adhesion contribute to muscle defects in DM1 (Table 1, Fig. 6).
Methods
Sample Preparation and Affymetrix Splicing Microarray Detection
RNA samples from the quadriceps muscle of individual 12–14-week-old male mice from the Mbnl1ΔE3/ΔE3, HSALR, and FVB inbred background lines (n = 4 for each group) were compared. To identify Mbnl1-dependent splicing events in heart (shown in Supplementary Table 2), RNA from quadriceps muscle and heart from Mbnl1ΔE3/ΔE3 mice and age-matched wild type mice in the C57BL/6J background were compared. RNA samples were processed for hybridization to Affymetrix “A-chip” oligonucleotide microarrays29 according to the standards of the manufacturer.
Microarray Data Analysis
Analysis was done according to ref. 29, with modifications indicated below. Preliminary analysis of data for the HSALR mice indicated that probes with any 7-mer comprised of CTG repeats reported significantly higher intensities than others in the same gene, likely reflecting cross-hybridization driven by the large mass of CUGexp RNA in the sample. We corrected for this by ignoring any gene with more than 4 probes containing the three 7-mer permutations of (CTG)n: CTGCTGC, GCTGCTG, or TGCTGCT.
The separation score method29 is illustrated for the Nfix(123) exon in Fig. 1a: when exon skipping (Skip) intensities are plotted against exon inclusion (Include) intensities, the wild type samples appear on the left side of the graph while the Mbnl1ΔE3/ΔE3 and HSALR samples appear in the lower right, indicating a shift from exon skipping in the wild type to inclusion in the mutants. The equation for sepscore is:
For each replicate set (HSALR, Mbnl1ΔE3/ΔE3, and wild type), we estimated the log2 ratio of skipping to inclusion using robust least squares analysis. We evaluated sepscore significance by permuting the assignments of data points to replicate sets, calculating the separation score for the permuted data, and estimating the likelihood that the observed data came from the permuted distribution. We estimated rates of overall gene expression according to probesets that measure constitutive features. We identified genes that are differentially expressed between replicate sets using SAM57.
RT-PCR and bioanalyzer analysis
Total RNA was reversed transcribed to cDNA with Superscript III (Invitrogen) and oligo-dT, following the manufacturer’s protocol. Then ~50 ng cDNA was used as template for PCR with primers to regions spanning the test cassette exons. PCR was carried out for 25–35 cycles using the Platinum Taq polymerase (Invitrogen). To measure splicing, 1 ul PCR products from above reaction were separated on the Agilent bioanalyzer, which then reported the size and concentrations of each splicing-derived product.
Quantitative RT-PCR
Quantitative RT-PCR was carried out in 30 ul reactions using Brilliant QRT-PCR Master Mix kits (SYBR-Green, Stratagene, La Jolla, CA) on a Bio-Rad iCycler. We used GAPDH for normalization, and the protocol of Livak et al.58 for calculating changes in threshold cycles.
Analysis of sequence motifs
We used Improbizer to search intronic sequences flanking each differentially-spliced exon. Background sequences were from alternative exons that were expressed but showed no changes in splicing. We searched for 4 copies of each motif during its search (maxOcc setting = 4). To map motifs, we searched in 40-base windows (offset by 5 bases) through the 150-base region immediately upstream and downstream of each alternative exon. We contrasted 55 Mbnl1-repressed exons (skipping lost in the mutant), 66 Mbnl1-induced exons (inclusion lost in the mutant), and 790 alternative exons expressed without splicing changes. We determined background motif frequencies by randomly sampling equivalent numbers of exons from the background 1000 times per sequence window. Mean background counts and the 95% confidence interval were determined through quartile analysis.
To identify sequence words associated with class II genes we contrasted the sequences of the 5′ and 3′ UTRs to those of all genes expressed in the experiment. We determine the number of times each k-mer appears in the selected sequences and in the background, for k = 5, 6, and 7. A public perl module (Statistics::ChisqIndep) for chi-squared evaluation of 2×2 contingency table data is used to determine the likelihood (p-value) that the observed enrichment is due to chance. A Bonferroni correction is applied by multiplying the p-value by the number of all possible k-mers (4k). This correction does not assume independence and ignores the fact that many k-mers only rarely appear in the genome. We consider k-mers to be significantly enriched in the test set relative to background if their false discovery rates are 0.1 or less (Supplementary Table 8).
Cloning, mutagenesis, cell culture and transfection
The human β-globin Dup33 minigene plasmid59 (gift of Doug Black) was used to create splicing reporters. The Vldlr(84) and Nfix(123) exons and associated intron sequences (~200 bp on each side) were amplified from mouse genomic DNA and inserted between the ApaI and BglII sites. Mutagenesis was performed using the QuikChange Site-Directed Mutagenesis Kit (Stratagene Corp.)
Mouse embryonic fibroblasts from Mbnl1ΔE3/ΔE3 (C57BL/6J background) were immortalized by transfection with pOT, a plasmid expressing SV40 T-antigen60 (gift of Xiang-Dong Fu). The cells were grown in antibiotic-free Dulbecco’s modified Eagle’s medium (DMEM) with 15% fetal bovine serum. To test the function of Mbnl1 and the motifs, 0.5 ug wild type or mutant minigenes were and 1 ug pEGFP-MBNL1 (ref. 28) or the control plasmid producing only GFP (pEGFPC1) were co-transfected using Lipofectamine 2000 reagent (Invitrogen). RNA was harvested 24 h after transfection.
Detection of enrichment of functional gene classes
We evaluated Gene Ontology term enrichment using GoMiner (Version 200904, April 2009; Application Build 246, ref. 38), relative to the set of all genes expressed in the experiment. Because of the use of Fisher’s exact test for scoring, enrichment of GO classes for which there are 5 or fewer genes must be considered tentative.
Supplementary Material
Acknowledgments
We thank Doug Black (UCLA) and Xiang-Dong Fu (UCSD) for gifts of plasmid and advice on mammalian cell culture. Thanks to Benoit Chabot and Jeremy Sanford for critical readings of the manuscript. This work was primarily supported by grant GM084317 to M.A., with additional support from GM040478 (to M.A.), grant AR046799 (to M.S.) and grants AR046806 and a Muscular Dystrophy Center grant U54NS048843 (to C.T.). M.H. acknowledges the California Institute of Regenerative Medicine for postdoctoral support.
Footnotes
Accession codes
Microarray data for this study has been deposited in GEO under accession number GSE17986.
Author contributions
M.A., H.D., M.S. and C.T. designed the experiments. H.D., R.O., D.T., and M.H. performed the experiments. T.C. provided materials and data analysis. M.C., J.D., and L.S. performed data analysis. M.A., H.D., and M.C. wrote the paper.
References
- 1.Mirkin SM. Expandable DNA repeats and human disease. Nature. 2007;447:932–40. doi: 10.1038/nature05977. [DOI] [PubMed] [Google Scholar]
- 2.Caskey CT, Pizzuti A, Fu YH, Fenwick RG, Jr, Nelson DL. Triplet repeat mutations in human disease. Science. 1992;256:784–9. doi: 10.1126/science.1589758. [DOI] [PubMed] [Google Scholar]
- 3.Buxton J, et al. Detection of an unstable fragment of DNA specific to individuals with myotonic dystrophy. Nature. 1992;355:547–8. doi: 10.1038/355547a0. [DOI] [PubMed] [Google Scholar]
- 4.Taneja KL, McCurrach M, Schalling M, Housman D, Singer RH. Foci of trinucleotide repeat transcripts in nuclei of myotonic dystrophy cells and tissues. J Cell Biol. 1995;128:995–1002. doi: 10.1083/jcb.128.6.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mankodi A, Lin X, Blaxall BC, Swanson MS, Thornton CA. Nuclear RNA foci in the heart in myotonic dystrophy. Circ Res. 2005;97:1152–5. doi: 10.1161/01.RES.0000193598.89753.e3. [DOI] [PubMed] [Google Scholar]
- 6.Orengo JP, et al. Expanded CTG repeats within the DMPK 3′ UTR causes severe skeletal muscle wasting in an inducible mouse model for myotonic dystrophy. Proc Natl Acad Sci U S A. 2008;105:2646–51. doi: 10.1073/pnas.0708519105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mankodi A, et al. Myotonic dystrophy in transgenic mice expressing an expanded CUG repeat. Science. 2000;289:1769–73. doi: 10.1126/science.289.5485.1769. [DOI] [PubMed] [Google Scholar]
- 8.Ebralidze A, Wang Y, Petkova V, Ebralidse K, Junghans RP. RNA leaching of transcription factors disrupts transcription in myotonic dystrophy. Science. 2004;303:383–7. doi: 10.1126/science.1088679. [DOI] [PubMed] [Google Scholar]
- 9.Krol J, et al. Ribonuclease dicer cleaves triplet repeat hairpins into shorter repeats that silence specific targets. Mol Cell. 2007;25:575–86. doi: 10.1016/j.molcel.2007.01.031. [DOI] [PubMed] [Google Scholar]
- 10.Kuyumcu-Martinez NM, Wang GS, Cooper TA. Increased steady-state levels of CUGBP1 in myotonic dystrophy 1 are due to PKC-mediated hyperphosphorylation. Mol Cell. 2007;28:68–78. doi: 10.1016/j.molcel.2007.07.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin X, et al. Failure of MBNL1-dependent post-natal splicing transitions in myotonic dystrophy. Hum Mol Genet. 2006;15:2087–97. doi: 10.1093/hmg/ddl132. [DOI] [PubMed] [Google Scholar]
- 12.Lueck JD, Mankodi A, Swanson MS, Thornton CA, Dirksen RT. Muscle chloride channel dysfunction in two mouse models of myotonic dystrophy. J Gen Physiol. 2007;129:79–94. doi: 10.1085/jgp.200609635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Charlet BN, et al. Loss of the muscle-specific chloride channel in type 1 myotonic dystrophy due to misregulated alternative splicing. Mol Cell. 2002;10:45–53. doi: 10.1016/s1097-2765(02)00572-5. [DOI] [PubMed] [Google Scholar]
- 14.Lueck JD, et al. Chloride channelopathy in myotonic dystrophy resulting from loss of posttranscriptional regulation for CLCN1. Am J Physiol Cell Physiol. 2007;292:C1291–7. doi: 10.1152/ajpcell.00336.2006. [DOI] [PubMed] [Google Scholar]
- 15.Wheeler TM, Lueck JD, Swanson MS, Dirksen RT, Thornton CA. Correction of ClC-1 splicing eliminates chloride channelopathy and myotonia in mouse models of myotonic dystrophy. J Clin Invest. 2007;117:3952–7. doi: 10.1172/JCI33355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Savkur RS, Philips AV, Cooper TA. Aberrant regulation of insulin receptor alternative splicing is associated with insulin resistance in myotonic dystrophy. Nat Genet. 2001;29:40–7. doi: 10.1038/ng704. [DOI] [PubMed] [Google Scholar]
- 17.Miller JW, et al. Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy. Embo J. 2000;19:4439–48. doi: 10.1093/emboj/19.17.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Warf MB, Berglund JA. MBNL binds similar RNA structures in the CUG repeats of myotonic dystrophy and its pre-mRNA substrate cardiac troponin T. Rna. 2007;13:2238–51. doi: 10.1261/rna.610607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Begemann G, et al. muscleblind, a gene required for photoreceptor differentiation in Drosophila, encodes novel nuclear Cys3His-type zinc-finger-containing proteins. Development. 1997;124:4321–31. doi: 10.1242/dev.124.21.4321. [DOI] [PubMed] [Google Scholar]
- 20.Teplova M, Patel DJ. Structural insights into RNA recognition by the alternative-splicing regulator muscleblind-like MBNL1. Nat Struct Mol Biol. 2008;15:1343–51. doi: 10.1038/nsmb.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ho TH, et al. Muscleblind proteins regulate alternative splicing. Embo J. 2004;23:3103–12. doi: 10.1038/sj.emboj.7600300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mooers BH, Logue JS, Berglund JA. The structural basis of myotonic dystrophy from the crystal structure of CUG repeats. Proc Natl Acad Sci U S A. 2005;102:16626–31. doi: 10.1073/pnas.0505873102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paul S, et al. Interaction of muscleblind, CUG-BP1 and hnRNP H proteins in DM1-associated aberrant IR splicing. Embo J. 2006;25:4271–83. doi: 10.1038/sj.emboj.7601296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Timchenko NA, et al. RNA CUG repeats sequester CUGBP1 and alter protein levels and activity of CUGBP1. J Biol Chem. 2001;276:7820–6. doi: 10.1074/jbc.M005960200. [DOI] [PubMed] [Google Scholar]
- 25.Kalsotra A, et al. A postnatal switch of CELF and MBNL proteins reprograms alternative splicing in the developing heart. Proc Natl Acad Sci U S A. 2008;105:20333–8. doi: 10.1073/pnas.0809045105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim DH, et al. HnRNP H inhibits nuclear export of mRNA containing expanded CUG repeats and a distal branch point sequence. Nucleic Acids Res. 2005;33:3866–74. doi: 10.1093/nar/gki698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanadia RN, et al. A muscleblind knockout model for myotonic dystrophy. Science. 2003;302:1978–80. doi: 10.1126/science.1088583. [DOI] [PubMed] [Google Scholar]
- 28.Kanadia RN, et al. Reversal of RNA missplicing and myotonia after muscleblind overexpression in a mouse poly(CUG) model for myotonic dystrophy. Proc Natl Acad Sci U S A. 2006;103:11748–53. doi: 10.1073/pnas.0604970103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sugnet CW, et al. Unusual intron conservation near tissue-regulated exons found by splicing microarrays. PLoS Comput Biol. 2006;2:e4. doi: 10.1371/journal.pcbi.0020004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ni JZ, et al. Ultraconserved elements are associated with homeostatic control of splicing regulators by alternative splicing and nonsense-mediated decay. Genes Dev. 2007;21:708–18. doi: 10.1101/gad.1525507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fardaei M, et al. Three proteins, MBNL, MBLL and MBXL, co-localize in vivo with nuclear foci of expanded-repeat transcripts in DM1 and DM2 cells. Hum Mol Genet. 2002;11:805–14. doi: 10.1093/hmg/11.7.805. [DOI] [PubMed] [Google Scholar]
- 32.Holt I, et al. Muscleblind-like proteins: similarities and differences in normal and myotonic dystrophy muscle. Am J Pathol. 2009;174:216–27. doi: 10.2353/ajpath.2009.080520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hino S, et al. Molecular mechanisms responsible for aberrant splicing of SERCA1 in myotonic dystrophy type 1. Hum Mol Genet. 2007;16:2834–43. doi: 10.1093/hmg/ddm239. [DOI] [PubMed] [Google Scholar]
- 34.Yuan Y, et al. Muscleblind-like 1 interacts with RNA hairpins in splicing target and pathogenic RNAs. Nucleic Acids Res. 2007;35:5474–86. doi: 10.1093/nar/gkm601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hsu F, et al. The UCSC Known Genes. Bioinformatics. 2006;22:1036–46. doi: 10.1093/bioinformatics/btl048. [DOI] [PubMed] [Google Scholar]
- 36.Lejeune F, Maquat LE. Mechanistic links between nonsense-mediated mRNA decay and pre-mRNA splicing in mammalian cells. Curr Opin Cell Biol. 2005;17:309–15. doi: 10.1016/j.ceb.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 37.Wheeler TM, et al. Reversal of RNA dominance by displacement of protein sequestered on triplet repeat RNA. Science. 2009;325:336–9. doi: 10.1126/science.1173110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zeeberg BR, et al. High-Throughput GoMiner, an ‘industrial-strength’ integrative gene ontology tool for interpretation of multiple-microarray experiments, with application to studies of Common Variable Immune Deficiency (CVID) BMC Bioinformatics. 2005;6:168. doi: 10.1186/1471-2105-6-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.He F, et al. Solution structure of the RNA binding domain in the human muscleblind-like protein 2. Protein Sci. 2009;18:80–91. doi: 10.1002/pro.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Adereth Y, Dammai V, Kose N, Li R, Hsu T. RNA-dependent integrin alpha3 protein localization regulated by the Muscleblind-like protein MLP1. Nat Cell Biol. 2005;7:1240–7. doi: 10.1038/ncb1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jiang H, Mankodi A, Swanson MS, Moxley RT, Thornton CA. Myotonic dystrophy type 1 is associated with nuclear foci of mutant RNA, sequestration of muscleblind proteins and deregulated alternative splicing in neurons. Hum Mol Genet. 2004;13:3079–88. doi: 10.1093/hmg/ddh327. [DOI] [PubMed] [Google Scholar]
- 42.Hao M, et al. Muscleblind-like 2 (Mbnl2) -deficient mice as a model for myotonic dystrophy. Dev Dyn. 2008;237:403–10. doi: 10.1002/dvdy.21428. [DOI] [PubMed] [Google Scholar]
- 43.Mankodi A, et al. Expanded CUG repeats trigger aberrant splicing of ClC-1 chloride channel pre-mRNA and hyperexcitability of skeletal muscle in myotonic dystrophy. Mol Cell. 2002;10:35–44. doi: 10.1016/s1097-2765(02)00563-4. [DOI] [PubMed] [Google Scholar]
- 44.Xu X, et al. ASF/SF2-regulated CaMKIIdelta alternative splicing temporally reprograms excitation-contraction coupling in cardiac muscle. Cell. 2005;120:59–72. doi: 10.1016/j.cell.2004.11.036. [DOI] [PubMed] [Google Scholar]
- 45.Lee SJ, et al. Regulation of muscle growth by multiple ligands signaling through activin type II receptors. Proc Natl Acad Sci U S A. 2005;102:18117–22. doi: 10.1073/pnas.0505996102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kanagawa M, Toda T. The genetic and molecular basis of muscular dystrophy: roles of cell-matrix linkage in the pathogenesis. J Hum Genet. 2006;51:915–26. doi: 10.1007/s10038-006-0056-7. [DOI] [PubMed] [Google Scholar]
- 47.Jimenez-Mallebrera C, Brown SC, Sewry CA, Muntoni F. Congenital muscular dystrophy: molecular and cellular aspects. Cell Mol Life Sci. 2005;62:809–23. doi: 10.1007/s00018-004-4510-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schessl J, Zou Y, Bonnemann CG. Congenital muscular dystrophies and the extracellular matrix. Semin Pediatr Neurol. 2006;13:80–9. doi: 10.1016/j.spen.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 49.Engelbert RH, et al. Osteogenesis imperfecta in childhood: impairment and disability. A prospective study with 4-year follow-up. Arch Phys Med Rehabil. 2004;85:772–8. doi: 10.1016/j.apmr.2003.08.085. [DOI] [PubMed] [Google Scholar]
- 50.Ishikawa H, et al. Ullrich disease due to deficiency of collagen VI in the sarcolemma. Neurology. 2004;62:620–3. doi: 10.1212/01.wnl.0000113023.84421.00. [DOI] [PubMed] [Google Scholar]
- 51.Eklund L, et al. Lack of type XV collagen causes a skeletal myopathy and cardiovascular defects in mice. Proc Natl Acad Sci U S A. 2001;98:1194–9. doi: 10.1073/pnas.031444798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Behan WM, et al. Muscle fibrillin deficiency in Marfan’s syndrome myopathy. J Neurol Neurosurg Psychiatry. 2003;74:633–8. doi: 10.1136/jnnp.74.5.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Percheron G, et al. Muscle strength and body composition in adult women with Marfan syndrome. Rheumatology (Oxford) 2007;46:957–62. doi: 10.1093/rheumatology/kel450. [DOI] [PubMed] [Google Scholar]
- 54.Stone EM, et al. Missense variations in the fibulin 5 gene and age-related macular degeneration. N Engl J Med. 2004;351:346–53. doi: 10.1056/NEJMoa040833. [DOI] [PubMed] [Google Scholar]
- 55.Lotery AJ, et al. Reduced secretion of fibulin 5 in age-related macular degeneration and cutis laxa. Hum Mutat. 2006;27:568–74. doi: 10.1002/humu.20344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Morellini F, Schachner M. Enhanced novelty-induced activity, reduced anxiety, delayed resynchronization to daylight reversal and weaker muscle strength in tenascin-C-deficient mice. Eur J Neurosci. 2006;23:1255–68. doi: 10.1111/j.1460-9568.2006.04657.x. [DOI] [PubMed] [Google Scholar]
- 57.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001;98:5116–21. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 59.Dominski Z, Kole R. Selection of splice sites in preRNAs with short internal exons. Mol Cell Biol. 1991;11:6075–83. doi: 10.1128/mcb.11.12.6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hanahan D, Lane D, Lipsich L, Wigler M, Botchan M. Characteristics of an SV40-plasmid recombinant and its movement into and out of the genome of a murine cell. Cell. 1980;21:127–39. doi: 10.1016/0092-8674(80)90120-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.