Abstract
Small RNAs (smRNAs) including miRNAs and siRNAs are critical for gene regulation and plant development. Among the highly diverse siRNAs, trans-acting siRNAs (ta-siRNAs) have been shown to be plant-specific. In Arabidopsis, eight TAS loci belonging to four families (TAS1, TAS2, TAS3, and TAS4) have been identified, and bioinformatics analysis reveals that the sequence of TAS3 is highly conserved in plants. In this study, the function of TAS3 ta-siRNA (tasiR-ARF) has been revealed in rice (Oryza sativa L.) on polarity establishment and stage transition from vegetative to reproductive development by over-expressing Osta-siR2141. Osta-siR2141 replaced miR390 in the miR390 backbone for ectopic expression in rice, and overexpression of Osta-siR2141 caused disturbed vascular bundle development and adaxialization in polarity establishment. Transgenic lines also displayed abnormal shoot apical meristems (SAMs) and retarded growth at the vegetative stage. Molecular analysis revealed that overexpression of Osta-siR2141 resulted in the down-regulation of miR166 and the up-regulation of class III homeodomain-leucine zipper genes (HD-ZIPIIIs) in the vegetative stage but not in the reproductive stage. Moreover, overexpression of Osta-siR2141 in Arabidopsis disturbed polarity establishment and retarded stage transition, suggesting that tasiR-ARF was functionally conserved in rice and Arabidopsis.
Keywords: Adaxialization, HD-ZIPIII, polarity, tasiR-ARF
Introduction
SmRNAs are a large class of non-coding RNAs ranging from 20–30 nucleotides in length (Aravin et al., 2003). Through association with the RNA-induced silencing complex (RISC), smRNAs identify mRNA based on anti-sense complementarity and result in mRNA cleavage, translation repression, chromatin modification, and even influence genome integrity (Allshire, 2002; Mochizuki and Gorovsky, 2004; Zamore and Haley, 2005; Vaucheret, 2006; Liu, 2008; Mosher et al., 2008). MiRNAs and siRNAs are the two broad categories of smRNAs, and the machinery of smRNA regulation exists in a wide variety of organisms (Chapman and Carrington, 2007; Molnar et al., 2007; Zhao et al., 2007; Xie and Qi, 2008). SiRNAs were first identified because of their association with the post-transcriptional gene silencing (PTGS) in plants (Hamilton and Baulcombe, 1999). Besides mediating gene-specific silencing, siRNAs may also take part in DNA methylation and transcriptional silencing (Carrington and Ambros, 2003; Kidner and Martienssen, 2005). In fact, miRNAs and siRNAs are functionally related and act in the common pathway interchangeably, depending on the degree of complementarity with their targets (Aravin et al., 2003; Carrington and Ambros, 2003; Bartel, 2005). And activity of them might differ according to their biogenesis (Tretter et al., 2008).
Sets of siRNAs prove to be highly diverse, and new members are constantly being identified in various organisms which adds further depth and complexity to the siRNA world (Hamilton et al., 2002; Lippman and Martienssen, 2004; Borsani et al., 2005; Katiyar-Agarwal et al., 2006, 2007; Kasschau et al., 2007). Trans-acting siRNA (ta-siRNA) is a kind of siRNA specific to plants. Ta-siRNAs originate from defined genetic loci (named TAS loci) in the genome through a miRNA-dependent pathway (Allen et al., 2005; Axtell et al., 2006). Generally, a TAS locus produces a non-protein-coding transcript, a portion of which is then converted into double stranded-RNA (dsRNA), a process that is triggered by cleavage of the original transcript by the corresponding miRNA; then the dsRNA is cleaved into siRNAs of 21 nucleotides, among which are the mature ta-siRNAs (Fahlgren et al., 2006). Most TAS loci have miRNA complementary sites at which miRNA-directed cleavage defines one end of the dsRNA intermediate, and thereby sets the register of phased ta-siRNA production (Allen et al., 2005; Axtell et al., 2006). Due to the dependence of ta-siRNAs on miRNA cleavage, factors needed in the biogenesis of miRNAs are also required by ta-siRNAs (Peragine et al., 2004; Vazquez et al., 2004). Ta-siRNAs negatively regulate mRNAs with no sequence relevance with the TAS loci (Peragine et al., 2004; Allen et al., 2005; Yoshikawa et al., 2005; Axtell et al., 2006; Vaucheret, 2006).
Until now, eight TAS loci belonging to four families (TAS1, TAS2, TAS3, and TAS4) have been identified in Arabidopsis (Allen et al., 2005; Yoshikawa et al., 2005; Rajagopalan et al., 2006). MiRNA cleavages of TAS1, TAS2, and TAS4 transcripts occur at the 5′ sides of ta-siRNA generating regions, while that of TAS3 occurs at the 3′ sides, although the TAS3 transcript is flanked by dual miR390 sites. The biogenesis of TAS3 ta-siRNAs (tasiR-ARFs) is in tight association with the AGO7 protein, while that of TAS1 and TAS2 ta-siRNAs is not (Axtell et al., 2006; Howell et al., 2007; Montgomery et al., 2008). tasiR-ARFs have been proved to influence various aspects of leaf morphology, leaf polarity, developmental timing, and patterning by targeting Auxin Responsive Factor 3 (ARF3) and ARF4 (Adenot et al., 2006; Fahlgren et al., 2006; Garcia et al., 2006; Montgomery et al., 2008). TAS3 is highly conserved in plants (Allen et al., 2005; Axtell et al., 2006, 2007). In maize (Zea mays), tasiR-ARFs were found to function in the maintenance of leaf polarity along with miR166, which establishes the abaxial character through the negative regulation of the class III homeodomain zipper (HD-ZIPIII) transcription factor genes (Nogueira et al., 2007).
The biogenesis and function of tasiR-ARF have been studied in Arabidopsis; in monocotyledons, and especially in rice, only a preliminary study has been carried out (Yoshikawa et al., 2005; Adenot et al., 2006; Fahlgren et al., 2006; Garcia et al., 2006; Nogueira et al., 2007; Montgomery et al., 2008). To reveal the function of tasiR-ARF in rice, Osta-siR2141 was over-expressed through the miR390 backbone. Overexpression of Osta-siR2141 caused various developmental changes, such as a disturbance in vascular bundle development and adaxialization in polarity establishment, abnormal shoot apical meristems (SAMs), and growth retardation at the vegetative stage etc. In addition, ectopic expression of Osta-siR2141 down-regulated the expression of miR166 and up-regulated that of HD-ZIPIIIs; however, at the reproductive stage, HD-ZIPIII genes were not up-regulated, implying that the transition from vegetative to reproductive growth might be sensitive to the level of HD-ZIPIII expression. Moreover, the conserved role of Osta-siR2141 was revealed by over-expressing it in Arabidopsis.
Materials and methods
Plant materials
Oryza sativa L. subsp. japonica cv. Zhonghua No. 11 (abbreviated as ZH11) was used as the wild type. Transformants that had ceased at the vegetative stage (CVS) were kept in tubes by tissue culture. ZH11 and non-CVS transformants were planted in the greenhouse, with 16/8 h light/dark, with a planting management that accorded with standard greenhouse practice.
The study of Arabidopsis thaliana, Columbia ecotype was used as the wild type. Seeds were sown on MS medium, cold-treated for 3 d at 4 °C, and then transferred to controlled environment cabinets under SD (8/16 h light/dark) conditions with a fluence rate of 120 μmol m−2 s−1 of white light at 22 °C.
Construction of OsmiR-ARF(390)
Firstly miR390 backbone of 176 base pairs was cloned into pCAMBIA1301 (kindly provided by Professor Richard Jefferson) by PCR amplification, using primers miR390F and miR390R, and resulting in the construct p1301(390). Then the mature miR390-producing DNA region in p1301(390) was substituted by Osta-siR2141 using overlapping PCR (Schwab et al., 2006) with the combination of primers siARFF, siARFR, miR390F, and miR390R, resulting in OsmiR-ARF(390). In this study, Osta-siR2141 from OsTAS3a was used in OsmiR-ARF(390), and three nucleotide mismatches between ta-siR2141 and ta-siR2141* were introduced (Fig. 1C). The whole process of OsmiR-ARF(390) construction is outlined in Supplementary Fig. S1 at JXB online. Primer sequences are listed in Supplementary Table S1 at JXB online.
Fig. 1.
Analysis of the TAS3 gene and tasiR-ARF in rice. (A) OsTAS3a on chromosome 3 and OsTAS3b on chromosome 5, as marked in red; OsTAS3a-phased tasiR-ARFs were from the 5′D6(+) and 5′D7(+) positions and OsTAS3b-phased from the 5′D7(+) and 5′D8(+) positions. (B) Alignment of tasiR-ARFs in rice and Arabidopsis, shaded nucleotides indicating mismatches. (C) Complementarity of tasiR-ARF and OsARF3s. In OsmiR-ARF(390), Osta-siR2141 from OsTAS3a was used, three red asterisks between ta-siR2141* and ta-siR2141 indicating the introduced mismatches; red asterisks between the OsTAS3a ta-siR2141 and OsARF3-1 site A indicating matching of OstasiR-ARF and OsARF3s.
Genetic transformation of rice and Arabidopsis
Rice was transformed using Agrobacterium-mediated transformation (Hiei et al., 1994). Arabidopsis was also transformed using Agrobacterium-mediated transformation (Clough and Bent, 1998).
Small RNA Northern blot analysis
Total RNAs from rice and Arabidopsis tissues were extracted using the Trizol reagent (Invitrogen, Carlsbad, CA), and the concentration was measured using a Thermo Scientific NanoDrop*1000 Spectrophotometer. Northern blot analysis was carried out as follows: at least 20 μg of total RNA was loaded for SDS-PAGE (19% concentration) electrophoresis, and then transferred onto nylon membrane (Amersham Hybond N+) by electrophoretic transfer; prehybridization was carried out for 2 h at 35 °C. The probes anti-sense Osta-siR2141 and anti-sense OsmiR166a were radioactively labelled using the terminal labelling method, and hybridization was carried out at 41 °C overnight.
RT-PCR analysis
In the reverse transcription process, about 1 μg of the DNaseI-treated total RNA template and oligod(T) primer were used in synthesis of the first strand of cDNA, using M-MLV RTase (Toyobo, Japan). All the molecular manipulation followed the routine protocols. Gene-specific primers are listed in Supplementary Table S1 at JXB online.
Scanning electron microscope (SEM) analysis
Shoot apical meristems (SAMs) were decorticated under a light microscope and leaves were cut using a sharp knife. All samples were fixed quickly in 50% FAA at 4 °C overnight after vacuuming and then dehydrated through a graded alcohol series of 70%, 85%, and 90% ethanol once, and 100% ethanol twice, each for 10 min. Samples were critical point dried using liquid carbon dioxide and mounted on SEM stubs, then sputter coated with gold and palladium (4:1) and examined using a SEM (Hitachi S-2460, Japan) and pictures were taken.
Anatomical analysis
Leaves and roots were cut using a sharp knife and fixed in 50% FAA at 4 °C overnight after vacuuming. After serial dehydration in several concentrations of ethanol, samples were embedded in epoxide resin and cut into slices 2–3 μm thick; strips of these slices were spread at 42 °C on a hot platform overnight, stained using 0.5% toluidine blue O, and sealed for observation under the microscope (Olympus BX51 plus DP70).
In situ hybridization
SAM regions were fixed in 4% (w/v) paraformaldehyde and 0.25% glutaraldehyde in 0.1 M sodium phosphate buffer (pH 7.4) overnight at 4 °C, dehydrated through a graded ethanol and xylene series, and embedded in Paraplast Plus (Sigma). Microtome sections (8 μm thick) were applied to glass slides treated with polylysine. For RNA synthesis and labelling, an OsHB3 cDNA fragment was cloned into the pBluescript II KS vector using primers OsHB3IF and OsHB3IR (sequences listed in Supplementary Table S1 at JXB online). In situ hybridization of digoxigenin-labelled sense/anti-sense RNA was conducted as described by Coen et al. (1990).
Results
Analysis of OsTAS3 and tasiR-ARF in rice
In rice, there are two homologous TAS3 gene loci, OsTAS3a on chromosome 3 and OsTAS3b on chromosome 5; each locus bearing two miR390 complementary sites at the 3′ and 5′ sides, respectively (Fig. 1A). In-phase 21-nucleotide positions on the 5′ side of the miR390 cleavage site were coded as 5′D1(+), 5′D2(+), 5′D3(+), and so on (Fig. 1A). OsTAS3a-phased tasiR-ARFs were from the 5′D6(+) and 5′D7(+) positions, and OsTAS3b-phased from the 5′D7(+) and 5′D8(+) positions (Fig. 1A). Compared with those from Arabidopsis, tasiR-ARFs produced by OsTAS3a showed two nucleotide mismatches within the ta-siR2141 sequence and one nucleotide mismatch within the ta-siR2142 sequence; tasiR-ARFs produced by OsTAS3b showed one nucleotide mismatch within the ta-siR2141 sequence and two nucleotide mismatches within the ta-siR2142 sequence (Fig. 1B), suggesting a high degree of sequence conservation between rice and Arabidopsis.
There are four ARF3 gene homologies in rice, i.e. Os05g48870, Os05g43920, Os01g48060, and Os01g54990. They were tentatively named as OsARF3-1, OsARF3-2, OsARF3-3, and OsARF3-4 respectively. Each of the OsARF3s contained two tandem OstasiR-ARF complementary sites (site A and site B in Fig. 1C). OstasiR-ARFs showed a nearly perfect match with their targets (red asterisks between ta-siR2141 and OsARF3-1 in Fig. 1C). No ARF4 gene homology was found in rice; while, in Arabidopsis, both ARF3 and ARF4 genes were proved to be the targets of tasiR-ARFs (Fahlgren et al., 2006).
Construction of OsmiR-ARF(390) and phenotypes of the transformants
To study the function of tasiR-ARF in rice, a vector, OsmiR-ARF(390), to over-express Osta-siR2141 was constructed first. A genomic fragment of the miR390 backbone was amplified from the rice genome and cloned into pCAMBIA1301 between the ubi promoter and the nos terminator. The mature miR390 region was substituted by Osta-siR2141 using overlapping PCR (Schwab et al., 2006). In OsmiR-ARF(390), ta-siR2141 from OsTAS3a was used, and three nucleotide mismatches between ta-siR2141 and ta-siR2141* were introduced (Fig. 1C).
More than 800 transgenic lines were obtained; about 99% of them displayed growth that had terminated at the vegetative stage (CVS transformants) with a seedling height of about 3 cm (Fig. 2A). These CVS transformants showed thick and rough sheaths (Fig. 2A, B, C, D), and seriously deformed leaves which rolled adaxially (Fig. 2A, B, left). In some cases, leaves were thread-like (Fig. 2C, red arrow); also, in rare cases, there was no production of leaves (data not shown). The leaf-sheath structures was disordered (Fig. 2D), suggesting an abnormal leaf initiation and phyllotaxy. Roots seemed normal in appearance (Fig. 2B). About 1% of the transformants could continue development until the reproductive stage (non-CVS transformants) with distorted leaves and abnormal phyllotaxy (Fig. 2E). Non-CVS transformants could develop flowers, but the seeds became crimped and began to die at about 10 days after pollination (DAP) (Fig. 2H).
Fig. 2.
Phenotypes of the transformants. (A) CVS transformants were about 3 cm; there was a ruler on left; (B) Roots of the CVS transformants (left) and transformants from a void vector (right). (C) Thread-like leaves (red arrows) in one CVS transformant. (D) Abnormal phyllotaxy in one CVS transformant. (E) A non-CVS transformant showing twisted leaves and abnormal phyllotaxy. (F) Alternative phyllotaxy in ZH11. (G) Seeds of ZH11. (H) Seeds of the non-CVS transformant. Seeds in (G) and (H) were of 1, 3, 5, and 10 DAPs, respectively (left to right). Bars in (A), (C), and (D) were 0.5 cm, in (B), (E), and (F) were 1 cm, in (G) and (H) were 1 mm.
CVS transformants showed adaxialization in polarity establishment
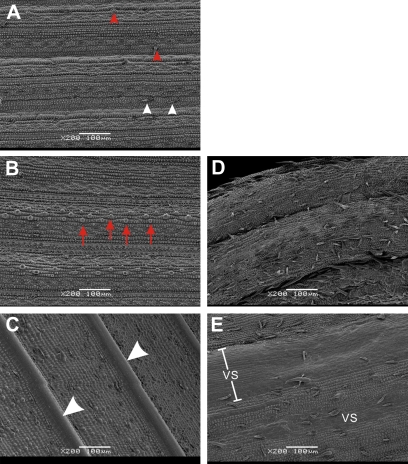
To characterize the cellular changes in the transformants, SEM analysis was performed. Since leaves of the CVS lines rolled up tightly, only the abaxial surface could be observed by SEM. In the wild-type, the adaxial epidermis was characterized by thorns (Fig. 3A, red arrowheads) and hairs (Fig. 3A, white arrowheads), and the abaxial epidermis was distinguished by water pores (Fig. 3B, red arrows). In the CVS transformants, fewer water pores on the abaxial epidermis were observed; while many thorns appreared (Fig. 3D), suggesting that some adaxial characters were converted to the abaxial side. On the sheath of the CVS lines, thorns and hairs seemed to be loosely connected to the surface (Fig. 3E); vascular bundles were confused and irregularly enlarged (Fig. 3E, ‘VS’), suggesting disturbance in vascular bundle development.
Fig. 3.
SEM analyses of leaves and sheaths of the CVS transformants. (A) Adaxial surface of a ZH11 leaf, white arrowheads indicating hairs and red arrowheads thorns. (B) Abaxial surface of a ZH11 leaf, red arrows showing water pores. (C) Outer surface of a ZH11 sheath, white arrowheads indicating the vascular bundles. (D) Abaxial surface of the CVS transformant leaf. (E) Outer surface of the CVS transformant sheath, ‘VS’ indicating the vascular bundles. ZH11 was at the five-leaf-stage.
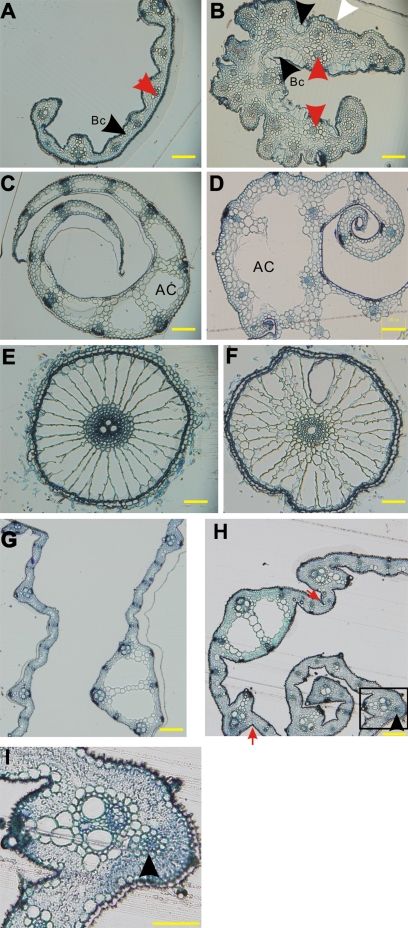
In addition, anatomical analyses of the leaves, sheaths, and roots were carried out. In the CVS transformants, the boundary of the leaf was relaxed, and the cells within were irregular and non-compact (Fig. 4B). Abaxial epidermal cells were exaggerated (Fig. 4B, white arrowhead). Bulliform cells, which are thin-walled cells specifically situated between two vascular bundles on the adaxial surface (black arrowhead with ‘Bc’ in Fig. 4A, B), were extraordinarily plump (Fig. 4B). Furthermore, there were bulliform-like cells on the abaxial surface (black arrowhead in Fig. 4B), suggesting an over-development of the adaxial characteristics. Vascular bundles in the wild type had already differentiated into phloem on the abaxial side and xylem on the adaxial side (Fig. 4A, red arrowhead), while those in the CVS transformants seemed to be radicalized, and increased in number (Fig. 4B), indicating abnormal vascular bundle development and polarity establishment. In addition, sheaths of the transformants also showed a rough boundary, undifferentiated and irregular vascular bundles, and irregular air cavities (Fig. 4D). Although the roots seemed normal in appearance, the vascular bundles within were devoid of sclerenchymatous cells (darker area in the central region), suggesting a reduction of the abaxial characters in the CVS tranformants (Fig. 4F).
Fig. 4.
Transverse sections of the leaves, sheaths, and roots of the transformants. (A) Half leaf of ZH11; (B) leaf of the CVS transformant, with a white arrowhead indicating the exaggerated epidermial cells, black arrowheads with ‘Bc’ in (A) and (B) indicating the bulliform cells, a black arrowhead indicating bulliform-like cells on the abaxial surface, and red arrowheads in (A) and (B) indicating vascular bundles. (C) ZH11 sheath; (D) sheath of the CVS transformant, with ‘AC’ in (C) and (D) indicating the air cavity. (E) ZH11 root; (F) root of the CVS transformants. (G) ZH11 leaves, left: half leaf blade without midrib, right: midrib with part of leaf blade. (H) Leaf of the non-CVS transformants with red arrows indicating bulliform-like cells on the abaxial surface, a black arrowhead indicating the ectopically formed vascular bundles. (I) Enlarged view of the rectangle in (H). Bars in (A)–(I) were 100 μm. ZH11 in (A), (C), (E) was at the five-leaf-stage, in (G) was at the booting stage.
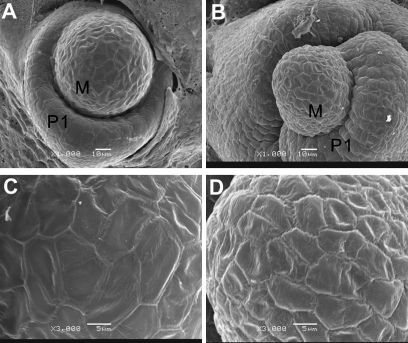
SAM is the centre for polarity establishment and plant development. SEM observation indicated that wild-type SAM at the five-leaf-stage was a regular orbicular tightly enwrapped by the newly formed leaf primordia (P1) (Fig. 5A). However, the CVS transformants displayed an elliptical SAM with detached leaf primordial (P1) (Fig. 5B). And wild-type SAM was smooth; while the transgenic SAM was coarse with a less-defined cell boundary (compare Fig. 5D with C). In addition, the non-CVS transformants also displayed abnormal polarity and vascular bundle development (Fig. 4H, I). These results indicated that over-expression of Osta-siR2141 resulted in over-development of the adaxial characteristics in polarity establishment.
Fig. 5.
SEM analyses of SAMs of the CVS transformants. (A) SAM of ZH11; (B) SAM of one CVS transformant; M in (A) and (B) indicate shoot apical meristem, P1 in (A) and (B) is the leaf primordia. (C) Enlargement of the P1 region in (A). (D) Enlargement of the P1 region in (B). ZH11 was at the five-leaf-stage.
Overexpression of Osta-siR2141 caused down-regulation of OsARF3s
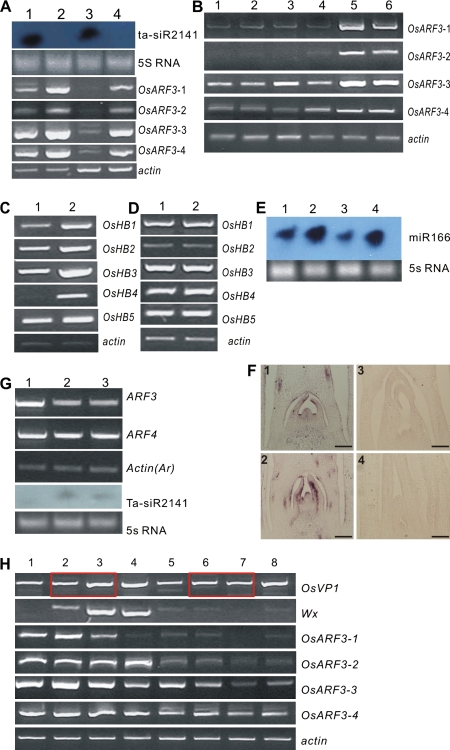
Molecular analysis revealed that, in the CVS transformants, tasiR-ARF was over-expressed in both the shoots and roots, and expression of all four OsARF3 genes was down-regulated simultaneously (Fig. 6A). In the non-CVS transformants, OsARF3s were also down-regulated, and no difference in degree was observed in the two kinds of transformants (Fig. 6B). Overexpression of Osta-siR2141 was closely associated with down-regulation of the OsARF3s, proving that OsARF3s were the functional targets of tasiR-ARF and, furthermore, the OsmiR-ARF(390) vector could work well to over-express siRNAs.
Fig. 6.
Molecular analyses of the rice and Arabidopsis transformants. (A) Osta-siR2141 Northern blot and RT-PCR analysis of the OsARF3s. (1, 2) leaves of the CVS transformants and ZH11, respectively; (3, 4) roots of the CVS transformants and ZH11, respectively; ZH11 was at the five-leaf-stage and the PCR was processed for 30 cycles for OsARF3s and 25 cycles for actin. (B) OsARF3s were down-regulated in leaves of the CVS and non-CVS transformants. (1, 2) Leaves of two respective CVS transformants, (3, 4) leaves of two respective non-CVS transformants, (5) leaves of the five-leaf-stage ZH11, (6) leaves of booting stage ZH11. Materials for PCR were sampled three times. PCRs were processed for 30 cycles for OsARF3s and 25 cycles for actin. (C) RT-PCR analysis of HD-ZIPIIIs in SAMs of the CVS transformants: (1) five-leaf-stage ZH11, (2) the CVS transformants. (D) RT-PCR analysis of HD-ZIPIIIs in IMs of the non-CVS transformants. (1) IMs of booting stage ZH11, (2) IMs of the non-CVS transformants; IMs were about 0.5 cm and materials for PCR were sampled three times. PCR was processed for 30 cycles for HD-ZIPIIIs and 25 cycles for actin. (E) Northern blot analysis of miR166 in SAMs/IMs of the transformants and anti-sense miR166a was used as probe. (1) SAMs of the CVS transformants, (2) SAMs of the five-leaf-stage ZH11, (3) IMs of the non-CVS transformants, (4) IMs of booting stage ZH11; IMs in (3) and (4) were about 0.5 cm. (F) In situ hybridization of the OsHB3 gene in SAMs of the CVS transformants. (1) ZH11, anti-sense probe; (2) the CVS transformants, anti-sense probe; (3) ZH11, sense probe; (4) the CVS transformants, sense probe. Bars in 1–4 were 100 μm. (G) Molecular analyses of the Arabidopsis transformants. (1) Seedling of Columbia; (2, 3) seedlings of two respective transformants; antisense Osta-siR2141 was used as the probe in the Northern blot; RT-PCR was processed for 36 cycles for ARF3 and ARF4, and for 24 cycles for actin (Ar). (H) Expression of Wx and OsVP1 genes in seeds of the non-CVS transformants. (1, 2, 3, 4) Seeds of ZH11 at about 1–2, 3–4, 5–6, >10 DAP, respectively; (5, 6, 7, 8) seeds of the non-CVS transformants at about 1–2, 3–4, 5–6, >10 DAP, respectively; Materials for PCRs were sampled three times. PCR was processed for 32 cycles for OsVP1, 29 cycles for Wx, 30 cycles for OsARF3s, and 25 cycles for actin.
HD-ZIPIII genes in SAMs of the CVS transformants were up-regulated
HD-ZIPIII genes have been proved to be pivotal in polarity establishment (Juarez et al., 2004). In rice, HD-ZIPIIIs contain five members, i.e. OsHB1 to OsHB5 (Zhong and Ye, 2004; Itoh et al., 2008). In the CVS transformants, HD-ZIPIII genes were greatly up-regulated (Fig. 6C) and miR166 down-regulated (Fig. 6E), while in the non-CVS transformants, expression of HD-ZIPIIIs showed no difference from that of the wild type (Fig. 6D) despite miR166 being similarly down-regulated (Fig. 6E). Furthermore, expression of OsHB3 was examined in the CVS transformants using in situ hybridization. In the wild type, OsHB3 was observed to be expressed mainly on the adaxial side of the leaf primordia, and the joint of the leaf primordia and the SAM (Fig. 6F1). However, in the CVS transformants, OsHB3 expression was greatly extended from the adaxial side to the abaxial region, and on the apex of the SAM (Fig. 6F2). These results indicated that the two kinds of transformants displayed over-expression of Osta-siR2141 and down-regulation of OsARF3s and miR166; while HD-ZIPIIIs were up-regulated in the CVS transformants but not in the non-CVS transformants.
The non-CVS transformants displayed arrested seed development
In the non-CVS transformants, most of the seeds developed until 10 DAP, but then began to shrink until death (Fig. 2H). Since starch synthesis is an important process during late development of the seed, and the Waxy (Wx) gene played a pivotal role in starch synthesis (Wang et al., 1990; Zhu et al., 2003), expression of the Wx gene was examined. In wild-type seeds, the Wx gene began to be expressed after 3 DAP and reached a climax at about 5 DAP which was maintained (Fig. 6H); but in the non-CVS transformants, expression of the Wx gene was hardly detectable at any DAPs (Fig. 6H), indicating the absence of starch synthesis. Similarly, four OsARF3s were down-regulated in the non-CVS lines (Fig. 6H). Expression of the OsVP1 gene, a B3 domain transcriptional factor important in seed development, was checked further (Hoecker et al., 1995; Fan et al., 2007). In the wild type, expression of the OsVP1 gene was low until 4 DAP and from 4–6 DAP it showed an obvious pyramiding; while in the non-CVS transformants, this pyramiding was not detected. Although OsVP1 expression could reach a similar level at 10 DAP, the timing was lost, so that abnormal OsVP1 gene expression might contribute to the failure in seed development, and if so, the timing of pyramiding at 4–6 DAP might be pivotal.
Osta-siR2141 overexpression in Arabidopsis revealed some function conservation
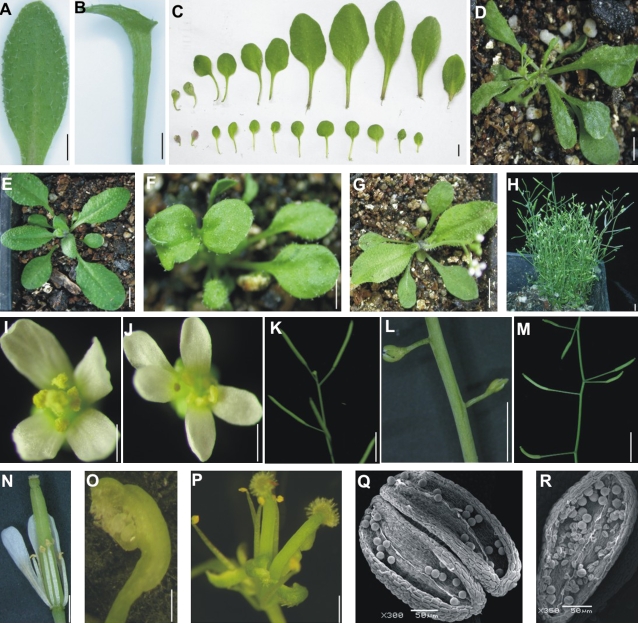
Alvarez et al. proved that synthesized miR-ARF could effectively targeting ARF2, ARF3 and ARF4 in Arabidopsis (Alvarez et al., 2006). In this study, OsmiR-ARF(390) was also transformed into Arabidopsis. The transformants developed various phenotypes, such as lotus leaves (Fig. 7B), indicating a possible polarity change. Judging from the leaf shape (Hunter et al., 2003), the transition from infancy to youth was seriously delayed (Fig. 7C), a process that was advanced in ago7 (Hunter et al., 2003), which functions in the biogenesis of tasiR-ARF (Adenot et al., 2006). Distortion appeared on some leaves (Fig. 7D), perhaps as a result of interference in vascular bundle development. Some petioles were fused (Fig. 7F). Some transformants were dwarf and clustered (Fig. 7H). Phyllotaxy in some transformants was opposite (Fig. 7G); the sequence of some siliques on the stalk was abnormal (Fig. 7L, M). In the reproductive stage, petals and the gynoecium were increased (Fig. 7J, P), the gynoecium swelled (Fig. 7O), and some pollen was infertile (Fig. 7L, R). Molecular analysis verified Osta-siR2141 over-expression and revealed the down-regulation of the ARF3 gene but not the ARF4 gene (Fig. 6G).
Fig. 7.
Phenotypes of the Arabidopsis transformants. (A) Leaf of Columbia. (B) Lotus leaf of the transformants. (C) Top and bottom: leaves of Columbia and one transformant arranged in a growth sequence, respectively. (D) Leaf curling and distortion in one transformant. (E) Columbia, showing leaf shape and phyllotaxy. (F) Fused-petiole in one transformant. (G) Opposite phyllotaxy in one transformant. (H) Transformant showing dwarf and cluster. (I) Flower of Columbia. (J) Five-petal flower in one transformant. (K) Silique in Columbia. (L) Infertile silique in one transformant. (M) Disordered silique of one transformant. (N) Gynoecium of Columbia. (O) Gynoecium in one transformant exaggerated. (P) Increased gynoecia in one transformant. (Q, R) SEM analysis of pollen in Columbia and one transformant, showing fertile pollen in (Q) and infertile pollen in (R). Bars in (A), (B), (C), (K), (L), (M), (N), (O), and (P) were 0.5 cm; in (D), (E), (F), (G), and (H) were 1 cm; in (I) and (J) were 1 mm. (This figure is available in colour at JXB online.)
Discussion
miRNA is a efficient vehicle for RNAi study
The functional knock-down of genes is the traditional way to study gene function and efficient methods were continually being developed to interfere with gene expression. Since its discovery, miRNA has been developed for the study of gene RNAi, mostly because the miRNA vector could produce siRNA more efficiently–specifically through its stem-loop structure. Since, in the process of ta-siRNA biogenesis, many unstable siRNAs were produced at the same time, the TAS gene itself was unsuitable for RNAi vector engineering; by contrast, the structure of pri-miRNA was stable. So, in this study, the miR390 backbone was used and successfully over-expressed Osta-siR2141 with four target genes being down-regulated simultaneously. The function of other ta-siRNAs or siRNAs could be studied through the miR390 backbone, and it is reasonable to suppose that other miRNAs could also be developed for use.
Conservation of tasiR-ARF in rice and Arabidopsis
Polarity is one of the basic events to be established in the leaf primordia, and the abaxial/adaxial polarity is of primary importance among the three polar axes (abaxial/adaxial, proximodistal, and mediolateral). Once established, abaxial/adaxial polarity is maintained throughout the development process to co-ordinate the proper growth and patterning of the leaf (McConnell and Barton, 1998). The sequence of tasiR-ARF has been found in various eudicots, monocots, and even gymnosperm (Axtell et al., 2006); in this study, overexpression of Osta-siR2141 in both rice and Arabidopsis disturbed vascular bundle development and polarity establishment, and retarded growth stage transition. The CVS transformants were kept at a young stage, and, in Arabidopsis transformants, the transition from juvenile to adult was seriously delayed. However, it seemed that rice was more sensitive to the level of tasiR-ARF during the vegetative stage, since most of the transformants could not develop into the reproductive stage; while most of the Arabidopsis transformants could. At the reproductive stage, the influence of tasiR-ARF on Arabidopsis was much more diverse, such as an increased number of floral organs (petals and gynoecium), infertile pollen, and the disorder of silique growth, etc (Fig. 7). In conclusion, although some divergence might occur during evolution, the function of tasiR-ARF was highly conserved between rice and Arabidopsis: tasiR-ARF and miR166/HD-ZIPIIIs function co-operatively in both dicotyledonous and monocotyledonous plants to co-ordinate polarity establishment.
The transition from vegetative to reproductive stage might be sensitive to the expressional level of HD-ZIPIII genes
The transition from vegetative to reproductive growth needs the co-ordination of many independently regulated processes to transform the SAMs from the indeterminate to the determinate state. In animals, homeobox genes control a vast array of developmental decisions and act as the molecular switch (Hayashi and Scott, 1990). In plants, HD-ZIPIII genes belong to a family of homeobox genes that has been proved to be a central regulator of crucial aspects of plant development, especially in leaf polarity and vascular development, SAM initiation, and embryo patterning (Nagasaki et al., 2007; Itoh et al., 2008). TasiR-ARF and miR165/166 negatively regulate ARF3 and HD-ZIPIII genes, respectively. In maize, tasiR-ARF and miR166 defined the opposing polarity of the abaxial/adaxial pattern (Nogueira et al., 2007). In rice, the possible relationship of tasiR-ARF/ARF3s and miR166/HD-ZIPIIIs had been discussed (Nagasaki et al., 2007). In this study, it was solidly proved that tasiR-ARF influenced leaf polarity establishment, vascular bundle and SAM development, and growth stage transition through co-operation with HD-ZIPIIIs. Furthermore, HD-ZIPIIIs were up-regulated in the CVS transformants but not in the non-CVS transformants, although miR166 was down-regulated similarly, so that HD-ZIPIII genes might act as a molecular switch in the transition from the vegetative to the reproductive stage in rice. If so, HD-ZIPIII genes should be modulated under a certain level and, if this level is breached, the transition could not be accomplished as in the CVS transformants. In the transition from the vegetative to the reproductive stage and during the reproductive growth, other important regulatory factors might be recruited to modulate HD-ZIPIIIs. A recent study showed that competitive inhibitors regulated HD-ZIPIII genes in SAM development in Arabidopsis. In mutant zpr3, no obvious abnormality could be observed during the vegetative stage, while in the reproductive stage, SAM was influenced and discrepancy occurred (Kim et al., 2008). This added a powerful possibility for our hypothesis.
Supplementary data
Supplementary data are available at JXB online.
Supplementary Fig. S1. Sketch map of plasmid OsmiR-ARF(390) construction.
Supplementary Table S1. Sequence of primers used in this study.
Supplementary Material
Acknowledgments
We would like to thank Professor Hai Huang for critical reading of the manuscript, and Professor Da Luo and Dr Jun Yang for instruction on in situ hybridization. This work was supported by grants from the National Natural Science Foundation of China (No. 30671113) the Ministry of Science and Technology of China (No. 2006AA10A102); the National Special Program on Research and Commercialization of Transgenic Plant (2008ZX08001-006); and the Scholarship Foundation from Shanghai Institutes for Biological Sciences (No. 2007KIP206).
Glossary
Abbreviations
- smRNA
small regulatory RNA
- ta-siRNA
trans-acting siRNA
- tasiR-ARF
TAS3 ta-siRNA
- CVS
ceased at vegetative stage
- non-CVS
not ceased at vegetative stage
- RISC
RNA-induced silencing complex
- dsRNA
double-stranded RNA
- HD-ZIPIII
class III homeodomain-leucine zipper gene
- ARF
auxin responsive factor
- ZH11
Japonica rice Zhonghua No. 11
- SEM
scanning electron microscope
- SAM
shoot apical meristem
- DAP
days after pollination
- MS
Murashige and Skoog medium
- SD
short day
References
- Adenot X, Elmayan T, Lauressergues D, Boutet S, Bouche N, Gasciolli V, Vaucheret H. DRB4-dependent TAS3 trans-acting siRNAs control leaf morphology through AGO7. Current Biology. 2006;16:927–932. doi: 10.1016/j.cub.2006.03.035. [DOI] [PubMed] [Google Scholar]
- Allen E, Xie Z, Gustafson AM, Carrington JC. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- Allshire R. Molecular biology. RNAi and heterochromatin: a hushed-up affair. Science. 2002;297:1818–1819. doi: 10.1126/science.1075874. [DOI] [PubMed] [Google Scholar]
- Alvarez JP, Pekker I, Goldshmidt A, Blum E, Amsellem Z, Eshed Y. Endogenous and synthetic microRNAs stimulate simultaneous, efficient, and localized regulation of multiple targets in diverse species. Plant Cell. 2006;18:1134–1151. doi: 10.1105/tpc.105.040725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin AA, Lagos-Quintana M, Yalcin A, Zavolan M, Marks D, Snyder B, Gaasterland T, Meyer J, Tuschl T. The small RNA profile during Drosophila melanogaster development. Developmental Cell. 2003;5:337–350. doi: 10.1016/s1534-5807(03)00228-4. [DOI] [PubMed] [Google Scholar]
- Axtell MJ, Jan C, Rajagopalan R, Bartel DP. A two-hit trigger for siRNA biogenesis in plants. Cell. 2006;127:565–577. doi: 10.1016/j.cell.2006.09.032. [DOI] [PubMed] [Google Scholar]
- Axtell MJ, Snyder JA, Bartel DP. Common functions for diverse small RNAs of land plants. The Plant Cell. 2007;19:1750–1769. doi: 10.1105/tpc.107.051706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel B. MicroRNAs directing siRNA biogenesis. Nature Structural Molecular Biology. 2005;12:569–571. doi: 10.1038/nsmb0705-569. [DOI] [PubMed] [Google Scholar]
- Borsani O, Zhu J, Verslues PE, Sunkar R, Zhu JK. Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell. 2005;123:1279–1291. doi: 10.1016/j.cell.2005.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington JC, Ambros V. Role of microRNAs in plant and animal development. Science. 2003;301:336–338. doi: 10.1126/science.1085242. [DOI] [PubMed] [Google Scholar]
- Chapman EJ, Carrington JC. Specialization and evolution of endogenous small RNA pathways. Nature Reviews Genetics. 2007;8:884–896. doi: 10.1038/nrg2179. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. The Plant Journal. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Coen ES, Romero JM, Doyle S, Elliott R, Murphy G, Carpenter R. floricaula: a homeotic gene required for flower development in Antirrhinum majus. Cell. 1990;63:1311–1322. doi: 10.1016/0092-8674(90)90426-f. [DOI] [PubMed] [Google Scholar]
- Fahlgren N, Montgomery TA, Howell MD, Allen E, Dvorak SK, Alexander AL, Carrington JC. Regulation of AUXIN RESPONSE FACTOR3 by TAS3 ta-siRNA affects developmental timing and patterning in Arabidopsis. Current Biology. 2006;16:939–944. doi: 10.1016/j.cub.2006.03.065. [DOI] [PubMed] [Google Scholar]
- Fan J, Niu X, Wang Y, Ren G, Zhuo T, Yang Y, Lu BR, Liu Y. Short, direct repeats (SDRs)-mediated post-transcriptional processing of a transcription factor gene OsVP1 in rice (Oryza sativa) Journal of Experimental Botany. 2007;58:3811–3817. doi: 10.1093/jxb/erm231. [DOI] [PubMed] [Google Scholar]
- Garcia D, Collier SA, Byrne ME, Martienssen RA. Specification of leaf polarity in Arabidopsis via the trans-acting siRNA pathway. Current Biology. 2006;16:933–938. doi: 10.1016/j.cub.2006.03.064. [DOI] [PubMed] [Google Scholar]
- Hamilton A, Voinnet O, Chappell L, Baulcombe D. Two classes of short interfering RNA in RNA silencing. EMBO Journal. 2002;21:4671–4679. doi: 10.1093/emboj/cdf464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton AJ, Baulcombe DC. A species of small antisense RNA in posttranscriptional gene silencing in plants. Science. 1999;286:950–952. doi: 10.1126/science.286.5441.950. [DOI] [PubMed] [Google Scholar]
- Hayashi S, Scott MP. What determines the specificity of action of Drosophila homeodomain proteins? Cell. 1990;63:883–894. doi: 10.1016/0092-8674(90)90492-w. [DOI] [PubMed] [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. The Plant Journal. 1994;6:271–282. doi: 10.1046/j.1365-313x.1994.6020271.x. [DOI] [PubMed] [Google Scholar]
- Hoecker U, Vasil IK, McCarty DR. Integrated control of seed maturation and germination programs by activator and repressor functions of Viviparous-1 of maize. Genes and Development. 1995;9:2459–2469. doi: 10.1101/gad.9.20.2459. [DOI] [PubMed] [Google Scholar]
- Howell MD, Fahlgren N, Chapman EJ, Cumbie JS, Sullivan CM, Givan SA, Kasschau KD, Carrington JC. Genome-wide analysis of the RNA-DEPENDENT RNA POLYMERASE6/DICER-LIKE4 pathway in Arabidopsis reveals dependency on miRNA- and tasiRNA-directed targeting. The Plant Cell. 2007;19:926–942. doi: 10.1105/tpc.107.050062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter C, Sun H, Poethig RS. The Arabidopsis heterochronic gene ZIPPY is an ARGONAUTE family member. Current Biology. 2003;13:1734–1739. doi: 10.1016/j.cub.2003.09.004. [DOI] [PubMed] [Google Scholar]
- Itoh J, Hibara K, Sato Y, Nagato Y. Developmental role and auxin responsiveness of Class III homeodomain leucine zipper gene family members in rice. Plant Physiology. 2008;147:1960–1975. doi: 10.1104/pp.108.118679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juarez MT, Kui JS, Thomas J, Heller BA, Timmermans MC. microRNA-mediated repression of rolled leaf1 specifies maize leaf polarity. Nature. 2004;428:84–88. doi: 10.1038/nature02363. [DOI] [PubMed] [Google Scholar]
- Kasschau KD, Fahlgren N, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Carrington JC. Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS Biology. 2007;5 doi: 10.1371/journal.pbio.0050057. e57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katiyar-Agarwal S, Gao S, Vivian-Smith A, Jin H. A novel class of bacteria-induced small RNAs in Arabidopsis. Genes and Development. 2007;21:3123–3134. doi: 10.1101/gad.1595107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katiyar-Agarwal S, Morgan R, Dahlbeck D, Borsani O, Villegas A, Jr, Zhu JK, Staskawicz BJ, Jin H. A pathogen-inducible endogenous siRNA in plant immunity. Proceedings of the National Academy of Sciences, USA. 2006;103:18002–18007. doi: 10.1073/pnas.0608258103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidner CA, Martienssen RA. The developmental role of microRNA in plants. Current Opinion in Plant Biology. 2005;8:38–44. doi: 10.1016/j.pbi.2004.11.008. [DOI] [PubMed] [Google Scholar]
- Kim YS, Kim SG, Lee M, et al. HD-ZIP III activity is modulated by competitive inhibitors via a feedback loop in Arabidopsis shoot apical meristem development. The Plant Cell. 2008;20:920–933. doi: 10.1105/tpc.107.057448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippman Z, Martienssen R. The role of RNA interference in heterochromatic silencing. Nature. 2004;431:364–370. doi: 10.1038/nature02875. [DOI] [PubMed] [Google Scholar]
- Liu J. Control of protein synthesis and mRNA degradation by microRNAs. Current Opinion in Cell Biology. 2008;20:214–221. doi: 10.1016/j.ceb.2008.01.006. [DOI] [PubMed] [Google Scholar]
- McConnell JR, Barton MK. Leaf polarity and meristem formation in Arabidopsis. Development. 1998;125:2935–2942. doi: 10.1242/dev.125.15.2935. [DOI] [PubMed] [Google Scholar]
- Mochizuki K, Gorovsky MA. Conjugation-specific small RNAs in Tetrahymena have predicted properties of scan (scn) RNAs involved in genome rearrangement. Genes and Development. 2004;18:2068–2073. doi: 10.1101/gad.1219904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnar A, Schwach F, Studholme DJ, Thuenemann EC, Baulcombe DC. miRNAs control gene expression in the single-cell alga Chlamydomonas reinhardtii. Nature. 2007;447:1126–1129. doi: 10.1038/nature05903. [DOI] [PubMed] [Google Scholar]
- Montgomery TA, Howell MD, Cuperus JT, Li D, Hansen JE, Alexander AL, Chapman EJ, Fahlgren N, Allen E, Carrington JC. Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell. 2008;133:128–141. doi: 10.1016/j.cell.2008.02.033. [DOI] [PubMed] [Google Scholar]
- Mosher RA, Schwach F, Studholme D, Baulcombe DC. PolIVb influences RNA-directed DNA methylation independently of its role in siRNA biogenesis. Proceedings of the National Academy of Sciences, USA. 2008;105:3145–3150. doi: 10.1073/pnas.0709632105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagasaki H, Itoh J, Hayashi K, et al. The small interfering RNA production pathway is required for shoot meristem initiation in rice. Proceedings of the National Academy of Sciences, USA. 2007;104:14867–14871. doi: 10.1073/pnas.0704339104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nogueira FT, Madi S, Chitwood DH, Juarez MT, Timmermans MC. Two small regulatory RNAs establish opposing fates of a developmental axis. Genes and Development. 2007;21:750–755. doi: 10.1101/gad.1528607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peragine A, Yoshikawa M, Wu G, Albrecht HL, Poethig RS. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes and Development. 2004;18:2368–2379. doi: 10.1101/gad.1231804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes and Development. 2006;20:3407–3425. doi: 10.1101/gad.1476406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D. Highly specific gene silencing by artificial microRNAs in Arabidopsis. The Plant Cell. 2006;18:1121–1133. doi: 10.1105/tpc.105.039834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tretter EM, Alvarez JP, Eshed Y, Bowman JL. Activity range of Arabidopsis small RNAs derived from different biogenesis pathways. Plant Physiol. 2008;147:58–62. doi: 10.1104/pp.108.117119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret H. Post-transcriptional small RNA pathways in plants: mechanisms and regulations. Genes and Development. 2006;20:759–771. doi: 10.1101/gad.1410506. [DOI] [PubMed] [Google Scholar]
- Vazquez F, Vaucheret H, Rajagopalan R, Lepers C, Gasciolli V, Mallory AC, Hilbert JL, Bartel DP, Crete P. Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Molecular Cell. 2004;16:69–79. doi: 10.1016/j.molcel.2004.09.028. [DOI] [PubMed] [Google Scholar]
- Wang ZY, Wu ZL, Xing YY, Zheng FG, Guo XL, Zhang WG, Hong MM. Nucleotide sequence of rice waxy gene. Nucleic Acids Research. 1990;18:5898. doi: 10.1093/nar/18.19.5898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Qi X. Diverse small RNA-directed silencing pathways in plants. Biochimica et Biophysica Acta. 2008;1779:720–724. doi: 10.1016/j.bbagrm.2008.02.009. [DOI] [PubMed] [Google Scholar]
- Yoshikawa M, Peragine A, Park MY, Poethig RS. A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes and Development. 2005;19:2164–2175. doi: 10.1101/gad.1352605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zamore PD, Haley B. Ribo-gnome: the big world of small RNAs. Science. 2005;309:1519–1524. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]
- Zhao T, Li G, Mi S, Li S, Hannon GJ, Wang XJ, Qi Y. A complex system of small RNAs in the unicellular green alga Chlamydomonas reinhardtii. Genes and Development. 2007;21:1190–1203. doi: 10.1101/gad.1543507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong R, Ye ZH. Amphivasal vascular bundle 1, a gain-of-function mutation of the IFL1/REV gene, is associated with alterations in the polarity of leaves, stems and carpels. Plant and Cell Physiology. 2004;45:369–385. doi: 10.1093/pcp/pch051. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Cai XL, Wang ZY, Hong MM. An interaction between a MYC protein and an EREBP protein is involved in transcriptional regulation of the rice Wx gene. Journal of Biological Chemistry. 2003;278:47803–47811. doi: 10.1074/jbc.M302806200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.