Abstract
Nedd4 E3 ligases are members of the HECT E3 ubiquitin ligase family and regulate ubiquitination-mediated protein degradation. In this report, we demonstrate that calcium releases the C2 domain-mediated auto-inhibition in both Nedd4-1 and Nedd4-2. Calcium disrupts binding of the C2 domain to the HECT domain. Consistent with this, calcium activates the E3 ubiquitin ligase activity of Nedd4. Elevation of intracellular calcium by ionomycin treatment, or activation of acetylcholine receptor or epidermal growth factor receptor by carbachol or epidermal growth factor stimulation induced activation of endogenous Nedd4 in vivo evaluated by assays of either Nedd4 E3 ligase activity or ubiquitination of Nedd4 substrate ENaC-β. The activation effect of calcium on Nedd4 E3 ligase activity was dramatically enhanced by a membrane-rich fraction, suggesting that calcium-mediated membrane translocation through the C2 domain might be an activation mechanism of Nedd4 in vivo. Our studies have revealed an activation mechanism of Nedd4 E3 ubiquitin ligases and established a connection of intracellular calcium signaling to regulation of protein ubiquitination.
Keywords: Calcium, E3 Ubiquitin Ligase, Enzyme Mechanisms, Phospholipid, Signal Transduction, Ubiquitination
Introduction
Protein ubiquitination is a major intracellular signaling event. E3 ubiquitin ligase (E3),3 including the HECT (homologous to E6-AP carboxyl terminus) domain containing and the RING (the really interesting new gene) domain containing E3 ligases, is the key enzyme that catalyzes ubiquitination and confers specificity of ubiquitination substrates (1–3). Nedd4 E3 ubiquitin ligases are members of the WW domain-containing HECT E3 ubiquitin ligase subfamily (4). There are two Nedd4 E3 ligases, Nedd4-1 and Nedd4-2, in mammalian cells (5). Human Nedd4-1 gene (Nedd4) is localized on chromosome 15, and Nedd4-2 gene (Nedd4L) is on chromosome 18 (5). Both Nedd4-1 and Nedd4-2 have the same domain structure, with the C2 domain at the N terminus, followed by four WW domains, and the HECT domain at the C terminus. The primary peptide sequences of human Nedd4-1 and Nedd4-2 are ∼65% identical. The most unconserved regions are located between the WW1 and the WW3 domains.
Four groups of Nedd4 substrates have been identified: 1) ion channels and membrane transporters; 2) membrane receptors; 3) tumor suppressors; and 4) endocytic regulation proteins. Accordingly, Nedd4 is involved in regulation of hypertension (6), neuronal signal transmission (7, 8), tumorigenesis and cell growth (9), cellular metabolism (10), receptor endocytosis and degradation (11), and viral endocytosis and budding (12, 13). Although Nedd4-1 and Nedd4-2 show partially redundant cellular functions (14), there seems to be a distinct preference for substrates between Nedd4-1 and Nedd4-2. For example, Nedd4-1 preferentially ubiquitinates tumor suppressors and endocytic proteins (15–18), whereas Nedd4-2 ubiquitinates ion channels and membrane transporters (19–23). It is not known how substrate preferences of Nedd4-1 and Nedd4-2 are determined. In addition, both Nedd4-1 and Nedd4-2 have multiple cellular substrates. How Nedd4 discriminates substrates in response to cellular signaling to execute corresponding cellular function is entirely unknown.
Regulation of Nedd4 activity in cells is poorly understood. Phosphorylation of Nedd4-2 by serum- and glucocorticoid-inducible kinase 1 (SGK1) is an only major finding in regulation of Nedd4-2 activity up to date (24). SGK1 was identified as a kinase causing hypertension through activation of epithelial sodium channel (ENaC) (25, 26). SGK1 phosphorylates Nedd4-2 on serine 444 (27), which is required for association of Nedd4-2 with 14-3-3, resulting in reduced Nedd4-mediated ubiquitination of ENaC and enhanced ENaC activity (26). In addition to ENaC, SGK1 is also involved in regulation of glucose transporter SGLT1, glutamate transporter EAAT1, glutamine transporter SN1, and cardiac sodium channel SCN5A through phosphorylation and inhibition of Nedd4-2 (28–31). Interestingly, SGK1 is activated by a serine/threonine kinase, WNK1 (with no lysine (K) kinase 1), which is a known causal molecule for pseudohypoaldosteronism type II hypertension (32). WNK1 activates SGK1, inhibiting Nedd4-2 and enhancing ENaC activity, which results in hypertension (33). Phosphorylation of Nedd4-2 by AMP-activated kinase, which causes inhibition of ENaC activity, was also observed (34). Phosphorylation of Nedd4 by AMP-activated kinase may be an important means for regulation of the E3 ligase activity or recognition of the substrate. A recent research report shows that Nedd4 is phosphorylated by G-protein-coupled receptor kinase 2 (35). The effect of phosphorylation by G-protein-coupled receptor kinase 2 on Nedd4 activity is not known.
The C2 domain was originally identified as a calcium-binding domain in protein kinase C, which is required for calcium-dependent activation of the kinase activity (36, 37). More than 100 proteins are now known to contain the C2 domain (38). The C2 domain of Nedd4 has been proposed as a calcium-dependent inositol phospholipid-binding module (39, 40). Early studies indicated that the C2 domain mediated the translocation of cytoplasmic Nedd4 to plasma membrane by binding to phospholipids (39). Subsequently, Annexin XIIIb was identified as a partner for the C2 domain of Nedd4 to bind to apical plasma membranes in epithelial cells (41). In yeast, the C2 domain of Rsp5, the homolog of Nedd4, is required for functional compartmentalization of Rsp5 and ubiquitination of its substrate (40). In mammalian cells the WW domains, but not the C2 domain, are required for Nedd4 to down-regulate ENaC (42). It was observed that the C2 domain truncation mutant of Nedd4 had stronger inhibition of ENaC activity than wild-type Nedd4 (42), suggesting that the C2 domain has an inhibitory role in Nedd4 function. A recent study has shown that the C2 domain of Smurf2 directly binds to the HECT domain and functions as an auto-inhibitory domain for E3 ligase activity (43).
In this report, we confirmed that the C2 domain of Nedd4 is the auto-inhibitory domain for its E3 ubiquitin ligase activity. Deletion of the C2 domain causes constitutive activation of Nedd4. Calcium activates Nedd4 E3 ubiquitin ligase activity both in vitro and in vivo by releasing the auto-inhibition. Signaling that elevates intracellular calcium activates Nedd4 E3 ubiquitin ligase activity. Thus, we propose that release of the C2 domain-mediated auto-inhibition by calcium ions or other cellular activators is a key mechanism for activation of Nedd4 E3 ubiquitin ligases in response to cellular signaling.
EXPERIMENTAL PROCEDURES
Materials
The human Nedd4-2b cDNA (IMAGE: 5528964) and partial human Nedd4-2a cDNA (IMAGE: 3604024) plasmids were purchased from Open Biosystems. Rat Nedd4-1 plasmid is a kind gift from Dr. Daniela Rotin at University of Toronto. Anti-ubiquitin antibody was purchased from Covance; anti-Nedd4 was purchased from UBI. Ubiquitin (U6253–5MG), ubiquitin activating enzyme (E1, UL758–25UG), UbcH7 (E2, U9132–100UG), and carbachol (C4382) were purchased from Sigma.
Construction of Tagged Nedd4s and the Mutants
pcDNA3 HA-human Nedd4-2b and pcDNA3 HA-rat Nedd4-1 were constructed by subcloning human Nedd4-2b and rat Nedd4-1 cDNAs into an HA-tagged pcDNA3 vector. The pcDNA3-HA-human Nedd4-2a was constructed by ligation of the partial human Nedd4-2a cDNA (IMAGE: 3604024) with the 5′-end of human Nedd4-2b cDNA. pcDNA3-HA-hNedd4-2aΔC2, pcDNA3-HA-hNedd4-2bΔC2, pcDNA3-HA-hNedd4-2bΔN, pcDNA3 HA-Nedd4-2bWW, pcDNA3 HA-rNedd4-1ΔC2, and pGEX-4T3-hNedd4-2b-C2 were constructed by PCR-directed mutagenesis. All the constructs were confirmed by nucleotide sequencing.
Culture and Transfection of Cells
HEK293T cells were maintained in Dulbecco's modified Eagle's medium plus 10% fetal bovine serum at 37 °C and 5% CO2. The cells were cultured to 90% confluence before transfection. For transfection, the plasmid DNA (1–2 μg/60 mm dish) was pre-mixed with ESCORTTM V transfection Reagent (Sigma, 1:4, w/v) in ESCORTTM V transfection reagent buffer (0.2 ml per 60-mm dish) and incubated at room temperature for 30 min. The cells were incubated in the plasmid DNA/ESCORTTM V mixture for 5 h. Then the mixture was replaced with the complete culture medium, and the cells were maintained under normal culture conditions up to 43 h before used for experiments.
Establishment of ENaC-β-GFP Stable Cell Line
The plasmids pEGFP-N1-ENaC-β and pMT-puro were transfected into HEK293T cells using Lipofectamine 2000 (Invitrogen). After 48 h, transfected cells were treated with 2 μg/ml puromycin to select transfectant colonies for 2 weeks. The positive colonies were determined by visualizing GFP fluorescence under a fluorescent microscope and immunoblotting the cell lysates with anti-GFP.
Immunoprecipitation and Immunoblotting
The cells were rinsed with phosphate-buffered saline once, then either lysed with the mammalian cell lysis buffer (40 mm Hepes, pH 7.4, 100 mm NaCl, 1% Triton X-100, 25 mm β-glycerophosphate, 1 mm sodium ortho-vanadate, 10 μg/ml leupeptin, and 10 μg/ml aprotinin) with rocking at 4 °C for 20 min, or homogenized in ubiquitin ligation reaction (ULR) buffer (25 mm Tris-HCl, pH 8.0, 100 mm NaCl, 2.0 mm MgCl2, and 1 mm dithiothreitol) using a 1-ml Kontes tissue grinder. The cell lysates or homogenates were collected into an Eppendorf tube and cleared by centrifugation at 14,000 rpm (20,000 × g) for 10 min in a microcentrifuge. The cleared lysates were transferred to a clean Eppendorf tube and incubated with primary antibody on ice for 30 min, then protein A or protein G beads were added, and the mixture was incubated at 4 °C for 2 h with rotation. The beads were washed either with lysis buffer two times followed by ULR buffer three times (for immunoprecipitation from cell lysates) or just with ULR buffer three times (for immunoprecipitation from homogenates). The immunoprecipitated Nedd4 was used for the E3 ubiquitin ligase activity assay. The immunoblot was performed as instructed by ECL immunoblot kits (Amersham Biosciences).
Preparation of Homogenate Fractions
For preparation of homogenate fractions, we cultured HEK293 cells to 90–100% confluence in 10-cm dishes, harvested cells by scraping the cells in 1 ml of pre-cooled ULR buffer for each dish, and homogenized the cells with a 2-ml homogenizer on ice. The cell debris, nuclei, and mitochondria were removed by centrifugation of the homogenates at 1,000 rpm (50 × g) for 15 min at 4 °C. The supernatant (50 × g S) was then spun at 3,000 × g for 15 min at 4 °C. The yielded supernatant and pellet were designated as 3Kxg S and 3Kxg P, respectively. The 3Kxg S was further spun at 400,000 × g for 30 min at 4 °C, resulting in a supernatant (400Kxg S) and pellet (400Kxg P). All the pellets were resuspended in equal volume of ULR buffer to the supernatant. In activation of Nedd4 E3 ubiquitin ligase assays, 5 μl of each fraction was added into the reaction mixture.
The in Vitro E3 Ubiquitin Ligase Activity Assay
The E3 ubiquitin ligase activity assay was based on the method described in a previous study (44) with minor modifications. The E3 ubiquitin ligase activity assay was performed by adding 15 μl of reaction mixture containing a final concentration of 100 nm E1, 0.5 μm UbcH7 (E2), 5 μm ubiquitin or K0-ubiquitin, and 2 μm ATP in ULR buffer (25 mm Tris-HCl, pH 8.0, 100 mm NaCl, 2.0 mm MgCl2, and 1 mm dithiothreitol) to immunoprecipitated Nedd4 (10 μl). For an activation assay of the E3 ubiquitin ligase activity, CaCl2 solution or the HEK293 cell homogenate fraction (5 μl/each sample) was added to the immunoprecipitated Nedd4 before addition of the reaction mixture. CaCl2 solution was buffered with EGTA, and the concentration of calcium was calculated according to WEBMAXCLITE. The reaction was initiated by adding the reaction mixture, carried out at 22 °C for 20 min, and stopped by addition of 25 μl of 2× SDS-PAGE sample buffer. The reaction mixtures were resolved by SDS-PAGE, transferred to polyvinylidene difluoride membrane, and subjected to immunoblot. The poly-ubiquitin products were visualized by ECL immunoblotting with an anti-ubiquitin antibody. The X-films yielded from ECL immunoblotting were scanned, and the arbitrary density counts of both poly-ubiquitin and Nedd4 bands were quantified by Kodak EDAS290 image system. The E3 ubiquitin ligase specific activity is defined as the ratio of density of poly-ubiquitin products to Nedd4.
ENaC-β in Vivo Ubiquitination Assay
For ionomycin treatment: pcDNA3-HA-Nedd4-2a, pcDNA3-HA-Nedd4-2a-ΔC2, or vector was transfected into the HEK293T-ENaβ-GFP cell line for 36 h followed by 12-h serum starvation. The medium was replaced with normal cell culture medium plus 1.25 mm CaCl2 and 10 μm MG-132 with or without 1 μm ionomycin for 6 h. Ubiquitinated ENaC-β-GFP was precipitated with a GST-ACK-Uba pulldown assay and detected by immunoblotting with anti-GFP antibody. For carbachol or EGF treatment, ENaβ-GFP stably transfected cells were serum-starved for 12 h followed by treatment with MG-132 (10 μm) for 6 h. The cells then stimulated with carbachol (5 μm) or EGF (50 ng/ml) at the indicated time. Ubiquitinated ENaC-β was precipitated by a GST-ACK1-Uba pulldown assay and detected by immunoblotting with anti-GFP antibody.
Expression and Purification of His-T7-tagged Nedd4 Proteins
His-T7-tagged E3 ligase Nedd4-2a, Nedd4-2a-ΔC2, Nedd4-2a C922A, and Nedd4-2a-HECT were expressed in Escherichia coli BL21 cell by isopropyl 1-thio-β-d-galactopyranoside induction. The cells were lysed by sonication in His lysis buffer (25 mm Tris-HCl (pH 8.0), 50 mm NaCl, 0.5% Triton X-100, 10% glycerol, 10 mm imidazole). The soluble fraction of the lysate was incubated with nickel-nitrilotriacetic acid-agarose beads at 4 °C for 2 h. The nickel beads were washed three times with lysis buffer containing 60 mm imidazole. The bound fractions were eluted with the same buffer containing 250 mm imidazole.
Expression and Purification of GST-Nedd4-2b C2 Domain and the GST-Nedd4-2b C2 Domain Pulldown Assay
Expression and purification of the GST-Nedd4-2b C2 domain were performed as described previously (45). The GST-Nedd4-2b C2 domain was bound to glutathione-conjugated agarose beads, resuspended in the bacteria lysis buffer (40 mm Tris/HCl, pH 8.0, 100 mm NaCl, 0.5% Triton X-100, 1 mm EDTA, 1 mm EGTA, and 10 μg/ml aprotinin and leupeptin) and ready for pulldown assays.
For GST-Nedd4-2b C2 domain pulldown assay, the GST-Nedd4-2b C2 domain-bound beads were washed two times with the mammalian cell lysis buffer and incubated with the purified His-T7-Nedd4-2a-HECT protein, or HA-Nedd4-2bΔN- and HA-Nedd4-2b-WW-expressed mammalian cell lysates with either 1 mm EGTA or the indicated concentrations of EGTA-buffered free calcium (calculated by WEBMAXCLITE) at 4 °C for 2 h with rotation. The beads were washed three times with the mammalian cell lysis buffer plus either 1 mm EGTA or the indicated concentrations of EGTA-buffered free calcium (calculated by WEBMAXCLITE), and resuspended in 2× SDS-PAGE sample buffer for SDS-PAGE. The proteins precipitated by the GST fusion protein were immunoblotted with either anti-T7 or anti-HA antibody.
RESULTS
The C2 Domain of Nedd4 Is an Auto-inhibitory Domain for E3 Ubiquitin Ligase Activity
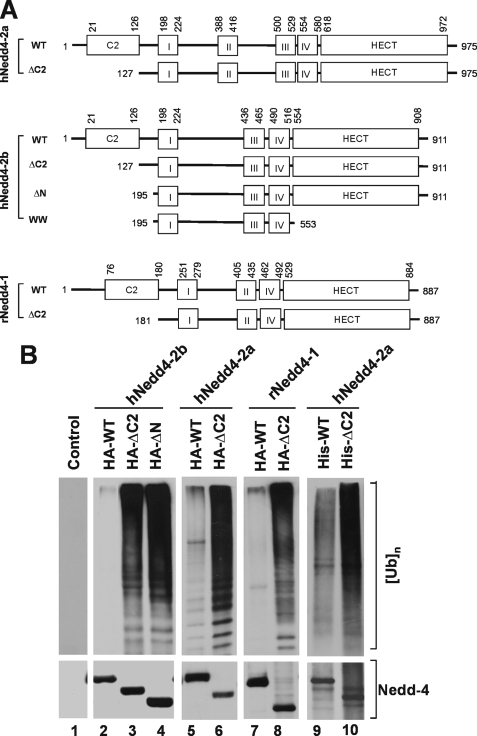
To search for the activation mechanism of the E3 ubiquitin ligase Nedd4, a series of truncation mutants of human Nedd4-2b (also Nedd4L) were constructed. These included the C2 domain truncation mutant ΔC2 and the N terminus truncation mutant ΔN (Fig. 1A). The mutants were HA-tagged, expressed in HEK293 cells by transient transfection, and immunoprecipitated for determination of E3 ubiquitin ligase activity. The activity was measured by an in vitro ubiquitin ligase activity assay (see “Experimental Procedures”), in which the amount of polyubiquitin formed from mono-ubiquitin (substrate) is used to evaluate the ligase activity (top panels, Fig. 1B). To control auto-ubiquitination of Nedd4 in the assay, the ubiquitination reaction was taken for 20 min at 22 °C. Within this reaction time, we observed little auto-ubiquitination of Nedd4 (data not shown). As shown in lanes 1–4 of Fig. 1B, truncation of the C2 domain enhanced the ligase activity of Nedd4-2b by 30-fold, suggesting that the C2 domain inhibits the E3 ubiquitin ligase activity. Nedd4-2b is an alternative splicing form of Nedd4-2, in which the WW2 domain is missing (Fig. 1A). To confirm that the C2 domain in full-length Nedd4-2 or Nedd4-1 has the same effect, the C2 domain truncation mutants of human Nedd4-2a (full-length) and rat Nedd4-1 were constructed (Fig. 1A). Similar to Nedd4-2b, truncation of the C2 domain of Nedd4-1 or Nedd4-2a markedly activated the E3 ubiquitin ligase (lanes 5–8, Fig. 1B). To exclude the possibility that increased activity in the C2 domain truncation mutants resulted from other cellular proteins co-immunoprecipitated with Nedd4 from HEK293 cell lysates, we used purified his-tagged Nedd4-2a expressed in bacteria to perform the in vitro E3 ubiquitin ligase activity assay. As shown in lanes 9 and 10 in Fig. 1B, the E3 ubiquitin ligase activity of the C2 domain truncation mutant was significantly higher than wild type Nedd4-2a, indicating that the activation is directly attributable to deletion of the C2 domain. These data demonstrate that the C2 domain is auto-inhibitory for Nedd4 E3 ubiquitin ligases, consistent with auto-inhibitory function of the C2 domain in Smurf2 (43).
FIGURE 1.
Truncation of the C2 domain activates the E3 ubiquitin ligase activity of Nedd4. A, schematic representation of the C2 domain deletion mutants of Nedd4-1 and Nedd4-2. The roman numbers I–IV refer to the WW domains 1 (WW1), 2 (WW2), 3 (WW3), and 4 (WW4). Human Nedd4-2a and Nedd4-2b (hNedd4-2a and hNedd4-2b) are Nedd4-2 splicing forms. The Nedd4-2b lacks the WW2 domain in Nedd4-2a. Rat Nedd4-1 (rNedd4-1) has only three WW domains. The positions for the deletions and the boundaries of the domains are labeled with the amino acid residue position numbers in the peptides. B, truncation of the C2 domain activates Nedd4. Lanes 1–8, HA-tagged wild-type or the C2 domain deletion mutant of Nedd4-1 and Nedd4-2 was transfected into HEK293 cells. Nedd4 protein was immunoprecipitated with anti-HA antibody and used for the E3 ubiquitin ligase activity assay. Lanes 9 and 10, His-tagged Nedd4-2a or its C2 domain deletion mutant was expressed in E. coli and purified with affinity beads. Purified His-tagged Nedd4 and the mutant (100 ng) were used for the ubiquitin ligase activity assay. In the assay, mono-ubiquitin was used as the substrate. The ubiquitin ligation products (poly-ubiquitins) were detected by immunoblotting with anti-ubiquitin antibody. Top panels, poly-ubiquitin products; bottom panels, HA- or his-tagged wild-type and deletion mutants of Nedd4s.
Calcium Dissociates Binding of the C2 Domain to the HECT Domain
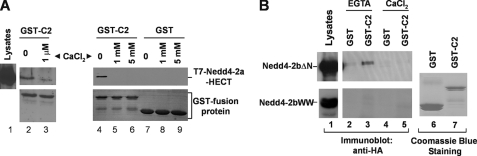
It has been demonstrated that the C2 domain directly interacts with the HECT domain in Smurf2 (43). Given that the C2 domain is a known calcium-binding domain (36, 37), we hypothesized that calcium regulates binding of the C2 domain to the HECT domain in Nedd4 and activates E3 ligase activity of Nedd4. To test this hypothesis, we constructed GST-Nedd4-2-C2 domain fusion protein and performed GST fusion protein pulldown assays with purified T7-tagged Nedd4-2 HECT domain or HEK293 cell-expressed Nedd4-2bΔN in the presence or absence of calcium. As shown in Fig. 2A, the GST-C2 domain precipitated the HECT domain in the absence of calcium (lane 1, top panel), but little in the presence of calcium, even at 1 μm of calcium (lanes 2–6, top panel), indicating that calcium disrupts association of the C2 domain with the HECT domain. This was confirmed by GST-Nedd4-2b C2 domain pulldown assays with HEK293 cell lysates expressing Nedd4-2bΔN, in which the C2 domain is deleted. In the absence of calcium (i.e. EGTA), the GST-C2 domain precipitated HA-tagged Nedd4-2bΔN from the cell lysates (lane 3, upper panel, Fig. 2B). Addition of calcium eliminated binding of the C2 domain to HA-Nedd4-2bΔN (lane 4, upper panel). As a control, we also performed the GST-C2 domain pulldown assay with HA-tagged Nedd4-2bWW that was generated by deleting the HECT domain in Nedd4-2bΔN. No binding of the C2 domain to Nedd4-2bWW was observed (lanes 1–5, bottom panel, Fig. 2B), demonstrating that GST-C2 domain co-precipitation of Nedd4-2bΔN required the HECT domain. These data indicate that calcium dissociates binding of the C2 domain to the HECT domain. Given that the binding of the C2 domain to the HECT domain inhibits the E3 ubiquitin ligase, calcium may activate the E3 ubiquitin ligase activity of Nedd4 by releasing the C2 domain-mediated inhibition of the E3 ubiquitin ligase activity.
FIGURE 2.
The C2 domain of Nedd4 interacts with its HECT domain in the absence of calcium. GST-C2 domain pulldown assay for binding of the C2 domain to purified T7-tagged HECT domain of Nedd4-2a (A), or HA-tagged Nedd4-2bΔN (B). The C2 domain of Nedd4-2 was subcloned into a GST fusion protein expression vector, expressed in E. coli, and purified by affinity purification with glutathione-conjugated agarose beads. The beads with GST (control) or GST-Nedd4-2b C2 domain fusion protein were incubated with purified T7-tagged HECT domain of Nedd4-2a in indicated calcium concentrations, or with HA-Nedd4-2bΔN- or HA-Nedd4-2bWW-expressed HEK293 cell lysates in the presence of 5 mm CaCl2 or 1 mm EGTA. The GST-C2 domain-precipitated T7-Nedd4-2a-HECT, HA-Nedd4-2bΔN, or HA-Nedd4-2bWW was detected by immunoblotting with anti-T7 antibody or anti-HA antibody. The GST and GST fusion proteins used for the pulldown assay are shown by Coomassie Blue staining. Lane 1 in A and B shows one-tenth the total input of the indicated proteins in the pulldown assay.
Calcium Activates Nedd4 E3 Ubiquitin Ligase Activity
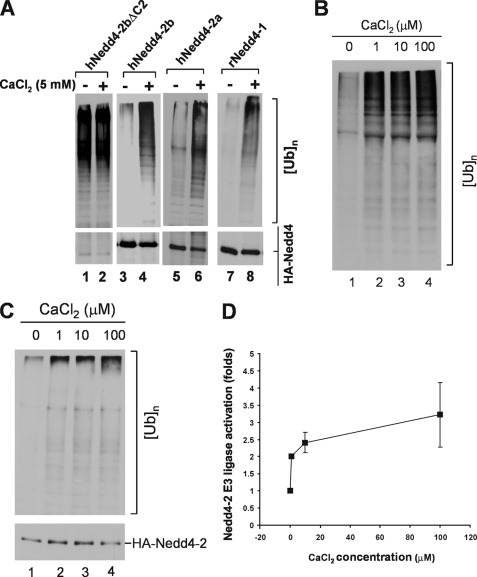
To test if calcium activates Nedd4 E3 ubiquitin ligase activity, we transfected and expressed human Nedd4-2a, -2b, -2bΔC2, or rat Nedd4-1 in HEK293 cells, immunoprecipitated the proteins, and determined the E3 ligase activity in the presence or absence of calcium by the in vitro ubiquitination assays. As expected, the E3 ubiquitin ligase activity of wild-type Nedd4-2a, -2b, and rat Nedd4-1 was markedly activated by 5 mm calcium (lanes 4, 6, and 8, Fig. 3A). Deletion of the C2 domain caused constitutive activation of Nedd4-2b ligase activity and abolished the response to calcium (lanes 1 and 2, Fig. 3A), indicating that the C2 domain is required for calcium-mediated activation of Nedd4 ligase activity. Together, these data suggest a mechanism in which calcium disrupts binding of the C2 domain to the HECT domain, which releases the auto-inhibition of the C2 domain and activates E3 ubiquitin ligase activity.
FIGURE 3.
Calcium activates Nedd4 E3 ubiquitin ligase activity. A, HA-tagged human Nedd4-2a, Nedd4-2b, Nedd4-2bΔC2, or rat Nedd4-1 was transfected into HEK293 cells, immunoprecipitated from the cell lysates, and used for the E3 ubiquitin ligase activity assay with or without addition of 5 mm CaCl2. The polyubiquitin products were detected by immunoblotting with anti-ubiquitin antibody (top panels), and the Nedd4 protein amount used in the ligase assay was detected by immunoblotting with anti-HA antibody (bottom panels). B, calcium activation assayed with purified Nedd4-2. His-tagged Nedd4-2a was expressed in E. coli and purified with affinity beads. Purified his-tagged Nedd4-2a (100 ng) was used for the ubiquitin ligase activity assay in the presence of indicated concentration of CaCl2. C and D, calcium activates immunoprecipitated Nedd4 E3 ubiquitin ligase from HEK293 cells. HA-tagged Nedd4-2a was transfected into HEK293 cells and immunoprecipitated with anti-HA antibody from cell homogenates. The E3 ubiquitin ligase activity of Nedd4-2a was assayed in presence of CaCl2 at indicated concentrations. C, a representative immunoblotting data; D, quantification of immunoblotting data from two independent assays of calcium-induced activation of Nedd4-2a E3 ubiquitin ligase activity by Kodak EDAS290 image system. The activation fold is defined as ([UB]n/[HA-Nedd4-2a])/([[UB]n]0/[HA-Nedd4-2a]0), where [UB]n stands for the quantity of poly-ubiquitin products at any concentration of calcium and [HA-Nedd4-2a] for the quantity of the corresponding HA-Nedd4-2a used in the assay; [[UB]n]0 for the quantity of poly-ubiquitin products with no calcium and [HA-Nedd4-2a]0 for the quantity of HA-Nedd4-2a used in the assay with no calcium.
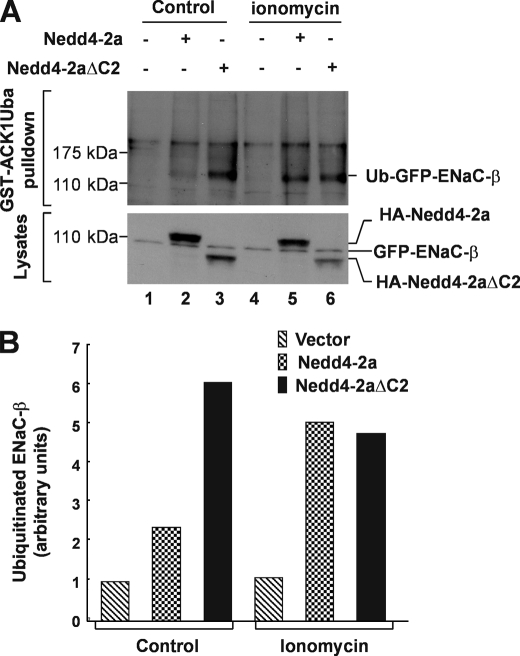
We further determined activation of Nedd4 E3 ligase activity by micromolar concentrations of calcium. We measured in vitro ubiquitination activity of purified Nedd4-2a expressed in bacteria in the presence of 1–100 μm calcium. As shown in Fig. 3B, maximal activation of Nedd4-2a was achieved by 1 μm calcium (lane 2). Similar results were obtained using Nedd4-2a expressed in HEK293 cells (Fig. 3, C and D). Previous studies have shown that peak concentrations of cytosolic calcium released by inositol 1,4,5-trisphosphate stimulation is ∼0.5–1 μm (46–49), which is within the concentration range we observed for activation of Nedd4. Intracellular calcium could reach even higher concentrations in microdomains (50, 51). We noticed that calcium activated the purified Nedd4-2 expressed in bacteria better than the immunoprecipitated Nedd4-2 expressed in HEK293 cells (Fig. 3, B and C). This might be due to co-immunoprecipitation of some endogenous Nedd4-2 activators in HEK293 cells that could elevate the basal activity of Nedd4-2, thus reducing the in vitro activation by calcium. Nevertheless, the data in Fig. 3 demonstrate that Nedd4 could be activated by physiological concentrations of calcium. To demonstrate that calcium activates Nedd4 in cells, we stably expressed the GFP-tagged ENaC-β subunit (GFP-ENaC-β), a known Nedd4 substrate (52), in HEK293 cells. We then transiently transfected HA-tagged Nedd4-2a or Nedd4-2aΔC2 into the GFP-ENaC-β cell line. We used the calcium ionophore ionomycin to introduce extracellular calcium into the cells and the proteasome inhibitor MG-132 for accumulating ubiquitinated ENaC-β. The ubiquitination of GFP-ENaC-β was assayed by GST-ACK1Uba pulldown, in which the Uba domain of ACK1, a known ubiquitin-binding domain (53), was fused with GST and used for precipitation of ubiquitinated GFP-ENaC-β from cell lysates. Precipitated ubiquitinated ENaC-β was detected by immunoblotting with anti-GFP. As shown in Fig. 4, without ionomycin treatment, ubiquitination of ENaC-β was slightly increased in cells overexpressing wild-type Nedd4-2a (lane 2, top panel, Fig. 4, A and B), but was substantially increased in cells overexpressing Nedd4-2aΔC2, the constitutively active mutant. With ionomycin treatment, ubiquitination of ENaC-β was robustly enhanced in cells overexpressing wild-type Nedd4-2a (lane 5, top panel, Fig. 4, A and B). However, treatment with ionomycin did not increase ubiquitination of ENaC-β in cells overexpressing Nedd4-2aΔC2, indicating that ubiquitination of ENaC-β by Nedd4-2aΔC2 was not sensitive to calcium. These data are consistent with the in vitro E3 ubiquitin ligase activity assay in Fig. 3A (lanes 1 and 2) and provide support for the calcium-dependent activation of Nedd4 E3 ubiquitin ligase in vivo.
FIGURE 4.
Influx of extracellular calcium by ionomycin enhances ubiquitination of ENaC-β subunit, a specific Nedd4 substrate. A, a stably GFP-ENaC-β-expressed HEK293 cells transfected 36 h with HA-Nedd4-2a, Nedd4-2aΔC2, or vector were serum-starved for 12 h followed by incubation in calcium buffer (20 mm Hepes, pH 7.4, 140 mm NaCl, 6 mm KCl, 1 mm MgCl2, 1.25 mm CaCl2, 0.1 mm EDTA, and 20 mm glucose) along with or without 1 μm ionomycin for 6 h in presence of 10 μm MG-132, a proteasome inhibitor. The ubiquitinated GFP-ENaC-β was precipitated by GST-ACK1-Uba and detected by immunoblotting with anti-GFP antibody (top panel). The expression level of GFP-ENaC-β, HA-Nedd4-2a, or HA-Nedd4-2aΔC2 was determined by immunoblotting of the cell lysates with anti-GFP and anti-HA, respectively (bottom panel). B, the quantification of ubiquitinated GFP-ENaC-β in (A) with GE-Gel Logic100 Imaging system.
Receptor Signals Activate Nedd4 E3 Ubiquitin Ligase Activity
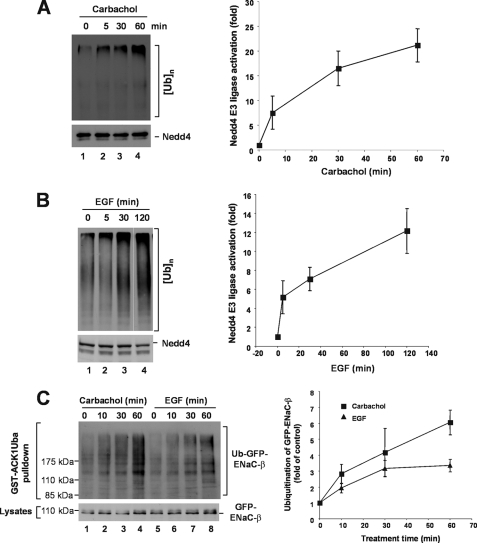
To define cellular signaling for activation of Nedd4, we determined effects of carbachol, a ligand for acetylcholine receptor, and epidermal growth factor (EGF), a ligand for epidermal growth factor receptor (EGFR), on activation of endogenous Nedd4 in HEK293 and MDA-MB-231 cells. Both the acetylcholine receptor and EGFR are known to activate phospholipase C and elevate intracellular calcium levels (54–56). Both carbachol and EGF activated Nedd4, measured by in vitro E3 ubiquitin ligase activity assay with immunoprecipitated endogenous Nedd4, by 15- to 20-fold upon treatment for 60–120 min (Fig. 5, A and B). Because immunoprecipitated Nedd4 could be contaminated with other E3 ubiquitin ligases, we further determined ubiquitination of ENaC-β, the Nedd4 substrate, in response to carbachol or EGF stimulation in HEK293 cells stably expressing GFP-ENaC-β. As shown in Fig. 5C, stimulation of the cells with carbachol or EGF significantly enhanced ubiquitination of ENaC-β. These data confirmed that carbachol or EGF stimulated Nedd4 activity, presumably through elevation of intracellular calcium level.
FIGURE 5.
In vivo activation of Nedd4 E3 ubiquitin ligase activity by stimulation of acetylcholine receptor and EGFR. A, HEK293 cells were starved in serum-free Dulbecco's modified Eagle's medium for 2 h, then stimulated with 5 mm carbachol at the indicated time. B, human breast cancer cell MDA-MB-231 cells were serum-starved overnight followed by stimulation with EGF (100 ng/ml) at indicated time. In both A and B, endogenous Nedd4 was immunoprecipitated with an anti-Nedd4 antibody from 20,000 × g cleared cell homogenates. The E3 ubiquitin ligase activity was assayed. The poly-ubiquitin products were determined by immunoblotting and quantified by using a Kodak EDAS290 imaging system for two independent experiments. C, stimulation of acetylcholine receptor by carbachol or EGFR by EGF enhances ubiquitination of Nedd4 substrate ENaC-β subunit. GFP-ENaC-β-expressed HEK293 cells were serum-starved for 12 h, then stimulated with 5 μm carbachol or 100 ng/ml EGF at the indicated times in the presence of 10 μm MG-132. The ubiquitinated GFP-ENaC-β was precipitated by GST-ACK1-Uba and detected by immunoblotting with anti-GFP (top panel). The expression level of GFP-ENaC-β was determined by immunoblotting of the cell lysates with anti-GFP (bottom panel). Ubiquitinated GFP-ENaC-β was quantified with normalized GFP-ENaC-β by using a GE-Gel Logic100 imaging system for two independent experiments.
A Heat-sensitive Membrane Component Enhances Calcium-dependent Activation of Nedd4
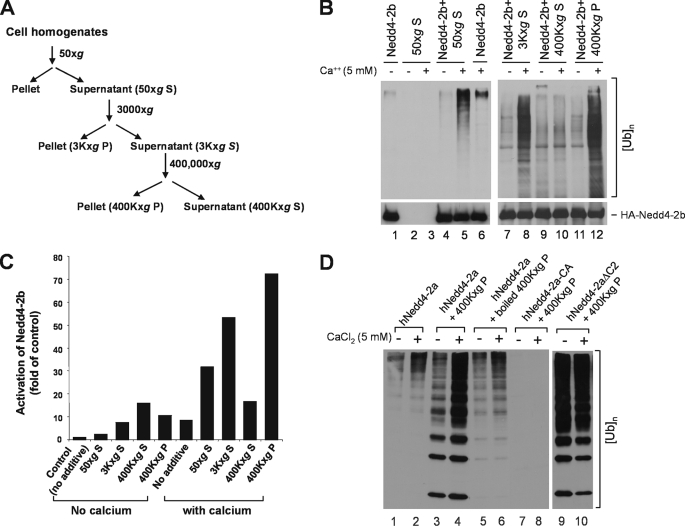
We noticed that activation of Nedd4 E3 ligase activity by calcium (∼4-fold) was significantly less than the activation by truncation of the C2 domain (∼30-fold) or the activation by carbachol or EGF signaling (10- to 20-fold). This suggests that additional component in cells is needed for calcium to fully activate Nedd4. The C2 domain has been proposed as a membrane-targeting domain for Nedd4 and that the C2 domain-mediated membrane translocation is required for cellular function of Nedd4 (39, 40). To investigate how Nedd4 is activated in cells, we searched for endogenous activators of Nedd4 in cells. We fractionated HEK293 cell homogenates by centrifugation and prepared supernatant and particulate fractions as outlined in Fig. 6A. An aliquot of each fraction was added to E3 ubiquitin ligase assays with Nedd4-2b immunoprecipitated from transfected HEK293 cell lysates. As shown in Fig. 6 (B and C), in the absence of cell fractions, calcium activated Nedd4-2b ∼8-fold (lanes 1 and 6, Fig. 6B). With addition of the 50 × g S fraction, activation of Nedd4-2b by calcium was enhanced to 30-fold (lanes 4 and 5, Fig. 6B). This activation was not due to endogenous E3 ubiquitin ligase activity in the 50 × g S fraction, because no E3 ubiquitin ligase activity was detected in this fraction (lanes 2 and 3, Fig. 6B). The Nedd4 activation component was mainly in the 400,000 × g particulate fraction (lanes 9–12, Fig. 6B). With addition of this fraction, calcium activated Nedd4-2b by 70-fold (Fig. 6C).
FIGURE 6.
One or more heat-sensitive components enhance calcium-induced activation of Nedd4 E3 ubiquitin ligases. A, schematic procedures for preparation of cell homogenate fractions by centrifugation. B, the HA-tagged Nedd4-2b was transfected in HEK293 cells and immunoprecipitated with an anti-HA antibody for the E3-ubiquitin ligase activity assay. The preparation of the homogenate fractions of HEK293 cells and the assay of E3 ligase activity were performed as described under “Experimental Procedures.” To examine the effect of the homogenate fractions on the E3 ubiquitin ligase activity of HA-Nedd4-2b, 5 μl of the homogenate fraction was added to the reaction mix during the ubiquitin ligase activity assay. The top panels represent poly-ubiquitin products generated from the E3 ubiquitin ligase assay; the bottom panels show amounts of immunoprecipitated HA-tagged Nedd4-2b used in the assay. S50, the 50 × g supernatant; S3K, the 3,000 × g supernatant; P3K, the 3,000 × g pellet; S96K, the 96,000 × g supernatant; P96K, the 96,000 × g pellet; S400K, 400,000 × g supernatant; and P400K, the 400,000 × g pellet. C, the poly-ubiquitin product in B was quantified by using the GE-Gel Logic100 Imaging system and used for calculation of Nedd4-2b activation. D, purified Nedd4-2a is activated by the component in the 400,000 × g pellet. His-tagged Nedd4-2a was expressed in E. coli and purified with affinity beads. Purified His-tagged Nedd4-2a (100 ng), active mutant Nedd4-2aΔC2 (100 ng), or ligase-dead mutant Nedd4-2a-CA (100 ng) was used for the ubiquitin ligase activity assay in the presence or absence of CaCl2 with addition of 5 μl of indicated homogenate fraction. The poly-ubiquitin product was detected by immunoblotting with anti-ubiquitin antibody.
To further characterize the active component in the 400,000 × g P fraction, we used purified human Nedd4-2a expressed in bacteria. As shown in Fig. 6D, calcium produced higher Nedd4-2a activity in the presence of the 400,000 × g P fraction (compare lane 4 with lane 2). The enhanced calcium-dependent activation of Nedd4 was eliminated by heating the 400,000 × g P fraction for 10 min at 100 °C (lane 6), indicating a heat-sensitive component. To control for endogenous E3 ubiquitin ligase activity in the 400,000 × g P fraction, this fraction was assayed for the E3 ubiquitin ligase activity with purified ligase-dead Nedd4-2a mutant, Nedd4-2a-CA. No poly-ubiquitin products were detected (lanes 7 and 8), indicating that the 400,000 × g P fraction has no detectable E3 ubiquitin ligase activity. Enhancement of calcium-dependent activation of Nedd4 by the 400,000 × g P fraction was mediated by release of the C2 domain auto-inhibition, because 400,000 × g P fraction had little effect on activity of the C2 domain truncation mutant Nedd4-2aΔC2 (lanes 9 and 10, Fig. 6D). Thus, we conclude that the Nedd4 activator in the 400,000 × g P fraction is a heat-sensitive component that enhances calcium-dependent release of the C2-domain-mediated auto-inhibition of Nedd4 E3 ubiquitin ligase activity.
There was also a calcium-independent activation of Nedd4 caused by addition of either the supernatant or the particulate fraction (lanes 4, 7, 9, and 11 in Fig. 6B, lanes 3 in Fig. 6D). This activation might not be through release of the C2 domain-mediated auto-inhibition, because the activity of Nedd4-2aΔC2 was much higher than that of Nedd4-2a with addition of 400,000 × g P in the absence of calcium (compare lane 9 with lane 3 in Fig. 6D). It has been observed that the WW domain-binding protein NDFIP activated ITCH E3 ligase activity (57). Such a protein in the supernatant or the particulate fraction could mediate calcium-independent activation of Nedd4 through interaction with the WW domains of Nedd4.
DISCUSSION
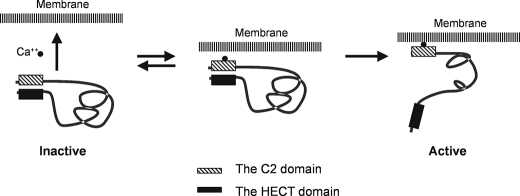
We have shown by mutational analysis and GST-Nedd4-C2 domain pulldown assays that the C2 domain of Nedd4 functions as an auto-inhibitory domain through binding to the HECT domain. Weisner et al. also reported that the C2 domain of Smurf2 directly binds to the HECT domain and functions as an auto-inhibition module to the E3 ligase activity (43). They proposed that Nedd4 has a similar structure to Smurf2 and showed that truncation of the C2 domain activates Nedd4 E3 ubiquitin ligase activity in an in vitro ubiquitin ligase activity assay (43). Thus, the auto-inhibition of the E3 ligase activity by the C2 domain is a key feature of Nedd4. Currently we do not know whether the auto-inhibition is formed through the intra- or the intermolecular interaction between the C2 and the HECT domains. We speculate that the both interactions could form the auto-inhibition. We hypothesized that activation of Nedd4 E3 ligase activity is directly related to releasing the auto-inhibition. Our data show that this auto-inhibitory mechanism is regulated by calcium. Calcium activates Nedd4 by binding to the C2 domain and releasing the C2 domain-mediated auto-inhibition. The membrane-rich fraction (400Kxg fraction) enhances calcium to bind to the C2 domain; together calcium and the membrane-rich fraction generate a robust activation of Nedd4. We also show signal-dependent activation of endogenous Nedd4 in cells. Finally, we also show that calcium-mediated activation of Nedd4 is enhanced by a heat-sensitive membrane fraction. Based on these results, we propose a model for activation of Nedd4 E3 ubiquitin ligase activity shown in Fig. 7. In this model, Nedd4 is kept inactive by either the intra- or intermolecular interaction (the intramolecular interaction depicted in Fig. 7) between the C2 and the HECT domains in calcium-free or low calcium environment. Cellular signaling elevates intracellular calcium concentration and results in binding of calcium to the C2 domain of Nedd4. The calcium binding enhances the affinity of the C2 domain to the membranes and promotes Nedd4 translocation to the membrane fraction, which releases the auto-inhibition and activates Nedd4 E3 ubiquitin ligase activity. We speculate that binding of calcium to the C2 domain (intermediate status) partially disrupts interaction of the C2 domain with the HECT domain and makes the C2 domain accessible to membranes. This is based on our finding that calcium alone only partially activates Nedd4 E3 ubiquitin ligase activity (Figs. 3 and 5). Binding to the membranes stabilizes the active conformation of Nedd4.
FIGURE 7.
A model for activation of Nedd4 in cells. Cellular signaling elevates the cytoplasmic concentration of calcium, which binds to the C2 domain and enables the C2 domain to interact with the cellular membrane. Calcium-dependent binding of the C2 domain to the membrane results in dissociation of the C2 domain from the HECT domain, and release of the C2 domain-mediated auto-inhibition thus activates Nedd4 E3 ligase activity.
The C2 domain of Nedd4 has been demonstrated to bind to inositol phospholipids and proposed as an inositol phospholipid-binding module (39, 40). The C2 domain also binds to phosphatidylcholine or/and phosphatidylserine in a calcium-promoting manner (39). Previous studies have shown that translocation of Nedd4 to plasma membrane occurred in response to elevation of intracellular calcium in cells (39, 41). We speculate that calcium-dependent membrane translocation of Nedd4 is a pivotal step for activation in response to intracellular signaling. We observed a dramatic enhancement in calcium-mediated activation of Nedd4 by a 400,000 × g particulate fraction. This enhancement was eliminated by heating the particulate fraction for 10 min at 100 °C, suggesting that the component in the particulate fraction that collaborates with calcium to activate Nedd4 might be a protein. This component could be an endogenous activator of Nedd4 that activates Nedd4 by augmenting or/and stabilizing the activation effect of calcium by releasing the C2 domain-mediated auto-inhibition. Identification of this component would help us further to understand the mechanism of Nedd4 ligase activation and delineate signaling pathways of Nedd4 ligases.
Although Smurf2, similar to Nedd4, also has the C2 domain-mediated auto-inhibitory mode for regulation of the E3 ubiquitin ligase activity, ITCH (AIP4) and WWP1, the two other members of the HECT E3 ubiquitin ligase family that are close to Nedd4, seem not to have such a regulatory mechanism. We observed no activation effect with the C2 domain truncation of ITCH and WWP1 (data not shown). It has been shown that JNK-mediated phosphorylation plays an important role in activation of ITCH (44). The phosphorylation site was located between the C2 domain and the WW1 domain. It was proposed that the region executes auto-inhibition by interaction with the HECT domain, while phosphorylation releases the auto-inhibition (44). Protein phosphorylation may also be involved in activation of Nedd4. It was observed that AMP activated kinase-phosphorylated Nedd4-2, and the phosphorylation enhanced binding of Nedd4-2 to its substrate ENaC, thus down-regulation of ENaC activity (34).
Although the catalytic mechanism of Nedd4 E3 ubiquitin ligases has been extensively studied (1–3), activation of the E3 ubiquitin ligase activity of Nedd4 in response to cellular signaling is poorly understood. Here we show that the C2 domain of Nedd4 E3 ubiquitin ligases functions as an auto-inhibitory domain to the ligase activity, and that the auto-inhibition is released by calcium. This molecular mechanism may serve as a switch for activation/inactivation of Nedd4 in cells and couple cellular signals, such as EGFR and acetylcholine receptor, to protein ubiquitination, degradation, and trafficking. It has been shown that Nedd4-1 ubiquitinates tumor suppressor proteins and may be oncogenic (15–18). Nedd4-2 ubiquitinates various ion channels and membrane transporters and determines their degradation (19–23). Thus, Nedd4 plays important roles in cell growth, survival, differentiation, and homeostasis. The C2/calcium switch model for the ligase activity will provide novel insight into how Nedd4 regulates biological and pathological processes in response to cellular signaling.
Acknowledgment
We thank Dr. Daniela Rotin at University of Toronto for sending us rat Nedd4-1 cDNA plasmid.
This work was supported by American Cancer Society Grant ACS-RSG TBE-110602 (to W. Y.).
- E3
- ubiquitin ligase
- E2
- ubiquitin-conjugating enzyme
- E1
- ubiquitin-activating enzyme
- SGK1
- serum- and glucocorticoid-inducible kinase 1
- ENaC
- epithelial sodium channel
- GFP
- green fluorescent protein
- ULR
- ubiquitin ligation reaction
- GST
- glutathione S-transferase
- HA
- hemagglutinin
- EGF
- epidermal growth factor
- EGFR
- EGF receptor
- JNK
- c-Jun amino-terminal kinase.
REFERENCES
- 1.Pickart C. M. (2001) Annu. Rev. Biochem. 70, 503–533 [DOI] [PubMed] [Google Scholar]
- 2.Jackson P. K., Eldridge A. G., Freed E., Furstenthal L., Hsu J. Y., Kaiser B. K., Reimann J. D. (2000) Trends Cell Biol. 10, 429–439 [DOI] [PubMed] [Google Scholar]
- 3.Passmore L. A., Barford D. (2004) Biochem. J. 379, 513–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shearwin-Whyatt L., Dalton H. E., Foot N., Kumar S. (2006) BioEssays 28, 617–628 [DOI] [PubMed] [Google Scholar]
- 5.Chen H., Ross C. A., Wang N., Huo Y., MacKinnon D. F., Potash J. B., Simpson S. G., McMahon F. J., DePaulo J. R., Jr., McInnis M. G. (2001) Eur. J. Hum. Genet. 9, 922–930 [DOI] [PubMed] [Google Scholar]
- 6.Russo C. J., Melista E., Cui J., DeStefano A. L., Bakris G. L., Manolis A. J., Gavras H., Baldwin C. T. (2005) Hypertension 46, 488–491 [DOI] [PubMed] [Google Scholar]
- 7.Arévalo J. C., Waite J., Rajagopal R., Beyna M., Chen Z. Y., Lee F. S., Chao M. V. (2006) Neuron 50, 549–559 [DOI] [PubMed] [Google Scholar]
- 8.Fotia A. B., Ekberg J., Adams D. J., Cook D. I., Poronnik P., Kumar S. (2004) J. Biol. Chem. 279, 28930–28935 [DOI] [PubMed] [Google Scholar]
- 9.Xu L. L., Shi Y., Petrovics G., Sun C., Makarem M., Zhang W., Sesterhenn I. A., McLeod D. G., Sun L., Moul J. W., Srivastava S. (2003) Cancer Res. 63, 4299–4304 [PubMed] [Google Scholar]
- 10.Lang F., Böhmer C., Palmada M., Seebohm G., Strutz-Seebohm N., Vallon V. (2006) Physiol. Rev. 86, 1151–1178 [DOI] [PubMed] [Google Scholar]
- 11.Marmor M. D., Yarden Y. (2004) Oncogene 23, 2057–2070 [DOI] [PubMed] [Google Scholar]
- 12.Vana M. L., Tang Y., Chen A., Medina G., Carter C., Leis J. (2004) J. Virol. 78, 13943–13953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Segura-Morales C., Pescia C., Chatellard-Causse C., Sadoul R., Bertrand E., Basyuk E. (2005) J. Biol. Chem. 280, 27004–27012 [DOI] [PubMed] [Google Scholar]
- 14.Henry P. C., Kanelis V., O'Brien M. C., Kim B., Gautschi I., Forman-Kay J., Schild L., Rotin D. (2003) J. Biol. Chem. 278, 20019–20028 [DOI] [PubMed] [Google Scholar]
- 15.Magnifico A., Ettenberg S., Yang C., Mariano J., Tiwari S., Fang S., Lipkowitz S., Weissman A. M. (2003) J. Biol. Chem. 278, 43169–43177 [DOI] [PubMed] [Google Scholar]
- 16.Woelk T., Oldrini B., Maspero E., Confalonieri S., Cavallaro E., Di Fiore P. P., Polo S. (2006) Nat. Cell Biol. 8, 1246–1254 [DOI] [PubMed] [Google Scholar]
- 17.Katz M., Shtiegman K., Tal-Or P., Yakir L., Mosesson Y., Harari D., Machluf Y., Asao H., Jovin T., Sugamura K., Yarden Y. (2002) Traffic 3, 740–751 [DOI] [PubMed] [Google Scholar]
- 18.Aoh Q. L., Castle A. M., Hubbard C. H., Katsumata O., Castle J. D. (2009) Mol. Biol. Cell 20, 1816–1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Snyder P. M., Steines J. C., Olson D. R. (2004) J. Biol. Chem. 279, 5042–5046 [DOI] [PubMed] [Google Scholar]
- 20.van Bemmelen M. X., Rougier J. S., Gavillet B., Apothéloz F., Daidié D., Tateyama M., Rivolta I., Thomas M. A., Kass R. S., Staub O., Abriel H. (2004) Circ. Res. 95, 284–291 [DOI] [PubMed] [Google Scholar]
- 21.Zhou R., Patel S. V., Snyder P. M. (2007) J. Biol. Chem. 282, 20207–20212 [DOI] [PubMed] [Google Scholar]
- 22.Ekberg J., Schuetz F., Boase N. A., Conroy S. J., Manning J., Kumar S., Poronnik P., Adams D. J. (2007) J. Biol. Chem. 282, 12135–12142 [DOI] [PubMed] [Google Scholar]
- 23.Sorkina T., Miranda M., Dionne K. R., Hoover B. R., Zahniser N. R., Sorkin A. (2006) J. Neurosci. 26, 8195–8205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Debonneville C., Flores S. Y., Kamynina E., Plant P. J., Tauxe C., Thomas M. A., Münster C., Chraïbi A., Pratt J. H., Horisberger J. D., Pearce D., Loffing J., Staub O. (2001) EMBO J. 20, 7052–7059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Busjahn A., Aydin A., Uhlmann R., Krasko C., Bähring S., Szelestei T., Feng Y., Dahm S., Sharma A. M., Luft F. C., Lang F. (2002) Hypertension 40, 256–260 [DOI] [PubMed] [Google Scholar]
- 26.Pearce D. (2001) Trends Endocrinol. Metab. 12, 341–347 [DOI] [PubMed] [Google Scholar]
- 27.Bhalla V., Daidié D., Li H., Pao A. C., LaGrange L. P., Wang J., Vandewalle A., Stockand J. D., Staub O., Pearce D. (2005) Mol. Endocrinol. 19, 3073–3084 [DOI] [PubMed] [Google Scholar]
- 28.Dieter M., Palmada M., Rajamanickam J., Aydin A., Busjahn A., Boehmer C., Luft F. C., Lang F. (2004) Obes. Res. 12, 862–870 [DOI] [PubMed] [Google Scholar]
- 29.Boehmer C., Henke G., Schniepp R., Palmada M., Rothstein J. D., Bröer S., Lang F. (2003) J. Neurochem. 86, 1181–1188 [DOI] [PubMed] [Google Scholar]
- 30.Boehmer C., Okur F., Setiawan I., Bröer S., Lang F. (2003) Biochem. Biophys. Res. Commun. 306, 156–162 [DOI] [PubMed] [Google Scholar]
- 31.Boehmer C., Wilhelm V., Palmada M., Wallisch S., Henke G., Brinkmeier H., Cohen P., Pieske B., Lang F. (2003) Cardiovasc. Res. 57, 1079–1084 [DOI] [PubMed] [Google Scholar]
- 32.Choate K. A., Kahle K. T., Wilson F. H., Nelson-Williams C., Lifton R. P. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 663–668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu B. E., Stippec S., Chu P. Y., Lazrak A., Li X. J., Lee B. H., English J. M., Ortega B., Huang C. L., Cobb M. H. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 10315–10320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bhalla V., Oyster N. M., Fitch A. C., Wijngaarden M. A., Neumann D., Schlattner U., Pearce D., Hallows K. R. (2006) J. Biol. Chem. 281, 26159–26169 [DOI] [PubMed] [Google Scholar]
- 35.Sanchez-Perez A., Kumar S., Cook D. I. (2007) Biochem. Biophys. Res. Commun. 359, 611–615 [DOI] [PubMed] [Google Scholar]
- 36.Luo J. H., Weinstein I. B. (1993) J. Biol. Chem. 268, 23580–23584 [PubMed] [Google Scholar]
- 37.Medkova M., Cho W. (1998) J. Biol. Chem. 273, 17544–17552 [DOI] [PubMed] [Google Scholar]
- 38.Nalefski E. A., Falke J. J. (1996) Protein Sci. 5, 2375–23790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Plant P. J., Yeger H., Staub O., Howard P., Rotin D. (1997) J. Biol. Chem. 272, 32329–32336 [DOI] [PubMed] [Google Scholar]
- 40.Dunn R., Klos D. A., Adler A. S., Hicke L. (2004) J. Cell Biol. 165, 135–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Plant P. J., Lafont F., Lecat S., Verkade P., Simons K., Rotin D. (2000) J. Cell Biol. 149, 1473–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Snyder P. M., Olson D. R., McDonald F. J., Bucher D. B. (2001) J. Biol. Chem. 276, 28321–28326 [DOI] [PubMed] [Google Scholar]
- 43.Wiesner S., Ogunjimi A. A., Wang H. R., Rotin D., Sicheri F., Wrana J. L., Forman-Kay J. D. (2007) Cell 130, 651–662 [DOI] [PubMed] [Google Scholar]
- 44.Gallagher E., Gao M., Liu Y. C., Karin M. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 1717–1722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang W., Lo C. G., Despenza T., Cerione R. A. (2001) J. Biol. Chem. 276, 17468–17473 [DOI] [PubMed] [Google Scholar]
- 46.Prentki M., Corkey B. E., Matschinsky F. M. (1985) J. Biol. Chem. 260, 9185–9190 [PubMed] [Google Scholar]
- 47.Prentki M., Wollheim C. B., Lew P. D. (1984) J. Biol. Chem. 259, 13777–13782 [PubMed] [Google Scholar]
- 48.Keizer J., Li Y. X., Stojilković S., Rinzel J. (1995) Mol. Biol. Cell 6, 945–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Thomas A. P., Bird G. S., Hajnóczky G., Robb-Gaspers L. D., Putney J. W., Jr. (1996) FASEB J. 10, 1505–1517 [PubMed] [Google Scholar]
- 50.Parekh A. B. (2008) J. Physiol. 586, 3043–3054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Berridge M. J. (2006) Cell Calcium 40, 405–412 [DOI] [PubMed] [Google Scholar]
- 52.Staub O., Dho S., Henry P., Correa J., Ishikawa T., McGlade J., Rotin D. (1996) EMBO J. 15, 2371–2380 [PMC free article] [PubMed] [Google Scholar]
- 53.Shen F., Lin Q., Gu Y., Childress C., Yang W. (2007) Mol. Biol. Cell 18, 732–742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Margolis B., Rhee S. G., Felder S., Mervic M., Lyall R., Levitzki A., Ullrich A., Zilberstein A., Schlessinger J. (1989) Cell 57, 1101–1107 [DOI] [PubMed] [Google Scholar]
- 55.Qian N. X., Winitz S., Johnson G. L. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 4077–4081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Seuwen K., Kahan C., Hartmann T., Pouyssegur J. (1990) J. Biol. Chem. 265, 22292–22299 [PubMed] [Google Scholar]
- 57.Mund T., Pelham H. R. (2009) EMBO Rep. 10, 501–507 [DOI] [PMC free article] [PubMed] [Google Scholar]