Abstract
Chromosomal and plasmid DNA molecules in bacterial cells are maintained under torsional tension and are therefore supercoiled. With the exception of extreme thermophiles, supercoiling has a negative sign, which means that the torsional tension diminishes the DNA helicity and facilitates strand separation. In consequence, negative supercoiling aids such processes as DNA replication or transcription that require global- or local-strand separation. In extreme thermophiles, DNA is positively supercoiled which protects it from thermal denaturation. While the role of DNA supercoiling connected to the control of DNA stability, is thoroughly researched and subject of many reviews, a less known role of DNA supercoiling emerges and consists of aiding DNA topoisomerases in DNA decatenation and unknotting. Although DNA catenanes are natural intermediates in the process of DNA replication of circular DNA molecules, it is necessary that they become very efficiently decatenated, as otherwise the segregation of freshly replicated DNA molecules would be blocked. DNA knots arise as by-products of topoisomerase-mediated intramolecular passages that are needed to facilitate general DNA metabolism, including DNA replication, transcription or recombination. The formed knots are, however, very harmful for cells if not removed efficiently. Here, we overview the role of DNA supercoiling in DNA unknotting and decatenation.
INTRODUCTION
DNA supercoiling
To present basic features of DNA supercoiling and its consequences for DNA properties, let us imagine that we could manipulate a linear fragment of DNA. We would see this fragment behaving like an elastic rod, i.e. we could bend it and the energetic costs of bending would increase with the square of the deflection angle. Under very strong bending deformations, the DNA would exit the elastic regime forming kinks that would facilitate further bending (1). However, it is questionable whether bending deformations in vivo reach the level at which kinking occurs (2,3). We could also impose a torque on the DNA, and would see that the energy of torsional deformation also initially increases with the square of the induced rotation angle between the two ends (4). Interestingly, despite the fact that the DNA structure is right-handed, the energetic costs for right- and left-handed torsional deformations are practically identical for small torsional deformations (5). However, for larger torsional deformations, there is an interval where a torque acting in the direction that unwinds the DNA helix leads to the separation of the DNA strands, while a similar torque, but acting in the direction that winds up the DNA helix just stabilizes the DNA helix (5,6).
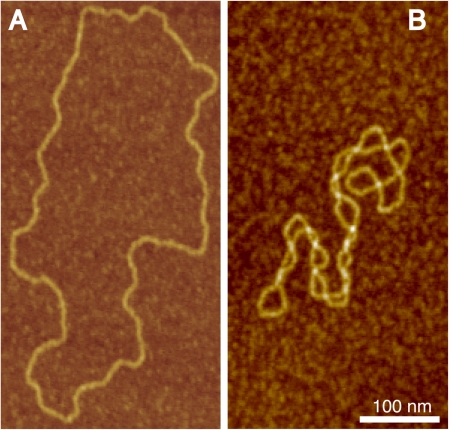
Let us now imagine that we close into a circle the manipulated DNA fragment. We could close the DNA without imposing any torsional deformation or just a minimal one needed to juxtapose ends of both strands for covalent bond formation. Such covalently closed DNA molecules would be torsionally relaxed, and, for plasmid size molecules, would adopt the shape of unconstrained freely fluctuating rings, that may adsorb without any intramolecular crossings when deposited on a surface for atomic force microscopy (AFM) imaging (Figure 1A). We could also close the DNA fragment into a circle after having introduced sufficient torsional deformation to remove, for example, one helical turn of the DNA for each 200 bp. If both DNA strands of such a DNA were ligated and then the molecule let free to relax, it would minimize its elastic energy by adopting the structure of an interwound supercoiled molecule (Figure 1B).
Figure 1.
Atomic force microscopy (AFM) visualization of torsionally relaxed (A) and negatively supercoiled (B) bacterial plasmids pBR322. The deposition of the 4361 bp-long DNA chains is made on APTES modified mica (82).
Negative supercoiling has an important biological function of facilitating local- and global-strand separation of DNA molecules such as these occurring during transcription and replication, respectively (7–9). Figure 2 explains why this is the case. Strand separation relaxes the torsional stress in negatively supercoiled DNA (10). Therefore, it is energetically less costly to separate strands in negatively supercoiled than in relaxed DNA, and the energy difference is furnished by DNA relaxation. Of course, there is no free lunch even for a bacteria, and the energetic cost of the torsional constraint has to be introduced by the ATP hydrolysis-driven action of DNA gyrase (11).
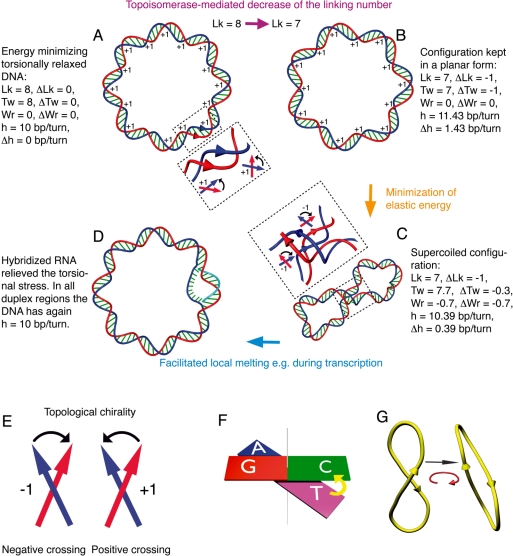
Figure 2.
Topology and mechanics of DNA supercoiling. (A) Energy minimizing, torsionally relaxed, covalently closed DNA molecule with Lk = 8, Tw = 8, Wr = 0 and having the intrinsic DNA helicity of 10 bp/turn. The blow-up shows that interstrand crossings in the right-handed DNA helix are positive, since for topological considerations one assumes parallel orientation of DNA strands [see (E) for the topological convention of crossings’ signs]. (B) As a result of a combined action of DNA gyrase that decreased the linking number by two followed by the relaxation reaction of topo I, that increased the linking number by 1, the molecule presented in (A) has changed its linking number to Lk = 7. If that molecule were maintained in a planar configuration by ionic interaction with a charged surface, for example, its Tw would also change to 7, while Wr would remain unchanged. The change of DNA twist introduces a significant torsional tension into the elastic structure of the DNA as its helical repeat would need to change to 11.43 bp/turn, while its minimal torsional energy is achieved for a helix with 10 bp/turn. (C) The molecule presented in (B) has detached from a charged surface and minimized its elastic energy by adopting a supercoiled form with a Wr ≈−0.7 which permitted the molecule to greatly diminish its torsional tension as its twist has changed to ≈7.7, which is close to torsionally relaxed state. Due to the quadratic dependence of torsional and bending energies on the respective elastic deformations, it is energetically favourable to repartition the elastic stress due to the deficit of linking ΔLk into torsional and bending deformations. Usually ∼70% of the ΔLk are compensated by the acquired writhe (41,83). The blow-up shows that the inter-helix crossings between the two strands in negatively supercoiled DNA have negative sign as opposed to intra-helix crossings that have positive signs. (D) Opening of 10 bp by hybridization with nascent RNA, for example, is energetically more favourable in an unwound chain C than in the covalently closed, torsionally relaxed form A. The twist value is lower than in the torsionally relaxed DNA shown in (A). However, this causes no torsional stress as this twist is realized over a shorter region of pairing between the DNA strands, which re-establishes there the helicity of 10 bp/turn, while the open region is stabilized by the interaction with the hybridized RNA. (E) The topological sign convention. To determine the sign of individual crossings of two oriented curves, one checks in which sense one should turn the orientation vector of the overlying segment to have it pointing in the same direction as the vector of the underlying segment, while the rotation can not exceed 180°. If that rotation is clockwise, the crossing is negative and it is positive otherwise. (F) The concept of twist. Twist of DNA molecules is the sum of all the twists angles between the consecutive base pairs. The twist units are 360° rotations. (G) The concept of writhe. The same 3D curve, representing the axis of a given DNA molecule is observed from two different directions. The score provided by segment crossings can vary between the two cases, explaining why as writhe one takes the average value of crossings scores over all directions equisampling the sphere enclosing the 3D curve.
Figure 2 helps to introduce topological and geometrical descriptors of various DNA configurations. Topological descriptors define the topology of a DNA molecule, and for a given covalently closed DNA molecule, their values do not change during continuous structural deformations, i.e. deformations which do not require breaking and resealing of covalent chemical bonds of the DNA backbone. One of these descriptors is the linking number (Lk) that tells us how many times and with what handedness the two single strands of a given DNA molecule are linked with each other. The absolute value of the DNA linking number corresponds to the minimal number of passages of one strand through the other, needed to completely separate in space the two strands as it is required during replication and segregation of circular DNA molecules in living cells. Such inter-strand passages are non-continuous structural deformations of DNA that can be catalysed by type I DNA topoisomerases. The positive sign of Lk indicates that the two strands of the molecule wind around each other in a right-handed sense as it is the case of native B-DNA structure. The mathematical way of determining the linking number of two closed curves in space is to score all the crossings of one curve with the other when the two curves are observed from any given direction (8) (see Figure 2B–D). Positive crossings score as +1 and negative ones as −1, and are defined as presented in Figure 2. The sum of scored values, but not the number of visible crossings is a topological invariant, i.e. it does not change with the direction of viewing and is independent of any continuous deformation of covalently closed DNA molecules. The value of Lk corresponds to half of the sum obtained after adding all the values for the perceived crossings of the two strands, and is always an integer. Figure 2B–D illustrates the invariant nature of Lk showing that it does remain unchanged after folding or after partial strand separation in a covalently closed DNA molecule. It is important to remember that, for practical reasons, the linking number for right-handed DNA, such as in a native B-DNA structure, was set to be positive and thus, for topological considerations, the two complementary DNA strands are considered as having a parallel orientation despite their anti-parallel chemical polarity (8).
Geometric descriptors of the DNA structure include the twist (Tw) and the writhe (Wr). Tw is a measure of the helical rotation of DNA around its axis and corresponds to the integrated Tw angle (expressed in number of 360° rotations) between all successive base pairs in an entire DNA molecule (Figure 2F). The Tw value is usually not an integer, however, it takes integer values when the axial path of the entire covalently closed DNA molecule is planar or when it lies on the surface of a perfect sphere (12). For right-handed DNA the Tw is positive. The Tw value can vary with the continuous elastic deformation of a covalently closed DNA molecule.
The second geometric descriptor, the Wr, is a measure of the winding of the axial trajectory of the DNA molecule around itself. Torsionally relaxed DNA molecules, such as shown in Figures 1A and 2A, do not wind around themselves to minimize their elastic energy and have their time averaged Wr close to zero. Negatively supercoiled molecules, such as shown in Figures 1B and 2C wind around themselves and thus have a significant Wr. To calculate the Wr one needs to give a consistent direction to the entire axial trajectory of a given DNA molecule to be able to determine the sign of perceived self-crossings. Similar to Lk, the right-handed crossings score as +1 and left-handed crossings as −1. However, to calculate the Wr, the signed sum value is not divided by two, contrary to Lk. In a further contrast to Lk, the signed sum over perceived right- and left-handed crossings may change with the direction of viewing, and therefore the Wr value is obtained after averaging the signed sum score over all directions of viewing, equisampling an imaginary sphere that encloses the trajectory for which the Wr is calculated (Figure 2G). The Wr values are negative for negatively supercoiled DNA and positive for positively supercoiled molecules. In the case shown in Figure 2D the Wr value has increased by ∼0.7 (from ∼−0.7 to 0) when negatively supercoiled DNA got torsionally relaxed as a result of strand separation extending over a region of ∼10 bp, as would be the case if an individual RNA polymerase started a transcription.
Figure 2 illustrates the mathematical relationship between Lk, Tw and Wr valid for any given covalently closed DNA molecule. This relationship is expressed by the formula Lk = Tw + Wr (8,12–14). One of the consequences of this relationship is that, when a given covalently closed DNA molecule undergoes continuous deformation, its Lk remains constant while changes of Tw and Wr are coupled in such a way that their sum is constant and equal to Lk (Figure 2).
In biochemical experiments, however, it is hardly possible to determine the Lk of a given DNA molecule. It is relatively easy, though, to determine by gel electrophoresis the linking number difference between different DNA topoisomers, i.e. DNA molecules with the same size and sequence but which differ in their linking number (8). The extent of DNA supercoiling of a given topoisomer is usually expressed by its ΔLk = (Lk−Lk0), i.e. the difference between its actual linking number (Lk) and a hypothetical linking number (Lk0) this molecule would have had under the same environmental conditions if it were permitted to completely release its torsional stress by a free rotation. Lk0 is equal to the sum of time-averaged twist (Tw0) and time averaged writhe (Wr0) of the nicked form of a given DNA molecule. The Tw0 value is related to the native DNA helical repeat (h) by the formula Tw0 = N/h, where N is the number of base pairs in the molecule. The helical repeat value is not universal for all DNA molecules but varies with a particular sequence of base pairs, temperature, the concentration of various solutes etc. (15–17). The Wr0 value is usually close to zero but it may significantly deviate from it for particular sequences that induce for example a helicoidal path of the DNA axis (18). In contrast to integer value of Lk of real DNA topoisomers, the value of Lk0 does not need to be an integer as it does not correspond to a realisable topoisomer but simply describes the reference point from which the torsional stress starts to arise in the considered DNA molecules.
The axial line of a covalently closed DNA molecule is usually isomorphic to a circle i.e. it can be continuously deformed to a circle without breaking and resealing of DNA strands. All molecules for which this is the case are unknotted. In contrast, if the axial line of a given molecule is not isomorphic to a circle then the molecule is knotted and may form one of infinitely many types of knots, although the most frequently observed DNA knots are simple knots like trefoils or figure-of-eight knots (19). The axial lines of two or more DNA molecules may also be topologically entangled (i.e. it is not possible to disentangle them without cutting one of the molecules). In that case, the molecules form catenanes. The most frequently studied DNA catenanes formed in vivo are multiply interlinked torus type of catenanes arising during replication of circular DNA molecules (20,21).
Supercoiling, knotting and catenation are not mutually exclusive, i.e. DNA molecules that are supercoiled might be in addition knotted or catenated or even knotted and catenated. We will discuss later the interplay between supercoiling and knotting or catenation and how this interplay helps DNA topoisomerases to unknot and decatenate DNA molecules.
Bacterial topoisomerases
DNA decatenation and unknotting can not happen without DNA topoisomerases that are the enzymes that can change DNA topology and as such allow non-continuous deformations of DNA molecules involving transient opening and sealing of covalent bonds in the DNA backbone. Since our survey concentrates on bacterial systems, we will limit our presentation to bacterial topoisomerases. For the processes of DNA supercoiling, decatenation and unknotting, the most important are type II DNA topoisomerases that transiently cut both strands of the DNA, keep the cut ends at a small distance during the passage of another double-stranded region through the cut, and finally reseal the cut strands (22,23). The double-stranded region that passes through the cut may belong to the same or to another DNA molecule. The two currently known type II DNA topoisomerases in eubacteria are DNA gyrase and topoisomerase IV. The typical action of DNA gyrase involves the right-handed wrapping of a ∼140 bp-long DNA fragment around the enzyme and the ensuing intersegmental passage of one of the DNA arms flanking the wrap through the transiently cut central region of the wrapped portion of the DNA (24). Each catalytic cycle of DNA gyrase diminishes by two the linking number of covalently closed DNA molecules (25). DNA supercoiling is energetically costly, and the energy for it is provided by the ATPase activity of DNA gyrase (11,26). When the DNA is negatively supercoiled to a physiological level, its torsional stress limits further supercoiling action of DNA gyrase and this contributes to setting the steady state level of DNA supercoiling in living cells (27,28).
In the case of Topo IV, DNA segments passing through the transient cuts are those that have found themselves close to the entry gate of the topoisomerase and were correctly positioned for the passage (29). The chain passages may involve pairs of segments belonging to two different DNA molecules or to the same one, and represent necessary events for decatenation and unknotting, respectively. The catalytic cycle of Topo IV, similarly to that of DNA gyrase, is coupled to ATP hydrolysis. However, the obtained energy is not used to increase the elastic energy of individual DNA molecules but rather to actively diminish the level of knotting or catenation below the topological equilibrium (see below). In consequence, the free energy of the system increases above the equilibrium value obtained in an ideal system made of phantom chains passing trough each other (26,30). It is important to add here that a cycle of cleavage and resealing per se is isoenergetic and does not require an energy input. For this reason, type I topoisomerases that cut and reseal single strands without coupling ATP hydrolysis to DNA passages can only catalyse strand-passage reactions that go down the free energy gradient.
In bacterial cells, there are two type I topoisomerases known as Topo I and Topo III, and their specific function significantly differ from each other although they both belong to class A of type I topoisomerases (22). The main function of Topo I is to survey the level of DNA torsional tension resulting from negative supercoiling by DNA gyrase or locally generated behind the progressing RNA polymerase (27,31). If that level becomes too high, uncontrolled melting could occur, and that is prevented by stepwise relaxation of the DNA by Topo I, until a safe level of negative supercoiling is attained. To fulfil this function, Topo I is activated when the DNA is strongly negatively supercoiled. Topo I's complex action involves local melting of the DNA followed by transient cutting of one strand and the passage of the opposing strand through the opening (32). During each catalytic cycle of Topo I the linking number of DNA increases by 1, diminishing the level of negative supercoiling (27). Topo I may also have other functions related to its activity on nicked DNA. Topo I preferentially interacts with nick sites and transiently opens the continuous strand facing the nick, upon which it can permit a duplex segment from the same or other DNA molecules to pass through the generated opening (32). Such a reaction can lead to unknotting or decatenation of nicked double-stranded DNA molecules. A very similar mechanism can operate during Topo III-mediated diminution of the catenation level or even complete decatenation of replication intermediates where the unfinished or freshly replicated DNA molecules are not yet converted to covalently closed circular DNA molecules and have short single-stranded regions or nicks (33,34).
DNA KNOTTING AND SUPECOILING
Knotting equilibrium
An important question related to the interplay of DNA supercoiling and DNA knotting is how the knotting equilibrium is affected by DNA supercoiling. To explain the concept of knotting equilibrium, let us first consider an imaginary situation, where torsionally relaxed, highly diluted circular DNA molecules of a given size undergo thermal fluctuations during which ideal type II DNA topoisomerases permit intersegmental passages whenever two DNA segments collide with each other. Such a reaction will reach knotting equilibrium when new rounds of passages will be as likely to form new DNA knots as to undo them, and as the reaction proceeds further from this point, the fraction of molecules that are knotted will remain fairly constant. Since DNA–DNA passages mediated by DNA topoisomerases are in reality non-random, the knotting equilibrium was usually achieved in experiments where linear DNA molecules were undergoing a slow cyclization reaction due to the annealing of their long cohesive ends. Upon such annealing, some molecules become knotted and the fraction of knotted molecules is believed to be the same as that obtained at knotting equilibrium due to free passages (35,36). Several biochemical and numerical studies confirmed the intuitive idea that the fraction of knots at knotting equilibrium increases with the chain size of DNA molecules and decreases with increasing electrostatic repulsion between DNA segments, which in turn depends on the counterions’ concentration (35–37). Let us now consider, how the knotting equilibrium changes when the studied DNA is actively maintained in a supercoiled form, as it is the case in living bacterial cells. We consider a hypothetical, idealized situation where bacterial Topo IV performs intersegmental passages that may lead to knotting or unknotting whenever two DNA segments collide with each other (in reality, as discussed below, the passages mediated by Topo IV are selective and depend on the spatial arrangement of the DNA segments). In addition, we consider that DNA gyrase and Topo I compensate for the possible decrease or increase of DNA torsional stress due to knot formation by re-establishing the initial level of torsional stress typical for native DNA supercoiling. Will the steady-state knotting equilibrium under such conditions result in a higher or lower fraction of knotted DNA molecules than at the knotting equilibrium involving torsionally relaxed DNA molecules?.
Studies of in vivo formed knots in Escherichia coli revealed that pBR322 plasmids grown in E. coli strains with defective DNA gyrase were up to 10 times more frequently knotted than plasmids grown in wild-type strains (38). Since DNA gyrase is responsible for maintaining the DNA in a supercoiled form and is not the unknotting topoisomerase per se, as Topo IV is (39), this result provides a strong indication that DNA supercoiling is helping Topo IV to keep the DNA unknotted in living bacterial cells. The effect of supercoiling on unknotting was also investigated in vitro by comparing unknotting of supercoiled and non-supercoiled DNA molecules by Topo IV (39), i.e. the enzyme that is believed to be responsible for DNA unknotting and decatenation of DNA in E. coli. The authors of that study arrived to the conclusion that DNA supercoiling does not appreciably increase the unknotting rate by Topo IV (39). However, a closer look at the presented data clearly reveals that, while complex DNA knots seemed to be equally quickly transformed into simpler knots by Topo IV irrespectively whether they were supercoiled or not, the unknotting of less complex knots, as trefoil knots, was clearly stimulated by DNA supercoiling, while trefoil knots were accumulating when Topo IV was acting on a mixture of complex DNA knots that were not supercoiled [see Figure 7 in ref. (39)]. A likely explanation of this behaviour is that the presence of simple knots in non-supercoiled DNA molecules does not produce a sufficient energy gradient that could lead to unknotting, and therefore, this gradient needs to be enhanced by DNA supercoiling. For complex knots, however, the energy gain due to a strand passage leading to knots’ simplification is sufficiently large to direct these passages even in the absence of DNA supercoiling.
Energetic components of the interplay between DNA knotting and supercoiling
As discussed above, in vivo and in vitro studies support the notion that DNA supercoiling inhibits DNA knotting. However, this inhibition may not be related to a change of the free energy gradient introduced by DNA supercoiling, but rather be the result of ATP consuming Topo IV action that does not need to follow the free energy gradient of DNA molecules. More precisely, DNA supercoiling could help Topo IV to choose the passages leading to unknotting even if these were not following the free energy gradient of the system. Therefore, the studies discussed above do not really tell us whether DNA supercoiling changes the energy gradient in a way that disfavours DNA knotting. However, simulation studies, where one can control the energy and the topology of the modelled DNA molecules, and where one can also impose the condition that passages occur irrespectively of a particular spatial arrangement of colliding segments, could give us this information.
Intriguingly, two different simulation studies investigating the effect of DNA supercoiling on DNA knotting arrived at the opposite conclusions. While Podtelezhnikov et al. (11) concluded that DNA supercoiling stimulated DNA knotting, Burnier et al. (40) concluded the contrary. Let us investigate the reasons of that discrepancy. Both studies used a very similar Metropolis–Monte Carlo simulation method that accounts for DNA elastic properties and DNA effective diameter (4). During the Metropolis–Monte Carlo simulation procedure, the modelled molecules constantly change their shape and explore the available configuration space. The latter can be seen as the ensemble of all possible configurations of the elastic chain under the influence of thermal agitation, and of course, in that ensemble, energetically favourable configurations will appear more often. Therefore, if knotting of supercoiled molecules would be energetically favourable, one should observe a strong tendency of supercoiled DNA molecules to become knotted. Thermal motion by itself cannot pass two DNA segments through each other. Therefore, to account for the action of DNA topoisomerases, during the simulation one has to permit segment moves leading to strand passages, and thus possibly to knotting or unknotting. However, if intersegmental passages are permitted without any restriction regarding the topological state of the molecules, the simulated supercoiled DNA molecules quickly relax. This invalidates the simulations aiming to test the effect of maintaining the DNA in supercoiled form on the tendency of DNA molecules to become knotted or unknotted.
Several earlier simulation studies showed that, upon setting a fixed ΔLk (the difference between the actual Lk of the modelled molecules and their Lk0), and upon permitting intersegmental passages that did not result in formation of knots, the simulated supercoiled DNA molecules maintained their original supercoiling and even sampled in a more efficient way the configuration space as compared to simulations in which intersegmental passages were not allowed (4). For example, with this technique, simulated configurations of supercoiled DNA molecules looked like those observed by electron microscopy (41). Podtelezhnikov et al. (42), in their simulations investigating the interplay between DNA supercoiling and DNA knotting, permitted also these moves that resulted in passages from unknots to knots and vice versa while keeping the ΔLk fixed. With that setting, the simulated supercoiled DNA molecules were becoming progressively knotted, forming left-handed torus knots with increasing complexity. On this basis, the authors concluded that DNA molecules that keep the ΔLk typical for negatively supercoiled DNA molecules would have the tendency to undergo passages leading to formation of left-handed torus knots.
Indeed that would have been the case if Topo II-mediated passages were not changing the linking number of DNA molecules upon each intramolecular passage. We know however that topo II mediated passage from unknotted DNA molecules to left-handed trefoil knots necessitate the decrease of the linking number by 2. So for example, if one starts with a supercoiled molecule with ΔLk = −5 and would perform a necessary passage leading to formation of a left-handed trefoil knot, the ΔLk of that molecule would change to −7 (Figure 3A). Consequently, a real passage would be energetically not favourable as opposed to an imaginary topo II-mediated passage with unrealistic property of maintaining the ΔLk constant (Figure 3A). In addition, even if the realistic passage has happened, then afterwards DNA gyrase would act to re-establish the previous level of torsional stress, which would push the molecule to the state with the ΔLk oscillating ∼−8.41. (Figure 3A) This is the consequence of the formula: Lk = Tw + Wr and the fact that the Wr of a torsionally relaxed left-handed trefoil knot is of ∼−3.41 (43). Therefore, the Lk0 in torsionally relaxed left-handed trefoil knots is smaller than in torsionally relaxed unknotted DNA molecules. To account for the change of Lk0 due to knotting, Burnier et al. (40) introduced the concept of the effective ΔLk (ΔLke), which is the linking number difference between the Lk0 of a torsionally relaxed knot of a given type including unknot and the actual Lk of the modelled DNA forming the same knot type. When modelled molecules are permitted to change the knot type but keep the same ΔLke, their torsional tension is roughly conserved. Simulations performed under such conditions revealed that supercoiling directs intersegmental passages toward unknotting since the lowest energy-state of DNA molecules with a given ΔLke is attained for unknotted DNA molecules, and this is independent of whether the formed knots are left- or right-handed (40). However, it needs to be stated here that simulations performed under conditions maintaining the same ΔLke before and after passages leading to knot formation are based on the unrealistic assumption that the torsional tension is re-established instantly after the passage, while in reality it will take some time until the homeostatic mechanisms controlling the level of torsional stress will re-establish the original level of torsional stress (27). The second assumption is that topoisomerase-mediated passages are driven by the free energy gradient, while in reality type II DNA topoisomerases couple the energy of ATP hydrolysis to the performed passage and can act against the free energy gradient (26). However, these assumptions are needed to evaluate the importance of energetic considerations in the study of the steady-state knotting equilibrium in the presence of homeostatic mechanisms that maintain a quasi-constant level of DNA torsional stress in supercoiled DNA.
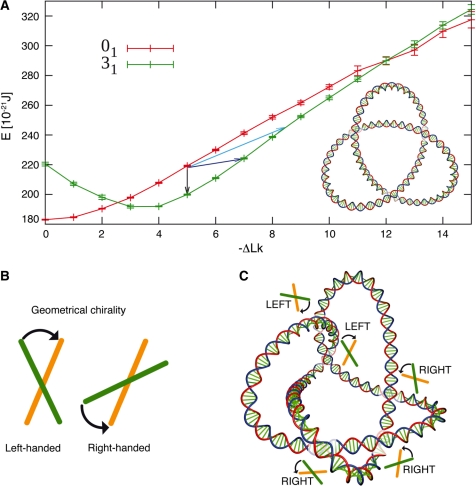
Figure 3.
DNA knots, supercoiling and the geometric chirality. (A) Comparison of the elastic energies of simulated negatively supercoiled DNA molecules with 3000 bp that were either unknotted or formed left-handed trefoil knots [the energy graph is reproduced from ref. (40)]. ΔLk refers to the difference between the actual linking number of knotted or unknotted DNA molecules and that of torsionally relaxed unknotted DNA molecules. The light blue arrow indicates the energy difference between unknotted and knotted DNA molecules with the same ΔLke. The dark blue arrow indicates the energy difference between unknotted and knotted DNA molecules resulting from one round of a Topo II-like action. The black arrow indicates the energy difference between unknotted and knotted DNA molecules under the unrealistic assumption that topoisomerase II could mediate an intramolecular passage reaction without changing the linking number. Note that left-handed trefoil knots reach their minimal energy state for ΔLk ≈3.5 and that this closely corresponds to the average writhe of torsionally relaxed left-handed trefoil knots (43) (shown in the inset). For the convenience of the presentation the figure shows three different energetic consequences of formation of left-handed trefoil knot starting from unknotted DNA molecules that were kept at their supercoiling equilibrium at ΔLk ≈ −5. However, natural supercoiling of 3000 bp-long DNA would rather keep the unknotted molecules at ΔLk ≈−15. Therefore, at physiological level of negative supercoiling the passages leading to knot formation will be much stronger opposed by the free energy gradient than for the case analysed here. (B) The concept of geometrical chirality. To determine the geometrical chirality of a crossing one looks what is the smallest rotation of the overlying segment that brings it parallel with the underlying one. If that rotation is clockwise the crossing is left-handed and it is right-handed otherwise. (C) Knot 52L with indicated geometrical chirality of their crossings. An intersegmental passage at any of left-handed crossing will unknot the knot, while a passage at any of right-handed crossings will convert the 52L knot into 31L knot.
Geometric components of the interplay between DNA supercoiling and DNA knotting
The free energy gradient provided by DNA supercoiling helps to guide DNA topoisomerases towards efficient unknotting, but this is not sufficient to explain the complexity of the interplay between DNA supercoiling, knotting and catenation. If DNA topoisomerases were simply catalysing reactions along the free energy gradient, then supercoiled DNA molecules in living bacterial cells would be subject to topoisomerase-mediated reactions leading to their rapid torsional relaxation. So for example, bacterial type I DNA topoisomerases, which perform passages of one strand through the other, would relax the torsional stress in supercoiled DNA. However, as the negative torsional stress is required for normal functioning of bacterial DNA, a metabolic short circuit would appear in this case, with type I topoisomerases constantly relaxing negatively supercoiled DNA molecules and with many molecules of DNA gyrase using ATP to re-establish the original level of negative supercoiling. Such a metabolic short-circuit would exhaust the cellular pool of ATP, and is in reality avoided trough molecular mechanisms that evolved to ensure that bacterial type I topoisomerases are only activated by an excessive torsional stress of the DNA or by the appearance of atypical DNA structures (27). The excessive torsional stress can be directly sensed at the local DNA level by the facility of strand separation, for example, and this provides the molecular basis of the mechanism protecting normally supercoiled DNA from the action of type I topoisomerases (27).
However, it seems more difficult to imagine molecular mechanisms that make naturally supercoiled DNA very good substrate for Topo IV-mediated passages leading to decatenation and unknotting but not to relaxation of negative supercoils (39). Since DNA topoisomerases are relatively small compared to overall dimensions of plasmid or chromosomal DNA, they can only distinguish between some local features that are characteristic for unperturbed negatively supercoiled DNA and these that appear in DNA molecules that are knotted and catenated. If there are such structural differences, then type II DNA topoisomerases could have acquired during the evolution the capacity to act preferentially on the DNA–DNA juxtapositions arising due to knotting or catenation, but not on those arising due to negative supercoiling.
Single molecule studies investigating the action of bacterial topoisomerase IV on braided DNA molecules revealed that it acts preferentially on left-handed braids having a local segment juxtaposition geometry similar to that of positively supercoiled DNA, while it leaves right-handed braids, comparable to negative supercoils, practically untouched (see Figure 3 for the explanation of the geometrical chirality of DNA–DNA juxtapositions) (44–46). These observations made a perfect sense as they explained why DNA molecules that are negatively supercoiled are practically immune to Topo IV relaxation, while positive supercoiling that arises during DNA replication can be efficiently relaxed by Topo IV, permitting further progression of the replication forks (47). Comparing DNA–DNA juxtapositions in positively- and negatively-supercoiled DNA, one notices that both types have a hooked character (48) but can be distinguished by the opposite geometric chirality of winding of juxtaposing segments (45,46).
It is important to mention here the difference between topological and geometric chirality. To determine the topological chirality one needs to operate with the concept of oriented curves and be aware in which direction the imaginary orientation vectors are pointing when two segments of DNA contact each other. Clearly, enzymes cannot recognize imaginary vectors. However, enzymes can recognize geometrical chirality of crossings if their DNA binding sites are arranged in such a way that, when the first DNA molecule is bound, the second can be bound on top of it but only at a certain relative inclination angle with respect to the already bound DNA segment. Depending on that angle, the enzyme may preferentially interact with DNA segments that wind around each other in left- or right-handed way (49,50). In negatively supercoiled DNA the topological sign of perceived intramolecular crossings is negative, however, the opposing DNA segments of negatively supercoiled DNA molecules wind around each other in a right-handed way. The left-handed direction of winding is then characteristic for the juxtapositions in positively supercoiled DNA molecules (Figure 4) and those are preferentially recognized by Topo IV.
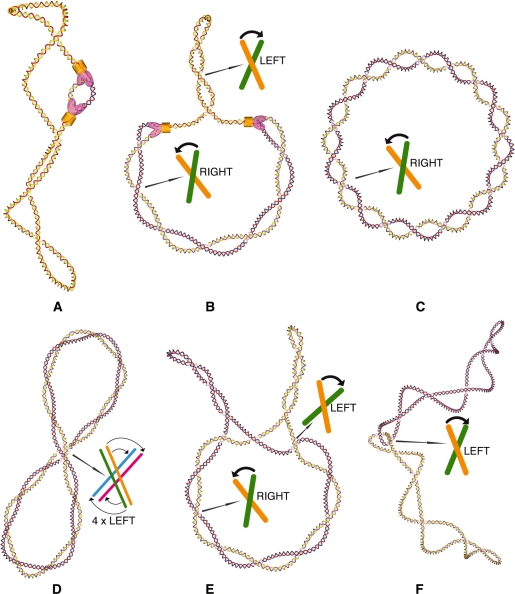
Figure 4.
Topological transitions during replication of circular DNA. (A) Supercoiled DNA that just started its process of DNA replication. The molecule is still negatively supercoiled and shows right-handed interwinding. The negative supercoiling helps to initiate the process of DNA replication (7). (B) Partially replicated DNA molecule with positive torsional stress causing formation of left-handed interwinding in the unreplicated portion and of right-handed interwinding of precatenanes in the replicated portion. The left-handed interwinding can be easily relaxed by DNA gyrase or by topoisomerase IV. Right-handed interwinding is a poor substrate for the relaxation reaction by Topo IV. (C) Standard representation of freshly replicated molecules forming multiply interlinked DNA catenanes. All crossings in this representation have right-handed chirality but this representation is not the equilibrium form of multiply interlinked DNA catenanes. (D) Schematic presentation of the equilibrium form of multiply interlinked DNA catenanes. Minimization of the elastic energy of multiply interlinked catenanes leads to the formation of a left-handed folding of the entire catenanes (21,78). This folding leads to the apparition of left-handed DNA–DNA juxtapositions that are very good substrates for Topo IV-mediated passages that lead to decrease of DNA catenation. (E) Catenanes with decreasing number of interlinks. Individual rings get supercoiled by DNA gyrase, and left-handed crossings may form in the region deformed by the extrusion of supercoils. (F) Singly interlinked catenanes have a complete freedom to form left-handed juxtapositions. Supercoiling provides the necessary ‘pressure’ leading to unlinking. The representations of DNA molecules at different steps of DNA replication are not shown at the same scale but their size was chosen in order to make important points clear. However, all the drawings correspond to sequential stages of DNA replication of a molecule with 800 bp.
The geometry of positively supercoiled DNA favours not only left-handed crossings of the formed DNA–DNA juxtapositions but also hooking of contacting DNA segments. Interestingly, the preferential action of type II DNA topoisomerases on hooked juxtapositions was also proposed as the mechanism that explained the seminal experiment by Rybenkov et al. (30,51). This experiment demonstrated that type II DNA topoisomerases acting in vivo on torsionally relaxed DNA keep the level of DNA knotting and catenation much below the topological equilibrium that would have arisen if the passages were simply driven by the free energy gradient (30). Type II DNA topoisomerases couple their DNA–DNA passage reaction to ATP hydrolysis and therefore there is no violation of thermodynamic principles connected to the fact that action of type II DNA topoisomerases on a statistical set of plasmid DNA molecules can push this set out of its lowest free-energy state. However, it was very intriguing how DNA topoisomerases, that can only sense local information, may get hints whether performing a DNA–DNA passage in a given place will rather unknot the DNA than lead to DNA knotting.
Several complex mechanisms were proposed over the years to explain how DNA topoisomerases could get these hints: the active sliding model (30), the kinetic proofreading (52) or the three-site interaction (53). While some of these models were incompatible with the known properties of type II topoisomerases (active sliding model), the other models seemed to be unnecessarily complex (kinetic proofreading, three site interaction). In 2002 Vologodskii et al. (51) proposed a model according to which type II DNA topoisomerases were actively bending the DNA in the so called G region (G stands for the gate as that DNA region will be later cut and will serve as a gate for the passage of transferred segment known as T segment). According to that model, the DNA topoisomerase was placing itself in the bend in such a way that the T segment could only be transferred if it approached the G region from the inside of the bend. As a consequence, the topoisomerase was providing directionality for the passage of the T segment from the inside to the outside of the strongly bent DNA loop with the G site in its centre. Simulations performed to test this model revealed that this relatively simple mechanism permits a 10-fold reduction of the knotting level below the topological equilibrium (51). However, biochemical experiments using Topo IV from E. coli showed ∼50-fold reduction of the knotting level below the topological equilibrium and therefore the mechanism proposed by Vologodskii et al. was not sufficient to explain the observed effect.
In 2004, a theoretical paper by Buck and Zechiedrich proposed that type II topoisomerases can keep a very low steady-state level of DNA knotting and of DNA catenation by preferentially acting on inter-hooked juxtapositions and performing passages there (48,54). The idea behind that model was that, in freely fluctuating, torsionally relaxed DNA, the interhooked juxtapositions form preferentially in the region where DNA is knotted or catenated (48,54). For entropic reasons the knotted domains in freely fluctuating knotted polymers tend to be rather tight (55,56) and this favours that hooked juxtapositions form there. Indeed, simulation studies using polygons confined to a cubic lattice confirmed that preferential action on hooked juxtapositions can decrease the level of knotting by ∼50 times as compared to random passages (57). Also, simulations performed using polygonal chains off-lattice revealed that selection of interhooked juxtapositions can result in a reduction of the knotting level as strong as that observed experimentally (58).
The hooked juxtapositions proposed by Buck and Zechiedrich (48) and tested in simulations by Liu et al. (57,59) were not assumed to wind in a right- or left-handed way but were modelled as achiral, interhooked juxtapositions where the planes of hooking were perpendicular to each other. Such achiral hooked juxtapositions were sufficient to explain the experiment by Rybenkov et al. (30) where the tested DNA was not supercoiled. However, even for unsupercoiled DNA there were other experiments requiring a chirality criteria to explain the observed results. Mann et al. (60) reported that human topoisomerase IIα has the very interesting ability to unknot left-handed five crossing twist knots (with the topological notation 52L) in just one passage. These results were puzzling since, even if DNA topoisomerases could specifically recognize hooked juxtapositions resulting from tight knotting of 52 knot, there are possibilities of forming five such juxtapositions and only passages occurring at two of them could lead to a direct conversion of the 52 knot into an unknot (61). The question arose then what structural difference could exist between DNA–DNA juxtapositions at these two different types of crossings in freely fluctuating DNA forming 52L knots?
Burnier et al. (58) used numerical simulations to test whether, by recognizing and performing passages within inter-hooked juxtapositions in which opposing segments wind around each other in a left-handed way (Figure 3), the topoisomerases could unknot 52L knots by performing just one passage. The simulations showed that this criteria was indeed very efficient in favouring unknotting of the 52L knot by a single topo II-mediated passage (58). In addition, the preferential action on inter-hooked juxtapositions with left-handed character explains also the mechanism by which human topoisomerase IIα relaxes positively supercoiled DNA molecules at least 10 times faster than negatively supercoiled ones (62).
Additional support for the hooking model was provided recently by crystallographic analysis of Topo IV DNA complexes (63). In these complexes, the DNA to which the topoisomerase is bound (G segment) is strongly bent in the direction consistent with the models proposing that Topo IV induces DNA bends (51,64) or that it preferably binds hooked juxtapositions (48,54). The observation mentioned earlier that plasmids isolated from bacterial strains with defective gyrase are frequently knotted (38) is consistent with the interpretation that DNA supercoiling may help Topo IV to distinguish better DNA juxtapositions that are due to knotting from other DNA juxtapositions.
As mentioned earlier, bacterial topoisomerases evolved in such a way that negatively supercoiled DNA molecules are protected against their relaxing activity. In the case of Topo IV, this protection is ensured by the chirality sensing mechanism explained above (44–46): right-handed juxtaposition present in negatively supercoiled DNA are practically immune to its action, whereas left-handed juxtapositions that are predominant in positively supercoiled DNA are very efficiently recognized by Topo IV and are substrate of processive passage reactions (44–46,65). Presumably, when knots are present in negatively supercoiled DNA molecules they promote formation of DNA–DNA juxtapositions with geometries that are somewhat different than the typical geometry of DNA–DNA juxtapositions in negatively supercoiled DNA. This is certainly the case of DNA molecules forming left-handed trefoil knots whose crossings have a natural tendency to form left-handed dsDNA–dsDNA juxtapositions. However, also DNA molecules forming right-handed trefoil knots, despite their preference to form right-handed dsDNA–dsDNA crossings, may in fact promote formation of juxtapositions that are more hooked or have somewhat different inclination angle than these normally present in negatively supercoiled DNA. In such a case also these knots may be efficiently unknotted by Topo IV action while regular DNA–DNA juxtapositions due to negative supercoiling would not serve as substrates of Topo IV.
DNA CATENATION AND SUPERCOILING
Formation of DNA precatenanes: gyrase paradox
During DNA replication, complementary strands of parental molecules become progressively separated, and this necessitates gradual untwisting of the double helix. In the case of covalently closed circular DNA, the two strands are topologically linked, and the action of DNA topoisomerases is required to reduce the linking number to exactly zero, in order to complete the segregation of newly replicated DNA molecules. As already mentioned, the negative supercoiling facilitates initial separation of DNA strands. However, as the strand separation in negatively supercoiled DNA molecules progresses beyond the point of DNA relaxation (Figure 2), it induces positive torsional stress. The mechanical torsional stress in elastic DNA molecules tends to equilibrate between replicated and non-replicated portions of partially replicated DNA molecules (Figure 4B). In non-replicated portions, the stress causes formation of interwound positive superhelices, in which the opposing segments wind around each other in a left-handed sense. In replicated portions, however, the stress causes formation of precatenanes, in which the opposing segments wind around each other in a right-handed sense. It is quite contra-intuitive that the geometric direction of winding changes between replicated and non-replicated portions of the same molecule minimizing its elastic energy. This phenomenon is due to the fact that in the non-replicated portion, the DNA is folded back on itself, which changes the topological orientation of segments wrapping. If that DNA were not allowed to fold back on itself it would adopt the solenoidal form of supercoiling, and form a right-handed helix (14) just like in precatenane regions of partially replicated DNA molecules (47).
The positive torsional stress opposes DNA strand separation, and induces replication fork reversal (66). Therefore, that torsional stress would eventually stop the progression of the replication forks if it were not continuously relieved by the action DNA topoisomerases. In vivo and in vitro experiments provided the evidence that the torsional stress is released by the activity of DNA gyrase and Topo IV acting on positive superhelices formed in the unreplicated portion of the DNA (67). In principle, the torsional stress could be also relieved by Topo IV action involving passages between precatenated portions of freshly replicated DNA. However, as already mentioned, Topo IV hardly acts on DNA duplexes that wrap around each other in a right-handed sense as otherwise it would constantly relax negatively supercoiled DNA, in which the opposing segments also wind around each other in a right-handed sense. This is most likely the reason why right-handed interwinding of freshly replicated regions is relatively stable (68). We propose below that the right-handed interwinding of freshly replicated DNA regions can be crucial for saving cells from gyrase induced exhaustion of cellular ATP.
Replicated portions of partially replicated DNA molecules are torsionally relaxed since newly synthesized strands are not covalently-closed (Figure 4B). As already discussed, bacterial cells are equipped with very efficient homeostatic mechanisms that keep cellular DNA at a quasi-constant level of negative supercoiling (27). As a consequence, if the torsional tension of DNA diminishes, it is very efficiently re-established by the action of DNA gyrase. However, any action of DNA gyrase on partially replicated portions will be useless, as it will only drive non-productive rotation of the DNA while every catalytic cycle will use two molecules of ATP. As shown by Gore et al. (69) gyrase can act very efficiently on linear nicked DNA and such enzymatic reaction could go practically forever using all available ATP. In growing bacterial cells partially replicated DNA molecules are practically omnipresent and the length of replicated regions may easily exceed that of non-replicated regions. Therefore, some mechanisms had to evolve to protect cells from spending their ATP on non-productive DNA rotations in partially replicated portions of DNA molecules as even if individual gyrase molecules show a relatively low ATPase rate of 1/s (70), there are ∼500–1000 gyrase molecules per cell (71).
Already early papers studying DNA gyrase have established that ATPase activity of DNA gyrase is ∼4–7 times lower in the presence of negatively supercoiled DNA than in the presence of relaxed DNA (72,73). Those early observations make a good sense since gyrase should stop hydrolysing ATP when the DNA is already sufficiently supercoiled, as otherwise a lot of ATP would be continuously wasted. We discussed already that Topo IV can discriminate between DNA juxtapositions with different geometrical chirality. It seems logical to propose that a similar mechanism could also operate in the case of DNA gyrase. Such a chiral discrimination of DNA substrates by DNA gyrase could protect the partially replicated portions of DNA from unproductive action of DNA gyrase. Since partially replicated portions of the DNA are naturally kept in the form of right-handed precatenanes by the positive torsional stress induced by progressing replication forks, this makes them looking like negative supercoils. Protection of partially replicated portions of DNA from action of DNA gyrase would make a good sense since it would importantly improve the efficiency of cell energetics. However, the proposal that the DNA that is strongly interwound in the right-handed sense is protected against further action of DNA gyrase should reconcile early reports that ATPase activity of DNA gyrase is partially inhibited by DNA supercoiling (72,73) with newer in vitro experiments showing that usage of ATP does not stop after gyrase has supercoiled the DNA to its thermodynamic limits (28).
The resolution of this apparent paradox may be provided by the fact that in vivo negative supercoiling promotes DNA interaction with bacterial structural proteins such as FIS and HU proteins that constrain negative supercoiling and form bacterial chromatin (74–76). These proteins are capable of wrapping the DNA into toroidal coils (74). Since binding of these proteins to DNA is favoured by negative supercoiling (76,77) and there is enough of these proteins to cover the DNA quite densely, it is reasonable to propose that upon reaching the physiological level of DNA supercoiling these proteins may displace DNA gyrase from the DNA and this will stop the ATPase activity of DNA gyrase in vivo. These proteins were not present in the in vitro experiments discussed above (28). Since it is most likely the right-handed wrapping of DNA that promotes the binding of HU proteins onto supercoiled DNA (77), then also right-handed precatenanes composed of torsionally relaxed DNA could enjoy HU protection against futile action of DNA gyrase.
Formation of DNA catenanes: Topo IV paradox
As the replication of circular DNA is nearly finished and the two replication forks converge together, less and less space remains for DNA topoisomerases to act on the unreplicated portion of the DNA. Replication forks are complex protein conglomerates where DNA helicases, single-strand binding proteins and other proteins interact with the DNA ahead of DNA polymerases. Therefore, as soon as the distance between the two approaching forks becomes smaller than few hundreds base pairs, it becomes very difficult for DNA gyrase or any other topoisomerase, to gain the access to the remaining unreplicated portion of the DNA and to entirely remove the linking of parental strands before their complete replication (Figure 4). As a consequence, in case of plasmid DNA molecules, for example, one observes formation of right-handed DNA catenanes, where the freshly replicated DNA molecules are wound around each other up to 40 times (21). That high interlinking number results from the fact that the number of turns of pre-existing precatenanes simply adds to the linking number of the terminal region where the replication proceeded without unlinking of DNA strands.
Several studies have shown that it is the bacterial Topo IV that is responsible for rapid and complete decatenation of freshly replicated sister duplexes (67). This decatenation is a vital process and any dysfunction of it is lethal. How then could Topo IV decatenate so efficiently right-handed catenanes that seem to have an overall structure very similar to negatively supercoiled DNA, which is hardly a substrate for Topo IV action? This problem is known in the literature as Topo IV paradox (45). We will try to argument here that it is DNA supercoiling (or rather higher order DNA coiling, in general) that provides crucial elements of solution of topo IV paradox. We believe that there are two different elements to the solution of Topo IV paradox: one of them works at early stages of DNA decatenation, and the second at late stages of DNA decatenation.
Early stages of DNA decatenation: second-order supercoiling of torus type catenanes
Just after the replication of circular DNA molecules is finished, the formed catenanes have a very high interlinking number that may exceed 40 (21). Two independent simulation studies investigated the structure of highly interlinked catenanes (21,78). It was observed that despite the fact that the composing rings were torsionally relaxed, the catenanes adopted supercoiled form in which the imaginary torus around which the two molecules wind is folded on itself in a left-handed way (Figure 4D). This left-handed higher-order winding permits the involved molecules to decrease their bending energy (21,78,79). As a consequence of the left-handed, higher order winding of the entire catenanes, the DNA segments contacting each other along the contact line of the interwound torus form left-handed juxtapositions (Figure 4D). Left-handed DNA–DNA juxtapositions have suitable-geometry for efficient action of Topo IV that could decrease the interlinking number between the two catenated DNA molecules. In fact, single molecule studies using braided DNA with increasing extent of braiding, established that right-handed braids were essentially immune to Topo IV action until the point where the braid folded on itself forming left-handed ‘super braid’ (45,46). From this point on, Topo IV-mediated passages were easily occurring in the folded braid and were reducing the interlinking of the braided duplexes. It should be clear that the structure of catenanes with high interlinking number is analogous to that described by Stone et al. (46) and Charvin et al. (45), and that Topo IV-mediated passages occurring in this structure will lead to partial decatenation of freshly replicated DNA molecules.
Late stages of DNA decatenation
As the decatenation progresses, the mechanical tension within the torus type catenanes decreases progressively, eliminating the second-order supercoils together with the left-handed juxtapositions that could be used by Topo IV for the decatenation (44–46). However, as the individual molecules become less constrained, they acquire an increasing level of DNA supercoiling due to the action of DNA gyrase, and this leads to an overall change of the structure of the catenanes (21). The region of mutual interlinking of catenated molecules shrinks and the individual molecules starts to wind on themselves forming typical interwound supercoils (Figure 4E). The regions where the interwound superhelices extrude from the axis of the torus show high bending and can possibly form inter-molecular juxtapositions with the geometry suitable for Topo IV recognition and the ensuing passage (G. Witz, in preparation). Also Stone et al. (46) proposed that in right-handed catenanes with small number of interlinks the left-handed juxtapositions can form, from time to time, by loose portions of the DNA. This would be possible if the weakly catenated molecules were less constrained and thus had enough freedom to contact each other in a way permitting formation of left-handed juxtapositions. However, the proposal of Stone et al. (46) neglected the fact that catenated molecules become supercoiled before they get completely decatenated, which effectively eliminates loose portions in the DNA catenanes.
We discussed above that DNA supercoiling favours DNA decatenation, by facilitating formation of Topo IV-recognizable DNA–DNA juxtapositions. But as in the case of knots, we can also consider the energetic influence of supercoiling on the equilibrium catenation level. If decatenation were proceeding without the concomitant supercoiling of the catenanes, the energetic tendency of molecules to separate would progressively decrease because the energy gradient would become flatter. However, this is not the case when the component molecules become progressively supercoiled as the decatenation proceeds (80). Under such conditions, supercoiling progressively replaces catenation to maintain a steep gradient creating a ‘pressure’ pushing towards complete decatenation (21,80).
CONCLUSIONS
We discussed the fact that the free energy of DNA supercoiling favours topoisomerase-mediated unknotting and decatenation reactions. We also discussed how DNA supercoiling can change the structure of knotted and catenated DNA molecules so that Topo IV could specifically unknot and decatenate supercoiled DNA molecules without inducing relaxation of negatively-supercoiled DNA molecules. The realization that DNA supercoiling can play an active role in the process of DNA decatenation and unknotting is relatively new and more research is needed to understand better all mechanisms implicated in very efficient decatenation of freshly replicated bacterial chromosomes and bacterial plasmids.
Open questions
There are many open problems concerning the interplay of DNA supercoiling with unknotting and decatenation. There is for example still no numerical study simulating the realistic case where the torsional level of knots or catenanes is maintained around a steady-state. For the moment, only the approximate solutions presented above exist. Also, one interesting question is why DNA gyrase uses ATP to relax positive supercoils that arise during ongoing DNA replication. Relaxation of positively supercoiled DNA is a downhill reaction and it could go without ATP hydrolysis. Millions of ATP molecules are needed for relaxation of chromosomal DNA during each replication round. Therefore, it seems unreasonable that during evolution this energetic wastefulness was not eliminated. Interestingly, early studies of bacterial topoisomerases revealed that one of the subunits of gyrase easily undergoes a proteolytic cleavage in vivo and the resulting enzyme can then relax positively supercoiled DNA without using ATP (81). Could it be that this cleavage variant of DNA gyrase indeed participates in the relaxation of positive supercoils generated during DNA replication, as it was originally proposed by Brown et al. (81)?
FUNDING
Swiss National Science Foundation grant (3100A0-116275 to A.S.).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Giovanni Dietler for many helpful discussions on DNA topology. All DNA graphics were realized with the software Blender, freely available at www.blender.org.
REFERENCES
- 1.Du Q, Kotlyar A, Vologodskii A. Kinking the double helix by bending deformation. Nucleic Acids Res. 2008;36:1120–1128. doi: 10.1093/nar/gkm1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cloutier TE, Widom J. Spontaneous sharp bending of double-stranded DNA. Mol. Cell. 2004;14:355–362. doi: 10.1016/s1097-2765(04)00210-2. [DOI] [PubMed] [Google Scholar]
- 3.Demurtas D, Amzallag A, Rawdon EJ, Maddocks JH, Dubochet J, Stasiak A. Bending modes of DNA directly addressed by cryo-electron microscopy of DNA minicircles. Nucleic Acids Res. 2009;37:2882–2893. doi: 10.1093/nar/gkp137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vologodskii AV, Levene SD, Klenin KV, Frank-Kamenetskii M, Cozzarelli NR. Conformational and thermodynamic properties of supercoiled DNA. J. Mol. Biol. 1992;227:1224–1243. doi: 10.1016/0022-2836(92)90533-p. [DOI] [PubMed] [Google Scholar]
- 5.Strick TR, Allemand JF, Bensimon D, Bensimon A, Croquette V. The elasticity of a single supercoiled DNA molecule. Science. 1996;271:1835–1837. doi: 10.1126/science.271.5257.1835. [DOI] [PubMed] [Google Scholar]
- 6.Perugino G, Valenti A, D'A;maro A, Rossi M, Ciaramella M. Reverse gyrase and genome stability in hyperthermophilic organisms. Biochem. Soc. Trans. 2009;37:69–73. doi: 10.1042/BST0370069. [DOI] [PubMed] [Google Scholar]
- 7.Marians KJ, Ikeda JE, Schlagman S, Hurwitz J. Role of DNA gyrase in phiX replicative-form replication in vitro. Proc. Natl Acad. Sci. USA. 1977;74:1965–1968. doi: 10.1073/pnas.74.5.1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bates AD, Maxwell A. DNA Topology. Oxford: Oxford University Press; 2005. [Google Scholar]
- 9.Nollmann M, Crisona NJ, Arimondo PB. Thirty years of Escherichia coli DNA gyrase: from in vivo function to single-molecule mechanism. Biochimie. 2007;89:490–499. doi: 10.1016/j.biochi.2007.02.012. [DOI] [PubMed] [Google Scholar]
- 10.Kasamatsu H, Vinograd J. Replication of circular DNA in eukaryotic cells. Annu. Rev. Biochem. 1974;43:695–719. doi: 10.1146/annurev.bi.43.070174.003403. [DOI] [PubMed] [Google Scholar]
- 11.Gellert M, Mizuuchi K, O'D;ea MH, Nash HA. DNA gyrase: an enzyme that introduces superhelical turns into DNA. Proc. Natl Acad. Sci. USA. 1976;73:3872–3876. doi: 10.1073/pnas.73.11.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fuller FB. The writhing number of a space curve. Proc. Natl Acad. Sci. USA. 1971;68:815–819. doi: 10.1073/pnas.68.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Călugăreanu G. Sur les classes d'i;sotopie des noeuds tridimensionnels et leurs invariants. Czech. Math. J. 1959;11:588–625. [Google Scholar]
- 14.Bauer WR, Crick FH, White JH. Supercoiled DNA. Sci. Am. 1980;243:100–113. [PubMed] [Google Scholar]
- 15.Peck LJ, Wang JC. Sequence dependence of the helical repeat of DNA in solution. Nature. 1981;292:375–378. doi: 10.1038/292375a0. [DOI] [PubMed] [Google Scholar]
- 16.Depew DE, Wang JC. Conformational fluctuations of DNA helix. Proc. Natl Acad. Sci. USA. 1975;72:4275–4279. doi: 10.1073/pnas.72.11.4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bauer WR. Structure and reactions of closed duplex DNA. Annu. Rev. Biophys. Bioeng. 1978;7:287–313. doi: 10.1146/annurev.bb.07.060178.001443. [DOI] [PubMed] [Google Scholar]
- 18.Dubochet J, Bednar J, Furrer P, Stasiak AZ, Stasiak A, Bolshoy AA. Determination of the DNA helical repeat by cryo-electron microscopy. Nat. Struct. Biol. 1994;1:361–363. doi: 10.1038/nsb0694-361. [DOI] [PubMed] [Google Scholar]
- 19.Dean FB, Stasiak A, Koller T, Cozzarelli NR. Duplex DNA knots produced by Escherichia Coli Topoisomerase-I—structure and requirements for formation. J. Biol. Chem. 1985;260:4975–4983. [PubMed] [Google Scholar]
- 20.Sundin O, Varshavsky A. Terminal stages of SV40 DNA replication proceed via multiply intertwined catenated dimers. Cell. 1980;21:103–114. doi: 10.1016/0092-8674(80)90118-x. [DOI] [PubMed] [Google Scholar]
- 21.Martinez-Robles ML, Witz G, Hernandez P, Schvartzman JB, Stasiak A, Krimer DB. Interplay of DNA supercoiling and catenation during the segregation of sister duplexes. Nucleic Acids Res. 2009;37:2882–2893. doi: 10.1093/nar/gkp530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schoeffler AJ, Berger JM. DNA topoisomerases: harnessing and constraining energy to govern chromosome topology. Q. Rev. Biophys. 2008;41:41–101. doi: 10.1017/S003358350800468X. [DOI] [PubMed] [Google Scholar]
- 23.Collins TR, Hammes GG, Hsieh TS. Analysis of the eukaryotic topoisomerase II DNA gate: a single-molecule FRET and structural perspective. Nucleic Acids Res. 2009;37:712–720. doi: 10.1093/nar/gkn1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Morrison A, Cozzarelli NR. Contacts between DNA gyrase and its binding site on DNA: features of symmetry and asymmetry revealed by protection from nucleases. Proc. Natl Acad. Sci. USA. 1981;78:1416–1420. doi: 10.1073/pnas.78.3.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brown PO, Cozzarelli NR. A sign inversion mechanism for enzymatic supercoiling of DNA. Science. 1979;206:1081–1083. doi: 10.1126/science.227059. [DOI] [PubMed] [Google Scholar]
- 26.Bates AD, Maxwell A. Energy coupling in type II topoisomerases: why do they hydrolyze ATP? Biochemistry. 2007;46:7929–7941. doi: 10.1021/bi700789g. [DOI] [PubMed] [Google Scholar]
- 27.Drlica K. Control of bacterial DNA supercoiling. Mol. Microbiol. 1992;6:425–433. doi: 10.1111/j.1365-2958.1992.tb01486.x. [DOI] [PubMed] [Google Scholar]
- 28.Bates AD, O'D;ea MH, Gellert M. Energy coupling in Escherichia coli DNA gyrase: the relationship between nucleotide binding, strand passage, and DNA supercoiling. Biochemistry. 1996;35:1408–1416. doi: 10.1021/bi952433y. [DOI] [PubMed] [Google Scholar]
- 29.Roca J, Berger JM, Harrison SC, Wang JC. DNA transport by a type II topoisomerase: direct evidence for a two-gate mechanism. Proc. Natl Acad. Sci. USA. 1996;93:4057–4062. doi: 10.1073/pnas.93.9.4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rybenkov VV, Ullsperger C, Vologodskii AV, Cozzarelli NR. Simplification of DNA topology below equilibrium values by type II topoisomerases. Science. 1997;277:690–693. doi: 10.1126/science.277.5326.690. [DOI] [PubMed] [Google Scholar]
- 31.Zechiedrich EL, Khodursky AB, Bachellier S, Schneider R, Chen D, Lilley DM, Cozzarelli NR. Roles of topoisomerases in maintaining steady-state DNA supercoiling in Escherichia coli. J. Biol. Chem. 2000;275:8103–8113. doi: 10.1074/jbc.275.11.8103. [DOI] [PubMed] [Google Scholar]
- 32.Brown PO, Cozzarelli NR. Catenation and knotting of duplex DNA by type 1 topoisomerases: a mechanistic parallel with type 2 topoisomerases. Proc. Natl Acad. Sci. USA. 1981;78:843–847. doi: 10.1073/pnas.78.2.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hiasa H, DiGate RJ, Marians KJ. Decatenating activity of Escherichia coli DNA gyrase and topoisomerases I and III during oriC and pBR322 DNA replication in vitro. J. Biol. Chem. 1994;269:2093–2099. [PubMed] [Google Scholar]
- 34.DiGate RJ, Marians KJ. Identification of a potent decatenating enzyme from Escherichia coli. J. Biol. Chem. 1988;263:13366–13373. [PubMed] [Google Scholar]
- 35.Shaw SY, Wang JC. Knotting of a DNA chain during ring closure. Science. 1993;260:533–536. doi: 10.1126/science.8475384. [DOI] [PubMed] [Google Scholar]
- 36.Rybenkov VV, Cozzarelli NR, Vologodskii AV. Probability of DNA knotting and the effective diameter of the DNA double helix. Proc. Natl Acad. Sci. USA. 1993;90:5307–5311. doi: 10.1073/pnas.90.11.5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deguchi T, Tsurusaki K. Universality of random knotting. Phys. Rev. E. 1997;55:6245. [Google Scholar]
- 38.Shishido K, Komiyama N, Ikawa S. Increased production of a knotted form of plasmid pBR322 DNA in Escherichia coli DNA topoisomerase mutants. J. Mol. Biol. 1987;195:215–218. doi: 10.1016/0022-2836(87)90338-x. [DOI] [PubMed] [Google Scholar]
- 39.Ullsperger C, Cozzarelli NR. Contrasting enzymatic activities of topoisomerase IV and DNA gyrase from Escherichia coli. J. Biol. Chem. 1996;271:31549–31555. doi: 10.1074/jbc.271.49.31549. [DOI] [PubMed] [Google Scholar]
- 40.Burnier Y, Dorier J, Stasiak A. DNA supercoiling inhibits DNA knotting. Nucleic Acids Res. 2008;36:4956–4963. doi: 10.1093/nar/gkn467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Boles TC, White JH, Cozzarelli NR. Structure of plectonemically supercoiled DNA. J. Mol. Biol. 1990;213:931–951. doi: 10.1016/S0022-2836(05)80272-4. [DOI] [PubMed] [Google Scholar]
- 42.Podtelezhnikov AA, Cozzarelli NR, Vologodskii AV. Equilibrium distributions of topological states in circular DNA: interplay of supercoiling and knotting. Proc. Natl Acad. Sci. USA. 1999;96:12974–12979. doi: 10.1073/pnas.96.23.12974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Katritch V, Bednar J, Michoud D, Scharein RG, Dubochet J, Stasiak A. Geometry and physics of knots. Nature. 1996;384:142–145. doi: 10.1038/384122a0. [DOI] [PubMed] [Google Scholar]
- 44.Crisona NJ, Strick TR, Bensimon D, Croquette V, Cozzarelli NR. Preferential relaxation of positively supercoiled DNA by E. coli topoisomerase IV in single-molecule and ensemble measurements. Genes Dev. 2000;14:2881–2892. doi: 10.1101/gad.838900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Charvin G, Bensimon D, Croquette V. Single-molecule study of DNA unlinking by eukaryotic and prokaryotic type-II topoisomerases. Proc. Natl Acad. Sci. USA. 2003;100:9820–9825. doi: 10.1073/pnas.1631550100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stone MD, Bryant Z, Crisona NJ, Smith SB, Vologodskii A, Bustamante C, Cozzarelli NR. Chirality sensing by Escherichia coli topoisomerase IV and the mechanism of type II topoisomerases. Proc. Natl Acad. Sci. USA. 2003;100:8654–8659. doi: 10.1073/pnas.1133178100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schvartzman JB, Stasiak A. A topological view of the replicon. EMBO Rep. 2004;5:256–261. doi: 10.1038/sj.embor.7400101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Buck GR, Zechiedrich EL. DNA disentangling by type-2 topoisomerases. J. Mol. Biol. 2004;340:933–939. doi: 10.1016/j.jmb.2004.05.034. [DOI] [PubMed] [Google Scholar]
- 49.Corbett KD, Schoeffler AJ, Thomsen ND, Berger JM. The structural basis for substrate specificity in DNA topoisomerase IV. J. Mol. Biol. 2005;351:545–561. doi: 10.1016/j.jmb.2005.06.029. [DOI] [PubMed] [Google Scholar]
- 50.McClendon AK, Dickey JS, Osheroff N. Ability of viral topoisomerase II to discern the handedness of supercoiled DNA: bimodal recognition of DNA geometry by type II enzymes. Biochemistry. 2006;45:11674–11680. doi: 10.1021/bi0520838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vologodskii AV, Zhang W, Rybenkov VV, Podtelezhnikov AA, Subramanian D, Griffith JD, Cozzarelli NR. Mechanism of topology simplification by type II DNA topoisomerases. Proc. Natl Acad. Sci. USA. 2001;98:3045–3049. doi: 10.1073/pnas.061029098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yan J, Magnasco MO, Marko JF. A kinetic proofreading mechanism for disentanglement of DNA by topoisomerases. Nature. 1999;401:932–935. doi: 10.1038/44872. [DOI] [PubMed] [Google Scholar]
- 53.Trigueros S, Salceda J, Bermudez I, Fernandez X, Roca J. Asymmetric removal of supercoils suggests how topoisomerase II simplifies DNA topology. J. Mol. Biol. 2004;335:723–731. doi: 10.1016/j.jmb.2003.11.011. [DOI] [PubMed] [Google Scholar]
- 54.Liu Z, Deibler RW, Chan HS, Zechiedrich L. The why and how of DNA unlinking. Nucleic Acids Res. 2009;37:661–671. doi: 10.1093/nar/gkp041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Katritch V, Olson WK, Vologodskii A, Dubochet J, Stasiak A. Tightness of random knotting. Phys. Rev. E Stat. Phys. Plasmas Fluids Relat. Interdiscip. Topics. 2000;61:5545–5549. doi: 10.1103/physreve.61.5545. [DOI] [PubMed] [Google Scholar]
- 56.Ercolini E, Valle F, Adamcik J, Witz G, Metzler R, De Los Rios P, Roca J, Dietler G. Fractal dimension and localization of DNA knots. Phys. Rev. Lett. 2007;98:058102. doi: 10.1103/PhysRevLett.98.058102. [DOI] [PubMed] [Google Scholar]
- 57.Liu Z, Mann JK, Zechiedrich EL, Chan HS. Topological information embodied in local juxtaposition geometry provides a statistical mechanical basis for unknotting by type-2 DNA topoisomerases. J. Mol. Biol. 2006;361:268–285. doi: 10.1016/j.jmb.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 58.Burnier Y, Weber C, Flammini A, Stasiak A. Local selection rules that can determine specific pathways of DNA unknotting by type II DNA topoisomerases. Nucleic Acids Res. 2007;35:5223–5231. doi: 10.1093/nar/gkm532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Liu Z, Zechiedrich EL, Chan HS. Inferring global topology from local juxtaposition geometry: interlinking polymer rings and ramifications for topoisomerase action. Biophys. J. 2006;90:2344–2355. doi: 10.1529/biophysj.105.076778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mann JK, Deibler RW, Sumners DW, Zechiedrich EL. Unknotting by type II topoisomerases. Abstr. Papers Presented Am. Math. Soc. 2004;25 994; 992–187. [Google Scholar]
- 61.Flammini A, Maritan A, Stasiak A. Simulations of action of DNA topoisomerases to investigate boundaries and shapes of spaces of knots. Biophys. J. 2004;87:2968–2975. doi: 10.1529/biophysj.104.045864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.McClendon AK, Rodriguez AC, Osheroff N. Human topoisomerase IIalpha rapidly relaxes positively supercoiled DNA: implications for enzyme action ahead of replication forks. J. Biol. Chem. 2005;280:39337–39345. doi: 10.1074/jbc.M503320200. [DOI] [PubMed] [Google Scholar]
- 63.Dong KC, Berger JM. Structural basis for gate-DNA recognition and bending by type IIA topoisomerases. Nature. 2007;450:1201–1205. doi: 10.1038/nature06396. [DOI] [PubMed] [Google Scholar]
- 64.Vologodskii A. Theoretical models of DNA topology simplification by type IIA DNA topoisomerases. Nucleic Acids Res. 2009;37:3125–3133. doi: 10.1093/nar/gkp250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Neuman KC, Charvin G, Bensimon D, Croquette V. Mechanisms of chiral discrimination by topoisomerase IV. Proc. Natl Acad. Sci. USA. 2009;106:6986–6991. doi: 10.1073/pnas.0900574106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Fierro-Fernandez M, Hernandez P, Krimer DB, Stasiak A, Schvartzman JB. Topological locking restrains replication fork reversal. Proc. Natl Acad. Sci. USA. 2007;104:1500–1505. doi: 10.1073/pnas.0609204104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Espeli O, Marians KJ. Untangling intracellular DNA topology. Mol. Microbiol. 2004;52:925–931. doi: 10.1111/j.1365-2958.2004.04047.x. [DOI] [PubMed] [Google Scholar]
- 68.Wang X, Reyes-Lamothe R, Sherratt DJ. Modulation of Escherichia coli sister chromosome cohesion by topoisomerase IV. Genes Dev. 2008;22:2426–2433. doi: 10.1101/gad.487508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gore J, Bryant Z, Stone MD, Nollmann M, Cozzarelli NR, Bustamante C. Mechanochemical analysis of DNA gyrase using rotor bead tracking. Nature. 2006;439:100–104. doi: 10.1038/nature04319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Maxwell A, Gellert M. The DNA dependence of the ATPase activity of DNA gyrase. J. Biol. Chem. 1984;259:14472–14480. [PubMed] [Google Scholar]
- 71.Condemine G, Smith CL. Transcription regulates oxolinic acid-induced DNA gyrase cleavage at specific sites on the E. coli chromosome. Nucleic Acids Res. 1990;18:7389–7396. doi: 10.1093/nar/18.24.7389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Sugino A, Cozzarelli NR. The intrinsic ATPase of DNA gyrase. J. Biol. Chem. 1980;255:6299–6306. [PubMed] [Google Scholar]
- 73.Mizuuchi K, O'D;ea MH, Gellert M. DNA gyrase: subunit structure and ATPase activity of the purified enzyme. Proc. Natl Acad. Sci. USA. 1978;75:5960–5963. doi: 10.1073/pnas.75.12.5960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Drlica K, Rouviere-Yaniv J. Histonelike proteins of bacteria. Microbiol. Rev. 1987;51:301–319. doi: 10.1128/mr.51.3.301-319.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.White SW, Appelt K, Wilson KS, Tanaka I. A protein structural motif that bends DNA. Proteins. 1989;5:281–288. doi: 10.1002/prot.340050405. [DOI] [PubMed] [Google Scholar]
- 76.Schneider R, Lurz R, Luder G, Tolksdorf C, Travers A, Muskhelishvili G. An architectural role of the Escherichia coli chromatin protein FIS in organising DNA. Nucleic Acids Res. 2001;29:5107–5114. doi: 10.1093/nar/29.24.5107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kobryn K, Lavoie BD, Chaconas G. Supercoiling-dependent site-specific binding of HU to naked Mu DNA. J. Mol. Biol. 1999;289:777–784. doi: 10.1006/jmbi.1999.2805. [DOI] [PubMed] [Google Scholar]
- 78.Vologodskii AV, Cozzarelli NR. Monte Carlo analysis of the conformation of DNA catenanes. J. Mol. Biol. 1993;232:1130–1140. doi: 10.1006/jmbi.1993.1465. [DOI] [PubMed] [Google Scholar]
- 79.Marko JF. Supercoiled and braided DNA under tension. Phys. Rev. E. 1997;55:1758. [Google Scholar]
- 80.Marko JF. Coupling of intramolecular and intermolecular linkage complexity of two DNAs. Phys. Rev. E. 1999;59:900–912. [Google Scholar]
- 81.Brown PO, Peebles CL, Cozzarelli NR. A topoisomerase from Escherichia coli related to DNA gyrase. Proc. Natl Acad. Sci. USA. 1979;76:6110–6114. doi: 10.1073/pnas.76.12.6110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Adamcik J, Valle F, Witz G, Rechendorff K, Dietler G. The promotion of secondary structures in single-stranded DNA by drugs that bind to duplex DNA: an atomic force microscopy study. Nanotechnology. 2008;19 doi: 10.1088/0957-4484/19/38/384016. Article ID 384016. [DOI] [PubMed] [Google Scholar]
- 83.Adrian M, ten Heggeler-Bordier B, Wahli W, Stasiak AZ, Stasiak A, Dubochet J. Direct visualization of supercoiled DNA molecules in solution. Embo J. 1990;9:4551–4554. doi: 10.1002/j.1460-2075.1990.tb07907.x. [DOI] [PMC free article] [PubMed] [Google Scholar]