Abstract
Muscleblind-like 1 (MBNL1) regulates alternative splicing and is a key player in the disease mechanism of myotonic dystrophy (DM). In DM, MBNL1 becomes sequestered to expanded CUG/CCUG repeat RNAs resulting in splicing defects, which lead to disease symptoms. In order to understand MBNL1’s role in both the disease mechanism of DM and alternative splicing regulation, we sought to identify its RNA-binding motif. A doped SELEX was performed on a known MBNL1-binding site. After five rounds of SELEX, MBNL1 selected pyrimidine-rich RNAs containing YGCY motifs. Insertion of multiple YGCY motifs into a normally MBNL1-independent splicing reporter was sufficient to promote regulation by MBNL1. MBNL1 was also shown to regulate the splicing of exon 22 in the ATP2A1 pre-mRNA, an exon mis-spliced in DM, via YGCY motifs. A search for YGCY motifs in 24 pre-mRNA transcripts that are mis-spliced in DM1 patients revealed an interesting pattern relative to the regulated exon. The intronic regions upstream of exons that are excluded in normal tissues relative to DM1, are enriched in YGCY motifs. Meanwhile, the intronic regions downstream of exons that are included in normal tissues relative to DM1, are enriched in YGCY motifs.
INTRODUCTION
Alternative splicing is essential for creating a diverse and functional proteome as well as for establishing tissue and developmentally specific repertoires of mRNAs. It has been shown that ∼90% of human pre-mRNAs are alternatively spliced (1). Several proteins [e.g. NOVA1, CUGBP1, MBNL1, A2BP1 (also known as Fox-1) and their related paralogs] have been shown to play important roles in the regulation of alternative splicing (2–4). NOVA1 is a neuron-specific regulator of alternative splicing that binds YCAY clusters in or near alternatively spliced exons and promotes exon inclusion or exclusion, depending on the location of the binding site (3,5). A2BP1 is expressed in brain, heart and skeletal muscle and binds the UGCAUG RNA motif (6). Based on hundreds of predicted A2BP1-binding sites, A2BP1 binding upstream of the regulated exon promotes exclusion while binding downstream promotes inclusion (4). Although much less is known about MBNL1-binding sites, a similar model of alternative splicing regulation has been proposed for the MBNL1 proteins (2,7).
The original member of the muscleblind family of proteins, muscleblind (Mbl) was identified in Drosophila and found to be important in photoreceptor and muscle differentiation (8,9). The orthologous proteins, muscleblind-like 1–3 (MBNL1, MBNL2 and MBNL3) were discovered in humans as the proteins sequestered to the toxic CUG and CCUG repeats that cause mytonic dystrophies 1 and 2 (DM1 and DM2), respectively (10–13). The muscleblind proteins are generally highly conserved, especially in the zinc finger domains, and bind RNA in a specific fashion through these domains (7,14–17). The sequestration of MBNL results in its lack of binding to normal pre-mRNA targets. This lack of binding by MBNL causes important developmentally specific transcripts to become mis-spliced and leads to symptoms of DM (for reviews see 13,18,19). For example, insulin receptor (INSR) and chloride ion channel (CLCN1) pre-mRNAs are mis-spliced in DM1 patients leading to inappropriate expression of fetal isoforms and/or degradation of the transcript (20–22). The lack of appropriate INSR and CLCN1 splice isoforms in DM1 patients is thought to lead to the symptoms of insulin resistance and myotonia, respectively.
MBNL1 promotes the exclusion of exon 5 in the TNNT2 (also known as cTNT) pre-mRNA, which produces a splice product found in adult tissue. However, in DM1, exon 5 is included aberrantly, thus producing a splice product normally found in fetal tissue (23). It has now been shown that the sequestration of MBNL1 and MBNL2 is responsible for this mis-splicing (24). MBNL1 binds a 32-nucleotide region upstream of exon 5 and regulates splicing through this site (7,24,25). In addition to TNNT2, several other pre-mRNA transcripts are regulated by MBNL1, including ATP2A1 (also known as SERCA1), and auto-regulation of MBNL1 and MBNL2 pre-mRNAs (for reviews see 13,19). The only previously characterized MBNL1-binding site in a human pre-mRNA is the 32-nucleotide TNNT2 site. The identification of additional MBNL1 RNA-binding sites would allow for a deeper understanding of MBNL1’s RNA-binding specificity. This will help determine pre-mRNA targets regulated by MBNL1 and to predict MBNL1-binding sites within these targets.
We performed a doped SELEX (Systematic Evolution of Ligands by Exponential Enrichment) experiment, using the TNNT2-binding site as a template, to identify sequences that MBNL1 binds with high affinity. The doping incorporated the endogenous binding site nucleotides at a rate of 51% for each of the 32 residue positions. The majority of sequences recovered after five rounds of selection bound MBNL1 with an affinity similar to MBNL1’s affinity for the native TNNT2 site. We observed that there was a general selection for pyrimidines and a specific selection for the motif YGCY. We showed that placing multiple YGCY motifs in a splicing reporter minigene, which was not previously regulated by MBNL1, was sufficient for MBNL1-dependent splicing regulation. We analyzed the sequences surrounding cassette exons in 24 pre-mRNAs that are mis-regulated in DM1 patients and identified over 100 YGCY potential MBNL1-binding sites. We then demonstrated that MBNL1 binds with high affinity to RNA oligomers derived from several of these endogenous YGCY motifs. We also showed that multiple YGCY instances significantly contribute to MBNL1 regulated splicing of an ATP2A1 exon known to be mis-spliced in DM1. And finally, we observed that YGCY motifs are enriched in the intronic regions upstream of cassette exons that are normally excluded (aberrantly included in DM1), and, conversely, are enriched downstream of exons that are normally included (aberrantly excluded in DM1). Together, these results aid in defining MBNL1’s RNA-binding specificity, which will ultimately help lead to the identification of more MBNL1 endogenous binding sites and provide a better understanding of the mechanisms by which MBNL1 regulates splicing.
MATERIALS AND METHODS
SELEX
The MBNL1 expression construct included residues 1–260 and contains an N-terminal GST tag. This construct, termed MBNL1 throughout the article, binds RNA as tightly as the full-length version (7). MBNL1 was expressed and purified as described earlier (7). The DNA template used for the SELEX (26) experiment was the following 81-mer oligonucleotide: 5′-GGGAATGGATCCACATCTACGAATTC(CCTGTCTCGCTTTTCCCCTCCGCTGCGGCCAC)AAGACTCGATACGTGACGAACCT-3.′ The oligonucleotide depicted here contains the 32-nucleotide MBNL1-binding site in TNNT2 (in parenthesis) flanked by two constant regions. To create SELEX pool 0, the nucleotides in parenthesis were randomized; 51% of the time the original nucleotide was kept at each position and the other 49% of the time each position was varied to incorporate an equal mix of the other three nucleotides. The forward primer for the SELEX was 5′-GATAATACGACTCACTATAGGGAATGGATCCACATCTACGA-3′ (including a T7 RNA polymerase site for in vitro transcription) and the reverse was 5′-AGGTTCGTCACGTATCGAGTCTT-3′. The initial PCR reaction was amplified by eight rounds of PCR and 1×1014 DNA molecules from the PCR reaction were used for the initial transcription reaction. The transcription reactions were done under the following conditions: 500 ng/µl template, 40 mM Tris pH 7.9, 26 mM MgCl2, 2 mM spermidine, 10 mM NaCl, 5 mM of each nucleotide, 0.1 µg/100 µl yeast pyrophosphatase, ∼2 mg/ml T7 polymerase and 40 mM DTT. The transcription reactions were performed with trace amounts of [α-P32] cytidine triphosphate (CTP) for monitoring purposes. After transcription, reactions were treated with DNase for 1 h at 37°C, gel purified and ethanol precipitated. For each round, the appropriate concentration of MBNL1 (see below) was bound to 20 µl of glutathione-agarose beads at 4°C for 15 min, then washed once with 200μl of SELEX-binding buffer (100 mM NaCl, 20 mM Tris pH 7.5, 5 mM MgCl2, 0.02% Triton X-100 and 5 mM DTT added the day of use). All washes were done in a similar manner. The RNA was heated in SELEX-binding buffer at 95°C for 3 min and placed immediately on ice for 10 min. The MBNL1-bound beads were incubated with the annealed RNA at 25°C for 20 min. The beads were washed once with 200 µl of SELEX binding buffer at 25°C and RNA was released and collected from the beads by a phenol–chloroform extraction and subsequent ethanol precipitation. The binding conditions were chosen such that 5–10% of the RNA was retained after each round. The collected RNA was reverse transcribed using Avian Myeloblastosis Virus (AMV) reverse transcriptase, 1× AMV buffer, 8.8 µM reverse primer, and two-third of the RNA isolated after the round for 1 h at 42°C. After the reverse transcription was complete, the DNA was amplified by 11 PCR cycles. The DNA was transcribed and the SELEX cycle repeated a total of five times with the concentration of RNA and MBNL1 varying. The concentrations of RNA and MBNL1 used were as follows: Round 1 (20 µM RNA, 20 µM MBNL1), Round 2 (9.8 µM RNA, 4.9 µM MBNL1), Round 3 (8 µM RNA, 2 µM MBNL1), Round 4 (2 µM RNA, 0.33 µM MBNL1), Round 5 (1 µM RNA, 0.13 µM MBNL1). After five rounds were completed, individual clones were isolated by TOPO cloning (Invitrogen) and sequenced.
Labeling of RNAs for gel mobility shift assays
SELEX RNA oligonucleotides were transcribed from DNA templates using T7 RNA polymerase and [α-P32] CTP. Commercially purchased RNA oligonucleotides were kinased using Polynucleotide Kinase and [γ-P32] ATP.
Gel mobility shift assay
Gel mobility shift assays and Kd calculations were performed as described earlier (7), with the following modifications. RNA (8 µl) was snap annealed in (75 mM NaCl, 5 mM MgCl2, 15 mM Tris pH 7.5, 0.25 mM β-ME), then mixed with 2 µl of protein for final reaction conditions of 175 mM NaCl, 5 mM MgCl2, 20 mM Tris pH 7.5, 1.25 mM β-ME, 10% glycerol, 2 mg/ml BSA, 0.1 mg/ml heparin and trace amounts of bromophenol blue. The binding reactions were incubated for 10–25 min at room temperature and 2–4 µl were loaded onto a 6% acrylamide gel (6% 37.5: 1 acrylamide:bisacrylamide, 0.5 × TB). Gels were run for 35 min at 170 V at 4°C. The Kd values were calculated based on a minimum of three gel mobility shift assays for each RNA, with the exception of F06 UCCA and (CUGCUU)6 RNAs, which were calculated based on two gel mobility shift assays.
Single nucleotide composition of selected RNAs
In order to examine biases in the single nucleotide composition that resulted during selection, we compared the total count for each nucleotide within the randomized region for Round 5 versus those observed in Round 0. To evaluate the statistical significance of the differences observed, we calculated a confidence interval (α = 0.05) for the binomial distribution using the previously described method (27) for a population that is the size of the sequences recovered (82 oligos × 32 nt length = 2624). The expected probabilities used in this estimate were those derived from the frequencies observed within the randomized regions of the Round 0 sequences.
K-mer enrichment
All K-mers from 4 to 6 nt were counted in the final 82 SELEX sequences, including the random sequence plus 5 nt of the constant flanking sequence. The inclusion of the flank allows one to analyze K-mers that overlap with the edges of the constant sequence. To calculate a Z-score for the observed versus expected occurrences we used Monte–Carlo simulation to estimate the mean (μ) and standard deviation (σ) for the occurrence of all K-mers in 1000 independent populations of 82 sequences constructed using the same random biases used to synthesize the oligos used as input in the SELEX experiment. The Z-score (Z) for each K-mer was calculated according to Equation 1 where μ is the mean observed over the random samples, σ is the standard deviation for the K-mer occurrence within the random samples and x is the observed occurrence within the population derived from SELEX.
| (1) |
Construction of splicing reporter constructs
The construction of the MBNL–eGFP plasmid was previously described and obtained from the laboratory of Maury Swanson (28). The DMPK-CUG960 plasmid was obtained from the laboratory of Thomas Cooper (24). The wild-type PLEKHH2 minigene (PLEKHH2 WT) was constructed by amplifying regions of the PLEKHH2 gene from HeLa genomic DNA using PCR primers containing unique restriction sites. Introns 20 and 21 are 2.4 and 1.7 Kb, respectively, so both were truncated using primers directed against the introns. The construct was built in three segments. The front segment contains exon 20 and the first 922 nt of intron 20 flanked by KpnI and BamHI sites (sense primer: 5'-CGGGGTACCAAATGCTGCAGTTGACTCTCC-3′ and anti-sense primer: 5′-CGCGGATCCCTGTGGCTAACAGGCAGTCA-3′). The middle segment is flanked by BamHI and NotI sites and contains the last 194 nt of intron 20, exon 21 and the first 297 nt of intron 21(sense primer: 5′-CGCGGATCCGGATCATAGATCTGACCCAATG and anti-sense primer: 5′-AGGAACATAGCGGCCGCTTGAATGAACACCCACTAATGC). The final segment is flanked by NotI and XhoI sites and contains the last 958 nt of intron 21 and exon 22 (sense primer: 5′-AGGAACATAGCGGCCGCATCTGCCTACAGGGCACTTG-3 and anti-sense primer: 5′-CCGCTCGAGCCATTCATGAAGTGCACAGG-3). The ligated segments were inserted between KpnI and XhoI sites of the pcDNA3 plasmid (Invitrogen), which contains the hCMV promoter/enhancer and a bovine growth hormone (BGH) poly(A) signal after the multiple cloning site.
The PLEKHH2-SM1F, PLEKHH2-SM1R, PLEKHH2-SM2, PLEKHH2-SM3 and PLEKHH2-SM4 constructs were all created from the PLEKHH2 WT template. To create PLEKHH2-SM1F, six, tandem copies of the SELEX motif 5′-CUGCUU-3′ were inserted into intron 20 of the PLEKHH2 minigene using PCR and standard cloning techniques. The 36-nt SELEX motif repeat, flanked by ClaI and PacI unique restriction sites (a total of 50 nt), was inserted 40 nt upstream of the 3′ splice site of intron 20, and replaced 36-nt (positions –76 to –41 relative to the 3′ splice site) of the intron to ensure that the relative size of the intron remains fixed. PLEKHH2-SM1R was created in the same manner except the six SELEX motifs were inserted into the intron backwards, 5′-UUCGUC–3′, as a negative control. PLEKHH2-SM2, PLEKHH2-SM3 and PLEKHH2-SM4 were also created in the same manner, but contain six tandem copies of 5′-CUGCCU-3′, 5′-CCGCUU-3′, and 5′-CCGCCU–3′ motifs, respectively.
Construction of ATP2A1 splicing minigene
The wild-type ATP2A1 minigene was constructed by amplifying the region of the ATP2A1 gene from HeLa genomic DNA containing exon 21, intron 21, exon 22, intron 22 and exon 23 using PCR primers (sense primer: 5′-GGGGTACCACCTCACCCAGTGGCTCATG-3′ and antisense primer: 5′-CGGGATCCCACAGCT CTGCCTGAAGATG-3′) containing unique KpnI and BamHI restriction sites. The PCR product was purified, digested and ligated into the pcDNA3 plasmid.
The ATP2A1 deletion minigene was created by deleting 151 residues of intron 22 (positions –117 to –267) using standard PCR techniques. The deletion minigene was created in two segments; the front portion (antisense primer: 5′-TGCTTACAATTGACGGCTCCAGGTGGAGCTGCGAGCACAAGTG-3′) contains exon 21, intron 21, exon 22, and the first 116 nt of intron 22 flanked by KpnI and Mfe1 restriction sites. The second section (sense primer: 5′-TGCTTACAATTGGGGCTGCAGTGGGGGGGGGCGGG-3′) contains the last 171 nt of intron 22 and exon 23 flanked by MfeI and BamHI sites. The digested fragments were ligated in the KpnI and BamHI sites of pcDNA3.
Cell culture and transfection
HeLa cells were routinely cultured as a monolayer in DMEM + GLUTAMAX media (Invitrogen) supplemented with 10% Fetal Bovine Serum (Gibco) at 37°C under 5% CO2. Prior to transfection, cells were plated in six-well plates at a density of 1.8 × 106 cells/well. Cells were transfected 18–24 h later at ∼80% confluency. Plasmid (1 µg/well) and when applicable antisense oligonucleotide (ASO) (100 pmol/well) were transfected into each well with 5 µl Lipofectamine2000 (Invitrogen) following manufacturer’s protocol. All ASOs contained a 2′-O-Methyl modification at every base and phosphorothioate backbones. The sequences of ASOs used herein to target intron 22 of the ATP2A1 minigene are as follows: ASO1 (5′-GGGCAAGAAGGGGGTGATACCTGTG-3′), ASO2 (5′-GGCGCGGGTGGCAGGGGCACAGCA-3′), ASO3 (5′-TTGACGGCTCCAGGTGGAGCTGCG-3′), and ASO4 (5′-GGCAGGCGGCAGGAGGGCAGCGAG-3′). In double transfection experiments, 500 ng of each plasmid was transfected into a single well; however, in single transfection experiments 500 ng of empty pcDNA3 vector was used to normalize plasmid concentration between wells. Cells were harvested 18–24 h after transfection using TriplE (GIBCO) and then pelleted by centrifugation. RNA was isolated from the cell pellets using an RNeasy kit (QIAGEN).
In vivo splicing: RT-PCR
Splicing assays were conducted as described earlier (29). Briefly, isolated RNA (500 ng) was incubated with 1 unit of RQI DNase (Promega) in a 10 µl reaction for 1 h at 37°C. DNased RNA [2 µl (100 ng)] was reverse transcribed in a 10 µl reaction (1: 5 dilution) using Superscript II (Invitrogen), according to manufacturer's; protocols, with the exception that we used half the recommended amount of Superscript II. All PLEKHH2 reporters and the ATP2A1 minigenes were reverse transcribed using an antisense primer (5′-AGCATTTAGGTGACACTATAGAATAGGG-3′) designed to the Sp6 promoter site of the pcDNA3 plasmid. The minus reverse transcription (−RT) reactions were treated identically to plus reverse transcription (+RT) reactions except the Superscript II was replaced with water. All reverse transcription reactions (2 µl) were subjected to 22 rounds of PCR amplification, which was found to be within the linear range for all primers used (data not shown), in a 20 µl reaction (1: 10 dilution). PCR was conducted to analyze the splice products of all PLEKHH2 reporters using the sense cloning primer designed to exon 20 and the antisense cloning primer designed to exon 22 described earlier. The ATP2A1 WT sense and antisense cloning primers designed to exon 21 and 23, respectively, were used for PCR of the ATP2A1 minigene. For quantification of the PLEKHH2 reporters, PCR was conducted using radiolabeled sense cloning primer. For the ATP2A1 minigene splicing, radiolabeled antisense primer was used in the PCR reaction for quantification. The resulting PCR products were resolved by electrophoresis on 8% (19:1) polyacrylamide native gels. The gels were dried and exposed overnight on a phosphorimager screen. Quantification of the radioactive bands corresponding to splice products was performed using ImageQuant software (Molecular Dynamics). The percent of exon inclusion was calculated by dividing the amount of the band corresponding to inclusion splice product by the total amount of splice product (calculated by adding the inclusion splice product band to the exclusion splice product band). Percentage of exon exclusion was computed in the same manner, except the exclusion product was divided by the total amount.
Calculation of the enrichment of YGCY motifs in the regions flanking exons mis-spliced in DM1
The identities of 24 exons that are mis-spliced in human DM1 tissues were gleaned from a search of literature (Supplementary Table S2). The sequences for the exons and up to 200 nt of the flanking introns were recovered from public databases. If the intron was <400 nt in length only the adjacent half was evaluated. Each exon was categorized as ‘excluded’ (E) or ‘included’ (I) if it was generally excluded or included, respectively, in normal tissues relative to DM1 tissues. The frequency of YGCY motifs in the flanking intronic regions was calculated according to Equation 2 where n equals the number of YGCY motifs observed, l equals the length of each intronic sequence evaluated and k indicates the number of introns evaluated. The denominator effectively represents the total number of positions that could contain the motif within the set of sequences evaluated.
| (2) |
The background frequency of YGCY in the upstream and downstream intronic flanks for all human introns (according to the UCSC hg18 annotation) was also calculated. The actual numbers are contained in Supplementary Table S3. The upper and lower confidence intervals (α = 0.01) for the frequencies expected in a randomly drawn sample that is the size of the regions of interest was calculated using the background frequencies, the sample sizes, and the method of Agresti et al. (27).
RESULTS
Doped SELEX with an MBNL1 RNA-binding site
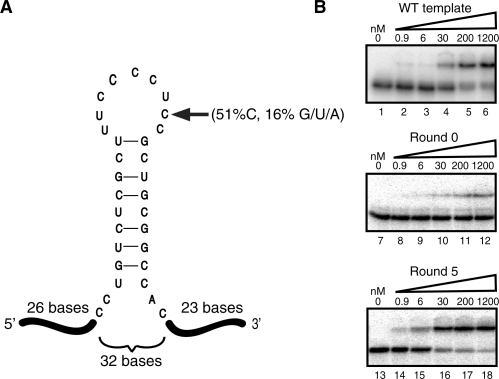
MBNL1 binds just upstream of exon 5 in the TNNT2 pre-mRNA (7,24). It has been previously shown that a 32-nt RNA corresponding to this region binds MBNL1 with high affinity and that mutations that disrupt MBNL1 binding also disrupt MBNL1-regulated exclusion of the downstream exon (7). In order to identify the nucleotides that contribute most to recognition by MBNL1, we performed a doped SELEX experiment (30). Unlike traditional SELEX, which starts with a pool of uniformly random RNAs, doped SELEX begins with a population of RNAs synthesized such that they are biased toward a specific starting sequence, but each position is still allowed to vary. For this particular experiment we began with a population of 32-nt RNAs synthesized such that each position was 51% likely to be the equivalent base found in the 32-nt TNNT2 MBNL1-binding site. Each of the other nucleotides had a 16.3% probability of being used. For example, the first position in the 32-nt region was cytidine, so in 51% of the RNAs it remained a cytidine, while the other 49% of the time the other three nucleotides were equally incorporated at that position (Figure 1A). This allowed MBNL1 to sample other possible residues at each position with a bias toward the endogenous residue. The 32 endogenous nucleotides were flanked on each side by constant regions, 26 nt on the 5′ side and 23 nt on the 3′ side, that are necessary for PCR and transcription. The MBNL1 construct included residues 1–260 and an N-terminal GST tag. This protein, termed MBNL1 throughout the article, bound RNA as tightly as the full-length version of MBNL1 (7).
Figure 1.
Round 5 SELEX RNAs bind significantly tighter than Round 0 RNAs. (A) The TNNT2 template used to create the pool 0 RNA for SELEX. The dark lines indicate the constant regions, while the central 32 bases denote the doped, variable region. The arrow indicates an example of the doping. For that particular position, the C was maintained in 51% of the sequences, while G, U and A were incorporated equally in 49% of the other sequences. The structure of the TNNT2 32mer was previously determined (7). (B) Gel mobility shift assays of MBNL1 with the 81 nucleotide wild-type TNNT2 template (top), Round 0 RNAs (middle), and Round 5 RNAs (bottom).
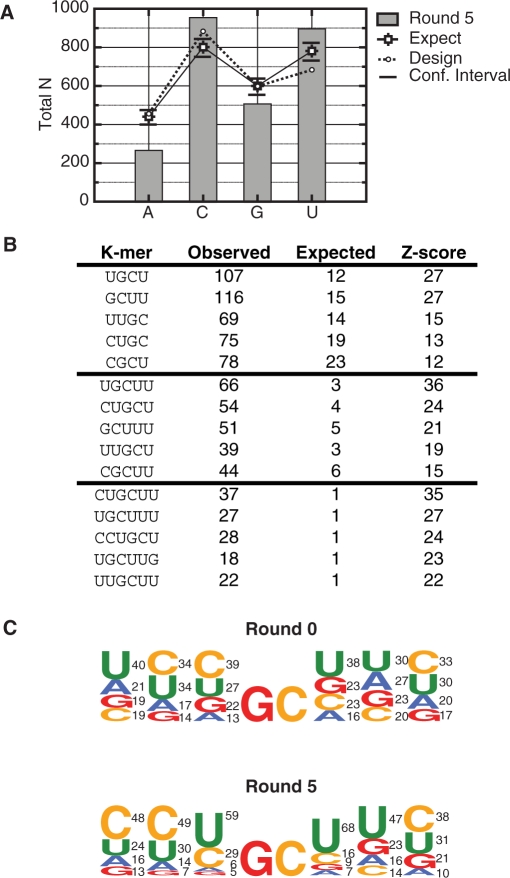
After five rounds of selection, the binding affinities of the recovered pools of RNAs were tested using a gel mobility shift assay. The binding affinity of MBNL1 was low for pool zero (Kd of >1.2 µM) and high for pool five (Figure 1B). Pool 5 RNA-bound MBNL1 with slightly higher affinity compared to the endogenous TNNT2-binding site template (Figure 1B, compare lanes 1–6 and lanes 13–18), which demonstrated that we were successful in selecting RNAs with high affinity toward MBNL1. Eighty-two sequences were obtained from Round 5, and there were no duplicates, indicating that over-selection did not take place. The sequence most closely related to the endogenous TNNT2 template contained six bases that differed from the original TNNT2 site. This result was not surprising, as pool 0 started out with only a 1 in 2 × 109 chance that a sequence would be identical to the endogenous template. Pool zero was sequenced (96 individual sequences) to insure that there was no bias in the initial pool, other than the designed doping. Analysis of these sequences demonstrated that pool 0 sequences had compositions similar to that expected according to the doping design. However, the pool 0 RNAs did show a slight enrichment in the number of uridines and a corresponding slight decrease in the number of cytidines expected according to the doping design (Figure 2A).
Figure 2.
Nucleotide composition, K-mer analysis and the frequency of nucleotides flanking GC dimers confirm a high occurrence of pyrimidines and YGCY motifs within SELEX sequences. (A) Single nucleotide counts for the randomized regions derived from Round 5 RNAs are compared to those expected (labeled ‘Expect’) given the frequencies observed in the starting pool (derived from the counts observed in Round 0). Upper and lower confidence intervals (α = 0.05) are indicated above and below each of the expected values (see ‘Materials and Methods’ section for the calculation). Also shown are the values expected according to the original doping design (labeled ‘Design’). (B) The K-mer analysis reveals highly enriched 4-, 5- and 6-mers (p < 0.01) found within the SELEX sequences from Round 5. The 15 K-mers listed have the highest Z-scores out of each K-mer size category (4–6). Observed is the total number of occurrences of the K-mer within the sample of 82 SELEX sequences. The expected is the mean number of the K-mer within 1000 sets of 82 random sequences, taking the doping into account. (C) The nucleotide frequencies flanking all GC dimers found within the SELEX Round 5 randomized regions were determined. The frequencies are represented as sequence-logos with the size of the nucleotide corresponding to frequency (numbers to the right of each nucleotide in percentages). The nucleotides are ordered top to bottom, highest frequency to lowest frequency.
The majority of SELEX-derived RNAs bind MBNL1 with high affinity and are highly enriched in YGCY motifs
MBNL1’s endogenous TNNT2-binding site is pyrimidine rich (28% U, 50% C) and purine poor (19% G, 3% A). In order to determine if there was an overall selection for pyrimidines over purines during the SELEX process, we analyzed the single nucleotide frequencies for the randomized region from the population of sequences recovered during Round 5 and compared these to the sequences from Round 0 (Figure 2A). This analysis demonstrated that there was a statistically significant enrichment in the overall uridine and cytidine content in the Round 5 RNAs when compared to Round 0 RNAs, suggesting that MBNL1 has a preference for pyrimidines over purines.
An analysis of the K-mer (4–6 nt) composition of the Round 5 RNAs relative to the composition expected by chance (based upon the starting template and doping regime, see ‘Materials and Methods’ section) revealed that the Round 5 RNAs are enriched in specific K-mers (Figure 2B and Supplemental Table S1). Meanwhile, the same analysis identified no significantly enriched K-mers within the Round 0 RNA sequences (data not shown). All of the most highly enriched K-mers contain a GC-dimer. Of these, the most significantly enriched contain UGCU (107 occurrences compared to 12 expected by chance, Z = 27). Closer inspection revealed that CGCU (Z = 12) and UGCC (Z = 3) are also enriched, but at lower levels (Figure 2B and Supplementary Table S1).
We aligned all GC-dimer instances in the Round 5 sequences, plus three nucleotides on either flank, and compared these to GC-dimers found in Round 0 (Figure 2C). This analysis revealed a selection toward pyrimidines in the three positions flanking either side of the central GC-dimer. This, and the previous analysis, suggests that MBNL1 has a higher affinity toward UGCU; however, since cytidines are also tolerated we chose to represent the general MBNL1 binding site as YGCY.
The majority of SELEX sequences contain two to six YGCY motifs (Class I, Figure 3). Over half of the sequences contain YGCY in the same locations as the wild type TNNT2 CGCU motifs. However, many of the sequences show that MBNL1 selected YGCY motifs that are shifted with respect to the positions of the YGCY motifs in the wild-type TNNT2. In some cases, this change is a slight shift of the YGCY position in comparison to the location of the wild-type motifs, whereas in other sequences a whole new second, third or forth additional motif appeared nearby or in a different region.
Figure 3.
SELEX sequences are highly enriched in YGCY motifs. The YGCY motifs highlighted in all 82 Round 5 SELEX sequences. Class I RNAs contain two or more YGCY motifs and bind MBNL1 with high affinity, while Class II contain zero or one motif and do not bind MBNL1 with high affinity. The wild-type TNNT2 sequence contains two YGCY motifs and is shown at the top of each column for reference.
Binding studies were performed on 12 of the SELEX sequences to determine the affinities of these sequences to MBNL1. Almost all of the sequences tested have binding affinities roughly equal to or higher than the endogenous TNNT2-binding site (Table 1). The SELEX sequences were grouped into two classes. Class I contains sequences with two or more YGCY motifs and Kds between 1 and 25 nM (of the RNAs tested). Class II sequences contain zero or one YGCY motif and have Kds > 1.2 µM (of the RNAs tested). Interestingly, two sequences in Class II that contain only one motif (C12 and B08), don’t bind MBNL1, suggesting an important role for multiple YGCY motifs for MBNL1 binding (Figure 3, Table 1).
Table 1.
Binding affinities of MBNL1 for selected SELEX sequences
 |
aGray shading refers to SELEX RNA oligomers in Class II, no shading refers to Class I.
bCA refers to GC to CA mutations in the 5'-YGCY-3' of the SELEX RNA.
To determine if the constant (non-randomized) regions were playing a role in the binding of MBNL1 to the SELEX sequences, the constant regions were removed from several SELEX sequences and binding studies were performed (Supplementary Figure S1). The addition of the constant regions to the TNNT2 sequence only slightly weakened binding (Kd of 30 nM), compared to a Kd of 20 nM for the 32mer endogenous TNNT2 site. Similar studies were performed on three of the SELEX sequences (F06, B07, A04) to determine the effects of removing the constant regions for these RNAs. The F06 32mer showed similar binding to the equivalent 81mer (Supplementary Figure S1 and Table 1). The B07 32mer bound MBNL1 with high affinity (Kd of 0.66 nM), which was ∼10-fold tighter than the B07 81mer. The A04 32mer bound much weaker to MBNL1 with a Kd of 180 nM compared to 6.0 nM for the 81mer RNA. Although these studies show that the constant regions can influence MBNL1 binding, in general the randomized regions appear to contain the necessary elements for MBNL1 high-affinity binding.
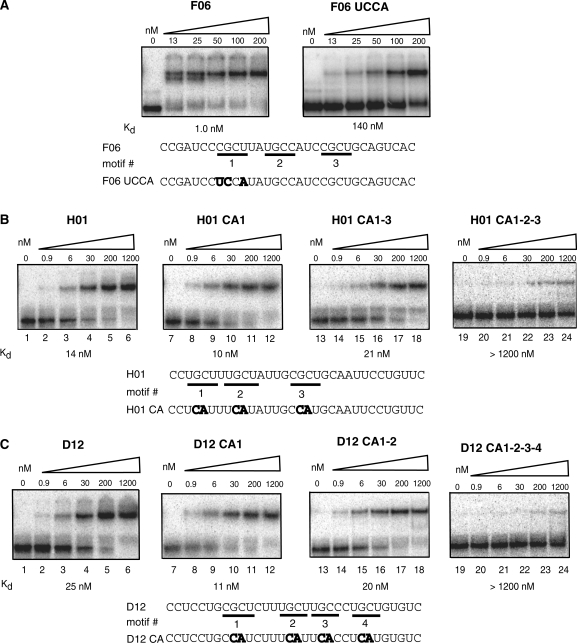
Mutations in YGCY motifs in SELEX sequences decrease MBNL1’s binding affinity
To determine the importance of the YGCY motif for MBNL1 recognition, this motif was mutated in three different 81mer SELEX sequences (F06, H01 and D12). The F06 sequence contains three YGCY motifs (Figure 4A). Motif #1 in F06 was mutated from CGCU to UCCA, which caused a 140-fold decrease in binding affinity to MBNL1. The H01 sequence contains three YGCY motifs, which were mutated from GC to CA individually (H01 CA1, HO1 CA2 and HO1 CA3), in pairs (H01 CA1-2, HO1 CA1-3, HO1 CA2-3) or all three at once (H01 CA1-2-3) (Figure 4B, Table 1). The H01 mutants with one or two motifs mutated bound with similar affinity to H01 (Kds of 2–10 nM), with the exception of H01 CA1-2, which bound with 12-fold lower affinity to MBNL1 compared to H01. When all three motifs were mutated (H01 CA1-2-3), binding was greatly weakened, with a Kd > 1.2 µM (Figure 4B, lanes 19–24). Together, these results demonstrate that, for this particular RNA, only one motif is required for MBNL1 binding. However, in the case of two SELEX sequences from Class II, C12 and B08, MBNL1 was unable to bind them although each contains one YGCY motif (Figure 3 and Table 1). This indicates that MBNL1 binding is context dependent and in some cases, requires more than one YGCY motif.
Figure 4.
Mutations in YGCY motifs decrease binding to SELEX RNAs. (A) SELEX RNA F06 has three YGCY motifs, indicated by motif #1, #2 and #3. Pictured are gel mobility shift assays to F06 and mutated F06 (F06 UCCA) to MBNL1, with the Kds listed below. The residues in bold indicate mutated residues. Notice the concentrations of MBNL1 used in the F06 gels are different than the concentrations used in 4B and 4C. (B) RNA H01 contains three YGCY motifs and binds MBNL1 with a Kd of 14 nM, indicated by the gel mobility shift assay labeled H01. Mutations were made within each motif individually and in combination. Representative gel mobility shift assays and Kds of MBNL1 binding to one of each type of mutation are shown. Mutations relative to H01 are shown in bold. H01 CA1 indicates the GC in motif #1 was mutated to CA, H01 CA1-3 indicates motifs #1 and #3 were mutated (but not motif #2). H01 CA1-2-3 has all three motifs mutated. All RNAs are 81-nt long. (C) D12 contains four YGCY motifs and binds MBNL1 with a Kd of 25 nM, indicated by the gel mobility shift assay labeled D12. D12 CA1, D12 CA1-2 and D12 CA1-2-3-4 refer to mutations in the corresponding motifs shown in the lower portion of the figure. Representative gel mobility shift assays and Kds, one from each class of mutation, are shown for D12 mutant RNAs bound to MBNL1.
The D12 RNA contains four YGCY motifs (Figure 4C). These were mutated from GC dimers to CA dimers one at a time, two at a time, three at a time or all four at once (Figure 4C, Table 1). All single, double or triple mutant combinations of D12 RNAs have an affinity within a 10-fold range of the native sequence (Kd of 25 nM). In contrast, when all four motifs were mutated (D12 CA1-2-3-4, Table 1) binding was drastically reduced (Kd > 1.2 µM). Certain motifs within these RNAs may play a more significant role in MBNL1 binding, such as the H01 motifs #1 and #2 compared to HO1 motif #3, but in general, MBNL1 does not appear to have a preference for specific YGCY motifs in these RNAs (Table 1). In conclusion, mutations in these SELEX sequences indicate that MBNL1 requires at least one YGCY for high-affinity binding.
YGCY motifs are sufficient for MBNL1 to regulate splicing in a PLEKHH2 minigene reporter
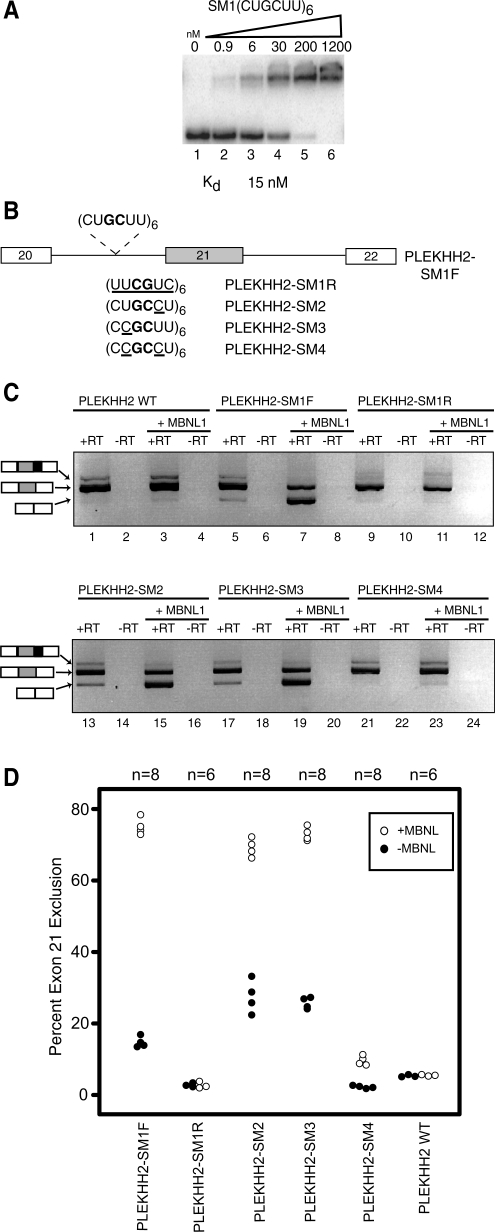
In order to determine if YGCY is sufficient for MBNL1 to regulate splicing, we created an artificial binding site using the SELEX CUGCUU K-mer (SELEX motif 1, SM1) because it had the highest Z-score in the K-mer analysis of 6-mers enriched in the SELEX sequences (Figure 2B). In addition, CUGCUU is found in several SELEX sequences that MBNL1 binds with high affinity (E01, A04, H01). MBNL1 binds (CUGCUU)6 RNA with high affinity in a gel mobility shift assay (Figure 5A).
Figure 5.
Multiple YGCY motifs embedded in pyrimidines are sufficient for MBNL1 regulated splicing of a PLEKHH2 reporter minigene. (A) Gel mobility shift assay of MBNL1 to SELEX motif 1(CUGCUU)6 RNA. (B) A diagram of the PLEKHH2 minigene reporter, PLEKHH2-SM1F, containing six copies of the SELEX motif 1 (CUGCUU). Additional reporters, PLEKHH2-SM1R, PLEKHH2-SM2, PLEKHH2- SM3 and PLEKHH2-SM4 were constructed similarly and only vary from PLEKHH2-SM1F in the sequence of the inserted motif, (UUCGUC), (CUGCCU), (CCGCUU) or (CCGCCU), respectively. Nucleotide variations in the motif around the GC core (bold) are underlined. The gray exon 21 refers to the MBNL1 regulated exon. (C) HeLa cells were transfected with either wild-type PLEKHH2 splicing reporter plasmid (PLEKHH2 WT), PLEKHH2-SM1F, PLEKHH2-SM1R, PLEKHH2-SM2, PLEKHH2-SM3 or PLEKHH2-SM4 with or without MBNL1 over-expression (+MBNL1). The RNA was harvested and RT-PCR performed to determine the splice products. Representative agarose gels are shown. A diagram of splice products including exon 20 or 22 (white boxes), exon 21 (gray boxes), the alternative 5′ splice site (downstream of exon 21) (black boxes) are shown to the left of their corresponding bands in the gel. (D) Quantification of the splicing of the PLEKHH2 reporters with (open circles) and without MBNL1 (solid circles) co-expression using radiolabeled RT-PCR.
We designed several splicing reporter minigenes based on the PLEKHH2 gene transcript. This transcript was chosen because MBNL1 does not normally regulate the splicing of this minigene. We designed several minigenes that include exons 20–22 and the intervening introns (Figure 5B). We replaced 36 nt (positions –76 to –40) with a 50-nt sequence containing six repeats of either CUGCUU (PLEKHH2-SM1F) or, as a negative control, UUCGUC (PLEKHH2-SM1R), the reverse of SM1. We also wanted to explore the effect of C versus U in the positions immediately flanking the GC core of the motif. To this end, we created three additional splicing reporter constructs in the same manner as PLEKHH2-SM1F using variations of SELEX motif 1; PLEKHH2-SM2, PLEKHH2-SM3 and PLEKHH2-SM4. PLEKHH2-SM2 contains six repeats of CUGCCU (the changed nucleotide is underlined). This reporter was designed to assess the effects of a C, instead of a U, immediately 3′ of the GC. Similarly, PLEKHH2-SM3 contains (CCGCUU)6, to evaluate the effects of a C at the position immediately 5′ of the GC. PLEKHH2-SM4 contains a C at both positions flanking the GC core to give (CCGCCU)6 (Figure 5B). In all cases the motifs are positioned ∼10 nt upstream of the putative branch site and of the poly-pyrimidine tract. It is important to note that the reversed sequence (PLEKHH2-SM1R) contains the same overall pyrimidine content and the same number of guanosines as in SELEX motif 1; however, in the reversed sequence the UGCU motif is converted to UCGU. All three constructs were transfected into HeLa cells with or without an MBNL1 protein expression vector (Figure 5C). Splice products were observed via harvesting of HeLa RNA and subsequent RT-PCR using primers to PLEKHH2.
Several splicing products were observed for the PLEKHH2 WT construct (Figure 5C, lane 1). The major splice product (Figure 5C, lane 1, middle band) corresponds to inclusion of exon 21. We also observed a smaller amount of product that arose from usage of an alternative 5′ splice site lying downstream of the normal 5' splice site (Figure 5C, lane 1, top band). Very little product (5%) corresponding to the skipping of exon 21 (Figure 5C, lane 1, bottom band) was observed in the WT construct. The splicing pattern for PLEKHH2 WT was completely unaffected by expression of MBNL1 (Figure 5C, lane 3 versus lane 1 and 5D), which demonstrates that the WT splicing pattern is MBNL1 independent.
The overall splicing pattern observed for PLEKHH2-SM1F, without expression of MBNL1, was very similar to that observed for the PLEKHH2 WT construct (Figure 5C, lane 5); however, exon 21 exclusion is increased to 15% for this reporter (Figure 5D). In sharp contrast to the effect seen for PLEKHH2 WT, when MBNL1 was co-expressed with the PLEKHH2-SM1 reporter, 75% of the splicing product observed corresponded to the skipped isoform (Figure 5C, lane 7 and 5D) demonstrating that MBNL1 robustly suppresses inclusion of exon 21 in this synthetic construct. HeLa cells contain a low level of endogenous MBNL1; this could explain the low level of the exon 21 skipped isoform seen when expressing PLEKHH2-SM1without co-expression of MBNL1 (Figure 5C, lane 5 versus lane 1).
The splicing pattern for PLEKHH2-SM1R (containing SELEX motif 1 reversed), like the other constructs, consists primarily of the exon 21 inclusion isoform (Figure 5C, lane 9), with only 2% exon 21 exclusion. However, in addition to the small amount of transcript derived from usage of the downstream alternative 5′ splice site, there is a minimal amount of higher molecular weight product corresponding to a cryptic 3′ splice site. Importantly, and in sharp contrast to PLEKHH2-SM1F, when MBNL1 is co-expressed with PLEKHH2-SM1R there is no corresponding increase in skipping of exon 21 observed and the overall percentage of exon 21 exclusion remains unchanged at 2% (Figure 5C, lane 11 versus lane 9 and 5D).
The splicing patterns for PLEKHH2-SM2 and PLEKHH2-SM3 were very similar to PLEKHH2-SM1F, with exon 21 exclusion of 28 and 26%, respectively (Figure 5C lanes 13 and 17 and 5D). Also, similar splicing changes were observed in both reporters with MBNL1 co-expression leading to strong MBNL1-induced increases in exon 21 exclusion to 70 and 73%, respectively (Figure 5C, lanes 15 and 19 and 5D). These findings suggest that MBNL1 recognizes a C upstream or downstream of the GC core equally efficiently when the other Y position in the YGCY motif is a U.
Splicing of the PLEKHH2-SM4 construct gave 2% exon 21 exclusion (Figure 5C lane 21). Surprisingly, however, only a slight increase in exon 21 exclusion to 10% was observed upon co-expression of MBNL1 (Figure 5C, lane 23 and 5D). This observation suggests that MBNL1 recognizes CGCC less efficiently than UGCU, UGCC or CGCU, which suggests that MBNL1 requires at least one U flanking the GC core for high-affinity recognition.
Together, these results demonstrate that, at least in the case of PLEKHH2, insertion of multiple YGCY motifs is sufficient for conferring MBNL1-dependent alternative splicing upon an exon that is normally constitutively included and does not normally require MBNL1 for splicing regulation. The observation that the reversed sequence is incapable of conferring MBNL1-dependent splicing demonstrates that a GC step (rather than a CG or GU step) is necessary for MBNL1-specific regulation. This distinction differentiates the MBNL1-binding site from the compositionally similar ETR-3-binding site: UGUU (31). Additionally, the observation that the exon 21 exclusion ratio in PLEKHH2-SM1F, PLEKHH2-SM2 and PLEKHH2-SM3 reporters are all robustly increased with MBNL1 co-expression suggests that MBNL1 recognizes a C or U flanking the GC core of the motif (i.e. CGCU or UGCC) with similar efficiency. Importantly however, MBNL1 co-expression with the PLEKHH2-SM4 reporter was much less efficient at inducing exon 21 exclusion in the reporter. This observation suggests that MBNL1 tolerates a C at either Y position only as long as the other position remains a U, therefore MBNL1 does not appear to recognize CGCC as well as it recognizes UGCU, UGCC or CGCU.
Novel putative MBNL1-binding sites identified in pre-mRNAs mis-spliced in DM1
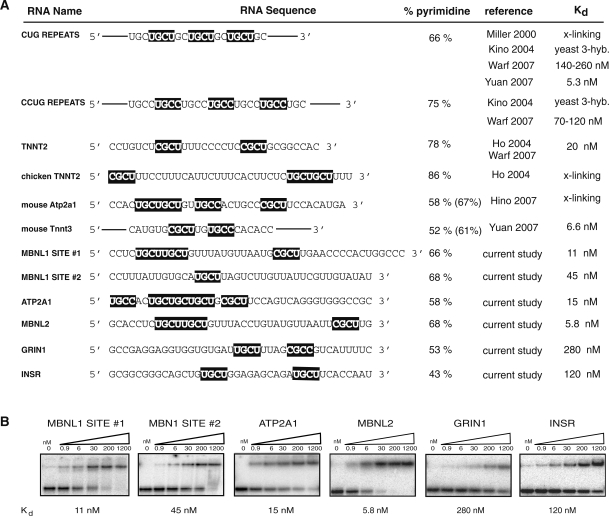
We predicted potential endogenous MBNL1-binding sites by identifying YGCY motifs clustered within introns flanking five exons mis-spliced in DM1. We evaluated these sites in vitro to determine if MBNL1 can bind these potential sites with similar affinity to that of the known TNNT2 site. Two sites in the MBNL1 pre-mRNA upstream of exon 7, one site downstream of ATP2A1 exon 22, one site in the MBNL2 pre-mRNA upstream of exon 7, one site downstream of exon 11 in INSR and one site upstream of exon 5 in GRIN1 were tested for MBNL1 binding (Figure 6). MBNL1 site #1 contains upstream residues 35–79 (45mer) and has three YGCY motifs. MBNL1 site #2 contains upstream residues 154–193 (40mer) and has one YGCY motif. The potential MBNL1-binding site found in ATP2A1 pre-mRNA has five motifs and is located downstream and contains residues 117–156 (40mer). The site in the MBNL2 pre-mRNA is between upstream residues 50–89 (40mer) and contains three motifs. The Kds of MBNL1 to the potential RNA-binding sites; MBNL1 site #1, MBNL1 site #2, ATP2A1 and MBNL2 sites were measured as 11, 45, 15 and 5.8 nM, respectively (Figure 6B), well within the range of MBNL1’s RNA-binding affinity to TNNT2 (Kd of 20 nM). MBNL1 bound the 40 nt GRIN1 and INSR RNAs, each containing two YGCY motifs, but did so with lower affinity (Kds of 280 and 120 nM, respectively). This observation suggests that aspects other than the YGCY motifs, such as pyrimidine content of adjacent regions of RNA, number of motifs, and RNA structure may also play a role in MBNL1’s RNA-binding affinity. Although these other aspects almost certainly play a role in MBNL1 binding, simply searching for YGCY occurrences has proven a fruitful method for predicting potential MBNL1-binding sites.
Figure 6.
YGCY motifs are found in multiple MBNL1 binding sites. (A) Shown are 12 RNAs that MBNL1 binds. The Kds on the right are reported in the literature (reference) or in the current study. Three studies used crosslinking (‘x-link’) to determine that MBNL1 binds specific RNAs, but did not identify affinities (12,24,33). Kino et al. used a yeast three-hybrid (‘yeast 3-hyb.’) assay to determine relative affinities of MBNL1 for CUG repeats, however no Kds were determined (15). YGCY motifs are highlighted in black. The lengths of the RNAs used for binding studies were 40 nt for MBNL1 site #2, ATP2A1, MBNL2, GRIN1, INSR, 45 nt for MBNL1 site #1, and 32 nt for human TNNT2. The two CUG repeat Kds reported were calculated under different binding conditions. The Kd reported for human TNNT2 was from this study (measured via gel mobility shift assays). The percent pyrimidine was calculated based on the sequences that the various binding studies were performed with. The percent pyrimidine calculations in parenthesis were calculated based on the sequences shown in the figure (binding studies were performed on larger regions of RNA in some cases). (B) Representative gel mobility shift assays and Kds to MBNL1 are shown for all binding sites identified in this study. The concentration (nM) of MBNL1 is indicated above each gel.
Multiple YGCY motifs contribute to MBNL1-regulated splicing of the ATP2A1 exon
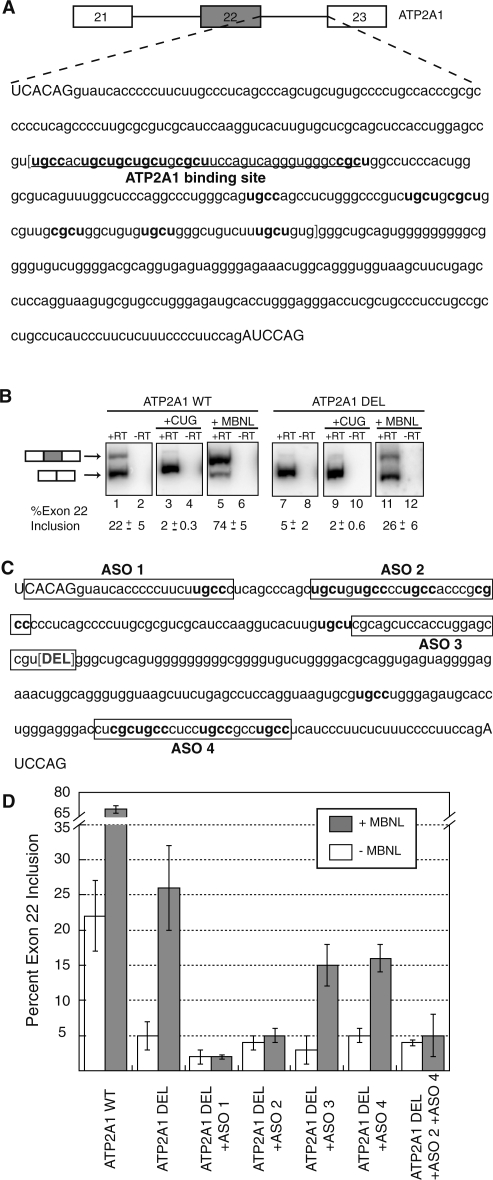
In order to validate the functionality of YGCY as the MBNL1 recognition motif in an endogenous target we created a splicing reporter minigene of the ATP2A1 transcript. Our minigene contains exons 21, 22 and 23 of the ATP2A1 transcript and the intervening introns (Figure 7A). In DM1 patients, this exon is aberrantly excluded and thus is potentially regulated by MBNL1, as shown in mouse Atp2a1 (32,33). As expected, our ATP2A1 reporter gave two major splicing product bands, corresponding to exon 22 inclusion (Figure 7B, lane 1, top band) and exon 22 exclusion (Figure 7B, lane 1, bottom band). For the ATP2A1 minigene we observed 22% exon 22 inclusion when spliced in HeLa cells (Figure 7B, lane 1 and 7D). As previously mentioned HeLa cells express some endogenous MBNL1, so the exon inclusion observed for the reporter is most likely due to exon inclusion induced by the endogenous MBNL1. In DM1, it is thought that MBNL1 is sequestered away from the ATP2A1 transcript and unable to regulate exon 22 inclusion, resulting in very low inclusion and an aberrant transcript (32). As expected, when our minigene is spliced in the presence of 960 CUG repeats, the inclusion of exon 22 drops to 2% (Figure 7B, lane 3). Importantly, splicing of the ATP2A1 reporter in combination with over-expression of MBNL1 lead to an increase in exon 22 inclusion to 74% (Figure 7B, lane 5 and 7D). In combination, these observations suggest that our ATP2A1 minigene is a good reporter and is amenable to studying MBNL1-mediated splicing effects in a DM1-related transcript.
Figure 7.
Evaluation of MBNL1 recognition of YGCY motifs in an endogenous target, ATP2A1. (A) Diagram of the ATP2A1 minigene, with white boxes indicating exons 21 and 23, lines to represent intervening introns, and a gray box representing the alternatively spliced exon 22, the inclusion of which is regulated by MBNL1. Sequence of intron 22 is shown with high-affinity MBNL1-binding site (ATP2A1-binding site) underlined. The intronic sequence contained within the red brackets indicates the portion of the intron (151 nt) that was deleted to create the ATP2A1 deletion (ATP2A1 DEL) minigene in B. Capitalized nucleotides represent exonic sequence, lower case nucleotides represent intronic sequence. YGCY motifs contained within the deletion region are in bold. (B) HeLa cells were transfected with either ATP2A1 WT or ATP2A1 DEL splicing reporter minigenes alone, or with either MBNL (+MBNL) or DMPK-CUG960 (+CUG) co-expression. Splice products are diagramed. The top band corresponds to the splice product containing exon 22 (gray box), whereas the bottom band corresponds to the splice product wherein exon 22 is excluded. The resulting splicing products were quantified using radiolabeled RT-PCR and percent exon 22 inclusion is reported below representative acrylamide gels. The values reported are an average of at least four independent experiments. (C) Sequence of intron 22 after the 151 nt deletion (red bracketed DEL). Target sequences of ASOs boxed and YGCY motifs contained within the ATP2A1 DEL minigene are indicated in bold. (D) HeLa cells were transfected with the ATP2A1 DEL minigene in combination with the ASOs diagrammed in C, both with (gray columns) or without (white columns) MBNL co-expression, and the percentage of exon 22 was quantified as in B. Inclusion values reported are the result of at least three independent experiments.
Multiple YGCY motifs are found in the intron downstream of ATP2A1 exon 22. We demonstrated that MBNL1 binds to an RNA oligonucleotide derived from this region (Figure 6B), making this site a candidate for an endogenous MBNL1-binding site responsible for regulation of exon 22. To test the importance of this site and the surrounding YGCY motifs on splicing regulation we deleted 151 nt of intron 22 in the splicing minigene to create an ATP2A1 deletion minigene (ATP2A1 DEL) that lacks both the high-affinity ATP2A1-binding site and six YGCY motifs located downstream of the ATP2A1 site (Figure 7C). When the ATP2A1 DEL minigene was tested under the same splicing conditions as the WT ATP2A1 minigene we observed that deletion of the high-affinity site lead to a decrease in exon 22 inclusion from 22 to 5% (Figure 7C, compare lane 7 to lane 1). This observation could imply that removal of the high-affinity site created a minigene that endogenous MBNL1 recognizes much less efficiently. Alternatively this decrease to 5% inclusion could be due to the loss of intronic enhancer elements that operate independently of MBNL1. Inclusion of exon 22 of the ATP2A1 DEL minigene was also reduced to 2% upon co-expression of CUG repeats (Figure 7B, lane 9). This is an important observation because it implies that although MBNL1-mediated regulation of the ATP2A1 DEL minigene is impaired significantly, it isn’t completely eliminated because MBNL1 sequestration is still capable of reducing the inclusion ratio slightly. Following this line of reasoning, splicing of the ATP2A1 DEL minigene in combination with over-expression of MBNL1 caused exon 22 inclusion to increase to 26% (Figure 7B, lane 11). This observation also suggests that the deletion was successful in reducing the MBNL1-mediated splicing effects of the ATP2A1 minigene, but did not eliminate MBNL1’s ability to regulate exon 22 inclusion. Taken together, these findings imply that deletion of the high-affinity ATP2A1 site from intron 22 of the minigene is not sufficient to eliminate all of the functional MBNL1-binding sites in the ATP2A1 transcript.
Upon further examination of the ATP2A1 DEL minigene, we observed two remaining regions of intron 22 containing clusters of YGCY motifs (Figure 7C). If these potential MBNL1-binding sites are functional in regulation of the ATP2A1 transcript, then they may be responsible for maintaining MBNL1-mediated regulation of exon 22 in the context of the deletion. Next, we used ASO (Figure 7C) to block the remaining motifs (see ‘Materials and Methods’ section) in combination with the deletion to determine if removing additional YCGY motifs is sufficient to completely eliminate MBNL1-mediated splicing effects on ATP2A1 exon 22. ASOs have previously been shown to affect splicing in vitro and in tissue culture (34,35). Splicing of the ATP2A1 DEL minigene in the presence of each ASO was evaluated both with and without co-expression of MBNL1 and percentage of exon 22 inclusion was quantified (Figure 7D). ASO1 was designed as a control to show that blocking the 5′splice site of the intron with an ASO directed against that region is sufficient to block the splicing machinery from recognizing the 5′ splice site, leading to a robust change in the splicing pattern of the minigene that is independent of MBNL1 over-expression. As expected, blocking the 5′ splice site with ASO1 resulted in a reduction in exon 22 inclusion to 2%, an effect that was MBNL1 independent (Figure 7D). ASO3 was designed as a control to show that not all ASO binding events alter splicing regulation. In the context of ATP2A1 DEL minigene and MBNL1 over-expression, ASO3 had little effect on MBNL1’s ability to regulate exon 22 inclusion (Figure 7D).
ASO2 targets a section of the ATP2A1 DEL that contains four YGCY motifs (Figure 7C) and is a logical candidate for an endogenous MBNL1-recognition site because it is located within the 5′ end of the intron, near the alternatively regulated 5′ splice junction. Blocking the potential MBNL1-binding site of the ATP2A1 DEL minigene with ASO2 caused a decrease in exon 22 inclusion to 4% (Figure 7D). Interestingly, over-expression of MBNL1 in combination with ASO2 treatment showed no significant increase in exon 22 inclusion. This observation suggests that blocking the four YCGY motifs immediately downstream of the 5′ splice junction via ASO2 is sufficient to eliminate MBNL1-mediated regulation of ATP2A1 exon 22 in the context of the deletion.
Finally, ASO4 was designed to a region at the 3′ end of the intron that also contains four YGCY motifs and is therefore another potential MBNL1-binding site. ASO4 treatment in combination with ATP2A1 DEL reduced exon 22 inclusion to 3%, similar to the effect observed upon ASO2 treatment. Although ASO4 treatment alone is capable of suppressing the ATP2A1 DEL minigene further than the deletion alone, in combination with over-expression of MBNL1, the inclusion was increased to 15% (Figure 7D). This observation suggests that blocking these four YGCY motifs found in ASO4 is not sufficient to eliminate MBNL1’s ability to regulate the inclusion of this exon. Not surprisingly however, when the ATP2A1 DEL minigene is spliced with ASO2 in combination with ASO4 the amount of exon 22 inclusion was decreased to 4%. When this minigene is subjected to splicing with ASO2, ASO4 and MBNL1 over-expression, no significant change in exon inclusion is observed. Therefore, the decrease in exon 22 inclusion with the combination of ASO2 and ASO4 is MBNL1-independent, suggesting that the combination of ASO2 and ASO4 is also sufficient to completely eliminate MBNL1-mediated regulation of ATP2A1 exon 22 in the context of the deletion.
These experiments indicate that the ATP2A1 transcript, an endogenous target of MBNL1, contains multiple functional and, potentially high-affinity, MBNL1-binding sites. It is important to note that in this particular case, simply eliminating one YGCY motif or high-affinity binding site is not sufficient to eliminate MBNL1-mediated splicing effects. Rather, multiple YGCY motifs, spanning much of the regulated intron, must be removed or blocked to fully remove MBNL1 regulation.
YGCY motifs are enriched in the intronic regions flanking DM1 mis-spliced exons
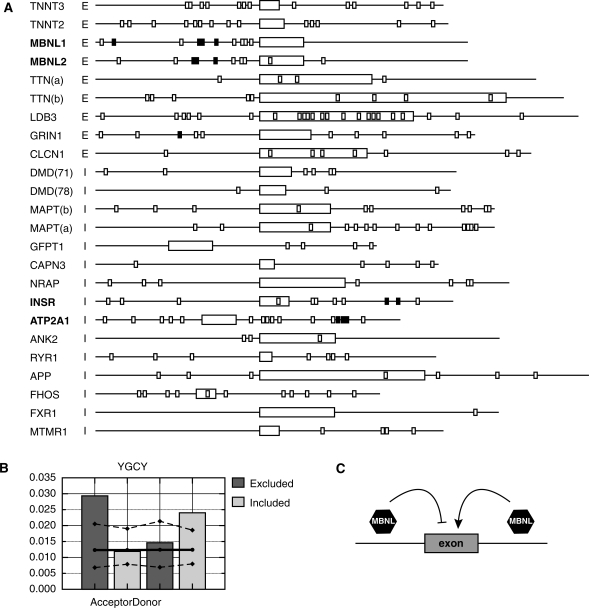
To identify new MBNL1 sites, we analyzed the intronic regions encompassing 24 human exons known to be mis-spliced in DM1 (for literature references see Supplementary Table S2). We identified the locations of all instances of YGCY within the last 200 nt of the upstream intron and the first 200 nt of the downstream intron (if either intron was <400 nt in length only half of the intron was included) (Figure 8A). Although we do not know if MBNL1 directly regulates the splicing of all of these exons, all 24 of these exons have one or more instances of YGCY in the adjacent intronic sequences and several contain instances within the exon as well. Considering both the small size and the degeneracy of this motif this observation by itself is not surprising. We carried out several statistical tests to determine if there is a significant correlation between the occurrence of the YGCY motif within these regions and the mis-regulation of splicing in DM1.
Figure 8.
(A) Schematic representation of exons and flanking regions that are mis-spliced in DM1 with instances of YGCY motifs. Two hundred bases (unless the intron is <400 bases, then only half of the intron was used) upstream and downstream of the mis-spliced exon are represented for each gene. All regions are drawn to scale. Exons are indicated as boxes and introns as lines. The gene names are those that appear in the UCSC genome browser (http://genome.ucsc.edu/). The ‘I’ or ‘E’ after each gene indicates that the exon is generally included or excluded in normal individuals (according to literature references, Supplementary Table S2). The smaller open boxes denote YGCY motifs. The filled boxes indicate YGCY motifs that are within regions tested for binding to MBNL1 in Figure 6. (B) YGCY motifs are enriched in the upstream acceptor flank of exons that are normally excluded and are enriched in the downstream donor flanks of exons that are normally included. The bars indicate the frequency of YGCY motifs in the upstream (acceptor) intronic flanks and downstream (donor) intronic flanks for the exons shown in A. For each bar, the background frequency for YGCY is indicated as a solid line. The upper and lower dashed lines represent the upper and lower 99% confidence intervals that define the range of frequencies expected if the same number of intronic flanks were randomly drawn from the population of all intronic flanks. These were calculated as described in the ‘Materials and Methods’ section. (C) Model of how MBNL1 binding correlates to exon regulation. When MBNL1 binds upstream of an alternatively spliced exon, MBNL1 suppresses inclusion of that exon. Conversely, when MBNL1 binds downstream of an alternatively spliced exon, MBNL1 enhances exon inclusion.
A correlation between binding-site location and enhancer or silencer activity has been observed for several alternative splicing regulators such as NOVA1 and A2BP1 (4,5). In order to determine if a similar correlation is observed for instances of YGCY and DM1 related mis-regulation, we evaluated the association between the frequency of YGCY in the upstream (acceptor) and downstream (donor) intronic flanks versus the type of mis-splicing seen in DM1. We observed that, if the exon is included (represented by an I in Figure 8A) in normal individuals (excluded in DM1), occurrences of YGCY are biased toward the donor region (Figure 8B). Conversely, if the exon is excluded (represented by an E in Figure 8A) in normal individuals, the motifs are biased toward the acceptor side (Figure 8B). The chi-squared test for association confirmed the significance of this bias within this dataset (P = 7.4 × 10–7).
In order to determine whether or not YGCY motifs are enriched within these regions relative to human intronic flanks in general, we determined the background frequency of YGCY within the corresponding regions of all human intronic flanks (solid line, Figure 8B) and then calculated a 99% confidence interval for the range of frequencies of YGCY expected within sample sizes equivalent to those used in this analysis (dashed lines, Figure 8B, and ‘Materials and Methods’ section). This test revealed that, despite the small size and degeneracy of the putative MBNL1-binding site and the small sample size of putative MBNL1 regulated exons currently available, YGCY is significantly enriched above background levels in the acceptor intronic flanks of exons that are excluded in normal individuals and in the donor intronic flanks of exons that are normally included (Figure 8B). Meanwhile the frequency of YGCY in the alternate flank is essentially the level expected by chance. To further demonstrate that this association is a phenomenon of YGCY, we performed identical analysis using several motifs similar to YGCY but which contain specific substitutions: YGUY, YCGY and RGCR (Supplementary Figure S2). We observed that, of the motifs tested, only YGCY shows a significant and biased enrichment correlated with the inclusion/exclusion of the adjacent exon.
Although YGCY motifs are often located both upstream and downstream of regulated exons (Figure 8A) it is likely that only subsets of these are functionally relevant MBNL1-binding sites. Additional features that we have not yet fully explored are likely to play important roles in creating physiologically functional sites, such as pyrimidine content and structure. However, the data presented here supports the model that MBNL1-binding upstream of an exon is likely to cause silencing of the downstream splice site while MBNL1-binding downstream of an exon is likely to enhance usage of the upstream splice site (Figure 8C).
DISCUSSION
The data presented here have provided insights into MBNL1’s RNA-binding specificity. The SELEX experiment recovered many RNAs that bind MBNL1 with high affinity. Analysis of the selected RNAs suggests that MBNL1 has an overall preference for pyrimidines and requires the motif YGCY for high-affinity binding. This motif is quite similar to the previously reported motif of YGCUU/GY, which was identified by comparing four MBNL1-binding sites, two in chicken TNNT2 and two in human TNNT2 (24). All nine RNAs that MBNL1 has been shown to bind, including CUG repeats, chicken and human TNNT2, mouse Atp2a1, mouse Tnnt3 and the four new putative sites identified in this study (those with Kds < 50 nM are being considered putative sites), have the common motif of YGCY (Figure 6). The analysis of nucleotide frequencies of regions flanking GC dimers suggests the Y positions in YGCY have a strong bias toward uridine (Figure 2C). Interestingly, the four known MBNL1-binding sites (human and chicken TNNT2, mouse Atp2a1 and mouse Tnnt3, Figure 6A) all contain multiple CGCU, UGCU or UGCU motifs and do not contain any CGCC motifs. GRIN1 is the only RNA in Figure 6A that contains CGCC and it binds MBNL1 with lower affinity (Figure 6A). In addition, our work on MBNL1-dependent PLEKHH2 minigene reporter splicing indicates that MBNL1 requires at least one U in either Y position in the YGCY motif to regulate splicing (Figure 5). The CCUG repeats have also been shown to be high-affinity binding sites for MBNL1 (7,15) and the common motif in these repeats is UGCC not CGCC (Figure 6A). Taken together, these results strongly suggest that MBNL1 requires multiple GC steps flanked by at least one U and a pyrimidine on the opposite flank.
Several lines of evidence suggest that local pyrimidine content influences MBNL1 binding: (i) this SELEX experiment demonstrated overall selection for uridines; (ii) the native TNNT2-binding site and most of the high-affinity sites adjacent to regulated exons (this study) are generally pyrimidine rich; (iii) CUG and CCUG repeat RNAs are highly pyrimidine rich; and (iv) The sequences inserted in PLEKHH2-SM1F, PLEKHH2-SM2 and PLEKHH2-SM3 that confer MBNL1-splicing regulation are 83% pyrimidine. The mouse Tnnt3 and the mouse Atp2a1 sites contain lower levels of pyrimidines in general compared to the other known MBNL1-binding sites (Figure 6). However, the regions of these transcripts proposed to bind MBNL1 (an 18 nt stem-loop and two YGCY motifs within 12 nt of each other, respectively) contain higher levels of pyrimidines (33,36). In other words, for these specific transcripts the regions of the transcripts proposed to contain an MBNL1-binding site have higher, local pyrimidine content than the overall transcript. Therefore, when only these regions of the transcripts are considered the mouse Tnnt3 and the mouse Atp2a1 transcripts are similar in pyrimidine composition to the other sites. This supports the model that MBNL1 prefers to bind YGCY motifs when they are embedded within pyrimidines. The GRIN1 and INSR RNAs bind more weakly to MBNL1 compared to the other RNAs, and, interestingly, both of these RNAs contain a lower pyrimidine content compared to the other RNAs (Figure 6). This observation also supports the model that high-affinity MBNL1-binding sites will be found within pyrimidine-rich sequences. Although MBNL1 bound the GRIN1 and INSR sites more weakly, it is still possible that MBNL1 interacts with these sites and regulates splicing through them, especially if other protein factors enhance the binding of MBNL1 to these sites.
Another commonality among the RNAs that MBNL1 binds with high affinity is that they contain multiple YGCY motifs. A significant fraction of the SELEX RNAs contains two or more YGCY motifs (Figure 3). The CUG repeats, CCUG repeats, TNNT2, ATP2A1 and several potential MBNL1 sites identified in this study also contain multiple copies of the YGCY motif. These results suggest that high-affinity MBNL1 sites will be both pyrimidine rich and contain multiple copies of the YGCY motif. It will be important to follow up the identification of these putative new MBNL1 sites with functional studies to determine if the YGCY motifs regulate the splicing of these exons and if their effect on regulating the inclusion and exclusion of these exons is MBNL1 dependent. We showed that in the case of ATP2A1 exon 22 regulation, MBNL1 utilizes multiple YGCY motifs, spanning a large portion of intron 22 for proper splicing regulation. This finding is in agreement with the model that MBNL1 requires multiple YGCY motifs for binding and splicing regulation.
MBNL1 has been shown to bind to several different types of RNA targets. The first type is the toxic CUG and CCUG repeat RNA (12,15), specifically the expanded CUG and CCUG repeats which form long, A-form stem-loops containing pyrimidine–pyrimidine mismatches in between G-C and C-G base pairs (37–40). Similarly, MBNL1’s binding sites in TNNT2 and mouse Tnnt3 pre-mRNAs are stem-loops containing pyrimidine–pyrimidine mismatches (7,36), suggesting MBNL1 is recognizing all of these sites through a common mode of recognition. A recent crystal structure of Zn fingers 3–4 from human MBNL1 in complex with a 6-mer RNA (CGCUGU) shows that the GC core of the YGCY motif is recognized via its Watson–Crick face, indicating that the GC core is not able to simultaneously bind MBNL1 and form Watson–Crick base-pairs in a stem-loop structure (17). This structure, in combination with the data presented here showing that MBNL1 prefers YGCY motifs embedded in pyrimidine-rich RNAs, suggests MBNL1 binds this motif when it is partially or fully unfolded. It is possible that RNA structure in the remainder of the RNA enhances binding to MBNL1. A similar combination of primary sequence and secondary structure has been shown to be important for NOVA1 binding in which its site can be presented in the loop portion of a stem-loop (41).
We used Sfold (website: http://sfold.wadsworth.org) to predict structures of the six potential MBNL1-binding sites identified in this study (40–45 nt) (data not shown) (42,43). Interestingly, there is a mix of strong stem-loops and weaker stem-loops and no strong correlation between MBNL1’s binding affinity and the stability of the predicted structure. The analysis of the RNAs from the SELEX using Sfold also did not indicate a correlation between predicted RNA structure and MBNL1 binding. It does not appear that within this set of RNAs, a specific RNA structure is required for MBNL1 binding. Structure may be one aspect influencing how MBNL1 regulates splicing and another influence could be binding site location.
The creation of the first ‘synthetic’ MBNL1 regulated exon (PLEKHH2-SM1F) demonstrates that six copies of the motif (CUGCUU) are all that is necessary and sufficient for MBNL1 to block inclusion of an exon. The minigene containing the reverse of the motif in PLEKHH2-SMIF (PLEKHH2-SM1R) shows that the GC core is critical for this control by MBNL1 (Figure 5). We chose to place the MBNL1 motifs 10 nt upstream of the putative branch site and poly-pyrimidine tract in intron 20 so as to avoid direct steric competition with the splicing factors recognizing these elements. This region upstream of the 3′ splice site has been shown by us and others to frequently contain splicing enhancers and repressors (29,44). Therefore, we felt this was an appropriate location for placement of the six CUGCUU motifs. It is likely that MBNL1 is repressing the use of the PLEKHH2-SM1F 3′ splice site through a different mechanism than the blocking of U2AF65 binding at the poly-pyrimidine tract, which is how MBNL1 negatively regulates exon 5 of the TNNT2 pre-mRNA (25). In the future it will be interesting to determine the multiple mechanisms through which MBNL1 acts to negatively and positively regulate splicing.
Multiple alternative splicing regulators have been shown to have positional biases with respect to where their binding site is relative to the regulated exon. For example, A2BP1 has been shown to function as a repressor when bound upstream of a regulated exon and as an enhancer when bound downstream (4). NOVA and the CELF family of proteins have also been shown to have a position-dependent mode of splicing regulation (2,5). Although there has not been a comprehensive analysis of MBNL1-binding sites, several pieces of evidence suggest MBNL1 may share common splicing mechanisms with these other factors. The characterization of MBNL1’s binding site in TNNT2 confirms that MBNL1 acts as a repressor when binding upstream of exon 5 (7,24). In addition, the presence of a branch point sequence and a poly-pyrimidine tract near the MBNL1 site suggests that MBNL1 blocks recognition of those sequences by inhibiting the binding of the splicing factor U2AF65 (25). In the case of the PLEKHH2 transcript, placing MBNL1-binding sites upstream of exon 21 also causes MBNL1-dependent repression of exon 21, similar to how MBNL1 regulates TNNT2 exon 5 splicing (Figure 5). In regards to positive regulation, YGCY sites downstream of exon 22 in the human ATP2A1 pre-mRNA function as positive regulators of exon inclusion through MBNL1 binding (Figure 7). And finally, statistical analysis of introns flanking DM1 mis-spliced exons revealed that YGCY sites are enriched upstream of exons that are normally excluded (relative to DM levels) and are enriched downstream of exons that are normally included (relative to DM levels). These results support the model (Figure 8C) that MBNL1 acts as a negative regulator when bound upstream of a regulated exon and as a positive regulator when bound downstream.
Identification of additional MBNL1-binding sites in pre-mRNA targets will be very useful toward understanding MBNL1’s alternative splicing mechanisms and to identify the full list of pre-mRNA targets MBNL1 regulates. However, with a potential structural aspect as well as the sequence aspects, MBNL1’s binding sites may be degenerate and challenging to recognize in a sea of intronic sequence. This doped SELEX has provided a useful set of criteria for identifying potential MBNL1-binding sites; YGCY motifs embedded in pyrimidines. We have shown that sites created (in the case of PLEKHH2 reporters) or identified (in the case of ATP2A1) using these criteria have proven to be sufficient for MBNL1-dependent splicing regulation. In the future these criteria will aid in the identification of additional MBNL1 targets and currently has furthered the understanding of MBNL1’s mode of splicing regulation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (AR053903 to J.A.B.). Funding for open access charges: National Institutes of Health.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank the Berglund lab for helpful feedback on the manuscript and Ralf Krahe and Linda Bachiniski (MD Anderson Cancer Center) for their help in identifying exons regulated by MBNL1 in the literature.
REFERENCES
- 1.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kalsotra A, Xiao X, Ward AJ, Castle JC, Johnson JM, Burge CB, Cooper TA. A postnatal switch of CELF and MBNL proteins reprograms alternative splicing in the developing heart. Proc. Natl Acad. Sci. USA. 2008;105:20333–20338. doi: 10.1073/pnas.0809045105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ule J, Jensen KB, Ruggiu M, Mele A, Ule A, Darnell RB. CLIP identifies Nova-regulated RNA networks in the brain. Science. 2003;302:, 1212–1215. doi: 10.1126/science.1090095. [DOI] [PubMed] [Google Scholar]
- 4.Zhang C, Zhang Z, Castle J, Sun S, Johnson J, Krainer AR, Zhang MQ. Defining the regulatory network of the tissue-specific splicing factors Fox-1 and Fox-2. Genes Dev. 2008;22:2550–2563. doi: 10.1101/gad.1703108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ule J, Stefani G, Mele A, Ruggiu M, Wang X, Taneri B, Gaasterland T, Blencowe BJ, Darnell RB. An RNA map predicting Nova-dependent splicing regulation. Nature. 2006;444:580–586. doi: 10.1038/nature05304. [DOI] [PubMed] [Google Scholar]
- 6.Jin Y, Suzuki H, Maegawa S, Endo H, Sugano S, Hashimoto K, Yasuda K, Inoue K. A vertebrate RNA-binding protein Fox-1 regulates tissue-specific splicing via the pentanucleotide GCAUG. EMBO J. 2003;22:905–912. doi: 10.1093/emboj/cdg089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Warf MB, Berglund JA. MBNL binds similar RNA structures in the CUG repeats of myotonic dystrophy and its pre-mRNA substrate cardiac troponin T. RNA. 2007;13:2238–2251. doi: 10.1261/rna.610607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Artero R, Prokop A, Paricio N, Begemann G, Pueyo I, Mlodzik M, Perez-Alonso M, Baylies MK. The muscleblind gene participates in the organization of Z-bands and epidermal attachments of Drosophila muscles and is regulated by Dmef2. Dev. Biol. 1998;195:131–143. doi: 10.1006/dbio.1997.8833. [DOI] [PubMed] [Google Scholar]
- 9.Begemann G, Paricio N, Artero R, Kiss I, Perez-Alonso M, Mlodzik M. Muscleblind, a gene required for photoreceptor differentiation in Drosophila, encodes novel nuclear Cys3His-type zinc-finger-containing proteins. Development. 1997;124:4321–4331. doi: 10.1242/dev.124.21.4321. [DOI] [PubMed] [Google Scholar]
- 10.Fardaei M, Larkin K, Brook JD, Hamshere MG. In vivo co-localisation of MBNL protein with DMPK expanded-repeat transcripts. Nucleic Acids Res. 2001;29:2766–2771. doi: 10.1093/nar/29.13.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mankodi A, Urbinati CR, Yuan QP, Moxley RT, Sansone V, Krym M, Henderson D, Schalling M, Swanson MS, Thornton CA. Muscleblind localizes to nuclear foci of aberrant RNA in myotonic dystrophy types 1 and 2. Hum. Mol. Genet. 2001;10:2165–2170. doi: 10.1093/hmg/10.19.2165. [DOI] [PubMed] [Google Scholar]
- 12.Miller JW, Urbinati CR, Teng-Umnuay P, Stenberg MG, Byrne BJ, Thornton CA, Swanson MS. Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy. EMBO J. 2000;19:4439–4448. doi: 10.1093/emboj/19.17.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Osborne RJ, Thornton CA. RNA-dominant diseases. Hum. Mol. Genet. 2006;15 Spec No 2:R162–169. doi: 10.1093/hmg/ddl181. [DOI] [PubMed] [Google Scholar]
- 14.Goers ES, Voelker RB, Gates DP, Berglund JA. RNA binding specificity of Drosophila muscleblind. Biochemistry. 2008;47:7284–7294. doi: 10.1021/bi702252d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kino Y, Mori D, Oma Y, Takeshita Y, Sasagawa N, Ishiura S. Muscleblind protein, MBNL1/EXP, binds specifically to CHHG repeats. Hum. Mol. Genet. 2004;13:495–507. doi: 10.1093/hmg/ddh056. [DOI] [PubMed] [Google Scholar]
- 16.Pascual M, Vicente M, Monferrer L, Artero R. The muscleblind family of proteins: an emerging class of regulators of developmentally programmed alternative splicing. Differentiation; research in biological diversity. 2006;74:65–80. doi: 10.1111/j.1432-0436.2006.00060.x. [DOI] [PubMed] [Google Scholar]
- 17.Teplova M, Patel DJ. Structural insights into RNA recognition by the alternative-splicing regulator muscleblind-like MBNL1. Nat. Struct. Mol. Biol. 2008;15:1343–1351. doi: 10.1038/nsmb.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cho DH, Tapscott SJ. Myotonic dystrophy: emerging mechanisms for DM1 and DM2. Biochim. Biophys. Acta. 2007;1772:195–204. doi: 10.1016/j.bbadis.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 19.Ranum LP, Cooper TA. RNA-mediated neuromuscular disorders. Annu. Rev. Neurosci. 2006;29:259–277. doi: 10.1146/annurev.neuro.29.051605.113014. [DOI] [PubMed] [Google Scholar]
- 20.Charlet BN, Savkur RS, Singh G, Philips AV, Grice EA, Cooper TA. Loss of the muscle-specific chloride channel in type 1 myotonic dystrophy due to misregulated alternative splicing. Mol. Cell. 2002;10:45–53. doi: 10.1016/s1097-2765(02)00572-5. [DOI] [PubMed] [Google Scholar]
- 21.Mankodi A, Takahashi MP, Jiang H, Beck CL, Bowers WJ, Moxley RT, Cannon SC, Thornton CA. Expanded CUG repeats trigger aberrant splicing of ClC-1 chloride channel pre-mRNA and hyperexcitability of skeletal muscle in myotonic dystrophy. Mol. Cell. 2002;10:35–44. doi: 10.1016/s1097-2765(02)00563-4. [DOI] [PubMed] [Google Scholar]
- 22.Savkur RS, Philips AV, Cooper TA. Aberrant regulation of insulin receptor alternative splicing is associated with insulin resistance in myotonic dystrophy. Nat. Genet. 2001;29:40–47. doi: 10.1038/ng704. [DOI] [PubMed] [Google Scholar]
- 23.Philips AV, Timchenko LT, Cooper TA. Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science. 1998;280:737–741. doi: 10.1126/science.280.5364.737. [DOI] [PubMed] [Google Scholar]
- 24.Ho TH, Charlet BN, Poulos MG, Singh G, Swanson MS, Cooper TA. Muscleblind proteins regulate alternative splicing. EMBO J. 2004;23:3103–3112. doi: 10.1038/sj.emboj.7600300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Warf MB, Diegel JV, von Hippel PH, Berglund JA. The protein factors MBNL1 and U2AF65 bind alternative RNA structures to regulate splicing. Proc. Natl Acad. Sci. USA. 2009;106:9203–9208. doi: 10.1073/pnas.0900342106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Conrad RC, Bruck FM, Bell S, Ellington AD. RNA: Protein Interactions. Oxford: Oxford University Press; 1998. [Google Scholar]
- 27.Agresti A, Coull B. Approximate is better than “exact” for interval estimation of binomial proportions. Am. Stat. 1998;52:119–126. [Google Scholar]
- 28.Ho TH, Savkur RS, Poulos MG, Mancini MA, Swanson MS, Cooper TA. Colocalization of muscleblind with RNA foci is separable from mis-regulation of alternative splicing in myotonic dystrophy. J. Cell. Sci. 2005;118:2923–2933. doi: 10.1242/jcs.02404. [DOI] [PubMed] [Google Scholar]
- 29.Murray JI, Voelker RB, Henscheid KL, Warf MB, Berglund JA. Identification of motifs that function in the splicing of non-canonical introns. Genome Biol. 2008;9:R97. doi: 10.1186/gb-2008-9-6-r97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bartel DP, Zapp ML, Green MR, Szostak JW. HIV-1 Rev regulation involves recognition of non-Watson-Crick base pairs in viral RNA. Cell. 1991;67:529–536. doi: 10.1016/0092-8674(91)90527-6. [DOI] [PubMed] [Google Scholar]
- 31.Faustino NA, Cooper TA. Identification of putative new splicing targets for ETR-3 using sequences identified by systematic evolution of ligands by exponential enrichment. Mol. Cell. Biol. 2005;25:879–887. doi: 10.1128/MCB.25.3.879-887.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kimura T, Nakamori M, Lueck JD, Pouliquin P, Aoike F, Fujimura H, Dirksen RT, Takahashi MP, Dulhunty AF, Sakoda S. Altered mRNA splicing of the skeletal muscle ryanodine receptor and sarcoplasmic/endoplasmic reticulum Ca2+-ATPase in myotonic dystrophy type 1. Hum. Mol. Genet. 2005;14:2189–2200. doi: 10.1093/hmg/ddi223. [DOI] [PubMed] [Google Scholar]
- 33.Hino S, Kondo S, Sekiya H, Saito A, Kanemoto S, Murakami T, Chihara K, Aoki Y, Nakamori M, Takahashi MP, et al. Molecular mechanisms responsible for aberrant splicing of SERCA1 in myotonic dystrophy type 1. Hum. Mol. Genet. 2007;16:2834–2843. doi: 10.1093/hmg/ddm239. [DOI] [PubMed] [Google Scholar]
- 34.Dominski Z, Kole R. Restoration of correct splicing in thalassemic pre-mRNA by antisense oligonucleotides. Proc. Natl Acad. Sci. USA. 1993;90:8673–8677. doi: 10.1073/pnas.90.18.8673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sierakowska H, Sambade MJ, Agrawal S, Kole R. Repair of thalassemic human beta-globin mRNA in mammalian cells by antisense oligonucleotides. Proc. Natl Acad. Sci. USA. 1996;93:12840–12844. doi: 10.1073/pnas.93.23.12840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yuan Y, Compton SA, Sobczak K, Stenberg MG, Thornton CA, Griffith JD, Swanson MS. Muscleblind-like 1 interacts with RNA hairpins in splicing target and pathogenic RNAs. Nucleic Acids Res. 2007;35:5474–5486. doi: 10.1093/nar/gkm601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Michalowski S, Miller JW, Urbinati CR, Paliouras M, Swanson MS, Griffith J. Visualization of double-stranded RNAs from the myotonic dystrophy protein kinase gene and interactions with CUG-binding protein. Nucleic Acids Res. 1999;27:3534–3542. doi: 10.1093/nar/27.17.3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mooers BH, Logue JS, Berglund JA. The structural basis of myotonic dystrophy from the crystal structure of CUG repeats. Proc. Natl Acad. Sci. USA. 2005;102:16626–16631. doi: 10.1073/pnas.0505873102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Napierala M, Krzyzosiak WJ. CUG repeats present in myotonin kinase RNA form metastable “slippery” hairpins. J. Biol. Chem. 1997;272:31079–31085. doi: 10.1074/jbc.272.49.31079. [DOI] [PubMed] [Google Scholar]
- 40.Sobczak K, de Mezer M, Michlewski G, Krol J, Krzyzosiak WJ. RNA structure of trinucleotide repeats associated with human neurological diseases. Nucleic Acids Res. 2003;31:5469–5482. doi: 10.1093/nar/gkg766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jensen KB, Musunuru K, Lewis HA, Burley SK, Darnell RB. The tetranucleotide UCAY directs the specific recognition of RNA by the Nova K-homology 3 domain. Proc. Natl Acad. Sci. USA. 2000;97:5740–5745. doi: 10.1073/pnas.090553997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ding Y, Chan CY, Lawrence CE. RNA secondary structure prediction by centroids in a Boltzmann weighted ensemble. RNA. 2005;11:1157–1166. doi: 10.1261/rna.2500605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ding Y, Lawrence CE. A statistical sampling algorithm for RNA secondary structure prediction. Nucleic Acids Res. 2003;31:7280–7301. doi: 10.1093/nar/gkg938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ladd AN, Cooper TA. Finding signals that regulate alternative splicing in the post-genomic era. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-11-reviews0008. reviews0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.