Abstract
Endogenous retroviruses (ERVs) are an inherited part of the eukaryotic genomes, and represent ∼400 000 loci in the human genome. Human endogenous retroviruses (HERVs) can be divided into distinct families, composed of phylogenetically related but structurally heterogeneous elements. The majority of HERVs are silent in most physiological contexts, whereas a significant expression is observed in pathological contexts, such as cancers. Owing to their repetitive nature, few of the active HERV elements have been accurately identified. In addition, there are no criteria defining the active promoters among HERV long-terminal repeats (LTRs). Hence, it is difficult to understand the HERV (de)regulation mechanisms and their implication on the physiopathology of the host. We developed a microarray to specifically detect the LTR-containing transcripts from the HERV-H, HERV-E, HERV-W and HERV-K(HML-2) families. HERV transcriptome was analyzed in the placenta and seven normal/tumoral match-pair samples. We identified six HERV-W loci overexpressed in testicular cancer, including a usually placenta-restricted transcript of ERVWE1. For each locus, specific overexpression was confirmed by quantitative RT-PCR, and comparison of the activity of U3 versus U5 regions suggested a U3-promoted transcription coupled with 5′R initiation. The analysis of DNA from tumoral versus normal tissue revealed that hypomethylation of U3 promoters in tumors is a prerequisite for their activation.
INTRODUCTION
Integral genome sequencing data from various species have highlighted the significant numbers of endogenous retroviral elements contained within the cellular genomes. In humans, retrovirus-like elements represent ∼8% of the euchromatin (1). There are at least 31 families of human endogenous retroviruses (HERVs), each derived as a result of independent infections of the germline by an infectious virus during the evolution of the human lineage. Each family encompasses tens to thousands of loci (2), essentially resulting from reinfection by replication-competent elements and intracellular retrotransposition of the transcriptionally active copies. All contemporary copies are found to be defective for viral replication owing to genetic drift, and are consequently transmitted in a Mendelian fashion. HERVs were initially considered as DNA parasites, but are now suggested to contribute to biological activities. For example, the Syncytin-1 envelope glycoprotein, encoded by the ERVWE1 locus, which is a full-length proviral member of the HERV-W family, is thought to contribute to human placentation (3–5), autoimmunity (6) and epithelial cancers (7–9). Furthermore, HERV long-terminal repeats (LTRs), whose original function is retroviral-expression control, may provide transcriptional regulatory elements for cellular genes consisting in promoter (10), enhancer (11), antisense transcripts negative modulators (12) or polyadenylation signal functions (13). Thus, HERV proteins and LTRs appear to be potential contributors to both human physiology and pathology.
Nevertheless, the vast majority of the data reflecting HERV behavior mainly rely on the evidence of transcriptional activity. Male and female reproductive tissues as well as embryonic tissues such as the placenta are the major sites of HERV expression. This suggests: (i) a hormone-dependent relationship between HERV and reproductive tract [reviewed in ref. (14)] and (ii) that at least some LTRs are hypomethylated in these tissues, as recently demonstrated for the ERVWE1 locus (Syncytin-1) expressed in the placenta (15,16), and several HERV-E LTRs that function as alternative gene promoter in the placenta (17). In somatic cells, HERVs are globally silent. This silencing is thought to be the result of the methylation of the LTRs (5′LTR, 3′LTR, or solo LTR) that contain the regulatory elements including the retroviral promoter in the LTR U3-subregion. Conversely, expression of HERV has been recurrently observed during cellular transformation processes in both cell culture and in vivo. Thus, approaches for analyzing the activity of HERVs may help to elucidate transcriptional regulation mechanisms, address physiopathological functions and eventually prove useful for diagnosis and therapy.
On the basis of functional hypotheses associated with candidate loci, the majority of HERV quantitative expression studies are based on RT-PCR (18–22). Global methods were also developed, essentially based on the relative conservation of the pol genes, a process known as pan-retrovirus amplification. For example, HERV-taxon-specific real-time RT-PCR assays using degenerate primers, was design to quantify the RNA expression of some gammaretroviral HERV families (23). Alternatively, PCR amplification using relatively complex degenerate primer mixtures, combined with low-density microarrays, were developed to detect and identify the expression of HERV families (24,25). Although these strategies aim to analyze the overall transcription of each family, each associated method is a compromise between quantitation and exhaustiveness, and it thus appears that none of the existing methods are exhaustive (26). Moreover, as designed, these methods do not permit to follow the expression of each locus within each family.
Thus, several global approaches were developed to try to analyze the expression of each individual locus. The first one, called ‘digital expression’, compared human EST data with prototypes loci of four families and showed the trends concerning family tropism of the expression, but failed to positively assign the locus to the EST (27). An improvement of such a method was recently published, based on HML-2 analysis (28). A second approach, based on the utilization of PCR with two primer pairs targeting gag and env regions of the HML-2 family and subsequent cloning and sequencing, detected the original transcriptionally active proviruses, but strictly depended on the primer design and associated putative bias (29,30). Although informative, these two methods cannot provide any information related to autonomous (U3 driven) versus non-autonomous (e.g. readthrough) expression of HERVs. Moreover, as 85% of HERV sequences consist of solitary LTR (1), namely without gag, pol, or env internal sequences, a major part of the HERV transcriptome remains hidden. The recently described genome repeat expression monitoring (GREM) technique, terminating at the cloning and sequencing steps, allowed identifying HML-2 human-specific solitary LTRs acting in vivo as promoters (31,32). In this study, we developed a high-density microarray to detect HERV transcriptional activities in a locus-specific way. More particularly, independent probesets for the U3 and U5 regions of every HERV LTR were designed from four HERV families [HERV-K(HML-2), HERV-W, HERV-H, HERV-E]. Additional groups of probesets were dedicated to the detection of the 16 known env genes, some env-related splice transcripts and putative gag ORFs. We used this microarray to explore the differential activity of HERV loci in tumoral versus normal RNA pairs from different tumor types and in the placenta. We found a specific up-regulation of a cluster of HERV-W U5 probesets in the tumoral testis (TT). Detailed analysis of the probesets provided additional information relative to alternative splicing and LTR self-activation, which in turn, resulted in a hypothesis concerning an epigenetic control of these loci. Both the levels of control were experimentally addressed on several patient samples.
MATERIALS AND METHODS
RNA and DNA samples
RNA samples, namely breast, colon, lung, ovary, prostate, testis and uterus First_Choice Human Tumor/Normal Adjacent Tissue RNA and normal placenta First_Choice Human Tissue RNA were purchased from Ambion (Austin, TX, USA). An RNA and DNA pair sample of human testis Tumoral/Normal Adjacent Tissue was purchased from Clinisciences/Biochain.
Constitution of HERV genomic sequence library
First, GenBank sequences (release 128) containing homologs for the families HERV-K(HML-2), HERV-W, HERV-H and HERV-E were identified by Blast procedure with default parameters (33). The accession numbers for the full-length proviral sequences used as queries are presented in Table 1. Subsequently, a screening of the identified GenBank sequences containing HERVs was performed using RepeatMasker (Smit, AFA and Green, P RepeatMasker at http://repeatmasker.genome.washington.edu/cgi-bin/RM2_req.pl) at a 20% divergence threshold with a library containing prototype HERV proviruses cut out into functional parts (LTR, gag, pol, env). Finally, the resulting sequences were collected and their structures were annotated.
Table 1.
Reference sequences used to constitute the HERV genomic library and microrarray probesets
| Family | Reference sequence |
Probeset | |
|---|---|---|---|
| Designation | Genomic position | Nomenclature | |
| HERV-K(HML-2) | HERV-K(C7)a,b | (−) chr7: 4588583–4598054 | H2'n;umber'.'region' |
| envK1c | (−) chr12: 57008430–57010526 | envk1 | |
| envK2c | (+) chr7: 4589506–4591605 | envk2 | |
| envK3c | (−) chr19: 32821333–32824298 | envk3 | |
| envK4c | (−) chr6: 78484295–78486392 | envk4 | |
| envK5c | (−) chr19: 32821286–32823386 | envk5 | |
| envK6c | (−) chr8: 7343729–7345828 | envk6 | |
| np9d | (+) chr22: 17312661–17314316 | sp.np9 | |
| cORF/RECd | (+) chr7: 4589627–4591360 | sp.corf | |
| HERV-W | ERVWE1a | (−) chr7: 91935220–91945441 | HW'n;umber'.'region' |
| envWc | (−) chr7: 91936015–91937571 | envw | |
| ERVWE1/Syncytin-1d | (−) chr7: 91936164–91945269 | sp.ervwe1 | |
| HERV-H | HERV-H19a | (+) chr2: 166272309–166280954 | HH'n;umber'.'region' |
| envH1c | (−) chr2: 166272946–166274699 | envh1 | |
| envH2c | (−) chr3: 168023715–168025406 | envh2 | |
| envH3c | (+) chr2: 155436978–155438645 | envh3 | |
| HERV-E | HERV-E4.1a | (−) chr19: 20721466–20730278 | HE'n;umber'.'region' |
| HERV-T | envTc | (+) chr19: 20341241–20343121 | envt |
| HERV-FRD | envFRDc | (+) chr6: 11211913–11213529 | envfrd |
| ERV3/HERV-R | envRc | (+) chr7: 64088729–64097156 | envr |
| HERV-R(b) | envR(b)c | (+) chr3: 16786814–16788359 | envr(b) |
| HERV-F(c) | envF(c)1c | (+) chrX: 96988918–96990890 | envf(c)1 |
| envF(c)2c | (−) chr7: 152737681–152739128 | envf(c)2 | |
Family affiliation of the reference sequences is indicated in the first column. The reference sequences were used to generate
aLTR probesets.
bgag probesets.
cenv probesets.
dprobesets at the splice junctions.
The position in accession number correspond to the whole HERV provirus in a and b, and to the probeset area in c and d. ‘Number’ in probeset nomenclature refers to the locus accession number in the HERV library. ‘Region’ in probeset nomenclature is ‘L5.’s ‘L3.’ or ‘LS.’ for 5′LTRs, 3′LTRs or soloLTRs, respectively, followed by ‘U3’ or ‘U5’ in function of the region targetted in the LTR and ‘GAG’ for HERV-K(HML-2) putative full-length ORFs.
Custom HERV GeneChip microarray
For each HERV family, probes were designed using the criteria of sequence uniqueness among the genomic sequence collection of the whole concerned HERV family. All potential 25-mer sequences were generated from the targeted regions. All the 25-mers were then compared with the others to identify those occurring uniquely. For each functional domain, a probeset comprising unique sequences was picked out to ensure a homogeneous spread over the targeted region. Thus, any solo or proviral LTR may consist of two probesets, one for the U3 region and one for the U5 region. Therefore, for a full-length provirus, the maximum number of probesets was four, i.e. targeting independently U3 5′LTR, U5 5′LTR, U3 3′LTR and U5 3′LTR. It is essential to note that, for a given provirus, divergence occurring between 5′ and 3′ LTRs since the proviral integration event made it possible to select distinct probes for the 5′ and 3′LTRs sub regions. In fact, owing to the uniqueness filter, some LTR or LTR subregions were not represented by any probe. Additionally, probesets for the 16 known HERV full-length envelope genes, envH 1–3, envK 1–6, envT, envW, envFRD, envR, envR(b), and envF(c) 1 and 2 [Table 1; (18)] were built. In addition to the envW_at probeset detecting the ERVWE1 env region, sp.ervwe1_at, a probeset specific for ERVWE1/Syncytin-1 mRNA was designed, consisting of probes located at the env splice junction (34) and at the ERVWE1 signature (35). Likewise, probesets for the HERV-K(HML-2) np9 and cORF/REC encoding spliced mRNA were built using type 1 HERV-K101 (AF164609) and type 2 HERV-K(HML-2.HOM) (AC072054) proviral copies as queries, respectively. The probes for np9 and cORF mRNA were located at the splice junction excluding the env region (36,37). Finally, the probesets were designed for five putative full-length gag HERV-K(HML-2) ORFs (2001 nt) identified in the HERV genomic sequence library. Thus, a total of 5100 HERV probesets were generated.
In addition to the HERV probes, the standard Affymetrix control probes for unbiased amplification (5′ and 3′ GAPDH and HSA probesets) and hybridization quality (bioB, bioC, bioD and cre probesets) were included in the microarray. MM mismatch standard Affymetrix control probes, including one central mismatch, were not included because of the probable frequent existence of the corresponding HERV paralogous element, and also because it was shown that such controls could be inefficient (38). A set of dedicated control probes containing two or three transversions were designed to evaluate intrafamily putative cross-hybridization (Supplementary Data 1A; .cdf file and basic annotation file are available upon request).
Target amplification/labeling and microarray hybridization
cDNA synthesis and transcriptional amplification were performed using 400 ng of RNA following the Whole Transcripts Amplification protocol (WTA, Affymetrix). Briefly, dsDNA synthesis was performed using random primers with a T7 promoter tail (T7-N6) and reverse transcriptase/RNAseH mix. An antisense RNA (cRNA) was produced by adding T7 polymerase. The cRNA amplification products were cleaned on RNeasy columns (RNeasy mini kit, Qiagen) and used as template for dsDNA synthesis. Amplified dsDNA products were purified using the QIAquick purification kit (Qiagen) and their quality was assessed with a Bioanalyzer capillary electrophoresis device (Agilent, Palo Alto, USA). Sixteen micrograms of purified dsDNA were fragmented into 50–200 bp fragments using DNAseI treatment and labeled at the 3′OH termini by introducing biotinylated dNTPs using terminal transferase. The 16-µg fragmented dsDNA were mixed with standard hybridization controls and oligo B2 following the recommendations of the supplier. This hybridization cocktail was heat-denatured at 90°C for 10 min, incubated at 50°C for 5 min and centrifuged at 16 000g for 2 min to pellet the residual salts. At the same time, the HERV GeneChip microarray was prehybridized using 200 µl of hybridization buffer in an oven at 60°C, under agitation at 60 r.p.m. for at least 10 min. The hybridization buffer was then replaced by the denatured hybridization cocktail containing labeled targets. Hybridization was performed at 50°C for 16 h in a hybridization oven with constant rotation (60 r.p.m.). Washing and staining were carried out according to the protocol supplied by the manufacturer, using a fluidic station (GeneChip fluidic station 450, Affymetrix). The probe arrays were scanned using a fluorometric scanner (GeneChip scanner, Affymetrix). The experiments were performed in triplicate, that is three independent amplifications and hybridizations were performed for each clinical sample.
Expression analysis
Expression analysis was performed using R statistical software (39) and packages from the Bioconductor project (40).
The quality of the microarrays was assessed with standard Affymetrix quality controls extracted from GCOS report (GeneChip® Expression Analysis Data Analysis Fundamentals, Affymetrix). All the microarrays fulfilled the criteria. The images produced by the scanner of the log-transformed intensities were visually inspected, and distribution homogeneity of the array intensities was also checked with boxplots and histograms of log2 intensities.
Robust multi-array averaging (RMA) method (41) was used for background correction (RMA), normalization (quantiles method) and summarization (median polish method) of the microarray data.
The expression data were filtered by removing the low-expressed probesets. The maximum expression of each probeset across the arrays was computed. Probesets with a maximum expression lower than the median (i.e. 70) of the distribution of maximum values were considered to be low-expressed and were removed.
Identification of differentially expressed probesets among the samples was performed with a multiclass significance analysis of microarrays (SAM) based on a fisher test (42). SAM provides permutation-based estimations of the false-discovery rate (FDR) associated with the lists of probesets for which the null hypothesis is rejected. The null hypothesis states that ‘the intensity associated with a probeset remains the same in all samples’. An FDR cutoff of 1% was applied (cel files are available upon request).
Real-time RT-PCR
A set of locus-specific PCR primer pairs was obtained using the Primer3 software, and the best primer pair was selected using BLAT and in silico PCR at UCSC genome browser (http://genome.ucsc.edu/cgi-bin/hgPcr) to optimize specificity (Supplementary Table S1). Primers targeting ERVWE1-spliced env mRNA were designed as previously described for overlapping splice junction (43). All the RT-PCR products were sequenced and mapped onto the genome (UCSC genome March 2006 version 36.1) to check whether each retained primer pair actually matched only one specific locus. One microgram of each total RNA was reverse-transcribed in a volume of 20 µl containing 1X first-strand buffer, 0.01 M of DTT, 40 U of RNaseOUT, 0.5 mM of dNTP-Mix, 250 ng of random primers (Promega, Madison, USA) and 200 U of SuperScriptII (Invitrogen, Carlsbad, USA) according to the manufacturers’ recommendations. Reverse-transcriptase-free reactions were carried out to verify the absence of contaminating genomic DNA.
The cDNA were used for real-time PCR based on SYBR green fluorescence. After 100-fold dilution, 10 µl of cDNA were mixed with 1 µl of 2.5 µM forward primer, 1 µl of 2.5 µM reverse primer (Supplementary Table S1), 0.5 µl of water and 12.5 µl of Brilliant® qPCRMaster Mix. The volume of the reaction mixture was 25 µl. PCR amplification was carried out in eight-well strips using an M×3005P thermal cycler (Stratagene). The housekeeping genes (G6PD, GAPDH and HPRT) were analyzed in the same experiment as the target transcripts. Amplifications of cDNAs, including those of RT-minus cDNA synthesis reactions, were performed as follows: a 10-min denaturation step at 95°C, followed by 40 or 46 cycles at 95°C for 30 s, at 55°C for 1 min, and at 72°C for 1 min. An additional cycle at 95°C for 1 min, at 60°C for 30 s and at 95°C for 30 min was performed at the end of the run to establish the dissociation curves. Each sample was analyzed at least twice to assess assay reproducibility. Quantification of each targeted transcript was performed using an external standard calibration curve, designed using serial dilutions of a reference sample. The reference samples in the cloned PCR-amplified genomic regions or cDNA were subsequently re-amplified using primers targeting the pCR2.1 topo cloning vector, and the amplification products were used as templates for calibration curves. G6PD that appeared to be the most stable housekeeping gene between the tumoral and the normal adjacent tissues was chosen to normalize the experiments.
5′ Rapid amplification of cDNA ends
For the identified differentially expressed transcripts, 5′ Rapid amplification of cDNA end (RACE) was performed using the SMARTTM RACE cDNA amplification Kit (Clontech). First-strand 5′RACE cDNA synthesis was performed on 1 μg of total RNA with the 5′-CDS primer, following the manufacturer’s instructions. Amplification of the transcripts at the 5′ end was performed on the resulting 5′-RACE-Ready cDNA, with the real-time qRT-PCR ‘U5’ reverse primers as the gene-specific primers (Supplementary Table S1). The recommended TFR-gene 5′-RACE control and gene-of-interest amplification control (using U5 forward and reverse primers) were carried out in the same experiments. Reverse primers were located maximally at ∼550 bp downstream from the theoretical retroviral initiation site. Therefore, the elongation time was fixed to 3 min, enabling amplification of up to 4-kb fragments. The initial cycle numbers were fixed at 25, and were increased up to 30 or 35 cycles when necessary. Alternatively, nested 5′RACE was carried out. The 5′-RACE products were loaded onto a 1.2% agarose gel, and excised and purified using NucleoSpin Extract II Kit (Macherey Nagel). The RACE products were then cloned using the TOPO TA cloning kit (Invitrogen) and sequenced for 5′-end characterization.
Sodium bisulfite modification of the genomic DNA and methylation analyses
The methylation status of LTR U3 region was determined using Bisulfite Sequencing PCR. Bisulfite treatment converts all, except the methylated (protected) cytosines, to uracil. Uracils are further read as thymines by the polymerase during the PCR step. Subsequently, sequencing allows for the identification of methylated cytosine that is not converted to thymidine. The genomic DNA (1 µg) from the testicular tumor and normal adjacent testicular tissue was modified by bisulfite using EZ DNA methylation kit (Zymo Research), according to the manufacturer’s instructions, except for the conversion time. The incubation time with the CT conversion reagent (Zymo Research), was reduced to 9 h, to obtain bisulfite-unconversion signatures discriminating the amplified genomic molecules without generating false-positive unmethylated CpGs (optimizations done in our laboratory are not presented). The DNA was eluted in 30 µl of sterile water.
PCR amplifications were performed using strand-specific primers (Supplementary Table S2) designed to specifically amplify the HERV of interest. The primers for the first and nested PCR are listed in the Supplementary Table S2. For the first PCR, 3 µl of modified DNA was mixed with 0.66 µM of reverse and forward primers, 0.2 mM of each dNTP, 1 U of Platinium Taq polymerase (Invitrogen), 1 ×of Mg-free buffer (Invitrogen) and 1.5 mM of MgCl2 (Invitrogen), reaching a final volume of 50 µl. The reactions were performed in an Eppendorf Thermocycler (Eppendorf). The reaction conditions were as follows: an initial denaturation step at 94°C for 5 min, followed by 40 cycles at 94°C for 1 min, at an annealing temperature depending on the primer for 1 min, at 72°C for 1 min, and a final extension step at 72°C for 10 min.
The nested PCR (if necessary) was performed with 1 µl of first PCR products under the same mixture and amplification conditions for 30 cycles (primer and annealing temperatures used are presented in Supplementary Table S2). The amplification products were purified using Nucleospin® Extract II (Machery Nagel), following extraction on 1% agarose gel. The amplicons were cloned using TOPO TA Cloning kits (Invitrogen). Positive clones were selected by LacZ gene reporter control and EcoRI restriction. The clones were then sequenced using ABI Prism 3100 Genetic Analyzer (Applied Biosystems). Each sequence was aligned and compared with the in silico modified sequence to determine the methylation status of each CpG, the rate of conversion, and the errors in cytosine conversion, which distinguish the molecularly independent clones.
RESULTS
Identification of differentially expressed HERV loci by DNA microarray and qRT-PCR validation
To investigate the HERV transcriptome in a locus-specific manner, we developed an HERV-dedicated high-density microarray. This microarray comprised probesets for the U3 and the U5 regions of HERV LTRs from four families, namely HERV-E, HERV-H, HML-2 (HERV-K superfamily) and HERV-W (Supplementary Figure S1), and probesets for selected HERV genes, namely gag, env, rec and np9 genes. The control probes containing two to three transversions were included in order to evaluate the risk of cross-reactions of the phylogenetically related loci. Chip validation was performed (i) by comparing the wild-type individual probes and counterpart control signals as well as (ii) by using a well-documented cellular model: fusion-induced BeWo cells that show transcriptional activation of specific HERV loci in response to forskolin treatment. The total RNA samples from non-induced and induced BeWo cells, normal placenta and seven pairs of tumor and normal adjacent tissue were analyzed on the custom HERV microarray. The raw data were corrected for noise, and normalized and summarized using the RMA method (‘Materials and Methods’ section). Briefly, although signals observed for the control probes could be considered as the background currently admitted for Affymetrix technology, some exceptions showed that a risk of cross-hybridization cannot be excluded when the divergence between the two probes/loci results from two adjacent bases and/or when hybridization signals are extremely high (see Supplementary Data 1A). This implies that in the absence of a systematic counterpart set of control probes in this chip version, identification of overexpressed loci should be confirmed by qRT-PCR. Indeed, the observed correlation between DNA chip and qRT-PCR analyzes of HERV candidate loci in the BeWo model indicates the potential of the chip approach to analyze the HERV transcriptome (see Supplementary Data 1B).
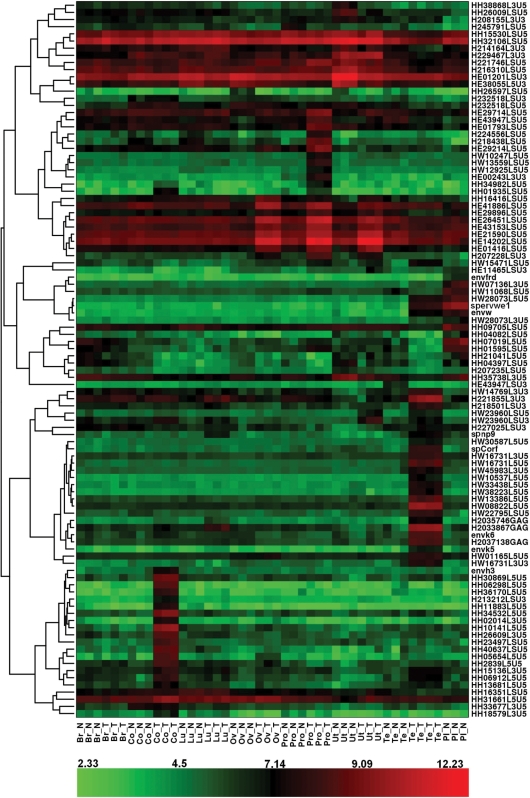
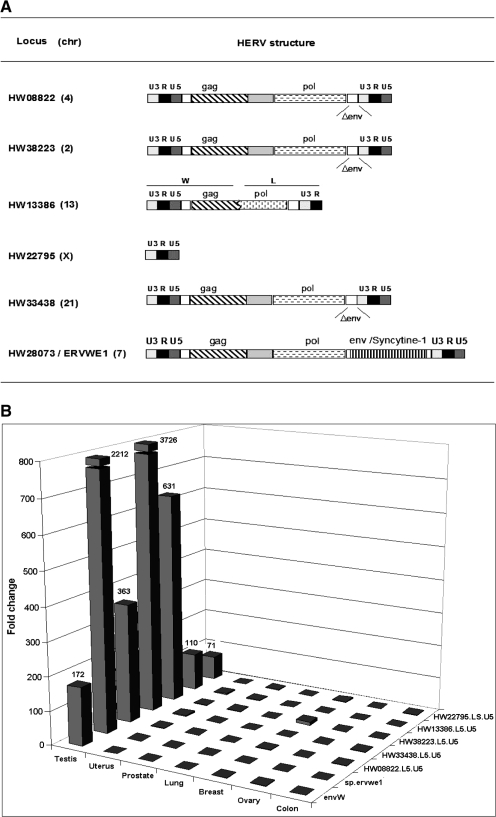
Total RNA samples from normal placenta and from pairs of tumor and normal adjacent tissue were subsequently analyzed on the custom HERV microarray. After removal of the low expressed probesets, differentially expressed probesets among the samples were identified using the SAM method. The FDR was fixed at 1%. Owing to putative intrafamily cross-reactivity, it should be noted that the mention of an expressed probeset reflects a global family, a family subset or a single locus reactivation. The undoubtedly tagged specific locus will be supported either by the control probeset behavior or qRT-PCR validation. In a first attempt, the expression of the 100th most significant differentially expressed probesets was visualized with a heatmap (Figure 1). In addition, hierarchical clustering was performed to generate a tree (dendrogram) that groups together probesets with similar expression profiles (Figure 1). First, this hierarchical clustering allowed us to observe one group of HERVs principally active in the placenta, such as ERVWE1 and ERVFRDE1 loci. The probesets detecting the ERVWE1 provirus associated mRNAs were localized in the 5′LTR U5 region (HW28073.L5.U5_at), env region (envW_at) and specific spliced env mRNA (sp.ervwe1_at), in addition the probeset detecting the ERVFRDE1 provirus associated mRNAs was localized in the env gene region (envfrd_at). The intensity fold change for these probesets in normal placenta, when compared with the mean intensity in other normal samples, is reported in Table 2. Second analyses showed tumor-specific overexpressions of HERV probeset groups in different tumors. We observed the signal expression of several probesets of HERV-H family in colorectal cancer; a set of HERV-E probesets activated in prostate, ovary and uterus cancers; and probesets of the HERV-W and HML-2 families in testicular cancer. Interestingly, we also observed a heterogeneous behavior of the HERV-E probesets in ovary, uterus and prostate tissues, that is, 14 probesets were overexpressed in all the three tumoral tissues, two probesets were overexpressed only in ovary and prostate tumors, seven probesets were overexpressed only in prostate tumor, five probesets were down-regulated in the uterus tumor, and finally, one probeset was highly and similarly expressed in the prostate and ovary tissues that were normal or tumoral. Nevertheless, an unambiguous identification of HERV-E and HERV-H loci was difficult owing to the inefficient design of the probes within those families. Expression detected by these probesets may result from the expression of different HERV-E elements. On the contrary, one probeset for the HERV-H family, HH34532.L5.U5_at, strictly corresponded to a unique locus whose overexpression in colon cancer (Table 2) was confirmed by qRT-PCR (data not shown). Furthermore, the envH3 sequence corresponding to an HERV-H full-length Env ORF behaved similarly. In testicular cancer, the expression of gag, env, cOrf and np9 regions of HML-2 loci was evidenced directly by their dedicated probesets, although only three loci were unambiguously identified (Table 2). Similarly, overexpression of six HERV-W loci in the testicular cancer was evidenced by their respective U5 LTR probesets, namely, HW28073.L5.U5_at, HW08822.L5.U5_at, HW33438.L5.U5_at, HW38223.L5.U5_at, HW13386.L5.U5_at and HW22795.LS.U5_at (fold change ranging from 5.90 to 12.80; Table 2). These six loci are localized on different chromosomes, showing that they do not represent a regional cluster of reactivation. In addition, they present heterogeneous structures. Thus, these six probesets correspond to one full-length provirus, three env-deleted proviruses, one chimeric provirus composed of an HERV-W (5′LTR-gag) and an HERV-L (pol-3′LTR) elements and one solitary LTR (Figure 2A). Notably, the full-length provirus is the ERVWE1 locus whose expression in testicular tumor is also identified by the env region (envW_at) and specific spliced env mRNA (sp.ervwe1_at) probesets.
Figure 1.
HERV expression heat-map visualization. Figure shows a heat-map visualization based on the expression of the 100 most significant probesets for all samples and obtained by hierarchical clustering of probeset expression. The hierarchical clustering is represented by the dendrogram on the left-hand side, and is based on the correlation distance between the probeset expression profiles. The color assigned to each point in the heat-map grid is based on the level of expression (log2 scale) of one probeset (rows) in one sample (columns). Red indicates a high expression and green indicates a low expression. Br, breast; Co, colon; Lu, lung; Ov, ovary; Pro, prostate; Ut, uterus; Te, testis; Pl, placenta; N, normal tissue; T, tumoral tissue.
Table 2.
Microarray-based differential expression of the most significant HERV probesetsa
| Probesetsb | Familyb | Copy positionc | Global analysisd |
Pairwise comparaisone |
|||
|---|---|---|---|---|---|---|---|
| raw p-value | q-value | expression levels | fold change | ||||
| Placenta | Others | Placenta | |||||
| HW28073.L5.U5_at | HERV-W | (−) chr7: 91935211–91945442 | 0.00E+00 | 0.00E+00 | 38.4 | 393.9 | 10.25 |
| envw_at | HERV-W | (–) chr7: 91935211–91945442 | 0.00E+00 | 0.00E+00 | 25 | 273.8 | 10.95 |
| sp.ervwe1_at | HERV-W | (–) chr7: 91935211–91945442 | 0.00E+00 | 0.00E+00 | 33.2 | 1014.8 | 30.57 |
| envfrd_at | HERV–FRD | (–) chr6: 11210982–11215545 | 1.16E–06 | 7.43E–06 | 10.7 | 95.4 | 8.92 |
| envR_at | ERV3/HERV–R | (–) chr7: 64088729–64097156 | 6.77E–05 | 4.14E–05 | 56.9 | 127.8 | 2.25 |
| Tumoral colon | Colon N | Colon T | |||||
| HH34532.L5.U5_at | HERV–H | (+) chr1: 68624274–68630265 | 9.70E–07 | 7.39E–06 | 9.9 | 14.1 | 1.42 |
| envH3_at | HERV–H | (+) chr2: 155436773–155438869 | 6.01E–06 | 1.54E–05 | 32.7 | 181.2 | 5.54 |
| Tumoral testis | Testis N | Testis T | |||||
| HW28073.L5.U5_at | HERV–W | (–) chr7: 91935211–91945442 | 0.00E+00 | 0.00E+00 | 29.9 | 202.6 | 6.78 |
| envw_at | HERV–W | (–) chr7: 91935211–91945442 | 0.00E+00 | 0.00E+00 | 12.1 | 154.9 | 12.80 |
| sp.ervwe1_at | HERV–W | (–) chr7: 91935211–91945442 | 0.00E+00 | 0.00E+00 | 21.1 | 268.6 | 12.73 |
| HW08822.L5.U5_at | HERV-W | (–) chr4: 41982210–41989670 | 0.00E+00 | 0.00E+00 | 76.9 | 984 | 12.80 |
| HW13386.L5.U5_at | HERV–W | (+) chr13: 68693758–68697469 | 3.88E–07 | 3.67E–06 | 54.9 | 428.4 | 7.80 |
| HW38223.L5.U5_at | HERV–W | (+) chr2: 17383688–17391462 | 3.88E–07 | 3.67E–06 | 18.3 | 211.4 | 11.55 |
| HW33438.L5.U5_at | HERV–W | (+) chr21: 27148626–27156169 | 3.88E–07 | 3.67E–06 | 27.1 | 203.9 | 7.52 |
| HW22795.LS.U5_at | HERV–W | (+) chrX: 113026617–113027400 | 2.91E–06 | 1.33E–05 | 49.5 | 292.2 | 5.90 |
| envk5_at | HML-2 | (–) chr5: 156018263–156019973 | 3.10E–06 | 1.33E–05 | 14.6 | 105.3 | 7.21 |
| envk6_at | HML-2 | (–) chr8: 7343775–7351309 | 3.10E−06 | 1.33E−05 | 47.3 | 411.7 | 8.70 |
| H2035746.GAG_at | HML-2 | (–) chr12: 57008477–57015997 | 3.88E−07 | 3.67E−06 | 18.4 | 164.6 | 8.95 |
| Tumoral prostate | Prostate N | Prostate T | |||||
| envR_at | ERV3/HERV-R | (–) chr7: 64088729–64097156 | 6.77E–05 | 4.14E–05 | 41 | 99 | 2.41 |
aOnly the most significant probesets identified by global SAM analysis are presented. Probesets are classified according to the site in which the overexpression is observed.
bThe names of the probeset and HERV family are indicated.
cThe orientation of the copy onto the DNA strand, the chromosome number, and the positions of the first and terminal bases of the copy on the chromosome are indicated (UCSC genome March 2006 version 36.1).
dRaw p– and q–value summarized the results of the global analysis among all the samples. The raw p–value was obtained by SAM multiclass test and q–value was the lowest False Discovery Rate at which the probeset was considered significant.
eFor pairwise comparison, the reference conditions are defined by either the corresponding normal tissue for tumoral site of overexpression, or by all non–placenta normal tissues for placenta. The expression levels are summarized by the median values. The degree of differential expression between reference and overexpressed sites is expressed as fold change.
Figure 2.
Identification of HERV-W loci specifically up-regulated in testicular tumor. (A) Structure and genomic position of the identified HERV-W loci. For all the identified loci, the HERV genomic structures are detailed: U3, R and U5 are the functional regions of the LTRs; gag, pol and env are the proviral genes; and Δenv indicates that the envelope gene is deleted. The family type of 5′ and 3′ parts of the HW13386 chimeric locus are indicated. (B) Fold change between normal and tumoral counterpart. Differential RNA expression between normal and tumoral counterpart is indicated for each tissue (testis, uterus, prostate, lung, breast, ovary and colon) for the six overexpressed loci (HW08822, HW13386, HW22795, HW33438, HW38223 and ERVWE1/Wenv). The RNA samples were similar to those used in the microarray experiments. The qRT-PCR results were normalized with G6PD housekeeping gene expression, which had an averaged copy number of 1000.
To validate the results obtained by the microarray experiments, the mRNA expression levels of the newly identified HERV-W loci were analyzed by qRT-PCR in the same normal/tumoral samples. Locus-specific primer pairs localized in the same regions as the probesets were used for real-time quantitative PCR. Thus, amplification was conducted in the LTR U5 region for HW08822.L5.U5, HW33438.L5.U5, HW38223.L5.U5, HW13386.L5.U5 and HW22795.LS.U5. HW28073/ERVWE1 amplification was carried out with two primer sets localized in the env region and at the env-specific mRNA splice junction, respectively. All the loci were overexpressed in the testicular tumor when compared with the normal adjacent section, with fold changes ranging from 71 for HW22795 up to 3226 for HW33438 (Figure 2B). In addition, an absence of differential expression in other tumoral/normal tissues was observed. Altogether, these results validate the microarray data for testicular-tumor-specific reactivation of the U5 or env regions for all the six loci.
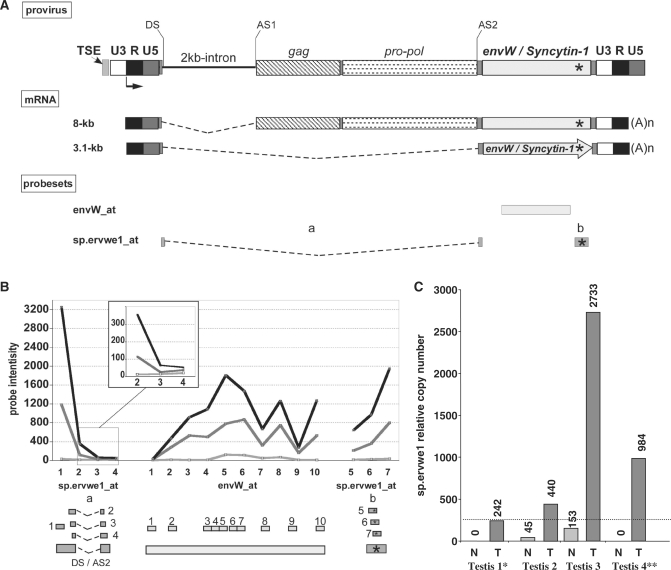
Identification of the splicing strategy: the particular case of ERVWE1 locus
The envW_at and the sp.ervwe1_at probesets, used to trace ERVWE1 proviral activity, could detect all env-containing transcripts (8 and 3.1 kb) and only the env-encoding transcript (3.1 kb), respectively (Figure 3A). The data generated from the microarray indicated that the locus did not present any significant activity in the normal testis sample, as opposed to the significant activity observed in placenta and TT (Figure 3B). Nevertheless, as only one probe (overlapping splice junction) of the sp.ervwe1_at probeset exhibited a significant intensity, which supports the env-specific splicing strategy, these results were confirmed by qRT-PCR. To reduce putative sample bias, four testicular sample pairs were investigated using two primer pairs (Supplementary Table S1) that amplified the 8 and 3.1-kb mRNAs in the env region (envW) and only the 3.1-kb mRNA (sp.ervwe1), respectively. All RT-PCR data were normalized on G6PD, which exhibited an average copy number among the experiments of 1000 copies per 5 ng of total RNA (250 cells). A significant activity was evidenced in three normal testicular samples (from 1700 to 5800 copies), except the one previously analyzed with the microarray. Conversely, no significant Syncytin-1-specific spliced mRNA expression was observed in all the normal testis samples. In all the four TT samples, the ERVWE1 locus expression level was found to be up-regulated (from 15 000 to 52 000 copies). Moreover, Syncytin-1 mRNA production appeared to be a common but heterogeneous attribute of TT samples (from 242 to 2733 copies) (Figure 3C), suggesting a change in ERVWE1 splicing strategy in the tumor.
Figure 3.
ERVWE1 locus splicing strategy in tumoral testis. (A) ERVWE1 genomic structure and transcripts. The transcription start site at the U3 promoter/R boundary of the 5′ LTR is represented by an arrow, and the splice donor site (DS) and the splice acceptor sites (AS1 and AS2) leading to the two major transcripts are indicated. ERVWE1-specific sequence signature is located by a star. The use of DS in combination with AS1 gives rise to an 8-kb transcript. mRNA (8-kb) contains gag, pol and env regions, and does not code for protein. The use of SD in combination with AS2 gives rise to a 3.1-kb transcript. The 3.1-kb mRNA contains the env ORF exclusively, and is responsible for the production of the Syncytin-1 protein. (B) ERVWE1 probesets. Location of the probesets designed for the detection of ERVWE1 overall transcription (envW_at) and Synyctin-1-specific mRNA (sp.ervwe1_at) are indicated by boxes under ERVWE1 mRNAs illustration. sp.ervwe1_at is composed of probes located at the SD/SA2 junction and probes located at the locus signature (boxes with star into). Position of each probe within each probeset is detailed below, and for each probeset, signal intensity in normal (light gray) and in related tumoral (dark gray) testis, and in placenta (black) samples are illustrated probe-by-probe. (C) qRT-PCR quantification of ERVWE1-spliced Syncytin-1 transcript in normal and tumoral testis. Syncytin-1-specific mRNA (sp.ervwe1) was evaluated by qRT-PCR in four pairs of normal and tumoral related testicular samples. Expressions are normalized with the expression of G6PD housekeeping gene having an averaged number of 1000 copies per 5 ng of total RNA in 250 cells. An arbitrary cutoff at 250 is indicated by a dotted line, mimicking one copy per cell. The star indicates the pair sample used in the microarray experiment.
Transcription of the reactivated HERV-W loci is essentially autonomous
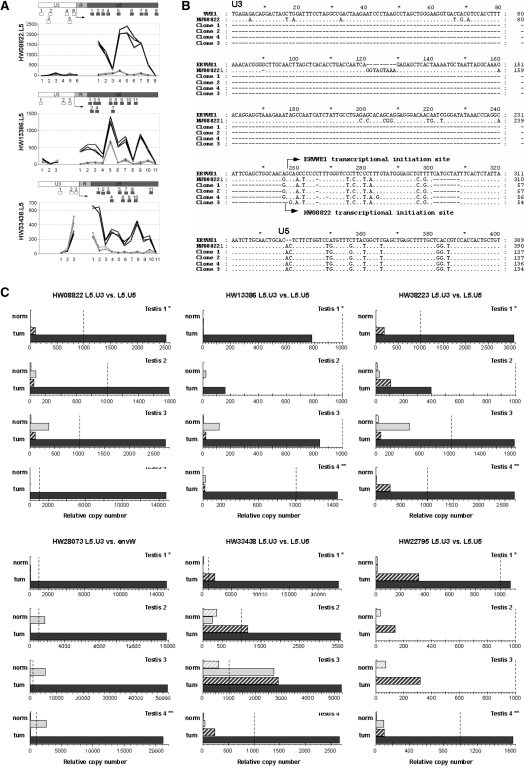
It is notable that among the 100 most differentially expressed probesets, the U5 regions are overrepresented when compared with the microarray composition (Supplementary Figure S1 and Figure 1). The U3, R and U5 juxtaposed regions of LTRs exhibit distinct putative functions. Conventionally, in proviral 5′LTR (and solo LTRs), U3 acts as a promoter and the R-U5 region is transcribed. In proviral 3′LTR (and putatively solo LTR), R acts as a polyadenylation signal and U3-R is transcribed. Thus, for a complete provirus, the larger transcript consists of RU5–gag–pol–env–U3R. As noted in the Materials and Methods section, divergence occurring between 5′ and 3′ LTRs since the proviral integration made it possible to select distinct probes and amplification primers for the 5′ and 3′LTRs sub regions. In line with this, all the HERV-W loci reactivated in testicular tumor are observed to possess full-length LTRs (5′ or solo), and only the U5 regions have differentially expressed probesets. Associated U3 probesets targeting 5′LTRs for HW08822, HW13386 and HW38223 and included in the microarray are poorly and not differentially expressed in the normal and tumoral testicular samples (Figure 4A). The divergence in microarray intensities between the U3 and U5 regions for each individual LTR could reflect an autonomous (U3-mediated) activation of these loci. Such information is not available for ERVWE1, HW22795 and HW38223 as U3 probesets could not be designed.\
Figure 4.
Identification of autonomous versus non-autonomous LTR activity. (A) Microarray data on U3 versus U5 activity. The dichotomic expressions observed between the U3 and U5 regions of the LTR are represented probe-by-probe for the three identified loci with U3 and U5 parts present on the microarray. Dark gray represents tumoral-testis intensity of the probes, and light gray shows the normal-testis intensity. The positions of all probes along the LTR are schematized below by boxes. The putative initiation site is indicated by an arrow. (B) 5′-RACE identification of autonomous transcriptional induction for the locus HW08822. Sequence alignment of ERVWE1 5′LTR, HW08822 5′LTR and clones obtained by 5′ RACE are represented. The transcriptional initiation site of ERVWE1 is indicated by an arrow as well as the initiation site of HW08822 observed in the three clones. TT1/TN1 match pair was the sample used as in microarray experiment. (C) U3- and U5-specific qRT-PCR s in four testicular normal/tumoral pairs. Expressions of the loci identified as overexpressed in the testicular tumors are represented by dendrograms. Expressions in LTR U3 part are in hatched gray boxes and expressions in LTR U5 part are in filled boxes. Expressions in normal RNA are in light gray and those in tumoral RNA are in dark gray. Expressions are normalized with the expression of G6PD housekeeping gene, which had an averaged copy number of 1000. The values for 1000 copies of G6PD gene are represented by a dotted line. The normal/tumoral sample pair used in the microarray experiment is Testis 1 (*).
We investigated the HERV-W LTRs transcription start by 5′-RACE experiments on the tumoral testicular mRNA. The results were conclusive for one locus, namely HW08822. The identified cap site of HW08822 mRNA correlated with the one previously described for ERVWE1 at the U3/R boundary (Figure 4B), thus confirming the promoter action of the U3 region in the testicular tumor transcriptional induction. Alternatively, using qPCR, we performed comparative assays on the transcriptional levels of the U3 regions versus the U5 regions for the six overexpressed HERV-W loci in the four normal/tumoral testicular sample pairs. In this assay, an autonomous driving was reflected by the U5-only transcription, and three main transcriptional profiles were observed (Figure 4C).
First, we observed a clear dichotomic expression between U3 and U5 regions for the loci HW08822, HW13386, HW38223 and HW28073/ERVWE1. The U3 regions were not transcribed in the normal testicular tissues and were absent or poorly expressed in the tumoral section. On the contrary, the tumoral transcriptional increase of the U5 region was confirmed in all the four sample pairs, even if activation (absolute copy number) varied essentially as a function of the LTR. In addition, in normal tissues, it is noticeable that the apparent level of U5 expression of these four loci varied among the samples, from a non-significant expression (<450 copies) for HW08822, HW13386 and HW38223 to a significant expression in three samples (from 1730 to 5000 copies) for ERVWE1. Thus, transcriptional profiles of HW08822, HW13386, HW38223 and ERVWE1 clearly suggest a transcriptional induction through the activation of the U3 promoter.
Second, the HW33438 locus roughly presented the same dichotomic expression, that is, a relatively low expression of the U3 region when compared with a high expression of the U5 region in all the four tumoral samples. Noticeably, an overexpression in the U3 region was observed in all the tumoral samples within a range between 1160 and 2870. The expression level of the U5 region, which exhibited intersample variations similar to those described for the loci of the first group, varied from 3500 to 26 700 copies in the tumoral samples and from 25 to 2700 copies in the normal counterpart.
Third, for the HW22795 locus, the situation was much more complex. The U3–U5 dichotomy described earlier was observed in T1 and T4 samples, that is, a non-significant expression of U3 region in the normal and tumoral tissues versus U5 expression restricted to tumoral tissues. Conversely, an exclusive U3 expression was evidenced in T2 and T3 samples. In these samples, U3 expression was consistently higher in the tumoral part of the sample, but remained faint, reaching a maximum of 347 copies.
Overall, these results suggest the existence of different transcriptional regulation. Nevertheless, these results highly suggest that the testicular tumoral expression of at least five out of the six HERV-W loci is driven by the HERV locus-specific promoter located in the U3 region of the LTR.
Global amount of methylation in differentially expressed HERV-W loci and control HERVs between normal and tumoral testicular samples
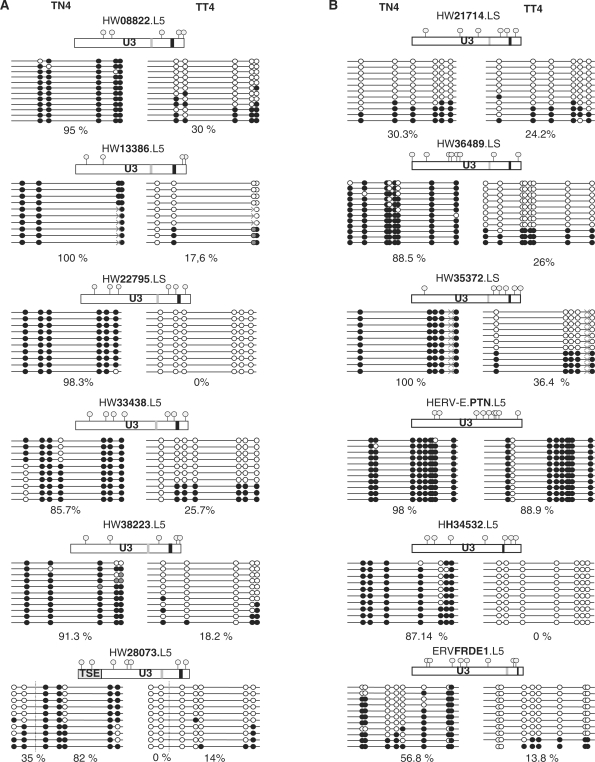
Among the possible regulatory changes that could lead to the observed up-regulation, methylation appears to be most frequently associated with tumoral contexts. To validate this hypothesis, we analyzed the methylation status of the LTR promoter part in the normal/tumoral DNA related to the normal/tumoral testicular mRNA sample number 4. Sequenced individual clones resulting from the bisulfite-PCR-sequencing method are shown in Figure 5. We observed a difference in the methylation status between the tumoral and the normal DNA for all the reactivated loci. The global amount of methylated CpGs varied from 82 to 100% in normal DNA. In contrast, in tumoral DNA, it varied from 0 to 30%. Furthermore, in the tumoral DNA, 42–100% of the sequences were completely unmethylated as illustrated in Figure 5A, whereas none was found unmethylated in the normal counterpart. Intriguingly, the ERVWE1 (placental) enhancer sequence, TSE, was found completely unmethylated in half of the sequences of this normal section, although the apparent increase in the frequency of unmethylated sequences between normal and tumoral tissue was observed to be consistent with the increase in the expression of the 6 HERV-W loci. This suggests that DNA methylation could be involved in the control of expression of HERV-W elements in this tumoral context.
Figure 5.
U3 promoter global methylation comparison in normal versus tumoral testis. (A) Global methylation comparison in the differentially expressed HERV-W LTR. (B) Global methylation comparison in the HERV control LTR. Comparison of CpG methylation status in normal (TN4) and tumoral (TT4) testicular cancer sample of the U3 part of the six differentially expressed HERV-W LTRs (column A) and six HERV LTR controls, namely, three of the HERV-W family and one of each of the HERV-E, HERV-H and HERV-FRD families (column B). All U3 parts are represented and CpG positions are noted by gray circles; CAAT boxes are noted in light gray; and TATAA boxes in darker gray. Highly mutated CAAT and TATAA boxes, in comparison with the reference sequence, are not represented (e.g. in the HERV-E.PTN LTR). Methylated CpG are represented in solid circles and unmethylated CpG are in open circles, undetermined methylation status is in gray circles. Identified SNPs are represented by gray crosses. The percentage of U3 global methylation is indicated for each LTR. For ERVWE1 locus, both the 5′ LTR U3 region and the upstream placental enhancer (TSE) are represented, and the percentage of methylation is indicated for each region.
To assess whether the hypomethylation of HERV-W elements in testicular cancer is restricted to these loci, we analyzed the methylation status of three additional HERV-W LTRs that appeared unexpressed in all the RNA samples. One locus (HW21714) was found to be phylogenetically related to active LTRs, whereas two (HW35372 and HW36489) were related to inactive LTRs as previously defined [(44), S. Prudhomme, unpublished data]. We observed two cases of methylation behavior, that is, either a methylation decrease appeared in the tumoral tissue or a low methylation was observed in both the tissues (normal and tumoral) (Figure 5B). Precisely, the global amount of methylation was over 88.5% in the normal DNA and lower than 36.4% in the tumoral DNA for the loci HW36489 and HW35372. Furthermore, in the tumoral DNA, up to 63.6% of the sequences were fully unmethylated, as illustrated in Figure 5A, whereas none was found unmethylated in the normal counterpart. For the locus HW21714, the percentage of methylation observed was 30.3% and 24.2% in normal and tumoral DNA, respectively, with a majority of fully unmethylated sequences in both the cases. To assess whether the testicular tumor-associated hypomethylation of HERV is restricted to the family W, we analyzed the HERV loci from other families, namely, HERV-H, HERV-E and HERV-FRD, that is, the loci HH34532 overexpressed in colorectal cancer, and HERV-E.PTN and ERVFRDE1 expressed in the placenta. Again, we observed two cases of methylation behavior, either a methylation decrease that appeared in the tumoral tissue or a high methylation observed in both the tissues (normal and tumoral). In the first case (ERVFRDE1 and HERV-H/HH34532), the amount of methylation was over 56.9%, with no fully unmethylated sequences in the normal tissue, and was lower than 13.8% in the tumoral tissue, with 80–100% of fully unmethylated sequences. In the second case (HERV-E.PTN), the amount of methylation was stable and over 88.9% in both the tissues.
Altogether, these results suggest that the methylation deregulation in this tumoral context concerns neither the HERVs as a whole nor the differentially expressed HERV-W loci alone.
DISCUSSION
HERV transcriptional activities have been evidenced in several physiological and pathological contexts. However, the multicopy nature of these repeated elements leads to substantial difficulties to identify which locus (loci) is (are) related to the observed activities, and whether such locus (loci) expression resulted from self-activation or readthrough. As a consequence, this often limits the range of the results and the determination of the physiological or pathological contribution that could have one or several HERV loci within a family. Thus, several research groups are currently developing specific tools to analyze HERV transcriptome. This study is the first attempt to elaborate a high-density microarray designed to specifically detect the activity of HERV individual sequences.
First, the probes were designed following the most simple/basic criteria excluding all other criteria (i.e. uniqueness), indicating that theoretically, each single sequence belonging to one locus and presenting at least one mutation, as referred to all other loci, is selected onto the microarray. In the case of HERV families, with a divergence between the loci below 20%, these criteria were relatively stringent, leading to several regions without any unique probe. In addition, despite being unique, a probe dedicated to a particular locus may not be specific under the hybridization conditions on the chip. Second, owing to the limited number of features on the chip and the redundancy of information about the neighboring overlapping probes, a probe selection process was developed. The choice between the neighboring probes was realized to preferentially select the probes with the informative mismatches located in their center part. These criteria of selection, which was preferred to the probe’s quality or probeset composition, as theoretically favoring specificity, led to two main adverse effects: (i) the exclusion of good quality probes and (ii) the creation of poorly representative probesets with few probes. An optimized solution would take into account all three parameters in the selection algorithm. Overall, this process of selection may explain the observed cross-reactivities, and thus, the difficulty to trace back the original loci, although clear differential expression may be attributed to the given probesets. In addition, exhaustivity and redundancy problems of GenBank release 128, when compared with the present assembled version of the human genome (release 171), in particular for repetitive elements, suggest a bias in the data used for this chip design. Hence, with respect to the number of identified active loci, this version of the microarray seems less efficient than other global approaches such as PCR coupled with sequencing (30) or GREM technique using PCR and terminating at the sequencing step (31). Conversely, the use of several probesets onto a locus provided more information concerning transcriptional regulation, than the PCR-based process did when targeting a single internal domain (29). Signals from independent probesets for U3 and U5 regions may reflect a promoter activity (U3 negative, U5 positive), a polyadenylation role (U3 positive, U5 negative), or a readthrough phenomena (U3 and U5 positive). Thus, the over-representation of the U5 regions for the 100 most differentially expressed LTR loci suggests that activation of HERVs, predominantly found in tumors, is essentially autonomous. More precisely, U3 negative versus U5 positive signals for HW08822, HW13386 and HW33438 loci, overexpressed in one TT sample, suggest an autonomous transcription, controlled by their respective U3 promoter. Nevertheless, we cannot formally exclude that some of the U5 observed activations are related to HERV pseudogenes lacking U3 promoter (45). In addition, the detection of high or significant signal intensities (i) in the placenta and TT for sp.ervwe1_at probeset targeting the Syncytin-1-specific splice junction and (ii) in testicular tumors for probesets detecting HML-2 double-spliced transcripts, np9 and cORF, indicate that the use of such microarray technology makes it possible to identify tissue-associated HERV splicing strategy, as previously described for conventional genes (46).
As an obligatory bridge between mRNA targets in samples and probes onto the chip, the amplification process employed is theoretically unbiased. Because of the use of random primers, this concerns the relative amplification efficiency of the 5′ and 3′ ends of mRNAs and the equivalency of amplification of distant versus closely related sequences, as opposed to gag–pol region-based PCR methods (30) or restriction enzyme coupled with the PCR-based method (31). This advantage is clearly illustrated by the efficient detection of 5′ terminal U5 regions, notably for the 100 most differentially expressed LTR loci that consist of both 5′ long proviral LTRs and short solo LTR. This is also illustrated by the simultaneous analysis of HERV-W, HML-2, HERV-E and HERV-H families, as primarily limited to the HML-2 family in other methods (30,32). Moreover, the relative abundance of all HERVs mRNA has been theoretically preserved using linear transcriptional amplification. The use of triplicates demonstrated remarkable robustness of the overall process (e.g. conserving relative abundance of distinct targets). The median-intensities correlation among the triplicates varied from 0.911 to 0.991 among all the samples (median = 0.979), highlighting the linearity of the relationship and the infrequency of the outliers. Moreover, by using raw data for each probe with a signal higher than 70, the median CV ranged from 11.3 to 24.7% (median 15.3), which is similar to the PCR-based target amplification coupled with hybridization onto a solid surface (25). In addition, the overall process was found to be less prone to ex vivo recombination, as observed with RT coupled with PCR exponential process (29).
Although the observed specificity problems limit the actual extent of the interpretable results from this version of the chip, several published data indicate that the microarray is reliable enough to reveal previously unidentified events. Yet, as a general concern regarding the biological significance of differential expression, it should be noted that further investigations will be required, as so-called normal tissue adjacent to the tumor is not necessarily the most appropriate control since (i) the cancer precursor cell is not necessarily neighboring the tumor depending on the type of cancer and conversely (ii) the adjacent tissue may represent a pre-cancerous state. Thus, all the samples evaluated (with the exception of placenta and ovary samples) with the chip technology were similar to those previously analyzed with a quantitative multiplex degenerate PCR coupled with an oligosorbent assay (MD-PCR OLISA), based on the relative conservation of the pol genes and by targeting nine families (25,26). The initial observations focused on the significant signals derived from the probesets of ERVWE1, ERVFRDE1 and ERV3/HERV-R loci, which are known to be highly expressed in the placenta (34,47–49). It can be noted that the p-value for the envR probeset differential intensity in the placenta was not as significant as that for ERVWE1 and HERV-FRD probesets, in accordance with the published envR expression in several other normal tissues, as opposed to ERVWE1 and ERVFRDE1 (18). As ERVWE1 probesets proved useful in analyzing placenta mRNA, the absence of signals in the breast tumoral sample may reflect a specific behavior of this particular sample, with respect to the report of ERVWE1 activation in 30% of breast cancer samples (7). Conversely, the absence of HERV-W family overexpression in the ovarian cancer sample, as previously described (50), may be owing to the lack of exhaustiveness of the HERV-W family onto the chip. The identification of several HERV-W loci in TT correlates with the initial observation of the overexpression of the HERV-W family in the same match-pair sample (25). The overexpression of at least five loci was confirmed by qRT-PCR on three additional seminomal samples. Thus, these HERV-W loci represent novel candidate cancer markers that need to be validated on larger sample panels.
However, we did not observe the previously described overexpressions of HERV-K(HML-2) types 1 and 2 in breast cancer (51–53). HERV-K(HML-2) expression in normal and tumoral breast samples is highly variable, and similar levels of expression are often found between the tumor and the adjacent normal breast tissues. However, the breast sample pair was previously investigated using MD-PCR OLISA that displayed more than 250-fold increase in HML-2 pol transcribed sequences (25). Although there is no definite explanation, the previously observed overexpressions may have resulted from the accumulation of a faint transcription of different and numerous loci, undetectable by the overall chip process based on statistical significance. Alternatively, because of the sequence uniqueness criterion used to design the probes, the HERV-K(HML-2) family may not be represented exhaustively on the microarray, as suggested earlier for the HERV-W family. Nevertheless, HML-2 5′LTR U5 regions, gag and env regions, as well as np9 and cORF probesets’ differential expressions, were found in the testicular tumor, in accordance with several reports on the presence of these HML-2 transcripts in testicular tumors (37). Moreover, the HML-2 overexpression previously observed in lung tumor sample (25) was limited to type 2 elements, as np9 but not Rec expression was detected.
The high intensity observed in HERV-H 5′LTR U5 regions and HERV-H envh3 gene probesets is in accordance with the report on HERV-H activity in colon tumor (25,27,54).
Finally, the differential expression presented by a probeset cluster from the HERV-E4.1 family in prostate, as previously described (21,25), and/or ovary and/or uterus tumors illustrates the heterogeneity of the transcriptional regulation process among the elements of this family. The fact that most of the corresponding probesets derived mainly from solo LTR (83%) may explain why such an expression have not been observed in tumoral uterus using the pol-based MD-PCR OLISA system (26). More precisely, these results putatively illustrate the existence of at least five HERV-E subgroups, namely, (i) overexpressed in prostate–ovary–uterus tumors; (ii) in prostate–ovary tumors; (iii) in prostate tumors only; (iv) down-regulated in tumoral uterus; and (v) highly expressed in both prostate and uterus tissues, irrespectively of their tumoral status. A preliminary phylogenetic analysis shows that all these elements are away from the placentally expressed subgroup consisting of HERV-E.PTN, HERV-E.EBR and HERV-E.APOC1 loci (10,55).
Analyses of the transcriptional regulation features of the several HERV-W loci, primarily identified as overexpressed in seminoma by the microarray whole procedure and confirmed using dedicated methods, are of general interest concerning the mechanisms leading to HERV contribution to the biological activities, essentially in human pathology.
First, a post-transcriptional alteration was illustrated by the ERVWE1 locus behavior in the tumoral section of four patients, evidenced using the microarray on one patient and confirmed by qRT-PCR on four samples. It has been originally described that the ERVWE1 locus can produce two major large-splice transcripts, a non-coding 8-kb mRNA and the Syncytin-1-related 3.1-kb mRNA, and a smaller largely-spliced 1.5-kb mRNA, at least in placenta (34). The 8-kb transcript was detectable in three out of four normal testicular samples, as formerly described (56). Interestingly, in the four seminomas, the overexpression of ERVWE1 was associated with an induction of the 3.1-kb mRNA production. However, whether the appearance of this spliced form reflects an overall increase in the transcription of the locus or results from a modification of the pool of cellular factors affecting the splicing strategy, needs further investigations. Most intriguingly, the presence of ERVWE1 3.1-kb transcript is presumed to lead to the production of the Syncytin-1 protein in seminoma, as observed quite frequently in other cancers (7–9).
Second, and of more general interest, the regulation of LTR expression in tumoral tissues was addressed by analyzing the six identified HERV-W elements overexpressed in seminoma. On one hand, it remains generally unclear whether transcribed HERV mRNA are LTR-promoted, LTR-polyadenylated, or readthrough products initiated from the neighboring conventional (non-retroviral) promoters. On the other hand, transcriptional re-activation in cancer could be attributable to several mechanisms, such as tumor-associated changes in the transcription factors (putatively altering transcription initiation), alterations in chromatin-histone code, or modifications in DNA methylation.
Using U3- and U5-specific primer pairs in real-time PCR comparative assays, we confirmed that all six HERV-W-loci-derived mRNA were U3-promoted. This mode of control of expression was exclusive and preserved for HW08822, HW13386 and HW38223 and ERVWE1/HW28073 elements in all patients. Coupled with this U3-driven expression, a classical 5′-R-dependent initiation of transcription has been suggested by the RACE analysis of the HW08822 locus in tumoral tissue. It is similar to the one identified with the domesticated ERVWE1 locus (34), and is different from the 3′-R-transcription start point of human-specific HML-2 elements (57). Nevertheless, whether this reflects a particular functionality or is a signature of the HERV-W family remains to be deciphered. U3-promoted transcription was also found to be a major but not an exclusive phenomenon for the HW33438 locus that additionally exhibited a low transcription of the U3 region. This observation may be owing to the generation of two independent transcripts, with the minor transcript resulting from an upstream initiation of transcription in the promoter domain (data not shown), possibly owing to an altered/inefficient TATAA box (58). Such a generation of two independent transcripts for the HW22795 locus, each containing either the U3 or the U5 LTR region, was also observed in two out of four patients. This kind of profile was previously reported by Vinogradova et al. (59) for HERV-K LTRs in the T-cell leukemia Jurkat cell line following heat-shock, as well as in seminoma. An overrepresentation of the U5 signal was found to indicate that the main mechanism is the U3-promoted sense-oriented transcription mechanism. Strikingly, such U3-containing mRNA was also detected in two other testis tumoral samples that lacked U5 expression. U3 expression of both HW22795 and HW33438 LTR may either reflect a transcriptional terminator function (U3-R-polyA) or an antisense transcript, as HERV-W LTR was shown to possess a bi-directional promoter activity (60). Alternatively, such features may reflect the heterogeneity of the tumor, leading to a combination of transcription factors targeting heterogeneously and independently in the subpopulation, HW22795 and HW33438 LTRs. In such situation, expression of HW08822, HW13386 and HW38223, as well as ERVWE1/HW28073 LTRs would not be affected by heterogeneity. Although speculative, it is worth noting that the activation of these HERVs may alter the expression of the adjacent genes (Supplementary Table S3); for instance, retroviral-induced production of GATAD1-interfering antisense transcripts owing to ERVWE1 reactivation (tail-to-tail elements) as previously described (12), and promoter competition between HW33438 and ADAMTS1 owing to the LTR-observed antisense activity (head-to-head elements), as described earlier (61).
Bisulfite-PCR sequencing of the six reactivated HERV-W loci initially showed that their promoter domains were highly methylated in the normal adjacent tissue, which is in agreement with the methylation-dependent silencing of the HERVs in most somatic cells (15–17). Conversely, in testicular cancer, a strong DNA hypomethylation of these activated LTR 5′ U3 promoter domains was observed. Nevertheless, analyses of three additional HERV-W loci that were not expressed showed that hypomethylation was tumor-associated in two of them and was an intrinsic attribute in one of them. In DNA isolated from a true normal testis tissue (fatal accident), CpG methylation of HW8822 active LTR and HW35372 inactive LTR were similar to the one observed in adjacent tissue (data not shown). Thus, analyses of this short collection of nine HERV-W LTRs suggest that the HERV-W intrafamily hypomethylation seems homogeneous in the tumoral section, irrespective of the transcriptional activation of the LTR copy. First, it is noticeable that LTR activity cannot be correlated with the age of the elements, as all active elements belong to the most recent HERV-W subfamily 3 and inactive LTR belongs both to the subfamily 3 and 2 (62). Thus, the difference in HERV-W elements’ transcriptional activity could be explained by the differences in the ability of the present transcriptional factors to bind onto each LTR. Second, taken as a whole, the median of methylation of those nine HERV LTRs drops from 91% in normal testis to 21% in the tumor, which is in agreement with methylation deregulations observed in seminoma, with the residual level of LTR methylation appearing higher than CpG methylation measured by immunohistochemistry (63). This suggests that demethylation in cancer does not equally affect all the components of the genome. More precisely, this suggests that the observed (de)methylation relies upon a (sub)family-dependent targeting, putatively based on sequence homology, which would have a direct impact on the risk and frequency of ectopic pairing and recombination among the homologous elements. It should be noted that seminoma was proposed to evolve from primordial germ cells (PGCs) (63,64), in which genome methylation is mostly erased before being reprogrammed along the spermatogenesis pathway (65). Therefore the observed homogeneous hypomethylation of the HERV-W LTRs in seminoma could at least partly reflect a physiological methylation state corresponding to PGCs. However, we cannot exclude that some HERV-W elements remain hypermethylated as proposed for malignant ovarian tissue, where a global hypomethylation of the HERV-W family was reported (50). Overall, the control of the (de)methylation process appears to be different in this pathological situation than that observed in the placenta physiological context, where (hypo)methylation of HERV sequences appeared limited to the functional domesticated elements (16,17). As opposed to the HERV-W elements, prototype LTRs of HERV-E, HERV-H and HERV-FRD families showed extremely different levels of methylation in the tumor. In the context of the PGCs scheme, this would indicate a distinct and apparently sequence-dependant remethylation process of HERV families (or at least particular elements) during spermatogenesis. Interestingly, the HERV-E LTR that remains highly methylated in the tumor is the element whose environment comprises scanty repeated elements and abundant genes (Supplementary Table S3). Nevertheless, environment is not the only factor influencing methylation, as the HW21714 LTR that is effectively hypomethylated in normal tissue is located within a gene-rich environment (Supplementary Table S3). Furthermore, the domesticated ERVFRDE1 locus, located in a region rich in repeated elements and genes (Supplementary Table S3), exhibits an unusually low level of methylation in the normal tissue, including almost completely unmethylated copies, when compared with the other HERV elements. Taken together, these observations suggest a multifactor process modulating methylation of HERVs in cancer, including preferably sequence (family), then chromosomal environment and possibly locus domestication/function. This is in agreement with the recent data showing that 5–8% of repetitive elements demonstrate cancer-related DNA methylation alteration, and that the loss of DNA methylation is most pronounced for certain members of the HERV families (66).
CONCLUDING REMARKS
This study is the first attempt to identify individual locus HERV expression using high density microarray format. When compared with the sequencing-based methods, the clearly identified advantages of the microarray format to decipher HERV transcriptome consist of (i) simultaneous detection of the differential activity of several, if not all, HERV families and (ii) simultaneous but independent examination of the different regions for each individual locus, including U3 and U5 parts of LTR, internal gag or env regions, and spliced junctions as well, without any a priori knowledge related to the functionality of the HERV sequence. Although the presented results represent the proof of concept of such method, the current version of the chip is far from this ideal situation. The microarray is currently being upgraded by improving the probes’ design and, in parallel, by increasing the exhaustiveness of HERV families. We are thus moving from the initial chosen criteria, uniqueness, to a second-generation one, namely, specificity. In addition, curated and assembled genome-based database, when compared with the release used for this chip version, may facilitate such a design. This first-generation HERV microarray allows for the confirmation of several previous independent reports on HERV expression in different contexts. Moreover, modifications of ERVWE1 splicing strategy in testicular tumor suggest that the implication of Syncytin-1 envelope has to be further investigated. Indeed, owing to its fusogenic, differentiation and anti-apoptotic properties, it is presumed to have a role of an oncogene or tumor suppressor in (testicular) cancer. Finally, the tumor-associated hypomethylation of the U3 LTR part of various HERV loci, whether reactivated or not, implies that alteration in methylation is a critical step toward HERV activation in cancers, and that the involvement of DNA methyltransferases in HERV targeting and regulation alterations in cancers should be addressed precisely.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Advanced Diagnostics for New Therapeutic Approaches (ADNA), a program dedicated to personalized medicine, coordinated by Mérieux Alliance and supported by the French public agency, OSEO. J.G. and C.M. are supported by a doctoral fellowship from bioMérieux; J.Ph.P. was supported by grants from ‘L’Association Nationale de la Recherche Technique (ANRT)’ and ‘L’Association pour la Recherche sur le Cancer (ARC)’; B.B. was supported by a doctoral fellowship from bioMérieux and Centre National de la Recherche Scientifique (CNRS) and a grant from ‘La fondation pour la recherche médicale (FRM)’. Funding for open access charge: bioMérieux SA.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 2.Katzourakis A, Rambaut A, Pybus OG. The evolutionary dynamics of endogenous retroviruses. Trends Microbiol. 2005;13:463–468. doi: 10.1016/j.tim.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 3.Blond JL, Lavillette D, Cheynet V, Bouton O, Oriol G, Chapel-Fernandes S, Mandrand B, Mallet F, Cosset FL. An envelope glycoprotein of the human endogenous retrovirus HERV-W is expressed in the human placenta and fuses cells expressing the type D mammalian retrovirus receptor. J. Virol. 2000;74:3321–3329. doi: 10.1128/jvi.74.7.3321-3329.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frendo JL, Olivier D, Cheynet V, Blond JL, Bouton O, Vidaud M, Rabreau M, Evain-Brion D, Mallet F. Direct involvement of HERV-W Env glycoprotein in human trophoblast cell fusion and differentiation. Mol. Cell Biol. 2003;23:3566–3574. doi: 10.1128/MCB.23.10.3566-3574.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mallet F, Bouton O, Prudhomme S, Cheynet V, Oriol G, Bonnaud B, Lucotte G, Duret L, Mandrand B. The endogenous retroviral locus ERVWE1 is a bona fide gene involved in hominoid placental physiology. Proc. Natl Acad. Sci. USA. 2004;101:1731–1736. doi: 10.1073/pnas.0305763101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Antony JM, van Marle G, Opii W, Butterfield DA, Mallet F, Yong VW, Wallace JL, Deacon RM, Warren K, Power C. Human endogenous retrovirus glycoprotein-mediated induction of redox reactants causes oligodendrocyte death and demyelination. Nat. Neurosci. 2004;7:1088–1095. doi: 10.1038/nn1319. [DOI] [PubMed] [Google Scholar]
- 7.Bjerregaard B, Holck S, Christensen IJ, Larsson LI. Syncytin is involved in breast cancer-endothelial cell fusions. Cell Mol. Life Sci. 2006;63:1906–1911. doi: 10.1007/s00018-006-6201-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Strick R, Ackermann S, Langbein M, Swiatek J, Schubert SW, Hashemolhosseini S, Koscheck T, Fasching PA, Schild RL, Beckmann MW, et al. Proliferation and cell-cell fusion of endometrial carcinoma are induced by the human endogenous retroviral Syncytin-1 and regulated by TGF-beta. J. Mol. Med. 2007;85:23–38. doi: 10.1007/s00109-006-0104-y. [DOI] [PubMed] [Google Scholar]
- 9.Larsen JM, Christensen IJ, Nielsen HJ, Hansen U, Bjerregaard B, Talts JF, Larsson LI. Syncytin immunoreactivity in colorectal cancer: potential prognostic impact. Cancer Lett. 2009;280:44–49. doi: 10.1016/j.canlet.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 10.Schulte AM, Lai S, Kurtz A, Czubayko F, Riegel AT, Wellstein A. Human trophoblast and choriocarcinoma expression of the growth factor pleiotrophin attributable to germ-line insertion of an endogenous retrovirus. Proc. Natl Acad. Sci. USA. 1996;93:14759–14764. doi: 10.1073/pnas.93.25.14759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ruda VM, Akopov SB, Trubetskoy DO, Manuylov NL, Vetchinova AS, Zavalova LL, Nikolaev LG, Sverdlov ED. Tissue specificity of enhancer and promoter activities of a HERV-K(HML-2) LTR. Virus Res. 2004;104:11–16. doi: 10.1016/j.virusres.2004.02.036. [DOI] [PubMed] [Google Scholar]
- 12.Gogvadze E, Stukacheva E, Buzdin A, Sverdlov E. Human-specific modulation of transcriptional activity provided by endogenous retroviral insertions. J. Virol. 2009;83:6098–6105. doi: 10.1128/JVI.00123-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mager DL, Hunter DG, Schertzer M, Freeman JD. Endogenous retroviruses provide the primary polyadenylation signal for two new human genes (HHLA2 and HHLA3) Genomics. 1999;59:255–263. doi: 10.1006/geno.1999.5877. [DOI] [PubMed] [Google Scholar]
- 14.Prudhomme S, Bonnaud B, Mallet F. Endogenous retroviruses and animal reproduction. Cytogenet. Genome Res. 2005;110:353–364. doi: 10.1159/000084967. [DOI] [PubMed] [Google Scholar]
- 15.Matouskova M, Blazkova J, Pajer P, Pavlicek A, Hejnar J. CpG methylation suppresses transcriptional activity of human syncytin-1 in non-placental tissues. Exp. Cell Res. 2006;312:1011–1020. doi: 10.1016/j.yexcr.2005.12.010. [DOI] [PubMed] [Google Scholar]
- 16.Gimenez J, Montgiraud C, Oriol G, Pichon JP, Ruel K, Tsatsaris V, Gerbaud P, Frendo JL, Evain-Brion D, Mallet F. Comparative Methylation of ERVWE1/Syncytin-1 and Other Human Endogenous Retrovirus LTRs in Placenta Tissues. DNA Res. 2009;16:195–211. doi: 10.1093/dnares/dsp011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reiss D, Zhang Y, Mager DL. Widely variable endogenous retroviral methylation levels in human placenta. Nucleic Acids Res. 2007;35:4743–4754. doi: 10.1093/nar/gkm455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.de Parseval N, Lazar V, Casella JF, Benit L, Heidmann T. Survey of human genes of retroviral origin: identification and transcriptome of the genes with coding capacity for complete envelope proteins. J. Virol. 2003;77:10414–10422. doi: 10.1128/JVI.77.19.10414-10422.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okahara G, Matsubara S, Oda T, Sugimoto J, Jinno Y, Kanaya F. Expression analyses of human endogenous retroviruses (HERVs): tissue-specific and developmental stage-dependent expression of HERVs. Genomics. 2004;84:982–990. doi: 10.1016/j.ygeno.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 20.Buscher K, Trefzer U, Hofmann M, Sterry W, Kurth R, Denner J. Expression of human endogenous retrovirus K in melanomas and melanoma cell lines. Cancer Res. 2005;65:4172–4180. doi: 10.1158/0008-5472.CAN-04-2983. [DOI] [PubMed] [Google Scholar]
- 21.Wang-Johanning F, Frost AR, Jian B, Azerou R, Lu DW, Chen DT, Johanning GL. Detecting the expression of human endogenous retrovirus E envelope transcripts in human prostate adenocarcinoma. Cancer. 2003;98:187–197. doi: 10.1002/cncr.11451. [DOI] [PubMed] [Google Scholar]
- 22.Smallwood A, Papageorghiou A, Nicolaides K, Alley MK, Jim A, Nargund G, Ojha K, Campbell S, Banerjee S. Temporal regulation of the expression of syncytin (HERV-W), maternally imprinted PEG10, and SGCE in human placenta. Biol. Reprod. 2003;69:286–293. doi: 10.1095/biolreprod.102.013078. [DOI] [PubMed] [Google Scholar]
- 23.Forsman A, Yun Z, Hu L, Uzhameckis D, Jern P, Blomberg J. Development of broadly targeted human endogenous gammaretroviral pol-based real time PCRs Quantitation of RNA expression in human tissues. J. Virol. Methods. 2005;129:16–30. doi: 10.1016/j.jviromet.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 24.Seifarth W, Frank O, Zeilfelder U, Spiess B, Greenwood AD, Hehlmann R, Leib-Mosch C. Comprehensive analysis of human endogenous retrovirus transcriptional activity in human tissues with a retrovirus-specific microarray. J. Virol. 2005;79:341–352. doi: 10.1128/JVI.79.1.341-352.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pichon JP, Bonnaud B, Cleuziat P, Mallet F. Multiplex degenerate PCR coupled with an oligo sorbent array for human endogenous retrovirus expression profiling. Nucleic Acids Res. 2006;34:e46. doi: 10.1093/nar/gkl086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pichon JP, Bonnaud B, Mallet F. Quantitative multiplex degenerate PCR for human endogenous retrovirus expression profiling. Nat. Protoc. 2006;1:2831–2838. doi: 10.1038/nprot.2006.475. [DOI] [PubMed] [Google Scholar]
- 27.Stauffer Y, Theiler G, Sperisen P, Lebedev Y, Jongeneel CV. Digital expression profiles of human endogenous retroviral families in normal and cancerous tissues. Cancer Immun. 2004;4:2. [PubMed] [Google Scholar]
- 28.Oja M, Peltonen J, Blomberg J, Kaski S. Methods for estimating human endogenous retrovirus activities from EST databases. BMC Bioinformatics. 2007;8(Suppl 2):S11. doi: 10.1186/1471-2105-8-S2-S11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Flockerzi A, Maydt J, Frank O, Ruggieri A, Maldener E, Seifarth W, Medstrand P, Lengauer T, Meyerhans A, Leib-Mosch C, et al. Expression pattern analysis of transcribed HERV sequences is complicated by ex vivo recombination. Retrovirology. 2007;4:39. doi: 10.1186/1742-4690-4-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Flockerzi A, Ruggieri A, Frank O, Sauter M, Maldener E, Kopper B, Wullich B, Seifarth W, Muller-Lantzsch N, Leib-Mosch C, et al. Expression patterns of transcribed human endogenous retrovirus HERV-K(HML-2) loci in human tissues and the need for a HERV Transcriptome Project. BMC Genomics. 2008;9:354. doi: 10.1186/1471-2164-9-354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Buzdin A, Kovalskaya-Alexandrova E, Gogvadze E, Sverdlov E. GREM, a technique for genome-wide isolation and quantitative analysis of promoter active repeats. Nucleic Acids Res. 2006;34:e67. doi: 10.1093/nar/gkl335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buzdin A, Kovalskaya-Alexandrova E, Gogvadze E, Sverdlov E. At least 50% of human-specific HERV-K (HML-2) long terminal repeats serve in vivo as active promoters for host nonrepetitive DNA transcription. J. Virol. 2006;80:10752–10762. doi: 10.1128/JVI.00871-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 34.Blond JL, Beseme F, Duret L, Bouton O, Bedin F, Perron H, Mandrand B, Mallet F. Molecular characterization and placental expression of HERV-W, a new human endogenous retrovirus family. J. Virol. 1999;73:1175–1185. doi: 10.1128/jvi.73.2.1175-1185.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bonnaud B, Bouton O, Oriol G, Cheynet V, Duret L, Mallet F. Evidence of selection on the domesticated ERVWE1 env retroviral element involved in placentation. Mol. Biol. Evol. 2004;21:1895–1901. doi: 10.1093/molbev/msh206. [DOI] [PubMed] [Google Scholar]
- 36.Lower R, Tonjes RR, Korbmacher C, Kurth R, Lower J. Identification of a Rev-related protein by analysis of spliced transcripts of the human endogenous retroviruses HTDV/HERV-K. J. Virol. 1995;69:141–149. doi: 10.1128/jvi.69.1.141-149.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Armbruester V, Sauter M, Krautkraemer E, Meese E, Kleiman A, Best B, Roemer K, Mueller-Lantzsch N. A novel gene from the human endogenous retrovirus K expressed in transformed cells. Clin. Cancer Res. 2002;8:1800–1807. [PubMed] [Google Scholar]
- 38.Naef F, Lim DA, Patil N, Magnasco M. DNA hybridization to mismatched templates: a chip study. Phys. Rev. E Stat. Nonlin. Soft Matter Phys. 2002;65:040902. doi: 10.1103/PhysRevE.65.040902. [DOI] [PubMed] [Google Scholar]
- 39.Team R DC. 2008. R: a language and environment for statistical computing. Ref Type: Computer Program. [Google Scholar]
- 40.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 42.Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl Acad. Sci. USA. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mallet F. Characterization of RNA using Continuous RT-PCR coupled with ELOSA. In: Rapley R, editor. The nucleic Acids Protocol Handdbook. Totowa, USA: Humana Press; 2000. pp. 219–228. [Google Scholar]
- 44.Schon U, Seifarth W, Baust C, Hohenadl C, Erfle V, Leib-Mosch C. Cell type-specific expression and promoter activity of human endogenous retroviral long terminal repeats. Virology. 2001;279:280–291. doi: 10.1006/viro.2000.0712. [DOI] [PubMed] [Google Scholar]
- 45.Pavlicek A, Paces J, Zika R, Hejnar J. Length distribution of long interspersed nucleotide elements (LINEs) and processed pseudogenes of human endogenous retroviruses: implications for retrotransposition and pseudogene detection. Gene. 2002;300:189–194. doi: 10.1016/s0378-1119(02)01047-8. [DOI] [PubMed] [Google Scholar]
- 46.Fehlbaum P, Guihal C, Bracco L, Cochet O. A microarray configuration to quantify expression levels and relative abundance of splice variants. Nucleic Acids Res. 2005;33:e47. doi: 10.1093/nar/gni047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gimenez J, Mallet F. ERVWE1 (Endogenous Retroviral family W, Env(C7), member 1) Atlas Genet Cytogenet Oncol Haematol. 2007 web: Ref Type: Electronic Citation. [Google Scholar]
- 48.Blaise S, de Parseval N, Benit L, Heidmann T. Genomewide screening for fusogenic human endogenous retrovirus envelopes identifies syncytin 2, a gene conserved on primate evolution. Proc. Natl Acad. Sci. USA. 2003;100:13013–13018. doi: 10.1073/pnas.2132646100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kato N, Pfeifer-Ohlsson S, Kato M, Larsson E, Rydnert J, Ohlsson R, Cohen M. Tissue-specific expression of human provirus ERV3 mRNA in human placenta: two of the three ERV3 mRNAs contain human cellular sequences. J. Virol. 1987;61:2182–2191. doi: 10.1128/jvi.61.7.2182-2191.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Menendez L, Benigno BB, McDonald JF. L1 and HERV-W retrotransposons are hypomethylated in human ovarian carcinomas. Mol. Cancer. 2004;3:12. doi: 10.1186/1476-4598-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Frank O, Verbeke C, Schwarz N, Mayer J, Fabarius A, Hehlmann R, Leib-Mosch C, Seifarth W. Variable transcriptional activity of endogenous retroviruses in human breast cancer. J. Virol. 2008;82:1808–1818. doi: 10.1128/JVI.02115-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang-Johanning F, Frost AR, Johanning GL, Khazaeli MB, LoBuglio AF, Shaw DR, Strong TV. Expression of human endogenous retrovirus k envelope transcripts in human breast cancer. Clin. Cancer Res. 2001;7:1553–1560. [PubMed] [Google Scholar]
- 53.Wang-Johanning F, Frost AR, Jian B, Epp L, Lu DW, Johanning GL. Quantitation of HERV-K env gene expression and splicing in human breast cancer. Oncogene. 2003;22:1528–1535. doi: 10.1038/sj.onc.1206241. [DOI] [PubMed] [Google Scholar]
- 54.Wentzensen N, Coy JF, Knaebel HP, Linnebacher M, Wilz B, Gebert J, von Knebel Doeberitz M. Expression of an endogenous retroviral sequence from the HERV-H group in gastrointestinal cancers. Int. J. Cancer. 2007;121:1417–1423. doi: 10.1002/ijc.22826. [DOI] [PubMed] [Google Scholar]
- 55.Medstrand P, Landry JR, Mager DL. Long terminal repeats are used as alternative promoters for the endothelin B receptor and apolipoprotein C-I genes in humans. J. Biol. Chem. 2001;276:1896–1903. doi: 10.1074/jbc.M006557200. [DOI] [PubMed] [Google Scholar]
- 56.Mi S, Lee X, Li X, Veldman GM, Finnerty H, Racie L, LaVallie E, Tang XY, Edouard P, Howes S, et al. Syncytin is a captive retroviral envelope protein involved in human placental morphogenesis. Nature. 2000;403:785–789. doi: 10.1038/35001608. [DOI] [PubMed] [Google Scholar]
- 57.Kovalskaya E, Buzdin A, Gogvadze E, Vinogradova T, Sverdlov E. Functional human endogenous retroviral LTR transcription start sites are located between the R and U5 regions. Virology. 2006;346:373–378. doi: 10.1016/j.virol.2005.11.007. [DOI] [PubMed] [Google Scholar]
- 58.Blake MC, Jambou RC, Swick AG, Kahn JW, Azizkhan JC. Transcriptional initiation is controlled by upstream GC-box interactions in a TATAA-less promoter. Mol. Cell Biol. 1990;10:6632–6641. doi: 10.1128/mcb.10.12.6632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vinogradova TV, Leppik LP, Nikolaev LG, Akopov SB, Kleiman AM, Senyuta NB, Sverdlov ED. Solitary human endogenous retroviruses-K LTRs retain transcriptional activity in vivo, the mode of which is different in different cell types. Virology. 2001;290:83–90. doi: 10.1006/viro.2001.1134. [DOI] [PubMed] [Google Scholar]
- 60.Lee WJ, Kwun HJ, Jang KL. Analysis of transcriptional regulatory sequences in the human endogenous retrovirus W long terminal repeat. J. Gen. Virol. 2003;84:2229–2235. doi: 10.1099/vir.0.19076-0. [DOI] [PubMed] [Google Scholar]
- 61.Conte C, Dastugue B, Vaury C. Promoter competition as a mechanism of transcriptional interference mediated by retrotransposons. Embo J. 2002;21:3908–3916. doi: 10.1093/emboj/cdf367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Costas J. Characterization of the intragenomic spread of the human endogenous retrovirus family HERV-W. Mol. Biol. Evol. 2002;19:526–533. doi: 10.1093/oxfordjournals.molbev.a004108. [DOI] [PubMed] [Google Scholar]
- 63.Netto GJ, Nakai Y, Nakayama M, Jadallah S, Toubaji A, Nonomura N, Albadine R, Hicks JL, Epstein JI, Yegnasubramanian S, et al. Global DNA hypomethylation in intratubular germ cell neoplasia and seminoma, but not in nonseminomatous male germ cell tumors. Mod. Pathol. 2008;21:1337–1344. doi: 10.1038/modpathol.2008.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sonne SB, Almstrup K, Dalgaard M, Juncker AS, Edsgard D, Ruban L, Harrison NJ, Schwager C, Abdollahi A, Huber PE, et al. Analysis of gene expression profiles of microdissected cell populations indicates that testicular carcinoma in situ is an arrested gonocyte. Cancer Res. 2009;69:5241–5250. doi: 10.1158/0008-5472.CAN-08-4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Morgan HD, Santos F, Green K, Dean W, Reik W. Epigenetic reprogramming in mammals. Hum. Mol. Genet. 2005;14(Spec No 1):R47–R58. doi: 10.1093/hmg/ddi114. [DOI] [PubMed] [Google Scholar]
- 66.Szpakowski S, Sun X, Lage JM, Dyer A, Rubinstein J, Kowalski D, Sasaki C, Costa J, Lizardi PM. Loss of epigenetic silencing in tumors preferentially affects primate-specific retroelements. Gene. 2009;448:151–167. doi: 10.1016/j.gene.2009.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.