Abstract
Background:
There is an important need to develop noninvasive biomarkers to detect disease in premature neonates. Our objective was to determine if salivary genomic analysis provides novel information about neonatal developmental gene expression.
Methods:
Saliva (50-200 μL) was prospectively collected from five premature infants at five time points: before, starting, and advancing enteral nutrition, at introduction of oral feeds, and at advanced oral feeds. Salivary RNA was extracted, amplified, and hybridized onto whole genomic microarrays.
Results:
Bioinformatic analyses identified 9,286 gene transcripts that showed statistically significant gene expression changes across subjects over time. Of these, 3,522 (37.9%) genes were down-regulated and 5,764 (62.1%) genes were up-regulated. Gene expression changes were highly associated with developmental pathways. Statistically significantly down-regulated expression was seen in embryonic development, connective tissue development and function, hematological system development and function, and survival of the organism (10−14 < p <10−3). Conversely, genes associated with behavior, nervous system development, tissue development, organ development, and digestive system development were statistically significantly up-regulated (10−11 < p < 10−2).
Discussion:
Comparative genomic salivary analyses provide robust, comprehensive, real-time information regarding nearly all organs and tissues in the developing preterm infant. This innovative and noninvasive technique represents a new approach for monitoring health, disease, and development in this vulnerable patient population. By comparing these data in healthy infants to those who develop medical complications, we expect to identify new biomarkers that will ultimately improve newborn care.
Keywords: Salivary genomics, neonatology, development, gene expression
Introduction
An estimated 540,000 infants are born prematurely in the United States each year, resulting in $12.6 billion annual health care costs. This neonatal population has unique and often severe medical sequelae, a consequence of the disruption of normal organ and tissue development (1). In particular, medical complications affecting the nervous system (i.e. developmental delay, cerebral palsy) (2, 3) and the gastrointestinal system (i.e. short gut syndrome following necrotizing enterocolitis) (4, 5) can result in lifelong morbidities. The ability to determine predisposing risk factors for these complications remains limited in neonates (6). Therefore, there is an important need to develop noninvasive biomarkers to detect disease early in order to initiate treatment (7).
Saliva is a body fluid that can be obtained noninvasively and repeatedly. Filtered and processed in the salivary glands from blood, saliva has been described as the ‘mirror of the body’ and the ‘perfect medium to be explored for health and disease surveillance’ (8). It is a rich source of nucleic acids, and recent technological advances allow stabilization of salivary RNA for downstream genomic applications (9). While genomic microarray analysis of adult saliva has been performed (10), no study to date has applied this technology to premature neonates.
We hypothesized that comparative microarray analyses of salivary RNA obtained serially from premature infants could provide novel information regarding their development and health, particularly with regard to their developing gastrointestinal and nervous systems. This discovery-driven approach could lead to a better understanding of the normal and abnormal developmental processes that occur in the premature infant and would potentially provide novel noninvasive biomarkers for assessment and diagnosis of this patient population, thus facilitating earlier treatment.
Materials and Methods
Study Subjects
This study was approved by the Tufts Medical Center Institutional Review Board. Infants (n=5) born between 28 and 32 weeks' gestation, without known genetic diseases or congenital anomalies, admitted to the Tufts Medical Center Neonatal Intensive Care Unit, were enrolled in the study with parental informed consent. Demographic and relevant clinical information regarding the subjects are shown in Table 1. Of note, there is one set of fraternal twins, and one set of identical twins from a set of quadruplets within this data set. Every attempt was made to control for medical complications and drug exposure among subjects (Table 1).
Table 1.
Clinical information regarding study subjects
| Subject | Gender | Gestational age at birth |
Birth Weight (grams) |
Medications (During Study) |
Medical Complications |
Nutrition |
|---|---|---|---|---|---|---|
| 1 | Male | 29 0/7 | 1389 |
|
|
Formula |
| 2 | Female Fraternal twin |
28 3/7 | 942 |
|
|
Breast Milk |
| 3 | Male Fraternal twin |
28 3/7 | 1123 |
|
|
Breast Milk |
| 4 | Female Identical twin from a set of quadruplets |
32 0/7 | 1683 |
|
|
Formula |
| 5 | Female Identical twin from a set of quadruplets |
32 0/7 | 1379 |
|
|
Formula |
Salivary Acquisition
Approximately 50-200 μL of saliva was collected from each enrolled neonate with a 1 mL syringe attached to low-wall suction. Salivary samples were collected at the following time points: 1) Prior to enteral nutrition (baseline); 2) At the start of enteral nutrition; 3) During the advancement of enteral nutrition; 4) At the start of oral feeding; and 5) At full or mostly full oral feeding. Saliva was collected prior to a scheduled feed to avoid milk contamination, and was stabilized in 1 mL of RNAProtect Saliva™ (Qiagen, Valencia, California) within 1 min of acquisition. Samples were briefly vortexed, placed immediately on ice, and then stored at 4°C for 48 to 96 h prior to RNA extraction.
Salivary RNA Extraction, Amplification, and Hybridization
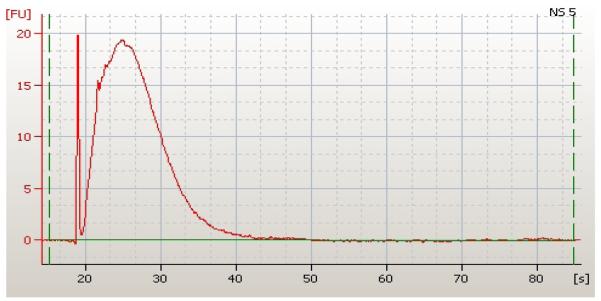
Total RNA extraction was performed with the RNeasy Protect Saliva Mini Kit (Qiagen, Valencia, California) as per the manufacturer's instructions. On-column DNase treatment occurred for each sample during RNA extraction. Eluted RNA was stored at −80°C prior to amplification with the WT-Ovation™ Pico RNA Amplification System (Nugen Technologies, San Carlos, California). The quantity and quality of amplified RNA was assessed with a Agilent™ Bioanalyzer 2100 (Agilent Technologies, Foster City, California) (Figure 1) prior to fragmentation and biotinylation with the FL-Ovation™ cDNA Biotin Module V2 (Nugen Technologies, San Carlos, California). For each sample, a standard 5 μg of amplified and labeled RNA was hybridized from each subject at each time point onto the Affymetrix™ HG U133 Plus 2.0 whole genomic microarray (total=25). Following hybridization, each array was washed and stained in the GeneChip® Fluidics Station 400 (Affymetrix, Santa Clara, California). Arrays were then scanned with the GeneArray Scanner (Affymetrix, Santa Clara, California) and initial analyses were performed using the GeneChip Microarray Suite 5.0 (Affymetrix, Santa Clara, California).
Figure 1.
Amplified neonatal salivary RNA as assessed by the Agilent™ Bioanalyzer. Time in seconds is on the x-axis and fluorescence is on the y-axis. Area under the curve represents quantity of RNA extracted from a salivary sample (849.11 ng/μL).
Bioinformatic and Computational Analyses
All calculations were done in R version 2.8.1 (11), Bioconductor version 2.3 (12) and lme4 (13). Probe sets were summarized and arrays were normalized using the RMA algorithm in the Bioconductor Affy package with default settings (14). For each probe set, we determined the influence of increasing postnatal age by fitting two statistical models. The first model fit a random individual subject effect. The second model fit a fixed linear age effect and random individual subject effect. Analysis of variance (ANOVA) testing was performed using the likelihood ratio test to compare the two models. P-values were adjusted using the BenjaminiHochberg procedure (15). Probe sets were identified as significantly differentially expressed for age when the false discovery rate (FDR) p-value was less than 0.05. T-scores were calculated to differentiate between those genes with significantly greater or lesser gene expression over time. To evaluate whether any one subject skewed the data, we also performed ANOVAs comparing subject and age interactions, and FDR p-values were calculated.
To determine the effect of feeding status as a result of gestational age on gene expression changes, we again compared two models. The first model fit age with a random subject effect; the second model accounted for whether the subject was orally fed (yes/no) and whether the subject was being fed via a nasogastric tube (yes/no). Analysis of variance (ANOVA) testing was performed using the likelihood ratio test to compare the two models. P-values were adjusted using the Benjamini-Hochberg procedure (15).
Subjects from Multiple Gestations
All five subjects in this data set were treated as individuals. To determine if the genetically related subjects (one fraternal twin set; one identical twin set from a set of quadruplets) could have led to biases caused by underestimating the variance between individuals, we performed an additional three regression analyses of expression as a function of postnatal age for each probe set: 1.) for all five subjects; 2.) for the two sets of related subjects; and 3.) for the identical twins only. The random individual subject effect was intentionally omitted so that any differences in subject groups would be reflected in the analysis. Mean square errors (MSE) were then calculated. To adjust for variability between probe sets, we took the log of the MSE from each of the additional three analyses, and for each gene subtracted the mean of the log MSE over each of the three regressions.
Data Analyses
Statistically significantly up or down-regulated gene transcripts were analyzed using the Ingenuity® software package to assess gene-gene relations, associated network functions, and physiological developmental systems. The top five significantly up-regulated and down-regulated physiological system development and function categories were further analyzed. For each of these categories, individual functions within a category that had 20 genes or more were assessed (Tables 2 and 3), and all individual genes were reviewed using EntrezGene to better understand the relevant physiological processes (Supplemental Data Tables 1 and 2).
Table 2.
Up-regulated gene expression categories and functions
| Category | Function (Functional Annotation) | # of genes | P Value |
|---|---|---|---|
| Behavior | Behavior | 147 | 1.2 × 10−11 |
| Learning (Learning) | 76 | 7.9 × 10−7 − 4.4 × 10−3 | |
| Cognition (Cognition) | 66 | 1.5 × 10−6 − 1.5 × 10−2 | |
| Psychological process | 68 | 9.2 × 10−6 | |
| Feeding (Feeding, feeding of organisms) | 80 | 1.2 × 10−5 − 8.7 × 10−3 | |
| Locomotion | 63 | 1.9 × 10−5 | |
| Memory | 32 | 2.0 × 10−4 | |
|
| |||
| Nervous System Development and Function |
Neurotransmission (Cells, normal cells, neurons, synapse) | 141 | 1.8 × 10−9 − 1.4 × 10−2 |
| Synaptic Transmission (Cells, normal cells) | 108 | 2.2 × 10−9 − 1.7 × 10−2 | |
| Differentiation (Neurons) | 70 | 4.9 × 10−7 | |
| Synaptogenesis | 21 | 5.0 × 10−7 | |
| Neurological Process (Cells, eukaryotic cells, normal cells, tissue, neurons, synapse, nervous tissue, nerves, brain, axons, cell lines, neuroglia) |
395 | 5.1 × 10−6 − 7.4 × 10−3 | |
| Development (Nervous system, neurons, central nervous system, nerves, dentate gyrus, brain, Schwann cells, olfactory receptor neurons, nervous tissue, forebrain, olfactory bulb, astrocytes, trigeminal ganglion neurons) |
319 | 1.1 × 10−4 − 1.7 × 10−2 | |
| Memory | 32 | 2.0 × 10−4 | |
| Branching (Dendrites, neurites) | 34 | 3.2 × 10−4 − 3.3 × 10−4 | |
| Neurogenesis | 101 | 9.6 × 10−4 | |
| Growth (Neurites) | 76 | 9.7 × 10−4 | |
| Quantity (Neurons, trigeminal ganglion neurons) | 47 | 1.3 × 10−3 − 1.1 × 10−2 | |
| Migration (Schwann cells, neurons) | 45 | 1.4 × 10−3 − 1.1 × 10−2 | |
| Outgrowth (Neurons) | 67 | 1.8 × 10−3 | |
| Myelination (myelination, nerves, normal cells) | 33 | 2.2 × 10−3 − 1.6 × 10−2 | |
| Survival (Trigeminal ganglion neurons, spinal cord cells, neurons, dorsal root ganglion cells, ganglion cells) |
61 | 2.7 × 10−3 − 1.2 × 10−2 | |
| Proliferation (Neurons, neuronal progenitor cells | 29 | 4.9 × 10−3 − 5.9 × 10−3 | |
|
| |||
| Tissue Development | Development (Tissue, cartilage tissue, nerves, placode, muscle, metanephros, epithelial tissue, fetal membranes, connective tissue, nervous tissue, allantois, bone marrow, olfactory bulb) |
394 | 4.6 × 10−7 − 1.7 × 10−2 |
| Developmental Process (Tissue, nerves, cartilage tissue, muscle, epithelial tissue, placode, nervous tissue, atrioventricular canal cushion, flexor muscle) |
363 | 4.5 × 10−5 − 1.7 × 10−2 | |
| Formation (Cortical bone, mammary placode 3, connective tissue, bone, tissue, trabecular bone) |
125 | 4.9 × 10−3 − 9.6 × 10−3 | |
|
| |||
| Organ Development | Development (Organ, testis, gonad, dentate gyrus, brain, ovarian follicle, heart, prostate gland, metanephros, kidney, ovary, forebrain, lung) |
454 | 6.0 × 10−6 − 1.6 × 10−2 |
| Organogenesis | 139 | 7.5 × 10−6 | |
| Morphogenesis | 41 | 2.4 × 10−3 | |
|
| |||
| Digestive System Development and Function |
Feeding (Feeding, feeding of organism) | 80 | 1.2 × 10−5 − 8.7 × 10−3 |
Table 3.
Down-regulated gene expression categories and functions
| Category | Function (Functional annotation) | # of genes | p value |
|---|---|---|---|
| Embryonic Development | Disease (Embryonic cell lines) | 86 | 7.2 × 10−14 |
| Infectious Disorder (Embryonic cell lines) | 84 | 3.9 × 10−13 | |
| Apoptosis (Embryonic cell lines) | 30 | 1.2 × 10−3 | |
| Cell Death (Embryonic cells lines) | 37 | 9.3 × 10−3 | |
|
| |||
| Connective Tissue Development and Function |
Proliferation (Fibroblast cell lines, fibroblasts) |
97 | 4.8 × 10−8 − 2.6 × 10−6 |
| Growth (Fibroblast cell lines, fibroblasts) | 75 | 3.8 × 10−6 − 1.3 × 10−4 | |
| G1 phase (Arrest in G1 phase of fibroblast cell lines; G1 phase of fibroblast cell lines) |
26 | 7.5 × 10−4 − 1.2 × 10−3 | |
|
| |||
| Hematological System Development and Function |
Differentiation (Blood cells, leukocytes, T lymphocytes, lymphocytes) |
272 | 1.0 × 10−7 − 7.3 × 10−4 |
| Hematopoiesis | 116 | 1.4 × 10−7 | |
| Hematological process | 129 | 2.4 × 10−6 | |
| Quantity (Lymphocytes, mononuclear leukocytes, blood cells, leukocytes, B and T lymphocytes) |
438 | 5.4 × 10−6 − 2.2 × 10−3 | |
| Development (Blood cells, leukocytes, lymphocytes, T lymphocytes) |
196 | 2.0 × 10−4 − 9.2 × 10−4 | |
| Proliferation (Leukocytes, lymphocytes, T lymphocytes) |
356 | 6.4 × 10−4 − 1.7 × 10−3 | |
|
| |||
| Hematopoiesis | Differentiation (Leukocytes, T lymphocytes, lymphocytes) |
178 | 1.1 × 10−7 − 7.3 × 10−4 |
| Hematopoiesis | 116 | 1.3 × 10−7 | |
| Development (Leukocytes, lymphocytes, T lymphocytes) |
135 | 9.2 × 10−4 | |
|
| |||
| Organismal Survival | Death (mammalia, animal) | 503 | 1.4 × 10−6 − 1.2 × 10−3 |
Results
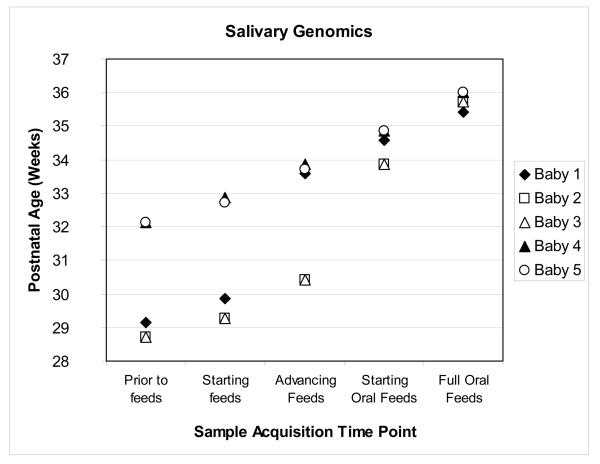
For each infant, five gene expression arrays were analyzed from each of the five previously described time points (total=25 arrays). There were 24 days of postnatal age separating the youngest and oldest infants at the time of the first salivary collection. At the time of the final salivary sample acquisition, when the infants were successfully orally feeding, these same infants were separated by only two postnatal days (Figure 2). Bioinformatic analyses revealed that gene expression was more statistically significantly affected by postnatal age rather than feeding status. There were no statistically significant changes in gene expression based upon feeding status with a FDR p-value <0.05. Conversely, of the 54,675 probes on the array, 9,286 (17%) gene transcripts showed significant changes in gene expression over time with a FDR p-value of <0.05. Calculated T-scores for the gene transcripts revealed that 3,522 genes (37.9%) were significantly down-regulated and 5,764 genes (62.1%) were significantly up-regulated. FDR p values for all interactions comparing subject and age were >0.20, suggesting that our results were not driven by any one subject.
Figure 2.
Scatter-plot depicting postnatal age of subjects at time of sample acquisition. Feeding milestone is on the x axis; postnatal age in weeks, on the y axis.
Significantly Up-Regulated Gene Transcripts over Time
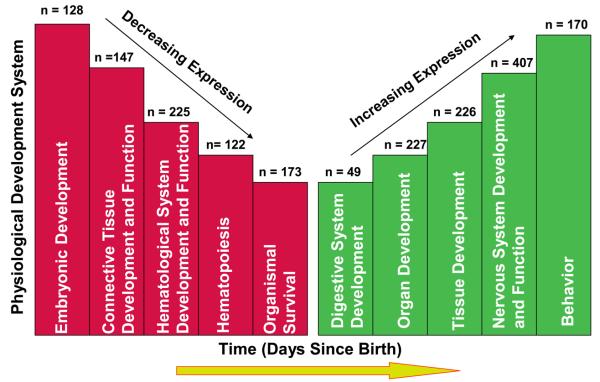
The top five up-regulated physiological systems that were statistically significantly affected by postnatal age were behavior (10−11 < p <10−2), and development of the nervous system (10−9 < p < 10−7), tissues (10−7 < p < 10−2), organs (10−6 < p < 10−2), and the digestive system (10−5 < p < 10−2) (Figure 3). Functional descriptions within each category, the number of genes within a specific function, and respective p-values are shown in Table 2. The complete list of up-regulated genes and their functional descriptions for each category are in Supplemental Data Table 1. Individual genes may be listed in more than one category.
Figure 3.
Schematic depiction of the five most significantly up-regulated and down-regulated physiological developmental systems (10−14 < p< 10−2) within this data set. Days since birth (x axis) represents real-time gene expression changes occurring with increasing postnatal age. While all categories were highly significant, the height of an individual column represents the degree of significance within a group (i.e. the higher the column the more statistically significant the system). Values at the top of each column represent the number of genes within the given category. Certain genes were considered significant in more than one category. Note that the figure is not drawn to scale.
Significantly Down-Regulated Gene Transcripts over Time
The top five down-regulated physiological systems were embryonic development (10−14 < p < 10−3), connective tissue development and function (10−8 < p < 10−3), hematological system development and function (10−7 < p < 10−3), hematopoiesis (10−7 < p < 10−4) and survival of the organism (10−7 < p <10−3) (Figure 3). Functional descriptions within each category, the number of genes within a function and the respective p values are shown in Table 3. The complete list of down-regulated genes and their functional descriptions for each category are provided in Supplemental Data Table 2. Individual genes may be listed in more than one category.
Analysis of Subjects from Multiple Gestations
The (natural) log MSE varied from −6.3 to 2.3 across probe sets, with quartiles at −1.8 and −0.5 (Supplemental Data Figure 1). The mean log MSE for all five subjects after subtracting the mean across the cases was 0.015 (A); the mean for the two sets of twins was −0.011 (B); the mean for the identical twins was −0.003 (C). The differences were small and were not in the hypothesized order (i.e.C < B < A). Thus, it is unlikely that our results were biased by including subjects that are genetically related to each other.
Discussion
Our work is the first to demonstrate the potential diagnostic and clinical utility of transcriptional salivary analysis in premature neonates. It lays the foundation for prospective clinical studies that develop hypotheses regarding abnormal gene expression in neonatal pathophysiology. We demonstrate here that comparative genomic analyses of salivary mRNA transcripts obtained from premature neonates during the first weeks of postnatal life provide novel dynamic information regarding nearly all developing organs and tissues.
Despite the limited quantities of saliva obtained for analysis in this study, the assay used here provides a comprehensive genomic analysis of both neonatal salivary cells and supernatant. This approach is novel compared to previously published reports on the cell-free adult salivary transcriptome obtained from supernatant (10). While the cellular source of the salivary samples is currently unknown, it likely contributes to the RNA pool in these genomic analyses. Future studies will be required to elucidate cell source and the respective contributions of RNA from both the supernatant and cellular layers.
There were 9,286 genes identified with statistically significant gene expression changes that occurred over time. While the achievement of oral feeding and advancing postnatal age are inherently linked, our analysis revealed that advancing postnatal age, rather than attainment of a feeding milestone, resulted in significant gene expression changes. The convergence of the postnatal ages of the subjects with the acquisition of successful oral feeding skills likely contributed to this finding (Figure 2). Although there were two sets of genetically related subjects within this data set, our additional statistical analysis suggested that no bias was introduced into our analysis by their inclusion.
While a comprehensive physiological and functional analysis of the complete gene list is beyond the scope this report, the data demonstrate important changes in pathways associated with neurodevelopment and digestion. The up-regulation of digestive system developmental transcripts revealed enzymatic genes necessary for the proper processing of enteral nutrition, neuronal genes regulating satiety and food consumption, and structural genes associated with normal dentition formation. Examples of these up-regulated genes include LALBA, a principal milk protein that enables lactose production; CCKAR, a major physiologic mediator of pancreatic enzyme secretion and smooth muscle contraction of the gallbladder and stomach; HCRTR2, involved in stimulation of food intake; MCHR1, involved in neuronal regulation of food consumption; and DMP1, an extracellular matrix protein crucial for proper mineralization of bone and dentin (Supplemental Data Table 2).
Within the ‘Nervous System Development and Function’ category, nearly every functional aspect of the developing brain, and peripheral and central nervous systems was represented by the salivary gene list, including neuronal development, myelination, synaptic formation, and neurogenesis. These findings also coincide with the major ‘burst’ of active brain growth that occurs in the last half of human gestation (16). Interestingly, genes associated with cranial nerve V (the trigeminal nerve) function were specifically highlighted by this analysis. Though primarily involved in facial sensation, the trigeminal nerve has associated motor functions that include biting, chewing and swallowing. One of the most important and difficult neurological tasks facing the premature neonate is the successful coordination of sucking and swallowing to facilitate oral feeding. In most neonatal intensive care units, the determination of an infant's readiness to feed is largely subjective. We speculate that salivary monitoring of gene expression data related to trigeminal nerve development may provide clear and objective evidence of a premature infant's ability to successfully feed orally.
We acknowledge that our work represents an early proof of principle study. These findings will need to be validated by an independent cohort, and with independent technologies such as reverse-transcriptase polymerase chain reaction (RT-PCR). As with any human subject study, each infant in this cohort had a unique clinical course. However, every attempt was made to control for similar drug exposure and outcome. It is unlikely that the small clinical between-infant variation observed in this population contributed to the findings of this study. Rather, a major strength of our salivary genomic analysis is the strong clinical correlation between the identified statistically significantly up-regulated and down-regulated physiological systems and expected neonatal physiology. The most significantly down-regulated physiological system was embryonic development. Over time, genes involved in neurogenesis during embryonic development, such as AES, and genes involved in the arrangement of three-dimensional tissue structure and angiogenesis, such as CEACAM1, were actively suppressed as the infants matured. Simultaneously, genes involved in lung development, including SFTPB, surfactant protein B, and tissue development, such as FREM2, which is required for maintaining skin epithelium, were up-regulated over time and were detectable in neonatal saliva. These findings highlight important normal physiological processes occurring in multiple developing organs within this population. Thus, transcripts identified in the saliva of these relatively healthy premature neonates can serve as a comparison transcriptome to gestationally-age matched infants who suffer from severe neonatal sequelae involving the lungs (i.e. bronchopulmonary dysplasia), the gastrointestinal system (i.e. necrotizing enterocolitis), the eyes (i.e. retinopathy of prematurity), and the immune system (i.e. sepsis).
While we acknowledge that serial neonatal salivary microarray analyses may be cost prohibitive for many research centers, the technique described here for salivary acquisition, stabilization, and RNA extraction is feasible, reproducible, and cost effective (approximately U.S. $11/sample). Alternative down-stream applications, including RT-PCR for specific genes of interest identified in this paper, could be used for large scale, international studies.
In summary, salivary genomic analyses provide a noninvasive means of assessing developmental progression in the premature neonate. This technique provides a large amount of data from a single sample. In particular, we demonstrated the dynamic nature of genes expressed as part of the neurodevelopmental and digestive systems in the first few weeks of postnatal life. By comparing these data in healthy preterm infants to those who develop medical complications, we expect to develop new noninvasive biomarkers that will ultimately improve newborn care.
Supplementary Material
Acknowledgments
We would like to thank the families who graciously participated in this research, and the nursing and medical staff of the Tufts Medical Center Neonatal Intensive Care Unit. We would also like to thank Dr. Donna Slonim for her bioinformatic contributions, and Helene Stroh for laboratory support.
Human Genes
- LALBA
lactalbumin, alpha
- CCKAR
cholecystokinin A receptor
- HCRTR2
hypocretin receptor 2
- MCHR1
melanin-concentrating hormone receptor 1
- DMP1
dentin matrix acidic phosphoprotein 1
- AES
amino terminal enhancer of spit
- CEACAM1
carcinoembryonic antigen-related cell adhesion molecule 1
- SFTPB
surfactant protein B
- FREM2
FRAS1 related extracellular matrix protein 2
References
- 1.Ward RM, Beachy JC. Neonatal complications following preterm birth. BJOG. 2003;110:8–16. doi: 10.1016/s1470-0328(03)00012-0. [DOI] [PubMed] [Google Scholar]
- 2.Kinney HC. The near-term (late preterm) human brain and risk for periventricular leukomalacia: A review. Semin Perinatol. 2006;30:81–8. doi: 10.1053/j.semperi.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 3.Adams-Chapman I. Insults to the developing brain and impact on neurodevelopmental outcome. J Commun Disord. 2009;42:256–62. doi: 10.1016/j.jcomdis.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 4.Vennarecci G, Kato T, Misiakos EP, Neto AB, Verzaro R, Pinna A, et al. Intestinal transplantation for short gut syndrome attributable to necrotizing enterocolitis. Pediatrics. 2000;105:E25. doi: 10.1542/peds.105.2.e25. [DOI] [PubMed] [Google Scholar]
- 5.Guner YS, Chokshi N, Petrosyan M, Upperman JS, Ford HR, Grikscheit TC. Necrotizing enterocolitis—bench to bedside: novel and emerging strategies. Semin Pediatr Surg. 2008;17:255–65. doi: 10.1053/j.sempedsurg.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 6.Bassler K, Stoll BJ, Schmidt B, Asztalos EV, Roberts RS, Robertson CM, et al. Using a count of neonatal morbidities to predict poor outcome in extremely low birth weight infants: added role of neonatal infection. Pediatrics. 2009;123:313–8. doi: 10.1542/peds.2008-0377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maresso A, Broeckel U. The role of genomics in the neonatal ICU. Clin Perinatol. 2009;36:189–204. doi: 10.1016/j.clp.2008.09.015. [DOI] [PubMed] [Google Scholar]
- 8.Segal A, Wong DT. Salivary diagnostics: enhancing disease detection and making medicine better. Eur J Dent Educ. 2008;12:22–9. doi: 10.1111/j.1600-0579.2007.00477.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zimmermann BG, Park NJ, Wong DT. Genomic targets in saliva. Ann N Y Acad Sci. 2007;1098:184–91. doi: 10.1196/annals.1384.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Y, Zhou X, John MA, Wong DT. RNA profiling of cell-free saliva using microarray technology. J Dent Res. 2004;83:199–203. doi: 10.1177/154405910408300303. [DOI] [PubMed] [Google Scholar]
- 11.R foundation for Statistical Computing R: A language and environment for statistical computing. http://www.R-project.org (Accessed September 2009)
- 12.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoits S, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Package lme4. lme4: linear mixed-effects models using S4 classes. http://cran.r-project.org. (Accessed September 2009)
- 14.Gautier L, Cope L, Bostad BM, Irizarry RA. Affy--analysis of affymetrix genechip data set at the probe level. Bioinformatics. 2004;20:307–15. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 15.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc (Ser A) 1995;B75:290–300. [Google Scholar]
- 16.Guihard-Costa A-M, Larroche J-C. Differential growth between the fetal brain and its infratentorial part. Early Hum Dev. 1990;23:27–40. doi: 10.1016/0378-3782(90)90126-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.