Abstract
Expression of the respiratory apparatus depends on both nuclear and mitochondrial genes. Although these genes are sequestered in distinct cellular organelles, their transcription relies on nucleus-encoded factors. Certain of these factors are directed to the mitochondria, where they sponsor the bi-directional transcription of mitochondrial DNA. Others act on nuclear genes that encode the majority of the respiratory subunits and many other gene products required for the assembly and function of the respiratory chain. The nuclear respiratory factors, NRF-1 and NRF-2, contribute to the expression of respiratory subunits and mitochondrial transcription factors and thus have been implicated in nucleo-mitochondrial interactions. In addition, coactivators of the PGC-1 family serve as mediators between the environment and the transcriptional machinery governing mitochondrial biogenesis. One family member, peroxisome proliferator-activated receptor γ coactivator PGC-1-related coactivator (PRC), is an immediate early gene product that is rapidly induced by mitogenic signals in the absence of de novo protein synthesis. Like other PGC-1 family members, PRC binds NRF-1 and activates NRF-1 target genes. In addition, PRC complexes with NRF-2 and HCF-1 (host cell factor-1) in the activation of NRF-2-dependent promoters. HCF-1 functions in cell-cycle progression and has been identified as an NRF-2 coactivator. The association of these factors with PRC is suggestive of a role for the complex in cell growth. Finally, shRNA-mediated knock down of PRC expression results in a complex phenotype that includes the inhibition of respiratory growth on galactose and the loss of respiratory complexes. Thus, PRC may help integrate the expression of the respiratory apparatus with the cell proliferative program.
Keywords: mitochondria, nuclear respiratory factors, PRC coactivator, respiratory chain, transcription, regulation, cell proliferation, gene expression, metabolism, PGC-1
Introduction
The energy-transducing systems of mitochondria produce the bulk of cellular energy through the oxidation of pyruvate and fatty acids. The electron transport chain of the mitochondrial inner membrane uses reducing equivalents derived from chemical bond energy to establish an electrochemical proton gradient. This gradient is dissipated by the adenosine 5′-triphosphate (ATP) synthase to drive the synthesis of ATP or by natural uncouplers to generate heat.1,2 Mitochondria contain their own genetic system based on a multicopy mitochondrial DNA (mtDNA) genome which, in vertebrates, is a covalently closed circular molecule of approximately 16.5 kilobases. The entire protein-coding capacity of mtDNA is devoted to 13 essential subunits of respiratory complexes I, III, IV, and V. The only other mitochondrial gene products are the 22 tRNAs and 2 rRNAs required for translation of the respiratory subunit mRNAs within the mitochondrial matrix.3–5 Because the coding capacity of mtDNA is limited, nuclear genes specify most of the numerous gene products required for the molecular architecture and biochemical functions of the organelle.6,7 These include the majority of respiratory proteins, all of the protein constituents of the mitochondrial translation system, and all of the gene products required for the transcription and replication of mtDNA.
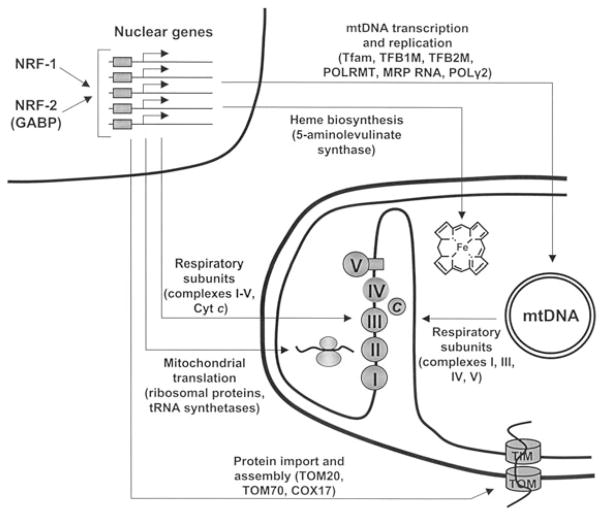
Two distinct classes of regulatory proteins govern nucleo-mitochondrial interactions at the transcriptional level in mammalian systems. The first class comprises transcription factors that bind the promoter regions of nuclear and mitochondrial genes. A small number of nucleus-encoded factors, including mitochondrial transcription Factor A (Tfam) and the mitochondrial transcription Factor B (mtTFB) isoforms TFB1M and TFB2M, direct transcription from divergent heavy- and light-strand promoters within the mitochondrial D-loop regulatory region.5,8 These factors work in conjunction with the mitochondrial RNA polymerase (POLRMT) to confer promoter specificity and to enhance the rate of transcription initiation. In addition, a second group of transcription factors act on the majority of nuclear genes whose products are required for respiratory chain expression and biological function.7 Among these are the nuclear respiratory factors NRF-1 and NRF-2 (GA binding protein [GABP]), which were originally identified as activators of cytochrome c9,10 and cytochrome oxidase11 genes, respectively. As depicted in Figure 1, these factors have subsequently been implicated in the expression of many other genes whose products contribute essential mitochondrial functions related to the respiratory apparatus.6,7 Both factors have also been linked to the control of genes essential to cell proliferation.12,13 This association is consistent with the early embryonic lethality of targeted disruptions of NRF-114 or NRF-2(GABP)15 in mice.
Figure 1.
Nuclear respiratory factors (NRF-1 and NRF-2) in the expression of nuclear genes governing mitochondrial respiratory function. NRFs act on the majority of nuclear genes that specify subunits of the five respiratory complexes of the mitochondrial inner membrane. In addition, they act on many other genes whose products direct the expression and assembly of the respiratory apparatus. Promoters for most of the nuclear genes encoding mtDNA transcription and replication factors have functional recognition sites for NRF-1, NRF-2, or both. These factors are required for the expression of respiratory subunits from complexes I, III, IV, and V encoded by mtDNA. Similarly, genes for mitochondrial translational components—including ribosomal proteins and tRNA synthetases as well as heme biosynthetic enzymes localized to the mitochondrial matrix—are NRF-dependent. Increasing evidence also suggests that a number of genes specifying proteins of the mitochondrial protein import and assembly machinery are NRF targets, including subunits of the import receptor complexes and COX assembly factors. Thus, NRF-1 and NRF-2 are part of a unifying mechanism for the coordinate transcriptional control of respiratory chain expression. MRP = mitochondrial RNA processing; POL = polymerase; TOM = translocase of the outer mitochondrial membrane; cytc = cytochrome c.
It is now apparent that a relatively small number of nuclear factors serve to coordinate the transcriptional expression of nuclear and mitochondrial respiratory proteins. In addition to the NRFs, stimulatory protein 1 (Sp1), estrogen related receptor α (ERRα), and yin yang 1 transcription factor (YY1) have also been linked to many genes required for respiratory chain expression and function.6,7,16 However, these factors are not universal in their control of all mitochondrial oxidative pathways, as exemplified by peroxisome proliferator-activated receptor (PPAR)α and δ in the expression of the fatty acid oxidation pathway.17,18 It is also likely that NRF-independent mechanisms are responsible for establishing and maintaining the molecular architecture of the organelle.19 This would mean that higher-order controls operate to integrate the actions of diverse transcription factors into a program of mitochondrial biogenesis. The PGC-1 family of regulated coactivators supplies such an integrative function by communicating physiological changes in the environment to a diverse array of transcription factors. The three family members, PGC-1α, PGC-1β, and PGC-1-related coactivator (PRC), are differentially regulated by environmental cues governing pathways of thermogenesis, gluconeogenesis, muscle differentiation, and cell growth.20–22 The coactivators, in turn, implement programs of gene expression through direct interaction with transcription factor targets or through their indirect effects on transcription factor expression. This paradigm is best exemplified by the control of thermogenesis in brown fat by PGC-1α, the founding member of the family.23 PGC-1α trans-activates the expression of the thermogenic uncoupling protein UCP-1 through its direct interaction with the nuclear hormone receptors PPARγ and thyroid receptor β (TRβ). It also targets NRF-1 and NRF-2 in orchestrating a program of mitochondrial biogenesis, an important part of the thermogenic response.24 Moreover, the protein exerts tissue-specific effects through other transcription factors to help drive gluconeogenesis in liver or fiber type switching in muscle.21,22 In addition to its regulated expression, PGC-1α also controls respiratory genes in response to post-translational modifications that serve to modulate protein–protein interactions.25–27 Finally, a closely related family member, PGC-1β, complements PGC-1α in differentiation-induced mitochondrial biogenesis in brown fat28 and directs the biogenesis of highly oxidative muscle fibers.29 Here, the focus is on PRC. This PGC-1 family coactivator has the properties of a growth regulator30,31 and may serve to integrate the expression of the respiratory apparatus with the genetic program controlling cell proliferation.
Nuclear Respiratory Factors in Nucleo-Mitochondrial Interactions
The gene encoding cytochrome c, a dissociable electron carrier that shuttles electrons between complexes III and IV, was characterized as the first vertebrate model for investigating the transcriptional control of respiratory gene expression.32 NRF-1 was identified during a systematic analysis of the cytochrome c promoter as a protein factor that binds a GC-rich palindrome.9,33 Subsequently, NRF-1 recognition sites were found in the promoters of a number of nuclear genes required for respiratory chain expression and function.10,34 NRF-1 has a unique DNA-binding domain, binds its recognition site as a homodimer, and functions as a positive regulator of transcription.35,36 It has a carboxy-terminal transcriptional activation domain composed of glutamine-containing clusters of hydrophobic amino acid residues that are essential for maximal activity.37 In proliferating mammalian cells, NRF-1 is serine phosphorylated within a concise amino-terminal domain, and phosphorylation within this domain has been associated with enhancement of both its DNA-binding activity36 and trans-activation functions.38 NRF-1 relatives have not been identified in mammalian genomes, although the factor is related through its DNA-binding domain to developmental regulatory proteins in sea urchins,39 Drosophila,40 and zebra fish.41
Investigations from a number of laboratories have identified NRF-1 as a key regulator of nucleus-encoded subunits of all five respiratory complexes.6,42,43 In addition, NRF-1 has a potential integrative function in coordinating bi-genomic respiratory subunit expression through its control over major regulators of mitochondrial transcription, including Tfam44 and both mtTFB isoform genes.45 Moreover, NRF-1 acts on a number of nuclear genes whose products play an indirect role in specifying respiratory function (Fig. 1). These include genes encoding key enzymes of the heme biosynthetic pathway46,47 and a cytochrome oxidase assembly factor.48 Finally, a number of physiological studies have established a role for NRF-1 in mediating in vivo changes in mitochondrial biogenesis and function.6,7 These are consistent with the fact that a targeted disruption of the NRF-1 gene in mice results in early embryonic lethality accompanied by severe depletion of mtDNA.14
A second nuclear respiratory factor, NRF-2, was identified by its specific binding to essential cis-acting elements in the cytochrome oxidase subunit IV (COXIV) promoter.11,49 Core binding sites for NRF-2 contain the GGAA motif that is common to the recognition sites of the E2G-specific (ETS)-domain family of transcription factors.50 Tandem direct repeats of the GGAA core sequence overlap multiple transcription initiation sites within the mouse COXIV promoter.49,51 Purification of NRF-2 to homogeneity from HeLa cell nuclear extracts led to the identification of five subunits designated α, β1, β2, γ1, and γ2. The α subunit binds DNA, whereas the others associate with α to form heterodimeric and heterotetrameric complexes.11 Molecular cloning of the five NRF-2 subunits52 revealed that NRF-2 is the human homologue of mouse GABP, a three-subunit (α, β1, β2), ETS-domain transcription factor that that was identified based on its activation of herpes virus immediate early genes.53 The human NRF-2β2 and γ2 subunits correspond to GABPβ1 and β2, respectively, whereas the two additional NRF-2 subunits, β1 and γ1, are minor splice variants.52 The NRF-2β subunits, corresponding to GABPβ1, have a dimerization domain that facilitates high-affinity binding of a heterotetrameric complex to tandem binding sites.52,54 In solution, GABP exists as a heterodimer but is induced to form the heterotetramer α2β2 by DNA containing two or more binding sites.55 A transcriptional activation domain composed of glutamine-containing hydrophobic clusters reminiscent of those found in NRF-1 is adjacent to the carboxy-terminus in all four NRF-2β and γ subunits.37
The first known cellular function for NRF-2(GABP) is the activation of COX subunit expression.11 Although initially recognized for its role in the expression of COXIV49,51 and COXVb,11,56 recent experiments have implicated the factor in the expression of all 10 nucleus-encoded cytochrome oxidase subunits.57 NRF-2 associates with COX subunit promoters in vivo.57 Moreover, a dominant negative NRF-2 allele reduced COX expression and an siRNA directed against NRF-2α reduced expression of all 10 subunits.58 However, a mitochondrial role for NRF-2 is not restricted to COX expression (Fig. 1). Like NRF-1, functional NRF-2 sites are present in the promoters of many genes whose products are required for respiratory chain expression.6,42 These include genes encoding the mitochondrial transcription factors Tfam44 and the TFB isoforms45 as well as three of the four human succinate dehydrogenase (complex II) subunit genes.59–61 Chromatin immunoprecipitations (ChIP) reveal NRF-1 and NRF-2α occupancy of TFB1M and 2M promoters in vivo.45 In addition, both NRF-1 and NRF-2 have been implicated in the expression of subunits of the membrane receptor complexes required for the import of the thousands of proteins that contribute to diverse mitochondrial functions.62–65 NRF control over key components of the protein import and assembly machinery is suggestive of a broad role for these factors in orchestrating mitochondrial biogenesis.
Importantly, both NRFs act on a broad spectrum of target genes and are not restricted to those involved in mitochondrial respiratory function. A screen for NRF-1 binding sites in mammalian promoters revealed a number of primate and rodent genes whose functions are not linked directly to mitochondrial biogenesis.35 The collection comprises genes that encode metabolic enzymes, signaling molecules, and gene products required for chromosome maintenance and nucleic acid metabolism among others. Notably, NRF-1 is one of seven transcription factors whose recognition sites most frequently occur in the proximal promoters of ubiquitously expressed genes.66 ChIP coupled with microarray assay (ChIP-on-chip) has revealed a collection of human promoters that are occupied by NRF-1 in vivo.12 Of approximately 13,000 human promoters surveyed by ChIP-on-chip analysis,67 691 are bound by NRF-1 in living cells.12 As expected, genes involved in mitochondrial biogenesis and metabolism represent a major subset, including many that had not been previously identified as NRF-1 targets. Among these are mitochondrial ribosomal protein and tRNA synthetase genes (Fig. 1). In addition, significant overlap was found between the NRF-1 target genes and those bound by E2F, a transcription factor family that participates in cell growth control. This is suggestive of a role for NRF-1 in the regulation of a subset of E2F-responsive genes. This subset was enriched in genes required for DNA replication, mitosis, and cytokinesis, and the expression of several of these E2F targets was repressed by an NRF-1 siRNA. It is of interest in this context that NRF-1 is dephosphorylated in quiescent fibroblasts and becomes phosphorylated upon cell cycle entry. The enhancement of NRF-1 transcriptional activity by phosphorylation may control the functional state of the DNA-bound factor.36,38 This, along with de-repression by release of E2F factors from a subset of NRF-1 target genes, may help promote cell proliferation. The participation of NRF-1 in the regulation of cell cycle progression may account for the early embryonic lethality of NRF-1 knockout embryos.14 Interestingly, a recent study has implicated NRF-2(GABP) in a D-cyclin-independent pathway of entry to the cell cycle.13 These findings are suggestive of an important link between the NRF-dependent transcriptional expression related to respiratory function and the cell proliferative cycle.
PRC: Molecular Interactions and Biological Functions
PRC was identified as a large cDNA with significant sequence similarities to PGC-1α within the carboxy-terminal RS domain and RNA recognition motif.30 The two coactivators also share a conserved amino-terminal activation domain, a central proline-rich region that is expanded in PRC, as well as a conserved LXXLL coactivator signature and DHDY host cell factor-1 (HCF-1) binding motifs (Fig. 2). The conserved spatial arrangement of these domains in otherwise dissimilar molecules is suggestive of related function. PRC shares with PGC-1α and β the ability to bind NRF-1 both in vitro and in vivo and to use NRF-1 for the trans-activation of NRF-1 target genes.30,45 NRF-1-dependent trans-activation requires the PRC activation domain, suggesting that this domain shares function with PGC-1α in recruiting chromatin-remodeling cofactors that drive transcription.68 PRC and PGC-1α are indistinguishable in trans-activating promoters for cytochrome c, 5-aminolevulinate synthase, and both of the TFB isoforms, suggesting that PRC may participate in the expression of the respiratory chain.30,45 Maximal trans-activation by both coactivators requires the NRF-1 and NRF-2 binding sites within the proximal promoters of these genes. Although similar to PGC-1α in these basic transcriptional properties, PRC mRNA is not enriched in brown versus white fat and is only slightly elevated in brown fat upon cold exposure, arguing against a major role for PRC in adaptive thermogenesis.30
Figure 2.
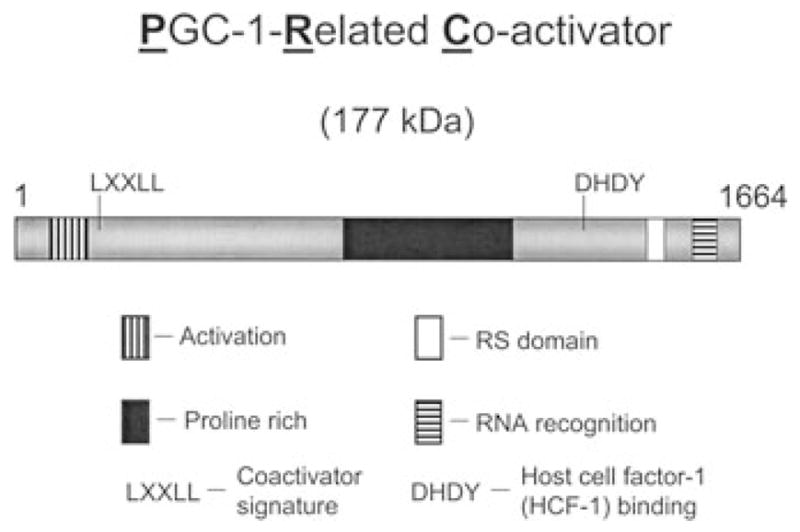
Arrangement of conserved sequence motifs in PGC-1-related coactivator (PRC). PRC is the largest member of the PGC-1 coactivator family. Sequence similarities between PRC and the other PGC-1 family coactivators are confined to distinct sequence blocks that are spatially conserved among the three family members. These include a potent amino-terminal transcriptional activation domain (vertical hatched box), an expanded proline-rich region (filled box), an arginine/serine (R/S) rich domain (open box) and an RNA recognition motif (horizontal hatched box). In addition, PRC has the LXXLL coactivator signature sequence and the DHDY host cell factor 1 (HCF-1) binding site present in the other family members.
Analysis of PRC expression in cultured fibroblasts revealed that PRC levels correlate with the cell proliferative cycle. The steady-state expression of PRC mRNA and protein is high in growing cells but markedly diminished upon exit from the cell cycle as a consequence of contact inhibition or serum withdrawal.30 PRC is also rapidly induced upon serum stimulation of quiescent fibroblasts. As shown in Table 1, this induction is correlated with a profile of gene expression that resembles that directed by PGC-1α.45 The table presents a comparison of relative mRNA expression in three systems undergoing mitochondrial biogenesis, including serum-stimulated fibroblasts (3 and 12 h following serum addition); C2C12 cells, where PGC-1α is overexpressed (compared with a green fluorescent protein (GFP) control);24 and differentiated L1 adipocytes (compared with undifferentiated cells).69 Relative mRNA expression for nuclear coactivators, nuclear transcription factors, mitochondrial transcription and replication factors, and both nuclear and mitochondrial respiratory subunits was assessed by quantitative real-time PCR; these represent regulatory and structural genes required for respiratory chain expression.
TABLE 1.
Expression of Regulatory and Structural Genes Contributing to the Expression of the Respiratory Chain in Systems Undergoing Mitochondrial Biogenesis
| Respiratory proteins |
||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nuclear coactivators |
Nuclear transcription factors |
Mitochondrial transcription and replication factors |
Nuclear DNA |
mtDNA |
||||||||||||||||||||||
| PRCd |
PGC-1α |
NRF-1 |
NRF2α |
NRF-2β |
Tfam |
TFB1M |
TFB2M |
POLRMT |
POLγ2 |
Cytc |
COXIV |
COXII |
||||||||||||||
| Serum stimulationa | 3 h | 12 h | 3 h | 12 h | 3 h | 12 h | 3 h | 12 h | 3 h | 12 h | 3 h | 12 h | 3 h | 12 h | 3 h | 12 h | 3 h | 12 h | 3 h | 12 h | 3 h | 12 h | 3 h | 12 h | 3 h | 12 h |
| Average | 6.4 | 5.8 | 1.2 | 0.9 | 2.5 | 2.9 | 2.1 | 3.3 | 6.3 | 4.7 | 4.7 | 5.2 | 0.3 | 2.6 | 4.0 | 5.9 | 1.7 | 3.6 | 1.2 | 2.0 | 4.5 | 10.0 | 1.4 | 1.7 | 2.5 | 3.2 |
| S.E.M. | 1.2 | 1.0 | 0.2 | 0.4 | 0.5 | 0.9 | 0.3 | 0.8 | 0.5 | 1.2 | 0.8 | 1.0 | 0.1 | 0.6 | 0.5 | 1.4 | 0.7 | 1.3 | 0.3 | 0.6 | 0.6 | 2.2 | 0.3 | 0.5 | 0.2 | 0.8 |
| Adeno PGC-1αb | 1.2 ± 0.2 | High | 1.5 ± 0.1 | 3.3 ± 0.2 | 1.7 ± 0.4 | 3.5 ± 0.6 | 2.4 ± 0.2 | 3.4 ± 0.3 | 2.3 ± 0.5 | 1.1 ± 0.1 | 16.1 ± 2.3 | 4.1 ± 0.3 | 3.3 ± 0.4 | |||||||||||||
| Adipocyte differentiationc | 0.9 ± 0.2 | 4.9 ± 1.6 | 1.4 ± 0.3 | 3.5 ± 0.6 | 2.6 ± 0.3 | 2.3 ± 0.6 | 5.3 ± 0.8 | 5.0 ± 0.8 | 5.8 ± 0.5 | 2.4 ± 0.1 | 17.3 ± 3.1 | 6.9 ± 2.0 | 15.2 ± 2.7 | |||||||||||||
Steady-state mRNA levels were measured by quantitative real-time PCR at 3 h and 12 h following serum stimulation of quiescent BALB/3T3 fibroblasts. Values represent the average fold increase in expression over that measured in serum-starved cells.
Steady-state mRNA levels were measured by quantitative real-time PCR in C2C12 myoblasts infected with an adenovirus expressing either PGC-1α or GFP (a gift from D. P. Kelly, Washington University). Values represent the average fold difference in expression in the PGC-1α expressing cells relative to the GFP expressing controls.
Steady-state mRNA levels were measured by quantitative real-time PCR in 3T3-L1 fibroblasts before and after their differentiation to adipocytes. Values represent the fold difference in expression in adipocytes relative to fibroblasts.
Steady-state mRNA levels were normalized to rRNA and represent the average of at least 3 separate determinations ± SEM.
Notably, PRC mRNA is not induced upon adenovirus overexpression of PGC-1α or during adipocyte differentiation. By contrast, PGC-1α mRNA is nearly undetectable in cultured fibroblasts and not induced by serum stimulation. However, with a few exceptions, it is evident that the pattern of gene expression is similar among the three systems. In general, NRF-2 and the mitochondrial transcriptional machinery, including the transcription factors (Tfam, TFB1M, TFB2M) and RNA polymerase (POLRMT), are upregulated in all three. Although the respiratory protein mRNAs for cytochrome c and COXII are induced in all three, COXIV shows only modest induction by serum. It is possible that an alternative COXIV isoform is induced, as has been observed in response to hypoxic conditions.70 Paradoxically, cyclin D1 appears to suppress NRF-1 function and mitochondrial content through a mechanism involving NRF-1 phosphorylation by cyclin D1-dependent kinase.71 This seems at odds with the results in Table 1 showing a clear induction of several groups of genes that are essential to the synthesis of the mitochondrial respiratory chain upon entry to the cell cycle. Interestingly, elevations in PRC have been observed in other systems where mitochondrial content is increased. PRC levels are elevated along with NRF-1 and Tfam in thyroid oncocytomas—thyroid tumors characterized by high mitochondrial density but lacking PGC-1α.72 The increased PRC expression is accompanied by elevated cytochrome oxidase activity and mtDNA content. PRC has also been implicated in the early adaptive changes in skeletal muscle gene expression in response to exercise training.73 Recent loss-of-function experiments support a direct role for PRC in respiratory chain expression.31
The rapid induction of PRC by serum in quiescent fibroblasts is reminiscent of the immediate early genes whose expression is induced by serum growth factors in the absence of de novo protein synthesis.74 The induction of this class of genes is thought to be a key event in initiating the genetic program leading to cell proliferation. Like other genes in this class, PRC mRNA induction by serum is insensitive to the protein synthesis inhibitor, cycloheximide, indicating that it occurs through the use of preexisting factors.31 PRC also resembles the immediate early genes in that its mRNA is super-induced and markedly stabilized by cycloheximide, probably because of a requirement for protein synthesis for mRNA turnover. The immediate early class comprises many growth regulators, including transcription factors, chemokines, growth factors, proto-oncogenes, and serine-threonine kinases, among others.75 Although certain coactivators are post-transcriptionally modified as part of the immediate early response,76 to our knowledge, serum-inducible transcriptional coactivators have not been described.
The regulated expression of PRC in accordance with the cell proliferative cycle may contribute to the integrated function of diverse transcription factors during cell growth. This possibility has been investigated using cytochrome c expression as a model. Cytochrome c is induced by serum during the G0-to-G1 transition in quiescent mouse fibroblasts (Table 1), leading to enhanced mitochondrial respiration.38 Maximal induction of cytochrome c mRNA requires both NRF-1 and cyclic AMP response element binding protein (CREB) recognition sites within its promoter.38 As discussed, NRF-1 has been associated with many genes required for DNA replication, cytokinesis, and mitosis,12 and CREB is a well-known target of mitogenic signaling pathways.77 Both proteins are phosphorylated upon entry to the cell cycle.38 As summarized in Figure 3, recent mapping experiments demonstrate that NRF-1 and CREB are indistinguishable in binding PRC both in vitro and in vivo and that both transcription factors interact with identical sites on PRC.31 PRC also binds both factors through their DNA-binding domains (Fig. 3), a property reminiscent of the interaction between the PPARγ DNA-binding domain and PGC-1α.24 Moreover, ChIP show that PRC, NRF-1, and CREB are bound to the cytochrome c promoter in vivo and that PRC occupancy, along with CREB phosphorylation, increases in response to serum stimulation.31 Taken together, the results are consistent with the notion that PRC can target both NRF-1 and CREB in response to mitogenic signals (Fig. 4). This is supported further by the observation that expression of the PRC NRF-1/CREB binding domain in trans from a lentivirus vector leads to dominant negative inhibition of cell growth on galactose as a carbon source.31 Mitochondrial respiratory function is required for growth on galactose,78 and the growth inhibition under these conditions suggests that disruption of the interaction between PRC and NRF-1/CREB is inhibitory to respiratory growth.
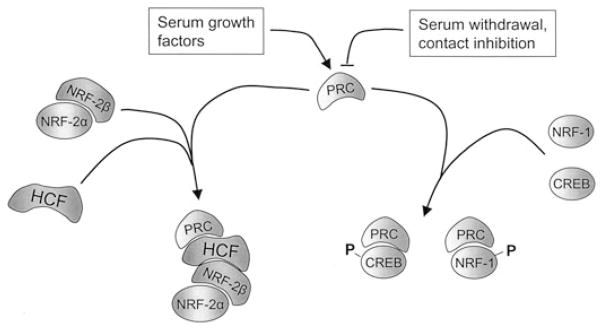
Figure 3.
Summary of molecular interactions between PRC and its cognate transcription factors, NRF-1 and CREB. NRF-1 and CREB engage in specific in vitro and in vivo interactions with PRC, as evidenced by pull-down assays and co-immunoprecipitations (see text for references). Deletion fine mapping revealed that both transcription factors share distinct binding sites located between the activation domain and the proline-rich region and within the R/S domain. A fragment containing the upstream binding site inhibits cytochrome c promoter activity and respiratory growth on galactose when expressed in trans. As observed for the interaction between PPARγ and PGC-1α, the PRC-transcription factor interactions occur through the DNA-binding domains (cross hatched boxes) of both NRF-1 and CREB.
Figure 4.
Summary of PRC-transcription factor complexes. The expression of PRC mRNA and protein is induced in quiescent fibroblasts by serum growth factors and downregulated upon exit from the cell cycle brought about either by serum withdrawal or contact inhibition. PRC engages in a direct interaction with NRF-1 and CREB in vitro and exists in a complex with each factor in cell extracts. PRC occupancy of the NRF-1- and CREB-dependent cytochrome c promoter also increases upon serum stimulation, along with transcription factor phosphorylation. By contrast, PRC does not bind any of the NRF-2 subunits in vitro. However, antibodies directed against PRC can immunoprecipitate both NRF-2β and HCF-1 from cell extracts. This, combined with the fact that HCF-1 engages in direct interactions with both PRC and NRF-2β suggests that all three proteins exist in a complex in vivo. This is supported by the observation that mutations in the DHDY HCF-1 binding site on PRC, or in key hydrophobic residues in the NRF-2β activation domain, that are required for interaction with HCF-1 inhibit PRC trans-activation through NRF-2.
New observations provide additional links between PRC and the program of cell growth. Recent experiments establish that PRC binds HCF-1, an abundant chromatin-associated protein required for progression through G1 of the cell cycle.79 HCF-1 was first identified as part of the molecular switch leading to the expression of herpes virus immediate early genes and has subsequently been found to associate with a number of transcription factors and chromatin remodeling activities.80 HCF-1 also functions as an NRF-2(GABP) coactivator81 and, as summarized in Figure 4, our recent experiments demonstrate that PRC exists in a complex with both HCF-1 and NRF-2β(GABPβ). This is supported both by in vitro binding assays and by co-immunoprecipitations from cell extracts.84 In fact, the same molecular determinants necessary for the interaction between PRC and HCF-1 and for that between HCF-1 and NRF-2β(GABPβ) are required for PRC trans-activation through NRF-2. These include the DHDY consensus HCF-1 binding motif in PRC and the NRF-2 activation domain localized to the NRF-2β subunit.52 The results suggest that HCF-1 may serve as a platform for PRC–transcription factor interactions in promoting the expression of genes necessary for cell growth, including the many NRF-2-dependent genes required for mitochondrial respiratory function. Interestingly, HCF-1 has also been implicated in the interaction between PGC-1α and NRF-2β(GABPβ) in the expression of genes encoding the neuromuscular junction in muscle.82 In this case, phosphorylation of both PGC-1α and NRF-2β(GABPβ) was required for the interaction with HCF-1. These modifications are thought to contribute to the specificity of PGC-1α in directing this pathway. HCF-1 also functions as a coactivator of PGC-1β-dependent transcription.83 Combined, these results argue that the PGC-1 family coactivators mediate at least some of their effects through their association with HCF-1-containing chromatin complexes.
A role for PRC in the expression of mitochondrial respiratory function is supported by the recent characterization of a U2 osteosarcoma cells (U2OS) cell line in which PRC expression is ablated by shRNAs expressed from a transduced lentivirus.84 The steady-state expression of PRC protein was reduced to undetectable levels, resulting in marked inhibition of growth on galactose media. The shRNA-mediated growth inhibition was greater than that observed with the PRC dominant negative allele derived from the NRF-1/CREB binding site31 confirming an important role for PRC in supporting respiratory growth. Moreover, the reduced respiratory growth rate on galactose was accompanied by the reduced expression of several mitochondrial respiratory subunits. Steady-state levels of the 20kDa NADH dehydrogenase subunit 6 (ND6) of complex I and core 2 of complex III were reduced to undetectable levels, whereas COXII of complex IV and succinate dehydrogenase B subunit (SDHB) of complex II were diminished significantly. The affected subunits were from both nuclear (core 2, SDHB) and mitochondrial (COXII and ND6) genes, indicating that the loss of PRC leads to a global defect in respiratory gene expression. The respiratory defect may be partly explained by the observation that mRNA encoding the mitochondrial transcriptional activator, TFB2M, is reduced in the PRC shRNA transductant, along with mitochondrial transcripts encoding COXII and cytochrome b. A defect in mitochondrial transcription may result in a failure to express optimal levels of key respiratory complexes. This possibility is supported by the fact that the reduction in COXII mRNA expression is accompanied by diminished cytochrome oxidase activity. Notably, the respiratory effects of PRC ablation were observed in the absence of detectable levels of PGC-1α. These results are consistent with the conclusion that PRC contributes to nucleo-mitochondrial interactions by controlling the expression of respiratory subunits encoded by both genomes.
Conclusion
In summary, PRC is a growth-regulated member of the PGC-1 family of inducible coactivators and has the characteristics of an immediate early gene. It associates with transcription factors, including NRF-1 and NRF-2, that have been implicated in the expression of the respiratory chain. These factors also act on genes involved in key aspects of cell growth, including DNA replication, mitosis, and cytokinesis, and thus have the potential to link the expression of the respiratory chain with the program of cell proliferation. PRC may serve to integrate mitogenic signals through its specific interaction with the transcriptional machinery specifying these functions. An additional link between PRC and the proliferative program comes from its association with HCF-1, a chromatin-bound coactivator that is required for progression through G1 of the cell cycle. HCF-1 serves as an intermediary between PRC and NRF-2 by binding both the transcription factor and the coactivator. These associations are functionally significant because mutations that disrupt the binding of PRC to HCF-1 or the binding of HCF-1 to NRF-2 inhibit trans-activation of target promoters by PRC. In addition, PRC loss of function results in the inhibition of respiratory growth and reduced expression of mitochondrial respiratory complexes. It is not clear why PRC binds factors such as NRF-1 and CREB directly, whereas it binds NRF-2 indirectly through its association with HCF-1. This may contribute to temporal patterns of expression or may act as a spatial requirement for targeting specific promoter contexts. Further studies will elucidate the unique contributions of PRC to the spectrum of activities assigned to the PGC-1 coactivator family.
Acknowledgments
I thank Natalie Gleyzer for excellent technical assistance. Work in the author’s laboratory was supported by United States Public Health Service Grant GM32525-25 from the National Institutes of Health.
Footnotes
Conflicts of Interest
The author declares no conflicts of interest.
References
- 1.Hatefi Y. The mitochondrial electron transport chain and oxidative phosphorylation system. Annu Rev Biochem. 1985;54:1015–1069. doi: 10.1146/annurev.bi.54.070185.005055. [DOI] [PubMed] [Google Scholar]
- 2.Cannon B, Nedergaard J. Brown adipose tissue: Function and physiological significance. Physiol Rev. 2004;84:277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- 3.Scarpulla RC. Molecular biology of the OXPHOS system. In: Smeitink JAM, Sengers RCA, Trij JMF, editors. Oxidative Phosphorylation in Health and Disease. Landes Bioscience; New York, NY: 2004. pp. 28–42. [Google Scholar]
- 4.Wallace DC. A mitochondrial paradigm of metabolic and degenerative diseases, aging, and cancer: A dawn for evolutionary medicine. Annu Rev Genet. 2005;39:359–407. doi: 10.1146/annurev.genet.39.110304.095751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonawitz ND, Clayton DA, Shadel GS. Initiation and beyond: Multiple functions of the human mitochondrial transcription machinery. Mol Cell. 2006;24:813–825. doi: 10.1016/j.molcel.2006.11.024. [DOI] [PubMed] [Google Scholar]
- 6.Kelly DP, Scarpulla RC. Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes Dev. 2004;18:357–368. doi: 10.1101/gad.1177604. [DOI] [PubMed] [Google Scholar]
- 7.Scarpulla RC. Nuclear control of respiratory gene expression in mammalian cells. J Cell Biochem. 2006;97:673–683. doi: 10.1002/jcb.20743. [DOI] [PubMed] [Google Scholar]
- 8.Gaspari M, Larsson NG, Gustafsson CM. The transcription machinery in mammalian mitochondria. Biochim Biophys Acta Bio-Energetics. 2004;1659:148–152. doi: 10.1016/j.bbabio.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 9.Evans MJ, Scarpulla RC. Interaction of nuclear factors with multiple sites in the somatic cytochrome c promoter. Characterization of upstream NRF-1, ATF and intron Sp1 recognition sites. J Biol Chem. 1989;264:14361–14368. [PubMed] [Google Scholar]
- 10.Evans MJ, Scarpulla RC. NRF-1: A trans-activator of nuclear-encoded respiratory genes in animal cells. Genes Dev. 1990;4:1023–1034. doi: 10.1101/gad.4.6.1023. [DOI] [PubMed] [Google Scholar]
- 11.Virbasius JV, Virbasius CA, Scarpulla RC. Identity of GABP with NRF-2, a multisubunit activator of cytochrome oxidase expression, reveals a cellular role for an ETS domain activator of viral promoters. Genes Dev. 1993;7:380–392. doi: 10.1101/gad.7.3.380. [DOI] [PubMed] [Google Scholar]
- 12.Cam H, Balciunaite E, Blias A, et al. A common set of gene regulatory networks links metabolism and growth inhibition. Mol Cell. 2004;16:399–411. doi: 10.1016/j.molcel.2004.09.037. [DOI] [PubMed] [Google Scholar]
- 13.Yang ZF, Mott S, Rosmarin AG. The Ets transcription factor GABP is required for cell-cycle progression. Nat Cell Biol. 2007;9:339–346. doi: 10.1038/ncb1548. [DOI] [PubMed] [Google Scholar]
- 14.Huo L, Scarpulla RC. Mitochondrial DNA instability and peri-implantation lethality associated with targeted disruption of nuclear respiratory factor 1 in mice. Mol Cell Biol. 2001;21:644–654. doi: 10.1128/MCB.21.2.644-654.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ristevski S, O’leary DA, Thornell AP, et al. The ETS transcription factor GABPα is essential for early embryogenesis. Mol Cell Biol. 2004;24:5844–5849. doi: 10.1128/MCB.24.13.5844-5849.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goffart S, Wiesner RJ. Regulation and co-ordination of nuclear gene expression during mitochondrial biogenesis. Exp Physiol. 2003;88:33–40. doi: 10.1113/eph8802500. [DOI] [PubMed] [Google Scholar]
- 17.Gulick T, Cresci S, Caira T, et al. The peroxisome proliferator-activated receptor regulates mitochondrial fatty acid oxidative enzyme gene expression. Proc Natl Acad Sci USA. 1994;91:11012–11016. doi: 10.1073/pnas.91.23.11012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barish GD, V, Narkar A, Evans RM. PPAR delta: A dagger in the heart of the metabolic syndrome. J Clin Invest. 2006;116:590–597. doi: 10.1172/JCI27955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mercy L, Pauw A, Payen L, et al. Mitochondrial biogenesis in mtDNA-depleted cells involves a Ca2+-dependent pathway and a reduced mitochondrial protein import. FEBS J. 2005;272:5031–5055. doi: 10.1111/j.1742-4658.2005.04913.x. [DOI] [PubMed] [Google Scholar]
- 20.Lin J, Handschin C, Spiegelman BM. Metabolic control through the PGC-1 family of transcriptional coactivators. Cell Metab. 2005;1:361–370. doi: 10.1016/j.cmet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 21.Handschin C, Spiegelman BM. Peroxisome proliferator-activated receptor gamma coactivator 1 coactivators, energy homeostasis, and metabolism. Endocr Rev. 2006;27:728–735. doi: 10.1210/er.2006-0037. [DOI] [PubMed] [Google Scholar]
- 22.Finck BN, Kelly DP. PGC-1 coactivators: Inducible regulators of energy metabolism in health and disease. J Clin Invest. 2006;116:615–622. doi: 10.1172/JCI27794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Puigserver P, Wu Z, Park CW, et al. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/s0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 24.Wu Z, Puigserver P, Andersson U, et al. Mechanisms controlling mitochondrial biogenesis and function through the thermogenic coactivator PGC-1. Cell. 1999;98:115–124. doi: 10.1016/S0092-8674(00)80611-X. [DOI] [PubMed] [Google Scholar]
- 25.Knutti D, Kressler D, Kralli A. Regulation of the transcriptional coactivator PGC-1 via MAPK-sensitive interaction with a repressor. Proc Natl Acad Sci USA. 2001;98:9713–9718. doi: 10.1073/pnas.171184698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fan M, Rhee J, St Pierre J, et al. Suppression of mitochondrial respiration through recruitment of p160 myb binding protein to PGC-1α: Modulation by p38 MAPK. Genes Dev. 2004;18:278–289. doi: 10.1101/gad.1152204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Teyssier C, Ma H, Emter R, et al. Activation of nuclear receptor coactivator PGC-1α by arginine methylation. Genes Dev. 2005;19:1466–1473. doi: 10.1101/gad.1295005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uldry M, Yang W, St-Pierre J, et al. Complementary action of the PGC-1 coactivators in mitochondrial biogenesis and brown fat differentiation. Cell Metab. 2006;3:333–341. doi: 10.1016/j.cmet.2006.04.002. [DOI] [PubMed] [Google Scholar]
- 29.Arany Z, Lebrasseur N, Morris C, et al. The transcriptional coactivator PGC-1β drives the formation of oxidative type IIX fibers in skeletal muscle. Cell Metab. 2007;5:35–46. doi: 10.1016/j.cmet.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 30.Andersson U, Scarpulla RC. PGC-1-related coactivator, a novel, serum-inducible coactivator of nuclear respiratory factor 1-dependent transcription in mammalian cells. Mol Cell Biol. 2001;21:3738–3749. doi: 10.1128/MCB.21.11.3738-3749.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vercauteren K, Pasko RA, Gleyzer N, et al. PGC-1-related coactivator (PRC): Immediate early expression and characterization of a CREB/NRF-1 binding domain associated with cytochrome c promoter occupancy and respiratory growth. Mol Cell Biol. 2006;26:7409–7419. doi: 10.1128/MCB.00585-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scarpulla RC, Wu R. Nonallelic members of the cytochrome c multigene family of the rat may arise through different messenger RNAs. Cell. 1983;32:473–482. doi: 10.1016/0092-8674(83)90467-1. [DOI] [PubMed] [Google Scholar]
- 33.Evans MJ, Scarpulla RC. Both upstream and intron sequence elements are required for elevated expression of the rat somatic cytochrome c gene in COS-1 cells. Mol Cell Biol. 1988;8:35–41. doi: 10.1128/mcb.8.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chau CA, Evans MJ, Scarpulla RC. Nuclear respiratory factor 1 activation sites in genes encoding the gamma-subunit of ATP synthase, eukaryotic initiation factor 2α, and tyrosine aminotransferase. Specific interaction of purified NRF-1 with multiple target genes. J Biol Chem. 1992;267:6999–7006. [PubMed] [Google Scholar]
- 35.Virbasius CA, Virbasius JV, Scarpulla RC. NRF-1, an activator involved in nuclear-mitochondrial interactions, utilizes a new DNA-binding domain conserved in a family of developmental regulators. Genes Dev. 1993;7:2431–2445. doi: 10.1101/gad.7.12a.2431. [DOI] [PubMed] [Google Scholar]
- 36.Gugneja S, Scarpulla RC. Serine phosphorylation within a concise amino-terminal domain in nuclear respiratory factor 1 enhances DNA binding. J Biol Chem. 1997;272:18732–18739. doi: 10.1074/jbc.272.30.18732. [DOI] [PubMed] [Google Scholar]
- 37.Gugneja S, Virbasius CA, Scarpulla RC. Nuclear respiratory factors 1 and 2 utilize similar glutamine-containing clusters of hydrophobic residues to activate transcription. Mol Cell Biol. 1996;16:5708–5716. doi: 10.1128/mcb.16.10.5708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Herzig RP, Scacco S, Scarpulla RC. Sequential serum-dependent activation of CREB and NRF-1 leads to enhanced mitochondrial respiration through the induction of cytochrome c. J Biol Chem. 2000;275:13134–13141. doi: 10.1074/jbc.275.17.13134. [DOI] [PubMed] [Google Scholar]
- 39.Calzone FJ, Hoog C, Teplow DB, et al. Gene regulatory factors of the sea urchin embryo. I. Purification by affinity chromatography and cloning of P3A2, a novel DNA binding protein. Development. 1991;112:335–350. doi: 10.1242/dev.112.1.335. [DOI] [PubMed] [Google Scholar]
- 40.Desimone SM, White K. The Drosophila erect wing gene, which is important for both neuronal and muscle development, encodes a protein which is similar to the sea urchin P3A2 DNA binding protein. Mol Cell Biol. 1993;13:3641–3949. doi: 10.1128/mcb.13.6.3641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Becker TS, Burgess SM, Amsterdam AH, et al. Not really finished is crucial for development of the zebrafish outer retina and encodes a transcription factor highly homologous to human nuclear respiratory factor 1 and avian initiation binding repressor. Development. 1998;124:4369–4378. doi: 10.1242/dev.125.22.4369. [DOI] [PubMed] [Google Scholar]
- 42.Scarpulla RC. Nuclear activators and coactivators in mammalian mitochondrial biogenesis. Biochem Biophys Acta. 2002;1576:1–14. doi: 10.1016/s0167-4781(02)00343-3. [DOI] [PubMed] [Google Scholar]
- 43.Scarpulla RC. Transcriptional activators and coactivators in the nuclear control of mitochondrial function in mammalian cells. Gene. 2002;2:81–89. doi: 10.1016/s0378-1119(01)00809-5. [DOI] [PubMed] [Google Scholar]
- 44.Virbasius JV, Scarpulla RC. Activation of the human mitochondrial transcription factor A gene by nuclear respiratory factors: A potential regulatory link between nuclear and mitochondrial gene expression in organelle biogenesis. Proc Natl Acad Sci USA. 1994;91:1309–1313. doi: 10.1073/pnas.91.4.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gleyzer N, Vercauteren K, Scarpulla RC. Control of mitochondrial transcription specificity factors (TFB1M and TFB2M) by nuclear respiratory factors (NRF-1 and NRF-2) and PGC-1 family coactivators. Mol Cell Biol. 2005;25:1354–1366. doi: 10.1128/MCB.25.4.1354-1366.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Braidotti G, I, Borthwick A, May BK. Identification of regulatory sequences in the gene for 5-aminolevulinate synthase from rat. J Biol Chem. 1993;268:1109–1117. [PubMed] [Google Scholar]
- 47.Aizencang GI, Bishop DF, Forrest D, et al. Uroporphyrinogen III synthase. An alternative promoter controls erythroid-specific expression in the murine gene. J Biol Chem. 2000;275:2295–2304. doi: 10.1074/jbc.275.4.2295. [DOI] [PubMed] [Google Scholar]
- 48.Takahashi Y, Kako K, Arai H, et al. Characterization and identification of promoter elements in the mouse COX17 gene. Biochem Biophys Acta. 2002;1574:359–364. doi: 10.1016/s0167-4781(01)00374-8. [DOI] [PubMed] [Google Scholar]
- 49.Virbasius JV, Scarpulla RC. Transcriptional activation through ETS domain binding sites in the cytochrome c oxidase subunit IV gene. Mol Cell Biol. 1991;11:5631–5638. doi: 10.1128/mcb.11.11.5631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Graves BJ. Inner workings of a transcription factor partnership. Science. 1998;279:1000–1002. doi: 10.1126/science.279.5353.1000. [DOI] [PubMed] [Google Scholar]
- 51.Carter RS, Bhat NK, Basu A, Avadhani NG. The basal promoter elements of murine cytochrome c oxidase subunit IV gene consist of tandemly duplicated ets motifs that bind to GABP-related transcription factors. J Biol Chem. 1992;267:23418–23426. [PubMed] [Google Scholar]
- 52.Gugneja S, Virbasius JV, Scarpulla RC. Four structurally distinct, non-DNA-binding subunits of human nuclear respiratory factor 2 share a conserved transcriptional activation domain. Mol Cell Biol. 1995;15:102–111. doi: 10.1128/mcb.15.1.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lamarco KL, Mcknight SL. Purification of a set of cellular polypeptides that bind to the purine-rich cis-regulatory element of herpes simplex virus immediate early genes. Genes Dev. 1989;3:1372–1383. doi: 10.1101/gad.3.9.1372. [DOI] [PubMed] [Google Scholar]
- 54.Thompson CC, Brown TA, Mcknight SL. Convergence of Ets- and notch-related structural motifs in a heteromeric DNA binding complex. Science. 1991;253:762–768. doi: 10.1126/science.1876833. [DOI] [PubMed] [Google Scholar]
- 55.Chinenov Y, Henzl M, Martin ME. The α and β subunits of the GA-binding protein form a stable heterodimer in solution. J Biol Chem. 2000;275:7749–7756. doi: 10.1074/jbc.275.11.7749. [DOI] [PubMed] [Google Scholar]
- 56.Sucharov C, Basu A, Carter RS, Avadhani NG. A novel transcriptional initiator activity of the GABP factor binding ets sequence repeat from the murine cytochrome c oxidase Vb gene. Gene Expr. 1995;5:93–111. [PMC free article] [PubMed] [Google Scholar]
- 57.Ongwijitwat S, Wong-Riley MT. Is nuclear respiratory factor 2 a master transcriptional coordinator for all ten nuclear-encoded cytochrome c oxidase subunits in neurons? Gene. 2005;360:65–77. doi: 10.1016/j.gene.2005.06.015. [DOI] [PubMed] [Google Scholar]
- 58.Ongwijitwat S, Liang HL, Graboyes EM, Wong-Riley MT. Nuclear respiratory factor 2 senses changing cellular energy demands and its silencing down-regulates cytochrome oxidase and other target gene mRNAs. Gene. 2006;374:39–49. doi: 10.1016/j.gene.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 59.Au HC, Scheffler IE. Promoter analysis of the human succinate dehydrogenase iron-protein gene. Both nuclear respiratory factors NRF-1 and NRF-2 are required. Eur J Biochem. 1998;251:164–174. doi: 10.1046/j.1432-1327.1998.2510164.x. [DOI] [PubMed] [Google Scholar]
- 60.Elbehti-Green A, Au HC, Mascarello JT, et al. Characterization of the human SDHC gene encoding one of the integral membrane proteins of succinate-quinone oxidoreductase in mitochondria. Gene. 1998;213:133–140. doi: 10.1016/s0378-1119(98)00186-3. [DOI] [PubMed] [Google Scholar]
- 61.Hirawake H, Taniwaki M, Tamura A, et al. Characterization of the human SDHD gene encoding the small subunit of cytochrome b (cybS) in mitochondrial succinate-ubiquinone oxidoreductase. Biochim Biophys Acta. 1999;1412:295–300. doi: 10.1016/s0005-2728(99)00071-7. [DOI] [PubMed] [Google Scholar]
- 62.Joseph AM, Rungi AA, Robinson BH, Hood DA. Compensatory responses of protein import and transcription factor expression in mitochondrial DNA defects. Am J Physiol Cell Physiol. 2004;286:C867–C875. doi: 10.1152/ajpcell.00191.2003. [DOI] [PubMed] [Google Scholar]
- 63.Blesa JR, Hernandez JM, Hernandez-Yago J. NRF-2 transcription factor is essential in promoting human Tomm70 gene expression. Mitochondrion. 2004;3:251–259. doi: 10.1016/j.mito.2004.02.001. [DOI] [PubMed] [Google Scholar]
- 64.Blesa JR, Hernandez-Yago J. Distinct functional contributions of 2 GABP-NRF-2 recognition sites within the context of the human TOMM70 promoter. Biochem Cell Biol. 2006;84:813–822. doi: 10.1139/o06-064. [DOI] [PubMed] [Google Scholar]
- 65.Blesa JR, Prieto-Ruiz JA, Hernandez JM, Hernandez-Yago J. NRF-2 transcription factor is required for human TOMM20 gene expression. Gene. 2007;391:198–208. doi: 10.1016/j.gene.2006.12.024. [DOI] [PubMed] [Google Scholar]
- 66.Fitzgerald PC, Shlyakhtenko A, Mir AA, Vinson C. Clustering of DNA sequences in human promoters. Genome Res. 2004;14:1562–1574. doi: 10.1101/gr.1953904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ren B, Dynlacht BD. Use of chromatin immunoprecipitation assays in genome-wide location analysis of mammalian transcription factors. Methods Enzymol. 2004;376:304–315. doi: 10.1016/S0076-6879(03)76020-0. [DOI] [PubMed] [Google Scholar]
- 68.Wallberg AE, Yamamura S, Malik S, et al. Coordination of p300-mediated chromatin remodeling and TRAP/mediator function through coactivator PGC-1α. Mol Cell. 2003;12:1137–1149. doi: 10.1016/s1097-2765(03)00391-5. [DOI] [PubMed] [Google Scholar]
- 69.Wilson-Fritch L, Burkart A, Bell G, et al. Mitochondrial biogenesis and remodeling during adipogenesis and in response to the insulin sensitizer rosiglitazone. Mol Cell Biol. 2003;23:1085–1094. doi: 10.1128/MCB.23.3.1085-1094.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Semenza GL. Oxygen-dependent regulation of mitochondrial respiration by hypoxia-inducible factor 1. Biochem J. 2007;405:1–9. doi: 10.1042/BJ20070389. [DOI] [PubMed] [Google Scholar]
- 71.Wang C, Li Z, Lu Y, et al. Cyclin D1 repression of nuclear respiratory factor 1 integrates nuclear DNA synthesis and mitochondrial function. Proc Natl Acad Sci USA. 2006;103:11567–11572. doi: 10.1073/pnas.0603363103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Savagner F, Mirebeau D, Jacques C, et al. PGC-1-related coactivator and targets are upregulated in thyroid oncocytoma. Biochem Biophys Res Commun. 2003;310:779–784. doi: 10.1016/j.bbrc.2003.09.076. [DOI] [PubMed] [Google Scholar]
- 73.Russell AP, Hesselink MKC, Lo SK, Schrauwen P. Regulation of metabolic transcriptional co-activators and transcription factors with acute exercise. FASEB J. 2005;19:NIL467–NIL486. doi: 10.1096/fj.04-3168fje. [DOI] [PubMed] [Google Scholar]
- 74.Herschman HR. Primary response genes induced by growth factors and tumor promoters. Annu Rev Biochem. 1991;60:281–319. doi: 10.1146/annurev.bi.60.070191.001433. [DOI] [PubMed] [Google Scholar]
- 75.Winkles JA. Serum- and polypeptide growth factor-inducible gene expression in mouse fibroblasts. Prog Nucleic Acid Res Mol Biol. 1998;58:41–78. doi: 10.1016/s0079-6603(08)60033-1. [DOI] [PubMed] [Google Scholar]
- 76.Cen B, Selvaraj A, Prywes R. Myocardin/MKL family of SRF coactivators: Key regulators of immediate early and muscle specific gene expression. J Cell Biochem. 2004;93:74–82. doi: 10.1002/jcb.20199. [DOI] [PubMed] [Google Scholar]
- 77.Mayr B, Montminy M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat Rev Mol Cell Biol. 2001;2:599–609. doi: 10.1038/35085068. [DOI] [PubMed] [Google Scholar]
- 78.Robinson BH. Use of fibroblast and lymphoblast cultures for detection of respiratory chain defects. Methods Enzymol. 1996;264:454–464. doi: 10.1016/s0076-6879(96)64041-5. [DOI] [PubMed] [Google Scholar]
- 79.Goto H, Motomura S, Wilson AC, et al. A single point mutation in HCF causes temperature-sensitive cell-cycle arrest and disrupts VP16 function. Genes Dev. 1997;11:726–737. doi: 10.1101/gad.11.6.726. [DOI] [PubMed] [Google Scholar]
- 80.Wysocka J, Herr W. The herpes simplex virus VP16-induced complex: The makings of a regulatory switch. Trends Biochem Sci. 2003;28:294–304. doi: 10.1016/S0968-0004(03)00088-4. [DOI] [PubMed] [Google Scholar]
- 81.Vogel JL, Kristie TM. The novel coactivator C1 (HCF) coordinates multiprotein enhancer formation and mediates transcription activation by GABP. EMBO J. 2000;19:683–690. doi: 10.1093/emboj/19.4.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Handschin C, Kobayashi YM, Chin S, et al. PGC-1alpha regulates the neuromuscular junction program and ameliorates Duchenne muscular dystrophy. Genes Dev. 2007;21:770–783. doi: 10.1101/gad.1525107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lin J, Puigserver P, Donovan J, et al. PGC-1β: A novel PGC-1-related transcription coactivator associated with host cell factor. J Biol Chem. 2002;277:1645–1648. doi: 10.1074/jbc.C100631200. [DOI] [PubMed] [Google Scholar]
- 84.Vercauteren K, Gleyzer N, Scarpula RC. PGC-1-related coactivator complexes with HCF-1 and NRF-2B ion mediating MRF-2 (GABP)-dependent respiratory gene expression. J Biol Chem. 2008;283:12102–12111. doi: 10.1074/jbc.M710150200. [DOI] [PMC free article] [PubMed] [Google Scholar]