Abstract
A newly described heterodimeric cytokine, interleukin-23 (IL-23) is emerging as a key player in both the innate and the adaptive T helper (Th)17 driven immune response as well as an initiator of several autoimmune diseases. The rate-limiting element of IL-23 production is believed to be driven by expression of the unique p19 subunit encoded by IL23A. We set out to perform comprehensive DNA sequencing of this previously under-studied gene in 96 individuals from two evolutionary distinct human population groups, Southern African Bantu and European. We observed a total of 33 different DNA variants within these two groups, 22 (67%) of which are currently not reported in any available database. We further demonstrate both inter-population and intra-species sequence conservation within the coding and known regulatory regions of IL23A, supporting a critical physiological role for IL-23. We conclude that IL23A may have undergone positive selection pressure directed towards conservation, suggesting that functional genetic variants within IL23A will have a significant impact on the host immune response.
Keywords: IL-23, IL23A, novel variants, genetic conservation
Interleukin (IL)-23 is a heterodimeric cytokine, comprised of a p40 subunit, which is common with IL-12, and a unique p19 subunit. Despite structural similarities between IL-12 and IL-23, they have vastly different immunological roles. The IL-23 complex is primarily secreted by monocyte-derived dendritic cells and macrophages1 and is reported to play a critical role in survival of T helper (Th)17 cells and possibly the formation of Th17 memory cells.2 Th17 cells have been implicated in resistance to fungal and extracellular bacterial pathogens as well as induction of various types of organ specific autoimmunity.3 In addition to the potential role in adaptive immunity, recent reviews have described IL-23 as being ‘primitive in origin’ and have proposed a primary role in the evolutionary older innate immune system,4 particularly in mediating inflammatory attacks against pathogens of the intestine.5–9 Such pleiotropic cytokines may prove useful in determining an evolutionary link between the innate and adaptive immune pathways. Consistent with the innate immune system evolving prior to the adaptive immune response, it is reasonable to assume that molecules involved in this system may have evolved more rapidly and be genetically conserved across a broad spectrum of evolutionary distant species.
Since the discovery of IL-23, investigators have queried previous reports that target the shared p40 subunit to implicate IL-12 in several common autoimmune diseases.10–12 Subsequent studies have described a role for IL-23 (and in some instances exonerated IL-12) in the pathogenesis of many of these diseases.5–9
The gene responsible for encoding the unique IL-23 p19 protein, later termed IL23A, was first described by Oppmann et al.1 It is generally believed that the rate-limiting step of IL-23 production is expression of the p19 transcript.13 In light of the important role, IL-23 plays in both the innate and the adaptive immune response reportedly by enhancing the production of IL-17,14 it is within reason to hypothesize that regulation of IL23A expression, and therefore genetic variants within this gene, may be critical in determining the host immune response, as well as the development of autoimmune disease.
In this study, we determine the extent of IL23A sequence variation in two ethnically distinct human populations, including 48 Europeans from Australia and 48 Bantu from Southern Africa. Participants were randomly selected, with no known disease status. Our linguistically mixed Southern African Bantu population (including Xhosa, Tswana and Sotho) more closely represents the origin of anatomically modern humans.15 We use the observed mutational spectrum and intra-species comparisons to determine the degree of evolutionary conservation and make predictions regarding functional implications.
Direct Sanger sequence analysis of IL23A (including ∼2.9 kb 5′ and 1.9 kb 3′ of the transcription start and stop sites, respectively) in each of the two human populations, revealed a total of 33 different DNA variants present within the 6.36-kb region screened. All DNA variants underwent dual independent observations either as a result of bidirectional sequence validation and/or locality in overlapping fragments. With the exception of one variant at −1752 bp from the translation start site (−1752C>G; rs73324334), all variants observed were population specific (Table 1). The obvious distinction between variants in the two groups is important to note for investigating diseases with a strong correlation to ethnicity, particularly in this case, autoimmune-related diseases.16–18
Table 1.
Variant distribution of human IL23A in 96 individuals of Bantu and European ancestry
| Population | RS number (if known) | Gene position | Variant ID | Variant frequency (n/48 samples) |
|
|---|---|---|---|---|---|
| Heterozygous | Homozygous | ||||
| Bantu (n = 48) | Promoter | −3009G>T | 3 | 0 | |
| Promoter | −2186C>T | 1 | 0 | ||
| Promoter | −2572A>Ga | 7 | 1 | ||
| Promoter | −2493T>G | 2 | 0 | ||
| Promoter | −2192C>Ta | 7 | 1 | ||
| Promoter | −1982C>T | 3 | 0 | ||
| rs11835054 | Promoter | −1774C>G | 10 | 0 | |
| rs73324334 | Promoter | −1752C>G | 20 | 18 | |
| Promoter | −1538G>Ta | 7 | 1 | ||
| Promoter | −1316C>T | 3 | 0 | ||
| Promoter | −1263C>T | 1 | 0 | ||
| rs11465743 | Promoter | −914C>T | 3 | 0 | |
| Promoter | −854A>G | 1 | 0 | ||
| rs59094153 | Promoter | −204A>G | 3 | 0 | |
| 5′UTR/exon 1 | −42G>A | 1 | 0 | ||
| rs11465744 | 5′UTR/exon 1 | −35G>A | 2 | 0 | |
| Exon 4 | 955A>G (Pro155Pro) | 2 | 0 | ||
| Downstream | 1792T>C | 3 | 0 | ||
| rs11575253 | Downstream | 1945G>A | 1 | 0 | |
| Downstream | 2136G>A | 3 | 0 | ||
| rs7977692 | Downstream | 2421A>G | 29 | 7 | |
| Downstream | 3010T>G | 1 | 0 | ||
| Downstream | 3176C>T | 1 | 0 | ||
| European (n = 48) | Promoter | −2799C>T | 1 | 0 | |
| Promoter | −2730T>C | 1 | 0 | ||
| rs73131077 | Promoter | −2142G>A | 1 | 0 | |
| Promoter | −1956G>C | 2 | 0 | ||
| rs73324334 | Promoter | −1752C>Gb | 5 | 0 | |
| Promoter | −1527T>C | 1 | 0 | ||
| Promoter | −1517T>C | 1 | 0 | ||
| 5′UTR/Exon 1 | −138G>A | 1 | 0 | ||
| rs11171806 | Exon 3 | 703G>A (Ser106Ser)b | 5 | 0 | |
| rs11575254 | Downstream | 1863C>T | 1 | 0 | |
| rs11575248 | Downstream | 3162G>Tb | 5 | 0 | |
Variants within a 6357-bp IL23A region were identified in both Bantu and Europeans for 48 individuals each, using direct Sanger sequencing (primer information and amplification conditions for the 10 overlapping fragments available upon request).
aVariants in complete LD within the Bantu population.
bVariants in complete LD within the European population.
In addition to demonstrating the ethnic uniqueness of IL23A, we also highlight the limited genetic analysis of this gene to date. A total of 23 different genetic variants were identified in the Bantu, 16 (70%) of which had not previously been reported. Novel variants were also observed in the European population, and represented 7 (64%) of the 11 variants observed in this group. The increased observation of novel variation in the Southern African Bantu population collective can be explained by the exclusion of this largely diverse population grouping from current whole genome sequencing efforts defining current SNP database content. A recent report defines the Xhosa Bantu-speaking Southern Africans as genetically distinct from the peoples of Western African Yoruba.19
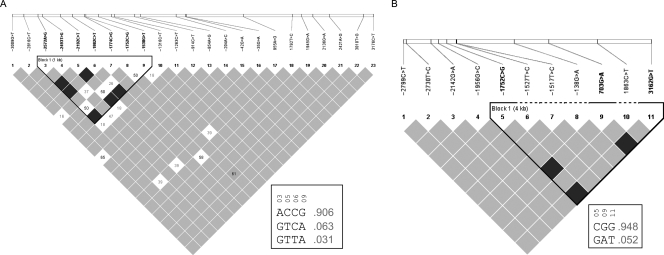
Haplotype predictive tools are not only important for identifying tag single-nucleotide polymorphisms (SNPs; relevant for disease association studies), but also for investigating the rate of gene evolution. As a result of population-specific variant distribution, linkage disequilibrium (LD) and haplotype analysis were performed on each the Bantu and European groups separately. Pairwise LD analysis on the Bantu samples, predicted a single haplotype block encompassing a 1-kb region towards the 5′ end of the gene, which showed strong LD between four variants, −2572A>G, −2192C>T, −1982C>T and −1538G>A (Fig. 1A). Three different combinations were predicted with the wild-type haplotype represented in 90.6% of the population (Fig. 1A). Overall, there were 25 allelic combinations predicted for the 23 variants identified in the Bantu (Supplementary Table S1). In Europeans, a single haplotype block within a 4-kb region towards the 3′ end of the gene was determined, including three variants in complete LD, −1752C>G (rs73324334), 703G>A (rs11171806, Ser106Ser) and 3162G>T (rs11575248) (Fig. 1B). The wild-type alleles were predicted to represent 94.8% of haplotypes (Fig. 1B). Frequency calculations for all 10 haplotype combinations identified in the Europeans demonstrate a high level of conservation within this group, with the wild-type alleles present in 85.4% of individuals (Supplementary Table S1). In accordance with genetic evolutionary data, the Southern African Bantu populations are among the genetically oldest populations in the world in comparison to the relatively young European populations,20 thus the evolutionary time-line allows for more genetic variation (as depicted in this study) as well as more recombination events to have occurred. Although only a single haplotype block is depicted in each population (Fig. 1), the genetic distance within the Bantu haplotype (1 kb) is 4-fold smaller than the European haplotype block (4 kb). The importance of the identified variants and haplotypes in disease susceptibility studies would require further validation.
Figure 1.
Pairwise LD plots and haplotype combinations of Bantu (A) and European (B) variants. (A) Variants 2572A>G, 2192C>T, 1982C>T and 1538G>T show strong LD within a 1-kb block in the Bantu population. Predicted haplotype frequencies for block 1 variants, depict the most common haplotype present in over 90% of the Bantu sequenced. (B) Variants 1752C>G, 703G>A and 3162G>T show strong LD within a 4-kb block in the Europeans. Predicted haplotype frequencies for block 1 reveal the most common haplotype is present in close to 95% of the Europeans sequenced. LD and haplotype information was generated using Haploview v4.1.
The majority of the 33 variants identified in this study occur outside the transcribed gene region. Of the five (15.6%) variants within the transcribed region, two are coding (both synonymous), and three occur in the 5′ untranslated region (UTR) of exon 1. Similarly, of the 21 SNPs described on one or both EntrezSNP (http://www.ncbi.nlm.nih.gov/snp/) and SNPper (http://snpper.chip.org) SNP databases (nine of which are reported in this study), only four are located within the transcribed region of IL23A, three synonymous and one intronic. Only one of these previously reported transcribed SNPs was identified in our study. We conclude that the amino acid coding region of IL23A is highly conserved not only in the recently migrated (out of Africa) European population, but also the genetically diverse Southern African Bantu. Genomic regions, which exhibit a high degree of conservation, beyond neutral expectations, are often reported as having important functional roles.21 It may further be postulated that variants which might affect the function of IL23A may be found in the regulatory regions, rather than coding regions of this gene. A recent study, which used a genome wide scan to identify psoriasis susceptibility loci in a European cohort, was the first to implicate a genetic variant within the IL23A locus to autoimmune disease susceptibility.22 This variant, rs2066808, is located 3.7 kb 3′ of IL23A (not within the 6.4-kb region of our analysis); however, any functional effect of this variant remains to be elucidated.
In this study, a total of 20 variants were observed 5′ of the transcription start site. Initially, we assessed whether the variants we observed were located within known transcription factor binding sites (TFBSs), namely NF-κB, ATF-2 and SMAD-3.23,24 We observed, no variants within these published sites. Assessing the entire 5′ region using MatInspector (Genomatix) for the wild-type and variant promoter sequences for known IL23A TFBSs NF-κB, ATF-2, SMAD-3 and Stat3,25 no difference in transcription factor binding was predicted. Lack of direct impact on protein function or transcriptional activity is suggestive that IL23A variation defined in this study may be as a direct result of neutral contribution and that functionally important variants would have undergone selection pressure as a result of reduced fitness. Concluding that the regulatory region of IL23A is highly conserved is in concordance with the proposed hypothesis of conservation of regulatory regions of genes involved in highly complex, adaptive physiological processes.26 As such genes are often not constitutively expressed, regulatory regions are considered more fundamental to maintaining an adequate response.
To determine the level of evolutionary conservation of IL23A, we performed sequence alignment of the transcribed region across the current content of vertebrates within the UCSC Genome Browser (http://genome.ucsc.edu). Comparisons revealed a high level of genetic conservation within the coding region of IL23A across all species presenting with a human IL23A gene homolog (32 of 44) from the platypus to the chimpanzee (Supplementary Fig. S1). Of note is the large degree of conservation across the transcribed region within the extended primate family.
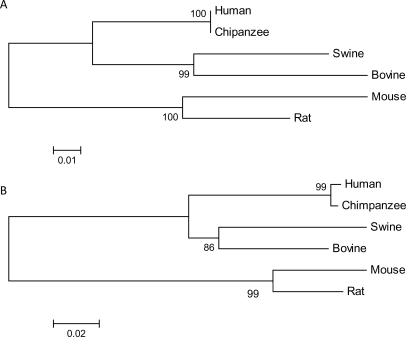
To further investigate intra-species conservation, the evolutionary relationship of IL23A between several mammalian species was predicted using Molecular Evolutionary Genetic Analysis version 4 (MEGA4) software.27 All publicly available (http://www.ncbi.nlm.nih.gov/) IL23A mRNA sequences, including Homo sapien (Human, GenBank no. NM_016584), Pan troglodytes (Chimpanzee, XM_522436), Sus scrofa (Swine, NM_001130236), Bos taurus (Bovine, XM_588269), Rattus norvegicus (Norwegian rat, NM_130410) and Mus musculus (common house mouse, NM_031252), were aligned using ClustalW, and a phylogenetic tree was constructed using the neighbour-joining and maximum likelihood methods on the basis of nucleotide differences (Fig. 2A). This analysis revealed complete homology of the IL23A mRNA transcript between humans and the closely related chimpanzee indicating that in the estimated 5–7 million years since evolutionary divergence of these two species,28 the transcribed sequence of IL23A has remained unaltered. In contrast, phylogenetic analysis of the gene, which encodes the p40 subunit of IL-23 and the binding partner of IL-23 p19, IL12B, revealed some (although minor) evolutionary divergence between the common transcript for both humans and chimpanzees (Fig. 2B). Similarly, the evolutionary distance between the human IL23A mRNA sequence and that of the remaining species was ∼2-fold less than that observed between the human IL12B mRNA sequence and the alternative species (Fig. 2A scale bar represents 0.01 base substitutions per site compared with 0.02 in Fig. 2B). These results further illustrate the exceptional level of evolutionary conservation of IL23A compared with another closely related member of this cytokine family.
Figure 2.
Phylogenetic tree representing the evolutionary relationship of six mammalian IL23A (A) and IL12B (B) mRNA sequences. Sequences were aligned using ClustalW and a neighbour-joining phylogenetic tree was generated using the maximum composite likelihood method and MEGA4. The tree is drawn to scale and the scale bar represents the same units as the evolutionary distance used to infer the phylogentic tree; base substitutions per site. Reliability of the tree was assessed by bootstrapping, using 1000 bootstrap replications. Numbers at branch nodes represent the confidence level of 1000 bootstrap replications. (A) The human IL23A transcript (H. sapien; NM_016584) is genetically identical to the predicted transcript of the chimpanzee (P. troglodytes; XM_522436). The next closest evolutionary related species is the swine (S. scrofa; NM_001130236), followed by the bovine (B. taurus; XM_588269), rat (R. norvegicus; NM_130410) and finally the mouse (M. musculus; NM_031252). The scale bar represents a genetic distance calculated by the number of base substitutions per site of 0.01. (B) The human IL12B transcript (NM_002187) is highly similar to the predicted transcript of the chimpanzee (XM_527101). The next closest evolutionary-related species is the swine (NM_214013), followed by the bovine (NM_174356), rat (NM_022611) and finally the mouse (NM_008352). The scale bar represents a genetic distance calculated by the number of base substitutions per site of 0.02.
Genes that display limited intra- and inter-species variation are thought to have undergone positive selection pressure directed towards a functional conservation and may have played a pivotal role in species evolution.28 Comparative genomics has revealed some of the most rapidly evolving genes are involved in reproduction and immune defense pathways.21 Genes which encode proteins with antibacterial, antiviral and antifungal activity are generally believed to be driven by co-evolution of host and pathogen and therefore pathogenic agents provide some of the strongest selective pressures on human evolution.29 A proposed role for IL-23 in mucosal immunity10 thus provides some explanation for the conservation of IL23A.
It has been suggested that when the immune response maintained by IL-23 persists in an uncontrolled manner, this protein may switch from being an integral part of the host defense to being a pro-inflammatory contributor to autoimmune disease.4 Thus, as a key component of the IL-23 complex, we suggest that IL23A may be vitally important to maintaining both innate and autoimmune responses. Although in this study we identify novel DNA variation at the IL23A locus, we also demonstrate the highly conserved nature of IL23A, at both amino acid and predicted regulatory levels. This study therefore highlights the potential significance of genetic variants found within this gene locus. Identifying functional variants, whether they are common disease associated variants or rare non-synonymous mutations, may provide key information in determining a role for IL23A in disease progression.
Supplementary data
Supplementary data are available at www.dnaresearch.oxfordjournals.org.
Funding
Funding sources include Cancer Institute New South Wales (CINSW) career development and support fellowship to V.M.H., CINSW scholar award and Australian Rotary Health PhD scholarship to E.A.T.
Acknowledgements
We would like to thank Dr Desiree Petersen and Prof. Web Miller for helpful communications. Ethics approval for this study was provided by the Human Ethics Research Committee of the University of New South Wales (HREC #08244).
Footnotes
Edited by Minoru Ko
References
- 1.Oppmann B., Lesley R., Blom B., et al. Novel p19 protein engages IL-12p40 to form a cytokine, IL-23, with biological activities similar as well as distinct from IL-12. Immunity. 2000;13:715–25. doi: 10.1016/s1074-7613(00)00070-4. [DOI] [PubMed] [Google Scholar]
- 2.McGeachy M.J., Cua D.J. Th17 cell differentiation: the long and winding road. Immunity. 2008;28:445–53. doi: 10.1016/j.immuni.2008.03.001. [DOI] [PubMed] [Google Scholar]
- 3.Boniface K., Blom B., Liu Y.J., de Waal Malefyt R. From interleukin-23 to T-helper 17 cells: human T-helper cell differentiation revisited. Immunol. Rev. 2008;226:132–46. doi: 10.1111/j.1600-065X.2008.00714.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tato C.M., Cua D.J. Reconciling id, ego, and superego within interleukin-23. Immunol. Rev. 2008;226:103–11. doi: 10.1111/j.1600-065X.2008.00715.x. [DOI] [PubMed] [Google Scholar]
- 5.Hue S., Ahern P., Buonocore S., et al. Interleukin-23 drives innate and T cell-mediated intestinal inflammation. J. Exp. Med. 2006;203:2473–83. doi: 10.1084/jem.20061099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kullberg M.C., Jankovic D., Feng C.G., et al. IL-23 plays a key role in Helicobacter hepaticus-induced T cell-dependent colitis. J. Exp. Med. 2006;203:2485–94. doi: 10.1084/jem.20061082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Uhlig H.H., McKenzie B.S., Hue S., et al. Differential activity of IL-12 and IL-23 in mucosal and systemic innate immune pathology. Immunity. 2006;25:309–18. doi: 10.1016/j.immuni.2006.05.017. [DOI] [PubMed] [Google Scholar]
- 8.Yen D., Cheung J., Scheerens H., et al. IL-23 is essential for T cell-mediated colitis and promotes inflammation via IL-17 and IL-6. J. Clin. Invest. 2006;116:1310–6. doi: 10.1172/JCI21404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cua D.J., Sherlock J., Chen Y., et al. Interleukin-23 rather than interleukin-12 is the critical cytokine for autoimmune inflammation of the brain. Nature. 2003;421:744–8. doi: 10.1038/nature01355. [DOI] [PubMed] [Google Scholar]
- 10.McGovern D., Powrie F. The IL23 axis plays a key role in the pathogenesis of IBD. Gut. 2007;56:1333–6. doi: 10.1136/gut.2006.115402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Torti D.C., Feldman S.R. Interleukin-12, interleukin-23, and psoriasis: current prospects. J. Am. Acad. Dermatol. 2007;57:1059–68. doi: 10.1016/j.jaad.2007.07.016. [DOI] [PubMed] [Google Scholar]
- 12.Furuzawa-Carballeda J., Vargas-Rojas M.I., Cabral A.R. Autoimmune inflammation from the Th17 perspective. Autoimmun. Rev. 2007;6:169–75. doi: 10.1016/j.autrev.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 13.Lyakh L., Trinchieri G., Provezza L., Carra G., Gerosa F. Regulation of interleukin-12/interleukin-23 production and the T-helper 17 response in humans. Immunol. Rev. 2008;226:112–31. doi: 10.1111/j.1600-065X.2008.00700.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sutton C.E., Lalor S.J., Sweeney C.M., Brereton C.F., Lavelle E.C., Mills K.H. Interleukin-1 and IL-23 induce innate IL-17 production from gammadelta T cells, amplifying Th17 responses and autoimmunity. Immunity. 2009;31:331–41. doi: 10.1016/j.immuni.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 15.Tishkoff S.A., Reed F.A., Friedlaender F.R., et al. The genetic structure and history of Africans and African Americans. Science. 2009;324:1035–44. doi: 10.1126/science.1172257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boster A.L., Endress C.F., Hreha S.A., Caon C., Perumal J.S., Khan O.A. Pediatric-onset multiple sclerosis in African-American black and European-origin white patients. Pediatr. Neurol. 2009;40:31–3. doi: 10.1016/j.pediatrneurol.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 17.Lee H.S., Korman B.D., Le J.M., et al. Genetic risk factors for rheumatoid arthritis differ in Caucasian and Korean populations. Arthritis Rheum. 2009;60:364–71. doi: 10.1002/art.24245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.White J.M., O'Connor S., Winter H.S., et al. Inflammatory bowel disease in African American children compared with other racial/ethnic groups in a multicenter registry. Clin. Gastroenterol. Hepatol. 2008;6:1361–9. doi: 10.1016/j.cgh.2008.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Patterson N., Petersen D.C., van der Ross R.E., et al. Genetic structure of a unique admixed population: implications for medical research. Hum. Mol. Genet. 2009;19:411–9. doi: 10.1093/hmg/ddp505. [DOI] [PubMed] [Google Scholar]
- 20.Campbell M.C., Tishkoff S.A. African genetic diversity: implications for human demographic history, modern human origins, and complex disease mapping. Annu. Rev. Genomics Hum. Genet. 2008;9:403–33. doi: 10.1146/annurev.genom.9.081307.164258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ellegren H. Comparative genomics and the study of evolution by natural selection. Mol. Ecol. 2008;17:4586–96. doi: 10.1111/j.1365-294X.2008.03954.x. [DOI] [PubMed] [Google Scholar]
- 22.Nair R.P., Duffin K.C., Helms C., et al. Genome-wide scan reveals association of psoriasis with IL-23 and NF-kappaB pathways. Nat. Genet. 2009;41:199–204. doi: 10.1038/ng.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Al-Salleeh F., Petro T.M. Promoter analysis reveals critical roles for SMAD-3 and ATF-2 in expression of IL-23 p19 in macrophages. J. Immunol. 2008;181:4523–33. doi: 10.4049/jimmunol.181.7.4523. [DOI] [PubMed] [Google Scholar]
- 24.Mise-Omata S., Kuroda E., Niikura J., Yamashita U., Obata Y., Doi T.S. A proximal kappaB site in the IL-23 p19 promoter is responsible for RelA- and c-Rel-dependent transcription. J. Immunol. 2007;179:6596–603. doi: 10.4049/jimmunol.179.10.6596. [DOI] [PubMed] [Google Scholar]
- 25.Kortylewski M., Xin H., Kujawski M., et al. Regulation of the IL-23 and IL-12 balance by Stat3 signaling in the tumor microenvironment. Cancer Cell. 2009;15:114–23. doi: 10.1016/j.ccr.2008.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee S., Kohane I., Kasif S. Genes involved in complex adaptive processes tend to have highly conserved upstream regions in mammalian genomes. BMC Genomics. 2005;6:168. doi: 10.1186/1471-2164-6-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tamura K., Dudley J., Nei M., Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007;24:1596–9. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 28.Kehrer-Sawatzki H., Cooper D.N. Understanding the recent evolution of the human genome: insights from human-chimpanzee genome comparisons. Hum. Mutat. 2007;28:99–130. doi: 10.1002/humu.20420. [DOI] [PubMed] [Google Scholar]
- 29.Nakajima T., Ohtani H., Satta Y., et al. Natural selection in the TLR-related genes in the course of primate evolution. Immunogenetics. 2008;60:727–35. doi: 10.1007/s00251-008-0332-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.