Abstract
Methods used for lipid analysis in embryos and oocytes usually involve selective lipid extraction from a pool of many samples followed by chemical manipulation, separation and characterization of individual components by chromatographic techniques. Herein we report direct analysis by matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS) of single and intact embryos or oocytes from various species. Biological samples were simply moisturized with the matrix solution and characteristic lipid (represented by phosphatidylcholines, sphingomyelins and triacylglycerols) profiles were obtained via MALDI-MS. As representative examples, human, bovine, sheep and fish oocytes, as well as bovine and insect embryos were analyzed. MALDI-MS is shown to be capable of providing characteristic lipid profiles of gametes and embryos and also to respond to modifications due to developmental stages and in vitro culture conditions of bovine embryos. Investigation in developmental biology of the biological roles of structural and reserve lipids in embryos and oocytes should therefore benefit from these rapid MALDI-MS profiles from single and intact species.
Keywords: oocyte, embryo, mass spectrometry, phospholipid, triacylglicerol, lipid fingerprinting
The double molecular layer of polar lipids is a marvelous architectural feature of exquisite biological engineering in cell membranes. Specific functions and variations of the various phospholipids (PL), the most abundant lipids in eukaryotic cell membranes, are, however, still poorly understood (1). A diversity of PL in a finely balanced equilibrium is used by cells to construct stable and functional membranes, and PL composition determines most of the physico-chemical cell membrane properties such as fluidity, permeability and thermal phase behavior (2).
Knowledge of the function of lipids within the cell has benefited from the development of increasingly sensitive and selective analytical techniques, particularly those based on mass spectrometry (3, 4). Among these techniques, matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS) (5) has been very successful in studies of the compositions of lipids and other crucial biological molecules. MALDI-MS has allowed direct analysis of complex and unfractionated samples, such as the study of peptide profiles below the level of a single cell (6). In lipidomics, MALDI-MS has provided fast and simple acquisition of mass spectra with lipid profiles of cells, tissues and body fluids (7).
MALDI-MS lipid fingerprinting can, for example, help studies aimed at understanding the effect of membrane lipid composition on cell membrane behavior after temperature changes. This knowledge is essential for cryopreservation studies of a variety of cells, including oocytes from patients undergoing cancer treatment (8); gametes and embryos for conservation of animal genetic resources; and to facilitate the international transit of genetic material (9). Cytoplasm lipid accumulation has been shown to affect the rate of post-thaw development of bovine embryos (10–12), which suggests that the embryo culture process causes lipid metabolism changes that affect the properties and stability of cell membranes (13). In laboratories for assisted reproduction for humans, embryos and oocytes are routinely cryopreserved, but there is still considerable need of research aiming to improve knowledge of the role of lipids and the efficiency of protocols used (14–16).
To date, changes in PL composition profiles of mammalian embryos and oocytes are poorly understood. Gas chromatography (GC) has been applied to study triacylglycerol (TAG) composition in human and bovine oocytes and embryos (17–19), but this approach requires chemical transformation via saponification and derivatization, providing free fatty acyl residue profiles but not TAG or PL identification. A main drawback for GC analyses is the need of a large pool of samples (more than 10 oocytes or embryos for each lipid analysis), especially when human embryos or oocytes are investigated (19).
MALDI-MS offers, however, the advantage of detecting intact lipids, and due to its unmatched high spatial resolution, allows targeting single species of microscopic dimensions (100–200 μm) such as mammalian oocytes and embryos. Herein we show that direct analysis by MALDI-MS can contribute significantly to embryo and oocyte lipidomic studies. With no solvent extraction and no chemical manipulation, direct MALDI-MS of single and intact embryos or oocytes of different species (from invertebrates to mammalian) is shown to provide reproducible lipid profiles. Besides species recognition, the method described herein is also shown to be sensitive and, hence, to reveal important changes in lipid profiles during bovine preimplantation development and in bovine embryo in vitro culture conditions.
MATERIALS AND METHODS
Reagents
Unless mentioned, chemicals and culture media for handling bovine and ovine oocytes were purchased from Sigma (St. Louis, USA). Media used for human oocytes handling (modified Human Tubal Fluid [HTF]) was purchased from Irvine Scientific (Santa Ana, CA, USA). Phosphate buffer saline (PBS) solution was supplied by Nutricell (Campinas, SP, Brazil).
Methanol (ACS/HPLC) grade was purchased from Burdick and Jackson (Muskegon, MI, USA) and 2,5-dihydroxybenzoic acid (DHB) was purchased from ICN Biomedicals (Aurora, OH, USA). Ultrapure water, purified by a Direct-Q water system (Millipore, Bedford, MA, USA) was used for the preparation of solvents.
Sample collection and storage for analysis
The care and use of animal samples (bovine, sheep, fish and fire ant) was approved by the Institutional Committee for Ethics in Animal Research of the State University of Campinas (UNICAMP), which follows the Ethical Principles of Animal Research established by the Brazilian College for Animal Experimentation (COBEA) under protocol number 1752-1. The use of human unfertilized oocytes was approved by the São Paulo Federal University (UNIFESP) Institutional Committee for Ethics under the protocol number 0411/07.
Immature bovine (Bos taurus) and sheep (Ovis Aries) oocytes were obtained by post mortem follicular aspiration of ovaries from cows and ewes slaughtered at commercial slaughterhouses. Ovaries were transported in 0.9% (w/v) saline solution at 25–30°C to the laboratory and follicles were aspirated using an 18-gauge needle attached to a 20-ml syringe. Cumulus oocyte complexes with at least three layers of cumulus cells and homogeneous cytoplasm were denuded of cumulus cells by gentle pipetting in 0.5% hyaluronidase.
Bovine in vivo–derived embryos were obtained from superovulated heifers kept in pasture (Brachiaria decumbens). The emergence of the animal's follicular wave was synchronized by one intramuscular (i.m.) injection of 2.0 mg estradiol benzoate (Estrogin, Farmavet, São Paulo, Brazil) and insertion of an intravaginal controlled internal drug release device (CIDR), containing 1.9 g progesterone (Pfizer, Hamilton, New Zealand) on day 0. On day 4.5, the superstimulatory treatments were initiated (follicle stimulating hormone [FSH]; Folltropin-V, Bioniche Animal Health, Belleville, Canada) and given in decreasing doses of 28, 21, 14 and 7 mg FSH twice daily, over a 4-day period, for a total dose of 70 mg. At the time of the fifth and sixth injections of FSH, 25 mg of dinoprost tromethamine (Lutalyse, Pfizer, Paulinia, Brazil) was injected intramuscularly. The CIDR was removed at the time of the seventh superstimulatory injection. Ovulation was induced with an i.m. injection of 0.05 mg gonadotrophin releasing hormone (GnRH; Gestran Plus; ARSA S.R.L., Buenos Aires, Argentina) 12 h after the last superstimulatory injection. All heifers were artificially inseminated (AI) with frozen/thawed semen from the same bull 12 and 24 h after GnRH injection. Seven days after the first AI, embryos/ova were recovered using a nonsurgical uterine flushing technique (20).
For bovine in vitro embryo production, oocytes obtained by post mortem follicular aspiration were in vitro fertilized and cultured as described previously (21), but with changes in culture medium supplementation and incubator atmosphere. After IVF, embryos were cultured in four different in vitro conditions: 5% O2 and BSA (Group 1); 20% O2 and BSA (Group 2); 20% O2 and FCS (Group 3); and 5% O2 and FCS (Group 4).
Human unfertilized oocytes from women submitted to transvaginal oocyte retrieval were provided by the Human Reproduction Service of São Paulo Federal University (UNIFESP) in Brazil. These samples would normally be discarded.
Mullet ova (Mugil spp.) were collected from fresh fish in the City Fish Market of Santos-SP, Brazil, and transported at 4°C to the laboratory.
Fire ant (Solenopsis spp.) eggs were collected in the field after species identification and were immediately transported to the laboratory.
Oocyte and embryo samples were stored in microtubes containing 100 μl of a 50% aqueous methanol solution at -80°C until analysis. Sample preparation involved placing each oocyte, egg or embryo in a given spot of the target plate under the stereomicroscope. Samples were allowed to dry at room temperature, and their location was recorded in order to place the laser at the correct location during analysis. Just before analysis, 1 μl of 1.0 mol/l 2,5-dihydroxybenzoic acid (DHB) in methanol was placed in each target spot and allowed to dry at room temperature.
Lipid analysis by MALDI-MS
MALDI-MS and MALDI-MS/MS spectra were acquired in the positive ion and reflectron modes using a Q-ToF Premier mass spectrometer (Waters, Manchester, UK) equipped with a 200-Hz solid-state laser in the m/z range of 700–950. The principal operating condition used was 10 V (sample plate), and laser irradiation consisted of diverse shots during the time of 60–90 s in the region were the sample had been placed on the target plate, until signals in the region of interest were observed and disappeared due to the consumption of the microscopic sample (Fig. 1). MALDI-MS/MS were manually acquired by increasing the collision energy until extensive dissociation of the precursor ion was observed. Argon was used as the collision gas. Spectra were centered and aligned using the MassLynx 4.0 software (Waters, Manchester, UK). From each spectrum, after the exclusion of isotopic peaks, the fifty most intense ions were considered as the starting point for searching m/z values corresponding to lipids. After attribution, only the m/z values which were clearly distinct from noise level in the spectra were included in the principal component analysis (PCA), which was performed using Pirouette v.3.11 (Infometrix Inc., Woodinville, WA, USA).
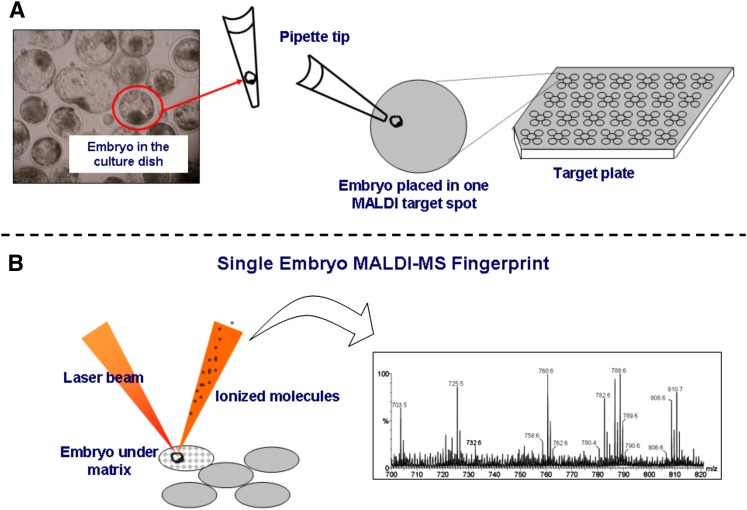
Fig. 1.
Schematic of sample preparation and spectrum acquisition for MALDI-MS analysis of a single embryo or oocyte. (A) Individual samples are removed from the culture dish, washed, transferred to a spot on a MALDI target plate, dried and moistened with the matrix solution. (B) The laser is focused on the single unit and the MALDI-MS data is collected. MALDI plates typically allow for the simultaneous loading of either 96 or 384 samples in individual spots, and full spectra acquisition requires a few milliseconds of spectra accumulation.
RESULTS
As proof-of-principle cases, we investigated the potential of MALDI-MS fingerprinting to provide characteristic lipid profiles from a representative set of single embryos and oocytes from mammals and from two distantly related species: insect and fish. We then evaluated the ability of the technique to reveal changes in the lipid profiles during embryo development and under different in vitro culture conditions for bovines as a representative biological model.
Lipid fingerprinting in different species
MALDI-MS analysis and sample preparation (described in detail in the “Material and Methods” section) involved no lipid extraction and no chemical manipulation which, as already mentioned, would require substantial sample pooling of these highly valuable and sometimes unavailable (particularly in large numbers) microscopic structures. An intact single embryo or oocyte was collected and then placed in one of the many spots of the MALDI target plate placed under a stereomicroscope (Fig. 1A). Then, the target plate spot was moistened with the MALDI matrix before spectra acquisition (Fig. 1B).
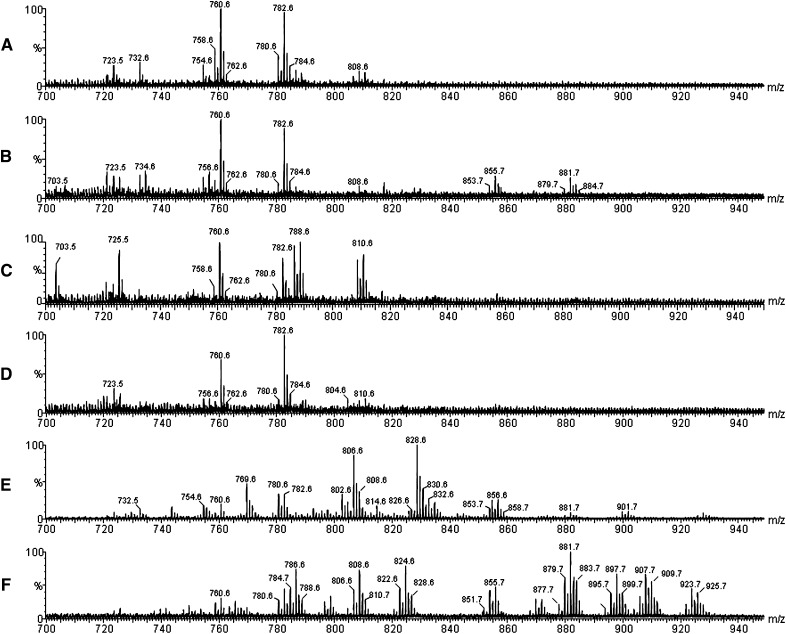
Fig. 2 displays representative MALDI-MS spectra from single embryos or oocytes from the five species investigated in this work. The spectra were acquired in the m/z 700–950 range, where most PL and TAG should be detected by MALDI(+)-MS mainly as either their protonated [M + H]+ or sodiated molecules [M + Na]+, or both (22). Note that the MALDI-MS profiles are rather characteristic, and in some cases, very distinctive with unique PC, SM and TAG ions (see below) with contrasting abundances or ratios. Group comparison by principal component analysis (PCA; see below) was also found to allow their prompt characterization.
Fig. 2.
MALDI-MS in the positive ion mode for a single and intact oocyte or embryo: (A) human (Homo sapiens) oocyte; (B) bovine (Bos taurus) oocyte; (C) bovine embryo; (D) sheep (Ovis aries) oocyte; (E) fish (Mugil spp.) oocyte; and (F) ant (Solenopsis spp.) egg.
The PL species were first attributed (Table 1) based on previous lipid profile studies by MALDI-MS and are described by the class abbreviation followed by the total number of carbons and double bounds in the acyl residues attached to the glycerol backbone (in parenthesis). The MALDI-MS of the human oocyte is characterized mainly by two clusters of ions, in which those of m/z 760.6, assigned as [PC (34:1) + H]+, and m/z 782.6, [PC (36:4) + H]+ or [PC (34:1) + Na]+ or both, predominate. These are also seen as major ions in the other mammalian samples: bovine oocytes (Fig. 2B), in vivo–produced bovine embryos (Fig. 2C) and sheep oocytes (Fig. 2D).
TABLE 1.
Phospholipids (PL) and triacylglycerols (TAG) identified via MALDI(+)-MS of single human oocytes, bovine oocytes, bovine embryos, sheep oocytes, fish oocyte or ant eggs
| m/z | Lipid Ion (carbons:unsaturations) | Reference |
|---|---|---|
| 703.5 | [SM (16:0) + H]+ | 26, 30 |
| 723.5 | [PC (34:1) + Na]+ and loss of N(CH3)3 | 29 |
| 725.5 | [SM (16:0) + Na]+ | 28, 30 |
| 731.5 | [SM (18:0) + H]+ | 26 |
| 732.5 | [PC (32:1) + H]+ | 26 |
| 734.6 | [PC(32:0) + H]+ | 26 |
| 753.6 | [SM (18:0) + Na]+ | 26 |
| 754.6 | [PC (32:1) + Na]+ | 26 |
| 756.6 | [PC (32:0) + Na]+ | 29, 30 |
| 758.6 | [PC (34:2) + H]+ | 26, 27, 30 |
| 760.6 | [PC (34:1) + H]+ | 28 |
| 762.6 | [PC (34:0) + H]+ | 27 |
| 780.6 | [PC (34:2) + Na]+, [PC (36:5) + H]+ | 26, 27, 30 |
| 782.6 | [PC (36:4) + H]+, [PC (34:1) + Na]+ | 27–30 |
| 784.6 | [PC (34:0) + Na]+ | 30 |
| 786.6 | [PC (36:2) + H]+ | 26, 28, 30 |
| 788.6 | [PC (36:1) + H]+ | 27, 28 |
| 802.6 | [PC 36:5 + Na]+ | a |
| 804.6 | [PC (38:7) + H]+, [PC36:4 + Na]+ | 30, a |
| 806.6 | [PC (38:6) + H]+, [PC36:3 + Na]+ | a |
| 808.6 | [PC (38:5) + H]+, [PC (36:2) + Na]+ | 26–8, 30 |
| 810.6 | [PC (38:4) + H]+, [PC (36:1) + Na]+ | 26–28, 30 |
| 828.6 | [PC (38:6) + Na]+ | a |
| 830.6 | [PC (38:5) + Na]+ | 28 |
| 832.6 | [PC (38:4) + Na]+ | 26, 30 |
| 834.6 | [PC (40:6) + H]+ | 27 |
| 836.6 | [PC (40:5) + H]+ | 28 |
| 851.7 | PPLn (50:3) + Na+ | 31 |
| 853.7 | PPL (50:2)+ Na+ | 31 |
| 855.7 | PPO (50:1) + Na+ | 31 |
| 856.6 | [PC (40:6) + Na]+ | 29 |
| 877.7 | PLL (52:4) + Na+ | 31 |
| 879.7 | PLO (52:3) + Na+ | 31 |
| 881.7 | POO (52:2) + Na+ | 31 |
| 883.7 | POS (52:1) + Na+ | 31 |
| 895.7 | TAG (54:9) + Na+ | a |
| 897.7 | TAG (54:8) + Na+ | a |
| 899.7 | LLLn (54:7) + Na+ | 31 |
| 901.7 | LLL (54:6) + Na+ | 31 |
| 903.7 | LLO, OOLn (54:5) + Na+ | 31 |
| 905.7 | OOL, LLS (54:4) + Na+ | 31 |
| 907.7 | OOO, SOL (54:3) + Na+ | 31 |
| 909.7 | OOS, SSL (54:2) + Na+ | 31 |
| 911.7 | SSO (54:1) + Na+ | 31 |
| 919.7 | LLO, OOLn (54:5) + K+ | 31 |
| 921.7 | OOL, LLS (54:4) + K+ | 31 |
| 923.7 | OOO, SOL (54:3) + K+ | 31 |
| 925.7 | OOS, SSL (54:2) + K+ | 31 |
Identification is based on earlier studies and MS/MS data. L, linoleic acid; Ln, linolenic acid; O, oleic acid; P, palmitic acid; PC, phosphatidylcholines; S, stearic acid; SM, sphingomyelin.
Attribution performed in this work.
In vivo bovine embryo [compare bovine oocyte (Fig. 2B) with the bovine embryo (Fig. 2C)], development is indeed found to produce considerable changes in the MALDI-MS lipid profiles as seen by the greater abundance of ions of m/z 786.6 [PC (36:2) + H]+, and the appearance of abundant ions of m/z 788.6 [PC (36:1) + H]+, 808.6 [PC (36:2) + Na]+ or [PC (38:5) + H]+ and 810.6 [PC (38:4) + H]+ or [PC (36:1) + Na]+. In the MALDI-MS fingerprint of unique fish oocyte (Fig. 2E), PC ions with higher degrees of unsaturation in the fatty acyl residues, such as those of m/z 806.6 [PC (38:6) + H]+ or [PC(36:3) + Na]+, as well as TAG ions of m/z 853.7 [PPL (50:2)+ Na]+ and 855.7 [PPO (50:1) + Na]+ are noticeable, as expected from fish samples (23).
The MALDI-MS of the insect embryo (Fig. 2F) is even more distinct and characteristic, displaying a much diverse and broader (in m/z range) set of abundant PC, SM and TAG ions. Indeed, about 30–40% of the dry weight of insect eggs are known to correspond to lipids, mainly of TAG (24–26).
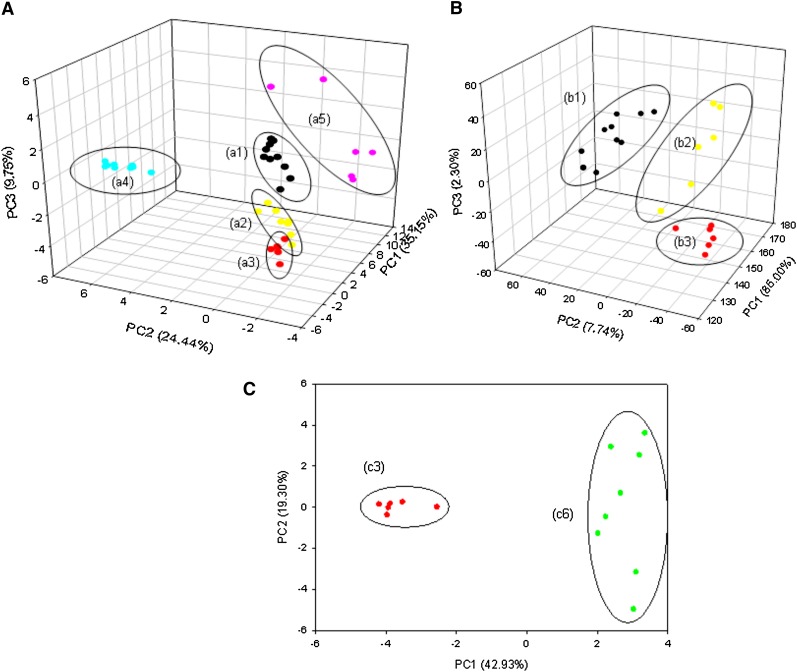
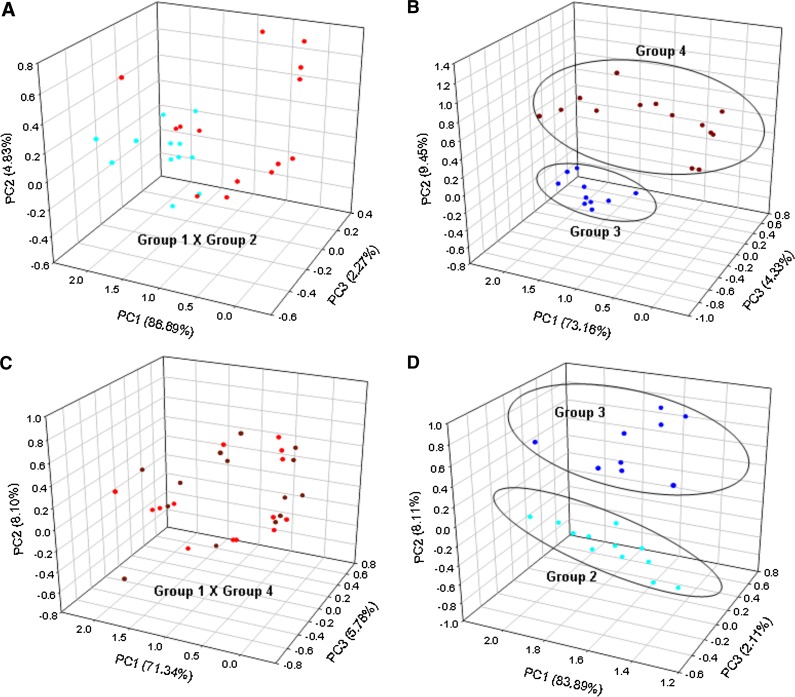
PCA analysis (Fig. 3A) shows that all classes of samples can be resolved via their MALDI-MS lipid profiles. As could be expected, the mammalian samples [human (a1), bovine (a2) and sheep (a3)] are placed far away from the fish (a4) and ant (a5) samples, but they form a group located close together in the 3D PCA domain. They are, however, clearly resolved and grouped properly by species with little overlap.
Fig. 3.
PCA plot for the MALDI-MS data of single embryos and oocytes of (A) all samples from the five species: (a1) human (Homo sapiens) oocytes (N = 10), (a2) sheep (Ovis Aries) oocytes (N = 6), (a3) bovine (Bos taurus) oocytes (N = 6), (a4) fish (Mugil spp.) oocytes (N = 9), and (a5) insect (Solenopsis spp.) eggs (N = 6); (B) of the three mammalian species: (b1) human, (b2) sheep and (b3) bovine; and (C) of only (c3) bovine oocytes (N = 6) and (c6) bovine embryos (N = 8).
When the MALDI-MS data for the mammalian samples were analyzed separately (Fig. 3B), group individualization in the 3D PCA plot was even more pronounced, with no overlaps. The fish and insect (ant) samples, as expected from their more distinct lipid distributions, form the best-resolved groups with a greater dispersion for the insect samples, probably due to the much greater diversity of the lipid ions detected in their MALDI-MS.
A 2D PCA (Fig. 3C) compared separately lipid profiles from bovine embryos and oocytes. Both types were very well resolved by proper grouping, which suggests an interesting application of single embryo and oocyte MALDI-MS fingerprinting, that is, monitoring changes in PL and TAG profiles during different developmental stages.
To investigate whether MALDI-MS of single embryos could monitor lipid changes owing to in vitro culture conditions, bovine embryos were cultured using four different in vitro conditions regarding incubator atmosphere and culture medium supplementation, respectively: 5% O2 and BSA (Group 1); 20% O2 and BSA (Group 2); 20% O2 and FCS (Group 3); and 5% O2 and FCS (Group 4).
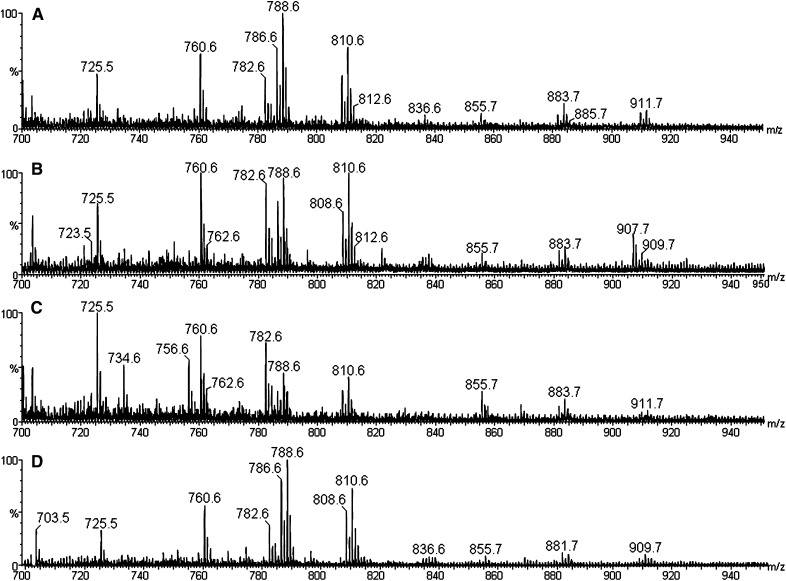
Comparisons were performed considering the culture medium supplementation (Group 1 versus Group 2 and Group 3 versus Group 4) and atmosphere influence (Group 1 versus Group 4 and Group 2 versus Group 3). Regarding culture medium supplementation (Fig. 4A, B and Fig. 5A), most embryos from BSA at low oxygen concentration presented less intense ions of m/z 760.6 and 782.6 [protonated and sodiated forms of PC 34:1, respectively] and of m/z 725.5 (sodiated SM 16:0) compared with higher oxygen concentration. The FCS culture medium supplementation effect is clearly noted at different oxygen concentrations (Fig. 4C–D and Fig. 5B), where the embryos cultured in high oxygen concentration present less intense PC 36:1 ions and higher PC 34:1 and SM 16:0 ions compared with lower oxygen concentration. Regarding the atmosphere influence, at low oxygen concentration, no influence of BSA or FCS was observed (Fig. 4A, D and Fig. 5C). At higher oxygen concentration, PCA clearly separates BSA from FCS-supplemented embryos (Fig. 4C, D and Fig. 5D). The principal difference is that embryos from Group 1 presented more intense PC 36:1 ions than embryos from Group 4, which presented more intense PC 34:1 and the ion of m/z 725.6. Together with GC data on the fatty acyl residue composition of bovine oocytes (18, 27), these results indicate that low oxygen concentration in the incubator and BSA as a medium supplement favor accumulation of PC containing 18:1 (PC 36:1) and not 16:0 (PC 34:1 and SM 16:0).
Fig. 4.
MALDI-MS in the positive ion mode for single and intact bovine embryos cultured using four different in vitro conditions regarding incubator atmosphere and culture medium supplementation, respectively: (A) 5% O2 and BSA (Group 1; N = 14); (B) 20% O2 and BSA (Group 2; N = 12); (C) 20% O2 and FCS (Group 3; N = 10); and (D) 5% O2 and FCS (Group 4; N = 14).
Fig. 5.
3D PCA using MALDI-MS lipid data from bovine embryos cultured in four different in vitro conditions regarding culture medium supplementation: (A) 5% O2 and BSA (Group 1; red dots) versus 20% O2 and BSA (Group 2; light blue dots), and (B) 20% O2 and FCS (Group 3; dark blue dots) versus 5% O2 and FCS (Group 4; brown dots), and incubator atmosphere: (C) 5% O2 and BSA (Group 1; red dots) versus 5% O2 and FCS (Group 4; brown dots); and (D) 20% O2 and BSA (Group 2; light blue dots) versus 20% O2 and FCS (Group 3; dark blue dots).
Characterization of lipids in oocytes and embryos
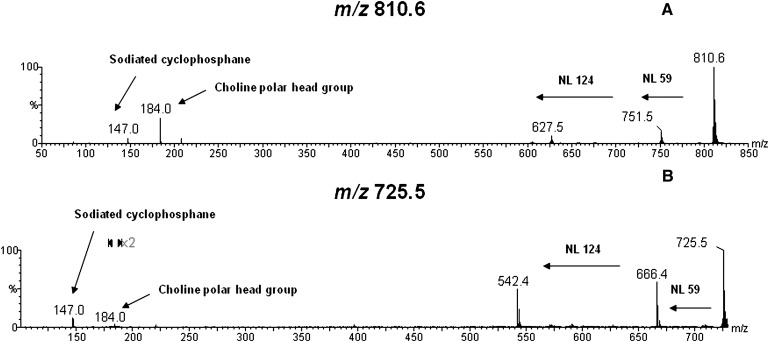
Table 1 summarizes the attribution of PL and TAG from the MALDI-MS data. To substantiate such attributions, major PL or TAG ions were subjected to MALDI-MS/MS. Dissociation patterns compatible with either PC, SM or TAG ions (28–33) were observed. Representative examples (Fig. 6A, B)show the MALDI-MS/MS of the PC ions of m/z 810.6 [PC (38:4) + H]+ or [PC (36:1) + Na]+ and m/z 725.5 [SM (16:0) + Na]+, respectively. The loss (NL) of 59 Da is due to neutral trimethylamine [(N(CH3)3], whereas 124 Da involves loss of the cyclophosphane ring (C2H5O4P). The fragment ion of m/z 147 corresponds to the sodiated cyclophosphane, and that of m/z 184 to the monoprotonated dihydrogenphosphate choline or the choline head fragment (C5H15PO4N). Supplementary Table I presents a list of fragment ions diagnostic of PC and SM species. Supplementary Table II lists m/z values for lipid ions followed by minimal and maximal relative ion abundances, as well as the percentage of samples containing the specific lipid in the five different species, as individual variation is expected to occur in biological samples.
Fig. 6.
Single bovine embryo MALDI-MS/MS for the ions of: (A) m/z 810.6 [PC (38:4) + H]+ or [PC (36:1) + Na]+ and (B) m/z 725.5 [SM (16:0) + Na]+. Both spectra display characteristic NL of 59 Da (trimethylamine) and of 124 Da from the loss of the cyclophosphane ring. Also, the sodiated cyclophosphane (m/z 147.0) and the choline polar head group (m/z 184.0) ion can be observed.
DISCUSSION
This article has described a practical and novel approach for embryo and oocyte lipid studies. Reproducible and characteristic MALDI-MS fingerprints can be obtained from a single oocyte or preimplantation intact embryo sample in a procedure involving no extraction, no chemical manipulation and no preseparation. MALDI-MS is a powerful tool in lipidomics (34), generating PL and TAG profiles due to detection of singly charged molecular ions, which greatly facilitates data interpretation. We estimate about 10–15 pmol as the sensitivity for lipids from single embryo or oocyte MALDI-MS analyses as each bovine oocyte is known to contain this amount of PL (18). Oocytes from other mammals have similar sizes, and embryos have greater amounts of PL due to their higher amount of cells.
Research on the biological role of lipids in cells requires the determination, usually from numerous samples, of detailed lipid composition and changes during organism development. Methods traditionally used in oocyte and embryo lipid analysis, such as GC (17–19, 27) and radioactive labeling (35), always demand the pooling of 10–400 (17, 18, 27) of these microscopic structures for lipid extraction with organic solvents, followed by chemical manipulation (hydrolysis/derivatization), separation and characterization with chromatographic techniques. Lipids with similar structural and chemical properties may also not be resolved. Mass spectrometry, especially when high accuracy m/z measurements or MS/MS techniques are applied, overcomes these failings, since the components of a complex lipid mixture are resolved by mass and identified in very low concentrations (typically pico- or fentomol) by the characteristic m/z values of their corresponding molecular ions and structurally diagnostic dissociation in MS/MS experiments.
Reports of fatty acyl residue composition in oocytes describe the palmitic (16:0), stearic (18:0) and oleic (18:1) groups as the predominant fatty acyl residues in immature bovine oocytes, in unfertilized human oocytes (17, 18) and in sheep oocytes (27). Herein, we have reported the structural composition of some PL species containing these fatty acyl residues, and we show that embryo and oocyte lipid fingerprinting by MALDI-MS allows differentiation among species. Also, developmental stage differences in PC profiles between bovine oocytes and embryos (at the developmental stage of blastocyst) were indicated to occur. It is known that blastocyst formation involves cell differentiation into trophoectoderm and the inner cell mass. Metabolomic, gene expression and proteomic studies in bovine blastocysts have been performed (36–38). Nonetheless, membrane PC and SM changes and their roles during embryo development have not been reported. Changes in lipid patterns during oocyte development have been observed in Bufo arenarum oogenesis and early embryo development (39, 40). Although used as a model for understanding oogenesis and embryogenesis, due to their high numbers of gametes and external fecundation, frog reproduction differs significantly from that of mammalians species. MALDI-MS monitoring of single samples seems to be able to provide an important tool for studies of mammalian samples.
Fatty acyl residues have previously been shown to be associated with oocyte maturation and fertility (18) as well as with embryo development (19, 35, 41, 42) in mammalian species. As an example, storage and membrane lipid content of oocytes or embryos influence their cryosensitivity (43, 44). Oocytes of all species are especially sensitive to low temperatures (45). Adding linoleic acid to bovine embryo culture medium was shown to enhance cryosurvival of embryos (42), increasing interest in studies involving the role of lipid composition in embryos with different cryosurvival rates.
In vitro culture conditions of bovine embryos are recognized to influence embryo developmental success, as well as embryo cryosensitivity (46–48). We evaluated the influence of the culture medium supplement (BSA or FCS) and incubator atmosphere (high oxygen concentration: 20% O2, and low oxygen concentration: 5% O2) on the MALDI-MS lipid profile (represented by SM, PC and TAG) of single bovine embryos. FCS is widely used as a supplement for embryo in vitro culture, but it alters embryo metabolism, stimulating lipid accumulation, as observed by staining methods and electron microscopy (47). Regarding incubator oxygen concentration, it has been suggested that lower oxygen concentrations (5% O2) mimic physiological conditions and significantly decrease oxidative stress in embryos (48).
Our results indicate that low incubator oxygen concentration and BSA as a culture medium supplement favor PC containing 18:1 (PC 36:1) and not 16:0 (PC 34:1 and SM 16:0). These results correlate well with studies showing the beneficial effects of low oxygen concentrations and serum-free medium (46, 48) and the negative effect of palmitic acid (16:0) supplementation on embryo cryosurvival (49).
The high quality and selectivity of the first single embryo MALDI-MS data presented herein point already to the suitability of the technique. Note that other direct desorption/ionization ambient techniques could be envisaged for single embryo MS analysis such as desorption electrospray ionization (DESI) and direct analysis in real time [DART (50)], but they lack the desirable high spatial resolution characteristic of MALDI-MS, which is a vital feature for microscopic samples. Besides, MALDI-MS is known to present the high sensitivity needed to detect the few picomols of lipids present in these samples.
We believe that the figures of merit presented for MALDI-MS of single embryo analysis can still be improved. Altering the laser source (nitrogen or a Nd:YAG laser source) and particularly the MALDI matrix may allow more selective or broader detection of different lipids according to the specific biological question investigated. The use of other matrices, such as para- nitroaniline (51), 2-(2-aminoethylamino)-5-nitropyridine (52), and 9-aminoacridine (53), has been reported for PL analysis. DHB (2,5-dihydroxybenzoic acid), although widely used as a matrix for lipid analysis, is not optimal for PL detection in the negative mode due to its acidic character and more effectively detects PC and SM in the positive ion mode (29). Recently, the use of 9-aminoacridine with improved preparation protocols for MALDI-MS allowed low background signals for the detection of low MW species (54) and various lipid classes (53), paving the way for practical lipidomic studies based on MALDI-MS.
The proof-of-principle data presented herein with characteristic spectra for five species at different developmental stages confirms the broad and successful application of direct single embryo lipid fingerprinting by MALDI-MS.
CONCLUSIONS
Single embryo and oocyte MALDI-MS of intact samples allows nearly direct lipid profiling (represented herein by SM, PC and TAG species) with no extraction, chemical manipulation or pre-separation steps. Mass measurements are made with accuracy, high reproducibility and high sample throughput. Typification and monitoring of some lipid changes during the developmental stages were shown to be dependent on species specificity and were related to the environment. MALDI-MS of intact embryos and oocytes is also open to further improvements as other lasers and matrices may boost lipid detection. MALDI-MS, by providing lipid profiles for single embryos and oocytes from many different species, should contribute to improve cryopreservation and in vitro culture conditions of embryos and oocytes in general.
Supplementary Material
Acknowledgments
The authors are grateful to Prof. Carol Hollingworth Collins for English review, and to Amadeu Hoshi Iglesias, Juliana Hayashi Tannura, Dr. Aguinaldo Pereira Cedenho, Amanda Begatti Victorino and Juliana Stevanato for technical assistance during sample collection and spectra acquisition.
Footnotes
Abbreviations:
- DHB
- 2,5-dihydroxybenzoic acid
- FCS
- fetal calf serum
- GC
- gas chromatography
- L
- linoleic acid
- Ln
- linolenic acid
- MALDI-MS
- matrix-assisted laser desorption/ionization mass spectrometry
- NL
- neutral loss
- O
- oleic acid
- P
- palmitic acid
- PC
- phosphatidylcholines
- PCA
- principal component analysis
- PL
- phospholipids
- S
- stearic acid
- SM
- sphingomyelin
- TAG
- triacylglycerol
This work was supported by the Brazilian research foundations FAPESP (Grant 2008/10756-7) and CNPq.
The online version of this article (available at http://www.jlr.org)contains supplementary data in the form of two tables.
REFERENCES
- 1.van Meer G., Voelker D. R., Feigenson G. W. 2008. Membrane lipids: where they are and how they behave. Nat. Rev. Mol. Cell Biol. 9: 112–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Edidin M. 2003. Lipids on the frontier: a century of cell-membrane bilayers. Nat. Rev. Mol. Cell Biol. 4: 414–418. [DOI] [PubMed] [Google Scholar]
- 3.Roberts L. D., McCombie G., Titman C. M., Griffin J. L. 2008. A matter of fat: an introduction to lipidomic profiling methods. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 871: 174–181. [DOI] [PubMed] [Google Scholar]
- 4.Ejsing C. S., Sampaio J. L., Surendranath V., Duchoslav E., Ekroos K., Klemm R. W., Simons K., Shevchenko A. 2009. Global analysis of the yeast lipidome by quantitative shotgun mass spectrometry. Proc. Natl. Acad. Sci. USA. 106: 2136–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karas M., Hillenkamp F. 1988. Laser desorption ionization of proteins with molecular masses exceeding 10,000 daltons. Anal. Chem. 60: 2299–2301. [DOI] [PubMed] [Google Scholar]
- 6.Li L., Garden R. W., Sweedler J. V. 2000. Single-cell MALDI: a new tool for direct peptide profiling. Trends Biotechnol. 18: 151–160. [DOI] [PubMed] [Google Scholar]
- 7.Fuchs B., Schiller J. 2008. MALDI-TOF MS analysis of lipids from cells, tissues and body fluids. Subcell. Biochem. 49: 541–565. [DOI] [PubMed] [Google Scholar]
- 8.Tao T., Del Valle A. 2008. Human oocyte and ovarian tissue cryopreservation and its application. J. Assist. Reprod. Genet. 25: 287–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pereira R. M., Marques C. C. 2008. Animal oocyte and embryo cryopreservation. Cell Tissue Bank. 9: 267–277. [DOI] [PubMed] [Google Scholar]
- 10.Abe H., Yamashita S., Satoh T., Hoshi H. 2002. Accumulation of cytoplasmic lipid droplets in bovine embryos and cryotolerance of embryos developed in different culture systems using serum-free or serum-containing media. Mol. Reprod. Dev. 61: 57–66. [DOI] [PubMed] [Google Scholar]
- 11.Barcelo-Fimbres M., Seidel G. E., Jr. 2007. Effects of either glucose or fructose and metabolic regulators on bovine embryo development and lipid accumulation in vitro. Mol. Reprod. Dev. 74: 1406–1418. [DOI] [PubMed] [Google Scholar]
- 12.George F., Daniaux C., Genicot G., Verhaeghe B., Lambert P., Donnay I. 2008. Set up of a serum-free culture system for bovine embryos: embryo development and quality before and after transient transfer. Theriogenology. 69: 612–623. [DOI] [PubMed] [Google Scholar]
- 13.Dinnyes A., Nedambale T. L. 2009. Cryopreservation of manipulated embryos: tackling the double jeopardy. Reprod. Fertil. Dev. 21: 45–59. [DOI] [PubMed] [Google Scholar]
- 14.Desai N., Blackmon H., Szeptycki J., Goldfarb J. 2007. Cryoloop vitrification of human day 3 cleavage-stage embryos: post-vitrification development, pregnancy outcomes and live births. Reprod. Biomed. Online. 14: 208–213. [DOI] [PubMed] [Google Scholar]
- 15.Cobo A., Kuwayama M., Perez S., Ruiz A., Pellicer A., Remohi J. 2008. Comparison of concomitant outcome achieved with fresh and cryopreserved donor oocytes vitrified by the Cryotop method. Fertil. Steril. 89: 1657–1664. [DOI] [PubMed] [Google Scholar]
- 16.Hiraoka K., Fuchiwaki M., Horiuchi T., Okano S., Kinutani M., Kinutani K. 2008. Vitrified human day-7 blastocyst transfer: 11 cases. Reprod. Biomed. Online. 17: 689–694. [DOI] [PubMed] [Google Scholar]
- 17.Matorras R., Ruiz J. I., Mendoza R., Ruiz N., Sanjurjo P., Rodriguez-Escudero F. J. 1998. Fatty acid composition of fertilization-failed human oocytes. Hum. Reprod. 13: 2227–2230. [DOI] [PubMed] [Google Scholar]
- 18.Kim J. Y., Kinoshita M., Ohnishi M., Fukui Y. 2001. Lipid and fatty acid analysis of fresh and frozen-thawed immature and in vitro matured bovine oocytes. Reproduction. 122: 131–138. [PubMed] [Google Scholar]
- 19.Haggarty P., Wood M., Ferguson E., Hoad G., Srikantharajah A., Milne E., Hamilton M., Bhattacharya S. 2006. Fatty acid metabolism in human preimplantation embryos. Hum. Reprod. 21: 766–773. [DOI] [PubMed] [Google Scholar]
- 20.Neto A. S. C., Sanches B. V., Binelli M., Seneda M. M., Perri S. H., Garcia J. F. 2005. Improvement in embryo recovery using double uterine flushing. Theriogenology. 63: 1249–1255. [DOI] [PubMed] [Google Scholar]
- 21.Ferreira C. R., Souza G. H. M. F., Riccio M. F., Catharino R. R., Pontes J. H. F., Basso A. C., Junior J. C. E., Perecin F., Eberlin M. N. 2009. Mass spectrometry fingerprinting of media used for in vitro production of bovine embryos. Rapid Commun. Mass Spectrom. 23: 1313–1320. [DOI] [PubMed] [Google Scholar]
- 22.Schiller J., Arnhold J., Benard S., Muller M., Reichl S., Arnold K. 1999. Lipid analysis by matrix-assisted laser desorption and ionization mass spectrometry: a methodological approach. Anal. Biochem. 267: 46–56. [DOI] [PubMed] [Google Scholar]
- 23.Recks M. A., Seaborn G. T. 2008. Variation in fatty acid composition among nine forage species from a southeastern US estuarine and nearshore coastal ecosystem. Fish Physiol. Biochem. 34: 275–287. [DOI] [PubMed] [Google Scholar]
- 24.Troy S., Anderson W. A., Spielman A. 1975. Lipid content of maturing ovaries of Aedes aegypti mosquitoes. Comp. Biochem. Physiol. B. 50: 457–461. [DOI] [PubMed] [Google Scholar]
- 25.Briegel H. 1990. Metabolic relationship between female body size, reserves, and fecundity of Aedes aegypti. J. Insect Physiol. 36: 165–172. [Google Scholar]
- 26.Kawooya J. K., Osir E. O., Law J. H. 1988. Uptake of the major hemolymph lipoprotein and its transformation in the insect egg. J. Biol. Chem. 263: 8740–8747. [PubMed] [Google Scholar]
- 27.McEvoy T. G., Coull G. D., Broadbent P. J., Hutchinson J. S., Speake B. K. 2000. Fatty acid composition of lipids in immature cattle, pig and sheep oocytes with intact zona pellucida. J. Reprod. Fertil. 118: 163–170. [PubMed] [Google Scholar]
- 28.Brugger B., Erben G., Sandhoff R., Wieland F. T., Lehmann W. D. 1997. Quantitative analysis of biological membrane lipids at the low picomole level by nano-electrospray ionization tandem mass spectrometry. Proc. Natl. Acad. Sci. USA. 94: 2339–2344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Petkovic M., Schiller J., Muller M., Benard S., Reichl S., Arnold K., Arnhold J. 2001. Detection of individual phospholipids in lipid mixtures by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry: phosphatidylcholine prevents the detection of further species. Anal. Biochem. 289: 202–216. [DOI] [PubMed] [Google Scholar]
- 30.Fuchs B., Jakop U., Goritz F., Hermes R., Hildebrandt T., Schiller J., Muller K. 2009. MALDI-TOF “fingerprint” phospholipid mass spectra allow the differentiation between ruminantia and feloideae spermatozoa. Theriogenology. 71: 568–575. [DOI] [PubMed] [Google Scholar]
- 31.Hayasaka T., Goto-Inoue N., Sugiura Y., Zaima N., Nakanishi H., Ohishi K., Nakanishi S., Naito T., Taguchi R., Setou M. 2008. Matrix-assisted laser desorption/ionization quadrupole ion trap time-of-flight (MALDI-QIT-TOF)-based imaging mass spectrometry reveals a layered distribution of phospholipid molecular species in the mouse retina. Rapid Commun. Mass Spectrom. 22: 3415–3426. [DOI] [PubMed] [Google Scholar]
- 32.Burnum K. E., Cornett D. S., Puolitaival S. M., Milne S. B., Myers D. S., Tranguch S., Brown H. A., Dey S. K., Caprioli R. M. 2009. Spatial and temporal alterations of phospholipids determined by mass spectrometry during mouse embryo implantation. J. Lipid Res. 50: 2290–2298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saraiva S. A., Cabral E. C., Eberlin M. N., Catharino R. R. 2009. Amazonian vegetable oils and fats: fast typification and quality control via triacylglycerol (TAG) profiles from dry matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) mass spectrometry fingerprinting. J. Agric. Food Chem. 57: 4030–4034. [DOI] [PubMed] [Google Scholar]
- 34.Schiller J., Suss R., Arnhold J., Fuchs B., Lessig J., Muller M., Petkovic M., Spalteholz H., Zschornig O., Arnold K. 2004. Matrix-assisted laser desorption and ionization time-of-flight (MALDI-TOF) mass spectrometry in lipid and phospholipid research. Prog. Lipid Res. 43: 449–488. [DOI] [PubMed] [Google Scholar]
- 35.Waterman R. A., Wall R. J. 1988. Lipid interactions with in vitro development of mammalian zygotes. Gamete Res. 21: 243–254. [DOI] [PubMed] [Google Scholar]
- 36.Wrenzycki C., Herrmann D., Niemann H. 2007. Messenger RNA in oocytes and embryos in relation to embryo viability. Theriogenology. 68(Suppl. 1): S77–S83. [DOI] [PubMed] [Google Scholar]
- 37.Dominguez F., Gadea B., Esteban F. J., Horcajadas J. A., Pellicer A., Simon C. 2008. Comparative protein-profile analysis of implanted versus non-implanted human blastocysts. Hum. Reprod. 23: 1993–2000. [DOI] [PubMed] [Google Scholar]
- 38.Urbanski J. P., Johnson M. T., Craig D. D., Potter D. L., Gardner D. K., Thorsen T. 2008. Noninvasive metabolic profiling using microfluidics for analysis of single preimplantation embryos. Anal. Chem. 80: 6500–6507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alonso T. S., Bonini de Romanelli I. C., Bazan N. G. 1982. Membrane lipids composition and metabolism during early embryonic development. Phospholipid subcellular distribution and 32P labeling. Biochim. Biophys. Acta. 688: 145–151. [DOI] [PubMed] [Google Scholar]
- 40.Bruzzone A., Buschiazzo J., Alonso T. S. 2003. Lipids during Bufo arenarum oogenesis. Zygote. 11: 95–100. [DOI] [PubMed] [Google Scholar]
- 41.Nonogaki T., Noda Y., Goto Y., Kishi J., Mori T. 1994. Developmental blockage of mouse embryos caused by fatty acids. J. Assist. Reprod. Genet. 11: 482–488. [DOI] [PubMed] [Google Scholar]
- 42.Pereira R. M., Baptista M. C., Vasques M. I., Horta A. E., Portugal P. V., Bessa R. J., Silva J. C., Pereira M. S., Marques C. C. 2007. Cryosurvival of bovine blastocysts is enhanced by culture with trans-10 cis-12 conjugated linoleic acid (10t,12c CLA). Anim. Reprod. Sci. 98: 293–301. [DOI] [PubMed] [Google Scholar]
- 43.Otoi T., Yamamoto K., Koyama N., Tachikawa S., Murakami M., Kikkawa Y., Suzuki T. 1997. Cryopreservation of mature bovine oocytes following centrifugation treatment. Cryobiology. 34: 36–41. [DOI] [PubMed] [Google Scholar]
- 44.Abe H., Sata R., Tsujii H., Hoshi H. 2008. Effects of bovine plasma lipoproteins on accumulation of cytoplasmic lipid droplets and mitochondrial morphology in bovine embryos. Reprod. Fertil. Dev. 20: 140. [Google Scholar]
- 45.Leibo S. P. 2008. Cryopreservation of oocytes and embryos: optimization by theoretical versus empirical analysis. Theriogenology. 69: 37–47. [DOI] [PubMed] [Google Scholar]
- 46.Seidel G. E. 2006. Modifying oocytes and embryos to improve their cryopreservation. Theriogenology. 65: 228–235. [DOI] [PubMed] [Google Scholar]
- 47.Abe H., Hoshi H. 2003. Evaluation of bovine embryos produced in high performance serum-free media. J. Reprod. Dev. 49: 193–202. [DOI] [PubMed] [Google Scholar]
- 48.Orsi N. M., Leese H. J. 2004. Amino acid metabolism of preimplantation bovine embryos cultured with bovine serum albumin or polyvinyl alcohol. Theriogenology. 61: 561–572. [DOI] [PubMed] [Google Scholar]
- 49.Shehab-El-Deen M. A., Leroy J. L., Maes D., Van Soom A. 2009. Cryotolerance of bovine blastocysts is affected by oocyte maturation in media containing palmitic or stearic acid. Reprod. Domest. Anim. 44: 140–142. [DOI] [PubMed] [Google Scholar]
- 50.Ifa D. R., Jackson A. U., Paglia G., Cooks R. G. 2009. Forensic applications of ambient ionization mass spectrometry. Anal. Bioanal. Chem. 394: 1995–2008. [DOI] [PubMed] [Google Scholar]
- 51.Estrada R., Yappert M. C. 2004. Alternative approaches for the detection of various phospholipid classes by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. J. Mass Spectrom. 39: 412–422. [DOI] [PubMed] [Google Scholar]
- 52.Lorkiewicz P., Yappert M. C. 2009. 2-(2-Aminoethylamino)-5-nitropyridine as a basic matrix for negative-mode matrix-assisted laser desorption/ionization analysis of phospholipids. J. Mass Spectrom. 44: 137–143. [DOI] [PubMed] [Google Scholar]
- 53.Sun G., Yang K., Zhao Z., Guan S., Han X., Gross R. W. 2008. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometric analysis of cellular glycerophospholipids enabled by multiplexed solvent dependent analyte-matrix interactions. Anal. Chem. 80: 7576–7585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Shroff R., Muck A., Svatos A. 2007. Analysis of low molecular weight acids by negative mode matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 21: 3295–3300. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.