The ER/Golgi protein p23/Tmp21 acts as a C1 domain-docking protein that mediates perinuclear translocation of β-chimaerin. C1 domains from PKC isozymes can also interact with p23/Tmp21. Our study highlights the relevance of C1 domains in protein-protein interactions in addition to their well-established lipid-binding properties.
Abstract
The C1 domains in protein kinase C (PKC) isozymes and other signaling molecules are responsible for binding the lipid second messenger diacylglycerol and phorbol esters, and for mediating translocation to membranes. Previous studies revealed that the C1 domain in α- and β-chimaerins, diacylglycerol-regulated Rac-GAPs, interacts with the endoplasmic reticulum/Golgi protein p23/Tmp21. Here, we found that p23/Tmp21 acts as a C1 domain-docking protein that mediates perinuclear translocation of β2-chimaerin. Glu227 and Leu248 in the β2-chimaerin C1 domain are crucial for binding p23/Tmp21 and perinuclear targeting. Interestingly, isolated C1 domains from individual PKC isozymes differentially interact with p23/Tmp21. For PKCε, it interacts with p23/Tmp21 specifically via its C1b domain; however, this association is lost in response to phorbol esters. These results demonstrate that p23/Tmp21 acts as an anchor that distinctively modulates compartmentalization of C1 domain-containing proteins, and it plays an essential role in β2-chimaerin relocalization. Our study also highlights the relevance of C1 domains in protein–protein interactions in addition to their well-established lipid-binding properties.
INTRODUCTION
C1 domains are 50–51 amino acid-long cysteine-rich motifs originally identified in protein kinase C (PKC) as the binding sites for the lipid second messenger diacylglycerol (DAG) and the phorbol ester tumor promoters. These domains contain the characteristic motif HX12CX2CX13/14CX2CX4HX2CX7C, where H is His, C is Cys, and X is any other amino acid. X-ray crystallography analysis revealed that C1 domains are compact globular structures coordinated through binding of two Zn2+ ions to conserved Cys and His residues. Although this motif is duplicated in tandem (C1a and C1b domains) in classical PKCs (cPKCα, βI, βII, and γ) and novel PKCs (nPKCδ, ε, η, and θ), a single copy is present in phorbol ester/DAG unresponsive atypical PKC isozymes (aPKCζ, ι/λ) (Newton, 1995; Mellor and Parker, 1998). C1 domains capable of binding phorbol esters and DAG are also present in other protein kinases such as protein kinase D isozymes (PKDs) and myotonic dystrophy kinase-related Cdc42-binding kinase, lipid kinases (DAG-kinases), GTPase activating proteins (α- and β-chimaerin Rac-GAPs), guanine nucleotide exchange factors (RasGRP guanine nucleotide exchange factors), and scaffolding proteins (Munc-13s) (Hall et al., 1990; Ahmed et al., 1991; Maruyama and Brenner, 1991; Valverde et al., 1994; Caloca et al., 1997; Betz et al., 1998; Ebinu et al., 1998; Shindo et al., 2003; Choi et al., 2008).
A distinctive feature of most phorbol ester receptors with C1 domains is their ability to redistribute to membranes in response to stimulation of receptors that couple to DAG generation or phorbol esters. A continuous hydrophobic surface generated by the phorbol ester or DAG facilitates the insertion of the C1 domain into lipid bilayers, which in cPKCs and nPKCs is followed by a conformational rearrangement that leads to kinase activation (Zhang et al., 1995; Csukai and Mochly-Rosen, 1999; Ron and Kazanietz, 1999). It is noteworthy that upon activation PKCs relocalize not only to plasma membrane but also to other intracellular compartments, including the nuclear membrane (Murray et al., 1994), perinuclear structures (Hu and Exton, 2004), and mitochondria (Majumder et al., 2000). A high degree of isozyme selectivity for translocation to different intracellular compartments seems to exist (Mochly-Rosen and Gordon, 1998). For example, early studies established that PKCε localizes to the Golgi complex in NIH 3T3 cells via the C1 domain (Lehel et al., 1994, 1995). Moreover, a recent study in neuroblastoma SK-N-BE(2)C cells revealed that mutation of specific residues in the PKCε C1b domain impairs its perinuclear localization without affecting its translocation to the plasma membrane in response to the C1 domain ligand phorbol 12-myristate 13-acetate (PMA) (Schultz et al., 2004).
We have established previously that α- and β-chimaerins, C1 domain-containing proteins with Rac-GAP activity, translocate both to the plasma membrane and the perinuclear region in response to PMA or DAG analogues (Caloca et al., 2001; Wang and Kazanietz, 2002; Wang et al., 2006). Deletion of the C1 domain or mutations of key amino acids implicated in phorbol ester binding impairs both the peripheral and perinuclear translocation of chimaerins, thus arguing that the C1 domain is essential for chimaerin intracellular targeting (Caloca et al., 1997, 1999, 2001; Wang and Kazanietz, 2002; Wang et al., 2006). Other “nonkinase” phorbol ester/DAG receptors such as RasGRP1/3 and Munc-13 also translocate to the plasma membrane and Golgi in response to PMA via their C1 domains (Song et al., 1999; Caloca et al., 2003). The molecular basis for the translocation of proteins with C1 domains is only partially understood. Speculation has been that protein–protein interactions are key factors for determining their selective intracellular relocalization, and studies have indeed identified numerous PKC interactors that dictate compartmentalization through binding to unique motifs present in individual isozymes, such as the receptors for activated C-kinases (Csukai et al., 1997). Interactions may require a conformational rearrangement that exposes the protein binding domain (Mochly-Rosen et al., 1991). Although these mechanisms have been extensively studied for PKC isozymes, the involvement of protein partners in targeting chimaerin Rac-GAPs has not been established yet.
In a yeast two-hybrid screening, we identified the Golgi/endoplasmic reticulum (ER) protein p23/Tmp21 (p24δ) as a chimaerin-interacting protein (Wang and Kazanietz, 2002). p23/Tmp21, a type I transmembrane protein, belongs to the p24 protein family that has been widely implicated in trafficking from the intermediate compartment to the Golgi (Blum et al., 1996, 1999; Sohn et al., 1996). A deletional analysis in α- and β-chimaerins revealed the C1 domain as the p23/Tmp21-interacting motif (Wang and Kazanietz, 2002). Although biochemical and imaging studies strongly support the formation of a chimaerin-p23/Tmp21 complex in cells (Wang and Kazanietz, 2002), it is yet unknown whether p23/Tmp21 actually serves as a perinuclear anchoring protein for chimaerin Rac-GAPs. It also remains to be determined whether p23/Tmp21 associates with other proteins containing C1 domains to drive their perinuclear translocation. This is relevant because proteins with C1 domains, such as PKCε, have been shown to localize at the Golgi.
In this study, we demonstrate that p23/Tmp21 is required for the perinuclear translocation of β2-chimaerin. This Rac-GAP indeed fails to redistribute to the perinuclear compartment in p23/Tmp21-deficient cells. We have also identified key residues in the β2-chimaerin C1 domain that specifically mediate its association with p23/Tmp21 and translocation to the perinuclear compartment. Interestingly, p23/Tmp21 also interacts with other C1 domains in isolation, including C1 domains from PKC isozymes. Our results support the notion that C1 domains act not only as lipid binding motifs but can also mediate protein–protein interactions that determine selective intracellular compartmentalization.
MATERIALS AND METHODS
Materials
PMA and GF109203X were purchased from LC Laboratories (Woburn, MA). Cell culture reagents were obtained from Invitrogen (Carlsbad, CA). Reagents for the expression and purification of recombinant glutathione transferase (GST)-fusion proteins and glutathione-Sepharose 4B beads were purchased from GE Healthcare (Little Chalfont, Buckinghamshire, United Kingdom). COS-1 and HeLa cells were obtained from the American Type Culture Collection (Manassas, VA). Yeast strain EGY48, and yeast culture reagents and media were obtained from Clontech (Mountain View, CA). O-Nitrophenyl-β-d-galactopyranoside (ONPG) and MISSION Lentiviral Transduction short hairpin RNA (shRNA) particles were obtained from Sigma-Aldrich (St. Louis, MO). The following primary antibodies were used: anti-pLexA (Santa Cruz Biotechnology, Santa Cruz, CA), anti-β-actin (Sigma-Aldrich), anti-V5 (Invitrogen), anti-p23/Tmp21 (ProSci, Poway, CA), anti-GST, anti-hemagglutinin (HA), anti-green fluorescent protein (GFP) (Covance, Emeryville, CA), anti-Rac1 (Millipore, Billerica, MA), anti-PKCε (Cell Signaling Technology, Danvers, MA), and anti-GS28 (BD Biosciences, San Jose, CA).
Plasmid Construction
Generation of pB42AD-p23/Tmp21 (amino acid [aa] 108-219), pEBG-p23/Tmp21 (aa 1-219), and pEBG-p23/Tmp21 (aa 108-219) were described previously (Wang and Kazanietz, 2002). Truncated p23/Tmp21 mutants were generated by polymerase chain reaction (PCR) and subcloned into EcoRI-XhoI sites in pB42AD-HA vector or BamHI-SpeI sites in pEBG vector. C1 regions from PKCε, PKCζ, and β2-chimaerin were isolated by PCR and fragments subcloned into EcoRI-BamHI sites in pLexA to generate pLexA-C1εa-b (aa 134-309), pLexA-C1ζ (aa 95-197), and pLexA-C1β2-ch (aa 179-281), respectively. C1 domain fragments were also subcloned into EcoRI-BamHI sites in pEGFP-C1 to generate pEGFP-C1εa-b, pEGFP-C1ζ, and pEGFP-C1β2-ch. Individual C1a and C1b domains from PKCε (comprising aa 168-222 and aa 241-294, respectively) were subcloned into EcoRI-BamHI sites in pEGFP-C1 and pLexA to generate pEGFP-C1εa, pEGFP-C1εb, pLexA-C1εa, and pLexA-C1εb. The primers used for PCR cloning are listed in Supplemental Table S1. All constructs were confirmed by sequencing.
Site-directed Mutagenesis
For PCR-based mutagenesis, we used the QuikChange XL site-directed mutagenesis kit (Strategene, La Jolla, CA), using GFP-β2-chimaerin, GFP-C1β2-ch, or pLexA-C1β2-ch as templates. For the mutant E227G-β2-chimaerin, we used the following primers (mutated nucleotides are underlined): forward, 5′-CGAGGCCCACACTGGTGTGGATATTGTGCCAATTTCATG; and reverse, 5′-CATGAAATTGGCACAATATCCACACCAGTGTGGGCCTCG. For the mutant L248A-β2-chimaerin, we used forward, 5′-GTCCGGTGCTCAGACTGTGGAGCTAACGTACACAAACAG and reverse, 5′-CTGTTTGTGTACGTTAGCTCCACAGTCTGAGCACCGGAC. For PKCε (E257G), we used forward, 5′-GGTCCCCACGTTCTGTGGCCACTGTGGGTCCCTGC and reverse, 5′-GCAGGGACCCACAGTGGCCACAGAACGTGGGGACC. For PKCε (M278G), we used: forward, 5′-GCAGTGTAAAGTCTGCAAAGGGAATGTTCACCGTCGATGTG and reverse, 5′-CACATCGACGGTGAACATTCCCTTTGCAGACTTTACACTGC.
Cell Culture, Transfections, and Adenoviral Infections
HeLa and COS-1 cells were cultured in DMEM (Invitrogen) supplemented with 10% fetal bovine serum (HyClone Laboratories, Logan, UT), 100 U/ml penicillin, and 100 μg/ml streptomycin in a humidified 5% CO2 atmosphere at 37°C. Cells in 6-well plates at ∼50% confluence were transfected with different mammalian expression vectors (1 μg) by using Lipofectamine 2000 (Invitrogen) following the manufacturer's protocol. Adenoviral infections were carried out essentially as described previously (Wang et al., 2006).
Generation of p23/Tmp21-depleted Cell Lines by Using shRNA Lentiviruses
HeLa cells were infected with 3 different MISSION Lentiviral Transduction particles encoding p23/Tmp21 shRNAs. p23/Tmp21 shRNA target sequences were as follows: #1, CCGGGAGATTCACAAGGACCTGCTACTCGAGTAGCAGGTCCTTGTGAATCTCTTTTT; #2, CCGGCCAACTCGTGATCCTAGACATCTCGAGATGTCTAGGATCACGAGTTGGTTTTT; and #3, CCGGCGCTTCTTCAAGGCCAAGAAACTCGAGTTTCTTGGCCTTGAAGAAGCGTTTTT. Stable cell lines (pools) were generated by selection with puromycin (1 μg/ml).
Yeast Two-hybrid Assay
pLexA expression vectors, which contain a His marker, were cotransformed into the yeast strain EGY48 together with pB42AD-HA–tagged expression vectors, which have a Trp marker, and the p8OP-LacZ reporter vector (Ura marker). Transformants were plated on yeast dropout medium lacking Trp, Ura, and His, thereby selecting for the plasmids encoding proteins capable of two-hybrid interaction as evidenced by transactivation of the LacZ reporter gene (Wang and Kazanietz, 2002).
For β-galactosidase liquid assays, yeast was cultured in galactose/raffinose/-His/-Ura/-Trp liquid SD selection medium until the cells were in mid-log phase (OD600 = 0.5–0.8). Cells were pelleted at 14,000 × g for 30 s and resuspended in 300 μl of a buffer, pH 7.0, containing 60 mM Na2HPO4, 40 mM NaH2PO4, · H2O, 10 mM KCl, 1 mM MgSO4, and 0.27% (vol/vol) β-mercaptoethanol. One hundred microliters of the cell suspension were then frozen and thawed three times in liquid nitrogen and a 37°C water bath, respectively, and an additional 700 μl of resuspension buffer was added. ONPG was then added (final concentration, 670 μg/ml), and the reaction was initiated by addition of Na2CO3 (final concentration, 300 μg/ml). β-Galactosidase activity was determined as described previously (Wang and Kazanietz, 2002). One unit of β-galactosidase activity is defined as the amount that hydrolyzes 1 μmol of ONPG to o-nitrophenol and d-galactose per minute and per cell.
Immunostaining and Confocal Microscopy
Plasmids encoding for full-length PKCε or β2-chimaerin together with p23/Tmp21 in V5 epitope-tagged pcDNA3.1 were cotransfected into HeLa cells by using Lipofectamine 2000. After 24 h, cells were treated with PMA for 30 min, washed twice with phosphate-buffered saline (PBS), and fixed with 4% paraformaldehyde for 20 min at room temperature. After washing once with PBS containing 0.5% SDS and 5% β-mercaptoethanol (at 37°C for 30 min), and twice with PBS alone, cells were incubated with an anti-V5 monoclonal antibody (1:500). A donkey anti-mouse antibody conjugated with Cy3 was used as secondary antibody (1:1000). Slides were mounted using Fluoromount-G (Southern Biotechnology Associates, Birmingham, AL) and viewed with an LSM 710 laser scanning microscope (Carl Zeiss, Thornwood, NY). The confocal images were processed with LSM Image Browser. All the images shown are individual middle sections of projected Z-series mounting. For quantification, images (RGB) from red and green channels were converted into 8-bits images in ImageJ (National Institutes of Health, Bethesda, MD), and the Pearson's r (Rr) was calculated using this software.
Time-Lapse Microscopy
HeLa cells were seeded into glass-bottomed culture dishes (MatTek, Ashland, MA) for 20 h. pEGFP-PKCε or pEGFP-β2-chimaerin plasmids (wild type or mutants) were transfected using Lipofectamine 2000 according to the manufacturer's protocols. Cells were cultured for 20 h in phenol red-free RPMI 1640 medium containing 10% fetal bovine serum and 5 mM HEPES. Cells were monitored under a fluorescence microscope (Eclipse TE2000U;) Nikon, Tokyo, Japan) at a 488 nm excitation wavelength with a 515 nm long pass barrier filter at 25°C.
Coprecipitation by Using Glutathione-Sepharose 4B Beads
COS-1 cells at ∼50% confluence were cotransfected with pEBG-p23/Tmp21 (full-length) or empty vector (pEBG). After 24 h, cells were washed twice with cold PBS and then lysed for 10 min at 4°C in 400 μl of a lysis buffer containing 50 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1% Nonidet P-40, 0.5% deoxycholate, 0.1% SDS, and protease inhibitor cocktail (Sigma-Aldrich). Ten microliters of glutathione-Sepharose 4B beads were added to the lysate and incubated for 1 h at 4°C. The beads were extensively washed in lysis buffer and boiled. Samples were resolved in a 12% SDS-polyacrylamide gel and transferred to a polyvinylidene difluoride membrane (Millipore) for Western blot analysis.
In Vitro Protein-Protein Binding Assay
Cell lysates were prepared from COS-1 cells expressing pEBG, pEBG-p23/Tmp21 (aa 1-219), pEBG-p23/Tmp21 (aa 1-208), pEBG-p23/Tmp21 (aa 108-219), or pEBG-p23/Tmp21 (aa 1-185). A fixed amount of GST or GST-fused p23/Tmp21 (wild type or truncated mutants) was incubated with lysates of COS-1 cells expressing HA-β2-chimaerin at 4°C for 2 h and then incubated with glutathione-Sepharose 4B beads for 1 h. After extensive washing, the beads were boiled in loading buffer and subjected to Western blot with an anti-HA antibody.
Determination of Rac-GTP Levels
Experiments were carried out as described previously (Wang and Kazanietz, 2002). In brief, cells were lysed in a buffer containing 8 μg of GST-PBD (p21 binding domain), 20 mM Tris-HCl, pH 7.5, 1 mM dithiothreitol, 5 mM MgCl2, 150 mM NaCl, 0.5% Nonidet P-40, 5 mM β-glycerophosphate, and protease inhibitors cocktail (Sigma-Aldrich). Lysates were centrifuged at 14,000 × g (at 4°C for 10 min) and then incubated with glutathione-Sepharose 4B beads (at 4°C for 1 h). After extensive washing, the beads were boiled in loading buffer and subject to Western blot analysis using an anti-Rac1 antibody.
Western Blot
Nonspecific binding in membranes was blocked by incubation with 5% nonfat milk or 5% bovine serum albumin for 2 h. Membranes were then incubated with primary antibodies for 2 h at room temperature, followed by incubation with peroxidase-conjugated goat anti-mouse or anti-rabbit immunoglobulin G (1:3000 or 1:10,000) for 1 h at room temperature. Immunoreactivity was visualized with an LAS3000 image reader (Fujifilm, Tokyo, Japan) by using an enhanced chemoluminescence detection kit (GE Healthcare).
RESULTS
The C1 Domain of β2-Chimaerin and C1b Domain of PKCε Interact with p23/Tmp21
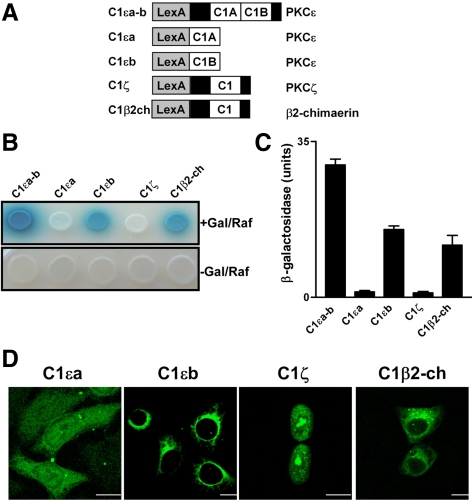
Previous studies have identified p23/Tmp21, a type I transmembrane protein highly enriched in the ER and Golgi, as an α- and β-chimaerin–interacting protein. A deletional analysis established that interaction with p23/Tmp21 occurs through a region that encompasses the chimaerin C1 domain (Wang and Kazanietz, 2002). Because PKCε was shown to localize at the perinuclear region via its C1 domain region (Schultz et al., 2004), we speculated that C1a and/or C1b domains in PKCε may interact with p23/Tmp21. To address this issue, we first examined whether the C1 region of PKCε, which include both C1a and C1b domains (C1εa-b), interacts with p23/Tmp21 in a yeast two-hybrid system. A pLexA construct encoding C1εa-b was generated (Figure 1A) and cotransformed with pB42AD-p23/Tmp21 (HA-tagged, aa 108-208, a fragment that interacts with chimaerins) into EGY48 yeast containing p8OP-LacZ vector (Wang and Kazanietz, 2002). The triple vectors cotransformants were selected by plating the yeast into SD/-Ura/-His/-Trp dropout plates. pLexA-fused proteins were expressed in both galactose/raffinose (+Gal/Raf) plates and glucose plates (−Gal/Raf), whereas HA-tagged pB42AD-p23/Tmp21 (aa 108-208) was only expressed in galactose/raffinose induction plates (data not shown). Figure 1B shows that both the β2-chimaerin C1 domain (C1β2-ch) and C1εa-b strongly interact with p23/Tmp21, as revealed by the induction of the LacZ reporter (blue color). These interactions were also detected using a liquid β-galactosidase assay (Figure 1C).
Figure 1.
Differential interaction of C1 domains with p23/Tmp21. (A) Schematic representation of C1εa-b, C1εa, C1εb, C1ζ, or C1β2-ch domain fused to pLexA. (B) EGY48 yeast (containing 8op-LacZ vector) was cotransformed with pLexA encoding C1εa-b, C1εa, C1εb, C1ζ, or C1β2-ch domain, and pB42AD-HA-tagged p23/Tmp21 (aa 108-208). Assay of β-galactosidase activity on induction (top) or no-induction (bottom) plates was carried out 72 h after transformation. Gal/Raf, galactosidase/raffinose. (C) Assay of β-galactosidase activity in liquid cultures using ONPG as a substrate. Results were expressed as mean ± SD (n = 3). (D) GFP-PKCεC1b domain localizes in the perinuclear region. HeLa cells were transfected with pEGFP-C1εa, C1εb, C1ζ, or C1β2-ch. Forty-eight hours later, cells were fixed and localization examined by confocal microscopy. Bar, 10 μm. All experiments have been performed at least three times with similar results.
Studies have determined that C1εb but not C1εa is essential for Golgi localization in neuroblastoma SK-N-BE(2)C cells (Schultz et al., 2004), suggesting a differential involvement of each domain in perinuclear targeting. We therefore generated pLexA-fused C1εa and C1εb constructs (Figure 1A), cotransformed each of them with pB42AD-p23/Tmp21 into the EGY48 (p8OP-LacZ) yeast, and expressed both proteins in +Gal/Raf plates. Interestingly, a strong association was observed with C1εb, whereas C1εa failed to interact with p23/Tmp21 (Figure 1, B and C). To further establish whether C1 domain specificity exists, we generated pLexA constructs encoding C1 domains of aPKCζ (C1ζ), PKCα (C1α), PKCδ (C1δ), and RasGRP1, although C1ζ failed to interact with p23/Tmp21 in the yeast two-hybrid assay (Figure 1B). In contrast, association was detected for C1α and C1δ, and the C1 domain of the Ras/Rap1 exchange factor RasGRP1 weakly interacted with p23/Tmp21 (Supplemental Figure S1, A–C).
Next, we determined the intracellular localization of C1β2-ch, C1εa, and C1εb domains in mammalian cells. C1 domains were expressed as GFP-fusion proteins in HeLa cells, and localization was examined by confocal microscopy (Figure 1D). Remarkably, individual C1 domains exhibited distinct intracellular localization, although GFP-C1εa distributed throughout the cell and had no obvious perinuclear localization, as reported previously in SK-N-BE(2)C cells (Schultz et al., 2004), GFP-C1εb and GFP-C1β2-ch showed a characteristic perinuclear localization. GFP-C1ζ displayed a strong nuclear localization when expressed in HeLa cells but no obvious perinuclear staining. Thus, C1 domains, when expressed in isolation, have unique localization properties.
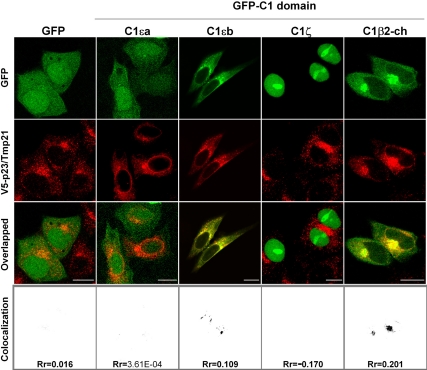
Next, we examined whether different C1 domains colocalize with p23/Tmp21. HeLa cells were cotransfected with pcDNA3-V5-p23/Tmp21 together with plasmids encoding different C1 domains fused to GFP, and colocalization determined by confocal microscopy. As shown in Figure 2, both GFP-C1εb and GFP-C1β2-ch colocalized with p23/Tmp21, as judged by the yellow color observed in the overlapped images. Quantification using ImageJ and analysis using a Pearson's r (Rr) confirmed these results. In contrast, neither GFP-C1εa nor GFP-C1ζ showed any obvious colocalization with p23/Tmp21. GFP alone also failed to colocalize with p23/Tmp21 in HeLa cells. We also found that full-length PKCα, PKCδ, and RasGRP1 have some degree of colocalization with Tmp21/p23 (Supplemental Figure S1D). These results are in agreement with those observed in the yeast two-hybrid analysis and reveal unique patterns of intracellular localization and protein interactions for discrete C1 domains.
Figure 2.
Colocalization of GFP-fused PKCε C1b and β2-chimaerin C1 domains with p23/Tmp21. HeLa cells were cotransfected with pEGFP-fused C1εa, C1εb, C1ζ, or C1β2-ch (or empty vector) and V5-tagged full-length pcDNA3-p23/Tmp21. Forty-eight hours later, cells were fixed and stained with an anti-V5 antibody, and localization was examined by confocal microscopy. Colocalization images and Pearson's r (Rr) were generated by ImageJ. Similar results were observed at least in three independent experiments. Bar, 10 μm.
Colocalization of Full-Length PKCε and p23/Tmp21
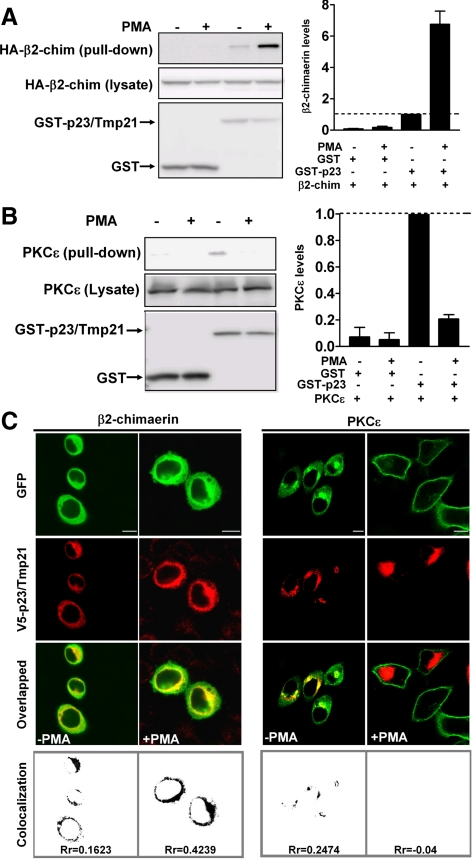
Experiments carried out with isolated C1 domains established proof-of-principle for differential targeting, but may or may not reflect what occurs with intact proteins in cells. Therefore, we next decided to investigate the association of full-length β2-chimaerin and PKCε with p23/Tmp21 in mammalian cells. COS-1 cells were transfected with pEBG control vector (which encodes GST alone) (Wang and Kazanietz, 2002) or pEBG-p23/Tmp21. After 24 h, cells were infected with adenoviruses for either full-length PKCε or full-length β2-chimaerin, and 24 h later subject to GST pull-down by using glutathione Sepharose 4B beads. As shown in Figure 3A (left), and in agreement with our previous study (Wang and Kazanietz, 2002), β2-chimaerin was detected in complex with GST-p23/Tmp21 but not with GST. The β2-chimaerin was shown to translocate to the perinuclear compartment in response to C1 domain ligands such as PMA. This effect was lost when key residues in the C1 domain are mutated but was not affected by the pan-PKC inhibitor GF 109203X, suggesting that the PMA effect was not mediated by PKCs (Caloca et al., 2001; Wang and Kazanietz, 2002). Figure 3A (left) also shows that the association of β2-chimaerin with GST-p23/Tmp21 was markedly enhanced by PMA. A densitometric analysis revealed that PMA caused an approximate sevenfold increase in the association of β2-chimaerin with p23/Tmp21 (Figure 3A, right). The association between β2-chimaerin and p23/Tmp21 can also be enhanced in cells growing in serum and in response to EGF treatment, although in this last case to a lower extent than that observed with PMA (Supplemental Figure S2). Interestingly, PKCε can be readily detected in GST-p23/Tmp21 precipitates, whereas it cannot be pulled down by GST alone, an indication that PKCε and p23/Tmp21 exist as a complex. However, in contrast to β2-chimaerin, PKCε dissociated from p23/Tmp21 when cells were treated with PMA (Figure 3B).
Figure 3.
Differential interaction of PKCε and β2-chimaerin with p23/Tmp21. (A and B) COS-1 cells were transfected with either pEBG (empty vector) or pEBG-p23/Tmp21. Twenty-four hours later, cells were infected with either HA-β2-chimaerin adenovirus (multiplicity of infection [MOI], 10 plaque-forming units [pfu]/cell) (A) or PKCε adenovirus (MOI = 3 pfu/cell) (B). After 24 h, cells were treated with PMA (1 μM) or vehicle for 30 min in the presence of the PKC inhibitor GF109203X (5 μM) and lysed. GST or GST-p23/Tmp21 proteins were precipitated with glutathione-Sepharose 4B beads, and associated HA-β2-chimaerin was detected by Western blot using an anti-HA antibody. Left, representative experiments. Right, densitometric analysis of three individual experiments, expressed as fold change relative to GST-p23/Tmp21 in the absence (A) or presence (B) of PMA. (C) HeLa cells were cotransfected with either pEGFP-PKCε or pEGFP-β2-chimaerin and pcDNA3.1/V5-p23/Tmp21 (full length). Forty-eight hours later, cells were treated with PMA (1 μM) or vehicle for 30 min, fixed, and stained with an anti-V5 antibody, and localization examined by confocal microscopy. Top, green fluorescence from GFP-PKCε or GFP-β2-chimaerin; middle, red fluorescence from pcDNA3.1/V5-p23/Tmp21; bottom, overlapped images. Colocalization images and Pearson's r (Rr) were generated by ImageJ. Similar results were obtained in three additional experiments. Bar, 10 μm.
The distinct association pattern of PKCε and β2-chimaerin with p23/Tmp21 prompted us to examine whether they differentially relocalize in response to phorbol ester treatment. HeLa cells expressing GFP-PKCε or GFP-β2-chimaerin (full-length) were treated with either PMA or vehicle, and localization examined by confocal microscopy. Like β2-chimaerin, PKCε displayed some degree of colocalization with p23/Tmp21 in the perinuclear region of vehicle-treated cells (Rr = 0.25). However, a remarkably distinct pattern of translocation for each protein was observed in response to PMA: whereas β2-chimaerin redistributed primarily to the perinuclear compartment and colocalized with p23/Tmp21 in response to PMA (Rr increases from 0.16 to 0.42) (plasma membrane localization also can be detected; see Caloca et al., 2001; Wang and Kazanietz, 2002), PKCε fully translocated to the cell periphery. Coincidentally, no perinuclear PKCε or colocalization with p23/Tmp21 could be observed in PMA-treated cells (Rr = −0.04) (Figure 3C). These results argue for a differential interaction of PKCε and β2-chimaerin with p23/Tmp21 in response to stimuli. Moreover, they also suggest that domain(s) in PKCε other than the C1b domain must have prominent targeting roles.
p23/Tmp21 Depletion Impairs Perinuclear Translocation of β2-Chimaerin
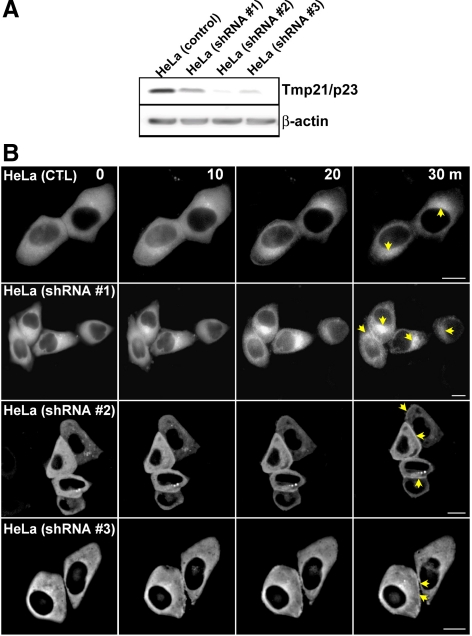
To determine the requirement of p23/Tmp21 for β2-chimaerin translocation, we established p23/Tmp21-depleted HeLa cell lines by using lentiviral shRNAs. Three different lentiviruses were used, and stable cell lines were generated after selection with puromycin. Figure 4A shows that a significant depletion (>80%) was observed with shRNA p23/Tmp21 lentiviruses #2 and #3, whereas shRNA lentivirus #1 was less effective. GFP-β2-chimaerin was expressed in the different stable cell lines, and its localization in response to PMA was monitored using real-time microscopy. Figure 4B revealed that GFP-β2-chimaerin efficiently translocated to the perinuclear region (marked with an arrow at 30-min time point) in control cells, as expected. Likewise, perinuclear translocation was readily detected in cells infected with p23/Tmp21 shRNA lentivirus #1, in which depletion was minimal. In contrast, perinuclear translocation of GFP-β2-chimaerin was essentially lost in those cell lines in which p23/Tmp21 has been markedly depleted. GFP-β2-chimaerin colocalized with the cis-Golgi marker GS-28 in the resting state, and PMA treatment enhanced colocalization (Supplemental Figure S3). Translocation of PKCε to the cell periphery by PMA was not affected by shRNA p23/Tmp21 depletion (data not shown). These results suggest that p23/Tmp21 is indispensable for the translocation of β2-chimaerin to the perinuclear region.
Figure 4.
p23/Tmp21 RNAi depletion impairs perinuclear β2-chimaerin translocation. (A) Expression of p23/Tmp21 in HeLa cells stably expressing different p23/Tmp21 shRNAs (shRNA#1, shRNA#2, and shRNA#3) or control cells. (B) Cells were transfected with pEGFP-β2-chimaerin and 48 h later treated with PMA (3 μM) in the presence of GF109203X (5 μM). Time-lapse images of β2-chimaerin translocation in living cells were captured at different times after PMA treatment. Perinuclear and periphery translocation were marked with arrows. Similar results were observed in three individual experiments. Bar, 10 μm.
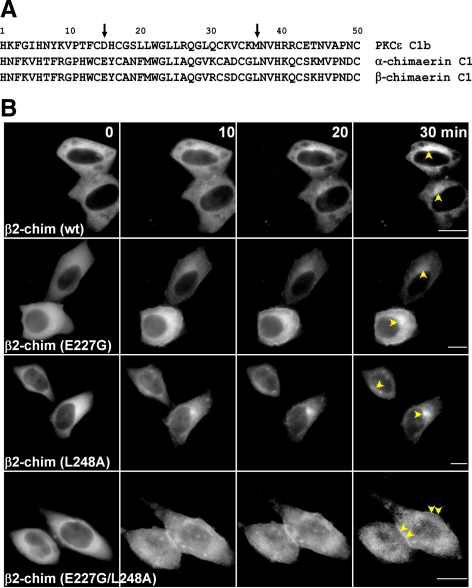
Glu227 and Leu248 in the β2-Chimaerin C1 Domain Are Critical for Translocation to the Perinuclear Compartment
In a recent study, Schultz et al. (2004) showed that mutation of Asp257 and Met278 in the PKCε C1b domain (amino acids 15 and 36 in the motif, respectively) abolished the perinuclear localization of PKCε or its PKCε C1b domain in neuroblastoma cells. We observed that when we mutated both Asp257 and Met278 to Gly in PKCε, the resulting mutant localized to the plasma membrane even in the absence of PMA stimulation and does not colocalize with p23/Tmp21 (Supplemental Fig. S4). Alignment of C1εb with C1 domains from both α- and β-chimaerins revealed that an acidic amino acid in position 15 and a lipophilic amino acid in position 36 of the motif were conserved (Figure 5A). We speculated that Glu227 and Leu248 in β2-chimaerin might be implicated in perinuclear translocation. Mutants in those positions in β2-chimaerin (E227G, L248A, and the double mutant E227G/L248A) were generated, expressed in HeLa cells as GFP-fused proteins, and their localization in real-time in response to PMA analyzed by microscopy. Single mutants E227G- and L248A-β2-chimaerin showed slightly higher translocation to the plasma membrane compared with wild-type β2-chimaerin. However, perinuclear translocation after PMA treatment can still be observed. Conversely, no perinuclear translocation could be observed for the double mutant E227G/L248A-β2-chimaerin, whereas plasma membrane fluorescence was readily detected (Figure 5B; videos presented in Supplemental Figure S5). Therefore, residues in the β2-chimaerin C1 domain homologous to those in C1εb play a significant role in targeting β2-chimaerin to the perinuclear compartment.
Figure 5.
Glu227 and Leu248 in the β2-chimaerin C1 domain are required for perinuclear translocation. (A) Alignment of PKCε C1b and α- and β-chimaerin C1 domains. Positions 15 and 36 are indicated with an arrow. (B) HeLa cells were transfected with pEGFP-β2-chimaerin (wt), pEGFP-β2-chimaerin (E227G), pEGFP-β2-chimaerin (L248A), or pEGFP-β2-chimaerin (E227G/L248A). Forty-eight hours later, cells were treated with PMA (3 μM) in the presence of GF109203X (5 μM). Time-lapse images of translocation of GFP-β2-chimaerin or its mutants in living cells were captured by fluorescence microscopy at different times after PMA treatment. Perinuclear and periphery translocation were marked with arrows. Similar results were observed in five independent experiments. Bar, 10 μm.
The Double Mutant E227G/L248A-β2-Chimaerin Fails to Interact with p23/Tmp21
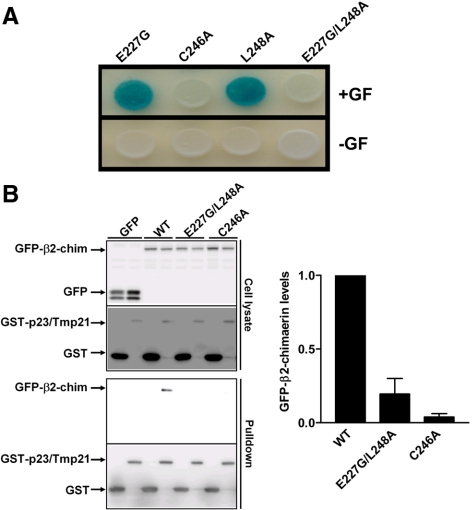
We speculated that the lack of perinuclear translocation of the double mutant E227G/L248A-β2-chimaerin by PMA was due to its inability to bind p23/Tmp21. We tested this hypothesis using a yeast two-hybrid assay. pLexA-fused constructs for E227G-, L248A-, and E227G/L248A-β2-chimaerin C1 domain mutants were generated (E227G-C1β2-ch, L248A-C1β2-ch, and E227G/L248A-C1β2-ch respectively). The pLexA plasmids were cotransformed with pB42AD-p23/Tmp21 into EGY48 (p8OP-LacZ) yeast. As shown in Figure 6A, single mutants E227G-C1β2-ch and L248A-C1β2-ch retained their ability to bind p23/Tmp21, as revealed by the induction of the LacZ reporter gene (blue). In contrast, the double mutant E227G/L248-C1β2-ch was unable to interact with p23/Tmp21. A mutant with a Cys essential for C1 domain folding mutated to Ala (C246A-C1β2-ch) also failed to interact with p23/Tmp21, consistent with the lack of translocation of C246A-β2-chimaerin in PMA-treated cells (Caloca et al., 2001; Wang and Kazanietz, 2002).
Figure 6.
Glu227 and Leu248 residues in the β2-chimaerin C1 domain are required for the interaction with p23/Tmp21. (A) EGY48 yeast (containing 8op-LacZ vector) was cotransformed with pLexA-β2-chim-C1 (E227G), pLexA-β2-chim-C1 (L248A), pLexA-β2-chim-C1 (E227G/L248A), or pLexA-β2-chim-C1 (C246A), and pB42AD-HA-tagged p23/Tmp21 (aa 108-208). Assay of β-galactosidase activity on induction (top) or no-induction (bottom) plates was carried out 72 h after transformation. Gal/Raf, galactosidase/raffinose. (B) COS-1 cells were cotransfected with either pEBG (empty vector) or pEBG-p23/Tmp21 and GFP vector, GFP-β2-chimaerin (wt), GFP-β2-chimaerin (E227G/L248A) or GFP-β2-chimaerin (C246A). After 24 h, cells were lysed. GST or GST-p23/Tmp21 proteins were precipitated with glutathione-Sepharose 4B beads and associated GFP-fused proteins detected by Western blot using an anti-GFP antibody. Left, representative experiments. Right, densitometric analysis of three individual experiments, expressed as fold change relative to GST-p23/Tmp21 bound β2-chimaerin (wt).
In the next experiments, we determined the association of GFP-β2-chimaerin mutants with p23/Tmp21 in COS-1 cells by using a coprecipitation approach. Cells were cotransfected with pEBG (empty vector) or pEBG-p23/Tmp21, together with GFP-β2-chimaerin (wt), GFP-β2-chimaerin (E227G/L248A) or GFP-β2-chimaerin (C246A). After 36 h, cells were subject to a pull-down assay using glutathione-Sepharose 4B beads. As shown in Figure 6B (left), whereas β2-chimaerin (wt) was readily detected in complex with GST-p23/Tmp21, no coprecipitation was observed for β2-chimaerin (E227G/L248A) or β2-chimaerin (C246A). A quantitative analysis of multiple experiments is presented in Figure 6B (right).
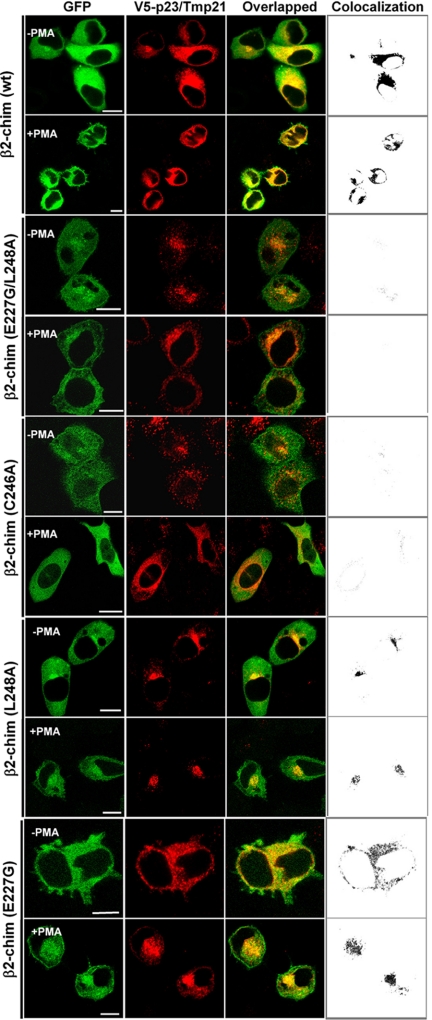
Furthermore, imaging studies using confocal microscopy showed that although perinuclear translocation and colocalization with p23/Tmp21 in response to PMA could still be observed for full-length GFP-β2-chimaerin in which Glu227 and Leu248 have been mutated individually, the double mutant GFP-E227G/L248A-β2-chimaerin was unable to redistribute to the perinuclear compartment or to colocalize with p23/Tmp21 in cells (Figure 7). As in Figure 5B, significant plasma membrane translocation could be detected with the double mutant. In agreement with previous studies (Caloca et al., 2001; Wang and Kazanietz, 2002), the PMA-unresponsive mutant C246A-β2-chimaerin was unable to translocate in response to the phorbol ester.
Figure 7.
Colocalization studies of β2-chimaerin mutants and p23/Tmp-21. HeLa cells were cotransfected with pEGFP-β2-chim (wt), pEGFP-β2-chim (C246A), pEGFP-β2-chim (E227G), pEGFP-β2-chim (L248A), or pEGFP-β2-chim (E227G/L248A) and V5-tagged pcDNA-p23/Tmp21. Forty-eight hours later, cells were treated with PMA (3 μM) or vehicle for 30 min in the presence of GF 109203X (5 μM). Cells were then washed and visualized by confocal microscopy. Left, green fluorescence from GFP-β2-chimaerin (wild-type or mutants); middle, red fluorescence from pcDNA3.1/V5-p23/Tmp21; right, overlapped images. Far right, colocalization images generated by ImageJ. Similar results were obtained in three different experiments. Bar, 10 μm.
Identification of the Chimaerin Binding Domain in p23/Tmp21
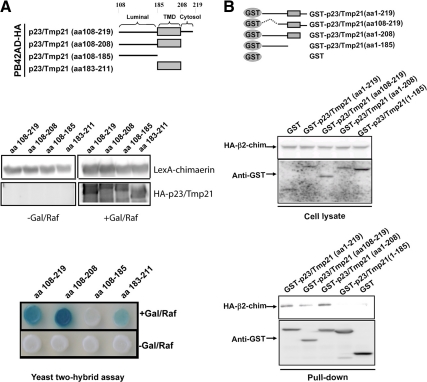
In the next set of experiments, we mapped the domain in p23/Tmp21 that interacts with the C1 domain in chimaerins by using a yeast two-hybrid approach. p23/Tmp21 comprises a luminal domain and a short cytoplasmic tail linked by a transmembrane domain. Deletions for each domain were generated, and the corresponding deleted mutants subcloned into pB42AD-HA as shown in Figure 8A (top). EGY48 (p8OP-LacZ) yeast was cotransformed with either mutant together with pLexA-α-chimaerin (aa 1-147) (it has similar interacting properties as the β-isoforms) and proteins expressed in Gal/Raf plates (Figure 8A, middle). A strong interaction could be detected even when the cytosolic tail (Δ208-219) was deleted. In contrast, deletion of amino acids 185-208, which correspond to the transmembrane domain, impaired the association. Expression of the transmembrane domain alone was sufficient to observe interaction (Figure 8A, bottom). A β-galactosidase liquid assay also revealed that the transmembrane domain alone interacts with chimaerin (data not shown). These results were confirmed using a coprecipitation approach. Deletion mutants were generated and subcloned into pEBG vector (Figure 8B, top). GST or GST-fused p23/Tmp21 mutants were mixed with COS-1 cell lysates expressing HA-β2-chimaerin (Figure 8B, middle). Although association of GST-p23/Tmp21 (aa 1-219; aa 108-219; aa 1-208) with HA-β2-chimaerin was readily detected, GST-p23/Tmp21 (aa 1-185), which has the transmembrane domain deleted, failed to associate with β2-chimaerin (Figure 8B, bottom). These results suggest that p23/Tmp21 transmembrane domain is implicated in the interaction.
Figure 8.
Identification of the β2-chimaerin C1 domain interacting region in p23/Tmp21. EGY48 yeast (containing 8op-LacZ vector) was cotransformed with pLexA-fused α1-chimaerin (aa 1–147) (Wang and Kazanietz, 2002) and pB42AD-HA–tagged p23/Tmp21 truncated mutants. (A) Top, schematic representation of p23/Tmp21 constructs used in the yeast two-hybrid assay. Middle, chimaerin expression in yeast lysates, as determined by Western blot using an anti-pLexA antibody; and expression of p23/Tmp21 truncated proteins in yeast lysates using an anti-HA antibody. Bottom, assay of β-galactosidase activity on induction or no-induction plates, carried out 72 h after transformation. Gal/Raf, galactosidase/raffinose. (B) COS-1 cells were transfected with either pEBG (empty vector) or pEBG-p23/Tmp21, and infected with a HA-β2-chimaerin adenovirus (multiplicity of infection [MOI] = 10 plaque-forming units [pfu]/cell). Thirty-six hours later, GST or GST-p23/Tmp21 proteins were precipitated with glutathione-Sepharose 4B beads and associated HA-β2-chimaerin detected by Western blot using an anti-HA antibody. Top, schematic representation of GST-p23/Tmp21 constructs used in the coprecipitation assays. Middle, expression of GST-p23/Tmp21 or its mutants and HA-β2-chimaerin in cell lysates. Bottom, associated HA-β2-chimaerin and GST-p23/Tmp21 or its mutants in pull-down assay were detected by Western blot using anti-HA and anti-GST antibody respectively. Similar results were observed in two additional experiments.
Disruption of the β2-Chimaerin-p23/Tmp21 Complex Leads to Enhanced β2-Chimaerin Rac-GAP Activity
We have shown previously that the expression levels of p23/Tmp21 may influence the ability of β2-chimaerin to regulate Rac-GTP (active) levels (Wang and Kazanietz, 2002). Conceivably, when complexed with p23/Tmp21, β2-chimaerin cannot access the plasma membrane to inactivate Rac. It is still elusive whether disruption of the β2-chimaerin-p23/Tmp21 complex affects β2-chimaerin Rac-GAP activity. We predicted that dissociation of this complex should result in enhanced availability of β2-chimaerin for Rac inactivation. To address this issue, we compared the Rac-GAP activity of β2-chimaerin (wild type) and E227G/L248A-β2-chimaerin in COS-1 cells by using a PBD pull-down assay. As shown in Figure 9, the double mutant is more active as a Rac-GAP than wild-type-β2-chimaerin is when expressed at comparable levels. PMA significantly enhanced Rac-GAP activity of wild-type β2-chimaerin. These results suggest that binding of β2-chimaerin to p23/Tmp21 limits the availability of this Rac-GAP for inhibiting Rac activity.
Figure 9.
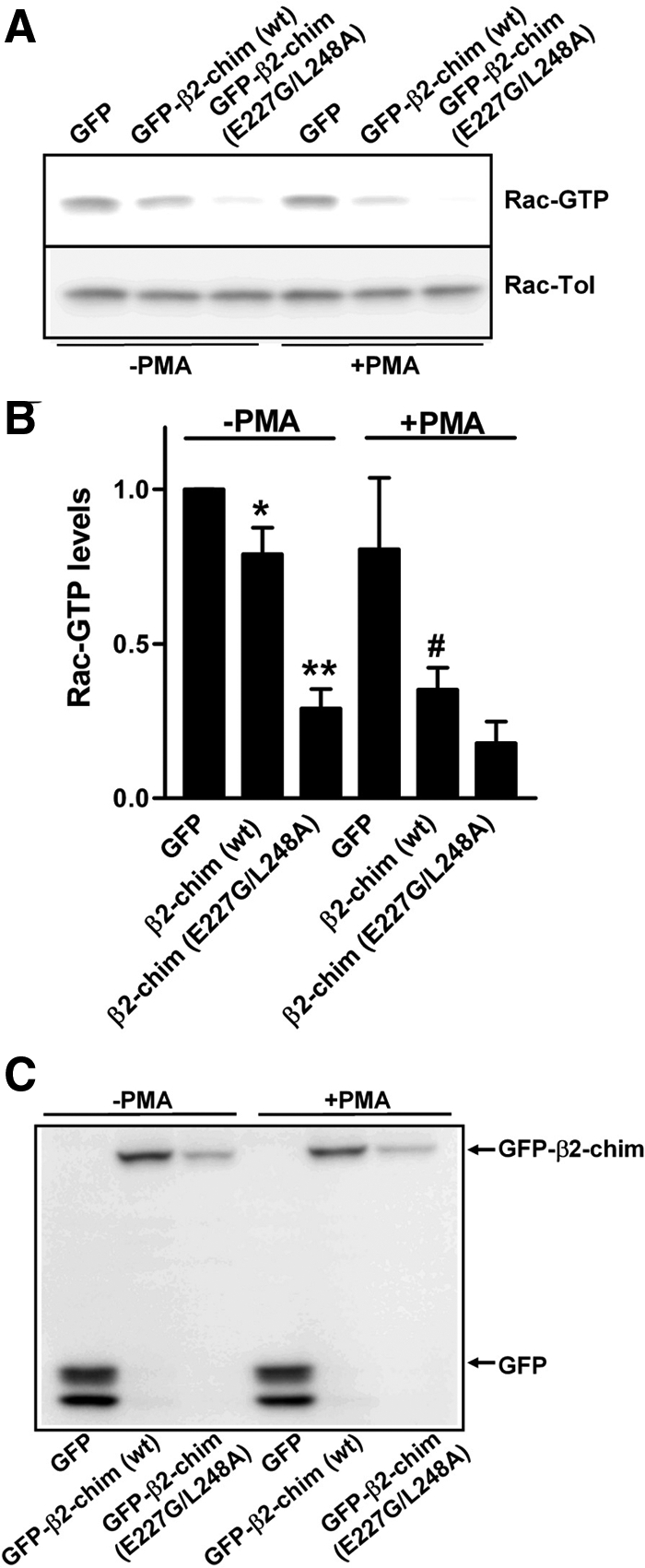
Disruption of β2-chimaerin-p23/Tmp21 interaction leads to enhanced β2-chimaerin Rac-GAP activity. (A) COS-1 cells were transfected with pEGFP, pEGFP-β2-chimaerin (wt), or pEGFP-β2-chimaerin (E227G/L248A). Forty-eight hours later, Rac-GTP levels were assayed using a GST-PBD pull-down assay. (B) Densitometric analysis of Rac-GTP levels relative to control (GFP alone). Data are expressed as mean ± SE of five independent experiments. *p < 0.05 between GFP versus GFP-β2-chim (wt); **p < 0.01 between GFP-β2-chim (wt) versus GFP-β2-chim (E227D/L248A). #p < 0.01 between GFP-β2-chim (wt) (−PMA) versus GFP-β2-chim(wt) (+PMA). (C) Western blots show GFP, GFP-β2-chim (wt) and GFP-β2-chim (E227D/L248A) protein expression.
DISCUSSION
In the present study, we demonstrated that p23/Tmp21 is an anchoring protein for the Rac-GAP β2-chimaerin and is required for its translocation to the perinuclear compartment. The interaction is mediated through the β2-chimaerin C1 domain. Interestingly, we found that C1 domains from other proteins, such as the second C1 domain of PKCε (C1εb), have the ability to interact with p23/Tmp21. However, a comparative analysis between β2-chimaerin and PKCε suggests that p23/Tmp21 plays differential roles in targeting C1 domain containing proteins, as only the former interacts with p23/Tmp21 in response to phorbol ester activation. We also identified key amino acids in the β2-chimaerin C1 domain required for the interaction that when mutated prevent the formation of the complex.
C1 Domains As Targeting Modules
It is well established that C1 domains play a fundamental role in targeting PKC isozymes and other molecules from the cytosol to membranes both in response to phorbol esters and DAG generated upon receptor activation (Colon-Gonzalez and Kazanietz, 2006). For classical and novel PKCs, this membrane targeting is essential for allosteric activation. C1 domains in other phorbol ester/DAG receptors also play key roles in translocation, because it has been extensively demonstrated for PKDs, RasGRPs, and chimaerin Rac-GAPs. X-Ray crystallography studies of the C1b domain of PKCδ established that it is constituted of two β sheets and a small α helix at the end of the C terminus. The β sheets form a pocket where phorbol esters and DAG bind (Zhang et al., 1995). Modeling analysis revealed that all C1 domains including those in chimaerins, have remarkable structural resemblance (Caloca et al., 1999). The C1 domain in α- and β-chimaerins binds phorbol esters and DAG analogs with affinities similar to PKC C1 domains and uses similar mechanisms for insertion into lipid bilayers (Caloca et al., 1999). It is remarkable, however, that individual C1 domains from phorbol ester/DAG receptors have quite distinct properties for ligand recognition and localize to entirely different compartments when expressed in cells. Various examples of differential ligand affinities have been reported for C1a and C1b domains in PKC isozymes. For example, a study showed that whereas in PKCδ the C1a domain preferentially binds DAG, the C1b domain has higher affinity for phorbol esters (Stahelin et al., 2004). Site-directed mutagenesis of individual C1 domains in PKCs has also established distinct roles in translocation, and it was suggested that this disparity relates to a differential exposure of C1a and C1b domains (Pu et al., 2009). The nonequivalency of C1 domains in PKCs is also exemplified by their differential ability to localize to the Golgi complex: whereas isolated C1a domains generally localize throughout the cell rather than specifically in the perinuclear region, C1b domains of cPKCs and nPKCs show colocalization with a Golgi marker (Schultz et al., 2004), as we have also demonstrated for full-length β2-chimaerin and its isolated C1 domain (Caloca et al., 2001), and brefeldin can disrupt their localization (Maissel et al., 2006). In PKDs, a family of PKC-related kinases, C1a and C1b domains also have differential biochemical and targeting properties (Maeda et al., 2001; Chen et al., 2008). Altogether, this suggests that mechanisms alternative or in addition to ligand binding may account for the selective intracellular compartmentalization.
Growing evidence suggests that C1 domains in PKCs and PKDs act as protein interaction modules that regulate their cellular localization and/or activation. For example, the C1b domain of PKCα interacts with the cell matrix protein fascin, and disruption of this interaction affects cell motility (Anilkumar et al., 2003). 14-3-3τ binding sites have been identified within the C1b domain of PKCγ (Nguyen et al., 2004). The association between the PKD1 C1 domain and 14-3-3τ in T cells negatively regulates PKD1 (Hausser et al., 1999). The recent identification of a 20 amino acid module in PKCε that directs localization to cell–cell contacts via protein–protein interaction is another remarkable example (Diouf et al., 2009). The PKCε C1b binds to peripherin to induce its aggregation, leading to apoptosis in neuroblastoma cells (Sunesson et al., 2008). Localization of PKCβII to centrosomes is mediated by the scaffolding protein pericentrin and involves the PKCβII C1a domain, and dissociation of this complex impairs spindle formation and cell division (Chen et al., 2004). Newton and coworkers identified a novel E3 ubiquitin ligase that interacts with the C1a domain of PKCβII (Chen et al., 2007).
The identification of p23/Tmp21 as a β2-chimaerin interactor allowed us to postulate an anchoring role for this Golgi protein. In this study, we showed that RNA interference (RNAi) depletion of p23/Tmp21 prevents the translocation of β2-chimaerin to the perinuclear region, an indication that binding to p23/Tmp21 is a requisite for relocalization. Deletion of the C1 domain in β2-chimaerin prevents β2-chimaerin interaction with p23/Tmp21 (Wang and Kazanietz, 2002), and this is also supported by the failure of the C1 domain mutant C246A-β2-chimaerin to associate with p23/Tmp21 observed in the present study. We also investigated the role of two highly conserved amino acids in the C1b domains of PKCs. Previous studies by Schultz et al. (2004) showed that Asp257 and Met278 in PKCε (positions 15 and 36 in the C1 domain consensus) or the equivalent residues in PKCθ (Glu246 and Met267) are required for Golgi targeting. Like PKC C1b domains, the C1 domain in β2-chimaerin possesses an acidic residue in position 15 (Glu227) and a hydrophobic residue in position 36 (Leu248). We found that mutation of both residues abolishes perinuclear translocation of β2-chimaerin. It is unclear why single mutants were not as effective, but Shultz et al. (2004) also described similar results for single mutants in PKCθ. It is interesting that, unlike β2-chimaerin, PKCε losses its perinuclear localization in response to PMA and mobilize to a peripheral compartment. Most likely, the forces that drive membrane translocation are sufficiently strong to overcome the PKCε–p23/Tmp21 interaction. The C1a domain in PKCε may suffice to drive plasma membrane localization, as the tandem C1a-C1b domain mobilizes to the plasma membrane in response to PMA (data not shown). In addition to its perinuclear relocalization, β2-chimaerin translocates to the plasma membrane in response to stimuli (Wang et al., 2006). It remains to be determined whether signals could lead to the dissociation of the complex and release β2-chimaerin to make it available for membrane translocation or whether different intracellular pools that respond differentially to stimuli exist. The ability of chimaerins to interact with other proteins independently of the C1 domain (Colon-Gonzalez and Kazanietz, unpublished data) clearly suggests complex regulatory mechanisms controlling relocalization and activation of this family of Rac-GAPs.
A key finding in this paper was the identification of amino acids 15 and 36 in the C1 domain consensus as essential for the interaction with p23/Tmp21. Mutation of both residues in β2-chimaerin impairs the interaction with p23/Tmp21 in a yeast two-hybrid assay. Likewise, mutation of amino acids Asp257 and Met278 in PKCε leads to dissociation of this kinase from the perinuclear compartment (see Supplemental Figure S4). The differential interaction of PKCε C1a and C1b domains with p23/Tmp21 may also reflect the differences in amino acids present in those positions (Ser and Cys in C1a). Although at the present time there is no structural information on the C1 domain-p23/Tmp21 interaction, three-dimensional studies clearly showed that amino acids in position 15 and 36 are in proximity and possibly not inserted into the membrane bilayer (Zhang et al., 1995; Schultz et al., 2004), thus making them available for interactions with other partners.
p23/Tmp21 As an Anchoring Protein for β2-Chimaerin
p23/Tmp21 is a type I protein belonging to the p24 family that has a receptor-like luminal domain and a short cytoplasmic tail. Members of the p24 family have been widely implicated as coat protein (COP) vesicle cargo receptors, and they participate in COP vesicle budding and the organization of the Golgi apparatus. The cytoplasmic tail of p23/Tmp21 carries motifs that bind to COPI, and this association is crucial for the retention and retrieval of cargo proteins in the early secretory pathway (Gommel et al., 2001). Our deletional analysis revealed that this C-terminal tail is not involved in binding to chimaerins. Despite multiple studies implicating p24 proteins in vesicle trafficking, insights into the actual function of p23/Tmp21 remain elusive. Recent studies have established that p23/Tmp21 is a component of the presenilin complex that modulates γ-secretase activity (Chen et al., 2006). p23/Tmp21 can also localized in post-Golgi compartments and even traffic to the plasma membrane (Blum and Lepier, 2008). In addition, emerging evidence suggests that p23/Tmp21 is implicated in the regulation of small GTPase function. Studies identified a mechanism of recruitment of the small GTPase ARF1 (ADP-ribosylation factor 1) to the Golgi via p23/Tmp21 (Gommel et al., 2001; Majoul et al., 2001). More recently, studies found that ARF1-dependent assembly of actin in the Golgi apparatus may involve Cdc42/Rac and is dependent on p23/Tmp21 (Fucini et al., 2002). Although early studies showed that Rac1 is present in the perinuclear region, primarily in an inactive guanosine diphosphate-bound state (Kraynov et al., 2000), functional studies on perinuclear Rac are scarce. Moreover, it is unclear what the relative contribution of this perinuclear-associated Rac is to the pool of active plasma membrane Rac generated in response to stimuli. It may be possible that recruitment of chimaerin Rac-GAPs via p23/Tmp21 contributes to the maintenance of the perinuclear Rac inactive pool. There is a precedent for the interaction of endogenous Rac and a Rac-GAP protein, OCRL1, at the trans-Golgi network; however, the mechanisms involved in targeting these proteins to the perinuclear region are yet to be established.
CONCLUSIONS
In summary, we identified p23/Tmp21 as an anchoring protein for β2-chimaerin via its C1 domain. Depletion of p23/Tmp21 from cells leads to reduced perinuclear translocation of β2-chimaerin, suggesting that when complexed with p23/Tmp21 its availability to mobilize to the plasma membrane and inactivate Rac is limited. Our studies also provide evidence for a marked selectivity of C1 domains for binding to p23/Tmp21, because some C1 domains from PKC isozymes and potentially other phorbol ester/DAG receptors have the ability to bind p23/Tmp21. In PKCε, it seems that p23/Tmp21 anchors this kinase via the C1b domain and that the PKCε-Tmp21 complex dissociates in response to stimuli. Whether this is also true for other PKC isozymes or phorbol ester/DAG receptors such as PKD and RasGRP isozymes remains to be determined. The ability of C1 domains to dictate intracellular localization via protein–protein interactions expands our view that these domains act solely as lipid binding motifs and highlight the complexity of DAG signaling.
Supplementary Material
ACKNOWLEDGMENTS
This work is supported by National Institutes of Health grant R01-CA74197 (to M.G.K.). We also thank Dr. Michael S. Marks (University of Pennsylvania) for supplying GS-28 antibody.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-08-0735) on February 17, 2010.
REFERENCES
- Ahmed S., Kozma R., Lee J., Monfries C., Harden N., Lim L. The cysteine-rich domain of human proteins, neuronal chimaerin, protein kinase C and diacylglycerol kinase binds zinc. Evidence for the involvement of a zinc-dependent structure in phorbol ester binding. Biochem. J. 1991;280:233–241. doi: 10.1042/bj2800233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anilkumar N., Parsons M., Monk R., Ng T., Adams J. C. Interaction of fascin and protein kinase Calpha: a novel intersection in cell adhesion and motility. EMBO J. 2003;22:5390–5402. doi: 10.1093/emboj/cdg521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betz A., Ashery U., Rickmann M., Augustin I., Neher E., Sudhof T. C., Rettig J., Brose N. Munc13-1 is a presynaptic phorbol ester receptor that enhances neurotransmitter release. Neuron. 1998;21:123–136. doi: 10.1016/s0896-6273(00)80520-6. [DOI] [PubMed] [Google Scholar]
- Blum R., Feick P., Puype M., Vandekerckhove J., Klengel R., Nastainczyk W., Schulz I. Tmp21 and p24A, two type I proteins enriched in pancreatic microsomal membranes, are members of a protein family involved in vesicular trafficking. J. Biol. Chem. 1996;271:17183–17189. doi: 10.1074/jbc.271.29.17183. [DOI] [PubMed] [Google Scholar]
- Blum R., Lepier A. The luminal domain of p23 (Tmp21) plays a critical role in p23 cell surface trafficking. Traffic. 2008;9:1530–1550. doi: 10.1111/j.1600-0854.2008.00784.x. [DOI] [PubMed] [Google Scholar]
- Blum R., Pfeiffer F., Feick P., Nastainczyk W., Kohler B., Schafer K. H., Schulz I. Intracellular localization and in vivo trafficking of p24A and p23. J. Cell Sci. 1999;112:537–548. doi: 10.1242/jcs.112.4.537. [DOI] [PubMed] [Google Scholar]
- Caloca M. J., Fernandez N., Lewin N. E., Ching D., Modali R., Blumberg P. M., Kazanietz M. G. Beta2-chimaerin is a high affinity receptor for the phorbol ester tumor promoters. J. Biol. Chem. 1997;272:26488–26496. doi: 10.1074/jbc.272.42.26488. [DOI] [PubMed] [Google Scholar]
- Caloca M. J., et al. beta2-chimaerin is a novel target for diacylglycerol: binding properties and changes in subcellular localization mediated by ligand binding to its C1 domain. Proc. Natl. Acad. Sci. USA. 1999;96:11854–11859. doi: 10.1073/pnas.96.21.11854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caloca M. J., Wang H., Delemos A., Wang S., Kazanietz M. G. Phorbol esters and related analogs regulate the subcellular localization of beta 2-chimaerin, a non-protein kinase C phorbol ester receptor. J. Biol. Chem. 2001;276:18303–18312. doi: 10.1074/jbc.M011368200. [DOI] [PubMed] [Google Scholar]
- Caloca M. J., Zugaza J. L., Bustelo X. R. Exchange factors of the RasGRP family mediate Ras activation in the Golgi. J. Biol. Chem. 2003;278:33465–33473. doi: 10.1074/jbc.M302807200. [DOI] [PubMed] [Google Scholar]
- Chen D., Gould C., Garza R., Gao T., Hampton R. Y., Newton A. C. Amplitude control of protein kinase C by RINCK, a novel E3 ubiquitin ligase. J. Biol. Chem. 2007;282:33776–33787. doi: 10.1074/jbc.M703320200. [DOI] [PubMed] [Google Scholar]
- Chen D., Purohit A., Halilovic E., Doxsey S. J., Newton A. C. Centrosomal anchoring of protein kinase C betaII by pericentrin controls microtubule organization, spindle function, and cytokinesis. J. Biol. Chem. 2004;279:4829–4839. doi: 10.1074/jbc.M311196200. [DOI] [PubMed] [Google Scholar]
- Chen F., et al. TMP21 is a presenilin complex component that modulates gamma-secretase but not epsilon-secretase activity. Nature. 2006;440:1208–1212. doi: 10.1038/nature04667. [DOI] [PubMed] [Google Scholar]
- Chen J., Deng F., Li J., Wang Q. J. Selective binding of phorbol esters and diacylglycerol by individual C1 domains of the PKD family. Biochem. J. 2008;411:333–342. doi: 10.1042/BJ20071334. [DOI] [PubMed] [Google Scholar]
- Choi S. H., Czifra G., Kedei N., Lewin N. E., Lazar J., Pu Y., Marquez V. E., Blumberg P. M. Characterization of the interaction of phorbol esters with the C1 domain of MRCK (myotonic dystrophy kinase-related Cdc42 binding kinase) alpha/beta. J. Biol. Chem. 2008;283:10543–10549. doi: 10.1074/jbc.M707463200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colon-Gonzalez F., Kazanietz M. G. C1 domains exposed: from diacylglycerol binding to protein-protein interactions. Biochim. Biophys. Acta. 2006;1761:827–837. doi: 10.1016/j.bbalip.2006.05.001. [DOI] [PubMed] [Google Scholar]
- Csukai M., Chen C. H., De Matteis M. A., Mochly-Rosen D. The coatomer protein beta'-COP, a selective binding protein (RACK) for protein kinase Cepsilon. J. Biol. Chem. 1997;272:29200–29206. doi: 10.1074/jbc.272.46.29200. [DOI] [PubMed] [Google Scholar]
- Csukai M., Mochly-Rosen D. Pharmacologic modulation of protein kinase C isozymes: the role of RACKs and subcellular localisation. Pharmacol. Res. 1999;39:253–259. doi: 10.1006/phrs.1998.0418. [DOI] [PubMed] [Google Scholar]
- Diouf B., et al. A 20-amino acid module of protein kinase C{epsilon} involved in translocation and selective targeting at cell-cell contacts. J. Biol. Chem. 2009;284:18808–18815. doi: 10.1074/jbc.M109.004614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebinu J. O., Bottorff D. A., Chan E. Y., Stang S. L., Dunn R. J., Stone J. C. RasGRP, a Ras guanyl nucleotide-releasing protein with calcium- and diacylglycerol-binding motifs. Science. 1998;280:1082–1086. doi: 10.1126/science.280.5366.1082. [DOI] [PubMed] [Google Scholar]
- Fucini R. V., Chen J. L., Sharma C., Kessels M. M., Stamnes M. Golgi vesicle proteins are linked to the assembly of an actin complex defined by mAbp1. Mol. Biol. Cell. 2002;13:621–631. doi: 10.1091/mbc.01-11-0547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gommel D. U., Memon A. R., Heiss A., Lottspeich F., Pfannstiel J., Lechner J., Reinhard C., Helms J. B., Nickel W., Wieland F. T. Recruitment to Golgi membranes of ADP-ribosylation factor 1 is mediated by the cytoplasmic domain of p23. EMBO J. 2001;20:6751–6760. doi: 10.1093/emboj/20.23.6751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall C., Monfries C., Smith P., Lim H. H., Kozma R., Ahmed S., Vanniasingham V., Leung T., Lim L. Novel human brain cDNA encoding a 34,000 Mr protein n-chimaerin, related to both the regulatory domain of protein kinase C and BCR, the product of the breakpoint cluster region gene. J. Mol. Biol. 1990;211:11–16. doi: 10.1016/0022-2836(90)90006-8. [DOI] [PubMed] [Google Scholar]
- Hausser A., Storz P., Link G., Stoll H., Liu Y. C., Altman A., Pfizenmaier K., Johannes F. J. Protein kinase C mu is negatively regulated by 14–3-3 signal transduction proteins. J. Biol. Chem. 1999;274:9258–9264. doi: 10.1074/jbc.274.14.9258. [DOI] [PubMed] [Google Scholar]
- Hu T., Exton J. H. Protein kinase Calpha translocates to the perinuclear region to activate phospholipase D1. J. Biol. Chem. 2004;279:35702–35708. doi: 10.1074/jbc.M402372200. [DOI] [PubMed] [Google Scholar]
- Kraynov V. S., Chamberlain C., Bokoch G. M., Schwartz M. A., Slabaugh S., Hahn K. M. Localized Rac activation dynamics visualized in living cells. Science. 2000;290:333–337. doi: 10.1126/science.290.5490.333. [DOI] [PubMed] [Google Scholar]
- Lehel C., Olah Z., Jakab G., Anderson W. B. Protein kinase C epsilon is localized to the Golgi via its zinc-finger domain and modulates Golgi function. Proc. Natl. Acad. Sci. USA. 1995;92:1406–1410. doi: 10.1073/pnas.92.5.1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehel C., Olah Z., Mischak H., Mushinski J. F., Anderson W. B. Overexpressed protein kinase C-delta and -epsilon subtypes in NIH 3T3 cells exhibit differential subcellular localization and differential regulation of sodium-dependent phosphate uptake. J. Biol. Chem. 1994;269:4761–4766. [PubMed] [Google Scholar]
- Maeda Y., Beznoussenko G. V., Van Lint J., Mironov A. A., Malhotra V. Recruitment of protein kinase D to the trans-Golgi network via the first cysteine-rich domain. EMBO J. 2001;20:5982–5990. doi: 10.1093/emboj/20.21.5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maissel A., Marom M., Shtutman M., Shahaf G., Livneh E. PKCeta is localized in the Golgi, ER and nuclear envelope and translocates to the nuclear envelope upon PMA activation and serum-starvation: C1b domain and the pseudosubstrate containing fragment target PKCeta to the Golgi and the nuclear envelope. Cell Signal. 2006;18:1127–1139. doi: 10.1016/j.cellsig.2005.09.003. [DOI] [PubMed] [Google Scholar]
- Majoul I., Straub M., Hell S. W., Duden R., Soling H. D. KDEL-cargo regulates interactions between proteins involved in COPI vesicle traffic: measurements in living cells using FRET. Dev. Cell. 2001;1:139–153. doi: 10.1016/s1534-5807(01)00004-1. [DOI] [PubMed] [Google Scholar]
- Majumder P. K., Pandey P., Sun X., Cheng K., Datta R., Saxena S., Kharbanda S., Kufe D. Mitochondrial translocation of protein kinase C delta in phorbol ester-induced cytochrome c release and apoptosis. J. Biol. Chem. 2000;275:21793–21796. doi: 10.1074/jbc.C000048200. [DOI] [PubMed] [Google Scholar]
- Maruyama I. N., Brenner S. A phorbol ester/diacylglycerol-binding protein encoded by the unc-13 gene of Caenorhabditis elegans. Proc. Natl. Acad. Sci. USA. 1991;88:5729–5733. doi: 10.1073/pnas.88.13.5729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellor H., Parker P. J. The extended protein kinase C superfamily. Biochem. J. 1998;332:281–292. doi: 10.1042/bj3320281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochly-Rosen D., Gordon A. S. Anchoring proteins for protein kinase C: a means for isozyme selectivity. FASEB J. 1998;12:35–42. [PubMed] [Google Scholar]
- Mochly-Rosen D., Khaner H., Lopez J. Identification of intracellular receptor proteins for activated protein kinase C. Proc. Natl. Acad. Sci. USA. 1991;88:3997–4000. doi: 10.1073/pnas.88.9.3997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray N. R., Burns D. J., Fields A. P. Presence of a beta II protein kinase C-selective nuclear membrane activation factor in human leukemia cells. J. Biol. Chem. 1994;269:21385–21390. [PubMed] [Google Scholar]
- Newton A. C. Protein kinase C: structure, function, and regulation. J. Biol. Chem. 1995;270:28495–28498. doi: 10.1074/jbc.270.48.28495. [DOI] [PubMed] [Google Scholar]
- Nguyen T. A., Takemoto L. J., Takemoto D. J. Inhibition of gap junction activity through the release of the C1B domain of protein kinase Cgamma (PKCgamma) from 14-3-3, identification of PKCgamma-binding sites. J. Biol. Chem. 2004;279:52714–52725. doi: 10.1074/jbc.M403040200. [DOI] [PubMed] [Google Scholar]
- Pu Y., Garfield S. H., Kedei N., Blumberg P. M. Characterization of the differential roles of the twin C1a and C1b domains of protein kinase C-delta. J. Biol. Chem. 2009;284:1302–1312. doi: 10.1074/jbc.M804796200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ron D., Kazanietz M. G. New insights into the regulation of protein kinase C and novel phorbol ester receptors. FASEB J. 1999;13:1658–1676. [PubMed] [Google Scholar]
- Schultz A., Ling M., Larsson C. Identification of an amino acid residue in the protein kinase C C1b domain crucial for its localization to the Golgi network. J. Biol. Chem. 2004;279:31750–31760. doi: 10.1074/jbc.M313017200. [DOI] [PubMed] [Google Scholar]
- Shindo M., Irie K., Masuda A., Ohigashi H., Shirai Y., Miyasaka K., Saito N. Synthesis and phorbol ester binding of the cysteine-rich domains of diacylglycerol kinase (DGK) isozymes. DGKgamma and DGKbeta are new targets of tumor-promoting phorbol esters. J. Biol. Chem. 2003;278:18448–18454. doi: 10.1074/jbc.M300400200. [DOI] [PubMed] [Google Scholar]
- Sohn K., Orci L., Ravazzola M., Amherdt M., Bremser M., Lottspeich F., Fiedler K., Helms J. B., Wieland F. T. A major transmembrane protein of Golgi-derived COPI-coated vesicles involved in coatomer binding. J. Cell Biol. 1996;135:1239–1248. doi: 10.1083/jcb.135.5.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y., Ailenberg M., Silverman M. Human munc13 is a diacylglycerol receptor that induces apoptosis and may contribute to renal cell injury in hyperglycemia. Mol. Biol. Cell. 1999;10:1609–1619. doi: 10.1091/mbc.10.5.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahelin R. V., Digman M. A., Medkova M., Ananthanarayanan B., Rafter J. D., Melowic H. R., Cho W. Mechanism of diacylglycerol-induced membrane targeting and activation of protein kinase Cdelta. J. Biol. Chem. 2004;279:29501–29512. doi: 10.1074/jbc.M403191200. [DOI] [PubMed] [Google Scholar]
- Sunesson L., Hellman U., Larsson C. Protein kinase Cepsilon binds peripherin and induces its aggregation, which is accompanied by apoptosis of neuroblastoma cells. J. Biol. Chem. 2008;283:16653–16664. doi: 10.1074/jbc.M710436200. [DOI] [PubMed] [Google Scholar]
- Valverde A. M., Sinnett-Smith J., Van Lint J., Rozengurt E. Molecular cloning and characterization of protein kinase D: a target for diacylglycerol and phorbol esters with a distinctive catalytic domain. Proc. Natl. Acad. Sci. USA. 1994;91:8572–8576. doi: 10.1073/pnas.91.18.8572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Kazanietz M. G. Chimaerins, novel non-protein kinase C phorbol ester receptors, associate with Tmp21-I (p23): evidence for a novel anchoring mechanism involving the chimaerin C1 domain. J. Biol. Chem. 2002;277:4541–4550. doi: 10.1074/jbc.M107150200. [DOI] [PubMed] [Google Scholar]
- Wang H., Yang C., Leskow F. C., Sun J., Canagarajah B., Hurley J. H., Kazanietz M. G. Phospholipase Cgamma/diacylglycerol-dependent activation of beta2-chimaerin restricts EGF-induced Rac signaling. EMBO J. 2006;25:2062–2074. doi: 10.1038/sj.emboj.7601098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G., Kazanietz M. G., Blumberg P. M., Hurley J. H. Crystal structure of the cys2 activator-binding domain of protein kinase C delta in complex with phorbol ester. Cell. 1995;81:917–924. doi: 10.1016/0092-8674(95)90011-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.