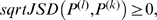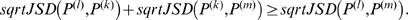Abstract
Background
Alzheimer's disease (AD) is characterized by a neurodegenerative progression that alters cognition. On a phenotypical level, cognition is evaluated by means of the MiniMental State Examination (MMSE) and the post-morten examination of Neurofibrillary Tangle count (NFT) helps to confirm an AD diagnostic. The MMSE evaluates different aspects of cognition including orientation, short-term memory (retention and recall), attention and language. As there is a normal cognitive decline with aging, and death is the final state on which NFT can be counted, the identification of brain gene expression biomarkers from these phenotypical measures has been elusive.
Methodology/Principal Findings
We have reanalysed a microarray dataset contributed in 2004 by Blalock et al. of 31 samples corresponding to hippocampus gene expression from 22 AD subjects of varying degree of severity and 9 controls. Instead of only relying on correlations of gene expression with the associated MMSE and NFT measures, and by using modern bioinformatics methods based on information theory and combinatorial optimization, we uncovered a 1,372-probe gene expression signature that presents a high-consensus with established markers of progression in AD. The signature reveals alterations in calcium, insulin, phosphatidylinositol and wnt-signalling. Among the most correlated gene probes with AD severity we found those linked to synaptic function, neurofilament bundle assembly and neuronal plasticity.
Conclusions/Significance
A transcription factors analysis of 1,372-probe signature reveals significant associations with the EGR/KROX family of proteins, MAZ, and E2F1. The gene homologous of EGR1, zif268, Egr-1 or Zenk, together with other members of the EGR family, are consolidating a key role in the neuronal plasticity in the brain. These results indicate a degree of commonality between putative genes involved in AD and prion-induced neurodegenerative processes that warrants further investigation.
Introduction
Gomez Ravetti and Moscato have recently shown that the abundance of five proteins, within a panel that also measured other 115 cytokines and growth factors, can be used to predict the development of clinical Alzheimer's Disease (AD) [1]. The biomarker molecular signature is composed of IL-1a, TNF-a, IL-3, EGF and G-CSF and has the same level of specificity and sensitivity as the original 18-protein signature proposed by Ray et al. [2] in late 2007, who introduced this important dataset in the literature. In the original work, Ray et al. had employed the abundance of 120 signalling proteins in plasma to obtain their 18-protein signature set. They used a training set of 83 samples to identify patients that progressed to AD in two to six years. The proposed 5-protein signature has an average of 96% accuracy in predicting clinical AD but it is still linked to the joint measurement of 120 protein abundances.
In this paper, we are revisiting the quest of finding biomarkers of AD. However, this time we aim at finding biomarkers in hippocampus tissue samples which would complement the results of the previous studies on plasma biomarkers. This study will now give a different perspective on the progression of the disease, keeping a systems biology and functional genomics approach. Towards this end, we have chosen to rely on an informative experimental design and dataset contributed by Blalock et al. [3]. We believe that their dataset may help us to locate, either directly or indirectly, other biomarkers of interest that could eventually be detectable in plasma.
Blalock et al. analysed samples from 35 patients with four different levels of AD severity: control, incipient, moderate and severe; for this paper we used only 31 samples for which information is available online. The label assigned to each sample (its “level of severity”) was decided after considering two important scores, those provided by the MiniMental State Examination (MMSE) and the Neurofibrillary Tangle count (NFT). The MMSE score is based on a questionnaire that aims at measuring the level of cognitive impairment of a patient. The questions are aimed at evaluating different aspects of cognition, such as orientation, short-term memory (retention and recall), attention and language. A normal score can range from 24 to 30, mild cognitive impairment on the interval 20 to 23, moderate AD between 10 to 19, and the rest (from 0 to 9) are all considered severe AD cases.
As previously mentioned, Blalock et al. [3] also used the NFT score to assign a severity label to each sample. The NFT score is a well established method for the neuropathological diagnosis of AD [4]. The score is usually based on the average counts of neurofibrilary tangles considering different regions of the brain. A NFT score is a recognised indicator of AD, nevertheless, it is not completely effective as there is evidence that NFTs were also identified in healthy aging brains [5], [6], [7], [8].
The analysis by Blalock et al. [3] focused on the identification of AD-related genes (ADG) and incipient ADG (IADG) using a methodology based on the correlation of the genes with NFT and MMSE scores. In turn, they identified putative biological processes and signalling pathways that are significantly present in those gene lists. Our analysis takes a different direction. While still based on the same dataset, we are attempting to map the progression of the disease, finding biomarkers linked to disease severity, by identifying the genes associated with the divergence of the gene expression profile of a sample with the gene expression average profile of the “Control” group. Analogously, we are interested in identifying the genes that seem to best correlate with the “convergence” to the average profile of the “AD Severe” group of samples. The difference between Blalock et al.'s [3] methodological approach to data analysis and ours is very important. We aim to uncover genes that correlate with the divergence of the gene expression profiles, instead of relying only on correlations with the NFT and MMSE values.
Our objective is to uncover genes which are highly correlated to the progression of the disease. With this objective in mind, we will concentrate the first part of our analysis on the two most extremely separated classes, the sets of samples that have been labelled as “Control” and those labelled “AD Severe”. This important initial decision was made based on the fact that the four classes are, in some sense, arbitrarily defined as specific thresholds for the MMSE and NFT scores that were decided ad hoc. Therefore, we decided to first focus on the transitional patterns that can be identified from a “normally aging” to an “AD-severe” gene expression profile in hippocampus. With this approach, we also avoid selecting genes that diverge from the normal-aged profile by causes other than AD, as we expect that the severity scale in AD has a higher probability of being correct in the “Severe AD” cases (since they have high values of NFT and low MMSE scores, clearly a joint combination highly appreciated as a disease hallmark). This approach has an additional advantage. Using this particular dataset and with focus on the effects of incorrect diagnoses, two publications indentify four possible misdiagnoses between control and incipient AD [9], [10]. In our case, the samples that have been labelled either “Incipient AD” or “Moderate AD” play the role of a “Test set”, as they are not used to select probes for establishing a molecular signature, thus avoiding misdiagnoses problems.
Results
The results have been obtained using four steps in tandem: 1) abundance quantization of gene expression values and filtering of probes (this step is supervised by using the samples labelled either “Control” or “Severe AD”); 2) a feature selection algorithm to refine the probe selection based on numerical solution of a combinatorial optimization problem (the (alpha,beta)-k-Feature Set methodology); 3) a correlation analysis (that requires the computation of Jensen-Shannon divergences). Finally, a fourth step involves the pathway and Gene Ontology analysis of the results.
The first two steps only used the samples labelled either “Control” or “Severe AD”. The third step requires several procedures and uses all of the samples. We first compute an average gene expression profile for the classes “Control” and “Severe AD”. This step is followed by the computation of the square root of the Jensen-Shannon divergence [11] of the gene expression profile of each sample with the average profiles of the classes “Control” and “Severe AD”. Finally, we perform a correlation analysis of each gene expression profile (now across all samples) with the results of the square root of the Jensen-Shannon divergence (we do it twice, one for the “Control” and the other for the “Severe AD” case). With this information, and using state-of-the-art pathway analysis and text mining tools, as a result of our final analysis step, we provide a comprehensive list of results of the differentially regulated genes, patterns of up (down)-regulation and the pathways that seem to be implicated in the progression of AD. We refer to the Methods section for a completely reproducible and in-depth explanation of our methodology.
Probe selection and Jensen-Shannon divergence computations based on class information
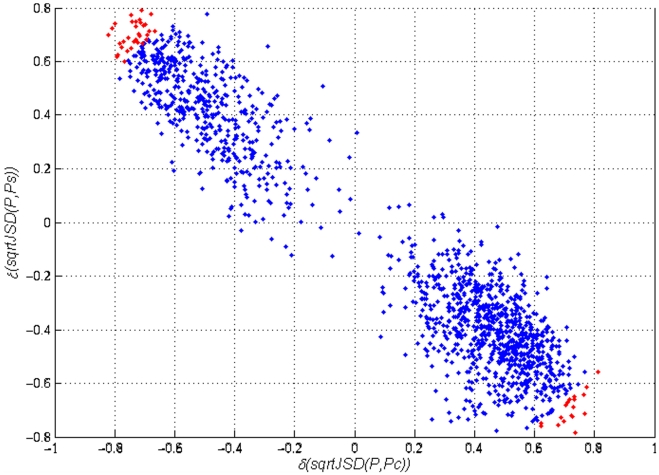
We start our analysis with a baseline comparison, which we have chosen to include for illustrative purposes. Figure 1 provides an example of the importance of performing an initial probe/gene selection step. The example serves as an argument for the necessity of the first two steps of our method. We have normalized each individual gene expression profile, and we have computed the average gene expression profile for classes “Control” and “Severe AD” (following the same procedure we will use in the third step of our method, but in this case using all probes in the array).
Figure 1. This plot illustrates that the third step of our methodology, the use of the Jensen-Shannon divergence, does not appear to give an interesting separation of the samples in the absence of a previous feature selection step.
For this graph, all 22,215 genes were considered in the calculation of the average profile of the samples in the “Control” and “Severe AD” classes. The square root of the Jensen-Shannon divergences to the “Control” and “Severe AD” average profile are computed, respectively giving, for each sample, its x and y coordinates in this plot. Observe that most of the “Control” samples have values lower than 0.12, with two exceptions. This result is expected, as the probability distribution function of the “Control” class was used. However, most of the samples from AD patients (having either “Incipient AD”, “Moderate” or “Severe” labels), show a divergence with the Control average gene expression profile. Figure 2 shows the important contribution provided by the feature selection step.
We have used the square root of the Jensen-Shannon divergence of a pair of samples (a pair of gene expression profiles) as our measure of “dissimilarity” between them. The square root of the Jensen-Shannon divergence quantifies the difference between two probability distribution functions (PDFs) and it is a metric (we refer the reader to the Methods section for a mathematical definition and a discussion of its properties). Figure 1 plots the divergence of each sample with the average expression profile of the classes ‘Control’ and ‘Severe AD’; sqrtJSD(P,
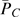 ) denotes the square root of the Jensen-Shannon divergence between sample P and the average profile on the ‘Control’ class
) denotes the square root of the Jensen-Shannon divergence between sample P and the average profile on the ‘Control’ class 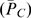 . Analogously, sqrtJSD(P ,
. Analogously, sqrtJSD(P ,
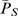 ) denotes the square root of the Jensen-Shannon divergence between sample P and the average profile on the ‘Severe AD’ class
) denotes the square root of the Jensen-Shannon divergence between sample P and the average profile on the ‘Severe AD’ class 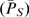 . The advantage of using the probe/gene selection steps, which reduces the number of genes to the most informative ones, will be evident when we later compare Figure 1 with Figure 2. However, Figure 1 already shows some interesting patterns. For instance, we can observe that a high percentage of the samples from AD patients (having either ‘Incipient AD’, ‘Moderate’ or ‘Severe’ labels) show sqrtJSD(P,
. The advantage of using the probe/gene selection steps, which reduces the number of genes to the most informative ones, will be evident when we later compare Figure 1 with Figure 2. However, Figure 1 already shows some interesting patterns. For instance, we can observe that a high percentage of the samples from AD patients (having either ‘Incipient AD’, ‘Moderate’ or ‘Severe’ labels) show sqrtJSD(P,
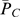 ) values greater than 0.115, which indicates measurable divergence with the Control average gene expression profile.
) values greater than 0.115, which indicates measurable divergence with the Control average gene expression profile.
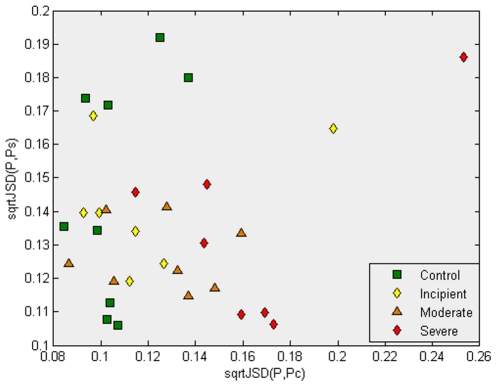
Figure 2. This plot illustrates that after application of the feature selection steps, followed by the computation of the gene expression profile's average profile of the samples in the “Control” and “Severe AD” classes (now on a set of 1,372 probes), the samples are now more clearly separated.
Here, all “Control” samples have the square root of the Jensen-Shannon divergences to the average gene expression of the “Control” samples (x-coordinate) smaller than 0.12 (almost all severe AD have x-coordinates greater than 0.15). In addition to that, most samples labelled “Severe AD” are located on the same region. Both results are expected. However, it is interesting that in this (x,y)-plot most samples that are labelled “Incipient AD” or “Moderate AD” seem to “bridge” between the regions that have most of the “Control” samples and the region that have most of the “Severe AD” group. This result is interesting as no samples from “Incipient AD” nor “Moderate AD” have been used in the first three steps of our methodology. In essence, the work is a “test set” indicating that it is reasonable to expect that some genes in the genetic signature of 1,372 probes have information about a putative “progression” trend of the disease, from the “Control” to the “Severe AD” profile. In what follows, correlations across all the samples with these divergences are used as a method to try to identify those gene profiles that are most correlated with the progression from “Control” to “Severe AD”.
Figure 2 presents the same procedure, but only after the feature selection step has significantly reduced the number of probes fom 22,215 to 1,372. We refer to the Methods section for details. In Figure 2, an arguably more coherent arrangement can be observed. As expected, the group of control samples (in green) have lower values of sqrtJSD(P,
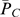 ) and higher values of sqrtJSD(P,
) and higher values of sqrtJSD(P,
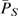 ). Obviously, the opposite behaviour is observed for the samples belonging to the severe cases. What cannot be expected, however, is a layout of the samples that could provide evidence of a continuous “progression” of the disease. The Figure shows that the samples of ‘Incipient AD’ are close to the control group and the ‘Moderate AD’ samples are closer to them and also link to severe AD. A priori, since those samples had not been used for probe selection, they could have been in any position in the (sqrtJSD(P ,
). Obviously, the opposite behaviour is observed for the samples belonging to the severe cases. What cannot be expected, however, is a layout of the samples that could provide evidence of a continuous “progression” of the disease. The Figure shows that the samples of ‘Incipient AD’ are close to the control group and the ‘Moderate AD’ samples are closer to them and also link to severe AD. A priori, since those samples had not been used for probe selection, they could have been in any position in the (sqrtJSD(P ,
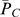 ), sqrtJSD(P,
), sqrtJSD(P,
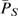 ) plane.
) plane.
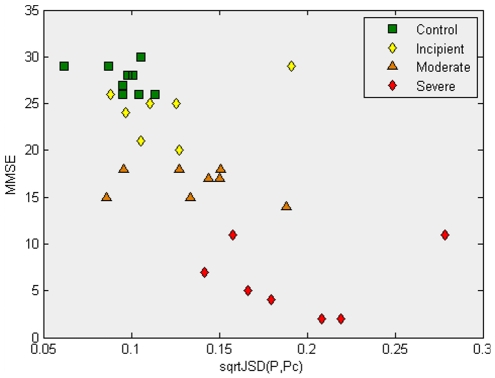
Finally, Figure 3 presents the results of the MMSE score as a function of the sqrtJSD(P ,
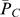 ), showing an inverse correlation between them. A similar situation happens between MMSE and sqrtJSD(P,
), showing an inverse correlation between them. A similar situation happens between MMSE and sqrtJSD(P,
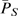 ), but in this case low MMSE scores correspond to low values of sqrtJSD(P,Ps), giving a positive correlation. It is this interplay between positive and negative correlations that has enabled us to find interesting biomarkers. In the next subsection, we explain how these correlations were used to identify probes that “diverge from” their values in the “Control” group and “converge to” the values in the “Severe AD” group.
), but in this case low MMSE scores correspond to low values of sqrtJSD(P,Ps), giving a positive correlation. It is this interplay between positive and negative correlations that has enabled us to find interesting biomarkers. In the next subsection, we explain how these correlations were used to identify probes that “diverge from” their values in the “Control” group and “converge to” the values in the “Severe AD” group.
Figure 3. This plot shows the MMSE scores as a function of the square root of the Jensen-Shannon divergences to the average gene expression of the “Control” samples.
‘Incipient AD’ samples, although having a lower value for their MMSE score, still do not show a dramatic change in their x-coordinates compared to the ‘Control’ samples. ‘Moderate AD’ samples appear to be more scattered, with some of them already having a significant divergence from the ‘Control’ average profile.
Gene correlation analysis
The third step employs a correlation analysis to select the group of probes that are the most strongly correlated. Intuitively, the idea is fairly straightforward as illustrated in the following “Gedankenexperiment” (a thought experiment). Assume, for argument's sake, that the MMSE of each patient P is not actually phenotypical information assigned to each sample. Instead, assume that the MMSE values are the microarray probe expression of some gene. In this “thought experiment”, let MMSE(P) be the expression of this hypothetical gene probe on sample P, and fDataset be the set of values it has for each sample. The correlation of the sample-ordered set of values {MMSE(P)} with the set of sample-ordered values {sqrtJSD(P,
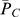 )} is negative, indicating that, in general, this hypothetical MMSE probe reduces its values as the whole gene expression profile of sample P diverges from the average “Control” profile (Figure 3). Analogously, there exists a positive correlation of the set of values {MMSE(P)} with the values of the set {sqrtJSD(P,
)} is negative, indicating that, in general, this hypothetical MMSE probe reduces its values as the whole gene expression profile of sample P diverges from the average “Control” profile (Figure 3). Analogously, there exists a positive correlation of the set of values {MMSE(P)} with the values of the set {sqrtJSD(P,
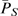 )}. This indicates that the values of MMSE tend to be reduced as the profile of sample P “converges to” the average profile of samples in the “Severe AD” group. We have computed these correlations for all probes in the signature, which are given in the supplementary material (File S2 sheet ‘correlation Analysis’) and are the basis for our analysis.
)}. This indicates that the values of MMSE tend to be reduced as the profile of sample P “converges to” the average profile of samples in the “Severe AD” group. We have computed these correlations for all probes in the signature, which are given in the supplementary material (File S2 sheet ‘correlation Analysis’) and are the basis for our analysis.
We also refer the reader to Figure 4, which presents the computed correlations. Tables 1 and 2 present the one hundred most correlated probes (in absolute values). In the supplementary material (File S2 sheet ‘correlation Analysis’), the correlation of each of the 1,372 probes that were selected by our method is given (and annotated, including Affymetrix and Stanford's Source outputs) to facilitate further analyses.
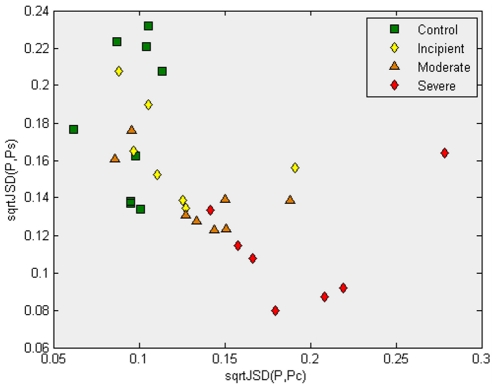
Figure 4. Correlation of the expression profiles of 1,372 probes (across samples) with the sqrtJSD of the samples of two reference groups (“Control” and “Severe AD”, represented by the average expression profile in the group).
The 50 probes in red are those most distant from the origin of this system of coordinates. Those probes have expression-value variations that are correlated with the divergences of the average “Control” profile and at the same time with the “Severe AD”.
Table 1. For each sample, we have calculated the sample's Jensen-Shannon divergence with the average Control gene expression profile.
| Gene symbol | Probe | Spearman rank correlation | |
| 1 | CSF1 | 211839_s_at | 0.79388 |
| 2 | MCL1 | 214057_at | 0.75484 |
| 3 | PSMC3IP | 205956_x_at | 0.74816 |
| 4 | ZHX3 | 217367_s_at | 0.74416 |
| 5 | C10orf76 | 55662_at | 0.74093 |
| 6 | FCAR | 211307_s_at | 0.72002 |
| 7 | TUBD1 | 210389_x_at | 0.71835 |
| 8 | AW974666 | 222365_at | 0.71835 |
| 9 | LRP10 | 201412_at | 0.71079 |
| 10 | SERTAD2 | 202656_s_at | 0.70679 |
| 11 | ITGB5 | 201125_s_at | 0.7059 |
| 12 | CDC2L6 | 212899_at | 0.70412 |
| 13 | RNF19A | 220483_s_at | 0.70367 |
| 14 | TTN | 208195_at | 0.70278 |
| 15 | DHFR | 202534_x_at | 0.69844 |
| 16 | FYCO1 | 218204_s_at | 0.69655 |
| 17 | HBEGF | 38037_at | 0.69388 |
| 18 | ZBTB20 | 205383_s_at | 0.69121 |
| 19 | KCNK5 | 219615_s_at | 0.69121 |
| 20 | KLHL20 | 204177_s_at | 0.68988 |
| 21 | DLG5 | 201681_s_at | 0.68899 |
| 22 | CHD2 | 203461_at | 0.68821 |
| 23 | TUG1 | 222244_s_at | 0.68721 |
| 24 | ZNF500 | 213641_at | 0.68454 |
| 25 | N58524 | 222332_at | 0.68276 |
| 26 | KIR2DL5A | 211410_x_at | 0.68165 |
| 27 | CYBRD1 | 217889_s_at | 0.67964 |
| 28 | DLG1 | 217208_s_at | 0.67831 |
| 29 | IL15 | 205992_s_at | 0.67731 |
| 30 | RND2 | 214393_at | 0.67508 |
| 31 | TNS1 | 221748_s_at | 0.67253 |
| 32 | CTBP2 | 210835_s_at | 0.6703 |
| 33 | AL050204 | 213929_at | 0.66852 |
| 34 | YES1 | 202933_s_at | 0.66763 |
| 35 | MYBL1 | 213906_at | 0.66719 |
| 36 | No gene associated | 213256_at | 0.66363 |
| 37 | C5orf4 | 48031_r_at | 0.66363 |
| 38 | FOXO1 | 202724_s_at | 0.66318 |
| 39 | UPF1 | 211168_s_at | 0.66096 |
| 40 | STAG3L1 | 221191_at | 0.66007 |
| 41 | SLC12A7 | 218066_at | 0.65784 |
| 42 | CYP3A4 | 205999_x_at | 0.65695 |
| 43 | KRCC1 | 218303_x_at | 0.65562 |
| 44 | P53AIP1 | 220402_at | 0.65462 |
| 45 | TLE3 | 212769_at | 0.6535 |
| 46 | ZNF669 | 220215_at | 0.65206 |
| 47 | CFLAR | 214486_x_at | 0.65206 |
| 48 | PAK4 | 203154_s_at | 0.65028 |
| 49 | M78162 | 217536_x_at | 0.6485 |
| 50 | MMP11 | 203876_s_at | 0.6485 |
| 51 | RGS7 | 206290_s_at | −0.67475 |
| 52 | ASTN1 | 213197_at | −0.67653 |
| 53 | TMSB10 | 217733_s_at | −0.67653 |
| 54 | SUPT4H1 | 201484_at | −0.67731 |
| 55 | COX6B1 | 201441_at | −0.67742 |
| 56 | WASF1 | 204165_at | −0.67742 |
| 57 | RALYL | 213967_at | −0.67786 |
| 58 | BBS7 | 219688_at | −0.67875 |
| 59 | SEC31A | 200945_s_at | −0.68009 |
| 60 | DDX1 | 201241_at | −0.68009 |
| 61 | RP11-336K24.9 | 218291_at | −0.68098 |
| 62 | GABBR2 | 209990_s_at | −0.68231 |
| 63 | SLC25A12 | 203340_s_at | −0.68454 |
| 64 | ATP5C1 | 205711_x_at | −0.68587 |
| 65 | NEFL | 221805_at | −0.68632 |
| 66 | NDUFB8 | 201226_at | −0.68854 |
| 67 | OPA1 | 212214_at | −0.69255 |
| 68 | KPNA2 | 201088_at | −0.69522 |
| 69 | PPIA | 211765_x_at | −0.69566 |
| 70 | CYP26B1 | 219825_at | −0.69566 |
| 71 | COX7AP2 | 217249_x_at | −0.69878 |
| 72 | VSNL1 | 203798_s_at | −0.69878 |
| 73 | ATP6V1D | 208898_at | −0.70145 |
| 74 | ATP5C1 | 213366_x_at | −0.70234 |
| 75 | NRXN1 | 209915_s_at | −0.7059 |
| 76 | PCSK2 | 204870_s_at | −0.70901 |
| 77 | AI708767 | 211978_x_at | −0.71034 |
| 78 | UGCGL2 | 218801_at | −0.71257 |
| 79 | KIAA0528 | 212943_at | −0.7139 |
| 80 | SERPINI1 | 205352_at | −0.71657 |
| 81 | LZTS1 | 219042_at | −0.71835 |
| 82 | NEFM | 205113_at | −0.71835 |
| 83 | FRY | 204072_s_at | −0.71924 |
| 84 | CSPG5 | 205344_at | −0.72291 |
| 85 | COX6A1 | 200925_at | −0.7277 |
| 86 | COX4I1 | 202698_x_at | −0.73037 |
| 87 | KIAA0368 | 212428_at | −0.73126 |
| 88 | MYT1L | 210016_at | −0.73304 |
| 89 | PPP3CA | 202457_s_at | −0.74194 |
| 90 | LOC100131599 | 213222_at | −0.74549 |
| 91 | CACNG3 | 206384_at | −0.75484 |
| 92 | PPP3R1 | 204506_at | −0.75573 |
| 93 | MAN1A1 | 221760_at | −0.75929 |
| 94 | NETO2 | 218888_s_at | −0.76819 |
| 95 | LPHN1 | 219145_at | −0.76852 |
| 96 | CAPRIN2 | 218456_at | −0.76997 |
| 97 | CAMK1G | 215161_at | −0.77041 |
| 98 | LDB2 | 206481_s_at | −0.7802 |
| 99 | TRIM36 | 219736_at | −0.79622 |
| 100 | LDHA | 200650_s_at | −0.80245 |
These values are then correlated with the individual expression profiles of each probe across the set of samples samples. We list here the 100 probes that have the highest Spearman correlation (absolute value, computed over all samples) between the expression of the probe and the square root of the Jensen-Shannon divergence of the sample with the average Control gene expression profile. Rows in boldface indicate the cases for which a putative relationship exist in the published literature between the gene and AD. A probe that has a positive correlation with the square root of the Jensen-Shannon divergence with the average Control gene expression profile roughly indicates, in this case, a probe that, over all samples in the set, tends to increase its expression from their values in the “Control” group to the “Severe AD”.
Table 2. List of the 100 probes with the highest Spearman correlation (absolute value, computed over all samples) between the expression of the probe and the square root of the Jensen-Shannon divergence of all samples with the average Severe AD gene expression profile.
| Gene symbol | Probe | Spearman rank correlation | |
| 1 | NEFM | 205113_at | 0.84472 |
| 2 | NRG1 | 206343_s_at | 0.83003 |
| 3 | VSNL1 | 203798_s_at | 0.80156 |
| 4 | NEFL | 221805_at | 0.79889 |
| 5 | SLC25A12 | 203340_s_at | 0.79666 |
| 6 | BCL11A | 219497_s_at | 0.79266 |
| 7 | RALYL | 213967_at | 0.78776 |
| 8 | SERPINI1 | 205352_at | 0.78242 |
| 9 | ATP2B2 | 204685_s_at | 0.78154 |
| 10 | LDB2 | 206481_s_at | 0.7802 |
| 11 | ENSA | 202596_at | 0.77931 |
| 12 | NDUFV2 | 202941_at | 0.77753 |
| 13 | KIAA0319 | 206017_at | 0.76418 |
| 14 | ATP5C1 | 213366_x_at | 0.7584 |
| 15 | TAGLN3 | 204743_at | 0.75617 |
| 16 | SV2B | 205551_at | 0.75484 |
| 17 | DOPEY1 | 213271_s_at | 0.75439 |
| 18 | FAR2 | 220615_s_at | 0.75395 |
| 19 | SNRK | 209481_at | 0.7535 |
| 20 | TRIM36 | 219736_at | 0.74994 |
| 21 | NRXN1 | 209915_s_at | 0.74772 |
| 22 | PKP4 | 214874_at | 0.74461 |
| 23 | CALM3 | 200622_x_at | 0.74149 |
| 24 | PIP4K2C | 218942_at | 0.73971 |
| 25 | CRYM | 205489_at | 0.73437 |
| 26 | SCFD1 | 215548_s_at | 0.73037 |
| 27 | COX6A1 | 200925_at | 0.72992 |
| 28 | OPA1 | 212214_at | 0.7277 |
| 29 | ATP5C1 | 205711_x_at | 0.72414 |
| 30 | LETMD1 | 207170_s_at | 0.71969 |
| 31 | PPP2R2B | 213849_s_at | 0.71657 |
| 32 | UQCRQ | 201568_at | 0.71301 |
| 33 | FKBP3 | 218003_s_at | 0.71268 |
| 34 | PBX1 | 212148_at | 0.71123 |
| 35 | CACNG3 | 206384_at | 0.71079 |
| 36 | TMSB10 | 217733_s_at | 0.70812 |
| 37 | KIAA1467 | 213234_at | 0.70812 |
| 38 | INA | 204465_s_at | 0.7059 |
| 39 | ARF5 | 201526_at | 0.70545 |
| 40 | CD200 | 209582_s_at | 0.70456 |
| 41 | CAMK1G | 215161_at | 0.70367 |
| 42 | TUBG2 | 203894_at | 0.70234 |
| 43 | LDHA | 200650_s_at | 0.70189 |
| 44 | LOC100131599 | 213222_at | 0.70056 |
| 45 | DIMT1L | 210802_s_at | 0.697 |
| 46 | RGS4 | 204339_s_at | 0.69655 |
| 47 | CAMKK2 | 212252_at | 0.69611 |
| 48 | BE731738 | 212661_x_at | 0.69477 |
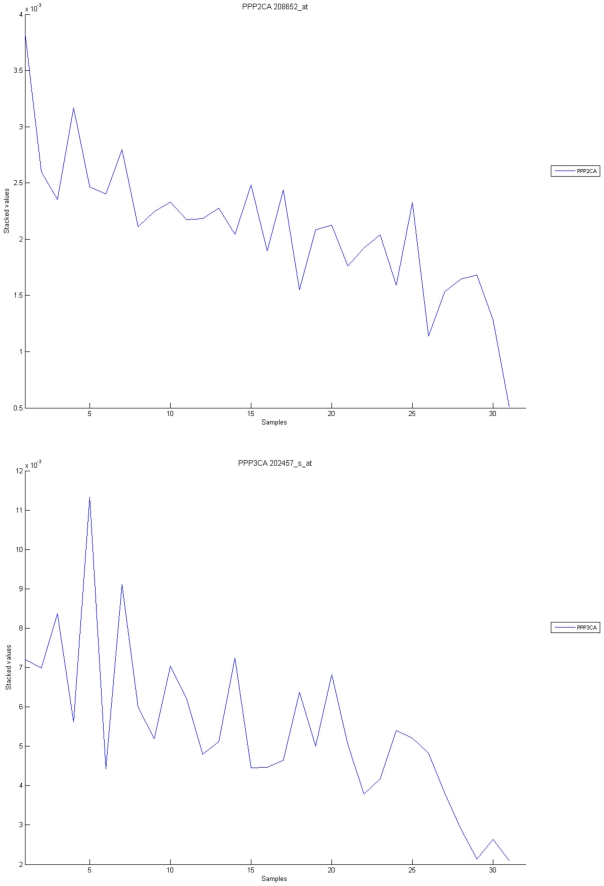
| 49 | PPP2CA | 208652_at | 0.69388 |
| 50 | SRD5A1 | 211056_s_at | 0.69388 |
| 51 | DMN | 212730_at | −0.68409 |
| 52 | AW974666 | 222365_at | −0.68721 |
| 53 | SLC33A1 | 203164_at | −0.68899 |
| 54 | SYNC1 | 221276_s_at | −0.68954 |
| 55 | ITGB5 | 201125_s_at | −0.69299 |
| 56 | CNOT6 | 217970_s_at | −0.69655 |
| 57 | DYNLT1 | 201999_s_at | −0.697 |
| 58 | ZMYND8 | 214795_at | −0.697 |
| 59 | TBL1X | 213400_s_at | −0.69967 |
| 60 | RND2 | 214393_a | −0.70378 |
| 61 | LRP10 | 201412_at | −0.70545 |
| 62 | GMPR | 204187_at | −0.70768 |
| 63 | LTF | 202018_s_at | −0.70812 |
| 64 | CSNK1A1 | 208865_at | −0.70812 |
| 65 | NBPF12 | 213612_x_at | −0.70901 |
| 66 | ZFP36L2 | 201368_at | −0.70945 |
| 67 | AV712577 | 201305_x_at | −0.71212 |
| 68 | FDFT1 | 208647_at | −0.71257 |
| 69 | ADARB2 | 220648_at | −0.71301 |
| 70 | CPT2 | 204264_at | −0.7139 |
| 71 | ADD3 | 201753_s_at | −0.71524 |
| 72 | 37681 | 213256_at | −0.71613 |
| 73 | ITGB8 | 205816_at | −0.71924 |
| 74 | RBM19 | 205115_s_at | −0.71969 |
| 75 | HIST1H1C | 209398_at | −0.72058 |
| 76 | NM_018612 | 220882_at | −0.73037 |
| 77 | CD68 | 203507_at | −0.73259 |
| 78 | GTF2A1L | 213413_at | −0.73348 |
| 79 | FAM114A1 | 213455_at | −0.73571 |
| 80 | FOXO1 | 202724_s_at | −0.73749 |
| 81 | C6orf145 | 212923_s_at | −0.73882 |
| 82 | KRCC1 | 218303_x_at | −0.74149 |
| 83 | TGFBR3 | 204731_at | −0.74372 |
| 84 | ZHX3 | 217367_s_at | −0.74594 |
| 85 | TSPO | 202096_s_at | −0.74816 |
| 86 | STAT5A | 203010_at | −0.74994 |
| 87 | AFF1 | 201924_at | −0.75039 |
| 88 | RASL12 | 219167_at | −0.75217 |
| 89 | AL359052 | 214927_at | −0.75528 |
| 90 | ALDH3A2 | 202054_s_at | −0.75706 |
| 91 | C1S | 208747_s_at | −0.76062 |
| 92 | AV700298 | 217523_at | −0.76062 |
| 93 | HBEGF | 38037_at | −0.76819 |
| 94 | BG251521 | 213156_at | −0.77086 |
| 95 | ZBTB20 | 205383_s_at | −0.77353 |
| 96 | AL049443 | 215306_at | −0.78109 |
| 97 | PTTG1IP | 200677_at | −0.78154 |
| 98 | FYCO1 | 218204_s_at | −0.78598 |
| 99 | ATP6V0E1 | 214150_x_at | −0.802 |
| 100 | SERTAD2 | 202656_s_at | −0.84338 |
We listed the top fifty probes with positive and negative correlation. Rows in boldface indicate the cases for which a putative relationship exist in the published literature between the gene and AD. A probe that has a positive correlation with the square root of the Jensen-Shannon divergence with the average Control gene expression profile roughly indicates a probe that, over all samples in the set, tends to increase its expression from their values in the “Control” group to the “Severe AD”.
As the objective is to detect the probes correlated with the progression of AD, we will select those probes with high absolute correlations values with both groups, an indication of a divergence of the average control profile together with a convergence to the severe AD profile; these correlations computed over all sample types. We need to check both groups according to their correlations to the average profile. The first group of probes we are interested in are those that have a positive correlation with the sqrtJSD(P,
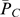 ) and a negative correlation with sqrtJSD(P,
) and a negative correlation with sqrtJSD(P,
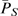 ). The probes in this group are those probes with under-expression in the non-disease sample but are over-expressed in the severe AD cases. The second group has the opposite behaviour, the probes' expression values have a negative correlation with sqrtJSD(P,
). The probes in this group are those probes with under-expression in the non-disease sample but are over-expressed in the severe AD cases. The second group has the opposite behaviour, the probes' expression values have a negative correlation with sqrtJSD(P,
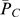 ) and a positive correlation with sqrtJSD(P,
) and a positive correlation with sqrtJSD(P,
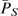 ). This pattern can be visualised in Figure 4, where the elliptical shape of the dispersion of the probes in this scatter plot indicates that our methodology has preserved all the significant probes for both classes and that there are no probes (after the filter) presenting a high correlation simultaneously with the control and severe AD profiles.
). This pattern can be visualised in Figure 4, where the elliptical shape of the dispersion of the probes in this scatter plot indicates that our methodology has preserved all the significant probes for both classes and that there are no probes (after the filter) presenting a high correlation simultaneously with the control and severe AD profiles.
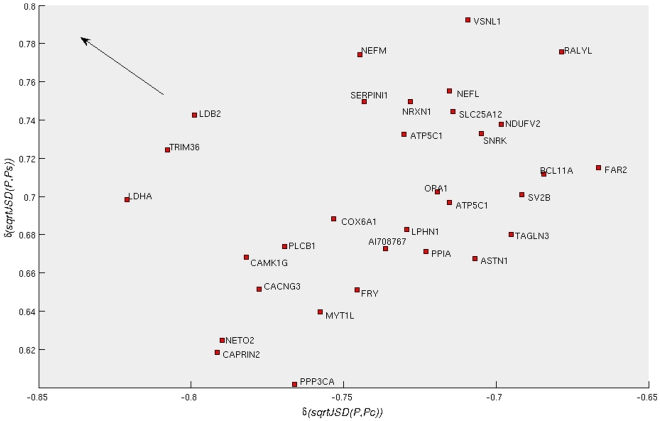
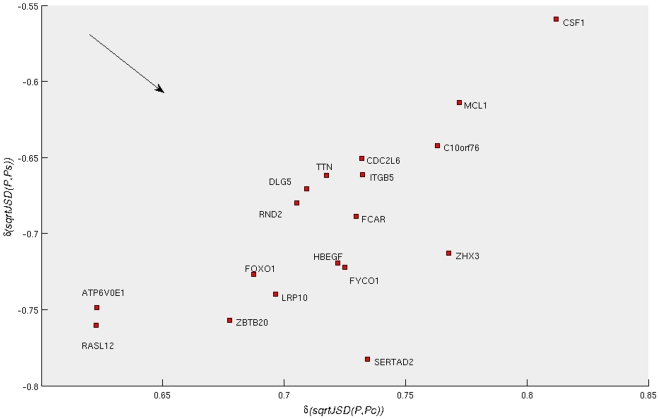
On these values a new selection criterion is applied, as we wanted to identify the group of probes that have strong correlations to both groups in absolute value. This symmetry of our argument stems from the interest in understanding the biology of the progression of the disease. For identifying disease biomarkers we may just concentrate in finding the probes that present an upregulation trend when progressing from “Control” to “Disease”. However, here we would also like to identify those probes that become increasingly downregulated, which, in turn, would help us to identify significantly dysregulated biological pathways (as members of the pathway will be either up or downregulated). Towards this end, we rank the probes in the order given by their Euclidean distance from the origin of coordinates in Figure 4. We selected an arbitrary cut-off value of fifty probes (the selected probes are marked in red). These fifty probes are also identified by their Gene Symbols in Figures 5 and 6.
Figure 5. Zoom of Figure 4, identifying the most distant probes from the origin with negative correlation with the control profile, 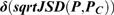 and positive correlation with the severe profile,
and positive correlation with the severe profile, 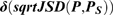 .
.
Figure 6. Zoom of Figure 4, identifying the most distant probes from the origin with positive correlation with the control profile, 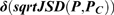 and negative correlation with the severe profile,
and negative correlation with the severe profile, 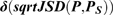 .
.
Calculating the distance of each probe to the origin, on the sqrtJSD system of coordinates, we further selected the 50 most distant probes and analysed their behaviour. Table 3 presents the 50 probes (corresponding to 48 genes), their correlation to each group and their distance to the origin of coordinates. File S2 sheet ‘correlation Analysis’ column ‘E - Distance’ of the supplementary material presents the distance to the origin of the 1,372 probes analysed. In Table 3, it can be seen which genes have some putative annotation that links them to AD (17 genes out of 48).
Table 3. The 50 genes most distant to the origin of the coordinates space 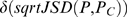 ×
×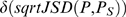 .
.
| Probe Set ID | Gene Symbol | Gene Title | δ(sqrtJSD (P,Pc)) | δ(sqrtJSD (P,Ps)) | Dist O | Ref (ADG) |
| 206481_s_at | LDB2 | LIM domain binding 2 | −0.7988 | 0.7427 | 1.0907 | |
| 219736_at | TRIM36 | tripartite motif-containing 36 | −0.8077 | 0.7242 | 1.0848 | |
| 200650_s_at | LDHA | lactate dehydrogenase A | −0.8210 | 0.6984 | 1.0778 | |
| 205113_at | NEFM | neurofilament, medium polypeptide 150kDa | −0.7448 | 0.7742 | 1.0743 | [379], [649], [672], [673], [674], [675], [676], [677], [678], [679], [680], [681], [682], [683], [684], [685], [686], [687], [688], [689], [690], [691], [692], [693], [694], [695], [696], [697], [698], [699], [700], [701], [702] |
| 202656_s_at | SERTAD2 | SERTA domain containing 2 | 0.7343 | −0.7827 | 1.0732 | |
| 203798_s_at | VSNL1 | visinin-like 1 | −0.7093 | 0.7923 | 1.0634 | [565], [566], [638], [641], [642], [703], [704], [705] |
| 205352_at | SERPINI1 | serpin peptidase inhibitor, clade I (neuroserpin), member 1 | −0.7432 | 0.7496 | 1.0555 | [706], [707], [708], [709], [710], [711], [712], [713], [714] |
| 217367_s_at | ZHX3 | zinc fingers and homeoboxes 3 | 0.7677 | −0.7129 | 1.0477 | |
| 209915_s_at | NRXN1 | neurexin 1 | −0.7282 | 0.7496 | 1.0451 | |
| 221805_at | NEFL | neurofilament, light polypeptide 68kDa | −0.7153 | 0.7552 | 1.0402 | [715], [716], [717] |
| 213366_x_at | ATP5C1 | ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 | −0.7302 | 0.7327 | 1.0344 | |
| 203340_s_at | SLC25A12 | solute carrier family 25 (mitochondrial carrier, Aralar), member 12 | −0.7141 | 0.7444 | 1.0315 | [718] |
| 213967_at | RALYL | RALY RNA binding protein-like | −0.6786 | 0.7758 | 1.0307 | |
| 215161_at | CAMK1G | calcium/calmodulin-dependent protein kinase IG | −0.7819 | 0.6682 | 1.0285 | |
| 218204_s_at | FYCO1 | FYVE and coiled-coil domain containing 1 | 0.7250 | −0.7222 | 1.0233 | |
| 213222_at | PLCB1 | phospholipase C, beta 1 (phosphoinositide-specific) | −0.7694 | 0.6738 | 1.0227 | [719], [720], [721], [722], [723], [724] |
| 200925_at | COX6A1 | cytochrome c oxidase subunit VIa polypeptide 1 | −0.7532 | 0.6883 | 1.0204 | |
| 38037_at | HBEGF | heparin-binding EGF-like growth factor | 0.7222 | −0.7194 | 1.0193 | [725] |
| 209481_at | SNRK | SNF related kinase | −0.7048 | 0.7331 | 1.0169 | |
| 201412_at | LRP10 | low density lipoprotein receptor-related protein 10 | 0.6964 | −0.7399 | 1.0161 | |
| 202941_at | NDUFV2 | NADH dehydrogenase (ubiquinone) flavoprotein 2, 24kDa | −0.6984 | 0.7379 | 1.0160 | |
| 205383_s_at | ZBTB20 | zinc finger and BTB domain containing 20 | 0.6774 | −0.7569 | 1.0157 | [726], [727] |
| 206384_at | CACNG3 | calcium channel, voltage-dependent, gamma subunit 3 | −0.7778 | 0.6516 | 1.0147 | |
| 218888_s_at | NETO2 | neuropilin (NRP) and tolloid (TLL)-like 2 | −0.7899 | 0.6246 | 1.0070 | |
| 212214_at | OPA1 | optic atrophy 1 (autosomal dominant) | −0.7194 | 0.7024 | 1.0054 | [728], [729], [730], [731], [732], [733], [734], [735], [736], [737], [738], [739], [740], [741], [742] |
| 218456_at | CAPRIN2 | caprin family member 2 | −0.7915 | 0.6186 | 1.0046 | |
| 211307_s_at | FCAR | Fc fragment of IgA, receptor for | 0.7297 | −0.6886 | 1.0033 | |
| 202724_s_at | FOXO1 | forkhead box O1 | 0.6875 | −0.7270 | 1.0006 | [743], [744], [745] |
| 219145_at | LPHN1 | latrophilin 1 | −0.7293 | 0.6826 | 0.9989 | [746] |
| 205711_x_at | ATP5C1 | ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1 | −0.7153 | 0.6968 | 0.9986 | |
| 55662_at | C10orf76 | chromosome 10 open reading frame 76 | 0.7632 | −0.6420 | 0.9973 | |
| 211978_x_at | PPIA | peptidylprolyl isomerase A (cyclophilin A) | −0.7363 | 0.6726 | 0.9972 | [747], [748], [749], [750] |
| 210016_at | MYT1L | myelin transcription factor 1-like /// hypothetical protein LOC100134306 | −0.7577 | 0.6395 | 0.9915 | |
| 204072_s_at | FRY | furry homolog (Drosophila) | −0.7456 | 0.6512 | 0.9899 | |
| 219497_s_at | BCL11A | B-cell CLL/lymphoma 11A (zinc finger protein) | −0.6843 | 0.7117 | 0.9873 | |
| 201125_s_at | ITGB5 | integrin, beta 5 | 0.7323 | −0.6613 | 0.9867 | |
| 211765_x_at | PPIA | peptidylprolyl isomerase A (cyclophilin A) | −0.7230 | 0.6714 | 0.9866 | [747], [748], [749], [750] |
| 214057_at | MCL1 | Myeloid cell leukemia sequence 1 (BCL2-related) | 0.7722 | −0.6137 | 0.9864 | [751] |
| 211839_s_at | CSF1 | colony stimulating factor 1 (macrophage) | 0.8120 | −0.5590 | 0.9858 | [752], [753], [754], [755], [756], [757], [758], [759], [760], [761], [762], [763], [764], [765], [766], [767], [768], [769], [770], [771], [772], [773], [774], [775], [776], [777], [778], [779], [780], [781], [782] |
| 205551_at | SV2B | synaptic vesicle glycoprotein 2B | −0.6915 | 0.7008 | 0.9846 | [66] |
| 219167_at | RASL12 | RAS-like, family 12 | 0.6226 | −0.7605 | 0.9828 | |
| 214393_at | RND2 | Rho family GTPase 2 | 0.7051 | −0.6799 | 0.9796 | |
| 212899_at | CDC2L6 | cell division cycle 2-like 6 (CDK8-like) | 0.7319 | −0.6504 | 0.9791 | |
| 220615_s_at | MLSTD1 | male sterility domain containing 1 | −0.6665 | 0.7149 | 0.9774 | |
| 201681_s_at | DLG5 | discs, large homolog 5 (Drosophila) | 0.7093 | −0.6706 | 0.9761 | |
| 208195_at | TTN | titin | 0.7173 | −0.6617 | 0.9759 | [783] |
| 202457_s_at | PPP3CA | protein phosphatase 3 (formerly 2B), catalytic subunit, alpha isoform | −0.7661 | 0.6016 | 0.9741 | |
| 214150_x_at | ATP6V0E1 | ATPase, H+ transporting, lysosomal 9kDa, V0 subunit e1 | 0.6230 | −0.7488 | 0.9741 | |
| 204743_at | TAGLN3 | transgelin 3 | −0.6952 | 0.6802 | 0.9726 | |
| 213197_at | ASTN1 | astrotactin 1 | −0.7069 | 0.6673 | 0.9721 |
The column “Dist O” shows the Euclidean distance from the origin for each gene. If the gene has a known relation with AD (ADG), the reference's codes are display in column “Ref ADG”.
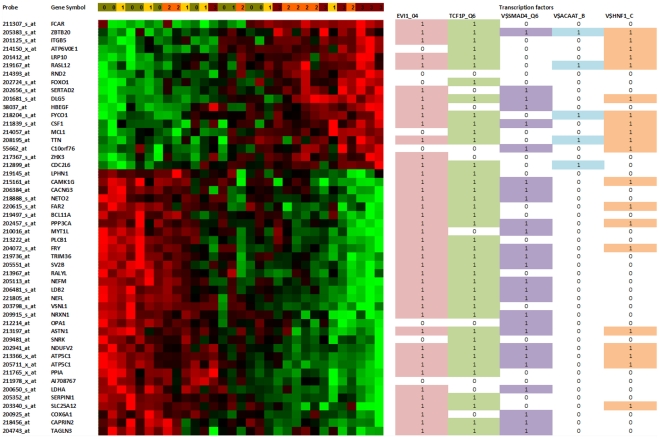
Figure 7 shows the heat map of the 50-probe signature, where the probes and patient samples are ordered by considering the similarity of their gene-expression values only. It can be observed that the Memetic Algorithm (MA), a high performance combinatorial optimization ordering method [12] for microarray datasets introduced in 2007, ordered most of the patients with or without an incipient level of AD on the left and the more severe cases on the right. When ordering the probes' gene expression, the MA perfectly sorted the groups previously described. We refer to [12], [13] for details of the MA. The supplementary material (File S2 ‘1372 norm. +heat map+GO’) presents the heat map of the 1,372 gene-probes, with samples and probes sorted by the MA.
Figure 7. Heat map of the 50-probe signature and the transcription factors with best p-values, for the whole set of 50 probes and for the two groups considered.
The samples and probes were sorted using the memetic algorithm given in [12], using the Euclidean distance. The transcription factors were obtained using Chang and Nevins' GATHER system to interpret genomic signatures [634]. The coloured cell and the number 1 indicate that the transcription factor has a binding motif with the gene for that row. The levels of severity as defined by Blalock et al. [635] are indicated in the first line: (0) Control, (1) Incipient AD, (2) Moderate AD and (3) Severe AD.
Transcription factors analysis of most correlated probes
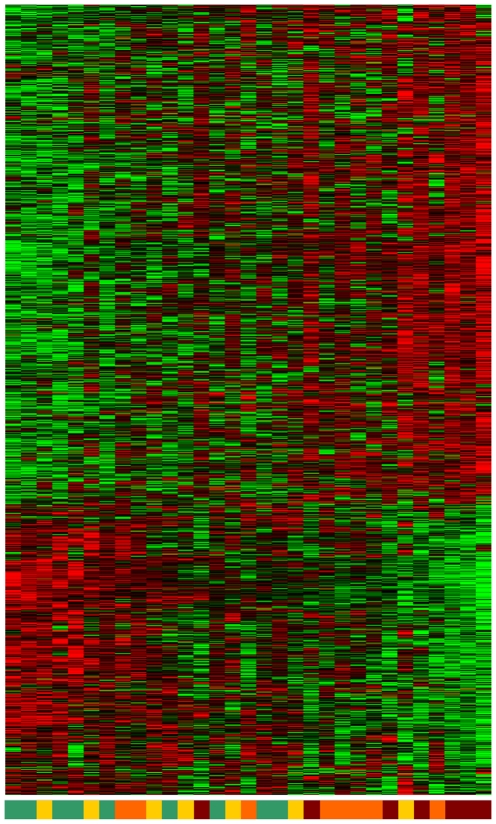
The signature of 50 probes we present in Figure 7 has 48 different genes (some probes are related to the same gene). The two repeated genes in this 50-probe list are ATP5C1 (ATP synthase, H+ transporting, mitochondrial F1 complex, gamma polypeptide 1) and PPIA (peptidylprolyl isomerase A (cyclophilin A)) [14], [15], [16], [17], a calcineurin regulatory protein. A recent study that used RT-PCR to examine tissue from 90 AD and 81 control human brains reports that cyclophilin is reduced in AD (both for females and males as compared with their gender-matched groups) [18]. We note here that the cutoff of 50 probes circumscribes the initial description a little, but most of the later discussion uses information from the whole signature to identify dysregulated pathways. Figure 8 presents the heat map of the 1,372-probe signature. The probes were sorted with the MA but the samples remain in the same position as obtained previously with the 50-probe signature.
Figure 8. Heat map of 1,372-probe signature.
The probes were sorted using the memetic algorithm but the samples remain in the same order than the 50-probe signature.
We analysed this list of genes using GATHER [19], an online tool for annotating signatures. Forty-one genes out of fifty have a motif for EVI1 (ecotropic viral integration site 1) and thirty-nine of them have a binding motif with V$TCF1P_Q6 (TCF1: transcription factor 1, hepatic; LF-B1, hepatic nuclear factor (HNF1), albumin proximal factor). The same analysis can be done if we divide the set of genes in two groups. The first group has positive correlation with the control profile and are overexpressed in AD; the second group has a positive correlation with the severe profile, and tend towards being underexpressed in AD (see Table 3). Table 4 presents the overrepresented motifs. We note, however, that we believe that the best results to identify putative overrepresented regulatory motifs can be obtained using the whole signature of 1,372 probes, and we will present the results of this investigation after presenting the case of the most correlated probes.
Table 4. Binding factors related to two groups of genes.
| Transcription Factors | Description | P value |
| First group | ||
| V$EVI1_04 | Ectopic viral integration site 1 encoded factor | 0.00069 |
| V$SMAD4_Q6 | SMAD family member 4 | 0.0033 |
| Second Group | ||
| V$HNF1_C | Hepatic nuclear factor 1 | 0.0022 |
| V$ACAAT_B | Avian C-type CCAAT box | 0.0015 |
The second group has the opposite behaviour, that is, positive correlation with the severe profile.
The first group has positive correlation with the control profile.
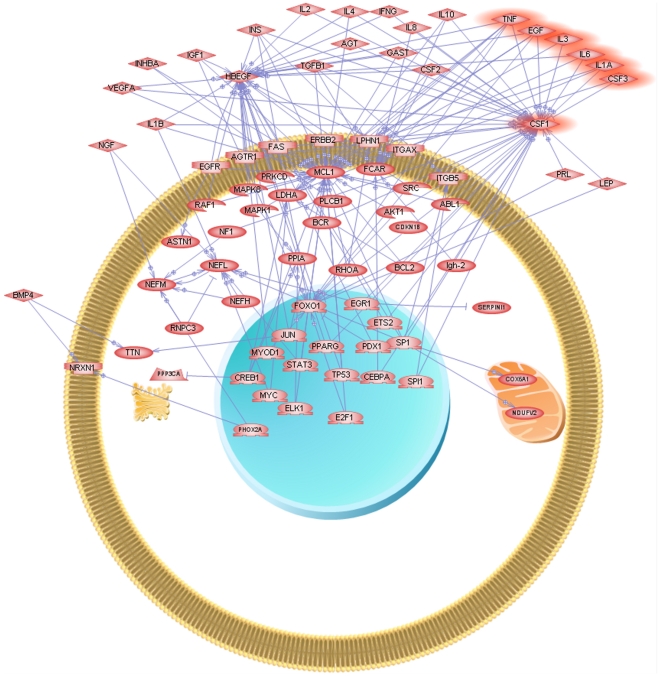
Another interesting pattern emerged when analysing the KEGG Pathways of the 50-probe signature using GATHER and PATHWAY Studio [20]. Using GATHER, three KEGG Pathways appear significantly represented, Amyotrophic lateral sclerosis (ALS), Oxidative phosphorylation and ATP synthesis. Using PATHWAY Studio, we automatically built the “common-regulators” diagram by selecting a filter that only considers protein interactions and binding. The resulting diagram is presented in Figure 9. As can be seen from the figure, we have chosen a circular membrane layout and our previously uncovered 5-protein signature [1] (IL1-a, TNF-a, IL-3, EGF and G-CSF) in plasma (plus IL-6) appears to have a strong relationship with CSF1 (colony stimulating factor 1 (macrophage)), the most positive correlated gene with the control profile (see Table 1). It is also worth mentioning, that CSF1 was found differentially expressed in blood of AD and Control subjects and belongs to the 18-protein signature uncovered by Ray et al. [2] in 2007.
Figure 9. ‘Common-regulators’ 50-probes’ signature.
The figure was obtained using Pathway Studio [569]. The program received as input the 50-probes displayed in Fig. 7 and automatically searched all the known putative common regulators relationships. The highlighted proteins are the 5-protein signature (IL1- α, TNF-α, IL-3, EGF and GCSF) of [1]. We have also highlighted IL-6 (discussed in [1] in the context of results of classifiers that also use it) and CSF1, Colony-stimulating factor 1, (macrophage).
Five of the 50 most correlated probes correspond to genes already mapped to KEGGs Alzheimer's disease Pathway KEGG:05010 and together with LDHA they link to impaired metabolism and the “novel glucocorticoid hypothesis”
We have observed that five genes, which are the most correlated probes with our putative signature for disease severity, can be mapped to the AD pathway of the Kyoto Encyclopaedia of Genes KEGG:05010. They are ATP5C1, COX6A1 [21], [22], NDUFV2 [23], [24], [25], [26], [27], [28], [29], [30], PLCB1[31], [32], [33], [34], and PPP3CA (protein phosphatase 3 (formerly 2B), catalytic subunit, alpha isoform), the last one also known as Calmodulin-dependent calcineurin A subunit alpha isoform. In all cases, the probes showed a reduction of expression with AD severity, which may indicate a sign of impaired mitochondrial functions and energy uptake [35], [36].
In addition to these five, we observed the reduced expression of the glycolytic enzyme LDHA, which may also indicate another challenge for energy metabolism in these neurons. Although glucose is generally considered to be the only substrate for brain energy metabolism, moncarboxylates have also been hypotheised as alternative substrates [37]. Laughton et al. report segregation in the hippocampus, with LDHA present in astrocytes and not in neurons. Instead, it is pyruvate dehydrogenase that is present in neurons but not in astrocytes and as a consequence of this study they support the argument that a metabolic compartmentalization exists in the human cortex and hippocampus where lactate produced by astrocytes could be oxidized by neurons [37]. We have also observed a reduction in expression of a probe that corresponds to PDHA1 (Pyruvate dehydrogenase (lipoamide) alpha 1, 200980_s_at) with increasing AD severity. The reduction of PDH expression, and the concurrent increase in pyruvate carboxylase gene expression, was discussed by Landfield et al. [38], who argue that: “These changes suggest that reduced pyruvate flux through PDH and decreased oxidative metabolism of glucose may develop early in AD. Interestingly, the inactivation of PDH is also a major pathway through which glucocorticoid activity acts to conserve glucose, and apparently, to induce insulin resistance [65], [66] . Thus, our data are consistent with the possibility that GC effects on this and other important target pathways in brain are enhanced in both aging and AD. If so, such alterations in glucocorticoid efficacy may have implications for AD pathogenesis as well as for the increased risk of AD associated with normal aging.” Our results seem to indicate that LDHA might also be discussed within the extended metabolic pathways that serve as the basic framework of this novel, more complex hypothesis [38], [39], [40], [41], [42], [43], [44], [45], [46], [47], [48], [49], [50], [51], [52], [53], [54], [55].
Four of the 50 most correlated gene probes are linked to synaptic function and neurofilament bundle assembly and also have reduced expressions with AD severity
NEFM, NRXN1, SV2B, and NEFL all have a similar pattern of reduced gene expression with AD severity. Experiments with mice depleted of the NEFL have been previously reported in the literature. Dubois et al state that this procedure: “mimics the reduced NFL mRNA levels seen in amyotrophic lateral sclerosis and causes perikaryal accumulation of neurofilament proteins and axonal hypotrophy in motoneurons. NFL−/− mice was evaluated for regional brain metabolism by means of quantitative histochemical estimation of cytochrome oxidase activity.” [56]. Mutations in the NEFL gene [56], [57], [58], [59], [60], [61], [62] and in the NEFM [63] have been linked to Charcot-Marie-Tooth disease. We will discuss the loss of expression of NRXN1 (Neurexin 1) later, when we comment on its presence in a panel of putative genes linked to prion-induced neurodegeneration [64]. However, we note here that both NRXN1 and NEFL appeared to be downregulated on a transcriptional profiling study of prion infection in mice [65].
The loss of expression of SV2B is also interesting. In 2001, Heese et al. [66] reported “a new transcript of SV2B (SV2Bb) mRNA that is up-regulated at mRNA level in neurons by amyloid beta peptide (Abeta) fragment (1–42). In comparison to SV2B this new mRNA encodes for the same protein but it has an elongated 3′-untranslated region (3′UTR) that contains several AU-rich (AUR) cis-acting elements which are probably involved in posttranscriptional regulating of SV2Bb translation. In conclusion, alteration of SV2B(b) expression appears to be involved in processes of neuronal degeneration” (see also [67]). We note that SV2B is only expressed in vesicles that undergo calcium-regulated exocytosis [68] and is a regulator of synaptotagmin 1 [69], which is a synaptic calcium sensor with a role in neurotransmitter release previously studied in AD [70], [71], [72], [73], [74], [75]. We present a number of genes related to synaptic function and neuronal plasticity which are increasingly down/up regulated later on the manuscript and on the supplementary material (File S3 Sheet ‘Synapse’).
Analysis of the 1,372-probe signature reveals alterations in calcium and insulin signalling
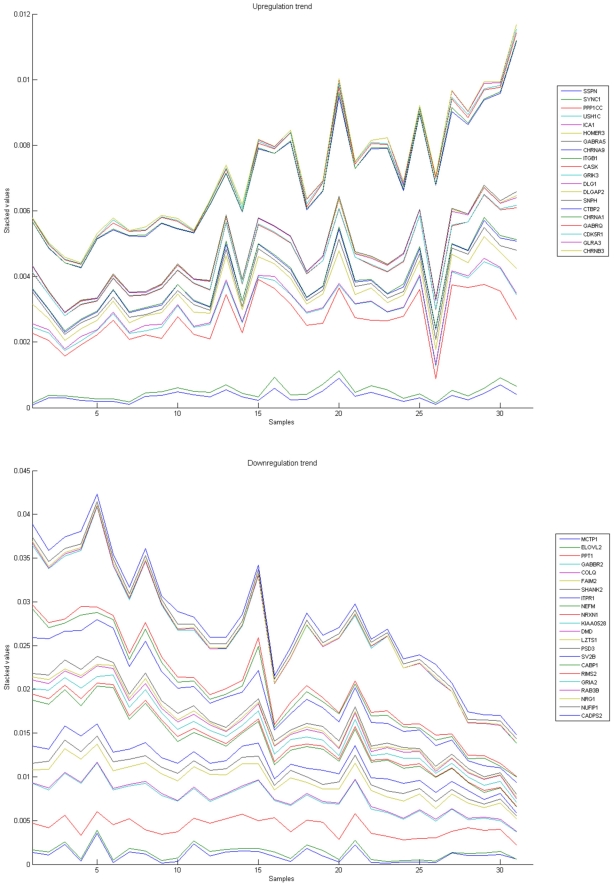
Using GATHER, we have identified 32 genes in the Calcium signalling pathway http://www.genome.jp/dbget-bin/show_pathway?hsa04020 (p-value<0.009). They are ADCY2, ADORA2B, AGTR1, ATP2A3, ATP2B1, ATP2B2, ATP2B4, AVPR1A, CALM1, CALM3, CREBBP, GNA14, GNAS, GRM5, HTR2A, ITPR1, ITPR2, LHCGR, NFATC1, PHKA2, PLCB1, PLCE1, PPP3CA, PPP3R1, PRKCB1, PTAFR, SLC25A6, SLC8A2, SYK, TBXA2R, TNNC2, and TTN. We cannot do enough justice in this manuscript to the several different hypotheses that point at imbalances/deregulation in calcium signalling and AD pathology. Instead, we contribute to these interesting discussions with our findings of genes related to this pathway within this group of 32 genes. The gene symbols in boldface can be mapped to the KEGG Pathway hsa04080, Neuroactive ligand-receptor interaction; those in italics can be mapped to KEGG Pathway hsa04310, Wnt Signalling. Being aware of the existing interest on Wnt Signalling and AD, we went back to the list of genes present in our (alpha,beta)-k-feature set signature and we identified others that can also be linked to Wnt signalling, like CSNK1G3, CSNK2A2, FRAT1[76], [77], [78], [79], [80], [81], [82], [83], [84], [85], [86], [87], [88], [89], FZD5[89], [90], [91], MDFIC, PIAS4, SOX2 [92], [93], [94], [95], [96], TCF7L1/TCF3[89], [97], [98], TCF7L2/TCF4[99], [100], [101], [102], [103], [104], [105], [106], and TLE3[106], [107], [108], [109].
In addition, most of the remaining 32 genes in the Calcium signalling pathway can be mapped to KEGG Pathway hsa04070, Phosphatidylinositol signalling system (CALM1, CALM3, ITPR1, ITPR2, PLCB1, PLCE1, PRKCB1), and Gap Junction (ADCY2, GNA14, GNAS, GRM5, HTR2A, ITPR1, ITPR2, PLCB1, PRKCB1).
This fact suggested that we should check how many genes were mapped to these pathways. We found that Phosphatidylinositol signalling system was indeed the third pathway with most “hits” in our signature, and also with other 12 genes (CDIPT, CSNK1G3 PIK3C3, PIK3R1, PIK3R4, PI4KB, PIP5K1A, PIP5K1C, PIP4K2C, PTEN, SKIP and TTK) which brings the total number to 19. We have also found (CCND3, CSNK1A1, CSNK2A2, CTBP1, CTBP2, FRAT1, FZD5, PPARD, PPP2CA, PPP2R2B, RBX1, SMAD3, TBL1X, TCF7L1, TCF7L2, VANGL1) bringing the total to 22 genes. We refer the reader to the supplementary material (File S3 Sheet ‘Phosphatidylinositol signalling’) for inspection of the individual pattern of expression of all these genes.
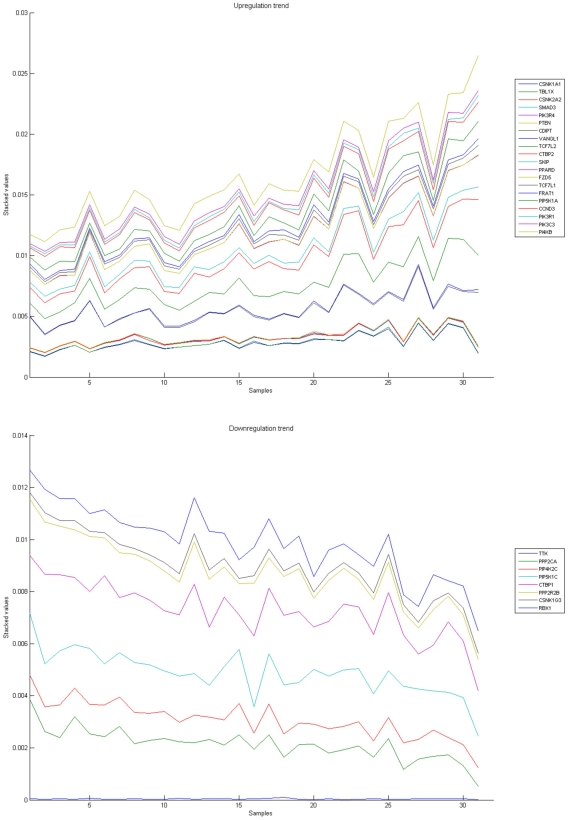
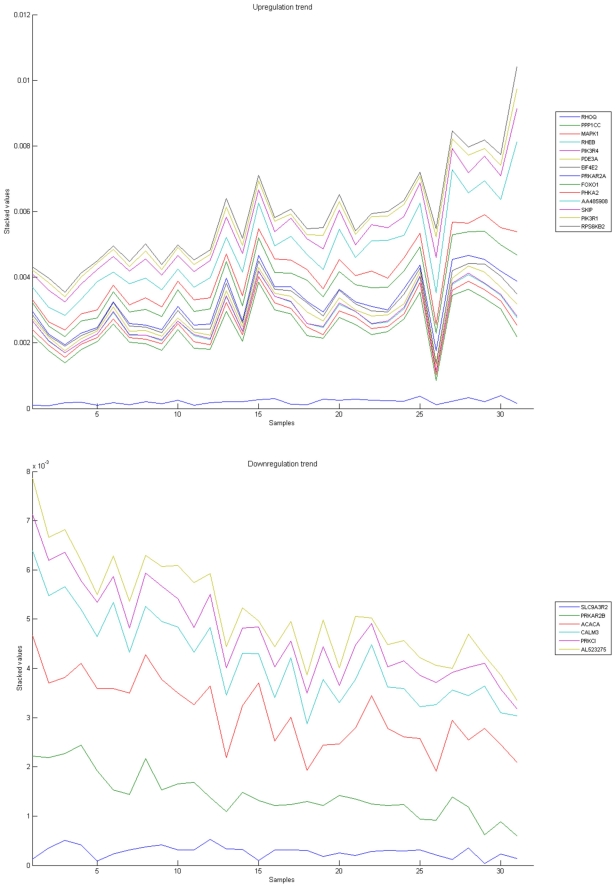
Together with the 20 genes mapped to the Insulin signalling pathway KEGG hsa04910 (ACACA, CALM1, CALM3, EIF4E2, FOXO1A, INSR [110], [111], [112], [113], [114], [115], [116], [117], MAPK1, PDE3A, PHKA2, PIK3R1, PIK3R4, PPP1CC, PRKAR2A, PRKAR2B, PRKCI, RHEB, RHOQ, RPS6KB2, SKIP, and TSC2), our results seem to give some support to the hypothesis of altered calcium dynamics [35], [118], [119], [120], [121], [122], [123], [124], [125], [126], [127], deregulation of insulin signalling [36], [41], [113], [114], [115], [116], [128], [129], [130], [131], [132], [133], [134], [135], [136], [137], [138], [139], [140], [141], [142], [143], [144], [145], [146], [147], [148], [149], [150], [151], [152], [153], [154], [155], [156], [157], [158], [159], [160], [161], [162], [163], [164], [165] and the implication of the Wnt pathway [166], [167], [168], [169], [170], [171], [172], [173], [174], [175], [176], [177], [178], [179], [180], [181], [182], [183], [184], [185], [186], [187], [188], [189], [190], [191], [192], [193], [194], [195], [196], [197], [198], [199] in AD pathogenesis.
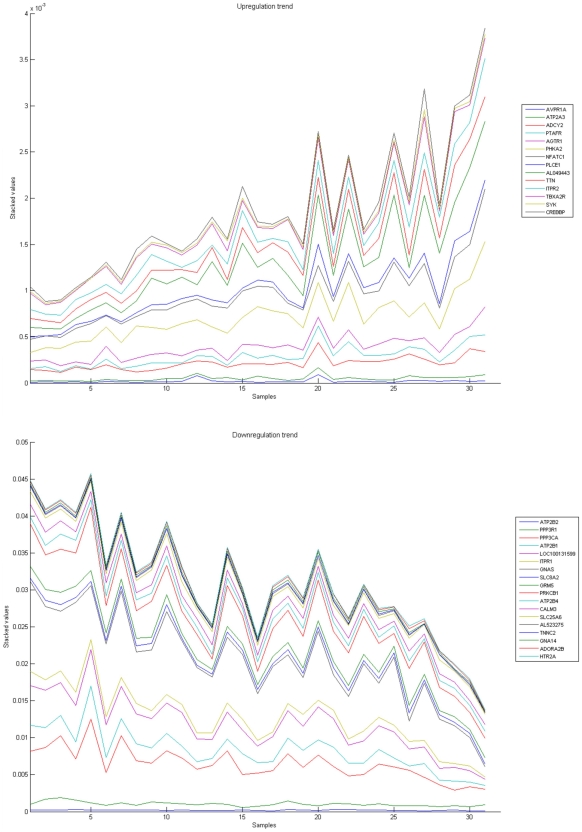
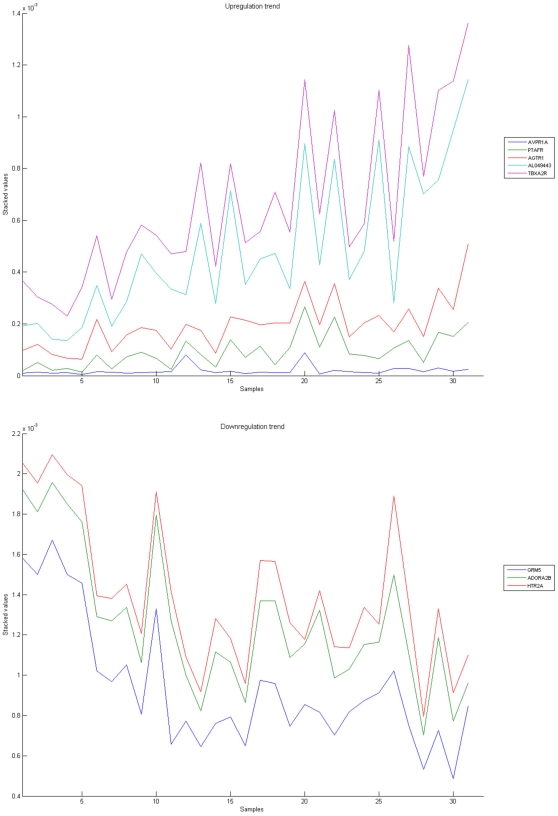
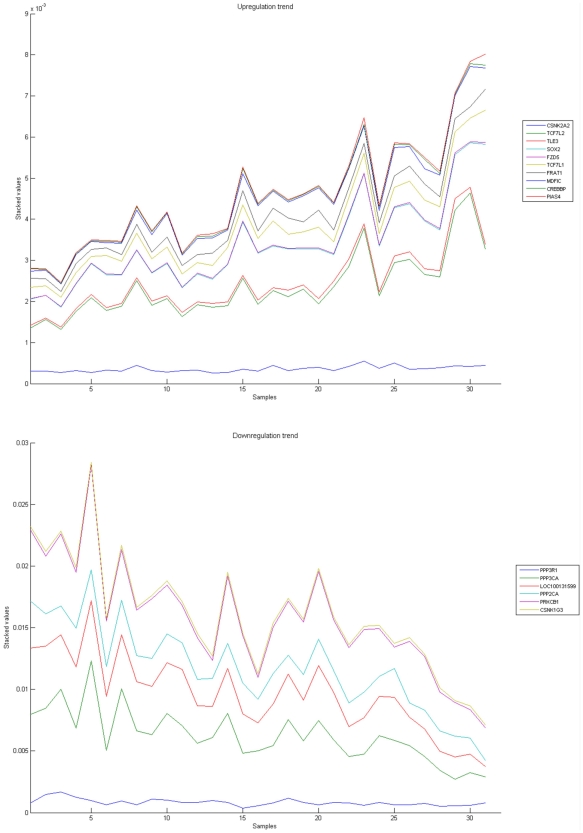
Figures 10, 11, 12, 13, and 14 illustrate down(up)-regulation of genes in these signalling pathways (Calcium signalling, Neuroactive ligand receptor pathway, WNT, Phosphatidylinositol and Insulin signalling, respectively). Figure 15 shows the expression of probes corresponding to genes for which there are known associations to synaptic function and neuronal plasticity. We refer the reader to the supplementary material (File S3) for more searchable information.
Figure 10. Calcium signaling pathway.
The upper graph presents the stacked normalized expression values of all the probes involved in the Calcium signaling with an upregulation trend. The lower graph analyses the genes involved in the pathway with a downregulation tendency. In the supplementary material (File S3 sheet ‘Calcium signalling pathway’), the reader will find all the individual gene expression values, normalised and not normalised.
Figure 11. Neuroactive ligand-receptor interaction pathway.
The upper graph presents the stacked normalized expression values of all the probes involved in the pathway with an upregulation trend. The lower graph analyses the genes involved in the pathway with a downregulation tendency. In the supplementary material (File S3 sheet ‘Neuroactive ligand-receptor’), the reader will find all the individual gene expression values, normalised and not normalised.
Figure 12. WNT signaling pathway.
The upper graph presents the stacked normalized expression values of all the probes involved in the pathway with an upregulation trend. The lower graph analyses the genes involved in the pathway with a downregulation tendency. In the supplementary material (File S3 sheet ‘Wnt Signalling’), the reader will find all the individual gene expression values, normalised and not normalised.
Figure 13. Phosphatidylinositol signaling pathway.
The upper graph presents the stacked normalized expression values of all the probes involved in the pathway with an upregulation trend. The lower graph analyses the genes involved in the pathway with a downregulation tendency. In the supplementary material (File S3 sheet ‘Phosphatidylinositol signalling’), the reader will find all the individual gene expression values, normalised and not normalised.
Figure 14. Insulin signaling pathway.
The upper graph presents the stacked normalized expression values of all the probes involved in the pathway with an upregulation trend. The lower graph analyses the genes involved in the pathway with a downregulation tendency. In the supplementary material (File S3 sheet ‘Insulin signalling’), the reader will find all the individual gene expression values, normalised and not normalised.
Figure 15. Genes related to synapse and neuronal plasticity.
The upper graph presents the stacked normalized expression values of all the related probes with an upregulation trend. The lower graph analyses the genes involved with a downregulation inclination. In the supplementary material (File S3, Sheet ‘Synapse’), the reader will find all the individual gene expression values, normalised and not normalised.
Transcription factors analysis of 1,372-probe signature reveals significant associations with the EGR/KROX family of proteins, MAZ, and E2F1
The analysis of the 1,372-probe signature indicates that they can be linked to putative transcription factors that have been previously implicated in AD and other neurodegenerative diseases. Using GATHER, we have observed that there is a strong association with motif V$KROX_Q6 (p-value<0.0004) with 719 out of 1294 genes in our signature; V$MAZ_Q6 (p-value<0.001, with 1003 genes); and V$E2F1_Q6_01 and V$E2F1_Q3_01 (with p-values which are smaller than 0.002 and 0.009 respectively). Of the 1294 genes associated with the 1,372 probes (by GATHER), more than half of them (656) have a motif for V$E2F1_Q6_01 and 603 have a motif for V$E2F1_Q3_01.
MAZ (MYC-associated zinc finger protein (purine-binding transcription factor)) , also known as ZF87 and Cys2His2-type zinc finger transcription factor serum amyloid A activating factor 1 [200], has been previously implicated in Alzheimer's disease [201] and as a blood biomarker in schizophrenia [202]. MAZ interacts with DCC, the receptor for netrin-1, a neuronal survival factor [203]. Deregulation of cyclin-dependent kinases and abnormal patterns of E2F1 regulation have also been linked with Alzheimer's disease [204], [205], [206], [207], [208], neurodegeneration [205], [207], [209], [210], [211], [212], [213], [214], [215], and neuronal apoptosis [216], [217], [218], [219], [220].
The involvement of the EGR/KROX (immediate early genes) family of proteins in the pathogenesis of Alzheimer's disease was first suggested in [221]. Studies of the behavioural consequences of stress have shown a link between the activation of the glucocorticoid receptor mediated response and EGR1, one of the members of this family [222]. It has been recently proposed that different members of the EGR/KROX family have different roles in learning and memory and cognitive functions [223], [224], [225], [226], [227], [228]. Mutant mice experiments showed that EGR1/KROX24 is required for the consolidation of long-term memory, while it is EGR3 the one linked to short-term memory [229], with EGR2 having perhaps other type of phenotypic characteristics not yet mapped [230]. In rat hippocampus, EGR1 decreases with aging [231]. In a recent study, it has been shown that initial playbacks of novel songs transiently increase EGR1 but that the observed response selectively habituates after repetition of the stimulus, with a different expression profile after one day [232] (see [233] and also [234] in which the homolog of NEFM, one of our biomarkers of reduced expression with increasing ‘AD severity’ called NF-M, is showed to be involved in the development and/or maturation of the oscine song control system).
We found the following connection between EGR/KROX, E2F1 and MAZ transcription factors that makes their concurrent finding notable. A recent study of microRNA signature of prion-induced neurodegeneration [64] has shown that EGR1, E2F1 and MAZ might be also implicated in the putative deregulation of immune response related genes by miRNAs via modulation of transcriptional regulators in scrapie-infected mice. We leave these findings for the next section of the manuscript where we will discuss them and present a list of common differentially expressed genes in these two neurodegenerative processes.
The 1,372-probe signature contains a significant number of genes differentially expressed that are linked to synaptic function and neuronal plasticity
The existence of several genes among the most correlated ones (NRXN1, SV2B, NEFM, etc.,) motivated us to try to identify which genes were present in the 1,372-probe signature that are also related to synaptic function and neuronal plasticity. We have identified 42 probes that can be divided into two groups, those that seem to be increasingly downregulated with AD severity (CABP1 [235], [236], [237], [238], [239], [240], [241], [242], [243], CADPS2 [244], [245], [246], [247], [248], [249], COLQ [250], DMD [251], [252], [253], [254], [255], [256], ELOVL2 [257], FAIM2/LFG [258], [259], [260], [261], GABBR2 [262], [263], [264], [265], GRIA2/GLUR2 [266], [267], [268], [269], [270], [271], [272], [273], [274], [275], [276], [277], ITPR1 [278], [279], [280], [281], [282], [283], KIAA0528, LZTS1/FEZ1 [284], [285], NEFM, NRG1, NRXN1, NUFIP1 [286], [287], [288], PPT1 [289], [290], [291], [292], [293], [294], [295], [296], [297], [298], [299], [300], [301], PSD3, RAB3B [302], [303], [304], [305], [306], [307], [308], [309], [310], [311], [312], [313], [314], [315], [316], [317], RIMS2 [318], [319], [320], [321], SHANK2 [322], [323], [324], [325], [326], [327], [328], [329], [330], [331], [332], [333], [334], [335], [336], [337], [338], [339], [340], SV2B [68], [69], [341], [342], [343], [344], [345], [346], [347], [348], [349], [350], [351], [352], [353], [354], [355], [356], [357], [358], [359]) and those that present an upregulation pattern (CASK [360], [361], [362], CDK5R1 [363], [364], [365], [366], [367], [368], [369], [370], [371], [372], [373], [374], [375], [376], [377], [378], [379], CHRNA1, CHRNA9, CHRNB3, CTBP2, DLG1/SAP97 [380], [381], [382], [383], [384], [385], [386], [387], [388], DLGAP2, GABRA5 [389], [390], [391], [392], [393], [394], GABRQ [395], GLRA3 [396], [397], [398], GRIK3/GLUR7 [399], HOMER3 [400], ICA1 [401], ITGB1 [402], [403], MCTP1 [404], [405], PPP1CC [406], SNPH [407], [408], [409], [410], [411], [412], [413], [414], SSPN [415], SYNC1, and USH1C [416], [417], [418]). The reader can consult the supplementary material (File S2) for the individual expression patterns of these genes. If, in agreement with Klemmer et al. [362], consider synapses as the most complex cellular organelle, with approximately 1500 proteins interacting in an activity dependent manner, we can argue that we must be inclusive with our list of references to help other researchers map the literature of their functions. Our aim is that experts can use this information to find ways of building novel testable hypotheses of AD neuronal plasticity impairment in the hippocampus. Our approach here has been to map what is currently known, and link it with the current biomedical literature, to facilitate experts that understand processes in detail.
We have already discussed some of the increasingly downregulated genes, another important candidate for further study is NRG1 (Neuregulin 1), a gene that has already been linked to several neuronal diseases. It is a candidate for susceptibility to schizophrenia and bipolar disorder (see [419], [420], [421], [422], [423], [424], [425], [426], [427], [428], [429], [430], [431], [432], [433], [434] and references therein). There have been reported links of NRG1 with AD. BACE1 (beta-Site APP-cleaving enzyme) is necessary for the cleavage of the amyloid-beta precursor protein, and BACE1 participates in the proteolytic processing of NRG1 [435], [436], and there exists some concerns about BACE1 inhibition as a potential therapeutic intervention due to its interaction with NRG1 and potential effects on remyelination [437]. In particular, NRG1 has been reported as a possible biomarker in cerebral spinal fluid, since its levels have been reported to be significantly increased in AD. Pankonin et al. suggest that: “While (NRG1) is not detected in human serum, a novel neuregulin antagonist activity was identified in human serum that could have prevented its detection. These results suggest that human neuregulin is selectively targeted from cortical neurons to white matter extracellular matrix where it exists in steady-state equilibrium with cerebral spinal fluid where it has the potential to serve as a biological marker in human neuronal disorders” [438]. NRG1 seems to collaborate with the ERBB4 receptor, and Li et al. propose that together they control glutamatergic synapse maturation and plasticity [439]. A single nucleotide polymorphism in NRG1 has also been associated as a risk factor to positive symptoms of psychosis in a proportion of late-onset AD [440]. With this evidence it is clear that NGR1 [439], [441], [442], [443], [444], [445], [446], [447] as well as the whole panel presented here are excellent candidates for further studies due to their well supported role in synaptic function in health and disease states.
Other biomarkers of interest
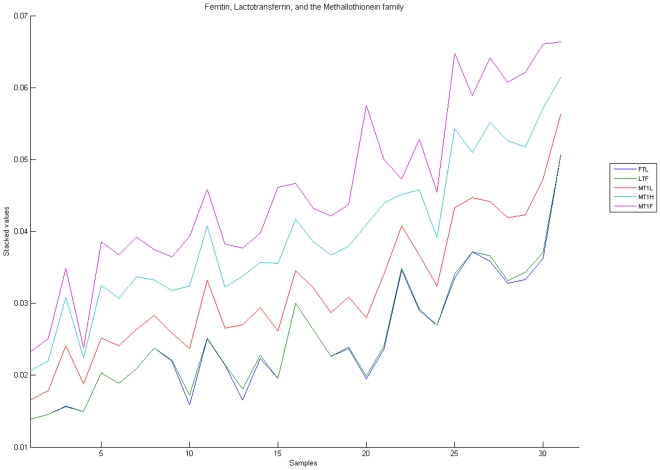
We should also mention some other biomarkers that could be interesting for further studies, including imaging purposes, like TSPO/PBR (translocator protein (18kDa)) also known as Mitochondrial Benzodiazepine Receptor (peripheral), thus supporting its current role as a putative imaging biomarker for AD [448], [449], [450], [451], [452], [453], [454], [455], C1S (complement component 1, s subcomponent) [456], [457], [458], [459], [460], [461], FDFT1 (the squalene synthase gene), which is critical for cholesterol synthesis [462], [463], BMP4 [92], [96], [464], [465], CD68 (as marker of enhanced lysosomal activity) [450], [466], [467], [468], [469], [470], [471], [472], SERTAD2/TRIP-Br2 [473], [474], [475], LTF (Lactotransferrin) [476], [477], [478], FTL (ferritin, light polypeptide; Ferritin light chain) [479], [480], [481], [482], MTF1 (Metal-regulatory transcription factor 1) [483], [484], [485], GSTA3 (Glutathione S-transferase A3), GSTM4 (Glutathione S-transferase M4), MT1L (Metallothionein 1L (gene/pseudogene) [486] (a human-specific truncated protein which may have changed its function or suppressed it [487]), MT1H (Metallothionein 1H) [488], MT1F (Metallothionein 1F) [488], [489] (Figure 16). These last three upregulated genes need to be put in concert with other reports on methallothioneins in AD brains [490], [491], [492]. Figure 16 shows the upregulation of Lactotransferrin, FTL (ferritin, light polypeptide; Ferritin light chain), and the Metallothionein family with increasing AD severity.
Figure 16. Metallothionein family.
Stacked line graph of the probes related to the Metallothionein family in the 1372-probe signature.
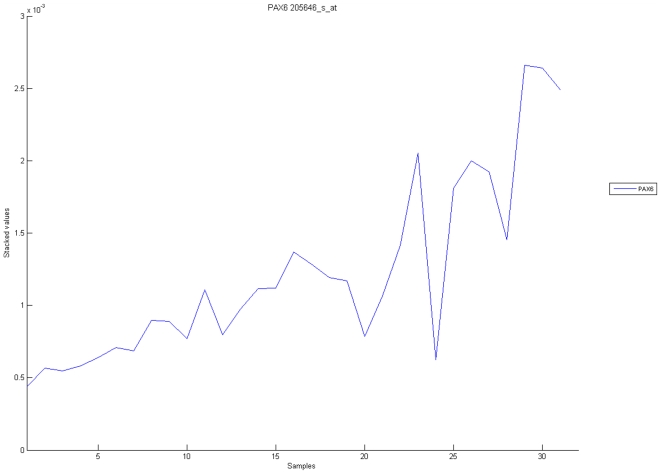
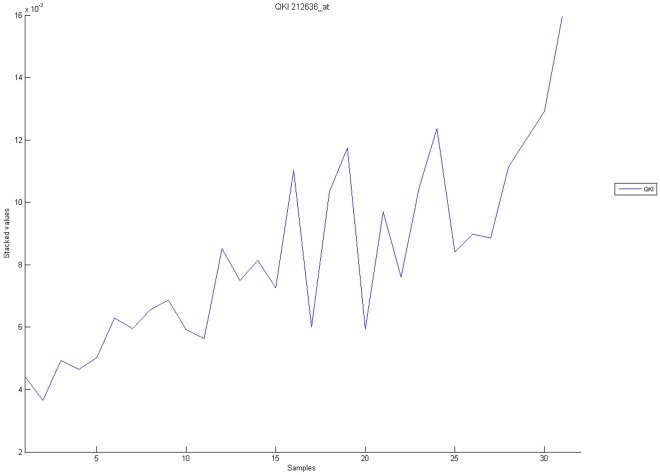
Other probes which present an upregulation trend that we would like to highlight are BCL2 [493], [494], FYCO1 [495], [496], PAX6 [111], [497], [498], [499] (Figure 17), and QKI [500] (Figure 18). The increase of expression of these probes, together with SOX2, is intriguing as they are related to differentiation from stem cells and are considered critical in neurogenesis [501], [502], [503], [504], [505], [506], [507], [508], [509], [510]. Our results support the combined use of them in tracking AD progression in this tissue. In addition, we have previously mentioned the relevance of EGR1 in coordinating a large number of genes that seem to be differentially expressed in this study. EGR1 also appears with a marked upregulation in severe AD patients (we refer to the supplementary material File S2 Sheet ‘1372 norm. +heat map+GO' for its gene expression profile). We found that this link is very important, as the homologues of EGR1, zif268, Egr-1 or ZENK, together with other members of the EGR family, are consolidating a key role in the neuronal plasticity in the brain [226], [230], [511], [512], [513], [514], [515], [516], [517], [518], [519], [520], [521], [522], [523], [524], [525], [526], [527], [528], [529], [530], [531], [532], [533], [534], [535], [536], [537], [538], [539], [540], [541], [542], [543], [544], [545], [546], [547], [548], [549] and links with AD and cognitive decline progression are starting to be reported [514], [515], [550], [551], [552], [553], [554].
Figure 17. Stacked line graph of the probe expression of Ferritin Light Chain, Lactotransferrin, and the Methallothionein family, in the 1,372-probe signature, that shows an increasing upregualtion with AD severity.
The expression of a PAX6 probe shows increasing upregualtion with AD severity.
Figure 18. The expression of a QKI probe, like PAX6, also shows increasing upregualtion with AD severity.
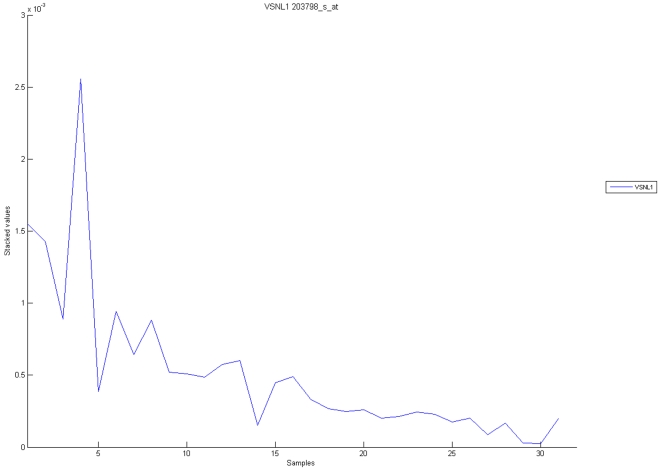
At the same time, prospective studies should encompass some other genes which appear downregulated with increasing AD severity. Top of the list is perhaps LDB2/CLIM1 (LIM domain binding 2), recently pointed as a marker (with LMO4 [555], [556]) of the control program of the development of neuronal subtype diversity of the cerebral cortex [557]. TRIM36 is another interesting candidate for further studies [558]. A gene that shares the same trend of dowregulation is CAMK1G (calcium/calmodulin-dependent protein kinase IG) [559], [560], [561], [562], [563], [564]. When analysing prefrontal cortical tissue from mice with inducible deletions of BDNF (Brain-derived neurotrofic factor), Glorioso et al. employed microarray gene expression profiling to show that there were alterations to early-immediate genes (including EGR1) and CAMK1G [563]. They conclude their manuscript stating that: “while altered BDNF expression may not represent the primary disturbance in AD, changed expression of, or altered responsiveness to BDNF (and subsequently decreased SST levels) may represent a critical feature of Alzheimer's disease progression.” VSNL1 (Visinin-like protein 1) [565], a CA++ sensor protein is also down-regulated (see Figure 19), a finding which is paralleled in the work of Youn et al. [566], who found similar changes in hippocampus.
Figure 19. The expression of a probe for VSNL1 (Visinin-like protein-1) shows increasing downregualtion with AD severity.
VSNL1, a neuronal calcium sensor that has received recent attention in AD [636], [637], [638], [639] has also been linked to model systems of schizophrenia, where it has been found upregulated in hippocampus [640]. A previous result by Schnurra et al. raised the possibility that the redution of VSNL1 expressing neurons indicate a selective vulnerabilty of these cells, since they observed that VSNL1 expression enhanced hyperphosphorylation of tau protein (in contrast with nontransfected or calbindin-D28K-transfected cells) [641]. In 2001, Braunewell et al. had already reported the reduction of VSNL1-immunoactive neurons in the temporal cortex of AD patients as compared with controls [642].
Discussion
Putative common genes involved in Alzheimer's disease and prion-induced neurodegenerative processes
In late 2008, a paper was published in PLoS ONE, shortly after the publication of our signature for prediction of clinical symptoms of AD [1] appeared online [64]. In this other contribution, Saba et al. present a microRNA signature of prion induced neurodegeneration [64]. By examination of the promoter regions of putative microRNA targets, they found that some transcription factor motifs were significantly enriched, E2F-1 (p-value = 6.01×10−14), KROX (p-value = 9.34×10−14), MAZ (p-value = 2.23×10−11) and PAX6 (p-value = 1.76×10−9). Our identification of EGR1/KROX-24 and PAX-6 as upregulated with AD progression, and the identification of motif V$KROX_Q6, V$MAZ_Q6, V$E2F1_Q6_01, V$E2F1_Q3_01 as enriched in our signature were two contributing factors that motivated us to explore any further similarities that we could find.
In [64], an analysis of the predicted target genes of their microRNA signature, linked with differentially expressed genes in scrapie-infected mice [65] as well as two other publications [567], [568], led Saba et al. [64] to identify a network of de-regulated immune response-related genes. Additionally, they identified the putative transcription regulator genes that are targets of miRNAs similarly de-regulated. In essence, a possible hierarchy of deregulations of microRNAs, which, deregulated transcription factors that then, modify 1282 target genes. A Gene Ontology analysis also indicated that the “data sets were found to be in the significant enrichment for genes involved in cell death, regulation of the cell cycle, nervous system development and function and cell signalling pathways.”
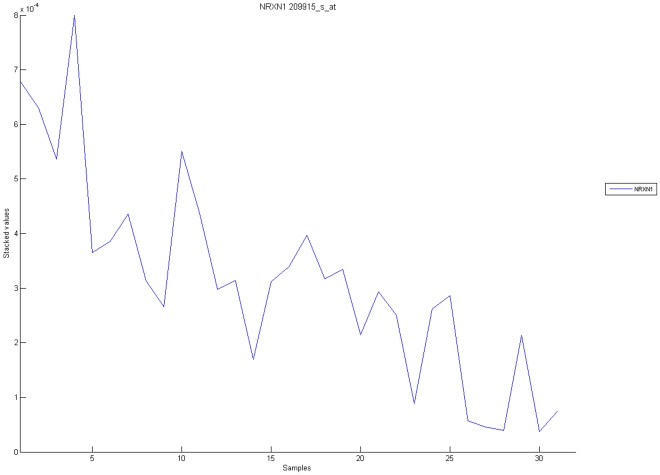
As a consequence, we have investigated if some of the 1,282 putative target genes of the miRNA signature of prion induced neurodegeneration also appear in our lists. Of those 1,282 genes we immediately noticed that there were 9 genes listed in our list of the 50 most correlated genes (Table 3). These genes are BCL11A, CSF1, DLG5, FOXO1, HBEGF, NRXN1, SERTAD2, SNRK and ZBTB20. Two of these genes, CSF1 (colony stimulating factor 1 (macrophage)) and HBEGF (heparin-binding EGF-like growth factor) appear to be conspicuous mediators of cytokine and growth factor signalling as Figure 9 illustrates (we obtained this network using Pathway Studio [569] as described in the previous section), and CSF1 and HBEGF seems to be increasing with AD severity. In opposition, the probe corresponding to NRXN1 (Neurexin 1, 209915_s_at) has decreasing expression (Figure 20). Although no connection has been found between NRXN1 and AD yet, this gene has been implicated in autism [570], [571], [572], [573], [574], [575], [576], schizophrenia [577], [578], [579], [580], [581], nicotine and alcoholism dependence [582], [583], [584], and mental retardation [585]. SERTAD2 (SERTA domain containing 2), mentioned in the previous section, is also known as Transcriptional regulator interacting with the PHD-bromodomain 2, TRIP-Br2, a member of the TRIP-Br family of transcriptional regulators, required for the transduction of mitogenic signals and the execution of serum-inducible E2F-mediated cell cycle progression [473]. In our data, the probe for SERTAD2 is increasing with AD severity. It has also been reported that overexpression of SERTAD2 is sufficient to transform murine fibroblasts and promotes tumorigenesis in athymic nude mice due to the deregulation of the E2F/DP-transcriptional pathway thanks to the upregulation of the key E2F-responsive genes [474]. FOXO1 (Forkhead box O1) also appears upregulated with increasing AD severity, and has been reported as a negative regulator of EGR1 expression via the activation of the PI3K/Akt/Forkhead pathway [586]. The expression of FOXO1 is also induced by E2F1 [587]. The product of this gene has also been reported as a survival factor in deprivation-induced neuronal cell death [588], [589] (see also the review in [590]). Although FOXO1 has not been previously implicated in AD, an exception may exist. van Der Heide et al. describe in [591] how the Forkhead transcription factors are involved in insulin signalling. The “PI3K route” is a name given to common signal transduction cascade that links neuronal survival, synaptic plasticity (and, as a consequence, learning and memory) [592]. This “PI3K-Akt-FOXO1 mechanism” and its role in neurons warrant the current intensive investigation [593], [594], [595], [596], [597], [598], [599], [600]. From this group of 9 genes, seven of them (NRX1, SERTAD2, SNRK, HBGEF, FOXO1, CSF1, BCL11A) and QKI have been predicted to be targeted by mmu-mir128 by two or more microRNA prediction tools. We found this to be a connection that is worth exploring. Lukiw and Pogue have reported that following metal-induced reactive oxygen species production (by iron and aluminium-sulfate at nanomolar concentrations) upregulates miR-128 in human neural cells in primary culture [601]. They also report that, together with miR-9, mir-125a, mir-128 is upregulated in AD brain. In the previously cited reference Lukiw reported that: “miR-9, miR-124a, miR-125b, miR-128, miR-132 and miR-219 are abundantly represented in fetal hippocampus, are differentially regulated in aged brain, and an alteration in specific micro-RNA complexity occurs in Alzheimer hippocampus.”
Figure 20. It is possible to observe that one of the probes for NRXN1 (Neurexin 1, 209915_s_at) has decreasing expression with increasing AD severity.
We have found no previous evidence of a connection of NRXN1 and AD, but this gene has been previously implicated in autism [570], [571], [572], [573], [574], [575], [576], schizophrenia [577], [578], [579], [580], [581], nicotine and alcoholism dependence [582], [583], [584], and mental retardation [585].
The expression of probes corresponding to PP2A and PP2B catalytic subunits (i.e. PPP2CA, Protein phosphatase 2 (formerly 2A), catalytic subunit, alpha isoform, and PPP3CA, Protein phosphatase 3 (formerly 2B), catalytic subunit, alpha isoform, Calcineurin A1) shows increasing downregualtion with the progression of AD., see Figure 21. This finding supports a role for downregulation of PPP2CA, PPP3CA in AD pathology [619]–[647].
Figure 21. The expression of two probes for PPP2CA (Protein phosphatase 2 (formerly 2A), catalytic subunit, alpha isoform,) and PPP3CA (Protein phosphatase 3 (formerly 2B), catalytic subunit, alpha isoform, Calcineurin A1) show increasing downregualtion with AD severity.
A similar plot exists for PPP3R1 (protein phosphatase 3 (formerly 2B), regulatory subunit B, alpha isoform, Calcineurin subunit B type 1). This result supports a role for downregulation of PPP2CA, PPP3CA in AD pathology [643], [644], [645], [646], [647], [648], [649], [650], [651], [652], [653], [654], [655], [656], [657], [658], [659], [660], [661], [662], [663], [664], [665], [666], [667], [668], [669], [670], [671].
Finally, in addition to the presence of hyperphosphorylated tau, the accumulation of Amyloid-beta (Abeta) peptide in brain tissue is a hallmark of AD [602]. The identification of the genes involved in the proteolytic processing of APP (beta-amyloid precursor protein), which in turn produces Abeta, is a subject of intense research. Researchers are currently looking at the alterations of APP cellular localization and endocytic trafficking as one mechanism that can modify the processing of APP to Abeta. LRPs are known to regulate APP's endocytic trafficking [603], [604], [605], [606], and seem to be a hub of a number of mounting evidences on processes that link to cholesterol metabolism and atherosclerosis [607]. In our selected panel of 50 proteins we have one member of this family, LRP10 (low density lipoprotein receptor-related protein 10), as one of the most correlated gene expression profiles. In our list of 1372 gene probe signature we also have another member of this family, LRP1B (low density lipoprotein-related protein 1B (deleted in tumors))[608], While LRP10 appears to be positively upregulated with cognitive decline an inverse relationship is observed for LRP1B.
LRPs are also known to linked to APP via a mechanism that involves the alternative splicing of APBB3/Fe65L2 [609], [610], [611]. Tanahashi and Tabira have proposed that the splicing of APBB3/Fe65L2 alters the ability to bind with APP and low-density-lipoprotein-receptor-related protein. They propose that the secretion of beta-amyloid peptide Abeta40 and Abeta42 is increased following the overexpression of APBB3, but there are no visible changes of half-life and maturation of APP, or the secretion of secreted APP [612]. In our dataset, we observe APBB3 expression being upregulated with the increasing cognitive decline, following the same pattern of LRP10.
Polymorphisms on these genes have previously been linked to AD. Tanahashi, Asada and Tabira have reported an association between a polymorphism in APBB3/Fe65L2 and early-onset AD [612] (the link between APBB3 and AD is being increasingly explored, we refer to [613], [614], [615], [616] for further references). Using 500K SNP microarray technology, Poduslo, Huang and Spiro have identified haplotypes in LRP1B as significant for successful aging without cognitive decline in a study involving individuals that were 85 years old or older, had MMSE scores greater than 26, no history of dementia in their families, and no major illnesses (i.e. no cardiovascular problems, diabetes, obesity, or major cancer diseases) and most of them had normal cholesterol levels. Their genome-wide association screening compared these individuals with those that have late-onset AD [617]. Poduslo et al. have suggested that if the decreased production of Abeta42 in successful aging is due to the haplotypes they describe, then LRP1B may be a new target for treatment of AD [608], [617], Taken together these results indicate that integrative bioinformatics analytic approaches will be needed to elicit the interactome of LRPs and their role in AD.
Conclusions
This re-analysis of the microarray dataset hippocampal gene expression contributed by Blalock et al. has shown that there exist a relatively large number of probes (1,372) that present a clear pattern of either up or down regulation with increasing AD severity. The signature reveals alterations in calcium, insulin, phosphatidylinositol and Wnt-signalling. Among the group of most correlated gene probes with AD severity we found some linked to synaptic function, neurofilament bundle assembly, neuronal plasticity and inflammation.
A transcription factors analysis of 1,372-probe gene expression signature reveals significant associations with the EGR/KROX family of proteins, MAZ, and E2F1. The gene homologous of EGR1, zif268, Egr-1 or ZENK, together with other members of the EGR family, are consolidating as key players in short and long-term memory and neuronal plasticity in the brain. We have also uncovered a large consensus of this gene expression signature with current genes putatively involved in AD progression. Our results also indicate a degree of commonality between putative genes involved in AD and prion-induced neurodegenerative processes that warrants further investigation.
Materials and Methods
Dataset
In this contribution, we have used a MIAME compliant, Affymetrix gene expression dataset that is public available and was contributed by Blalock et al [3] in 2004. We thank the authors of that publication for making this useful dataset available to the research community at large allowing further exploration and reanalysis.
The dataset is available from GEO Dataset Browser, accession number GDS1297 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE1297). The Affymetrix human GeneChip, HG-U133A, containing 22,283 targets was used. The dataset is de-identified and the methods for disease classification, based on MMSE and NFT scores, are described in full detail by Blalock et al. in Ref. [3].
The hippocampal samples used by Blalock et al. were obtained from the autopsy of 31 subjects through the Brain Bank of the University of Kentucky Alzheimer's Disease Research Center (ADRC), Sanders-Brown Center on Aging, University of Kentucky. The ADRC was established in 1985 and in operation since 1989 a pool of research volunteers that have agreed in principle to be research participants. Participants were asked questions based on NINCDS/ADRDA criteria [618] to establish their physical and mental condition to determine if their were eligible for the study. When a mutual agreement existed, the individuals were visted in their homes to review and sign the informed-consent document (which was approved by the University of Kentucky Institutional Review Board). Participants also signed a donor card, and the visit also aimed to establish their baseline mental-status testing. Elegibility for the purpose of the study included having a Mini-Mental State Exam score above 24 [619], passing a series of cognitive tests, and a previous history of absence of neurological disease [620], as well as neither substance abuse nor major psychiatric illnesses. All eligible volunteers were 60 years of age or older and satisfactorily performed normal activities of daily living. The Wechsler Adult Intelligence Scale (Vocabulary) was also applied to exclude significant other medical diseases that could affect cognition and elegible participants must had no previous history of head injury with loss of consciousness.
The research participants that were deemed eligible also signed a form (in addition to the consent document) indicating their agreement to donate their brain to the Sanders-Brown Center on Aging. A full description of the methods used can be found in Brain Donation in Normal Aging Procedures, Motivations, and Donor Characteristics from the Biologically Resilient Adults in Neurological Studies (BRAiNS) Project [621].
Blalock et al. [3] categorized the samples in four groups, with a labelling that indicates different “levels of severity”. These labels were decided based on the MiniMental State Examination (MMSE) and the Neurofibrillary Tangle count (NFT) of each sample [622]. Samples are then separated in the types ‘Control’, ‘Incipient AD’, ‘Moderate AD’ and ‘Severe AD’. Table 1 of Blalock et al. shows the mean values of MMSE and NFT for each one of these groups. In addition, they give the mean Braak stage [623], [624], [625] for each one of the groups (2.1 for ‘Control’, 5 for ‘Incipient’, 5.6 for ‘Moderate’ and 5.9 for ‘Severe’). We are grateful to Dr. Blalock who has kindly given us these values of the Braak stage for each sample in the dataset. Together with the individual values of MMSE, NFT, the Braak stage of each sample is included in the Supplementary Material (File S2 sheet ‘Braak’) section of this publication.
Methodology
Our analysis method consisted of four steps: abundance quantization and filtering of probes; a feature selection algorithm to refine the probe selection; a Jensen-Shannon divergence computation; and finally, a correlation analysis. Each of these steps is described below.
As mentioned in the Results section, we only used the samples labelled as “Control” or “Severe AD” for feature selection, thus we have a two-class probe/gene selection task. We did not use the samples labelled as “Incipient AD” or “Moderate AD” for the probe selection steps. Those samples were only used in the final step, at the time of computing the correlation of the gene profile, across all samples, with the Jensen-Shannon divergences computed for the “Control” and “Severe” classes as explained later in this section.
For the first step, the quantization of the expression values, as well as for the initial data pruning, we used Fayyad and Irani's algorithm [626]. The heuristic algorithm minimises the feature-class entropy and discards genes according to the Minimum Description Length principle. The application of Fayyad and Irani's algorithm not only filters several thousand genes, it also provides thresholds for each probe remaining in the dataset. These quantized values of gene expression leave us with an instance of a combinatorial optimization problem, the (α, β)-k-Feature Set problem [13], [627], [628].
The (α, β)-k-Feature Set problem is a combinatorial optimisation problem introduced by Cotta, Sloper and Moscato[628] in 2004 to address the problem of feature selection in high-dimensional datasets. We solve an instance of this problem numerically using an integer programming formulation. This approach has been previously employed to obtain molecular biomarker signatures in Alzheimer's Disease [1], [629], models of Parkinson disease [630], prostate cancer [631], electrode selection in EEGs [632], and elsewhere. To obtain mathematically proven optimal solutions of the integer programming formulation, the CPLEX commercial optimization solver was used. As in previous contributions of our group, we found gene expression signatures corresponding to values of α maximum and β maximal [1], [13], [627], [628], [633]. We refer the reader to these previous contributions for a detailed explanation of the methodology.
At this point, we have a selection of 1,372 probes, a set which we denote as Ω. For each sample m and probe  , let fim be its expression value. We now define a probability distribution function (PDF) for each sample. For sample, m its PDF
, let fim be its expression value. We now define a probability distribution function (PDF) for each sample. For sample, m its PDF 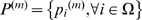 , is given by
, is given by
We can now compute an average PDF profile for samples in the “Control” and “Severe AD” groups, denoted by 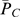 and
and  respectively. Let C and S be the set of samples with the labels “Control” and “Severe AD” respectively. The average profile
respectively. Let C and S be the set of samples with the labels “Control” and “Severe AD” respectively. The average profile 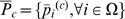 , is then:
, is then:
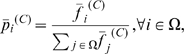 |
where
where  represents the number of samples in class C.
represents the number of samples in class C.  is analogously defined.
is analogously defined.
The Jensen-Shannon divergence between two sample PDFs, i.e. samples l and k (P(l) and P(k)) is defined as
where
S[P] is the Shannon Entropy for a specific PDF sample with N states. It is well known that the square root of the JSD (sqrtJSD) is a metric, which means that for a given set of PDFs the following four properties are satisfied:
Once the sqrtJSD between each patient and the two average profiles ( and
and 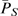 ) has been computed, the genes most correlated with these metrics can be uncovered. We used the Spearman rank correlation, which is a well-known non-parametric method, and can thus be used even when the data does not satisfy assumptions about normality, homoscedasticity and linearity.
) has been computed, the genes most correlated with these metrics can be uncovered. We used the Spearman rank correlation, which is a well-known non-parametric method, and can thus be used even when the data does not satisfy assumptions about normality, homoscedasticity and linearity.
Supplementary Material
Supplementary ‘File S1’ provides a glossary of each gene referenced in this paper including synoms and refrences to iHOP (http://www.ihop-net.org/).
The results referenced in this manuscript are provided in supplementary ‘File S2’ and ‘File S3’ in Microsoft Excel format.
Supporting Information
IHop Glossary of Genes.
(0.15 MB DOC)
Supplementary Data 1.
(4.10 MB XLS)
Supplementary Data 2.
(1.26 MB XLS)
Acknowledgments
The authors wish to thank the late Dr. William Markesbry, Dr. Erik Blalock and through them the whole team of the University of Kentucky's Sanders-Brown Center on Aging who have contributed a very valuable dataset.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: The authors acknowledge partial support from the Australian Research Council (ARC) Centre for Bioinformatics, Australia. OAR acknowledges partial support from the Consejo Nacional de Investigaciones Cientificias y Tecnicas (CONICET), Argentina. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Gomez Ravetti M, Moscato P. Identification of a 5-Protein Biomarker Molecular Signature for Predicting Alzheimer's Disease. PLoS One. 2008;3:e3111. doi: 10.1371/journal.pone.0003111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ray S, Britschgi M, Herbert C, Takeda-Uchimura Y, Boxer A, et al. Classification and prediction of clinical Alzheimer's diagnosis based on plasma signaling proteins. Nat Med. 2007;13:1359–1362. doi: 10.1038/nm1653. [DOI] [PubMed] [Google Scholar]
- 3.Blalock EM, Geddes JW, Chen KC, Porter NM, Markesbery WR, et al. Incipient Alzheimer's disease: Microarray correlation analyses reveal major transcriptional and tumor suppressor responses. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:2173–2178. doi: 10.1073/pnas.0308512100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmitt FA, Davis DG, Wekstein DR, Smith CD, Ashford JW, et al. “Preclinical” AD revisited: Neuropathology of cognitively normal older adults. Neurology. 2000;55:370–376. doi: 10.1212/wnl.55.3.370. [DOI] [PubMed] [Google Scholar]
- 5.Haroutunian V, Purohit DP, Perl DP, Marin D, Khan K, et al. Neurofibrillary Tangles in Nondemented Elderly Subjects and Mild Alzheimer Disease. Arch Neurol. 1999;56:713–718. doi: 10.1001/archneur.56.6.713. [DOI] [PubMed] [Google Scholar]
- 6.Price DL, Sisodia SS. Mutant genes in familial Alzheimer's disease and transgenic models. Annual Review of Neuroscience. 1998;21:479–505. doi: 10.1146/annurev.neuro.21.1.479. [DOI] [PubMed] [Google Scholar]
- 7.Price JL. The relationship between tangle and plaque formation during healthy aging and mild dementia. Neurobiology of Aging. 1993;14:661–663. doi: 10.1016/0197-4580(93)90062-g. [DOI] [PubMed] [Google Scholar]
- 8.Price JL, Davis PB, Morris JC, White DL. The distribution of tangles, plaques and related immunohistochemical markers in healthy aging and Alzheimer's disease. Neurobiology of Aging. 1991;12:295–312. doi: 10.1016/0197-4580(91)90006-6. [DOI] [PubMed] [Google Scholar]
- 9.Robbins K, Sandeep J, Zhang W, Rekaya R. Classification of incipient Alzheimer patients using gene expression data: Dealing with potential misdiagnosis. Online Journal of Bioinformatics. 2006;7:9. [Google Scholar]
- 10.Sandeep J, Robbins K, Zhang W, Rekaya R. Effects of Misdiagnosis in Input Data on the Identification of Differential Expression Genes in Incipient Alzheimer Patients. In Silico Biology. 2008;8:9. [PubMed] [Google Scholar]
- 11.Grosse I, Bernaola-Galvan P, Carpena P, Roman-Roldan R, Oliver J, et al. Analysis of symbolic sequences using the Jensen-Shannon divergence. Phys Rev E Stat Nonlin Soft Matter Phys. 2002;65:041905. doi: 10.1103/PhysRevE.65.041905. [DOI] [PubMed] [Google Scholar]
- 12.Moscato P, Mendes A, Berretta R. Benchmarking a memetic algorithm for ordering microarray data. Biosystems. 2007;88:56–75. doi: 10.1016/j.biosystems.2006.04.005. [DOI] [PubMed] [Google Scholar]
- 13.Berretta R, Costa W, Moscato P. Combinatorial Optimization Models for Finding Genetic Signatures from Gene Expression Datasets. In: Keith JM, editor. Bioinformatics. Humana Press; 2008. pp. 363–377. [DOI] [PubMed] [Google Scholar]
- 14.Gubern C, Hurtado O, Rodriguez R, Morales JR, Romera VG, et al. Validation of housekeeping genes for quantitative real-time PCR in in-vivo and in-vitro models of cerebral ischaemia. BMC Mol Biol. 2009;10:57. doi: 10.1186/1471-2199-10-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Santos AR, Duarte CB. Validation of internal control genes for expression studies: effects of the neurotrophin BDNF on hippocampal neurons. J Neurosci Res. 2008;86:3684–3692. doi: 10.1002/jnr.21796. [DOI] [PubMed] [Google Scholar]
- 16.Johansson S, Fuchs A, Okvist A, Karimi M, Harper C, et al. Validation of endogenous controls for quantitative gene expression analysis: application on brain cortices of human chronic alcoholics. Brain Res. 2007;1132:20–28. doi: 10.1016/j.brainres.2006.11.026. [DOI] [PubMed] [Google Scholar]
- 17.Hughes V, Smith S, Garcia-Sanchez A, Sales J, Stevenson K. Proteomic comparison of Mycobacterium avium subspecies paratuberculosis grown in vitro and isolated from clinical cases of ovine paratuberculosis. Microbiology. 2007;153:196–205. doi: 10.1099/mic.0.29129-0. [DOI] [PubMed] [Google Scholar]
- 18.Preece P, Cairns NJ. Quantifying mRNA in postmortem human brain: influence of gender, age at death, postmortem interval, brain pH, agonal state and inter-lobe mRNA variance. Brain Res Mol Brain Res. 2003;118:60–71. doi: 10.1016/s0169-328x(03)00337-1. [DOI] [PubMed] [Google Scholar]
- 19.Chang JT, Nevins JR. GATHER: a systems approach to interpreting genomic signatures. Bioinformatics. 2006;22:2926–2933. doi: 10.1093/bioinformatics/btl483. [DOI] [PubMed] [Google Scholar]
- 20.Ariadne Genomics I. 2007. Pathway Studio™. 5.0 ed.
- 21.Ongwijitwat S, Wong-Riley MT. Functional analysis of the rat cytochrome c oxidase subunit 6A1 promoter in primary neurons. Gene. 2004;337:163–171. doi: 10.1016/j.gene.2004.04.024. [DOI] [PubMed] [Google Scholar]
- 22.Wong-Riley M, Guo A, Bachman NJ, Lomax MI. Human COX6A1 gene: promoter analysis, cDNA isolation and expression in the monkey brain. Gene. 2000;247:63–75. doi: 10.1016/s0378-1119(00)00121-9. [DOI] [PubMed] [Google Scholar]
- 23.Ben-Shachar D, Karry R. Neuroanatomical pattern of mitochondrial complex I pathology varies between schizophrenia, bipolar disorder and major depression. PLoS One. 2008;3:e3676. doi: 10.1371/journal.pone.0003676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ji B, La Y, Gao L, Zhu H, Tian N, et al. A Comparative Proteomics Analysis of Rat Mitochondria from the Cerebral Cortex and Hippocampus in Response to Antipsychotic Medications. J Proteome Res. 2009;8:3633–3641. doi: 10.1021/pr800876z. [DOI] [PubMed] [Google Scholar]
- 25.Zhang J, Li X, Wang Y, Ji J, Yang F, et al. Association study on the mitochondrial gene NDUFV2 and bipolar disorder in the Chinese Han population. J Neural Transm. 2009;116:357–361. doi: 10.1007/s00702-009-0185-1. [DOI] [PubMed] [Google Scholar]
- 26.Washizuka S, Iwamoto K, Kakiuchi C, Bundo M, Kato T. Expression of mitochondrial complex I subunit gene NDUFV2 in the lymphoblastoid cells derived from patients with bipolar disorder and schizophrenia. Neurosci Res. 2009;63:199–204. doi: 10.1016/j.neures.2008.12.004. [DOI] [PubMed] [Google Scholar]
- 27.Xu C, Li PP, Kennedy JL, Green M, Hughes B, et al. Further support for association of the mitochondrial complex I subunit gene NDUFV2 with bipolar disorder. Bipolar Disord. 2008;10:105–110. doi: 10.1111/j.1399-5618.2008.00535.x. [DOI] [PubMed] [Google Scholar]
- 28.Ben-Shachar D, Karry R. Sp1 expression is disrupted in schizophrenia; a possible mechanism for the abnormal expression of mitochondrial complex I genes, NDUFV1 and NDUFV2. PLoS One. 2007;2:e817. doi: 10.1371/journal.pone.0000817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lazarou M, McKenzie M, Ohtake A, Thorburn DR, Ryan MT. Analysis of the assembly profiles for mitochondrial- and nuclear-DNA-encoded subunits into complex I. Mol Cell Biol. 2007;27:4228–4237. doi: 10.1128/MCB.00074-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakatani N, Hattori E, Ohnishi T, Dean B, Iwayama Y, et al. Genome-wide expression analysis detects eight genes with robust alterations specific to bipolar I disorder: relevance to neuronal network perturbation. Hum Mol Genet. 2006;15:1949–1962. doi: 10.1093/hmg/ddl118. [DOI] [PubMed] [Google Scholar]
- 31.Spires TL, Molnar Z, Kind PC, Cordery PM, Upton AL, et al. Activity-dependent regulation of synapse and dendritic spine morphology in developing barrel cortex requires phospholipase C-beta1 signalling. Cereb Cortex. 2005;15:385–393. doi: 10.1093/cercor/bhh141. [DOI] [PubMed] [Google Scholar]
- 32.Litosch I. Novel mechanisms for feedback regulation of phospholipase C-beta activity. IUBMB Life. 2002;54:253–260. doi: 10.1080/15216540215673. [DOI] [PubMed] [Google Scholar]
- 33.Bohm D, Schwegler H, Kotthaus L, Nayernia K, Rickmann M, et al. Disruption of PLC-beta 1-mediated signal transduction in mutant mice causes age-dependent hippocampal mossy fiber sprouting and neurodegeneration. Mol Cell Neurosci. 2002;21:584–601. doi: 10.1006/mcne.2002.1199. [DOI] [PubMed] [Google Scholar]
- 34.Hannan AJ, Blakemore C, Katsnelson A, Vitalis T, Huber KM, et al. PLC-beta1, activated via mGluRs, mediates activity-dependent differentiation in cerebral cortex. Nat Neurosci. 2001;4:282–288. doi: 10.1038/85132. [DOI] [PubMed] [Google Scholar]
- 35.Tiveci S, Akin A, Cakir T, Saybasili H, Ulgen K. Modelling of calcium dynamics in brain energy metabolism and Alzheimer's disease. Comput Biol Chem. 2005;29:151–162. doi: 10.1016/j.compbiolchem.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 36.Moreira PI, Santos MS, Seica R, Oliveira CR. Brain mitochondrial dysfunction as a link between Alzheimer's disease and diabetes. J Neurol Sci. 2007;257:206–214. doi: 10.1016/j.jns.2007.01.017. [DOI] [PubMed] [Google Scholar]
- 37.Laughton JD, Bittar P, Charnay Y, Pellerin L, Kovari E, et al. Metabolic compartmentalization in the human cortex and hippocampus: evidence for a cell- and region-specific localization of lactate dehydrogenase 5 and pyruvate dehydrogenase. BMC Neurosci. 2007;8:35. doi: 10.1186/1471-2202-8-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Landfield PW, Blalock EM, Chen KC, Porter NM. A new glucocorticoid hypothesis of brain aging: implications for Alzheimer's disease. Curr Alzheimer Res. 2007;4:205–212. doi: 10.2174/156720507780362083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Elgh E, Lindqvist Astot A, Fagerlund M, Eriksson S, Olsson T, et al. Cognitive dysfunction, hippocampal atrophy and glucocorticoid feedback in Alzheimer's disease. Biol Psychiatry. 2006;59:155–161. doi: 10.1016/j.biopsych.2005.06.017. [DOI] [PubMed] [Google Scholar]
- 40.Bao AM, Meynen G, Swaab DF. The stress system in depression and neurodegeneration: focus on the human hypothalamus. Brain Res Rev. 2008;57:531–553. doi: 10.1016/j.brainresrev.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 41.Lee HK, Kumar P, Fu Q, Rosen KM, Querfurth HW. The insulin/Akt signaling pathway is targeted by intracellular beta-amyloid. Mol Biol Cell. 2009;20:1533–1544. doi: 10.1091/mbc.E08-07-0777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Escribano L, Simon AM, Perez-Mediavilla A, Salazar-Colocho P, Del Rio J, et al. Rosiglitazone reverses memory decline and hippocampal glucocorticoid receptor down-regulation in an Alzheimer's disease mouse model. Biochem Biophys Res Commun. 2009;379:406–410. doi: 10.1016/j.bbrc.2008.12.071. [DOI] [PubMed] [Google Scholar]
- 43.Bonomo SM, Rigamonti AE, Giunta M, Galimberti D, Guaita A, et al. Menopausal transition: a possible risk factor for brain pathologic events. Neurobiol Aging. 2009;30:71–80. doi: 10.1016/j.neurobiolaging.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 44.Aisa B, Gil-Bea FJ, Marcos B, Tordera R, Lasheras B, et al. Neonatal stress affects vulnerability of cholinergic neurons and cognition in the rat: Involvement of the HPA axis. Psychoneuroendocrinology. 2009 doi: 10.1016/j.psyneuen.2009.05.003. [DOI] [PubMed] [Google Scholar]
- 45.Sotiropoulos I, Cerqueira JJ, Catania C, Takashima A, Sousa N, et al. Stress and glucocorticoid footprints in the brain-the path from depression to Alzheimer's disease. Neurosci Biobehav Rev. 2008;32:1161–1173. doi: 10.1016/j.neubiorev.2008.05.007. [DOI] [PubMed] [Google Scholar]
- 46.Jing H, Iwasaki Y, Nishiyama M, Taguchi T, Tsugita M, et al. Multisignal regulation of the rat NMDA1 receptor subunit gene–a pivotal role of glucocorticoid-dependent transcription. Life Sci. 2008;82:1137–1141. doi: 10.1016/j.lfs.2008.03.022. [DOI] [PubMed] [Google Scholar]
- 47.White LD, Cory-Slechta DA, Gilbert ME, Tiffany-Castiglioni E, Zawia NH, et al. New and evolving concepts in the neurotoxicology of lead. Toxicol Appl Pharmacol. 2007;225:1–27. doi: 10.1016/j.taap.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 48.Green KN, Billings LM, Roozendaal B, McGaugh JL, LaFerla FM. Glucocorticoids increase amyloid-beta and tau pathology in a mouse model of Alzheimer's disease. J Neurosci. 2006;26:9047–9056. doi: 10.1523/JNEUROSCI.2797-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de Quervain DJ, Poirier R, Wollmer MA, Grimaldi LM, Tsolaki M, et al. Glucocorticoid-related genetic susceptibility for Alzheimer's disease. Hum Mol Genet. 2004;13:47–52. doi: 10.1093/hmg/ddg361. [DOI] [PubMed] [Google Scholar]
- 50.Dai J, Buijs R, Swaab D. Glucocorticoid hormone (cortisol) affects axonal transport in human cortex neurons but shows resistance in Alzheimer's disease. Br J Pharmacol. 2004;143:606–610. doi: 10.1038/sj.bjp.0705995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Polleri A, Gianelli MV, Murialdo G. Dementia: a neuroendocrine perspective. J Endocrinol Invest. 2002;25:73–83. doi: 10.1007/BF03343964. [DOI] [PubMed] [Google Scholar]
- 52.Chen H, Sun S, Mei Y, Liu C, Liu A, et al. The effect of beta-amyloid on neurons and the influence of glucocorticoid and age on such effect. J Huazhong Univ Sci Technolog Med Sci. 2002;22:250–252. doi: 10.1007/BF02828194. [DOI] [PubMed] [Google Scholar]
- 53.Aisen PS. The potential of anti-inflammatory drugs for the treatment of Alzheimer's disease. Lancet Neurol. 2002;1:279–284. doi: 10.1016/s1474-4422(02)00133-3. [DOI] [PubMed] [Google Scholar]
- 54.Rasmuson S, Andrew R, Nasman B, Seckl JR, Walker BR, et al. Increased glucocorticoid production and altered cortisol metabolism in women with mild to moderate Alzheimer's disease. Biol Psychiatry. 2001;49:547–552. doi: 10.1016/s0006-3223(00)01015-5. [DOI] [PubMed] [Google Scholar]
- 55.Pedersen WA, McCullers D, Culmsee C, Haughey NJ, Herman JP, et al. Corticotropin-releasing hormone protects neurons against insults relevant to the pathogenesis of Alzheimer's disease. Neurobiol Dis. 2001;8:492–503. doi: 10.1006/nbdi.2001.0395. [DOI] [PubMed] [Google Scholar]
- 56.Dubois M, Lalonde R, Julien JP, Strazielle C. Mice with the deleted neurofilament of low-molecular-weight (Nefl) gene: 1. Effects on regional brain metabolism. J Neurosci Res. 2005;80:741–750. doi: 10.1002/jnr.20449. [DOI] [PubMed] [Google Scholar]
- 57.Jordanova A, De Jonghe P, Boerkoel CF, Takashima H, De Vriendt E, et al. Mutations in the neurofilament light chain gene (NEFL) cause early onset severe Charcot-Marie-Tooth disease. Brain. 2003;126:590–597. doi: 10.1093/brain/awg059. [DOI] [PubMed] [Google Scholar]
- 58.Abe A, Numakura C, Saito K, Koide H, Oka N, et al. Neurofilament light chain polypeptide gene mutations in Charcot-Marie-Tooth disease: nonsense mutation probably causes a recessive phenotype. J Hum Genet. 2009;54:94–97. doi: 10.1038/jhg.2008.13. [DOI] [PubMed] [Google Scholar]
- 59.Tradewell ML, Durham HD, Mushynski WE, Gentil BJ. Mitochondrial and axonal abnormalities precede disruption of the neurofilament network in a model of charcot-marie-tooth disease type 2E and are prevented by heat shock proteins in a mutant-specific fashion. J Neuropathol Exp Neurol. 2009;68:642–652. doi: 10.1097/NEN.0b013e3181a5deeb. [DOI] [PubMed] [Google Scholar]
- 60.Evgrafov OV, Mersiyanova I, Irobi J, Van Den Bosch L, Dierick I, et al. Mutant small heat-shock protein 27 causes axonal Charcot-Marie-Tooth disease and distal hereditary motor neuropathy. Nat Genet. 2004;36:602–606. doi: 10.1038/ng1354. [DOI] [PubMed] [Google Scholar]
- 61.Nakagawa M, Takashima H. [Update on hereditary neuropathy]. Rinsho Shinkeigaku. 2004;44:991–994. [PubMed] [Google Scholar]
- 62.Takashima H. [Molecular genetics of inherited neuropathies]. Rinsho Shinkeigaku. 2006;46:1–18. [PubMed] [Google Scholar]
- 63.Lus G, Nelis E, Jordanova A, Lofgren A, Cavallaro T, et al. Charcot-Marie-Tooth disease with giant axons: a clinicopathological and genetic entity. Neurology. 2003;61:988–990. doi: 10.1212/wnl.61.7.988. [DOI] [PubMed] [Google Scholar]
- 64.Saba R, Goodman CD, Huzarewich RL, Robertson C, Booth SA. A miRNA signature of prion induced neurodegeneration. PLoS One. 2008;3:e3652. doi: 10.1371/journal.pone.0003652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sorensen G, Medina S, Parchaliuk D, Phillipson C, Robertson C, et al. Comprehensive transcriptional profiling of prion infection in mouse models reveals networks of responsive genes. BMC Genomics. 2008;9:114. doi: 10.1186/1471-2164-9-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Heese K, Nagai Y, Sawada T. Identification of a new synaptic vesicle protein 2B mRNA transcript which is up-regulated in neurons by amyloid beta peptide fragment (1–42). Biochem Biophys Res Commun. 2001;289:924–928. doi: 10.1006/bbrc.2001.5932. [DOI] [PubMed] [Google Scholar]
- 67.Heese K, Nagai Y, Sawada T. The 3′ untranslated region of the new rat synaptic vesicle protein 2B mRNA transcript inhibits translational efficiency. Brain Res Mol Brain Res. 2002;104:127–131. doi: 10.1016/s0169-328x(02)00326-1. [DOI] [PubMed] [Google Scholar]
- 68.Morgans CW, Kensel-Hammes P, Hurley JB, Burton K, Idzerda R, et al. Loss of the Synaptic Vesicle Protein SV2B results in reduced neurotransmission and altered synaptic vesicle protein expression in the retina. PLoS One. 2009;4:e5230. doi: 10.1371/journal.pone.0005230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lazzell DR, Belizaire R, Thakur P, Sherry DM, Janz R. SV2B regulates synaptotagmin 1 by direct interaction. J Biol Chem. 2004;279:52124–52131. doi: 10.1074/jbc.M407502200. [DOI] [PubMed] [Google Scholar]
- 70.Veinbergs I, Mante M, Jung MW, Van Uden E, Masliah E. Synaptotagmin and synaptic transmission alterations in apolipoprotein E-deficient mice. Prog Neuropsychopharmacol Biol Psychiatry. 1999;23:519–531. doi: 10.1016/s0278-5846(99)00013-5. [DOI] [PubMed] [Google Scholar]
- 71.Davidsson P, Blennow K. Neurochemical dissection of synaptic pathology in Alzheimer's disease. Int Psychogeriatr. 1998;10:11–23. doi: 10.1017/s1041610298005110. [DOI] [PubMed] [Google Scholar]
- 72.Ferrer I, Marti E, Tortosa A, Blasi J. Dystrophic neurites of senile plaques are defective in proteins involved in exocytosis and neurotransmission. J Neuropathol Exp Neurol. 1998;57:218–225. doi: 10.1097/00005072-199803000-00002. [DOI] [PubMed] [Google Scholar]
- 73.Shimohama S, Kamiya S, Taniguchi T, Akagawa K, Kimura J. Differential involvement of synaptic vesicle and presynaptic plasma membrane proteins in Alzheimer's disease. Biochem Biophys Res Commun. 1997;236:239–242. doi: 10.1006/bbrc.1997.6940. [DOI] [PubMed] [Google Scholar]
- 74.Davidsson P, Jahn R, Bergquist J, Ekman R, Blennow K. Synaptotagmin, a synaptic vesicle protein, is present in human cerebrospinal fluid: a new biochemical marker for synaptic pathology in Alzheimer disease? Mol Chem Neuropathol. 1996;27:195–210. doi: 10.1007/BF02815094. [DOI] [PubMed] [Google Scholar]
- 75.Reddy PH, Mani G, Park BS, Jacques J, Murdoch G, et al. Differential loss of synaptic proteins in Alzheimer's disease: implications for synaptic dysfunction. J Alzheimers Dis. 2005;7:103–117. doi: 10.3233/jad-2005-7203. discussion 173–180. [DOI] [PubMed] [Google Scholar]
- 76.Kirikoshi H, Katoh M. Expression of WNT7A in human normal tissues and cancer, and regulation of WNT7A and WNT7B in human cancer. Int J Oncol. 2002;21:895–900. [PubMed] [Google Scholar]
- 77.Wang Y, Liu S, Zhu H, Zhang W, Zhang G, et al. FRAT1 overexpression leads to aberrant activation of beta-catenin/TCF pathway in esophageal squamous cell carcinoma. Int J Cancer. 2008;123:561–568. doi: 10.1002/ijc.23600. [DOI] [PubMed] [Google Scholar]
- 78.Hongisto V, Vainio JC, Thompson R, Courtney MJ, Coffey ET. The Wnt pool of glycogen synthase kinase 3beta is critical for trophic-deprivation-induced neuronal death. Mol Cell Biol. 2008;28:1515–1527. doi: 10.1128/MCB.02227-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hagen T, Cross DA, Culbert AA, West A, Frame S, et al. FRAT1, a substrate-specific regulator of glycogen synthase kinase-3 activity, is a cellular substrate of protein kinase A. J Biol Chem. 2006;281:35021–35029. doi: 10.1074/jbc.M607003200. [DOI] [PubMed] [Google Scholar]
- 80.Hongisto V, Smeds N, Brecht S, Herdegen T, Courtney MJ, et al. Lithium blocks the c-Jun stress response and protects neurons via its action on glycogen synthase kinase 3. Mol Cell Biol. 2003;23:6027–6036. doi: 10.1128/MCB.23.17.6027-6036.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hino S, Michiue T, Asashima M, Kikuchi A. Casein kinase I epsilon enhances the binding of Dvl-1 to Frat-1 and is essential for Wnt-3a-induced accumulation of beta-catenin. J Biol Chem. 2003;278:14066–14073. doi: 10.1074/jbc.M213265200. [DOI] [PubMed] [Google Scholar]
- 82.Hay E, Faucheu C, Suc-Royer I, Touitou R, Stiot V, et al. Interaction between LRP5 and Frat1 mediates the activation of the Wnt canonical pathway. J Biol Chem. 2005;280:13616–13623. doi: 10.1074/jbc.M411999200. [DOI] [PubMed] [Google Scholar]
- 83.Fraser E, Young N, Dajani R, Franca-Koh J, Ryves J, et al. Identification of the Axin and Frat binding region of glycogen synthase kinase-3. J Biol Chem. 2002;277:2176–2185. doi: 10.1074/jbc.M109462200. [DOI] [PubMed] [Google Scholar]
- 84.Franca-Koh J, Yeo M, Fraser E, Young N, Dale TC. The regulation of glycogen synthase kinase-3 nuclear export by Frat/GBP. J Biol Chem. 2002;277:43844–43848. doi: 10.1074/jbc.M207265200. [DOI] [PubMed] [Google Scholar]
- 85.Saitoh T, Moriwaki J, Koike J, Takagi A, Miwa T, et al. Molecular cloning and characterization of FRAT2, encoding a positive regulator of the WNT signaling pathway. Biochem Biophys Res Commun. 2001;281:815–820. doi: 10.1006/bbrc.2001.4421. [DOI] [PubMed] [Google Scholar]
- 86.Killick R, Pollard CC, Asuni AA, Mudher AK, Richardson JC, et al. Presenilin 1 independently regulates beta-catenin stability and transcriptional activity. J Biol Chem. 2001;276:48554–48561. doi: 10.1074/jbc.M108332200. [DOI] [PubMed] [Google Scholar]
- 87.Culbert AA, Brown MJ, Frame S, Hagen T, Cross DA, et al. GSK-3 inhibition by adenoviral FRAT1 overexpression is neuroprotective and induces Tau dephosphorylation and beta-catenin stabilisation without elevation of glycogen synthase activity. FEBS Lett. 2001;507:288–294. doi: 10.1016/s0014-5793(01)02990-8. [DOI] [PubMed] [Google Scholar]
- 88.Crowder RJ, Freeman RS. Glycogen synthase kinase-3 beta activity is critical for neuronal death caused by inhibiting phosphatidylinositol 3-kinase or Akt but not for death caused by nerve growth factor withdrawal. J Biol Chem. 2000;275:34266–34271. doi: 10.1074/jbc.M006160200. [DOI] [PubMed] [Google Scholar]
- 89.Katoh M. Networking of WNT, FGF, Notch, BMP, and Hedgehog signaling pathways during carcinogenesis. Stem Cell Rev. 2007;3:30–38. doi: 10.1007/s12015-007-0006-6. [DOI] [PubMed] [Google Scholar]
- 90.Liu C, Wang Y, Smallwood PM, Nathans J. An essential role for Frizzled5 in neuronal survival in the parafascicular nucleus of the thalamus. J Neurosci. 2008;28:5641–5653. doi: 10.1523/JNEUROSCI.1056-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Carmon KS, Loose DS. Wnt7a interaction with Fzd5 and detection of signaling activation using a split eGFP. Biochem Biophys Res Commun. 2008;368:285–291. doi: 10.1016/j.bbrc.2008.01.088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Steventon B, Araya C, Linker C, Kuriyama S, Mayor R. Differential requirements of BMP and Wnt signalling during gastrulation and neurulation define two steps in neural crest induction. Development. 2009;136:771–779. doi: 10.1242/dev.029017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lee MY, Lim HW, Lee SH, Han HJ. Smad, PI3K/Akt, and Wnt-Dependent Signaling Pathways are Involved in BMP-4-Induced ES Cell Self-Renewal. Stem Cells. 2009 doi: 10.1002/stem.124. [DOI] [PubMed] [Google Scholar]
- 94.Kelberman D, de Castro SC, Huang S, Crolla JA, Palmer R, et al. SOX2 plays a critical role in the pituitary, forebrain, and eye during human embryonic development. J Clin Endocrinol Metab. 2008;93:1865–1873. doi: 10.1210/jc.2007-2337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Shi Y, Sun G, Zhao C, Stewart R. Neural stem cell self-renewal. Crit Rev Oncol Hematol. 2008;65:43–53. doi: 10.1016/j.critrevonc.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Linker C, Stern CD. Neural induction requires BMP inhibition only as a late step, and involves signals other than FGF and Wnt antagonists. Development. 2004;131:5671–5681. doi: 10.1242/dev.01445. [DOI] [PubMed] [Google Scholar]
- 97.Tam WL, Lim CY, Han J, Zhang J, Ang YS, et al. T-cell factor 3 regulates embryonic stem cell pluripotency and self-renewal by the transcriptional control of multiple lineage pathways. Stem Cells. 2008;26:2019–2031. doi: 10.1634/stemcells.2007-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ruzov A, Hackett JA, Prokhortchouk A, Reddington JP, Madej MJ, et al. The interaction of xKaiso with xTcf3: a revised model for integration of epigenetic and Wnt signalling pathways. Development. 2009;136:723–727. doi: 10.1242/dev.025577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fancy SP, Baranzini SE, Zhao C, Yuk DI, Irvine KA, et al. Dysregulation of the Wnt pathway inhibits timely myelination and remyelination in the mammalian CNS. Genes Dev. 2009;23:1571–1585. doi: 10.1101/gad.1806309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Koslowski MJ, Kubler I, Chamaillard M, Schaeffeler E, Reinisch W, et al. Genetic variants of Wnt transcription factor TCF-4 (TCF7L2) putative promoter region are associated with small intestinal Crohn's disease. PLoS ONE. 2009;4:e4496. doi: 10.1371/journal.pone.0004496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Schinner S. Wnt-signalling and the metabolic syndrome. Horm Metab Res. 2009;41:159–163. doi: 10.1055/s-0028-1119408. [DOI] [PubMed] [Google Scholar]
- 102.Nazwar TA, Glassmann A, Schilling K. Expression and molecular diversity of Tcf7l2 in the developing murine cerebellum and brain. J Neurosci Res. 2009;87:1532–1546. doi: 10.1002/jnr.21989. [DOI] [PubMed] [Google Scholar]
- 103.Ye F, Chen Y, Hoang T, Montgomery RL, Zhao XH, et al. HDAC1 and HDAC2 regulate oligodendrocyte differentiation by disrupting the beta-catenin-TCF interaction. Nat Neurosci. 2009;12:829–838. doi: 10.1038/nn.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Murray KD, Rubin CM, Jones EG, Chalupa LM. Molecular correlates of laminar differences in the macaque dorsal lateral geniculate nucleus. J Neurosci. 2008;28:12010–12022. doi: 10.1523/JNEUROSCI.3800-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Murray KD, Choudary PV, Jones EG. Nucleus- and cell-specific gene expression in monkey thalamus. Proc Natl Acad Sci U S A. 2007;104:1989–1994. doi: 10.1073/pnas.0610742104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Lukas J, Mazna P, Valenta T, Doubravska L, Pospichalova V, et al. Dazap2 modulates transcription driven by the Wnt effector TCF-4. Nucleic Acids Res. 2009;37:3007–3020. doi: 10.1093/nar/gkp179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Lepourcelet M, Shivdasani RA. Characterization of a novel mammalian Groucho isoform and its role in transcriptional regulation. J Biol Chem. 2002;277:47732–47740. doi: 10.1074/jbc.M208154200. [DOI] [PubMed] [Google Scholar]
- 108.Bachar-Dahan L, Goltzmann J, Yaniv A, Gazit A. Engrailed-1 negatively regulates beta-catenin transcriptional activity by destabilizing beta-catenin via a glycogen synthase kinase-3beta-independent pathway. Mol Biol Cell. 2006;17:2572–2580. doi: 10.1091/mbc.E06-01-0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Daniels DL, Weis WI. Beta-catenin directly displaces Groucho/TLE repressors from Tcf/Lef in Wnt-mediated transcription activation. Nat Struct Mol Biol. 2005;12:364–371. doi: 10.1038/nsmb912. [DOI] [PubMed] [Google Scholar]
- 110.Bauer J, Plaschke K, Martin E, Bardenheuer HJ, Hoyer S. Causes and consequences of neuronal energy deficit in sporadic Alzheimer's disease. Ann N Y Acad Sci. 1997;826:379–381. doi: 10.1111/j.1749-6632.1997.tb48487.x. [DOI] [PubMed] [Google Scholar]
- 111.Lu Y, He X, Zhong S. Cross-species microarray analysis with the OSCAR system suggests an INSR→Pax6→NQO1 neuro-protective pathway in aging and Alzheimer's disease. Nucleic Acids Res. 2007;35:W105–114. doi: 10.1093/nar/gkm408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Wang H, Wang R, Zhao Z, Ji Z, Xu S, et al. Coexistences of insulin signaling-related proteins and choline acetyltransferase in neurons. Brain Res. 2009;1249:237–243. doi: 10.1016/j.brainres.2008.10.046. [DOI] [PubMed] [Google Scholar]
- 113.Frolich L, Blum-Degen D, Riederer P, Hoyer S. A disturbance in the neuronal insulin receptor signal transduction in sporadic Alzheimer's disease. Ann N Y Acad Sci. 1999;893:290–293. doi: 10.1111/j.1749-6632.1999.tb07839.x. [DOI] [PubMed] [Google Scholar]
- 114.Hoyer S. Age as risk factor for sporadic dementia of the Alzheimer type? Ann N Y Acad Sci. 1994;719:248–256. doi: 10.1111/j.1749-6632.1994.tb56833.x. [DOI] [PubMed] [Google Scholar]
- 115.Hoyer S, Lannert H. Inhibition of the neuronal insulin receptor causes Alzheimer-like disturbances in oxidative/energy brain metabolism and in behavior in adult rats. Ann N Y Acad Sci. 1999;893:301–303. doi: 10.1111/j.1749-6632.1999.tb07842.x. [DOI] [PubMed] [Google Scholar]
- 116.Hoyer S, Lee SK, Loffler T, Schliebs R. Inhibition of the neuronal insulin receptor. An in vivo model for sporadic Alzheimer disease? Ann N Y Acad Sci. 2000;920:256–258. doi: 10.1111/j.1749-6632.2000.tb06932.x. [DOI] [PubMed] [Google Scholar]
- 117.Hoyer S. Oxidative metabolism deficiencies in brains of patients with Alzheimer's disease. Acta Neurol Scand. 1996;165(Suppl):18–24. doi: 10.1111/j.1600-0404.1996.tb05868.x. [DOI] [PubMed] [Google Scholar]
- 118.Verkhratsky A, Toescu EC. Endoplasmic reticulum Ca(2+) homeostasis and neuronal death. J Cell Mol Med. 2003;7:351–361. doi: 10.1111/j.1582-4934.2003.tb00238.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Norris CM, Kadish I, Blalock EM, Chen KC, Thibault V, et al. Calcineurin triggers reactive/inflammatory processes in astrocytes and is upregulated in aging and Alzheimer's models. J Neurosci. 2005;25:4649–4658. doi: 10.1523/JNEUROSCI.0365-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Bojarski L, Herms J, Kuznicki J. Calcium dysregulation in Alzheimer's disease. Neurochem Int. 2008;52:621–633. doi: 10.1016/j.neuint.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 121.Cowburn RF, Popescu BO, Ankarcrona M, Dehvari N, Cedazo-Minguez A. Presenilin-mediated signal transduction. Physiol Behav. 2007;92:93–97. doi: 10.1016/j.physbeh.2007.05.053. [DOI] [PubMed] [Google Scholar]
- 122.Giacomello M, Barbiero L, Zatti G, Squitti R, Binetti G, et al. Reduction of Ca2+ stores and capacitative Ca2+ entry is associated with the familial Alzheimer's disease presenilin-2 T122R mutation and anticipates the onset of dementia. Neurobiol Dis. 2005;18:638–648. doi: 10.1016/j.nbd.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 123.Peers C, Smith IF, Boyle JP, Pearson HA. Remodelling of Ca2+ homeostasis in type I cortical astrocytes by hypoxia: evidence for association with Alzheimer's disease. Biol Chem. 2004;385:285–289. doi: 10.1515/BC.2004.023. [DOI] [PubMed] [Google Scholar]
- 124.LaFerla FM. Calcium dyshomeostasis and intracellular signalling in Alzheimer's disease. Nat Rev Neurosci. 2002;3:862–872. doi: 10.1038/nrn960. [DOI] [PubMed] [Google Scholar]
- 125.Eckert A, Forstl H, Zerfass R, Hennerici M, Muller WE. Free intracellular calcium in peripheral cells in Alzheimer's disease. Neurobiol Aging. 1997;18:281–284. doi: 10.1016/s0197-4580(97)80308-9. [DOI] [PubMed] [Google Scholar]
- 126.Schubert P, Ogata T, Miyazaki H, Marchini C, Ferroni S, et al. Pathological immuno-reactions of glial cells in Alzheimer's disease and possible sites of interference. J Neural Transm. 1998;54(Suppl):167–174. doi: 10.1007/978-3-7091-7508-8_16. [DOI] [PubMed] [Google Scholar]
- 127.Eckert A, Forstl H, Zerfass R, Hartmann H, Muller WE. Lymphocytes and neutrophils as peripheral models to study the effect of beta-amyloid on cellular calcium signalling in Alzheimer's disease. Life Sci. 1996;59:499–510. doi: 10.1016/0024-3205(96)00329-3. [DOI] [PubMed] [Google Scholar]
- 128.Salkovic-Petrisic M, Hoyer S. Central insulin resistance as a trigger for sporadic Alzheimer-like pathology: an experimental approach. J Neural Transm. 2007;(Suppl):217–233. doi: 10.1007/978-3-211-73574-9_28. [DOI] [PubMed] [Google Scholar]
- 129.Qin W, Zhao W, Ho L, Wang J, Walsh K, et al. Regulation of forkhead transcription factor FoxO3a contributes to calorie restriction-induced prevention of Alzheimer's disease-type amyloid neuropathology and spatial memory deterioration. Ann N Y Acad Sci. 2008;1147:335–347. doi: 10.1196/annals.1427.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Carro E, Trejo JL, Spuch C, Bohl D, Heard JM, et al. Blockade of the insulin-like growth factor I receptor in the choroid plexus originates Alzheimer's-like neuropathology in rodents: new cues into the human disease? Neurobiol Aging. 2006;27:1618–1631. doi: 10.1016/j.neurobiolaging.2005.09.039. [DOI] [PubMed] [Google Scholar]
- 131.Rhein V, Eckert A. Effects of Alzheimer's amyloid-beta and tau protein on mitochondrial function – role of glucose metabolism and insulin signalling. Arch Physiol Biochem. 2007;113:131–141. doi: 10.1080/13813450701572288. [DOI] [PubMed] [Google Scholar]
- 132.Abbas T, Faivre E, Holscher C. Impairment of synaptic plasticity and memory formation in GLP-1 receptor KO mice: Interaction between type 2 diabetes and Alzheimer's disease. Behav Brain Res. 2009 doi: 10.1016/j.bbr.2009.06.035. [DOI] [PubMed] [Google Scholar]
- 133.Castri P, Iacovelli L, De Blasi A, Giubilei F, Moretti A, et al. Reduced insulin-induced phosphatidylinositol-3-kinase activation in peripheral blood mononuclear leucocytes from patients with Alzheimer's disease. Eur J Neurosci. 2007;26:2469–2472. doi: 10.1111/j.1460-9568.2007.05869.x. [DOI] [PubMed] [Google Scholar]
- 134.Li L, Holscher C. Common pathological processes in Alzheimer disease and type 2 diabetes: a review. Brain Res Rev. 2007;56:384–402. doi: 10.1016/j.brainresrev.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 135.Salkovic-Petrisic M, Tribl F, Schmidt M, Hoyer S, Riederer P. Alzheimer-like changes in protein kinase B and glycogen synthase kinase-3 in rat frontal cortex and hippocampus after damage to the insulin signalling pathway. J Neurochem. 2006;96:1005–1015. doi: 10.1111/j.1471-4159.2005.03637.x. [DOI] [PubMed] [Google Scholar]
- 136.Biessels GJ, Kappelle LJ. Increased risk of Alzheimer's disease in Type II diabetes: insulin resistance of the brain or insulin-induced amyloid pathology? Biochem Soc Trans. 2005;33:1041–1044. doi: 10.1042/BST0331041. [DOI] [PubMed] [Google Scholar]
- 137.Nelson TJ, Alkon DL. Insulin and cholesterol pathways in neuronal function, memory and neurodegeneration. Biochem Soc Trans. 2005;33:1033–1036. doi: 10.1042/BST20051033. [DOI] [PubMed] [Google Scholar]
- 138.Biessels GJ, Bravenboer B, Gispen WH. Glucose, insulin and the brain: modulation of cognition and synaptic plasticity in health and disease: a preface. Eur J Pharmacol. 2004;490:1–4. doi: 10.1016/j.ejphar.2004.02.057. [DOI] [PubMed] [Google Scholar]
- 139.Morgan C, Colombres M, Nunez MT, Inestrosa NC. Structure and function of amyloid in Alzheimer's disease. Prog Neurobiol. 2004;74:323–349. doi: 10.1016/j.pneurobio.2004.10.004. [DOI] [PubMed] [Google Scholar]
- 140.Zhao WQ, Townsend M. Insulin resistance and amyloidogenesis as common molecular foundation for type 2 diabetes and Alzheimer's disease. Biochim Biophys Acta. 2009;1792:482–496. doi: 10.1016/j.bbadis.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 141.De Felice FG, Vieira MN, Bomfim TR, Decker H, Velasco PT, et al. Protection of synapses against Alzheimer's-linked toxins: insulin signaling prevents the pathogenic binding of Abeta oligomers. Proc Natl Acad Sci U S A. 2009;106:1971–1976. doi: 10.1073/pnas.0809158106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Chen Y, Zhou K, Wang R, Liu Y, Kwak YD, et al. Antidiabetic drug metformin (GlucophageR) increases biogenesis of Alzheimer's amyloid peptides via up-regulating BACE1 transcription. Proc Natl Acad Sci U S A. 2009;106:3907–3912. doi: 10.1073/pnas.0807991106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Puglielli L. Aging of the brain, neurotrophin signaling, and Alzheimer's disease: is IGF1-R the common culprit? Neurobiol Aging. 2008;29:795–811. doi: 10.1016/j.neurobiolaging.2007.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Jolivalt CG, Lee CA, Beiswenger KK, Smith JL, Orlov M, et al. Defective insulin signaling pathway and increased glycogen synthase kinase-3 activity in the brain of diabetic mice: parallels with Alzheimer's disease and correction by insulin. J Neurosci Res. 2008;86:3265–3274. doi: 10.1002/jnr.21787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.de Tullio MB, Morelli L, Castano EM. The irreversible binding of amyloid peptide substrates to insulin-degrading enzyme: a biological perspective. Prion. 2008;2:51–56. doi: 10.4161/pri.2.2.6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Dore S, Kar S, Quirion R. Insulin-like growth factor I protects and rescues hippocampal neurons against beta-amyloid- and human amylin-induced toxicity. Proc Natl Acad Sci U S A. 1997;94:4772–4777. doi: 10.1073/pnas.94.9.4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Gandy S, Czernik AJ, Greengard P. Phosphorylation of Alzheimer disease amyloid precursor peptide by protein kinase C and Ca2+/calmodulin-dependent protein kinase II. Proc Natl Acad Sci U S A. 1988;85:6218–6221. doi: 10.1073/pnas.85.16.6218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Deutsch SI, Rosse RB, Lakshman RM. Dysregulation of tau phosphorylation is a hypothesized point of convergence in the pathogenesis of alzheimer's disease, frontotemporal dementia and schizophrenia with therapeutic implications. Prog Neuropsychopharmacol Biol Psychiatry. 2006;30:1369–1380. doi: 10.1016/j.pnpbp.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 149.Wickelgren I. Tracking insulin to the mind. Science. 1998;280:517–519. doi: 10.1126/science.280.5363.517. [DOI] [PubMed] [Google Scholar]
- 150.Jafferali S, Dumont Y, Sotty F, Robitaille Y, Quirion R, et al. Insulin-like growth factor-I and its receptor in the frontal cortex, hippocampus, and cerebellum of normal human and alzheimer disease brains. Synapse. 2000;38:450–459. doi: 10.1002/1098-2396(20001215)38:4<450::AID-SYN10>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 151.Brune S, Kolsch H, Ptok U, Majores M, Schulz A, et al. Polymorphism in the peroxisome proliferator-activated receptor alpha gene influences the risk for Alzheimer's disease. J Neural Transm. 2003;110:1041–1050. doi: 10.1007/s00702-003-0018-6. [DOI] [PubMed] [Google Scholar]
- 152.Frolich L, Blum-Degen D, Bernstein HG, Engelsberger S, Humrich J, et al. Brain insulin and insulin receptors in aging and sporadic Alzheimer's disease. J Neural Transm. 1998;105:423–438. doi: 10.1007/s007020050068. [DOI] [PubMed] [Google Scholar]
- 153.Grunblatt E, Hoyer S, Riederer P. Gene expression profile in streptozotocin rat model for sporadic Alzheimer's disease. J Neural Transm. 2004;111:367–386. doi: 10.1007/s00702-003-0030-x. [DOI] [PubMed] [Google Scholar]
- 154.Hoyer S. Is sporadic Alzheimer disease the brain type of non-insulin dependent diabetes mellitus? A challenging hypothesis. J Neural Transm. 1998;105:415–422. doi: 10.1007/s007020050067. [DOI] [PubMed] [Google Scholar]
- 155.Hoyer S. The aging brain. Changes in the neuronal insulin/insulin receptor signal transduction cascade trigger late-onset sporadic Alzheimer disease (SAD). A mini-review. J Neural Transm. 2002;109:991–1002. doi: 10.1007/s007020200082. [DOI] [PubMed] [Google Scholar]
- 156.Hoyer S. The brain insulin signal transduction system and sporadic (type II) Alzheimer disease: an update. J Neural Transm. 2002;109:341–360. doi: 10.1007/s007020200028. [DOI] [PubMed] [Google Scholar]
- 157.Hoyer S, Nitsch R. Cerebral excess release of neurotransmitter amino acids subsequent to reduced cerebral glucose metabolism in early-onset dementia of Alzheimer type. J Neural Transm. 1989;75:227–232. doi: 10.1007/BF01258634. [DOI] [PubMed] [Google Scholar]
- 158.Xu WH, Huber R, Riepe MW. Gender- and region-specific expression of insulin receptor protein in mouse brain: effect of mild inhibition of oxidative phosphorylation. J Neural Transm. 2007;114:373–377. doi: 10.1007/s00702-006-0588-1. [DOI] [PubMed] [Google Scholar]
- 159.Hoyer S, Nitsch R, Oesterreich K. Predominant abnormality in cerebral glucose utilization in late-onset dementia of the Alzheimer type: a cross-sectional comparison against advanced late-onset and incipient early-onset cases. J Neural Transm Park Dis Dement Sect. 1991;3:1–14. doi: 10.1007/BF02251132. [DOI] [PubMed] [Google Scholar]
- 160.Blum-Degen D, Frolich L, Hoyer S, Riederer P. Altered regulation of brain glucose metabolism as a cause of neurodegenerative disorders? J Neural Transm. 1995;46(Suppl):139–147. [PubMed] [Google Scholar]
- 161.Hoyer S. Models of Alzheimer's disease: cellular and molecular aspects. J Neural Transm. 1997;49(Suppl):11–21. doi: 10.1007/978-3-7091-6844-8_2. [DOI] [PubMed] [Google Scholar]
- 162.Hoyer S, Muller D, Plaschke K. Desensitization of brain insulin receptor. Effect on glucose/energy and related metabolism. J Neural Transm. 1994;44(Suppl):259–268. doi: 10.1007/978-3-7091-9350-1_20. [DOI] [PubMed] [Google Scholar]
- 163.Aguado-Llera D, Arilla-Ferreiro E, Campos-Barros A, Puebla-Jimenez L, Barrios V. Protective effects of insulin-like growth factor-I on the somatostatinergic system in the temporal cortex of beta-amyloid-treated rats. J Neurochem. 2005;92:607–615. doi: 10.1111/j.1471-4159.2004.02889.x. [DOI] [PubMed] [Google Scholar]
- 164.Crews FT, McElhaney R, Freund G, Ballinger WE, Jr, Raizada MK. Insulin-like growth factor I receptor binding in brains of Alzheimer's and alcoholic patients. J Neurochem. 1992;58:1205–1210. doi: 10.1111/j.1471-4159.1992.tb11330.x. [DOI] [PubMed] [Google Scholar]
- 165.Hoyer S. Somatostatin and Alzheimer's disease. J Neurol. 1987;234:266–267. doi: 10.1007/BF00618265. [DOI] [PubMed] [Google Scholar]
- 166.De Ferrari GV, Moon RT. The ups and downs of Wnt signaling in prevalent neurological disorders. Oncogene. 2006;25:7545–7553. doi: 10.1038/sj.onc.1210064. [DOI] [PubMed] [Google Scholar]
- 167.Balaraman Y, Limaye AR, Levey AI, Srinivasan S. Glycogen synthase kinase 3beta and Alzheimer's disease: pathophysiological and therapeutic significance. Cell Mol Life Sci. 2006;63:1226–1235. doi: 10.1007/s00018-005-5597-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Widelitz R. Wnt signaling through canonical and non-canonical pathways: recent progress. Growth Factors. 2005;23:111–116. doi: 10.1080/08977190500125746. [DOI] [PubMed] [Google Scholar]
- 169.Cerpa W, Dinamarca MC, Inestrosa NC. Structure-function implications in Alzheimer's disease: effect of Abeta oligomers at central synapses. Curr Alzheimer Res. 2008;5:233–243. doi: 10.2174/156720508784533321. [DOI] [PubMed] [Google Scholar]
- 170.Mercado-Gomez O, Hernandez-Fonseca K, Villavicencio-Queijeiro A, Massieu L, Chimal-Monroy J, et al. Inhibition of Wnt and PI3K signaling modulates GSK-3beta activity and induces morphological changes in cortical neurons: role of tau phosphorylation. Neurochem Res. 2008;33:1599–1609. doi: 10.1007/s11064-008-9714-9. [DOI] [PubMed] [Google Scholar]
- 171.Magdesian MH, Carvalho MM, Mendes FA, Saraiva LM, Juliano MA, et al. Amyloid-beta binds to the extracellular cysteine-rich domain of Frizzled and inhibits Wnt/beta-catenin signaling. J Biol Chem. 2008;283:9359–9368. doi: 10.1074/jbc.M707108200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Esposito G, Scuderi C, Lu J, Savani C, De Filippis D, et al. S100B induces tau protein hyperphosphorylation via Dickopff-1 up-regulation and disrupts the Wnt pathway in human neural stem cells. J Cell Mol Med. 2008;12:914–927. doi: 10.1111/j.1582-4934.2008.00159.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Ferrero A, Cereseto M, Sifonios L. [The relationship between the Wnt signaling and the psychiatric diseases]. Vertex. 2006;17:165–171. [PubMed] [Google Scholar]
- 174.De Ferrari GV, Papassotiropoulos A, Biechele T, Wavrant De-Vrieze F, Avila ME, et al. Common genetic variation within the low-density lipoprotein receptor-related protein 6 and late-onset Alzheimer's disease. Proc Natl Acad Sci U S A. 2007;104:9434–9439. doi: 10.1073/pnas.0603523104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Inestrosa NC, Varela-Nallar L, Grabowski CP, Colombres M. Synaptotoxicity in Alzheimer's disease: the Wnt signaling pathway as a molecular target. IUBMB Life. 2007;59:316–321. doi: 10.1080/15216540701242490. [DOI] [PubMed] [Google Scholar]
- 176.Maiese K, Chong ZZ, Shang YC. Mechanistic insights into diabetes mellitus and oxidative stress. Curr Med Chem. 2007;14:1729–1738. doi: 10.2174/092986707781058968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Repetto E, Yoon IS, Zheng H, Kang DE. Presenilin 1 regulates epidermal growth factor receptor turnover and signaling in the endosomal-lysosomal pathway. J Biol Chem. 2007;282:31504–31516. doi: 10.1074/jbc.M704273200. [DOI] [PubMed] [Google Scholar]
- 178.Fuentealba RA, Farias G, Scheu J, Bronfman M, Marzolo MP, et al. Signal transduction during amyloid-beta-peptide neurotoxicity: role in Alzheimer disease. Brain Res Brain Res Rev. 2004;47:275–289. doi: 10.1016/j.brainresrev.2004.07.018. [DOI] [PubMed] [Google Scholar]
- 179.Inestrosa NC, Urra S, Colombres M. Acetylcholinesterase (AChE)–amyloid-beta-peptide complexes in Alzheimer's disease. the Wnt signaling pathway. Curr Alzheimer Res. 2004;1:249–254. doi: 10.2174/1567205043332063. [DOI] [PubMed] [Google Scholar]
- 180.De Ferrari GV, Inestrosa NC. Wnt signaling function in Alzheimer's disease. Brain Res Brain Res Rev. 2000;33:1–12. doi: 10.1016/s0165-0173(00)00021-7. [DOI] [PubMed] [Google Scholar]
- 181.Inestrosa NC, Alvarez A, Godoy J, Reyes A, De Ferrari GV. Acetylcholinesterase-amyloid-beta-peptide interaction and Wnt signaling involvement in Abeta neurotoxicity. Acta Neurol Scand. 2000;176(Suppl):53–59. doi: 10.1034/j.1600-0404.2000.00308.x. [DOI] [PubMed] [Google Scholar]
- 182.Small DH. Alzheimer Symposium. Gamma-secretase, presenilins and WNT proteins. IDrugs. 2000;3:740–741. [PubMed] [Google Scholar]
- 183.De Strooper B, Annaert W. Where Notch and Wnt signaling meet. The presenilin hub. J Cell Biol. 2001;152:F17–20. doi: 10.1083/jcb.152.4.f17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Caricasole A, Copani A, Caruso A, Caraci F, Iacovelli L, et al. The Wnt pathway, cell-cycle activation and beta-amyloid: novel therapeutic strategies in Alzheimer's disease? Trends Pharmacol Sci. 2003;24:233–238. doi: 10.1016/s0165-6147(03)00100-7. [DOI] [PubMed] [Google Scholar]
- 185.De Ferrari GV, Chacon MA, Barria MI, Garrido JL, Godoy JA, et al. Activation of Wnt signaling rescues neurodegeneration and behavioral impairments induced by beta-amyloid fibrils. Mol Psychiatry. 2003;8:195–208. doi: 10.1038/sj.mp.4001208. [DOI] [PubMed] [Google Scholar]
- 186.Grilli M, Ferrari Toninelli G, Uberti D, Spano P, Memo M. Alzheimer's disease linking neurodegeneration with neurodevelopment. Funct Neurol. 2003;18:145–148. [PubMed] [Google Scholar]
- 187.Selkoe D, Kopan R. Notch and Presenilin: regulated intramembrane proteolysis links development and degeneration. Annu Rev Neurosci. 2003;26:565–597. doi: 10.1146/annurev.neuro.26.041002.131334. [DOI] [PubMed] [Google Scholar]
- 188.Alvarez AR, Godoy JA, Mullendorff K, Olivares GH, Bronfman M, et al. Wnt-3a overcomes beta-amyloid toxicity in rat hippocampal neurons. Exp Cell Res. 2004;297:186–196. doi: 10.1016/j.yexcr.2004.02.028. [DOI] [PubMed] [Google Scholar]
- 189.Busby V, Goossens S, Nowotny P, Hamilton G, Smemo S, et al. Alpha-T-catenin is expressed in human brain and interacts with the Wnt signaling pathway but is not responsible for linkage to chromosome 10 in Alzheimer's disease. Neuromolecular Med. 2004;5:133–146. doi: 10.1385/NMM:5:2:133. [DOI] [PubMed] [Google Scholar]
- 190.Caricasole A, Copani A, Caraci F, Aronica E, Rozemuller AJ, et al. Induction of Dickkopf-1, a negative modulator of the Wnt pathway, is associated with neuronal degeneration in Alzheimer's brain. J Neurosci. 2004;24:6021–6027. doi: 10.1523/JNEUROSCI.1381-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Chong ZZ, Maiese K. Targeting WNT, protein kinase B, and mitochondrial membrane integrity to foster cellular survival in the nervous system. Histol Histopathol. 2004;19:495–504. doi: 10.14670/hh-19.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Toledo EM, Colombres M, Inestrosa NC. Wnt signaling in neuroprotection and stem cell differentiation. Prog Neurobiol. 2008;86:281–296. doi: 10.1016/j.pneurobio.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 193.Boonen RA, van Tijn P, Zivkovic D. Wnt signaling in Alzheimer's disease: up or down, that is the question. Ageing Res Rev. 2009;8:71–82. doi: 10.1016/j.arr.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 194.Cairney CJ, Sanguinetti G, Ranghini E, Chantry AD, Nostro MC, et al. A systems biology approach to Down syndrome: identification of Notch/Wnt dysregulation in a model of stem cells aging. Biochim Biophys Acta. 2009;1792:353–363. doi: 10.1016/j.bbadis.2009.01.015. [DOI] [PubMed] [Google Scholar]
- 195.Caricasole A, Bakker A, Copani A, Nicoletti F, Gaviraghi G, et al. Two sides of the same coin: Wnt signaling in neurodegeneration and neuro-oncology. Biosci Rep. 2005;25:309–327. doi: 10.1007/s10540-005-2893-6. [DOI] [PubMed] [Google Scholar]
- 196.Lee EO, Shin YJ, Chong YH. Mechanisms involved in prostaglandin E2-mediated neuroprotection against TNF-alpha: possible involvement of multiple signal transduction and beta-catenin/T-cell factor. J Neuroimmunol. 2004;155:21–31. doi: 10.1016/j.jneuroim.2004.05.012. [DOI] [PubMed] [Google Scholar]
- 197.Doble BW, Woodgett JR. GSK-3: tricks of the trade for a multi-tasking kinase. J Cell Sci. 2003;116:1175–1186. doi: 10.1242/jcs.00384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Anderton BH, Dayanandan R, Killick R, Lovestone S. Does dysregulation of the Notch and wingless/Wnt pathways underlie the pathogenesis of Alzheimer's disease? Mol Med Today. 2000;6:54–59. doi: 10.1016/s1357-4310(99)01640-8. [DOI] [PubMed] [Google Scholar]
- 199.Weihl CC, Ghadge GD, Kennedy SG, Hay N, Miller RJ, et al. Mutant presenilin-1 induces apoptosis and downregulates Akt/PKB. J Neurosci. 1999;19:5360–5369. doi: 10.1523/JNEUROSCI.19-13-05360.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Ray A, Dhar S, Shakya A, Ray P, Okada Y, et al. SAF-3, a novel splice variant of the SAF-1/MAZ/Pur-1 family, is expressed during inflammation. FEBS J. 2009 doi: 10.1111/j.1742-4658.2009.07136.x. [DOI] [PubMed] [Google Scholar]
- 201.Jordan-Sciutto KL, Dragich JM, Caltagarone J, Hall DJ, Bowser R. Fetal Alz-50 clone 1 (FAC1) protein interacts with the Myc-associated zinc finger protein (ZF87/MAZ) and alters its transcriptional activity. Biochemistry. 2000;39:3206–3215. doi: 10.1021/bi992211q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Bowden NA, Weidenhofer J, Scott RJ, Schall U, Todd J, et al. Preliminary investigation of gene expression profiles in peripheral blood lymphocytes in schizophrenia. Schizophr Res. 2006;82:175–183. doi: 10.1016/j.schres.2005.11.012. [DOI] [PubMed] [Google Scholar]
- 203.Bataller L, Wade DF, Graus F, Rosenfeld MR, Dalmau J. The MAZ protein is an autoantigen of Hodgkin's disease and paraneoplastic cerebellar dysfunction. Ann Neurol. 2003;53:123–127. doi: 10.1002/ana.10434. [DOI] [PubMed] [Google Scholar]
- 204.Sajan FD, Martiniuk F, Marcus DL, Frey WH, 2nd, Hite R, et al. Apoptotic gene expression in Alzheimer's disease hippocampal tissue. Am J Alzheimers Dis Other Demen. 2007;22:319–328. doi: 10.1177/1533317507302447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Lim AC, Qi RZ. Cyclin-dependent kinases in neural development and degeneration. J Alzheimers Dis. 2003;5:329–335. doi: 10.3233/jad-2003-5409. [DOI] [PubMed] [Google Scholar]
- 206.Jordan-Sciutto KL, Malaiyandi LM, Bowser R. Altered distribution of cell cycle transcriptional regulators during Alzheimer disease. J Neuropathol Exp Neurol. 2002;61:358–367. doi: 10.1093/jnen/61.4.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Jordan-Sciutto K, Rhodes J, Bowser R. Altered subcellular distribution of transcriptional regulators in response to Abeta peptide and during Alzheimer's disease. Mech Ageing Dev. 2001;123:11–20. doi: 10.1016/s0047-6374(01)00334-7. [DOI] [PubMed] [Google Scholar]
- 208.Motonaga K, Itoh M, Hirayama A, Hirano S, Becker LE, et al. Up-regulation of E2F-1 in Down's syndrome brain exhibiting neuropathological features of Alzheimer-type dementia. Brain Res. 2001;905:250–253. doi: 10.1016/s0006-8993(01)02535-5. [DOI] [PubMed] [Google Scholar]
- 209.Putzer BM. Targeting E2F1 Death Signaling: Opposing Role in Cancer Control and Neurodegeneration. Discov Med. 2006;6:123–127. [PubMed] [Google Scholar]
- 210.Strachan GD, Koike MA, Siman R, Hall DJ, Jordan-Sciutto KL. E2F1 induces cell death, calpain activation, and MDMX degradation in a transcription independent manner implicating a novel role for E2F1 in neuronal loss in SIV encephalitis. J Cell Biochem. 2005;96:728–740. doi: 10.1002/jcb.20574. [DOI] [PubMed] [Google Scholar]
- 211.Fortin A, MacLaurin JG, Arbour N, Cregan SP, Kushwaha N, et al. The proapoptotic gene SIVA is a direct transcriptional target for the tumor suppressors p53 and E2F1. J Biol Chem. 2004;279:28706–28714. doi: 10.1074/jbc.M400376200. [DOI] [PubMed] [Google Scholar]
- 212.Jordan-Sciutto KL, Wang G, Murphey-Corb M, Wiley CA. Cell cycle proteins exhibit altered expression patterns in lentiviral-associated encephalitis. J Neurosci. 2002;22:2185–2195. doi: 10.1523/JNEUROSCI.22-06-02185.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Jordan-Sciutto KL, Murray Fenner BA, Wiley CA, Achim CL. Response of cell cycle proteins to neurotrophic factor and chemokine stimulation in human neuroglia. Exp Neurol. 2001;167:205–214. doi: 10.1006/exnr.2000.7594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Jordan-Sciutto KL, Wang G, Murphy-Corb M, Wiley CA. Induction of cell-cycle regulators in simian immunodeficiency virus encephalitis. Am J Pathol. 2000;157:497–507. doi: 10.1016/S0002-9440(10)64561-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Strachan GD, Kopp AS, Koike MA, Morgan KL, Jordan-Sciutto KL. Chemokine- and neurotrophic factor-induced changes in E2F1 localization and phosphorylation of the retinoblastoma susceptibility gene product (pRb) occur by distinct mechanisms in murine cortical cultures. Exp Neurol. 2005;193:455–468. doi: 10.1016/j.expneurol.2004.08.038. [DOI] [PubMed] [Google Scholar]
- 216.Verdaguer E, Susana Gde A, Clemens A, Pallas M, Camins A. Implication of the transcription factor E2F-1 in the modulation of neuronal apoptosis. Biomed Pharmacother. 2007;61:390–399. doi: 10.1016/j.biopha.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 217.Ranganathan S, Bowser R. Alterations in G(1) to S phase cell-cycle regulators during amyotrophic lateral sclerosis. Am J Pathol. 2003;162:823–835. doi: 10.1016/S0002-9440(10)63879-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.MacManus JP, Jian M, Preston E, Rasquinha I, Webster J, et al. Absence of the transcription factor E2F1 attenuates brain injury and improves behavior after focal ischemia in mice. J Cereb Blood Flow Metab. 2003;23:1020–1028. doi: 10.1097/01.WCB.0000084249.20114.FA. [DOI] [PubMed] [Google Scholar]
- 219.Konishi Y, Bonni A. The E2F-Cdc2 cell-cycle pathway specifically mediates activity deprivation-induced apoptosis of postmitotic neurons. J Neurosci. 2003;23:1649–1658. doi: 10.1523/JNEUROSCI.23-05-01649.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Cooper-Kuhn CM, Vroemen M, Brown J, Ye H, Thompson MA, et al. Impaired adult neurogenesis in mice lacking the transcription factor E2F1. Mol Cell Neurosci. 2002;21:312–323. doi: 10.1006/mcne.2002.1176. [DOI] [PubMed] [Google Scholar]
- 221.MacGibbon GA, Lawlor PA, Walton M, Sirimanne E, Faull RL, et al. Expression of Fos, Jun, and Krox family proteins in Alzheimer's disease. Exp Neurol. 1997;147:316–332. doi: 10.1006/exnr.1997.6600. [DOI] [PubMed] [Google Scholar]
- 222.Revest JM, Di Blasi F, Kitchener P, Rouge-Pont F, Desmedt A, et al. The MAPK pathway and Egr-1 mediate stress-related behavioral effects of glucocorticoids. Nat Neurosci. 2005;8:664–672. doi: 10.1038/nn1441. [DOI] [PubMed] [Google Scholar]
- 223.Illing RB, Michler SA, Kraus KS, Laszig R. Transcription factor modulation and expression in the rat auditory brainstem following electrical intracochlear stimulation. Exp Neurol. 2002;175:226–244. doi: 10.1006/exnr.2002.7895. [DOI] [PubMed] [Google Scholar]
- 224.Bozon B, Davis S, Laroche S. A requirement for the immediate early gene zif268 in reconsolidation of recognition memory after retrieval. Neuron. 2003;40:695–701. doi: 10.1016/s0896-6273(03)00674-3. [DOI] [PubMed] [Google Scholar]
- 225.Knapska E, Kaczmarek L. A gene for neuronal plasticity in the mammalian brain: Zif268/Egr-1/NGFI-A/Krox-24/TIS8/ZENK? Prog Neurobiol. 2004;74:183–211. doi: 10.1016/j.pneurobio.2004.05.007. [DOI] [PubMed] [Google Scholar]
- 226.Bozon B, Kelly A, Josselyn SA, Silva AJ, Davis S, et al. MAPK, CREB and zif268 are all required for the consolidation of recognition memory. Philos Trans R Soc Lond B Biol Sci. 2003;358:805–814. doi: 10.1098/rstb.2002.1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Alder J, Thakker-Varia S, Bangasser DA, Kuroiwa M, Plummer MR, et al. Brain-derived neurotrophic factor-induced gene expression reveals novel actions of VGF in hippocampal synaptic plasticity. J Neurosci. 2003;23:10800–10808. doi: 10.1523/JNEUROSCI.23-34-10800.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 228.Illing RB. Activity-dependent plasticity in the adult auditory brainstem. Audiol Neurootol. 2001;6:319–345. doi: 10.1159/000046844. [DOI] [PubMed] [Google Scholar]
- 229.Poirier R, Cheval H, Mailhes C, Garel S, Charnay P, et al. Distinct functions of egr gene family members in cognitive processes. Front Neurosci. 2008;2:47–55. doi: 10.3389/neuro.01.002.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Poirier R, Cheval H, Mailhes C, Charnay P, Davis S, et al. Paradoxical role of an egr transcription factor family member, egr2/krox20, in learning and memory. Front Behav Neurosci. 2007;1:6. doi: 10.3389/neuro.08.006.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Desjardins S, Mayo W, Vallee M, Hancock D, Le Moal M, et al. Effect of aging on the basal expression of c-Fos, c-Jun, and Egr-1 proteins in the hippocampus. Neurobiol Aging. 1997;18:37–44. doi: 10.1016/s0197-4580(96)00206-0. [DOI] [PubMed] [Google Scholar]
- 232.Dong S, Replogle KL, Hasadsri L, Imai BS, Yau PM, et al. Discrete molecular states in the brain accompany changing responses to a vocal signal. Proc Natl Acad Sci U S A. 2009;106:11364–11369. doi: 10.1073/pnas.0812998106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Velho TA, Mello CV. Synapsins are late activity-induced genes regulated by birdsong. J Neurosci. 2008;28:11871–11882. doi: 10.1523/JNEUROSCI.2307-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Velho TA, Lovell P, Mello CV. Enriched expression and developmental regulation of the middle-weight neurofilament (NF-M) gene in song control nuclei of the zebra finch. J Comp Neurol. 2007;500:477–497. doi: 10.1002/cne.21180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Zhu MX. Multiple roles of calmodulin and other Ca(2+)-binding proteins in the functional regulation of TRP channels. Pflugers Arch. 2005;451:105–115. doi: 10.1007/s00424-005-1427-1. [DOI] [PubMed] [Google Scholar]
- 236.Zhou H, Yu K, McCoy KL, Lee A. Molecular mechanism for divergent regulation of Cav1.2 Ca2+ channels by calmodulin and Ca2+-binding protein-1. J Biol Chem. 2005;280:29612–29619. doi: 10.1074/jbc.M504167200. [DOI] [PubMed] [Google Scholar]
- 237.Wingard JN, Chan J, Bosanac I, Haeseleer F, Palczewski K, et al. Structural analysis of Mg2+ and Ca2+ binding to CaBP1, a neuron-specific regulator of calcium channels. J Biol Chem. 2005;280:37461–37470. doi: 10.1074/jbc.M508541200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Li C, Chan J, Haeseleer F, Mikoshiba K, Palczewski K, et al. Structural insights into Ca2+-dependent regulation of inositol 1,4,5-trisphosphate receptors by CaBP1. J Biol Chem. 2009;284:2472–2481. doi: 10.1074/jbc.M806513200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Chen ML, Chen YC, Peng IW, Kang RL, Wu MP, et al. Ca2+ binding protein-1 inhibits Ca2+ currents and exocytosis in bovine chromaffin cells. J Biomed Sci. 2008;15:169–181. doi: 10.1007/s11373-007-9217-8. [DOI] [PubMed] [Google Scholar]
- 240.Tippens AL, Lee A. Caldendrin, a neuron-specific modulator of Cav/1.2 (L-type) Ca2+ channels. J Biol Chem. 2007;282:8464–8473. doi: 10.1074/jbc.M611384200. [DOI] [PubMed] [Google Scholar]
- 241.Haynes LP, Fitzgerald DJ, Wareing B, O'Callaghan DW, Morgan A, et al. Analysis of the interacting partners of the neuronal calcium-binding proteins L-CaBP1, hippocalcin, NCS-1 and neurocalcin delta. Proteomics. 2006;6:1822–1832. doi: 10.1002/pmic.200500489. [DOI] [PubMed] [Google Scholar]
- 242.Lautermilch NJ, Few AP, Scheuer T, Catterall WA. Modulation of CaV2.1 channels by the neuronal calcium-binding protein visinin-like protein-2. J Neurosci. 2005;25:7062–7070. doi: 10.1523/JNEUROSCI.0447-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Haeseleer F, Imanishi Y, Maeda T, Possin DE, Maeda A, et al. Essential role of Ca2+-binding protein 4, a Cav1.4 channel regulator, in photoreceptor synaptic function. Nat Neurosci. 2004;7:1079–1087. doi: 10.1038/nn1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Eran A, Graham KR, Vatalaro K, McCarthy J, Collins C, et al. Comment on “Autistic-like phenotypes in Cadps2-knockout mice and aberrant CADPS2 splicing in autistic patients”. J Clin Invest. 2009;119:679–680. doi: 10.1172/JCI38620. author reply 680–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Sadakata T, Furuichi T. Developmentally Regulated Ca(2+)-Dependent Activator Protein for Secretion 2 (CAPS2) is Involved in BDNF Secretion and is Associated with Autism Susceptibility. Cerebellum. 2009 doi: 10.1007/s12311-009-0097-5. [DOI] [PubMed] [Google Scholar]
- 246.Sadakata T, Washida M, Furuichi T. Alternative splicing variations in mouse CAPS2: differential expression and functional properties of splicing variants. BMC Neurosci. 2007;8:25. doi: 10.1186/1471-2202-8-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Sadakata T, Washida M, Iwayama Y, Shoji S, Sato Y, et al. Autistic-like phenotypes in Cadps2-knockout mice and aberrant CADPS2 splicing in autistic patients. J Clin Invest. 2007;117:931–943. doi: 10.1172/JCI29031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Sadakata T, Kakegawa W, Mizoguchi A, Washida M, Katoh-Semba R, et al. Impaired cerebellar development and function in mice lacking CAPS2, a protein involved in neurotrophin release. J Neurosci. 2007;27:2472–2482. doi: 10.1523/JNEUROSCI.2279-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Sadakata T, Washida M, Morita N, Furuichi T. Tissue distribution of Ca2+-dependent activator protein for secretion family members CAPS1 and CAPS2 in mice. J Histochem Cytochem. 2007;55:301–311. doi: 10.1369/jhc.6A7033.2006. [DOI] [PubMed] [Google Scholar]
- 250.Feng G, Krejci E, Molgo J, Cunningham JM, Massoulie J, et al. Genetic analysis of collagen Q: roles in acetylcholinesterase and butyrylcholinesterase assembly and in synaptic structure and function. J Cell Biol. 1999;144:1349–1360. doi: 10.1083/jcb.144.6.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Fritschy JM, Schweizer C, Brunig I, Luscher B. Pre- and post-synaptic mechanisms regulating the clustering of type A gamma-aminobutyric acid receptors (GABAA receptors). Biochem Soc Trans. 2003;31:889–892. doi: 10.1042/bst0310889. [DOI] [PubMed] [Google Scholar]
- 252.Banks GB, Chamberlain JS, Froehner SC. Truncated dystrophins can influence neuromuscular synapse structure. Mol Cell Neurosci. 2009;40:433–441. doi: 10.1016/j.mcn.2008.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Fradkin LG, Baines RA, van der Plas MC, Noordermeer JN. The dystrophin Dp186 isoform regulates neurotransmitter release at a central synapse in Drosophila. J Neurosci. 2008;28:5105–5114. doi: 10.1523/JNEUROSCI.4950-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Bogdanik L, Framery B, Frolich A, Franco B, Mornet D, et al. Muscle dystroglycan organizes the postsynapse and regulates presynaptic neurotransmitter release at the Drosophila neuromuscular junction. PLoS One. 2008;3:e2084. doi: 10.1371/journal.pone.0002084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Minatel E, Neto HS, Marques MJ. Acetylcholine receptor distribution and synapse elimination at the developing neuromuscular junction of mdx mice. Muscle Nerve. 2003;28:561–569. doi: 10.1002/mus.10416. [DOI] [PubMed] [Google Scholar]
- 256.Marchand S, Stetzkowski-Marden F, Cartaud J. Differential targeting of components of the dystrophin complex to the postsynaptic membrane. Eur J Neurosci. 2001;13:221–229. [PubMed] [Google Scholar]
- 257.Monroig O, Rotllant J, Sanchez E, Cerda-Reverter JM, Tocher DR. Expression of long-chain polyunsaturated fatty acid (LC-PUFA) biosynthesis genes during zebrafish Danio rerio early embryogenesis. Biochim Biophys Acta. 2009. [DOI] [PubMed]
- 258.Fernandez M, Segura MF, Sole C, Colino A, Comella JX, et al. Lifeguard/neuronal membrane protein 35 regulates Fas ligand-mediated apoptosis in neurons via microdomain recruitment. J Neurochem. 2007;103:190–203. doi: 10.1111/j.1471-4159.2007.04767.x. [DOI] [PubMed] [Google Scholar]
- 259.Reimers K, Choi CY, Mau-Thek E, Vogt PM. Sequence analysis shows that Lifeguard belongs to a new evolutionarily conserved cytoprotective family. Int J Mol Med. 2006;18:729–734. [PubMed] [Google Scholar]
- 260.Chen W, Lee J, Cho SY, Fine HA. Proteasome-mediated destruction of the cyclin a/cyclin-dependent kinase 2 complex suppresses tumor cell growth in vitro and in vivo. Cancer Res. 2004;64:3949–3957. doi: 10.1158/0008-5472.CAN-03-3906. [DOI] [PubMed] [Google Scholar]
- 261.Somia NV, Schmitt MJ, Vetter DE, Van Antwerp D, Heinemann SF, et al. LFG: an anti-apoptotic gene that provides protection from Fas-mediated cell death. Proc Natl Acad Sci U S A. 1999;96:12667–12672. doi: 10.1073/pnas.96.22.12667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Lopez-Bendito G, Shigemoto R, Kulik A, Paulsen O, Fairen A, et al. Expression and distribution of metabotropic GABA receptor subtypes GABABR1 and GABABR2 during rat neocortical development. Eur J Neurosci. 2002;15:1766–1778. doi: 10.1046/j.1460-9568.2002.02032.x. [DOI] [PubMed] [Google Scholar]
- 263.Fatemi SH, Folsom TD, Reutiman TJ, Thuras PD. Expression of GABA(B) receptors is altered in brains of subjects with autism. Cerebellum. 2009;8:64–69. doi: 10.1007/s12311-008-0075-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Correa SA, Munton R, Nishimune A, Fitzjohn S, Henley JM. Development of GABAB subunits and functional GABAB receptors in rat cultured hippocampal neurons. Neuropharmacology. 2004;47:475–484. doi: 10.1016/j.neuropharm.2004.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Straessle A, Loup F, Arabadzisz D, Ohning GV, Fritschy JM. Rapid and long-term alterations of hippocampal GABAB receptors in a mouse model of temporal lobe epilepsy. Eur J Neurosci. 2003;18:2213–2226. doi: 10.1046/j.1460-9568.2003.02964.x. [DOI] [PubMed] [Google Scholar]
- 266.Mead AN, Morris HV, Dixon CI, Rulten SL, Mayne LV, et al. AMPA receptor GluR2, but not GluR1, subunit deletion impairs emotional response conditioning in mice. Behav Neurosci. 2006;120:241–248. doi: 10.1037/0735-7044.120.2.241. [DOI] [PubMed] [Google Scholar]
- 267.Beveridge NJ, Tooney PA, Carroll AP, Gardiner E, Bowden N, et al. Dysregulation of miRNA 181b in the temporal cortex in schizophrenia. Hum Mol Genet. 2008;17:1156–1168. doi: 10.1093/hmg/ddn005. [DOI] [PubMed] [Google Scholar]
- 268.Le-Niculescu H, Balaraman Y, Patel S, Tan J, Sidhu K, et al. Towards understanding the schizophrenia code: an expanded convergent functional genomics approach. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:129–158. doi: 10.1002/ajmg.b.30481. [DOI] [PubMed] [Google Scholar]
- 269.Dhar SS, Liang HL, Wong-Riley MT. Nuclear respiratory factor 1 co-regulates AMPA glutamate receptor subunit 2 and cytochrome c oxidase: tight coupling of glutamatergic transmission and energy metabolism in neurons. J Neurochem. 2009;108:1595–1606. doi: 10.1111/j.1471-4159.2009.05929.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Mead AN, Stephens DN. Involvement of AMPA receptor GluR2 subunits in stimulus-reward learning: evidence from glutamate receptor gria2 knock-out mice. J Neurosci. 2003;23:9500–9507. doi: 10.1523/JNEUROSCI.23-29-09500.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Williams C, Mehrian Shai R, Wu Y, Hsu YH, Sitzer T, et al. Transcriptome analysis of synaptoneurosomes identifies neuroplasticity genes overexpressed in incipient Alzheimer's disease. PLoS One. 2009;4:e4936. doi: 10.1371/journal.pone.0004936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Perlis RH, Smoller JW, Ferreira MA, McQuillin A, Bass N, et al. A genomewide association study of response to lithium for prevention of recurrence in bipolar disorder. Am J Psychiatry. 2009;166:718–725. doi: 10.1176/appi.ajp.2009.08111633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Medvedev NI, Rodriguez-Arellano JJ, Popov VI, Davies HA, Tigaret CM, et al. The glutamate receptor 2 subunit controls post-synaptic density complexity and spine shape in the dentate gyrus. Eur J Neurosci. 2008;27:315–325. doi: 10.1111/j.1460-9568.2007.06005.x. [DOI] [PubMed] [Google Scholar]
- 274.Hou Q, Zhang D, Jarzylo L, Huganir RL, Man HY. Homeostatic regulation of AMPA receptor expression at single hippocampal synapses. Proc Natl Acad Sci U S A. 2008;105:775–780. doi: 10.1073/pnas.0706447105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Heine M, Thoumine O, Mondin M, Tessier B, Giannone G, et al. Activity-independent and subunit-specific recruitment of functional AMPA receptors at neurexin/neuroligin contacts. Proc Natl Acad Sci U S A. 2008;105:20947–20952. doi: 10.1073/pnas.0804007106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Bagal AA, Kao JP, Tang CM, Thompson SM. Long-term potentiation of exogenous glutamate responses at single dendritic spines. Proc Natl Acad Sci U S A. 2005;102:14434–14439. doi: 10.1073/pnas.0501956102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Passafaro M, Nakagawa T, Sala C, Sheng M. Induction of dendritic spines by an extracellular domain of AMPA receptor subunit GluR2. Nature. 2003;424:677–681. doi: 10.1038/nature01781. [DOI] [PubMed] [Google Scholar]
- 278.Tsim KW, Choi RC, Siow NL, Cheng AW, Ling KK, et al. ATP induces post-synaptic gene expressions in vertebrate skeletal neuromuscular junctions. J Neurocytol. 2003;32:603–617. doi: 10.1023/B:NEUR.0000020613.25367.78. [DOI] [PubMed] [Google Scholar]
- 279.Zhang GC, Mao LM, Liu XY, Parelkar NK, Arora A, et al. In vivo regulation of Homer1a expression in the striatum by cocaine. Mol Pharmacol. 2007;71:1148–1158. doi: 10.1124/mol.106.028399. [DOI] [PubMed] [Google Scholar]
- 280.Todd KJ, Auld DS, Robitaille R. Neurotrophins modulate neuron-glia interactions at a vertebrate synapse. Eur J Neurosci. 2007;25:1287–1296. doi: 10.1111/j.1460-9568.2007.05385.x. [DOI] [PubMed] [Google Scholar]
- 281.Mao L, Yang L, Tang Q, Samdani S, Zhang G, et al. The scaffold protein Homer1b/c links metabotropic glutamate receptor 5 to extracellular signal-regulated protein kinase cascades in neurons. J Neurosci. 2005;25:2741–2752. doi: 10.1523/JNEUROSCI.4360-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Ogasawara H, Doi T, Kawato M. Systems biology perspectives on cerebellar long-term depression. Neurosignals. 2008;16:300–317. doi: 10.1159/000123040. [DOI] [PubMed] [Google Scholar]
- 283.Faraut B, Barbier J, Ravel-Chapuis A, Doyennette MA, Jandrot-Perrus M, et al. Thrombin downregulates muscle acetylcholine receptors via an IP3 signaling pathway by activating its G-protein-coupled protease-activated receptor-1. J Cell Physiol. 2003;196:105–112. doi: 10.1002/jcp.10280. [DOI] [PubMed] [Google Scholar]
- 284.Sakae N, Yamasaki N, Kitaichi K, Fukuda T, Yamada M, et al. Mice lacking the schizophrenia-associated protein FEZ1 manifest hyperactivity and enhanced responsiveness to psychostimulants. Hum Mol Genet. 2008;17:3191–3203. doi: 10.1093/hmg/ddn215. [DOI] [PubMed] [Google Scholar]
- 285.Konno D, Ko JA, Usui S, Hori K, Maruoka H, et al. The postsynaptic density and dendritic raft localization of PSD-Zip70, which contains an N-myristoylation sequence and leucine-zipper motifs. J Cell Sci. 2002;115:4695–4706. doi: 10.1242/jcs.00127. [DOI] [PubMed] [Google Scholar]
- 286.Bardoni B, Giglio S, Schenck A, Rocchi M, Mandel JL. Assignment of NUFIP1 (nuclear FMRP interacting protein 1) gene to chromosome 13q14 and assignment of a pseudogene to chromosome 6q12. Cytogenet Cell Genet. 2000;89:11–13. doi: 10.1159/000015580. [DOI] [PubMed] [Google Scholar]
- 287.Bardoni B, Schenck A, Mandel JL. A novel RNA-binding nuclear protein that interacts with the fragile X mental retardation (FMR1) protein. Hum Mol Genet. 1999;8:2557–2566. doi: 10.1093/hmg/8.13.2557. [DOI] [PubMed] [Google Scholar]
- 288.Bardoni B, Willemsen R, Weiler IJ, Schenck A, Severijnen LA, et al. NUFIP1 (nuclear FMRP interacting protein 1) is a nucleocytoplasmic shuttling protein associated with active synaptoneurosomes. Exp Cell Res. 2003;289:95–107. doi: 10.1016/s0014-4827(03)00222-2. [DOI] [PubMed] [Google Scholar]
- 289.Macauley SL, Wozniak DF, Kielar C, Tan Y, Cooper JD, et al. Cerebellar pathology and motor deficits in the palmitoyl protein thioesterase 1-deficient mouse. Exp Neurol. 2009;217:124–135. doi: 10.1016/j.expneurol.2009.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Tardy C, Sabourdy F, Garcia V, Jalanko A, Therville N, et al. Palmitoyl protein thioesterase 1 modulates tumor necrosis factor alpha-induced apoptosis. Biochim Biophys Acta. 2009;1793:1250–1258. doi: 10.1016/j.bbamcr.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 291.Kim SJ, Zhang Z, Sarkar C, Tsai PC, Lee YC, et al. Palmitoyl protein thioesterase-1 deficiency impairs synaptic vesicle recycling at nerve terminals, contributing to neuropathology in humans and mice. J Clin Invest. 2008;118:3075–3086. doi: 10.1172/JCI33482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Zhang Z, Lee YC, Kim SJ, Choi MS, Tsai PC, et al. Production of lysophosphatidylcholine by cPLA2 in the brain of mice lacking PPT1 is a signal for phagocyte infiltration. Hum Mol Genet. 2007;16:837–847. doi: 10.1093/hmg/ddm029. [DOI] [PubMed] [Google Scholar]
- 293.Ramadan H, Al-Din AS, Ismail A, Balen F, Varma A, et al. Adult neuronal ceroid lipofuscinosis caused by deficiency in palmitoyl protein thioesterase 1. Neurology. 2007;68:387–388. doi: 10.1212/01.wnl.0000252825.85947.2f. [DOI] [PubMed] [Google Scholar]
- 294.Qiao X, Lu JY, Hofmann SL. Gene expression profiling in a mouse model of infantile neuronal ceroid lipofuscinosis reveals upregulation of immediate early genes and mediators of the inflammatory response. BMC Neurosci. 2007;8:95. doi: 10.1186/1471-2202-8-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Lyly A, von Schantz C, Salonen T, Kopra O, Saarela J, et al. Glycosylation, transport, and complex formation of palmitoyl protein thioesterase 1 (PPT1)–distinct characteristics in neurons. BMC Cell Biol. 2007;8:22. doi: 10.1186/1471-2121-8-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Kielar C, Maddox L, Bible E, Pontikis CC, Macauley SL, et al. Successive neuron loss in the thalamus and cortex in a mouse model of infantile neuronal ceroid lipofuscinosis. Neurobiol Dis. 2007;25:150–162. doi: 10.1016/j.nbd.2006.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Kalviainen R, Eriksson K, Losekoot M, Sorri I, Harvima I, et al. Juvenile-onset neuronal ceroid lipofuscinosis with infantile CLN1 mutation and palmitoyl-protein thioesterase deficiency. Eur J Neurol. 2007;14:369–372. doi: 10.1111/j.1468-1331.2007.01668.x. [DOI] [PubMed] [Google Scholar]
- 298.Ahtiainen L, Kolikova J, Mutka AL, Luiro K, Gentile M, et al. Palmitoyl protein thioesterase 1 (Ppt1)-deficient mouse neurons show alterations in cholesterol metabolism and calcium homeostasis prior to synaptic dysfunction. Neurobiol Dis. 2007;28:52–64. doi: 10.1016/j.nbd.2007.06.012. [DOI] [PubMed] [Google Scholar]
- 299.Zhang Z, Lee YC, Kim SJ, Choi MS, Tsai PC, et al. Palmitoyl-protein thioesterase-1 deficiency mediates the activation of the unfolded protein response and neuronal apoptosis in INCL. Hum Mol Genet. 2006;15:337–346. doi: 10.1093/hmg/ddi451. [DOI] [PubMed] [Google Scholar]
- 300.Bible E, Gupta P, Hofmann SL, Cooper JD. Regional and cellular neuropathology in the palmitoyl protein thioesterase-1 null mutant mouse model of infantile neuronal ceroid lipofuscinosis. Neurobiol Dis. 2004;16:346–359. doi: 10.1016/j.nbd.2004.02.010. [DOI] [PubMed] [Google Scholar]
- 301.Ahtiainen L, Van Diggelen OP, Jalanko A, Kopra O. Palmitoyl protein thioesterase 1 is targeted to the axons in neurons. J Comp Neurol. 2003;455:368–377. doi: 10.1002/cne.10492. [DOI] [PubMed] [Google Scholar]
- 302.Francis SC, Sunshine C, Kirk KL. Coordinate regulation of catecholamine uptake by rab3 and phosphoinositide 3-kinase. J Biol Chem. 2002;277:7816–7823. doi: 10.1074/jbc.M109743200. [DOI] [PubMed] [Google Scholar]
- 303.Schluter OM, Basu J, Sudhof TC, Rosenmund C. Rab3 superprimes synaptic vesicles for release: implications for short-term synaptic plasticity. J Neurosci. 2006;26:1239–1246. doi: 10.1523/JNEUROSCI.3553-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Sons MS, Plomp JJ. Rab3A deletion selectively reduces spontaneous neurotransmitter release at the mouse neuromuscular synapse. Brain Res. 2006;1089:126–134. doi: 10.1016/j.brainres.2006.03.055. [DOI] [PubMed] [Google Scholar]
- 305.Nishioka H, Haraoka J. Significance of immunohistochemical expression of Rab3B and SNAP-25 in growth hormone-producing pituitary adenomas. Acta Neuropathol. 2005;109:598–602. doi: 10.1007/s00401-005-1008-6. [DOI] [PubMed] [Google Scholar]
- 306.Schluter OM, Schmitz F, Jahn R, Rosenmund C, Sudhof TC. A complete genetic analysis of neuronal Rab3 function. J Neurosci. 2004;24:6629–6637. doi: 10.1523/JNEUROSCI.1610-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Matsuno A, Itoh J, Takekoshi S, Itoh Y, Ohsugi Y, et al. Dynamics of subcellular organelles, growth hormone, Rab3B, SNAP-25, and syntaxin in rat pituitary cells caused by growth hormone releasing hormone and somatostatin. Microsc Res Tech. 2003;62:232–239. doi: 10.1002/jemt.10364. [DOI] [PubMed] [Google Scholar]
- 308.Sidhu RS, Bhullar RP. Rab3B in human platelet is membrane bound and interacts with Ca(2+)/calmodulin. Biochem Biophys Res Commun. 2001;289:1039–1043. doi: 10.1006/bbrc.2001.6113. [DOI] [PubMed] [Google Scholar]
- 309.Masumoto N, Ikebuchi Y, Tahara M, Yokoi T, Tasaka K, et al. Expression of Rab3A in the cortical region in mouse metaphase II eggs. J Exp Zool. 1998;280:91–96. [PubMed] [Google Scholar]
- 310.Madison DL, Kruger WH, Kim T, Pfeiffer SE. Differential expression of rab3 isoforms in oligodendrocytes and astrocytes. J Neurosci Res. 1996;45:258–268. doi: 10.1002/(SICI)1097-4547(19960801)45:3<258::AID-JNR7>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 311.Grabs D, Bergmann M, Urban M, Post A, Gratzl M. Rab3 proteins and SNAP-25, essential components of the exocytosis machinery in conventional synapses, are absent from ribbon synapses of the mouse retina. Eur J Neurosci. 1996;8:162–168. doi: 10.1111/j.1460-9568.1996.tb01177.x. [DOI] [PubMed] [Google Scholar]
- 312.Stettler O, Nothias F, Tavitian B, Vernier P. Double in situ hybridization reveals overlapping neuronal populations expressing the low molecular weight GTPases Rab3a and Rab3b in Rat brain. Eur J Neurosci. 1995;7:702–713. doi: 10.1111/j.1460-9568.1995.tb00674.x. [DOI] [PubMed] [Google Scholar]
- 313.Redecker P, Cetin Y, Grube D. Differential distribution of synaptotagmin I and rab3 in the anterior pituitary of four mammalian species. Neuroendocrinology. 1995;62:101–110. doi: 10.1159/000126994. [DOI] [PubMed] [Google Scholar]
- 314.Fischer von Mollard G, Stahl B, Khokhlatchev A, Sudhof TC, Jahn R. Rab3C is a synaptic vesicle protein that dissociates from synaptic vesicles after stimulation of exocytosis. J Biol Chem. 1994;269:10971–10974. [PubMed] [Google Scholar]
- 315.Lledo PM, Vernier P, Vincent JD, Mason WT, Zorec R. Inhibition of Rab3B expression attenuates Ca(2+)-dependent exocytosis in rat anterior pituitary cells. Nature. 1993;364:540–544. doi: 10.1038/364540a0. [DOI] [PubMed] [Google Scholar]
- 316.Lledo PM, Johannes L, Vernier P, Henry JP, Vincent JD, et al. [Calcium-dependent regulated secretion is controlled by GTPase Rab3 in neuroendocrine cells]. C R Seances Soc Biol Fil. 1993;187:726–736. [PubMed] [Google Scholar]
- 317.Karniguian A, Zahraoui A, Tavitian A. Identification of small GTP-binding rab proteins in human platelets: thrombin-induced phosphorylation of rab3B, rab6, and rab8 proteins. Proc Natl Acad Sci U S A. 1993;90:7647–7651. doi: 10.1073/pnas.90.16.7647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Weidenhofer J, Scott RJ, Tooney PA. Investigation of the expression of genes affecting cytomatrix active zone function in the amygdala in schizophrenia: effects of antipsychotic drugs. J Psychiatr Res. 2009;43:282–290. doi: 10.1016/j.jpsychires.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 319.Fujimoto K, Shibasaki T, Yokoi N, Kashima Y, Matsumoto M, et al. Piccolo, a Ca2+ sensor in pancreatic beta-cells. Involvement of cAMP-GEFII.Rim2.Piccolo complex in cAMP-dependent exocytosis. J Biol Chem. 2002;277:50497–50502. doi: 10.1074/jbc.M210146200. [DOI] [PubMed] [Google Scholar]
- 320.Kashima Y, Miki T, Shibasaki T, Ozaki N, Miyazaki M, et al. Critical role of cAMP-GEFII–Rim2 complex in incretin-potentiated insulin secretion. J Biol Chem. 2001;276:46046–46053. doi: 10.1074/jbc.M108378200. [DOI] [PubMed] [Google Scholar]
- 321.Ozaki N, Shibasaki T, Kashima Y, Miki T, Takahashi K, et al. cAMP-GEFII is a direct target of cAMP in regulated exocytosis. Nat Cell Biol. 2000;2:805–811. doi: 10.1038/35041046. [DOI] [PubMed] [Google Scholar]
- 322.Brandstatter JH, Dick O, Boeckers TM. The postsynaptic scaffold proteins ProSAP1/Shank2 and Homer1 are associated with glutamate receptor complexes at rat retinal synapses. J Comp Neurol. 2004;475:551–563. doi: 10.1002/cne.20194. [DOI] [PubMed] [Google Scholar]
- 323.Uemura T, Mori H, Mishina M. Direct interaction of GluRdelta2 with Shank scaffold proteins in cerebellar Purkinje cells. Mol Cell Neurosci. 2004;26:330–341. doi: 10.1016/j.mcn.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 324.Bockers TM, Segger-Junius M, Iglauer P, Bockmann J, Gundelfinger ED, et al. Differential expression and dendritic transcript localization of Shank family members: identification of a dendritic targeting element in the 3′ untranslated region of Shank1 mRNA. Mol Cell Neurosci. 2004;26:182–190. doi: 10.1016/j.mcn.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 325.Qualmann B, Boeckers TM, Jeromin M, Gundelfinger ED, Kessels MM. Linkage of the actin cytoskeleton to the postsynaptic density via direct interactions of Abp1 with the ProSAP/Shank family. J Neurosci. 2004;24:2481–2495. doi: 10.1523/JNEUROSCI.5479-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 326.de Bartolomeis A, Fiore G. Postsynaptic density scaffolding proteins at excitatory synapse and disorders of synaptic plasticity: implications for human behavior pathologies. Int Rev Neurobiol. 2004;59:221–254. doi: 10.1016/S0074-7742(04)59009-8. [DOI] [PubMed] [Google Scholar]
- 327.Hwang JI, Kim HS, Lee JR, Kim E, Ryu SH, et al. The interaction of phospholipase C-beta3 with Shank2 regulates mGluR-mediated calcium signal. J Biol Chem. 2005;280:12467–12473. doi: 10.1074/jbc.M410740200. [DOI] [PubMed] [Google Scholar]
- 328.Bresler T, Shapira M, Boeckers T, Dresbach T, Futter M, et al. Postsynaptic density assembly is fundamentally different from presynaptic active zone assembly. J Neurosci. 2004;24:1507–1520. doi: 10.1523/JNEUROSCI.3819-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 329.Kim JY, Han W, Namkung W, Lee JH, Kim KH, et al. Inhibitory regulation of cystic fibrosis transmembrane conductance regulator anion-transporting activities by Shank2. J Biol Chem. 2004;279:10389–10396. doi: 10.1074/jbc.M312871200. [DOI] [PubMed] [Google Scholar]
- 330.Park E, Na M, Choi J, Kim S, Lee JR, et al. The Shank family of postsynaptic density proteins interacts with and promotes synaptic accumulation of the beta PIX guanine nucleotide exchange factor for Rac1 and Cdc42. J Biol Chem. 2003;278:19220–19229. doi: 10.1074/jbc.M301052200. [DOI] [PubMed] [Google Scholar]
- 331.Kreienkamp HJ, Soltau M, Richter D, Bockers T. Interaction of G-protein-coupled receptors with synaptic scaffolding proteins. Biochem Soc Trans. 2002;30:464–468. doi: 10.1042/bst0300464. [DOI] [PubMed] [Google Scholar]
- 332.Boeckers TM, Bockmann J, Kreutz MR, Gundelfinger ED. ProSAP/Shank proteins - a family of higher order organizing molecules of the postsynaptic density with an emerging role in human neurological disease. J Neurochem. 2002;81:903–910. doi: 10.1046/j.1471-4159.2002.00931.x. [DOI] [PubMed] [Google Scholar]
- 333.Ehlers MD. Molecular morphogens for dendritic spines. Trends Neurosci. 2002;25:64–67. doi: 10.1016/s0166-2236(02)02061-1. [DOI] [PubMed] [Google Scholar]
- 334.Okamoto PM, Gamby C, Wells D, Fallon J, Vallee RB. Dynamin isoform-specific interaction with the shank/ProSAP scaffolding proteins of the postsynaptic density and actin cytoskeleton. J Biol Chem. 2001;276:48458–48465. doi: 10.1074/jbc.M104927200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 335.Kreienkamp HJ, Zitzer H, Richter D. Identification of proteins interacting with the rat somatostatin receptor subtype 2. J Physiol Paris. 2000;94:193–198. doi: 10.1016/s0928-4257(00)00204-7. [DOI] [PubMed] [Google Scholar]
- 336.Tobaben S, Sudhof TC, Stahl B. The G protein-coupled receptor CL1 interacts directly with proteins of the Shank family. J Biol Chem. 2000;275:36204–36210. doi: 10.1074/jbc.M006448200. [DOI] [PubMed] [Google Scholar]
- 337.Sheng M, Kim E. The Shank family of scaffold proteins. J Cell Sci. 2000;113(Pt 11):1851–1856. doi: 10.1242/jcs.113.11.1851. [DOI] [PubMed] [Google Scholar]
- 338.Ehlers MD. Synapse structure: glutamate receptors connected by the shanks. Curr Biol. 1999;9:R848–850. doi: 10.1016/s0960-9822(00)80043-3. [DOI] [PubMed] [Google Scholar]
- 339.Boeckers TM, Winter C, Smalla KH, Kreutz MR, Bockmann J, et al. Proline-rich synapse-associated proteins ProSAP1 and ProSAP2 interact with synaptic proteins of the SAPAP/GKAP family. Biochem Biophys Res Commun. 1999;264:247–252. doi: 10.1006/bbrc.1999.1489. [DOI] [PubMed] [Google Scholar]
- 340.Lim S, Naisbitt S, Yoon J, Hwang JI, Suh PG, et al. Characterization of the Shank family of synaptic proteins. Multiple genes, alternative splicing, and differential expression in brain and development. J Biol Chem. 1999;274:29510–29518. doi: 10.1074/jbc.274.41.29510. [DOI] [PubMed] [Google Scholar]
- 341.Bajjalieh SM, Peterson K, Linial M, Scheller RH. Brain contains two forms of synaptic vesicle protein 2. Proc Natl Acad Sci U S A. 1993;90:2150–2154. doi: 10.1073/pnas.90.6.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 342.Lynch BA, Lambeng N, Nocka K, Kensel-Hammes P, Bajjalieh SM, et al. The synaptic vesicle protein SV2A is the binding site for the antiepileptic drug levetiracetam. Proc Natl Acad Sci U S A. 2004;101:9861–9866. doi: 10.1073/pnas.0308208101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Bindra PS, Knowles R, Buckley KM. Conservation of the amino acid sequence of SV2, a transmembrane transporter in synaptic vesicles and endocrine cells. Gene. 1993;137:299–302. doi: 10.1016/0378-1119(93)90024-w. [DOI] [PubMed] [Google Scholar]
- 344.Schivell AE, Batchelor RH, Bajjalieh SM. Isoform-specific, calcium-regulated interaction of the synaptic vesicle proteins SV2 and synaptotagmin. J Biol Chem. 1996;271:27770–27775. doi: 10.1074/jbc.271.44.27770. [DOI] [PubMed] [Google Scholar]
- 345.Yao J, Bajjalieh SM. Synaptic vesicle protein 2 binds adenine nucleotides. J Biol Chem. 2008;283:20628–20634. doi: 10.1074/jbc.M800738200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Iezzi M, Theander S, Janz R, Loze C, Wollheim CB. SV2A and SV2C are not vesicular Ca2+ transporters but control glucose-evoked granule recruitment. J Cell Sci. 2005;118:5647–5660. doi: 10.1242/jcs.02658. [DOI] [PubMed] [Google Scholar]
- 347.Lockhart ST, Mead JN, Pisano JM, Slonimsky JD, Birren SJ. Nerve growth factor collaborates with myocyte-derived factors to promote development of presynaptic sites in cultured sympathetic neurons. J Neurobiol. 2000;42:460–476. [PubMed] [Google Scholar]
- 348.Hayashi M, Yamamoto A, Yatsushiro S, Yamada H, Futai M, et al. Synaptic vesicle protein SV2B, but not SV2A, is predominantly expressed and associated with microvesicles in rat pinealocytes. J Neurochem. 1998;71:356–365. doi: 10.1046/j.1471-4159.1998.71010356.x. [DOI] [PubMed] [Google Scholar]
- 349.Scranton TW, Iwata M, Carlson SS. The SV2 protein of synaptic vesicles is a keratan sulfate proteoglycan. J Neurochem. 1993;61:29–44. doi: 10.1111/j.1471-4159.1993.tb03535.x. [DOI] [PubMed] [Google Scholar]
- 350.Bajjalieh SM, Frantz GD, Weimann JM, McConnell SK, Scheller RH. Differential expression of synaptic vesicle protein 2 (SV2) isoforms. J Neurosci. 1994;14:5223–5235. doi: 10.1523/JNEUROSCI.14-09-05223.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 351.Custer KL, Austin NS, Sullivan JM, Bajjalieh SM. Synaptic vesicle protein 2 enhances release probability at quiescent synapses. J Neurosci. 2006;26:1303–1313. doi: 10.1523/JNEUROSCI.2699-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 352.Janz R, Hofmann K, Sudhof TC. SVOP, an evolutionarily conserved synaptic vesicle protein, suggests novel transport functions of synaptic vesicles. J Neurosci. 1998;18:9269–9281. doi: 10.1523/JNEUROSCI.18-22-09269.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 353.Dong M, Liu H, Tepp WH, Johnson EA, Janz R, et al. Glycosylated SV2A and SV2B mediate the entry of botulinum neurotoxin E into neurons. Mol Biol Cell. 2008;19:5226–5237. doi: 10.1091/mbc.E08-07-0765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Schivell AE, Mochida S, Kensel-Hammes P, Custer KL, Bajjalieh SM. SV2A and SV2C contain a unique synaptotagmin-binding site. Mol Cell Neurosci. 2005;29:56–64. doi: 10.1016/j.mcn.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 355.Janz R, Goda Y, Geppert M, Missler M, Sudhof TC. SV2A and SV2B function as redundant Ca2+ regulators in neurotransmitter release. Neuron. 1999;24:1003–1016. doi: 10.1016/s0896-6273(00)81046-6. [DOI] [PubMed] [Google Scholar]
- 356.Lambeng N, Grossmann M, Chatelain P, Fuks B. Solubilization and immunopurification of rat brain synaptic vesicle protein 2A with maintained binding properties. Neurosci Lett. 2006;398:107–112. doi: 10.1016/j.neulet.2005.12.059. [DOI] [PubMed] [Google Scholar]
- 357.Janz R, Sudhof TC. SV2C is a synaptic vesicle protein with an unusually restricted localization: anatomy of a synaptic vesicle protein family. Neuroscience. 1999;94:1279–1290. doi: 10.1016/s0306-4522(99)00370-x. [DOI] [PubMed] [Google Scholar]
- 358.Clegg N, Ferguson C, True LD, Arnold H, Moorman A, et al. Molecular characterization of prostatic small-cell neuroendocrine carcinoma. Prostate. 2003;55:55–64. doi: 10.1002/pros.10217. [DOI] [PubMed] [Google Scholar]
- 359.Dong M, Yeh F, Tepp WH, Dean C, Johnson EA, et al. SV2 is the protein receptor for botulinum neurotoxin A. Science. 2006;312:592–596. doi: 10.1126/science.1123654. [DOI] [PubMed] [Google Scholar]
- 360.Leonoudakis D, Conti LR, Radeke CM, McGuire LM, Vandenberg CA. A multiprotein trafficking complex composed of SAP97, CASK, Veli, and Mint1 is associated with inward rectifier Kir2 potassium channels. J Biol Chem. 2004;279:19051–19063. doi: 10.1074/jbc.M400284200. [DOI] [PubMed] [Google Scholar]
- 361.Atasoy D, Schoch S, Ho A, Nadasy KA, Liu X, et al. Deletion of CASK in mice is lethal and impairs synaptic function. Proc Natl Acad Sci U S A. 2007;104:2525–2530. doi: 10.1073/pnas.0611003104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Klemmer P, Smit AB, Li KW. Proteomics analysis of immuno-precipitated synaptic protein complexes. J Proteomics. 2009;72:82–90. doi: 10.1016/j.jprot.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 363.Hernandez-Ortega K, Ferrera P, Arias C. Sequential expression of cell-cycle regulators and Alzheimer's disease-related proteins in entorhinal cortex after hippocampal excitotoxic damage. J Neurosci Res. 2007;85:1744–1751. doi: 10.1002/jnr.21301. [DOI] [PubMed] [Google Scholar]
- 364.Utreras E, Maccioni R, Gonzalez-Billault C. Cyclin-dependent kinase 5 activator p35 over-expression and amyloid beta synergism increase apoptosis in cultured neuronal cells. Neuroscience. 2009;161:978–987. doi: 10.1016/j.neuroscience.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 365.Ubeda M, Kemp DM, Habener JF. Glucose-induced expression of the cyclin-dependent protein kinase 5 activator p35 involved in Alzheimer's disease regulates insulin gene transcription in pancreatic beta-cells. Endocrinology. 2004;145:3023–3031. doi: 10.1210/en.2003-1522. [DOI] [PubMed] [Google Scholar]
- 366.Mateo I, Vazquez-Higuera JL, Sanchez-Juan P, Rodriguez-Rodriguez E, Infante J, et al. Epistasis between tau phosphorylation regulating genes (CDK5R1 and GSK-3beta) and Alzheimer's disease risk. Acta Neurol Scand. 2008 doi: 10.1111/j.1600-0404.2008.01128.x. [DOI] [PubMed] [Google Scholar]
- 367.Moncini S, Bevilacqua A, Venturin M, Fallini C, Ratti A, et al. The 3′ untranslated region of human Cyclin-Dependent Kinase 5 Regulatory subunit 1 contains regulatory elements affecting transcript stability. BMC Mol Biol. 2007;8:111. doi: 10.1186/1471-2199-8-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Rademakers R, Sleegers K, Theuns J, Van den Broeck M, Bel Kacem S, et al. Association of cyclin-dependent kinase 5 and neuronal activators p35 and p39 complex in early-onset Alzheimer's disease. Neurobiol Aging. 2005;26:1145–1151. doi: 10.1016/j.neurobiolaging.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 369.Lopes JP, Oliveira CR, Agostinho P. Role of cyclin-dependent kinase 5 in the neurodegenerative process triggered by amyloid-Beta and prion peptides: implications for Alzheimer's disease and prion-related encephalopathies. Cell Mol Neurobiol. 2007;27:943–957. doi: 10.1007/s10571-007-9224-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 370.Monaco EA., 3rd Recent evidence regarding a role for Cdk5 dysregulation in Alzheimer's disease. Curr Alzheimer Res. 2004;1:33–38. doi: 10.2174/1567205043480519. [DOI] [PubMed] [Google Scholar]
- 371.Maccioni RB, Otth C, Concha, Munoz JP. The protein kinase Cdk5. Structural aspects, roles in neurogenesis and involvement in Alzheimer's pathology. Eur J Biochem. 2001;268:1518–1527. doi: 10.1046/j.1432-1033.2001.02024.x. [DOI] [PubMed] [Google Scholar]
- 372.Sato S, Xu J, Okuyama S, Martinez LB, Walsh SM, et al. Spatial learning impairment, enhanced CDK5/p35 activity, and downregulation of NMDA receptor expression in transgenic mice expressing tau-tubulin kinase 1. J Neurosci. 2008;28:14511–14521. doi: 10.1523/JNEUROSCI.3417-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 373.Patrick GN, Zukerberg L, Nikolic M, de la Monte S, Dikkes P, et al. Conversion of p35 to p25 deregulates Cdk5 activity and promotes neurodegeneration. Nature. 1999;402:615–622. doi: 10.1038/45159. [DOI] [PubMed] [Google Scholar]
- 374.Orellana DI, Quintanilla RA, Maccioni RB. Neuroprotective effect of TNFalpha against the beta-amyloid neurotoxicity mediated by CDK5 kinase. Biochim Biophys Acta. 2007;1773:254–263. doi: 10.1016/j.bbamcr.2006.10.010. [DOI] [PubMed] [Google Scholar]
- 375.Ubeda M, Rukstalis JM, Habener JF. Inhibition of cyclin-dependent kinase 5 activity protects pancreatic beta cells from glucotoxicity. J Biol Chem. 2006;281:28858–28864. doi: 10.1074/jbc.M604690200. [DOI] [PubMed] [Google Scholar]
- 376.Camins A, Verdaguer E, Folch J, Canudas AM, Pallas M. The role of CDK5/P25 formation/inhibition in neurodegeneration. Drug News Perspect. 2006;19:453–460. doi: 10.1358/dnp.2006.19.8.1043961. [DOI] [PubMed] [Google Scholar]
- 377.Quintanilla RA, Orellana DI, Gonzalez-Billault C, Maccioni RB. Interleukin-6 induces Alzheimer-type phosphorylation of tau protein by deregulating the cdk5/p35 pathway. Exp Cell Res. 2004;295:245–257. doi: 10.1016/j.yexcr.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 378.Lee MS, Tsai LH. Cdk5: one of the links between senile plaques and neurofibrillary tangles? J Alzheimers Dis. 2003;5:127–137. doi: 10.3233/jad-2003-5207. [DOI] [PubMed] [Google Scholar]
- 379.Kesavapany S, Li BS, Pant HC. Cyclin-dependent kinase 5 in neurofilament function and regulation. Neurosignals. 2003;12:252–264. doi: 10.1159/000074627. [DOI] [PubMed] [Google Scholar]
- 380.Mauceri D, Gardoni F, Marcello E, Di Luca M. Dual role of CaMKII-dependent SAP97 phosphorylation in mediating trafficking and insertion of NMDA receptor subunit NR2A. J Neurochem. 2007;100:1032–1046. doi: 10.1111/j.1471-4159.2006.04267.x. [DOI] [PubMed] [Google Scholar]
- 381.Surena AL, de Faria GP, Studler JM, Peiretti F, Pidoux M, et al. DLG1/SAP97 modulates transforming growth factor alpha bioavailability. Biochim Biophys Acta. 2009;1793:264–272. doi: 10.1016/j.bbamcr.2008.09.005. [DOI] [PubMed] [Google Scholar]
- 382.Sato J, Shimazu D, Yamamoto N, Nishikawa T. An association analysis of synapse-associated protein 97 (SAP97) gene in schizophrenia. J Neural Transm. 2008;115:1355–1365. doi: 10.1007/s00702-008-0085-9. [DOI] [PubMed] [Google Scholar]
- 383.Cai C, Li H, Kangasniemi A, Pihlajamaa T, Von Ossowski L, et al. Somatostatin receptor subtype 1 is a PDZ ligand for synapse-associated protein 97 and a potential regulator of growth cone dynamics. Neuroscience. 2008;157:833–843. doi: 10.1016/j.neuroscience.2008.09.048. [DOI] [PubMed] [Google Scholar]
- 384.Migues PV, Cammarota M, Kavanagh J, Atkinson R, Powis DA, et al. Maturational changes in the subunit composition of AMPA receptors and the functional consequences of their activation in chicken forebrain. Dev Neurosci. 2007;29:232–240. doi: 10.1159/000096408. [DOI] [PubMed] [Google Scholar]
- 385.Cai C, Li H, Rivera C, Keinanen K. Interaction between SAP97 and PSD-95, two Maguk proteins involved in synaptic trafficking of AMPA receptors. J Biol Chem. 2006;281:4267–4273. doi: 10.1074/jbc.M505886200. [DOI] [PubMed] [Google Scholar]
- 386.Nash JE, Johnston TH, Collingridge GL, Garner CC, Brotchie JM. Subcellular redistribution of the synapse-associated proteins PSD-95 and SAP97 in animal models of Parkinson's disease and L-DOPA-induced dyskinesia. FASEB J. 2005;19:583–585. doi: 10.1096/fj.04-1854fje. [DOI] [PubMed] [Google Scholar]
- 387.Wakabayashi K, Narisawa-Saito M, Iwakura Y, Arai T, Ikeda K, et al. Phenotypic down-regulation of glutamate receptor subunit GluR1 in Alzheimer's disease. Neurobiol Aging. 1999;20:287–295. doi: 10.1016/s0197-4580(99)00035-4. [DOI] [PubMed] [Google Scholar]
- 388.Tavalin SJ. AKAP79 selectively enhances protein kinase C regulation of GluR1 at a Ca2+-calmodulin-dependent protein kinase II/protein kinase C site. J Biol Chem. 2008;283:11445–11452. doi: 10.1074/jbc.M709253200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 389.Gerdjikov TV, Rudolph U, Keist R, Mohler H, Feldon J, et al. Hippocampal alpha 5 subunit-containing GABA A receptors are involved in the development of the latent inhibition effect. Neurobiol Learn Mem. 2008;89:87–94. doi: 10.1016/j.nlm.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 390.Glykys J, Mann EO, Mody I. Which GABA(A) receptor subunits are necessary for tonic inhibition in the hippocampus? J Neurosci. 2008;28:1421–1426. doi: 10.1523/JNEUROSCI.4751-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 391.Bonin RP, Martin LJ, MacDonald JF, Orser BA. Alpha5GABAA receptors regulate the intrinsic excitability of mouse hippocampal pyramidal neurons. J Neurophysiol. 2007;98:2244–2254. doi: 10.1152/jn.00482.2007. [DOI] [PubMed] [Google Scholar]
- 392.Dawson GR, Maubach KA, Collinson N, Cobain M, Everitt BJ, et al. An inverse agonist selective for alpha5 subunit-containing GABAA receptors enhances cognition. J Pharmacol Exp Ther. 2006;316:1335–1345. doi: 10.1124/jpet.105.092320. [DOI] [PubMed] [Google Scholar]
- 393.Caraiscos VB, Newell JG, You-Ten KE, Elliott EM, Rosahl TW, et al. Selective enhancement of tonic GABAergic inhibition in murine hippocampal neurons by low concentrations of the volatile anesthetic isoflurane. J Neurosci. 2004;24:8454–8458. doi: 10.1523/JNEUROSCI.2063-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 394.Zarnowska ED, Keist R, Rudolph U, Pearce RA. GABAA receptor alpha5 subunits contribute to GABAA,slow synaptic inhibition in mouse hippocampus. J Neurophysiol. 2009;101:1179–1191. doi: 10.1152/jn.91203.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 395.Fernandez F, Esposito T, Lea RA, Colson NJ, Ciccodicola A, et al. Investigation of gamma-aminobutyric acid (GABA) A receptors genes and migraine susceptibility. BMC Med Genet. 2008;9:109. doi: 10.1186/1471-2350-9-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 396.Weiss J, O'Sullivan GA, Heinze L, Chen HX, Betz H, et al. Glycinergic input of small-field amacrine cells in the retinas of wildtype and glycine receptor deficient mice. Mol Cell Neurosci. 2008;37:40–55. doi: 10.1016/j.mcn.2007.08.012. [DOI] [PubMed] [Google Scholar]
- 397.Ramanathan S, Woodroffe A, Flodman PL, Mays LZ, Hanouni M, et al. A case of autism with an interstitial deletion on 4q leading to hemizygosity for genes encoding for glutamine and glycine neurotransmitter receptor sub-units (AMPA 2, GLRA3, GLRB) and neuropeptide receptors NPY1R, NPY5R. BMC Med Genet. 2004;5:10. doi: 10.1186/1471-2350-5-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Nikolic Z, Laube B, Weber RG, Lichter P, Kioschis P, et al. The human glycine receptor subunit alpha3. Glra3 gene structure, chromosomal localization, and functional characterization of alternative transcripts. J Biol Chem. 1998;273:19708–19714. doi: 10.1074/jbc.273.31.19708. [DOI] [PubMed] [Google Scholar]
- 399.Hunter RG, Bellani R, Bloss E, Costa A, McCarthy K, et al. Regulation of kainate receptor subunit mRNA by stress and corticosteroids in the rat hippocampus. PLoS One. 2009;4:e4328. doi: 10.1371/journal.pone.0004328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 400.Parisiadou L, Bethani I, Michaki V, Krousti K, Rapti G, et al. Homer2 and Homer3 interact with amyloid precursor protein and inhibit Abeta production. Neurobiol Dis. 2008;30:353–364. doi: 10.1016/j.nbd.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 401.Cao M, Xu J, Shen C, Kam C, Huganir RL, et al. PICK1-ICA69 heteromeric BAR domain complex regulates synaptic targeting and surface expression of AMPA receptors. J Neurosci. 2007;27:12945–12956. doi: 10.1523/JNEUROSCI.2040-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402.Huang Z, Shimazu K, Woo NH, Zang K, Muller U, et al. Distinct roles of the beta 1-class integrins at the developing and the mature hippocampal excitatory synapse. J Neurosci. 2006;26:11208–11219. doi: 10.1523/JNEUROSCI.3526-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Thomas U. Modulation of synaptic signalling complexes by Homer proteins. J Neurochem. 2002;81:407–413. doi: 10.1046/j.1471-4159.2002.00869.x. [DOI] [PubMed] [Google Scholar]
- 404.Scott LJ, Muglia P, Kong XQ, Guan W, Flickinger M, et al. Genome-wide association and meta-analysis of bipolar disorder in individuals of European ancestry. Proc Natl Acad Sci U S A. 2009;106:7501–7506. doi: 10.1073/pnas.0813386106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 405.Shin OH, Han W, Wang Y, Sudhof TC. Evolutionarily conserved multiple C2 domain proteins with two transmembrane regions (MCTPs) and unusual Ca2+ binding properties. J Biol Chem. 2005;280:1641–1651. doi: 10.1074/jbc.M407305200. [DOI] [PubMed] [Google Scholar]
- 406.Enz R, Croci C. Different binding motifs in metabotropic glutamate receptor type 7b for filamin A, protein phosphatase 1C, protein interacting with protein kinase C (PICK) 1 and syntenin allow the formation of multimeric protein complexes. Biochem J. 2003;372:183–191. doi: 10.1042/BJ20021750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 407.Boczan J, Leenders AG, Sheng ZH. Phosphorylation of syntaphilin by cAMP-dependent protein kinase modulates its interaction with syntaxin-1 and annuls its inhibitory effect on vesicle exocytosis. J Biol Chem. 2004;279:18911–18919. doi: 10.1074/jbc.M400496200. [DOI] [PubMed] [Google Scholar]
- 408.Kang JS, Tian JH, Pan PY, Zald P, Li C, et al. Docking of axonal mitochondria by syntaphilin controls their mobility and affects short-term facilitation. Cell. 2008;132:137–148. doi: 10.1016/j.cell.2007.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 409.Das S, Gerwin C, Sheng ZH. Syntaphilin binds to dynamin-1 and inhibits dynamin-dependent endocytosis. J Biol Chem. 2003;278:41221–41226. doi: 10.1074/jbc.M304851200. [DOI] [PubMed] [Google Scholar]
- 410.Das S, Boczan J, Gerwin C, Zald PB, Sheng ZH. Regional and developmental regulation of syntaphilin expression in the brain: a candidate molecular element of synaptic functional differentiation. Brain Res Mol Brain Res. 2003;116:38–49. doi: 10.1016/s0169-328x(03)00212-2. [DOI] [PubMed] [Google Scholar]
- 411.Lao G, Scheuss V, Gerwin CM, Su Q, Mochida S, et al. Syntaphilin: a syntaxin-1 clamp that controls SNARE assembly. Neuron. 2000;25:191–201. doi: 10.1016/s0896-6273(00)80882-x. [DOI] [PubMed] [Google Scholar]
- 412.Mahad DJ, Ziabreva I, Campbell G, Lax N, White K, et al. Mitochondrial changes within axons in multiple sclerosis. Brain. 2009;132:1161–1174. doi: 10.1093/brain/awp046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 413.Funakoshi E, Hamano A, Fukui M, Nishiyama N, Ogita K, et al. Molecular cloning of the m-Golsyn gene and its expression in the mouse brain. Gene Expr. 2006;13:27–40. doi: 10.3727/000000006783991917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Funakoshi E, Nakagawa KY, Hamano A, Hori T, Shimizu A, et al. Molecular cloning and characterization of gene for Golgi-localized syntaphilin-related protein on human chromosome 8q23. Gene. 2005;344:259–271. doi: 10.1016/j.gene.2004.10.024. [DOI] [PubMed] [Google Scholar]
- 415.Zhou D, Wang J, Zapala MA, Xue J, Schork NJ, et al. Gene expression in mouse brain following chronic hypoxia: role of sarcospan in glial cell death. Physiol Genomics. 2008;32:370–379. doi: 10.1152/physiolgenomics.00147.2007. [DOI] [PubMed] [Google Scholar]
- 416.Williams D, Aleman T, Lillo C, Lopes VS, Hughes LC, et al. Harmonin in the murine retina and the retinal phenotypes of Ush1c-mutant mice and human USH1C. Invest Ophthalmol Vis Sci. 2009 doi: 10.1167/iovs.08-3358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 417.Lillo C, Kitamoto J, Williams DS. Roles and interactions of usher 1 proteins in the outer retina. Adv Exp Med Biol. 2006;572:341–348. doi: 10.1007/0-387-32442-9_48. [DOI] [PubMed] [Google Scholar]
- 418.Reiners J, Nagel-Wolfrum K, Jurgens K, Marker T, Wolfrum U. Molecular basis of human Usher syndrome: deciphering the meshes of the Usher protein network provides insights into the pathomechanisms of the Usher disease. Exp Eye Res. 2006;83:97–119. doi: 10.1016/j.exer.2005.11.010. [DOI] [PubMed] [Google Scholar]
- 419.Keri S, Seres I, Kelemen O, Benedek G. The Relationship Among Neuregulin 1-Stimulated Phosphorylation of AKT, Psychosis Proneness, and Habituation of Arousal in Nonclinical Individuals. Schizophr Bull. 2009 doi: 10.1093/schbul/sbp063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 420.Le Strat Y, Ramoz N, Gorwood P. The role of genes involved in neuroplasticity and neurogenesis in the observation of a gene-environment interaction (GxE) in schizophrenia. Curr Mol Med. 2009;9:506–518. doi: 10.2174/156652409788167104. [DOI] [PubMed] [Google Scholar]
- 421.Schijndel JE, Loo KM, Zweeden MV, Djurovic S, Andreassen OA, et al. Three-cohort targeted gene screening reveals a non-synonymous TRKA polymorphism associated with schizophrenia. J Psychiatr Res. 2009 doi: 10.1016/j.jpsychires.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 422.So HC, Fong PY, Chen RY, Hui TC, Ng MY, et al. Identification of neuroglycan C and interacting partners as potential susceptibility genes for schizophrenia in a Southern Chinese population. Am J Med Genet B Neuropsychiatr Genet. 2009 doi: 10.1002/ajmg.b.30961. [DOI] [PubMed] [Google Scholar]
- 423.Wood JD, Bonath F, Kumar S, Ross CA, Cunliffe VT. Disrupted-in-schizophrenia 1 and neuregulin 1 are required for the specification of oligodendrocytes and neurones in the zebrafish brain. Hum Mol Genet. 2009;18:391–404. doi: 10.1093/hmg/ddn361. [DOI] [PubMed] [Google Scholar]
- 424.Wong J, Weickert CS. Transcriptional Interaction of an Estrogen Receptor Splice Variant and ErbB4 Suggests Convergence in Gene Susceptibility Pathways in Schizophrenia. J Biol Chem. 2009;284:18824–18832. doi: 10.1074/jbc.M109.013243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 425.Wang F, Jiang T, Sun Z, Teng SL, Luo X, et al. Neuregulin 1 genetic variation and anterior cingulum integrity in patients with schizophrenia and healthy controls. J Psychiatry Neurosci. 2009;34:181–186. [PMC free article] [PubMed] [Google Scholar]
- 426.Voineskos D, De Luca V, Macgregor S, Likhodi O, Miller L, et al. Neuregulin 1 and age of onset in the major psychoses. J Neural Transm. 2009;116:479–486. doi: 10.1007/s00702-008-0182-9. [DOI] [PubMed] [Google Scholar]
- 427.Tabares-Seisdedos R, Rubenstein JL. Chromosome 8p as a potential hub for developmental neuropsychiatric disorders: implications for schizophrenia, autism and cancer. Mol Psychiatry. 2009;14:563–589. doi: 10.1038/mp.2009.2. [DOI] [PubMed] [Google Scholar]
- 428.Reinhard S, Vela E, Bombara N, Devries GH, Raabe TD. Developmental regulation of Neuregulin1 isoforms and erbB receptor expression in intact rat dorsal root ganglia. Neurochem Res. 2009;34:17–22. doi: 10.1007/s11064-008-9732-7. [DOI] [PubMed] [Google Scholar]
- 429.Prata DP, Breen G, Osborne S, Munro J, St Clair D, et al. An association study of the neuregulin 1 gene, bipolar affective disorder and psychosis. Psychiatr Genet. 2009;19:113–116. doi: 10.1097/YPG.0b013e32832a4f69. [DOI] [PubMed] [Google Scholar]
- 430.Pedrosa E, Nolan KA, Stefanescu R, Hershcovitz P, Novak T, et al. Analysis of a Promoter Polymorphism in the SMDF Neuregulin 1 Isoform in Schizophrenia. Neuropsychobiology. 2009;59:205–212. doi: 10.1159/000223732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 431.Mata I, Perez-Iglesias R, Roiz-Santianez R, Tordesillas-Gutierrez D, Gonzalez-Mandly A, et al. A neuregulin 1 variant is associated with increased lateral ventricle volume in patients with first-episode schizophrenia. Biol Psychiatry. 2009;65:535–540. doi: 10.1016/j.biopsych.2008.10.020. [DOI] [PubMed] [Google Scholar]
- 432.Kircher T, Thienel R, Wagner M, Reske M, Habel U, et al. Neuregulin 1 ICE-single nucleotide polymorphism in first episode schizophrenia correlates with cerebral activation in fronto-temporal areas. Eur Arch Psychiatry Clin Neurosci. 2009;259:72–79. doi: 10.1007/s00406-008-0837-4. [DOI] [PubMed] [Google Scholar]
- 433.Kircher T, Krug A, Markov V, Whitney C, Krach S, et al. Genetic variation in the schizophrenia-risk gene neuregulin 1 correlates with brain activation and impaired speech production in a verbal fluency task in healthy individuals. Hum Brain Mapp. 2009 doi: 10.1002/hbm.20761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 434.Keri S, Seres I, Kelemen O, Benedek G. Neuregulin 1-stimulated phosphorylation of AKT in psychotic disorders and its relationship with neurocognitive functions. Neurochem Int. 2009 doi: 10.1016/j.neuint.2009.06.002. [DOI] [PubMed] [Google Scholar]
- 435.Willem M, Garratt AN, Novak B, Citron M, Kaufmann S, et al. Control of peripheral nerve myelination by the beta-secretase BACE1. Science. 2006;314:664–666. doi: 10.1126/science.1132341. [DOI] [PubMed] [Google Scholar]
- 436.Savonenko AV, Melnikova T, Laird FM, Stewart KA, Price DL, et al. Alteration of BACE1-dependent NRG1/ErbB4 signaling and schizophrenia-like phenotypes in BACE1-null mice. Proc Natl Acad Sci U S A. 2008;105:5585–5590. doi: 10.1073/pnas.0710373105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 437.Hu X, He W, Diaconu C, Tang X, Kidd GJ, et al. Genetic deletion of BACE1 in mice affects remyelination of sciatic nerves. FASEB J. 2008;22:2970–2980. doi: 10.1096/fj.08-106666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 438.Pankonin MS, Sohi J, Kamholz J, Loeb JA. Differential distribution of neuregulin in human brain and spinal fluid. Brain Res. 2009;1258:1–11. doi: 10.1016/j.brainres.2008.12.047. [DOI] [PubMed] [Google Scholar]
- 439.Li B, Woo RS, Mei L, Malinow R. The neuregulin-1 receptor erbB4 controls glutamatergic synapse maturation and plasticity. Neuron. 2007;54:583–597. doi: 10.1016/j.neuron.2007.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 440.Go RC, Perry RT, Wiener H, Bassett SS, Blacker D, et al. Neuregulin-1 polymorphism in late onset Alzheimer's disease families with psychoses. Am J Med Genet B Neuropsychiatr Genet. 2005;139B:28–32. doi: 10.1002/ajmg.b.30219. [DOI] [PubMed] [Google Scholar]
- 441.Gallagher JP, Orozco-Cabal LF, Liu J, Shinnick-Gallagher P. Synaptic physiology of central CRH system. Eur J Pharmacol. 2008;583:215–225. doi: 10.1016/j.ejphar.2007.11.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 442.Liu Y, Tao YM, Woo RS, Xiong WC, Mei L. Stimulated ErbB4 internalization is necessary for neuregulin signaling in neurons. Biochem Biophys Res Commun. 2007;354:505–510. doi: 10.1016/j.bbrc.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 443.MacDonald AW, 3rd, Chafee MV. Translational and developmental perspective on N-methyl-D-aspartate synaptic deficits in schizophrenia. Dev Psychopathol. 2006;18:853–876. [PubMed] [Google Scholar]
- 444.Jaworski A, Burden SJ. Neuromuscular synapse formation in mice lacking motor neuron- and skeletal muscle-derived Neuregulin-1. J Neurosci. 2006;26:655–661. doi: 10.1523/JNEUROSCI.4506-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 445.Jacobson C, Duggan D, Fischbach G. Neuregulin induces the expression of transcription factors and myosin heavy chains typical of muscle spindles in cultured human muscle. Proc Natl Acad Sci U S A. 2004;101:12218–12223. doi: 10.1073/pnas.0404240101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 446.Okada M, Corfas G. Neuregulin1 downregulates postsynaptic GABAA receptors at the hippocampal inhibitory synapse. Hippocampus. 2004;14:337–344. doi: 10.1002/hipo.10185. [DOI] [PubMed] [Google Scholar]
- 447.Bennett AOM. Dual constraints on synapse formation and regression in schizophrenia: neuregulin, neuroligin, dysbindin, DISC1, MuSK and agrin. Aust N Z J Psychiatry. 2008;42:662–677. doi: 10.1080/00048670802203467. [DOI] [PubMed] [Google Scholar]
- 448.Papadopoulos V, Lecanu L, Brown RC, Han Z, Yao ZX. Peripheral-type benzodiazepine receptor in neurosteroid biosynthesis, neuropathology and neurological disorders. Neuroscience. 2006;138:749–756. doi: 10.1016/j.neuroscience.2005.05.063. [DOI] [PubMed] [Google Scholar]
- 449.Hazell AS. Astrocytes and manganese neurotoxicity. Neurochem Int. 2002;41:271–277. doi: 10.1016/s0197-0186(02)00013-x. [DOI] [PubMed] [Google Scholar]
- 450.Roberts JC, Friel SL, Roman S, Perren M, Harper A, et al. Autoradiographical imaging of PPARgamma agonist effects on PBR/TSPO binding in TASTPM mice. Exp Neurol. 2009;216:459–470. doi: 10.1016/j.expneurol.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 451.Ji B, Maeda J, Sawada M, Ono M, Okauchi T, et al. Imaging of peripheral benzodiazepine receptor expression as biomarkers of detrimental versus beneficial glial responses in mouse models of Alzheimer's and other CNS pathologies. J Neurosci. 2008;28:12255–12267. doi: 10.1523/JNEUROSCI.2312-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 452.Gulyas B, Makkai B, Kasa P, Gulya K, Bakota L, et al. A comparative autoradiography study in post mortem whole hemisphere human brain slices taken from Alzheimer patients and age-matched controls using two radiolabelled DAA1106 analogues with high affinity to the peripheral benzodiazepine receptor (PBR) system. Neurochem Int. 2009;54:28–36. doi: 10.1016/j.neuint.2008.10.001. [DOI] [PubMed] [Google Scholar]
- 453.Wang M, Gao M, Hutchins GD, Zheng QH. Synthesis of [11C]FEDAA1106 as a new PET imaging probe of peripheral benzodiazepine receptor expression. Eur J Med Chem. 2009;44:2748–2753. doi: 10.1016/j.ejmech.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 454.Yasuno F, Ota M, Kosaka J, Ito H, Higuchi M, et al. Increased binding of peripheral benzodiazepine receptor in Alzheimer's disease measured by positron emission tomography with [11C]DAA1106. Biol Psychiatry. 2008;64:835–841. doi: 10.1016/j.biopsych.2008.04.021. [DOI] [PubMed] [Google Scholar]
- 455.Laquintana V, Denora N, Lopedota A, Suzuki H, Sawada M, et al. N-benzyl-2-(6,8-dichloro-2-(4-chlorophenyl)imidazo[1,2-a]pyridin-3-yl)-N-( 6-(7-nitrobenzo[c][1,2,5]oxadiazol-4-ylamino)hexyl)acetamide as a new fluorescent probe for peripheral benzodiazepine receptor and microglial cell visualization. Bioconjug Chem. 2007;18:1397–1407. doi: 10.1021/bc060393c. [DOI] [PubMed] [Google Scholar]
- 456.Walker DG, Dalsing-Hernandez JE, Lue LF. Human postmortem brain-derived cerebrovascular smooth muscle cells express all genes of the classical complement pathway: a potential mechanism for vascular damage in cerebral amyloid angiopathy and Alzheimer's disease. Microvasc Res. 2008;75:411–419. doi: 10.1016/j.mvr.2007.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 457.Veerhuis R, Janssen I, De Groot CJ, Van Muiswinkel FL, Hack CE, et al. Cytokines associated with amyloid plaques in Alzheimer's disease brain stimulate human glial and neuronal cell cultures to secrete early complement proteins, but not C1-inhibitor. Exp Neurol. 1999;160:289–299. doi: 10.1006/exnr.1999.7199. [DOI] [PubMed] [Google Scholar]
- 458.Yasojima K, McGeer EG, McGeer PL. Complement regulators C1 inhibitor and CD59 do not significantly inhibit complement activation in Alzheimer disease. Brain Res. 1999;833:297–301. doi: 10.1016/s0006-8993(99)01514-0. [DOI] [PubMed] [Google Scholar]
- 459.Bergamaschini L, Canziani S, Bottasso B, Cugno M, Braidotti P, et al. Alzheimer's beta-amyloid peptides can activate the early components of complement classical pathway in a C1q-independent manner. Clin Exp Immunol. 1999;115:526–533. doi: 10.1046/j.1365-2249.1999.00835.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 460.Yasojima K, Schwab C, McGeer EG, McGeer PL. Up-regulated production and activation of the complement system in Alzheimer's disease brain. Am J Pathol. 1999;154:927–936. doi: 10.1016/S0002-9440(10)65340-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 461.Terai K, Walker DG, McGeer EG, McGeer PL. Neurons express proteins of the classical complement pathway in Alzheimer disease. Brain Res. 1997;769:385–390. doi: 10.1016/s0006-8993(97)00849-4. [DOI] [PubMed] [Google Scholar]
- 462.Mori M, Sawashita J, Higuchi K. Functional polymorphisms of the Lss and Fdft1 genes in laboratory rats. Exp Anim. 2007;56:93–101. doi: 10.1538/expanim.56.93. [DOI] [PubMed] [Google Scholar]
- 463.Funfschilling U, Saher G, Xiao L, Mobius W, Nave KA. Survival of adult neurons lacking cholesterol synthesis in vivo. BMC Neurosci. 2007;8:1. doi: 10.1186/1471-2202-8-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 464.Tang J, Song M, Wang Y, Fan X, Xu H, et al. Noggin and BMP4 co-modulate adult hippocampal neurogenesis in the APP(swe)/PS1(DeltaE9) transgenic mouse model of Alzheimer's disease. Biochem Biophys Res Commun. 2009;385:341–345. doi: 10.1016/j.bbrc.2009.05.067. [DOI] [PubMed] [Google Scholar]
- 465.Li D, Tang J, Xu H, Fan X, Bai Y, et al. Decreased hippocampal cell proliferation correlates with increased expression of BMP4 in the APPswe/PS1DeltaE9 mouse model of Alzheimer's disease. Hippocampus. 2008;18:692–698. doi: 10.1002/hipo.20428. [DOI] [PubMed] [Google Scholar]
- 466.Malm TM, Magga J, Kuh GF, Vatanen T, Koistinaho M, et al. Minocycline reduces engraftment and activation of bone marrow-derived cells but sustains their phagocytic activity in a mouse model of Alzheimer's disease. Glia. 2008;56:1767–1779. doi: 10.1002/glia.20726. [DOI] [PubMed] [Google Scholar]
- 467.Kobayashi K, Hayashi M, Nakano H, Fukutani Y, Sasaki K, et al. Apoptosis of astrocytes with enhanced lysosomal activity and oligodendrocytes in white matter lesions in Alzheimer's disease. Neuropathol Appl Neurobiol. 2002;28:238–251. doi: 10.1046/j.1365-2990.2002.00390.x. [DOI] [PubMed] [Google Scholar]
- 468.Fiala M, Liu QN, Sayre J, Pop V, Brahmandam V, et al. Cyclooxygenase-2-positive macrophages infiltrate the Alzheimer's disease brain and damage the blood-brain barrier. Eur J Clin Invest. 2002;32:360–371. doi: 10.1046/j.1365-2362.2002.00994.x. [DOI] [PubMed] [Google Scholar]
- 469.Heneka MT, Wiesinger H, Dumitrescu-Ozimek L, Riederer P, Feinstein DL, et al. Neuronal and glial coexpression of argininosuccinate synthetase and inducible nitric oxide synthase in Alzheimer disease. J Neuropathol Exp Neurol. 2001;60:906–916. doi: 10.1093/jnen/60.9.906. [DOI] [PubMed] [Google Scholar]
- 470.Kobayashi K, Muramori F, Aoki T, Hayashi M, Miyazu K, et al. KP-1 is a marker for extraneuronal neurofibrillary tangles and senile plaques in Alzheimer diseased brains. Dement Geriatr Cogn Disord. 1998;9:13–19. doi: 10.1159/000017015. [DOI] [PubMed] [Google Scholar]
- 471.Reynolds WF, Rhees J, Maciejewski D, Paladino T, Sieburg H, et al. Myeloperoxidase polymorphism is associated with gender specific risk for Alzheimer's disease. Exp Neurol. 1999;155:31–41. doi: 10.1006/exnr.1998.6977. [DOI] [PubMed] [Google Scholar]
- 472.Damjanac M, Rioux Bilan A, Barrier L, Pontcharraud R, Anne C, et al. Fluoro-Jade B staining as useful tool to identify activated microglia and astrocytes in a mouse transgenic model of Alzheimer's disease. Brain Res. 2007;1128:40–49. doi: 10.1016/j.brainres.2006.05.050. [DOI] [PubMed] [Google Scholar]
- 473.Sim KG, Cheong JK, Hsu SI. The TRIP-Br family of transcriptional regulators is essential for the execution of cyclin E-mediated cell cycle progression. Cell Cycle. 2006;5:1111–1115. doi: 10.4161/cc.5.10.2797. [DOI] [PubMed] [Google Scholar]
- 474.Cheong JK, Gunaratnam L, Zang ZJ, Yang CM, Sun X, et al. TRIP-Br2 promotes oncogenesis in nude mice and is frequently overexpressed in multiple human tumors. J Transl Med. 2009;7:8. doi: 10.1186/1479-5876-7-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 475.Zang ZJ, Sim KG, Cheong JK, Yang CM, Yap CS, et al. Exploiting the TRIP-Br family of cell cycle regulatory proteins as chemotherapeutic drug targets in human cancer. Cancer Biol Ther. 2007;6:712–718. doi: 10.4161/cbt.6.5.3964. [DOI] [PubMed] [Google Scholar]
- 476.An L, Sato H, Konishi Y, Walker DG, Beach TG, et al. Expression and localization of lactotransferrin messenger RNA in the cortex of Alzheimer's disease. Neurosci Lett. 2009;452:277–280. doi: 10.1016/j.neulet.2009.01.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 477.Leveugle B, Spik G, Perl DP, Bouras C, Fillit HM, et al. The iron-binding protein lactotransferrin is present in pathologic lesions in a variety of neurodegenerative disorders: a comparative immunohistochemical analysis. Brain Res. 1994;650:20–31. doi: 10.1016/0006-8993(94)90202-x. [DOI] [PubMed] [Google Scholar]
- 478.Kawamata T, Tooyama I, Yamada T, Walker DG, McGeer PL. Lactotransferrin immunocytochemistry in Alzheimer and normal human brain. Am J Pathol. 1993;142:1574–1585. [PMC free article] [PubMed] [Google Scholar]
- 479.Morozova N, Khrapko K, Panee J, Liu W, Harney JW, et al. Glutathione depletion in hippocampal cells increases levels of H and L ferritin and glutathione S-transferase mRNAs. Genes Cells. 2007;12:561–567. doi: 10.1111/j.1365-2443.2007.01074.x. [DOI] [PubMed] [Google Scholar]
- 480.Connor JR, Snyder BS, Arosio P, Loeffler DA, LeWitt P. A quantitative analysis of isoferritins in select regions of aged, parkinsonian, and Alzheimer's diseased brains. J Neurochem. 1995;65:717–724. doi: 10.1046/j.1471-4159.1995.65020717.x. [DOI] [PubMed] [Google Scholar]
- 481.Vidal R, Miravalle L, Gao X, Barbeito AG, Baraibar MA, et al. Expression of a mutant form of the ferritin light chain gene induces neurodegeneration and iron overload in transgenic mice. J Neurosci. 2008;28:60–67. doi: 10.1523/JNEUROSCI.3962-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 482.Thomas M, Jankovic J. Neurodegenerative disease and iron storage in the brain. Curr Opin Neurol. 2004;17:437–442. doi: 10.1097/01.wco.0000137534.61244.d1. [DOI] [PubMed] [Google Scholar]
- 483.Wimmer U, Wang Y, Georgiev O, Schaffner W. Two major branches of anti-cadmium defense in the mouse: MTF-1/metallothioneins and glutathione. Nucleic Acids Res. 2005;33:5715–5727. doi: 10.1093/nar/gki881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 484.Bellingham SA, Coleman LA, Masters CL, Camakaris J, Hill AF. Regulation of prion gene expression by transcription factors SP1 and metal transcription factor-1. J Biol Chem. 2009;284:1291–1301. doi: 10.1074/jbc.M804755200. [DOI] [PubMed] [Google Scholar]
- 485.Colangelo V, Schurr J, Ball MJ, Pelaez RP, Bazan NG, et al. Gene expression profiling of 12633 genes in Alzheimer hippocampal CA1: transcription and neurotrophic factor down-regulation and up-regulation of apoptotic and pro-inflammatory signaling. J Neurosci Res. 2002;70:462–473. doi: 10.1002/jnr.10351. [DOI] [PubMed] [Google Scholar]
- 486.Holloway AF, Stennard FA, West AK. Human metallothionein gene MT1L mRNA is present in several human tissues but is unlikely to produce a metallothionein protein. FEBS Lett. 1997;404:41–44. doi: 10.1016/s0014-5793(97)00082-3. [DOI] [PubMed] [Google Scholar]
- 487.Hahn Y, Lee B. Human-specific nonsense mutations identified by genome sequence comparisons. Hum Genet. 2006;119:169–178. doi: 10.1007/s00439-005-0125-6. [DOI] [PubMed] [Google Scholar]
- 488.Choi KH, Elashoff M, Higgs BW, Song J, Kim S, et al. Putative psychosis genes in the prefrontal cortex: combined analysis of gene expression microarrays. BMC Psychiatry. 2008;8:87. doi: 10.1186/1471-244X-8-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 489.Goncalves I, Quintela T, Baltazar G, Almeida MR, Saraiva MJ, et al. Transthyretin interacts with metallothionein 2. Biochemistry. 2008;47:2244–2251. doi: 10.1021/bi7016377. [DOI] [PubMed] [Google Scholar]
- 490.Aschner M, West AK. The role of MT in neurological disorders. J Alzheimers Dis. 2005;8:139–145. doi: 10.3233/jad-2005-8206. discussion 209–115. [DOI] [PubMed] [Google Scholar]
- 491.Richarz AN, Bratter P. Speciation analysis of trace elements in the brains of individuals with Alzheimer's disease with special emphasis on metallothioneins. Anal Bioanal Chem. 2002;372:412–417. doi: 10.1007/s00216-001-1187-5. [DOI] [PubMed] [Google Scholar]
- 492.Irie Y, Keung WM. Metallothionein-III antagonizes the neurotoxic and neurotrophic effects of amyloid beta peptides. Biochem Biophys Res Commun. 2001;282:416–420. doi: 10.1006/bbrc.2001.4594. [DOI] [PubMed] [Google Scholar]
- 493.Kobayashi K, Hayashi M, Nakano H, Shimazaki M, Sugimori K, et al. Correlation between astrocyte apoptosis and Alzheimer changes in gray matter lesions in Alzheimer's disease. J Alzheimers Dis. 2004;6:623–632. doi: 10.3233/jad-2004-6606. discussion 673–681. [DOI] [PubMed] [Google Scholar]
- 494.Mattson MP. Glutamate and neurotrophic factors in neuronal plasticity and disease. Ann N Y Acad Sci. 2008;1144:97–112. doi: 10.1196/annals.1418.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 495.Stahelin RV, Long F, Diraviyam K, Bruzik KS, Murray D, et al. Phosphatidylinositol 3-phosphate induces the membrane penetration of the FYVE domains of Vps27p and Hrs. J Biol Chem. 2002;277:26379–26388. doi: 10.1074/jbc.M201106200. [DOI] [PubMed] [Google Scholar]
- 496.Falasca M, Maffucci T. Emerging roles of phosphatidylinositol 3-monophosphate as a dynamic lipid second messenger. Arch Physiol Biochem. 2006;112:274–284. doi: 10.1080/13813450601094664. [DOI] [PubMed] [Google Scholar]
- 497.Sansom SN, Griffiths DS, Faedo A, Kleinjan DJ, Ruan Y, et al. The level of the transcription factor Pax6 is essential for controlling the balance between neural stem cell self-renewal and neurogenesis. PLoS Genet. 2009;5:e1000511. doi: 10.1371/journal.pgen.1000511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 498.von Bohlen Und Halbach O. Immunohistological markers for staging neurogenesis in adult hippocampus. Cell Tissue Res. 2007;329:409–420. doi: 10.1007/s00441-007-0432-4. [DOI] [PubMed] [Google Scholar]
- 499.Kallur T, Gisler R, Lindvall O, Kokaia Z. Pax6 promotes neurogenesis in human neural stem cells. Mol Cell Neurosci. 2008;38:616–628. doi: 10.1016/j.mcn.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 500.Levers TE, Edgar JM, Price DJ. The fates of cells generated at the end of neurogenesis in developing mouse cortex. J Neurobiol. 2001;48:265–277. doi: 10.1002/neu.1056. [DOI] [PubMed] [Google Scholar]
- 501.Hardy RJ. QKI expression is regulated during neuron-glial cell fate decisions. J Neurosci Res. 1998;54:46–57. doi: 10.1002/(SICI)1097-4547(19981001)54:1<46::AID-JNR6>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 502.Bibel M, Richter J, Schrenk K, Tucker KL, Staiger V, et al. Differentiation of mouse embryonic stem cells into a defined neuronal lineage. Nat Neurosci. 2004;7:1003–1009. doi: 10.1038/nn1301. [DOI] [PubMed] [Google Scholar]
- 503.Baer K, Eriksson PS, Faull RL, Rees MI, Curtis MA. Sox-2 is expressed by glial and progenitor cells and Pax-6 is expressed by neuroblasts in the human subventricular zone. Exp Neurol. 2007;204:828–831. doi: 10.1016/j.expneurol.2006.12.008. [DOI] [PubMed] [Google Scholar]
- 504.Nacher J, Varea E, Blasco-Ibanez JM, Castillo-Gomez E, Crespo C, et al. Expression of the transcription factor Pax 6 in the adult rat dentate gyrus. J Neurosci Res. 2005;81:753–761. doi: 10.1002/jnr.20596. [DOI] [PubMed] [Google Scholar]
- 505.Malaterre J, Mantamadiotis T, Dworkin S, Lightowler S, Yang Q, et al. c-Myb is required for neural progenitor cell proliferation and maintenance of the neural stem cell niche in adult brain. Stem Cells. 2008;26:173–181. doi: 10.1634/stemcells.2007-0293. [DOI] [PubMed] [Google Scholar]
- 506.Lauriat TL, Shiue L, Haroutunian V, Verbitsky M, Ares M, Jr, et al. Developmental expression profile of quaking, a candidate gene for schizophrenia, and its target genes in human prefrontal cortex and hippocampus shows regional specificity. J Neurosci Res. 2008;86:785–796. doi: 10.1002/jnr.21534. [DOI] [PubMed] [Google Scholar]
- 507.Chen Y, Tian D, Ku L, Osterhout DJ, Feng Y. The selective RNA-binding protein quaking I (QKI) is necessary and sufficient for promoting oligodendroglia differentiation. J Biol Chem. 2007;282:23553–23560. doi: 10.1074/jbc.M702045200. [DOI] [PubMed] [Google Scholar]
- 508.Zhao L, Ku L, Chen Y, Xia M, LoPresti P, et al. QKI binds MAP1B mRNA and enhances MAP1B expression during oligodendrocyte development. Mol Biol Cell. 2006;17:4179–4186. doi: 10.1091/mbc.E06-04-0355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 509.Aberg K, Saetre P, Jareborg N, Jazin E. Human QKI, a potential regulator of mRNA expression of human oligodendrocyte-related genes involved in schizophrenia. Proc Natl Acad Sci U S A. 2006;103:7482–7487. doi: 10.1073/pnas.0601213103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 510.Galarneau A, Richard S. Target RNA motif and target mRNAs of the Quaking STAR protein. Nat Struct Mol Biol. 2005;12:691–698. doi: 10.1038/nsmb963. [DOI] [PubMed] [Google Scholar]
- 511.Valjent E, Aubier B, Corbille AG, Brami-Cherrier K, Caboche J, et al. Plasticity-associated gene Krox24/Zif268 is required for long-lasting behavioral effects of cocaine. J Neurosci. 2006;26:4956–4960. doi: 10.1523/JNEUROSCI.4601-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 512.Nikitin VP, Kozyrev SA. Effects of antisense oligonucleotides to mRNA for the early gene zif268 on the mechanisms of synapse-specific plasticity. Neurosci Behav Physiol. 2007;37:607–612. doi: 10.1007/s11055-007-0059-7. [DOI] [PubMed] [Google Scholar]
- 513.Poirier GL, Amin E, Aggleton JP. Qualitatively different hippocampal subfield engagement emerges with mastery of a spatial memory task by rats. J Neurosci. 2008;28:1034–1045. doi: 10.1523/JNEUROSCI.4607-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 514.Renaudineau S, Poucet B, Laroche S, Davis S, Save E. Impaired long-term stability of CA1 place cell representation in mice lacking the transcription factor zif268/egr1. Proc Natl Acad Sci U S A. 2009;106:11771–11775. doi: 10.1073/pnas.0900484106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 515.Romcy-Pereira RN, Erraji-Benchekroun L, Smyrniotopoulos P, Ogawa S, Mello CV, et al. Sleep-dependent gene expression in the hippocampus and prefrontal cortex following long-term potentiation. Physiol Behav. 2009;98:44–52. doi: 10.1016/j.physbeh.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 516.Bailey DJ, Wade J. Differential expression of the immediate early genes FOS and ZENK following auditory stimulation in the juvenile male and female zebra finch. Brain Res Mol Brain Res. 2003;116:147–154. doi: 10.1016/s0169-328x(03)00288-2. [DOI] [PubMed] [Google Scholar]
- 517.Eda-Fujiwara H, Satoh R, Bolhuis JJ, Kimura T. Neuronal activation in female budgerigars is localized and related to male song complexity. Eur J Neurosci. 2003;17:149–154. doi: 10.1046/j.1460-9568.2003.02414.x. [DOI] [PubMed] [Google Scholar]
- 518.Shimizu T, Bowers AN, Budzynski CA, Kahn MC, Bingman VP. What does a pigeon (Columba livia) brain look like during homing? selective examination of ZENK expression. Behav Neurosci. 2004;118:845–851. doi: 10.1037/0735-7044.118.4.845. [DOI] [PubMed] [Google Scholar]
- 519.Terpstra NJ, Bolhuis JJ, den Boer-Visser AM. An analysis of the neural representation of birdsong memory. J Neurosci. 2004;24:4971–4977. doi: 10.1523/JNEUROSCI.0570-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 520.Bailey DJ, Wade J. FOS and ZENK responses in 45-day-old zebra finches vary with auditory stimulus and brain region, but not sex. Behav Brain Res. 2005;162:108–115. doi: 10.1016/j.bbr.2005.03.016. [DOI] [PubMed] [Google Scholar]
- 521.Terpstra NJ, Bolhuis JJ, Den Boer-Visser AM, Ten Cate C. Neuronal activation related to auditory perception in the brain of a non-songbird, the ring dove. J Comp Neurol. 2005;488:342–351. doi: 10.1002/cne.20592. [DOI] [PubMed] [Google Scholar]
- 522.Bischofe HJ, Lieshoff C, Watanabe S. Spatial memory and hippocampal function in a non-foodstoring songbird, the zebra finch (Taeniopygia guttata). Rev Neurosci. 2006;17:43–52. doi: 10.1515/revneuro.2006.17.1-2.43. [DOI] [PubMed] [Google Scholar]
- 523.Brito I, Britto LR, Ferrari EA. Classical tone-shock conditioning induces Zenk expression in the pigeon (Columba livia) hippocampus. Behav Neurosci. 2006;120:353–361. doi: 10.1037/0735-7044.120.2.353. [DOI] [PubMed] [Google Scholar]
- 524.Terpstra NJ, Bolhuis JJ, Riebel K, van der Burg JM, den Boer-Visser AM. Localized brain activation specific to auditory memory in a female songbird. J Comp Neurol. 2006;494:784–791. doi: 10.1002/cne.20831. [DOI] [PubMed] [Google Scholar]
- 525.Vignal C, Bouchut C, Mathevon N. Sound-induced brain activity depends on stimulus subjective salience in female zebra finches. C R Biol. 2008;331:347–356. doi: 10.1016/j.crvi.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 526.Fischer AJ, McGuire JJ, Schaeffel F, Stell WK. Light- and focus-dependent expression of the transcription factor ZENK in the chick retina. Nat Neurosci. 1999;2:706–712. doi: 10.1038/11167. [DOI] [PubMed] [Google Scholar]
- 527.Mello CV, Vicario DS, Clayton DF. Song presentation induces gene expression in the songbird forebrain. Proc Natl Acad Sci U S A. 1992;89:6818–6822. doi: 10.1073/pnas.89.15.6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 528.Mello CV, Clayton DF. Song-induced ZENK gene expression in auditory pathways of songbird brain and its relation to the song control system. J Neurosci. 1994;14:6652–6666. doi: 10.1523/JNEUROSCI.14-11-06652.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 529.Mello C, Nottebohm F, Clayton D. Repeated exposure to one song leads to a rapid and persistent decline in an immediate early gene's response to that song in zebra finch telencephalon. J Neurosci. 1995;15:6919–6925. doi: 10.1523/JNEUROSCI.15-10-06919.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 530.Mello CV, Clayton DF. Differential induction of the ZENK gene in the avian forebrain and song control circuit after metrazole-induced depolarization. J Neurobiol. 1995;26:145–161. doi: 10.1002/neu.480260112. [DOI] [PubMed] [Google Scholar]
- 531.Mello CV, Ribeiro S. ZENK protein regulation by song in the brain of songbirds. J Comp Neurol. 1998;393:426–438. doi: 10.1002/(sici)1096-9861(19980420)393:4<426::aid-cne3>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 532.Ribeiro S, Cecchi GA, Magnasco MO, Mello CV. Toward a song code: evidence for a syllabic representation in the canary brain. Neuron. 1998;21:359–371. doi: 10.1016/s0896-6273(00)80545-0. [DOI] [PubMed] [Google Scholar]
- 533.Tischmeyer W, Grimm R. Activation of immediate early genes and memory formation. Cell Mol Life Sci. 1999;55:564–574. doi: 10.1007/s000180050315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 534.Nedivi E, Hevroni D, Naot D, Israeli D, Citri Y. Numerous candidate plasticity-related genes revealed by differential cDNA cloning. Nature. 1993;363:718–722. doi: 10.1038/363718a0. [DOI] [PubMed] [Google Scholar]
- 535.Li L, Yun SH, Keblesh J, Trommer BL, Xiong H, et al. Egr3, a synaptic activity regulated transcription factor that is essential for learning and memory. Mol Cell Neurosci. 2007;35:76–88. doi: 10.1016/j.mcn.2007.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 536.Soule J, Penke Z, Kanhema T, Alme MN, Laroche S, et al. Object-place recognition learning triggers rapid induction of plasticity-related immediate early genes and synaptic proteins in the rat dentate gyrus. Neural Plast. 2008;2008:269097. doi: 10.1155/2008/269097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 537.Takahata T, Higo N, Kaas JH, Yamamori T. Expression of immediate-early genes reveals functional compartments within ocular dominance columns after brief monocular inactivation. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0905092106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 538.Lam BY, Zhang W, Enticknap N, Haggis E, Cader MZ, et al. Inverse regulation of plasticity-related immediate early genes by calcineurin in hippocampal neurons. J Biol Chem. 2009;284:12562–12571. doi: 10.1074/jbc.M901121200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 539.Pfenning AR, Schwartz R, Barth AL. A comparative genomics approach to identifying the plasticity transcriptome. BMC Neurosci. 2007;8:20. doi: 10.1186/1471-2202-8-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 540.Nikitin VP. A new mechanism of synapse-specific neuronal plasticity. Neurosci Behav Physiol. 2007;37:559–570. doi: 10.1007/s11055-007-0053-0. [DOI] [PubMed] [Google Scholar]
- 541.Toscano CD, McGlothan JL, Guilarte TR. Experience-dependent regulation of zif268 gene expression and spatial learning. Exp Neurol. 2006;200:209–215. doi: 10.1016/j.expneurol.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 542.Jenkins TA, Amin E, Brown MW, Aggleton JP. Changes in immediate early gene expression in the rat brain after unilateral lesions of the hippocampus. Neuroscience. 2006;137:747–759. doi: 10.1016/j.neuroscience.2005.09.034. [DOI] [PubMed] [Google Scholar]
- 543.James AB, Conway AM, Morris BJ. Regulation of the neuronal proteasome by Zif268 (Egr1). J Neurosci. 2006;26:1624–1634. doi: 10.1523/JNEUROSCI.4199-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 544.Ihara H, Mie M, Funabashi H, Takahashi F, Sawasaki T, et al. In vitro selection of zinc finger DNA-binding proteins through ribosome display. Biochem Biophys Res Commun. 2006;345:1149–1154. doi: 10.1016/j.bbrc.2006.05.029. [DOI] [PubMed] [Google Scholar]
- 545.Hernandez PJ, Schiltz CA, Kelley AE. Dynamic shifts in corticostriatal expression patterns of the immediate early genes Homer 1a and Zif268 during early and late phases of instrumental training. Learn Mem. 2006;13:599–608. doi: 10.1101/lm.335006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 546.James AB, Conway AM, Morris BJ. Genomic profiling of the neuronal target genes of the plasticity-related transcription factor – Zif268. J Neurochem. 2005;95:796–810. doi: 10.1111/j.1471-4159.2005.03400.x. [DOI] [PubMed] [Google Scholar]
- 547.Lee JL, Everitt BJ, Thomas KL. Independent cellular processes for hippocampal memory consolidation and reconsolidation. Science. 2004;304:839–843. doi: 10.1126/science.1095760. [DOI] [PubMed] [Google Scholar]
- 548.Korzus E. The relation of transcription to memory formation. Acta Biochim Pol. 2003;50:775–782. [PubMed] [Google Scholar]
- 549.Davis S, Bozon B, Laroche S. How necessary is the activation of the immediate early gene zif268 in synaptic plasticity and learning? Behav Brain Res. 2003;142:17–30. doi: 10.1016/s0166-4328(02)00421-7. [DOI] [PubMed] [Google Scholar]
- 550.Blanchard J, Decorte L, Nogues X, Micheau J. Characterization of cognition alteration across the course of the disease in APP751SL mice with parallel estimation of cerebral Abeta deposition. Behav Brain Res. 2009;201:147–157. doi: 10.1016/j.bbr.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 551.Blanchard J, Martel G, Guillou JL, Nogues X, Micheau J. Impairment of spatial memory consolidation in APP(751SL) mice results in cue-guided response. Neurobiol Aging. 2008;29:1011–1021. doi: 10.1016/j.neurobiolaging.2007.02.010. [DOI] [PubMed] [Google Scholar]
- 552.Becker M, Lavie V, Solomon B. Stimulation of endogenous neurogenesis by anti-EFRH immunization in a transgenic mouse model of Alzheimer's disease. Proc Natl Acad Sci U S A. 2007;104:1691–1696. doi: 10.1073/pnas.0610180104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 553.Dickey CA, Loring JF, Montgomery J, Gordon MN, Eastman PS, et al. Selectively reduced expression of synaptic plasticity-related genes in amyloid precursor protein+presenilin-1 transgenic mice. J Neurosci. 2003;23:5219–5226. doi: 10.1523/JNEUROSCI.23-12-05219.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 554.Dickey CA, Gordon MN, Mason JE, Wilson NJ, Diamond DM, et al. Amyloid suppresses induction of genes critical for memory consolidation in APP+PS1 transgenic mice. J Neurochem. 2004;88:434–442. doi: 10.1111/j.1471-4159.2004.02185.x. [DOI] [PubMed] [Google Scholar]
- 555.Kashani AH, Qiu Z, Jurata L, Lee SK, Pfaff S, et al. Calcium activation of the LMO4 transcription complex and its role in the patterning of thalamocortical connections. J Neurosci. 2006;26:8398–8408. doi: 10.1523/JNEUROSCI.0618-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 556.Bulchand S, Subramanian L, Tole S. Dynamic spatiotemporal expression of LIM genes and cofactors in the embryonic and postnatal cerebral cortex. Dev Dyn. 2003;226:460–469. doi: 10.1002/dvdy.10235. [DOI] [PubMed] [Google Scholar]
- 557.Azim E, Shnider SJ, Cederquist GY, Sohur US, Macklis JD. Lmo4 and Clim1 progressively delineate cortical projection neuron subtypes during development. Cereb Cortex. 2009;19(Suppl 1):i62–69. doi: 10.1093/cercor/bhp030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 558.Miyajima N, Maruyama S, Nonomura K, Hatakeyama S. TRIM36 interacts with the kinetochore protein CENP-H and delays cell cycle progression. Biochem Biophys Res Commun. 2009;381:383–387. doi: 10.1016/j.bbrc.2009.02.059. [DOI] [PubMed] [Google Scholar]
- 559.Harrill JA, Li Z, Wright FA, Radio NM, Mundy WR, et al. Transcriptional response of rat frontal cortex following acute in vivo exposure to the pyrethroid insecticides permethrin and deltamethrin. BMC Genomics. 2008;9:546. doi: 10.1186/1471-2164-9-546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 560.Takemoto-Kimura S, Terai H, Takamoto M, Ohmae S, Kikumura S, et al. Molecular cloning and characterization of CLICK-III/CaMKIgamma, a novel membrane-anchored neuronal Ca2+/calmodulin-dependent protein kinase (CaMK). J Biol Chem. 2003;278:18597–18605. doi: 10.1074/jbc.M300578200. [DOI] [PubMed] [Google Scholar]
- 561.Takemoto-Kimura S, Ageta-Ishihara N, Nonaka M, Adachi-Morishima A, Mano T, et al. Regulation of dendritogenesis via a lipid-raft-associated Ca2+/calmodulin-dependent protein kinase CLICK-III/CaMKIgamma. Neuron. 2007;54:755–770. doi: 10.1016/j.neuron.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 562.Korostynski M, Piechota M, Kaminska D, Solecki W, Przewlocki R. Morphine effects on striatal transcriptome in mice. Genome Biol. 2007;8:R128. doi: 10.1186/gb-2007-8-6-r128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 563.Glorioso C, Sabatini M, Unger T, Hashimoto T, Monteggia LM, et al. Specificity and timing of neocortical transcriptome changes in response to BDNF gene ablation during embryogenesis or adulthood. Mol Psychiatry. 2006;11:633–648. doi: 10.1038/sj.mp.4001835. [DOI] [PubMed] [Google Scholar]
- 564.Nishimura H, Sakagami H, Uezu A, Fukunaga K, Watanabe M, et al. Cloning, characterization and expression of two alternatively splicing isoforms of Ca2+/calmodulin-dependent protein kinase I gamma in the rat brain. J Neurochem. 2003;85:1216–1227. doi: 10.1046/j.1471-4159.2003.01760.x. [DOI] [PubMed] [Google Scholar]
- 565.Zhao C, Braunewell KH. Expression of the neuronal calcium sensor visinin-like protein-1 in the rat hippocampus. Neuroscience. 2008;153:1202–1212. doi: 10.1016/j.neuroscience.2007.10.067. [DOI] [PubMed] [Google Scholar]
- 566.Youn H, Jeoung M, Koo Y, Ji H, Markesbery WR, et al. Kalirin is under-expressed in Alzheimer's disease hippocampus. J Alzheimers Dis. 2007;11:385–397. doi: 10.3233/jad-2007-11314. [DOI] [PubMed] [Google Scholar]
- 567.Huang JC, Babak T, Corson TW, Chua G, Khan S, et al. Using expression profiling data to identify human microRNA targets. Nat Methods. 2007;4:1045–1049. doi: 10.1038/nmeth1130. [DOI] [PubMed] [Google Scholar]
- 568.Liu T, Papagiannakopoulos T, Puskar K, Qi S, Santiago F, et al. Detection of a microRNA signal in an in vivo expression set of mRNAs. PLoS One. 2007;2:e804. doi: 10.1371/journal.pone.0000804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 569.Nikitin A, Egorov S, Daraselia N, Mazo I. Pathway studio–the analysis and navigation of molecular networks. Bioinformatics. 2003;19:2155–2157. doi: 10.1093/bioinformatics/btg290. [DOI] [PubMed] [Google Scholar]
- 570.Bucan M, Abrahams BS, Wang K, Glessner JT, Herman EI, et al. Genome-wide analyses of exonic copy number variants in a family-based study point to novel autism susceptibility genes. PLoS Genet. 2009;5:e1000536. doi: 10.1371/journal.pgen.1000536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 571.Bourgeron T. A synaptic trek to autism. Curr Opin Neurobiol. 2009 doi: 10.1016/j.conb.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 572.Glessner JT, Wang K, Cai G, Korvatska O, Kim CE, et al. Autism genome-wide copy number variation reveals ubiquitin and neuronal genes. Nature. 2009;459:569–573. doi: 10.1038/nature07953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 573.Kumar RA, Christian SL. Genetics of autism spectrum disorders. Curr Neurol Neurosci Rep. 2009;9:188–197. doi: 10.1007/s11910-009-0029-2. [DOI] [PubMed] [Google Scholar]
- 574.Bourgeron T. The possible interplay of synaptic and clock genes in autism spectrum disorders. Cold Spring Harb Symp Quant Biol. 2007;72:645–654. doi: 10.1101/sqb.2007.72.020. [DOI] [PubMed] [Google Scholar]
- 575.Marshall CR, Noor A, Vincent JB, Lionel AC, Feuk L, et al. Structural variation of chromosomes in autism spectrum disorder. Am J Hum Genet. 2008;82:477–488. doi: 10.1016/j.ajhg.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 576.Kim HG, Kishikawa S, Higgins AW, Seong IS, Donovan DJ, et al. Disruption of neurexin 1 associated with autism spectrum disorder. Am J Hum Genet. 2008;82:199–207. doi: 10.1016/j.ajhg.2007.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 577.Owen MJ, Williams HJ, O'Donovan MC. Schizophrenia genetics: advancing on two fronts. Curr Opin Genet Dev. 2009;19:266–270. doi: 10.1016/j.gde.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 578.Need AC, Ge D, Weale ME, Maia J, Feng S, et al. A genome-wide investigation of SNPs and CNVs in schizophrenia. PLoS Genet. 2009;5:e1000373. doi: 10.1371/journal.pgen.1000373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 579.Rujescu D, Ingason A, Cichon S, Pietilainen OP, Barnes MR, et al. Disruption of the neurexin 1 gene is associated with schizophrenia. Hum Mol Genet. 2009;18:988–996. doi: 10.1093/hmg/ddn351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 580.Vrijenhoek T, Buizer-Voskamp JE, van der Stelt I, Strengman E, Sabatti C, et al. Recurrent CNVs disrupt three candidate genes in schizophrenia patients. Am J Hum Genet. 2008;83:504–510. doi: 10.1016/j.ajhg.2008.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 581.Kirov G, Gumus D, Chen W, Norton N, Georgieva L, et al. Comparative genome hybridization suggests a role for NRXN1 and APBA2 in schizophrenia. Hum Mol Genet. 2008;17:458–465. doi: 10.1093/hmg/ddm323. [DOI] [PubMed] [Google Scholar]
- 582.Nussbaum J, Xu Q, Payne TJ, Ma JZ, Huang W, et al. Significant association of the neurexin-1 gene (NRXN1) with nicotine dependence in European- and African-American smokers. Hum Mol Genet. 2008;17:1569–1577. doi: 10.1093/hmg/ddn044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 583.Bierut LJ, Madden PA, Breslau N, Johnson EO, Hatsukami D, et al. Novel genes identified in a high-density genome wide association study for nicotine dependence. Hum Mol Genet. 2007;16:24–35. doi: 10.1093/hmg/ddl441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 584.Yang HC, Chang CC, Lin CY, Chen CL, Fann CS. A genome-wide scanning and fine mapping study of COGA data. BMC Genet. 2005;6(Suppl 1):S30. doi: 10.1186/1471-2156-6-S1-S30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 585.Zahir FR, Baross A, Delaney AD, Eydoux P, Fernandes ND, et al. A patient with vertebral, cognitive and behavioural abnormalities and a de novo deletion of NRXN1alpha. J Med Genet. 2008;45:239–243. doi: 10.1136/jmg.2007.054437. [DOI] [PubMed] [Google Scholar]
- 586.Cabodi S, Morello V, Masi A, Cicchi R, Broggio C, et al. Convergence of integrins and EGF receptor signaling via PI3K/Akt/FoxO pathway in early gene Egr-1 expression. J Cell Physiol. 2009;218:294–303. doi: 10.1002/jcp.21603. [DOI] [PubMed] [Google Scholar]
- 587.Nowak K, Killmer K, Gessner C, Lutz W. E2F-1 regulates expression of FOXO1 and FOXO3a. Biochim Biophys Acta. 2007;1769:244–252. doi: 10.1016/j.bbaexp.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 588.Yuan Z, Lehtinen MK, Merlo P, Villen J, Gygi S, et al. Regulation of neuronal cell death by MST1-FOXO1 signaling. J Biol Chem. 2009;284:11285–11292. doi: 10.1074/jbc.M900461200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 589.Yuan Z, Becker EB, Merlo P, Yamada T, DiBacco S, et al. Activation of FOXO1 by Cdk1 in cycling cells and postmitotic neurons. Science. 2008;319:1665–1668. doi: 10.1126/science.1152337. [DOI] [PubMed] [Google Scholar]
- 590.Kim AH, Bonni A. Cdk1-FOXO1: a mitotic signal takes center stage in post-mitotic neurons. Cell Cycle. 2008;7:3819–3822. doi: 10.4161/cc.7.24.7215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 591.van der Heide LP, Ramakers GM, Smidt MP. Insulin signaling in the central nervous system: learning to survive. Prog Neurobiol. 2006;79:205–221. doi: 10.1016/j.pneurobio.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 592.Horwood JM, Dufour F, Laroche S, Davis S. Signalling mechanisms mediated by the phosphoinositide 3-kinase/Akt cascade in synaptic plasticity and memory in the rat. Eur J Neurosci. 2006;23:3375–3384. doi: 10.1111/j.1460-9568.2006.04859.x. [DOI] [PubMed] [Google Scholar]
- 593.Polter A, Yang S, Zmijewska AA, van Groen T, Paik JH, et al. Forkhead box, class O transcription factors in brain: regulation and behavioral manifestation. Biol Psychiatry. 2009;65:150–159. doi: 10.1016/j.biopsych.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 594.Wu H, Lu D, Jiang H, Xiong Y, Qu C, et al. Increase in phosphorylation of Akt and its downstream signaling targets and suppression of apoptosis by simvastatin after traumatic brain injury. J Neurosurg. 2008;109:691–698. doi: 10.3171/JNS/2008/109/10/0691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 595.Sasaki T, Han F, Shioda N, Moriguchi S, Kasahara J, et al. Lithium-induced activation of Akt and CaM kinase II contributes to its neuroprotective action in a rat microsphere embolism model. Brain Res. 2006;1108:98–106. doi: 10.1016/j.brainres.2006.06.009. [DOI] [PubMed] [Google Scholar]
- 596.Hoekman MF, Jacobs FM, Smidt MP, Burbach JP. Spatial and temporal expression of FoxO transcription factors in the developing and adult murine brain. Gene Expr Patterns. 2006;6:134–140. doi: 10.1016/j.modgep.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 597.Samarin J, Cicha I, Goppelt-Struebe M. Cell type-specific regulation of CCN2 protein expression by PI3K-AKT-FoxO signaling. J Cell Commun Signal. 2009;3:79–84. doi: 10.1007/s12079-009-0055-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 598.Fukuda M, Jones JE, Olson D, Hill J, Lee CE, et al. Monitoring FoxO1 localization in chemically identified neurons. J Neurosci. 2008;28:13640–13648. doi: 10.1523/JNEUROSCI.4023-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 599.Tajes M, Yeste-Velasco M, Zhu X, Chou SP, Smith MA, et al. Activation of Akt by lithium: pro-survival pathways in aging. Mech Ageing Dev. 2009;130:253–261. doi: 10.1016/j.mad.2008.12.006. [DOI] [PubMed] [Google Scholar]
- 600.Chiang CW, Yan L, Yang E. Phosphatases and regulation of cell death. Methods Enzymol. 2008;446:237–257. doi: 10.1016/S0076-6879(08)01614-5. [DOI] [PubMed] [Google Scholar]
- 601.Lukiw WJ, Pogue AI. Induction of specific micro RNA (miRNA) species by ROS-generating metal sulfates in primary human brain cells. J Inorg Biochem. 2007;101:1265–1269. doi: 10.1016/j.jinorgbio.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 602.Alafuzoff I, Arzberger T, Al-Sarraj S, Bodi I, Bogdanovic N, et al. Staging of neurofibrillary pathology in Alzheimer's disease: a study of the BrainNet Europe Consortium. Brain Pathol. 2008;18:484–496. doi: 10.1111/j.1750-3639.2008.00147.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 603.Marzolo MP, Bu G. Lipoprotein receptors and cholesterol in APP trafficking and proteolytic processing, implications for Alzheimer's disease. Semin Cell Dev Biol. 2009;20:191–200. doi: 10.1016/j.semcdb.2008.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 604.Jaeger S, Pietrzik CU. Functional role of lipoprotein receptors in Alzheimer's disease. Curr Alzheimer Res. 2008;5:15–25. doi: 10.2174/156720508783884675. [DOI] [PubMed] [Google Scholar]
- 605.Cam JA, Bu G. Modulation of beta-amyloid precursor protein trafficking and processing by the low density lipoprotein receptor family. Mol Neurodegener. 2006;1:8. doi: 10.1186/1750-1326-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 606.Bu G, Cam J, Zerbinatti C. LRP in amyloid-beta production and metabolism. Ann N Y Acad Sci. 2006;1086:35–53. doi: 10.1196/annals.1377.005. [DOI] [PubMed] [Google Scholar]
- 607.Carter CJ. Convergence of genes implicated in Alzheimer's disease on the cerebral cholesterol shuttle: APP, cholesterol, lipoproteins, and atherosclerosis. Neurochem Int. 2007;50:12–38. doi: 10.1016/j.neuint.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 608.Cam JA, Zerbinatti CV, Knisely JM, Hecimovic S, Li Y, et al. The low density lipoprotein receptor-related protein 1B retains beta-amyloid precursor protein at the cell surface and reduces amyloid-beta peptide production. J Biol Chem. 2004;279:29639–29646. doi: 10.1074/jbc.M313893200. [DOI] [PubMed] [Google Scholar]
- 609.Duilio A, Faraonio R, Minopoli G, Zambrano N, Russo T. Fe65L2: a new member of the Fe65 protein family interacting with the intracellular domain of the Alzheimer's beta-amyloid precursor protein. Biochem J. 1998;330(Pt 1):513–519. doi: 10.1042/bj3300513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 610.Tanahashi H, Tabira T. Genome structure and chromosomal mapping of the gene for Fe65L2 interacting with Alzheimer's beta-amyloid precursor protein. Biochem Biophys Res Commun. 1999;258:385–389. doi: 10.1006/bbrc.1999.0639. [DOI] [PubMed] [Google Scholar]
- 611.Tanahashi H, Tabira T. Molecular cloning of human Fe65L2 and its interaction with the Alzheimer's beta-amyloid precursor protein. Neurosci Lett. 1999;261:143–146. doi: 10.1016/s0304-3940(98)00995-1. [DOI] [PubMed] [Google Scholar]
- 612.Tanahashi H, Asada T, Tabira T. c954C→T polymorphism in the Fe65L2 gene is associated with early-onset Alzheimer's disease. Ann Neurol. 2002;52:691–693. doi: 10.1002/ana.10368. [DOI] [PubMed] [Google Scholar]
- 613.Tamayev R, Zhou D, D'Adamio L. The interactome of the amyloid beta precursor protein family members is shaped by phosphorylation of their intracellular domains. Mol Neurodegener. 2009;4:28. doi: 10.1186/1750-1326-4-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 614.Zhou D, Zambrano N, Russo T, D'Adamio L. Phosphorylation of a tyrosine in the amyloid-beta protein precursor intracellular domain inhibits Fe65 binding and signaling. J Alzheimers Dis. 2009;16:301–307. doi: 10.3233/JAD-2009-0970. [DOI] [PubMed] [Google Scholar]
- 615.McLoughlin DM, Miller CC. The FE65 proteins and Alzheimer's disease. J Neurosci Res. 2008;86:744–754. doi: 10.1002/jnr.21532. [DOI] [PubMed] [Google Scholar]
- 616.Chang Y, Tesco G, Jeong WJ, Lindsley L, Eckman EA, et al. Generation of the beta-amyloid peptide and the amyloid precursor protein C-terminal fragment gamma are potentiated by FE65L1. J Biol Chem. 2003;278:51100–51107. doi: 10.1074/jbc.M309561200. [DOI] [PubMed] [Google Scholar]
- 617.Poduslo SE, Huang R, Spiro A., 3rd A genome screen of successful aging without cognitive decline identifies LRP1B by haplotype analysis. Am J Med Genet B Neuropsychiatr Geneti. 2010;153B doi: 10.1002/ajmg.b.30963. [DOI] [PubMed] [Google Scholar]
- 618.McKhann G, Drachman D, Folstein M, Katzman R, Price D, et al. Clinical diagnosis of Alzheimer's disease: Report of the NINCDS-ADRDA Work Group* under the auspices of Department of Health and Human Services Task Force on Alzheimer's Disease. Neurology. 1984;34:939. doi: 10.1212/wnl.34.7.939. [DOI] [PubMed] [Google Scholar]
- 619.Folstein MF, Folstein SE, McHugh PR. “Mini-mental state”: A practical method for grading the cognitive state of patients for the clinician. Journal of Psychiatric Research. 1975;12:189–198. doi: 10.1016/0022-3956(75)90026-6. [DOI] [PubMed] [Google Scholar]
- 620.Rosen W, Mohs R, Davis K. A new rating scale for Alzheimer's disease. Am J Psychiatry. 1984;141:1356–1364. doi: 10.1176/ajp.141.11.1356. [DOI] [PubMed] [Google Scholar]
- 621.Schmitt FA, Wetherby MMC, Wekstein DR, Dearth CMS, Markesbery WR. Brain Donation in Normal Aging: Procedures, Motivations, and Donor Characteristics From the Biologically Resilient Adults in Neurological Studies (BRAiNS) Project. Gerontologist. 2001;41:716–722. doi: 10.1093/geront/41.6.716. [DOI] [PubMed] [Google Scholar]
- 622.Geddes JW, Tekirian TL, Soultanian NS, Ashford JW, Davis DG, et al. Comparison of neuropathologic criteria for the diagnosis of Alzheimer's disease. Neurobiol Aging. 1997;18:S99–105. doi: 10.1016/s0197-4580(97)00063-8. [DOI] [PubMed] [Google Scholar]
- 623.Braak H, Braak E. Neuropathological stageing of Alzheimer-related changes. Acta Neuropathol. 1991;82:239–259. doi: 10.1007/BF00308809. [DOI] [PubMed] [Google Scholar]
- 624.Bancher C, Braak H, Fischer P, Jellinger KA. Neuropathological staging of Alzheimer lesions and intellectual status in Alzheimer's and Parkinson's disease patients. Neurosci Lett. 1993;162:179–182. doi: 10.1016/0304-3940(93)90590-h. [DOI] [PubMed] [Google Scholar]
- 625.Braak H, Braak E. Staging of Alzheimer's disease-related neurofibrillary changes. Neurobiol Aging. 1995;16:271–278. doi: 10.1016/0197-4580(95)00021-6. discussion 278–284. [DOI] [PubMed] [Google Scholar]
- 626.Fayyad UM, Irani KB, editors. Multi-Interval Discretization of Continuous-Valued Attributes for Classification Learning. Morgan Kaufmann; 1993. pp. 1022–1029. [Google Scholar]
- 627.Berretta R, Mendes A, Moscato P. Selection of Discriminative Genes in Microarray Experiments Using Mathematical Programming. Journal of Research and Practice in Information Technology. 2007;39:287–299. [Google Scholar]
- 628.Cotta C, Sloper C, Moscato P. Evolutionary Search of Thresholds for Robust Feature Set Selection: Application to the Analysis of Microarray Data. In: Raidl GR, Cagnoni S, Branke J, Corne DW, Drechsler R, et al., editors. Applications of Evolutionary Computing. Springer Berlin/Heidelberg; 2004. pp. 21–30. [Google Scholar]
- 629.Gómez Ravetti M, Berretta R, Moscato P. Novel Biomarkers for Prostate Cancer Revealed by (α,β)-k-Feature Sets. 2009. pp. 149–175. Foundations of Computational Intelligence Volume 5.
- 630.Hourani M, Berretta R, Mendes A, Moscato P. Genetic signatures for a rodent model of Parkinson's disease using combinatorial optimization methods. Methods Mol Biol. 2008;453:379–392. doi: 10.1007/978-1-60327-429-6_20. [DOI] [PubMed] [Google Scholar]
- 631.Mendes A, Scott RJ, Moscato P. Microarrays–identifying molecular portraits for prostate tumors with different Gleason patterns. Methods Mol Med. 2008;141:131–151. doi: 10.1007/978-1-60327-148-6_8. [DOI] [PubMed] [Google Scholar]
- 632.Rosso OA, Mendes A, Berretta R, Rostas JA, Hunter M, et al. Distinguishing childhood absence epilepsy patients from controls by the analysis of their background brain electrical activity (II): a combinatorial optimization approach for electrode selection. J Neurosci Methods. 2009;181:257–267. doi: 10.1016/j.jneumeth.2009.04.028. [DOI] [PubMed] [Google Scholar]
- 633.Cotta C, Langston MA, Moscato P. Combinatorial and Algorithmic Issues for Microarray Analysis. In: Gonzalez TF, editor. Handbook of Approximation Algorithms and Metaheuristics. Chapman & Hall/CRC; 2007. pp. 74.71–74.14. [Google Scholar]
- 634.Chang JT, Nevins JR. GATHER: a systems approach to interpreting genomic signatures. Bioinformatics. 2006;22:2926–2933. doi: 10.1093/bioinformatics/btl483. [DOI] [PubMed] [Google Scholar]
- 635.Blalock EM, Geddes JW, Chen KC, Porter NM, Markesbery WR, et al. Incipient Alzheimer's disease: microarray correlation analyses reveal major transcriptional and tumor suppressor responses. Proc Natl Acad Sci U S A. 2004;101:2173–2178. doi: 10.1073/pnas.0308512100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 636.Zhao CJ, Noack C, Brackmann M, Gloveli T, Maelicke A, et al. Neuronal Ca2+ sensor VILIP-1 leads to the upregulation of functional alpha4beta2 nicotinic acetylcholine receptors in hippocampal neurons. Mol Cell Neurosci. 2009;40:280–292. doi: 10.1016/j.mcn.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 637.Bernstein HG, Braunewell KH. Some notes on visinin-like protein 1 and Alzheimer disease. Clin Chem. 2009;55:1041–1043. doi: 10.1373/clinchem.2008.116970. [DOI] [PubMed] [Google Scholar]
- 638.Lee JM, Blennow K, Andreasen N, Laterza O, Modur V, et al. The brain injury biomarker VLP-1 is increased in the cerebrospinal fluid of Alzheimer disease patients. Clin Chem. 2008;54:1617–1623. doi: 10.1373/clinchem.2008.104497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 639.Zhao C, Anand R, Braunewell KH. Nicotine-induced Ca2+-myristoyl switch of neuronal Ca2+ sensor VILIP-1 in hippocampal neurons: a possible crosstalk mechanism for nicotinic receptors. Cell Mol Neurobiol. 2009;29:273–286. doi: 10.1007/s10571-008-9320-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 640.Gierke P, Zhao C, Bernstein HG, Noack C, Anand R, et al. Implication of neuronal Ca2+ -sensor protein VILIP-1 in the glutamate hypothesis of schizophrenia. Neurobiol Dis. 2008;32:162–175. doi: 10.1016/j.nbd.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 641.Schnurra I, Bernstein HG, Riederer P, Braunewell KH. The neuronal calcium sensor protein VILIP-1 is associated with amyloid plaques and extracellular tangles in Alzheimer's disease and promotes cell death and tau phosphorylation in vitro: a link between calcium sensors and Alzheimer's disease? Neurobiol Dis. 2001;8:900–909. doi: 10.1006/nbdi.2001.0432. [DOI] [PubMed] [Google Scholar]
- 642.Braunewell K, Riederer P, Spilker C, Gundelfinger ED, Bogerts B, et al. Abnormal localization of two neuronal calcium sensor proteins, visinin-like proteins (vilips)-1 and -3, in neocortical brain areas of Alzheimer disease patients. Dement Geriatr Cogn Disord. 2001;12:110–116. doi: 10.1159/000051244. [DOI] [PubMed] [Google Scholar]
- 643.Killick R, Scales G, Leroy K, Causevic M, Hooper C, et al. Deletion of Irs2 reduces amyloid deposition and rescues behavioural deficits in APP transgenic mice. Biochem Biophys Res Commun. 2009;386:257–262. doi: 10.1016/j.bbrc.2009.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 644.Bulbarelli A, Lonati E, Cazzaniga E, Gregori M, Masserini M. Pin1 affects Tau phosphorylation in response to Abeta oligomers. Mol Cell Neurosci. 2009;42:75–80. doi: 10.1016/j.mcn.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 645.Zhao WQ, Feng C, Alkon DL. Impairment of phosphatase 2A contributes to the prolonged MAP kinase phosphorylation in Alzheimer's disease fibroblasts. Neurobiol Dis. 2003;14:458–469. doi: 10.1016/s0969-9961(03)00124-4. [DOI] [PubMed] [Google Scholar]
- 646.Vafai SB, Stock JB. Protein phosphatase 2A methylation: a link between elevated plasma homocysteine and Alzheimer's Disease. FEBS Lett. 2002;518:1–4. doi: 10.1016/s0014-5793(02)02702-3. [DOI] [PubMed] [Google Scholar]
- 647.Wei Q, Holzer M, Brueckner MK, Liu Y, Arendt T. Dephosphorylation of tau protein by calcineurin triturated into neural living cells. Cell Mol Neurobiol. 2002;22:13–24. doi: 10.1023/A:1015385527187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 648.Tian Q, Wang J. Role of serine/threonine protein phosphatase in Alzheimer's disease. Neurosignals. 2002;11:262–269. doi: 10.1159/000067425. [DOI] [PubMed] [Google Scholar]
- 649.Gong CX, Wang JZ, Iqbal K, Grundke-Iqbal I. Inhibition of protein phosphatase 2A induces phosphorylation and accumulation of neurofilaments in metabolically active rat brain slices. Neurosci Lett. 2003;340:107–110. doi: 10.1016/s0304-3940(03)00096-x. [DOI] [PubMed] [Google Scholar]
- 650.Munch G, Kuhla B, Luth HJ, Arendt T, Robinson SR. Anti-AGEing defences against Alzheimer's disease. Biochem Soc Trans. 2003;31:1397–1399. doi: 10.1042/bst0311397. [DOI] [PubMed] [Google Scholar]
- 651.Sontag E, Luangpirom A, Hladik C, Mudrak I, Ogris E, et al. Altered expression levels of the protein phosphatase 2A ABalphaC enzyme are associated with Alzheimer disease pathology. J Neuropathol Exp Neurol. 2004;63:287–301. doi: 10.1093/jnen/63.4.287. [DOI] [PubMed] [Google Scholar]
- 652.Rametti A, Esclaire F, Yardin C, Terro F. Linking alterations in tau phosphorylation and cleavage during neuronal apoptosis. J Biol Chem. 2004;279:54518–54528. doi: 10.1074/jbc.M408186200. [DOI] [PubMed] [Google Scholar]
- 653.Iqbal K, Liu F, Gong CX, Alonso Adel C, Grundke-Iqbal I. Mechanisms of tau-induced neurodegeneration. Acta Neuropathol. 2009;118:53–69. doi: 10.1007/s00401-009-0486-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 654.Deters N, Ittner LM, Gotz J. Substrate-specific reduction of PP2A activity exaggerates tau pathology. Biochem Biophys Res Commun. 2009;379:400–405. doi: 10.1016/j.bbrc.2008.12.140. [DOI] [PubMed] [Google Scholar]
- 655.Liang Z, Liu F, Iqbal K, Grundke-Iqbal I, Wegiel J, et al. Decrease of protein phosphatase 2A and its association with accumulation and hyperphosphorylation of tau in Down syndrome. J Alzheimers Dis. 2008;13:295–302. doi: 10.3233/jad-2008-13307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 656.Chen S, Li B, Grundke-Iqbal I, Iqbal K. I1PP2A affects tau phosphorylation via association with the catalytic subunit of protein phosphatase 2A. J Biol Chem. 2008;283:10513–10521. doi: 10.1074/jbc.M709852200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 657.Liu R, Zhou XW, Tanila H, Bjorkdahl C, Wang JZ, et al. Phosphorylated PP2A (tyrosine 307) is associated with Alzheimer neurofibrillary pathology. J Cell Mol Med. 2008;12:241–257. doi: 10.1111/j.1582-4934.2008.00249.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 658.Meske V, Albert F, Ohm TG. Coupling of mammalian target of rapamycin with phosphoinositide 3-kinase signaling pathway regulates protein phosphatase 2A- and glycogen synthase kinase-3 -dependent phosphorylation of Tau. J Biol Chem. 2008;283:100–109. doi: 10.1074/jbc.M704292200. [DOI] [PubMed] [Google Scholar]
- 659.Walton JR. An aluminum-based rat model for Alzheimer's disease exhibits oxidative damage, inhibition of PP2A activity, hyperphosphorylated tau, and granulovacuolar degeneration. J Inorg Biochem. 2007;101:1275–1284. doi: 10.1016/j.jinorgbio.2007.06.001. [DOI] [PubMed] [Google Scholar]
- 660.Nowotny P, Bertelsen S, Hinrichs AL, Kauwe JS, Mayo K, et al. Association studies between common variants in prolyl isomerase Pin1 and the risk for late-onset Alzheimer's disease. Neurosci Lett. 2007;419:15–17. doi: 10.1016/j.neulet.2007.03.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 661.Chohan MO, Khatoon S, Iqbal IG, Iqbal K. Involvement of I2PP2A in the abnormal hyperphosphorylation of tau and its reversal by Memantine. FEBS Lett. 2006;580:3973–3979. doi: 10.1016/j.febslet.2006.06.021. [DOI] [PubMed] [Google Scholar]
- 662.Plattner F, Angelo M, Giese KP. The roles of cyclin-dependent kinase 5 and glycogen synthase kinase 3 in tau hyperphosphorylation. J Biol Chem. 2006;281:25457–25465. doi: 10.1074/jbc.M603469200. [DOI] [PubMed] [Google Scholar]
- 663.Kerr F, Rickle A, Nayeem N, Brandner S, Cowburn RF, et al. PTEN, a negative regulator of PI3 kinase signalling, alters tau phosphorylation in cells by mechanisms independent of GSK-3. FEBS Lett. 2006;580:3121–3128. doi: 10.1016/j.febslet.2006.04.064. [DOI] [PubMed] [Google Scholar]
- 664.Liu F, Grundke-Iqbal I, Iqbal K, Gong CX. Contributions of protein phosphatases PP1, PP2A, PP2B and PP5 to the regulation of tau phosphorylation. Eur J Neurosci. 2005;22:1942–1950. doi: 10.1111/j.1460-9568.2005.04391.x. [DOI] [PubMed] [Google Scholar]
- 665.Madeira A, Pommet JM, Prochiantz A, Allinquant B. SET protein (TAF1beta, I2PP2A) is involved in neuronal apoptosis induced by an amyloid precursor protein cytoplasmic subdomain. FASEB J. 2005;19:1905–1907. doi: 10.1096/fj.05-3839fje. [DOI] [PubMed] [Google Scholar]
- 666.Snyder EM, Nong Y, Almeida CG, Paul S, Moran T, et al. Regulation of NMDA receptor trafficking by amyloid-beta. Nat Neurosci. 2005;8:1051–1058. doi: 10.1038/nn1503. [DOI] [PubMed] [Google Scholar]
- 667.Rahman A, Grundke-Iqbal I, Iqbal K. PP2B isolated from human brain preferentially dephosphorylates Ser-262 and Ser-396 of the Alzheimer disease abnormally hyperphosphorylated tau. J Neural Transm. 2006;113:219–230. doi: 10.1007/s00702-005-0313-5. [DOI] [PubMed] [Google Scholar]
- 668.Rahman A, Grundke-Iqbal I, Iqbal K. Phosphothreonine-212 of Alzheimer abnormally hyperphosphorylated tau is a preferred substrate of protein phosphatase-1. Neurochem Res. 2005;30:277–287. doi: 10.1007/s11064-005-2483-9. [DOI] [PubMed] [Google Scholar]
- 669.Cheng LY, Wang JZ, Gong CX, Pei JJ, Zaidi T, et al. Multiple forms of phosphatase from human brain: isolation and partial characterization of affi-gel blue binding phosphatases. Neurochem Res. 2000;25:107–120. doi: 10.1023/a:1007547701518. [DOI] [PubMed] [Google Scholar]
- 670.Garver TD, Lehman RA, Billingsley ML. Microtubule assembly competence analysis of freshly-biopsied human tau, dephosphorylated tau, and Alzheimer tau. J Neurosci Res. 1996;44:12–20. doi: 10.1002/(SICI)1097-4547(19960401)44:1<12::AID-JNR2>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- 671.Matsuo ES, Shin RW, Billingsley ML, Van deVoorde A, O'Connor M, et al. Biopsy-derived adult human brain tau is phosphorylated at many of the same sites as Alzheimer's disease paired helical filament tau. Neuron. 1994;13:989–1002. doi: 10.1016/0896-6273(94)90264-x. [DOI] [PubMed] [Google Scholar]
- 672.Bajo M, Yoo BC, Cairns N, Gratzer M, Lubec G. Neurofilament proteins NF-L, NF-M and NF-H in brain of patients with Down syndrome and Alzheimer's disease. Amino Acids. 2001;21:293–301. doi: 10.1007/s007260170015. [DOI] [PubMed] [Google Scholar]
- 673.Blass JP, Markesbery WR, Ko LW, DeGiorgio L, Sheu KF, et al. Presence of neuronal proteins in serially cultured cells from autopsy human brain. J Neurol Sci. 1994;121:132–138. doi: 10.1016/0022-510x(94)90341-7. [DOI] [PubMed] [Google Scholar]
- 674.Cairns NJ, Uryu K, Bigio EH, Mackenzie IR, Gearing M, et al. alpha-Internexin aggregates are abundant in neuronal intermediate filament inclusion disease (NIFID) but rare in other neurodegenerative diseases. Acta Neuropathol. 2004;108:213–223. doi: 10.1007/s00401-004-0882-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 675.DeGiorgio LA, Sheu KF, Blass JP. Culture from human leptomeninges of cells containing neurofilament protein and neuron-specific enolase. J Neurol Sci. 1994;124:141–148. doi: 10.1016/0022-510x(94)90319-0. [DOI] [PubMed] [Google Scholar]
- 676.Deng Y, Li B, Liu F, Iqbal K, Grundke-Iqbal I, et al. Regulation between O-GlcNAcylation and phosphorylation of neurofilament-M and their dysregulation in Alzheimer disease. Faseb J. 2008;22:138–145. doi: 10.1096/fj.07-8309com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 677.Faigon M, Hadas E, Alroy G, Chapman J, Auerbach JM, et al. Monoclonal antibodies to the heavy neurofilament subunit (NF-H) of Torpedo cholinergic neurons. J Neurosci Res. 1991;29:490–498. doi: 10.1002/jnr.490290409. [DOI] [PubMed] [Google Scholar]
- 678.Hollosi M, Holly S, Majer Z, Laczko I, Fasman GD. Complexes of aluminium with peptide ligands: a Fourier transform IR spectroscopic study. Biopolymers. 1995;36:381–389. doi: 10.1002/bip.360360311. [DOI] [PubMed] [Google Scholar]
- 679.Hollosi M, Shen ZM, Perczel A, Fasman GD. Stable intrachain and interchain complexes of neurofilament peptides: a putative link between Al3+ and Alzheimer disease. Proc Natl Acad Sci U S A. 1994;91:4902–4906. doi: 10.1073/pnas.91.11.4902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 680.Hollosi M, Urge L, Perczel A, Kajtar J, Teplan I, et al. Metal ion-induced conformational changes of phosphorylated fragments of human neurofilament (NF-M) protein. J Mol Biol. 1992;223:673–682. doi: 10.1016/0022-2836(92)90983-q. [DOI] [PubMed] [Google Scholar]
- 681.Holly S, Laczko I, Fasman GD, Hollosi M. FT-IR spectroscopy indicates that Ca(2+)-binding to phosphorylated C-terminal fragments of the midsized neurofilament protein subunit results in beta-sheet formation and beta-aggregation. Biochem Biophys Res Commun. 1993;197:755–762. doi: 10.1006/bbrc.1993.2543. [DOI] [PubMed] [Google Scholar]
- 682.Hu YY, He SS, Wang XC, Duan QH, Khatoon S, et al. Elevated levels of phosphorylated neurofilament proteins in cerebrospinal fluid of Alzheimer disease patients. Neurosci Lett. 2002;320:156–160. doi: 10.1016/s0304-3940(02)00047-2. [DOI] [PubMed] [Google Scholar]
- 683.Julien JP, Mushynski WE. Neurofilaments in health and disease. Prog Nucleic Acid Res Mol Biol. 1998;61:1–23. doi: 10.1016/s0079-6603(08)60823-5. [DOI] [PubMed] [Google Scholar]
- 684.Kittur S, Hoh J, Endo H, Tourtellotte W, Weeks BS, et al. Cytoskeletal neurofilament gene expression in brain tissue from Alzheimer's disease patients. I. Decrease in NF-L and NF-M message. J Geriatr Psychiatry Neurol. 1994;7:153–158. doi: 10.1177/089198879400700305. [DOI] [PubMed] [Google Scholar]
- 685.Ksiezak-Reding H, Yen SH. Two monoclonal antibodies recognize Alzheimer's neurofibrillary tangles, neurofilament, and microtubule-associated proteins. J Neurochem. 1987;48:455–462. doi: 10.1111/j.1471-4159.1987.tb04114.x. [DOI] [PubMed] [Google Scholar]
- 686.Lee VM, Otvos L, Jr, Carden MJ, Hollosi M, Dietzschold B, et al. Identification of the major multiphosphorylation site in mammalian neurofilaments. Proc Natl Acad Sci U S A. 1988;85:1998–2002. doi: 10.1073/pnas.85.6.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 687.Li SP, Deng YQ, Wang XC, Wang YP, Wang JZ. Melatonin protects SH-SY5Y neuroblastoma cells from calyculin A-induced neurofilament impairment and neurotoxicity. J Pineal Res. 2004;36:186–191. doi: 10.1111/j.1600-079x.2004.00116.x. [DOI] [PubMed] [Google Scholar]
- 688.Miller CC, Brion JP, Calvert R, Chin TK, Eagles PA, et al. Alzheimer's paired helical filaments share epitopes with neurofilament side arms. Embo J. 1986;5:269–276. doi: 10.1002/j.1460-2075.1986.tb04209.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 689.Pollak D, Cairns N, Lubec G. Cytoskeleton derangement in brain of patients with Down syndrome, Alzheimer's disease and Pick's disease. J Neural Transm. 2003;(Suppl):149–158. doi: 10.1007/978-3-7091-6721-2_13. [DOI] [PubMed] [Google Scholar]
- 690.Roder HM, Ingram VM. Two novel kinases phosphorylate tau and the KSP site of heavy neurofilament subunits in high stoichiometric ratios. J Neurosci. 1991;11:3325–3343. doi: 10.1523/JNEUROSCI.11-11-03325.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 691.Save MP, Shetty VP, Shetty KT, Antia NH. Alterations in neurofilament protein(s) in human leprous nerves: morphology, immunohistochemistry and Western immunoblot correlative study. Neuropathol Appl Neurobiol. 2004;30:635–650. doi: 10.1111/j.1365-2990.2004.00578.x. [DOI] [PubMed] [Google Scholar]
- 692.Schmidt ML, Lee VM, Trojanowski JQ. Analysis of epitopes shared by Hirano bodies and neurofilament proteins in normal and Alzheimer's disease hippocampus. Lab Invest. 1989;60:513–522. [PubMed] [Google Scholar]
- 693.Schmidt ML, Lee VM, Trojanowski JQ. Comparative epitope analysis of neuronal cytoskeletal proteins in Alzheimer's disease senile plaque neurites and neuropil threads. Lab Invest. 1991;64:352–357. [PubMed] [Google Scholar]
- 694.Shen ZM, Perczel A, Hollosi M, Nagypal I, Fasman GD. Study of Al3+ binding and conformational properties of the alanine-substituted C-terminal domain of the NF-M protein and its relevance to Alzheimer's disease. Biochemistry. 1994;33:9627–9636. doi: 10.1021/bi00198a031. [DOI] [PubMed] [Google Scholar]
- 695.Trojanowski JQ, Schmidt ML, Otvos L, Jr, Gur RC, Gur RE, et al. Selective expression of epitopes in multiphosphorylation repeats of the high and middle molecular weight neurofilament proteins in Alzheimer neurofibrillary tangles. Ann Med. 1989;21:113–116. doi: 10.3109/07853898909149196. [DOI] [PubMed] [Google Scholar]
- 696.Troy CM, Greene LA, Shelanski ML. Neurite outgrowth in peripherin-depleted PC12 cells. J Cell Biol. 1992;117:1085–1092. doi: 10.1083/jcb.117.5.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 697.Wang J, Tung YC, Wang Y, Li XT, Iqbal K, et al. Hyperphosphorylation and accumulation of neurofilament proteins in Alzheimer disease brain and in okadaic acid-treated SY5Y cells. FEBS Lett. 2001;507:81–87. doi: 10.1016/s0014-5793(01)02944-1. [DOI] [PubMed] [Google Scholar]
- 698.Wang Y, Wang Q, Wang J. [Detection of level and mutation of neurofilament mRNA in Alzheimer's disease]. Zhonghua Yi Xue Za Zhi. 2002;82:519–522. [PubMed] [Google Scholar]
- 699.Wang YP, Wei ZL, Wang XC, Wang Q, Wang JZ. [Comparative study of the expression and phosphorylation of neurofilament proteins of brain gray matter in Alzheimer's disease]. Zhongguo Yi Xue Ke Xue Yuan Xue Bao. 2001;23:445–449. [PubMed] [Google Scholar]
- 700.Wurtman RJ, Ulus IH, Cansev M, Watkins CJ, Wang L, et al. Synaptic proteins and phospholipids are increased in gerbil brain by administering uridine plus docosahexaenoic acid orally. Brain Res. 2006;1088:83–92. doi: 10.1016/j.brainres.2006.03.019. [DOI] [PubMed] [Google Scholar]
- 701.Yang X, Yang Y, Luo Y, Li G, Wang J, et al. Hyperphosphorylation and Accumulation of Neurofilament Proteins in Transgenic Mice with Alzheimer Presenilin 1 Mutation. Cell Mol Neurobiol. 2009 doi: 10.1007/s10571-008-9341-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 702.Yao PJ, Coleman PD. Reduced O-glycosylated clathrin assembly protein AP180: implication for synaptic vesicle recycling dysfunction in Alzheimer's disease. Neurosci Lett. 1998;252:33–36. doi: 10.1016/s0304-3940(98)00547-3. [DOI] [PubMed] [Google Scholar]
- 703.Laterza OF, Modur VR, Crimmins DL, Olander JV, Landt Y, et al. Identification of novel brain biomarkers. Clin Chem. 2006;52:1713–1721. doi: 10.1373/clinchem.2006.070912. [DOI] [PubMed] [Google Scholar]
- 704.Zhao C, Anand R, Braunewell KH. Nicotine-induced Ca(2+)-myristoyl Switch of Neuronal Ca(2+) Sensor VILIP-1 in Hippocampal Neurons: A Possible Crosstalk Mechanism for Nicotinic Receptors. Cell Mol Neurobiol. 2008 doi: 10.1007/s10571-008-9320-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 705.Zhao C, Noack C, Brackmann M, Gloveli T, Maelicke A, et al. Neuronal Ca(2+) sensor VILIP-1 leads to the upregulation of functional alpha4beta2 nicotinic acetylcholine receptors in hippocampal neurons. Mol Cell Neurosci. 2008 doi: 10.1016/j.mcn.2008.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 706.Briand C, Kozlov SV, Sonderegger P, Grutter MG. Crystal structure of neuroserpin: a neuronal serpin involved in a conformational disease. FEBS Lett. 2001;505:18–22. doi: 10.1016/s0014-5793(01)02764-8. [DOI] [PubMed] [Google Scholar]
- 707.Bruno MA, Cuello AC. Activity-dependent release of precursor nerve growth factor, conversion to mature nerve growth factor, and its degradation by a protease cascade. Proc Natl Acad Sci U S A. 2006;103:6735–6740. doi: 10.1073/pnas.0510645103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 708.Carrell RW. Cell toxicity and conformational disease. Trends Cell Biol. 2005;15:574–580. doi: 10.1016/j.tcb.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 709.Crowther DC. Familial conformational diseases and dementias. Hum Mutat. 2002;20:1–14. doi: 10.1002/humu.10100. [DOI] [PubMed] [Google Scholar]
- 710.Dufour A, Corsini E, Gelati M, Massa G, Tarcic N, et al. Mutations in the neuroserpin gene are rare in familial dementia. French Alzheimer's Disease and Fronto-Temporal Dementia Genetics Study Groups. Ann Neurol. 2000;47:688. [PubMed] [Google Scholar]
- 711.Kinghorn KJ, Crowther DC, Sharp LK, Nerelius C, Davis RL, et al. Neuroserpin binds Abeta and is a neuroprotective component of amyloid plaques in Alzheimer disease. J Biol Chem. 2006;281:29268–29277. doi: 10.1074/jbc.M600690200. [DOI] [PubMed] [Google Scholar]
- 712.Nielsen HM, Minthon L, Londos E, Blennow K, Miranda E, et al. Plasma and CSF serpins in Alzheimer disease and dementia with Lewy bodies. Neurology. 2007;69:1569–1579. doi: 10.1212/01.wnl.0000271077.82508.a0. [DOI] [PubMed] [Google Scholar]
- 713.Tabira T. [Familial non-Alzheimer dementia]. Rinsho Shinkeigaku. 2003;43:775–778. [PubMed] [Google Scholar]
- 714.Yamasaki M, Li W, Johnson DJ, Huntington JA. Crystal structure of a stable dimer reveals the molecular basis of serpin polymerization. Nature. 2008;455:1255–1258. doi: 10.1038/nature07394. [DOI] [PubMed] [Google Scholar]
- 715.Opii WO, Joshi G, Head E, Milgram NW, Muggenburg BA, et al. Proteomic identification of brain proteins in the canine model of human aging following a long-term treatment with antioxidants and a program of behavioral enrichment: relevance to Alzheimer's disease. Neurobiol Aging. 2008;29:51–70. doi: 10.1016/j.neurobiolaging.2006.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 716.Poon HF, Farr SA, Thongboonkerd V, Lynn BC, Banks WA, et al. Proteomic analysis of specific brain proteins in aged SAMP8 mice treated with alpha-lipoic acid: implications for aging and age-related neurodegenerative disorders. Neurochem Int. 2005;46:159–168. doi: 10.1016/j.neuint.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 717.Somerville MJ, Percy ME, Bergeron C, Yoong LK, Grima EA, et al. Localization and quantitation of 68 kDa neurofilament and superoxide dismutase-1 mRNA in Alzheimer brains. Brain Res Mol Brain Res. 1991;9:1–8. doi: 10.1016/0169-328x(91)90123-f. [DOI] [PubMed] [Google Scholar]
- 718.Wishart TM, Paterson JM, Short DM, Meredith S, Robertson KA, et al. Differential proteomics analysis of synaptic proteins identifies potential cellular targets and protein mediators of synaptic neuroprotection conferred by the slow Wallerian degeneration (Wlds) gene. Mol Cell Proteomics. 2007;6:1318–1330. doi: 10.1074/mcp.M600457-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 719.Shen Y, Halperin JA, Lee CM. Complement-mediated neurotoxicity is regulated by homologous restriction. Brain Res. 1995;671:282–292. doi: 10.1016/0006-8993(94)01264-i. [DOI] [PubMed] [Google Scholar]
- 720.Wallace MA. Effects of Alzheimer's disease-related beta amyloid protein fragments on enzymes metabolizing phosphoinositides in brain. Biochim Biophys Acta. 1994;1227:183–187. doi: 10.1016/0925-4439(94)90093-0. [DOI] [PubMed] [Google Scholar]
- 721.Shimohama S, Matsushima H. Signal transduction mechanisms in Alzheimer disease. Alzheimer Dis Assoc Disord. 1995;9(Suppl 2):15–22. doi: 10.1097/00002093-199501002-00004. [DOI] [PubMed] [Google Scholar]
- 722.Sambamurti K, Sevlever D, Koothan T, Refolo LM, Pinnix I, et al. Glycosylphosphatidylinositol-anchored proteins play an important role in the biogenesis of the Alzheimer's amyloid beta-protein. J Biol Chem. 1999;274:26810–26814. doi: 10.1074/jbc.274.38.26810. [DOI] [PubMed] [Google Scholar]
- 723.Strosznajder JB, Zambrzycka A, Kacprzak MD, Strosznajder RP. Amyloid beta peptide 25–35 modulates hydrolysis of phosphoinositides by membrane phospholipase(s) C of adult brain cortex. J Mol Neurosci. 1999;12:101–109. doi: 10.1007/BF02736924. [DOI] [PubMed] [Google Scholar]
- 724.Zambrzycka A, Cakala M, Kaminska M. Transition metal ions significantly decrease phospholipase C activity degrading phosphatidylinositol-4,5-bisphosphate in the brain cortex. Pol J Pharmacol. 2003;55:915–917. [PubMed] [Google Scholar]
- 725.Merlos-Suarez A, Ruiz-Paz S, Baselga J, Arribas J. Metalloprotease-dependent protransforming growth factor-alpha ectodomain shedding in the absence of tumor necrosis factor-alpha-converting enzyme. J Biol Chem. 2001;276:48510–48517. doi: 10.1074/jbc.M103488200. [DOI] [PubMed] [Google Scholar]
- 726.Gilmore GC, Wenk HE, Naylor LA, Koss E. Motion perception and Alzheimer's disease. J Gerontol. 1994;49:P52–57. doi: 10.1093/geronj/49.2.p52. [DOI] [PubMed] [Google Scholar]
- 727.Hayes TL, Lewis DA. Nonphosphorylated neurofilament protein and calbindin immunoreactivity in layer III pyramidal neurons of human neocortex. Cereb Cortex. 1992;2:56–67. doi: 10.1093/cercor/2.1.56. [DOI] [PubMed] [Google Scholar]
- 728.Chin J, Palop JJ, Puolivali J, Massaro C, Bien-Ly N, et al. Fyn kinase induces synaptic and cognitive impairments in a transgenic mouse model of Alzheimer's disease. J Neurosci. 2005;25:9694–9703. doi: 10.1523/JNEUROSCI.2980-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 729.Falangola MF, Dyakin VV, Lee SP, Bogart A, Babb JS, et al. Quantitative MRI reveals aging-associated T2 changes in mouse models of Alzheimer's disease. NMR Biomed. 2007;20:343–351. doi: 10.1002/nbm.1163. [DOI] [PubMed] [Google Scholar]
- 730.Helpern JA, Lee SP, Falangola MF, Dyakin VV, Bogart A, et al. MRI assessment of neuropathology in a transgenic mouse model of Alzheimer's disease. Magn Reson Med. 2004;51:794–798. doi: 10.1002/mrm.20038. [DOI] [PubMed] [Google Scholar]
- 731.Lahiri DK, Chen D, Ge YW, Farlow M, Kotwal G, et al. Does nitric oxide synthase contribute to the pathogenesis of Alzheimer's disease?: effects of beta-amyloid deposition on NOS in transgenic mouse brain with AD pathology. Ann N Y Acad Sci. 2003;1010:639–642. doi: 10.1196/annals.1299.117. [DOI] [PubMed] [Google Scholar]
- 732.Levine S, Saltzman A, Levy E, Ginsberg SD. Systemic pathology in aged mouse models of Down's syndrome and Alzheimer's disease. Exp Mol Pathol. 2009;86:18–22. doi: 10.1016/j.yexmp.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 733.Neiva TJ, Fries DM, Monteiro HP, D'Amico EA, Chamone DA. Aluminum induces lipid peroxidation and aggregation of human blood platelets. Braz J Med Biol Res. 1997;30:599–604. doi: 10.1590/s0100-879x1997000500005. [DOI] [PubMed] [Google Scholar]
- 734.Parent A, Linden DJ, Sisodia SS, Borchelt DR. Synaptic transmission and hippocampal long-term potentiation in transgenic mice expressing FAD-linked presenilin 1. Neurobiol Dis. 1999;6:56–62. doi: 10.1006/nbdi.1998.0207. [DOI] [PubMed] [Google Scholar]
- 735.Perez SE, Lumayag S, Kovacs B, Mufson EJ, Xu S. Beta-amyloid deposition and functional impairment in the retina of the APPswe/PS1DeltaE9 transgenic mouse model of Alzheimer's disease. Invest Ophthalmol Vis Sci. 2009;50:793–800. doi: 10.1167/iovs.08-2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 736.Sadowski M, Pankiewicz J, Scholtzova H, Ji Y, Quartermain D, et al. Amyloid-beta deposition is associated with decreased hippocampal glucose metabolism and spatial memory impairment in APP/PS1 mice. J Neuropathol Exp Neurol. 2004;63:418–428. doi: 10.1093/jnen/63.5.418. [DOI] [PubMed] [Google Scholar]
- 737.Sahara N, Vega IE, Ishizawa T, Lewis J, McGowan E, et al. Phosphorylated p38MAPK specific antibodies cross-react with sarkosyl-insoluble hyperphosphorylated tau proteins. J Neurochem. 2004;90:829–838. doi: 10.1111/j.1471-4159.2004.02558.x. [DOI] [PubMed] [Google Scholar]
- 738.Wang CY, Shen YC, Lo FY, Su CH, Lee SH, et al. Normal tension glaucoma is not associated with the interleukin -1alpha (-889) genetic polymorphism. J Glaucoma. 2007;16:230–233. doi: 10.1097/IJG.0b013e3180300818. [DOI] [PubMed] [Google Scholar]
- 739.Wang CY, Shen YC, Su CH, Lo FY, Lee SH, et al. Investigation of the association between interleukin-1beta polymorphism and normal tension glaucoma. Mol Vis. 2007;13:719–723. [PMC free article] [PubMed] [Google Scholar]
- 740.Wang X, Su B, Siedlak SL, Moreira PI, Fujioka H, et al. Amyloid-beta overproduction causes abnormal mitochondrial dynamics via differential modulation of mitochondrial fission/fusion proteins. Proc Natl Acad Sci U S A. 2008;105:19318–19323. doi: 10.1073/pnas.0804871105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 741.Yu WH, Matsuoka Y, Sziraki I, Hashim A, Lafrancois J, et al. Increased dopaminergic neuron sensitivity to 1-methyl-4-phenyl-1,2,3,6-tetrahydropyridine (MPTP) in transgenic mice expressing mutant A53T alpha-synuclein. Neurochem Res. 2008;33:902–911. doi: 10.1007/s11064-007-9533-4. [DOI] [PubMed] [Google Scholar]
- 742.Zhuo JM, Prakasam A, Murray ME, Zhang HY, Baxter MG, et al. An increase in Abeta42 in the prefrontal cortex is associated with a reversal-learning impairment in Alzheimer's disease model Tg2576 APPsw mice. Curr Alzheimer Res. 2008;5:385–391. doi: 10.2174/156720508785132280. [DOI] [PubMed] [Google Scholar]
- 743.Freude S, Plum L, Schnitker J, Leeser U, Udelhoven M, et al. Peripheral hyperinsulinemia promotes tau phosphorylation in vivo. Diabetes. 2005;54:3343–3348. doi: 10.2337/diabetes.54.12.3343. [DOI] [PubMed] [Google Scholar]
- 744.Lowe XR, Lu X, Marchetti F, Wyrobek AJ. The expression of Troponin T1 gene is induced by ketamine in adult mouse brain. Brain Res. 2007;1174:7–17. doi: 10.1016/j.brainres.2007.07.039. [DOI] [PubMed] [Google Scholar]
- 745.Morris BJ. A forkhead in the road to longevity: the molecular basis of lifespan becomes clearer. J Hypertens. 2005;23:1285–1309. doi: 10.1097/01.hjh.0000173509.45363.dd. [DOI] [PubMed] [Google Scholar]
- 746.Nakagawa K, Kitazume S, Oka R, Maruyama K, Saido TC, et al. Sialylation enhances the secretion of neurotoxic amyloid-beta peptides. J Neurochem. 2006;96:924–933. doi: 10.1111/j.1471-4159.2005.03595.x. [DOI] [PubMed] [Google Scholar]
- 747.Lian Q, Ladner CJ, Magnuson D, Lee JM. Selective changes of calcineurin (protein phosphatase 2B) activity in Alzheimer's disease cerebral cortex. Exp Neurol. 2001;167:158–165. doi: 10.1006/exnr.2000.7534. [DOI] [PubMed] [Google Scholar]
- 748.Shibata N, Kobayashi M. [The role for oxidative stress in neurodegenerative diseases]. Brain Nerve. 2008;60:157–170. [PubMed] [Google Scholar]
- 749.Wang H, Dong K, Li G, Peng X, Zhu H. [Effect of yizhi jiannao granules on the expression of Pin1 and HMGB1 mRNA in the hippocampus of SAMP8 mice.]. Zhong Nan Da Xue Xue Bao Yi Xue Ban. 2009;34:63–66. [PubMed] [Google Scholar]
- 750.Yang G, Wang L, Zhu M, Xu D. Identification of non-Alzheimer's disease tauopathies-related proteins by proteomic analysis. Neurol Res. 2008;30:613–622. doi: 10.1179/174313208X284124. [DOI] [PubMed] [Google Scholar]
- 751.Desjardins P, Ledoux S. Expression of ced-3 and ced-9 homologs in Alzheimer's disease cerebral cortex. Neurosci Lett. 1998;244:69–72. doi: 10.1016/s0304-3940(98)00138-4. [DOI] [PubMed] [Google Scholar]
- 752.Akiyama H, Nishimura T, Kondo H, Ikeda K, Hayashi Y, et al. Expression of the receptor for macrophage colony stimulating factor by brain microglia and its upregulation in brains of patients with Alzheimer's disease and amyotrophic lateral sclerosis. Brain Res. 1994;639:171–174. doi: 10.1016/0006-8993(94)91779-5. [DOI] [PubMed] [Google Scholar]
- 753.Boissonneault V, Filali M, Lessard M, Relton J, Wong G, et al. Powerful beneficial effects of macrophage colony-stimulating factor on {beta}-amyloid deposition and cognitive impairment in Alzheimer's disease. Brain. 2009 doi: 10.1093/brain/awn331. [DOI] [PubMed] [Google Scholar]
- 754.Du Yan S, Zhu H, Fu J, Yan SF, Roher A, et al. Amyloid-beta peptide-receptor for advanced glycation endproduct interaction elicits neuronal expression of macrophage-colony stimulating factor: a proinflammatory pathway in Alzheimer disease. Proc Natl Acad Sci U S A. 1997;94:5296–5301. doi: 10.1073/pnas.94.10.5296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 755.Ebadi M, Bashir RM, Heidrick ML, Hamada FM, Refaey HE, et al. Neurotrophins and their receptors in nerve injury and repair. Neurochem Int. 1997;30:347–374. doi: 10.1016/s0197-0186(96)00071-x. [DOI] [PubMed] [Google Scholar]
- 756.Flanagan AM, Lader CS. Update on the biologic effects of macrophage colony-stimulating factor. Curr Opin Hematol. 1998;5:181–185. doi: 10.1097/00062752-199805000-00006. [DOI] [PubMed] [Google Scholar]
- 757.Georganopoulou DG, Chang L, Nam JM, Thaxton CS, Mufson EJ, et al. Nanoparticle-based detection in cerebral spinal fluid of a soluble pathogenic biomarker for Alzheimer's disease. Proc Natl Acad Sci U S A. 2005;102:2273–2276. doi: 10.1073/pnas.0409336102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 758.Hamilton JA, Whitty G, White AR, Jobling MF, Thompson A, et al. Alzheimer's disease amyloid beta and prion protein amyloidogenic peptides promote macrophage survival, DNA synthesis and enhanced proliferative response to CSF-1 (M-CSF). Brain Res. 2002;940:49–54. doi: 10.1016/s0006-8993(02)02589-1. [DOI] [PubMed] [Google Scholar]
- 759.Hasegawa Y, Sawada M, Ozaki N, Inagaki T, Suzumura A. Increased soluble tumor necrosis factor receptor levels in the serum of elderly people. Gerontology. 2000;46:185–188. doi: 10.1159/000022157. [DOI] [PubMed] [Google Scholar]
- 760.Ito S, Sawada M, Haneda M, Fujii S, Oh-Hashi K, et al. Amyloid-beta peptides induce cell proliferation and macrophage colony-stimulating factor expression via the PI3-kinase/Akt pathway in cultured Ra2 microglial cells. FEBS Lett. 2005;579:1995–2000. doi: 10.1016/j.febslet.2005.02.048. [DOI] [PubMed] [Google Scholar]
- 761.Kaku M, Tsutsui K, Motokawa M, Kawata T, Fujita T, et al. Amyloid beta protein deposition and neuron loss in osteopetrotic (op/op) mice. Brain Res Brain Res Protoc. 2003;12:104–108. doi: 10.1016/j.brainresprot.2003.09.001. [DOI] [PubMed] [Google Scholar]
- 762.Kawata T, Tsutsui K, Kohno S, Kaku M, Fujita T, et al. Amyloid beta protein deposition in osteopetrotic (op/op) mice is reduced by injections of macrophage colony stimulating factor. J Int Med Res. 2005;33:654–660. doi: 10.1177/147323000503300607. [DOI] [PubMed] [Google Scholar]
- 763.Kondo Y, Lemere CA, Seabrook TJ. Osteopetrotic (op/op) mice have reduced microglia, no Abeta deposition, and no changes in dopaminergic neurons. J Neuroinflammation. 2007;4:31. doi: 10.1186/1742-2094-4-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 764.Kong QL, Zhang JM, Zhang ZX, Ge PJ, Xu YJ, et al. [Serum levels of macrophage colony stimulating factor in the patients with Alzheimer's disease]. Zhongguo Yi Xue Ke Xue Yuan Xue Bao. 2002;24:298–301. [PubMed] [Google Scholar]
- 765.Kumar AP, Piedrafita FJ, Reynolds WF. Peroxisome proliferator-activated receptor gamma ligands regulate myeloperoxidase expression in macrophages by an estrogen-dependent mechanism involving the -463GA promoter polymorphism. J Biol Chem. 2004;279:8300–8315. doi: 10.1074/jbc.M311625200. [DOI] [PubMed] [Google Scholar]
- 766.Lee SC, Liu W, Brosnan CF, Dickson DW. GM-CSF promotes proliferation of human fetal and adult microglia in primary cultures. Glia. 1994;12:309–318. doi: 10.1002/glia.440120407. [DOI] [PubMed] [Google Scholar]
- 767.Li M, Pisalyaput K, Galvan M, Tenner AJ. Macrophage colony stimulatory factor and interferon-gamma trigger distinct mechanisms for augmentation of beta-amyloid-induced microglia-mediated neurotoxicity. J Neurochem. 2004;91:623–633. doi: 10.1111/j.1471-4159.2004.02765.x. [DOI] [PubMed] [Google Scholar]
- 768.Lue LF, Rydel R, Brigham EF, Yang LB, Hampel H, et al. Inflammatory repertoire of Alzheimer's disease and nondemented elderly microglia in vitro. Glia. 2001;35:72–79. doi: 10.1002/glia.1072. [DOI] [PubMed] [Google Scholar]
- 769.Lue LF, Walker DG, Brachova L, Beach TG, Rogers J, et al. Involvement of microglial receptor for advanced glycation endproducts (RAGE) in Alzheimer's disease: identification of a cellular activation mechanism. Exp Neurol. 2001;171:29–45. doi: 10.1006/exnr.2001.7732. [DOI] [PubMed] [Google Scholar]
- 770.Lue LF, Walker DG, Rogers J. Modeling microglial activation in Alzheimer's disease with human postmortem microglial cultures. Neurobiol Aging. 2001;22:945–956. doi: 10.1016/s0197-4580(01)00311-6. [DOI] [PubMed] [Google Scholar]
- 771.Majumdar A, Cruz D, Asamoah N, Buxbaum A, Sohar I, et al. Activation of microglia acidifies lysosomes and leads to degradation of Alzheimer amyloid fibrils. Mol Biol Cell. 2007;18:1490–1496. doi: 10.1091/mbc.E06-10-0975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 772.Mitrasinovic OM, Grattan A, Robinson CC, Lapustea NB, Poon C, et al. Microglia overexpressing the macrophage colony-stimulating factor receptor are neuroprotective in a microglial-hippocampal organotypic coculture system. J Neurosci. 2005;25:4442–4451. doi: 10.1523/JNEUROSCI.0514-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 773.Mitrasinovic OM, Murphy GM., Jr Microglial overexpression of the M-CSF receptor augments phagocytosis of opsonized Abeta. Neurobiol Aging. 2003;24:807–815. doi: 10.1016/s0197-4580(02)00237-3. [DOI] [PubMed] [Google Scholar]
- 774.Mitrasinovic OM, Perez GV, Zhao F, Lee YL, Poon C, et al. Overexpression of macrophage colony-stimulating factor receptor on microglial cells induces an inflammatory response. J Biol Chem. 2001;276:30142–30149. doi: 10.1074/jbc.M104265200. [DOI] [PubMed] [Google Scholar]
- 775.Mitrasinovic OM, Robinson CC, Tenen DG, Lee YL, Poon C, et al. Biolistic expression of the macrophage colony stimulating factor receptor in organotypic cultures induces an inflammatory response. J Neurosci Res. 2004;77:420–429. doi: 10.1002/jnr.20168. [DOI] [PubMed] [Google Scholar]
- 776.Mitrasinovic OM, Vincent VA, Simsek D, Murphy GM., Jr Macrophage colony stimulating factor promotes phagocytosis by murine microglia. Neurosci Lett. 2003;344:185–188. doi: 10.1016/s0304-3940(03)00474-9. [DOI] [PubMed] [Google Scholar]
- 777.Murphy GM, Jr, Zhao F, Yang L, Cordell B. Expression of macrophage colony-stimulating factor receptor is increased in the AbetaPP(V717F) transgenic mouse model of Alzheimer's disease. Am J Pathol. 2000;157:895–904. doi: 10.1016/s0002-9440(10)64603-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 778.Vincent VA, Robinson CC, Simsek D, Murphy GM. Macrophage colony stimulating factor prevents NMDA-induced neuronal death in hippocampal organotypic cultures. J Neurochem. 2002;82:1388–1397. doi: 10.1046/j.1471-4159.2002.01087.x. [DOI] [PubMed] [Google Scholar]
- 779.Vincent VA, Selwood SP, Murphy GM., Jr Proinflammatory effects of M-CSF and A beta in hippocampal organotypic cultures. Neurobiol Aging. 2002;23:349–362. doi: 10.1016/s0197-4580(01)00338-4. [DOI] [PubMed] [Google Scholar]
- 780.Wollmer MA, Nitsch RM, Hock C, Papassotiropoulos A. Genetic association study on colony-stimulating factor 1 in Alzheimer's disease. Neurodegener Dis. 2006;3:334–337. doi: 10.1159/000097302. [DOI] [PubMed] [Google Scholar]
- 781.Yan SD, Stern D, Kane MD, Kuo YM, Lampert HC, et al. RAGE-Abeta interactions in the pathophysiology of Alzheimer's disease. Restor Neurol Neurosci. 1998;12:167–173. [PubMed] [Google Scholar]
- 782.Zhang JM, Kong QL, Wang H, Qin C, He W. [Expression of macrophage colony stimulating factor in brains of PDAPPV717I transgenic mice]. Zhongguo Yi Xue Ke Xue Yuan Xue Bao. 2004;26:62–65. [PubMed] [Google Scholar]
- 783.Marsagishvili LG, Shpagina MD, Emel'ianenko VI, Podlubnaia ZA. [Sarcomeric proteins of the titin family form amyloids]. Biofizika. 2005;50:803–809. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
IHop Glossary of Genes.
(0.15 MB DOC)
Supplementary Data 1.
(4.10 MB XLS)
Supplementary Data 2.
(1.26 MB XLS)