Summary
It is widely recognized that biofuel production from lignocellulosic materials is limited by inadequate technology to efficiently and economically release fermentable sugars from the complex multi-polymeric raw materials. Therefore, endoglucanases, exoglucanase, pectate lyases, cutinase, swollenin, xylanase, acetyl xylan esterase, beta glucosidase and lipase genes from bacteria or fungi were expressed in E. coli or tobacco chloroplasts. A PCR based method was used to clone genes without introns from Trichoderma reesei genomic DNA. Homoplasmic transplastomic lines showed normal phenotype and were fertile. Based on observed expression levels, up to 49, 64 and 10,751 million units of pectate lyases or endoglucanase can be produced annually, per acre of tobacco. Plant production cost of endoglucanase is 3,100-fold and pectate lyase is 1,057 or 1,480 fold lower than the same recombinant enzymes sold commercially, produced via fermentation. Chloroplast-derived enzymes had higher temperature stability and wider pH optima than enzymes expressed in E. coli. Plant crude-extracts showed higher enzyme activity than E. coli with increasing protein concentration, demonstrating their direct utility without purification. Addition of E. coli extracts to the chloroplast-derived enzymes significantly decreased their activity. Chloroplast-derived crude-extract enzyme cocktails yielded more (up to 3,625%) glucose from filter paper, pine wood or citrus peel than commercial cocktails. Furthermore, pectate lyase transplastomic plants showed enhanced resistance to Erwina soft rot. This is the first report of using plant-derived enzyme cocktails for production of fermentable sugars from lignocellulosic biomass. Limitations of higher cost and lower production capacity of fermentation systems are addressed by chloroplast-derived enzyme cocktails.
Keywords: Biofuel, Renewable Energy, Cellulosic Ethanol, Cell Wall Degrading Enzymes, Fermentable Sugars, Lignocellulosic biomass
Introduction
Plant cell wall is the major component of lignocellulosic biomass which provides abundant renewable polysaccharides in nature. Because of changing global energy needs and finite petroleum reserves, there is an urgent need to develop technologies for harnessing renewable energy resources. The US Congress ‘Energy Independence and Security Act of 2007’ set the goal for annual production of 16 billion gallons of cellulosic ethanol by 2022. European Union's requirement that 10% of all transport fuels come from renewable sources makes this a global challenge (Robertson et al., 2008). The major biofuel in use today is corn-derived ethanol. In the US, 25 to 30% of corn production is currently used for ethanol production. Increasing infrastructure investment in grain ethanol production will consume a substantial portion of corn production (Robertson et al., 2008) raising prices of corn and other food/feed sources. Corn ethanol has been reported to produce more greenhouse gas emission than gasoline. On the other hand cellulosic ethanol from nonfood crops and from waste produce much less greenhouse gas emission, even less than electricity or hydrogen, the two energy sources that are thought to be important for solving the problem of greenhouse gas emission (Charles, 2009). Therefore, renewable lignocellulosic biomass from agricultural wastes and wood products is a very attractive feedstock for bioethanol production (US DOE, 2007).
Pectate lyases (EC 4.2.2.2) play an important role in degrading pectic polysaccharides that are important components of primary cell wall of plants (Carpita and Gibeaut, 1993). Pectate lyase randomly cleaves α-(1-4) linkages between galacturonosyl residues, generating 4,5-unsaturated oligogalacturonates by β-elimination (Yoder et al., 1993). They have been extensively studied in plant pathogens and the action of these enzymes results in the maceration of plant tissues leading to pathogenesis (Collmer and Keen, 1986; Crawford and Kolattukudy, 1987; Herron et al., 2000; Leitzke et al., 1994). Many pathogens like Erwinia, Fusarium, Clostridium and Bacillus produce pectate lyases that are involved in the degradation of pectic compounds. Pectic compounds of plant cell wall are primarily made of α-1,4 linked polygalactosyluronic acid residues interspersed with regions of alternating galactosyluronic acid and rhamnosyl residues. Pectin compounds form the key binding material between plant cells. Hydrolysis of pectin compound is an important step in the enzymatic hydrolysis of citrus peel because it has high pectin content (>30%, Yapo et al., 2007).
Pectate lyases play a major role in bacterial pathogenesis. After invading the host plant tissue, Erwinia produces a large amount of cell wall degrading enzymes, generating typical soft rot symptoms. Erwinia bacteria secrete several isoenzymatic forms of pectate lyase which degrade pectin into unsaturated oligogalacturonates (OG), the major virulence determinant of Erwinia, known to trigger plant defense responses (Ryan, 1988). Therefore, it has been shown that the expression of pectate lyase in potato enhanced resistance to Erwinia soft rot (Wegener, 2002). Different pectate lyases have been isolated from Fusarium solani f. sp. Pisi (Gonzalez-Candelas et al., 1992; Guo et al., 1995, 1996). These pectate lyases belong to polysaccharide lyase family 3 (http://www.cazy.org/fam/PL3.html). F. solani f. sp. pisi, is a causative agent of root rot disease in pea (Funnell et al., 2001) and chickpea plants (Bhatti and Kraft, 1992). Pectate lyases are also produced by other disease causing organisms like Erwinia, Bacillus, Aspergillus and many other plant pathogenic organisms. F. solani f. sp. pisi produces at least four pectate lyases of which PelA and PelD are inducible and PelB and PelC are constitutively expressed (Rogers et al., 2000). Therefore, in this study, PelB and PelD have been expressed in chloroplasts and used in enzyme cocktails for biomass hydrolysis or their role in enhanced resistance to Erwinia soft rot has been investigated.
Cellulosic biomass or lignocellulosic biomass is a heterogeneous complex of different polymers (Sticklen, 2008). Acid or alkaline pretreatment of wood biomass makes this substrate more accessible to enzymes and converts cellulosic polymers into fermentable sugars (Margeot et al., 2009; Merino and Cherry, 2007; Wyman et al., 2005). Likewise, citrus waste is rich in pectin, cellulose and hemicellulosic polysaccharides, which can be hydrolyzed into sugars and fermented into ethanol. Citrus processing plants in Florida annually yield about 5 million tons of wet waste, which has the potential to produce 200 million gallons of ethanol (www.nps.ars.usda.gov). Upon complete hydrolysis by enzymes, citrus waste should yield fermentable hexose sugars; monosaccharides including galacturonic acid, can be fermented to ethanol and acetic acid by the recombinant bacterium E. coli KO11 (Grohmann et al., 1994).
The conversion of cellulosic biomass into fermentable sugars is a complex process and involves enzymatic hydrolysis using three major classes of enzymes including endoglucanase, exoglucanase and beta glucosidase. Especially the endoglucanases (EC No. 3.2.1.4) constitute an important enzyme group for hydrolysis. Endoglucanases catalyze endo hydrolysis of 1,4-beta-D-glycosidic linkages in cellulose, cellulose derivatives (such as carboxy methyl cellulose and hydroxy ethyl cellulose), lichenin, beta-1,4 bonds in mixed beta-1,3 glucans such as cereal beta-D-glucans or xyloglucans and other plant materials containing cellulosic components. Several endoglucanases from various organisms including Acidothermus cellulolyticus, Syncephalastrum racemosum, Thermobifida fusca, Trichoderma viride, Aspergillus niger, Bacillus polymyxa and Clostridium thermocellum have been well characterized (Baird et al., 1990; Hasper et al., 2002; Irwin et al., 2004; Kwon et al., 1999; Ng and Zeikus, 1981; Wonganu et al., 2008).
Since the lignocellulosic wastes are composed of a complex of multiple intertwined polymers, simultaneous presence of multiple hydrolases that can increase the access of each other will be required to get efficient release of monomers. Thus, a mixture of enzymes like hemicellulases including xylanase, acetyl xylan esterase and ligninases, lipases, pectate lyases may be required for efficient hydrolysis depending upon the composition of cell walls as it varies depending on plant taxa, tissue, age and cell type (Sticklen, 2008). For example in orange peel which has high pectin content requires high dosage of pectinase whereas wood biomass requires high dosage of xylanase and its accessory enzymes like acetyl xylan esterase and ferulic acid esterase for efficient hydrolysis. Enzymes like expansins have been proposed to disrupt hydrogen bonding between cellulose microfibrils or between cellulose and other cell wall polysaccharides without having hydrolytic activity and including this enzyme for biomass hydrolysis enhance the access of other enzymes for hydrolysis (Saloheimo et al., 2002). In some biomass, such as citrus peel, a cutin layer is present in epidermal layer and therefore hydrolysis of this polymer by cutinase is likely to enhance the access of hydrolases underlying carbohydrate polymers. All these enzymes are produced naturally by a range of microbial species including bacteria and fungi. Many cell wall degrading enzymes have been isolated and characterized and many more are still not uncovered. Availabilty of genome sequences of Trichoderma reesei (Martinez et al., 2008) and other organisms (Rubin, 2008) have increased inventory of enzymes for biomass utilization. Expression of all different classes of cell wall degrading enzymes individually provides great opportunity for developing biomass specific enzyme cocktails and no such plant-derived has been reported so far in the literature.
Production of cellulosic ethanol is currently limited by the lack of technology, infrastructure and high cost of enzymes. Due to their complex structure, lignocellulosic biomass degradation requires different classes of enzymes in large quantities to efficiently release fermentable sugars. Bioethanol process would require about 11 million FPU (19 kg, 42 lbs) of cellulase to yield 84 gallons of ethanol (Himmel et al., 1997, 1999) or 15-25 kg cellulase per ton of biomass (Carroll and Somerville, 2009; TaylorII et al., 2008). Moreover, due to the different polymer compositions, it is necessary to produce different classes of enzymes individually and then create cocktails for hydrolysis of different types of biomass. Therefore, the first challenge in lignocellulosic biotechnology is to develop an efficient enzyme production system for rapid and less expensive biomass depolymerization. Because of the high cost and limited capacity for producing these enzymes through fermentation, in planta expression of biomass degrading enzymes should lower the cost of cellulosic ethanol.
Although several reports have investigated heterologous cellulase production in plants, most utilized nuclear transformation technology (Dai et al, 2000; Kawazu et al., 1999; TaylorII et al., 2008; Ziegelhoffer et al., 1999). So far, the beta 1,4 endoglucanase (E1), cellulases, xylanases, alpha glucosidase, amylases, mixed-linkage glucanases from a variety of bacteria and fungi have been investigated (Biswas et al., 2006; Dai et al., 2000; Kawazu et al., 1999; Montalvo-Rodriguez et al., 2000; Oraby et al., 2007; TaylorII et al., 2008; Xu et al., 2008; Ziegelhoffer et al., 1999, 2001). None of these studies have used real lignocellulosic biomass as substrate or determined the combination and concentration of enzymes for the development of enzyme cocktail for biomass hydrolysis. Production of enzymes via plant nuclear transformation has a few limitations including lower levels of expression (with a few exceptions), gene silencing and position effect (Verma and Daniell, 2007). In contrast, plastid transformation results in high levels of expression, with minimal concerns of transgene silencing or position effect (Bally et al., 2009; Daniell et al., 2001; DeCosa et al., 2001; Lee et al., 2003; Singh et al., 2008; Verma and Daniell, 2007). Compartmentalization of toxic proteins within chloroplasts protects transgenic plants from pleiotropic effects (Daniell et al., 2001; Lee et al., 2003). Most importantly, harvesting leaves before flowering offers nearly complete transgene containment, in addition to protection offered by maternal inheritance of transgenes, especially in tobacco (Daniell, 2007; Ruf et al., 2007; Svab and Maliga, 2007). Therefore, bacterial genes have been expressed via the tobacco plastid genome for biofuel enzyme production (Gray et al., 2008; Leelavathi et al., 2003; Yu et al., 2007). However, fungal genes have not yet been expressed in transgenic chloroplasts because of concerns of codon usage or appropriate post-translational modifications. Furthermore, efficacy of enzyme cocktails derived from plants for production of fermentable sugars from biomass has not yet been investigated (TaylorII et al., 2008).
In this study, we have used endoglucanase, exoglucanase or lipase from bacteria, pectate lyases, cutinase, endoglucanases, swollenin, xylanase, acetyl xylan esterase or beta glucosidase from fungi to create chloroplast vectors. A PCR based method was used to clone open reading frames without introns (up to 5) from Trichoderma reesei genomic DNA using An et al. (2007) protocol. Because chloroplast vectors function efficiently in E. coli (Brixey et al., 1997), it was possible to express enzymes in both systems. Enzyme cocktails were used for biomass degradation to produce fermentable sugars and for direct comparison of properties of enzymes produced via fermentation or in planta, using identical genes and regulatory sequences. Observed results indicate that plant-derived enzymes offer an inexpensive and efficient method to produce fermentable sugars from lignocellulosic biomass.
Results and Discussion
Assembly of chloroplast expression constructs
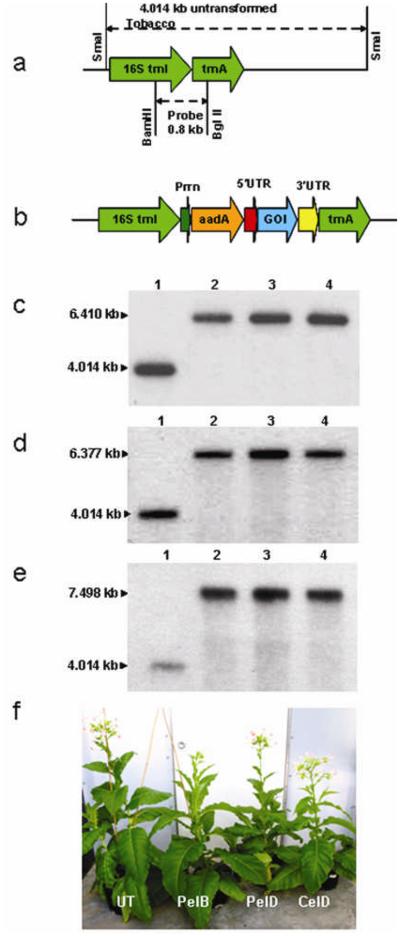
For the integration of transgenes, transcriptionally active spacer region between the trnI and trnA genes was used (Figure 1a). This region allows highly efficient transgene integration and expression (Arlen et al., 2008; Daniell et al., 2009; DeCosa et al., 2001; Koya et al., 2005; Lelivelt et al., 2005). PCR resulted in the amplification of various genes of interest (GOI) including endoglucanase (celD), exoglucanase (celO) from Clostridium thermocellum genomic DNA, lipase (lipY) from Mycobacterium tuberculosis genomic DNA, pectate lyases (pelA, pelB, pelD) and cutinase from Fusarium solani. Due to the existence of introns, low copy number of genes, high complexity of the eukaryotic genome and multiple steps involved in cDNA preparation, it is very challenging to clone full-length cDNA from fungi. Using a PCR based method (An et al., 2007), coding sequences of GOI including endoglucanases (egI), swollenin (swo1 similar to expansins), xylanase (xyn2), acetyl xylan esterase (axe1) and beta glucosidase (bgl1) were cloned without introns (ranging from 1 to 5) from Trichoderma reesei genomic DNA. This method can be used to isolate any gene from genomic DNA of an organism whose genomic sequence is available. Tobacco chloroplast transformation vectors were made with each GOI (Figure 1b). All chloroplast vectors included the 16S trnI/trnA flanking sequences for homologous recombination into the inverted repeat regions of the chloroplast genome and the aadA gene conferring resistance to spectinomycin. The origin of replication, oriA exists inside the trnI flanking region and might assist in replication of foreign vectors within chloroplasts (Daniell et al., 1990) thereby increasing the chances of transgene integration and reach homoplasmy even in the first round of selection (Guda et al., 2000). The aadA gene was driven by the constitutive rRNA operon promoter with GGAGG ribosome binding site. The GOI was driven by the psbA promoter and 5′ UTR in order to achieve high levels of expression. The 3′ UTR located at the 3′ end of the GOI conferred transcript stability. The aadA gene conferring spectinomycin resistance was used for selection.
Figure 1.
Regeneration and analysis of transplastomic lines (a) Schematic representation of the chloroplast 16S trnI/trnA region. Transgenes were inserted at the trnI/trnA spacer region in the tobacco chloroplast genome. (b) Schematic representation of the chloroplast transformation vectors. The gene of interest (GOI) is celD, celO, pelA, pelB, pelD, cutinase, lipY, egI, swo1, xyn2, axe1 or bgl1. Prrn, rRNA operon promoter; aadA, aminoglycoside 3′-adenylytransferase gene; 5′ UTR, promoter and 5′ untranslated region of psbA gene; 3′ UTR, 3′ untranslated region of psbA gene. (c) Evaluation of transgene integration and homoplasmy by Southern blot of pelB, (d) pelD and (e) celD transplastomic Petite Havana lines hybridized with the flanking sequence probe (1, untransformed; 2 to 4, transplastomic lines). (f) Phenotypes of untransformed (UT) and transplastomic lines (Petite Havana) grown in green house showing normal growth.
Generation and characterization of transplastomic tobacco expressing pectate lyases (PelB & PelD) and endoglucanase (CelD)
Transplastomic tobacco plants of experimental Petite Havana cultivar were obtained as described previously (Daniell et al., 2005; Verma et al., 2008). Southern blot analysis was performed to confirm site specific integration of the pLD-pelB, pLD-pelD and pLD-celD cassettes into the chloroplast genome and to determine homoplasmy. Digestion of total plant DNA with SmaI from untransformed and transplastomic lines generated a 4.014 kb fragment untransformed (UT) or 6.410 kb in pelB, 6.377 kb in pelD or 7.498 kb fragment in celD when hybridized with the [32P]-labeled trnI-trnA probe, confirming site specific integration of the transgenes into the spacer region between the trnI and trnA genes (Figure 1c-e). Furthermore, the absence of a 4.014 kb fragment in the transplastomic lines confirmed that homoplasmy was achieved (within the levels of detection). Transplastomic lines showed normal phenotype when compared to untransformed plants and were fertile (produced flowers, seeds Figure 1f).
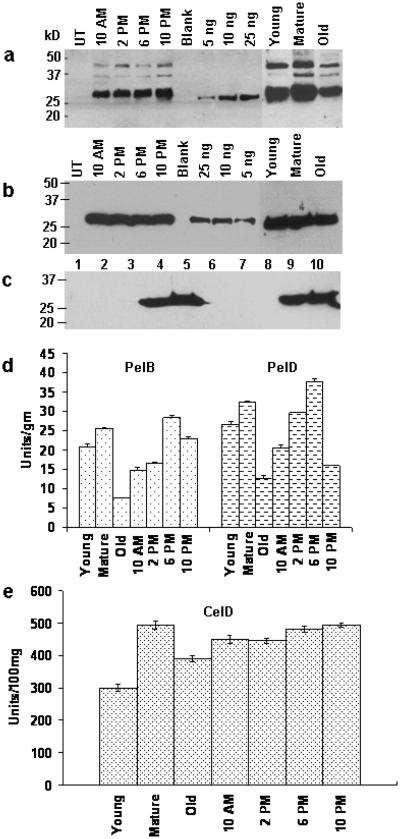
Immunoblots with antibodies raised against PelA and inhibition of pectate lyase activity in the presence of PelA antibody showed that PelB and PelD are immunologically related to PelA (Guo et al., 1995, 1996). Therefore, PelA antibody was used to detect the expression of PelB and PelD, although their affinity was variable in transplastomic lines. All transplastomic lines showed similar expression levels of PelB or PelD at different times of harvest, even though both transgenes were regulated by light (Figure 2a,b). This may be because of variable affinity between antigen epitopes of PelB, PelD and PelA antibody. Enzyme concentration slightly changed with leaf age and decreased in older leaves (Figure 2a,b). Western blots for expression of PelB and PelD from E. coli were performed using the His-tag antibody because PelA antibody cross reacted with too many proteins in E. coli cell extract, but not with any other protein in plant extract. Western blots show that PelB and PelD are expressed well in E. coli (Figure 2c). However, the His-tag antibody could not detect the chloroplast-derived PelB and PelD (Figure 2c). This could be due to the difference in folding of these proteins after formation of disulfide bonds in chloroplasts, making the His-tag inaccessible to the antibody. CelD western blots couldn't be done because of non-availability of this antibody.
Figure 2.
Western blot analysis and quantitation of transplastomic lines. Western blot of transplastomic lines expressing (a) PelB or (b) PelD. UT: untransformed, mature leaves harvested at 10 AM, 2 PM, 6 PM and 10 PM; 5ng, 10ng and 25ng: PelA purified protein, young, mature and old leaves. (c) Western blot analysis of PelB and PelD with His-tag antibody. Lane 1, protein marker, lane 2, untransformed plant extract; lane 3, PelD plant extract; lanes 4 & 5, PelD E. coli; lane 6, untransformed E. coli; lane 7, PelB plant extract; lane 8, blank; lanes 9 & 10, PelB E. coli. Enzyme units of PelB and PelD (d) or CelD (e) from one g or 100 mg leaf of different age or harvesting time.
Quantification of pectate lyases (PelB, PelD), and endoglucanase (CelD) at different harvesting time and leaf age
The activity of the enzyme varied significantly depending on the developmental stages and time of leaf harvest. Maximum enzyme activity was observed in mature leaves of PelB, PelD and CelD, with reduced activity in older leaves (Figure 2d,e). Mature leaves harvested at 6 PM showed maximum activity in both PelB and PelD whereas CelD showed maximum activity at 10 PM (Figure 2d,e). This may be due to increased stability of endoglucanase against proteases in plant extracts. Activity of cpCelD did not significantly decrease in plant crude extracts stored at room temperature, for more than thirty days (data not shown).
CelD enzyme activity was calculated using DNS reagent (Miller, 1959) according to the IUPAC protocol (Ghose, 1987). The specific activity of cpCelD using 2% CMC substrate was 493 units/mg total soluble protein (TSP) or 100 mg leaf tissue, in crude extracts prepared from mature leaves harvested at 10 PM. Using the glucose hexokinase assay, which is highly specific for glucose, the specific activity was 4.5 units/mg TSP and 6.28 units/mg TSP, when 5% avicel and sigmacell solution respectively was used as substrate (at pH 6.0, 60 °C).
CelD is known to have activity on microcrystalline substrate like Avicel and BMCC (Carrard et al., 2000; Fukumura et al., 1997; Kataeva et al., 1997). Endoglucanases randomly break down the β (1→4) glycosidic bonds existing between glucose molecules in cellulose. Some endoglucanases, though may not be active on cellobiose, hydrolyze microcrystalline substrate and release not only various lengths of cello-oligosaccharides but also individual glucose molecules. For example, CelT of Clostridium thermocellum is also an endoglucanase without carbohydrate binding domain (like CelD), hydrolyzes Avicel and releases individual molecules of glucose (Kurokawa et al., 2002). We did not find any detectable endogenous beta glucosidase activity in untransformed plant extracts, under our experimental conditions. Therefore, based on appropriate negative controls, authors are confident that the glucose released is indeed from the activity of CelD in transplastomic crude extracts.
Figures 2d and 2e show that approximately 26 units, 32 units and 4,930 units of PelB, PelD and CelD were obtained per gram fresh weight of mature leaves harvested at 6 PM or 10 PM. Thus, 2,048, 2,679 and 447,938 units of PelB, PelD and CelD can be harvested from each tobacco plant (experimental cultivar, Petite Havana). With 8,000 tobacco plants grown in one acre of land, 16, 21 and 3,584 million units of PelB, PelD or CelD can be obtained per single cutting (Table I). Based on three cuttings of tobacco in one year, up to 49, 64 and 10,751 million units of PelB, PelD or CelD can be harvested each year. The commercial cultivar yields 40 metric tons biomass of fresh leaves as opposed to 2.2 tons in experimental cultivar Petit Havana. Therefore, the commercial cultivar is expected to give 18 fold higher yields than the experimental cultivar.
Table I.
Enzyme yield in transplastomic tobacco plants
| Enzyme | Leaf age |
No of leaves /plant |
Avg. Wt (g)/leaf |
Units/ g in fresh leaf |
Units | Whole plant yield |
Units(millions)/ acre/ cutting |
Units(millions) /acre/year |
|
|---|---|---|---|---|---|---|---|---|---|
| Per leaf | Per age group |
||||||||
| PelB | Young | 3.5 | 2.5 | 20.82 | 52.05 | 182.18 | 8.89 % | 16.39 | 49.17 |
| Mature | 8.2 | 8.0 | 25.56 | 204.48 | 1,676.74 | 81.84% | |||
| Old | 4.5 | 5.6 | 7.54 | 42.22 | 190.00 | 9.27% | |||
| PelD | Young | 3.5 | 2.5 | 26.65 | 66.63 | 233.19 | 8.70% | 21.43 | 64.3 |
| Mature | 8.2 | 8.0 | 32.44 | 259.52 | 2,128.06 | 79.43% | |||
| Old | 4.5 | 5.6 | 12.62 | 70.67 | 318.02 | 11.87% | |||
| CelD | Young | 3.5 | 2.5 | 3,000 | 7,500 | 26,250 | 5.86% | 3,583.50 | 10751 |
| Mature | 8.2 | 8.0 | 4,930 | 39,440 | 323,408 | 72.20% | |||
| Old | 4.5 | 5.6 | 3,900 | 21,840 | 98,280 | 21.94% | |||
One unit of PelB and PelD enzyme is defined as the amount of enzyme which forms 1 μmol of unsaturated soluble oligogalacturonates per min with a molar extinction coefficient of 4,600 μmol−1 cm−1 in a 2.5 ml reaction containing 2.5 mg/ml polygalacturonic acid.
One unit of CelD enzyme is defined as the amount of enzyme that released 1 μmole glucose equivalents per minute in a 1ml reaction containing 2%CMC.
Effect of pH & temperature on pectate lyases (PelB & PelD) and endoglucanase (CelD) enzyme activity
The goal of this study is to use crude extracts and not purified enzymes to achieve low cost production. Therefore, the kinetic data provided in this manuscript are only operational parameters and such parameters are needed for development of enzyme cocktails (range of pH, functional temperature, etc.) and in the context of the applications of the results for production of soluble sugars from agricultural wastes. Further characterization of the expressed enzymes is not particularly useful for this application. Several laboratories have reported characterization of enzyme activities using crude plant extracts (Bae et al., 2008; Dai et al., 2000; Gray et al., 2008; Jin et al., 2003; Leelavathi et al., 2003; Montalvo-Rodriguez et al., 2000; Oraby et al., 2007; Sun et al., 2007; Yu et al., 2007; Zeigler et al., 2000; Zeigelhoffer et al., 1999; Ziegelhoffer et al., 2009).
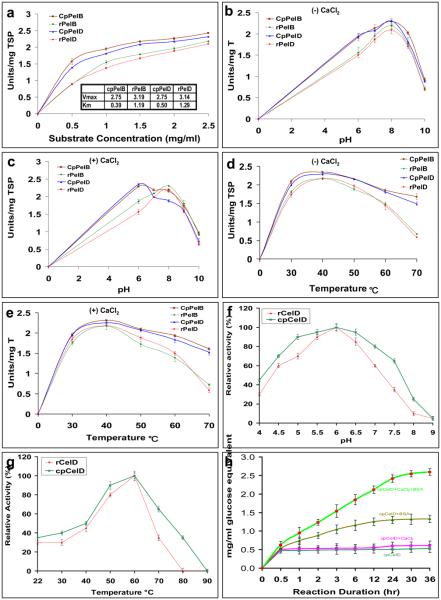
Both plant and E. coli extracts showed optimal pectate lyase activity at 2.5 mg/ml PGA (Figure 3a). Therefore, all enzyme characterization studies were performed at this substrate concentration. Kinetic studies carried out by using 4 μg of TSP, with increasing concentration of PGA (0 to 2.5 mg), under standard assay conditions gave Km values of 0.39 and 1.19 μg/ml in chloroplast (cp) and E. coli (r) PelB respectively, whereas values for chloroplast and E. coli PelD were 0.50 and 1.29 μg/ml respectively. The Vmax values obtained were 2.75, 3.19, 2.75 and 3.14 units/mg for cpPelB, rPelB, cpPelD and rPelD, respectively (Figure 3a).
Figure 3.
Effect of substrate, pH, temperature and cofactors on cpPelB, rPelB, cpPelD, rPelD, rCelD and cpCelD enzyme activity. (a) Effect of increasing PGA concentration on pectate lyases activity. (b) Effect of pH on pectate lyases activity in the absence of CaCl2 and (c) in the presence of CaCl2 (d) Effect of temperature (30 to 70°C) on enzyme activity at pH 8.0 in the absence of CaCl2 and (e) in the presence of CaCl2. (f) Optimization of pH and (g) effect of increasing temperature for cpCelD and rCelD enzyme activity (h) Enhancement of cpCelD (25 μg TSP/ml reaction) activity using 10 mM CaCl2 and 20 μg/ml BSA individually or in combination with 50 mM sodium acetate during the prolonged enzymatic hydrolysis. The hydrolysis was carried out up to 36 hours at 60°C, pH 6.0 in the presence of CMC (2%). Untransformed E. coli and leaf crude extracts did not yield any detectable level of unsaturated galacturonic acid or reducing sugar under these assay conditions.
The crude extract (4 to 5 μg TSP) from plant or E. coli was used to study the effect of pH and temperature on the activity of enzymes. The optimal pH for the E. coli derived pectate lyase in the presence and absence of 1mM CaCl2 under the standard assay conditions was 8.0 whereas plant derived pectate lyases showed a pH optimum of 6.0 in the presence of CaCl2 and 8.0 in the absence of Ca2+ ions (Figure 3b,c). The pH stability curve showed that the pectate lyase activity of chloroplast and E. coli derived enzyme in the absence of CaCl2 remained over 79% and 71% respectively in the buffers ranging from pH 6.0 to pH 9 (Figure 3b). However, increasing the pH to 10 resulted in decline of activity of chloroplast and E. coli derived enzyme to about 38% and 33% respectively. In the presence of CaCl2, pectate lyase B and D (both chloroplast and E. coli derived) retained over 76% and 67% activity respectively in the buffers ranging from pH 6.0 to pH 9 and retained 29-42% activity at pH 10 (Figure 3c). The pH stability curve for endoglucanase (CelD) showed that the endoglucanase activity of chloroplast and E. coli derived enzyme remained over 65% and 35% respectively in the buffers ranging from pH 4.5 to pH 7.5 (Figure 3f). However, at pH 9 complete loss in enzyme activity was observed irrespective of source of enzyme (Figure 3f).
The optimal temperature for the E. coli and chloroplast derived pectate lyase in the presence or absence of 1mM CaCl2 under the standard assay conditions was 40°C (Figure 3d,e). The chloroplast derived pectate lyases retained 65-76% activity at 70°C temperature whereas E. coli derived pectate lyase retained only 25-34% activity (Figure 3d, e). The temperature stability curve for endoglucanase (CelD) showed that the enzyme activity increased with increasing temperature up to 70°C in both E. coli and chloroplast-derived endoglucanase. Futher increase in temperature resulted in rapid decline of enzyme activity (Figure 3g). These data show that the chloroplast derived pectate lyases and endoglucanases are reasonably stable up to 70°C. Untransformed E. coli and leaf crude extracts did not yield any detectable amount of unsaturated galacturonic acid/glucose equivalents under standard assay conditions (data not shown).
Clostridium thermocellum CelD is structurally known to have affinity for CaCl2 ions and it also provides thermostability (Chauvaux et al., 1990). Even though 10 mM CaCl2 increased CelD activity in 2% CMC to 2 fold in E. coli crude extract, this was not apparent in chloroplast CelD crude extract during initial period of incubation. This may be due to optimum concentration of calcium ion present in plant cells. However, CaCl2 with 20 μg BSA yielded 5 fold increased activity at the end of 36 hour incubation for cpCelD crude extract (Figure 3h). Even though Ca2+ ions are required for pectate lyase activity it is unclear whether it binds to the enzyme (Crawford and Kolattukudy, 1987). However, Yoder et al (1993) found a putative binding site for Ca2+ on the outside of parallel β sheet of PelC pectate lyase of Erwinia. Ca2+ ions also play role in the cross linking of pectins during plant cell wall development and organization of plant cell wall polysaccharides (Carpita and Gibeaut, 1993; Wellner et al., 1998). Ca2+ ions also influenced the enzyme stability of PelB and PeD at higher temperature (40-55°C) compared to enzymes expressed in E.coli (Fig. 3d, e) or purified enzymes which had optimum temperature of 30 °C except for PelC which had 55°C as optimum temperature (Guo et al., 1995). These differences in enzyme properties from two different hosts may be due to their folding. This possibility was supported by the observation that it was possible to detect the E. coli enzyme with HIS-tag antibody but not the chloroplast enzyme (Figure 2c). It is well known that foreign proteins form disulfide bonds in chloroplasts (Arlen et al., 2007; Bally et al., 2008; Ruhlman et al., 2007) but not in E. coli when expressed in the cytoplasm. Both PelB and PelD enzymes have even number (12 or 14) cysteines that could form disulfide bonds (Guo et al., 1995).
E. coli vs. Chloroplast CelD, PelB & PelD
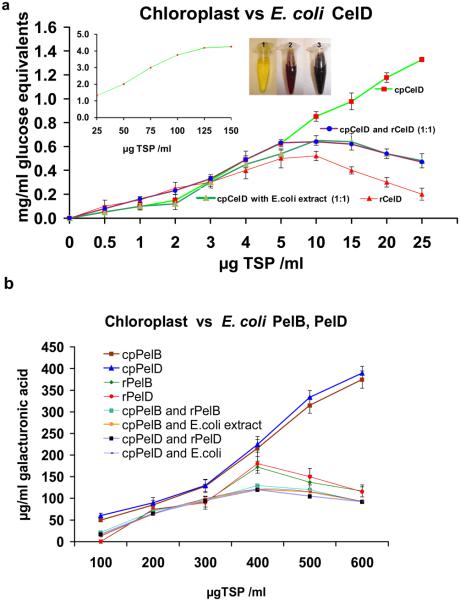
E. coli crude extract containing CelD enzyme showed decrease in enzyme activity when the reaction mixture contained more than 10 μg TSP, where as plant crude extract containing CelD released more reducing sugar with increasing protein concentration (Figure 4a). Chloroplast expressed CelD activity was saturated (in 2% CMC) at 150 μg TSP (Figure 4a inset) and there was no decrease in chloroplast CelD enzyme activity even up to 500 μg TSP as determined by end point assay. This finding is potentially of high practical significance because use of crude extracts eliminates the need for expensive purification steps. Large amounts of crude plant enzymes can be utilized in the cocktail as shown below without causing detrimental effect on enzyme activities, hydrolysis or yield of end products.
Figure 4.
E. coli vs. chloroplast derived enzymes at different protein concentrations of crude extracts. (a) Enzyme kinetics of cpCelD and rCelD using carboxymethyl cellulose (2%) substrate. The reaction mixture contained increasing concentration of cpCelD and rCelD TSP (μg/ml) with 10 mM CaCl2 and 50 mM sodium acetate buffer, pH 6.0. Enzyme hydrolysis was carried out for 30 minutes at 60°C. Figure inset shows enzyme kinetics saturation point for cpCelD TSP amount (μg/ml) towards CMC (2%). Eppendorf tubes with reaction mixture shown in inset represents, 1 untransformed plant, 2 and 3 rCelD and cpCelD 10μg TSP. (b) Effect of cpPelB, cpPelD, rPelB, and rPelD on hydrolysis of 5.0 mg/ml sodium polygalacturonate substrate. The reaction mixture contained increasing concentration of cpPelB, cpPelD, rPelB, and rPelD (μg/ml) in 20 mM Tris-HCl buffer (pH 8.0). Enzyme hydrolysis was carried out for 2 hour at 40°C on rotary shaker at 150 rpm.
Crude plant extracts containing cpCelD, cpPelB or cpPelD can be directly used for biomass degradation without any need for purification whereas E. coli extracts probably contain endoglucanase inhibitors. At higher protein concentrations, E. coli expressing rCelD, rPelB or rPelD showed reduced pectate lyase or endoglucanase activity and therefore prohibits high protein loading for higher hydrolysis whereas cpCelD, cpPelB or cpPelD continued to increase activity even up to 600 μg TSP. There may be inhibitors of enzyme activities in E. coli extracts, which are not present in plant crude extracts. Addition of E. coli crude cell extract from untransformed control or cells expressing CelD or pectate lyase inhibited CelD or pectate lyase activities in plant extracts. (Figure 4a, b). Crude extracts were used for assays without any dilution or concentration in both E. coli and tobacco. Therefore, molar ratio of protein and inhibitor was not changed under our experimental conditions. Inhibition observed in E. coli at higher protein concentrations is not due to expressed enzymes or products formed. Furthermore, product inhibition is not a possible explanation because higher products are formed in tobacco extracts than in E. coli.
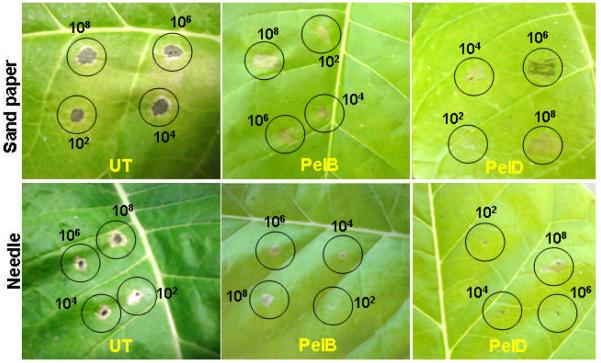
Resistance of transplastomic pectate lyase to Erwinia carotovora
The ability of pectate lyase to trigger plant defense responses was studied by investigating enhanced resistance to Erwinia soft rot either by using syringe or sand paper method. One day after inoculation with Erwinia, the first signs of damage were observed on leaves of untransformed plants in the region of the inoculated surface. On the 3rd day, virtually all inoculated untransformed leaf surfaces underwent necrosis whereas in leaves of transgenic plants, no damage zone was observed. Inoculation of potted plants with E. carotovora using a sandpaper technique and needle/syringe method resulted in areas of necrosis surrounding the point of inoculation in untransformed control for all cell densities, whereas transplastomic PelB and PelD mature leaves showed no areas of necrosis (Figure 5). Even inoculation of 108 cells resulted in no necrosis in mature transplastomic leaves. However, untransformed plants inoculated with 102 cells displayed necrosis. Similar results were obtained with bacteria inoculated using a syringe (Figure 5). Transplastomic mature leaves injected with E. carotovora showed a mild discoloration at the site of inoculation of 108 cells. These results support the hypothesis that expression of pectate lyase induces plant defense responses.
Figure 5.
In planta bioassays. Five- to 7-mm areas of untransformed, PelB and PelD transplastomic tobacco cv Petit Havana leaves were scraped with fine-grain sandpaper. Twenty microliters of 108, 106, 104 and 102_cells from an overnight culture of E. carotovora were inoculated to each prepared area. Photos were taken 5 d after inoculation.
Evaluation of enzyme activity for use in enzyme cocktail
Transplastomic lines were also generated expressing other biomass degrading enzymes for use in enzyme cocktails for hydrolysis of different lignocellulosic biomass. In order to demonstrate that individual enzymes used in the cocktails are indeed active, crude extract prepared from both E. coli and transplastomic plants were independently evaluated for enzyme activity using either model substrate (based on published data) or natural substrates (Table II). As reported above for pectate lyases or endoglucanases, enzyme activities should be measured at different developmental stages or leaves harvested at different times of the day and quantified using appropriate standards. As transplastomic plants expressing other enzymes were in different stages of development, we quantified enzyme activity with the available material. Chloroplast-derived pectate lyases and endoglucanase (CelD) had 1.1 to 1.41 fold higher activity when compared to E. coli crude extract. However, another endoglucanase (EgI) had 12.1 fold higher activity in plant extracts than E. coli. Maximum difference (24.55) in enzyme activity between E. coli and chloroplast-derived crude extract was observed in exoglucanase (CelO). Chloroplast-derived beta-glucosidase and xylanase also had several fold higher activity than E. coli derived crude extract. Enzymes expressed in transplastomic tobacco chloroplasts performed better than those expressed in E.coli. This could be due to several reasons including absence of disulfide bond and improper folding, formation of inclusion bodies (e.g., EG III, Okada et al., 1998; Sandgren et al., 2001) or certain unknown inhibitory substances present in the E. coli crude extract, as repeatedly observed in this study.
Table II.
Enzyme activity of cell wall degrading enzymes expressed in E. coli and chloroplast
| Enzyme | pH | °C | Substrate | Enzyme activity (units/mg) in crude total soluble protein |
Activity of transplastomic over E.coli (fold) |
|
|---|---|---|---|---|---|---|
| E.coli | Transplastomic | |||||
| CelD | 5.2 | 60 °C | CMC (2%) | 349 ±36 | 493 (±21) | 1.41 |
| EG1 | 5.2 | 50 °C | CMC (2%) | 28 ±7 | 339 (±12) | 12.10 |
| CelO | 5.2 | 60 °C | β-D-glucan (1%) | 18 ±2 | 442 (±19) | 24.55 |
| Bgl1 | 5.2 | 50 °C | p-nitrophenyl-β-D-glucopyranoside (4mM) | 2 ±0.02 | 14 (±2) | 7.0 |
| Xyn2 | 5.2 | 50 °C | Oat spelt xylan (1%) | 89 ±3 | 421 ±9 | 4.73 |
| PelB | 6 & 8 | 40 °C | Polygactouronic acid (0.25%) | 2.17 ±0.2 | 2.42 ±0.1 | 1.12 |
| PelD | 6 & 8 | 40 °C | Polygactouronic acid (0.25%) | 2.09 ±0.3 | 2.31 ±0.4 | 1.10 |
| PelA | 6 & 8 | 40 °C | Polygactouronic acid (0.25%) | 2.50 ±0.5 | 2.81 ±0.9 | 1.12 |
| Cutinase | 8.0 | 30 °C | p-nitrophenyl butyrate (0.03%) | 24 ±4 | 15 ±4 | <0.625 |
| SwoI | Swelling of cotton fiber was observed with E. coli and chloroplast-derived crude extract as described earlier (Saloheimo et al., 2002). |
|||||
| Axe1 | Color change with E. coli enzyme extract was observed using 1 mM α-naphthyl acetate as described earlier (Poutanen and Sundberg, 1988) |
|||||
Enzyme assays were done in triplicates and standard errors were calculated. Untransformed tobacco leaves and E.coli did not show detectable level of hydrolysis of any of the above substrates under conditions described in the materials and method.:
The primary goal of our investigations was to use low cost crude extracts from E. coli or transplastomic plants, we did not attempt any purification of enzymes. Pectate lyases are known to act synergistically in a variety of environment during degradation of its specific pectic compounds. Some of the reported pectate lyases also have activity on methylated pectin (Bartling et al., 1995; Guo et al., 1996). Therefore we used all three pectate lyases in the enzyme cocktails. Enzyme cocktails were prepared using crude soluble protein extracts from E. coli cultures or from transplastomic tobacco leaves, expressing the cell wall degrading enzymes.
Enzyme cocktail for hydrolysis of Filter paper
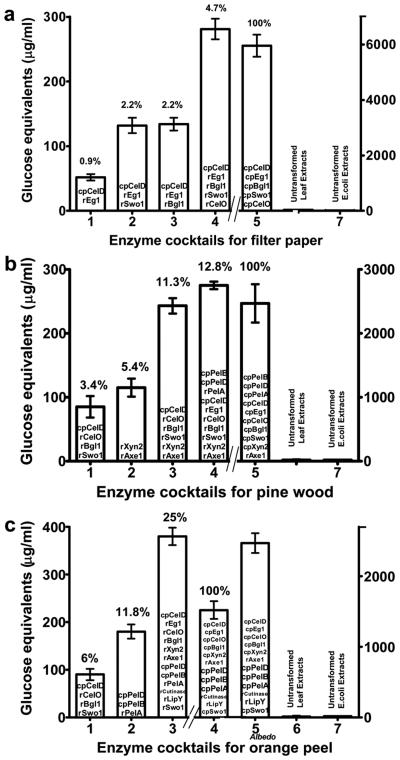
Before evaluation of enzyme cocktails, activity of each enzyme was tested independently with an appropriate substrate and enzyme units were calculated (Table II). Chloroplast or E. coli expressed endoglucanase (cpCelD or rEg1) alone did not release any detectable glucose from filter paper but when mixed together up to 0.9% of total hydrolysis was observed (Figure 6a, bar 1). The synergistic activity was further enhanced up to 2.2% when the endoglucanases (cpCelD and rEg1) were mixed with swollenin or beta-glucosidase (Figure 6a, bars 2,3). The increase in hydrolysis with swollenin (expansin like) could be due to the loosening or disruption of the packaging of the plant cell wall and polysaccharides. Similar increase in the yield of sugar was observed with Bacillus subtilis expansin when filter paper was incubated with a mixture of expansin and low dosage of cellulase (Kim et al., 2009). Addition of exoglucanase to this cocktail doubled the hydrolysis of filter paper (Figure 6a, bar 4). Observed synergism is probably due to the exo-mode of action of cellobiohydrolase (Zverlov et al., 2002) from reducing ends that were formed by random cuts in cellulose chains through endoglucanases, along with the action of swollenin and beta-glucosidase. It could also be due to the increased activity of exoglucanase in the presence of beta glucosidase, which reduces cellobiose, an exoglucanase inhibitor (Gupta and Lee, 2009). When we used same cocktail but comprising of chloroplast expressed enzymes, there was 21.3 fold increase in hydrolysis releasing maximum amount of reducing sugars (Figure 6a, bar 5). Addition of leaf extract from untransformed plants and E. coli to filter paper didn't yield any detectable glucose (Figure 6a, bars 6,7). Although there are reports on synergistic effect of cellulase on substrates like avicel, cotton fibers and filter paper (Gusakov et al., 2007; Irwin et al., 1993; Zhou and Ingram, 2000), they used purified recombinant enzymes expressed in bacteria or fungi but not crude extracts used in our study.
Figure 6.
Enzyme cocktails for filter paper, pine wood and citrus peel. (a) Enzyme cocktail activity on Whatman No1 filter paper (50 mg/ml). (b) Hydrolysis of pine wood sample (200 mg/5 ml reaction). (c) Hydrolysis of Valencia orange peel and albedo portion (200 mg/5 ml reaction).
Enzyme cocktail for hydrolysis of pine wood
The enzyme cocktail mentioned in figure 6a bar 4 for filter paper (except rEg1) resulted in 3.4% of total hydrolysis after 36 hours (Figure 6b, bar 1) when tested on pine wood. An enzyme cocktail of endoxylanase and acetyl xylan esterase showed 5.4% of total hydrolysis (Figure 6b, bar 2). Similar interaction between xylanase and acetyl xylan esterase was noticed confirming the strong synergistic relationship between these two enzymes (Kosugi et al., 2002; Selig et al., 2008). When the two cocktails (bar 1 and 2) were combined together, the hydrolysis increased up to 11.3% (Figure 6b, bar 3). Xylose removal probably enhanced cellulose accessibility and thus result in greater release of glucose. Supplementation of cellulase with xylanase enhanced glucose release from poplar pretreated solids and corn cell wall (Kumar and Wyman, 2009; Murashima et al., 2003). Pectin is the major structural component of plant cell wall of woody plants including pine trees (Hafren et al., 2000) along with cellulose and hemicellulose. Pectin is located mainly in the middle lamella and primary cell wall and functions as a matrix anchoring the cellulose and hemicellulose fibers (Carpita and Gibeaut, 1993). Therefore hydrolysis of pectin should result in loosening of cellulose and hemicellulosic fibers, resulting in enhanced glucose release by cellulases and hemicellulases. When pine wood substrate was first treated with pectate lyases, followed by the addition of the enzyme cocktail in bar 3, the overall hydrolysis was further enhanced up to 12.8% after 36 hour incubation (Figure 6b, bar 4). When we used same cocktail but comprising of chloroplast expressed enzymes (except rAxe1), there was 7.8 fold increase in hydrolysis releasing maximum amount of reducing sugars (Figure 6b bar 5). Addition of leaf extract from untransformed plants and E. coli to pine wood didn't yield any detectable glucose (Figure 6b, bars 6,7).
Enzyme cocktail for hydrolysis of citrus waste
The enzyme cocktail of endoglucanase (cpCelD), exoglucanase, swollenin and beta-glucosidase released up to 6% of total hydrolysis with citrus peel (Figure 6c, bar 1). When citrus peel was treated with pectate lyases (cpPelB, cpPelD and rPelA), hydrolysis was doubled (Figure 6c, bar 2) because of high pectin content (23%) in citrus peel (Yapo et al., 2007). Addition of endoxylanase, acetyl xylan esterase, cutinase and lipase to the both these cocktails released 2.1 fold more glucose equivalents (Figure 6c, bar 3). Enzymes like cutinase and lipase may have hydrolyzed oil bodies present in the citrus peel, providing greater access to endoglucanase, endoxylanase and pectate lyases for efficient hydrolysis of citrus peel. Using the same cocktail from chloroplast expressed enzymes (except rCutinase, rLipY and rAxe1) resulted in 1,520 μg/ml and 2,470 μg/ml (Figure 6c, bars 4,5) glucose equivalents from 200 mg of ground citrus peel and albedo portion of citrus peel respectively, after 36 hour incubation period. Addition of leaf extract from untransformed plants and E. coli to citrus waste didn't yield any detectable glucose (Figure 6c, bars 6,7). High amount of glucose released in albedo portion could be due to high content of cellulose and lack of oil bodies. Addition of other enzyme classes including the accessory enzymes produced in chloroplasts should further enhance yield of glucose in a cost effective manner.
Conclusions
Concerns over finite petroleum reserve require development of alternative energy resources. Lower emission of green house gases from alternative energy resources is also highly desirable. The fact that corn ethanol produces more green house gas emissions than gasoline and that cellulosic ethanol from non food crops produces less green house gas emissions than electricity or hydrogen, highly favors production of ethanol from cellulosic biomass. However, biofuel production from lignocellulosic materials is a challenging problem because of the multifaceted nature of raw materials and lack of technology to efficiently and economically release fermentable sugars from the complex multi-polymeric raw materials. The high costs of enzyme production and the tremendous amount of enzymes needed to hydrolyse pretreated biomass are often considered as key obstacles in the commercial lignocellulosic ethanol industry (Margeot et al., 2009). Therefore, in this study, we have used coding sequences from bacterial or fungal genomes to create chloroplast vectors. A PCR based method was used to clone ORFs without introns from fungal genomic DNA. E. coli expression system was used to evaluate functionality of each enzyme independently or their efficacy in enzyme cocktails before creating transgenic lines. The phenotype of homoplasmic transplastomic lines was normal and produced flowers & seeds. Based on three cuttings of tobacco in one year, 49, 64 and 10,751 million units of pectate lyase and endoglucanase activity can be obtained each year in an experimental cultivar. Commercial cultivars yield 40 metric tons biomass of fresh leaves as opposed to 2.2 tons in experimental cultivar Petit Havana. Therefore, the commercial cultivar is expected to give 18 fold higher yields than the experimental cultivar. Because most enzymes for hydrolysis of plant biomass are active at higher temperatures, it is feasible to harvest leaves and sun dry them, as reported previously for chloroplast derived xylanase (Leelavathi et al., 2003). Moreover, in our study, activity of cpCelD did not decrease significantly in plant crude extracts stored at room temperature for more than thirty days. This is the first study which directly compared properties of enzymes produced in E.coli or in planta using identical genes and regulatory sequences. Chloroplast-derived enzymes showed higher temperature stability, broad pH optima and higher enzyme activity with increasing protein concentration than enzymes expressed in E. coli indicating their direct use in biomass hydrolysis with higher protein loadings.
Expression of cell wall hydrolyzing enzymes in plant cells could confer useful agronomic traits, especially enhanced defense against plant pathogens. For example, the pectate lyase transplastomic plants showed enhance resistance to bacterial pathogen E. carotovora. These results support the hypothesis that expression of pectate lyase induces plant defense responses. An early recognition of the pathogenic attack is important for successful plant defense. The pectate lyase released from chloroplast should have initiated early recognition through the formation of oligogalacturonate (OG) elicitors from cell wall pectin. It has been known that when pectate lyase enzymes were liberated from cells get in contact with pectin results in the release of OGs, which confers plant defense responses (Wegener and Olsen, 2004). OGs in turn induce expression of several genes involved in plant defense (Casasoli et al., 2008; Ferrari et al., 2007) and may be regarded as host associated molecular patterns involved in the innate immunity (Stern et al., 2006; Taylor and Gallo, 2006).
The hydrolytic effectiveness of a multienzyme complex in the process of lignocellulose saccharification depends both on efficiency of individual enzymes and their ratio in the multienzyme cocktail. So far no model system of enzyme cocktails for the hydrolysis of lignocellulosic biomass has been developed that has the flexibility to optimize different enzyme classes. To the best of our knowledge, this is the first study using enzyme cocktails expressed in plants for hydrolysis of lignocellulosic biomass to produce fermentable sugars with greater flexibility to manipulate the cocktail depending upon the composition of the biomass. For example, in this study when we used xylanase and acetyl xylan esterase or pectate lyases for the hydrolysis of filter paper, no detectable sugar was released (data not shown). On the other hand, when xylan and acetyl xylan esterase or pectate lyases were used for pine wood and citrus peel hydrolysis respectively, detectable amount of glucose were released as wood and citrus peel has significant amounts of xylan or pectin. Our study has developed a new platform for creation of chloroplast-derived enzyme cocktails for digestion of different kinds of lignocellulosic biomass.
Because lignocellulosic wastes are composed of a complex of multiple intertwined polymers, simultaneous presence of multiple hydrolases that can increase the access of each other will be required to get efficient release of monomers. Majority of enzymatic hydrolysis studies on natural substrates like pretreated wood, corn stover or wheat straw have used commercially available enzymes (Merino and Cherry, 2007; Rosgaard et al., 2007a) or purified recombinant enzymes spiked with purified commercial enzymes (Gusakov et al., 2007; Selig et al., 2008). Accurate comparison of crude extract enzyme cocktails with commercial cocktails is not possible because of their unknown enzyme compositions. We used equivalent enzyme units based on CMC hydrolysis as a basis for general comparison and to approximately calibrate enzyme dose. Chloroplast-derived enzyme cocktail yielded 3,625% and 261% more glucose equivalent units for filter paper and citrus peel, respectively than Novozyme 188 cocktail with equivalent enzyme units; no glucose was released by Novozyme 188 from pine wood. Chloroplast-derived enzyme cocktail yielded 396%, 684% and 69% more glucose equivalent units for filter paper, pine wood and citrus peel, respectively than Celluclast 1.5L cocktail with equivalent enzyme units. A major drawback of submerged fermentation technology used in commercial cocktails is that the amount or composition of different enzymes can not be manipulated at will for hydrolysis of different biomass. According to Novozymes, a careful design of a combination of single component enzymes is necessary for rational utilization of these enzyme cocktails (Rosgaard et al., 2007b).
Commercial production of enzymes in fermentation systems is limited by both higher cost and lower production capacity. Both these concerns are addressed by chloroplast-derived enzyme cocktails. According to NC State University Burley Tobacco Guide 2009, the cost of production of Burley tobacco in 2008 was $3,506.35 per acre. Based on enzyme activity observed in plant crude extracts in this study, there is no need for purification. Therefore, enzymes could be produced as low as 0.003 cents for CelD, 0.007 cents for PelB and 0.005 cents for PelD per enzyme unit (as defined in the commercial source Megazyme). This is 3,100 fold and 1,057 to 1,480 fold less expensive than endoglucanase and for pectate lyase B & D respectively, when compared with current recombinant/purified commercial enzymes produced via fermentation cost (endoglucanase and pectate lyase from Megazyme). While this cost or yield comparison may not be the same for all chloroplast-derived enzymes, this concept provides a promising new platform for inexpensive enzyme cocktails to produce fermentable sugars from lignocellulosic biomass.
Experimental Procedures
Isolation of genes and construction of plastid transformation vectors
Genomic DNA of Clostridium thermocellum and Trichoderma reesei was obtained from ATCC and used as template for the amplification of different genes. Gene specific primers using a forward primer containing a NdeI site and a reverse primer containing a XbaI site for cloning in the pLD vector were designed for celD, celO and lipY genes. The mature region (without signal peptide) of cellulase genes celD (X04584) and celO (AJ275975) were amplified from genomic DNA of Clostridium thermocellum. LipY (NC_000962) was amplified from genomic DNA of Mycobacterium tuberculosis. Overlapping primers were designed for the amplification of various exons of egI (AB003694), swoI (AJ245918), axe1 (Z69256), xyn2 (X69574) and bgl1 (U09580) from genomic DNA of Trichoderma reesei. Full length cDNA of these genes was amplified from different exons by a PCR based method (An et al., 2007) using the forward of first exon and reverse of last exon containing a NdeI site and XbaI site respectively. Pectate lyase genes pelA, pelB & pelD from Fusarium solani with similar restriction sites including sequence for the His Tag were amplified using gene specific primers from pHILD2A, pHILD2B (Guo et al., 1995) and pHILD2D (Guo et al., 1996) respectively. A similar strategy was used to amplify cutinase gene (Soliday et al., 1984) from recombinant clone of Fusarium solani. All the full length amplified products were ligated to pCR Blunt II Topo vector (Invitrogen) and were subjected to DNA sequencing (Genewiz). Each gene cloned in Topo vector was digested with NdeI/XbaI and inserted into the pLD vector (Daniell et al., 1998, 2001) to make the tobacco chloroplast expression vector.
Regeneration of transplastomic plants and evaluation of transgene integration by PCR and Southern blot
Nicotiana tabacum var. Petite Havana was grown aseptically on hormone-free Murashige and Skoog (MS) agar medium containing 30 g/l sucrose. Sterile young leaves from plants at the 4 to 6 leaf stages were bombarded using gold particles coated with vectors pLD-PelB, pLD-PelD and pLD-CelD and transplastomic plants were regenerated as described previously (Daniell et al., 2005; Verma et al., 2008). Plant genomic DNA was isolated using Qiagen DNeasy plant mini kit from leaves. PCR analysis was performed to confirm transgene integration into the inverted repeat regions of the chloroplast genome using two sets of primers 3P/3M and 5P/2M, respectively (Daniell et al., 2001). The PCR reaction was performed as described previously (Daniell et al., 2001; Verma et al., 2008). Leaf from the PCR positive shoots were again cut into small pieces and transferred on RMOP (regeneration medium of plants) medium containing spectinomycin for another round of selection and subsequently moved to MSO (MS salts without vitamins and growth hormones) medium containing spectinomycin for another round of selection to generate homoplasmic lines. Southern blot analysis was performed to confirm homoplasmy according to lab protocol (Kumar and Daniell, 2004). In brief, total plant genomic DNA (1 to 2 μg) isolated from leaves was digested with SmaI and hybridized with 32P α[dCTP] labeled chloroplast flanking sequence probe (0.81 kb) containing the trnI-trnA genes. Hybridization was performed by using Stratagene QUICK-HYB hybridization solution and protocol.
Immunoblot analysis
Approximately 100 mg of leaf was ground in liquid nitrogen and used for immunoblot analysis as described previously (Kumar and Daniell, 2004). Protein concentration was determined by Bradford protein assay reagent kit (Bio-Rad). Equal amounts of total soluble protein were separated by SDS-PAGE and transferred to nitrocellulose membrane. The transgenic protein expression was detected using polyclonal serum raised against PelA in rabbit.
E. coli enzyme (crude) preparation
E. coli strain (XL-10 gold) harboring chloroplast expression vectors expressing rCelD, rEg1 (EC 3.2.1.4), rCelO (EC 3.2.1.91), rXyn2 (EC 3.2.1.8), rAxe1 (EC 3.1.1.72), rBgl1 (EC 3.2.1.21), rCutinase (EC 3.1.1.74), rLipY (lipase, EC 3.1.1.3), rPelA, rPelB, rPelD (EC 4.2.2.2) or rSwo1 was grown overnight at 37°C. Cells were harvested at 4°C and sonicated four times with 30s pulse in appropriate buffer (50 mM sodium acetate buffer with pH 5.5 for CelD, Eg1, CelO, Swo1, Xyn2, Axe1, Bgl1, 100 mM Tris-Cl with pH 7.0 for cutinase, lipase, PelA, PelB and PelD) containing protease inhibitor cocktail (Roche) and sodium azide (0.02%). Supernatant was collected after centrifugation at 16,000 × g for 10 minutes and protein concentration was determined.
Enzyme preparation from tobacco transplastomic leaf material
Fresh green leaves were collected and ground in liquid nitrogen. Total soluble protein was extracted in 50 mM sodium acetate buffer, pH 5.5 for cpCelD, cpXyn2 or 100 mM Tris-Cl buffer, pH 7.0 for PelD and PelB. Each enzyme used in this study (from E. coli and transplastomic plants) were tested for activity using suitable substrate(s). Different parameters and substrates used in these assays are given in Table II. Protein extraction from untransformed plants was also performed under similar conditions. All buffers contained protease inhibitor cocktail (Roche) and sodium azide (0.02%). Total soluble protein was filtered using 0.22 μm syringe filter. Protein concentration (mg/ml) in TSP was spectrophotometrically determined using Bradford method at 595 nm absorption, after subtracting the turbidity of extracts at 700 nm.
Enzyme assays for Pectate lyase B and Pectate lyase D
Pectate lyases B and D were assayed spectrophotometrically by measuring the increase in A235 (Crawford and Kolattukudy, 1987; Gonzalez-Candelas and Kolattukudy, 1992; Guo et al., 1995). Kinetics of the pectate lyase B and D were studied in a reaction mixtures contained 1ml of 50 mM Tris-HCl buffer (pH 8.0) with 1 mM CaCl2 (freshly prepared), 1ml of 0.0 to 2.5 mg/ml sodium polygalacturonate (Sigma) and 0.5 ml of suitably diluted enzyme solution. Measurements were carried out at 40°C. One unit of enzyme was defined as the amount of enzyme which forms 1 μmol of product per min with a molar extinction coefficient of 4,600 μmol−1 cm−1. Kinetic parameters (Km & Vmax) were calculated using non linear regression using Graphpad Prism 5.0. The initial slope of each substrate concentration was calculated, where as the velocity (units/mg/min) was defined through the release of unsaturated galacturonic acid. The temperature optimization for pectate lyase B and D activity was carried out in 50 mM Tris-HCl buffer, pH 8.0 with or without 1mM CaCl2 at different temperatures ranging from 30°C to 70°C. In each case, the substrate was pre-incubated at the desired temperature for 5 min. In order to study the thermal stability of the enzyme, buffered enzyme samples were incubated for fixed time period at different temperatures. The pH optimum of the pectate lyase B and D was measured at 40°C using different buffers like 50 mM phosphate buffer (pH 6 to 7), 50 mM Tris-HCl buffer (pH 8), 50 mM glycine/NaOH buffer (pH 9) or 50 mM CAPS buffer (pH 10.0) with 2.5 mg substrate and 4 μg of TSP of PelB and PelD from both plant and E. coli.
Enzyme assay for CelD and commercial cocktail (Celluclast 1.5L and Novozyme 188)
Cellulase enzyme activity of cpCelD was determined by incubating crude extract in 2% carboxylmethylcellulose, 5% avicel and 5% sigmacell (Sigma) as substrate according to IUPAC recommendations (Ghose, 1987) in 50 mM sodium acetate buffer pH 6.0, 10 mM CaCl2 and incubated at 60°C for 30 minutes for CMC and 2 hours for avicel and sigmacell. Similar conditions were used in determining the pH and temperature activity profile of cpCelD using 2% CMC. Relative activity (%) was measured with reference to maximum activity obtained with 25 μg/ml for cpCelD and 10 μg/ml for rCelD. Enzyme units of commercial cocktails Celluclast 1.5L and Novozyme 188 were determined using 2% CMC, under identical assay conditions. Reducing sugar amount was determined using 3,5-dinitrosalicylic acid (Miller, 1959). D-glucose and D-galacturonic acid were used as standard to measure release of glucose equivalents and unsaturated galacturonic acid molecules. One unit of enzyme was defined as the amount of enzyme that released 1 μmole glucose equivalents per minute/ml. Cellulase unit calculation for avicel and sigmacell was based on glucose hexokinase method according to the manufacturer's protocol (Sigma).
In planta assay for resistance to Erwinia soft rot
To verify the resistance of PelB and PelD, control and transplastomic leaves were inoculated with bacterial suspension culture. Erwinia carotovora strain was obtained from Dr. Jerry Bartz's laboratory (University of Florida, Gainesville) and grown for 24h at 25 °C in 5 ml of LB medium. Different dilutions of bacterial cells were prepared. Five to 7 mm areas of green house grown untransformed, PelB and PelD transplastomic tobacco leaves were scraped with fine-grain sandpaper and 20 μl of 108, 106 104 and 102 of Erwinia cells were inoculated to each prepared area. In a parallel study, 20 μl of the same dilutions of Erwinia cells were injected into leaves of untransformed, PelB and PelD transplastomic tobacco using a syringe with a precision glide needle. Photos were taken 5 d after inoculation.
Enzymatic hydrolysis of filter paper, pine wood and citrus peel
Enzyme assays were carried out either with one enzyme component or as cocktail on filter paper, pine wood and orange peel and released reducing sugar was determined using DNS method. Orange peel prepared from Valencia orange (Citrus sinensis cv Valencia) fruit and albedo portion was air dried overnight and ground in liquid nitrogen. Ground Valencia orange peel, albedo portion and pine wood biomass were washed several times in distilled water until no reducing sugar was detected by DNS reagent as well as by glucose hexokinase method.
For enzymatic digestion, 50 to 200 mg of filter paper, pine wood sample or ground orange peel was used. Enzyme catalytic function of chloroplast expressed enzymes like Eg1, CelO, Bgl1, Swo1, Xyn2, PelA, and cutinase were tested using known amount of crude extract TSP with appropriate substrates before making suitable enzyme cocktail. Filter paper activity was determined using Whatman No. 1 filter paper strip at pH 5.5 and 50°C. Different combinations of crude extracts contained TSP in the range of 25 to 200 μg/ml. rEg1 (100 μg/ml), rBgl1 (200 μg/ml), rSwo1 (120 μg/ml), rCelO (100 μg/ml), cpCelD (100 μg/ml), cpEg1 (100 μg/ml), cpBgl1 (200 μg/ml), cpSwo1 (25 μg/ml) and cpCelO (100 μg/ml) were used. Untransformed leaf and E. coli crude extracts contained 485 μg TSP. The samples were incubated in 50 mM sodium acetate buffer plus 10 mM CaCl2, 20 μg BSA for 36 hours. Hydrolysis of pine wood sample was carried (pH 5.5 to 8.0, 40°C to 50°C, 36 hours incubation) using a cocktail of crude extracts contained TSP in the range of 50 to 250 μg/5ml. cpPelB (250 μg), cpPelD (250 μg) (at pH 8.0,), cpCelD (200 μg), cpXyn2 (200 μg), rEg1 (100 μg), rBgl1(200 μg), rSwo1 (120 μg), rCelO (100μg), rAxe1 (100 μg), rPelA (200 μg), rCutinase (50 μg), rLipY (100 μg) were used in the cocktail. Untransformed leaf and E. coli crude extracts contained 1550 μg TSP/5ml. Hydrolysis of Valencia orange peel (200 mg/5 ml reaction, pH5.5 to 8.0, 40°C to 50°C, 36 hour incubation) was done using a cocktail of crude extracts contained TSP in the range of 50 to 250 μg/5ml. cpPelB (250 μg) , cpPelD (250 μg) cpCelD (100 μg) and cpXyn2 (100 μg), rEg1 (100 μg), rBgl1(200 μg), rSwo1(120 μg), rCelO (100μg) and cpCelD (100μg), rAxe2 (100 μg), rCutinase (50 μg), rLipY (100 μg), rPelA (200 μg) were used in the cocktail. Untransformed leaf and E. coli crude extracts contained 1670 μg TSP/5ml. All experiments were carried out under specified conditions in a rotary shaker at 150 rpm. Crude extracts containing enzymes from E. coli and plants were used in the cocktail for hydrolysis. End product reducing sugar was determined using DNS reagent (Miller, 1959) with D-glucose and D-galacturonic acid as standard. Ampicillin and kanamycin 100 μg/ml was added to prevent any microbial growth during the long durations of enzyme hydrolysis. Commercial enzyme cocktails Celluclast 1.5L and Novozyme 188 were tested for hydrolysis of citrus peel and pine wood in the same assay conditions used for enzyme cocktails from crude extracts. Enzyme units of Celluclast 1.5L and Novozyme 188 used for hydrolysis assays were equivalent to cpCelD enzyme units (based on CMC hydrolysis) present in cocktails of crude extracts. In all experiments control assays contained substrate without enzyme or enzyme without substrate. All experiments and assays were carried out in triplicate.
Acknowledgement
The investigations reported in this article were supported in part by grants from USDA 3611-21000-021-02S and NIH R01 GM 63879 to Henry Daniell. Authors are thankful to Dr. Velpula for optimization of pectate lyase assays and Dr. Jerry Bartz (University of Florida, Gainesville) for providing Erwinia carotovora culture.
References
- An X, Lu J, Huang J, Zhang B, Liu D, Zhang X, Chen J, Zhou Y, Tong Y. Rapid assembly of multiple-exon cDNA directly from genomic DNA. PLoS One. 2007;2(11):e1179. doi: 10.1371/journal.pone.0001179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlen PA, Falconer R, Cherukumilli S, Cole A, Cole AM, Oishi KK, Daniell H. Field production and functional evaluation of chloroplast-derived interferon-alpha2b. Plant Biotechnol. J. 2007;5:511–525. doi: 10.1111/j.1467-7652.2007.00258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arlen PA, Singleton M, Adamovicz JJ, Ding Y, Davoodi-Semiromi A, Daniell H. Effective plague vaccination via oral delivery of plant cells expressing F1-V antigens in chloroplasts. Infect. Immun. 2008;76:3640–3650. doi: 10.1128/IAI.00050-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae H-J, Kim HJ, Kim YS. Production of recombinant xylanase in plants and its potential for pulp biobleaching applications. Bioresour. Technol. 2008;99:3513–3519. doi: 10.1016/j.biortech.2007.07.064. [DOI] [PubMed] [Google Scholar]
- Baird SD, Johnson DA, Seligy VL. Molecular cloning, expression, and characterization of endo-3-1,4-glucanase genes from Bacillus polymyxa and Bacillus circulans. J. Bacteriol. 1990;172:1576–1586. doi: 10.1128/jb.172.3.1576-1586.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bally J, Nadai M, Vitel M, Rolland A, Dumain R, Dubald M. Plant physiological adaptations to the massive foreign protein synthesis occurring in recombinant chloroplasts. Plant Physiol. 2009 doi: 10.1104/pp.109.139816. DOI:10.1104/pp.109.139816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bally J, Paget E, Droux M, Job C, Job D, Dubald M. Both the stroma and thylakoid lumen of tobacco chloroplasts are competent for the formation of disulphide bonds in recombinant proteins. Plant Biotechnol. J. 2008;6:46–61. doi: 10.1111/j.1467-7652.2007.00298.x. [DOI] [PubMed] [Google Scholar]
- Bartling S, Wegener C, Olsen O. Synergism between Erwinia pectate lyase isoenzymes that depolymerize both pectate and pectin. Microbiology. 1995;141:873–881. doi: 10.1099/13500872-141-4-873. [DOI] [PubMed] [Google Scholar]
- Bhatti MA, Kraft JM. Influence of soil moisture on root rot and wilt of chickpea. Plant Disease. 1992;76:1259–1262. [Google Scholar]
- Biswas GCG, Ransom C, Sticklen M. Expression of biologically active Acidothermus cellulolyticus endoglucanase in transgenic maize plants. Plant Sci. 2006;171:617–623. [Google Scholar]
- Brixey PJ, Guda C, Daniell H. The chloroplast psbA promoter is more efficient in Escherichia coli than the T7 promoter for hyperexpression of a foreign protein. Biotechnol. Lett. 1997;19:395–400. [Google Scholar]
- Carrard G, Koivula A, Söderlund H, Béguin P. Cellulose-binding domains promote hydrolysis of different sites on crystalline cellulose. Proc. Natl. Acad. Sci. USA. 2000;97:10342–10347. doi: 10.1073/pnas.160216697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll A, Somerville C. Cellulosic biofuels. Annu. Rev. Plant Biol. 2009;60:165–182. doi: 10.1146/annurev.arplant.043008.092125. [DOI] [PubMed] [Google Scholar]
- Carpita NC, Gibeaut DM. Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J. 1993;3:1–30. doi: 10.1111/j.1365-313x.1993.tb00007.x. [DOI] [PubMed] [Google Scholar]
- Casasoli M, Spadoni S, Lilley KS, Cervone F, De Lorenzo G, Mattei B. Identification by 2D-DIGE of apoplastic proteins regulated by oligogalacturonides in Arabidopsis thaliana. Proteomics. 2008;8:1042–1054. doi: 10.1002/pmic.200700523. [DOI] [PubMed] [Google Scholar]
- Charles D. Corn-based ethanol flunks key test. Science. 2009;324:1055–1057. doi: 10.1126/science.324_587. [DOI] [PubMed] [Google Scholar]
- Chauvaux S, Beguin P, Aubert JP, Bhat KM, Gow LA, Wood TM, Bairoch A. Calcium-binding affinity and calcium-enhanced activity of Clostridium thermocellum endoglucanase D. Biochem. J. 1990;265:261–265. doi: 10.1042/bj2650261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collmer A, Keen NT. The role of pectic enzymes in plant pathogenesis. Annu. Rev. Phytopathol. 1986;24:383–409. [Google Scholar]
- Crawford MS, Kolattukudy PE. Pectate lyase from Fusarium solani f. sp. pisi: Purification, characterization, in vitro translation of the mRNA, and involvement in pathogenicity. Arch. Biochem. Biophys. 1987;258:196–205. doi: 10.1016/0003-9861(87)90336-5. [DOI] [PubMed] [Google Scholar]
- Dai Z, Hooker BS, Anderson DB, Thomas SR. Improved plant-based production of E1 endoglucanase using potato: expression optimization and tissue targeting. Mol. Breed. 2000;6:277–285. [Google Scholar]
- Daniell H. Transgene containment by maternal inheritance: Effective or elusive? Proc. Natl. Acad. Sci. USA. 2007;104:6879–6880. doi: 10.1073/pnas.0702219104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H, Datta R, Varma S, Gray S, Lee SB. Containment of herbicide resistance through genetic engineering of the chloroplast genome. Nat. Biotechnol. 1998;16:345–348. doi: 10.1038/nbt0498-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H, Lee SB, Panchal T, Wiebe PO. Expression of the native cholera toxin B subunit gene and assembly as functional oligomers in transgenic tobacco chloroplasts. J. Mol. Biol. 2001;311:1001–1009. doi: 10.1006/jmbi.2001.4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H, Ruiz G, Denes B, Sandberg L, Langridge W. Optimization of codon composition and regulatory elements for expression of human insulin like growth factor-1 in transgenic chloroplasts and evaluation of structural identity and function. BMC Biotechnol. 2009;9:33. doi: 10.1186/1472-6750-9-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniell H, Ruiz ON, Dhingra A. Chloroplast genetic engineering to improve agronomic traits. Methods Mol. Biol. 2005;286:111–138. doi: 10.1385/1-59259-827-7:111. [DOI] [PubMed] [Google Scholar]
- Daniell H, Vivekananda J, Nielsen BL, Ye GN, Tewari KK. Transient foreign gene expression in chloroplasts of cultured tobacco cells after biolistic delivery of chloroplast vectors. Proc. Natl. Acad. Sci. USA. 1990;87:88–92. doi: 10.1073/pnas.87.1.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCosa B, Moar W, Lee SB, Miller M, Daniell H. Overexpression of the Bt cry2Aa2 operon in chloroplasts leads to formation of insecticidal crystals. Nat. Biotechnol. 2001;19:71–74. doi: 10.1038/83559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari S, Galletti R, Denoux C, De Lorenzo G, Ausubel FM, Dewdney J. Resistance to Botrytis cinerea induced in Arabidopsis by elicitors is independent of salicylic acid, ethylene or jasmonate signaling but requires PAD3. Plant Physiol. 2007;144:367–379. doi: 10.1104/pp.107.095596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukumura M, Begum A, Kruus K, David Wu JH. Interactions and synergism between the recombinant CelD, an endoglucanase, and the cellulosome-integrating protein (CipA) of Clostridium thermocellum. J. Ferment. Bioeng. 1997;83:146–151. [Google Scholar]
- Funnell DL, Matthews PS, VanEtten HD. Breeding for highly fertile isolates of Nectria haematococca MPVI that are highly virulent on pea and in planta selection for virulent recombinants. Phytopathology. 2001;91:92–101. doi: 10.1094/PHYTO.2001.91.1.92. [DOI] [PubMed] [Google Scholar]
- Ghose TK. Measurement of cellulase activities. Pure Appl. Chem. 1987;59:257–268. [Google Scholar]
- Gonzalez-Candelas L, Kolattukudy PE. Isolation and analysis of a novel inducible pectate lyase gene from the phytopathogenic fungus Fusarium solani f. sp. pisi (Nectria haematococca, mating population VI) J. Bacteriol. 1992;174:6343–6349. doi: 10.1128/jb.174.20.6343-6349.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray BN, Ahner BA, Hanson MR. High-level bacterial cellulase accumulation in chloroplast-transformed tobacco mediated by downstream box fusions. Biotechnol. Bioeng. 2008;102:1045–1054. doi: 10.1002/bit.22156. [DOI] [PubMed] [Google Scholar]
- Grohmann K, Baldwin EA, Buslig BS, Ingram LO. Fermentation of galacturonic acid and other sugars in orange peel hydrolysates by the ethanologenic strain of Escherichia coli. Biotechnol. Lett. 1994;16:281–286. [Google Scholar]
- Guda C, Lee SB, Daniell H. Stable expression of a biodegradable protein-based polymer in tobacco chloroplasts. Plant Cell Rep. 2000;19:257–262. doi: 10.1007/s002990050008. [DOI] [PubMed] [Google Scholar]
- Guo W, Gonzalez-Candelas L, Kolattukudy PE. Cloning of a novel constitutively expressed pectate lyase gene pelB from Fusarium solani f. sp. pisi (Nectria haematococca, mating type VI) and characterization of the gene product expressed in Pichia pastoris. J. Bacteriol. 1995;177:7070–7077. doi: 10.1128/jb.177.24.7070-7077.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, González-Candelas L, Kolattukudy PE. Identification of a novel pelD gene expressed uniquely in planta by Fusarium solani f. sp. pisi (Nectria haematococca, mating Type VI) and characterization of its protein product as an endo-pectate lyase. Arch. Biochem. Biophys. 1996;332:305–312. doi: 10.1006/abbi.1996.0346. [DOI] [PubMed] [Google Scholar]
- Gupta R, Lee YY. Mechanism of cellulase reaction on pure cellulosic substrates. Biotechnol. Bioeng. 2009;102:1570–1581. doi: 10.1002/bit.22195. [DOI] [PubMed] [Google Scholar]
- Gusakov AV, Salanovich TN, Antonov AI, Ustinov BB, Okunev ON, Burlingame R, Emalfarb M, Baez M, Sinitsyn AP. Design of highly efficient cellulase mixtures for enzymatic hydrolysis of cellulose. Biotechnol. Bioeng. 2007;97:1028–1038. doi: 10.1002/bit.21329. [DOI] [PubMed] [Google Scholar]
- Hafren J, Daniel G, Westermark U. The distribution of acidic and esterified pectin in cambium, developing xylem and mature xylem of Pinus sylvestris. Iawa J. 2000;21:157–168. [Google Scholar]
- Hasper AA, Dekkers E, van Mil M, van de Vondervoort PJI, de Graaff LH. EglC, a new endoglucanase from Aspergillus niger with major activity towards xyloglucan. Appl. Environ. Microbiol. 2002;68:1556–1560. doi: 10.1128/AEM.68.4.1556-1560.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herron SR, Benen JA, Scavetta RD, Visser J, Jurnak F. Structure and function of pectic enzymes: virulence factors of plant pathogens. Proc. Natl. Acad. Sci. USA. 2000;97:8762–8769. doi: 10.1073/pnas.97.16.8762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himmel ME, Adney WS, Baker JO, Elander R, McMillan JD, Nieves RA, Sheehan JJ, Thomas SR, Vinzant TB, Zhang M. Advanced bioethanol production technologies: a prospective. American Chemical Society Washington, DC. In ACS Symposium. 1997;666:2–45. [Google Scholar]
- Himmel ME, Ruth MF, Wyman CE. Cellulase for commodity products from cellulosic biomass. Curr. Opin. Biotechnol. 1999;10:358–364. doi: 10.1016/S0958-1669(99)80065-2. [DOI] [PubMed] [Google Scholar]
- Irwin DC, Spezio M, Walker LP, Wilson DB. Activity studies of eight purified cellulases: Specificity, synergism, and binding domain effects. Biotechnol. Bioeng. 1993;42:1002–1013. doi: 10.1002/bit.260420811. [DOI] [PubMed] [Google Scholar]
- Jin R, Richter S, Zhong R, Lamppa GK. Expression and import of an active cellulase from a thermophilic bacterium into the chloroplast both in vitro and in vivo. Plant Mol. Biol. 2003;51:493–507. doi: 10.1023/a:1022354124741. [DOI] [PubMed] [Google Scholar]
- Kataeva I, Guglielmi G, Béguin P. Interaction between Clostridium thermocellum endoglucanase CelD and polypeptides derived from the cellulosome-integrating protein CipA: stoichiometry and cellulolytic activity of the complexes. Biochem J. 1997;326:617–624. doi: 10.1042/bj3260617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawazu T, Sun JL, Shibata M, Kimura T, Sakka K, Ohmiya K. Expression of a bacterial endoglucanase gene in tobacco increases digestibility of its cell wall fibers. J. Biosci. Bioeng. 1999;88:421–425. doi: 10.1016/s1389-1723(99)80220-5. [DOI] [PubMed] [Google Scholar]
- Kim ES, Lee HJ, Bang WG, Choi IG, Kim KH. Functional characterization of a bacterial expansin from Bacillus subtilis for enhanced enzymatic hydrolysis of cellulose. Biotechnol. Bioeng. 2009;102:1342–53. doi: 10.1002/bit.22193. [DOI] [PubMed] [Google Scholar]
- Kosugi A, Murashima K, Doi RH. Xylanase and acetyl xylan esterase activities of XynA, a key subunit of the Clostridium cellulovorans cellulosome for xylan degradation. Appl. Environ. Microbiol. 2002;68:6399–6402. doi: 10.1128/AEM.68.12.6399-6402.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koya V, Moayeri M, Leppla SH, Daniell H. Plant-based vaccine: mice immunized with chloroplast-derived anthrax protective antigen survive anthrax lethal toxin challenge. Infect Immun. 2005;73:8266–8274. doi: 10.1128/IAI.73.12.8266-8274.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar R, Wyman CE. Effects of cellulase and xylanase enzymes on the deconstruction of solids from pretreatment of poplar by leading technologies. Biotechnol. Prog. 2009;25:302–314. doi: 10.1002/btpr.102. [DOI] [PubMed] [Google Scholar]
- Kumar S, Daniell H. Engineering the chloroplast genome for hyperexpression of human therapeutic proteins and vaccine antigens. Methods Mol. Biol. 2004;267:365–383. doi: 10.1385/1-59259-774-2:365. [DOI] [PubMed] [Google Scholar]
- Kurokawa J, Hemjinda E, Arai T, Kimura T, Sakka K, Ohmiya K. Clostridium thermocellum cellulase CelT, a family 9 endoglucanase without an Ig-like domain or family 3c carbohydrate-binding module. Appl. Microbiol. Biotechnol. 2002;59:455–461. doi: 10.1007/s00253-002-1048-y. [DOI] [PubMed] [Google Scholar]
- Kwon I, Ekino K, Goto M, Furukawa K. Heterologous expression and characterization of endoglucanase I (EGI) from Trichoderma viride HK-75. Biosci. Biotechnol. Biochem. 1999;63:1714–1720. doi: 10.1271/bbb.63.1714. [DOI] [PubMed] [Google Scholar]
- Lee SB, Kwon HB, Kwon SJ, Park SC, Jeong MJ, Han SE, Byun MO, Daniell H. Accumulation of trehalose within transgenic chloroplasts confers drought tolerance. Mol. Breed. 2003;11:1–13. [Google Scholar]
- Leelavathi S, Gupta N, Maiti S, Ghosh A, Reddy VS. Overproduction of an alkali- and thermo-stable xylanase in tobacco chloroplasts and efficient recovery of the enzyme. Mol. Breed. 2003;11:59–67. [Google Scholar]
- Lelivelt CL, McCabe MS, Newell CA, Desnoo CB, van Dun KM, Birch-Machin I, Gray JC, Mills KH, Nugent JM. Stable plastid transformation in lettuce (Lactuca sativa L.) Plant Mol. Biol. 2005;58:763–74. doi: 10.1007/s11103-005-7704-8. [DOI] [PubMed] [Google Scholar]
- Lietzke SE, Yoder MD, Keen NT, Jurnak F. The three-dimensional structure of pectate lyase E, a plant virulence factor from Erwinia chrysanthemi. Plant Physiol. 1994;106:849–862. doi: 10.1104/pp.106.3.849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margeot A, Hahn-Hagerdal B, Edlund M, Slade R, Monot F. New improvements for lignocellulosic ethanol. Curr. Opin. Biotechnol. 2009;20:1–9. doi: 10.1016/j.copbio.2009.05.009. [DOI] [PubMed] [Google Scholar]
- Martinez D, Berka RM, Henrissat B, Saloheimo M, Arvas M, Baker SE, Chapman J, Chertkov O, Coutinho PM, Cullen D, Danchin EGJ, Grigoriev IV, Harris P, Jackson M, Kubicek CP, Han CS, Ho I, Larrondo LF, de Leon AL, Magnuson JK, Merino S, Misra M, Nelson B, Putnam N, Robbertse B, Salamov AA, Schmoll M, Terry A, Thayer N, Westerholm-Parvinen A, Schoch CL, Yao J, Barabote R, Nelson MA, Detter C, Bruce D, Kuske CR, Xie G, Richardson P, Rokhsar DS, Lucas SM, Rubin EM, Dunn-Coleman N, Ward M, Brettin TS. Genome sequencing and analysis of the biomass-degrading fungus Trichoderma reesei (syn. Hypocrea jecorina) Nat. Biotechnol. 2008;26:553–560. doi: 10.1038/nbt1403. [DOI] [PubMed] [Google Scholar]
- Merino ST, Cherry J. Progress and challenges in enzyme development for biomass utilization. Adv. Biochem. Eng. Biotechnol. 2007;108:95–120. doi: 10.1007/10_2007_066. [DOI] [PubMed] [Google Scholar]
- Miller GL. Use of Dinitrosalicylic acid reagent for determination of reducing sugar. Anal. Chem. 1959;31:426–428. [Google Scholar]
- Montalvo-Rodriguez R, Haseltine C, Huess-LaRossa K, Clemente T, Soto J, Staswick P, Blum P. Autohydrolysis of plant polysaccharides using transgenic hyperthermophilic enzymes. Biotechnol. Bioeng. 2000;70:151–159. doi: 10.1002/1097-0290(20001020)70:2<151::aid-bit4>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- Murashima K, Kosugi A, Doi RH. Synergistic effects of cellulosomal xylanase and cellulases from Clostridium cellulovorans on plant cell wall degradation. J. Bacteriol. 2003;185:1518–1524. doi: 10.1128/JB.185.5.1518-1524.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng TK, Zeikus JG. Purification and characterization of an endoglucanase (1,4-beta-D-glucan glucanohydrolase) from Clostridium thermocellum. Biochem J. 1981;199:341–350. doi: 10.1042/bj1990341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada H, Tada K, Sekiya T, Yokoyama K, Takahashi A, Tohda H, Kumagai H, Morikawa Y. Molecular characterization and heterologous expression of the gene encoding a low-molecular-mass endoglucanase from Trichoderma reesei QM9414. Appl. Environ. Microbiol. 1998;64:555–563. doi: 10.1128/aem.64.2.555-563.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oraby H, Venkatesh B, Dale B, Ahmad R, Ransom C, Oehmke J, Sticklen M. Enhanced conversion of plant biomass into glucose using transgenic rice-produced endoglucanase for cellulosic ethanol. Transgenic Res. 2007;16:739–749. doi: 10.1007/s11248-006-9064-9. [DOI] [PubMed] [Google Scholar]
- Poutanen K, Sundberg M. An acetyl esterase of Trichoderma reesei and its role in the hydrolysis of acetyl xylan. Appl. Microbiol. Biotechnol. 1988;28:419–424. [Google Scholar]
- Robertson GP, Dale VH, Doering OC, Hamburg SP, Melillo JM, Wander MM, Parton WJ, Adler PR, Barney JN, Cruse RM, Duke CS, Fearnside PM, Follett RF, Gibbs HK, Goldemberg J, Mladenoff DJ, Ojima D, Palmer MW, Sharpley A, Wallace L, Weathers KC, Wiens JA, Wilhelm WW. Sustainable biofuels redux. Science. 2008;322:49–50. doi: 10.1126/science.1161525. [DOI] [PubMed] [Google Scholar]
- Rogers LM, Kim YK, Guo W, González-Candelas L, Li D, Kolattukudy PE. Requirement for either a host- or pectin-induced pectate lyase for infection of Pisum sativum by Nectria hematococca. Proc. Natl. Acad. Sci. USA. 2000;97:9813–9818. doi: 10.1073/pnas.160271497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosgaard L, Pedersen S, Meyer AS. Comparison of different pretreatment strategies for enzymatic hydrolysis of wheat and barley straw. Appl. Biochem. Biotechnol. 2007a;143:284–296. doi: 10.1007/s12010-007-8001-6. [DOI] [PubMed] [Google Scholar]
- Rosgaard L, Pedersen S, Langston J, Akerhielm D, Cherry JR, Meyer AS. Evaluation of minimal Trichoderma reesei cellulase mixtures on differently pretreated Barley straw substrates. Biotechnol. Prog. 2007b;23:1270–1276. doi: 10.1021/bp070329p. [DOI] [PubMed] [Google Scholar]
- Rubin EM. Genomics of cellulosic biofuels. Nature. 2008;454:841–845. doi: 10.1038/nature07190. [DOI] [PubMed] [Google Scholar]
- Ruf S, Karcher D, Bock R. Determining the transgene containment level provided by chloroplast transformation. Proc. Natl. Acad. Sci. USA. 2007;104:6998–7002. doi: 10.1073/pnas.0700008104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruhlman T, Ahangari R, Devine A, Samsam M, Daniell H. Expression of cholera toxin B-proinsulin fusion protein in lettuce and tobacco chloroplasts--oral administration protects against development of insulitis in non-obese diabetic mice. Plant Biotechnol. J. 2007;5:495–510. doi: 10.1111/j.1467-7652.2007.00259.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan CA. Oligosaccharides as recognition signals for the expression of defensive genes in plants. Biochemistry. 1988;27:8879–8898. [Google Scholar]
- Saloheimo M, Paloheimo M, Hakola S, Pere J, Swanson B, Nyyssonen E, Bhatia A, Ward M, Penttila M. Swollenin, a Trichoderma reesei protein with sequence similarity to the plant expansins, exhibits disruption activity on cellulosic materials. Eur. J. Biochem. 2002;269:4202–4211. doi: 10.1046/j.1432-1033.2002.03095.x. [DOI] [PubMed] [Google Scholar]
- Sandgren M, Shaw A, Ropp TH, Wu S, Bott R, Cameron AD, Ståhlberg J, Mitchinson C, Jones TA. The X-ray crystal structure of the Trichoderma reesei family 12 endoglucanase 3, Cel12A, at 1.9 A resolution. J. Mol. Biol. 2001;308:295–310. doi: 10.1006/jmbi.2001.4583. [DOI] [PubMed] [Google Scholar]
- Selig MJ, Knoshaug EP, Adney WS, Himmel ME, Decker SR. Synergistic enhancement of cellobiohydrolase performance on pretreated corn stover by addition of xylanase and esterase activities. Bioresour. Technol. 2008;99:4997–5005. doi: 10.1016/j.biortech.2007.09.064. [DOI] [PubMed] [Google Scholar]
- Singh ND, Li M, Lee SB, Schnell D, Daniell H. Arabidopsis Tic40 expression in tobacco chloroplasts results in massive proliferation of the inner envelope membrane and upregulation of associated proteins. Plant Cell. 2008;20:3405–3417. doi: 10.1105/tpc.108.063172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soliday CL, Flurkey WH, Okita TW, Kolattukudy PE. Cloning and structure determination of cDNA for cutinase, an enzyme involved in fungal penetration of plants. Proc. Natl. Acad. Sci. USA. 1984;81:3939–3943. doi: 10.1073/pnas.81.13.3939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern R, Asari AA, Sugahara KN. Hyaluronan fragments: an information-rich system. Eur. J. Cell Biol. 2006;85:699–715. doi: 10.1016/j.ejcb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- Sticklen MB. Plant genetic engineering for biofuel production: towards affordable cellulosic ethanol. Nat. Rev. Genet. 2008;9:433–443. doi: 10.1038/nrg2336. [DOI] [PubMed] [Google Scholar]
- Sun Y, Cheng JJ, Himmel ME, Skory CD, Adney WS, Thomas SR, Tissert B, Nishimura Y, Yamamoto YT. Expression and characterization of Acidothermus cellulolyticus E1 endoglucanase in transgenic duckweed Lemna minor 8627. Bioresour. Technol. 2007;98:2866–2872. doi: 10.1016/j.biortech.2006.09.055. [DOI] [PubMed] [Google Scholar]
- Svab Z, Maliga P. Exceptional transmission of plastids and mitochondria from the transplastomic pollen parent and its impact on transgene containment. Proc. Natl. Acad. Sci. USA. 2007;104:7003–7008. doi: 10.1073/pnas.0700063104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor KR, Gallo RL. Glycosaminoglycans and their proteoglycans: host-associated molecular patterns for initiation and modulation of inflammation. FASEB J. 2006;20:9–22. doi: 10.1096/fj.05-4682rev. [DOI] [PubMed] [Google Scholar]
- TaylorII LE, Dai Z, Decker SR, Brunecky R, Adney WS, Ding SY, Himmel ME. Heterologous expression of glycosyl hydrolases in planta: a new departure for biofuels. Trends Biotechnol. 2008;26:413–424. doi: 10.1016/j.tibtech.2008.05.002. [DOI] [PubMed] [Google Scholar]
- Verma D, Daniell H. Chloroplast vector systems for biotechnology applications. Plant Physiol. 2007;145:1129–1143. doi: 10.1104/pp.107.106690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma D, Samson NP, Koya V, Daniell H. A protocol for expression of foreign genes in chloroplasts. Nat. Protocols. 2008;3:739–758. doi: 10.1038/nprot.2007.522. [DOI] [PubMed] [Google Scholar]
- Wegener GB. Induction of defence responses against Erwinia soft rot by an endogenous pectate lyase in potatoes. Physiol. Mol. Plant Pathol. 2002;60:91–100. [Google Scholar]
- Wegener CB, Olsen O. Heterologous pectate lyase isoenzymes are not different in their effects on soft rot resistance in transgenic potato. Physiol. Mol. Plant Pathol. 2004;65:59–66. [Google Scholar]
- Wellner N, Kauráková M, Malovíková A, Wilson RH, Belton PS. FT-IR study of pectate and pectinate gels formed by divalent cations. Carbohydrate Res. 1998;308:123–131. [Google Scholar]
- Wonganu B, Pootanakit K, Boonyapakron K, Champreda V, Tanapongpipat S, Eurwilaichitr L. Cloning, expression and characterization of a thermotolerant endoglucanase from Syncephalastrum racemosum ( BCC18080) in Pichia pastoris. Protein Expr. Purif. 2008;58:78–86. doi: 10.1016/j.pep.2007.10.022. [DOI] [PubMed] [Google Scholar]
- Wyman CE, Dale BE, Elander RT, Holtzapple M, Ladisch MR, Lee YY. Coordinated development of leading biomass pretreatment technologies. Bioresour. Technol. 2005;96:1959–1966. doi: 10.1016/j.biortech.2005.01.010. [DOI] [PubMed] [Google Scholar]
- Xu X, Fang J, Wang W, Guo J, Chen P, Cheng J, Shen Z. Expression of a bacterial alpha-amylase gene in transgenic rice seeds. Transgenic Res. 2008;17:645–650. doi: 10.1007/s11248-007-9144-5. [DOI] [PubMed] [Google Scholar]
- Yapo BM, Lerouge P, Thibault JF, Ralet MC. Pectins from citrus peel cell walls contain homogalacturonans homogenous with respect to molar mass, rhamnogalacturonan I and rhamnogalacturonan II. Carbohydr. Polym. 2007;69:426–435. [Google Scholar]
- Yoder MD, Keen NT, Jurnak F. New domain motif: the structure of pectate lyase C, a secreted plant virulence factor. Science. 1993;260:1503–1507. doi: 10.1126/science.8502994. [DOI] [PubMed] [Google Scholar]
- Yu LX, Gray BN, Rutzke CJ, Walker LP, Wilson DB, Hanson MR. Expression of thermostable microbial cellulases in the chloroplasts of nicotine-free tobacco. J. Biotechnol. 2007;131:362–369. doi: 10.1016/j.jbiotec.2007.07.942. [DOI] [PubMed] [Google Scholar]
- Zeigler MT, Thomas SR, Danna KJ. Accumulation of a thermostable endo-1,4-β-D-glucanase in the apoplast of Arabidopsis thaliana leaves. Mol. Breed. 2000;6:37–46. [Google Scholar]
- Zhou S, Ingram LO. Synergistic hydrolysis of carboxymethyl cellulose and acid-swollen cellulose by two endoglucanases (CelZ and CelY) from Erwinia chrysanthemi. J. Bacteriol. 2000;182:5676–5682. doi: 10.1128/jb.182.20.5676-5682.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegelhoffer T, Raasch JA, Austin-Phillips S. Dramatic effects of truncation and sub-cellular targeting on the accumulation of recombinant microbial cellulase in tobacco. Mol. Breed. 2001;8:147–158. [Google Scholar]
- Ziegelhoffer T, Raasch JA, Austin-Phillips S. Expression of Acidothermus cellulolyticus E1 endo- β-1,4-glucanase catalytic domain in transplastomic tobacco. Plant Biotechnol. J. 2009;7:527–536. doi: 10.1111/j.1467-7652.2009.00421.x. [DOI] [PubMed] [Google Scholar]
- Ziegelhoffer T, Will J, Austin-Phillips S. Expression of bacterial cellulase genes in transgenic alfalfa (Medicago sativa L.), potato (Solanum tuberosum L.) and tobacco (Nicotiana tabacum L.) Mol. Breed. 1999;5:309–318. [Google Scholar]
- Zverlov VV, Velikodvorskaya GA, Schwarz WH. A newly described cellulosomal cellobiohydrolase, CelO, from Clostridium thermocellum: investigation of the exo-mode of hydrolysis, and binding capacity to crystalline cellulose. Microbiology. 2002;148:247–255. doi: 10.1099/00221287-148-1-247. [DOI] [PubMed] [Google Scholar]