Abstract
Detection of multi-drug resistant tuberculosis (MDR-TB), a frequent cause of treatment failure, takes two or more weeks to identify by culture. Rifampicin (RIF) resistance is a hallmark of MDR-TB, and detection of mutations in the rpoB gene of Mycobacterium tuberculosis using molecular beacon probes with real-time quantitative PCR (qPCR) is a novel approach that takes ≤ 2 days. However, qPCR identification of resistant isolates, particularly for isolates with mixed RIF-susceptible and RIF-resistant bacteria, is reader-dependent and limits its clinical use. The aim of this study was to develop an objective, reader independent method to define rpoB mutants using beacon qPCR. This would facilitate the transition from a research protocol to the clinical setting, where high-throughput methods with objective interpretation are required. For this, DNAs from 107 M. tuberculosis clinical isolates with known susceptibility to RIF by culture-based methods were obtained from two regions where isolates have not previously been subjected to evaluation using molecular beacon qPCR: The Texas-Mexico border and Colombia. Using coded DNA specimens, mutations within an 81 bp hot-spot region of rpoB were established by qPCR with five beacons spanning this region. Visual and mathematical approaches were used to establish whether the qPCR cycle threshold (Ct) of the experimental isolate was significantly higher (mutant) compared to a reference wild-type isolate. Visual classification of the beacon qPCR required reader training for strains with a mixture of RIF-susceptible and resistant bacteria. Only then, the visual interpretation by an experienced reader had 100% sensitivity and 94.6% specificity versus RIF-resistance by culture phenotype, and 98.1% sensitivity and 100% specificity versus mutations based on DNA sequence. The mathematical approach was 98% sensitive and 94.5% specific versus culture, and 96.2% sensitive and 100% specific versus DNA sequence. Our findings indicate the mathematical approach has advantages over the visual reading, in that it uses a Microsoft Excel template to eliminate reader bias or inexperience, and allows objective interpretation from high-throughput analyses even in the presence of a mixture of RIF-resistant and RIF-susceptible isolates without the need for reader training.
Keywords: Mycobacterium, tuberculosis, molecular beacon, real-time polymerase chain reaction, MDR-TB, Texas-Mexico border, Colombia, rifampicin resistance
1. Introduction
Approximately 1/3 of the world's population is infected with Mycobacterium tuberculosis, with more than 2 million deaths among those who progress to active tuberculosis (TB) (Stop TB Partnership and World Health Organization, 2006). One of the most serious threats to the global control of TB is emergence of multidrug resistant tuberculosis (MDR-TB) isolates, resistant to at least isoniazid (INH) and rifampicin (RIF) (World Health Organization, 2008). Most of the 500,000 annual cases of MDR-TB are not detected or treated properly (World Health Organization, 2009). A major reason for treatment failure and fatal clinical outcome is resistance to RIF, an antibiotic with excellent early bactericidal effect on metabolically active M. tuberculosis, as well as late sterilizing action on semi-dormant organisms undergoing short bursts of metabolic activity. Most RIF-resistant isolates are also resistant to INH, also a first line bactericidal agent that upon activation produces a range of damaging reactive radical species. Therefore, detection of RIF-resistance predicts MDR-TB (Somoskovi et al., 2001).
Improvement of current TB control programs requires the development of simple and rapid methods for MDR-TB detection, enabling prompt initiation of appropriate TB treatment (Nikolayevskyy et al., 2007). The current “gold standard” to assess MDR-TB is based on culture phenotype: growth of M. tuberculosis on media containing the corresponding antibiotic. The advent of liquid media has expedited the turnaround time of culture methods, but results still take at least 11 days for MDR-TB detection (Robledo et al., 2008), and drug resistance culture-based testing is not always available in developing countries, particularly those classified as `high-burden' for TB by the World Health Organization (WHO, 2009).
Approximately 96% of the RIF-resistant isolates have a mutation in the 81 bp core region of the rpoB gene, facilitating detection of mutations associated with RIF resistance (Piatek et al., 2000; Piatek et al., 1998). Various molecular techniques have been proposed to detect these mutations. Some are limited to experimental use due to technical complexity, including conventional sequencing (Palomino, 2006), pyrosequencing (Jureen et al., 2006) and high-resolution thermal melt analysis (Hoek et al., 2008; McCammon et al., 2005). Solid-phase hybridization methods have been developed for diagnostic use (Genotype M. tuberculosis DR, Hain Lifescience, Nehren, Germany; and INNO-LiPa Rif TB assay, Innogenetics, Gent, Belgium). The increased simplicity of these assays depends on their capability to detect only the most common mutations worldwide, leading to variation in susceptibility between geographical regions (Ahmad et al., 2000; Ahmad and Mokaddas, 2005; Bartfai et al., 2001; Cavusoglu et al., 2002; Cooksey et al., 1997; Hirano et al., 1999; Hoek et al., 2008; Matsiota-Bernard et al., 1998; Nikolayevskyy et al., 2007; Ozkutuk et al., 2007). Another technique, real-time quantitative PCR (qPCR) using molecular beacons as probes has the potential for experimental and diagnostic use (Piatek et al., 2000; Piatek et al., 1998; Lin et al., 2004; Varma-Basil et al., 2004). Its use in research comes from its capacity to detect any mutation in the 81bp region of rpoB. For the purposes of diagnosis, molecular beacon qPCR can be used to screen TB patients for MDR-TB in the increasing number of laboratories worldwide with real time PCR capability. In settings where qPCR is not available, molecular beacons can still be used by amplification in enclosed tubes, and then measuring fluorescence with an appropriate light source in a fluorometer (Piatek et al., 1998; Tyagi and Kramer, 1996). Recent adaptations using a simpler, more operator-friendly platform have become available. (Cepheid, Sunnydale, CA) (Stop TB Partnership and World Health Organization, 2008).
The expanded use of beacon qPCR from a research tool to the clinical setting will require a systematic approach for its interpretation that is not operator-dependent. In this study we evaluated the performance of the method in M. tuberculosis isolates from two geographical sites not evaluated previously using this PCR-based method: The Texas-Mexico border and Medellin, Colombia. We observed that visual interpretation of the assay is affected by the PCR experience of the reader. We developed a mathematical approach to circumvent reader misinterpretation and implemented its use by designing a Microsoft Excel template to automatically classify isolates based on qPCR output. We compared the performance of visual reading and mathematical analysis, and highlight the observed advantages and potential applications of the mathematical approach.
2. Methods
2.1. Bacterial isolates
One hundred and seven M. tuberculosis clinical isolates with known susceptibility to RIF by microbiological methods (see below) were identified from The Texas-Mexico border and from Medellin, Colombia. These are two distant geographical regions where beacon PCR had not been used previously for assessment of RIF resistance. We selected isolates from patients from The Texas-Mexico border (n=53) that were previously characterized for MDR-TB using the rapid BACTEC-460TB radiometric system (Becton Dickinson, Paramus, N.J.) and had their rpoB gene sequenced (McCammon et al., 2005; Quitugua et al., 2002). Isolates from Colombia (n=54) were obtained during the routine isolation and drug-susceptibility testing of M. tuberculosis from clinical specimens submitted to the Corporación para Investigaciones Biológicas (CIB) in Medellin between 1997 and 2006. Conventional antibiotic susceptibility testing was performed at CIB using the Centers for Disease Control and Prevention version of the agar proportion method (Kent and Kubica, 1985). Resistance was defined as greater than 1% growth in the presence of 1 μg/ml of RIF.
2.2. DNA isolation, purification and quantification
DNA from The Texas-Mexico border isolates was extracted by boiling (Quitugua et al., 2002), and purified with phenol:chloroform:isoamyl alcohol (Eickbush and Moudrianakis, 1978). DNA from the Colombian isolates was isolated at CIB laboratories prior to shipping to Brownsville as described previously (van Soolingen et al., 1991). Mycobacterial DNA from both study sites was resuspended in 0.1x TE buffer (1 mM Tris-HCl, 0.1 mM EDTA, pH 8.0) for PCR. Mycobacterial DNA was quantified using Picogreen (Molecular Probes, Eugene, OR) and the mycobacterial DNA quantity and quality was assessed by absorbance using both 260:280 (between 1.8 – 2.2) and 260:230 (1.6 – 2.0) ratios (ND-1000 spectrophotometer; NanoDrop Tech, Wilmington, DE) prior to qPCR analysis.
2.3. Oligonucleotide sequences
The sequences of the primers and molecular beacon probes A thru D were described previously (El-Hajj et al., 2001; Piatek et al., 2000). The primers covered a 189 bp core of the rpoB gene from M. tuberculosis (accession no. Z95972). The molecular beacon MB531 (Beacon E in this study) was designed by Lin and collaborators (Lin et al., 2004). The combination of beacons A through E hybridize to an 83 bp wild-type region within the rpoB gene core (accession no. AE000516; locus tag MT0695; nucleotides 762,988–763,177), except for a gap in 5 nucleotides between beacons D and E (codons 447 and 448). A sixth molecular beacon for the 16S rRNA gene confirms the presence of M. tuberculosis (Piatek et al., 1998). All beacons were labeled with a reporter (6 – Carboxyfluorescein; 6-FAM) on the 5' end and quencher (Dabcyl) on the 3' end. Beacons were synthesized by Sigma-Genosys (Sigma-Aldrich, St. Louis, MO).
2.4. Assay conditions
Real time quantitative PCR (qPCR) assays were performed in 96-well plates (ABgene, Rochester, NY). All reactions were run in a 7900HT Sequence detector (Applied Biosystems, Foster City, CA), with each 25 μl reaction containing 10 mM Tris/HCl pH 8.0, 50 mM KCl, 200 μM dNTP mix, 400 nM each primer, 100 nM of the molecular beacon, 0.65 units of hot start JumpStart Taq DNA polymerase (Sigma-Aldrich, St. Louis, MO), ROX as a reference dye and either 4 mM MgCl2 for the rpoB beacons, and 5 mM MgCl2 for 16SrRNA. Conditions for amplification were 1 cycle at 95°C for 2 min, followed by 40 cycles at 95°C for 15 s for denaturation, 62°C for 30 s for primer and molecular beacon annealing and detection, and 72°C for 20 s for primer extension. Each run included experimental samples (103 copies/well), a no-DNA negative control (water instead of mycobacterial DNA to exclude contamination of the qPCR reagents) and a positive control (103 genomes of CDC1551 wild type strain, RIF-susceptible). Fluorescence was measured during each annealing step throughout the course of the 40 cycles. The spectral data were automatically analyzed using the SDS 2.1 software (Applied Biosystems, Foster City, CA) to determine the fluorescence intensity contributed by each beacon. The background fluorescence of each probe (calculated from thermal cycles 1 through 15) was subtracted. The threshold cycle (Ct) was then set up manually for all beacons by setting the threshold point at a ΔRn of 0.1 which was i) above the background from all beacons, ii) within the geometric phase of the amplification curve for all beacons for the reference CDC1551 strain.
2.5. DNA sequencing
The 83-bp region of rpoB was sequenced for all isolates from Colombia as had been previously done for all isolates from Texas. For this a standard PCR was run using assay conditions identical to those described for qPCR, except for absence of beacons, in a MJ Research PTC-200 thermocycler (Bio-Rad, Hercules, CA). Primers and unincorporated dNTPs were then removed from the amplicon by filtration (Montage PCR filter units, Millipore Corp, Billerica, MA). Amplicons were quantified at A260 (ND-1000 spectrophotometer; NanoDrop Tech, Wilmington, DE) and shipped to SeqWright DNA technology services (Houston, TX) where sequencing was performed with the same amplification primers in an ABI Prism 3730xl DNA sequencer (Applied Biosystems, Foster City, CA). Sequences were analyzed with CLUSTAL X Multiple Sequence Alignment Program (version 2.0.10) (Larkin et al., 2007).
2.6. Data analysis
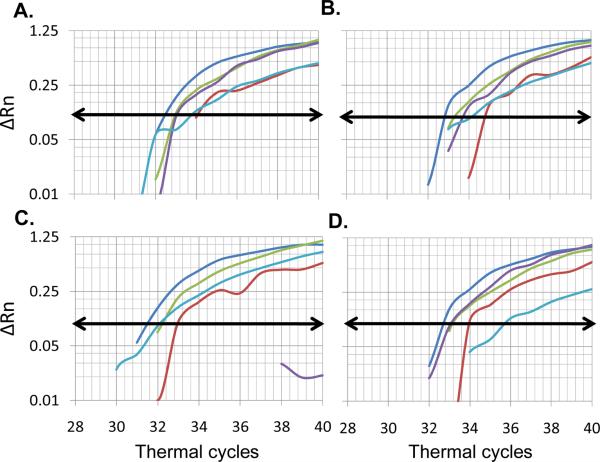
The number of cycles required for each amplicon to reach the Ct was recorded using SDS 2.1 (Applied Biosystems) (Heid et al., 1996). Mutations in the 83-bp rpoB region were established using two approaches. The first was visual inspection of the amplification plots, with mutations identified when there was either an absence or a compromise in amplification (subjective reading) of any of the five beacons (A–E) in a given isolate (Figs 1C–D). The second was a mathematical approach to objectively establish the Ct cut-off for defining mutations for each beacon after data on SDS 2.1 was exported into Microsoft Excel 2007. Details on the development of the second approach are provided in the Results section. Kappa statistic was conducted to establish the concordance between the standard drug-susceptibility characterization methods by culture or DNA sequencing, and the beacon approach. Descriptive statistics are provided. Positive predictive value (number of true positives/(number of true positives + number of false negatives), and negative predictive values (number of true negatives/(number of true negatives + number of false negatives) were calculated.
Figure 1.
Distribution of amplification plots for beacons A–E for CDC1551 and representative experimental isolates. The average delta Rn for duplicates of each beacon is shown for the 40 thermal cycles run for each isolate in a given plate. A, The signal efficiency was slightly variable between beacons for the reference strain CDC1551: strongest for beacon A (blue), followed by C (green), D (purple), B (red) and E (light blue). B, Similar beacon distribution was observed for the wild-type experimental isolate T138. C, Isolate MD465 showed similar distribution for beacons A, B, C, and E, but beacon D did not reach the threshold of fluorescence indicating a mutation in the sequence covered by that beacon. D, Isolate MD540 had a similar distribution for beacons A–D, but beacon E had a detectable but clearly higher Ct when compared to that for CDC1551, suggesting a mixed RIF-S and RIF-R population. Horizontal lines with arrows represent threshold set point at a delta Rn of 0.1.
3. Results
3.1. Study design
We adapted a molecular beacon qPCR assay and compared the performance of a visual and a mathematical analysis for prompt identification of RIF-resistance in isolates from two geographical regions not tested previously by this method: The Texas-Mexico border (n=32 RIF- susceptible and 21 RIF-resistant) and Medellin, Colombia (n=24 RIF-susceptible and 30 RIF-resistant). Our molecular team was blinded for the previous RIF-resistance. Total DNA from each M. tuberculosis isolate was quantified and 103 genomes per qPCR reaction were submitted to amplification with six molecular beacon assays: five spanning an 83-bp rpoB region (A, B, C, D, E) and one specific for M. tuberculosis 16SrRNA. Each 96-well plate was set up with DNA from CDC1551 and six experimental isolates, each run in duplicates (Fig S1). After amplification of M. tuberculosis DNA was confirmed with the 16S rRNA beacon, two strategies were used to determine the Ct cut-off for presence or absence of mutations in a beacon: visual or mathematical analysis. As a `gold-standard' reference we used the microbiological classification of drug resistance or the presence of DNA mutations in the rpoB target based on DNA sequence.
3.2. Characteristic distribution of amplification curves
The distribution of the amplification plots from all isolates revealed that classification of isolates as RIF-susceptible or –resistant required careful observation. First, despite placing “103” M. tuberculosis genomes per qPCR well, there were variations in the amplification signal efficiency between isolates that could lead to difficulties in comparing the amplification curves of each isolate with the reference strain. Second, the signal efficiency was slightly variable between the five beacons for the reference strain CDC1551: consistently stronger for beacon A, and weaker for B and E (Fig 1A and S2). Third, the amplification curves from the experimental isolates behaved in one of three ways when compared to CDC1551 (Fig. 1A): i) Ct similar to the reference strain (wild-type; Fig. 1B), ii) no Ct value for at least one beacon when the curve did not reach the threshold value (Fig. 1C), indicating a mutation in the sequence covered by that beacon in nearly 100% of the M. tuberculosis cells, or iii) at least one beacon with detectable but “clearly higher” Ct than that for CDC1551 (Fig. 1D), suggesting a possible mixture of wild type (mixed RIF-susceptible) and mutant (RIF-resistant) bacteria for the sequence covered by that molecular beacon. Twenty eight of the 51 mutant isolates fell into this category (data not shown). This was particularly common for beacon E, where 15/16 isolates with mutations in this region had a detectable Ct.
These observations led us to the realization that beacon interpretation by visual inspection could be compromised by inherent bias of the reader, particularly for isolates with higher but detectable Ct compared to the wild-type. This was supported after four volunteers (two with qPCR experience and two without) were given a guide to detect mutations using molecular beacon qPCR, and asked to record their interpretation of the amplification plots from all the isolates evaluated in this study (Table S1). All their misclassifications were for isolates that had at least one beacon with detectable Ct. The most frequent discordance between readers was for beacon E.
3.3. Visual detection of mutations
The varying characteristics of the amplification curves described in the previous section were the basis on which we designed a strategy for visual classification of RIF-resistant or –susceptible isolates. First, within each 96-well plate, the distribution of beacons A–E for strain CDC1551 were used as reference for the other five experimental isolates run in parallel, to avoid inter-plate variations. Second, taking into account the distribution of beacon signals for the wild type strain(s) (Fig 1A), we then evaluated each experimental isolate to determine whether any of the beacons had a Ct that was either undetectable or “clearly higher” (mutant) than the others with a lower Ct (wild-type regions). Since we noted that among the 51 isolates with nucleotide mutations, 50 had mutations that spanned up to two different beacons (Table 1), the amplification curves from the remaining three or four beacons that appeared to span wild-type sequences were used as internal Ct reference for each isolate. This intra-isolate comparison controlled for Ct variations between isolates, due to inherent differences in the amount of DNA loaded per well (even though 103 genomes were routinely loaded per qPCR reaction), and/or efficiency of amplification of each DNA preparation. Isolates with mutations in at least one beacon were classified as RIF-resistant. Using these rules, our qPCR operator interpreted the molecular beacons for each isolate without knowledge of culture phenotype, and then compared the results to RIF-resistance by culture as reference. Beacon qPCR was 100% sensitive and 94.6% specific, with 94.2% positive predictive value and 100% negative predictive value. Discordance was attributed to three isolates, RIF-susceptible by culture, but RIF-resistant by visual beacon classification (isolates 5514, 3300 and T106; Tables 1 and 2).
Table 1.
Mutations within the 83 bp region of rpoB from Colombian and Texan strains
| Beacon: | A |
B |
C |
D |
E |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Codon Noa | 428 | 429 | 430 | 433 | 435 | 436 | 441 | 445 | 447 | 450 | |
| Amino acid | Ser | Gln | Leu | Phe | Asp | Gln | Ser | His | Arg | Ser | |
| CDC1551 | AGC | CAG | CTG | TTC | GAC | CAG | TCG | CAC | CGC | TCG | |
| COLOMBIA (n=30) | MD17 | GTC | |||||||||
| MD24 | ACG | TTG | |||||||||
| MD31 | TTG | ||||||||||
| MD32 | TTG | ||||||||||
| MD43 | ATG | ACG | TTG | ||||||||
| MD44 | TAC | ||||||||||
| MD46 | TTG | ||||||||||
| MD48 | TTG | ||||||||||
| MD135 | TTG | ||||||||||
| MD141 | TTC | ||||||||||
| T102 | GAC | ||||||||||
| T106 | TAC | ||||||||||
| MD230 | TTG | ||||||||||
| MD232 | GTC | ||||||||||
| T126 | TTG | ||||||||||
| MD298 | GTC | ||||||||||
| MD315 | TTG | ||||||||||
| MD351 | TTG | ||||||||||
| MD390 | TTG | ||||||||||
| MD465 | GAC | ||||||||||
| MD506 | TTG | ||||||||||
| MD540 | TTG | ||||||||||
| MD561 | TTC | ||||||||||
| MD572 | AGG | TAC | |||||||||
| MD582 | TTG | ||||||||||
| T152 | AAC | TGG | |||||||||
| MD643 | TTG | ||||||||||
| MD794 | TTG | ||||||||||
| MD2101 | GTC | ||||||||||
| MD2160 | GTC | ||||||||||
| TEXAS-MEXICO BORDER (n=21) | 570 | CAC | TAC | ||||||||
| 870 | GAC | ||||||||||
| 2179 | CGC | ||||||||||
| 2220 | GAC | GCG | |||||||||
| 2267 | GGC | ||||||||||
| 2308 | ΔGAC-CAG | ||||||||||
| 2325 | TAC | ||||||||||
| 2608 | CGC | ||||||||||
| 3200 | CGC | ||||||||||
| 3300 | TTT | ||||||||||
| 3302 | GAC | ||||||||||
| 3573 | CGT | TTG | |||||||||
| 3765 | TAC | ||||||||||
| 4017 | GTC | ||||||||||
| 4384 | CGT | ||||||||||
| 4694 | CGC | ||||||||||
| 4727 | GAC | ||||||||||
| 5242 | GAG | AAC | |||||||||
| 5387 | GTC | ||||||||||
| 5514 | GAC | ||||||||||
| 5518 | GAC | ||||||||||
Codon numer based on M. tuberculosis rpoB sequence Accession no. AE000516.
Region: 761762–765298; CDC1551 sequence shown as a wild-type reference for nucleotide and amino acid sequence; bold nucleotide, mutation site; Δ, deletion; shaded codon presented discordant results between beacon interpretation and either culture phenotype or DNA sequence (summary in Table 1)
Table 2.
Performance of molecular beacons versus conventional culture or DNA sequencea
| Beacon evaluation method | Final classification | Culture phenotype |
DNA sequence |
||
|---|---|---|---|---|---|
| RIF-R (n=49) | RIF-S (n=56) | RIF-mutant (n=53) | RIF-wild type (n=52) | ||
| Visual | RIF-R | 49 (100.0%) | 3 (5.4%) | 52 (98.1%) | 0 |
| RIF-S | 0 | 53 (94.6%) | 1 (1.9%) | 52 (100.0%) | |
|
| |||||
| Mathematical | RIF-R | 48 (98.0%) | 3 (5.5%) | 51 (96.2%) | 0 |
| RIF-S | 1 (2.0%) | 53 (94.5%) | 2 (3.8%) | 52 (100.0%) | |
Details for classification of strains by each method are provided in the text; RIF-R, RIF-resistant; RIF-S, RIF-sensitive
To explore possible explanations for discordance between both techniques, and identify the nucleotide changes conferring RIF-resistance in isolates from each geographical region, analysis of the 83-bp rpoB DNA sequence was conducted. Mutations were more frequent in codon 450 for Colombian isolates (18/30), and codon 445 for the Texas-Mexico border isolates (12/21; Table 2). One Texas isolate (2308) had a 6 nucleotide (two codon) deletion. Three Texas isolates (3300, 3573, 4384) had silent mutations: two were RIF-susceptible, and one was RIF-R due to the presence of another mutation (Table 2; Table S2). Coincidently, the five nucleotide region containing these two silent mutations was within a 5 nucleotide gap between beacons D and E. The qPCR visual method had a sensitivity of 98.1% and specificity of 100% when compared to mutations by DNA sequence (Table 2). Discordance was attributed to one isolate with no detectable mutation with beacons but a nucleotide substitution (CAC→GAC; His→Asp) based on DNA sequence. This isolate (870) was reported as susceptible using traditional methods. DNA sequence analysis explained the discordance between visual beacon interpretation and culture phenotype for isolate 3300, which had a silent mutation, but not for isolates 5514 or T106, which contained mutations resulting in amino acid changes (Table 1).
3.4. Mathematical approach to define a Ct cut-off for mutant classification
Even though the visual interpretation of mutations provided sensitivities and specificities higher than 94%, we noted that classification of isolates with mixed populations of RIF-resistant and RIF-susceptible bacteria (Fig 1C) was somewhat subjective and varied depending on the reader training and qPCR experience (Table S1). Therefore, we explored the possibility of setting an automated cut-off Ct value for classification of mutants that would eliminate reader bias. The strategy was to design a set of rules to objectively determine the Ct cut-off for isolates evaluated in the 18 96-well plates of this study. First, beacon A had the lowest raw Ct (mean±SEM; 32.2±0.10), followed by C (32.8±0.10), D (33.3±0.13), B (33.7±0.14) and E (34.0±0.12; Fig. S1). Therefore, we adjusted the efficiency of detection of each beacon so the Ct for beacons A–E would be comparable. The adjustment factor was based on data from CDC1551 from each plate, to account for the inherent variation in the efficiency of amplification of each experiment (run in duplicates; Fig 2A). Beacon A was arbitrarily chosen as reference, and the conversion factor to adjust for efficiency of amplification of all other beacons was estimated by dividing the mean Ct for the duplicates of beacon A by the mean Ct of beacons B, C, D or E (e.g. Conversion factor for beacon B= mean Ct for beacon A/mean Ct for beacon B; Table 3, Conversion factor using A as reference). Then the raw Ct for each beacon from the experimental isolates in the same plate was adjusted by multiplying by its corresponding adjustment factor (Adjusted Ct in Table 3; Fig 2B).
Figure 2.
Adjustment of Ct based on variable signal efficiency between beacons A – E for isolates evaluated in one 96-well plate. A, Raw Ct for beacons A–E for CDC1551 and six experimental isolates run in the same plate. B, Adjusted Ct based on the amplification efficiency of beacons A–E from strain CDC1551 as reference. Bars indicate the mean of the duplicate Ct for each beacon from each isolate. The solid horizontal lines in panel B indicate the Ct cut-off (avg-Ct-adj plus 1.5 cycles for beacons A–D, and 0.5 cycles for beacon E) based on our mathematical approach.
Table 3.
Steps to identify the final Ct for each beacon from each strain
| Reference strain |
Experimental isolate in same 96-well plate |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Beacon | Mean raw Ct | Conversion factor using A as reference | Mean raw Ct | Adjusted Ct | Mean of all adjusted Ct's | Select three adjusted Ct's closest to mean-adjusted Ct (A–E)b | Mean Ct-adjusted | Add Ct to select final Ct threshold | Final Ct cut-off for this experimental isolate |
| A | R-CtA | R-CtA/R-CtA | E-CtA | R-CtA/R-CtA × E-CtA | Mean-adj-Ct (A–E) | R-CtA/R-CtA × E-CtA | Mean adj-Ct (A,C,D) | 1.5 | Mean Adj-Ct (A,C,D)+1.5 |
| B | R-CtB | R-CtA/R-CtB | E-CtB | R-CtA/R-CtB × E-CtB | 1.5 | Mean Adj-Ct (A,C,D)+1.5 | |||
| C | R-CtC | R-CtA/R-CtC | E-CtC | R-CtA/R-CtC × E-CtC | R-CtA/R-CtC × E-CtC | 1.5 | Mean Adj-Ct (A,C,D)+1.5 | ||
| D | R-CtD | R-CtA/R-CtD | E-CtD | R-CtA/R-CtD × E-CtD | R-CtA/R-CtD × E-CtD | 1.5 | Mean Adj-Ct (A,C,D)+1.5 | ||
| E | R-CtE | R-CtA/R-CtE | E-CtE | R-CtA/R-CtE × E-CtE | 0.5 | Mean Adj-Ct (A,C,D)+0.5 | |||
Details provided in section 3.4
In this table, the Ct's from beacons A, C and D are arbitrarily selected to illustrate example
R-Ct, CT for reference strain; E-Ct, Ct for experimental strain; adj, adjusted; subscript letters denote beacons;
Second, we established the cut-off Ct value for each isolate in a given plate, using as gold standard the 83 bp rpoB DNA sequence (and not culture phenotype), given the presence of silent mutations (Table S1). By visual inspection we noticed that 98% (50/51) of the RIF-resistant isolates had mutations that would be detected by one or two beacons (Table 1). Therefore, to estimate the average Ct that represented the average amplification of the wild-type region of rpoB within a given isolate, we identified three beacons within each isolate that were not spanning a mutant region. These would be used to determine the optimal Ct cut-off that distinguishes mutants from wild-type isolates. To do this, we first calculated the average of adjusted Ct for the five beacons in a given experimental isolate (Mean of all adjusted Ct's in Table 3), and selected the three beacons that had an adjusted Ct closest to this overall mean (beacons A, C and D as example in Table 3). Then we calculated the mean of the adjusted Ct from these three beacons (mean-Ct-adjusted). Using this mean-Ct-adjusted value as a reference, we evaluated the sensitivity and specificity of the assay for the five beacons from the 105 isolates evaluated in this study by adding either 0.5, 1.0, 1.5 or 2.0 additional cycles. Receiver-operator curves (ROC) were used to establish the cut-off providing the best sensitivity and specificity for most isolates. The selected thresholds comprised the addition of 1.5 cycles for beacons A–D, and 0.5 cycles for beacon E (Table 4). The formulas for the mathematical approach were inserted into a Microsoft Excel template. Since all experiments had an identical 96-well plate set-up, once a qPCR run was finished, data were directly downloaded from the ABI7900 SDS software to the Microsoft Excel template, followed by automated calculation of Ct threshold and final classification of each isolate. This mathematical approach was 98% sensitive and 94.5% specific versus culture phenotype, and 96.2% sensitive and 100% specific versus DNA sequence (Table 2). When compared to the visual analysis, the mathematical approach only failed to detect one RIF-resistant isolate by culture and DNA sequence (MD643).
Table 4.
Summary of beacon performance for selection of cut-off Ct values
| Cycles added to Avg-Ct-adj: | Performance | Beacon sensitivity and specificity (%) |
||||
|---|---|---|---|---|---|---|
| A | B | C | D | E | ||
| 0.5 | Sensitivity | 100 | 100 | 100 | 94.7 | 95.2 |
| Specificity | 90.3 | 73.9 | 99.0 | 94.2 | 100 | |
|
| ||||||
| 1 | Sensitivity | 100 | 100 | 100 | 94.7 | 81.0 |
| Specificity | 100 | 93.5 | 100 | 100 | 100 | |
|
| ||||||
| 1.5 | Sensitivity | 100 | 100 | 100 | 94.7 | 38.1 |
| Specificity | 100 | 100 | 100 | 100 | 100 | |
|
| ||||||
| 2 | Sensitivity | 50.0 | 92.3 | 100 | 94.7 | 10 |
| Specificity | 100 | 100 | 100 | 100 | 100 | |
Light shade, best performance for each beacon; Dark shade, final selected threshold
4. Discussion
We propose an objective, simple and reader-independent mathematical approach for a molecular beacon qPCR method for detection of rpoB mutations associated with RIF-resistant as a surrogate marker of MDR-TB. The mathematical method provided 98% sensitivity and 94.5% specificity versus traditional drug susceptibility by culture. Most importantly, this method did not require reader training, which is particularly critical for interpretation of isolates with mixed populations of RIF-resistant and RIF-susceptible bacteria, which accounted for 51% of the RIF-R specimens evaluated in this study. In the process, we also showed a sensitivity of 96.2% and specificity of 100% versus DNA sequence analysis among M. tuberculosis isolates from two geographical regions not tested previously using this beacon qPCR approach.
The final strategies for visual and mathematical interpretation of mutations proposed in this study involved evaluation of several approaches. For visual analysis, we compared the amplification curves for a given beacon (e.g. beacon A) for all the isolates tested in the project to identify those that would “stand out” with a higher Ct (mutant). However, this method was only suitable for comparisons for a given operator and required at least 10 plates to determine a reliable Ct threshold. A second visual analysis was based on comparison of isolates run in a single 96-well plate to adjust for operator variability, reagents and experimental conditions. Comparison of each experimental isolate, one beacon at a time, to the corresponding beacon from the CDC1551 reference was not reliable since there were differences in DNA concentration and/or efficiency of amplification for each isolate, despite attempts to load the same amount of DNA per qPCR well. These experiments, together with the observations of the distribution of the amplification curves (Fig 1), led us to establish a visual analysis method which compared the Ct of the five beacons from a given isolate, allowing for inherent variations in the efficiency of each beacon. In our hands this approach had nearly 100% sensitivity and specificity, but the method required substantial reader experience since there were marked differences between beacon curves, particularly with mixed isolates (Table S1).
We therefore developed several mathematical approaches before designing the final model described in this study. The ability to create a “universal” Ct for each beacon based on the mean Ct from the 18 plates from CDC1551 was limited by the inherent variations in amplification of each isolate, despite attempts to load the same number of genomes per qPCR well (estimated <10% variation; data not shown). Amplification efficiency was not associated with the DNA extraction method for isolates from both sites (data not shown). We also tried designating a threshold based on the area under the amplification curve by reading Rn or delta Rn (Rn adjusted for background fluorescence and amplification in the first 15 cycles) and taking into account differences in the signal intensity of each beacon (consistently higher for A). However, not all curves had the same shape, with notable variation between isolates, and therefore, differences in total fluorescence despite similar Ct values. Beacon E was particularly problematic due to the narrow difference between a “wild-type” versus “mutant” Ct. In fact, in the process of adapting the published assay conditions for the five beacons (Lin et al., 2004; El-Hajj et al., 2001; Piatek et al., 2000), optimal efficiency of amplification of all beacons, including E, required careful titration of MgCl2 and dimethyl sulphoxide (data not shown).
Our beacon qPCR results had a similar sensitivity and specificity to that reported for MDR-TB detection of strains from different geographical regions (Lin et al., 2004; Piatek et al., 2000; Varma-Basil et al., 2004). However, previous studies had not addressed the importance of reader training and experience, particularly for interpretation of amplification curves with detectable but clearly higher Cts. This is critical for transition of research protocols to the clinical setting. These limitations are overcome by the mathematical method we propose: it was reproducible regardless of date of assay, master mix batch or different operators. This approach is suitable for high-throughput screening of MDR-TB, and only limited by the number of plates that can be run simultaneously. Our proposed mathematical model could be tailored to other software platforms, a step required for semi-automation of molecular beacon technology. The capacity of taking qPCR to this level is illustrated by the recent development of a field-method for automated RIF-resistance detection using the GeneXpert device and Xpert M. tuberculosis cartridge (Cepheid, Sunnyvale, CA)(Stop TB Partnership and World Health Organization, 2008).
The most common rpoB mutations in the literature are in codon 450 (31–67%), followed by codon 445 (7–48%), with variations between geographical regions (Ahmad et al., 2000; Ahmad and Mokaddas, 2005; Bartfai et al., 2001; Cavusoglu et al., 2002; Cooksey et al., 1997; Hirano et al., 1999; Matsiota-Bernard et al., 1998; Nikolayevskyy et al., 2007; Ozkutuk et al., 2007). Our isolates from Colombia were randomly selected and followed this frequency distribution. That was not the case for the isolates from Texas, which were selected to include isolates with mutations that spanned the regions probed by all beacons. However, in a previous study with a larger collection of isolates from the Texas-Mexico border, the distribution of mutations was similar to those from Colombia and other regions of the world (McCammon et al., 2005). Where resources are limited, but distribution of mutations known, screening assays might be limited to beacons D and E, which would detect about 50–70% of the RIF-resistant (and MDR-TB) isolates.
Even though the ultimate goal for diagnosis is to determine resistance, the quantitative nature of the molecular beacon qPCR method provides an insight into the complex biology of drug resistance. It is possible that each patient carries a mixed population of susceptible and resistant isolates, and the proportion of these varies. Detection of small numbers of resistant organisms in a mixed population exposed to RIF might be used to predict resistance not yet detectable clinically or phenotypically. Among the 51 mutant isolates evaluated in this study, 28 (55%) had a detectable Ct (beacon A, n=1; beacon B, n=6; beacon C, n=2; beacon D, n=3; beacon E, n=16). As a preliminary approach to assess the potential for quantification of molecular beacon qPCR, we mixed different ratios of RIF-resistant and RIF-susceptible bacteria for beacon B, and were able to detect and quantify as few as 30% of RIF-resistant among a total of 103 bacterial genomes per qPCR well (Table S3). We anticipate a similar quantification potential for beacons A–D, but most likely the need to modify beacon E to ensure its Ct reflects the proportion of mutants. This opens up the possibility of following the dynamics of drug resistance during the course of treatment, including prediction of treatment failure.
A potential study limitation is that we assumed that the changes in the Ct of a given isolate were attributed to the presence of bacteria with mixed resistance. Microbiological confirmation of the proportion of resistant bacteria in each isolate would be ideal, but this was not considered necessary for several reasons: First, our findings when mixing known quantities of “resistant” and “sensitive” bacteria provided the expected, proportional change in the Ct of the wild-type beacon PCR output, as observed in patient specimens. Second, it is well-recognized that drug-resistance arises from random mutations in a given bacterium that eventually multiplies to become more frequent than the drug-susceptible bacteria- this is the basis for the classical “proportions” method for assessing drug resistance of M. tuberculosis. Third, changes in the Ct were consistent with mutations detected by DNA sequence. Another limitation of our proposed beacon qPCR is the 5-nucleotide gap between beacons D and E, with failure to detect mutations in this region. In our hands the previously described beacon “E” by Piatek and collaborators was problematic (Piatek et al., 2000), and hence, the development of a beacon for this region is needed.
In summary, we propose a simple and objective, Microsoft Excel-based automated approach to standardize the identification of mutations associated with RIF-resistance using molecular beacon qPCR. This method would ease the expansion of the beacon qPCR technique from a research tool towards a standardized assay for prompt MDR-TB screening in clinical reference laboratories. The next step will be to test the mathematical approach on an extended number of isolates from different geographical regions, and across laboratories. Finally, the quantitative nature of this technique could be further developed into an assay to monitor or predict MDR-TB development.
Supplementary Material
Acknowledgments
We thank Jessica Ballou, Chris Yan, Mary Walsh and Caroline Mullin for volunteering to interpret the qPCR beacon, and Dr. David Alland for technical and scientific advice.
This project was supported by grants NIH- R21 AI 064297-01A1 and by the Hispanic Health Research Center EXPORT grant NIHMHD P20 MD000170 020.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahmad S, Araj GF, Akbar PK, Fares E, Chugh TD, Mustafa AS. Characterization of rpoB mutations in rifampin-resistant Mycobacterium tuberculosis isolates from the Middle East. Diagn Microbiol Infect Dis. 2000;38:227–232. doi: 10.1016/s0732-8893(00)00200-5. [DOI] [PubMed] [Google Scholar]
- Ahmad S, Mokaddas E. The occurrence of rare rpoB mutations in rifampicin-resistant clinical Mycobacterium tuberculosis isolates from Kuwait. Int J Antimicrob Agents. 2005;26:205–212. doi: 10.1016/j.ijantimicag.2005.06.009. [DOI] [PubMed] [Google Scholar]
- Bartfai Z, Somoskovi A, Kodmon C, Szabo N, Puskas E, Kosztolanyi L, Farago E, Mester J, Parsons LM, Salfinger M. Molecular characterization of rifampin-resistant isolates of Mycobacterium tuberculosis from Hungary by DNA sequencing and the line probe assay. J Clin Microbiol. 2001;39:3736–3739. doi: 10.1128/JCM.39.10.3736-3739.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavusoglu C, Hilmioglu S, Guneri S, Bilgic A. Characterization of rpoB mutations in rifampin-resistant clinical isolates of Mycobacterium tuberculosis from Turkey by DNA sequencing and line probe assay. J Clin Microbiol. 2002;40:4435–4438. doi: 10.1128/JCM.40.12.4435-4438.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooksey RC, Morlock GP, Glickman S, Crawford JT. Evaluation of a line probe assay kit for characterization of rpoB mutations in rifampin-resistant Mycobacterium tuberculosis isolates from New York City. J Clin Microbiol. 1997;35:1281–1283. doi: 10.1128/jcm.35.5.1281-1283.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eickbush TH, Moudrianakis EN. The compaction of DNA helices into either continuous supercoils or folded-fiber rods and toroids. Cell. 1978;13:295–306. doi: 10.1016/0092-8674(78)90198-8. [DOI] [PubMed] [Google Scholar]
- El-Hajj HH, Marras SA, Tyagi S, Kramer FR, Alland D. Detection of rifampin resistance in Mycobacterium tuberculosis in a single tube with molecular beacons. J Clin Microbiol. 2001;39:4131–4137. doi: 10.1128/JCM.39.11.4131-4137.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heid CA, Stevens J, Livak KJ, Williams PM. Real time quantitative PCR. Genome Res. 1996;6:986–994. doi: 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- Hirano K, Abe C, Takahashi M. Mutations in the rpoB gene of rifampin-resistant Mycobacterium tuberculosis strains isolated mostly in Asian countries and their rapid detection by line probe assay. J Clin Microbiol. 1999;37:2663–2666. doi: 10.1128/jcm.37.8.2663-2666.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoek KG, Gey van Pittius NC, Moolman-Smook H, Carelse-Tofa K, Jordaan A, van der Spuy GD, Streicher E, Victor TC, van Helden PD, Warren RM. Fluorometric assay for testing rifampin susceptibility of Mycobacterium tuberculosis complex. J Clin Microbiol. 2008;46:1369–1373. doi: 10.1128/JCM.02343-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jureen P, Engstrand L, Eriksson S, Alderborn A, Krabbe M, Hoffner SE. Rapid detection of rifampin resistance in Mycobacterium tuberculosis by Pyrosequencing technology. J Clin Microbiol. 2006;44:1925–1929. doi: 10.1128/JCM.02210-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent PT, Kubica GP. Public Health Mycobacteriology: A Guide for the Level III Laboratory. Center for Disease Control and Prevention; Atlanta, GA: 1985. [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lin S-YG, Probert W, Lo M, Desmond E. Rapid detection of isoniazid and rifampin resistance mutations in Mycobacterium tuberculosis complex from cultures or smear-positive sputa by use of molecular beacons. J Clin Microbiol. 2004;42:4204–4208. doi: 10.1128/JCM.42.9.4204-4208.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsiota-Bernard P, Vrioni G, Marinis E. Characterization of rpoB mutations in rifampin-resistant clinical Mycobacterium tuberculosis isolates from Greece. J Clin Microbiol. 1998;36:20–23. doi: 10.1128/jcm.36.1.20-23.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCammon MT, Gillette JS, Thomas DP, Ramaswamy SV, Graviss EA, Kreiswirth BN, Vijg J, Quitugua TN. Detection of rpoB mutations associated with rifampin resistance in Mycobacterium tuberculosis using denaturing gradient gel electrophoresis. Antimicrob Agents Chemother. 2005;49:2200–2209. doi: 10.1128/AAC.49.6.2200-2209.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolayevskyy VV, Brown TJ, Bazhora YI, Asmolov AA, Balabanova YM, Drobniewski FA. Molecular epidemiology and prevalence of mutations conferring rifampicin and isoniazid resistance in Mycobacterium tuberculosis strains from the southern Ukraine. Clin Microbiol Infect. 2007;13:129–138. doi: 10.1111/j.1469-0691.2006.01583.x. [DOI] [PubMed] [Google Scholar]
- Ozkutuk N, Gazi H, Surucuoglu S, Gunduz A, Ozbakkaloglu B. Characterization of rpoB mutations by line probe assay in rifampicin-resistant mycobacterium tuberculosis clinical isolates from the aegean region in Turkey. Jpn J Infect Dis. 2007;60:211–213. [PubMed] [Google Scholar]
- Palomino JC. Newer diagnostics for tuberculosis and multi-drug resistant tuberculosis. Curr Opin Pulm Med. 2006;12:172–178. doi: 10.1097/01.mcp.0000219265.50310.9b. [DOI] [PubMed] [Google Scholar]
- Piatek AS, Telenti A, Murray MR, El-Hajj H, Jacobs WR, Kramer FR, Alland D. Genotypic analysis of Mycobacterium tuberculosis in two distinct populations using molecular beacons: implications for rapid susceptibility testing. Antimicrob Agents Chemother. 2000;44:103–110. doi: 10.1128/aac.44.1.103-110.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piatek AS, Tyagi S, Pol AC, Telenti A, Miller LP, Kramer FR, Alland D. Molecular beacon sequence analysis for detecting drug resistance in Mycobacterium tuberculosis. Nat Biotechnol. 1998;16:359–363. doi: 10.1038/nbt0498-359. [DOI] [PubMed] [Google Scholar]
- Quitugua TN, Seaworth BJ, Weis SE, Taylor JP, Gillette JS, Rosas II, Jost KC, Jr, Magee DM, Jr., Cox RA. Transmission of drug-resistant tuberculosis in Texas and Mexico. J Clin Microbiol. 2002;40:2716–2724. doi: 10.1128/JCM.40.8.2716-2724.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robledo J, Mejia GI, Paniagua L, Martin A, Guzman A. Rapid detection of rifampicin and isoniazid resistance in Mycobacterium tuberculosis by the direct thin-layer agar method. Int J Tuberc Lung Dis. 2008;12:1482–1484. [PubMed] [Google Scholar]
- Somoskovi A, Parsons LM, Salfinger M. The molecular basis of resistance to isoniazid, rifampin, and pyrazinamide in Mycobacterium tuberculosis. Respir Res. 2001;2:164–168. doi: 10.1186/rr54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stop TB Partnership and World Health Organization . The Global Plan to Stop TB, 2006–2015. Stop TB Partnership; World Health Organization; Geneva: 2006. www.stoptb.org/globalplan WHO/HTM/STB/2006.35. [Google Scholar]
- Stop TB Partnership and World Health Organization . New Laboratory Diagnostic Tools for Tuberculosis Control. 2008. www.stoptb.org/.../New%20tools%20for%20TB%20control%20-%20filling%20gaps,%20overcoming%20barriers.pdf WHO/HTM/STB/2008.49. [Google Scholar]
- Tyagi S, Kramer FR. Molecular beacons: probes that fluoresce upon hybridization. Nat Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- van Soolingen D, Hermans PW, de Haas PE, Soll DR, van Embden JD. Occurrence and stability of insertion sequences in Mycobacterium tuberculosis complex strains: evaluation of an insertion sequence-dependent DNA polymorphism as a tool in the epidemiology of tuberculosis. J Clin Microbiol. 1991;29:2578–2586. doi: 10.1128/jcm.29.11.2578-2586.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varma-Basil M, El-Hajj H, Colangeli R, Hazbon MH, Kumar S, Bose M, Bobadilla-del-Valle M, Garcia LG, Hernandez A, Kramer FR, Osornio JS, Ponce-De-Leon A, Alland D. Rapid detection of rifampin resistance in Mycobacterium tuberculosis isolates from India and Mexico by a molecular beacon assay. J Clin Microbiol. 2004;42:5512–5516. doi: 10.1128/JCM.42.12.5512-5516.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization Global Tuberculosis Control: surveillance, planning, financing. 2009 http://www.who.int/tb/publications/global_report/2009/en/index.html. 1-1-2003.
- World Health Organization Anti-tuberculosis drug resistance in the world. Fourth Global Report. 2008 www.who.int/tb/publications/2008/drs_report4_26feb08.pdf WHO/HTM/TB/2008.394.
- World Health Organization . Global tuberculosis control : epidemiology, strategy, financing : WHO report 2009. World Health Organization; Geneva: 2009. http://www.who.int/tb/publications/global_report/2009/en/index.html WHO/HTM/TB/2009.411. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.