Abstract
Transcriptional profiling of two isogenic models of transformation identifies a gene signature linking cancer with inflammatory and metabolic diseases. In accord with this common transcriptional program, many drugs used for treatment of diabetes and cardiovascular diseases inhibit transformation and tumor growth. Unexpectedly, lipid metabolism genes are important for transformation and are up-regulated in cancer tissues. As in atherosclerosis, oxidized LDL and its receptor OLR1 activate the inflammatory pathway through NF-κB, leading to transformation. OLR1 is important for maintaining the transformed state in developmentally diverse cancer cell lines and for tumor growth, suggesting a molecular connection between cancer and atherosclerosis. We suggest that the interplay between this common transcriptional program and cell-type-specific factors gives rise to phenotypically disparate human diseases.
Keywords: cancer, metabolic syndrome, inflammation, gene networks, cellular transformation
INTRODUCTION
Clinical and epidemiological studies have linked cancer and other chronic medical conditions. For example, patients diagnosed with metabolic syndrome, inflammatory diseases, and auto-immune conditions show increased incidence and aggressiveness of tumor formation (Giovannucci, 2007; Manatovani et al., 2008; Pischon et al., 2008). Conversely, diabetics treated with metformin to lower insulin levels have reduced levels of cancer in comparison to untreated individuals (Hsu et al., 2007; Larsson et al., 2007). Smoking is linked not only to lung cancer, but also to cardiovascular and other diseases. In general, the molecular bases of these links among diseases are poorly understood.
Inflammation is commonly associated with cancer formation and progression, and it is estimated that 15–20% of all cancer related deaths can be attributed to inflammation and underlying infections (Manatovani et al., 2008). Inflammatory molecules are elevated in many forms of cancer, and they provide growth signals that promote the proliferation of malignant cells (Balkwill and Mantovani, 2001; Karin, 2006; De Marzo et al., 2007; Naugler and Karin, 2008; Pierce et al., 2009). Constitutively active NF-κB, the key transcription factor that mediates the inflammatory response, occurs in many types of cancer, and mouse models provide evidence for a causative role of NF- κB in malignant conversion and progression (Luedde et al., 2007; Naugler and Karin, 2008; Sakurai et al., 2008).
Increased cancer risk is also associated with obesity, type II diabetes, high cholesterol, and atherosclerosis, which are components of a disease state known as metabolic syndrome. Mechanistically, the link between metabolic diseases and cancer is less understood than the connection to inflammation. However, a pathway consisting of AMP-activated protein kinase, an energy sensor (Hardie, 2008), Akt, and PI3 kinase plays a critical role in diabetes and other metabolic diseases, and AMPK activation requires LKB1, a protein kinase that is a tumor suppressor associated with Peutz-Jeghers syndrome (Shaw et al., 2004; Shaw et al., 2005). In addition, fatty acid synthase also plays an important role in cancer pathogenesis, and inhibitors against this enzyme are being tested as anti-cancer drugs (Kuhajda, 2006; Menendez and Lupu, 2007).
Transcriptional profiling has been a common way to identify genes and signaling pathways important for carcinogenesis. However, the various approaches have limitations with regard to identifying genes relevant to cancer from those affected for unrelated reasons. Clinical samples derive from a mixed pool of patients with different clinical characteristics and different cancer subtypes. Additionally, transcriptional profiles of primary tumor samples are often not compared to normal matched samples from the same patient that may be unavailable. Even when normal matched samples are available, they are usually derived from cells next to the tumor and may be affected by the tumor microenvironment and/or may have some of the genetic alterations as the tumor. Studies utilizing cell lines rarely involve isogenically matched normal and transformed cell lines, and we are unaware of a time-course analysis of the cellular transformation process. The current study uses a different approach to identify genes involved in cellular transformation and carcinogenesis by performing transcriptional profiling in two isogenic models of cellular transformation.
RESULTS
Identification of a cancer gene signature from expression profiling of two isogenic models of cellular transformation
To identify genes differentially regulated during the process of cellular transformation, we used two isogenic cellular models (Figure 1A) derived from different tissue types; i.e. for each model, the non-transformed and transformed states are genetically identical. One model involves normal mammary epithelial cells (MCF-10A)(Soule et al., 1990) containing ER-Src, a derivative of the Src kinase oncoprotein (v-Src) that is fused to the ligand-binding domain of the estrogen receptor. Treatment of such cells with tamoxifen rapidly induces Src, and morphological transformation is observed within 24–36 hours (Hirsch et al., 2009; Iliopoulos et al., 2009), thereby making it possible to kinetically follow the transition between normal and transformed cells. Transformation of these cells results in colony formation in soft agar, foci formation rather than contact inhibition in monolayers, formation of mammospheres in suspension, and tumors in nude mice (Hirsch et al., 2009; Iliopoulos et al., 2009).
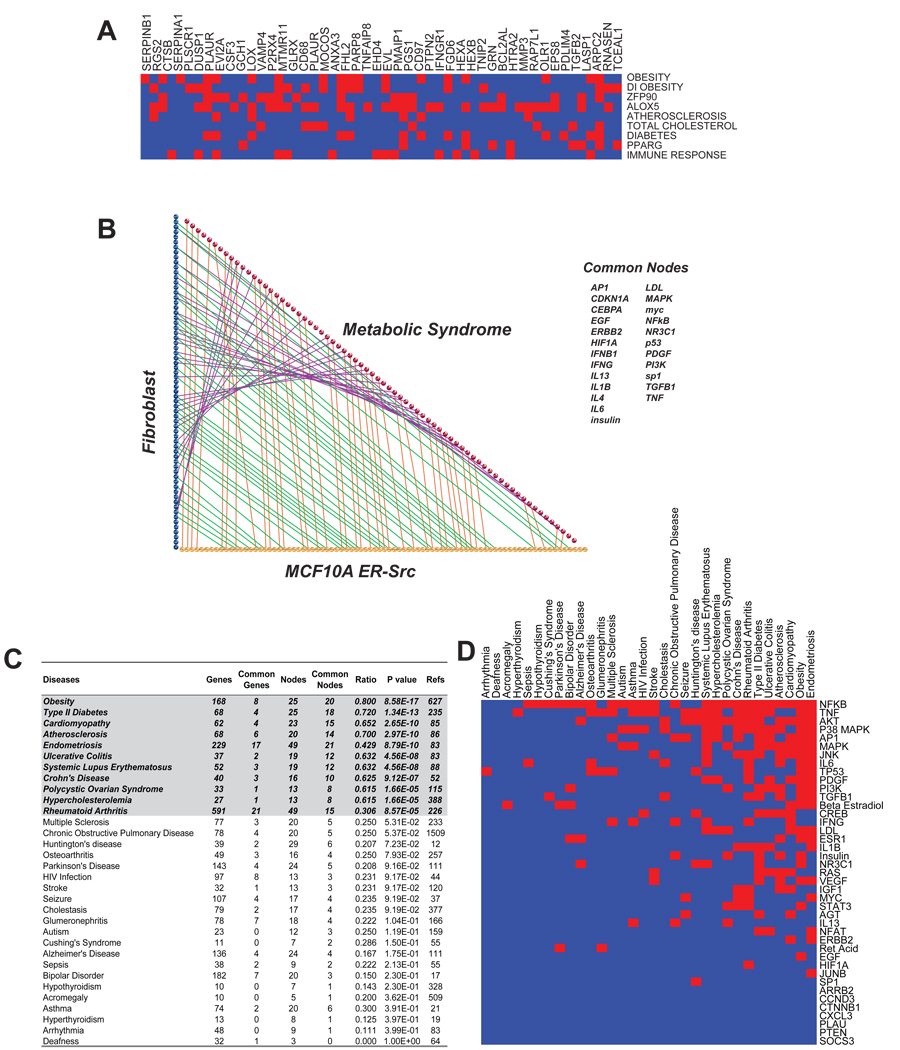
Figure 1. A 343-gene signature of cellular transformation.
(A) Phase contrast images (scale bars: 10µm) of morphology of non-transformed and transformed cells. MCF10A cells are transformed after Src induction by tamoxifen (TAM) treatment. BJ fibroblasts have stable integration of one (hTERT) or two (hTERT, SV40E) or three (hTERT, SV40E, HRAS-V12) genetic elements.
(B) Differentially expressed genes during MCF10A cell transformation at the indicated time points after TAM treatment using different (0.5, 1, 2 fold) log cut-offs.
(C) Differentially expressed genes in BJ fibroblasts were analyzed as two way comparisons in all combinations for the three cell types using different (0.5, 1, 2, 4-fold) log cut-offs.
(D) The 343-gene signature of cellular transformation defined by the overlap of differentially expressed genes in the two isogenic models.
(E) Transcription factor families whose DNA-binding motifs are enriched in the 2 kb or 10 kb regions flanking the transcriptional start site of up- or down-regulated genes within the indicated biofunctions and diseases.
(F) Relationship between the 343 gene set and the indicated diseases. The overlap counts and p-values are indicated.
The other model consists of three isogenic cell lines derived from primary fibroblasts in a serial manner (Hahn et al., 1999)(Figure 1A). The first is immortalized by overexpression of telomerase (hTERT), and exhibits normal fibroblast morphology. The second expresses hTERT along with both large and small T antigens of Simian virus 40, and it displays an altered morphology but is not transformed. The third cell line expresses hTERT, T antigens, and an oncogenic derivative of Ras (H-RasV12); it is morphologically transformed and has tumorigenic potential in soft agar and nude mice. These two isogenic models of cellular transformation differ with respect to cell type, oncogene, and mode of oncogenic transition (time course or staged cell lines), and hence should permit the identification of a gene signature common to the process of cellular transformation.
Using microarrays capable of assaying most protein-coding mRNAs, we performed transcriptional profiling of the transformation process in MCF-10A cells (8 time points from 1–36 hours after tamoxifen treatment) and in the three fibroblast cell lines. At a 1% false discovery rate by SAM analysis (Tusher et al., 2001), we identified 1201 genes differentially expressed at any time point in the ER-Src cells (Figure 1B; Supplemental Table 1) and 3208 genes (Figure 1C; Supplemental Tables 2) in any of the two-way comparisons of the fibroblast cell lines. In the ER-Src model, few genes were differentially expressed in the first time point (1h), more than 100 genes 4 hours post treatment and more than 700 genes 36 hours post induction. Interestingly, in the fibroblast model, most changes in gene expression are due to T antigens, not Ras, mirroring the morphology patterns but not transformation per se (Figure 1A), and suggesting that a viral oncogene can deregulate multiple pathways to create a pre-malignant phenotype.
The availability of transcriptional profiles for different isogenic models of cellular transformation makes it possible to distinguish between genes that play a relatively general role in transformation as opposed to those affected only by the specific experimental model. We therefore analyzed the combined datasets for differentially regulated genes that were either up-regulated or down-regulated in both experimental models. We define a gene signature of cellular transformation that contains 343 differentially regulated genes of which 238 are up-regulated and 105 are down-regulated (Figure 1D; Supplemental Table 3). This signature of differentially expressed genes, hereafter termed the cancer gene signature, will be the basis of bioinformatic approaches described below.
Identification of transcription factors linked to cellular transformation
The cancer gene signature is defined by coordinate regulation of gene expression during the process of cellular transformation, and such regulation must involve DNA-binding transcription factors. To identify such transcription factors, we used a computational approach (Warner et al., 2008) that asks whether DNA sequence motifs defined by comprehensive analysis of protein-DNA binding specificity in vitro are statistically overrepresented in candidate gene sets. We considered 2 kb or 10 kb (Supplemental Table 4) of sequence flanking each side of the transcriptional start site for genes included in the cancer gene signature as well as subsets of genes comprising specific biofunction categories. This analysis indicates that several families of transcription factors play a role in transformation including Myc/Max, IRF, Ets, Sox, Hox, Myb, KLF, GATA and Pou (Figure 1E). Interestingly, factors such as the SOX and Pou family members have been implicated in formation of cancer stem cells (Ben-Porath et al., 2008).
The gene signature of cellular transformation is strongly linked to diverse human cancers
To validate our combined experimental system of cellular transformation as a model of human cancer, we used two bioinformatic approaches. First, we performed intensive literature mining to correlate our gene signature with common 18 cancer types. We found that 208/343 genes are correlated with at least one cancer type (Supplemental Table 5), with a range of 22–88 genes correlated with each cancer type (Supplemental Figure 1). K-means clustering reveals 50 genes involved in most cancer types, including STAT3, IL1β, SOCS3, VEGF, HIF1α and TGFβ1, which play a significant role in inflammation. Second, we compared our cancer gene signature with array-based transcriptional profiles of specific cancer types. As shown in Figure 1F, the 343 common gene set significantly overlaps inflammatory (breast (Lerebours et al., 2008) and gastrointestinal (Ellmark et al., 2006)) and metabolism-related (thyroid (Delys et al., 2007) and pancreatic (Logsdon et al., 2003)) cancers (Supplemental Table 6). Thus, the gene signature defined by our isogenic models of cellular transformation is very strongly linked to diverse human cancers, thereby validating these experimental systems as models of oncogenesis.
Linkage of the cancer gene signature to metabolic diseases, including obesity, diabetes, and atherosclerosis
Using Ingenuity Pathway Analysis, we identified three groups of biofunctions and diseases that are significantly correlated (p < 10−5) with the cancer gene signature (Supplemental Table 7). The first group includes cancer-related biofunctions such as cellular growth and proliferation, cell cycle and cell death. The second group contains genes involved in inflammation and immune system function, as well as genes linked to inflammatory and gastrointestinal diseases. Many of these inflammatory genes have been linked to cancer in the literature, in accord with the multiple connections between cancer and inflammation. The third group of biofunctions includes lipid metabolism, metabolic disease, cardiovascular disease, and gastrointestinal disease. While certain aspects of metabolic disease have been linked to cancer through the Akt pathway, the identification of multiple genes involved in lipid metabolism was unexpected. For example, OLR1, SREBP-1, SNAP23, and VAMP4 are well described in studies of lipid metabolism, cholesterol biosynthesis, and atherosclerosis, but have not been discussed in terms of cancer.
As an independent method to confirm the linkage between cancer and metabolic diseases, we compared the cancer gene signature identified here to microarray transcriptional profiling studies from diseased individuals. There is significant overlap between the cancer gene signature and expression signatures found in obesity (Lee et al., 2005), atherosclerosis (Sluimer et al., 2007; Skogsberg et al., 2008) and metabolic syndrome (Chen et al., 2008) (Supplemental Table 7). In addition, among a group of 54 genes, we observe a significant correlation between cancer and various metabolic conditions (Figure 2A).
Figure 2. Linkage of the cancer gene signature to inflammatory and metabolic diseases.
(A) Heat-map representation of the 54 common genes between cancer and sub-categories of metabolic syndrome gene set.
(B) Common central nodes between gene networks derived from differentially expressed genes from MCF10A, BJ fibroblast and Metabolic Syndrome gene sets. Each sphere represents a central node of a gene network. The 24 common nodes are listed.
(C) Comparison of genes and central nodes between the cancer gene signature and gene sets of 32 diseases derived from literature (number of references indicated). For each disease, the number of genes and nodes are indicated along with the overlap with the cancer gene signature and the p-value for the significance of the overlap.
(D) Heat map representation of the relationship between common nodes of cellular transformation and the indicated diseases, with significant relationships indicated in red..
Gene network analysis identifies central players that link cellular transformation to metabolic diseases
Although transcriptional profiling is useful for uncovering common regulatory networks between disease states, it cannot account for the contribution of protein-protein interactions, post-translational modification, DNA-binding events, and subcellular localization. To address this issue, we organized the set of differentially expressed genes into networks with central nodes via Ingenuity Pathways analysis. A three-way comparison between networks derived from our 2 experimental datasets and from a gene set describing metabolic syndrome reveals a high overlap between the central nodes of cancer and metabolic syndrome (Figure 2B). Specifically, we identified 24 common central nodes, including inflammation-related nodes such as IFNγ, IL1β, IL6 and NF-κB, suggesting that inflammatory processes are important factors for both cancer and metabolic diseases. In addition, insulin and LDL appear as central nodes in cancer gene networks, suggesting the importance of protein and lipid metabolism in cellular transformation.
To address the relationship between our cancer gene signature and human diseases that lack transcriptional profiling data, we compiled gene sets based on literature-curated data, and organized them into networks to identify central nodes (Supplemental Table 8). Interestingly, 10 out of 32 diseases were highly correlated with cancer at the gene network level (Figure 2C, D and Supplemental Table 8). These diseases can be grouped as metabolic disorders (obesity, type II diabetes, atherosclerosis, hypercholesterolemia, polycystic ovarian syndrome) and auto-immune disorders (ulcerative colitis, Crohn’s disease, SLE).
Genes identified as central nodes are important for cellular transformation
We validated the functional importance of these central nodes for cellular transformation by a number of approaches: modulation of function through drugs; modulation of expression (siRNA or overexpression); modulation of cytokine levels (antibodies or addition of exogenous cytokines). We chose experimental conditions where the treatment did not significantly affect the growth of non-transformed cells. Tamoxifen- or non-tamoxifen-treated cells subjected to these perturbations were examined for cellular transformation either by morphology or by focus formation. Any perturbed node that decreased transformation of tamoxifen-treated cells or increased transformation in non-tamoxifen-treated cells by at least 25% was considered to have a contributing role to the transformation process (summarized in Supplemental Table 9. Importantly, all 23 nodes tested affected cellular transformation when their function was altered.
Drugs designed for treatment of metabolic diseases inhibit cellular transformation and tumor growth
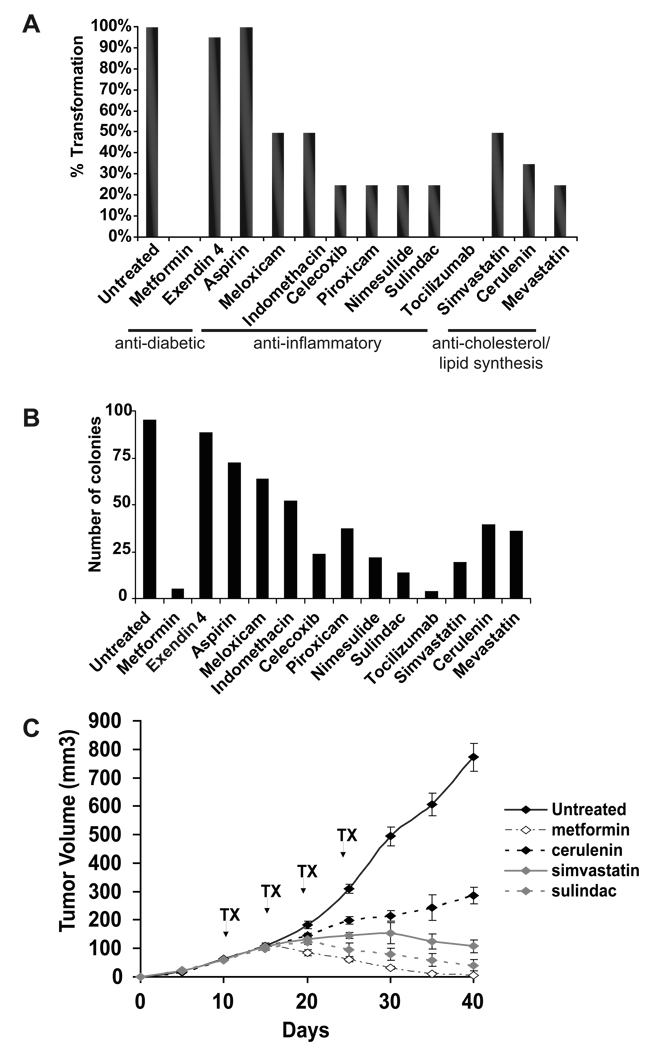
The molecular similarities among the various diseases predicts that drugs that designed or used for treatment of one disease may also help treat other diseases or in this case cancer. We therefore tested drugs that are used therapeutically for different diseases for their ability to inhibit oncogenic transformation and colony formation when plated in soft agar. Interestingly, 11 out of 13 drugs tested inhibit morphological transformation (Figure 3A), and 12 out of 13 drugs tested suppress colony formation (Figure 3B) to varying extents. The concentrations of the drugs used in these experiments do not significantly affect cell growth (Supplementary Figure 2), indicating that their effects on transformation and colony formation are not due to general cytotoxicity.
Figure 3. Many drugs used to treat non-cancer diseases block cellular transformation.
(A) Percentage of transformed cells (morphology assay) observed by treating TAM-induced MCF10 ER-Src cells with the indicated drugs.
(B) Soft agar colony assay of the effect of the indicated drugs on transformation.
(C) Tumor growth (mean ± SD) of ER-Src cells after 4 cycles of i.p treatments with the indicated drugs.
As metformin, sulindac, tocilizumab, simvastatin and cerulenin show the strongest effects on cellular transformation and tumorigenicity, we tested their ability to inhibit tumor growth in nude mice. To mimic conditions when patients are diagnosed with a tumor, we treated tumors that arose 10 days after injection of transformed ER-Src cells. Drugs were delivered by 5 cycles of intraperitoneal injections near the tumors every fifth day. Tumor growth is completely suppressed by metformin and sulindac and significantly delayed by cerulenin and simvastatin (Figure 3C). The effect of metformin is much stronger than we observed previously (Hirsch et al., 2009), presumably because drug concentrations employed here are 8-fold higher and treatment was for 5 cycles instead of 3. Thus, drugs designed to combat metabolic diseases can preferentially inhibit transformed cells, and hence may be useful in treating some types of cancer.
OLR1 (oxidized LDL receptor) and other lipid metabolic genes are important for cellular transformation
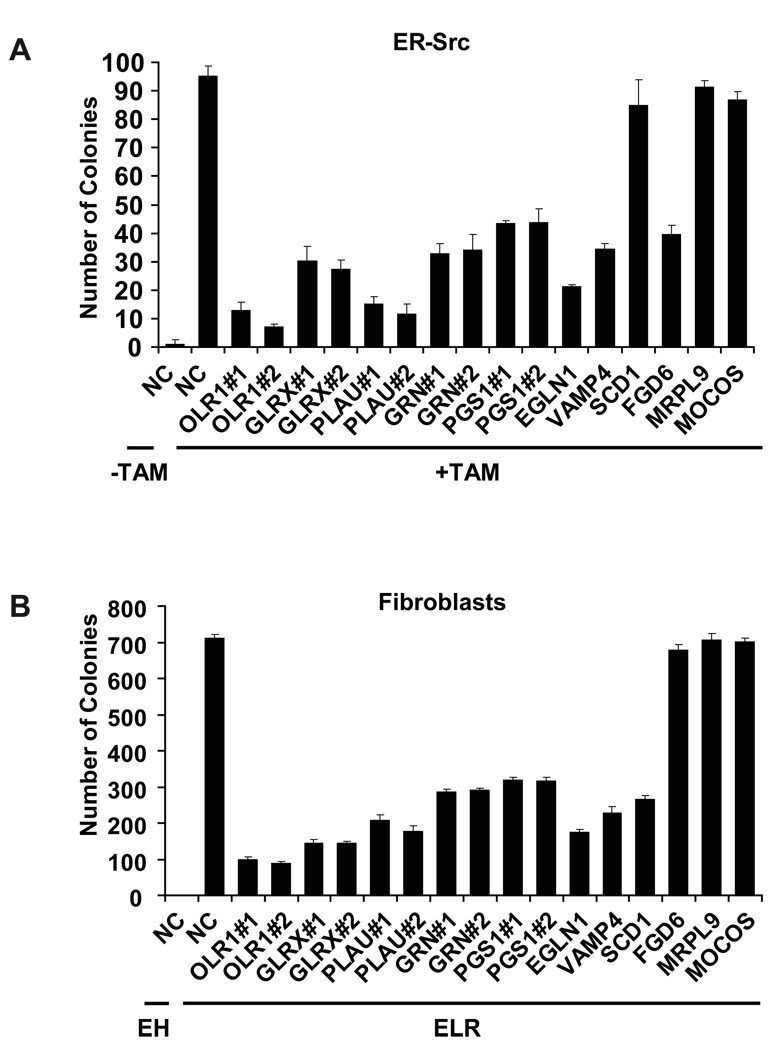
As mentioned above, the cancer gene signature includes a number of metabolic genes not previously linked to cancer. We used siRNA inhibition to examine the role of 11 such metabolic genes in cellular transformation. Efficient depletion of OLR1, GLRX, SNAP23, EGLN1, VAMP4, GRN and PGS1 (in most cases by two different siRNAs; Supplementary Figure 3) significantly reduced anchorage-independent growth in soft agar in both epithelial and fibroblast transformation models (Figure 4), but had no effect on growth of non-transformed MCF10A cells (Supplemental Figure 3C). Depletion of SCD1 reduced colony formation in the fibroblast model, depletion of FGD6 reduced colony formation in the MCF10A epithelial model, and depletion of MRLP9 and MOCOS had no effect. Thus, 9 out of 11 metabolic genes not previously linked to cancer tested are important for cellular transformation, and 7 of these are involved in both experimental models.
Figure 4. Metabolic genes affect the tumorigenicity of transformed cells.
(A) Number of colonies in soft agar (mean ± SD) of untreated and TAM-treated MCF10A ER-Src cells 24h post transfection with siRNAs against the indicated genes (NC indicates negative control siRNA). Number of colonies are presented as the mean ± SD of three experiments.
(B) Soft agar colony or foci assay in non-transformed (EH) and transformed (ELR) BJ fibroblasts 24h post transfection with siRNAs against the indicated genes (mean ± SD).
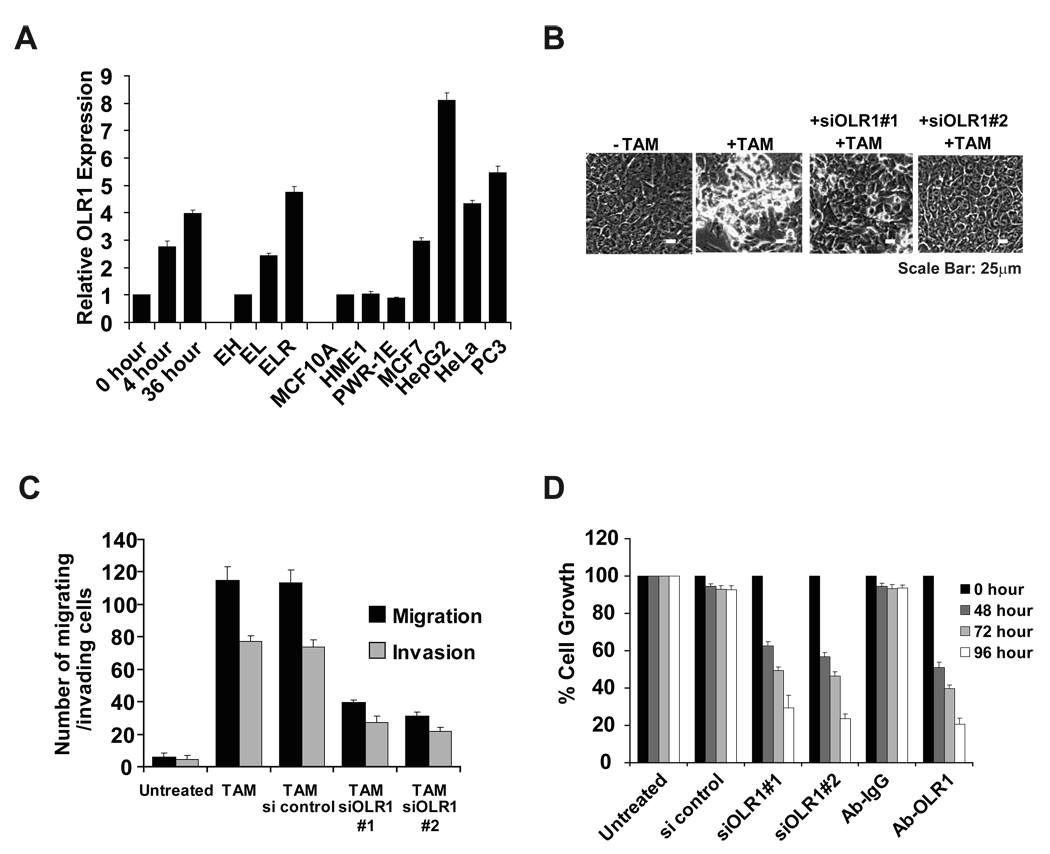
OLR1, SNAP23, VAMP4 and SCD1 are lipid-related genes, and LDL is a common hub between cancer and metabolic gene networks, suggesting the importance of lipid metabolism during cellular transformation. As OLR1 (oxidized LDL receptor) expression is induced in a transformation-dependent (Figure 5A) and depletion of OLR1 shows the strongest effects on anchorage-independent growth in both models, we further examined the role of OLR1 in the MCF-10A model. Depletion of OLR1 by two different siRNAs blocks morphological transformation (Figure 5B) and it inhibits cell motility (migration, invasion, and wound-healing assays; Figure 5C and Supplemental Figure 4). In addition, cell growth of transformed cells (treated with TAM for 48h) is blocked by inhibition of OLR1 expression via siRNAs or monoclonal antibody (Figure 5D). Thus, OLR1 and lipid metabolism are important for cellular transformation and maintenance of the transformed state.
Figure 5. OLR1 regulates transformation, cell growth and motility.
(A) OLR1 mRNA expression levels (mean ± SD) in untreated and TAM-treated MCF10A ER-Src cells and the EH, EL and ELR BJ fibroblasts.
(B) Representative phase-contrast images (scale bars: 25µm) of MCF10A ER-Src cells that were or were not treated with TAM together with two different siRNAs against OLR1.
(C) Migration and invasion assays in untreated and TAM-treated MCF10A ER-Src cells in the presence or absence of control or OLR1 siRNAs. For all panels, the data are presented as mean ± SD.
(D) Cell growth of MCF10A ER-Src TAM-treated cells (mean ± SD) after treatment with control or OLR1 siRNAs or with an OLR1 antibody and an IgG isotype control relative to untreated cells.
OLR1 is important for maintaining the transformed state in cell lines of diverse developmental origin
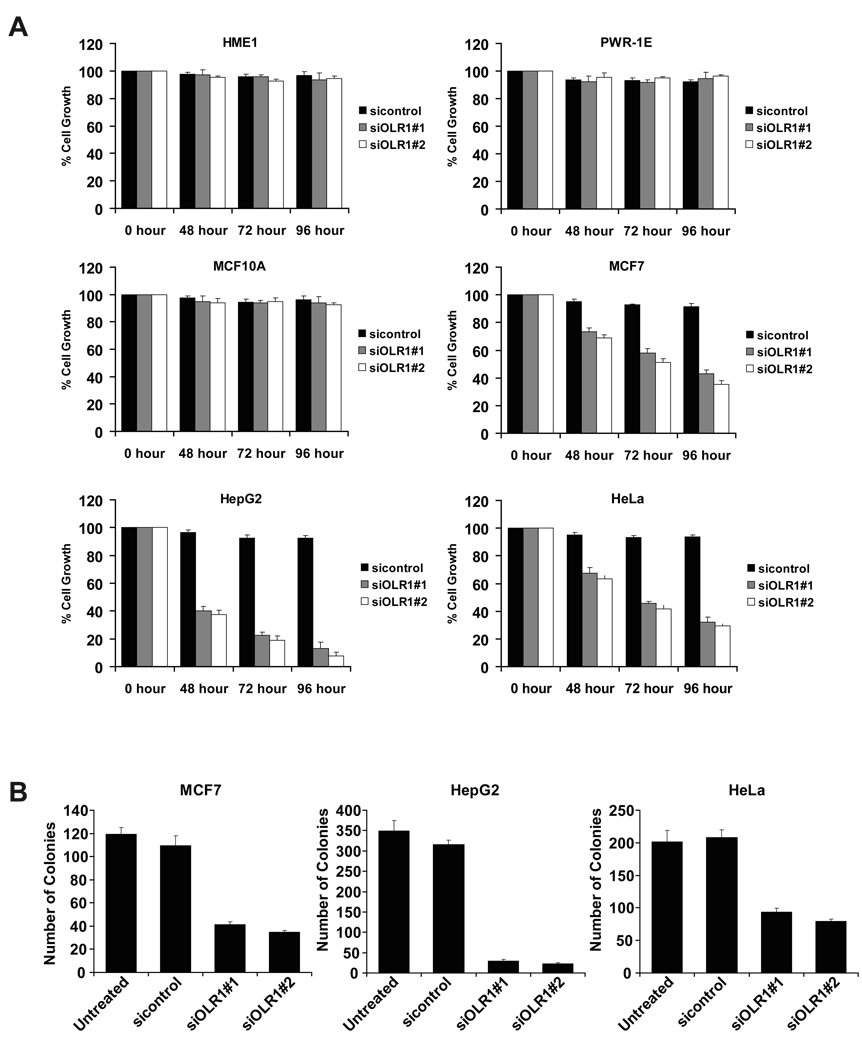
To generalize these results, we examined the role of OLR1 in developmentally diverse normal and cancer cell lines. siRNA-mediated inhibition of OLR1 expression does not affect the growth of non-transformed breast (MCF10A and HME1) or prostate (PWR-1E) cells. In contrast, inhibition of OLR1 suppresses growth of MCF7 (breast cancer), HepG2 (hepatocellular carcinoma), and HeLa (cervical cancer) cells (Figure 6A). In addition, inhibition of OLR1 reduces tumorigenicity (Figure 6B) of all three cancer cell lines. Thus, OLR1 is important for maintenance of the transformed state in a variety of developmentally unrelated cancer cell lines.
Figure 6. OLR1 regulates cell growth and tumorigenicity of cancer cells.
(A) Cell growth of normal (MCF10A, HME1, PWR-1E) and cancer (MCF7, HepG2, HeLa) cells after treatment with control or OLR1 siRNAs. Data are presented as mean ± SD.
(B) Soft agar colony assays for the cancer cell lines. The data are presented as mean ± SD.
OLR1-mediated activation of inflammatory and hypoxic pathways through NF-κB is important for cellular transformation
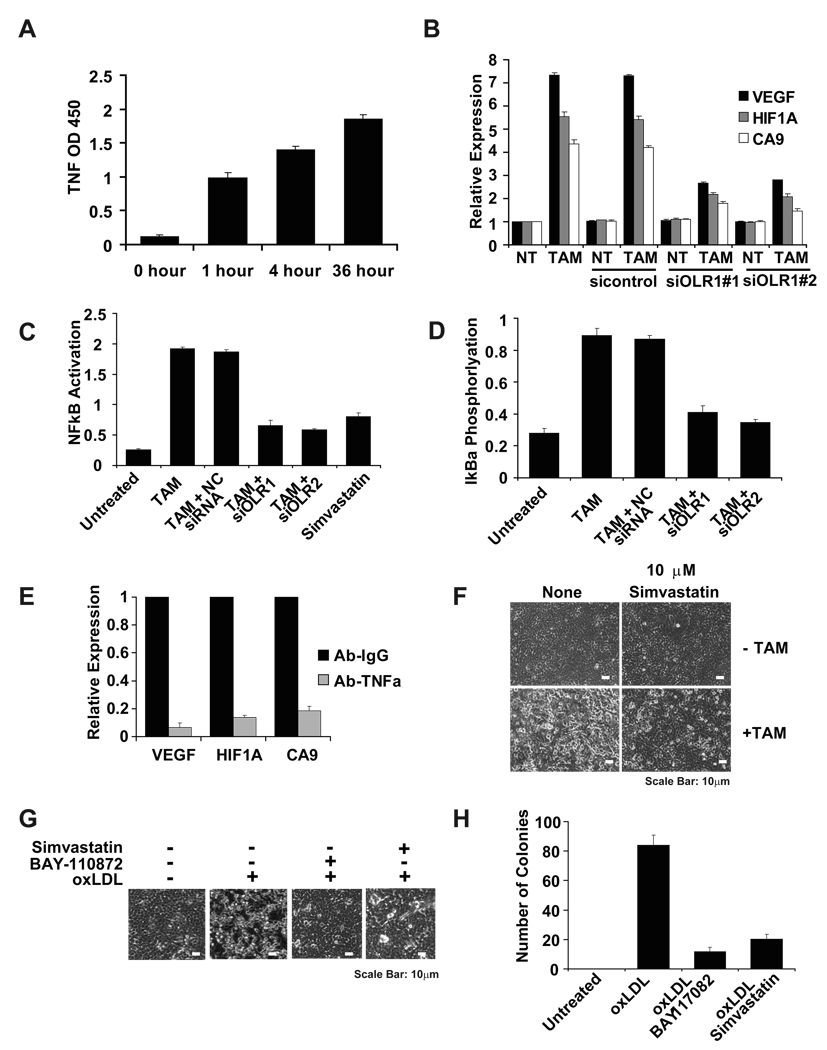
OLR1 is activated in response to oxidized LDL, angiotensin II, TNFα and other stress stimuli (Mehta et al., 2003), and TNFα production is increased very early (1–4h) during ER-Src transformation (Figure 7A). OLR1 is a marker of atherosclerosis, and it activates inflammatory and hypoxia pathways in vascular endothelial cells and macrophages. Similarly, genes involved in inflammation (e.g. IL1β, IL6, and IL8) and hypoxia (HIF1α, VEGF, CA9) are induced during the process of cellular transformation in the MCF-10A model. In accord with these observations, inhibition of OLR1 reduces the level of the inflammatory- and hypoxia-regulated genes (Figure 7B), and it also reduces NF-κB activation (Figure 7C) through inhibition of IκBα phosphorylation (Figure 7D). Furthermore, TNFα inhibition blocks the activation of HIF1α, VEGF, CA9, the downstream targets of OLR1 (Figure 7E). In addition, simvastatin, a lipid-lowering drug that inhibits OLR1 expression in endothelial cells, strongly inhibits cellular transformation (Figure 7F) in a manner associated with inhibition of NF-κB activity, but it does not affect growth of non-transformed MCF10A cells (data not shown). Conversely, treatment of MCF-10A cells with a low dose of oxidized LDL (oxLDL) induces cellular transformation and colony formation (Figure 7G, H and Supplemental Figure 5), with 40–50% transformation observed after 72 h and 85% observed after 120 h. Transformation mediated by oxLDL requires NF-κB, because inhibition of NF-kB (BAY-117082 treatment) blocks transformation (Figure 7G, H). Taken together, these observations suggest that OLR1 regulates the inflammatory and hypoxia responses during transformation in the MCF-10A model and that TNFα is a ligand for OLR1 activation.
Figure 7. OLR1 regulates transformation through NF-κB pathway.
(A) TNFα levels (mean ± SD) at the indicated time points during transformation.
(B) VEGF, HIF1A and CA9 mRNA levels (mean ±SD) assessed in non-treated (NT) and TAM-treated (36h) MCF10A ER-Src cells in the presence or absence of two different siRNAs against OLR1.
(C) NF-κB activity (ELISA assay; mean ± SD) in untreated and TAM-treated MCF10A ER-Src cells in the presence of the indicated siRNAs or 10 µM simvastatin.
(D) IκBα phosphorylation levels (ELISA assay; mean ± SD) in untreated and TAM-treated MCF10A ER-Src cells in the presence or absence of control or OLR1 siRNAs.
(E) VEGF, HIF1A and CA9 mRNA levels (mean ± SD) assessed in Ab-IgG or Ab-TNFα treated ER-Src cells.
(F) Representative phase contrast images (scale bars: 10µm) of untreated and TAM-treated MCF10A ER-Src cells in the presence or absence of simvastatin.
(G) Representative phase-contrast images (scale bars: 10µm) and (H) number of colonies of MCF10A cells treated with oxidized LDL (oxLDL) in the presence or absence of 5uM NF-κB inhibitor (BAY-117082). For all relevant panels, the data are presented as mean ± SD.
OLR1 is important for tumor growth in mice
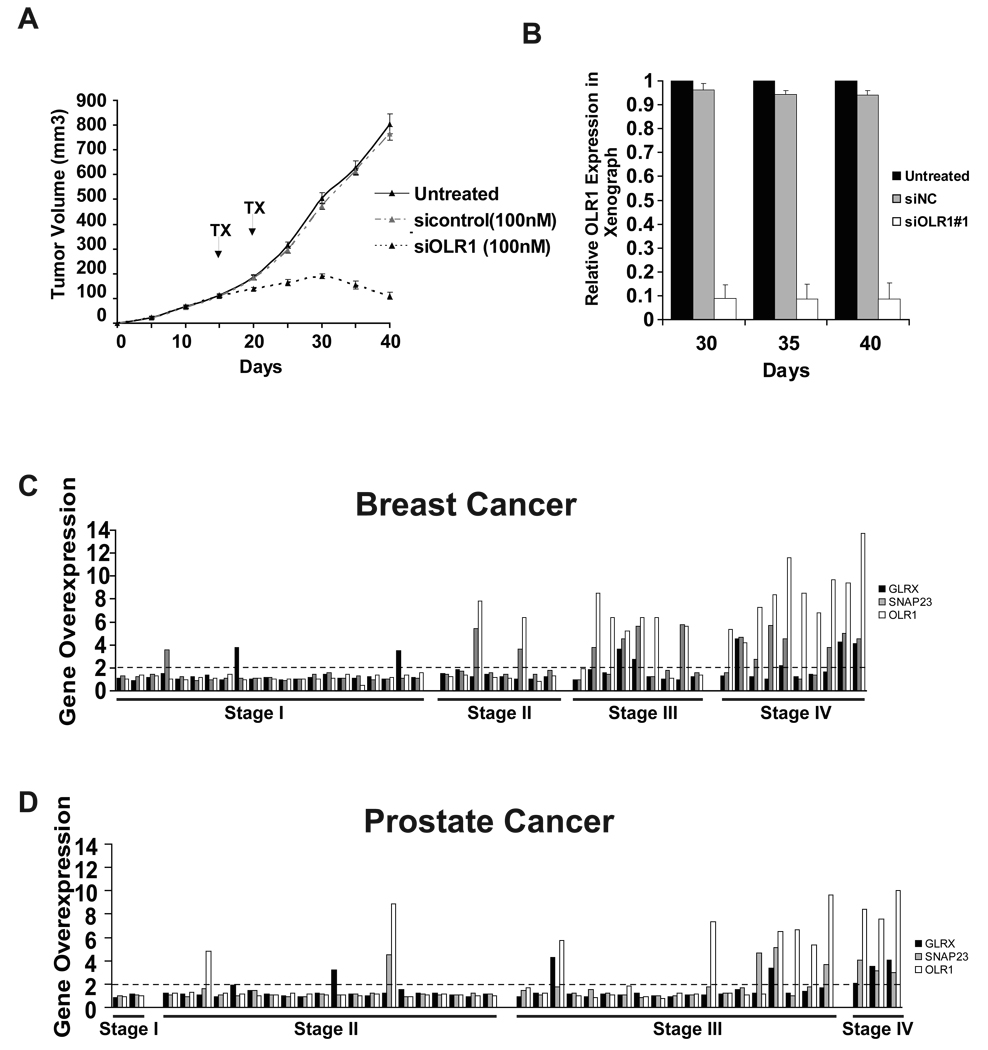
When injected as xenografts into nude mice, transformed MCF10A-ER-Src cells invariably cause tumors. Intraperitoneal injection of siRNA against OLR1 (4 cycles every 5 days) causes a marked reduction in tumor growth (Figure 8A), and along with a strong decrease in OLR1 expression levels (Figure 8B). In contrast, parallel injections of a control siRNA into the same cohort of tumor-containing mice has no effect on tumor growth or OLR1 expression. Thus, OLR1 is important for tumor growth in mouse xenografts.
Figure 8. OLR1 is important for tumor growth and overexpressed along with GLRX and SNAP23 in cancer tissues.
(A) Tumor volume (mean ± SD) of mice injected at time 0 with transformed MCF10A-ER-Src cells that were untreated, or treated by intraperitoneal injections every 5 days (4 cycles starting at day 15; arrows indicate the day or injections) with 100 nM siRNA against OLR11 or an siRNA control.
(B) OLR1 expression levels (mean ± SD) from tumors derived from the above experiment.
(C) Expression of OLR1, GLRX, and SNAP23 in breast cancer tissues separated by clinicopathological stage.
(D) Expression of OLR1, GLRX, and SNAP23 in prostate cancer tissues separated by clinicopathological stage.
Lipid metabolism genes OLR1, GLRX, and SNAP23 are often coordinately overexpressed in late stage breast and prostate cancer tissues
We examined whether expression of OLR1, GLRX and SNAP23 is altered in human cancers (Figure 8C, D). OLR1 is overexpressed in 37% of mammary adenocarcinomas and 25% of prostate adenocarcinomas, and in some cases 10-fold overexpression is observed. GLRX and SNAP23 are overexpressed in 17% and 29% of mammary adenocarcinomas and in 14% and 16% of prostate adenocarcinomas, although the magnitude of overexpression is less than observed for OLR1. When tumors are classified according to clinicopathological parameters, average mRNA expression levels of these genes increase with the stage grading of the tumor, with stage IV tumors showing high levels in most cases. In many, but not all, stage IV tumors, these three lipid metabolism genes are overexpressed, suggestive of coordinate regulation. Thus, lipid metabolism genes are often overexpressed in breast and prostate cancer tissues, and high levels of expression are associated with more aggressive, metastatic stage tumors.
DISCUSSION
Identification of a cancer gene signature
We define a cancer gene signature by a common set of genes that are differentially regulated in the same manner in two diverse models of cellular transformation. Several features of this approach are advantageous. First, isogenic models make it possible to identify genes involved in transformation without the complications of unknown and irrelevant genetic differences between non-transformed and transformed cells. Second, the two diverse model systems help to distinguish transformation-specific genes from genes that are regulated in a cell-type specific manner. In this regard, our cancer gene signature contains only 343 genes, whereas thousands of genes are differentially regulated in only one experimental model. This increased specificity is important for mechanistic understanding of transformation and for increasing the statistical significance of bioinformatics analyses. An important consequence of this specificity is our discovery of genes involved in lipid metabolism as contributing to cellular transformation. Third, the inducible Src system in MCF-10A cells offers the unique opportunity to kinetically follow the transcriptional program of cellular transformation in a manner similar to that used to study viral infection and other temporally ordered processes. Importantly, the transcriptional signature derived from the combined analysis of two isogenic, but biologically unrelated, models of cellular transcription is highly correlated with a wide variety of human cancers, thereby validating the clinical relevance of our experimental models.
A role for lipid metabolism in cellular transformation and a link between cancer and atherosclerosis
Although most genes in the cancer gene signature have been previously linked to cancer by some criterion, some have not. In support of their functional importance and relevance to cancer, 7 out of 11 such genes tested are important for cellular transformation in both experimental models (2 others in one model) and those tested are overexpressed in breast and prostate cancer tissues. Unexpectedly, genes involved in lipid metabolism are highly enriched in our cancer gene signature, particularly among those not previously linked to cancer. For example, OLR1, SNAP23, VAMP4, SCD1, and SREBP1 are lipid-related genes, and low-density lipoprotein (LDL) is a common hub between cancer and metabolic gene networks. Similarly, GALNT2 is associated with high cholesterol and triglyceride levels (Kathiresan et al., 2008), but not been linked with any type of cancer.
Strikingly, oxidized LDL can cause transformation of MCF-10A cells lacking ER-Src in a manner that depends on NF-κB. The receptor for oxidized LDL, OLR1, is a marker for atherosclerosis, and it activates inflammatory and hypoxia pathways in vascular endothelial cells and macrophages. In the MCF10A ER-Src model, inhibition of OLR1 also reduces NF-κB activation and the inflammatory and hypoxia pathways, suggesting a mechanistic connection between cellular transformation and atherosclerosis. Finally, OLR1 is important for maintaining the transformed state in cancer cell lines of diverse developmental origin, and for tumor growth in xenografts experiments. Thus, the importance of OLR1 and other genes involved in lipid metabolism in cellular transformation is relevant for, and a major determinant of, the connection between cancer and atherosclerosis. The finding of lipid metabolism genes in our cancer gene signature is noteworthy, because breast epithelial cells and primary fibroblasts are not major players in lipid biology and are functionally unrelated to cells involved in heart disease.
A common molecular signature for diverse human diseases
Clinical and epidemiological studies have linked cancer with inflammatory and metabolic diseases. In addition, specific molecular pathways involved in cancer are also involved in inflammatory diseases (e.g. NF-kB) and metabolic diseases (e.g. Akt). Our cancer gene signature and underlying regulatory networks significantly extend these observations by linking cancer with a variety of human diseases in a genome-wide manner that is based solely on experimental models of cellular transformation. These links between cancer gene signature and other diseases are robust, as they are based on 1) common biological functions, 2) correlations with literature-based annotations of individual human diseases, 3) similarity to transcriptional profiles of diseased patients, 4) identification and overlap of central nodes that define regulatory pathways. Furthermore, it is striking the studies of cellular transformation in breast epithelial cells and fibroblasts uncovered connections to diseases involving developmentally unrelated cell types and biological functions.
More importantly, our results indicate that many disease states share common molecular properties and biological programs. These similarities go beyond pairwise connections between cancer and a particular disease or regulatory pathway. Further, they do not simply reflect a stress response, because the transcriptional signature is not linked to any stress conditions. Instead, our results strongly argue that a core group of biological pathways is critical for normal cellular growth and behavior in a variety of cell types. Genetic or physiological disruption of these pathways leads to a transcriptional signature that is common to a diverse set of human diseases. As a consequence, genetic or environmental factors that affect the common genes or gene networks should predispose individuals to the development of multiple human diseases, and may contribute to the epidemiological connections between seemingly unrelated pathologic conditions. However, our results do not address whether the presence of one disease per se, increases the probability that another disease state may arise, because the different disease states involve different cell types and hence may arise independently. Conversely, the existence of a common transcriptional program and regulatory network for many diseases suggests that drugs used to treat one disease may be effective against other diseases. In this regard, 11 out of 13 drugs used to treat non-cancer diseases inhibit cellular transformation.
How does a common transcriptional program contribute to a diverse set of human diseases? We suggest that the interplay between cell-type specific transcription factors and the common transcriptional program leads to cell-type specific transcription profiles and phenotypes that are associated with specific disease states. This suggestion is in accord with, and indeed prompted by, the well-established principle that expression of mammalian genes requires combinations of transcriptional regulatory proteins bound to enhancers (Struhl, 1991). In this view, the combination of cell-type specific factors with a common disease program can either lead to inappropriate proliferation diagnostic of cancer or non-proliferative abnormalities that lead to other diseases such as diabetes or atherosclerosis.
MATERIALS AND METHODS
Cell lines and culture conditions
MCF-10A ER-Src and MCF-10A pBABE cells were grown in DMEM/F12 medium supplemented as described in (Debnath et al., 2003) with the addition of puromycin. The Src oncogene was induced by the addition of 1 uM Tamoxifen (Sigma) to confluent cell cultures for times indicated in the text. For testing the effect of drugs on transformation, each drug was titrated for optimum inhibition with minimal effects on non-transformed cells. All BJ fibroblast cell lines, described previously (Hahn et al., 1999), were cultured on KO-DMEM media, 10% FBS, Medium 199 glutamine and Pen/strep. Each drug used in the drug screen was titrated for optimum inhibition with minimal effects on non transformed cells. The following drugs were used in the following concentrations: metformin (0.1 mM), cerulenin (1 µg/ml), tocilizumab (2 µg/ml), Aspirin (0.1 mM), exendin4 (15 µm), sulindac (100 mM), Simvastatin (10 µM), meloxicam (30 µM), Indomethacin (30 µg/ml), celecoxib (10 µM), piroxicam (100 µM), Nimesulide (50 µM), Sulindac (100 µM), mevastatin (1 µM).
RNA preparation
RNA was extracted from all cell lines by Trizol method followed by RNeasy columns purification. These samples were hybridized on an Affymetrix U133 2.0A array at the Dana Farber Array Facility.
Gene expression analyses
All gene expression data was normalized and summarized with RMA algorithm (Irizarry et al., 2003) with an updated Entrez gene probeset definition (Dai et al., 2005). ComBat (Johnson et al., 2007) was used to remove non-biological experimental variation or batch effects between batches of microarray experiments. In order to detect differentially expressed genes, Significance Analysis of Microarrays (SAM) algorithm (Tusher et al., 2001) was used to calculate the q-values (False Discovery Rate) for genes in each time point. For ER-Src expression arrays, 7 samples were used as controls, including: Er-src_12EtOH (D1, D2, D3), Er-src_24EtOH (D1, D3), Er-src_0hr_TAM (D2, D3). A gene will be regarded as differentially expressed gene, only if 1) it was ‘present’, in terms Affymetrix MAS5 present/absent calls, in at least one time points and 2) q-value < 1 (either up-regulated or down-regulated) in at least one time points.
Disease gene sets
Gene sets were collected directly from previously published papers. These include the 1406 gene set for metabolic disorders (Chen et al., 2008), the 494 gene set for atherosclerosis (Sluimer et al., 2007), the 60 gene set for inflammatory breast cancer (Lerebours et al., 2008), the 28 gene set for inflammatory gastric cancer (Ellmark et al., 2006), the 687 gene set for thyroid cancer (Delys et al., 2007), and the 80 gene set for pancreatic cancer (Logsdon et al., 2003).
Lever algorithm analysis
The Lever algorithm was described previously (Warner et al., 2008). We incorporated the phylogenetic information from 12 mammals: mouse, rat, human, rabbit, chimp, macaque, cow, dog, armadillo, tenrec, opossum and elephant and used the MultiZ 17-way alignment as described in Supplementary Methods. The PBM data used in the study was for 104 TFs (transcription factors) from 22 structural classes (Badis et al., 2009) and for 178 TFs from the Homeodomain class (Berger et al., 2008). We used “Seed-and-Wobble” algorithm that has been described previously (Berger et al., 2006) in order to represent these data as position weight matrices (PWMs) for each TF. We used these PWMs in the Lever analysis.
Ingenuity pathway analysis
Ingenuity Pathways Analysis, (Ingenuity Systems, Mountain View, CA) is a robust and expertly curated database containing up-to-date information on over 20,000 mammalian genes and proteins, 1.4 million biological interactions, and one hundred canonical pathways incorporating over 6,000 discreet gene concepts. This information is integrated with other relevant databases such as EntrezGene and Gene Ontology. The experimental datasets were used to query the IPA and to compose a set of interactive networks taking into consideration canonical pathways, the relevant biological interactions, and the cellular and disease processes.
Statistics
The overlap count was computed by counting the number of genes in the intersection between two different gene sets. P values were calculated by Fisher Exact Test and Hypergeometric Probability Distribution Analysis in order to estimate the statistical significance of overlap between two gene sets.
Small interference RNA transfection experiments
MCF10A ER-Src cells were seeded in 6-well plates and were transfected with 100 nM siRNAs from Ambion Inc. against OLR1 (s9842 and s9843), GLRX (s5841 and s229668), PLAU (s10610 and s10612), GRN (s6149 and s6151), PGS1 (s18191 and s18192), SCD1 (s12505), FGD6 (s31504), MRPL9 (s35151), MOCOS (s230170), MYC (s1930), AKT (s659), SOCS3 (s17190), STAT3 (s744), HIF1A (s6539), NF-κB (s11914), IL6 (s7313), RAS (s806), VEGF (s460) using siPORT NeoFX transfection agent. SiPORT NeoFX is a lipid transfection agent consisting of a mixture of lipids that spontaneously complex small interference RNA and facilitates its transfer to the cells. Transfection with 100 nM siRNA (s4390846) was used as a control. No cell toxicity was detected due to the transfection agent.
Soft agar colony assay
Triplicate samples were mixed 4:1 (v/v) with 2.0% agarose in cell growth medium for a final concentration of 0.4% agarose. The cell mixture was plated on top of a solidified layer of 0.5% agarose in growth medium. Cells were fed every 6 to 7 days with growth medium containing 0.4% agarose. The number of colonies was counted after 15 days. The experiment was repeated thrice and the statistical significance was calculated using Student’s t test.
Cell migration, invasion, and wound healing assays
For the migration assay, 105 trypinized cells were added to the top chambers of the transwell (8 µm pore size; BD Bioscience, Bedford, MA), and assay medium was added to the bottom chambers. After overnight incubation, the migratory cells were fixed and stained with 0.1% crystal violet solution. The experiment was repeated thrice and the statistical significance was calculated using Student’s t test. Invasion of matrigel has conducted by using standardized conditions with BDBioCoat growth factor reduced MATRIGEL invasion chambers (PharMingen). Assays were conducted according to manufacturer’s protocol, by using 5% horse serum (GIBCO) and 20 ng/ml EGF as chemoattractants. Wound healing assays have been described previously (Hirsch et al., 2009).
RNA analysis
Equal amounts of purified RNA samples from untreated and TAM-induced (1, 2, 4, 8, 12, 16, 24, 36h) MCF10A ER-Src cells or from other cancer cell lines were reverse-transcribed to form cDNA, which was subjected to SYBR Green based real-time PCR analysis. To analyze patient samples, RNAs from 48 mammary adenocarcinoma tissues and 3 normal mammary tissues; 44 prostate adenocarcinoma and 3 normal tissues were purchased from Origene (Rockville, MD). The experiments have been performed in triplicate and data are presented as mean ± SD.
ELISA assays
The NF-kB/p65 ActivELISA Kit measured nuclear p65 levels in MCF10 ER-Src untreated or TAM-treated for 36h. The anti-p65 antibody coated plate captures free p65 and the amount of bound p65 is detected by adding a second anti-p65 antibody followed by alkaline phosphatase (AKP) -conjugated secondary antibody using colorimetric detection in an ELISA plate reader at absorbance 405nm. To detect IκBα phosphorylation status (serine 32), we used a solid phase sandwich enzyme-linked immunosorbent assay (cat no 7276, Cell Signaling) according to the manufacturer’s instructions. The magnitude of the absorbance (450 nm) is proportional to the quantity of bound target protein. To detect TNFα production, we used a TNF-alpha Quantikine ELISA Kit (cat no. DTA00C, R&D Systems) according to manufacturer’s instructions. For all ELIZA assays, each sample was loaded in triplicate and data are presented as mean ± SD.
Tumor growth in xenografts
To assess the role of OLR1, 5×106 transformed MCF10A ER-Src cells were injected into the right flank of 15 female nu/nu mice (Charles River Laboratories), all of which developed tumors in 15 days with size ~125mm3. The mice were randomly distributed into 3 groups that were untreated, or treated by intraperitoneal injections every 5 days (4 cycles) with 100 nM siRNA against OLR1 or a control siRNA. To assess the effects of metformin (20 mg/kg), cerulenin (40 mg/kb), simvastatin (20 mg/kg) and sulindac (15 mg/kg), the same procedure was followed except that drug treatment started 10 days of tumor formation (size ~60mm3). Tumor volume was measured at various times after the initial injection. All mouse experiments were approved by the Tufts University Institutional Animal Care and Use Committee.
Microarray data
Microarray data has been deposited at GEO, with an accession number of GSE17941.
SIGNIFICANCE
Although there are epidemiological and clinical connections between cancer and other diseases, the molecular bases of these connections are not well understood. mRNA expression profiling in two isogenic models of cellular transformation identifies a transcriptional signature and underlying gene regulatory networks that underlie diverse human diseases. In addition, it reveals the heretofore unappreciated importance of lipid metabolism to cellular transformation as well as the connection of cancer to atherosclerosis. These observations lead to the view that a variety of phenotypically diverse disease states are nevertheless linked through a common transcriptional program involving inflammatory and metabolic pathways.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Joan Brugge for the ER-Src and control cell lines, Bill Hahn for the fibroblast cell lines, and Konstantinos Drosatos for helpful discussions and suggestions regarding the involvement of lipid metabolism pathways in cancer. We thank members of the Bulyk lab, in particular Mike Berger, Andrew Gehrke, and Anthony Philippakis, and members of Tim Hughes’ lab, in particular Gwenael Badis and Shaheynoor Talukder, for sharing pre-publication PBM data on 104 mouse transcription factors. This project was supported by an NSF Postdoctoral Fellowship in Biological Informatics to S.J., a postdoctoral fellowship from the American Cancer Society to H.A.H., an Alfred P. Sloan Research Fellowship to X.S.L., and research grants from the National Institutes of Health to M.L.B (HG2966), P.N.T. (CA57486), X.S.L. (HG4069-02), and K.S. (CA107486).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Badis G, Berger MF, Philippakis AA, Talukder S, Gehrke AR, Jaeger SA, Chan ET, Metzler G, Vedenko A, Chen X, et al. Diversity and complexity in DNA recognition by transcription factors. Science. 2009;324:1720–1723. doi: 10.1126/science.1162327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balkwill F, Mantovani A. Inflammation and cancer: back to Virchow? Lancet. 2001;357:539–545. doi: 10.1016/S0140-6736(00)04046-0. [DOI] [PubMed] [Google Scholar]
- Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, Weinberg RA. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger MF, Badis G, Gehrke AR, Talukder S, Philippakis AA, Pena-Castillo L, Alleyne TM, Mnaimneh S, Botvinnik OB, Chan ET, et al. Variation in homeodomain DNA binding revealed by high-resolution analysis of sequence preferences. Cell. 2008;133:1266–1276. doi: 10.1016/j.cell.2008.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger MF, Philippakis AA, Qureshi AM, He FS, Estep PW, 3rd, Bulyk ML. Compact, universal DNA microarrays to comprehensively determine transcription-factor binding site specificities. Nat.Biotechnol. 2006;24:1429–1435. doi: 10.1038/nbt1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Zhu J, Lum PY, Yang X, Pinto S, MacNeil DJ, Zhang C, Lamb J, Edwards S, Sieberts SK, et al. Variations in DNA elucidate molecular networks that cause disease. Nature. 2008;452:429–435. doi: 10.1038/nature06757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai M, Wang P, Boyd AD, Kostov G, Athey B, Jones EG, Bunney WE, Myers RM, Speed TP, Akil H, et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucl. Acids Res. 2005;33:e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Marzo AM, Platz EA, Sutcliffe S, Xu J, Gronberg H, Drake CG, Nakai Y, Isaacs WB, Nelson WG. Inflammation in prostate carcinogenesis. Nat. Rev. Cancer. 2007;7:256–269. doi: 10.1038/nrc2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- Delys L, Detours V, Franc B, Thomas G, Bogdanova T, Tronko M, Libert F, Dumont JE, Maenhaut C. Gene expression and the biological phenotype of papillary thyroid carcinomas. Oncogene. 2007;26:7894–7903. doi: 10.1038/sj.onc.1210588. [DOI] [PubMed] [Google Scholar]
- Ellmark P, Ingvarsson J, Carlsson A, Lundin BS, Wingren C, Borrebaeck CA. Identification of protein expression signatures associated with Helicobacter pylori infection and gastric adenocarcinoma using recombinant antibody microarrays. Mol. Cell Proteomics. 2006;5:1638–1646. doi: 10.1074/mcp.M600170-MCP200. [DOI] [PubMed] [Google Scholar]
- Giovannucci E. Metabolic syndrome, hyperinsulinemia, and colon cancer: a review. Am. J. Clin.Nutr. 2007;86:836–842. doi: 10.1093/ajcn/86.3.836S. [DOI] [PubMed] [Google Scholar]
- Hahn WC, Counter CM, Lundberg AS, Beijersbergen RL, Brooks MW, Weinberg RA. Creation of human tumour cells with defined genetic elements. Nature. 1999;400:464–468. doi: 10.1038/22780. [DOI] [PubMed] [Google Scholar]
- Hardie DG. Role of AMP-activated protein kinase in the metabolic syndrome and in heart disease. FEBS Lett. 2008;582:81–89. doi: 10.1016/j.febslet.2007.11.018. [DOI] [PubMed] [Google Scholar]
- Hirsch HA, Iliopoulos D, Tsichlis PN, Struhl K. Metformin selectively targets cancer stem cells and acts together with chemotherapy to blocks tumor growth and prolong remission. Cancer Res. 2009;69:7507–7511. doi: 10.1158/0008-5472.CAN-09-2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu IR, Kim SP, Kabir M, Bergman RN. Metabolic syndrome, hyperinsulinemia, and cancer. Am. J. Clin.Nutr. 2007;86:867–871. doi: 10.1093/ajcn/86.3.867S. [DOI] [PubMed] [Google Scholar]
- Iliopoulos D, Hirsch HA, Struhl K. An epigenetic switch involving NF-kB, lin 28, let-7 microRNA, and IL6 links inflammation to cell transformation. Cell. 2009:693–706. doi: 10.1016/j.cell.2009.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- Karin M. Nuclear factor-kappaB in cancer development and progression. Nature. 2006;441:431–436. doi: 10.1038/nature04870. [DOI] [PubMed] [Google Scholar]
- Kathiresan S, Melander O, Guiducci C, Surti A, Burtt NP, Rieder MJ, Cooper GM, Roos C, Voight BF, Havulinna AS, et al. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat.Genet. 2008;40:189–197. doi: 10.1038/ng.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhajda FP. Fatty acid synthase and cancer: new application of an old pathway. Cancer Res. 2006;66:5977–5980. doi: 10.1158/0008-5472.CAN-05-4673. [DOI] [PubMed] [Google Scholar]
- Larsson SC, Mantzoros CS, Wolk A. Diabetes mellitus and risk of breast cancer: a meta-analysis. Int. J.Cancer. 2007;121:856–862. doi: 10.1002/ijc.22717. [DOI] [PubMed] [Google Scholar]
- Lee YH, Nair S, Rousseau E, Allison DB, Page GP, Tataranni PA, Bogardus C, Permana PA. Microarray profiling of isolated abdominal subcutaneous adipocytes from obese vs non-obese Pima Indians: increased expression of inflammation-related genes. Diabetologia. 2005;48:1776–1783. doi: 10.1007/s00125-005-1867-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerebours F, Vacher S, Andrieu C, Espie M, Marty M, Lidereau R, Bieche I. NF-kappa B genes have a major role in inflammatory breast cancer. BMC Cancer. 2008;8:41. doi: 10.1186/1471-2407-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logsdon CD, Simeone DM, Binkley C, Arumugam T, Greenson JK, Giordano TJ, Misek DE, Kuick R, Hanash S. Molecular profiling of pancreatic adenocarcinoma and chronic pancreatitis identifies multiple genes differentially regulated in pancreatic cancer. Cancer Res. 2003;63:2649–2657. [PubMed] [Google Scholar]
- Luedde T, Beraza N, Kotsikoris V, van Loo G, Nenci A, De Vos R, Roskams T, Trautwein C, Pasparakis M. Deletion of NEMO/IKKgamma in liver parenchymal cells causes steatohepatitis and hepatocellular carcinoma. Cancer Cell. 2007;11:119–132. doi: 10.1016/j.ccr.2006.12.016. [DOI] [PubMed] [Google Scholar]
- Manatovani A, Allavena P, Sica A, Balkwill F. Cancer-related inflammation. Nature. 2008;454:436–444. doi: 10.1038/nature07205. [DOI] [PubMed] [Google Scholar]
- Mehta JL, Hu B, Chen J, Li D. Pioglitazone inhibits LOX-1 expression in human coronary artery endothelial cells by reducing intracellular superoxide radical generation. Arterioscler. Thromb. Vasc. Biol. 2003;23:2203–2208. doi: 10.1161/01.ATV.0000094411.98127.5F. [DOI] [PubMed] [Google Scholar]
- Menendez JA, Lupu R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat. Rev.Cancer. 2007;7:763–777. doi: 10.1038/nrc2222. [DOI] [PubMed] [Google Scholar]
- Naugler WE, Karin M. NF-kappaB and cancer-identifying targets and mechanisms. Curr. Opin. Genet.Dev. 2008;18:19–26. doi: 10.1016/j.gde.2008.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce BL, Ballard-Barbash R, Bernstein L, Baumgartner RN, Neuhouser ML, Wener MH, Baumgartner KB, Gilliland FD, Sorensen BE, McTiernan A, et al. Elevated biomarkers of inflammation are associated with reduced survival among breast cancer patients. J. Clin.Oncol. 2009;27:3437–3444. doi: 10.1200/JCO.2008.18.9068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pischon T, Nothlings U, Boeing H. Obesity and Cancer. Proc. Nutr. Soc. 2008;67:128–145. doi: 10.1017/S0029665108006976. [DOI] [PubMed] [Google Scholar]
- Sakurai T, He G, Matsuzawa A, Yu GY, Maeda S, Hardiman G, Karin M. Hepatocyte necrosis induced by oxidative stress and IL-1 alpha release mediate carcinogen-induced compensatory proliferation and liver tumorigenesis. Cancer Cell. 2008;14:156–165. doi: 10.1016/j.ccr.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw RJ, Kosmatka M, Bardeesy N, Hurley RL, Witters LA, DePinho RA, Cantley LC. The tumor suppressor LKB1 kinase directly activates AMP-activated kinase and regulates apoptosis in response to energy stress. Proc. Natl. Acad. Sci.U.S.A. 2004;101:3329–3335. doi: 10.1073/pnas.0308061100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw RJ, Lamia KA, Vasquez D, Koo SH, Bardeesy N, Depinho RA, Montminy M, Cantley LC. The kinase LKB1 mediates glucose homeostasis in liver and therapeutic effects of metformin. Science. 2005;310:1642–1646. doi: 10.1126/science.1120781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skogsberg J, Lundstrom J, Kovacs A, Nilsson R, Noori P, Maleki S, Kohler M, Hamsten A, Tegner J, Bjorkegren J. Transcriptional profiling uncovers a network of cholesterol-responsive atherosclerosis target genes. PLoS Genet. 2008;4:e1000036. doi: 10.1371/journal.pgen.1000036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sluimer JC, Kisters N, Cleutjens KB, Volger OL, Horrevoets AJ, van den Akker LH, Bijnens AP, Daemen MJ. Dead or alive: gene expression profiles of advanced atherosclerotic plaques from autopsy and surgery. Physiol.Genomics. 2007;30:335–341. doi: 10.1152/physiolgenomics.00076.2007. [DOI] [PubMed] [Google Scholar]
- Soule HD, Maloney TM, Wolman SR, Peterson WD, Brenz R, McGrath CM, Russo J, Pauley RJ, Jones RF, Brooks SC. Isolation and characterization of a spontaneously immortallized human breast epithelial cell line, MCF10. Cancer Res. 1990;50:6075–6086. [PubMed] [Google Scholar]
- Struhl K. Mechanisms for diversity in gene expression patterns. Neuron. 1991;7:177–181. doi: 10.1016/0896-6273(91)90256-y. [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc. Natl. Acad. Sci.U.S.A. 2001;98:5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner JB, Philippakis AA, Jaeger SA, He FS, Lin J, Bulyk ML. Systematic identification of mammalian regulatory motifs' target genes and functions. Nat. Methods. 2008;5:347–353. doi: 10.1038/nmeth.1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.