Abstract
Gene duplication is a major mechanism facilitating adaptation to changing environments. From recent genomic analyses, the acquisition of zinc hypertolerance and hyperaccumulation characters discriminating Arabidopsis halleri from its zinc sensitive/non-accumulator closest relatives Arabidopsis lyrata and Arabidopsis thaliana was proposed to rely on duplication of genes controlling zinc transport or zinc tolerance. Metal Tolerance Protein 1 (MTP1) is one of these genes. It encodes a Zn2+/H+ antiporter involved in cytoplasmic zinc detoxification and thus in zinc tolerance. MTP1 was proposed to be triplicated in A. halleri, while it is present in single copy in A. thaliana and A. lyrata. Two of the three AhMTP1 paralogues were shown to co-segregate with zinc tolerance in a BC1 progeny from a cross between A. halleri and A. lyrata. In this work, the MTP1 family was characterized at both the genomic and functional levels in A. halleri. Five MTP1 paralogues were found to be present in A. halleri, AhMTP1-A1, -A2, -B, -C, and -D. Interestingly, one of the two newly identified AhMTP1 paralogues was not fixed at least in one A. halleri population. All MTP1s were expressed, but transcript accumulation of the paralogues co-segregating with zinc tolerance in the A. halleri X A. lyrata BC1 progeny was markedly higher than that of the other paralogues. All MTP1s displayed the ability to functionally complement a Saccharomyces cerevisiæ zinc hypersensitive mutant. However, the paralogue showing the least complementation of the yeast mutant phenotype was one of the paralogues co-segregating with zinc tolerance. From our results, the hypothesis that pentaplication of MTP1 could be a major basis of the zinc tolerance character in A. halleri is strongly counter-balanced by the fact that members of the MTP1 family are likely to experience different evolutionary fates, some of which not concurring to increase zinc tolerance.
Author Summary
Arabidopsis halleri has developed the characters of zinc hypertolerance and hyperaccumulation as compared to its close relatives Arabidopsis thaliana and Arabidopsis lyrata. Different candidate genes were proposed to account for the appearance of these characters in A. halleri. One of them is MTP1 (Metal Tolerance Protein 1), which is involved in cytoplasmic zinc detoxification. We found that A. halleri harbored five gene copies of MTP1, whereas A. thaliana and A. lyrata possess a single copy. It is thus tempting to associate the zinc hypertolerance character of A. halleri to the pentaplication of MTP1. However, we observed that one of the five MTP1 copies is not fixed in a population growing on metal contaminated soil. Also, the different MTP1 genes are markedly differentially expressed in A. halleri, two of them being poorly expressed and repressed in response to zinc constraint. AhMTP1 copies were also demonstrated to display a differential ability to complement the zinc hypersensitivity of a yeast mutant that is dysfunctional in vacuolar zinc transporters. Our findings suggest that different evolutionary fates, some of them not concurring to increase zinc tolerance, are likely to take place for the members of the MTP1 family in A. halleri.
Introduction
Adaptation of an organism to a challenging environment entails dramatic modifications in cellular, physiological, and regulatory processes. Gene duplications are postulated to be one of the main mechanisms providing raw genetic material for the origin of these adaptive modifications [1]. Conversely, the absence of gene duplications is thought to severely limit the plasticity for a genome or species in adapting to a challenging environment [2]. Plants are particular in the regard that they exhibit higher percentage of duplicated genes than other organisms [2]. Within the Plant kingdom, comparative genomic studies unravelled lineage-specific differential expansion of gene families in different plants species [3]–[5]. Much of the plant diversity may have arisen following the duplication and adaptive specialization of pre-existing genes rather than following the invention of new gene(s) [6]. After duplication and once fixed within species, three possible fates are typically envisaged for duplicated genes/paralogues [7]. Because selective constraints can be relaxed on duplicated genes initially underlying a same function, degenerative mutations can occur and result in the loss of function for one of the gene copies, therefore creating a pseudogene (non-functionalization). Alternatively, a new advantageous mutation can occur and confer a new function to one of the gene copy (neo-functionalization). Finally, rather than one gene duplicate retaining the original function and the others either degrading or evolving new functions, the original function of the single-copy gene may be partitioned among the duplicates (sub-functionalization). Thus, whereas orthologues in different species are usually expected to share similar functions, paralogues within a genome could have no or different functions.
Studying plant adaptation to extreme environments such as metal contaminated areas is an excellent way to characterise mechanisms underlying the evolution of a species, especially since there are only very few species that can survive and reproduce in these extreme environments [8]. In addition, metal tolerance is an important trait to investigate because its understanding can help to develop phytoremediation strategies for polluted soils [9]. Analysis of the natural adaptive evolution towards zinc hypertolerance in plants has attracted the attention of the scientific community due to the availability of diverse flora. Indeed, the same genus can host species showing zinc hypertolerance together with species showing zinc sensitivity. This is the case in the Arabidopsis genus: Arabidopsis halleri is a zinc hypertolerant species while its two closest relatives Arabidopsis thaliana and Arabidopsis lyrata are zinc sensitive. For example, A. halleri seedlings grown on 500 µM zinc appeared healthy and showed continued growth while A. thaliana and A. lyrata seedlings displayed dry and chlorotic leaves and stopped growing in presence of 100 µM zinc in the culture medium [10]. Zinc tolerance, together with zinc hyperaccumulation, are constitutive traits in A. halleri [11]. These traits have been acquired quite recently in A. halleri, since this species is estimated to have diverged from A. lyrata and A. thaliana ∼2 and ∼5 million years ago, respectively [12]. A significant level of quantitative variation of zinc tolerance was however observed among A. halleri populations [10], [11]. In particular, populations collected on metalliferous soils (known as metallicolous populations) display enhanced zinc tolerance compared to populations collected in metal uncontaminated sites (known as non metallicolous populations) [11]. Interestingly, a phylogeographic survey proposed that geographically distant metallicolous populations have been founded independently in distinct polluted areas [13] suggesting that the adaptive improvement of zinc tolerance may involve distinct genetic mechanisms in distinct metallicolous populations.
As already mentioned, A. halleri has experienced dramatic alterations of mechanisms involved in zinc tolerance. Gene duplications were proposed to underlie acquisition of this character in A. halleri [14], [15]. For instance ZIP3, ZIP6 and ZIP9, members of the zinc-regulated transporter/iron-regulated transporter-like proteins family [16], the HMA4 transporter controlling root to shoot zinc transport and zinc tolerance [15] and the MTP1 transporter controlling zinc transport into the vacuole [17] were shown to display increase in gene copy number in A. halleri compared to in A. thaliana or in A. lyrata. MTP1 (METAL TOLERANCE PROTEIN 1) is a Zn2+/H+ antiporter effluxing zinc out of the cytoplasm [17], [18]. When ectopically over-expressed in A. thaliana, AtMTP1 confers enhanced zinc tolerance [19]. MTP1 is present as single copy in A. thaliana. In A. halleri, three orthologues to the unique A. thaliana MTP1 were reported: AhMTP1-A, AhMTP1-B and AhMTP1-C [14]. Altogether, these AhMTP1s are over-expressed in A. halleri as compared to in A. thaliana. From the analysis of a backcross between the zinc hypertolerant A. halleri spp. halleri and the zinc sensitive A. lyrata spp. petraea, the three AhMTP1 paralogues were mapped to 3 different linkage groups. Two of the three paralogues, AhMTP1-A and AhMTP1-B, co-segregated with zinc tolerance QTLs controlling short-term root elongation in response to zinc constraint, while AhMTP1-C did not [20].
Until now detailed characterisation had not been performed for each of the AhMTP1 orthologues. This was a limiting step both for the further functional characterization of this gene sub-family in A. halleri and for the analysis of the genetic evolution of this family in relation to adaptation to zinc in A. halleri. In this work, cloning, sequencing and genetic mapping of all the A. halleri MTP1 paralogues revealed two new MTP1 paralogues. A. halleri was thus found to harbour five AhMTP1 paralogues, AhMTP1-A1, -A2, -B, -C and -D. The AhMTP1-D paralogue is not fixed in the metallicolous Auby population. Transcript accumulation studies together with functional characterizations in a heterologous yeast system enabled to propose that the different MTP1 duplications in A. halleri had different evolutionary fates including non-functionalization, sub-functionalization, and neo-functionalization. These results should provide a strong basis for further genetic diversity and linkage disequilibrium studies in A. halleri.
Results
Identification of all the MTP1 paralogues in A. halleri
In order to identify all the members of the MTP1 family in A. halleri, a BAC library [21] was screened with a labelled full length AhMTP1 probe obtained by PCR using primers designed from the published A. halleri MTP1 mRNA sequence (AJ556183 accession). Eight BAC clones, 3F23, 7G24, 16A6, 12L21, 1O21, 2B14, 3I23 and 1F18 were identified and confirmed by PCR sub-screening using the same primers.
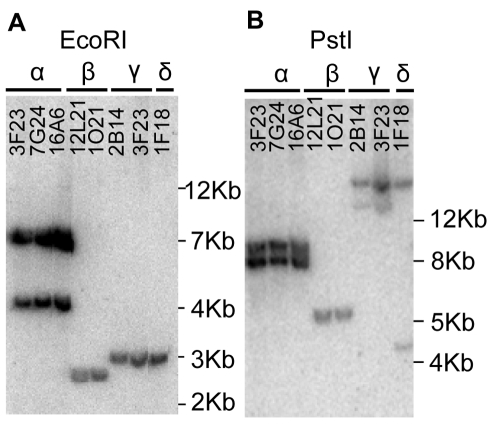
A. halleri was initially proposed to harbour three MTP1 paralogues [14]. With these data in mind, grouping of the eight BAC clones was attempted from the Southern hybridization profiles obtained using four different restriction enzymes, EcoRI, HindIII, NcoI and PstI (Figure 1 and data not shown). The eight BAC clones could be arranged into four groups, α, β, γ and δ displaying different profiles. γ and δ groups displayed however closely related profiles: their EcoRI profile was identical (Figure 1A) and their profile for other restriction enzymes shared a common band (Figure 1B and data not shown). Since A. halleri is an out-crossing species, BAC clones from groups γ and δ might harbour two allelic forms of a single locus. Alternatively, they may represent two similar but distinct loci harbouring AhMTP1 paralogues. These analyses thus suggested that A. halleri could harbour more than three MTP1 loci.
Figure 1. Southern hybridization of A. halleri MTP1 harbouring BAC clones.
BAC DNA digested with EcoRI (panel A) or PstI (panel B) was probed with a full length AhMTP1 probe. Names of the BAC clones are given at the top of the lanes. The α, β, γ, and δ symbols are the names given to the four distinct hybridisation profiles. Sizes of ladder are shown on the right of each panel.
Genetic mapping revealed the existence of a new MTP1 locus in A. halleri
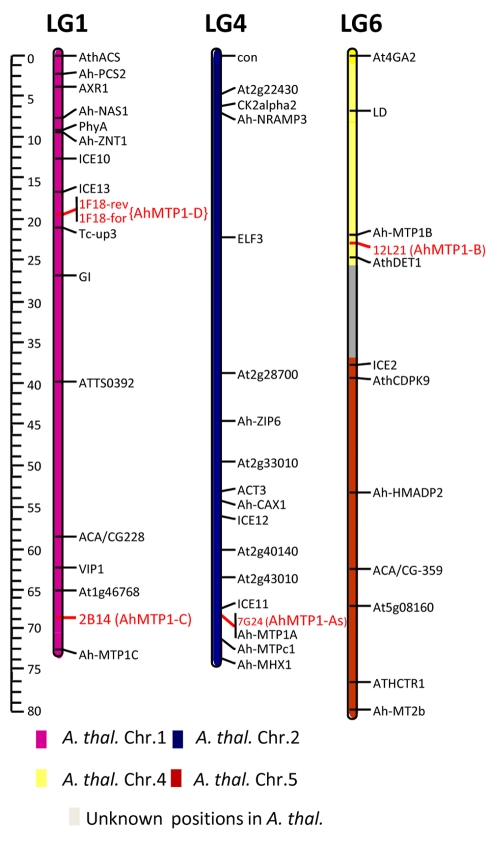
The three already described A. halleri MTP1 paralogues, AhMTP1-A, AhMTP1-B and AhMTP1-C, had been mapped to the bottom of linkage group 4, to the top of linkage group 6 and to the bottom of linkage group 1 on the A. halleri X A. lyrata linkage map, respectively [20]. To associate the 4 groups of BAC clones that we identified with the already described MTP1 paralogues, genetic mapping was performed using markers derived from selected BAC clones representing each group (Figure 2). The mapped positions of BAC clones 7G24, 12L21 and 2B14 representing the α, β, and γ groups, respectively, corresponded to the already mapped positions of AhMTP1-A, -B and -C, respectively. Thus the MTP1 copies characterising the α, β, and γ groups were considered to correspond to the AhMTP1-A, AhMTP1-B and AhMTP1-C paralogues, respectively. The BAC clone 1F18, which represents the δ group, was mapped to the upper part of linkage group 1 on the A. halleri X A. lyrata linkage map. The positioning was ascertained using 2 independent markers derived from each end of the BAC clone (Figure 2). No known MTP1 paralogue had already been mapped at that locus. Therefore, the MTP1 copy characterising the δ group was considered as a new A. halleri MTP1 paralogue and was named as AhMTP1-D.
Figure 2. Mapping of the AhMTP1 paralogues on the A. halleri × A. lyrata petraea BC1 linkage map.
The mapping was performed from the analysis of 199 plants and the four parents of an A. halleri × A. lyrata petraea BC1 population. Positions of BAC clones 7G24, 12L21, 2B14, and 1F18 that harbour the AhMTP1-A1 & -A2, AhMTP1-B, AhMTP1-C, and AhMTP1-D paralogues, respectively, are indicated in red letters (1F18rev and 1F18for are two independent markers). The other markers presented on the map have been previously described [20]. Only linkage groups (LG) 1, 4, and 6 of the A. halleri genome are shown. Colours of the bars representing the three linkage groups refer to the conserved synteny between the A. halleri and the A. thaliana genomes as inferred [22]. The regions showing no conserved synteny with A. thaliana are indicated in grey colour. The map was constructed using Joinmap 3.0. The scale to the left of figure represents centi-morgan distances.
The unique AtMTP1 gene (At2g46800) is located at the bottom of chromosome II of A. thaliana. Considering the shared synteny between the genome of A. thaliana and the genomes of other brassicaceae [22], position of the AtMTP1 locus corresponds to the position of the AhMTP1-A locus (Figure 2).
Sequence analyses unravel five AhMTP1 paralogues showing a significant diversity in non-coding regions
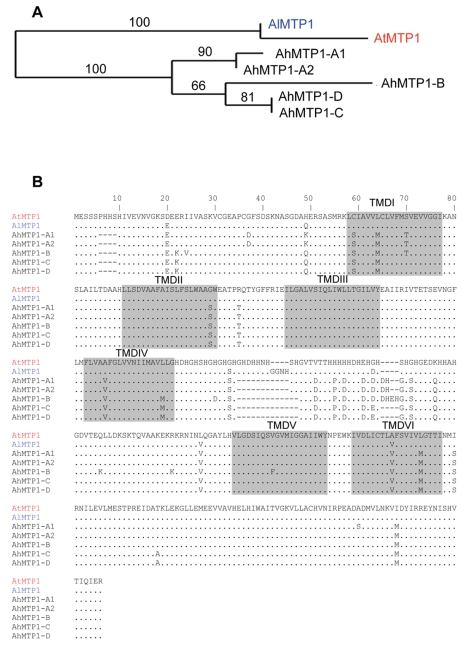
Genomic sequences of all the A. halleri MTP1 paralogues were obtained from the partial or complete sequencing of BAC clones. Analysis of the complete sequence of BAC clone 7G24 unravelled shared synteny between A. halleri and A. thaliana genomes in the A. thaliana region harbouring the sole A. thaliana MTP1 gene. This is in correspondence with the mapping results. Detailed sequence analysis revealed that the genetic structure (i.e. the gene order) is exactly the same in both species, except that the A. halleri genome displays a direct duplication of a 5 kbp region containing an MTP1 orthologue and a copy of a Retrovirus-related Pol polyprotein from transposon TNT 1-94. The complete sequence of BAC clone 7G24 thus revealed a fifth AhMTP1 paralogue. The two MTP1 paralogues arranged in tandem repeat on BAC clone 7G24 were named as AhMTP1-A1 and AhMTP1-A2. They displayed 100% identity in the promoter, 5′ UTR and 3′ UTR regions and differed by only two nucleotides in the coding DNA sequence. One of these differences resulted in an A365S substitution and the other one was silent (Figure 3B). The AhMTP1-A2 predicted protein showed 100% sequence identity with the already published full length AhMTP1 predicted protein [14]. The two nucleotidic differences discriminating AhMTP1-A2 from AhMTP1-A1 were shared with AtMTP1, AhMTP1-A2 thus being more similar to AtMTP1 than AhMTP1-A1 (Figure 3).
Figure 3. Phylogenetic analysis of MTP1 gene sub-family from A. thaliana, A. lyrata, and A. halleri.
(A) Maximum likelihood tree of MTP1 protein sequences from A. thaliana, A. lyrata and A. halleri. Bootstrap values are indicated in percentage (100 replicates). (B) Alignment of predicted amino acid sequences of MTP1s from A. thaliana, A. lyrata and A. halleri. Sequences are represented in 80 amino acids long blocks. The AtMTP1 protein sequence is from accession NP_850459 and the AlMTP1 one was extracted from scaffold 4 of the A. lyrata sequencing project available on http://genome.jgi-psf.org/cgi-bin/runAlignment?db=Araly1&advanced=1. Identities with AtMTP1 are represented by a dot and differences are written in alphabets. Dashes (–) signify deletions. Six transmembrane domains (TMDI to TMDVI) predicted using TMHMM server v. 2.0 [34] are shaded. The histidine rich loop is located between TMDIV and TMDV.
AhMTP1-B, AhMTP1-C and AhMTP1-D sequences were obtained from BAC clones 12L21, 2B14 and 1F18, respectively. They comprised at least 1.3 kb of putative promoter sequence, the complete 5′ UTR and coding DNA sequences, and at least 156 bp of the putative 3′ UTR and terminator sequences. At the protein level, the five AhMTP1 paralogues displayed on average 97.5% identity with each other (Figure 3B). The most divergent regions were the cytoplasmic N-terminus and the histidine rich loop between transmembrane domains IV and V; this loop has already been proposed to function as a zinc buffering pocket and a sensor of the zinc level at the cytoplasmic surface [18]. The five AhMTP1 paralogues shared only 91–93% identity with their A. thaliana and A. lyrata orthologues. Together with the result of the phylogenetic analysis of the MTP1 family in these species (Figure 3A), this suggests that pentaplication of MTP1 occurred recently in the A. halleri lineage. Such a conclusion is similar as the conclusion drawn for the zinc transporting HMA4 P1Btype-ATPase, which is present in one copy in A. thaliana and is triplicated in A. halleri [15].
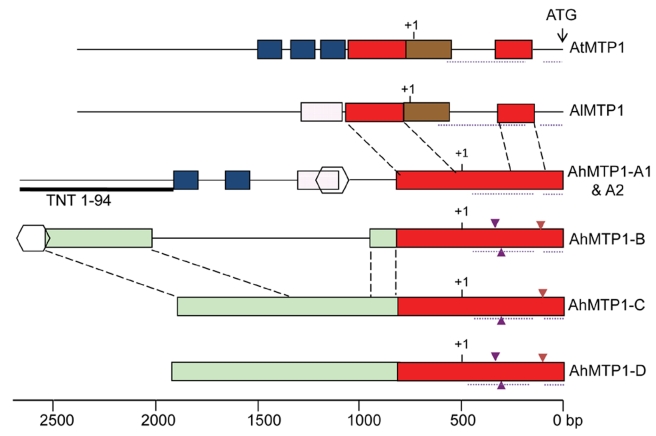
No intron was present in the region corresponding to the coding sequence for the five AhMTP1s, as for AtMTP1 and AlMTP1. In contrast, two introns were present in the 5′ UTR of all MTP1 orthologues (Figure 4), as revealed by the comparison between the genomic sequences and the published AhMTP1 and AtMTP1 mRNA sequences (AJ556183 and AF072858 accession numbers, respectively). Two 11 and 12 bp-long indel differences located in the first intron and a 17 bp-long indel difference located in the second intron discriminated the AhMTP1 paralogues (triangles in Figure 4). The proximal 800 bp region located upstream of the start codon were on average 95% identical among AhMTP1s (red boxes in Figure 4). In contrast, AhMTP1s showed on average 65–70% identities with AtMTP1 or AlMTP1 in that region. The more upstream parts of the MTP1 putative promoter regions were markedly divergent (Figure 4). At that location, AhMTP-A1 and -A2 displayed only few short regions in common with either AtMTP1/AlMTP1 or with the other AhMTP1s (blue or pink boxes in Figure 4). The AhMTP1-C and AhMTP1-D putative promoter regions shared 95% identity over their entire length and on average 97% identities with the AhMTP1-B promoter region only on distal sides (green boxes in Figure 4). Scanning of these MTP1 sequences was performed to identify transcription factor recognition motives that could be important in relation to zinc physiology. The “TGCACAC” conserved motif of metal response element b (MREb) [23] was found in all the MTP1s considered here but it was located in the coding DNA sequences. Apart from this MREb motif, no other metal responsive motives were found in the putative promoter regions of any of the AhMTP1s. The putative 3′ UTR regions of AhMTP1-A1, -A2, -C and -D were found to be 100% identical among themselves, and shared 97% identities with the 3′ UTR regions of either AtMTP1 or AlMTP1. In contrast, AhMTP1-B showed only 59% identities with other MTP1s in its 3′ UTR region.
Figure 4. Physical maps comparing the putative promoter plus 5′ UTR regions among A. thaliana, A. lyrata, and A. halleri MTP1 homologues.
Regions sharing >80% identity are shown by same coloured rectangular boxes or by same shapes. Dashed lines between different gene structures enable the relative positioning of the similar regions. Dotted lines below the gene structures indicate the position of introns. Small triangles present below or above the gene structures indicate 11 bp to 17 bp insertions (see text). Putative transcription start sites and translation start sites are indicated by +1 and ATG respectively. The Retrovirus-related Pol-polyprotein from transposon TNT 1–94 located in the putative promoter region of MTP1-A is represented by a thick line below the gene structure. Scale is shown at the bottom, relative to the ATG initiation codon.
Following the principle of parsimony an order of origin of AhMTP1 duplicates can be hypothesized based on genetic mapping, phylogenetic and sequences analyses. Because phylogenetically AtMTP1 is more closely related to AhMTP1-A1 and -A2 than to other AhMTP1s and because the AhMTP1-A locus was mapped to a region that shares conserved synteny with the A. thaliana region harbouring AtMTP1, it is therefore considered that AtMTP1 and AhMTP1-A have been derived from the MTP1 locus present in the common ancestor of A. thaliana and A. halleri. Thus, the AhMTP1-A locus harbours the parent MTP1 copy of other A. halleri MTP1s. Within this locus, it seems impossible to predict whether AhMTP1-A1 or -A2 is the parent MTP1 copy as they differ by only two nucleotides. From this parent copy it seems unlikely that AhMTP1-B, -C and -D have been independently derived because these three segmental duplicates show higher similarity to each other than to AhMTP1-A in coding as well as in putative promoter regions. AhMTP1-C or -D probably duplicated from one another because they share >95% identities over their entire length, but it seems impossible to predict which one of them was the first one to come into existence. AhMTP1-B is the most different from the AhMTP1-A. Its 3′ end in particular completely differs from the 3′ ends of other AhMTP1s that are 100% identical. Thus, AhMTP1-B is probably a segmental duplicate of either AhMTP1-C or -D.
The AhMTP1-D paralogue is not fixed in the A. halleri Auby population
In some experiments, the AhMTP1-D paralogue could not be found in a few A. halleri plants within the Auby population. In order to verify the presence of all the AhMTP1 gene copies in all the A. halleri plants within the Auby population, 188 plants were selected from this population and PCR was done using gene copy specific primer pairs (Table S1). The 188 plants were collected every ∼3 m along a 500 m-long transect starting from the least polluted zone at the periphery of the site towards the most polluted zone close to the centre of the site. This choice was made to check whether there could be a link between the possible presence of AhMTP1-D and zinc concentration in soil. In order to overcome possible allelic variation interfering with our analysis, two independent primer pairs enabling the specific detection of AhMTP1-D were designed. This strategy was chosen so that a lack of amplification by both primer pairs ascertains absence of the AhMTP1-D copy.
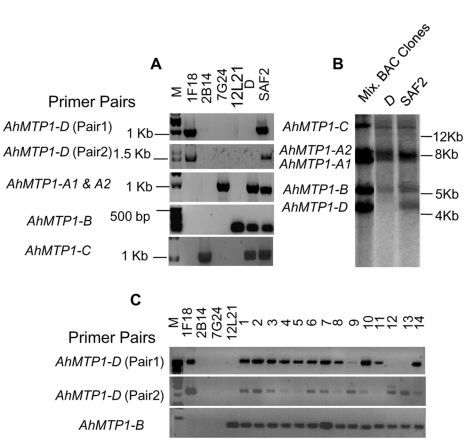
AhMTP1-A1 & A2, -B and -C amplicons were produced from all the plants (Figure S1 and data not shown). In contrast, 25% of the plants produced no amplicon for AhMTP1-D. This indicated that the AhMTP1-D gene is not fixed in the Auby population. No correlation was observed between the ability of a plant to produce AhMTP1-D amplicons and its position in the transect (data not shown). In order to ascertain that the AhMTP1-D paralogue was missing in some of the plants from the Auby accession, a Southern analysis was performed to analyse a plant producing no amplicon with AhMTP1-D specific primer pairs (D line) and another plant producing amplicons (SAF2 line). Comparison between the hybridization profiles of both plants and of a mix of the AhMTP1s harbouring BAC clones revealed that the bands specific to AhMTP1-D was only detected in the SAF2 line while the bands corresponding to AhMTP1-A1, -A2, -B and -C were detected in both lines (Figure 5A and 5B). These results confirmed that AhMTP1-D is not fixed in the Auby population.
Figure 5. Analysis of the presence of the MTP1-D paralogue in different A. halleri genotypes.
(A) PCR analysis of two plants of the A. halleri Auby accession (D and SAF2 plants) using AhMTP1-A, -B, -C, and -D gene specific primer pairs. DNA from BAC clones 1F18, 2B14, 7G24 and 12L21 are positive controls for demonstrating the specific amplification of AhMTP1-D, -C, -A, and -B, respectively. Corresponding sizes of amplicons are indicated on the right of each lane. (B) Southern analysis of the D and SAF2 plants of the Auby accession. Plant genomic DNAs and mixed BAC clone DNAs (positive control) were digested with PstI restriction enzyme. The probe was amplified from the AhMTP1-D harbouring BAC clone. Sizes deduced from a ladder are shown to right of the panel. (C) Production of AhMTP1-D specific amplicons from plants belonging to 14 different A. halleri populations. The 1 through 14 numbers above the lanes represent the following A. halleri populations, respectively: M Auby, M Sauerland, NM Regen, M Harz, NM CZ8-13, NM nord Tyrol, M Katowice-Weinowice, NM Zakopane, NM Appusenes, NM Fagaras Ro-12-6, NM Fagaras Ro-ovirensis, NM Southern Tyrol, M Lombardie and NM Tessin, where M = metallicolous and NM = non metallicolous. AhMTP1-B specific primer pairs were used as a control.
Presence of the AhMTP1-D copy was assayed by PCR in 14 different A. halleri populations representing the whole geographic distribution of the A. halleri species (Figure 5C). Interestingly at least one of the AhMTP1-D specific primer pairs produced amplicon for all the plants representing these populations. This indicates that the AhMTP1-D copy is present in all the analysed accessions. Our analysis cannot help to determine whether AhMTP1-D is fixed or not in all the A. halleri accessions. However, it can be concluded that AhMTP1-D is not in the process of being gained specifically in the A. halleri Auby population.
The AhMTP1 paralogues differentially complemented the zinc hypersensitivity of the zrc1 cot1 yeast mutant
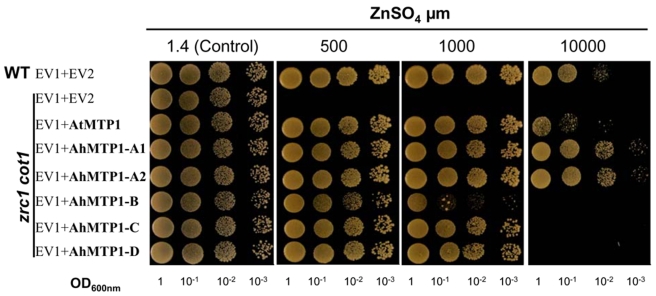
The five AhMTP1s and AtMTP1 were assayed for their ability to complement the zinc hypersensitivity of the yeast zrc1 cot1 double mutant, which is defective in vacuolar zinc transport [24]. Drop tests conducted on modified LSP medium supplemented with 500 µM zinc showed that the five AhMTP1s indeed induced functional complementation of the double zrc1 cot1 mutations (Figure 6). However, increasing zinc concentration in the medium up to 10 mM revealed the differential ability of the paralogues to complement zinc hypersensitivity of the zrc1 cot1 strain. AhMTP1-A1 and -A2 showed equal and highest complementation. They were slightly more efficient than AtMTP1. In contrast, AhMTP1-B was the least efficient among all of AhMTP1s in imparting complementation. Complementation imparted by AhMTP1-C and -D was equal and intermediate. Similar results were reproduced using independent clones and different media.
Figure 6. Functional complementation of the zinc-hypersensitivity of the zrc1 cot1 yeast mutant by AhMTP1s and AtMTP1.
Wild-type BY4741 (WT) and mutant zrc1 cot1 yeast strains harboured the empty pFL38H (EV1) that brought histidine autotrophy and either empty pYX212 vector (EV2) or pYX212 bearing one of MTP1s. Serial dilutions were spotted on modified selective LSP medium supplemented with different concentrations of ZnSO4 as indicated above the panels. Each spot was made with 10 µl of a yeast culture diluted at the OD600nm mentioned below the drops. Pictures were taken after 2 days for control and 4 days for other treatments.
AhMTP1 transcripts are differentially accumulated in planta
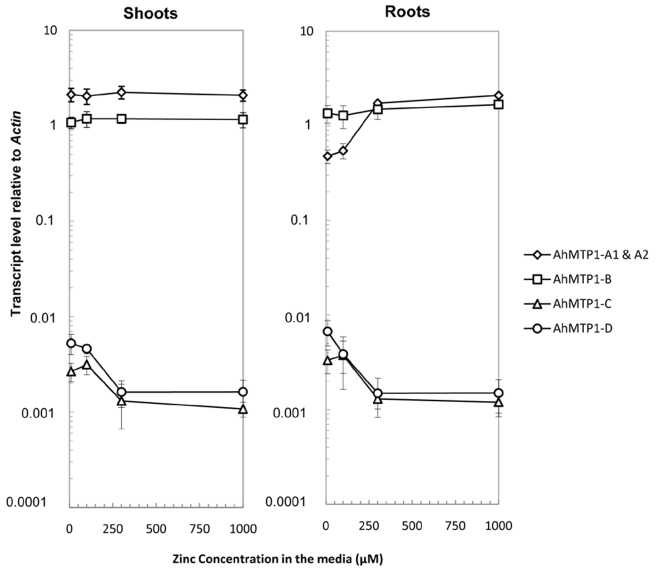
The transcript accumulation of the different AhMTP1 paralogues was analysed using Real-time quantitative RT-PCR in shoots and roots of individual mature plants grown in hydroponics on media supplemented with 10 (control), 100, 300 or 1000 µM ZnSO4 for 4 days (Table S2). The AhMTP1-A1 and -A2 genes sharing 99.9% identities in the whole gene including the promoter region could not be discriminated in that analysis. Otherwise, paralogue-specific primer pairs enabled to discriminate the AhMTP1-A, -B, -C and -D genes. In both roots and shoots, AhMTP1-A1 & -A2 and AhMTP1-B transcripts were much more abundant than AhMTP1-C and AhMTP1-D ones (Figure 7). On average, the relative abundances differed by nearly three orders of magnitude. In shoots steady-state AhMTP1-A1 & -A2 transcripts were more abundant than steady-state AhMTP1-B transcripts whereas it was the reverse in roots. In response to increasing zinc concentration in the culture medium, AhMTP1-A1 & -A2 transcripts as well as AhMTP1-B ones remained stable in shoots. AhMTP1-A1 & -A2 transcripts were induced in roots while AhMTP1-B ones remained stable (Figure 7). AhMTP1-C and AhMTP1-D transcript levels, which were very close to each other, decreased in shoots as well as in roots in response to increasing zinc concentration in the culture medium.
Figure 7. AhMTP1s transcript accumulation in plants submitted to different zinc treatments.
Roots and shoots were collected from plants of the A. halleri SAF2 genotype issued from the Auby accession that were exposed to 10 (control), 100, 300, or 1000 µM ZnSO4 for 4 days. Real-time quantitative RT–PCR was performed using gene copy specific primer pairs separately for shoots and roots. Data shown are transcript levels of AhMTP1s relative to Actin. Each data point in the graph is the average of three PCR repetitions for each of six biological replicates. Errors bars correspond to confidence intervals at the 0.05 threshold.
Discussion
With the aim to better understand the adaptive evolutionary processes leading to zinc hyperaccumulation and tolerance in A. halleri, we characterised the AhMTP1 gene family, which had been proposed to play a role in the control of these traits [14], [17]. Whereas, previous work identified three genetically unlinked MTP1 loci in A. halleri, the present study revealed the existence of five AhMTP1 paralogues located at 4 different loci. We consider that all the possible AhMTP1 paralogues have now been identified for the following two reasons. First, the paralogues were obtained from the screening of an A. halleri BAC library representing ∼4 equivalent genomes, meaning that any given A. halleri gene has a 0.986 probability to be present in at least one BAC clone of the library [21]. Then, the MTP1 hybridization profiles were identical between DNA mix of our BAC clones and different plant genomic DNAs coming from various accessions (Figure 5B, and compare Figure 1A of this work to Figure 4(a) from [16]).
From the genomic sequences that we obtained, the mechanism(s) responsible for the generation of MTP1 duplicates could not be identified. However, we could propose the most likely order of origin of theses duplications. Either of AhMTP1-A1 or -A2 is considered as the parent copy from which either of AhMTP1-C or -D was derived. AhMTP1-B would then have been derived from either of AhMTP1-C or -D.
Other genes involved in zinc tolerance or in zinc homeostasis display different degrees of multiplication in A. halleri as compared to in A. thaliana. These are for instance ZIP3, ZIP6 and ZIP9, members of the zinc-regulated transporter/iron-regulated transporter-like proteins family [16], the HMA4 transporter controlling root to shoot zinc transport [15] and type I defensins involved in cellular zinc tolerance [25]. The hypothesis that having more copies of genes involved in zinc homeostasis would be a general characteristic of A. halleri can however not be raised since other genes related to zinc tolerance or zinc homeostasis such as ZIP10, IRT3 or FRD3 are present as single copies in A. halleri as in A. thaliana [16]. In this context, the fact that five MTP1 copies are present in A. halleri may be the consequence of the fact that MTP1 plays a critical function with respect to zinc detoxification. This led us to study functional characteristics of the five paralogues.
The five AhMPT1 paralogues displayed a significant diversity in their ability to functionally complement the zinc hypersensitivity of the S. cerevisiae zrc1 cot1 mutant (Figure 6). While different causes can underlie this diversity, we favour the hypothesis that the differential functionality of the AhMTP1 paralogous proteins is linked to amino acid sequence differences. AhMTP1 predicted protein sequences displayed the greatest density of differences in a histidine rich loop located between transmembrane domains IV and V that has already been proposed to be a main regulatory domain of the protein [18]. Although it cannot be excluded that amino acid differences in other domains of the protein may also explain the functional differences between AhMTP1s, it seems likely that differences within the histidine rich loop are the determining factors for the differential ability of the AhMTP1s to complement the zinc hypersensitive phenotype of the mutant yeast.
The greatest functional difference discriminating the five AhMTP1 paralogues relates to their transcript levels, which varied by nearly three orders of magnitude (Figure 7). Our results are a bit different from previous ones [14] concerning the relative transcript abundances of the different AhMTP1 paralogues. We consider our data to be more accurate since we performed quantitative PCR and used gene specific probes made from clearly identified clones, which was not the case in the previous study. Alternatively, differences between our findings and previous ones might be due to different growing conditions in the two studies. One interesting novelty is that two of the AhMTP1 paralogues, AhMTP1-C and -D, displayed reduction in mRNA abundances following application of zinc. These paralogues were also the ones that showed a markedly lower transcript abundance compared to the three other ones in normal growing condition. This might be the sign that AhMTP1-C and -D play a completely different function from the other paralogues. Unsurprisingly, the differences between the transcript levels of the five AhMTP1 paralogues are in coherence with the differences between the corresponding promoter sequences (Figure 4 and Figure 7). For instance, the AhMPT1-D paralogue was found to display a transcript accumulation profile very similar to that of AhMTP1-C. This similarity can be related to the 95% identities observed between the putative promoter regions of the two paralogues. The extensive analysis of the relationship between AhMTP1 transcript levels and promoter sequences lead us to specific regions that may be responsible for the control of either the level of expression or the response to the zinc constraint. Among these, the 400 and 1000 bp long regions differentiating the putative AhMTP1-B promoter and the putative AhMTP1-C promoter should bring the most fruitful outcomes.
Among the genes proposed to be involved in zinc tolerance in A. halleri, only IRT3 and HMA4 have been fully characterised at both the genomic and functional levels [15], [26]. The situations characterising MTP1, IRT3 and HMA4 in A. halleri are completely different. First, AhIRT3 is in single copy while AhHMA4 is triplicated, with the three paralogues being present in tandem, and AhMTP1 is pentaplicated with both tandem and segmental duplicates. Transcripts of all three gene families were over-accumulated in A. halleri as compared to in A. thaliana [14], [15], [26]. However, analysing the relative contribution of different gene copies revealed completely different situations. For IRT3, transcript over-accumulation was attributable to the sole copy [26] while for HMA4, it was mainly due to the additive and equal contribution of the three paralogues [15]. In contrast, transcript over-accumulation could only be attributed to three of the five members of the AhMTP1 family (this work). The MTP1 family thus displays original characteristics in A. halleri. Remarkably, these original characteristics are not observed in another zinc hypertolerant and hyperaccumulating species, Thlaspi goesingense, as this species harbours only one MTP1 copy, which is over-expressed compared to in A. thaliana [27]. These characteristics displayed by AhMTP1 duplicates suggest that different evolutionary fates might take place for the duplicates in A. halleri.
As mentioned above, gene duplication together with mutations occurring in duplicates are promoting novelty in the evolutionary process [1]. Then, genes can be exposed to different kinds of evolutionary fates: sub-functionalization, neo-functionalization or non-functionalization [7]. Analysing the AhMTP1 gene family revealed asymmetric relationships among the AhMTP1 duplicates from the point of view of transcript accumulation patterns and protein function. Transcripts of AhMTP1-A1, -A2 and -B were found to be far more abundant than transcripts of AhMTP1-C and -D. At the same time, AhMTP1-B was less competent than AhMTP1-C and -D in complementing the zinc-hypersensitivity of the S. cerevisiae zrc1 cot1 double mutant. Since AhMTP1-A1, -A2 and -B were found to co-localize with previously described zinc-tolerance QTLs for short term root elongation whereas AhMTP1-C and -D did not, it appears that transcript abundance is a more important factor controlling the contribution of AhMTP1s to zinc tolerance in A. halleri than the ability of protein itself to confer zinc tolerance. In this context, the fate of AhMTP1s duplications could be sub-functionalization for the AhMTP1-A1, -A2 and -B copies. This hypothesis needs to be validated by systematic comparison of single, double and triple mutants for each gene copy to assess the functional redundancy of these paralogues. By contrast, the AhMTP1-C and -D duplicates are more difficult to characterise. Because AhMTP1-C and -D are neither expressed well nor present in the previously described zinc-tolerance QTLs, they may appear to be in the process of non-functionalization. This hypothesis would be supported by the additional observations that AhMTP1-D is not fixed in the metallicolous A. halleri Auby population, and that its occurrence in plants from this accession is unlinked to the zinc concentration in the soil, which reveals freedom from selective pressure. However, since the AhMTP1-C and -D proteins functionally complemented the yeast mutant and the corresponding genes are both still expressed in planta, another hypothesis can be raised. This hypothesis would be that changes in the regulatory profile of these copies would correspond to a neo-functionalization process [28] leading AhMTP1-C and -D to play another role than being involved in zinc tolerance for short term root elongation. In that situation, the interpretation of the non fixed nature of the AhMTP1-D in the Auby population could be that this copy is redundant with AhMTP1-C. Concluding on the actual evolutionary fate of the AhMTP1-C and -D copies would thus require an extensive analysis of the temporal and spatial expression patterns of all the AhMTP1 paralogues. Fates of duplicates were proposed to differ depending on the nature of the duplication [6]. Tandem duplication was proposed to provide a means of amplifying adaptatively important genes, particularly resistance genes, while segmental duplication was proposed to permit gene family diversification and long-term evolutionary plasticity. Our findings might be in agreement with this assumption. Indeed, the AhMTP1-A1 & -A2 tandem duplicates experience sub-functionalization, while two of the three segmental duplicates experience either neo- or non-functionalization.
In conclusion, based on genomics as well as functional approaches we propose that different evolutionary fates are likely to take place for AhMTP1 duplicates. Two paralogues do not appear to be under selective pressure for zinc tolerance, while three others appear to be. This study thus brings important outcomes to understand the mechanisms underlying the adaptation of A. halleri to zinc.
Materials and Methods
Plant material
Two different micropropagated lines from the A. halleri Auby population (the D and SAF2 lines) were used for Southern hybridization and/or transcript accumulation analyses. For the analysis of the presence of all the MTP1 paralogues in accessions representing the genetic diversity within the A. halleri species, one plant was taken at random from each of the following populations: M Auby from France, M Sauerland, NM Bavarian Forest and M Harz from Germany, NM CZ8-13 from Czech Republic, NM Nord Tyrol from Austria, M Katowice-Weinowice and NM Zakopane from Poland, NM Apuseni mountains, NM Fagaras Ro-12-6 and NM Fagaras Ro-ovirensis from Romania, NM Southern Tyrol, M Lombardie and NM Tessin from Italy, where M qualifies a metallicolous population and NM a non metallicolous one according to already described criteria [13].
BAC clone handling and Southern hybridisation
BAC clone identification was performed through the Southern screening of a BAC library made from an A. halleri plant from the Auby population, as described [21]. Then, BAC clone DNA was extracted from 3 ml of overnight culture grown in 2YT medium containing 12.5 mg/ml chloramphenicol, using the Nucleobond Plasmid DNA Purification kit (Macherey Nagel) but skipping the column purification step. AhMTP1-A1 and A2 sequences were obtained by full length sequencing of the 7G24 BAC clone (Genoscope, Evry, France). AhMTP1-B, -C and -D genomic sequences were obtained by partial sequencing of BAC clones 12L21, 2B14 and 1F18, respectively (GATC Biotech, Konstanz, Germany and Genoscreen, Lille, France). Sequences were deposited in the EMBL database. Accession numbers of the 7G24 BAC clone sequence and of the AhMTP1-B, -C and -D genomic sequences are (FN428855), (FN386317), (FN386316), and (FN386315), respectively. Sequences from which BAC clone specific genetic markers were designed are available under the accession numbers (FN386313) for BAC clone 2B14, (FN386314) and (FN428827) for BAC clone 1F18, (FN386317) for BAC clone 12L21 and (FN428855) for BAC clone 7G24.
For Southern analyses, 2 µg of BAC clone DNA or 10 µg of A. halleri genomic DNA were digested with 50 U of restriction enzyme at 37°C for 6–7 h and separated on a 0.8% (w/v) agarose gel in TEA1X buffer. Then, the agarose gel was submerged into 0.25 N HCl for 15 min and rinsed with water 2–3 times. DNA fragments were transferred onto a positively charged nylon membrane (Hybond-N+, Amersham Biosciences) by capillary action using 0.4 N NaOH for 8 h and then cross-linked onto the membrane for 80 sec under 254 nm UV light at 0.120 J.cm−2 with the Fluo-Link apparatus (Bioblock, Illkirch, France). For Southern analysis of BAC clones, the MTP1 probe was obtained from a PCR fragment produced from A. halleri genomic DNA using the 5′ -CGAGTCTTCAATTTCTGCAACT-3′ and 5′-AAACTTTATTGATTTATTGTTAA-3′ primers and purified using the Wizard SV Gel and PCR clean-up system (Promega). For the Southern analysis of genomic DNA, the probe was obtained from a PCR fragment produced from the 1F18 BAC clone using the 5′-TTTTCGGTTAAAGCGCACG-3′ and 5′-TGCAGAACTCGAAATCAAC-3′ primers. Fifty nanograms of purified PCR product were radioactively labelled by random priming (Prime-a-gene kit, Promega). The probe was then purified on illustra NICK columns (GE Healthcare). Prehybridization was carried out in Church buffer [29] for >2 h. Hybridization was carried out in the same buffer overnight at 65°C. Then the blots were washed. The final and more stringent wash was 0.5XSSC, 0.1% (w/v) SDS for BAC clones Southern blots and 0.1XSSC, 0.1% (w/v) SDS for genomic Southern blots for 20 min at 50°C. Blots were then placed against "Imaging Plate BAS-MS" screens, which were revealed using a BAS 5000 apparatus (Fujifilm, Japan).
Mapping of AhMTP1 paralogues on the A. halleri X A. lyrata petraea BC1 genetic map
Already available genomic DNA of the parents of the A. halleri X A. lyrata petraea BC1 population and of 199 plants from this population was used for genotyping, as described [30]. Mapping of the 7G24, 12L21, 2B14 and 1F18 BAC clones harbouring MTP1-A, -B, -C and -D, respectively, was performed by CAPS and SSCP analysis as described [20], using the markers described in Table S3. To make these markers, primer pairs were designed from sequences of the BAC clones and tested on genomic DNA of the A. halleri and A. lyrata petraea parents of the BC1 population. When no SSCP polymorphism could be detected, CAPS-type markers were made, as a result of assaying a set of different restriction enzymes on A. halleri and A. lyrata petraea amplicons.
Genotypes obtained in the BC1 population for the BAC-derived markers were combined with the data set used for the A. halleri X A. lyrata petraea linkage map construction [20], [30], using the Joinmap 3.0 program [31]. Individuals lacking information for more than 25% of all markers were excluded from the analysis. Linkage groups were obtained at a logarithm-of-odds (LOD) score threshold of 4. The best order of markers along each linkage group was determined using the sequential method implemented in Joinmap, comparing the goodness-of fit of the resulting map for each tested order using thresholds of 0.5 and 1.0 for the linkage groups and the loci, respectively. Translating recombination frequencies into map distances was made using Kosambi's mapping function [32].
Functional complementation in yeast
A. halleri and A. thaliana MTP1s are without any intron in the region corresponding to the coding DNA sequence. Thus their full length open reading frames were amplified directly from genomic or BAC DNAs using the proofreading Pfu DNA polymerase (Promega). A. halleri MTP1s were amplified using the forward 5′-AAAGAATTCATGGAGTCTTCAAGTCAC-3′ and reverse 5′-CCCCTCGAGTTAACGCTCGATTTGTATCG-3′ primers containing EcoRI and XhoI restriction sites, respectively (underlined sequences). A. thaliana MTP1 was amplified using the forward 5′-AAAGAATTCATGGAGTCTTCAAGTCCCCAC-3′ and reverse 5′-CCCCTCGAGTTAGCGCTCGATTTGTATCG-3′ primers also containing EcoRI and XhoI restriction sites, respectively. The PCR products were cloned downstream of the triose phosphate isomerase promoter in the pYX212 yeast expression vector at the EcoRI and XhoI restriction sites. A Saccharomyces cerevisiae zrc1 cot1 mutant (Mat a, zrc1::natMX3, cot1::kan-MX4, his3Δ1, leu2Δ0, met15Δ0, ura3Δ0) and its parental wild-type strain BY4741 were double transformed with the empty pFL38H (his+) vector and either empty pYX212 (ura+) or pYX212 (ura+) expressing AhMTP1-A1, -A2, -B, -C, -D or AtMTP1, using the lithium acetate/single-stranded carrier DNA/polyethylene glycol method [33]. Systematic co-transformation with pFL38H empty vector was done to avoid addition of histidine in the culture medium, as histidine is supposed to be a zinc chelator in growth medium. For drop assays, transformed yeast strains were grown overnight in 5 ml selective liquid YNB medium to early stationary phase. Yeast cells were then washed twice with ultrapure H2O and diluted in water to OD600nm = 1, 0.1, 0.01 and 0.001. Drop assays were performed on selective modified Low Sulphate/Phosphate medium [14] with pH adjusted to 4.7. The medium was supplemented with various concentrations of ZnSO4: 1.4 µM for control condition and from 100 µM to 10 mM for zinc treatments. At least four independent colonies were tested for each construct.
Gene expression analysis
For Real-Time Quantitative RT-PCR experiments, the A. halleri SAF2 genotype from the Auby accession was micropropagated in vitro on standard Murashige and Skoog culture medium with 0.8% (w/v) sucrose and 2% (w/v) agar. Four weeks later, rooted clones were transferred in hydroponics as described [25] and were let to acclimate themselves to this medium for 6 days. Then, individual clones were submitted for 4 days to different zinc treatments: 10 µM (control), 100 µM, 300 µM and 1000 µM ZnSO4. Six replicates were treated in parallel for each condition and analysed independently. Roots and shoots were harvested separately.
Total RNA was extracted (RNeasy kit; Qiagen, Hilden, Germany) then genomic DNA was removed using the RQ1 RNase-Free DNase kit (Promega). Four micrograms of total RNA were used as a template for first strand cDNA synthesis, which was performed using M-MLV Reverse Transcriptase, RNase H Minus, Point Mutant (Promega) and Oligo (dT)15 Primer (Promega) in a final volume of 100 µl, according to the instructions from the manufacturer.
Real-time RT-PCRs were performed in 384-well plates with the LightCycler 480 Real-Time PCR System (Roche diagnostics GmbH, USA) using SYBR Green to monitor cDNA amplification. Two microlitres of DNA sample were then used for PCR in a 10 µl reaction mixture containing 5 µl of LightCycler 480 SYBR Green I Master kit (Roche diagnostics GmbH) and 0.5 µM of each primer. The primer pairs used for the transcript accumulation analysis were designed in the specific regions of different AhMTP1s (Table 1). Actin was considered as an internal control. The primers used to analyse Actin have already been described [25]. The PCR program started with an initial 5 min-long treatment at 95°C. Then the samples were submitted to 45 PCR cycles composed of 10 sec at 95°C, 10 sec at respective annealing temperatures for each primer pair (Table 1) and 10 sec at 72°C. The specificity of the amplified PCR products was assessed for every sample by analysing the amplicon dissociation during the gradual increase of the temperature from 72°C to 95°C at the rate of 0.11°C/sec, using the Tm calling method proposed by the LightCycler 480 Software release 1.5.0. Six out of 750 PCR reactions showed unspecific amplification. The corresponding data were discarded.
Table 1. Gene-specific primer pairs used to characterise the different AhMTP1 paralogues in real-time quantitative RT–PCR analyses.
| Gene | Primer name | Primer sequence 5′--3′ | Annealing temperature (°C) | Average PCR efficiencies | |
| gDNA | cDNA | ||||
| Actin | ActinF | GGTAACATTGTGCTCAGTGGTGG | 65.9 | 1.87 | 1.90 |
| ActinR | AACGACCTTAATCTTCATGCTGC | ||||
| AhMTP1-A1 &A2 | ARTF | AGTGGTGAACATCATAATGGCTGTTC | 67.2 | 1.96 | 1.93 |
| ACRTR | TCGATTTGTCCAACAGTTGCTCAG | ||||
| AhMTP1-B | BRTF | TGGACGTGAAGTTATGGAGAC | 64.3 | 2.06 | 2.04 |
| BRTR | CTTCACATTTGGGCTATCACAG | ||||
| AhMTP1-C | CRTF | GGTGAACATCATAATGGCTGTTATGCTG | 67.8 | 1.98 | NP a |
| ACRTR | TCGATTTGTCCAACAGTTGCTCAG | ||||
| AhMTP1-D | DRTF | GTACACCCAGAGAGATTGACGCCG | 71.3 | 1.91 | NP a |
| DRTR | GACTAATGTTGTACTCCCTGCGGATG | ||||
a NP: Not possible to be done (see Materials and Methods).
For each primer pair specific to AhMTP1 paralogues, the PCR efficiency (E) was determined after the analysis of 5 serial 1∶10 dilutions of BAC clone DNA (Table 1) by using the equation E = (10−1/s), where “s” is the slope of the linear regression of the threshold cycle (Ct) values per the log10 values of the starting DNA copy numbers. When analysing the cDNA samples, the PCR efficiency was also evaluated from the analysis of 1∶3, 1∶12 and 1;48 dilutions of first strand cDNA (Table 1, Table S2) for Actin, AhMTP1-A1 and -A2 and AhMTP1-B. Transcript accumulation of the different genes was calculated using the respective experimentally determined PCR efficiency values for each primer pair. For the AhMTP1-C and -D genes, results from the 1∶12 and 1∶48 cDNA dilutions were unreliable because of surpassing the trustworthy detection limit of the real-time quantitative RT-PCR and were thus discarded (Table S2). For the AhMTP1-C and -D genes, it was thus impossible to calculate PCR efficiencies on cDNA samples; efficiencies measured on BAC clone DNA were thus used for calculations. Relative expression levels (REL) of AhMTP1s compared to the Actin were determined for every sample from the result of the 1∶3 dilution using the equation REL = [(E)-Ct]AhMTP1 / [(E)-Ct]Actin, where E and Ct are the PCR amplification efficiency and the threshold cycle, respectively, for the considered AhMTP1 and Actin. Six independent plant samples were considered for each condition.
Supporting Information
Analysis of the presence of the AhMTP1 paralogues in 44 plants from the Auby accession using gene copy specific primer pairs. Each of the horizontal panel show the amplification obtained from a primer pair specific to the MTP1 paralogue named at the left of the panel. Samples from BAC clones 1F18, 2B14, 7G24, and 12L21 were used as controls for specificity of the primer pairs. The lane M represents1kb invitrogen DNA ladder.
(0.09 MB PDF)
Sequences of gene-specific primer pairs and the corresponding annealing temperatures.
(0.03 MB DOC)
Threshold cycles (Ct) obtained in quantitative RT-PCR analyses performed on cDNAs coming from shoots and roots of A. halleri plants.
(0.06 MB XLS)
Markers used for genetic mapping of AhMTP1 harbouring BAC Clones.
(0.04 MB DOC)
Acknowledgments
We are thankful to A. Adiveze, H. Afonso, C. Baracco, H. Baudot, F. Bourgeois, C. Dasen, G. Doucet, C. Gagneré, S. Gélin, F. Lecocq, V. Rafin, G. Ruiz, and C. Zicler for technical and administrative supports.
Footnotes
The authors have declared that no competing interests exist.
ZS was supported by a scholarship from the Higher Education Commission, Pakistan. NR was supported by the Marie Curie intra-European Fellowship "Metolevol" (contract No.024683 MEIF-CT-2005-0224683). This work was in part supported by the "Fondation pour la Recherche en Biodiversité" (FRB; contract BRG No. 92), and from the INSU-CNRS program ACI ECCO (contract No. 04 2 9 FNS). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Ohno S. New York: Springer-Verlag.; 1970. Evolution by gene duplication. ISBN 0-04-575015-7. [Google Scholar]
- 2.Zhang JZ. Evolution by gene duplication: an update. Trends Ecol Evol. 2003;18:292–298. [Google Scholar]
- 3.Velasco R, Zharkikh A, Troggio M, Cartwright DA, Cestaro A, et al. A high quality draft consensus sequence of the genome of a heterozygous grapevine variety. PLoS ONE. 2007;2:e1326. doi: 10.1371/journal.pone.0001326. doi: 10.1371/journal.pone.0001326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ming R, Hou S, Feng Y, Yu Q, Dionne-Laporte A, et al. The draft genome of the transgenic tropical fruit tree papaya (Carica papaya Linnaeus). Nature. 2008;452:991–996. doi: 10.1038/nature06856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rensing SA, Lang D, Zimmer AD, Terry A, Salamov A, et al. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science. 2008;319:64–69. doi: 10.1126/science.1150646. [DOI] [PubMed] [Google Scholar]
- 6.Flagel LE, Wendel JF. Gene duplication and evolutionary novelty in plants. New Phytol. 2009;183:557–564. doi: 10.1111/j.1469-8137.2009.02923.x. [DOI] [PubMed] [Google Scholar]
- 7.Hurles M. Gene Duplication: The Genomic Trade in Spare Parts. PLoS Biol. 2004;2:e206. doi: 10.1371/journal.pbio.0020206. doi: 10.1371/journal.pbio.0020206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Antonovics J, Bradshaw AD, Turner RG. Heavy metal tolerance in plants. Adv Ecol Res. 1971;7:1–85. [Google Scholar]
- 9.Pilon-Smits E. Phytoremediation. Annu Rev Plant Biol. 2005;56:15–39. doi: 10.1146/annurev.arplant.56.032604.144214. [DOI] [PubMed] [Google Scholar]
- 10.Bert V, Macnair MR, Saumitou-Laprade P, De Laguerie P, Petit D. Zinc Tolerance and Accumulation in metallicolous and non-metallicolous populations of Arabidopsis halleri (Brassicaceae). New Phytol. 2000;146:225–233. doi: 10.1046/j.1469-8137.2000.00634.x. [DOI] [PubMed] [Google Scholar]
- 11.Pauwels M, Frerot H, Bonnin I, Saumitou-Laprade P. A broad-scale analysis of population differentiation for Zn tolerance in an emerging model species for tolerance study: Arabidopsis halleri (Brassicaceae). J Evol Biol. 2006;19:1838–1850. doi: 10.1111/j.1420-9101.2006.01178.x. [DOI] [PubMed] [Google Scholar]
- 12.Koch MA, Matschinger M. Evolution and genetic differentiation among relatives of Arabidopsis thaliana. Proc Natl Acad Sci USA. 2007;104:6272–6277. doi: 10.1073/pnas.0701338104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pauwels M, Willems G, Roosens NCJ, Frérot H, Saumitou-Laprade P. Merging methods in molecular and ecological genetics to study the adaptation of plants to anthropogenic metal-pollutedsites: implication for phytoremediation. Mol Ecol. 2008;17:108–119. doi: 10.1111/j.1365-294X.2007.03486.x. [DOI] [PubMed] [Google Scholar]
- 14.Dräger DB, Desbrosses-Fonrouge AG, Krach C, Chardonnens AN, Meyer RC, et al. Two genes encoding Arabidopsis halleri MTP1 metal transport proteins co-segregate with zinc tolerance and account for high MTP1 transcript levels. Plant J. 2004;39:425–439. doi: 10.1111/j.1365-313X.2004.02143.x. [DOI] [PubMed] [Google Scholar]
- 15.Hanikenne M, Talke IN, Haydon MJ, Lanz C, Nolte A, et al. Evolution of metal hyperaccumulation required cis-regulatory changes and triplication of HMA4. Nature. 2008;453:391–396. doi: 10.1038/nature06877. [DOI] [PubMed] [Google Scholar]
- 16.Talke IN, Hanikenne M, Krämer U. Zinc-dependent global transcriptional control, transcriptional deregulation, and higher gene copy number for genes in metal homeostasis of the hyperaccumulator Arabidopsis halleri. Plant Physiol. 2006;142:148–167. doi: 10.1104/pp.105.076232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Desbrosses-Fonrouge AG, Voigt K, Schröder A, Arrivault S, Thomine S, et al. Arabidopsis thaliana MTP1 is a Zn transporter in the vacuolar membrane which mediates Zn detoxification and drives leaf Zn accumulation. FEBS Lett. 2005;579:4165–4174. doi: 10.1016/j.febslet.2005.06.046. [DOI] [PubMed] [Google Scholar]
- 18.Kawachi M, Kobae Y, Mimura T, Maeshima M. Deletion of a histidine-rich loop of AtMTP1, a vacuolar Zn2+/H+ antiporter of Arabidopsis thaliana, stimulates the transport activity. J Biol Chem. 2008;283:8374–8383. doi: 10.1074/jbc.M707646200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van der Zaal BJ, Neuteboom LW, Pinas JE, Chardonnens AN, Schat H, et al. Overexpression of a novel Arabidopsis gene related to putative zinc-transporter genes from animals can lead to enhanced zinc resistance and accumulation. Plant Physiol. 1999;119:1047–1055. doi: 10.1104/pp.119.3.1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Willems G, Dräger DB, Courbot M, Godé C, Verbruggen N, et al. The genetic basis of zinc tolerance in the metallophyte Arabidopsis halleri ssp. halleri (Brassicaceae): an analysis of quantitative trait loci. Genetics. 2007;176:659–674. doi: 10.1534/genetics.106.064485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lacombe E, Cossegal M, Mirouze M, Adam T, Varoquaux F, et al. Construction and characterisation of a BAC library from Arabidopsis halleri: Evaluation of physical mapping based on conserved syntheny with Arabidopsis thaliana. Plant Sci. 2008;174:634–640. [Google Scholar]
- 22.Schranz ME, Lysak MA, Mitchell-Olds T. The ABC's of comparative genomics in the Brassicaceae: building blocks of crucifer genomes. Trends Plant Sci. 2006;11:535–542. doi: 10.1016/j.tplants.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 23.Koizumi S, Suzuki K, Ogra Y, Yamada H, Otsuka F. Transcriptional activity and regulatory protein binding of metal-responsive elements of the human metallothionein-IIA gene. Eur J Biochem. 1999;259:635–642. doi: 10.1046/j.1432-1327.1999.00069.x. [DOI] [PubMed] [Google Scholar]
- 24.MacDiarmid CW, Gaither AL, Eide D. Zinc transporters that regulate vacuolar zinc storage in Saccharomyces cerevisiae. EMBO J. 2000;19:2845–2855. doi: 10.1093/emboj/19.12.2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mirouze M, Sels J, Richard O, Czernic P, Loubet S, et al. A putative novel role for plant defensins: a defensin from the zinc hyperaccumulating plant, Arabidopsis halleri, confers zinc tolerance. Plant J. 2006;47:329–342. doi: 10.1111/j.1365-313X.2006.02788.x. [DOI] [PubMed] [Google Scholar]
- 26.Lin YF, Liang HM, Yang SY, Boch A, Clemens S, et al. Arabidopsis IRT3 is a zinc-regulated and plasma membrane localized zinc/iron transporter. New Phytol. 2009;182:392–404. doi: 10.1111/j.1469-8137.2009.02766.x. [DOI] [PubMed] [Google Scholar]
- 27.Persans MW, Nieman K, Salt DE. Functional activity and role of cation-efflux family members in Ni hyperaccumulation in Thlaspi goesingense. Proc Natl Acad Sci USA. 2001;98:9995–10000. doi: 10.1073/pnas.171039798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Force A, Lynch M, Pickett FB, Amores A, Yan Y-L, Postlethwait J. Preservation of duplicate genes by complementary, degenerative mutations. Genetics. 1999;151:1531–1545. doi: 10.1093/genetics/151.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Church GM, Gilbert W. Genomic sequencing. Proc Natl Acad Sci USA. 1984;81:1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roosens NCJ, Willems G, Godé C, Courseaux A, Saumitou-Laprade P. The use of comparative genome analysis and syntenic relationships allows extrapolating the position of Zn tolerance QTL regions from Arabidopsis halleri into Arabidopsis thaliana. Plant Soil. 2008;306:105–116. [Google Scholar]
- 31.Van Ooijen JW, Voorrips RE. Wageningen: Plant Research International; 2001. JoinMap® version 3.0: software for the calculation of genetic linkage maps. [Google Scholar]
- 32.Kosambi DD. The estimation of map distances from recombination values. Ann Eugen. 1944;12:172–175. [Google Scholar]
- 33.Gietz RD, Woods RA. Transformation of yeast by the LiAc/SS carrier DNA/PEG method. Methods Enzymol. 2002;350:87–96. doi: 10.1016/s0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- 34.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Analysis of the presence of the AhMTP1 paralogues in 44 plants from the Auby accession using gene copy specific primer pairs. Each of the horizontal panel show the amplification obtained from a primer pair specific to the MTP1 paralogue named at the left of the panel. Samples from BAC clones 1F18, 2B14, 7G24, and 12L21 were used as controls for specificity of the primer pairs. The lane M represents1kb invitrogen DNA ladder.
(0.09 MB PDF)
Sequences of gene-specific primer pairs and the corresponding annealing temperatures.
(0.03 MB DOC)
Threshold cycles (Ct) obtained in quantitative RT-PCR analyses performed on cDNAs coming from shoots and roots of A. halleri plants.
(0.06 MB XLS)
Markers used for genetic mapping of AhMTP1 harbouring BAC Clones.
(0.04 MB DOC)