Abstract
Our recent findings demonstrate that Astrocyte Elevated Gene-1 (AEG-1) is overexpressed in >90% of human hepatocellular carcinoma (HCC) samples and AEG-1 plays a central role in regulating development and progression of HCC. In the present manuscript, we elucidate a molecular mechanism of AEG-1-induced chemoresistance, an important characteristic of aggressive cancers. AEG-1 increases the expression of multidrug resistance gene 1 (MDR1) protein resulting in increased efflux and decreased accumulation of doxorubicin (DOX) promoting DOX-resistance. Suppression of MDR1, by siRNA or by chemical reagents, or inhibition of AEG-1 or a combination of both genes significantly increases in vitro sensitivity to DOX. In nude mice xenograft studies, a lentivirus expressing AEG-1 shRNA, in combination with DOX, profoundly inhibited growth of aggressive human HCC cells compared to either agent alone. We document that although AEG-1 does not affect MDR1 gene transcription, it facilitates association of MDR1 mRNA to polysomes resulting in increased translation and AEG-1 also inhibits ubiquitination and subsequent proteasome-mediated degradation of MDR1 protein. This study is the first documentation of a unique aspect of AEG-1 function, i.e., translational and post-translational regulation of proteins. Inhibition of AEG-1 might provide a means of more effectively using chemotherapy to treat HCC, which displays inherent chemoresistance with aggressive pathology.
Keywords: Astrocyte Elevated Gene-1 (AEG-1), doxorubicin, Multidrug resistance gene-1 (MDR1), translation, nude mice
Introduction
The observation that the expected incidence and mortality for hepatocellular carcinoma (HCC) runs closely parallel highlights the paucity of effective treatment modalities for this fatal disease (1). In the US, the estimated new cases of HCC for 2008 were 21,370 out of which 18,410 were expected to die (2). HCC is a tumor with rapid growth and early vascular invasion (3). Most HCC patients present with advanced symptomatic tumors with underlying cirrhotic changes that are not amenable to surgical resection or liver transplantation (4). Even after surgical resection, the recurrence rate is very high. Transarterial chemoembolization (TACE) and systemic therapy with doxorubicin alone or a combination of cisplatin, interferon, doxorubicin and 5-fluorouracil (PIAF) are being used for advanced disease with moderate improvement in overall survival duration varying between 6.8 months to 8.6 months (5–7). These dismal figures point to chemoresistance as an inherent trait of HCC. Identification of key molecules contributing to this chemoresistance and creating strategies to counteract this process will help augment the efficiency of current modalities of treatment and increase patient survival.
Anthracyclines are the most effective anticancer chemotherapeutics and Doxorubicin (DOX) is the first anthracycline isolated from Streptomyces peucetius early in the 1960s (8). DOX is the most common chemotherapeutic used for HCC (9). It is a component of TACE and systemic chemotherapy (6, 7). Although DOX is widely used in diverse cancer indications, development of resistance to DOX is a very common event. The most common mechanism of DOX resistance is the enhanced efflux of drug by cancer cells, including HCC cells, mediated by increases in drug transporter family known as adenosine triphosphate-binding cassette (ABC) transporters (10).
ABC transporters are ATP-dependent efflux pumps with broad drug specificity. As yet, 48 human ABC genes have been identified and divided into seven distinct subfamilies (ABCA to ABCG) on the basis of their sequence homology and domain organization (11). The most common member of this family is ABCB1 (also known as multidrug resistance gene1 or MDR1) that code for the protein P-glycoprotein (P-gp) (12, 13). MDR1 is a broad-spectrum multidrug efflux pump that has 12 transmembrane regions and two ATP-binding sites (14). Overexpression of MDR1 accompanied by a decrease in doxorubicin accumulation has been observed in HCC cell lines (15, 16). MDR1 expression level is an adverse prognostic factor correlated with reduced survival (16, 17). The molecular mechanism of MDR1 overexpression has not been fully elucidated. Epigenetic regulation by promoter demethylation leading to increased expression of MDR1 mRNA is one mechanism of MDR1 overexpression (18). In HCC cells the riboregulator H19 has been implicated in regulating this process (19).
Astrocyte Elevated Gene-1 (AEG-1) is an important gene regulating development and progression of multiple cancers including HCC (20, 21). Our recent results using immunohistochemical analysis of tissue microarray revealed that AEG-1 expression gradually increases with the stages and grades of HCC, while its expression is virtually undetected in normal liver (20). Among 109 HCC patients, >90% showed positive staining for AEG-1. In a subset of patients, the overexpression of AEG-1 mRNA and protein is caused by genomic amplification of the AEG-1 gene (20). Overexpression of AEG-1 in poorly aggressive HCC cells, such as HepG3, generated aggressive, highly vascular and metastatic tumors in nude mice while inhibition of AEG-1 by siRNA in highly aggressive HCC cells, such as QGY-7703, markedly inhibited growth of the tumors in nude mice (20). In HCC cells, AEG-1 activates multiple pro-survival signaling pathways including MEK/ERK, Akt, NF-κB and Wnt signaling (20). Affymetrix microarray confirmed that AEG-1 modulated specific genes regulating invasion, angiogenesis, chemoresistance and senescence thus playing a major role in HCC progression (20).
Chemoresistance is a major effect attributed to AEG-1 expression (22–24). We have shown that AEG-1 confers resistance to 5-fluorouracil (5-FU) by increasing the transcription factor LSF that induces thymidylate synthase, the substrate for 5-FU and by increasing the 5-FU catabolizing enzyme DPYD (22). In the present manuscript we document that AEG-1 also confers resistance to DOX in HCC cells. We elucidate a molecular mechanism by which AEG-1 confers DOX-resistance and demonstrate that inhibition of AEG-1 effectively reverses DOX-resistance. Thus, AEG-1 inhibition might provide an effective strategy to improve DOX-efficacy in HCC patients.
Materials and Methods
Cell lines, culture condition, viability assays and chemical reagents
HepG3 and QGY-7703 human HCC cells were cultured as described (20). Generation of Hep-AEG-1-14 and Hep-AEG-1-8 clones, HepG3 cells stably expressing AEG-1, and Hep-pc-4, HepG3 cells stably transduced with empty vector, has been described previously (20). Cell viability was determined by standard MTT assay as described (20). Doxorubicin was obtained from Sigma. MDR1 inhibitors Verapamil, C-4 and Cyclosporin A were obtained from Calbiochem and were used at a working concentration of 10 µM, 50 µM and 2.5 µM, respectively. MEK/ERK inhibitor PD98059, NF-κB inhibitor quinazoline and PI3 kinase inhibitor LY294002 (all obtained from Calbiochem) were used at a concentration of 10 µM, 1 µM and 10 µM, respectively. Proteosome inhibitor MG132 (Calbiochem) was used at a concentration of 5 µM.
Patient samples
Patient samples were obtained from the Liver Tissue Cell Distribution System (LTCDS), a National Institutes of Health (NIH) service contract to provide human liver and isolated hepatocytes from regional centers for distribution to scientific investigators throughout the United States (NIH contract# #N01-DK-7-0004 / HHSN267200700004C).
siRNAs and lentiviruses
Scrambled siRNA and siRNA for AEG-1 and MDR-1 were obtained from Santa Cruz Biotechnology and were used at a concentration of 20 nM. The 19-bp AEG-1 sequence used to generate AEG-1 shRNA is 5’ CAGAAGAAGAAGAACCGGA 3’. Detailed description of lentivirus vector production is described previously (22, 25).
Doxorubicin efflux and accumulation assays
Cells were treated with 3.44 µM Dox for 2 h, washed in PBS several times and incubated with DOX-free complete growth media for up to 120 min. Cell pellets were analyzed for intracellular DOX accumulation and the bathing media were used for efflux assay. The fluorescence images were obtained by an Olympus fluorescence microscope. DOX fluorescence intensity of intracellular retention and efflux was measured using λexcitation at 490 nm and λemission at 510–570 nm by using a fluorescence spectrophotometer (Turner Biosystems).
Preparation of Whole Cell Lysates and Western Blot Analyses
Preparation of whole cell lysates and Western blot analyses was performed as described (20). The primary antibodies used were anti-AEG-1 (1:5,000; chicken polyclonal; in house) (26), anti-CYP2B6 (1:2,000; mouse monoclonal; BD Biosciences), anti-AKR1C2 (1:1,000; rabbit polyclonal; Santa Cruz), anti-ABCC11 (1:1,000; mouse monoclonal; Abcam), anti-MDR1 (1:1,000; mouse monoclonal; Abcam), Anti-phospho-eIF4G (1:1,000; rabbit polyclonal; Cell Signaling), Anti-eIF4G (1:1,000; rabbit polyclonal; Cell Signaling), Anti-phospho-eIF4E (1:1,000; rabbit polyclonal; Cell Signaling), Anti-eIF4E (1:1,000; rabbit polyclonal; Cell Signaling), and Anti-phospho-4E-BP1 (1:1,000; rabbit polyclonal; Cell Signaling). Blots were stripped and normalized by re-probing with anti-β-tubulin (1:2,000; mouse monoclonal; Sigma). Densitometric analysis was performed using Image J software (http://rsbweb.nih.gov/ij/).
Pulse-chase analysis and immunoprecipitation
Hep-pc-4 and Hep-AEG-1-14 cells were cultured for 4 h in methionine-free medium and then 35S-methionine was added at a concentration of 50 µCi/ml. The cells were incubated for 0 to 12 h, and cell lysates were prepared and subjected to immunoprecipitation using anti-MDR1 antibody as described (27). The immunoprecipitates were run on a SDS-PAGE gel which was exposed to autoradiogram. Immunoprecipitation of ubiquitinated proteins were performed with anti-ubiquitin antibody (mouse monoclonal; Cell Signaling).
Immunofluorescence and immunohistochemical analyses
Immunofluorescence studies in cells and tumor sections and immunohistochemical studies in tumor sections were performed as described (20). For immunofluorescence the primary antibodies used were: anti-MDR1 and anti-Ki67 (1:200; mouse monoclonal; BD Bioscience). The secondary antibody was Alexa 488-conjugated anti-mouse IgG or anti-rabbit IgG (Molecular Probes). The samples were mounted in VectaShield fluorescence mounting medium containing 4,6 -diamidino-2- phenylindole (DAPI; Vector Laboratories). Images were analyzed by a Zeiss confocal laser scanning microscope. For immunohistochemistry anti-AEG-1 antibody was used at 1:200 dilution and anti-MDR1 antibody was used at 1:100 dilution. The sections were developed by avidin-biotin-peroxidase complexes with DAB substrate solution (Vector laboratories). The slides were co-stained with 10% Hematoxylin solution. The images were taken by an Olympus microscope.
Preparation of polysomes and Northern blot analysis
Polysomal fractions were purified essentially as described (28). Each fraction was taken at 500 µL monitoring the absorbance at 260 nm, and polysome fractions were identified (typically fractions 10–20). RNA was extracted from each fraction with Qiagen RNeasy mini kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol, and Northern blotting was done as described using radiolabeled human MDR1 cDNA probe (28).
Nude mice xenograft studies
QGY-7703 cells were transduced with either a lentivirus expressing control (scrambled) shRNA or a lentivirus expressing AEG-1 shRNA at a concentration of 2 moi per cell for 48 h. One million cells were subcutaneously implanted in the flanks of athymic nude mice. Doxorubicin was injected 5 times/week for 2 weeks at a dose of 4 mg/kg. Tumor diameter was measured with calipers at 2 weeks later after injection, and the tumor volume in mm3 was calculated by the formula: (Width)2 × length/2.
TUNEL assay
Apoptotic cell death was detected by the deoxynucleotidyl transferase-mediated dUTP nick end-labeling (TUNEL) method using ApoAlert DNA fragmentation assay kit (Clontech) according to the manufacturer’s protocol. Images were analyzed using an Olympus immunofluorescence microscope.
Statistical analysis
Data were represented as the mean ± Standard Error of Mean (S.E.M) and analyzed for statistical significance using one-way analysis of variance (ANOVA) followed by Newman-Keuls test as a post hoc test. A P value of < 0.05 was considered as significant.
Results
AEG-1 increases MDR-1 protein expression
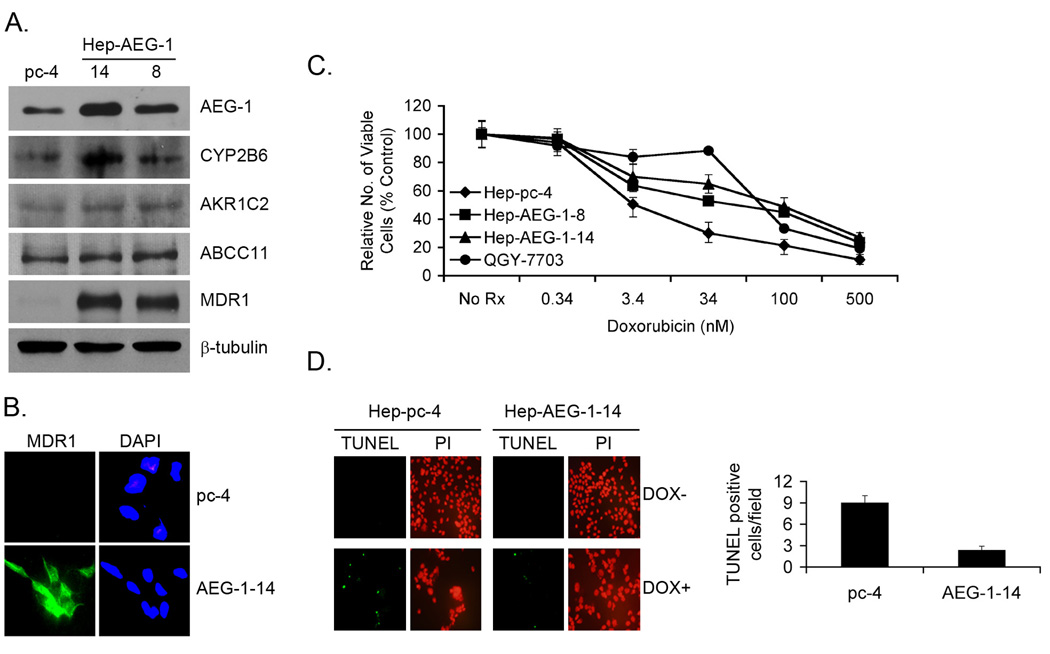
Human HCC cells HepG3 do not form tumors in nude mice and express low levels of AEG-1 (20). In contrast, QGY-7703 HCC cells form aggressive tumors in nude mice and express high levels of AEG-1 (20, 22). Stable overexpression of AEG-1 in HepG3 cells (Hep-AEG-1-14 and Hep-AEG-1-8) significantly increases invasion and anchorage-independent growth and generate highly aggressive, vascular tumors in nude mice (20). To identify AEG-1-downstream genes conferring its oncogenic activity, we performed Affymetrix cDNA microarray analysis between Hep-AEG-1-14 and Hep-pc-4 cells, the latter is HepG3 cells stably transformed with empty pcDNA3.1-hygro vector. A list of the modulated genes has been discussed previously (20). Of relevance to the present studies, we identified induction of several chemoresistance-associated genes by AEG-1 that include (i) dihydropyrimidine dehydrogenase (DPYD), principal enzyme inactivating 5-fluorouracil (5-FU); (ii) cytochrome P4502B6 (CYP2B6), involved in metabolism of multiple drugs; (iii) dihydrodiol dehydrogenase (AKR1C2), conferring resistance to doxorubicin and cisplatin; (iv) the transcription factor LSF which activates the transcription of thymidylate synthase, target of 5-FU; and (v) ABCC11/MRP8, a member of the ABC-family transporter (20, 22). We extensively characterized the consequence of AEG-1-mediated induction of DPYD and LSF that promotes resistance to 5-FU (22). Real-time PCR and Western blot analysis confirmed the AEG-1-mediated induction of CYP2B6 (Fig. 1A) (20). Since AKR1C2 and ABCC11 are known to mediate resistance to DOX we checked whether AEG-1 increases their expression at the protein level to confirm the microarray data. However, very modest increases were observed for AKR1C2 and ABCC11/MRP8 in Hep-AEG-1 clones compared to the Hep-pc-4 clone. Although MDR1 mRNA upregulation was not detected by microarray or real-time PCR (data not shown), robust increase in MDR1 protein level was detected in Hep-AEG-1-14 and Hep-AEG-1-8 clones compared to Hep-pc-4 clone (Fig. 1A). This increase in protein level was confirmed by immunofluorescence analysis (Fig. 1B). Comparable levels of AEG-1 and MDR1 proteins were detected in Hep-AEG-1-14 and QGY-7703 cells (Fig. S1A). We also checked AEG-1 and MDR1 protein expressions in sections of normal liver and matched HCC from five patients by immunohistochemistry. Significantly high level of AEG-1 and MDR1 was observed in HCC sections in comparison to normal liver in all five patients (representative result from one patient is shown in Fig. S1B).
Fig. 1.
AEG-1 induces MDR1 and confers resistance to doxorubicin (DOX). A. Analysis of expression of the indicated proteins in Hep-pc-4 (pc-4), Hep-AEG-1-14 and Hep-AEG-1-8 cells by Western blot analysis. B. Immunofluorescence analysis of MDR1 expression in Hep-pc-4 (pc-4) and Hep-AEG-1-14 (AEG-1-14) cells. C. Cell viability (MTT) assay in the indicated cells with the indicated concentrations of DOX at 7 days post-treatment. Data represents mean ± SEM. D. Analysis of apoptosis by TUNEL assay in Hep-pc-4 and Hep-AEG-1-14 cells treated or untreated with DOX. Left panel, immunofluorescence analysis. PI: propidium iodide. Right panel: graphical representation of the number of TUNEL positive cells per field in DOX-treated Hep-pc-4 and Hep-AEG-1-14 cells. Data represents mean ± SEM.
AEG-1 confers resistance to DOX
Since MDR1 overexpression is commonly associated with DOX resistance, we analyzed cell viability of Hep-pc-4 and Hep-AEG-1-8 and Hep-AEG-1-14 clones as well as QGY-7703 cells (naturally AEG-1-overexpressing HCC cell line) upon treatment with increasing concentrations of DOX. In a one-week assay, the IC50 of DOX for Hep-pc-4 was 3.4 nM, while that for Hep-AEG-1-8, Hep-AEG-1-14 and QGY-7703 cells was 60, 93 and 76 nM, respectively, demonstrating that AEG-1 overexpression results in profound resistance to DOX (Fig. 1C). Detection of apoptosis by TUNEL assay demonstrated a significant decrease in TUNEL-positive cells in the Hep-AEG-1-14 clone compared to the Hep-pc-4 clone upon DOX treatment (Fig. 1D). No TUNEL-positive cells were detected in either clone without DOX treatment (Fig. 1D, left panel). It should be noted that we also observed resistance of these cells towards cisplatin, supporting multidrug resistance, but we used DOX as a prototype drug to dissect the molecular mechanism of AEG-1-induced chemoresistance.
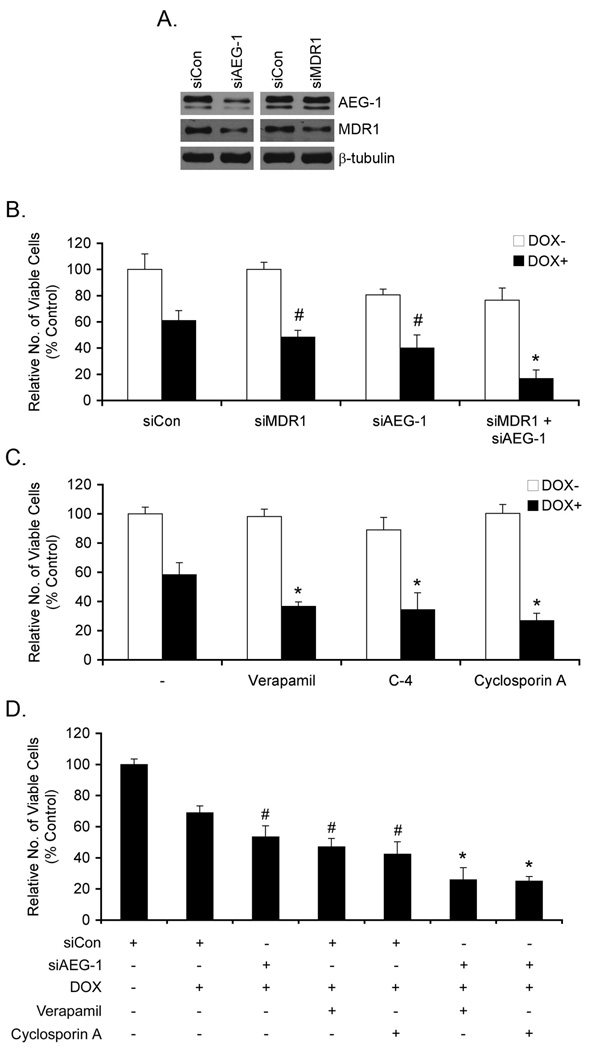
We explored the involvement of AEG-1 and MDR1 in mediating DOX-resistance through both loss-of-function (LOF) and gain-of-function (GOF) experiments. Inhibition of AEG-1 by siRNA significantly decreased the levels of both AEG-1 and MDR1 protein while MDR1 siRNA only decreased MDR1 protein level, but not AEG-1 protein level, indicating that MDR1 is downstream of AEG-1 (Fig. 2A). While MDR1 siRNA alone did not affect viability of Hep-AEG-1-14 cells, it significantly (although modestly) increased the sensitivity to DOX (Fig. 2B). AEG-1 siRNA alone resulted in a significant decrease in viability of Hep-AEG-1-14 cells, since AEG-1 regulates cell proliferation as shown previously (20), and increased sensitivity to DOX significantly (Fig. 2B). However, a combination of MDR1 siRNA and AEG-1 siRNA markedly increased sensitivity to DOX (Fig. 2B). Similar finding was also observed in QGY-7703 cells (Fig. S2). The observation that AEG-1 siRNA was more potent than MDR1 siRNA in conferring sensitivity to DOX might be explained by the regulation of other ABC transporters, such as ABCC11, and metabolic enzymes, such as AKR1C2, or other as yet identified genes by AEG-1 as shown in Fig. 1A. Chemical inhibitors of MDR1, such as Verapamil, C-4 and Cyclosporin A, also increased sensitivity of Hep-AEG-14 cells to DOX (Fig. 2C). A combination of AEG-1 siRNA with Verapamil or Cyclosporin A enhanced the sensitivity of Hep-AEG-1-14 cells to DOX (Fig. 2D).
Fig. 2.
Inhibition of MDR1 or AEG-1 reverses resistance to DOX. A. Hep-AEG-1-14 cells were transfected with control siRNA (siCon), AEG-1 siRNA (siAEG-1) or MDR1 siRNA (siMDR1) and expression of AEG-1, MDR1 and β-tubulin was analyzed by Western blot. B. Hep-AEG-1-14 cells were transfected with siCon, siAEG-1 or siMDR1 or a combination or siAEG-1 and siMDR1 and treated with 3.4 nM DOX. #: p<0.05 vs siCon; *: p<0.01 vs siCon. C. Hep-AEG-1-14 cells were treated with Verapamil (10 µM), C-4 (50 µM) or Cyclosporin A (2.5 µM) and treated with 3.4 nM DOX. *: p<0.01 vs DOX treatment only. D. Hep-AEG-1-14 cells were transfected with siCon or siAEG-1, treated with Verapamil (10 µM) or Cyclosporin A (2.5 µM) and treated with 3.4 nM DOX. #: p<0.05 vs siCon+DOX; *: p<0.01 vs siCon+DOX. B–D: Cell viability was analyzed 7 days post-treatment. Data represents mean ± SEM.
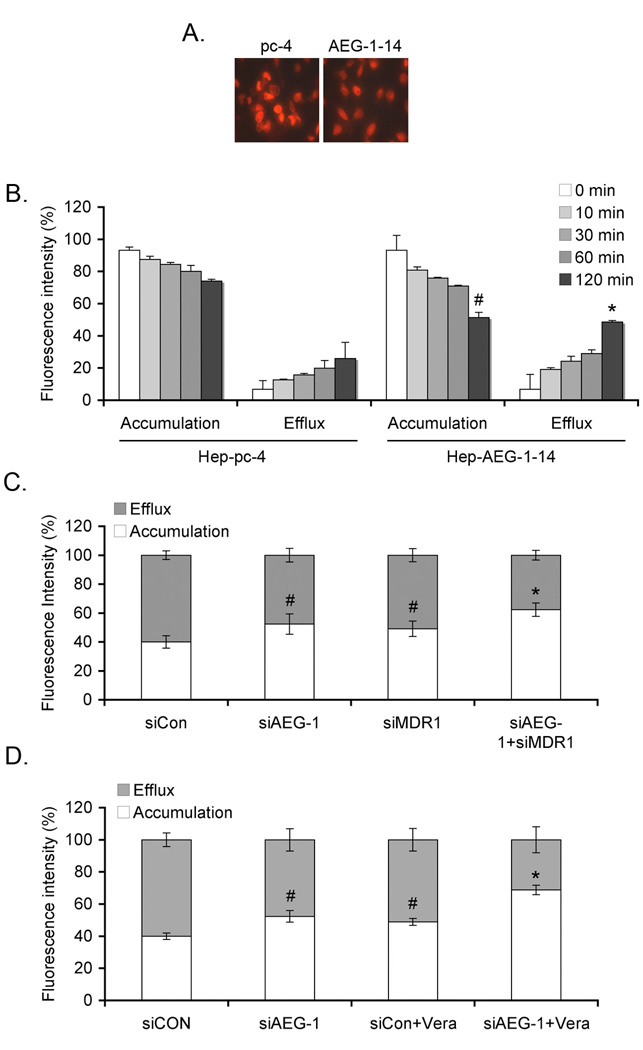
Since MDR1 functions by increasing drug efflux and decreasing drug accumulation we next determined the efflux and accumulation of DOX in Hep-pc-4 and Hep-AEG-1-14 cells. In a kinetic analysis, there was a significant increase in the efflux of DOX and a significant decrease in accumulation of DOX in Hep-AEG-1-14 cells when compared to Hep-pc-4 cells (Fig. 3A and 3B). In Hep-AEG-14 cells, DOX accumulation was significantly increased and DOX efflux was significantly decreased by treatment with AEG-1 siRNA or MDR1 siRNA and these effects were further augmented when treated with a combination of both siRNAs (Fig. 3C). Similarly, a combination of AEG-1 siRNA and verapamil also significantly increased DOX accumulation and decreased DOX efflux in Hep-AEG-1-14 cells, when compared to either agent used alone (Fig. 3D).
Fig. 3.
AEG-1 increases DOX efflux and decreases DOX accumulation. A. Hep-pc-4 and Hep-AEG-1-14 cells were treated with 3.44 µM Dox for 2 h, washed in PBS several times and incubated with DOX-free complete growth media for 2 h. Intracellular accumulation of DOX was determined by fluoroscence microscopy. B. Cells were treated as in A for the indicated time points. Cell pellets were analyzed for intracellular DOX accumulation and the bathing media were used for efflux assay as described in Materials and Methods. #: p<0.01 vs accumulation in Hep-pc-4 cells at 120 min; *: p<0.01 vs efflux in Hep-pc-4 cells at 120 min. C. Hep-AEG-1-14 cells were transfected with siCon, siAEG-1 or siMDR1 or a combination of siAEG-1 and siMDR1 and treated with 3.44 µM Dox for 2 h. DOX accumulation and efflux assays were performed as in B. #: p<0.05 vs siCon; *: p<0.01 vs siCon. D. Hep-AEG-1-14 cells were transfected with siCon or siAEG-1, treated with Verapamil (Vera; 10 µM) and treated with 3.44 µM Dox for 2 h. DOX accumulation and efflux assays were performed as in B. #: p<0.05 vs siCon; *: p<0.01 vs siCon. Data represents mean ± SEM.
AEG-1 increases translation of MDR-1 protein
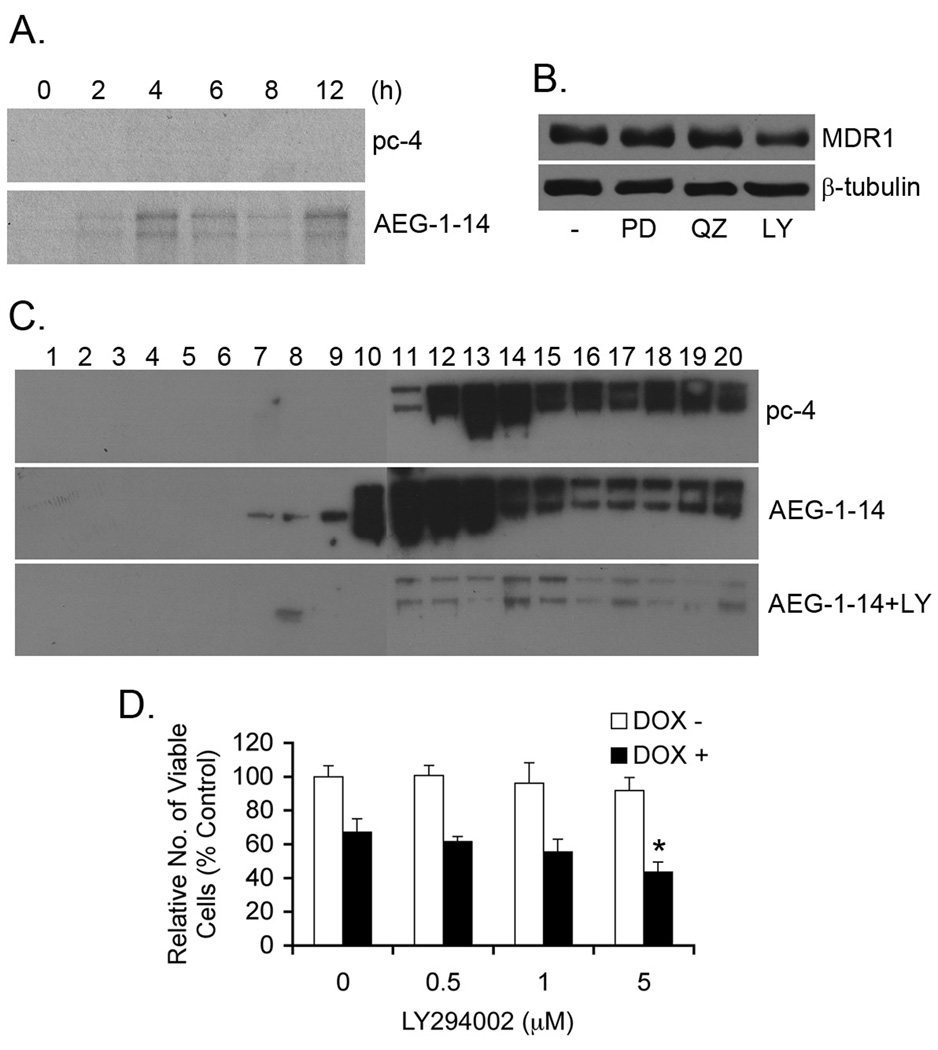
Based on our microarray data, it appears that AEG-1 increases MDR1 protein level without affecting MDR1 transcription. This increase in MDR1 protein by AEG-1 might be due to an increase in the translation of MDR1 protein or a decrease in ubiquitin-proteasome-mediated degradation of MDR1 protein. Pulse-chase experiment using 35S-labeled methionine clearly demonstrated increased translation of MDR1 protein in Hep-AEG-1-14 cells compared to Hep-pc-4 cells (Fig. 4A). AEG-1 is known to activate MEK/ERK, NF-κB and PI3K/Akt signaling pathways in HCC cells (20). Using chemical inhibitors of these 3 pathways, PD98059, Quinazoline and LY294002, respectively, it was observed that treatment with only LY294002 downregulated MDR1 protein level in the Hep-AEG-1-14 clone indicating that activation of PI3K/Akt pathway by AEG-1 contributes to an increase in MDR1 level in these cells (Fig. 4B). Analyzing the association of MDR1 mRNA with polysomes further corroborated the importance of PI3K/Akt pathway in mediating MDR1 translation by AEG-1. Polysomal fractions were isolated from Hep-pc-4 and Hep-AEG-1-14 cells as well as Hep-AEG-1-14 cells treated with LY294002 and the association of MDR1 mRNA to the polysomes was analyzed by Northern blot analysis. Increased association of MDR1 mRNA to polysomes was clearly observed in Hep-AEG-1-14 cells compared to Hep-pc-4 cells, which was profoundly inhibited by LY294002 (Fig. 4C). While treatment of Hep-AEG-1-14 cells with DOX alone decreased cell viability by ~30%, in combination with a non-cytotoxic concentration of LY294002 (5 µM) the decrease in cell viability was increased to ~57% providing further confirmation of the role of PI3/Akt pathway in mediating DOX-resistance (Fig. 4D).
Fig. 4.
AEG-1 increases translation of MDR-1 via PI3K/Akt pathway. A. Pulse-chase analysis of Hep-pc-4 (pc-4) and Hep-AEG-1-14 (AEG-1-14) cells as described in Materials and Methods. Immunoprecipitation was performed using anti-MDR1 antibody. B. Hep-AEG-14 cells were treated with PD98059 (PD; 10 µM), quinazoline (QZ; 1 µM) and LY294002 (LY; 10 µM) for 24 h and expression of MDR1 and β-tubulin was analyzed by Western blot. C. Polysomal fractions were purified from the cell lysates of Hep-pc-4 (pc-4), Hep-AEG-1-14 (AEG-1-14) cells or Hep-AEG-1-14 cells treated for 24 h with 10 µM LY294002 (AEG-1-14+LY) as described in Materials and Methods. Polysomal RNA was purified from each fraction and Northern blotting was done with a human MDR1 cDNA probe. The numbers represent fraction numbers, with fractions 10 to 20 representing the polysomes. D. Hep-AEG-1-14 cells were treated with the indicated concentrations of LY294002 with or without 3.4 nM DOX. Cell viability was analyzed 5 days post-treatment. Data represents mean ± SEM. *: p<0.01 vs DOX treatment alone.
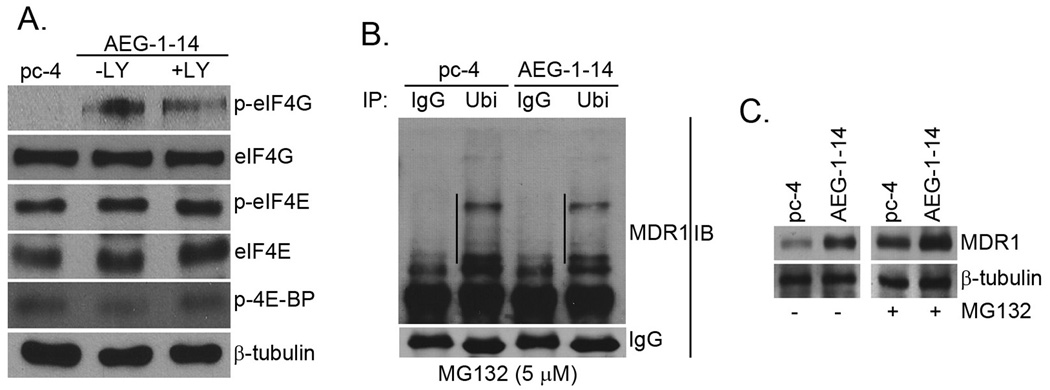
In addition to facilitating association of MDR1 mRNA to polysomes, we checked whether AEG-1 also affects the translational machinery. We checked the phosphorylation and activation status of key cap-dependent translational initiation factors, namely eukaryotic translation initiation factor 4E (eIF4E), eukaryotic translation initiation factor 4G (eIF4G) and 4E binding protein (4E-BP) in Hep-pc-4 cells and Hep-AEG-1-14 cells untreated or treated with LY294002. The reason to check these particular proteins is that they are modulated by mammalian target of rapamycin (mTOR) pathway, a PI3K/Akt-downstream signaling pathway, which plays a fundamental role in regulating translation initiation. Marked phosphorylation of eIF4G was observed in Hep-AEG-1-14 cells compared to Hep-pc-4 cells, which was not affected by LY294002 treatment that was confirmed by densitometric analysis (Fig. 5A). The phosphorylation of eIF4E and 4E-BP was not modulated by AEG-1.
Fig. 5.
AEG-1 augments translation initiation and decreases degradation of MDR-1. A. The expression of the indicated proteins were detected in Hep-pc-4 (pc-4) cells and Hep-AEG-1-14 (AEG-1-14) cells treated or untreated with 10 µM LY294002 for 24 h by Western blot analysis. B. Hep-pc-4 (pc-4) and Hep-AEG-1-14 (AEG-1-14) cells were treated with MG132 (5 µM) for 24 h and the cell lysates were subjected to immunoprecipitation by anti-ubiquitin antibody (Ubi). The immunoprecipitates were subjected to Western blot analysis using anti-MDR1 antibody. IgG: normal mouse IgG. The lines in the figure indicate ubiquitinated MDR1. C. Hep-pc-4 (pc-4) and Hep-AEG-1-14 (AEG-1-14) cells were treated with MG132 (5 µM) for 24 h and the expression of MDR1 and β-tubulin was analyzed by Western blot.
To check if AEG-1 modulates ubiquitination and degradation of MDR1 protein, the total level of ubiquitinated MDR1 was determined in Hep-pc-4 and Hep-AEG-1-14 cells. The cells were treated with the proteasome inhibitor MG132 and the cell lysates were subjected to immunoprecipitation with anti-ubiquitin antibody followed by immunoblot with anti-MDR1 antibody. The level of ubiquitinated MDR1 was significantly lower in the Hep-AEG-1-14 clone compared to the Hep-pc-4 clone (Fig. 5B). As a corollary, treatment of Hep-pc-4 and Hep-AEG-1-14 clones with MG132 significantly increased the protein level of MDR1 in both clones (Fig. 5C). MG132 treatment increased MDR1 protein level of Hep-pc-4 cells to a level that is comparable to basal AEG-1 level in Hep-AEG-1-14 cells (Fig. 5C). These findings indicate that AEG-1 might interfere with ubiquitination and proteasomal degradation of MDR1 protein, thus increasing the total MDR1 protein level.
Combination of AEG-1 shRNA and DOX significantly inhibit HCC xenograft growth in nude mice
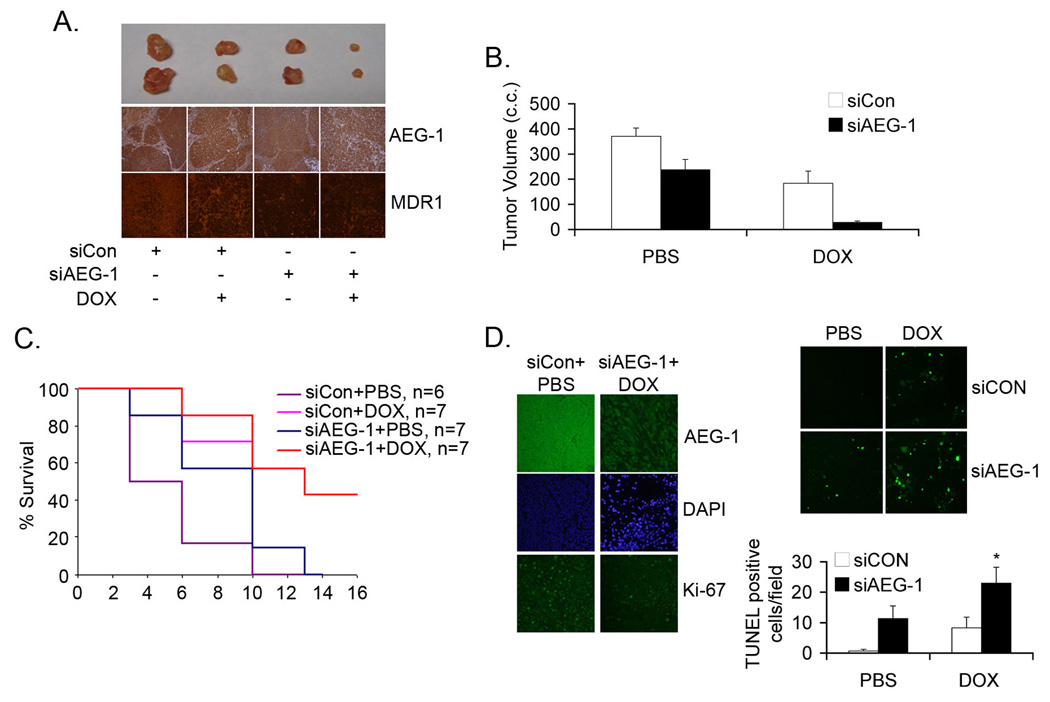
The in vitro findings that inhibition of AEG-1 might overcome DOX resistance were confirmed in in vivo nude mice xenograft studies. QGY-7703 cells were ex vivo transduced with a lentivirus expressing control siRNA (Lenti.siCon) or AEG-1 siRNA (Lenti.siAEG-1) and two days later the cells were subcutaneously implanted on the flanks of athymic nude mice. After establishment of the tumor (~100 mm3 requiring ~7 days) the animals received intraperitoneal injection of either PBS or DOX (4 mg/kg) 5 days/week for 2 weeks. Inhibition of AEG-1 alone resulted in significant inhibition of tumor growth (Fig. 6A–B). While DOX treatment alone also resulted in inhibition of tumor growth, the combination of AEG-1 inhibition and DOX treatment resulted in profound inhibition of tumor growth versus either agent alone (Fig. 6A–B). Analysis of tumor sections revealed that Lenti.siAEG-1 treatment resulted in profound downregulation of AEG-1 and MDR1 proteins compared to Lenti.siCon treatment in combination with PBS or DOX (Fig. 6A). Kaplan-Meier survival curves demonstrated that 100% siCon + PBS-treated animals died within 10 days of starting the treatment while treatment with either siAEG-1 or DOX alone resulted in 100% mortality within 14 days (Fig. 6C). Combinatorial treatment of siAEG-1 and DOX resulted in a significant increase in the survival of the animals (Fig. 6C). Treatment with siAEG-1 + DOX significantly downregulated Ki-67 staining, a marker for proliferation, in tumor samples (11.67 ± 3.5 Ki-67 positive cells/field) compared to siCon + PBS-treated tumors (41.33 ± 6.5 Ki-67 positive cells/field) (Fig. 6D, left panel). While siAEG-1 or DOX alone increased TUNEL-positive cells in the tumor section, the combination of both agents markedly increased TUNEL-positive cells indicating that the combination treatment decreases proliferation and increases apoptosis therefore resulting in profound inhibition of tumor growth (Fig. 6D, right panel).
Fig. 6.
Combination of AEG-1 inhibition and DOX inhibits growth of QGY-7703 cells in athymic nude mice. A. QGY-7703 cells were ex vivo transduced with a lentivirus expressing control siRNA (siCon) or AEG-1 siRNA (siAEG-1). Two days later the cells were subcutaneously implanted onto the flanks of athymic nude mice. After the establishment of the tumors the animals were treated with DOX 5 times/week for 2 weeks at a dose of 4 mg/kg. Top panel: photomicrograph of tumors of the different treatment groups at the end of the study. Middle panel: AEG-1 expression in the tumor sections at the end of the study. Bottom panel: MDR1 expression in the tumor sections at the end of the study. B. Tumor volume was measured at the end of the study (2 weeks after the initiation of DOX treatment). C. Kaplan-Meier survival curve of the different treatment groups. Data represents mean ± SEM. D. Left panel: Tumor sections from the indicated treatment groups were stained for AEG-1 (top), DAPI (middle) or Ki-67 (bottom) as described in the Materials and Methods. Top right panel: Tumor sections from the indicated treatment groups were subjected to TUNEL assay. Bottom right panel: Graphical representation of the number of TUNEL positive cells per field in the indicated groups. Data represents mean ± SEM. *: p<0.01 vs siAEG-1 or DOX alone.
Discussion
The ability of AEG-1 to confer chemoresistance represents an important attribute of this cancer-promoting gene. We have documented that AEG-1 activates a specific pathway involving LSF transcription factor and the catabolizing enzyme DPYD to confer resistance to 5-FU (22). A study using neuroblastoma cells demonstrate that AEG-1 mediates resistance to DOX and cisplatin, although the molecular mechanism of the resistance was not explored (24). Studies using breast cancer cells showed resistance to broad-spectrum chemotherapeutics conferred by AEG-1 that involves upregulation of aldehyde dehydrogenase 3 family, member A1 (ALDH3A1) and hepatocyte growth factor receptor (MET) (23). These two candidates were identified by microarray analysis looking for AEG-1 downstream genes. In the microarray analysis MDR1 mRNA upregulation was not observed since we now show that MDR1 upregulation by AEG-1 occurs at the protein level. DOX is the most common chemotherapeutic used for HCC. However, the clinical efficacy of DOX is not profound thus emphasizing an inherent resistance of HCC cells to DOX that has been attributed to overexpression of MDR1 (16, 17). Our observation that AEG-1 increases MDR1 protein level and contributes to DOX resistance has important potential clinical significance. Since AEG-1 confers a broad-spectrum resistance to chemotherapeutics and since AEG-1 is overexpressed in >90% of human HCC samples, inhibition of AEG-1, either by Lenti.siAEG-1 or by small molecules, might be considered as a component of any combinatorial therapeutic strategy for HCC.
Our findings indicate that AEG-1 increases MDR1 protein at multiple levels. AEG-1 facilitates association of MDR1 mRNA to polysomes thus increasing translation and inhibits polyubiquitination and subsequent degradation by proteasomes. Modulation of both processes by a single molecule is very intriguing. Indeed, AEG-1 is a distinctive molecule with pleiotropic properties. It resides on the cell surface facilitating adhesion to the endothelium, thus helping to facilitate metastasis (23, 29). It is also located in the nucleus where it functions as a transcriptional co-activator, increasing NF-κB activity, and inhibits transcriptional repression by PLZF thereby augmenting c-Myc transcription (27, 30, 31). In addition, AEG-1 is also located in the Endoplasmic Reticulum (ER) as well as in the cytoplasm (32). Our present studies indicate potential functions of the cytoplasmic AEG-1, i.e., regulation of translation and post-translational modification of target proteins.
How does AEG-1 affect translation? We document that by activating the PI3K/Akt pathway, AEG-1 increases MDR1 translation by augmenting the association of MDR1 mRNA to polysomes. However, the mechanism by which AEG-1 activates the PI3K/Akt pathway remains poorly defined. Additionally, the mechanism by which the activated PI3K/Akt pathway increases association of MDR1 mRNA to polysomes also needs clarification. We document that AEG-1 causes phosphorylation of translation initiation factor eIF4G and this phosphorylation was not affected by co-treatment with LY294002. On the other hand, we did not detect phosphorylation of eIF4E or 4E-BP by AEG-1. Hypophosphorylation or 4E-BP binds to the 5′ cap binding protein of eIF4E, thereby preventing its interaction with eIF4G and inhibiting translation (33). Phosphorylation of 4E-BP via the mTOR pathway, that lies downstream of the PI3K/Akt pathway, releases 4E-BP from eIF4E resulting in the recruitment of eIF4G to the 5’ cap and thereby allowing translation initiation to proceed (33). Since we did not detect 4E-BP phosphorylation by AEG-1, the augmentation of translation by AEG-1 might be independent of the mTOR pathway. Several studies reported alterations in eIF4E-eIF4G association that were independent of changes in 4E-BP1 binding to eIF4E and a potential mechanism for enhanced eIF4E to eIF4G assembly may involve phosphorylation of eIF4G (34–39). Increased phosphorylation of eIF4G correlates with conditions known to stimulate protein synthesis (38). Similarly, decreased eIF4G phosphorylation in the presence of mTOR activation is observed in skeletal muscle of septic rats, a condition that manifests resistance to stimulation of protein synthesis by insulin (40). Thus, eIF4G phosphorylation might occur by an mTOR-independent pathway. One possible mechanism by which AEG-1 facilitates translation initiation might be direct involvement of AEG-1 in the translational machinery. AEG-1 is a highly basic protein (26) that might function as a scaffold facilitating binding of nucleic acids, including mRNA. Our efforts aimed at defining AEG-1-interacting proteins have identified multiple proteins involved in translation initiation and elongation as well as ribosomal proteins as potential interacting partners of AEG-1 (B.K.Y. & D.S., unpublished data). Our ongoing studies aim at establishing relevance of these potential interactions and deciphering the role of AEG-1 in regulating translation initiation. The molecular mechanism by which AEG-1 interferes with ubiquitination and proteasomal degradation of MDR1 protein also requires further experimentation. Deciphering these molecular pathways will provide an in-depth understanding of the multitude of functions of AEG-1 and how it contributes to normal and abnormal cellular physiology.
In summary, the present manuscript reveals novel aspects of AEG-1 function, identifies a novel regulator of MDR1 and ushers in new ways of potentially overcoming chemoresistance in HCC and other cancers. Since AEG-1 is being identified as a key player and prognostic indicator in diverse histologically distinct cancers (21), efforts are justified in developing strategies to effectively inhibit AEG-1 as a potentially important cancer therapeutic.
Supplementary Material
Acknowledgements
The present study was supported in part by The Goldhirsh Foundation grant and The Dana Foundation grant (DS); NIH grants R01 CA134721 and R01 CA035675 (PBF). DS is the Harrison Endowed Scholar in Cancer Research. PBF holds the Thelma Newmeyer Corman Chair in Cancer Research and is a Samuel Waxman Cancer Research Foundation (SWCRF) Investigator.
References
- 1.El-Serag HB, Rudolph KL. Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology. 2007;132:2557–2576. doi: 10.1053/j.gastro.2007.04.061. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Ward E, et al. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 3.Pang RW, Joh JW, Johnson PJ, Monden M, Pawlik TM, Poon RT. Biology of hepatocellular carcinoma. Ann Surg Oncol. 2008;15:962–971. doi: 10.1245/s10434-007-9730-z. [DOI] [PubMed] [Google Scholar]
- 4.Poon RT, Fan ST, Lo CM, et al. Improving survival results after resection of hepatocellular carcinoma: a prospective study of 377 patients over 10 years. Ann Surg. 2001;234:63–70. doi: 10.1097/00000658-200107000-00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Llovet JM, Bruix J. Systematic review of randomized trials for unresectable hepatocellular carcinoma: Chemoembolization improves survival. Hepatology. 2003;37:429–442. doi: 10.1053/jhep.2003.50047. [DOI] [PubMed] [Google Scholar]
- 6.Llovet JM, Real MI, Montana X, et al. Arterial embolisation or chemoembolisation versus symptomatic treatment in patients with unresectable hepatocellular carcinoma: a randomised controlled trial. Lancet. 2002;359:1734–1739. doi: 10.1016/S0140-6736(02)08649-X. [DOI] [PubMed] [Google Scholar]
- 7.Patt YZ, Hoque A, Roh M, et al. Durable clinical and pathologic response of hepatocellular carcinoma to systemic and hepatic arterial administration of platinol, recombinant interferon alpha 2B, doxorubicin, and 5-fluorouracil: a communication. Am J Clin Oncol. 1999;22:209–213. doi: 10.1097/00000421-199904000-00024. [DOI] [PubMed] [Google Scholar]
- 8.Minotti G, Menna P, Salvatorelli E, Cairo G, Gianni L. Anthracyclines: molecular advances and pharmacologic developments in antitumor activity and cardiotoxicity. Pharmacol Rev. 2004;56:185–229. doi: 10.1124/pr.56.2.6. [DOI] [PubMed] [Google Scholar]
- 9.Nerenstone S, Friedman M. Medical treatment of hepatocellular carcinoma. Gastroenterol Clin North Am. 1987;16:603–612. [PubMed] [Google Scholar]
- 10.Gottesman MM, Fojo T, Bates SE. Multidrug resistance in cancer: role of ATP-dependent transporters. Nature Rev. 2002;2:48–58. doi: 10.1038/nrc706. [DOI] [PubMed] [Google Scholar]
- 11.Dean M, Rzhetsky A, Allikmets R. The human ATP-binding cassette (ABC) transporter superfamily. Genome Res. 2001;11:1156–1166. doi: 10.1101/gr.184901. [DOI] [PubMed] [Google Scholar]
- 12.Juliano RL, Ling V. A surface glycoprotein modulating drug permeability in Chinese hamster ovary cell mutants. Biochim Biophys Acta. 1976;455:152–162. doi: 10.1016/0005-2736(76)90160-7. [DOI] [PubMed] [Google Scholar]
- 13.Ueda K, Cardarelli C, Gottesman MM, Pastan I. Expression of a full-length cDNA for the human "MDR1" gene confers resistance to colchicine, doxorubicin, and vinblastine. Proc Natl Acad Sci U S A. 1987;84:3004–3008. doi: 10.1073/pnas.84.9.3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen CJ, Chin JE, Ueda K, et al. Internal duplication and homology with bacterial transport proteins in the mdr1 (P-glycoprotein) gene from multidrug-resistant human cells. Cell. 1986;47:381–389. doi: 10.1016/0092-8674(86)90595-7. [DOI] [PubMed] [Google Scholar]
- 15.Park JG, Lee SK, Hong IG, et al. MDR1 gene expression: its effect on drug resistance to doxorubicin in human hepatocellular carcinoma cell lines. J Natl Cancer Inst. 1994;86:700–705. doi: 10.1093/jnci/86.9.700. [DOI] [PubMed] [Google Scholar]
- 16.Ng IO, Liu CL, Fan ST, Ng M. Expression of P-glycoprotein in hepatocellular carcinoma. A determinant of chemotherapy response. Am J Clin Pathol. 2000;113:355–363. doi: 10.1309/AC1M-4TY4-U0TN-EN7T. [DOI] [PubMed] [Google Scholar]
- 17.Kato A, Miyazaki M, Ambiru S, et al. Multidrug resistance gene (MDR-1) expression as a useful prognostic factor in patients with human hepatocellular carcinoma after surgical resection. J Surg Oncol. 2001;78:110–115. doi: 10.1002/jso.1129. [DOI] [PubMed] [Google Scholar]
- 18.Kantharidis P, El-Osta A, deSilva M, et al. Altered methylation of the human MDR1 promoter is associated with acquired multidrug resistance. Clin Cancer Res. 1997;3:2025–2032. [PubMed] [Google Scholar]
- 19.Tsang WP, Kwok TT. Riboregulator H19 induction of MDR1-associated drug resistance in human hepatocellular carcinoma cells. Oncogene. 2007;26:4877–4881. doi: 10.1038/sj.onc.1210266. [DOI] [PubMed] [Google Scholar]
- 20.Yoo BK, Emdad L, Su ZZ, et al. Astrocyte elevated gene-1 regulates hepatocellular carcinoma development and progression. J Clin Invest. 2009;119:465–477. doi: 10.1172/JCI36460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sarkar D, Emdad L, Lee S-G, Yoo BK, Su Z-Z, Fisher PB. Astrocyte elevated gene-1 (AEG-1): far more than just a gene regulated in astrocytes. Cancer Res. 2009 doi: 10.1158/0008-5472.CAN-09-1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoo BK, Gredler R, Vozhilla N, et al. Identification of genes conferring resistance to 5-fluorouracil. Proc Natl Acad Sci U S A. 2009;106:12938–12943. doi: 10.1073/pnas.0901451106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu G, Chong RA, Yang Q, et al. MTDH activation by 8q22 genomic gain promotes chemoresistance and metastasis of poor-prognosis breast cancer. Cancer Cell. 2009;15:9–20. doi: 10.1016/j.ccr.2008.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu H, Song X, Liu C, Xie L, Wei L, Sun R. Knockdown of astrocyte elevated gene-1 inhibits proliferation and enhancing chemo-sensitivity to cisplatin or doxorubicin in neuroblastoma cells. J Exp Clin Cancer Res. 2009;28:19. doi: 10.1186/1756-9966-28-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kock N, Kasmieh R, Weissleder R, Shah K. Tumor therapy mediated by lentiviral expression of shBcl-2 and S-TRAIL. Neoplasia. 2007;9:435–442. doi: 10.1593/neo.07223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kang DC, Su ZZ, Sarkar D, Emdad L, Volsky DJ, Fisher PB. Cloning and characterization of HIV-1-inducible astrocyte elevated gene-1, AEG-1. Gene. 2005;353:8–15. doi: 10.1016/j.gene.2005.04.006. [DOI] [PubMed] [Google Scholar]
- 27.Sarkar D, Park ES, Emdad L, Lee SG, Su ZZ, Fisher PB. Molecular basis of nuclear factor-kappaB activation by astrocyte elevated gene-1. Cancer Res. 2008;68:1478–1484. doi: 10.1158/0008-5472.CAN-07-6164. [DOI] [PubMed] [Google Scholar]
- 28.Lebedeva IV, Sarkar D, Su ZZ, et al. Molecular target-based therapy of pancreatic cancer. Cancer Res. 2006;66:2403–2413. doi: 10.1158/0008-5472.CAN-05-3510. [DOI] [PubMed] [Google Scholar]
- 29.Brown DM, Ruoslahti E. Metadherin, a cell surface protein in breast tumors that mediates lung metastasis. Cancer Cell. 2004;5:365–374. doi: 10.1016/s1535-6108(04)00079-0. [DOI] [PubMed] [Google Scholar]
- 30.Emdad L, Sarkar D, Su ZZ, et al. Activation of the nuclear factor kappaB pathway by astrocyte elevated gene-1: implications for tumor progression and metastasis. Cancer Res. 2006;66:1509–1516. doi: 10.1158/0008-5472.CAN-05-3029. [DOI] [PubMed] [Google Scholar]
- 31.Thirkettle HJ, Mills IG, Whitaker HC, Neal DE. Nuclear LYRIC/AEG-1 interacts with PLZF and relieves PLZF-mediated repression. Oncogene. 2009;28:3663–3670. doi: 10.1038/onc.2009.223. [DOI] [PubMed] [Google Scholar]
- 32.Sutherland HG, Lam YW, Briers S, Lamond AI, Bickmore WA. 3D3/lyric: a novel transmembrane protein of the endoplasmic reticulum and nuclear envelope, which is also present in the nucleolus. Exp Cell Res. 2004;294:94–105. doi: 10.1016/j.yexcr.2003.11.020. [DOI] [PubMed] [Google Scholar]
- 33.Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol. 2009;10:307–318. doi: 10.1038/nrm2672. [DOI] [PubMed] [Google Scholar]
- 34.Vary TC, Jefferson LS, Kimball SR. Amino acid-induced stimulation of translation initiation in rat skeletal muscle. Am J Physiol. 1999;277:E1077–E1086. doi: 10.1152/ajpendo.1999.277.6.E1077. [DOI] [PubMed] [Google Scholar]
- 35.Fraser CS, Pain VM, Morley SJ. The association of initiation factor 4F with poly(A)-binding protein is enhanced in serum-stimulated Xenopus kidney cells. J Biol Chem. 1999;274:196–204. doi: 10.1074/jbc.274.1.196. [DOI] [PubMed] [Google Scholar]
- 36.Vary TC, Jefferson LS, Kimball SR. Role of eIF4E in stimulation of protein synthesis by IGF-I in perfused rat skeletal muscle. Am J Physiol Endocrinol Metab. 2000;278:E58–E64. doi: 10.1152/ajpendo.2000.278.1.E58. [DOI] [PubMed] [Google Scholar]
- 37.Morley SJ, Traugh JA. Differential stimulation of phosphorylation of initiation factors eIF-4F, eIF-4B, eIF-3, and ribosomal protein S6 by insulin and phorbol esters. J Biol Chem. 1990;265:10611–10616. [PubMed] [Google Scholar]
- 38.Raught B, Gingras AC, Gygi SP, et al. Serum-stimulated, rapamycin-sensitive phosphorylation sites in the eukaryotic translation initiation factor 4GI. Embo J. 2000;19:434–444. doi: 10.1093/emboj/19.3.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vary TC, Jefferson LS, Kimball SR. Insulin fails to stimulate muscle protein synthesis in sepsis despite unimpaired signaling to 4E-BP1 and S6K1. Am J Physiol Endocrinol Metab. 2001;281:E1045–E1053. doi: 10.1152/ajpendo.2001.281.5.E1045. [DOI] [PubMed] [Google Scholar]
- 40.Jurasinski C, Gray K, Vary TC. Modulation of skeletal muscle protein synthesis by amino acids and insulin during sepsis. Metabolism. 1995;44:1130–1138. doi: 10.1016/0026-0495(95)90005-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.