Summary
Fluorescent proteins (FP) have become essential tools in molecular and biological applications. Here, we present a novel fluorescent protein isolated from warm water coral, Cyphastrea microphthalma. The protein, which we named VFP (vivid Verde FP), matures readily at 37 °C and emits bright green light. Further characterizations revealed that VFP has a tendency to form dimers. By creating a homology model of VFP, based on the structure of red fluorescent protein DsRed, we were able to make mutations that alter the protein’s oligomerization state. We present two proteins, mVFP and mVFP1, that are both exclusively monomeric, and one, dVFP, which is dimeric. We characterized the spectroscopic properties of VFP and its variants in comparison with enhance green fluorescent protein (EGFP), a widely use variant of GFP. All the VFP variants are at least twice as bright as EGFP. Finally, we demonstrate the effectiveness of the VFP variants both in vitro and in vivo detection applications.
Keywords: fluorescent protein, relative brightness, oligomeric states, fluorescence correlation spectroscopy, detection marker
Introduction
Fluorescent proteins (FPs) have become ubiquitous tools in biological and biomedical research. Since the cloning and exogenous expression of green fluorescent protein (GFP) from the jellyfish Aequorea victoria, researchers have sought new variants of this protein as well as other FPs, with properties that are well-suited for a particular application [1–3]. Extensive mutagenesis has been performed on FPs to better tailor its properties to the needs of biologists [1, 2, 4, 5]. Of special interest are FPs with new excitation and emission wavelengths, FPs with increased brightness, FPs that are monomeric, and FPs that mature rapidly at 37°C.
GFP is a 238 amino acid protein, whose chromophore is formed by the post-translational rearrangement of an internal Ser-Tyr-Gly sequence to a 4-(p-hydroxybenzylidene)-imidazolidine-5-one structure [6]. Crystal structure of GFP revealed that the chromophore is buried in the center of a β-barrel structure [7, 8]. Amino acid mutagenesis and protein engineering were applied on GFP to improve its spectral characteristics, oligomeric state, and chromophore-maturation at 37°C [6, 9–12]. A broad range of GFP variants with fluorescence emission ranging from blue to yellow regions of the visible spectrum was created [1, 2]. Enhanced green fluorescent protein (EGFP) is a widely used variant of GFP, which has mutations at two positions: F64L and S65T [9, 10]. EGFP is brighter and matures rapidly at 37°C than wild-type GFP [1, 9]. Protein engineering of EGFP has yielded several green variants with improved characteristics such as Emerald. This Emerald FP has improved photostability and brightness than EGFP [11]. Another GFP variant is the “superfolder” GFP that is designed to fold faster at 37°C. This “superfolder” GFP is also brighter and more acid resistant than either EGFP or Emerald [12]. A weak tendency of GFP and its variants to dimerize was completely eliminated using point mutation at F223K, L221K, or A206K [13, 14].
Another fluorescent protein, DsRed from the sea anemone Discosoma striata, is also of great interest to researchers, because its intrinsic fluorescence is red rather than green [15, 16]. The chromophore of DsRed is closely related to that of GFP, being formed by rearrangement of an internal Gln-Tyr-Gly tripeptide [15]. The extended conjugation in the chromophore causes the red-shift observed in DsRed and other red FPs [4]. DsRed forms a strong tetramer both in solution and in crystal and its chromophore maturation is very slow [17–19]. Due to these limitations, DsRed has been a target of protein engineering and mutations to improve its chromophore maturation rate and to reduce oligomerization [20–22]. A directed evolution approach was performed on DsRed to make a monomeric version, mRFP1, which has a total of 33 amino acid mutations [21]. In addition to DsRed, there are many other FPs ranging from blue-, cyan-, green-, yellow- to red-emitting having different spectral properties, brightness, and stabilities isolated from reef corals and other Anthozoa species [1, 2]. Most of these FPs display higher degree of oligomerization which is detrimental for cellular labeling [17, 18, 23]. To overcome FP oligomerization, mutations must be made at the monomer-monomer interface. The exact nature of such interfaces varies depending on the nature and origin of the FP [2].
Many FPs either isolated from natural sources or engineered from GFP or DsRed are known and available [1, 2]. However, only few of the current FPs are widely used in various cell-imaging applications and most of them have certain limitations [1, 2, 24]. A continuing effort to improve the spectral characteristics and stabilities of the FPs, or alternatively, to search for new FPs with optimal properties must be done for maximum utility in cellular imaging.
The natural habitat of A. victoria is located in the cool waters off the northwest coast of Washington State. One might expect organisms that inhabit warmer waters to have evolved FPs that mature more rapidly at higher temperatures. Here we describe the characterization and modification of a novel FP that was isolated from Cyphastrea microphthalma, a scleractinian coral found in the warmer waters of the Australian Great Barrier Reef (Fig. 1). Several new fluorescent organisms were identified by diving at night with UV illumination and the FPs were cloned from these organisms, and expressed in E. coli [25, 26]. We found that the vast majority of proteins we characterized indeed mature robustly and rapidly at 37°C. Here we report the properties of one of the novel green-emitting FPs, VFP (vivid Verde FP), which exhibits useful properties. VFP is very bright, it matures rapidly at 37°C and we have engineered exclusively monomeric or dimeric variant of it. These properties are particularly well suited to a variety of molecular and biological applications.
Figure 1.
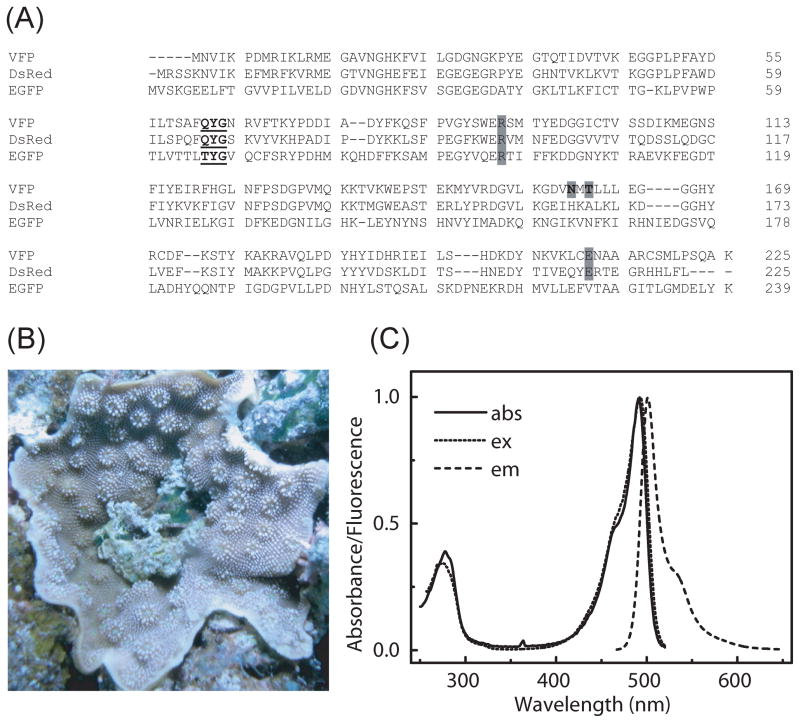
(A) Amino acid sequence alignment of VFP with DsRed and EGFP. The chromophore forming amino acid residues are highlighted in bold and underlined. Amino acid residues, N158 and T160 of VFP, where mutations were made are indicated by bold letter in gray background. The conserved Arg and Glu (corresponding to Arg96 and Glu222 of GFP) are highlighted in gray background. (B) A scleractinian coral, Cyphastrea microphthalma, collected in 1.2 m of water off Lizard Island on the Australian Great Barrier Reef. (C) Overlay of the absorption, fluorescence excitation and fluorescence emission spectra of VFP. The samples were excited at 450 nm and emission spectra were measured from 465 nm to 650 nm. Fluorescence excitation spectra were obtained from 250 nm to 515 nm by monitoring the emission at 530 nm. The spectra were normalized at the maximum peak.
Results
Sequence of the new fluorescent protein (FP) and relation to other known FP
A new fluorescent protein, VFP (vivid Verde FP), was isolated and cloned from the Cyphastrea microphthalma coral, collected in 1.2 m of water off Lizard Island on the Australian Great Barrier Reef [25, 26]. The alignment of the amino acid sequences of VFP, DsRed and EGFP is shown in Fig. 1A. The amino acid residues that form the chromophore are in bold and underlined. The chromophore residues at positions 66, 67 and 68, following the amino acid residues numbering in DsRed, are QYG in VFP, QYG in DsRed and TYG in EGFP. VFP shows greater sequence identity overall to DsRed than to EGFP with 53% sequence identity to DsRed and only 20% sequence identity to EGFP. Sequence alignment demonstrates the conservation of many positions in VFP, which are presumably structurally and/or functionally important. Arg96 and Glu222 of GFP, which was proposed to participate in chromophore maturation [27], is also conserved in DsRed and VFP. The VFP coding sequence was deposited at EMBL nucleotide sequence database under the accession number FN597286. Using the Swiss Institute of Bioinformatics BLAST Network Service, the VFP sequence has the highest sequence identity of 83% to a green fluorescent protein isolated from coral Montastraea cavernosa [28]. Sequence alignment also showed that there are several cyan, green, or red FPs and chromoproteins from coral with QYG chromophore forming amino acid residues similar to VFP.
VFP fluorescence protein exhibits maximum excitation and emission peaks at 491 and 503 nm, respectively as shown in Fig. 1C. These spectral properties are more similar to those of EGFP rather than those of DsRed, despite the fact that the sequence of VFP is more closely related to DsRed than to EGFP. The chromophore formation in GFP involves cyclization, oxidation and dehydration and in DsRed and other coral FPs, an additional oxidation step occurs [4, 29–31]. Previously, DsRed chromophore maturation has been shown to proceed through a green-emitting anionic GFP-like intermediate, which has excitation and emission peaks at 475 and 499 nm, respectively [17]. However, it has also been proposed that the red-emitting chromophore of DsRed and of related chromoproteins is produced from a blue-emitting neutral form of a GFP-like chromophore, the green anionic species being the dead-end product [32]. The GFP-like chromophore of VFP is stable and further conversion into red-emitting chromophore was not observed.
Two tryptophan residues at positions 93 and 143 of DsRed located in the immediate vicinity of the chromophore are conserved in VFP (corresponding to positions 89 and 139). Thus, absorption spectrum of VFP showed a peak at 280 nm (Fig. 1C) due to the presence of these Trp residues and excitation at 280 nm gave an emission peak at 503 nm.
Oligomeric state of VFP
For many applications, it is essential that the FP used to ‘tag’ another protein is monomeric [24]. If an FP is not monomeric, then its oligomerization may influence the behavior of the tagged protein, thus perturbing the system under study. We used gel filtration chromatography to assess the oligomeric state of VFP. To allow a direct comparison with a known protein, we also purified EGFP, which is monomeric at concentrations < 1 mg/ml [33]. Gel filtration chromatogram of VFP showed a major peak and a shoulder indicating a mixture of dimer and monomer species (Supplementary Fig. S1). We therefore sought to design mutations to shift the equilibrium to a fully monomeric state. With this goal, we aligned the sequences of DsRed and VFP and created a homology model for VFP.
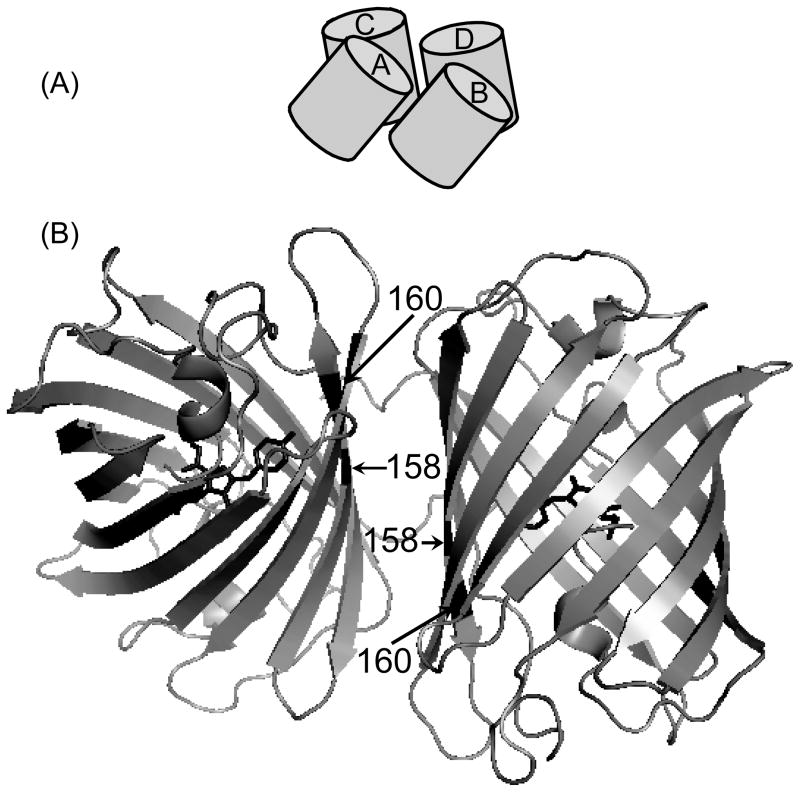
It is known that DsRed forms a strong tetramer, both in solution and in the crystal structure [17, 18]. An examination of the crystal structure of the DsRed tetramer shows that the monomers are arranged as a dimer of dimers, with AB (or CD) and AC (or BD) interfaces as illustrated in Fig. 2A. The AB interface is dominated by hydrophobic interactions, whereas the AC interface is comprised predominantly of salt bridges and hydrogen bonds [19]. Thus, the dimer formation of VFP could either be due to AB or AC interaction. Several point mutations such as I125R, H162K, A164R, and I180T on the surface of DsRed are documented in the literature, which convert the DsRed tight tetramer into a monomer [1, 21]. We compared the residues at these positions in VFP with those in DsRed to identify mutations in VFP that might shift the monomer-dimer equilibrium towards monomer. The corresponding amino acid residues in VFP are H121, N158, T160 and T176, allowing us to identify possible mutations in VFP as H121R, N158K and T160R. We focused on examining the N158K and T160R mutations. The location of these mutations in the AC interface are indicated in Fig. 2B. The rationale for N158K mutation is that it replaces a polar uncharged Asn with a positively charged Lys and this mutation should disrupt the AC dimerization interface. In DsRed, His162 of the A monomer is involved in a stacking interaction with His162 of the adjacent C monomer, whilst simultaneously making a electrostatic interactions with Glu176 of the C monomer, forming what appears to be an important part of the AC interface [19]. In VFP, residue 158 (corresponding to 162 in DsRed) is Asn and residue 172 (corresponding to 176 in DsRed) is Asp. On the other hand, in T160R mutations, the polar uncharged Thr was replaced with positively charged Arg. In DsRed, position 164 is occupied by Ala, which creates small hydrophobic patches in the AC interface and by replacing it with Arg, the AC interaction is disrupted. Also, previous studies showed that substituting hydrophilic or charged amino acids for hydrophobic and neutral residues of the FP tetrameric interfaces could generate the monomer form of the protein [13, 21].
Figure 2.
(A) A cartoon illustration of DsRed tetramer arranged as a dimer of dimers with AB (=CD) and AC (=BD) interfaces. (B) Structure of two of the four subunits of the tetrameric DsRed consisting of the AC polar interface. The positions of amino acid residues at 158 and 160 (corresponding to 162 and 164, respectively, in DsRed) where mutations were made are indicated by arrows. The chromophore at the center of the β-barrel structure is shown in black sticks. Protein Data Bank (PDB) code: 1GGX. [55]
The mutations were made individually, with the intention of combining any of them if individual mutation was insufficient to bring VFP to monomerize. We expressed and purified each VFP mutant (N158K or T160R) and assessed its oligomeric states using gel filtration chromatography. We found that either N158K or T160R mutation is sufficient to convert VFP into an exclusively monomeric species (Supplementary Fig. S1). We named these monomeric N158K and T160R mutants as mVFP1 and mVFP, respectively. In the course of the cloning, we also serendipitously isolated T160A mutant of VFP. Gel filtration chromatography revealed that T160A mutant is fully dimeric, with no evidence of the monomer-dimer equilibrium that we observed for VFP (Supplementary Fig. S1). Presumably, the introduction of small hydrophobic patches on the surface of the protein promotes strong dimer formation. We named this dimeric variant of VFP as dVFP.
Spectral properties of VFP and its variants
We proceeded with further characterizations of all four proteins: VFP and its variants mVFP1 (N158K), mVFP (T160R), and dVFP (T160A). The excitation and emission spectra for all four proteins are identical, with excitation maximum (Ex) of 491 nm, emission maximum (Em) of 503 nm and with a Stokes shift of 12 nm (Table 1, Fig. 1C and Supplementary Fig. S2). The measured extinction coefficient of VFP is 83,700 M−1cm−1, which is higher than that of EGFP (54,400 M−1cm−1). The extinction coefficients calculated for mVFP1 and mVFP are 80,400 and 85,000 M−1cm−1, respectively. However, a higher extinction coefficient value of 107, 000 M−1cm−1 was observed for dVFP. The increase in extinction coefficient of dVFP compared to VFP can be due to its tight dimer formation. Table 1 summarizes these data, alongside to the measured results for EGFP and Venus for comparison. Venus is a variant of yellow fluorescent protein with fast maturation and high brightness [34]. The results obtained for EGFP and Venus are consistent with the values reported in literature [34, 35]. Table 1 includes the list of selected green-emitting FPs that have relevant spectral properties to VFP variants for comparison purposes. We reported the fluorescence excitation (Ex) and emission (Em) wavelength peaks, molar extinction coefficient (EC), quantum yield (QY), oligomeric states, relative brightness, and photostability of these selected FPs.
Table 1. Spectral properties of VFP and its variants in comparison to EGFP, Venus and selected FPs.
The excitation (Ex) and emission (Em) wavelengths, molar extinction coefficient (EC), quantum yield (QY), oligomeric states, relative brightness and photostability are listed. The relative brightness was calculated from the product of EC and QY. Photostability was calculated based on 100% EGFP measured at the same time. nd is not determined.
| Protein | Ex (nm) | Em (nm) | EC × 10−3 (M−1cm−1) | QY | Oligomeric states | Relative brightness | Photo- stability (% EGFP) | Ref. |
|---|---|---|---|---|---|---|---|---|
| VFP | 491 | 503 | 83.7 | 1.0 | Weak Dimer | 84 | 33 | This work |
| mVFP | 491 | 503 | 85.0 | 0.86 | Monomer | 73 | 16 | This work |
| mVFP1 | 491 | 503 | 80.4 | 0.84 | Monomer | 68 | 11 | This work |
| dVFP | 491 | 503 | 107 | 1.0 | Dimer | 107 | 39 | This work |
| EGFP | 488 | 509 | 54.4 | 0.60 | Monomer | 33 | 100 | This work |
| Venus | 515 | 527 | 93.3 | 0.75 | Monomer | 70 | 27 | This work |
| Green FPs -Anthozoa | ||||||||
| AzamiGreen | 492 | 505 | 55.0 | 0.74 | Monomer | 41 | nd | [44] |
| mWasabi | 493 | 509 | 70.0 | 0.80 | Monomer | 56 | 53 | [45] |
| ZsGreen | 493 | 505 | 43.0 | 0.91 | Tetramer | 39 | nd | [15] |
| copGFP | 482 | 502 | 70.0 | 0.60 | Tetramer | 42 | nd | [46] |
| cmFP512 | 503 | 512 | 58.8 | 0.66 | Tetramer | 39 | 92 | [47] |
| aacuGFP2 | 502 | 513 | 93.9 | 0.71 | nd | 67 | nd | [48] |
| aeurGFP | 504 | 515 | 145.7 | 0.67 | nd | 98 | nd | [48] |
| eechGFP1 | 497 | 510 | 124.2 | 0.75 | nd | 93 | nd | [48] |
| efasGFP | 496 | 507 | 125.8 | 0.80 | nd | 101 | nd | [48] |
| gfasGFP | 492 | 506 | 102.5 | 0.73 | nd | 75 | nd | [48] |
| plamGFP | 502 | 514 | 98.6 | 0.96 | nd | 95 | nd | [48] |
| sarcGFP | 483 | 500 | 76.7 | 0.96 | nd | 74 | nd | [48] |
| GFP-Aequorea derivatives | ||||||||
| Superfolder | 485 | 510 | 83.3 | 0.65 | Monomer | 54 | 90 | [12] |
| mEmerald | 487 | 509 | 57.5 | 0.68 | Monomer | 39 | 58 | [11] |
| Cyan FPs-Anthozoa | ||||||||
| mTFP1 | 462 | 492 | 64 | 0.85 | Monomer | 54 | 63 | [49] |
| Photoconvertable FPs | ||||||||
| Kaede | 508 | 518 | 98.8 | 0.88 | Tetramer | 87 | 26 | [50] |
| dEos (G) | 506 | 516 | 84 | 0.66 | Dimer | 55 | 24 | [51] |
| mEos2 (G) | 506 | 519 | 56 | 0.84 | Monomer | 47 | 21 | [52] |
| Photoswitchable FPs | ||||||||
| Dronpa | 503 | 517 | 95 | 0.85 | Monomer | 81 | nd | [53] |
| mKikGR | 505 | 515 | 49 | 0.69 | Monomer | 34 | 7 | [54] |
| cerFP505 | 494 | 505 | 54 | 0.55 | Tetramer | 30 | 54 | [47] |
The extinction coefficient (EC) and quantum yield (QY) for each FP were determined and the product of these two parameters (EC × QY) provides the relative brightness (Table 1). We used the reported EGFP quantum yield of 0.60 [35] as a reference for calculating the quantum yield of VFP and its variants. The relative quantum yield values of VFP and its variants range from 0.84 to 1.0, which are higher than those of both EGFP and Venus. Thus, having high extinction coefficient and quantum yield, VFP and variants produced high relative brightness. It is evident that the dimeric form, VFP or dVFP variant, is brighter than the monomeric form of VFP. Either the mVFP1 or the mVFP variant is at least twice as bright as EGFP and the dVFP variant is much brighter than Venus. To our knowledge, there is no monomeric green-emitting FP available to date that is at least 2-fold brighter than EGFP, except for the photoswitchable Dronpa FP (Table 1).
We also investigated the pH dependence of VFP and its variants fluorescence emission at 503 nm upon excitation at 491 nm as shown in Supplementary Fig. S3. All, VFP and its variants, have similar pH stability profiles to EGFP between pH 6 to 10.
Fluorescence correlation spectroscopy (FCS) measurements of size and photostability
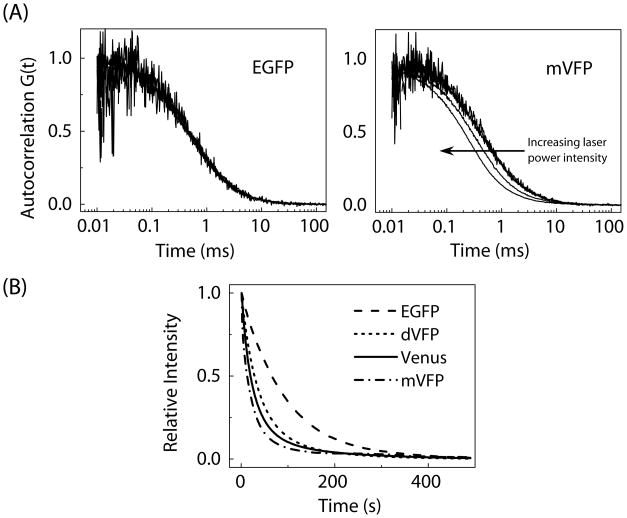
We used fluorescence correlation spectroscopy (FCS) to investigate the photobleaching, molecular brightness, and oligomeric states of the FPs in more detail. The traces of FCS autocorrelation curves obtained for EGFP and mVFP are shown in Fig. 3A. No shifts in the autocorrelation curves are observed for EGFP as a function of laser power intensity. However, the diffusion curves shifted to the left for VFP and its variants as the laser power intensity was increased (Fig. 3A and Supplementary Fig. S4). This shift in autocorrelation curve to the left, noted by shorter apparent diffusion times, is indicative of photobleaching.
Figure 3.
(A) Representative FCS autocorrelation curves of EGFP and mVFP taken at increasing laser power intensities from 0.25 to 5 μW. A shift in the autocorrelation curve to the left, to apparent shorter diffusion times, as a function of laser power intensity was observed for VFP and its variants. The autocorrelation curves are normalized to the number of molecules obtained from the fitting. (B) Photobleaching curves for the EGFP, Venus, mVFP and dVFP under mercury arc lamp illumination using wide-field microscope. The relative photostability of VFP and its variants are reported in Table 1.
The autocorrelation curves for each sample were fitted using single- or two-diffusion component equation. The best-fit curve was assessed based on the residual of the fitting. A detailed analysis of the other photophysical dynamics (e.g. triplet blinking) occurring at the submillisecond timescale is beyond the scope of this paper and will be presented elsewhere. The diffusion time (tD) and the average fluorescence intensity were determined from the fitting of the autocorrelation curves taken at 0.25 μW laser power as reported in Table 2. At this low laser power intensity, the effects of other photophysical processes were minimized. The relative molecular brightness of the FPs is calculated by dividing the average fluorescence intensity by the number of molecules within the illuminated region. The results that we obtained here supported our earlier findings that VFP and its variants were nearly 2-fold brighter than EGFP, based on the counts per molecules (kHz/molecule) in Table 2 and Supplementary Table S1.
Table 2. Summary of Fluorescence Correlation Spectroscopy Analysis.
The autocorrelation curves of each FP obtained at 0.25 μW laser power intensity were fitted using single diffusion component equation. The brightness expressed as counts per molecules was calculated by dividing the intensity by number of molecules.
| FPs | Diffusion time (ms) | Intensity (Hz) 1 × 104 | Counts per molecules (kHz/molecule) |
|---|---|---|---|
| EGFP | 0.486 ± 0.012 | 2.49 ± 0.05 | 0.262 ± 0.005 |
| Venus | 0.543 ± 0.019 | 3.25 ± 0.02 | 0.251 ± 0.002 |
| VFP | 0.646 ± 0.004 | 4.07 ± 0.14 | 1.76 ± 0.06 |
| mVFP1 | 0.460 ± 0.007 | 3.40 ± 0.19 | 0.485 ± 0.028 |
| mVFP | 0.472 ± 0.007 | 3.70 ± 0.14 | 0.476 ± 0.018 |
| dVFP | 0.763 ± 0.004 | 3.00 ± 0.11 | 1.55 ± 0.06 |
At 0.25 μW laser power intensity, the measured relative diffusion times (tD) of either mVFP1 or mVFP are comparable to that of the EGFP, indicating that both variants are monomeric. Furthermore, VFP and dVFP have tD values greater than that of EGFP indicating higher oligomeric states (Table 2). These results supported our findings on the oligomeric states of the VFP variants using gel filtration chromatography as described earlier.
Based on our FCS results, we noticed that photobleaching occurs in VFP and its variants. This observation prompted us to further investigate the rate of photobleaching of VFP and its variants in comparison to EGFP and Venus using wide-field microscope as described in the Materials and Methods. Figure 3B depicts the relative photobleaching curves of the EGFP, Venus, mVFP and dVFP from 0 to 500 seconds. We have determined the relative half time (t½) to photobleach the VFP samples, EGFP and Venus (Supplementary S5). Based on the t½ values, we calculated the percentage of photostability of the VFP and its variants relative to 100% EGFP. We also included in Table 1 the reported photostability of some FPs relative to 100% EGFP measured at the same time. The photostability data of other reported green-emitting FPs have not yet been reported or determined. The mVFP1 and mVFP variants having 11% and 16% photostability, respectively, are less photostable than the VFP and dVFP. However, the dVFP variant has 39% photostability, and thus exhibits greater photostability than Venus and other photoconvertable or photoswitchable FPs. For other imaging applications [24], the difference in photostability has no relevance. Even with this photostability, our VFP variants can be useful for numerous in vitro and in vivo detection applications.
Application of VFP variants as detection markers
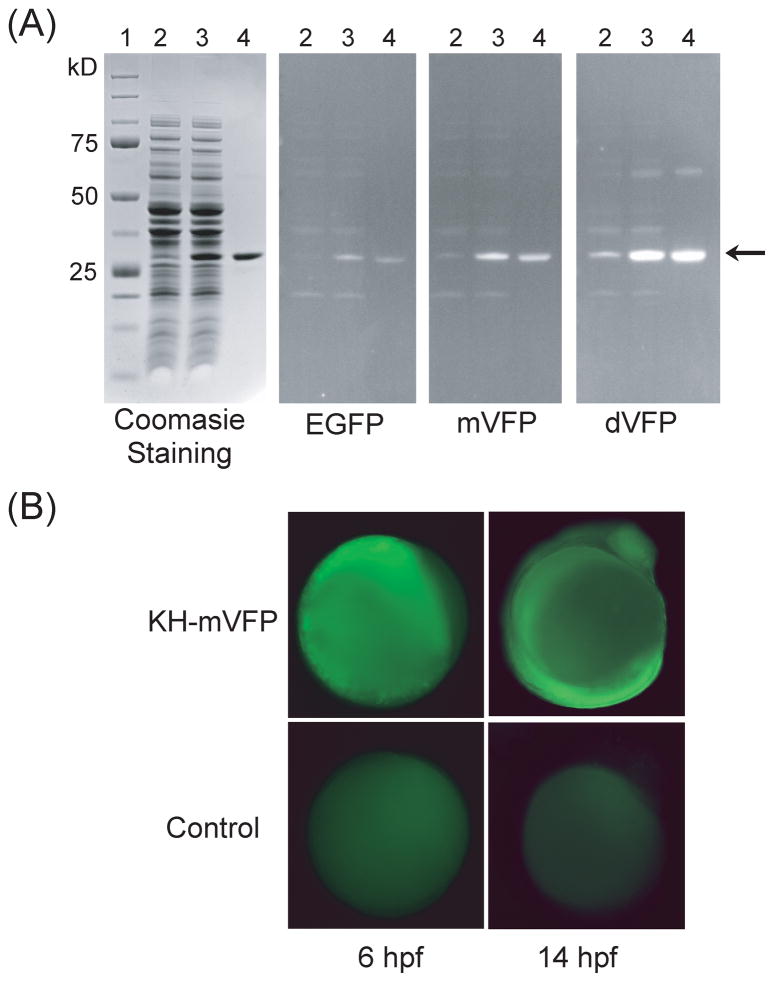
It has been shown previously in our laboratory that a protein recognition domain, tetratricopeptide repeats (TPR), fused to EGFP can be used to detect the protein-peptide interaction in a single step, completely eliminating the use of primary and secondary antibodies in Western blot analysis [36]. T-Mod (TPR-based recognition module) was demonstrated to bind specifically to MEEVF peptide fused to GST (Glutathione S-Transferase) protein [36]. FP-fusion to T-Mod can completely eliminate the need for any antibodies or developing procedures, which makes Western blotting faster, simpler, and less costly. We adapted this experiment to show the usefulness of mVFP and dVFP brightness in comparison to EGFP. We expressed and purified the T-Mod fused to EGFP, mVFP or dVFP. Following the SDS-PAGE electrophoresis of E. coli-expressing GST-MEEVF lysate, gels were transferred to PVDF membrane and processed as for Western blot. After blocking the membrane, we incubated the blots separately with different T-Mod-FPs for 1 hr at room temperature. The membrane was then visualized using a UV transilluminator at 302 nm as shown in Fig. 4A. The visible band indicated by arrow is the GST-MEEVF protein detected by the binding of T-Mod-FP. The intensity of the bands from T-Mod–mVFP or –dVFP are at least 2-fold brighter than that of the EGFP. Additional bands were visible in the membrane incubated with T-Mod-dVFP due to intense brightness of the dVFP protein. This result illustrates the advantage of having high brightness in terms of sensitivity in a practical detection application.
Figure 4.
(A) Comparison of the T-Mod fused to EGFP, mVFP, or dVFP as a replacement for antibodies in Western blot analysis. A duplicate SDS-PAGE gel used in Western blotting is stained with Coomassie Brilliant Blue. Lane (1) precision plus protein standard (BioRad); (2) lysate; (3) lysate supplemented with 1 mg/mL of purified GST-MEEVF protein; (4) purified GST-MEEVF. Arrow indicates the GST-MEEVF protein band. (B) Microinjection of KH-mVFP fusion mRNA into zebrafish embryos. The expression of KH-mVFP protein was monitored in the embryos at 6 hpf (hour post fertilization) and 14 hpf by fluorescence microscopy. Zebrafish embryos without RNA injections were used as a control.
Application of mVFP as in vivo marker
To demonstrate that our VFP can be used for in vivo labeling, we chose the monomeric form, mVFP and fused it to the KH domains of Fragile X Mental Retardation Protein (FMRP). We injected mRNA encoding the KH-mVFP fusion protein into zebrafish embryos at the 1-cell stage. Live embryos at 6-hour post fertilization (hpf) and at 14-hpf (10-somite) stages were mounted on glass slides and visualized using fluorescence microscope as shown in Fig. 4B. The fluorescence signal from zebrafish embryos with KH-mVFP are more intense compared to that of the control, which showed a faint cellular autofluorescence.
Discussion
We have described a detailed characterization of a new fluorescent protein from the warm water coral, Cyphastrea microphthalma, collected off Lizard Island on the Australian Great Barrier Reef. The protein, which we have named VFP (vivid Verde fluorescent protein) matures rapidly at 37°C and emits bright green fluorescence. VFP, as isolated, showed a propensity to form fairly weakly associating dimers. By creating a homology model of VFP, we were able to create surface mutations that convert VFP into either an exclusively monomeric species (N158K or T160R) – which we named mVFP1 and mVFP, respectively, or into an exclusively dimeric species (T160A) – which we named dVFP. This rational approach to creating monomeric variants can be used as a guide for re-engineering of other coral FPs having higher oligomeric forms.
These novel proteins have features that will be useful for a variety of applications. The mVFP1 and mVFP variants are both monomeric and fluoresce at least twice as brightly as EGFP. The dimeric dVFP is even brighter having at least 1.5 times as bright as Venus. For applications where oligomerization is not critical, the dVFP variant would be advantageous to use because of its high brightness. When a bright, monomeric protein is desired, mVFP1 or mVFP would be the protein of choice. Based on the list of reported FPs either wild-type or engineered (Table 1), there are no FPs that are monomeric and have at least 2-times brighter than EGFP except for photoswitchable Dronpa. The data we presented should allow investigators to choose which VFP variant is the most appropriate for their specific research application.
With regards to photostability, VFP and its variants photobleached at a faster rate than EGFP. The vast majority of reports in the literature describing green-emitting FPs isolated from corals do not include photostability measurements, which makes it difficult to assess the level of photostability of VFP variants in relation to other coral FPs [2, 24]. However, for many imaging applications, this photobleaching property will not be influential [24, 34]. In conclusion, the monomeric or dimeric form of VFP represent viable alternatives to widely used EGFP and Venus.
Materials and Methods
Plasmid constructions and mutations
The plasmids encoding VFP, EGFP and Venus with polyhistidine tags were constructed as previously described in [26, 37]. The VFP coding sequence was deposited at EMBL nucleotide sequence database under the accession number FN597286 and at UniProtKBT/TrEMBL protein sequence database with D1J6P8 accession number. Site-directed mutagenesis (QuikChange Site-Directed Mutagenesis Kit, Stratagene) was used to introduce the N158K and T160R mutations in VFP. Mutations were verified by DNA sequencing (W.M. Keck Foundation Facility, Yale University).
Sequence alignment and homology modeling
Sequence alignment of VFP with EGFP and DsRed was performed from ClustalW2 program (EMBL-EBI). Homology modeling was done using SWISS-MODEL program [38].
Recombinant Protein expression
The proteins were expressed in E. coli DH10β cells grown in Luria Bertani (LB) media for 24 hrs at 37°C. The cells were harvested by centrifugation and the pellets were resuspended in lysis buffer (50 mM Tris-HCl pH 7.4, 300 mM NaCl) supplemented with a tablet of complete EDTA-free protease inhibitor cocktail (Roche) and 5 mM β-mercaptoethanol. The lysate was sonicated, and then centrifuged. The supernatant solution was loaded into Ni-NTA agarose (Qiagen) and the pure protein was eluted with 50 mM Tris-HCl pH 7.4, 150 mM NaCl, 200 mM imidazole. The fractions containing the protein were pooled and dialyzed into 50 mM Tris-HCl, pH 7.4, 150 mM NaCl. The purity of the samples was determined by SDS-PAGE. The proteins were concentrated by centriprep YM-10 with 10,000 MWCO (Amicon) to about 100–200 mM then stored in aliquots at −20 °C. The buffer used in all spectroscopic analyses was 50 mM Tris-HCl, pH 7.4, 150 mM NaCl, unless otherwise noted.
Analytical gel filtration chromatography
The molecular size of the purified FPs were analyzed using a Superdex S200 10/30 gel filtration column (Amersham Pharmacia) in FPLC at room temperature. A 100 ml of < 0.01 mg/ml of each FP was injected into the column with a flowrate of 0.5 ml/min and absorbance was monitored at 280 nm. The oligomeric states of the VFP and mutants were determined based on EGFP elution time and protein standards (Bio-Rad).
Absorption Spectroscopy
Absorbance spectra of the FPs were recorded on a Hewlett Packard 845X UV-visible Chemstation. The extinction coefficients of the FPs were calculated based on the absorbance of the native and acid- or alkali-denatured proteins. The extinction coefficients of the GFP-like chromophores used in the calculation are 44,000 M−1 cm−1 at 447 nm in 1M NaOH [33] and 28,500 M−1 cm−1 at 382 nm in 1M HCl [39]. For yellow fluorescent protein, Venus, the extinction coefficient of the chromophore was back calculated using 22,000 M−1cm−1 at 280 in 10 mM Tris-HCl buffer.
Fluorescence Spectroscopy
Fluorescence excitation and emission measurements were performed using a PTI Quantamaster C-61 two channel fluorescence spectrophotometer. The samples were excited at 450 nm and emission spectra were measured from 465 nm to 650 nm with a 2 nm slit-width. Fluorescence excitation spectra were obtained from 250 nm to 515 nm by monitoring the emission at 530 nm with a 2 nm slit-width. The quantum yields (QY) of the VFP and its variants were determined relative to EGFP (QY=0.60 [35]). The pH dependence of VFP and its variants fluorescence emission at 503 nm was monitored upon excitation at 491 nm at room temperature. pH titrations were performed using a series of 100–200 mM citrate-phosphate buffer (pH 2.0 – 11.0), 150 mM NaCl.
Fluorescence Correlation Spectroscopy (FCS)
FCS measurements were made on a lab-built instrument based around an inverted microscope with a 488 nm DPSS laser for excitation as previously described [40, 41]. All measurements are carried out on approximately 100 nM FP samples with varying laser power intensities from 5 μW to 0.25 μW measured on the table before entering the microscope. The output of the detection channels was autocorrelated in a digital correlator (Correlator.com). Control measurements with Alexa 488 solutions were performed to ensure the proper alignment of the confocal optics and the absence of artifacts in the FCS. The autocorrelation curves were fitted using a single-or two-component equation as previously described [41]. The parameters extracted from the fittings are relative diffusion time, tD, number of molecules, and fluorescence intensities.
Photobleaching
Photobleaching measurements of purified FP samples were performed using a inverted wide-field microscope equipped with a 100W mercury arc lamp similar to those described in the literature [42]. The FP samples were mixed with mineral oil and about 5 μl of the mixture was sandwiched between a glass slide and a cover slip. Neutral density filter was used initially for sample alignment and then removed when the actual measurements were occurring. The FP samples were imaged with 50 ms exposure time with a frame rate of 1 image per second. The measurement was just taken in 600 seconds time span with continues illumination.
Western Blots Assay
The T-Mod (TPR-based recognition module)- fused to FP and GST-MEEVF constructs were prepared as previously described [36]. The FP fused to T-Mod was EGFP, mVFP, or dVFP. Each construct was transformed in E. coli BL21(DE3) cells and the protein was purified following the protocol previously described [36]. The GST-MEEVF lysate was obtained from 6-mL overnight culture cell pellet by adding 1-mL B-Per (Pierce) and shaking with occasional vortexing for 10 minutes. The lysates were supplemented either with or without 1 mg/mL of purified GST-MEEVF protein. The samples mixed with a reducing loading buffer were precisely loaded into 4–12% gradient SDS-polyacrylamide gels together with equivalent amount of purified GST-MEEVF protein. The gels were run at room temperature for 1 hour at a voltage of 120V using NuPAGE buffer (Invitrogen). One gel was stained with Coomassie Brilliant Blue while the other gels were transferred onto a PVDF membrane (Millipore). Membrane transferring was done at cold room for 3 hrs at a constant current of 380 mAmp. The transfer buffer used contains 24mM Tris-Base, 192 mM Glycine, 10% methanol and 0.01% SDS. The membranes were blocked in 5% nonfat milk in TBS-T (20 mM Tris-Base pH 8.0, 150 mM NaCl, 0.1% Tween-20) overnight at 4°C with shaking. The membranes were then incubated individually in each 5 μM T-Mod-FP fusion constructs in TBS-T with 0.1% nonfat milk for 1 hour at room temperature with shaking. The membranes were washed three times with TBS-T with 10-min interval each washing and then visualized using a UV transilluminator at 302 nm and the images were captured using a digital camera (Kodak).
mRNA microinjection assay
To assemble the KH-mVFP fusion construct, the delete KH domain of human Fragile X Mental Retardation Protein, hFMRP(KH1-KH2Δ) was fused with the N-terminal of mVFP and cloned into mammalian PCS2+ vector. The construct was sequenced (W.M. Keck Foundation Facility, Yale University) and named as KH-mVFP for simplicity. The in vitro synthesis of large amounts of capped RNA was prepared using mMESSAGE mMACHINE kit (Ambion) following the manufacturer’s protocols. The capped transcription reaction was prepared at room temperature and then incubated at 37°C for 2 hours. TURBODNase (Ambion) was added into the reaction and incubated at 37 °C for another 15 mins to remove the template DNA. The RNA was purified using RNeasy Mini kit (Qiagen). The concentration of the RNA was determine using UV-vis spectrometer and then stored at −80°C until ready for use. The RNA microinjections were performed at the 1-cell stage using standard protocols [43]. Injection solution consisted of 200 ng/μl KH-mVFP and 0.15% phenol Red in Danieau’s solution. Live embryos at 6 hpf (hour post fertilization) and 14 hpf stages were manually dechorionated and mounted in methylcellulose. In parallel, we also mounted embryos without RNA injections as a control. Fluorescent images were acquired on a Zeiss Axioskop microscope using 20x objective and FITC filter. Color adjustment of the fluorescent images was made equally for both KH-mVFP-injected and control zebrafish using ImageJ software.
Supplementary Material
Acknowledgments
We thank Dr. Joseph Wolenski of MCDB Imaging Facilities at Yale University for helping us with photobleaching experiments; and Dr. Scott Holley and Jamie Schwendinger-Schreck of MCDB at Yale University for RNA microinjection assay in zebrafish. We also thank the members of Regan laboratory for comments and suggestions on the manuscripts. This work is funded by HFSP (RGP44/2207 to L.R.), Leslie H. Warner Postdoctoral Fellowship (to R.P.I.), NIH (GM070348 to H-T.K. and Earthwatch Institute (Grant: “Luminous Life in the Great Barrier Reef” to V.A.P.)
Abbreviations
- FP
fluorescent protein
- GFP
green fluorescent protein
- EGFP
enhance GFP
- VFP
vivid verde fluorescent protein
- mVFP
monomeric VFP
- dVFP
dimeric VFP
- FCS
fluorescence correlation spectroscopy
- TPR
tetratricopeptide repeats
- T-Mod
TPR-based recognition module
- FMRP
fragile X mental retardation protein
Footnotes
Note: The nucleotide sequence data are available in the DDBJ/EMBL/GenBank databases under the accession number FN597286 and the protein sequence data are in UniProtKB/TrEMBL with D1J6P8 accession number.
This section includes the complete set of characterization of VFP and its variants in comparison to EGFP and Venus: 1. Gel-filtration chromatogram, 2. Absorption, fluorescence excitation and emission spectra, 3. pH dependence, 4. FCS autocorrelation curves, 5. Photobleaching curves, and 6. Table of fluorescence correlation spectroscopy (FCS) fitting results. The figures are named as Supplementary Figure S1 to S5 and the table is named as Table S1.
References
- 1.Shaner NC, Patterson GH, Davidson MW. Advances in fluorescent protein technology. J Cell Sci. 2007;120:4247–4260. doi: 10.1242/jcs.005801. [DOI] [PubMed] [Google Scholar]
- 2.Day RN, Davidson MW. The fluorescent protein palette: tools for cellular imaging. Chem Soc Rev. 2009;38:2887–2921. doi: 10.1039/b901966a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lippincott-Schwartz J, Patterson GH. Photoactivatable fluorescent proteins for diffraction-limited and super-resolution imaging. Trends Cell Biol. 2009;19:555–565. doi: 10.1016/j.tcb.2009.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nienhaus GU, Wiedenmann J. Structure, Dynamics and Optical Properties of Fluorescent Proteins: Perspectives for Marker Development. Chemphyschem. 2009;10:1369–1379. doi: 10.1002/cphc.200800839. [DOI] [PubMed] [Google Scholar]
- 5.Patterson GH, Lippincott-Schwartz J. A photoactivatable GFP for selective photolabeling of proteins and cells. Science. 2002;297:1873–1877. doi: 10.1126/science.1074952. [DOI] [PubMed] [Google Scholar]
- 6.Heim R, Prasher DC, Tsien RY. Wavelength mutations and posttranslational autoxidation of green fluorescent protein. Proc Natl Acad Sci U S A. 1994;91:12501–12504. doi: 10.1073/pnas.91.26.12501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ormo M, Cubitt AB, Kallio K, Gross LA, Tsien RY, Remington SJ. Crystal structure of the Aequorea victoria green fluorescent protein. Science. 1996;273:1392–1395. doi: 10.1126/science.273.5280.1392. [DOI] [PubMed] [Google Scholar]
- 8.Yang F, Moss LG, Phillips GN., Jr The molecular structure of green fluorescent protein. Nat Biotechnol. 1996;14:1246–1251. doi: 10.1038/nbt1096-1246. [DOI] [PubMed] [Google Scholar]
- 9.Heim R, Tsien RY. Engineering green fluorescent protein for improved brightness, longer wavelengths and fluorescence resonance energy transfer. Curr Biol. 1996;6:178–182. doi: 10.1016/s0960-9822(02)00450-5. [DOI] [PubMed] [Google Scholar]
- 10.Brejc K, Sixma TK, Kitts PA, Kain SR, Tsien RY, Ormo M, Remington SJ. Structural basis for dual excitation and photoisomerization of the Aequorea victoria green fluorescent protein. Proc Natl Acad Sci U S A. 1997;94:2306–2311. doi: 10.1073/pnas.94.6.2306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cubitt AB, Woollenweber LA, Heim R. Understanding structure-function relationships in the Aequorea victoria green fluorescent protein. Methods Cell Biol. 1999;58:19–30. doi: 10.1016/s0091-679x(08)61946-9. [DOI] [PubMed] [Google Scholar]
- 12.Pedelacq JD, Cabantous S, Tran T, Terwilliger TC, Waldo GS. Engineering and characterization of a superfolder green fluorescent protein. Nat Biotechnol. 2006;24:79–88. doi: 10.1038/nbt1172. [DOI] [PubMed] [Google Scholar]
- 13.Zacharias DA, Tsien RY. Molecular Biology and Mutation of Green Fluorescent Protein. In: Chalfie M, RKS, editors. Green Fluorescent Protein: Properties, Applications, and Protocols. John Wiley & Sons, Inc; New Jersey: 2006. pp. 83–120. [PubMed] [Google Scholar]
- 14.Zhang J, Campbell RE, Ting AY, Tsien RY. Creating new fluorescent probes for cell biology. Nat Rev Mol Cell Biol. 2002;3:906–918. doi: 10.1038/nrm976. [DOI] [PubMed] [Google Scholar]
- 15.Matz MV, Fradkov AF, Labas YA, Savitsky AP, Zaraisky AG, Markelov ML, Lukyanov SA. Fluorescent proteins from nonbioluminescent Anthozoa species. Nat Biotechnol. 1999;17:969–973. doi: 10.1038/13657. [DOI] [PubMed] [Google Scholar]
- 16.Davidson MW, Campbell RE. Engineered fluorescent proteins: innovations and applications. Nat Methods. 2009;6:713–717. doi: 10.1038/nmeth1009-713. [DOI] [PubMed] [Google Scholar]
- 17.Baird GS, Zacharias DA, Tsien RY. Biochemistry, mutagenesis, and oligomerization of DsRed, a red fluorescent protein from coral. Proc Natl Acad Sci U S A. 2000;97:11984–11989. doi: 10.1073/pnas.97.22.11984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gross LA, Baird GS, Hoffman RC, Baldridge KK, Tsien RY. The structure of the chromophore within DsRed, a red fluorescent protein from coral. Proc Natl Acad Sci U S A. 2000;97:11990–11995. doi: 10.1073/pnas.97.22.11990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yarbrough D, Wachter RM, Kallio K, Matz MV, Remington SJ. Refined crystal structure of DsRed, a red fluorescent protein from coral, at 2.0-A resolution. Proc Natl Acad Sci U S A. 2001;98:462–467. doi: 10.1073/pnas.98.2.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bevis BJ, Glick BS. Rapidly maturing variants of the Discosoma red fluorescent protein (DsRed) Nat Biotechnol. 2002;20:83–87. doi: 10.1038/nbt0102-83. [DOI] [PubMed] [Google Scholar]
- 21.Campbell RE, Tour O, Palmer AE, Steinbach PA, Baird GS, Zacharias DA, Tsien RY. A monomeric red fluorescent protein. Proc Natl Acad Sci U S A. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Strongin DE, Bevis B, Khuong N, Downing ME, Strack RL, Sundaram K, Glick BS, Keenan RJ. Structural rearrangements near the chromophore influence the maturation speed and brightness of DsRed variants. Protein Eng Des Sel. 2007;20:525–534. doi: 10.1093/protein/gzm046. [DOI] [PubMed] [Google Scholar]
- 23.Verkhusha VV, Lukyanov KA. The molecular properties and applications of Anthozoa fluorescent proteins and chromoproteins. Nat Biotechnol. 2004;22:289–296. doi: 10.1038/nbt943. [DOI] [PubMed] [Google Scholar]
- 24.Shaner NC, Steinbach PA, Tsien RY. A guide to choosing fluorescent proteins. Nat Methods. 2005;2:905–909. doi: 10.1038/nmeth819. [DOI] [PubMed] [Google Scholar]
- 25.Gruber DF, Desalle R, Lienau EK, Tchernov D, Pieribone VA, Kao HT. Novel Internal Regions of Fluorescent Proteins Undergo Divergent Evolutionary Patterns. Mol Biol Evol. 2009 doi: 10.1093/molbev/msp194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gruber DF, Kao HT, Janoschka S, Tsai J, Pieribone VA. Patterns of fluorescent protein expression in Scleractinian corals. Biol Bull. 2008;215:143–154. doi: 10.2307/25470695. [DOI] [PubMed] [Google Scholar]
- 27.Wood TI, Barondeau DP, Hitomi C, Kassmann CJ, Tainer JA, Getzoff ED. Defining the role of arginine 96 in green fluorescent protein fluorophore biosynthesis. Biochemistry. 2005;44:16211–16220. doi: 10.1021/bi051388j. [DOI] [PubMed] [Google Scholar]
- 28.Kelmanson IV, Matz MV. Molecular basis and evolutionary origins of color diversity in great star coral Montastraea cavernosa (Scleractinia: Faviida) Mol Biol Evol. 2003;20:1125–1133. doi: 10.1093/molbev/msg130. [DOI] [PubMed] [Google Scholar]
- 29.Craggs TD. Green fluorescent protein: structure, folding and chromophore maturation. Chem Soc Rev. 2009;38:2865–2875. doi: 10.1039/b903641p. [DOI] [PubMed] [Google Scholar]
- 30.Pakhomov AA, Martynov VI. GFP family: structural insights into spectral tuning. Chem Biol. 2008;15:755–764. doi: 10.1016/j.chembiol.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 31.Remington SJ. Fluorescent proteins: maturation, photochemistry and photophysics. Curr Opin Struct Biol. 2006;16:714–721. doi: 10.1016/j.sbi.2006.10.001. [DOI] [PubMed] [Google Scholar]
- 32.Verkhusha VV, Chudakov DM, Gurskaya NG, Lukyanov S, Lukyanov KA. Common pathway for the red chromophore formation in fluorescent proteins and chromoproteins. Chem Biol. 2004;11:845–854. doi: 10.1016/j.chembiol.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 33.Ward WW. Biochemical and Physical Properties of Green Fluorescent Protein. In: Chalfie M, RKS, editors. Green Fluorescent Protein: Properties, Applications, and Protocols. John Wiley & Sons, Inc; New Jersey: 2006. pp. 39–65. [Google Scholar]
- 34.Nagai T, Ibata K, Park ES, Kubota M, Mikoshiba K, Miyawaki A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- 35.Patterson GH, Knobel SM, Sharif WD, Kain SR, Piston DW. Use of the green fluorescent protein and its mutants in quantitative fluorescence microscopy. Biophys J. 1997;73:2782–2790. doi: 10.1016/S0006-3495(97)78307-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jackrel ME, Valverde R, Regan L. Redesign of a protein-peptide interaction: characterization and applications. Protein Sci. 2009;18:762–774. doi: 10.1002/pro.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kao HT, Sturgis S, DeSalle R, Tsai J, Davis D, Gruber DF, Pieribone VA. Dynamic regulation of fluorescent proteins from a single species of coral. Mar Biotechnol (NY) 2007;9:733–746. doi: 10.1007/s10126-007-9025-1. [DOI] [PubMed] [Google Scholar]
- 38.Bordoli L, Kiefer F, Arnold K, Benkert P, Battey J, Schwede T. Protein structure homology modeling using SWISS-MODEL workspace. Nat Protoc. 2009;4:1–13. doi: 10.1038/nprot.2008.197. [DOI] [PubMed] [Google Scholar]
- 39.Niwa H, Inouye S, Hirano T, Matsuno T, Kojima S, Kubota M, Ohashi M, Tsuji FI. Chemical nature of the light emitter of the Aequorea green fluorescent protein. Proc Natl Acad Sci U S A. 1996;93:13617–13622. doi: 10.1073/pnas.93.24.13617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trexler A, Rhoades E. a-Synuclein binds large unilamellar vesicles as an extended helix. Biochemistry. 2009;48:2304–2306. doi: 10.1021/bi900114z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen H, Rhoades E, Butler JS, Loh SN, Webb WW. Dynamics of equilibrium structural fluctuations of apomyoglobin measured by fluorescence correlation spectroscopy. Proc Natl Acad Sci U S A. 2007;104:10459–10464. doi: 10.1073/pnas.0704073104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Subach OM, Gundorov IS, Yoshimura M, Subach FV, Zhang J, Gruenwald D, Souslova EA, Chudakov DM, Verkhusha VV. Conversion of red fluorescent protein into a bright blue probe. Chem Biol. 2008;15:1116–1124. doi: 10.1016/j.chembiol.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gilmour DT, Jessen JR, Lin S. Manipulating gene expression in the zebrafish. In: Nusslein-Volhard C, Dahm R, editors. Zebrafish: A Practical Approach. Oxford University Press; New York: 2002. pp. 121–143. [Google Scholar]
- 44.Karasawa S, Araki T, Yamamoto-Hino M, Miyawaki A. A green-emitting fluorescent protein from Galaxeidae coral and its monomeric version for use in fluorescent labeling. J Biol Chem. 2003;278:34167–34171. doi: 10.1074/jbc.M304063200. [DOI] [PubMed] [Google Scholar]
- 45.Ai HW, Olenych SG, Wong P, Davidson MW, Campbell RE. Hue-shifted monomeric variants of Clavularia cyan fluorescent protein: identification of the molecular determinants of color and applications in fluorescence imaging. BMC Biol. 2008;6:13. doi: 10.1186/1741-7007-6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shagin DA, Barsova EV, Yanushevich YG, Fradkov AF, Lukyanov KA, Labas YA, Semenova TN, Ugalde JA, Meyers A, Nunez JM, et al. GFP-like proteins as ubiquitous metazoan superfamily: evolution of functional features and structural complexity. Mol Biol Evol. 2004;21:841–850. doi: 10.1093/molbev/msh079. [DOI] [PubMed] [Google Scholar]
- 47.Vogt A, D’Angelo C, Oswald F, Denzel A, Mazel CH, Matz MV, Ivanchenko S, Nienhaus GU, Wiedenmann J. A green fluorescent protein with photoswitchable emission from the deep sea. PLoS ONE. 2008;3:e3766. doi: 10.1371/journal.pone.0003766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alieva NO, Konzen KA, Field SF, Meleshkevitch EA, Hunt ME, Beltran-Ramirez V, Miller DJ, Wiedenmann J, Salih A, Matz MV. Diversity and evolution of coral fluorescent proteins. PLoS ONE. 2008;3:e2680. doi: 10.1371/journal.pone.0002680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ai HW, Henderson JN, Remington SJ, Campbell RE. Directed evolution of a monomeric, bright and photostable version of Clavularia cyan fluorescent protein: structural characterization and applications in fluorescence imaging. Biochem J. 2006;400:531–540. doi: 10.1042/BJ20060874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ando R, Hama H, Yamamoto-Hino M, Mizuno H, Miyawaki A. An optical marker based on the UV-induced green-to-red photoconversion of a fluorescent protein. Proc Natl Acad Sci U S A. 2002;99:12651–12656. doi: 10.1073/pnas.202320599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wiedenmann J, Ivanchenko S, Oswald F, Schmitt F, Rocker C, Salih A, Spindler KD, Nienhaus GU. EosFP, a fluorescent marker protein with UV-inducible green-to-red fluorescence conversion. Proc Natl Acad Sci U S A. 2004;101:15905–15910. doi: 10.1073/pnas.0403668101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McKinney SA, Murphy CS, Hazelwood KL, Davidson MW, Looger LL. A bright and photostable photoconvertible fluorescent protein. Nat Methods. 2009;6:131–133. doi: 10.1038/nmeth.1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ando R, Mizuno H, Miyawaki A. Regulated fast nucleocytoplasmic shuttling observed by reversible protein highlighting. Science. 2004;306:1370–1373. doi: 10.1126/science.1102506. [DOI] [PubMed] [Google Scholar]
- 54.Habuchi S, Tsutsui H, Kochaniak AB, Miyawaki A, van Oijen AM. mKikGR, a monomeric photoswitchable fluorescent protein. PLoS One. 2008;3:e3944. doi: 10.1371/journal.pone.0003944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wall MA, Socolich M, Ranganathan R. The structural basis for red fluorescence in the tetrameric GFP homolog DsRed. Nat Struct Biol. 2000;7:1133–1138. doi: 10.1038/81992. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.