Abstract
Proteasomes are large multisubunit complexes responsible for regulated protein degradation. Made of a core particle (20S) and regulatory caps (19S), proteasomal proteins are encoded by at least 33 genes, of which 12 have been shown to have testis-specific isoforms in Drosophila melanogaster. Pros28.1A (also known as Prosα4T1), a young retroduplicate copy of Pros28.1 (also known as Prosα4), is one of these isoforms. It is present in the D. melanogaster subgroup and was previously shown to be testis-specific in D. melanogaster. Here, we show its testis-specific transcription in all D. melanogaster subgroup species. Due to this conserved pattern of expression in the species harboring this insertion, we initially expected that a regulatory region common to these species evolved prior to the speciation event. We determined that the region driving testis expression in D. melanogaster is not far from the coding region (within 272 bp upstream of the ATG). However, different Transcription Start Sites (TSSs) are used in D. melanogaster and D. simulans, and a “broad” transcription start site is used in D. yakuba. These results suggest one of the following scenarios: 1) there is a conserved motif in the 5′ region of the gene that can be used as an upstream or downstream element or at different distance depending on the species; 2) different species evolved diverse regulatory sequences for the same pattern of expression (i.e., “TSS turnover”); or 3) the transcription start site can be broad or narrow depending on the species. This work reveals the difficulties of studying gene regulation in one species and extrapolating those findings to close relatives.
Keywords: retrogene, Drosophila, proteasome, testis expression, Pros28.1A, Prosα4T1, regulatory sequence
Introduction
Retroposition is a type of gene duplication that involves the creation of a new gene in a new genomic position via reverse transcription (RT).1 This reaction is likely catalyzed by reverse transcriptases of LINE-like transposable elements that mistakenly act on “host” gene transcripts; the resulting cDNAs are inserted into the genome by a process called Target Primed Reverse Transcription (TPRT).2, 3 Hallmarks of these new sequences include lack of introns, presence of poly-A tails and target site duplications. In Drosophila, the last two features are often lost in ancient retrogenes.4 It has long been known that for these retroposed copies to become functional genes, they must recruit either a “de novo” regulatory region, carry regulatory regions from the parental gene, insert in front of a region with regulatory capabilities or form chimeric genes. 5–7 It has also been suggested that gene expression can be facilitated by the surrounding chromatin context. 8 However, overall, little evidence supports any of these possibilities as a general mechanism by which the regulatory regions of retrogenes originate in Drosophila.5
Pros28.1, a gene located on the X chromosome of D. melanogaster, encodes the α4 subunit, a component of the proteasome core particle (CP). With two introns, it encodes a 248 amino acid-long protein that is expressed ubiquitously in D. melanogaster. 9 This is an expected pattern of expression, as most protein degradation in the cell is mediated by the proteasome through the attachment of ubiquitin to targeted proteins.10 While its conservation through evolution indicates its important housekeeping role in the proteasome, Pros28.1 has also given rise to two paralogs, both of which are located on autosomes with male-germline patterns of expression.11 These genes are examples of X-to-autosome duplication in Drosophila, which can occur to avoid X-chromosome inactivation during spermatogenesis, to increase the expression levels of X-linked genes or due to sexual antagonism4, 12–14 (also see discussion in Belote and Zhong 15). Of the two duplicates, Pros28.1B is the older, non-retroposed copy 11, 16 that has been shown to be transcribed during spermatid elongation 11, while Pros28.1A, the younger retroposed copy, is expressed primarily in spermatocytes and during the spermatid elongation stage.11 Pros28.1A is present only in the species of the D. melanogaster subgroup, suggesting that the retroposition event occurred 13–44 million years ago (Mya). 17 Pros28.1A was retroposed into the third intron of CG42322, a gene of unknown function that is conserved across Drosophila. 18
In Drosophila, the proteasome is encoded by 33 genes, of which 12 have been shown to have testis-specific isoforms.15 These duplicated genes are found primarily in the core particle (CP). Encoded by 14 genes (7 α and 7 β), the CP is composed of 4 heptameric rings organized in an α(1–7)β(1–7)β′(1–7)α′(1–7) fashion. Of these 14 genes, 6 have paralogs that show male-specific expression in Drosophila. The co-expression of these duplicated copies, along with functional data, supports the existence of a testis-specific proteasome 11, 15, 19, 20 whose function remains unknown. However, these expression data are based entirely on studies of D. melanogaster, and little information exists regarding the expression profiles in other species.
Gene regulation has been studied in detail for a number of genes, and a classic view has emerged. Gene regulation often encompasses a core promoter (located around the TSS) where transcription factors and RNA polymerase II assemble to initiate transcription, while other cis-regulatory modules (repressors and/or enhancers) farther away affect gene expression. 21 Several core promoter sequences have been described: some upstream of the TSS (e.g., the TATA box), some overlapping the TSS (e.g., the initiator, Inr) and some downstream of the TSS (e.g., the DPE and MTE).22–25 These motifs are not present in every promoter region, but they often occur in combination.25 In the case of testis-expressed genes, the regulatory regions have been shown to be in close vicinity of the TSS. 26–39 The only well-studied element that drives testis-specific expression is the β2 tubulin 14 bp element, which is present at identical positions (beginning at −51 bp from the TSS) in D. melanogaster and D. hydei. 39 This element has been shown to be associated with a 7 bp quantitative element and an Inr. 39, 40
In this work, we sought to describe the pattern of expression of Pros28.1A in different species to infer whether the testis-specific function is evolutionary conserved. We also studied the TSS of Pros28.1A in different species to characterize the potentially evolutionarily conserved regulatory region and determine its origin. Our data show that the testis-specific transcription is conserved in representative species of the entire D. melanogaster subgroup (i.e., D. melanogaster, D. simulans, D. yakuba and D. erecta) with different (up to 84 bp apart) TSSs in two species analyzed (D. melanogaster and D. simulans) and a broad TSS in D. yakuba. This difference was unexpected, since full-length transcripts usually start in a narrow window close to the TSS. 25 Thus, male-specific transcription in these species could be mediated by different regulatory sequences or by one that is common to all species, but that can either drive a broad or narrow start of transcription depending on the species. Alternately, this region may be capable of acting at different distances in some species, or as either a downstream regulatory region or upstream regulatory region. P element transformations using the upstream region of D. melanogaster Pros28.1A reveal that the testis-specific regulatory region in this species is within 272 bp upstream of the ATG. Although, not a lot can be inferred at present about its evolutionary origin, it is likely that transduced sequence at the time of insertion or additional reorganizations and nucleotide substitutions produced the current sequence and arrangement. In addition, this region does not resemble any known testis-specific regulatory sequence.
Results
Pros28.1A expression in different species
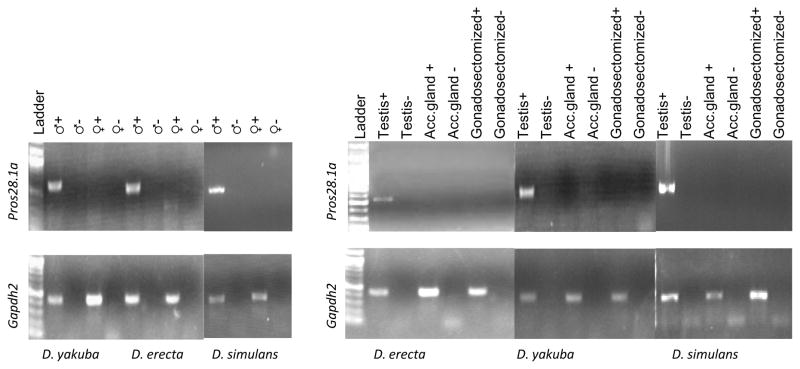
Expression of Pros28.1A was previously shown to be male specific in D. melanogaster pupae and adults, despite the ubiquitous expression of the parental copy.11 Our RT-PCR results also show the presence of Pros28.1A transcript only in males of D. simulans, D. yakuba and D. erecta. In addition, the presence of the transcript in males is limited to the testis, and no transcripts could be detected in the accessory glands or the gonadectomized body (Figure 1).
Figure 1.
RT-PCR results of Pros28.1A and Gapdh2 in D. simulans, D. yakuba and D. erecta are shown. Negative controls are PCR assays using DNA-digested total RNA where no reverse transcriptase was added.
Pros28.1A upstream region
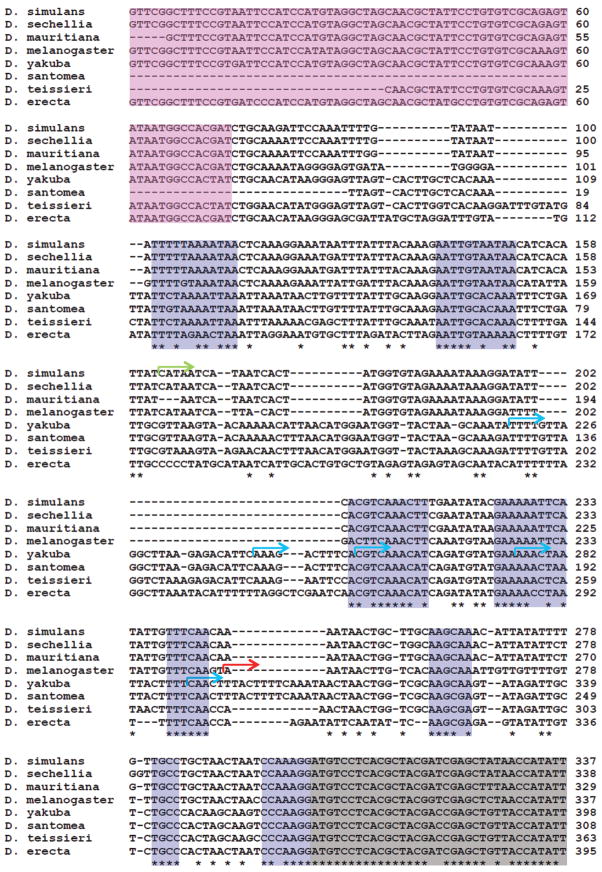
An alignment of the Pros28.1A upstream region from all the species in which the gene is present is shown in Figure 2. This alignment includes regions downloaded from FlyBase for D. melanogaster (gene CG17268 and upstream region), D. simulans (gene GD20037 and upstream region), D. sechellia (gene GM23164 and upstream region), D. yakuba (gene GE25043 and upstream region) and D. erecta (gene GG15180 and upstream region), and additional sequences obtained in this work (D. mauritiana, D. santomea, and D. teisseri). This alignment reveals several conserved regions (in blue) that could potentially act as regulatory sequences in all species given the conservation of the expression pattern in D. melanogaster, D. simulans, D. yakuba and D. erecta. As shown below, we found different TSSs in D. melanogaster, D. simulans and D. yakuba that challenged our original hypothesis.
Figure 2.
Alignment of the 5′ putative promoter region of Pros28.1A in D. melanogaster subgroup species. The beginning of the Pros28.1A coding sequence, which is inserted in the 3rd intron of CG42322, is light grey, and the 4th exon of CG42322 is pink. Note that CG42322 is encoded on the opposite strand from Pros28.1A. Blue boxes are conserved regions. Transcription start sites (TSSs) in each species are shown in different colors: The D. melanogaster TSS is shown in red, the D. simulans TSS is shown in green and the D. yakuba multiple TSSs are shown in blue.
Transcription start sites (TSSs)
Characterizing the regulatory regions of a gene requires knowledge of the TSS. Usually, regulatory motifs such as TATA 25 and the β2tubulin promoter (a known male-specific promoter) 39 are found upstream of the TSS. Therefore we performed 5′ Rapid Amplification of cDNA End (5′RACE) in D. melanogaster, D. simulans and D. yakuba using RNA from abdomens (see Materials and Methods), the results of which are shown in Figure 2. 5′RACE on Pros28.1A RNA from D. melanogaster demonstrated that the TSS is located 57 bp upstream of the CDS (i.e., the 5′UTR is 57 bp long). This 5′UTR is 1 bp longer than the annotated version in FlyBase, which is based on a full cDNA from the testis (AT30052). However, in D. simulans, the Pros28.1A TSS is located 140 bp upstream of the annotated CDS and 84 bp upstream of the D. melanogaster TSS. While we observed clear sequencing peaks in the D. melanogaster and D. simulans 5′RACE products, suggesting a single TSS, clear peaks were not observed in D. yakuba. This result is due to the presence of at least five different TSSs that are several base pairs (<74 bp) apart (Figure 2), an arrangement that can be classified as a broad TSS. 41 Those products were characterized after TA cloning and sequencing of the D. yakuba 5′RACE products.
As described in the Introduction, sequences surrounding the TSS often conform to a consensus sequence called the initiator sequence. Two types of initiators have been described in Drosophila: Inr (TCA(G/T)T(C/T))25 and Inr1 (TCATTCG)42, neither of which were found in the sequences surrounding the TSSs described in this work.
In contrast to our initial hypothesis, and despite the consistency in the pattern of transcription of Pros28.1A in D. melanogaster subgroup species, we observed different TSSs in the species studied. Therefore, we performed BLAST sequence analyses (http://blast.ncbi.nlm.nih.gov/Blast.cgi) 43 to compare the upstream regions (i.e., upstream of ATG) of Pros28.1A in D. melanogaster, D. simulans and D. yakuba. We used the default parameters but set the word size to 7 bp. Supplementary data file 1 and 2 show the results. These results reveal the following: 1) there are high levels of similarity between the sequences, as shown in Figure 2; 2) there is no region of similarity between the D. melanogaster and D. simulans TSS regions; and 3) there are no repeated motifs in either strand in D. simulans or D. yakuba, apart from some AT-rich short sequences that do not co-occur with the described TSSs. This result suggests that different but functionally equivalent regulatory sequences may have evolved in these lineages and highlights the interspersed nature of TSSs44 in some species. Alternatively, it is possible that a shared motif is present that could be used at different distances in some species, or that could act as an upstream motif in some and as a downstream regulatory region in others.
EGFP expression in transformed flies
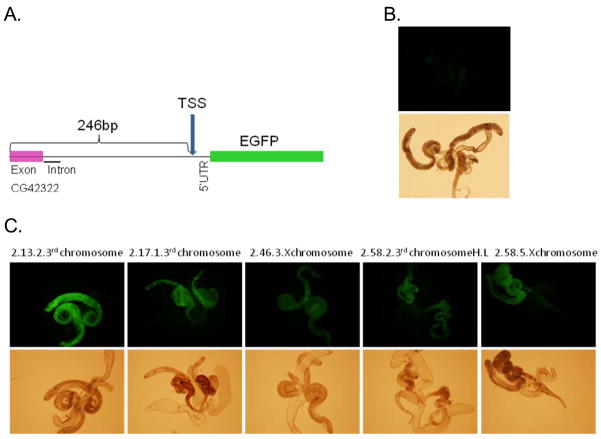
The sequence 246 bp upstream of D. melanogaster TSS and part of the 5′UTR (26 bp) were used to drive EGFP expression in transformed flies. Testis expression was observed in nine independent insertions, five of which are shown in Figure 3. Despite the intensity differences (possibly due to the position effects, depending on whether the insertion is homozygous lethal, in which case only heterozygotes were screened, or whether the insertion is located on the X chromosome35) all transformants showed EGFP expression in testis when compared to the injected strain (w1118). This pattern of expression is consistent with the previous observation of Yuan et al. 11, who reported expression of a Pros28.1A-lacZ reporter gene in primary spermatocytes during the spermatid elongation stage.11 Their transformant flies harbored ~3.5 kb of upstream sequence and ~2 kb of downstream sequence from Pros28.1A. However, we found that a much shorter region (272 bp) is sufficient to drive the same pattern of expression in the testis.
Figure 3.
Overview of the insert of the transformed construct is shown in Panel A. Panel B shows level of green fluorescence in testes in the control strain (w1118). Panel C shows EGFP florescence driven by the 272-bp region (246 bp upstream of the gene and 26 of 5′UTR) in the testis of five different transformants.
Microarray data in FlyBase (Gauhar et al. 2008.10.3). D. melanogaster life-cycle gene expression dataset and microarray normalization protocols; FlyBase personal communication) suggest that Pros28.1A is transcribed in early embryo. However, we did not observe embryonic (0–5 hours) EGFP expression in our transformant lines or transcript in our RT-PCR in D. melanogaster (Supplementary figures 1 and 2). This result could be explained by the microarray data being cross-hybridization with the parental gene. Since the platform used by Gauhar et al. were spotted amplicons in the coding region of Pros28.1A, the results could come from cross-hybridization with the parental gene (Pros28.1).
We previously studied the presence of known regulatory motifs or novel cis-regulatory modules (CRM) in D. melanogaster retrogenes. 45 A short sequence in the coding region of CG42322 upstream of Pros28.1A was proposed as a potential CRM associated with testes expression (TA1) because of its overrepresentation in testis-expressed retrogenes. However, this region was not included in our construct, and yet we were able to drive testis-specific EGFP expression; thus, we conclude that this motif does not drive testis expression in this gene, in agreement with what we postulated previously 45, and the TA1 found in coding region of CG42322 was a false positive.
Origin of the regulatory region
As described above, the fluorescence observed in transformant flies shows that in D. melanogaster, 246 bp upstream of the TSS and 26 bp of the 5′UTR is sufficient to drive expression of Pros28.1A in the male germline. While the particular motif driving expression of this retrogene in different species is still being explored, we can investigate the origin of this 272-bp region. As shown in Figure 2 and Supplementary data files 1 and 2, this region is conserved between the species that contain the Pros28.1A insertion but it is very different from the 5′UTR or upstream region of the parental gene (see Supplementary data file 3). A single transcript of the parental gene Pros28.1-RA has been annotated with a 146-bp 5′UTR (FlyBase annotation). Supplementary data file 3 shows the output of a BLAST 2 sequences analysis (http://blast.ncbi.nlm.nih.gov/Blast.cgi) 43 using the upstream regions of Pros28.1A (272 bp) and Pros28.1 (275 bp) from D. melanogaster. We used the BLAST default parameters but set the word size at 7 bp. No long stretches of similarity or statistically significant short matches were found. Only 9 short [7–11 bp] matches (Expected value (E-value) ≥ 1.1) were found in D. melanogaster between these regions of Pros28.1A and Pros28.1. The E-value represents the number of times you can expect to see the alignment with the same score or better purely by chance when searching against a database of the same size. 43 In BLAST 2 sequences searches, the database is the Subject sequence (http://www.ncbi.nlm.nih.gov/blast/producttable.shtml#blastn). E-values approach P values when they are small. 46 Therefore, we do not consider a hit significant unless E-value is smaller than the standard 0.05. This is in agreement with our previously published results 5, where we looked at sequence similarity between parental and retrogene upstream regions of all annotated retrogenes and found no conservation, even for retrogenes younger than Pros28.1A. So far, we have no evidence of sequence similarity upstream of the TSS between the parental gene and retrogene for any retrogene in Drosophila.
We also looked at the similarity of the upstream region between Pros28.1A (272 bp) and Pros28.1B (275 bp; another testis-specific member of the gene family; see Introduction). The results are shown in Supplementary data file 4. Again, no statistically significant matches were found using BLAST 2 sequences (E-value ≥0.087). 43 Although there were several matches found between the upstream regions of Pros28.1A and Pros28.1B, none of the hits between these two sequences occurred in a region conserved between the species that have the retrogene, precluding us from suggesting the existence of a conserved element in Pros28.1A that resembles a Pros28.1B element.
To look outside of the gene family, we blasted the 272-bp region of the D. melanogaster Pros28.1A gene against the whole genomes of all the species in the D. melanogaster group. No hit with an E-value smaller than 0.17 or a long similarity that could reveal the origin of this region was found.
Additionally, this region does not seem to belong to the original intron of CG42322. As revealed in Figure 4, the intron was likely small at the time of insertion, ranging from 57 to 74 bp in all species except in D. grimshawi (see Supplementary data file 5). In addition, little to no sequence similarity could be found in the intron between the species that have the insertion (Pros28.1A) and those without (Figure 4). Thus, we infer that the intron present at the time of insertion, which was likely short, did not give rise to the more than 250-bp region upstream of the CDS (Figure 2).
Figure 4.
Alignment of exons 3 and 4 and intron 3 of CG42322 in several Drosophila species. Grey corresponds to part of the exons. Blue arrows point to the approximate position where Pros28.1A and likely additional sequence at its 5′ and 3′ end have been inserted (not shown). See supplementary data file 2 for the complete alignment. D. grimshawi is not shown because it has an unusually long intron.
According to the phylogenetic distribution, the Pros28.1A duplication event could be 12–44 My old.17 Using sequence divergence information at nonsynonymous sites (see Materials and Methods), we dated the duplication event to ~ 39.6 Mya. This finding supports the hypothesis that the duplication is relatively old, and thus there has been plenty of time for the upstream region of Pros28.1A to diverge from the initial sequence (nucleotide substitutions and indels), resulting in sequences that are quite different. Hence, almost no information remains to make an inference about the origin of this 272-bp sequence.
Discussion
It is often expected that functional regulatory regions will be under purifying selection, and that orthologous genes maintaining the same pattern of expression will use the same regulatory regions/conserved motifs in similar way39, 47. However, this scenario is not supported by our present study.
In this work, we studied the expression of a young (12–44 My old) Drosophila retrogene (Pros28.1A) in several species and revealed that the pattern of expression has been conserved for more than 12 My. The gene has a male testis-specific transcription pattern in all the species studied. As described in the Introduction, the regulatory regions of many testis-expressed genes lie in close vicinity to the TSS. 26–39 In particular, β2 tubulin, the well-studied element that drives testis-specific expression and acts as a promoter and enhancer in both D. melanogaster and D. hydei, is present at identical positions in both species (i.e., beginning at −51 bp from the TSS 39 18). Thus, we expected to observe conservation of the Pros28.1A TSS in the species harboring this gene. However, as described earlier, our 5′ RACE analyses of Pros28.1A in three different species (D. melanogaster, D. simulans and D. yakuba) revealed significantly different TSSs (84 bp apart); additionally, a broad TSS was observed in D. yakuba, with multiple TSSs separated by up to 74 bp. This result is in disagreement with our initial expectation of conserved locations and sequences of cis-regulatory sequences, given the conserved pattern of expression of this gene. This finding can be explained in at least three different ways. First, it is possible that the same regulatory motif is used in all species but acts as downstream element in some species and as an upstream element in others. To our knowledge, this has been described before for enhancers but never for elements with promoter properties. Usually, if a motif is described to be an upstream or downstream promoter motif, it is used similarly in other species.23, 24, 39
The same regulatory motif could also function at different distances from the TSS in different species, but the broadening of the TSS in D. yakuba would remain to be explained. The presence of motifs potentially leading to a broadening of the TSS in this species could be ocurring.44 In some cases, the transcription of genes with the same regulatory motif(s) does not start in the same position in different genes or species due to other motifs or additional cis- and trans-regulatory changes.48 In addition, some species (D. yakuba in this case) show interspersed TSSs more often than others, and this could even be characteristic of some species. This change in regulatory region shape is not expected from what has been observed in mammals, as shape classes between orthologous mouse and human TSSs are quite conserved 41. In D. melanogaster, a single TSS peak or broad start of transcription correlates with the type of core promoter motif (i.e., TATA, Inr, DPE and MTE vs. Ohler 1, DRE, Ohler 6 and Ohler 7) and GC content.49 More data from across species and genes will be needed to determine the level of purifying selection or evolvability of this peak and broad TSS in Drosophila, as well as any species-specific characteristic that might be related to the effective population size of the species. 50
A third possibility is that different independently evolved regulatory regions (also called “TSS turnover”) drive the expression of Pros28.1A in the same tissue of different species.51, 52 Turnover has been defined as a TSS distance bigger than 100 bp 53, but this is an arbitrary definition, and motifs driving the expression of Pros28.1A in these species remain to be explored to determine whether TSS turnover has occurred. However, while this is a possibility, the low levels of sequence divergence argues against this hypothesis (see Figure 2). Detailed experimental analyses of the motifs driving the transcription of Pros28.1A in every species and of TSSs in different Drosophila species (using, for example, comparative Drosophila CAGE analyses for particular tissues) should help to answer these questions.
The features of the testis-specific regulatory region are also of interest. Manual and computational screening for the presence of previously described motifs 25, 39, 42 in the transformed 272-bp region failed to detect any similarity to any known motif responsible for transcription of Pros28.1A in D. melanogaster (this work and Bai et al. 45). Of this 246 bp, 78 bp encodes part of the third protein-coding exon of CG42322, which is unlikely to harbor regulatory regions due to functional constraints on the coding sequences. We previously studied the presence of a novel cis-regulatory module (CRM) in D. melanogaster retrogenes. 45 A short sequence in the coding region of CG42322 farther upstream of Pros28.1A was proposed as a potential CRM associated with testes expression (TA1) because of its overrepresentation in testis-expressed retrogenes. Since this region was not included in our construct we conclude that this motif does not drive testis expression in this gene. Experiments are underway to characterize the testis-specific regulatory motifs in the different species.
Here, we attempted to determine the origin of the testis-specific regulatory region of Pros28.1A. This region could be part of the original CG42322 intron or part of the parental transcript (5′UTR or upstream region in an aberrantly long parental transcript; see also 5). With no evidence of sequence similarity between parental and retrogene upstream regions and the small intron of CG42322, we concluded that neither one made a major contribution to the evolution of this testis-specific regulatory region. We also considered the possibility that the region could be a transduced sequence that appeared at the time of insertion. There are reports suggesting the ability of the LINE element to jump from one template to another, thus introducing a piece of DNA, such as extra pieces of the LINE element or its flanking sequences, originally absent in the transcript.54 To determine whether this was the origin of our 272-bp region, we performed BLAST on this sequence but found only sporadic insignificant hits against the sequenced genomes. Based on these findings, we conclude that the Pros28.1A duplication, which we estimate to be ~39.6 My old, is too old to reveal ancestral similarities. However, the region is too big to be derived only from the 5′ of ancestral intron (~25 bp) plus the parental 5′UTR (146 bp), suggesting that transduced sequence at the time of insertion or additional reorganizations and nucleotide substitutions likely produced the current sequence and arrangement.
Materials and Methods
Strains used
The following isofemale lines were used in this work: D. melanogaster (Besançon; from P. Gilbert), D. simulans (Florida; from J. Coyne), D. mauritiana (Synthetic; from P. Michalak), D. santomea (Sto10; from M. Long), D. teisseri (118.2 55), D. yakuba (115 55), and D. erecta (154.155). The strains were grown in standard corn media at 25°C.
Expression analysis
Transcription of Pros28.1A was studied in both male and female tissues. Tissues were homogenized in a glass homogenizer and total RNA was extracted using the RNeasy kit according to the manufacturer′s instructions (Qiagen, Valencia, CA) from ~30 males and virgin females. Mature males (1–5 days old) were dissected in saline solution to separate the testes, accessory glands and carcass (gonadectomized body). The tissues were preserved in RNA-later solution (Applied Biosystems/Ambion, Austin, TX), soaked at 4°C overnight and then stored at −80°C until processing. RNA was extracted from 20 gonadectomized males, 100 testes and 100 accessory glands of D. simulans, D. yakuba and D. erecta. RNA was also extracted from ~100 embryos (0–5 hours old) of D. melanogaster (Besancon strain).
RT-PCR was conducted on total RNA from males, virgin females, gonadectomized males, testes and accessory glands. Analysis of expression of intronless genes (such as Pros28.1A) is challenging because genomic contamination can produce a band of the same size as that of expected from the cDNA. Therefore, we digested possible contaminating DNA from the total RNA (DNase I amplification grade; Invitrogen, Carlsbad, CA) and ran controls including DNA-digested total RNA without reverse transcriptase. Single strand complementary DNA (cDNA) was synthesized using Superscript (Invitrogen, Carlsbad, CA) and oligo-dT (Promega, Madison, WI). RT-PCR was carried out using specific primers 5′-GTTCGTGGAGGCAATTGTGTG-3′ and 5′-GTACGCCCAGGAAGCTGTTC-3′ for amplification of Pros28.1A in D. yakuba and D. erecta, and 5′-TGCCTGCTAACTAACCCAAAG-3′ and 5′-AACTGGGTTAACCTCGAGAAGG-3′ in D. simulans and D. melanogaster. The Gapdh2 gene was used as positive control for the RT reaction, using primers 5′-CAAACGAACATGGGAGCATC-3′ and 5′-TCAGCCATCAGAGTCGATTC-5′.
DNA samples and sequencing
Sequences of the CG42322 intron that in some species contains Pros28.1A and its flanking exons were obtained from FlyBase for the available species.18 For species in which this sequence was not available (D. santomea, D. teisseri, and D. mauritiana), genomic DNA was extracted from single female flies using the Puregene kit, and Pros28.1A and its flanking sequence was PCR amplified using the primers 5′TTAGGGTTCGGCTTTCCGTA3′, 5′ACCTGCTATCCTGGGTGATC3′ and 5′CAACGCTATCCTGTGTCGC3′. PCR products were then sequenced directly after purification (Qiagen kit, Qiagen, Valencia, CA) on an ABI automated DNA sequencer and fluorescent DyeDeoxy terminator reagents (Applied Biosystems, Foster City, CA). Sequences were obtained from both strands to confirm every position and contigs were made using Sequencher 4.5 (Gene Codes Corporation, Ann Arbor, MI). Sequences were aligned by means of Clustal W.56 New sequences have been submitted to GenBank with accession numbers GU391592-GU391594.
Transcription start site (TSS) description
The full-length 5′ end sequence of the D. melanogaster, D. simulans and D. yakuba Pros28.1A transcripts were obtained by 5′ RACE experiments using the First choice RLM-RACE kit from Ambion (Applied Biosystems/Ambion, Austin, TX). Around 1 μg of total RNA obtained from male abdominal halves was phosphatase treated to remove the 5′ phosphate of degraded mRNA, rRNA, tRNA, and genomic DNA, leaving the capped mRNA intact. The capped mRNAs were then treated to remove the cap and ligated to an adaptor. Single strand cDNA was synthesized from mRNA using reverse transcriptase using random decamers as primers. PCRs were conducted to amplify the target transcript using the primer 5′-GCGAGCACAGAATTAATACGACT-3′ provided in the kit specific to the adapter and the following primers specific to the Pros28.1A of D. melanogaster, D. simulans and D. yakuba, respectively: 5′-AGGGTCACCTGGTTTTCGAAG-3′, 5′-GGTCACCGGTTTGTCGAAG-3′, 5′-GACCTGCCCTCGATTTATTAGGATC-3′. Since one round of PCR usually did not yield any product, these products were further amplified using the following nested primers: 5′-CAGCACCACACAATTGGCTCCA-3′, specific to D. melanogaster and D. simulans, and 5′-GTGATCTTGCGCACCGTTCGGTA-3′, specific to D. yakuba along, with the RACE inner primer 5′-CGCGGATCCGAATTAATACGACTCACTATAGG-3′ provided in the kit. The products were then sequenced. While clear sequences indicate single TSS, double peak overlaps could indicate more than one TSS. In such cases, the PCR product was cloned into the TOPO TA cloning kit (Invitrogen, Carlsbad, CA), and 10 colonies with insert were sequenced to characterize the ends.
Strains and clones for transformation
Genomic DNA extracted from single D. melanogaster fly was used to amplify the 5′ upstream region (i.e., upstream of the CDS) of Pros28.1A, which likely contains the putative promoter, using the primers 5′-CCGCGGATTACTCACCCTAAAC-3′ and 5′-ACCGGTCAACAATTTGCTTGTGACAAG-3′. High fidelity Taq polymerase (Finnzymes, Espoo, Finland) was used to prevent the introduction of sequence changes by PCR amplification. The PCR product was TA cloned using the TOPO TA cloning kit (Invitrogen, Carlsbad, CA), digested with SacII (New England Biolabs, Ipswich, MA) and AgeI (New England Biolabs, Ipswich, MA) and ligated directionally to EGFP vector (U55761; Clontech, Mountain View, CA) using T4ligase (New England Biolabs, Ipswich, MA). The ligated plasmid was then transformed into XL-blue super competent cells (Stratagene/Agilent Technologies, Cedar Creek, TX). After PCR screening and sequencing the plasmid to confirm the integrity of the insert, the purified plasmid (Qiagen; Valencia, CA) was digested by AflII (New England Biolabs, Ipswich, MA), blunt ended using Mung Bean nuclease (New England Biolabs, Ipswich, MA) and digested by SacII (New England Biolabs, Ipswich, MA). This insert was then ligated into a pCaSpeR4 vector (X81645) containing a blunt end and a SacII site produced in a similar way. The transformed and sequenced plasmid was injected into 30-minute-old embryos of the w1118 strain along with Turbo transposase plasmid to produce insertion of the construct in the genome.57 Injected individuals were backcrossed to individuals of the opposite sex of the w1118 strain. At least one individual from the crosses that had individuals with orange eyes were kept to map the P element insertion and then fix the insertion using balancer chromosomes. Genomic DNA from these transformed flies was used to amplify a shorter region. High fidelity Taq polymerase (Finnzymes, Espoo, Finland) was again used to prevent the introduction of sequence changes in our PCR amplifications. The primers used to make the final shorter construct used in the analyses were as follows: 5′-CTGCAGTTCGGCTTTCCGTAATTC-3′ in the 5′ region and 5′-CTGCAGTGATGAGTTTGGACAAAC-3′ in the P element, including the EGFP gene and termination signal. The amplified region included only 246 bp of the Pros28.1A 5′ upstream region and 26 bp of the 5′UTR upstream of EGFP. This region was cloned using the TOPO TA cloning kit (Invitrogen, Carlsbad, CA), then digested with KpnI and XhoI and ligated into the pCaSpeR4 vector (X81645) digested with the same enzymes. These were then transformed into XL1Blue competent cells (Stratagene/Agilent Technologies, Cedar Creek, TX). The plasmids were purified and were injected into w1118 embryos by Genetic Services Inc. (Cambridge, MA). Injected individuals were backcrossed to w1118 strain individuals of the opposite sex. At least one individual from the crosses that had individuals with orange eyes were kept to map the P element insertion and then fix the insertion using balancer chromosomes. Expression of EGFP was then screened in the transformants fixed for the insertion.
Age of the duplication
Age of the duplication was calculated using the method described in Li 58 that assumes that parental gene and retrogenes are evolving at different but constant rates. MEGA software was used to calculate the nonsynonymous distances between all sequences using the Pamilo-Bianchi-Li method (see Supplementary data file 6). Divergence between D. melanogaster/D. simulans/D. sechellia and D. mauritiana and D. yakuba were used to estimate the rate of evolution of parental and retrogene separately. Divergence time between D. melanogaster/D. simulans/D. sechellia and D. mauritiana and D. yakuba was set at 12.8 My.59 The average divergence between parental genes and retrogenes was estimated from all possible comparisons with the available data (see Supplementary data file 3).
Supplementary Material
BLAST 2 sequence output of the 272-bp region of D. melanogaster Pros28.1A aligned with the 5′UTR and upstream region of D. simulans Pros28.1A.
BLAST 2 sequence output of the 272-bp region of D. melanogaster Pros28.1A aligned with 5′UTR and upstream region of D. yakuba Pros28.1A.
BLAST 2 sequence output of the 272-bp region of D. melanogaster Pros28.1A aligned with 5′UTR and upstream region of D. melanogaster Pros28.1.
BLAST 2 sequence output of the 272-bp region of D. melanogaster Pros28.1A aligned with 5′UTR and upstream region of D. melanogaster Pros28.1B.
Sequence alignment of the Pros28.1A and flanking region, including the CG42322 flanking coding exons (exons 3 and 4 of the transcript encoded on the other strand) in the species where it is present and the same intron in the species where the Pros28.1A gene is not present. Grey corresponds to CG42322 exons and orthologs. Blue shows the CDS of Pros28.1A.
Shown is the nonsynonymous distance matrix between all parental and all retrogenes, created using the Pamilo-Bianchi-Li method. Divergence values between either D. melanogaster, D. simulans, D. sechellia, or D. mauritiana and D. yakuba were used to estimate the average rate of evolution of parental gene and retrogene separately (shown in bold). The average divergence between parental genes and retrogenes was estimated from all possible comparisons with the available data (shown in italics).
Panel a. shows level of green fluorescence in embryos in the control strain (w1118). Panel b. shows lack of EGFP florescence driven by the 272-bp region (246 bp upstream of the gene and 26 of 5′UTR) in the embryos of five different transformants.
RT-PCR results of Pros28.1A and Gapdh2 in 0–5 hour embryos of D. melanogaster (Besancon strain). Lane 1 shows ladder. Lanes 2–4 show RT+, RT− and PCR negative of Pros28.1A. Lanes Lane 5–7 show RT+, RT− and PCR negative of Gapdh2.
Acknowledgments
We would like to thank members of the Betrán lab for comments and help on this work. We also thank Jerry Coyne, Patricia Gilbert, Françoise Lemeunier, Manyuan Long, and Pawel Michalak for providing stocks. Research was supported by UTA startup funds and grant GM071813 from National Institutes of Health, USA to EB.
Abbreviations and Acronyms
- TSS
Transcription start site
- RT
reverse transcription
- My
million years
References
- 1.Brosius J. RNAs from all categories generate retrosequences that may be exapted as novel genes or regulatory elements. Gene. 1999;238:115–34. doi: 10.1016/s0378-1119(99)00227-9. [DOI] [PubMed] [Google Scholar]
- 2.Esnault C, Maestre J, Heidmann T. Human LINE retrotransposons generate processed pseudogenes. Nat Genet. 2000;24:363–7. doi: 10.1038/74184. [DOI] [PubMed] [Google Scholar]
- 3.Kazazian HH., Jr Mobile elements: drivers of genome evolution. Science. 2004;303:1626–32. doi: 10.1126/science.1089670. [DOI] [PubMed] [Google Scholar]
- 4.Betran E, Thornton K, Long M. Retroposed New Genes Out of the X in Drosophila. Genome Res. 2002;12:1854–9. doi: 10.1101/gr.604902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bai Y, Casola C, Betrán E. Evolutionary origin of regulatory regions of retrogenes in Drosophila. BMC Genomics. 2008;9:241. doi: 10.1186/1471-2164-9-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Long M, Langley CH. Natural selection and the origin of jingwei, a chimeric processed functional gene in Drosophila. Science. 1993;260:91–5. doi: 10.1126/science.7682012. [DOI] [PubMed] [Google Scholar]
- 7.McCarrey JR. Evolution of tissue-specific gene expression in mammals: How a new phosphoglycerate kinase was formed and refined. BioScience. 1994;44:20–7. [Google Scholar]
- 8.Kalmykova AI, Nurminsky DI, Ryzhov DV, Shevelyov YY. Regulated chromatin domain comprising cluster of co-expressed genes in Drosophila melanogaster. Nucleic Acids Res. 2005;33:1435–44. doi: 10.1093/nar/gki281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haass C, Pesold-Hurt B, Multhaup G, Beyreuther K, Kloetzel PM. The Drosophila PROS-28.1 gene is a member of the proteasome gene family. Gene. 1990;90:235–41. doi: 10.1016/0378-1119(90)90185-t. [DOI] [PubMed] [Google Scholar]
- 10.Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiol Rev. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- 11.Yuan X, Miller M, Belote JM. Duplicated proteasome subunit genes in Drosophila melanogaster encoding testes-specific isoforms. Genetics. 1996;144:147–57. doi: 10.1093/genetics/144.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parisi M, Nuttall R, Naiman D, Bouffard G, Malley J, Andrews J, et al. Paucity of genes on the Drosophila X chromosome showing male-biased expression. Science. 2003;299:697–700. doi: 10.1126/science.1079190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ranz JM, Castillo-Davis CI, Meiklejohn CD, Hartl DL. Sex-dependent gene expression and evolution of the Drosophila transcriptome. Science. 2003;300:1742–5. doi: 10.1126/science.1085881. [DOI] [PubMed] [Google Scholar]
- 14.Vicoso B, Charlesworth B. The deficit of male-biased genes on the D. melanogaster X chromosome is expression-dependent: a consequence of dosage compensation? J Mol Evol. 2009;68:576–83. doi: 10.1007/s00239-009-9235-4. [DOI] [PubMed] [Google Scholar]
- 15.Belote JM, Zhong D. Duplicated proteasome subunit genes in Drosophila and their roles in spermatogenesis. Heredity. 2009;103:23–31. doi: 10.1038/hdy.2009.23. [DOI] [PubMed] [Google Scholar]
- 16.Belote JM, Miller M, Smyth KA. Evolutionary conservation of a testes-specific proteasome subunit gene in Drosophila. Gene. 1998;215:93–100. doi: 10.1016/s0378-1119(98)00256-x. [DOI] [PubMed] [Google Scholar]
- 17.Bai Y, Casola C, Feschotte C, Betran E. Comparative Genomics Reveals a Constant Rate of Origination and Convergent Acquisition of Functional Retrogenes in Drosophila. Genome Biology. 2007;8:R11. doi: 10.1186/gb-2007-8-1-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilson RJ, Goodman JL, Strelets VB, Consortium F. FlyBase: integration and improvements to query tools. Nucleic Acids Research. 2008;36:D588–D93. doi: 10.1093/nar/gkm930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma J, Katz E, Belote JM. Expression of proteasome subunit isoforms during spermatogenesis in Drosophila melanogaster. Insect Mol Biol. 2002;11:627–39. doi: 10.1046/j.1365-2583.2002.00374.x. [DOI] [PubMed] [Google Scholar]
- 20.Zhong L, Belote JM. The testis-specific proteasome subunit Prosalpha6T of D. melanogaster is required for individualization and nuclear maturation during spermatogenesis. Development. 2007;134:3517–25. doi: 10.1242/dev.004770. [DOI] [PubMed] [Google Scholar]
- 21.Lemos B, Landry CR, Fontanillas P, Renn SCP, Kulathinal R, Brown KM, et al. Evolution of Genomic Expression. In: Pagel M, Pomiankowski A, editors. Evolutionary Genomics and Proteomics. Sunderland: Sinauer Associates; 2008. pp. 81–118. [Google Scholar]
- 22.Down TA, Bergman CM, Su J, Hubbard TJ. Large-Scale Discovery of Promoter Motifs in Drosophila melanogaster. PLoS Comput Biol. 2007;3:e7. doi: 10.1371/journal.pcbi.0030007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kadonaga JT. The DPE, a core promoter element for transcription by RNA polymerase II. Exp Mol Med. 2002;34:259–64. doi: 10.1038/emm.2002.36. [DOI] [PubMed] [Google Scholar]
- 24.Lim CY, Santoso B, Boulay T, Dong E, Ohler U, Kadonaga JT. The MTE, a new core promoter element for transcription by RNA polymerase II. Genes Dev. 2004;18:1606–17. doi: 10.1101/gad.1193404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ohler U, Liao GC, Niemann H, Rubin GM. Computational analysis of core promoters in the Drosophila genome. Genome Biol. 2002;3:RESEARCH0087. doi: 10.1186/gb-2002-3-12-research0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Robinson MO, McCarrey JR, Simon MI. Transcriptional regulatory regions of testis-specific PGK2 defined in transgenic mice. Proc Natl Acad Sci U S A. 1989;86:8437–41. doi: 10.1073/pnas.86.21.8437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nurminsky DI, Nurminskaya MV, De Aguiar D, Hartl DL. Selective sweep of a newly evolved sperm-specific gene in Drosophila. Nature. 1998;396:572–5. doi: 10.1038/25126. [DOI] [PubMed] [Google Scholar]
- 28.Han SY, Xie W, Kim SH, Yue L, DeJong J. A short core promoter drives expression of the ALF transcription factor in reproductive tissues of male and female mice. Biology of Reproduction. 2004;71:933–41. doi: 10.1095/biolreprod.104.030247. [DOI] [PubMed] [Google Scholar]
- 29.Kuhn R, Schafer U, Schafer M. Cis-acting regions sufficient for spermatocyte-specific transcriptional and spermatid-specific translational control of the Drosophila melanogaster gene mst(3)gl-9. EMBO J. 1988;7:447–54. doi: 10.1002/j.1460-2075.1988.tb02832.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hempel LU, Rathke C, Raja SJ, Renkawitz-Pohl R. In Drosophila, don juan and don juan like encode proteins of the spermatid nucleus and the flagellum and both are regulated at the transcriptional level by the TAF II80 cannonball while translational repression is achieved by distinct elements. Dev Dyn. 2006;235:1053–64. doi: 10.1002/dvdy.20698. [DOI] [PubMed] [Google Scholar]
- 31.Di Cara F, Morra R, Cavaliere D, Sorrentino A, De Simone A, Polito CL, et al. Structure and expression of a novel gene family showing male germline specific expression in Drosophila melanogaster. Insect Mol Biol. 2006;15:813–22. doi: 10.1111/j.1365-2583.2006.00688.x. [DOI] [PubMed] [Google Scholar]
- 32.Blumer N, Schreiter K, Hempel L, Santel A, Hollmann M, Schafer MA, et al. A new translational repression element and unusual transcriptional control regulate expression of don juan during Drosophila spermatogenesis. Mech Dev. 2002;110:97–112. doi: 10.1016/s0925-4773(01)00577-9. [DOI] [PubMed] [Google Scholar]
- 33.Kuhn R, Kuhn C, Borsch D, Glatzer KH, Schafer U, Schafer M. A cluster of four genes selectively expressed in the male germ line of Drosophila melanogaster. Mech Dev. 1991;35:143–51. doi: 10.1016/0925-4773(91)90064-d. [DOI] [PubMed] [Google Scholar]
- 34.Gigliotti S, Balz V, Malva C, Schafer MA. Organisation of regulatory elements in two closely spaced Drosophila genes with common expression characteristics. Mech Dev. 1997;68:101–13. doi: 10.1016/s0925-4773(97)00137-8. [DOI] [PubMed] [Google Scholar]
- 35.Hense W, Baines JF, Parsch J. X chromosome inactivation during Drosophila spermatogenesis. PLoS Biol. 2007;5:e273. doi: 10.1371/journal.pbio.0050273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Somboonthum P, Ohta H, Yamada S, Onishi M, Ike A, Nishimune Y, et al. cAMP-responsive element in TATA-less core promoter is essential for haploid-specific gene expression in mouse testis. Nucleic Acids Res. 2005;33:3401–11. doi: 10.1093/nar/gki652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schulz RA, Xie XL, Miksch JL. cis-acting sequences required for the germ line expression of the Drosophila gonadal gene. Dev Biol. 1990;140:455–8. doi: 10.1016/0012-1606(90)90094-y. [DOI] [PubMed] [Google Scholar]
- 38.Yanicostas C, Lepesant JA. Transcriptional and translational cis-regulatory sequences of the spermatocyte-specific Drosophila janusB gene are located in the 3′ exonic region of the overlapping janusA gene. Mol Gen Genet. 1990;224:450–8. doi: 10.1007/BF00262440. [DOI] [PubMed] [Google Scholar]
- 39.Michiels F, Gasch A, Kaltschmidt B, Renkawitz-Pohl R. A 14 bp promoter element directs the testis specificity of the Drosophila beta 2 tubulin gene. Embo J. 1989;8:1559–65. doi: 10.1002/j.1460-2075.1989.tb03540.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Santel A, Kaufmann J, Hyland R, Renkawitz-Pohl R. The initiator element of the Drosophila beta2 tubulin gene core promoter contributes to gene expression in vivo but is not required for male germ-cell specific expression. Nucleic Acids Res. 2000;28:1439–46. doi: 10.1093/nar/28.6.1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carninci P, Sandelin A, Lenhard B, Katayama S, Shimokawa K, Ponjavic J, et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat Genet. 2006;38:626–35. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- 42.Fitzgerald PC, Sturgill D, Shyakhtenko A, Oliver B, Vinson C. Comparative genomics of Drosophila and human core promoters. Genome Biol. 2006;7:R53. doi: 10.1186/gb-2006-7-7-r53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tatusova TA, Madden TL. BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol Lett. 1999;174:247–50. doi: 10.1111/j.1574-6968.1999.tb13575.x. [DOI] [PubMed] [Google Scholar]
- 44.Okamura K, Nakai K. Retroposition as a source of new promoters. Mol Biol Evol. 2008;25:1231–8. doi: 10.1093/molbev/msn071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bai Y, Casola C, Betran E. Quality of regulatory elements in Drosophila retrogenes. Genomics. 2009;93:83–9. doi: 10.1016/j.ygeno.2008.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pevsner J. Bioinformatics and Functional Genomics. Wiley & Sons; 2003. [Google Scholar]
- 47.Stark A, Lin MF, Kheradpour P, Pedersen JS, Parts L, Carlson JW, et al. Discovery of functional elements in 12 Drosophila genomes using evolutionary signatures. Nature. 2007;450:219–32. doi: 10.1038/nature06340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Megraw M, Pereira F, Jensen ST, Ohler U, Hatzigeorgiou AG. A transcription factor affinity-based code for mammalian transcription initiation. Genome Res. 2009;19:644–56. doi: 10.1101/gr.085449.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rach EA, Yuan HY, Majoros WH, Tomancak P, Ohler U. Motif composition, conservation and condition-specificity of single and alternative transcription start sites in the Drosophila genome. Genome Biol. 2009;10:R73. doi: 10.1186/gb-2009-10-7-r73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Keightley PD, Lercher MJ, Eyre-Walker A. Evidence for widespread degradation of gene control regions in hominid genomes. PLoS Biol. 2005;3:e42. doi: 10.1371/journal.pbio.0030042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Costas J, Casares F, Vieira J. Turnover of binding sites for transcription factors involved in early Drosophila development. Gene. 2003;310:215–20. doi: 10.1016/s0378-1119(03)00556-0. [DOI] [PubMed] [Google Scholar]
- 52.Ludwig MZ, Bergman C, Patel NH, Kreitman M. Evidence for stabilizing selection in a eukaryotic enhancer element. Nature. 2000;403:564–7. doi: 10.1038/35000615. [DOI] [PubMed] [Google Scholar]
- 53.Frith MC, Ponjavic J, Fredman D, Kai C, Kawai J, Carninci P, et al. Evolutionary turnover of mammalian transcription start sites. Genome Res. 2006;16:713–22. doi: 10.1101/gr.5031006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Babushok DV, Ostertag EM, Courtney CE, Choi JM, Kazazian HH., Jr L1 integration in a transgenic mouse model. Genome Res. 2006;16:240–50. doi: 10.1101/gr.4571606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lemeunier F, Ashburner MA. Relationships within the melanogaster species subgroup of the genus Drosophila (Sophophora). II. Phylogenetic relationships between six species based upon polytene chromosome banding sequences. Proc R Soc Lond B Biol Sci. 1976;193:275–94. doi: 10.1098/rspb.1976.0046. [DOI] [PubMed] [Google Scholar]
- 56.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–80. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rubin GM, Spradling AC. Genetic transformation of Drosophila with transposable element vectors. Science. 1982;218:348–53. doi: 10.1126/science.6289436. [DOI] [PubMed] [Google Scholar]
- 58.Li W-H. Molecular Evolution. Sunderland, MA: Sinauer Associates, Inc; 1997. [Google Scholar]
- 59.Tamura K, Subramanian S, Kumar S. Temporal patterns of fruit fly (Drosophila) evolution revealed by mutation clocks. Mol Biol Evol. 2004;21:36–44. doi: 10.1093/molbev/msg236. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
BLAST 2 sequence output of the 272-bp region of D. melanogaster Pros28.1A aligned with the 5′UTR and upstream region of D. simulans Pros28.1A.
BLAST 2 sequence output of the 272-bp region of D. melanogaster Pros28.1A aligned with 5′UTR and upstream region of D. yakuba Pros28.1A.
BLAST 2 sequence output of the 272-bp region of D. melanogaster Pros28.1A aligned with 5′UTR and upstream region of D. melanogaster Pros28.1.
BLAST 2 sequence output of the 272-bp region of D. melanogaster Pros28.1A aligned with 5′UTR and upstream region of D. melanogaster Pros28.1B.
Sequence alignment of the Pros28.1A and flanking region, including the CG42322 flanking coding exons (exons 3 and 4 of the transcript encoded on the other strand) in the species where it is present and the same intron in the species where the Pros28.1A gene is not present. Grey corresponds to CG42322 exons and orthologs. Blue shows the CDS of Pros28.1A.
Shown is the nonsynonymous distance matrix between all parental and all retrogenes, created using the Pamilo-Bianchi-Li method. Divergence values between either D. melanogaster, D. simulans, D. sechellia, or D. mauritiana and D. yakuba were used to estimate the average rate of evolution of parental gene and retrogene separately (shown in bold). The average divergence between parental genes and retrogenes was estimated from all possible comparisons with the available data (shown in italics).
Panel a. shows level of green fluorescence in embryos in the control strain (w1118). Panel b. shows lack of EGFP florescence driven by the 272-bp region (246 bp upstream of the gene and 26 of 5′UTR) in the embryos of five different transformants.
RT-PCR results of Pros28.1A and Gapdh2 in 0–5 hour embryos of D. melanogaster (Besancon strain). Lane 1 shows ladder. Lanes 2–4 show RT+, RT− and PCR negative of Pros28.1A. Lanes Lane 5–7 show RT+, RT− and PCR negative of Gapdh2.