Abstract
Fibrotic tissue is characterized by an overabundance of myofibroblasts. Thus, understanding the factors that induce myofibroblast differentiation is paramount to preventing fibrotic healing. Previous studies have shown that mechanical stress derived from the integrin-mediated interaction between extracellular matrix and the cytoskeleton promotes myofibroblast differentiation. Integrin α11β1 is a collagen receptor on fibroblasts. To determine whether α11β1 can act as a mechanosensor to promote the myofibroblast phenotype, mouse embryonic fibroblasts and human corneal fibroblasts were utilized. We found that α11 mRNA and protein levels were up-regulated in mouse embryonic fibroblasts grown in attached three-dimensional collagen gels and conversely down-regulated in cells grown in floating gels. α11 up-regulation could be prevented by manually detaching the collagen gels or by cytochalasin D treatment. Furthermore, SB-431542, an inhibitor of signaling via ALK4, ALK5, and ALK7, prevented the up-regulation of α11 and the concomitant phosphorylation of Smad3 under attached conditions. In attached gels, TGF-β1 was secreted in its inactive form but surprisingly not further activated, thus not influencing α11 regulation. However, inhibition of activin A attenuated the up-regulation of α11. To determine the role of α11 in myofibroblast differentiation, human corneal fibroblasts were transfected with small interfering RNA to α11, which decreased α-smooth muscle actin expression and myofibroblast differentiation. Our data suggest that α11β1 is regulated by cell/matrix stress involving activin A and Smad3 and that α11β1 regulates myofibroblast differentiation.
Keywords: Cell/Fibroblast, Cell/Adhesion, Cell/Differentiation, Extracellular Matrix/Collagen, Extracellular Matrix/Integrin, Organisms/Mouse
Introduction
A major function of fibroblasts is to take part in wound closure during wound healing (1). During this process, fibroblasts assume a more activated contractile phenotype and differentiate into myofibroblasts under the influence of mechanical tension (2) and soluble factors, such as TGF-β1 (3). Similar activation of fibroblasts occurs under pathological conditions, such as fibrosis and in the desmoplastic reaction in the tumor stroma (4, 5). In the latter instance, the tumor fibroblasts are called cancer-associated fibroblasts. Molecularly, myofibroblasts and cancer-associated fibroblasts are characterized by the expression of α-smooth muscle actin (α-SMA)2 and a splice variant of fibronectin (FN-EDA) (1). In addition to soluble factors, cell adhesion receptors of the integrin family play important roles during myofibroblast differentiation (3, 6). Integrins are heterodimeric cell adhesion receptors composed of non-covalently associated α- and β-chains. Nine of the 24 integrin αβ heterodimers are characterized by the presence of an inserted and interactive domain in the α-chain, known as the αI-domain, which is present in the four collagen receptors, α1β1, α2β1, α10β1, and α11β1 (7). We have recently determined that the integrin α11β1 is the major collagen receptor on cultured mouse embryonic fibroblasts (MEFs) and that it is uniquely needed during tooth eruption on periodontal ligament fibroblasts in vivo (8). In an experimental model for lung cancer, recent data suggest that α11β1 acts in tumor stroma fibroblasts by regulating the autocrine secretion of IGF-II (9).
Integrins have previously been shown to play a role in TGF-β activation. The groundbreaking work in the 1990s demonstrating that the RGD-binding αv integrins play an important role in the activation of TGF-β resulted in a new paradigm for integrins as master effectors of TGF-β activation (10). In this context, the αvβ6 integrin appears to be central in creating a conformational change in the latency-associated protein complex, leading to activation of TGF-β. The αvβ8 integrin can activate the complex by inducing the matrix metalloproteinase MMP-14, which in turn cleaves latency-associated protein to activate TGF-β (11). Generation of mice with a mutation of the RGD sequence in latency-associated protein to RGE results in a phenotype reminiscent of TGF-β1-deficient animals (12). More recently, this phenotype has also been mimicked by functionally inactivating αvβ6 and αvβ8 (13). Myofibroblasts have been shown to activate TGF-β by contracting the extracellular matrix (ECM) and the latency-associated protein complex as part of a mechanism suggested to restrict myofibroblast differentiation to stiffened matrices (3). In these experiments, αvβ5 has been shown to play a major role, but a role for β1 integrins has also been indicated. When bioavailable, TGF-β can initiate different signaling pathways involving the canonical Smad-dependent pathway with Smad phosphorylation and nuclear transport of Smads leading to transcriptional activation of fibrosis-related genes (14). A recent paper by Liu et al. (15) convincingly demonstrates the central role of fibroblast β1 integrins in fibrosis. Relatively little is known about the role of integrins in cancer-associated fibroblasts in the tumor stroma except for the data suggesting that matrix stiffness might regulate tumor growth (16, 17), in turn indicating that fibroblast-driven reorganization of the tumor stroma might be an important factor regulating tumorigenesis.
Two of the collagen-binding integrins, α1β1 (18) and α2β1 (19), influence myofibroblast differentiation under some conditions in vitro, whereas the role of α11β1 in this process is unknown. To better understand α11 expression and function in fibroblasts within a three-dimensional microenvironment, we have used as a model MEFs immortalized with the T large antigen of the SV40 virus (SV40 MEFs) and primary MEFs. Our findings demonstrate that α11β1 is regulated by a mechanical strain-driven mechanism that involves activin A. Upon finding that α-SMA and α11 are similarly regulated, we could furthermore establish that α11 is involved in collagen I-mediated myofibroblast differentiation in human corneal fibroblasts (HCFs), suggesting that α11β1 might be an important effector of collagen reorganization mediated by myofibroblasts.
EXPERIMENTAL PROCEDURES
Antibodies, Reagents, and Growth Factors
Rabbit polyclonal antisera to mouse and human α11 have been described previously (20). The polyclonal rabbit antibody to mouse integrin α2 was used as described (21). Monoclonal antibodies to α-tubulin, α-smooth muscle actin (α-SMA), and β-actin, clones DM1A, 1A4, and AC15 respectively, were purchased from Sigma. Monoclonal antibody to TGF-β1, β2, and β3 (clone 1D11) was acquired from R&D Systems (Oxon, UK). The monoclonal antibody to activin A (clone 69403) was purchased from R&D Systems. Rabbit polyclonal antiserum to phospho-Smad2 (22) was kindly donated by Aris Moustakas (Ludwig Institute for Cancer Research, Uppsala, Sweden). Rabbit polyclonal antiserum to phospho-Smad3 (pSpS423/425) was purchased from Acris (Hiddenhausen, Germany). Both horseradish peroxidase-conjugated secondary antibodies, goat anti-rabbit and goat anti-mouse IgGs, were obtained from Santa Cruz Biotechnology (Heidelberg, Germany). Goat anti-mouse and goat anti-rabbit Cy2- and Cy3-conjugated IgGs (multiple labeling grade) were obtained from Jackson ImmunoResearch Laboratories (Fornebu, Norway). Cytochalasin-D and SB-431542 were both purchased from Sigma. Recombinant mouse follistatin (FS288), recombinant human activin A, and recombinant FGF-2 (FGF-basic 157 amino acids) were bought from R&D Systems. Recombinant human TGF-β1 was bought from PeproTech (London, UK).
Cell Culture
To generate SV40 MEFs, primary mouse embryonic fibroblasts were isolated as described previously (8) and infected at +37 °C for 2 h with recombinant retroviruses containing the SV40 T large antigen (23) as described earlier (24). Cells were cultured in DMEM (Invitrogen) supplemented with 10% fetal calf serum (FBS) (PAA Laboratories, Pasching, Austria) and 1% penicillin and streptomycin (Pest) (Sigma). Corneal fibroblasts from adult human corneas were collected with the approval of the Ethical Committee of the Medical Faculty (Umeå University, Sweden), following informed consent and in accordance with the tenets of the Declaration of Helsinki of 1975. Small pieces of corneal stroma were cut from a corneal button and initially placed in cell culture flasks in 1:1 DMEM/Ham's F-12 medium (Invitrogen) supplemented with 10% FBS and 72 μg/ml benzylpenicillin. Upon cell growth, culture medium was replaced with standard DMEM, 10% FBS, 1% Pest, and cells were expanded and frozen at −150 °C at passage 3, with a splitting ratio of 1:3. Smad3−/− dermal fibroblasts and control wild type cells were kindly provided by K. Flanders (NCI, National Institutes of Health, Bethesda, MD) and were isolated as described previously (25). Dermal fibroblasts were isolated as described previously (26). Immortalization was achieved by ∼10 passages at the permissive temperature for large T expression (33 °C) in DMEM (Sigma) supplemented with 10% FBS and 20 units/ml interferon γ (Sigma) (27).
Collagen Type I Gels
Collagen I gels were mixed as previously described with some modifications (28). Briefly, each ml of collagen I solution contained 500 μl of 2× DMEM with 5 × 105 cells, 10 μl of 200 mm glutamine (MedProbe AS, St. Hanshaugen, Norway), antibiotics, 100 μl of 0.2 m HEPES (Sigma), pH 8.0, and 400 μl of 3 mg/ml collagen type I (PureCol, bovine skin collagen I, 3.0 mg/ml stock solution, Nutacon BV, Leimuiden, Netherlands). 450 μl of this mixture were added into each well of a 24-well plate (Nunc, Roskilde, Denmark). Gels attached to the wall and bottom of the wells during polymerization (∼90 min). Cells remained within these attached gels and cultured at 37 °C in the presence of 5% CO2 until collected. For floating conditions, lattices were either formed in wells previously coated overnight with 5% bovine serum albumin (Roche Applied Science) in sterile phosphate-buffered solution (PBS) (Invitrogen) or manually detached with a spatula after gels had polymerized and attached to the wall and bottom of uncoated wells. Once gels had formed, DMEM supplemented with 1% Pest and 10% FBS was applied on top of each gel. Cells were extracted from lattices at each time point by digesting gels with collagenase I (400 units/ml, CLS I, Worthington) at 37 °C for ∼15 min. Subsequently, cold PBS containing protease inhibitor (complete mini-EDTA-free, Roche Applied Science) was added, and cells were centrifuged. Upon discarding supernatants, pellets were further lysed in SDS-sample buffer without reducing reagent (Bio-Rad) and subjected to Western blotting.
Western Blotting
Cells cultured in monolayer were washed three times with PBS (Invitrogen) and trypsinized. They were further centrifuged, and after discarding supernatants, the resulting pellets were lysed in SDS-sample buffer without reducing agent (Bio-Rad), sonicated, and subjected to a SDS-PAGE on 6% gels. Separated proteins were transferred (100 V, 90 min) onto nitrocellulose membranes (GE Healthcare). Membranes were blocked for 1 h at room temperature with 5% nonfat dry milk (Marvel, UK) in Tris-buffered solution containing 0.05% Tween 20 (TBS-T) (Sigma), incubated with the primary antibodies to α11 or α2 (both diluted 1:500), α-SMA (1:1000), α-tubulin (1:2000), Psmad2 (1:1000), Psmad3 (1:1000), or β-actin (1:5000), in TBS-T, 1% bovine serum albumin overnight at +4 °C. Upon washing in TBS-T three times for 10 min, membranes were further incubated with goat anti-mouse and goat anti-rabbit horseradish peroxidase-conjugated secondary IgGs (both diluted 1:5000, 50 min at room temperature). Membranes were developed using the ECL Western blotting detection kit (GE Healthcare) and photographed using the ChemiDoc XRS device and the Quantity One 1-D analysis software (Bio-Rad). Each result shown by Western blot was repeated in independent assays a minimum of two times. Band intensities were quantified with Image J software (available on the National Institutes of Health Web site).
Immunofluorescence and Bright Field Microscopy
For bright field microscopy, 1.5 × 104 SV40 MEFs were seeded on 12-well plates (Nunc) and treated with Cytochalasin-D (Sigma) for 72 h. For immunofluorescence studies and except for Fig. 8D (where 8 × 103 cells were seeded), 1.5 × 104 cells were grown on 100 μg/ml collagen I-coated coverslips overnight and fixed the next day with methanol for 5 min at −20 °C. Following rehydration in PBS, cells were further permeabilized with 0.2% Triton X-100 (Sigma) in PBS for 15 min and blocked with 10% goat serum in PBS containing 0.05% Tween 20 (PBS-T) for 45 min at 37 °C. Fixed cells were next incubated with monoclonal mouse anti-α-SMA and polyclonal rabbit antisera to mouse or human α11 (diluted 1:400 and 1:1000, respectively) for 45 min at 37 °C, washed three times for 10 min with PBS-T, and incubated in goat anti-mouse Cy2- and goat anti-rabbit Cy3-conjugated secondary IgGs (diluted 1:50 and 1:500, respectively) for 45 min at 37 °C. Cells were visualized under a Zeiss Axioscope fluorescence microscope (Oslo, Norway), and pictures were acquired with a digital AxioCam MRm camera. Corneal fibroblasts were processed in the same way except that, when needed, they were treated with 5 ng/ml TGF-β or 2 nm activin A along with α11-specific siRNA or non-targeting mock siRNA at 100 nm for 3 days prior to fixation (see siRNA transfection procedure below).
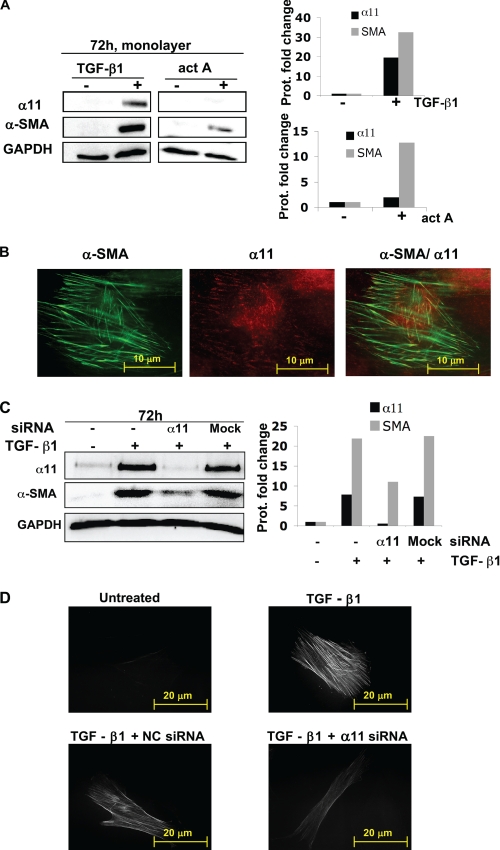
FIGURE 8.
α11 influences myofibroblast differentiation in human corneal fibroblasts. A, α11 and α-SMA protein levels in corneal fibroblasts that remained unstimulated (−) or were treated (+) with 5 ng/ml of TGF-β1 or 2 nm activin A (act A) for 3 days. B, Immunolocalization of α11 (red) at focal adhesions and α-SMA (green) at stress fibers in corneal fibroblasts treated with 5 ng/ml TGF-β1 for 3 days. C, α11 and α-SMA protein levels in corneal fibroblasts that remained unstimulated (−) or were treated (+) for 3 days with 5 ng/ml TGF-β1 and siRNA (100 nm) to α11 or an off-target siRNA (mock) as a control. D, immunolocalization of α-SMA in corneal fibroblasts that remained untreated (upper left), were stimulated with 5 ng/ml TGF-β1 only (upper right), or were treated with 5 ng/ml TGF-β1 and siRNA (100 nm) to α11 (lower right) or an off-target siRNA (negative control (NC); lower left). The exposure time when acquiring pictures was identical in all conditions. In A and C, band intensities were quantified, normalized to β-actin, and calibrated to the normalized value corresponding to untreated cells. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Enzyme-linked Immunosorbent Assay
The enzyme-linked immunosorbent assay kit Quantikine® (R&D Systems) was used according to the manufacturer's instructions to detect relative concentrations of mouse TGF-β1 present in conditioned media from SV40 MEF-populated attached collagen gels at either 24 or 96 h after casting the gels. Bars in the graph (see Fig. 5B) show the average value of three independent experiments, whereas the error bars represent the S.D. for the corresponding average value.
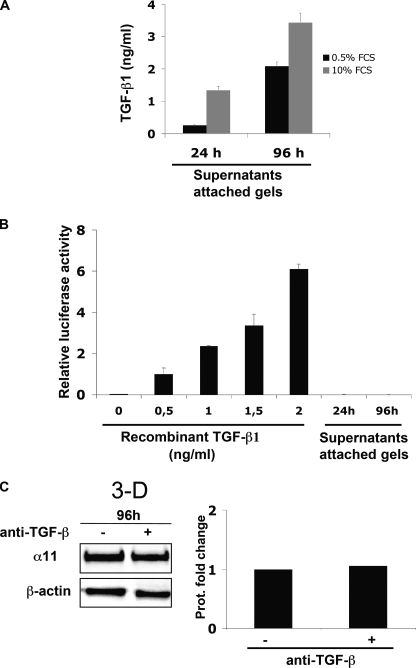
FIGURE 5.
TGF-β1 is secreted but not activated by SV40 MEFs inside a collagen gel. A, amount of TGF-β in supernatants from attached collagen lattices containing SV40 MEFs determined by ELISA at the indicated time points. The analysis was performed in the presence of two different concentrations of serum. The bars show the average value for three independent experiments, whereas the error bars represent the S.D. for the corresponding average value. B, luciferase activity normalized to β-galactosidase activity in transiently transfected SV40 MEFs (see “Experimental Procedures”) cultured with supernatants similar to those used in A. Recombinant active TGF-β1 exogenously added was used as a control. Luciferase activities were calibrated to the normalized value for cells stimulated with 0.5 ng/ml recombinant active TGF-β1 exogenously added. C, α11 protein levels in SV40 MEFs seeded in attached collagen gels in the presence or absence of a 10 μg/ml concentration of a function-blocking antibody to TGF-β. Band intensities were quantified, normalized to β-actin, and calibrated to the normalized value corresponding to the sample obtained from cells without anti-TGF-β added.
TGF-β Activity (Luciferase Assay)
2 × 105 SV40-immortalized MEFs were seeded on each well of a 12-well plate and cultured overnight in DMEM, 10% FBS, 1% Pest at 37 °C, 5% CO2. The next day, cells were co-transfected with 1 μg of a plasmid containing the TGF-β-responsive element linked to the luciferase reporter gene p(GACA)12-lux (29), and 10 ng of pCG-β-gal was used as an internal control (provided by C. Svensson (Uppsala, Sweden)), using FuGene transfection reagent (Roche R&D, Oslo Pharma) for 3 h according to the manufacturer's instructions. The medium containing plasmids and transfection reagent was then removed and replaced by fresh DMEM, 10% FBS, 1% Pest, and either the corresponding amounts of recombinant TGF-β1 or conditioned medium from SV40 immortalized MEFs seeded in attached gels were added. After 24 h, luciferase and β-gal activities of duplicate samples for each condition were measured in two independent experiments as reported previously (30). Transfected cells that survived in selection medium (29) for 14 days were considered as stably transfected, further grown under selection medium, and seeded within collagen gels (supplemental Fig. S1). Cells were extracted from collagen gels in the same manner as the untransfected cells until the step where cell pellets were obtained (see “Collagen Type I Gels”). From this step, cells were processed as cells cultured in monolayer and subjected to the luciferase assay (30).
siRNA Transfection
Upon thawing and overnight culture of low passage primary human corneal fibroblasts, cells were seeded on a 12-well plate (1.0 × 105 cells/well) and left in culture until the next day. 6 μl of Hi Perfect transfection reagent (Qiagen, Oslo, Norway) together with 100 nm ON-TARGET plus siRNA to human α11 (J-008000–10; target sequence, GGACUCAGACGGUAGCAUU; Dharmacon (Northumberland, UK)) or 100 nm non-targeting mock siRNA (D-001810-02-05; Dharmacon) were mixed and incubated with plain DMEM for ∼20 min at room temperature before adding the siRNA mixtures and 5 ng/ml TGF-β1 into the corresponding wells with cultured cells and incubating for 3 days. Corneal fibroblasts were then collected for Western blotting or for immunofluorescence analysis as described above.
Quantitative PCR (qPCR)
Total RNA was isolated from α11+/+ SV40-immortalized MEFs using the RNeasy minikit (Qiagen) according to the manufacturer's instructions. cDNA was generated from 1 μg of total RNA using murine leukemia virus reverse transcriptase (Fermentas, Helsingborg, Sweden) and oligo(dT)18 primer (Thermo Scientific, Oslo, Norway). For each sample, 100 ng of amplified cDNA were used in triplicates as template in the PCRs using iQ SYBR Green Supermix reagent according to the manufacturer's instructions (Bio-Rad). The qPCRs were performed in a LightCycler® 480 Instrument II (Roche Applied Science). The mRNA expression for each gene was measured using the LightCycler® 480 software version 1.5 (Roche Applied Science). The -fold change in mRNA expression for each gene was calculated using the 2−ΔΔCt method (31). mRNA quantifications of each target gene under each condition were normalized to glyceraldehyde-3-phosphate dehydrogenase. Quantifications of mRNA levels for each target gene in control cells seeded in monolayer were used for calibration/reference. The primers and annealing temperatures used are listed in supplemental Table S1.
RESULTS
α11 Integrin Levels in Mouse Embryonic Fibroblasts Cultured in Floating Versus Attached Collagen Lattices
We have previously shown that α11β1 is the major collagen receptor on MEFs cultured in vitro (8). Because fibroblasts are present in a three-dimensional collagen-rich environment in vivo, and culture within a three-dimensional matrix has been shown to influence the levels of another major collagen receptor, α2β1 (32), we wanted to determine whether α11 expression and function was affected by culturing MEFs inside attached or floating collagen gels. A remarkable difference between these two conditions is the freedom of cells to contract the diameter of floating but not attached gels. Both conditions (described under “Experimental Procedures”) were used to address whether matrix stiffness has an influence on the expression of the α11 integrin subunit in SV40 MEFs.
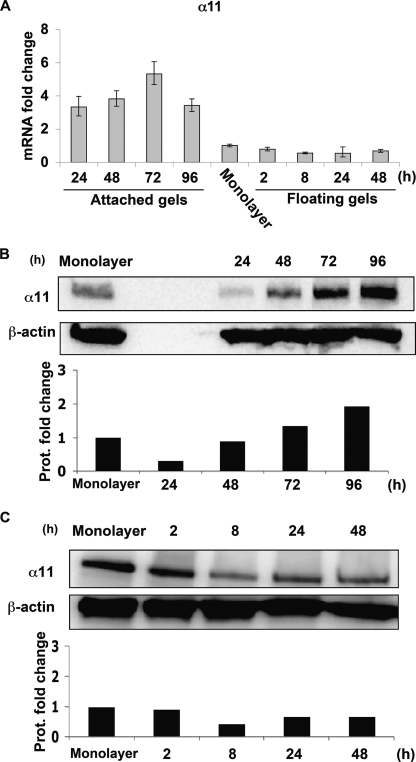
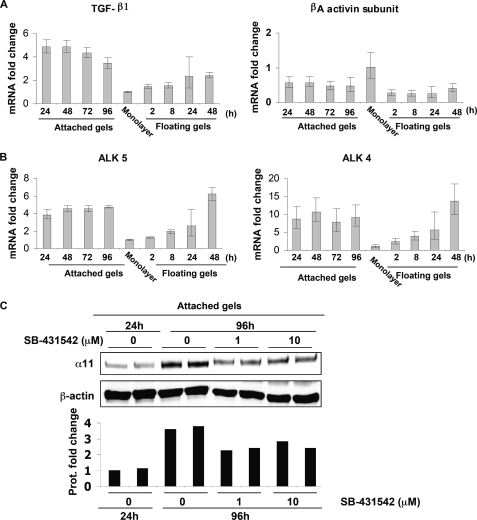
In attached gels, these cells up-regulated α11 RNA and protein levels in a time-dependent manner (Fig. 1, A and B, respectively). The lack of specific reactivity of preimmune serum is shown in supplemental Fig. S2. Whereas RNA levels increased in attached gels to peak at 72 h (Fig. 1A), protein levels at 24 h dropped when compared with levels in monolayers, to later increase at 72 and 96 h. In contrast, within floating lattices, SV40 MEFs down-regulated α11 transcript and protein levels already at 8 h (Fig. 1, A and C, respectively). Later time points were not considered in floating gels because α11 RNA levels did not decrease beyond 48 h (data not shown). These data suggest that the mechanical properties of collagen lattices regulate the expression of α11.
FIGURE 1.
α11 levels are dynamically regulated inside collagen lattices. A, quantifications of α11 mRNA levels in wild type SV40 MEFs seeded in attached and floating collagen gels at different time points. mRNA quantifications under each condition were normalized to glyceraldehyde-3-phosphate dehydrogenase. Quantifications of mRNA levels in control cells (seeded in monolayer) were used for calibration/reference. The error bars represent asymmetric S.D. values for the respective conditions and were calculated as previously described (31). The specific values for each error bar are provided in supplemental Table S2. B, α11 protein levels in SV40 MEFs seeded in attached collagen gels at different time points. C, α11 protein levels in SV40 MEFs seeded in floating collagen gels at different time points. Band intensities in B and C were quantified, normalized to β-actin, and calibrated to the normalized value for cells in monolayer.
An Attached Collagen Lattice and an Intact Actin Cytoskeleton Are Both Necessary for Increased Expression of α11
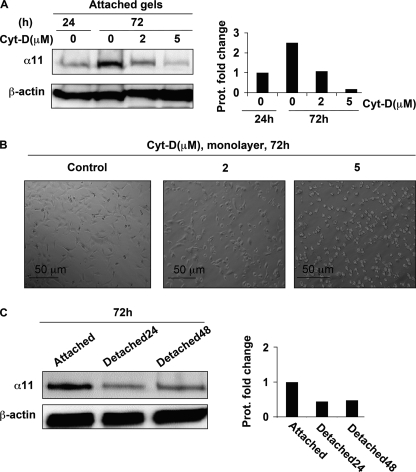
Because cells develop mechanical stress with time when cultured within attached gels (33), we hypothesized that mechanical tension could be involved in regulating the expression of α11. Consistent with our theory, the up-regulation of α11 could be prevented in a dose-dependent manner when cytochalasin D, an actin cytoskeleton-disrupting drug, was added for up to 72 h (Fig. 2A). Despite a change in morphology, cells treated with cytochalasin D on two-dimensional substrates did not detach from the plates or show any signs of toxic effects (Fig. 2B). In addition, α11 protein levels were measured in cells cultured in collagen gels that either remained attached at all times or were manually detached (thus becoming floating) 24 or 48 h before collecting cells for protein levels analyses. In these three different situations, SV40 MEFs remained for a total of 72 h within collagen lattices, but only cells cultured in gels that remained attached at all times were able to express relatively high levels of α11 (Fig. 2C). These data suggest that both an intact actin cytoskeleton and a continuously attached collagen lattice are needed for high expression of α11 in three-dimensional collagen gels.
FIGURE 2.
α11 levels inside attached collagen lattices depend on an intact actin cytoskeleton. A, effect of cytochalasin D at different concentrations on α11 protein levels in SV40 MEFs seeded in attached collagen lattices at 24 and 72 h. B, SV40 MEFs seeded in monolayer and treated with different concentrations of cytochalasin D (Cyt-D) for 72 h. C, α11 protein levels at 72 h in SV40 MEFs seeded in collagen lattices that were attached at all times (attached) or manually detached at 24 or 48 h in order to obtain floating conditions (Detached24 and Detached48, respectively). Band intensities in Western blots were quantified, normalized to β-actin, and calibrated to the normalized value corresponding to untreated cells at 24 h (A) or cells populating permanently attached gels (C).
The Up-regulation of α11 in Mouse Embryonic Fibroblasts Cultured in Attached Collagen Gels Involves the TGF-β/Activin/Nodal Signaling Pathway
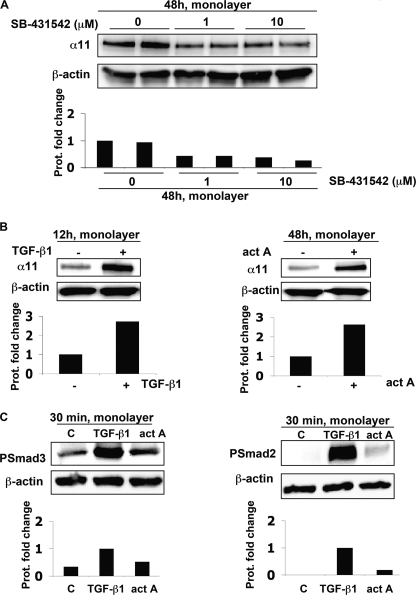
Given that a number of cell types transcriptionally up-regulate members of the TGF-β superfamily under attached conditions (34, 35) and that TGF-β has previously been reported to regulate other integrins, such as α2β1 (36), we hypothesized that TGF-β1 could be involved in the regulation of α11 in cells cultured in stressed gels. qPCR showed increased mRNA levels of TGF-β1 in cells cultured in attached lattices compared with cells in floating lattices, whereas βA activin levels were lower inside floating and attached gels compared with cells cultured on a monolayer (Fig. 3A). The mRNA levels of the corresponding activin-like receptors, ALK5 and ALK4, increased quickly in attached gels, whereas in floating gels, there was a delay in their increased expression (Fig. 3B). In addition, attached lattices populated by SV40 MEFs were treated with SB-431542, an inhibitor for signaling triggered by ALK4, ALK5, and ALK7 (37), which bind activin A, TGF-β1, and nodal, respectively (35). The addition of SB-431542 attenuated the up-regulation of α11 in SV40 MEFs cultured within attached gels (Fig. 3C), thus suggesting the involvement of at least one of the activin-like receptors in α11 up-regulation. Moreover, treatment with SB-431542 for 48 h also down-regulated the expression of α11 in SV40-MEFs seeded on planar two-dimensional substrates and in the absence of serum (Fig. 4A). Given (a) the inhibition produced by SB-431542 (Fig. 3C), (b) that we failed to detect any regulation of α11 upon treating cells in monolayer with nodal for different time periods, and (c) that we could barely detect mRNA levels for this ligand in SV40 MEFs within attached lattices (data not shown), we focused our interest on TGF-β1 and activin A as the potential ligands responsible for the up-regulation of α11 in attached collagen gels. The exogenous addition of either of these factors up-regulated the expression of α11 in SV40 MEFs seeded in monolayer, although with different kinetics (Fig. 4B), suggesting that ALK4 and ALK5 are functional and can mediate signaling affecting α11 levels in the presence of their corresponding bioactive ligands. Furthermore, phosphorylation of Smad2 and Smad3 was induced 30 min after the addition of TGF-β1 or activin A. However, TGF-β1 produced a more prominent phosphorylation of both Smads than activin A in these cells (Fig. 4C). Thus, considering these results and those obtained through qPCR (Fig. 3, A and B), TGF-β1 was at this point our main candidate potentially responsible for the up-regulation of the α11 integrin subunit in cells within attached collagen gels via Smad signaling. Because SB-431542 attenuated α11 expression in cells on a two-dimensional collagen substrate under serum-free conditions (Fig. 4A), it appears that the expression of α11 depends on an autocrine loop involving the TGF-β superfamily.
FIGURE 3.
Involvement of the TGF-β superfamily in the regulation of α11 in attached collagen gels. A and B, quantifications of the mRNA levels for TGF-β superfamily ligands TGF-β1 and βA activin (A) and their respective receptors, ALK5 and ALK4 (B), at different time points in SV40 MEFs seeded in attached or floating collagen gels. mRNA quantifications for each target gene under each condition were normalized to glyceraldehyde-3-phosphate dehydrogenase. Quantifications of mRNA levels for each target gene in control cells (seeded in monolayer) were used for calibration/reference. The error bars represent asymmetric S.D. values for the respective conditions and were calculated as described previously (31). The specific value for each error bar is provided in supplemental Fig. S2. C, effect of the indicated concentrations of SB-431542 on α11 protein levels in SV40 MEFs seeded in attached gels at 96 h. Band intensities were quantified, normalized to β-actin, and calibrated to the normalized value corresponding to untreated cells at 24 h.
FIGURE 4.
α11 levels in SV40 MEFs in monolayer are regulated by SB-431542, TGF-β1, and activin A. A, effect of the indicated concentrations of SB-431542 on α11 protein levels in SV40 MEFs seeded in monolayer at 48 h under serum free conditions. B, α11 protein levels in SV40 MEFs seeded in monolayer were serum-starved when stimulated (+) for the indicated time with 5 ng/ml of TGF-β1 or activin A (act A; 2 nm). C, phospho-Smad2 (PSmad2) and phospho-Smad3 (PSmad3) protein levels in SV40 MEFs seeded in monolayer and stimulated (+) for 30 min with 5 ng/ml TGF-β1 or activin A (act A; 2 nm). In A and B band intensities were quantified, normalized to β-actin, and calibrated to the normalized value for untreated cells. In C, the calibration was relative to the normalized band intensities corresponding to cells treated with TGF-β1.
TGF-β1 Is Not Involved in the Up-regulation of α11 in Mouse Embryonic Fibroblasts within Attached Lattices
Although TGF-β1 is detected extracellularly in its inactive form, this cytokine needs to be activated in order to be able to bind its receptor, ALK5, and trigger downstream events. Such activation can occur in several ways (6). Once we confirmed by ELISA that, consistent with the RNA data (Fig. 3A), SV40 MEFs secreted TGF-β1 in the range of 1–3 ng/ml when cultured in attached collagen gels (Fig. 5A), we proceeded to verify the activation of TGF-β1 by these cells. Supernatants from cells cultured in attached gels for 24 or 96 h were added to SV40 MEFs in monolayer that had been transiently transfected with a vector containing a TGF-β-responsive element linked to the firefly luciferase reporter gene (29). Given that (a) α11 was induced when we added recombinant active TGF-β to SV40 MEFs in monolayer (Fig. 4B) and that (b) these cells secrete relatively high amounts of TGF-β1 when grown in attached gels (Fig. 5A), we were expecting to detect luciferase activity when SV40 MEFs transfected with the TGF-β-responsive element were exposed to conditioned media from attached gels. Detecting luciferase activity in this scenario would have indicated that the TGF-β1 secreted by SV40 MEFs within attached gels had been activated and therefore could potentially be responsible for the up-regulation of α11 in attached gels. Surprisingly, no luciferase activity was detected in this case (Fig. 5B), which suggests that the amount of TGF-β1 secreted by these cells was not further activated. We added active recombinant TGF-β1 to the same transfected cells as a positive control for the luciferase assay (Fig. 5B). In order to exclude local activation of TGF-β that may have never reached the supernatants used to measure luciferase activity, the reporter plasmid was stably transfected into cells that were further seeded within collagen lattices, but no activity was observed in this instance either (supplemental Fig. S1). Importantly, exogenously added activin A did not activate the reporter plasmid (supplemental Fig. S1). In addition, a pan-TGF-β function-blocking antibody failed to block the up-regulation of α11 when added to cells in attached gels (Fig. 5C), whereas it efficiently blocked the up-regulation of α11 induced by recombinant active TGF-β1 that had been exogenously added (supplemental Fig. S3). These data suggest that the TGF-β1 secreted by SV40 MEFs within the attached gels is not further activated and, therefore, is not involved in the up-regulation of α11 in these cells and under these conditions.
Activin A Is Involved in the Up-regulation of α11 in Mouse Embryonic Fibroblasts within Attached Lattices
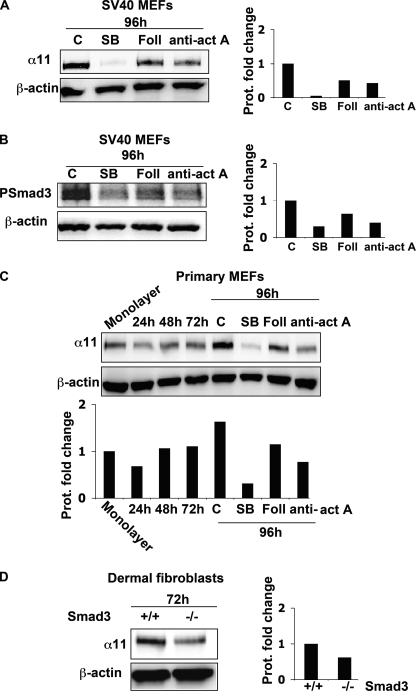
Given that (a) SB-431542 inhibited the up-regulation of α11 in the attached gels (Fig. 3C), (b) exogenously added recombinant activin A induced α11 in monolayer (Fig. 4B), and (c) TGF-β was not involved in the regulation of α11 in SV40 MEFs (Fig. 5, B and C, and supplemental Fig. S1), we proceeded to address whether activin A was taking part in up-regulating α11 levels in collagen gels in these cells. In attached gels, cells were treated with follistatin, a molecule that binds activin A and neutralizes its bioactivity (38), or with a function-blocking antibody to activin A. In both instances, the up-regulation of α11 was attenuated in SV40 MEFs (Fig. 6A). Moreover, Smad3 phosphorylation was also partially inhibited in these cells following follistatin or anti-activin A treatment (Fig. 6B). To ascertain that the identified mechanism was not restricted to SV40 immortalized MEFs, we confirmed that α11 up-regulation in an attached three-dimensional gel also involved activin A in primary MEFs (Fig. 6C). Consistent with a role for Smad3 in mediating the up-regulation of α11, Smad3−/− dermal fibroblasts seeded in attached collagen gels for 72 h showed decreased α11 protein levels compared with wild type dermal fibroblasts (Fig. 6D). Because we failed to detect phospho-Smad2 levels under these conditions (data not shown), and the levels of phospho-Smad2 were clearly detectable only when cells were treated with recombinant TGF-β1 (Fig. 4C), it appears that, in the attached gels, the regulation of α11 in SV40 MEFs involves activin A and Smad3 as the principal receptor-associated Smad mediating this event.
FIGURE 6.
Activin A regulates α11 and Smad3 levels in MEFs cultured in attached collagen gels. A, α11 levels in SV40 MEFs seeded in attached collagen gels in the presence or absence of SB-431542 (SB; 10 μm), follistatin (Foll; 5 nm), or activin A function blocking antibody (anti-act A; 25 μg/ml). B, phospho-Smad3 (PSmad3) levels in SV40 MEFs seeded in attached collagen gels in the presence or absence of SB-431542 (10 μm), follistatin (5 nm), or activin A function-blocking antibody (25 μg/ml). C, α11 levels in primary MEFs seeded in attached collagen gels in the presence or absence of SB-431542 (10 μm), follistatin (5 nm), or activin A function-blocking antibody (25 μg/ml). D, α11 protein levels in wild type or Smad3−/− dermal fibroblasts seeded in attached lattices at the indicated time point. Band intensities were quantified and normalized to β-actin. Normalized bands were calibrated to the value corresponding to untreated cells (A and B), cells in monolayer (C), or Smad3+/+ cells (D).
α11 Is Part of the Myofibroblast Phenotype of Mouse Fibroblasts
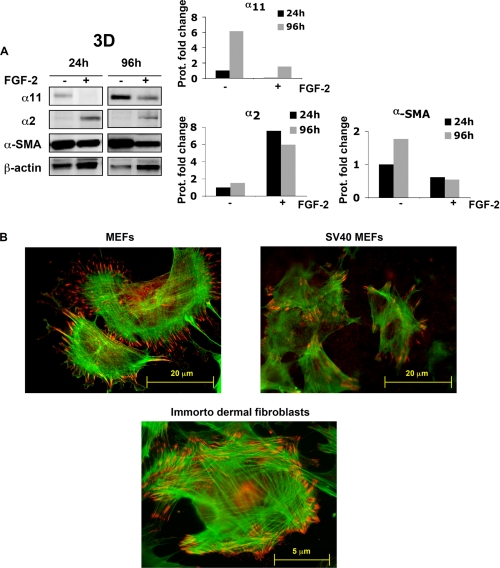
In addition to up-regulating α11, both activin A (39, 40) and mechanical tension (41) have been shown to be involved in the up-regulation of α- smooth muscle actin (α-SMA). This isoform of actin is a classical marker for myofibroblasts, a cell type playing a key role in wound healing (1) and pathological conditions, such as fibrosis (1) and cancer (42). Moreover, both α-SMA (43) and α11 (44) are also down-regulated by FGF-2 in some cell types. α-SMA and α11 were also concomitantly regulated by mechanical tension and FGF-2 in SV40 MEFs within attached gels, whereas α2 levels were increased by FGF-2 treatment (Fig. 7A). Thus, since α11 and α-SMA appeared similarly regulated, we hypothesized that α11 could be increased in myofibroblasts. Double immunofluorescence microscopy performed on primary and SV40 MEFs seeded on collagen I-coated surfaces allowed visualization of cells expressing α-SMA stress fibers terminating at α11-expressing focal adhesions (Fig. 7B). These data demonstrate that α11 is expressed and is functional in myofibroblasts. In addition, immunofluorescent labeling of primary MEFS and immortalized dermal fibroblasts (see “Experimental Procedures”) seeded in monolayer demonstrates that α11 is not only expressed in myofibroblasts derived from SV40 MEFs (Fig. 7B).
FIGURE 7.
α11 levels and regulation correlate with a myofibroblastic phenotype. A, α11, α2, and α-SMA protein levels in SV40 MEFs in attached gels in the presence (+) or absence (−) of 20 ng/ml FGF-2 at the indicated time points. Band intensities were quantified, normalized to β-actin, and calibrated to the normalized value corresponding to untreated cells at 24 h. B, immunolocalization of α11 (red) at focal adhesions and α-SMA (green) in stress fibers (red) in primary MEFs, SV40 MEFs, and dermal fibroblasts isolated from an immortalized mouse (see “Experimental Procedures”).
α11 Is Necessary for Myofibroblast Differentiation in Corneal Fibroblasts
Following the observation that the up-regulation of α11 in MEFs was linked to the myofibroblast phenotype, we wondered whether α11 could affect the differentiation process itself. Because SV40 MEFs, primary MEFs, and dermal fibroblasts all already appeared to express significant amounts of α-SMA in culture at early time points (Fig. 7), we initially tried to reduce these levels. Serum starvation failed to down-regulate α-SMA, and cultures of cells in floating collagen lattices combined with FGF-2 treatment (44, 45) appeared to disturb α11 regulation (data not shown).
Thus, we instead used primary HCFs, which express relatively modest levels of α-SMA and α11 at early passages (46). Because we were unable to reproducibly retrieve viable HCFs from the three-dimensional collagen lattice at the times needed to appreciate myofibroblast differentiation, we seeded these cells on a two-dimensional collagen I-coated surface. Upon treating HCFs cells with either activin A or active TGF-β for 3 days, α-SMA and hence the differentiation of fibroblasts into myofibroblasts could be induced. Here α-SMA and α11 were also concomitantly up-regulated, as previously observed for α11 in SV40 MEFs cultured inside an attached three-dimensional collagen matrix. However, stimulation with TGF-β1 led to a much more prominent up-regulation of α-SMA and α11 in corneal fibroblasts cultured on collagen I than that caused by activin A treatment at the concentrations used (Fig. 8A). In fact, Western blot membranes containing samples corresponding to cells stimulated with activin A had to be exposed for much longer to appreciate α11 up-regulation in these cells (supplemental Fig. S4). For this reason, only TGF-β was used in further experiments. TGF-β1-stimulated α-SMA induction was reduced when siRNA to α11 was included at the beginning of the experiment (Fig. 8, C and D, and supplemental Fig. S5). These data suggest that α11β1 is necessary for TGF-β-induced myofibroblast differentiation on a collagen I substrate.
DISCUSSION
α11β1 Levels Are Regulated by Mechanical Strain
In recent years, the field of ECM biology has attracted increased interest with the realization that the ECM plays a central role in maintaining the homeostasis of the cellular microenvironment (47, 48). In addition to its traditional role as a structural framework, it has become clear that the ECM regulates a number of dynamic events involving cell adhesion, cell migration, paracrine signaling, and tissue remodeling. Cell surface receptors, including members of the integrin family and matrix proteases, are central in mediating these effects.
Fibroblasts are connective tissue cells that play a central role in pathological events, such as fibrosis and tumorigenesis (49). In addition to the well known communication between tumor and stroma cells (42), a recent study suggests that fibroblasts and fibroblast integrins are involved in generating the migration pathways for metastasizing tumor cells (50).
We have previously shown that α11β1 is the major collagen receptor on mouse embryonic fibroblasts (8). In the current study, we have used mainly SV40 MEFs as a model to examine basic mechanisms of α11β1 regulation in a three-dimensional matrix. Although both α2β1 and α11β1 have been shown to mediate matrix reorganization in a three-dimensional collagen matrix (20, 32), careful analysis of the levels of α2β1 and α11β1 reveals that during matrix reorganization, the levels of these integrins are dynamically regulated. Other data demonstrate that some integrin functions involve a certain amount of cross-talk and that a hierarchical control exists within the integrin family (51–53). Recently, α2β1 was shown to negatively regulate α1β1 integrin when present on the same cells (53). These factors need to be taken into account when studying conditions that regulate α11 levels. Although the three-dimensional environment of certain fibroblasts is enough to up-regulate α2β1 levels (32, 54), that of α11β1 seems to further require the gel being mechanically strained.
The stiffness of the ECM has been shown to influence fibroblast adhesion and migration (55). Accumulating data have shown that mechanical strain is an important factor stimulating myofibroblast differentiation (2, 56–58). The influence of matrix stiffness on cell behavior is thus important to consider when analyzing fibroblasts inside a floating or attached three-dimensional collagen lattice (59).
Previous studies have indicated that cells within an attached collagen gel up-regulate transcription of a number of genes, including those encoding collagen I, periostin, collagen XII, and tenascin-C (60–64). Our data suggest that the α11 integrin subunit should be added to this group of mechanoregulated genes.
Role of TGF-β Family Members in α11 Expression
In the case of both collagen α1(I) (63) and periostin (64), an autocrine loop involving TGF-β1 has been implied in the mechanosensitive up-regulation of protein synthesis. Regarding tenascin-C, the mechanical strain-induced synthesis occurs in a manner dependent on integrin β1 and ILK (65). The fact that SB-431542 almost completely blocked α11 up-regulation indicates that TGF-β family signaling is a major mechanism to regulate α11 levels within a three-dimensional collagen gel. Recent analysis of the human ITGA11 proximal promoter has revealed a functional Smad3-binding site, which, together with a Smad2-dependent Sp1 site, seems to mediate responsiveness of ITGA11 to TGF-β1 (66). However, in the present study, the amount of TGF-β1 secreted by SV40 MEFs in attached lattices appeared not to be activated and therefore not to be responsible for the up-regulation of α11 in these cells. It will be interesting to determine if activin A also can mediate its effects on α11 levels via these or other sites in the promoter. Binding of SRF to CArG-boxes in genes responding to mechanical strain has previously been demonstrated in fibroblasts (67, 68). The fact that an intact cytoskeleton is needed for the α11 up-regulation might indicate that such a mechanical stress-sensitive element also is present in the ITGA11 promoter. No obvious conserved canonical CArG-box is present in the 3-kb part of the promoter (30), so the involvement of CArG-boxes in the regulation of α11 is currently unclear.
Activin A Stimulates α11 Synthesis
The reason for the lack of activation of TGF-β1 secreted by SV40 MEFs might be related to deregulated signaling caused by the immortalization process (69) or the origin/species differences of the fibroblasts. We are confident that the lack of effect of antibodies to TGF-β in the collagen gel is not a general diffusion/accessibility problem because antibodies to activin A have an effect on cells in the collagen gel. Finally, although we have gone relatively far to prove that TGF-β1 is not activated in stressed gels in this case, the possibility that activation of TGF-β1 cannot be detected by the methods used here should not be discarded.
In the present study, the lack of detectable TGF-β1 activation instead allowed us to identify activin A as an autocrine factor up-regulating α11β1 and in turn stimulating myofibroblast differentiation. Interestingly, βA activin mRNA levels were highest in monolayers (the condition where mechanical tension is highest) and lower in attached gels and barely detectable in floating gels. The results suggest that activin A in these cells may be regulated by mechanical strain. Interestingly, mechanical strain effects in embryonic stem cells have been found to be mediated by activin A and TGF-β (35). In our study, follistatin and antibodies to activin A partially inhibited the up-regulation of α11 inside the three-dimensional collagen gel in SV40 MEFs and, importantly, also in primary MEFs. The fact that activin A is regulated by follistatin adds another level of control in the cytokine-integrin response to strain. Although the Smad phosphorylation in two-dimensional conditions was readily monitored upon the addition of exogenous TGF-β1 or activin A, the signal appeared low in three-dimensional gels, and only low levels of phospho-Smad3 were detected. In a previous study using HaCaT cells, it was similarly observed that activin A stimulated cells via Smad3, whereas TGF-β1 stimulated cells in a Smad2- and Smad3-dependent manner (70). We have no good explanation for the lower effectiveness of anti-activin A and follistatin in comparison with the effect of SB-431542 in regulating α11 synthesis within an attached collagen gel. It is possible that the lower effect of anti-activin A antibodies is related to affinity issues. The modest increase in βA activin mRNA as judged by qPCR data does not lend support to a model where activin A is the main driver of strain-sensitive responses. It is possible that activin A instead plays a maintenance role, and a more important mechanosensitive ligand remains to be identified.
The in vivo implications of our findings are that α11 expression might be regulated by mechanical strain-dependent mechanisms during normal tissue homeostasis, during wound healing, and in fibrosis. In vivo, the integrin subunit α11 is highly expressed in fibroblasts of ectomesenchymal origin in the head and in fibroblasts derived from different types of mesoderm (24). Consistent with the data presented here, a common finding is that the highest α11 expression is noted at sites of mechanical tension, such as the periodontal ligament, tendons, ligaments, periosteum, and perichondrium and in the intervertebral discs (20, 24). Activin A in the developing embryo has been reported to be highly expressed in the craniofacial mesenchyme as well as in perichondrium and intervertebral discs (71). TGF-β1 shows a wide expression pattern (72) but is also expressed at sites of mechanical tension. Our data suggest that a signaling mechanism involving activin A and TGF-β1 might regulate α11 expression at these sites in vivo.
Role for α11 in Myofibroblast Differentiation
Members of the TGF-β family might thus be involved in restricting the in vivo expression pattern of α11 to different sets of fibroblasts. The recent finding that α11 is also expressed in dermal fibroblasts (26, 74) together with the expression of α11 in skin myofibroblasts shown here underscores the importance of examining a role for α11β1 in myofibroblasts, which are key cells mediating wound contraction (75).
Recent reports show that cell-generated mechanical tension can activate TGF-β1 in an strain-dependent manner (3). These assays and other assays demonstrating a role for αv-integrins (76) have been performed with cells cultured in serum, thus favoring RGD-binding integrins as the receptors mediating an effect on myofibroblast differentiation. Indirect binding to collagen fibrils can also occur when cells are grown in collagen gels in the presence of serum or alternatively cultured for extended periods of time, allowing synthesis of non-collagen integrin ligands (77). Binding of αv-ligands to collagen most likely mediates the αvβ3-dependent rapid collagen remodeling that has been observed upon release of attached collagen lattices (78).
In vivo, fibroblasts are surrounded by a matrix dominated by fibrillar collagens. Hence, the role of collagen-binding integrins during myofibroblast differentiation might have high biological relevance. In support of such a role for β1 integrins in myofibroblasts, a recent study demonstrates that a lack of β1 integrins in dermal fibroblasts protects mice from bleomycin-induced skin fibrosis (15). It will be important to determine the nature of the αβ1 integrin heterodimer involved in regulating the fibrotic response in dermal myofibroblasts and determine if this β1 integrin-dependent mechanism involves a TGF-β1 activating role or whether the integrin acts as a downstream effector (79).
Due to the high endogenous expression of α-SMA in MEFs at early passages, we could not use these cells to analyze the role of α11 during myofibroblast differentiation. We have previously noted a high expression of α11 in developing mouse and human cornea (20, 24) and have determined that α11β1 is a major collagen-binding integrin on cultured HCFs of low passage number (46). We chose HCFs for functional studies of the role of α11 in myofibroblast differentiation because these cells appeared to express low α-SMA at early passages. In order to promote the cell-collagen interaction over the cell-fibronectin interactions, we opted for analysis in the three-dimensional collagen matrix, which would also have best fit our analysis in MEFs. However, due to technical problems, maybe related to poor cell survival of the primary cells in the gels, we had to resort to inducing differentiation on collagen I-coated surfaces. Our results showing that both activin A and TGF-β stimulate myofibroblast differentiation of corneal fibroblasts are in agreement with previous work (40). However, HCFs displayed a relatively weak up-regulation of α11 and in our hands showed a lower induction of α-SMA in response to exogenously added activin A when compared with the induction produced by TGF-β1.
Thus, our failure to demonstrate a role for α11 in activin A-mediated myofibroblast differentiation in HCFs might be related to the low level of myofibroblast differentiation induced by activin A under our experimental conditions. Previous studies have indicated active activin A synthesis in fibroblasts, and activin A is one secreted growth factor from fibroblast feeder cell layers that maintains embryonic stem cell pluripotency (80). In the skin, a role for autocrine and paracrine activin A signaling has been suggested in post-burn scars (81). Finally, ex vivo experiments with scar fibroblasts implicate activin A in Akt-mediated signaling in myofibroblast differentiation (81). In summary, we believe that activin A is a good candidate to regulate α11 levels in various fibroblasts in vivo.
Despite the problems regarding activin A-induced myofibroblast differentiation in the HCFs, the cells were useful to demonstrate a role for α11β1 in TGF-β1-stimulated myofibroblast differentiation. A recent study suggests that α1β1 is important in a variety of human myofibroblasts in vivo (82), whereas a separate study suggests that α2β1 regulates myofibroblast differentiation inside three-dimensional collagen matrices (19). The studies involving α2β1 were performed prior to the existing knowledge about α11, and it will thus be important to analyze these two collagen receptors with similar techniques under similar conditions to estimate their relative contribution to the myofibroblast differentiation process. Because, in addition to being expressed on fibroblasts, α1β1 (83) and α2β1 (26) both are present on endothelial cells, it can be difficult to separate their role on myofibroblasts from their angiogenic effect during pathological processes in tissues. This is a complication that is not likely to be observed with α11, which is restricted to fibroblasts.
From the above discussion, we speculate that different collagen-binding integrins have different roles in different types of fibroblasts and that the type of collagen-binding integrin that influences myofibroblast differentiation might be tissue- and context-dependent. This is similar to the situation with αv integrins, where αvβ3 and αvβ5 have been reported to influence myofibroblast differentiation in cells isolated from the mouth and skin, whereas αvβ5 alone seems to have this property in kidney fibroblasts (77). In future studies, it will be important to investigate the role of TGF-β1 and activin A in α11 expression and function in fibroblasts/myofibroblasts of different origins (73).
In summary, we show here that α11β1 is regulated by mechanical strain and members of the TGF-β superfamily. It is tempting to suggest that the striking expression of α11 at sites of mechanical tension reflects inherent structural properties of α11β1, making it particularly suitable for consolidating cell-collagen interactions. The finding that α11β1 regulates myofibroblast differentiation warrants further studies of its role in pathological conditions involving fibroblasts.
Supplementary Material
Acknowledgments
We are grateful to Fatima Pedrosa-Domellöf and Anders Behndig (both at University of Umeå, Sweden) for providing human keratocytes.
This work was supported by European Union Marie Curie Early Stage Training Contract MEST-CT-2004-514483 (to S. C.), the Strategic Research Program at Helse Bergen (to R. J.), a bilateral German-Norwegian Deutsche Akademische Austauschdienst, Program des Projektbezogenen Personenaustausch grant from the Research Council of Norway (to D. G.), Helse Vest, Kreftforeningen Grant PR 2007-0103 (to D. G.), and Research Council of Norway Grant 172330 V40 (to D. G.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1–S4.
- α-SMA
- α-smooth muscle actin
- MEF
- mouse embryonic fibroblast
- TGF
- transforming growth factor
- HCF
- human corneal fibroblast
- FGF
- fibroblast growth factor
- DMEM
- Dulbecco's modified Eagle's medium
- Pest
- penicillin and streptomycin
- FBS
- fetal bovine serum
- PBS
- phosphate-buffered saline
- qPCR
- quantitative PCR
- ECM
- extracellular matrix
- siRNA
- small interfering RNA.
REFERENCES
- 1.Hinz B. (2007) J. Invest. Dermatol. 127, 526–537 [DOI] [PubMed] [Google Scholar]
- 2.Tomasek J. J., Gabbiani G., Hinz B., Chaponnier C., Brown R. A. (2002) Nat. Rev. Mol. Cell Biol. 3, 349–363 [DOI] [PubMed] [Google Scholar]
- 3.Wipff P. J., Rifkin D. B., Meister J. J., Hinz B. (2007) J. Cell Biol. 179, 1311–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Desmoulière A., Guyot C., Gabbiani G. (2004) Int. J. Dev. Biol. 48, 509–517 [DOI] [PubMed] [Google Scholar]
- 5.Hinz B., Dugina V., Ballestrem C., Wehrle-Haller B., Chaponnier C. (2003) Mol. Biol. Cell 14, 2508–2519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wipff P. J., Hinz B. (2008) Eur. J. Cell Biol. 87, 601–615 [DOI] [PubMed] [Google Scholar]
- 7.Popova S. N., Lundgren-Akerlund E., Wiig H., Gullberg D. (2007) Acta Physiol. 190, 179–187 [DOI] [PubMed] [Google Scholar]
- 8.Popova S. N., Barczyk M., Tiger C. F., Beertsen W., Zigrino P., Aszodi A., Miosge N., Forsberg E., Gullberg D. (2007) Mol. Cell. Biol. 27, 4306–4316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhu C. Q., Popova S. N., Brown E. R., Barsyte-Lovejoy D., Navab R., Shih W., Li M., Lu M., Jurisica I., Penn L. Z., Gullberg D., Tsao M. S. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 11754–11759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Munger J. S., Huang X., Kawakatsu H., Griffiths M. J., Dalton S. L., Wu J., Pittet J. F., Kaminski N., Garat C., Matthay M. A., Rifkin D. B., Sheppard D. (1999) Cell 96, 319–328 [DOI] [PubMed] [Google Scholar]
- 11.Mu D., Cambier S., Fjellbirkeland L., Baron J. L., Munger J. S., Kawakatsu H., Sheppard D., Broaddus V. C., Nishimura S. L. (2002) J. Cell Biol. 157, 493–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang Z., Mu Z., Dabovic B., Jurukovski V., Yu D., Sung J., Xiong X., Munger J. S. (2007) J. Cell Biol. 176, 787–793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aluwihare P., Mu Z., Zhao Z., Yu D., Weinreb P. H., Horan G. S., Violette S. M., Munger J. S. (2009) J. Cell Sci. 122, 227–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moustakas A., Pardali K., Gaal A., Heldin C. H. (2002) Immunol. Lett. 82, 85–91 [DOI] [PubMed] [Google Scholar]
- 15.Liu S., Kapoor M., Denton C. P., Abraham D. J., Leask A. (2009) Arthritis Rheum. 60, 2817–2821 [DOI] [PubMed] [Google Scholar]
- 16.Paszek M. J., Zahir N., Johnson K. R., Lakins J. N., Rozenberg G. I., Gefen A., Reinhart-King C. A., Margulies S. S., Dembo M., Boettiger D., Hammer D. A., Weaver V. M. (2005) Cancer Cell 8, 241–254 [DOI] [PubMed] [Google Scholar]
- 17.Levental K. R., Yu H., Kass L., Lakins J. N., Egeblad M., Erler J. T., Fong S. F., Csiszar K., Giaccia A., Weninger W., Yamauchi M., Gasser D. L., Weaver V. M. (2009) Cell 139, 891–906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ng C. P., Hinz B., Swartz M. A. (2005) J. Cell Sci. 118, 4731–4739 [DOI] [PubMed] [Google Scholar]
- 19.Arora P. D., Narani N., McCulloch C. A. (1999) Am. J. Pathol. 154, 871–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tiger C. F., Fougerousse F., Grundström G., Velling T., Gullberg D. (2001) Dev. Biol. 237, 116–129 [DOI] [PubMed] [Google Scholar]
- 21.Zhang Z. G., Bothe I., Hirche F., Zweers M., Gullberg D., Pfitzer G., Krieg T., Eckes B., Aumailley M. (2006) J. Cell Sci. 119, 1886–1895 [DOI] [PubMed] [Google Scholar]
- 22.Persson U., Izumi H., Souchelnytskyi S., Itoh S., Grimsby S., Engström U., Heldin C. H., Funa K., ten Dijke P. (1998) FEBS Lett. 434, 83–87 [DOI] [PubMed] [Google Scholar]
- 23.Jat P. S., Cepko C. L., Mulligan R. C., Sharp P. A. (1986) Mol. Cell. Biol. 6, 1204–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Popova S. N., Rodriguez-Sánchez B., Lidén A., Betsholtz C., Van Den Bos T., Gullberg D. (2004) Dev. Biol. 270, 427–442 [DOI] [PubMed] [Google Scholar]
- 25.Arany P. R., Flanders K. C., Kobayashi T., Kuo C. K., Stuelten C., Desai K. V., Tuan R., Rennard S. I., Roberts A. B. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 9250–9255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zweers M. C., Davidson J. M., Pozzi A., Hallinger R., Janz K., Quondamatteo F., Leutgeb B., Krieg T., Eckes B. (2007) J. Invest. Dermatol. 127, 467–478 [DOI] [PubMed] [Google Scholar]
- 27.Whitehead R. H., Joseph J. L. (1994) Epithelial Cell Biol. 3, 119–125 [PubMed] [Google Scholar]
- 28.Gullberg D., Tingström A., Thuresson A. C., Olsson L., Terracio L., Borg T. K., Rubin K. (1990) Exp. Cell Res. 186, 264–272 [DOI] [PubMed] [Google Scholar]
- 29.Abe M., Harpel J. G., Metz C. N., Nunes I., Loskutoff D. J., Rifkin D. B. (1994) Anal. Biochem. 216, 276–284 [DOI] [PubMed] [Google Scholar]
- 30.Lu N., Heuchel R., Barczyk M., Zhang W. M., Gullberg D. (2006) Matrix Biol. 25, 118–129 [DOI] [PubMed] [Google Scholar]
- 31.Livak K. J., Schmittgen T. D. (2001) Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 32.Klein C. E., Dressel D., Steinmayer T., Mauch C., Eckes B., Krieg T., Bankert R. B., Weber L. (1991) J. Cell Biol. 115, 1427–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Delvoye P., Wiliquet P., Levêque J. L., Nusgens B. V., Lapière C. M. (1991) J. Invest. Dermatol. 97, 898–902 [DOI] [PubMed] [Google Scholar]
- 34.Chiquet M., Renedo A. S., Huber F., Flück M. (2003) Matrix Biol. 22, 73–80 [DOI] [PubMed] [Google Scholar]
- 35.Saha S., Ji L., de Pablo J. J., Palecek S. P. (2008) Biophys. J. 94, 4123–4133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heino J., Ignotz R. A., Hemler M. E., Crouse C., Massagué J. (1989) J. Biol. Chem. 264, 380–388 [PubMed] [Google Scholar]
- 37.DaCosta Byfield S., Major C., Laping N. J., Roberts A. B. (2004) Mol. Pharmacol. 65, 744–752 [DOI] [PubMed] [Google Scholar]
- 38.de Winter J. P., ten Dijke P., de Vries C. J., van Achterberg T. A., Sugino H., de Waele P., Huylebroeck D., Verschueren K., van den Eijnden-van Raaij A. J. (1996) Mol. Cell. Endocrinol. 116, 105–114 [DOI] [PubMed] [Google Scholar]
- 39.Ohga E., Matsuse T., Teramoto S., Katayama H., Nagase T., Fukuchi Y., Ouchi Y. (1996) Biochem. Biophys. Res. Commun. 228, 391–396 [DOI] [PubMed] [Google Scholar]
- 40.You L., Kruse F. E. (2002) Invest. Ophthalmol. Vis. Sci. 43, 72–81 [PubMed] [Google Scholar]
- 41.Chipev C. C., Simon M. (2002) BMC Dermatol. 2, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.De Wever O., Demetter P., Mareel M., Bracke M. (2008) Int. J. Cancer 123, 2229–2238 [DOI] [PubMed] [Google Scholar]
- 43.Papetti M., Shujath J., Riley K. N., Herman I. M. (2003) Invest. Ophthalmol. Vis. Sci. 44, 4994–5005 [DOI] [PubMed] [Google Scholar]
- 44.Varas L., Ohlsson L. B., Honeth G., Olsson A., Bengtsson T., Wiberg C., Bockermann R., Järnum S., Richter J., Pennington D., Johnstone B., Lundgren-Akerlund E., Kjellman C. (2007) Stem Cells Dev. 16, 965–978 [DOI] [PubMed] [Google Scholar]
- 45.Maltseva O., Folger P., Zekaria D., Petridou S., Masur S. K. (2001) Invest. Ophthalmol. Vis. Sci. 42, 2490–2495 [PubMed] [Google Scholar]
- 46.Bystrom B., Carracedo S., Behndig A. B., Gullberg D., Pedrosa-Domellof F. (2009) Invest. Ophthalmol. Vis. Sci. 50, 5044–5053 [DOI] [PubMed] [Google Scholar]
- 47.Johnson K. R., Leight J. L., Weaver V. M. (2007) Methods Cell Biol. 83, 547–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Marastoni S., Ligresti G., Lorenzon E., Colombatti A., Mongiat M. (2008) Connect. Tissue Res. 49, 203–206 [DOI] [PubMed] [Google Scholar]
- 49.Kalluri R., Zeisberg M. (2006) Nat. Rev. Cancer 6, 392–401 [DOI] [PubMed] [Google Scholar]
- 50.Gaggioli C., Hooper S., Hidalgo-Carcedo C., Grosse R., Marshall J. F., Harrington K., Sahai E. (2007) Nat. Cell Biol. 9, 1392–1400 [DOI] [PubMed] [Google Scholar]
- 51.Schwartz M. A., Ginsberg M. H. (2002) Nat. Cell Biol. 4, E65–68 [DOI] [PubMed] [Google Scholar]
- 52.Orr A. W., Ginsberg M. H., Shattil S. J., Deckmyn H., Schwartz M. A. (2006) Mol. Biol. Cell 17, 4686–4697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Abair T. D., Sundaramoorthy M., Chen D., Heino J., Ivaska J., Hudson B. G., Sanders C. R., Pozzi A., Zent R. (2008) Exp. Cell Res. 314, 3593–3604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eckes B., Zweers M. C., Zhang Z. G., Hallinger R., Mauch C., Aumailley M., Krieg T. (2006) J. Investig. Dermatol. Symp. Proc. 11, 66–72 [DOI] [PubMed] [Google Scholar]
- 55.Pelham R. J., Jr., Wang Y. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 13661–13665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hinz B. (2009) Curr. Rheumatol. Rep. 11, 120–126 [DOI] [PubMed] [Google Scholar]
- 57.Patwari P., Lee R. T. (2008) Circ. Res. 103, 234–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wipff P. J., Hinz B. (2009) J. Body Mov. Ther. 13, 121–127 [DOI] [PubMed] [Google Scholar]
- 59.Solon J., Levental I., Sengupta K., Georges P. C., Janmey P. A. (2007) Biophys. J. 93, 4453–4461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chiquet-Ehrismann R., Tannheimer M., Koch M., Brunner A., Spring J., Martin D., Baumgartner S., Chiquet M. (1994) J. Cell Biol. 127, 2093–2101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Flück M., Giraud M. N., Tunç V., Chiquet M. (2003) Biochim. Biophys. Acta 1593, 239–248 [DOI] [PubMed] [Google Scholar]
- 62.Kessler D., Dethlefsen S., Haase I., Plomann M., Hirche F., Krieg T., Eckes B. (2001) J. Biol. Chem. 276, 36575–36585 [DOI] [PubMed] [Google Scholar]
- 63.Lindahl G. E., Chambers R. C., Papakrivopoulou J., Dawson S. J., Jacobsen M. C., Bishop J. E., Laurent G. J. (2002) J. Biol. Chem. 277, 6153–6161 [DOI] [PubMed] [Google Scholar]
- 64.Rios H. F., Ma D., Xie Y., Giannobile W. V., Bonewald L. F., Conway S. J., Feng J. Q. (2008) J. Periodontol. 79, 1480–1490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Maier S., Lutz R., Gelman L., Sarasa-Renedo A., Schenk S., Grashoff C., Chiquet M. (2008) Biochim. Biophys. Acta 1783, 1150–1162 [DOI] [PubMed] [Google Scholar]
- 66.Lu N., Carracedo S., Ranta J., Heuchel R., Soininen R., Gullberg D. (2009) Matrix Biol., in press [DOI] [PubMed] [Google Scholar]
- 67.Miano J. M., Long X., Fujiwara K. (2007) Am. J. Physiol. Cell Physiol 292, C70–C81 [DOI] [PubMed] [Google Scholar]
- 68.Chiquet M., Gelman L., Lutz R., Maier S. (2009) Biochim. Biophys. Acta 1793, 911–920 [DOI] [PubMed] [Google Scholar]
- 69.Lamar J. M., Pumiglia K. M., DiPersio C. M. (2008) Cancer Res. 68, 7371–7379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shimizu A., Kato M., Nakao A., Imamura T., ten Dijke P., Heldin C. H., Kawabata M., Shimada S., Miyazono K. (1998) Genes Cells 3, 125–134 [DOI] [PubMed] [Google Scholar]
- 71.Feijen A., Goumans M. J., van den Eijnden-van Raaij A. J. (1994) Development 120, 3621–3637 [DOI] [PubMed] [Google Scholar]
- 72.Lehnert S. A., Akhurst R. J. (1988) Development 104, 263–273 [DOI] [PubMed] [Google Scholar]
- 73.Hinz B., Phan S. H., Thannickal V. J., Galli A., Bochaton-Piallat M. L., Gabbiani G. (2007) Am. J. Pathol. 170, 1807–1816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Svendsen Ø. S., Barczyk M. M., Popova S. N., Lidén A., Gullberg D., Wiig H. (2009) Arterioscler. Thromb. Vasc. Biol. 29, 1864–1870 [DOI] [PubMed] [Google Scholar]
- 75.Eckes B., Zigrino P., Kessler D., Holtkötter O., Shephard P., Mauch C., Krieg T. (2000) Matrix Biol. 19, 325–332 [DOI] [PubMed] [Google Scholar]
- 76.Asano Y., Ihn H., Yamane K., Jinnin M., Tamaki K. (2006) Am. J. Pathol. 168, 499–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lygoe K. A., Norman J. T., Marshall J. F., Lewis M. P. (2004) Wound Repair Regen. 12, 461–470 [DOI] [PubMed] [Google Scholar]
- 78.Cooke M. E., Sakai T., Mosher D. F. (2000) J. Cell Sci. 113, 2375–2383 [DOI] [PubMed] [Google Scholar]
- 79.Gullberg D. (2009) Matrix Biol. 28, 383. [DOI] [PubMed] [Google Scholar]
- 80.Beattie G. M., Lopez A. D., Bucay N., Hinton A., Firpo M. T., King C. C., Hayek A. (2005) Stem Cells 23, 489–495 [DOI] [PubMed] [Google Scholar]
- 81.Fumagalli M., Musso T., Vermi W., Scutera S., Daniele R., Alotto D., Cambieri I., Ostorero A., Gentili F., Caposio P., Zucca M., Sozzani S., Stella M., Castagnoli C. (2007) Exp. Dermatol. 16, 600–610 [DOI] [PubMed] [Google Scholar]
- 82.Rodriguez A., Karen J., Gardner H., Gerdin B., Rubin K., Sundberg C. (2009) J. Cell. Mol. Med. 13, 3449–3462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pozzi A., Wary K. K., Giancotti F. G., Gardner H. A. (1998) J. Cell Biol. 142, 587–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.