Abstract
Coprinopsis cinerea is a model organism for fruiting body development in homobasidiomycetes. Here, we focused on N-linked oligosaccharides (NLO) of cell wall proteins in the hyphae of two developmental stages, vegetative mycelium and fruiting body. High mannose-type glycans were the most commonly found structures. In addition, we observed a novel glycan, predominantly present in fruiting body. This oligosaccharide structure was of the high mannose type with at least five mannoses and a bisecting α1–4 N-acetylglucosamine (GlcNAc) at the β-mannose of the N-glycan core. The transferase responsible for this modification, CcGnt1 (C. cinerea GlcNAc transferase 1), was identified and expressed in insect cells. In vitro activity of CcGnt1 was demonstrated. This novel glycosyltransferase belongs to the glycosyltransferase family 8 (GT8) and is predicted to be a type II membrane protein. Expression of the CcGnt1 locus was up-regulated in fruiting body, but down-regulation of expression by means of RNAi decreased the level of bisected NLO; however had no apparent effect on fruiting body formation.
Keywords: Carbohydrate/Biosynthesis, Carbohydrate/Processing, Carbohydrate/Structure, Enzymes, Glycoproteins/Biosynthesis, Glycosylation, Organisms/Fungi, Protein/Post-translational Modification
Introduction
N-Linked protein glycosylation is present in all domains of life and has two characteristics in common: the oligosaccharide is preassembled on a lipid carrier and is then transferred en bloc to an asparagine residue within the consensus sequence (N-X-(S/T)) of the protein. The composition of the oligosaccharide differs between bacteria and eukaryotes. Eukaryotic glycans are built on a chitobiose core (two N-acetylglucosamine, GlcNAc)3 that is further modified with mannoses (1). Higher eukaryotes share a conserved structure with two GlcNAc, nine mannoses, and three glucoses. After the initial trimming of the N-linked oligosaccharide (NLO) in the endoplasmic reticulum (ER), the structure and function of the glycans diverge in different eukaryotes. In animals, extensive remodeling by glycosylhydrolases (GH) and glycosyltransferases (GT) can lead to three classes of N-glycan: high mannose-, hybrid-, and complex-type. The combination of various monosaccharides, such as GlcNAc, galactose, fucose, and sialic acid, differently substituted and linked yields hundreds of protein-linked glycoforms. Plants do also have highly modified glycans, less complex and without sialic acid, but with the characteristic β1–2 xylose modification at the core mannose and α1–3 fucose at the innermost GlcNAc of the chitobiose core (2).
The glycan processing in the Golgi apparatus of fungi is less well studied but seems to be simpler. In the ascomycetous yeast Saccharomyces cerevisiae, high mannose-type oligosaccharides are not trimmed but elongated on a newly introduced branch with up to 200 mannoses (3, 4). In filamentous ascomycetes this outer chain mannosylation is less pronounced; instead galactofuranose is found attached to the middle branch of the high mannose sugar and α1–2 mannoses can be trimmed (5).
In basidiomycetes, investigation of N-glycan modification in Golgi was only recently approached using bioinformatic tools. Surprisingly, no homologs to equivalent Golgi-glycosyltransferases present in other eukaryotes were found to be encoded in four genomes of homobasidiomycetes, Coprinopsis cinerea, Phanerochaete chrysosporium, Laccaria bicolor, and Schizophyllum commune (6) and one heterobasidiomycete, Ustilago maydis (7). On the other hand, up to seven putative Golgi-localized mannosidases were identified in the genome of the homobasidiomycetes, and mannosidase activity was identified in S. commune (6).
In this study, we characterized N-glycans of cell wall proteins from vegetative mycelium and young fruiting bodies of the homobasidiomycete C. cinerea. High mannose-type structures with five to nine mannoses were identified predominantly, but an additional glycan, characterized as a high mannose-type glycan with a bisecting α1–4 GlcNAc at the β-mannose was isolated from fruiting bodies. The GlcNAc transferase responsible for this modification was identified, termed CcGnt1, and its activity was confirmed by both in vivo and in vitro experiments.
EXPERIMENTAL PROCEDURES
Strains, Growth, and Transformation Conditions
Escherichia coli strain DH5α was used for cloning and amplification of plasmids. Transformation-competent cells were prepared as described previously (8). Plasmid-containing bacteria were selected at 37 °C on Luria broth containing 100 mg of ampicillin/liter. S. cerevisiae laboratory strain W303–1A (MATa ura3–1 trp1–1 his3–11, 15 leu2–3, 112 ade2–1 can1–100) was used for cloning by homologous recombination. Transformants generated by the LiOAc method (9) were selected at 30 °C on synthetic complete medium without uracil (10). C. cinerea strain AmutBmut (A43mut B43mut pab1.2) (11, 12) was used for all Coprinopsis experiments. The transformation of mononucleate asexual C. cinerea spores (oidia) with the RNAi constructs was described previously (13). Selection for transformants was done on minimal medium (MM). Primordia (referred to as fruiting bodies in this report) of strain AmutBmut were produced by precultivating vegetative mycelium on cellophane disks on YMG plates for 4 days (triple inocula) at 37 °C in darkness (in ventilated closed black boxes) and subsequent transferral to 25 °C in a 12-h light/dark regime for another 3 days (resulting in secondary hyphal knots and primordia of up to 2 mm in diameter) and up to 10 days, respectively. The size of harvested primordia was 4–10 mm. Oidia for transformation and DNA isolation were produced by transferring vegetative mycelium grown on YMG (AmutBmut) without cellophane disks at 37 °C for 3 days in darkness (triple inocula) into constant white light (20 to 25 μE per m2 and s; emission spectrum of 275 to 780 nm) and incubation at 37 °C for another 4 and 5 days, respectively.
Cloning and Plasmid Construction
CcGnt1 was amplified from genomic DNA of AmutBmut, because the coding sequence is not interrupted by any introns. The sequence in AmutBmut was identical to the one of the strain okayama7 no. 130 and can be retrieved from GenBankTM (AACS01000037.1 286227-287411). The primer pair NTF_GnT1fwd/NTF_GnT1rev was used for PCR amplification, the product was cloned into pGem-T Easy Vector System (Promega), amplified in E. coli and sequenced. The DNA encoding the open reading frame without the start codon was subcloned into pFASTBAC vector (Invitrogen) for insect cell expression cleaved with the restriction endonucleases NarI and HindIII. The ATG start codon and an N-terminal sequence encoding for the FLAG tag were included on the plasmid.
The RNAi construct was built by homologous recombination in yeast with linearized plasmid (no. 359, Ref. 14) and the two PCR fragments (sense and antisense) amplified with primer pair RNAi_GnT1sfwd/RNAi_GnT1srev1 and RNAi_GnT1asfwd1/RNAi_GnT1asrev as described in Ref. 14. NTF_GnT1fwd (GGCGCCTGGCCGACCAATACTAGACCG); NTF_GnT1rev (AAGCTTTCACCTCGTCGGCCAAC); RNAi_GnT1sfwd (GTATCACCAGTCTAACATCCCGCGGTGGGCCGACCAATACTAGAC); RNAi_GnT1srev1 (TCTCTTGAATTCTCTTGAATTTGGAACGCCATCATCTTCAAG); RNAi_GnT1asfwd1 (TTCAAGAGAATTCAAGAGATTTGGAACGCCATCATCTTCAAG); RNAi_GnT1asrev (GGGGGAGCAATCCATGGACACTAGTTGGGCCGACCAATACTAGAC).
N-Glycan Isolation
50 mg of lyophilized material (either vegetative mycelium or fruiting body primordia) was powdered with glass beads in the Fast-Prep machine (Bio 101 Savant; Savant Instruments, Inc., Holbrook, NY) and the protocol established by Schulz and Aebi (15) was followed to enrich for cell wall-linked proteins. N-Glycans were then released with 500 units of either PNGase F (NEB; 500 units/μl) or Endo H (NEB; 500 units/μl) overnight at 37 °C. The supernatant and two consecutive washes of the pellet were pooled, subjected to cleanup, and 2-AB labeling (see below) (16).
Carbohydrate Cleanup and 2-AB Labeling
The extract was passed over C18- (Sep-Pak Cartridges C18; Waters) and graphitized carbon columns (Supelclean ENVI-Carb SPE bulk packing; Supelco) for purification: the two columns were first washed twice with 1 ml of acetonitrile, then equilibrated twice with 2% acetonitrile in H2O. The extract was then applied to the C18 column, the tube was washed with 2% acetonitrile, and passed through the column. The flow-through was loaded onto the carbon column and washed three times with 2% acetonitrile. NLO were eluted twice with 200 μl of 50% acetonitrile in H2O, dried in a Speedvac (Savant AES 1000) and subjected to 2-AB-labeling (2-AB: 2-aminobenzamide (Aldrich; anthranilamide)(17). Excess label was removed using Ultrafree-MC centrifugal filters (UFC30LH25, Millipore) supported by two paper discs (3 mm Chr, chromatography paper; Whatman). The columns were sequentially pretreated with twice 450 μl of 30% glacial acetic acid (J. T. Baker), ddH2O, 100% acetonitrile (J.T. Baker), and 95% acetonitrile. The labeling reaction was stopped by addition of acetonitrile to a final concentration of 95%. The reaction was loaded onto the column, and the columns were washed eight times with 95% acetonitrile in H2O. Elution was by centrifugation with ddH2O.
Glycosidase Treatment
Glycans were resuspended in 50 mm sodium citrate (pH 5) for α-mannosidase (Jack Bean, Sigma M7257; ∼20 units/mg) treatment, in 50 mm sodium citrate (pH 4.5) for β-HexNAc'ase (Jack Bean, Sigma; ∼50 units/mg) treatment, in 50 mm sodium citrate (pH 5) for double digest with α-mannosidase and β-HexNAc'ase and in 50 mm sodium acetate (pH 5.2) for α1–2 mannosidase treatment (Trichoderma reesei, a gift from R. Contreras, Gent). 1 μl of enzyme was used in a total volume of 20 μl. The reaction was incubated overnight at 37 °C. Glycans were desalted and purified from enzymes with a small scale carbon column as described below.
Insect Cell Culture and in Vitro Assay
SF9 cells (Invitrogen) were cultured in Grace's insect cell culture medium (Invitrogen) supplemented with 10% fetal calf serum in 6-well plates (TPP, Switzerland) at 28 °C. Two million cells were infected with recombinant baculovirus either containing pFASTBAC vector control (Invitrogen) or pFASTBAC-N-terminal FLAG-CcGnt1 according to the manufacturer's instructions and grown for 3 days. Cells were then lysed with shaking (4 °C, 15 min) in 150 μl of Tris-buffered saline (TBS, pH 7.4) containing 2% (w/v) Triton X-100 and protease inhibitor mixture (Roche, Complete EDTA-free). The lysis mixture was centrifuged (2000 × g, 5 min), the postnuclear supernatant was recovered (raw extract), and used for all further enzymatic studies.
Enzymatic activity toward appropriate carbohydrates was assessed using 2 μl of raw extract in a 5-μl final volume of MES buffer (pH 6.5, 40 μm) containing manganese(II) chloride (10 μm), UDP-GlcNAc (1 mm) and the acceptor glycan, prepared from RNAseB (Sigma) by release of the glycan with PNGase (NEB), followed by digestion with α1–2-mannosidase. Glycosylation reactions were typically run for 16 h at room temperature. For donor specificity analysis, UDP-GlcNAc was replaced by equal concentrations of UDP-GalNAc. For cofactor specificity analysis, MnCl2 was replaced by equal concentrations of MgCl2 or Na2EDTA. Products were desalted and purified from proteins with a small scale carbon column as described below. Analysis of the reaction product was performed by MALDI-TOF mass spectrometry.
Mass Spectrometry
For MS analysis, samples were purified by an additional carbon column. The small scale, self-made column was prepared as follows: a 200-μl pipette tip was filled with glass wool, and 100 μl of carbon suspension (Supelco, in methanol) was added. The column was washed twice with 200 μl of acetonitrile, equilibrated twice with 200 μl of 2% acetonitrile. The sample was loaded, and the column was washed twice with 200 μl of 2% acetonitrile before eluting twice with 50% acetonitrile. The eluate was dried and resuspended in 0.1% trifluoroacetic acid, spotted on a MALDI plate and covered with matrix (10 mg/ml 2,5-dihydroxybenzoic acid (Sigma), 70% acetonitrile, and 0.1% trifluoroacetic acid). MALDI-TOF-MS and MS/MS were performed with ABI 4800 MALDI TOF/TOFTM (Applied Biosystems Inc.).
qRT-PCR
Real-time PCR was carried out by using a SensiMixTMPlus SYBR® (QUANTACE) with 0.9 μm forward and reverse primer concentrations each and a variable amount of cDNA (20–0.02 ng per reaction) in a final reaction volume of 20 μl. Thermocycling was performed in a Rotor-Gene 3000 Real-Time Thermal Cycler (Corbett Research, Sydney, Australia) initiated by a 14-min incubation at 95 °C, followed by 40 cycles of 15 s at 95 °C, 30 s at 60 °C, and 30 s at 72 °C. Fluorescence data were acquired during the elongation step in every cycle. Each run was completed with a melting curve analysis to confirm specificity of amplification and absence of primer dimers. The amplification of genomic DNA was prevented by DNase digestion during RNA extraction and monitored by including a cDNA control, where reverse transcriptase was omitted. benA (tubulin) expression was detected with the primers 5′- GTCATGTCCGGTATCACCAC-3′ and 5′-GGGAAAGGAACCATGTTGA-3′ served as a reference gene for the relative quantification of CcGnt1 and CcGnt2, which were detected with the primers CcGnt1-fwd 5′-ACCAAAATGGATGCTTCACC-3′, CcGnt1-rev 5′-GATTGAGAGGCCTGAGATGC-3′ and CcGnt2-fwd 5′-GAGGCGACGCTTACTACTGG-3′, CcGnt2-rev 5′- GGTCCGAGTCCATAACGATG- 3′, respectively. PCR efficiencies were determined with serial dilutions of cDNA. Transcript quantification of individual samples was based on measurements in triplicate and analysis using the mathematical model of Pfaffl (18).
RESULTS
N-Glycans in C. cinerea
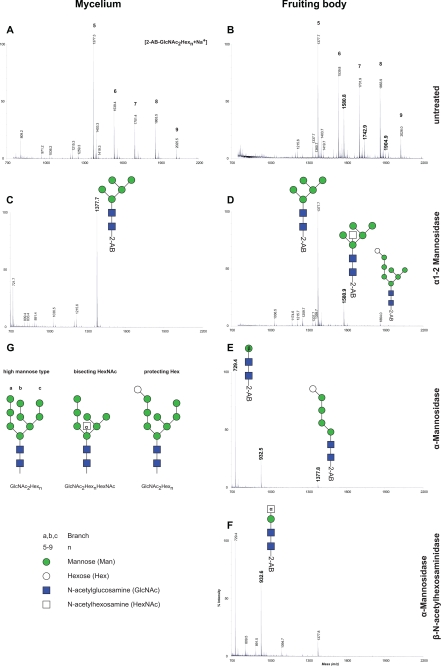
To describe the NLO in C. cinerea, the cell wall was chosen as the source enriched in glycosylated proteins (15). The fact that cell wall proteins can be covalently attached to the polysaccharide scaffold was exploited to enrich for glycoproteins prior to enzymatic release of the NLO by peptide: N-glycosidase F (PNGase F). Purified and 2-aminobenzamide (2-AB)-derivatized glycan were subsequently analyzed by MALDI-MS. In both, mycelium and fruiting body, we identified glycans with masses corresponding to two N-acetylhexosamine (HexNAc) and five to nine Hex units (Fig. 1, A and B; m/z: 1377, 1539, 1701, 1863, 2025). Treating the preparation with α1–2 mannosidase reduced the complexity of the spectra, and we observed a predominant glycan in mycelium-derived samples with a mass of m/z = 1377 (Fig. 1C). Based on the knowledge concerning biosynthesis and early processing of N-glycans in the ER (19) we concluded that these masses correspond to high mannose-type glycans (Fig. 1G). The different glycans detected were probably generated by the trimming of the ER oligosaccharide GlcNAc2Man8–9 to GlcNAc2Man5 by Golgi α1–2 mannosidases, as found to be the case in other filamentous fungi like Aspergillus fumigatus (20). In fruiting body-derived preparations, an additional series of oligosaccharides, differing from the high mannose series by the mass of one additional HexNAc unit appeared in the spectra (Fig. 1B; m/z: 1580, 1742, 1904). Digestion of glycans by α1–2 mannosidase again reduced complexity of the spectra and gave the GlcNAc2Man5 oligosaccharide (m/z 1377) as in the mycelium sample, but in addition two oligosaccharides appeared: a putative GlcNAc2Man5HexNAc and a putative GlcNAc2Hex8 oligosaccharide. Additional treatment of the sample by α-mannosidase resulted in a further trimming and yielded a GlcNAc2Man (m/z 729; derived from GlcNAc2Man5), a GlcNAc2ManHexNAc (m/z 932) and a GlcNAc2Hex5 (m/z 1377; Fig. 1E). These structures were resistant to treatment by β-N-acetylhexosaminidase (β-HexNAc'ase; Fig. 1F). We confirmed that β-N-acetylhexosaminidase was fully active under the conditions used because commercially available hybrid-type glycans with bisecting β-GlcNAc was processed by the enzyme (supplemental Fig. S1). We concluded that the novel series of glycans appearing in the fruiting body were modified with an α-HexNAc bisecting the structure at the core mannose and that oligosaccharides with (glucosylated) A-branch were also present in the preparations from fruiting bodies (Fig. 1G). A hexose attached to the A-branch protected the two α-mannoses from degradation by the enzyme. Based on biosynthetic information, we speculated that this hexose was glucose that was not removed by the processing glucosidase II in the ER.
FIGURE 1.
N-Glycan profile of C. cinerea cell wall proteins. N-Linked glycans were enzymatically released by PNGase F from cell wall proteins of C. cinerea, purified, derivatized, and analyzed by MALDI-TOF-MS. The spectra in A and B were obtained from analyzing extracts of mycelium and fruiting body, respectively. Vertical numbers indicate the peak mass, and selected peaks are labeled with an index (n) in bold, representing high mannose-type glycans of the composition 2-AB-GlcNAc2Hexn. Bold masses in B emphasize peaks appearing in fruiting body that were not detected in mycelium sample. In C and D, samples derived from mycelium and fruiting body, respectively, were digested with α1–2 mannosidase from T. reesei. To characterize the glycan profile further, fruiting body derived extract was either treated with Jack Bean α-mannosidases (E) or with α-mannosidases in combination with β-N-acetylhexosaminidase (β-HexNAc'ase) (F). Putative structures of N-linked glycans present in cell wall in G.
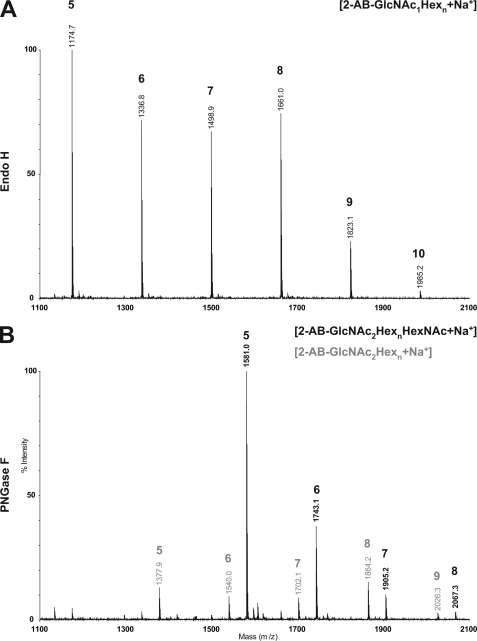
Importantly, the high mannose oligosaccharide with an additional HexNAc were not obtained by treating the cell wall preparation of fruiting bodies with Endo H (Fig. 2A), but subsequent treatment of the cell wall preparation by PNGase F released the GlcNAc-containing oligosaccharides. We concluded that the fruiting body specific bisecting α-linked GlcNAc modification inhibited Endo H activity. Such an inhibition does not occur for glycans that are bisected with β-GlcNAc, structures commonly found on mammalian N-glycoproteins such as ovalbumin (21).
FIGURE 2.
Novel glycan structure is resistant to Endo H. Cell wall protein from fruiting body extract was consecutively treated with Endo H, followed by PNGase F. The released N-glycans were purified, labeled with 2-AB, and analyzed with MALDI-MS. In A, the MALDI-TOF spectrum of glycans released by Endo H is shown. Peaks are labeled with mass (vertical) and indices n (bold) belonging to 2-AB-GlcNAc1Hexn. B shows the spectrum for PNGase F-released glycans from cell wall subsequent to Endo H treatment. In black, masses and indices correspond to 2-AB-GlcNAc2HexnHexNAc; in gray to 2-AB-GlcNAc2Hexn.
Identification and Characterization of the GlcNAc Transferase
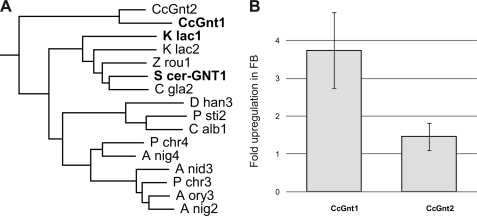
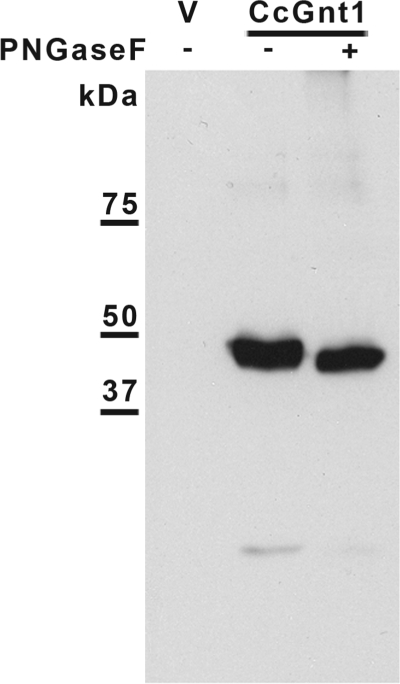
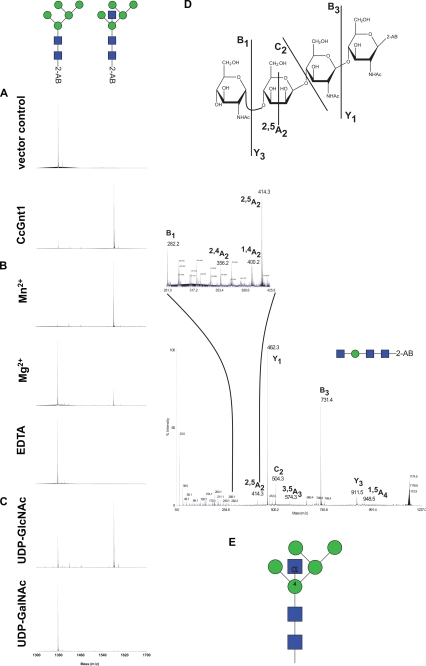
Genes homologous to the GlcNAc-transferases of higher eukaryotes (GnT) (22) were not identified using homology searches (by the BLAST algorithm) against the Coprinopsis genome (Broad Institute Coprinopsis Database). Instead five loci related to the GlcNAc transferases from the ascomycetous yeasts S. cerevisiae (24) and Kluyveromyces lactis (25) were found. This glycosyltransferase was first identified in K. lactis in a screen for mutant strains with altered mannan biosynthesis (26). In this report, a terminal α1–2 GlcNAc was found on the outer chain of mannoproteins. The genes, called Gnt1, code for glycosyltransferases of the family 8 (GT8), as classified by the CaZY data base (27). GT8 members are retaining transferases that add various monosaccharides from nucleotide activated substrates in α-conformation to various acceptors (28). Aligning all fungal GT8 proteins listed in the CaZY data base, together with the five orthologs found in C. cinerea, and performing a phylogenetic analysis allowed us to identify a group of putative glycosyltransferases that included the yeast Gnt1 as well as putative orthologs from ascomycetes. Interestingly, two closely related putative proteins in C. cinerea, denominated CcGnt1 and CcGnt2 (C. cinerea GlcNAc transferase 1 and 2), were present in this group (Fig. 3A). The two putative C. cinerea Gnt proteins consist of 394 and 324 amino acid residues, respectively, and are 51% identical in sequence. Like the yeast ortholog, they are predicted to be type II membrane protein, typical for Golgi glycosyltransferases. To evaluate the hypothesis that one of these transferases was responsible for the N-glycan modification observed in fruiting body cell wall protein, transcript levels of CcGnt1 and CcGnt2 were determined by quantitative real-time PCR (qRT-PCR) with RNA extracted from either vegetative mycelium or fruiting body. CcGnt1 was found to be up-regulated in fruiting body, whereas CcGnt2 mRNA levels were found to be the same in both developmental stages (Fig. 3B). Therefore, CcGnt1 was amplified from genomic DNA and the corresponding protein, marked with an N-terminal FLAG-tag, was expressed in a baculovirus expression system. Extracts derived from insect cells containing the expression construct revealed a clear signal in immunoblots obtained after SDS/PAGE and subsequent probing with anti-FLAG antiserum (Fig. 4, lane 2). Treatment of the extract with PNGase F prior to SDS/PAGE resulted in a slightly increased mobility of the tagged protein, consistent with the presence of a single N-glycosylation site in the protein (Fig. 4, lane 3). To evaluate glycosyltransferase activity of CcGnt1, crude extracts of CcGnt1-expressing and control insect cells were tested. 2-AB-labeled GlcNAc2Man5 oligosaccharide and UDP- GlcNAc were used as substrates. Product analysis was done by MALDI-TOF mass spectrometry. Crude extract of insect cells expressing CcGnt1 but not the control extract was able to transfer GlcNAc from UDP-GlcNAc to 2-AB-GlcNAc2Man5 in the presence of Mn2+ (Fig. 5A). The addition of Mn2+ as cofactor in the reaction was crucial for the transfer, because in the presence of Mg2+ ions, the substrate turnover was incomplete, and the reaction was totally inhibited by depleting divalent cations from the extract with EDTA (Fig. 5B). Using UDP-GalNAc as alternative donor did not result in any transfer (Fig. 5C), supporting the conclusion that CcGnt1 indeed is a GlcNAc transferase. We analyzed the in vitro reaction product in more detail. MS/MS fragmentation of permethylated in vitro produced glycan further provided strong evidence for addition of the GlcNAc to the 4 position on the β-mannose. The ions relevant for structure interpretation are found in the spectrum in Fig. 5D and are schematically represented in the Haworth projection. The Y1 ion, one GlcNAc with the 2-AB label, indicated that the reducing-end GlcNAc was not further substituted. The C2 ion revealed the presence of a GlcNAc molecule attached to the mannose. Together, those two ions provided good evidence for the additional GlcNAc to be linked to the β-mannose rather than to the chitobiose core. Because position 3 and 6 of the β-mannose are normally substituted by mannoses, only position 2 and 4 remained for the GlcNAc modification. The cross ring fragment 2,4A2 ion was indicative for the 1–4 linkage.
FIGURE 3.
Yeast Gnt1 homologues as candidates for the transferase activity in fruiting body. A, phylogenetic analysis of fungal members of the GT8-family found in CaZY data base (27). Shown here, the clade of the yeast Gnt1's (24, 25) and two homologs of C. cinerea. Aspergillus niger: A nig2 (CAK44598.1), A nig3 (CAK43246.1), A nig4 (CAK97109.1); Aspergillus oryzae: A ory3 (BAE63310.1); Candida albicans: C alb1 (AAV04198.1); Candida glabrata: C gla2 (CAG60616.1); C. cinerea: CcGnt1 (CC1G_14119.2), CcGnt2 (CC1G_14164.2) (sequences can be retrieved from the Broad Institute Coprinopsis Database); Debaryomyces hansenii: D han3 (CAG86035.1); Kluyveromyces lactis: K lac1 (AAD25740.1), K lac2 (CAH01694.1); Penicillium chrysogenum: P chr3 (CAP92752.1), P chr4 (CAP98553.1); Pichia stipitis: P sti2 (ABN66322.1); S. cerevisiae: S cer-GNT1 (CAA62175.1); Zygosaccharomyces rouxii: Z rou1 (CAR28822.1). B, relative mRNA levels of CcGnt1 and CcGnt2 in vegetative mycelium and fruiting body. mRNA isolated from mycelium and fruiting body of C. cinerea was used for quantitative real-time PCR (qRT-PCR) analysis of these two developmental states. CcGnt1 is differentially expressed, but CcGnt2 is not. The error bars represent the standard deviation of three experimental replicates.
FIGURE 4.
Immunoblot analysis of CcGnt1 expression in insect cells. Protein crude extract of SF9 cells was separated by SDS-PAGE and blotted on a nitrocellulose membrane. FLAG-tagged CcGnt1 was detected using monoclonal anti-FLAG antibody. Protein extract of cells transfected with the vector control (lane 1) and of cells transfected with CcGnt1-FLAG (lanes 2 and 3) was loaded. In lane 3, protein extract of cells transfected with CcGnt1-FLAG was treated with PNGase F prior SDS-PAGE. Bands from protein marker are indicated on the left.
FIGURE 5.
Characterization of CcGnt1 (A–C) and its glycan product in vitro (D). A–C, in vitro reaction with variable conditions were performed and the qualitative glycan educt to product conversion was analyzed by MALDI-TOF-MS. Glycans with the mass of 1377 Da (educt) and 1580 Da (product) are indicated schematically above the spectra. A, standard conditions with crude extract from cells transfected with vector control or with CcGnt1, respectively, were compared. B, influence of divalent cations on the enzyme activity. The reaction was performed with either Mn2+ (standard), Mg2+ or EDTA. C, to determine the transferase specificity, either UDP-GlcNAc or UDP-GalNAc was used as sugar donor. D, product was analyzed in more detail. In vitro reaction product was digested with α-mannosidase, purified, and permethylated. MS/MS spectrum of the resulting glycans is shown with peaks corresponding to characteristic ions. These are labeled and illustrated in the Haworth representation above the spectrum. E, scheme summarizing the findings concerning the structure of the reaction product.
GlcNAc2Man9 oligosaccharide was accepted as a substrate by the GlcNAc-transferase CcGnt1 (data not shown), supporting the view that the GlcNAc modification in Coprinopsis can occur, independent of Golgi α1–2 mannosidase trimming. This is different in mammalian cells where the α1–2 mannoses have to be removed by Golgi α1–2 mannosidases (29) before GnT I (30) and GnT III (31) can act to form hybrid- and complex-type glycans.
Depletion of CcGnt1 Does Not Affect Fruiting Body Formation
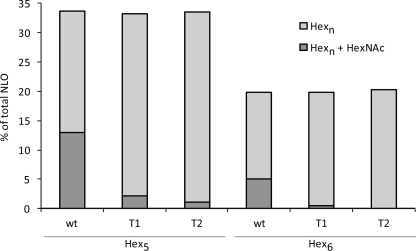
To address the role of Gnt1 in fruiting body formation, RNA silencing of CcGnt1 was done to down-regulate the transcript levels of the locus. Fruiting body formation was not affected in two representative transformants, but we observed a strong reduction of GlcNAc modification in N-linked glycans due to the expression of RNAi (Fig. 6), while the overall relative amounts of Man5 and Man6 containing NLO, respectively, were not affected.
FIGURE 6.
Effect of CcGnt1 down-regulation on N-linked glycan. Fruiting bodies of wild type (wt) and of two independent RNAi transformants (T1, T2) were collected, and NLO of cell wall proteins were analyzed by MALDI-MS. Signal to noise (s/n) ratios of all NLO peaks within a spectrum were summed and divided by the s/n ratio of a glycan HexNAc2Hexn either with (dark gray bar) or without (light gray bar) additional GlcNAc.
DISCUSSION
Our analysis of mature N-glycan from C. cinerea reflected the low diversity of NLO modification in fungal Golgi compartment. Mainly high mannose-type oligosaccharides were described so far in fungi, occasionally decorated with galactose, as it has been shown in Schizosaccharomyces pombe (4) and Aspergillus spp. (32). The addition of GlcNAc described in this study is the first Golgi modification reported for basidiomycetes. The position of the modification resembles a bisected-hybrid-type glycan as found in higher eukaryotes. There, the same position of the β-mannose is substituted with GlcNAc, however in a β conformation. GnT III, the transferase responsible for this modification is proposed to be a regulator of cell adhesion. Bisected glycans promote E-cadherin-mediated cell-cell adhesion and prevent in a feedback loop integrin-mediated cell migration (33). In the context of fungal cell wall, hyphal adhesion and fruiting body formation can be seen as analogous to cell adhesion in animals. It is also in that developmental process where CcGnt1 was up-regulated and the modified N-glycan were found on cell wall proteins. However, we did not observe a defect in fruiting body development. This result can be interpreted in that this N-glycan modification was not essential for fruiting body formation. Alternatively, the residual enzyme activity after RNAi treatment or the redundant activity of CcGnt2 may be sufficient to mask a possible defect. Complete knock-out, overexpression of CcGnt1 and enzymatic characterization of the homolog CcGnt2 need to be performed to elucidate the function of the fungal bisecting GlcNAc.
It is interesting to note that animals, plant and fungi can substitute the β-mannose of the core N-glycan in the Golgi compartment. However, this modification seems to be specific for the different kingdoms (β-GlcNAc in animals, β-xylose in plants, and α-GlcNAc in fungi).
The transferase responsible for the novel α1–4-GlcNAc modification belongs to the retaining glycosyltransferases from the family 8, as classified by B. Henrissat and co-workers in the CaZY data base. Phylogenetic analysis of human and fungal members of that family allowed a further division into at least five subfamilies, where one clade includes relatives of these fungal GlcNAc-transferases, a second and third the homologues of galactinol synthases and glycogenin, respectively. The fourth subgroup contains mainly human members, Large being among them (34). Little is known about general function and characteristics of Large, but loss of function leads to muscle dystrophy caused by aberrant glycosylation of a single protein, α-Dystroglycan, which impairs binding to its ligand laminin (35). The ortholog Large2 has a distinct expression pattern but functional redundancy could be shown in cell culture experiments (36).
The fifth clade unites fungal glycosyltransferases with unknown function. One of them, Mug136 (meiosis up-regulated gene; SPBC4C3.08) from S. pombe, was found to be differentially regulated but its role in meiosis is not yet clear (23). Other members of that subfamily are found in the basidiomycetous yeast Cryptococcus neoformans, where an additional pentose could be identified on N-glycan of cell wall protein.4
The fact that only a few fungal Golgi modifications on N-glycan are described does not necessarily mean that they do not exist. Amounts might be below the detection limit of our analytical methods, and these processes might be spatially or temporally regulated, so that these minor modifications escape our attention. Nevertheless, the complexity of N-glycan processing in the Golgi compartment seems to be low.
Supplementary Material
Acknowledgments
We thank Ben Schulz for inspiring discussions. We thank Yao-Yun (Jessy) Fan, Peter Gehrig, Bernd Roschitzki, and the Functional Genomic Center Zürich for support with mass spectrometry, Alex Titz and Belinda Schegg for help with the insect cell handling, and R. Contreras and N. Callewaert for α1–2 mannosidase.
This work was supported in part by ETH Zürich and by an ESKAS Swiss Federal Scholarship (to Z. L.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
R. Buser and M. Aebi, unpublished results.
- GlcNAc
- N-acetylglucosamine
- NLO
- N-linked oligosaccharide(s)
- MES
- 4-morpholineethanesulfonic acid
- HexNAc
- N-acetylhexosamine
- MALDI-MS
- matrix-assisted laser desorption/ionization-mass spectrometry
- ER
- endoplasmic reticulum.
REFERENCES
- 1.Burda P., Aebi M. (1999) Biochim. Biophys. Acta 1426, 239–257 [DOI] [PubMed] [Google Scholar]
- 2.Stanley P., Schachter H., Taniguchi N. (2009) in Essentials of Glycobiology (Varki A. ed) 2nd Ed., pp. 101–114, Cold Spring Harbor Laboratory Press, New York: [PubMed] [Google Scholar]
- 3.Dean N. (1999) Biochim. Biophys. Acta 1426, 309–322 [DOI] [PubMed] [Google Scholar]
- 4.Gemmill T. R., Trimble R. B. (1999) Biochim. Biophys. Acta 1426, 227–237 [DOI] [PubMed] [Google Scholar]
- 5.Wallis G. L., Easton R. L., Jolly K., Hemming F. W., Peberdy J. F. (2001) Eur. J. Biochem. 268, 4134–4143 [DOI] [PubMed] [Google Scholar]
- 6.Berends E., Ohm R. A., de Jong J. F., Rouwendal G., Wosten H. A., Lugones L. G., Bosch D. (2009) Appl. Environ. Microbiol. 75, 4648–4652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Deshpande N., Wilkins M. R., Packer N., Nevalainen H. (2008) Glycobiology 18, 626–637 [DOI] [PubMed] [Google Scholar]
- 8.Inoue H., Nojima H., Okayama H. (1990) Gene 96, 23–28 [DOI] [PubMed] [Google Scholar]
- 9.Ito H., Fukuda Y., Murata K., Kimura A. (1983) J. Bacteriol. 153, 163–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kaiser C., Michaelis S., Mitchell A. (1994) Methods in Yeast Genetics a Cold Spring Harbor Laboratory Course Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 11.May G., Le Chevanton L., Pukkila P. J. (1991) Genetics 128, 529–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Swamy S., Uno I., Ishikawa T. (1984) J. Gen. Microbiol. 130, 3219–3224 [Google Scholar]
- 13.Granado J. D., Kertesz-Chaloupková K., Aebi M., Kües U. (1997) Mol. Gen. Genet. 256, 28–36 [DOI] [PubMed] [Google Scholar]
- 14.Wälti M. A., Villalba C., Buser R. M., Grünler A., Aebi M., Künzler M. (2006) Eukaryot. Cell 5, 732–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schulz B. L., Aebi M. (2009) Mol. Cell. Proteomics 8, 357–364 [DOI] [PubMed] [Google Scholar]
- 16.Grubenmann C. E. (2004) Molecular and Biochemical Characterization of Four Novel Types of Congenital Disorders of Glycosylation. Ph.D. thesis, University of Zürich, Zürich, Switzerland [Google Scholar]
- 17.Bigge J. C., Patel T. P., Bruce J. A., Goulding P. N., Charles S. M., Parekh R. B. (1995) Anal. Biochem. 230, 229–238 [DOI] [PubMed] [Google Scholar]
- 18.Pfaffl M. W. (2001) Nucleic Acids Res. 29, e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Helenius A., Aebi M. (2004) Annu. Rev. Biochem. 73, 1019–1049 [DOI] [PubMed] [Google Scholar]
- 20.Herscovics A. (2001) Biochimie 83, 757–762 [DOI] [PubMed] [Google Scholar]
- 21.Tai T., Yamashita K., Ito S., Kobata A. (1977) J. Biol. Chem. 252, 6687–6694 [PubMed] [Google Scholar]
- 22.Taniguchi N., Honke K., Fukuda M. (2002) Handbook of Glycosyltransferases and Related Genes, Springer, Tokyo, New York [Google Scholar]
- 23.Mata J., Lyne R., Burns G., Bähler J. (2002) Nat. Genet. 32, 143–147 [DOI] [PubMed] [Google Scholar]
- 24.Yoko-o T., Wiggins C. A., Stolz J., Peak-Chew S. Y., Munro S. (2003) Glycobiology 13, 581–589 [DOI] [PubMed] [Google Scholar]
- 25.Guillen E., Abeijon C., Hirschberg C. B. (1999) J. Biol. Chem. 274, 6641–6646 [DOI] [PubMed] [Google Scholar]
- 26.Smith W. L., Nakajima T., Ballou C. E. (1975) J. Biol. Chem. 250, 3426–3435 [PubMed] [Google Scholar]
- 27.Cantarel B. L., Coutinho P. M., Rancurel C., Bernard T., Lombard V., Henrissat B. (2009) Nucleic Acids Res. 37, D233–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gibson R. P., Turkenburg J. P., Charnock S. J., Lloyd R., Davies G. J. (2002) Chem. Biol. 9, 1337–1346 [DOI] [PubMed] [Google Scholar]
- 29.Herscovics A. (1999) Biochim. Biophys. Acta 1473, 96–107 [DOI] [PubMed] [Google Scholar]
- 30.Kumar R., Yang J., Larsen R. D., Stanley P. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 9948–9952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Narasimhan S. (1982) J. Biol. Chem. 257, 10235–10242 [PubMed] [Google Scholar]
- 32.Morelle W., Bernard M., Debeaupuis J. P., Buitrago M., Tabouret M., Latgé J. P. (2005) Eukaryot. Cell 4, 1308–1316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gu J., Sato Y., Kariya Y., Isaji T., Taniguchi N., Fukuda T. (2009) J. Proteome Res. 8, 431–435 [DOI] [PubMed] [Google Scholar]
- 34.Peyrard M., Seroussi E., Sandberg-Nordqvist A. C., Xie Y. G., Han F. Y., Fransson I., Collins J., Dunham I., Kost-Alimova M., Imreh S., Dumanski J. P. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 598–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Patnaik S. K., Stanley P. (2005) J. Biol. Chem. 280, 20851–20859 [DOI] [PubMed] [Google Scholar]
- 36.Grewal P. K., McLaughlan J. M., Moore C. J., Browning C. A., Hewitt J. E. (2005) Glycobiology 15, 912–923 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.