Abstract
Mutations in leucine-rich repeat kinase 2 (LRRK2) are prevalent causes of late-onset Parkinson’s disease (PD). Here, we show that LRRK2 binds to mitogen-activated protein kinase (MAPK) kinases MKK3, 6, and 7, and that LRRK2 is able to phosphorylate MKK3, 6 and 7. Over-expression of LRRK2 and MKK6 increased the steady state levels of each protein beyond that observed with over-expression of either protein alone. Co-expression increased levels of MKK6 in the membrane more than in the cytoplasm. The increased expression of LRRK2 and MKK6 requires MKK6 activity. The disease-linked LRRK2 mutations, G2019S, R1441C and I2020T, enhance binding of LRRK2 to MKK6. This interaction was further supported by in vivo studies in C. elegans. RNAi knockdown in C. elegans of the endogenous orthologs for MKK6 or p38, sek-1 and pmk-1, abolishes LRRK2-mediated protection against mitochondrial stress. These results were confirmed by deletion of sek-1 in C. elegans. These data demonstrate that MKKs and LRRK2 function in similar biological pathways, and support a role for LRRK2 in modulating the cellular stress response.
Keywords: MAP kinase, phosphorylation, C. elegans, JNK, p38, membrane
INTRODUCTION
Parkinson’s disease (PD) is the most common neurodegenerative movement disorder (Olanow et al. 2009). Although the majority of PD cases are sporadic, the identification of genes responsible for familial parkinsonism has enhanced our understanding of the pathogenesis of PD. Autosomal-dominant mutations in leucine-rich repeat kinase 2 (LRRK2) are particularly interesting because they are associated with sporadic PD and a familial form of PD that resembles sporadic PD (Zimprich et al. 2004). The majority of LRRK2 cases exhibit pathological findings consisting of α-synuclein inclusions, which is the pathological hallmark of PD (Zimprich et al. 2004). However, some cases with LRRK2 mutations exhibit pleiomorphic pathology, including tau inclusions, or no inclusions (Zimprich et al. 2004).
The LRRK2 protein has several conserved domains including: a leucine-rich domain (LRR), a Roc GTPase domain, a COR (carboxy-terminal of Ras) domain, a kinase domain, and a WD40 domain at the carboxy-terminal (Zimprich et al. 2004, Greggio & Cookson 2009). The G2019S familial mutation leads to increased LRRK2 autophosphorylation and kinase activity (West et al. 2005, Smith et al. 2005, Smith et al. 2006, Greggio et al. 2006). Other mutations appear to affect kinase activity to a lesser extent (West et al. 2005, Smith et al. 2005, Smith et al. 2006, Gloeckner et al. 2009, Jaleel et al. 2007). The over-expression of some of these mutants lead to increased caspase-dependent toxicity in primary neurons (Macleod et al. 2006, Iaccarino et al. 2007, Ho et al. 2009). Missense mutations causing disruption in LRRK2 GTPase activity have also been shown to lead to cell death (Ito et al. 2007, Lewis et al. 2007, Li et al. 2007). In addition, LRRK2 appears to regulate neurite morphology in rodents, and in human neuronal cell lines (Macleod et al. 2006, Plowey et al. 2008). However, the exact mechanism by which LRRK2 lead to the pathogenesis of PD remains a critical unanswered question.
The kinase domain of LRRK2 shares homology with receptor interacting protein kinases (RIPs) and mixed lineage kinases (MLKs) (Greggio & Cookson 2009). RIP kinases, such as RIP-1, mediate signaling through death receptors, and activate stress kinases (JNK and p38) as well as NF-kB (Festjens et al. 2007). The RIP kinase family is particularly interesting in light of recent observations that LRRK2 mediates signaling through the death receptor protein Fas binding protein (Ho et al. 2009). MLKs are a family of serine/threonine protein kinases that function upstream in the MAPK signaling cascades (Silva et al. 2005). The MLK family of kinases activate the c-jun amino-terminal kinase (JNK) pathway by phosphorylating MAPK kinase 4 and 7 (MKK4 and MKK7), and the p38 pathway by phosphorylating MAPK kinase 3 and 6 (MKK3 and MKK6). These MKKs are then recruited into a multi-protein complex by scaffold proteins JNK Interacting Proteins (JIPs) 1–3 (Gallo & Johnson 2002). The homology between LRRK2, RIPs and MLKs lead us to hypothesize that LRRK2 might interact with the MKKs in a manner similar to these kinases.
Our data demonstrate interactions between LRRK2 and MKK 3, 6, and 7. We observed that LRRK2 binds to these MKKs through the COR and kinase domains, and phosphorylates the MKKs. The G2019S, R1441C and I2020T LRRK2 mutations increase the association of LRRK2 with MKK6. Binding of LRRK2 to MKK6 is associated with increased levels of both proteins in the plasma membrane and cytoplasm, and this change is dependent on the activity of MKK6. LRRK2 is able to phosphorylate MKK6. We validated the importance of MKK6 for LRRK2 function during mitochondrial stress by using RNAi knockdown or deletion in transgenic LRRK2 C. elegans. Our data suggest an intriguing mechanism by which MKK6 might regulate the function of LRRK2, and suggests a role for LRRK2 in the stress response.
MATERIALS AND METHODS
Constructs
Wild-type (WT) and mutant LRRK2 V5-His tagged constructs were generated as described in Greggio et al. (Greggio et al. 2006). All the pathogenic, kinase dead, and deletion LRRK2 constructs utilized in this study are illustrated schematically in Fig. S1. pRSV-Flag-MKK3, dominant negative pRSV-Flag-MKK3 (Ala), pcDNA3-Flag-MKK6, dominant negative pcDNA3-Flag-MKK6 (K82A), pcDNA3-Flag-MKK7, and dominant negative pcDNA3-Flag-MKK7 (K82A) were generous gifts from Dr. Roger Davis (University of Massachusetts). Recombinant MKK3, 6 and 7 were purchased from R&D Systems.
Cell culture
HEK-293 and HEK-293FT cells were maintained in DMEM (Invitrogen) supplemented with 10% FBS (Atlanta Biologicals), 1× non-essential amino acids, penicillin (100 units/ml), and streptomycin (100 µg/ml). Transfections were performed using FuGENE 6.0 reagent (Roche Applied Science) for HEK-293 cells and Lipofectamine 2000 (Invitrogen) for HEK-293FT cells.
Quantitative real-time PCR assay
HEK-293 cells were transfected with LRRK2 constructs and RNA was extracted 8 hours post-transfection using Qiagene RNeasy mini kit. Intact RNA was checked by running in 1.0% agarose gel and quantified spectrometrically (NanoDrop ND-1000) before proceeding to subsequent steps. 1 µg of total RNA was reverse-transcribed using iScript cDNA synthesis kit (Bio-Rad) according to manufacturer’s instructions. Real-time PCR was performed on a MyiQ™ Detection System (Bio-Rad) using iQ SYBR Green supermix. Expression levels for each target gene were calculated by the 2-ΔΔCT method (Livak & Schmittgen 2001). All analyses were performed in triplicates. Primers used for c-jun were 5'-GGAACAGGTGGCACAGCTTAAACA-3' (forward) and 5'-TTGCAACTGCTGCGTTAGCATGAG-3' (reverse) and for GAPDH were 5'TGGCCAAGGTCATCCATGACAACT-3' (forward) and 5'-CACAGTCTTCTGGGTGGCAGTGAT-3' (reverse).
Co-immunoprecipitation
HEK-293FT cells were co-transfected with V5-His tagged LRRK2 and Flag-tagged MKK constructs, and harvested in immunoprecipitation (IP) buffer consisted of 150 mM NaCl, 20 mM Hepes (pH = 7.4), 50 mM β-glycerophosphate, 5 mM sodium fluoride, 1% Triton X-100, 10% glycerol, 2 mM EGTA, 1 mM sodium orthovanadate, 1× protease inhibitor cocktail (Sigma) for LRRK2-MKK3 co-immunoprecipitation and 0.5% Triton-X, 1 mM EDTA in 1X PBS and 1X protease inhibitor cocktail (Sigma) for all other co-immunoprecipitations. The lysates were rotated at 4 °C for 30 minutes followed by centrifugation at 13,000g for 10 minutes. The supernatant was pre-cleared with Protein G Sepharose 4 Fast Flow (Amersham) followed with addition of the appropriate antibody and rotated overnight at 4 °C. Protein G Sepharose was added to pull down the antibody pre-complex. The Sepharose beads were stringently washed three times with IP buffer and once with 50 mM Tris (pH = 8) wash buffer. The immunoprecipitation proteins were eluted in lithium dodecyl sulphate (LDS) sample buffer (Invitrogen) by heating at 95 °C for 3 minutes. Immunoprecipitates were resolved by 3–8% NuPAGE Tris-Acetate or 4–20% Tris-Glycine gels (Invitrogen) and analyzed by western blot. Antibodies were purchased from Sigma (V5, Flag, c-Myc, actin), Calbiochem (Calnexin), and Biodesign (GAPDH).
For immunoaffinity chromatography, lysates (5 mg) were generated using the IP buffer, loaded on a column containing 400 µl anti-V5-agarose (Sigma), washed with IP buffer followed by 50 mM Tris (pH 8) wash buffer, and then eluted with V5-peptide (1 mg/ml, Sigma). Fractions were then analyzed by immunoblot as described above.
Subcellular fractionation
HEK-293FT cells were transfected with the appropriate constructs for 48 hours, washed with cold 1× PBS, and lysed with relaxation buffer consisted of 100 mM KCl, 3 mM NaCl, 3.5 mM MgCl2, 1.25 mM EGTA, 10 mM PIPES, and 1× protease inhibitor cocktail. The lysates were centrifuged at 500g for 5 minutes to eliminate cellular debris, and the supernatant was further centrifuged at 100,000g for 1 hour at 4 °C to separate the cytosolic fraction. The pellets were solublized in 200 µl of relaxation buffer to obtain the membrane fraction. Equal amounts of lysate were loaded and resolved by 3–8% NuPAGE Tris-Acetate or 4–20% Tris-Glycine gels for western blotting.
Kinase assays
Kinase assays were performed as described by Gloeckner et al (Gloeckner et al. 2009, Greggio et al. 2007). Recombinant LRRK2 was obtained from Invitrogen. The MKKs were generated by transfecting 293FT cells with Flag-MKK constructs, immunopurifying the Flag-MKK with anti-Flag M2 agarose and eluting with Flag peptide (25 µg peptide/run, using a 10 cm dish for each purification). 25 nM of recombinant LRRK2 (Invitrogen) was incubated with ~ 1µM of Flag-tagged MKK substrate protein in 30 µL assay buffer (25 mM Tris–HCl pH 7.5, 5 mM beta-glycerophosphate, 2 mM DTT, 0.1 mM Na3VO4, 10 mM MgCl2, Cell Signaling) supplemented with 5 µCi γ33P-ATP (3000 Ci/mmol; PerkinElmer Life Sciences). The reaction mix was incubated for 1 h at 30°C. The reaction was stopped by addition of 10 µL 5 × Laemmli buffer and samples were incubated for 5 min at 95°C prior to SDS gel-electrophoresis with subsequent immunoblotting, as needed.
Immunocytochemistry
HEK-293FT cells plated on glass coverslips were transfected with Lipofectamine 2000 at 50% confluency. After 48 hours, the transfected cells were fixed with 4% paraformaldehyde in 1× PBS for 10 minutes at room temperature. The cells were washed 3 times with PBS at 5 minutes each and were then incubated with 0.3% triton X-100 in PBS with 1% bovine serum albumin (BSA) for 1 hour at room temperature. The cells were incubated with 1:1000 dilution anti-V5 and anti-FLAG antibodies overnight at room temperature. After washing the cells 3 times with PBS at 5 minutes each, the cells were incubated with 1:750 dilution FITC-conjugated goat anti-rabbit antibody (Jackson ImmunoResearch) in 1× PBS. The washes were repeated with 1× PBS and the cells were incubated with 1:750 dilution Texas Red conjugated donkey anti-mouse antibody (Jackson ImmunoResearch). After washing the cells 3 times with 1× PBS at 5 minutes each, they were embedded in Antifade DAPI gel mounting medium (Invitrogen). Antibody against N-terminus APP (22C11, Calbiochem) was used as a marker for cell membranes.
Microscopy
The cells were visualized by three-dimensional multiple wavelength fluorescence microscopy using Olympus IX70 microscope equipped with SoftWor× 3.3.6, Deltavision 90000 (Applied Precision) for DAPI, FITC and Texas Red fluorescence at room temperature using an oil immersion 60× Olympus PlanApo objective (aperture set at 1.40). The images were captured using HP CoolSNAP camera, deconvolved using SoftWor× software (Applied Precision), and examined as either single sections or projections of an entire stack of optical sections. All images were processed using Adobe Photoshop CS2.
C. elegans
Transgenic LRRK2 C. elegans lines were generated as described by Saha et al (Saha et al. 2009). The WT LRRK2 wlzIs2 line was crossed with the sek-1 [km4] line and bred to homozygocity (Tanaka-Hino et al. 2002). The presence of LRRK2 and the km4 deletion were validated by PCR. RNAi knockdown was performed according to Kamath et al. (Kamath et al. 2001). Non-transgenic Bristol N2 and WT LRRK2 worms were allowed to lay eggs for 3 hours on NGM plates containing OP50 bacteria. After 3 days, synchronized young adult nematodes were moved to NGM plates to feed on HT115 bacteria containing either empty vector PL4440 or double-stranded RNAi expressing plasmid for 2 additional days. 25 adult worms were then moved to triplicate plates containing 25 µM rotenone and HT115 bacteria. NGM plates were made with 1 mM IPTG to keep the constant induction of RNAs. Nose-poke assays were performed to identify and quantify live worms over 4 days.
Statistical analysis
All data were expressed as mean ± SEM and graphed using Excel or Prism (GraphPad). One-way ANOVA and Tukey’s post hoc tests were performed to identify statistically significant differences using InStat (GraphPad).
RESULTS
LRRK2 binds to MKK3 and 6
HEK-293FT cells were transiently transfected with V5-His tagged WT LRRK2 and Flag-tagged MKK3 or MKK6. Protein complexes containing MKK6 (Fig. 1 A) or MKK3 (SI Fig. S2 A) were evident upon co-immunoprecipitation of LRRK2 or of the MKKs. Interestingly, co-transfecting LRRK2 and MKK6 increased the total levels of MKK6, as shown in the input lanes in Fig. 1A & C. Analysis of LRRK2 deletion mutants demonstrated that LRRK2 COR and kinase domains were essential and sufficient for binding to MKK6 (Fig. 1 B) and MKK3 (Fig. S2 B). In contrast, the LRR domain was not required for binding. Selectivity was also apparent in the study of MKK mutants, because a dominant negative MKK mutation K82A strongly reduced binding of LRRK2 to MKK6 (Fig. 1 C) but not MKK3 (Fig. S2 D). Finally, we also examined LRRK2 associated with endogenous MKK6. HEK-293 FT cells were transfected with empty vector or V5-tagged wild-type LRRK2. Lysates were collected in co-immunoprecipitation buffer, passed over an anti-V5 column and eluted with V5-peptide. Endogenous MKK6 was readily apparent in the eluate from V5-LRRK2 transfected lysates and absent from vector-transfected lysates (Fig.1D). Immuboblots of the cell lysates without immunoprecipitation also showed an increase in endogenous MKK6 levels upon transfection with LRRK2 (Fig. 1D), which was similar to the increase observed in Figures 1A & C. These results suggest that LRRK2 interacts with MKK6 as well as MKK3.
Fig. 1.
LRRK2 binds to MKK6, and pathogenic mutations in LRRK2 increase the binding of LRRK2 to MKK6. LRRK2 activates the p38 pathway. (A) LRRK2 co-immunoprecipitates with MKK6 in HEK-293FT cells. (B) LRRK2 kinase and COR domains are essential and sufficient for its interaction with MKK6. (C) Dominant negative K82A mutation in MKK6 reduces MKK6 binding to LRRK2. All experiments have been performed three times. D.) Eluate from anti-V5 column shows the association of endogenous MKK6 with LRRK2 (V5). Levels of each protein in the lysates are shown below.
LRRK2 binds to MKK7 but not MKK4
Because the JNK pathway is regulated via MKK4 and MKK7, we hypothesize that LRRK2 may also interact with these two MKKs. Co-immunoprecipitation experiments demonstrated that LRRK2 binds to MKK7 but not MKK4 (Fig. 2 A). The LRRK2 kinase and COR domains were essential and sufficient for interaction with MKK7 (Fig. 2 B), similar to that observed with MKK6 and MKK3. The dominant negative K82A mutation completely abolished the binding of MKK7 to LRRK2 (Fig. 2 C), suggesting that similar to the interaction between MKK6 and LRRK2, the LRRK2/MKK7 interaction is dependent on the activation of MKK7. We also observed that co-transfecting LRRK2 and MKK4 or 7 was associated with an increase in MKK levels similar to that seen with MKK6 (Fig. 2A & C).
Fig. 2.
LRRK2 binds to MKK7 but not MKK4 and activates the JNK pathway. (A) LRRK2 co-immunoprecipitates with MKK7 but not MKK4 in HEK-293FT cells. (B) LRRK2 kinase and COR domains are essential and sufficient for its interaction with MKK7. (C) Dominant negative K82A mutation in MKK7 reduces MKK7 binding to LRRK2. All experiments have been performed three times.
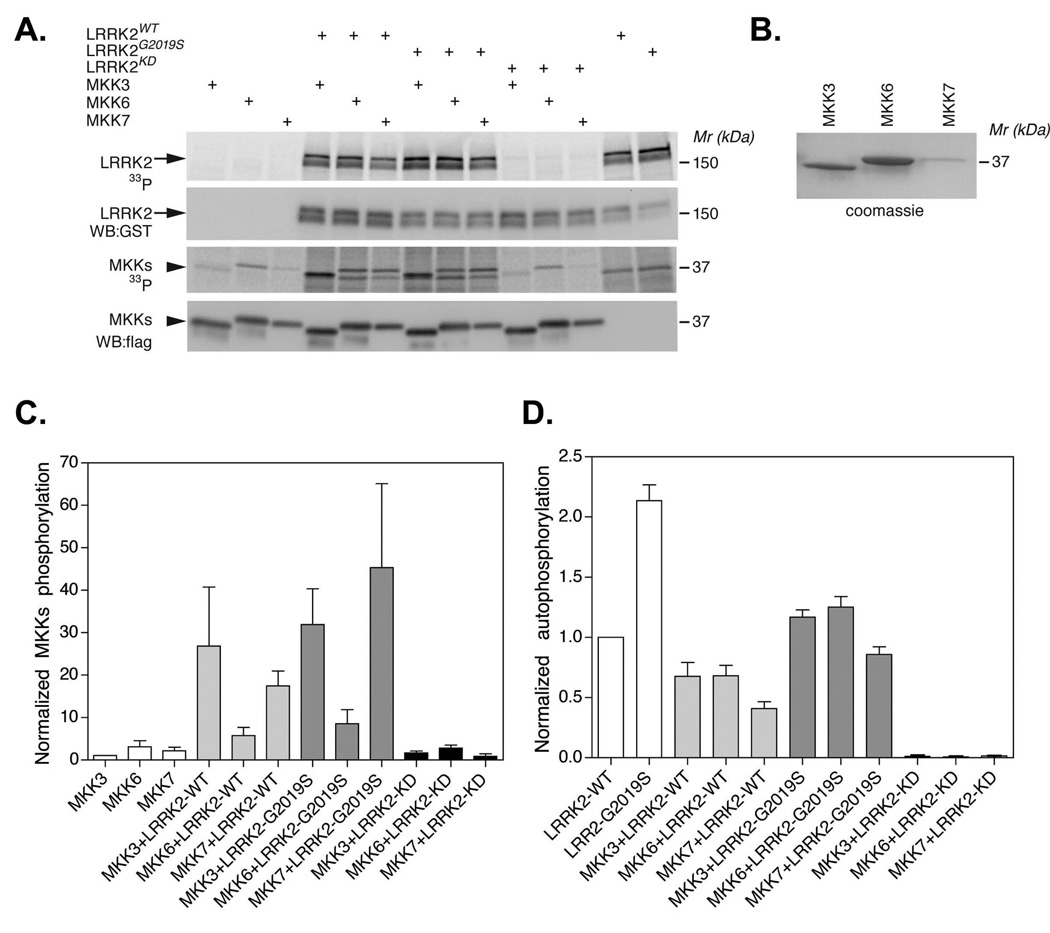
LRRK2 weakly phosphorylates MKKs
The association of LRRK2 with the MKKs led us to hypothesize that these two families of kinases could potentially be mutual substrates. Recent studies indicate that LRRK2 phosphorylates recombinant MKKs (Gloeckner et al. 2009). To test this hypothesis, radioactive kinase assays were performed using Flag-tagged MKK3, 6 or 7 immunoprecipitated from HEK-293FT cells and recombinant LRRK2 (fig. 3A & B). Phosphorylation measured using 32P-ATP (fig. 3A, C & D). Incubating WT or G2019S LRRK2 with MKK3, 6 or 7 moderately stimulated phosphorylation of MKK3 and MKK7, but had only marginal effects on MKK6 (fig. 3C). Phosphorylation required LRRK2 kinase activity because kinase dead LRRK2 was not able to stimulate MKK phosphorylation (fig. 3A & C). Interestingly, each of the MKKs appeared to reduce autophosphorylation of G2019S LRRK2 (fig. 3D). These data provide evidence that LRRK2 can modify MKK function, and vice-versa. The amount of phosphorylation was modest, and no changes in phosphorylation of the downstream kinases, JNK or p38, was observed upon over-expressing WT or G2019S LRRK2 (data not shown), which is consistent with prior reports (Liou et al. 2008).
Fig. 3.
LRRK2 stimulates MKK phosphorylation. A. Recombinant MKK3, 6 or 7 (Flag-tagged) was incubated ± LRRK2 (WT, G2019S or KD, GST-tagged), in the presence of kinase buffer and 32P-ATP. B. Coomassie gel showing the purity of the recombinant MKK3, 6 and 7. C & D. Quantification of MKK phosphorylation (C) and LRRK2 autophosphorylation (D) by densitometry. Error bars show the SEM from three independent experiments.
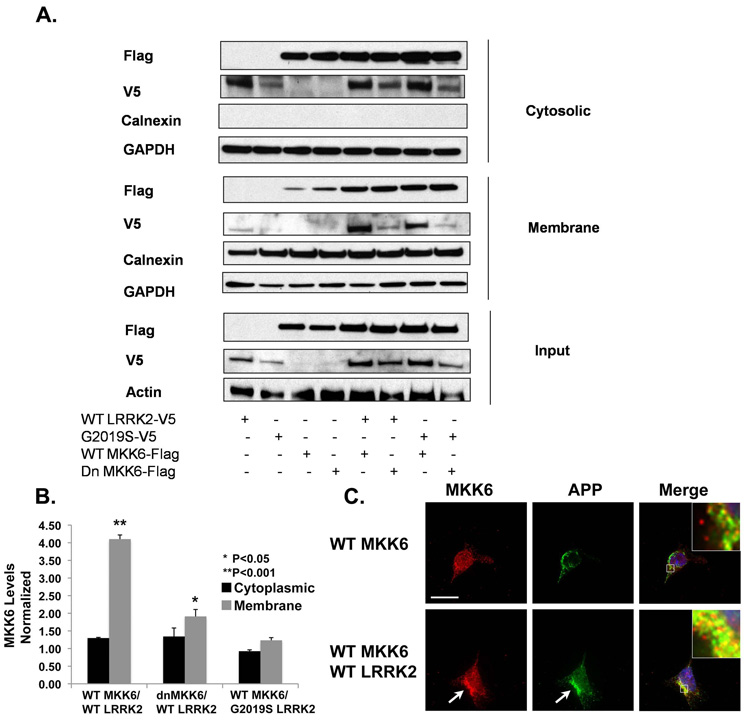
Co-expressing LRRK2 and MKK6 increases levels of MKK6 in the membrane more than in the cytoplasm
The activity of the MAPK cascades is also regulated by factors other than phosphorylation. The regulation of MAPK specificity frequently requires the tethering of appropriate kinases to their scaffold proteins to form a stimuli-specific module, and such module is then targeted to appropriate subcellular compartment for the activation of downstream signals (Garrington & Johnson 1999). We hypothesized that LRRK2 might also act as a scaffold protein to recruit MKKs to specific subcellular compartments. To test this hypothesis, we investigated whether co-expression of LRRK2 altered the distribution of MKK6. HEK-293FT cells were transfected with WT LRRK2 and WT or dominant negative MKK6, and the lysates fractionated into cytosolic and membrane fractions (Fig. 4A). Expressing WT LRRK2 increased levels of MKK6 in the membrane fraction approximately 4-fold (Fig. 4 A & B). Total MKK6 levels were also increased by co-expression with LRRK2, but the increase (~50%) was less than the change seen in the membrane fraction (Fig. 4A). LRRK2 expression appeared to be somewhat reduced when co-expressed with dominant negative MKK6 compared to WT MKK6, but this occurred in both the cytoplasm and membrane fractions (fig. 4A). To examine subcellular localization of the MKKs, we focused on the interaction with MKK6 using imaging, and performed immunocytochemistry by utilizing an antibody against N-terminus of amyloid precursor protein (22C11) to label the membrane. Co-localization of MKK6 with the membrane marker (amyloid precursor protein) increased in cells co-transfected with LRRK2 and MKK6, compared to cells expressing MKK6 alone (fig. 4C); the pattern of reactivity suggested localization at the plasma membrane.
Fig. 4.
Expressing LRRK2 increases the level of MKK6 in the membrane fraction. (A) Both WT and G2019S LRRK2 increased the level of membrane MKK6. (B) Quantification indicated that the level of membrane MKK6 increased about 4-fold when expressed along with LRRK2 (*p<0.05; N=3, compared to WT MKK6). There was no significant difference in the level of membrane MKK6 between the WT or G2019S LRRK2-transfected samples. The level of MKK6 in the membrane fraction was normalized to the level of MKK6 in the total lysate, and compared between samples transfected ± LRRK2. Experiments were repeated three times in HEK-293FT cells. (C) Co-expression of WT and G2019S LRRK2 with WT MKK6 increased levels of MKK6 present at the plasma membrane, as shown by co-localization with amyloid precursor protein (APP). The arrows point to LRRK2/APP co-localization. Bar, 15 µm. All experiments have been performed three times.
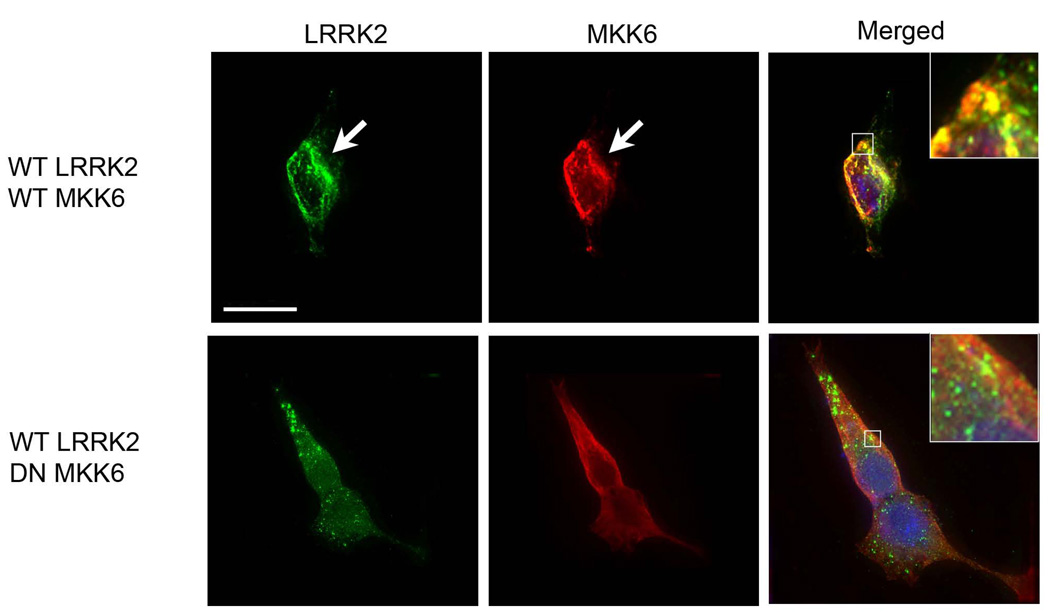
We also examined whether co-expression with MKK6 changed the subcellular localization of LRRK2. The fractionation studies showed increases in both cytoplasmic and membrane levels of LRRK2 (fig. 4A). We proceeded to examine this question by Immunocytochemistry. Cells that co-expressed LRRK2 and MKK6 showed a pattern of reactivity with a more distinct “rim” of reactivity, compared to cells expressing LRRK2 cells expressing dnMKK6 (fig. 5). These data are consistent with increased membrane binding, however since MKK6 also increases total levels of LRRK2, microscopy alone cannot rule-out that the changes result simply from increased expression.
Fig. 5.
Co-expression of LRRK2 and MKK6 leads to co-localization. HEK 293 cells were co-transfected with LRRK2 (WT or G2019S) ± MKK6 (WT or dominant negative). The proteins were subsequently identified by immunocytochemistry. WT and G2019S LRRK2 co-localize with WT MKK6. This co-localization is reduced with dominant negative MKK6, suggesting the importance of MKK6 function for its interaction with LRRK2. The arrows point to areas of LRRK2 membrane localization. Bar, 15 µm. All experiments have been performed three times.
MKK6 and p38 orthologs in C. elegans are required for LRRK2 function
To gain insight into the role of MKK6 in LRRK2 function in vivo and to examine whether endogenous MKKs regulated LRRK2 function in neurons, we utilized transgenic C. elegans lines that express human WT LRRK2 driven by the synaptobrevin promoter, which is a neuron-specific promoter. We recently generated lines of C. elegans expressing LRRK2 and showed that LRRK2 complements the function of its C. elegans ortholog, lrk-1 in a rotenone-protection assay (Saha et al. 2009). Nematode lines expressing WT LRRK2 were strongly protected against toxicity of rotenone, a mitochondrial complex I inhibitor (Saha et al. 2009). The strong phenotype associated with LRRK2 expression provides a robust assay to test the genetic pathways mediating LRRK2 function.
RNAi was used to knock down the C. elegans MKK6/3 ortholog sek-1 and its downstream target, the p38 MAPK ortholog pmk-1. Non-transgenic Bristol N2 or WT LRRK2 C. elegans were exposed to RNAi feeding vectors coding for sek-1 or pmk-1 and then exposed to 25 µM rotenone for 4 days while maintaining RNAi treatment. We observed that nematodes expressing LRRK2 were protected against rotenone (Fig. 6), which is consistent with our prior results (Saha et al. 2009). Knockdown of sek-1 (Fig. 6A) and pmk-1 (Fig. 6B) completely abolished the ability of WT LRRK2 to protect the C. elegans against rotenone, while control empty vector had no effect. Analysis of mRNA levels by real time PCR indicated a 60 ± 9% (P < 0.01, N=3) decrease in expression for the sek-1 transcript and a similar decrease for pmk-1. The role of MKK3/6 (sek-1) was confirmed by deleting the sek-1 gene from the WT LRRK2 wlzIs2 by crossing with the sek-1 [km4] C. elegans line, which carries a deletion of sek-1 exons 4–6 (www.wormbase.org), and breeding to homozygocity (Tanaka-Hino et al. 2002). The effects on rotenone toxicity were then examined. The LRRK2 wlzIs2 line showed strong resistance to rotenone toxicity, while the LRRK2/sek-1 [km4] line showed no protection (Fig. 6C). Finally, we examined whether endogenous lrk-1 was required for protection by sek1 or pmk-1. To determine whether sek-1 and pmk-1 required lrk-1 to modify rotenone toxicity, we examined the effects of sek-1 and pmk-1 knockdown on the lrk-1 [km17] C. elegans line that has a deletion in lrk-1. The lrk-1 [km17] line was highly sensitive to rotenone (25 µM, 4 days), as expected. In addition, pmk-1 knockdown provided no protection and the effect of sek-1 knockdown was much less than observed in the N2 line that contains active lrk-1 (fig. 6D for the lrk-1[km17] line, vs. fig. 6A&B for the lines containing lrk-1). Taken together these data suggest that lrk-1 and LRRK2 require sek-1 and pmk-1 for action (the C. elegans homologues of MKK6 and p38), and vice versa.
Fig. 6.
Knockdown of C. elegans MKK6/3 ortholog sek-1 (A) and p38 ortholog pmk-1 (B) sensitize WT LRRK2 C. elegans while protecting N2 worms against rotenone. (*p<0.001; #p<0.001 compared to Day 0 N2/empty vector). (C) Crossing LRRK2 to Sek-1 [km4] abolishes protection against rotenone seen with LRRK2 alone (*P<0.0001, LRRK2, day 2 vs. LRRK2/Sek-1 [km4], day 2). All experiments have been performed three times. (D) The lrk-1 [km17] nematode line, which have a deletion in lrk-1, are highly sensitive to rotenone. Knockdown of pmk-1 does not protect against toxicity, and knockdown of sek-1 protects only marginally (N=12). These effects of sek-1 and pmk-1 knockdown are much less than observed with the N2 line.
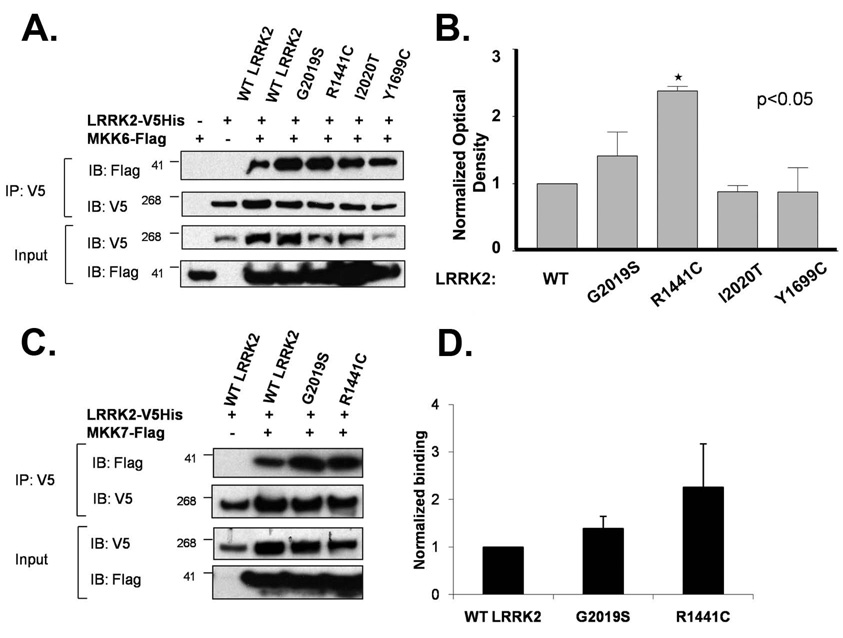
Disease-linked mutations in LRRK2 modify binding
Finally, we tested whether mutations in LRRK2 that are associated with disease modified the interaction of LRRK2 with the MKKs. HEK 293 cells were transfected with LRRK2 (WT, G2019S, R1441C, Y1699C or I2020T) and MKK6. LRRK2 was then immunoprecipitated and binding to MKK6 quantified by immunoblot (Fig. 7A & B). The G2019S, R1441C and I2020T mutations each enhanced binding of LRRK2 to MKK6, but the Y1699C mutation did not alter binding (Fig. 7 A & B). The enhanced binding to the mutant LRRK2 exerted stronger effects on MKK6 than on MKK3 or 7 binding; the R1441C and G2019S mutations did not show statistically significant increases in binding of LRRK2 to MKK7 (Fig. 7 C & D) or to MKK3 (Fig. S2 C).
Fig. 7.
The G2019S, R1441C and I2020T LRRK2 mutations increase binding to MKK6 (A) Comparison of binding of WT and mutant LRRK2 constructs (G2019S, R1441C, I2020T and Y1699C) to MKK6 by immunoprecipitation. (B) Quantification of binding of LRRK2 constructs to MKK6 (*p<0.01, N=3). Experiments were repeated four times in HEK-293FT cells. Binding to MKK6 was normalized to levels of LRRK2 in each pull-down. (C) Pathogenic mutations in LRRK2 did not significantly increase the binding of MKK7 to LRRK2. (D) Quantification of binding of LRRK2 constructs to MKK7 (*p<0.01, N=3). Experiments were repeated three times in HEK-293FT cells. All experiments have been performed three times.
DISCUSSION
Our results indicate that LRRK2 interacts with MKK3, 6 and 7. We provide multiple independent lines of investigation supporting the hypothesis that LRRK2 interacts with the MKKs. LRRK2 binds to MKK3, 6, and 7, and the R1441C LRRK2 mutation increases binding to MKK6. LRRK2 phosphorylates MKK3 and MKK7 in vitro but has a relatively minor effect on MKK6 phosphorylation. Co-expression of LRRK2 and MKK6 increased levels of each protein beyond that observed with expression of either protein alone. Co-expression increased levels of LRRK2 and MKK6 more in the membrane than in the cytoplasm. Membrane localization of LRRK2 and MKK6 is evident using biochemical fractionation, as well as with immunocytochemical studies. Finally, these cellular studies are complemented by studies showing that knockdown or deletion of MKK6 (sek-1) and p38 (pmk-1) in C. elegans abrogates LRRK2-mediated protection during rotenone toxicity, which demonstrates that endogenous MKK6 is required for LRRK2 function. Together, these data point to a role for the MKKs and JNK/p38 pathways in LRRK2 function. Our data are consistent with a hypothesis that the interaction between MKKs and LRRK2 affects two different biochemical processes, protein translation (or degradation) and membrane association.
We observed robust binding of LRRK2 to MKK3, 6 and 7, but not to MKK4. The structures of MKK4 and other MKK2 differ at the N-termini, which are not conserved, and in three short sequences (MKK4 amino acids 193-9, 278–283 and 329-35) that are conserved for MKK 3, 6 and 7 but not for MKK4. Future studies will determine whether any of these domains contribute to specificity for LRRK2 binding. The domains on LRRK2 that regulate binding were identified through deletion studies showed similar binding patterns for MKK3, 6 and 7. Each of the MKKs bound to LRRK2 deletion constructs containing the COR and kinase domains, but not to the LRR domain of LRRK2. Other studies also highlight the importance of the ROC and COR domains. The ROC domain of LRRK2 contributes to its homo-dimerization (Greggio et al. 2008), although sequences outside of the ROC-COR bi-domain are also required for dimer formation (Klein et al. 2009, Greggio et al. 2008). LRRK2 binds CHIP, a ubiquitin ligase that binds Hsp-70, and this interaction occurs via the COR and ROC domains (Ko et al. 2009). The ROC and COR domains (as well as the kinase domain) are also necessary for phosphorylation of moesin (Jaleel et al. 2007). Several other putative LRRK2 binding proteins that have also been identified, including Fas binding protein, Hsp90, 4E-BP and JIP1-4, however none of these studies analyzed the domain requirements for binding (Ho et al. 2009, Dachsel et al. 2007, Imai et al. 2008, Hsu et al. 2009). Thus, the ROC and COR domains appear to play particularly important roles in the association of LRRK2 with other proteins.
Our study identifies a new group of proteins that regulate both the levels of LRRK2 and the membrane association of LRRK2. Co-expressing LRRK2 with the MKKs increased total levels of LRRK2 and MKKs (fig. 1, 2 & 4). We hypothesize that these changes are regulated at the level of translation or degradation because expression was increased for both the transgenic proteins (LRRK2 and MKKs), which are regulated by the constitutive CMV promoter, and for endogenous MKK6 (fig. 1). Prior studies indicate that LRRK2 binds the protein 4E-BP, which inhibits protein translation, and that stimulating 4E-BP with rapamycin can suppress phenotypes associated with PD (Imai et al. 2008, Tain et al. 2009). We have recently confirmed that LRRK2 phosphorylates 4E-BP, although phosphorylation was only observed in vitro, and not in cell systems (Kumar et al. 2009). Interestingly, we found that p38, a potential effector of MKK activity is a more convincing kinase than LRRK2 for 4EBP. Whether direct or mediated through intermediate kinases, the interaction of LRRK2 with 4E-BP provides a link between LRRK2 and protein translation that could contribute to the changes in LRRK2 levels associated with co-expressing MKKs. Further studies will be needed to clarify whether the LRRK2 complex includes both MKKs and 4E-BP or other proteins linked to protein translation.
LRRK2 localizes with membranes but the significance of this localization is poorly understood (West et al. 2005, Biskup et al. 2006, Higashi et al. 2007). Our studies suggest that co-transfecting LRRK2 with MKK6 (and possibly other MKKs) increases membrane association of MKK6. Whether there is also an increase in LRRK2 membrane localization remains ambiguous because the increase in total LRRK2 associated with MKK co-transfection could account for any apparent increase in LRRK2 membrane localization. Although the hypothesis that the association of LRRK2 with membranes might regulate localization of other proteins is new, multiple prior studies provide clear evidence for localization of LRRK2 with membranes. Studies from Hatana et al. (Hatano et al. 2007) indicate that LRRK2 localizes in the plasma membrane and lipid rafts, while studies by several groups demonstrate that LRRK2 associates with organelle membranes (West et al. 2005, Biskup et al. 2006, Higashi et al. 2007). The observation that LRRK2 regulates MKK localization also complements recent studies from our laboratory indicating that LRRK2 binds to JIP1 – 4, which are scaffold proteins that bring together MKKs with MAPKs (e.g., JNK and p38) (Hsu et al. 2009). JIPs appear to function in directing the MAP kinase signaling complex to different sites within the cells (Whitmarsh 2006). We hypothesize that LRRK2 acts as part of this complex to facilitate localization of the MKK/MAPK proteins to sites of stress.
The actions of LRRK2 appear to differ from that of RIPs and MLKs, despite moderate homology with these proteins. The RIPs and MLKs are thought to function in a multi-tiered system by phosphorylating an appropriate downstream MAPKK, which in turn phosphorylates and activates JNK or p38 to regulate specific transcription factors and cellular responses. This system contrasts with the relatively modest phosphorylation of the MKKs by LRRK2. We were able to repeat the observations of Gloeckner et al showing that LRRK2 phosphorylates MKKs 3, 6 and 7. However, the concentration of MKKs needed to elicit phosphorylation of MKKs is quite high (40-fold greater than that of LRRK2), which suggests only a small fraction of the MKK is actually phosphorylated by LRRK2, reminiscent of recent observations of a low level of 4EBP phosphorylation by LRRK2. The low level of phosphorylation might explain why no changes in MKK, JNK or p38 phosphorylation were evident upon co-transfection of the two proteins.
The modest effects of the kinase domain are consistent with results observed in C. elegans and Drosophila (Saha et al. 2009, Wang et al. 2008). In C. elegans, we found that kinase dead LRRK2 was also functional and protected against toxicity induced by rotenone, although less so than WT LRRK2 (Saha et al. 2009). Similar results were observed in Drosophila, where Wang and colleagues observed that deletion of the kinase domain from the Drosophila LRRK2 homolog protected against toxicity induced by rotenone or paraquat (Wang et al. 2008). These studies suggest that LRRK2 exhibits multiple physiological actions, some of which are independent of kinase activity. This type of kinase-independent regulation is similar to that observed with RIP kinases, which are the closest homologues of LRRK2 (Festjens et al. 2007). RIP-1 activates NF-kB in response to DNA damage through a mechanism that does not require RIP-1 kinase activity (Festjens et al. 2007). However, other pathways regulated by RIP-1, such as the TNF-receptor/MEK-1/ERK cascade, require RIP-1 kinase activity (Festjens et al. 2007).
Studies of LRRK2 are beginning to illuminate the functional role of LRRK2. Experiments in mammalian cells indicate that LRRK2 potentiates peroxide-induced neurotoxicity in a kinase dependent manner (West et al. 2007). The studies in vivo support a role for LRRK2 in the response to stress, but differ in the details (Saha et al. 2009, Wang et al. 2008). We observed that WT LRRK2 protects against toxicity induced by rotenone or paraquat in C. elegans, both of which inhibit complex I of the electron transport chain (Saha et al. 2009). The G2019S and R1441C LRRK2 mutants show reduced protection against mitochondrial inhibition and actually increase degeneration of dopaminergic neurons under basal conditions and during mitochondrial inhibition (Saha et al. 2009). LRRK2 expression in C. elegans complements the function of lrk-1, the homolog of LRRK2 in C. elegans, and knockdown or deletion of lrk-1 potentiates rotenone toxicity (Saha et al. 2009, Sakaguchi-Nakashima et al. 2007). Wang and colleagues observed similar results, although their study pointed to a selective role for LRRK2 in protecting against toxicity induced by hydrogen peroxide (Wang et al. 2008). The selective toxicity that we observe for mutant LRRK2 is consistent with studies in transgenic mice, rats and in neuronal SY5Y cells which all suggest that mutant forms of LRRK2 are much more toxic than WT LRRK2 (Li et al. 2009, Greggio et al. 2006, Greggio et al. 2007, Macleod et al. 2006, Smith et al. 2005). In the current manuscript, we extend these observations by demonstrating that the LRRK2-mediated stress response requires the activity of the MKK6/sek-1 and p38/pmk-1 pathway, because blocking this pathway abrogated protection by LRRK2 in C. elegans. Knockdown of MKK6/sek-1 or p38/pmk-1 protects against rotenone-mediated toxicity in non-transgenic nematodes, and brings protection to a level equal to that seen with LRRK2 expression. Conversely, knockdown of MKK6/sek-1 or p38/pmk-1 in LRRK2 nematodes effectively abrogates the protection by LRRK2 and brings protection down to the level of non-transgenic nematodes. These data are consistent with a hypothesis that LRRK2 acts through a mechanism involving MKK6/sek-1 and p38/pmk-1.
Whether the MAPK cascade contributes to disease remains unclear. In the current study, the G2019S, R1441C and I2020T LRRK2 mutant constructs all showed increased binding to MKKs, although the Y1699C LRRK2 mutation did not show a statistically significant increase in binding. Binding to MKK3 and 7 did not show statistically significant changes, although the apparent mutation-linked differences raises the possibility that future studies using larger sample sizes could identify significant effects. These results parallel most other binding studies, which do not identify specific binding interaction that is altered by a large panel of disease-linked mutations (Imai et al. 2008, Ko et al. 2009, Ho et al. 2009, Dachsel et al. 2007, Jaleel et al. 2007); (Hsu et al. 2009). Studies of phosphorylation and of inclusion formation also do not show effects that are consistent across all disease-linked mutations (Greggio et al. 2006, Greggio et al. 2007, Smith et al. 2005, Gloeckner et al. 2009, Greggio et al. 2008, Ito et al. 2007, West et al. 2005). The notable exception to this trend is binding of LRRK2 to Fas-associated protein, which does show robust increases in binding with a panel of four different LRRK2 mutations (Ho et al. 2009). The Fas-associated protein utilizes a signal transduction pathway mediated by RIP kinases and also mediated by MKKs (Festjens et al. 2007). Thus, a Fas-mediated pathway involving MKKs might contribute to the pathophysiology of PD, but much more information is needed before such a conclusion can be stated with strong confidence.
The increasing evidence linking LRRK2 function to the stress response, and in particular to the MAPK pathways, illuminates its biological function and might ultimately reveal biological mechanisms linked to PD. Many studies have shown that p38 and JNK actions are crucial for the regulation of cell death in PD models. CHOP/Gadd153, a transcription factor downstream of p38, was up-regulated by both 6-OHDA and 1-methyl-4-phenylpyridinium (MPP+) treatments in a dopaminergic cell line. Manganese and 6-OHDA treatments also strongly activate p38 and JNK pathways by increasing phosphorylated p38 and c-jun (Holtz & O'Malley 2003). c-Jun is activated in dopaminergic neurons from PD patients and in 1-methyl-4-phenyl-1,2,4,6 tetrahydropyridine (MPTP) mouse model of PD. JNK2 and JNK3, but not JNK 1, are required for such MPTP-induced c-jun activation because the elimination of JNK2 and JNK3 prevent neurodegeneration and improve motor function of MPTP-treated mice (Hunot et al. 2004). Inhibitors of JNK and MLK such as CEP1347 have been shown to be protective in various in vitro and in vivo PD models (Roux et al. 2002, Silva et al. 2005). Our data suggest a role of LRRK2 in the p38 and JNK cascades and the response to stresses that are related to PD.
Supplementary Material
Fig. S1. Schematic of LRRK2 structure and constructs utilized in this study.
Fig. S2. LRRK2 binds to MKK3. (A) LRRK2 co-immunoprecipitates with MKK3 in HEK-293FT cells. (B) LRRK2 kinase and COR domains are essential and sufficient for its interaction with MKK3. (C) Pathogenic mutations in LRRK2 did not increase the binding of MKK3 to LRRK2. Experiments were repeated three times in HEK-293FT cells. (D) WT and dominant negative MKK3 showed similar degrees of binding to LRRK2.
Acknowledgements
We thank Roger Davis for his generosity in sharing the MKK plasmids, and Leonard Petrucelli for his valuable advice. We also thank Shelley Russek and Ramona Faris for their assistance with the luciferase assays, and Andrew Ferree with help in editing the manuscript. This work is supported by NIEHS ES15567, NINDS NS41786, the Alzheimer Association and the Michael J. Fox Foundation (B.W.). This research was supported in part by the Intramural Research Program of the NIH, National Institute on Aging, Z01AG000948 (M.R.C.).
ABBREVIATIONS
- COR
carboxy-terminal of Ras
- JIP
c-Jun interacting protein
- JNK
c-Jun kinase
- LRRK2
leucine-rich repeat kinase 2
- MAPK
mitogen-activating protein kinase, MAPK
- MKK
MAPK kinase
- MLK
mixed-lineage kinase
- PD
Parkinson’s disease
- RIP
Receptor Interacting Protein kinase
- ROC
Ras of complex
REFERENCES
- Biskup S, Moore DJ, Celsi F, et al. Localization of LRRK2 to membranous and vesicular structures in mammalian brain. Ann Neurol. 2006;60:557–569. doi: 10.1002/ana.21019. [DOI] [PubMed] [Google Scholar]
- Dachsel JC, Taylor JP, Mok SS, et al. Identification of potential protein interactors of Lrrk2. Parkinsonism Relat Disord. 2007;13 doi: 10.1016/j.parkreldis.2007.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Festjens N, Vanden Berghe T, Cornelis S, Vandenabeele P. RIP1, a kinase on the crossroads of a cell's decision to live or die. Cell Death Differ. 2007;14:400–410. doi: 10.1038/sj.cdd.4402085. [DOI] [PubMed] [Google Scholar]
- Gallo KA, Johnson GL. Mixed-lineage kinase control of JNK and p38 MAPK pathways. Nat Rev Mol Cell Biol. 2002;3:663–672. doi: 10.1038/nrm906. [DOI] [PubMed] [Google Scholar]
- Garrington TP, Johnson GL. Organization and regulation of mitogen-activated protein kinase signaling pathways. Curr Opin Cell Biol. 1999;11:211–218. doi: 10.1016/s0955-0674(99)80028-3. [DOI] [PubMed] [Google Scholar]
- Gloeckner CJ, Schumacher A, Boldt K, Ueffing M. The Parkinson disease-associated protein kinase LRRK2 exhibits MAPKKK activity and phosphorylates MKK3/6 and MKK4/7, in vitro. J Neurochem. 2009;109:959–968. doi: 10.1111/j.1471-4159.2009.06024.x. [DOI] [PubMed] [Google Scholar]
- Greggio E, Cookson MR. Leucine-rich repeat kinase 2 mutations and Parkinson's disease: three questions. ASN Neuro. 2009;1 doi: 10.1042/AN20090007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greggio E, Jain S, Kingsbury A, et al. Kinase activity is required for the toxic effects of mutant LRRK2/dardarin. Neurobiol Dis. 2006;23:329–341. doi: 10.1016/j.nbd.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Greggio E, Lewis PA, van der Brug MP, Ahmad R, Kaganovich A, Ding J, Beilina A, Baker AK, Cookson MR. Mutations in LRRK2/dardarin associated with Parkinson disease are more toxic than equivalent mutations in the homologous kinase LRRK1. J Neurochem. 2007;102:93–102. doi: 10.1111/j.1471-4159.2007.04523.x. [DOI] [PubMed] [Google Scholar]
- Greggio E, Zambrano I, Kaganovich A, et al. The Parkinson disease-associated leucine-rich repeat kinase 2 (LRRK2) is a dimer that undergoes intramolecular autophosphorylation. J Biol Chem. 2008;283:16906–16914. doi: 10.1074/jbc.M708718200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatano T, Kubo S, Imai S, Maeda M, Ishikawa K, Mizuno Y, Hattori N. Leucine-rich repeat kinase 2 associates with lipid rafts. Hum Mol Genet. 2007;16:678–690. doi: 10.1093/hmg/ddm013. [DOI] [PubMed] [Google Scholar]
- Higashi S, Moore DJ, Colebrooke RE, Biskup S, Dawson VL, Arai H, Dawson TM, Emson PC. Expression and localization of Parkinson's disease-associated leucine-rich repeat kinase 2 in the mouse brain. J Neurochem. 2007;100:368–381. doi: 10.1111/j.1471-4159.2006.04246.x. [DOI] [PubMed] [Google Scholar]
- Ho CC, Rideout HJ, Ribe E, Troy CM, Dauer WT. The Parkinson disease protein leucine-rich repeat kinase 2 transduces death signals via Fas-associated protein with death domain and caspase-8 in a cellular model of neurodegeneration. J Neurosci. 2009;29:1011–1016. doi: 10.1523/JNEUROSCI.5175-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtz WA, O'Malley KL. Parkinsonian mimetics induce aspects of unfolded protein response in death of dopaminergic neurons. J Biol Chem. 2003;278:19367–19377. doi: 10.1074/jbc.M211821200. [DOI] [PubMed] [Google Scholar]
- Hsu C, Chan D, Wolozin B. LRRK2 and the Stress Response: Interaction with MKKs and JIPs. Neurodegener Dis. 2009 doi: 10.1159/000285509. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunot S, Vila M, Teismann P, Davis RJ, Hirsch EC, Przedborski S, Rakic P, Flavell RA. JNK-mediated induction of cyclooxygenase 2 is required for neurodegeneration in a mouse model of Parkinson's disease. Proc Natl Acad Sci U S A. 2004;101:665–670. doi: 10.1073/pnas.0307453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iaccarino C, Crosio C, Vitale C, Sanna G, Carri MT, Barone P. Apoptotic mechanisms in mutant LRRK2-mediated cell death. Hum Mol Genet. 2007;16:1319–1326. doi: 10.1093/hmg/ddm080. [DOI] [PubMed] [Google Scholar]
- Imai Y, Gehrke S, Wang HQ, Takahashi R, Hasegawa K, Oota E, Lu B. Phosphorylation of 4E-BP by LRRK2 affects the maintenance of dopaminergic neurons in Drosophila. Embo J. 2008;27:2432–2443. doi: 10.1038/emboj.2008.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito G, Okai T, Fujino G, Takeda K, Ichijo H, Katada T, Iwatsubo T. GTP Binding Is Essential to the Protein Kinase Activity of LRRK2, a Causative Gene Product for Familial Parkinson's Disease. Biochemistry. 2007;46:1380–1388. doi: 10.1021/bi061960m. [DOI] [PubMed] [Google Scholar]
- Jaleel M, Nichols RJ, Deak M, Campbell DG, Gillardon F, Knebel A, Alessi DR. LRRK2 phosphorylates moesin at threonine-558: characterization of how Parkinson's disease mutants affect kinase activity. Biochem J. 2007;405:307–317. doi: 10.1042/BJ20070209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamath RS, Martinez-Campos M, Zipperlen P, Fraser AG, Ahringer J. Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2001;2 doi: 10.1186/gb-2000-2-1-research0002. RESEARCH0002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein CL, Rovelli G, Springer W, Schall C, Gasser T, Kahle PJ. Homo- and heterodimerization of ROCO kinases: LRRK2 kinase inhibition by the LRRK2 ROCO fragment. J Neurochem. 2009;111:703–715. doi: 10.1111/j.1471-4159.2009.06358.x. [DOI] [PubMed] [Google Scholar]
- Ko HS, Bailey R, Smith WW, et al. CHIP regulates leucine-rich repeat kinase-2 ubiquitination, degradation, and toxicity. Proc Natl Acad Sci U S A. 2009;106:2897–2902. doi: 10.1073/pnas.0810123106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Greggio E, Beilina A, Kaganovich A, Chan D, Wolozin B, Cookson M. The Parkinson’s disease associated LRRK2 exhibits weaker in vitro phosphorylation of 4E-BP compared to autophosphorylation. PLoS One. 2009 doi: 10.1371/journal.pone.0008730. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis PA, Greggio E, Beilina A, Jain S, Baker A, Cookson MR. The R1441C mutation of LRRK2 disrupts GTP hydrolysis. Biochem Biophys Res Commun. 2007;357:668–671. doi: 10.1016/j.bbrc.2007.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Tan YC, Poulose S, Olanow CW, Huang XY, Yue Z. Leucine-rich repeat kinase 2 (LRRK2)/PARK8 possesses GTPase activity that is altered in familial Parkinson's disease R1441C/G mutants. J Neurochem. 2007;103:238–247. doi: 10.1111/j.1471-4159.2007.04743.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Liu W, Oo TF, et al. Mutant LRRK2(R1441G) BAC transgenic mice recapitulate cardinal features of Parkinson's disease. Nat Neurosci. 2009 doi: 10.1038/nn.2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou AK, Leak RK, Li L, Zigmond MJ. Wild-type LRRK2 but not its mutant attenuates stress-induced cell death via ERK pathway. Neurobiol Dis. 2008;32:116–124. doi: 10.1016/j.nbd.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Macleod D, Dowman J, Hammond R, Leete T, Inoue K, Abeliovich A. The Familial Parkinsonism Gene LRRK2 Regulates Neurite Process Morphology. Neuron. 2006;52:587–593. doi: 10.1016/j.neuron.2006.10.008. [DOI] [PubMed] [Google Scholar]
- Olanow CW, Stern MB, Sethi K. The scientific and clinical basis for the treatment of Parkinson disease (2009) Neurology. 2009;72:S1–S136. doi: 10.1212/WNL.0b013e3181a1d44c. [DOI] [PubMed] [Google Scholar]
- Plowey ED, Cherra SJ, 3rd, Liu YJ, Chu CT. Role of autophagy in G2019S-LRRK2-associated neurite shortening in differentiated SH-SY5Y cells. J Neurochem. 2008;105:1048–1056. doi: 10.1111/j.1471-4159.2008.05217.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux PP, Dorval G, Boudreau M, Angers-Loustau A, Morris SJ, Makkerh J, Barker PA. K252a and CEP1347 are neuroprotective compounds that inhibit mixed-lineage kinase-3 and induce activation of Akt and ERK. J Biol Chem. 2002;277:49473–49480. doi: 10.1074/jbc.M203428200. [DOI] [PubMed] [Google Scholar]
- Saha S, Guillily M, Ferree A, et al. LRRK2 modulates vulnerability to mitochondrial dysfunction in C. elegans. J. Neuroscience. 2009 doi: 10.1523/JNEUROSCI.2281-09.2009. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakaguchi-Nakashima A, Meir JY, Jin Y, Matsumoto K, Hisamoto N. LRK-1, a C. elegans PARK8-related kinase, regulates axonal-dendritic polarity of SV proteins. Curr Biol. 2007;17:592–598. doi: 10.1016/j.cub.2007.01.074. [DOI] [PubMed] [Google Scholar]
- Silva RM, Kuan CY, Rakic P, Burke RE. Mixed lineage kinase-c-jun N-terminal kinase signaling pathway: a new therapeutic target in Parkinson's disease. Mov Disord. 2005;20:653–664. doi: 10.1002/mds.20390. [DOI] [PubMed] [Google Scholar]
- Smith WW, Pei Z, Jiang H, Dawson VL, Dawson TM, Ross CA. Kinase activity of mutant LRRK2 mediates neuronal toxicity. Nat Neurosci. 2006;9:1231–1233. doi: 10.1038/nn1776. [DOI] [PubMed] [Google Scholar]
- Smith WW, Pei Z, Jiang H, Moore DJ, Liang Y, West AB, Dawson VL, Dawson TM, Ross CA. Leucine-rich repeat kinase 2 (LRRK2) interacts with parkin and mutant LRRK2 induces neuronal degeneration. Proc Natl Acad Sci U S A. 2005;102:18676–18681. doi: 10.1073/pnas.0508052102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tain LS, Mortiboys H, Tao RN, Ziviani E, Bandmann O, Whitworth AJ. Rapamycin activation of 4E-BP prevents parkinsonian dopaminergic neuron loss. Nat Neurosci. 2009;12:1129–1135. doi: 10.1038/nn.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka-Hino M, Sagasti A, Hisamoto N, Kawasaki M, Nakano S, Ninomiya-Tsuji J, Bargmann CI, Matsumoto K. SEK-1 MAPKK mediates Ca2+ signaling to determine neuronal asymmetric development in Caenorhabditis elegans. EMBO Rep. 2002;3:56–62. doi: 10.1093/embo-reports/kvf001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D, Tang B, Zhao G, Pan Q, Xia K, Bodmer R, Zhang Z. Dispensable role of Drosophila ortholog of LRRK2 kinase activity in survival of dopaminergic neurons. Mol Neurodegener. 2008;3:3. doi: 10.1186/1750-1326-3-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AB, Moore DJ, Biskup S, Bugayenko A, Smith WW, Ross CA, Dawson VL, Dawson TM. Parkinson's disease-associated mutations in leucine-rich repeat kinase 2 augment kinase activity. Proc Natl Acad Sci U S A. 2005;102:16842–16847. doi: 10.1073/pnas.0507360102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West AB, Moore DJ, Choi C, et al. Parkinson's disease-associated mutations in LRRK2 link enhanced GTP-binding and kinase activities to neuronal toxicity. Hum Mol Genet. 2007;16:223–232. doi: 10.1093/hmg/ddl471. [DOI] [PubMed] [Google Scholar]
- Whitmarsh AJ. The JIP family of MAPK scaffold proteins. Biochem Soc Trans. 2006;34:828–832. doi: 10.1042/BST0340828. [DOI] [PubMed] [Google Scholar]
- Zimprich A, Biskup S, Leitner P, et al. Mutations in LRRK2 Cause Autosomal-Dominant Parkinsonism with Pleomorphic Pathology. Neuron. 2004;44:601–607. doi: 10.1016/j.neuron.2004.11.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Schematic of LRRK2 structure and constructs utilized in this study.
Fig. S2. LRRK2 binds to MKK3. (A) LRRK2 co-immunoprecipitates with MKK3 in HEK-293FT cells. (B) LRRK2 kinase and COR domains are essential and sufficient for its interaction with MKK3. (C) Pathogenic mutations in LRRK2 did not increase the binding of MKK3 to LRRK2. Experiments were repeated three times in HEK-293FT cells. (D) WT and dominant negative MKK3 showed similar degrees of binding to LRRK2.