Abstract
Ehrlichia chaffeensis is an obligately intracellular bacterium that exhibits tropism for mononuclear phagocytes and survives by reprogramming the host cell. Here we review new information regarding the newly characterized effector molecules and the complex network of molecular host-pathogen interactions that the organism exploits enabling it to thrive and persist intracellularly.
Keywords: Ehrlichia, tandem repeat proteins, ankyrin proteins, effector, host-pathogen interaction
1. Introduction
Ehrlichia chaffeensis is an obligately intracellular Gram-negative bacterium and the etiologic agent of human monocytotropic ehrlichiosis (HME), an emerging life-threatening tick-borne zoonosis [1]. HME is a systemic disease resembling sepsis or toxic shock syndrome with symptoms that most commonly include fever, malaise, myalgia, and headache, and is frequently accompanied by hematologic abnormalities including leucopenia, thrombocytopenia, and anemia, and elevations in serum hepatic aminotransferases. Hospitalization is required in 40 to 60% of HME cases and approximately 3% of cases are fatal [1].
E. chaffeensis exhibits tropism for mononuclear phagocytes, replicates within cytoplasmic vacuoles that have early endosomal characteristics, and survives by evading and/or suppressing the activation of innate and adaptive host defenses [2]. A primary strategy utilized by Ehrlichia to escape destruction is to interfere with immune activating signals produced by T cells rather than by inhibiting antigen presentation or T-cell activation [3]. Escape from phagocyte killing and intracellular persistence involves modulation of numerous host cell processes, including gene transcription, membrane trafficking, cell differentiation, activation and suppression of tyrosine and MAP kinase activity, downregulation of toll-like receptors and transcription factors, apoptosis, superoxide generation, lysosomal fusion, endosomal maturation, and transferrin receptor gene expression [4;5]. The inhibition of host MAP kinases by E. chaffeensis has been linked to the downregulation of transcription factors and corresponding target genes related to host defense [6]. After entry, E. chaffeensis blocks tyrosine phosphorylation of Janus kinase (Jak) and signal transducer and activator of transcription (Stat) signaling, inhibiting the anti-ehrlichial activity of IFN-γ [7]. Although many host cell processes are modulated by ehrlichiae, the effector proteins and host targets involved in the cellular reprogramming strategy to create a permissive host have been undefined. However, recent studies are providing new insight into the ehrlichial effector molecules responsible for modulating these important host-pathogen interactions.
Previous reviews have summarized the known cellular processes that are affected during Ehrlichia infection [2;5]. In this review, we focus on the newly defined E. chaffeensis effector proteins, 47-kDa tandem-repeat protein (TRP47) and 200-kDa ankyrin-repeat protein (Ank200) and their potential roles in pathobiology and disease pathogenesis. Molecular interactions between these effectors and specific host cell targets have been identified that are involved in controlling cellular processes such as gene transcription, cell signaling, cytoskeleton and vesicle trafficking, ATPase activity, apoptosis, and IFN-γ signaling, providing insights into the molecular mechanisms that Ehrlichia modulates to subvert host defenses and persist within mononuclear phagocytes.
2. Genome insights into Ehrlichia host-pathogen interactions
Complete genomes from three Ehrlichia species (E. canis, E. chaffeensis, and E. ruminantium) have been recently sequenced [8–10]. Ehrlichia have relatively small genome sizes (~1–1.5 Mb) with a high degree of genomic synteny, low G+C content (~30%) and one of the smallest genome coding ratios that is attributed to long non-coding regions and numerous long tandemly repeated sequences (TRs) [11]. The TRs appear to be actively created and deleted through a mechanism compatible with DNA slippage, but the mechanisms involved in this process are still unknown; however the generation of TRs by Ehrlichia appears to be a host adaptation mechanism [12]. TRs of different Ehrlichia species have no phylogenetic relationships and suggest that duplication occurred after diversification of the repeat-encoding DNA [12].
Several key genome features associated with host-pathogen interactions have been identified in Ehrlichia, including genes that encode tandem and ankyrin repeat containing proteins, actin polymerization proteins, a multigene family encoding outer membrane proteins, and a group of poly(G-C) tract (short sequence repeats) containing proteins [8]. In addition, mechanisms for the delivery of effector proteins have been identified, including many of the known type IV secretion system (T4SS) components [8–10]. The Sec-dependent and Sec-independent protein export pathways for secretion of protein across the inner membrane as well as a putative type I secretion system have also been identified. The Ehrlichia genomes also have genes that encode three response regulator two-component systems (TCS), a family of signal sensor, transduction, and response regulatory systems, composed of a pair of a sensor histidine kinase and a response regulator, that allows bacteria to sense signals and respond to changes in their environment through specific gene activation or repression [9].
3. TRP and Ank proteins and the host immune response
E. chaffeensis and E. canis have a small subset of major immunoreactive proteins that react strongly with antibodies in sera from infected humans or dogs, and most of these proteins have recently been molecularly characterized [13]. Many of the molecularly characterized E. chaffeensis immunoreactive proteins contain tandem or ankyrin repeats [14–17]. Major continuous species-specific antibody epitope(s) have been mapped to the acidic serine-rich TRs of E. chaffeensis, TRP120, TRP47, and TRP32 [14–16], and in the TRs of E. canis orthologs TRP140, TRP36, and TRP19, respectively [14;16;18]. In addition, multiple species-specific antibody eptiopes have been mapped to acidic terminal domains of the Ank200s [17;19].
4. Outer membrane proteins and immune evasion
A unique superfamily of immunorective outer membrane proteins has been identified in the family Anaplasmataceae. E. chaffeensis has a paralogous family of 22 major outer membrane proteins (OMP-1/p28) arranged in single locus upstream from the secA gene and downstream from a hypothetical transcriptional regulator gene [20]. Although recombination of the major outer membrane proteins of closely related Anaplasma spp. occurs to create antigenic diversity, there is no evidence that recombination of the Ehrlichia OMP-1 family occurs. Differential expression of the OMP-1 genes in ticks and animal hosts has been reported, suggesting that they play a role in host adaptation [21]. Expression of only one OMP-1 gene (OMP-1B) has been reported in ticks and tick cell lines that appear to involve a temperature sensitive regulation mechanism [21;22]. In contrast, all OMP-1 family members are expressed in mammalian hosts and cells and antibodies against all OMP-1 proteins have been detected in experimentally infected dogs [21;23]. Although the role of the proteins in antigenic variation and immune evasion is still uncertain, other characteristics have been identified for the OMP-1 proteins, including porin-like structural features, suggesting that they may facilitate nutrient acquisition [24].
5. TRP and Ank expression and secretion
E. chaffeensis exhibits two ultrastructural cell types, a small dense-cored (DC) form characterized by a dense nucleoid, and large replicating form, the reticulate cell (RC) that has uniformly dispersed nucleoid filaments. Ehrlichiae typically reside as microcolonies of bacteria within cytoplasmic vacuoles (morulae) derived from early endosomes [25]. The ultrastructural forms (DC and RC) can be distinguished phenotypically by the expression of two TRPs (TRP47 and TRP120) that are differentially expressed by DC ehrlichiae [14;26]. Immunoelectron microscopy has identified these TRPs, in addition to a non-differentially expressed TRP32, extracellularly associated with morular fibrillar matrix and the morula membrane, indicating that these proteins are secreted [14;15;26]. The expression of E. chaffeensis TRP120 and TRP47 by DC ehrlichiae suggests that one of their functions is related to attachment and entry, and some evidence of this function has been demonstrated with the TRP120 [26] as well as the TRP47 ortholog in E. ruminantium (Erum1110) [27]. Several effector TRPs have been reported to be secreted by the type III secretion system (T3SS), but the secretion mechanism utilized by ehrlichial TRPs is currently unknown. Recent study in our laboratory has indicated that E. chaffeensis TRP120, TRP47, TRP32, and Ank200 appear not to be secreted by VirB/ViD4 dependent Type IV secretion system (T4SS) (Wakeel and McBride, unpublished data). However, an E. chaffeensis Ank200 ortholog in A. phagocytophilum, AnkA, was recently reported to be secreted by a T4SS [28]. Although E. chaffeensis and A. phagocytophilum are closely related, they are different in many aspects such as tropism for different cell types and residence in different cytoplasmic compartments [29]. In addition, and A. phagocytophilum VirD4, the T4S substrate coupling protein, exhibits a higher identity with Agrobacterium tumefaciens VirD4 than E. chaffeensis [9;30].
6. Characteristics of TRP and Ank proteins
All of the characterized E. chaffeensis TRPs (TRP120, TRP47, and TRP32) and their orthologs exhibit similar biophysical characteristics including an acidic nature due to the predominance of acidic amino acid residues primarily in the TR region, abnormal electrophoretic mobilities, and a high frequency of polar amino acids, such as serine, particularly within TRs [14–16]. The comparison of TR amino acid usage has demonstrated that in spite of amino acid sequence variation of the TR among different ehrlichial species, conservation of amino acid usage is consistent. A total of 10 amino acids are used in all of the repeats, with a particularly high frequency of serine, threonine, alanine, proline, valine, aspartate, and glutamate [14–16]. Recent studies using mass spectrometry have determined that abnormal migration of E. chaffeensis TRPs during electrophoresis is attributed to the highly acidic TR regions and is not as result of large post translational modifications such as glycosylation, but rather the acidic nature of these repeat domains [15;16].
TRPs in pathogenic bacteria have been associated with host-pathogen interactions such as adhesion, actin nucleation and immune evasion. Examination of the E. chaffeensis TRPs has identified a relationship with several functional protein domains and motifs. The E. chaffeensis TRP47 contains seven 19-mer (ASVSEGDAVVNAVSQETPA) TRs that dominate the C-terminal region of the protein, and approximately half of the TRP47 is represented by the TR domain [31]. The TRP47 TR region exhibits homology with eukaryotic proteins including renin receptor/ATP6AP2/CAPER protein, DNA polymerase III subunits gamma and tau-conserved domain, and ribonuclease E suggesting similar functional characteristics [31]. Furthermore, new evidence indicates that the TRP47 is phosphorylated based on predicted tyrosine phosphorylation sites, mass spectrometry data consistent with addition of phosphate, and the fact that TRP47 can be immunoprecipitated with anti-phosphotyrosine antibodies (Wakeel and McBride, unpublished data). Other TRPs may be phosphorylated, such as TRP32, which has an unusually high frequency of tyrosine residues (20%) in the C-terminal tail [15].
Ehrlichia spp. are among only a few prokaryotes that are known to have ankryin repeat (Ank)-containing proteins. Ank is a ubiquitous eukaryote motif that mediates protein-protein interactions, and they are found in proteins that modulate many cellular functions such as transcriptional regulation, cell cycle, cytoskeleton organization, developmental regulation, signal transduction, toxicity, and the inflammatory response. The ankyrin structural repeat unit, Ank motif, is 33 amino acids long and contains two antiparallel helices and a beta-hairpin (helix-turn-helix). The Ank may occur in combinations with other types of domains and cooperatively fold into structures that mediate molecular recognition via protein-protein interactions. The most extensively studied Ank protein in E. chaffeensis is a 200 kDa protein (Ank200) that has central domain that has 19 Anks flanked by acidic (pI 4 to 5) C- and N-terminal domains that have a predominance of glutamate and aspartate residues [17]. In addition, like the TRPs, E. canis and E. chaffeensis Ank200s have a high proportion of polar amino acids, including serine and threonine [17;19].
7. Host cell gene expression during E. chaffeensis infection
E. chaffeensis significantly alters the transcriptional levels of approximately 5% of host genes within 24 hr of infection [4]. Genes that are modulated include those coding for apoptosis inhibitors, regulation of cell cycle and differentiation, signal transduction, proinflammatory cytokines, biosynthetic and metabolic proteins, and membrane trafficking proteins. This transcriptional profile has provided new information on host cell processes targeted by Ehrlichia and revealed key themes in disease pathogenesis. Furthermore, the unique Ehrlichia-host interaction is illustrated by the fact that among intracellular bacteria only a relatively few host genes were found to be commonly induced during E. chaffeensis infection, while no genes were commonly repressed, suggesting that ehrlichial survival mechanisms have evolved distinctly from other intracellular pathogens [4].
Microarray analysis during infection has shown that E. chaffeensis appears to manipulate genes related to three primary areas of the host response. First, E. chaffeensis represses the transcription of cytokines involved in the early innate immune response and cell-mediated immune response to intracellular microbes, including host cell cytokines that modulate innate and adaptive immunity to intracellular bacteria such as IL-12, IL-15, and IL-18, which are repressed. These cytokines play fundamental roles in stimulating NK cells and T helper 1 cells to produce gamma interferon (IFN-γ), which then activates macrophages to kill phagocytized bacteria. IL-12 and IL-15 also activate NK cells and cytotoxic T lymphocytes to kill cells infected with intracellular bacteria. Thus, repression of IL-12, IL-15, and IL-18 suggests that modulating these cytokines is critical to the survival of E. chaffeensis.
Second, E. chaffeensis up-regulates NF-κB and apoptosis inhibitors, which may enhance host cell survival. Apoptosis is an innate mechanism of host defense used to prevent proliferation of internalized bacteria. E. chaffeensis infection induce apoptosis inhibitors such as IER3 (immediately early response 3), BirC3 (baculoviral IAP repeat-containing protein 3), and BCL2, but inhibits apoptosis inducers such as BIK (BCL2-interacting killer) and BNIP3L (BCL2/adenovirus E1B 19-kDa interacting protein 3-like) during the early stage of infection, thus impairing host cell apoptosis and maintaining a prolonged growth opportunity for ehrlichiae.
Third, E. chaffeensis inhibits the transcription of genes involved in membrane trafficking. E. chaffeensis lives in an early endosome and inhibits the maturation of the endosome to evade destruction by lysosomal enzymes [29]. E. chaffeensis represses the production of Rab5, SNAP23, and STX16 (syntaxin 16) during infection, most dramatically during the first hour of infection. E. chaffeensis induces the production of vimentin, a reservoir for SNAP23 [32]. Thus, E. chaffeensis appears to modulate phagosome-lysosome fusion by apregulating expression of Rab5 and SNAPs in the macrophage. A current model of vesicle fusion is explained by the SNARE hypothesis which proposes that the docking and fusion of vesicles with the plasma membrane are mediated by the specific interaction of vesicle proteins (v-SNARE and SNAR receptor) with the target plasma membrane protein (t-SNARE). Among the proteins implicated are syntaxins, which have at least 16 members (synaptosome-associated proteins; SNAPs). These proteins form a complex that juxtaposes the two membranes to be fused, and this interaction is regulated by Rab5, a small GTPase of the Rab family. Depletion of Rab5 inhibits the fusion of the phagosome containing Listeria monocytogenes with lysosomes [33].
8. E. chaffeensis TRP47 network of host interactions
Progress in understanding the specific interactions of TRP47 with the host has been advanced by a recent study demonstrating interactions between E. chaffeensis TRP47 and multiple host proteins including polycomb group ring finger 5 (PCGF5), Src protein tyrosine kinase FYN (FYN), protein tyrosine phosphatase non-receptor type 2 (PTPN2), adenylate cyclase-associated protein 1 (CAP1), and immunoglobulin lambda-like polypeptide 1 (IGLL1) with distinct cellular functions associated with signaling, transcriptional regulation, vesicle trafficking, and cellular proliferation and differentiation [31] (Figure 1). Furthermore, the potential importance of the TRP47 in pathobiology is supported by the recent studies demonstrating that the TRP47 gene is the most highly expressed gene in the E. chaffeensis transcriptome (Kuriakose and McBride, unpublished data). Although, the relevance of these ehrlichiae-host molecular interactions in the context of ehrlichial pathobiology remains to be determined, the host targets identified suggest that TRP47 is a multifunctional effector that plays an important role in establishing bacterial infection and promoting intracellular survival.
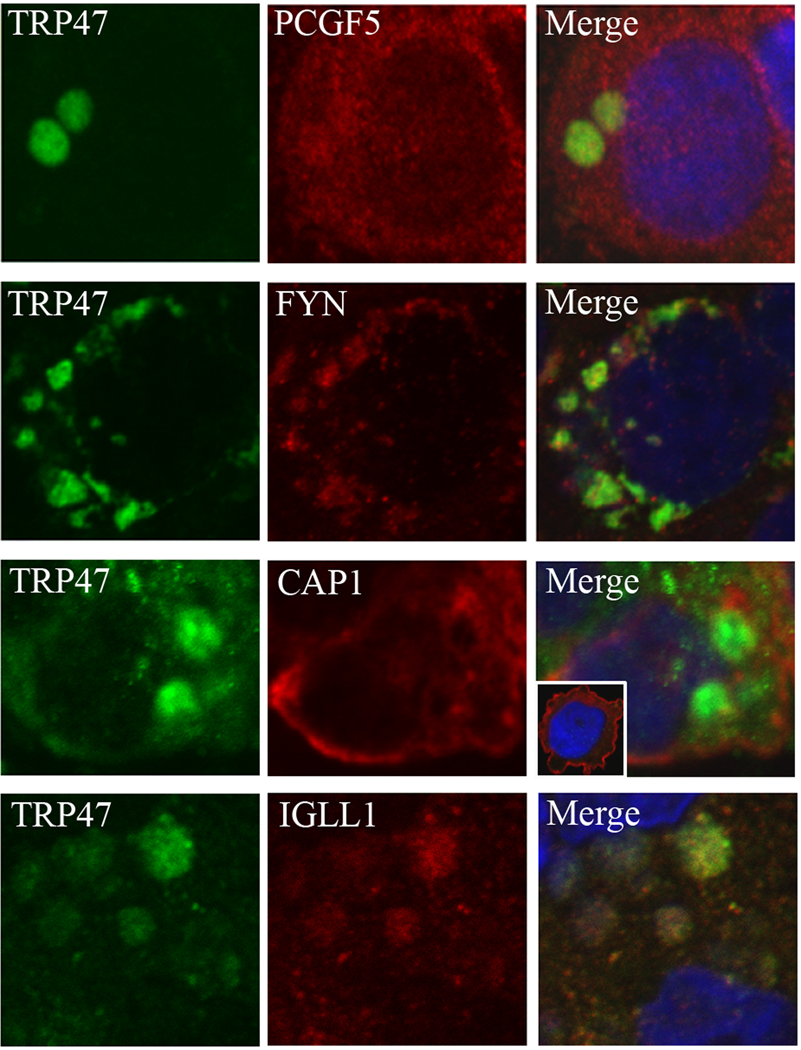
Fig. 1.
Colocalization of E. chaffeensis TRP47 with PCGF5, FYN, CAP1, and IGLL1 in E. chaffeensis-infected THP-1 cells. THP-1 cells were infected with E. chaffeensis and 3 days postinfection were dually labeled and examined by confocal microscopy. The panels on the left were labeled with TRP47 (green), middle panels labeled either with PCGF5, FYN, CAP1, or IGLL1 (red) and the panels on the right are merged images. PCGF5, FYN, CAP1, and IGLL1 colocalize with E. chaffeensis TRP47-labeled morulae (right panels, merged images). In the inset (TRP1 + CAP1 merged panel), a normal uninfected THP-1 cell reveals that CAP1 is mainly associated with plasma membrane, while in the E. chaffeensis-infected THP-1 cell, CAP1 is distributed in cytoplasm and associated with E. chaffeensis-containing morulae.
8.1 Host cell signaling and TRP47
Tyrosine phosphorylation of host and/or bacterial proteins has been implicated in signaling pathways triggering the entry of many intracellular pathogens. Tyrosine kinases are known to be involved in ehrlichial entry; however, the specific kinases involved have not been determined [34]. The association of TRP47 with tyrosine kinase FYN and the intracellular DC form of E. chaffeensis suggests that it may be recruited by TRP47 to facilitate the entry process. FYN specifically phosphorylates caveolin-1 and is required for coxsackievirus internalization and infection via caveolin-associated vesicles to polarized epithelial cells [35]. FYN was not observed associated with E. chaffeensis RCs demonstrating a selective association with DC ehrlichiae that express TRP47. E. chaffeensis TRP47 is strikingly similar to Chlamydia trachomatis serovars L2, Tarp, which is an immunoreactive, secreted (T3SS), highly acidic (pI 4) protein that migrates abnormally on SDS-PAGE, contains six near-identical tandem repeats, and is tyrosine phosphorylated by the Abl kinase at the site of entry and associated with recruitment of actin [36].
Another TRP47 interacting protein, PTPN2, is a protein tyrosine phosphatase (PTP) also known as T cell PTP (TC-PTP) that catalyzes the dephosphorylation of phosphotyrosine peptides and regulates phosphotyrosine levels in signal transduction pathways. PTPs are known to regulate a variety of cellular processes including cell growth, differentiation, mitotic cycle, and oncogenic transformation [37;38]. Multiple substrates of PTPN2 include CSF-1R, EGFR, PDGFR, IR, p52Shc, Jak1, Jak3, Stat1, Stat3, Stat5a/b, and Stat6 [38]. The in vivo and in vitro analyses indicate that PTPN2 could control cytokine signaling events by its negative action on the Jak/Stat pathway. The loss of PTPN2 results in Stat5 hyper-activation, increased production of gamma interferon (IFN-γ), tumor necrosis factor alpha (TNF-α), interleukin (IL)-12, and inducible nitric oxide synthase (iNOS), increased tyrosine phosphorylation, recruitment of a Grb2/Gab2/Shp2 complex to the CSF-1 receptor, and enhanced activation of ERK, and may affect transcription factor PU.1 signaling.
PTPN2 is ubiquitously expressed with particularly high expression in hematopoietic tissues. It is remarkable, not only by the fact that it appears to broadly influence hematopoietic cell development, but recent findings also demonstrate a role in several human diseases from autoimmune disease to cancer [37]. The Jak/Stat pathway is inhibited by monocytotropic E. chaffeensis [7], and supports the possibility that TRP47 may not only be involved in the inhibition of IFN-γ-induced tyrosine phosphorylation of Stat1, Jak1, and Jak2 by interacting with PTPN2, but also in the regulation of cellular development.
8.2 Vesicle trafficking, apoptosis and TRP47
Membrane trafficking is a cellular process that has been identified as an Ehrlichia target by multiple studies. A specific interaction between TRP47 and the multifunctional protein CAP1 has been defined that appears to occur at the morula membrane interface, where CAP1 localizes with the DC morulae adjacent to the morula boundaries (membrane) [31]. The distribution pattern of CAP1 was remarkably different in E. chaffeensis-infected cells as compared to uninfected cells, where it is primarily associated with the plasma membrane, indicating that distribution of this protein is altered as a result of E. chaffeensis infection [31]. CAPs were originally identified in yeast as a component of the adenylyl cyclase complex, and yeast cells deficient in CAPs are defective in cytoskeleton organization. Although CAPs do not regulate cAMP in animal cells, their role in regulation of actin remodeling in response to cellular signals is widely conserved. CAP1 is a highly conserved monomeric actin binding protein that contains actin (C-terminal), adenylyl cyclase and cofilin (N-terminal), and Src homology 3 (SH3) and profilin (central region) binding domains and plays an active role in actin turnover [39]. Genetic studies in yeast have implicated CAP1 in vesicle trafficking and endocytosis. In mammalian cells, CAP1 is associated with SH3 domain-dependent mAbp1-dynamin complex involved in receptor-mediated endocytosis [40]. Many intracellular bacteria including Listeria, Rickettsia, Burkholderia, Shigella and Mycobacterium species, subvert cellular actin dynamics to facilitate their movement within the host cytosol and to infect neighboring cells while evading host immune surveillance and promoting their intracellular survival [41]. Thus, in an effort to survive in the intracellular niche Ehrlichia may manipulate the mononuclear phagocyte cytoskeleton components such as actin by modulating CAP1.
Ehrlichiae, like chlamydiae, inhibit apoptosis early in infection, but while chlamydiae induce cell death at the end of the infection cycle, ehrlichial exit mechanisms remain undefined [42;43]. Interestingly, CAP1 has also been implicated in promoting apoptosis by functioning as an actin shuttle to mitochondria. Similar to cofilin, BAD, and BAX, CAP1 rapidly translocates to mitochondria independent of caspase activation where it promotes apoptosis [44]. Associations between ehrlichial morulae and mitochrondria have been consistently observed [25]. Thus, the TRP47 and CAP1 interaction may serve a dual function by facilitating endocytosis and vesicle trafficking, and promoting apoptosis in the late stages of infection.
8.3 Immunoglobulin lambda-like protein 1 and TRP47
IGLL1 is also interacts with TRP47 and shows colocalization on the surface of morulae. IGLL1 gene encodes one of the surrogate light chain subunits and is a member of the immunoglobulin gene superfamily and does not undergo rearrangement [45]. The preB cell receptor is composed of a membrane-bound Ig Mu heavy chain in association with a heterodimeric surrogate light chain (IGLL1) and is found on the surface of proB and preB cells [45]. The preB cell receptor is involved in transduction of signals for cellular proliferation, differentiation from the proB cell to the preB cell stage, allelic exclusion at the Ig heavy chain gene locus, and promotion of Ig light chain gene rearrangements [45]. Thus, the significance of the interaction between TRP47 and IGLL1 might involve signaling and development, but suggests a novel role for IGLL1 in the macrophage and one that will require further study to understand.
8.4 Gene silencing complex and TRP47
The TRP47 interacting partner, PCGF5 has been associated with DNA-dependent regulation of transcription, metal ion binding, and protein-protein interactions. It has a specialized Zn-finger domain consisting of 40 to 60 residues that binds two atoms of zinc, is defined by the 'cross-brace' motif involved in protein-protein interactions [46;47]. PCGF5 is related to the polycomb group proteins (transcriptional repressors) Bmi-1/PCGF4 and Mel-18/PCGF2 that play important role in the regulation of Hox gene expression, X-chromosome inactivation, tumorigenesis, and self-renewal, maintenance of pluripotency of stem cells, and stimulation of E3 ubiquitin ligase activity. Thus, it appears that TRP47-expressing DC ehrlichiae may recruit PCGF5 in an effort to modulate host cell gene expression to favor survival. Altered gene expression in E. chaffeensis-infected cells has been reported, but the mechanisms involved are largely unknown [4]. However, a recent study [48] has demonstrated that E. chaffeensis Ank200 targets host cell genes related to apoptosis, ATPase activity, and transcription and may modulate transcription.
9. Ehrlichia chaffeensis Ank200
The E. chaffeensis Ank 200 is a large immunoreactive protein that contains 19 ankyrin repeats in a centralized domain flanked by acidic terminal domains [17;48]. E. chaffeensis Ank200 is a nuclear translocated protein that lacks a classic nuclear localization signal (NLS); however, its specific association with host cell DNA motifs suggests that it plays an important direct role in modulating host cell gene transcription. Nuclear effector proteins have been reported recently in several intracellular human bacterial pathogens including Ehrlichia, Anaplasma, Shigella and Yersinia. It is well documented that E. chaffeensis modulates host cell gene transcription, and this can occur through multiple pathways and host-pathogen interactions. However, directly targeting genes in the host cell nucleus identifies an interesting new and relatively unexplored mechanism that pathogens, including Ehrlichia, utilize to modulate host gene transcription.
9.1 Ank 200 interaction with host Alu elements
E. chaffeensis Ank200 is translocated to the nuclei of Ehrlichia-infected mononuclear phagocytes where it interacts with an adenine-rich motif in promoter and intronic Alu elements, [48]. Alu elements are short interspersed mobile DNA elements distributed in a nonrandom manner that comprise approximately 5–10% of the human genome and are thought to be involved in transcriptional regulation as a carrier of cis regulatory elements [49;50]. Alu elements have known transcription factor binding sites including all MEF2 family members, HNF1.03, OC.2, BARX2 and PAX4 [51]. The association of Ank200 with Alu elements suggests that Ank200 could affect gene transcription globally through Alu-mediated transcriptional control mechanisms. The global analysis of binding sites of Ank200 demonstrated that this protein binds to multiple regions distributed on nearly every chromosome via direct DNA interaction or with other DNA-binding proteins.
9.2 Ank200 targets apoptosis, ATPase and transcriptional regulatory genes
Chromatin immunoprecipitation (ChIP) and microarrays (ChIP-chip) analysis has identified a subset of Ank200 target genes that have been classified into three gene ontology (GO) databases (biological processes, molecular function and cell structure). These included genes associated with transcriptional regulation (DNA and RNA), apoptosis, ATPase activity, and structural associations with the nucleus [48]. Interestingly, the most targeted genes are associated with cellular processes that are known to be modulated by E. chaffeensis, including apoptosis, ATPase activity, and regulation of gene transcription. In addition, Ank200 also appears to bind genes associated with transcription, and there is evidence that a large number of genes associated with transcription are modulated during E. chaffeensis infection [4]. ATPase activity is present in ehrlichial inclusions, and genes associated with ATPase activity also appear to be targets of E. chaffeensis Ank200 [48].
9.3 Ank200 modulation genes associated with pathobiology
Analysis of Ank200 gene targets identified a number of genes that have been linked to pathogenesis and immune evasion. Specific Ank200 target genes of potential importance that are significantly upregulated or silenced during infection are TNF-α, Jak2 and CD48. Although TNF-α expression is not induced early in infection (<48 hr) [52;53], TNF-α expression is upregulated approximately 30-fold by day 5 post infection [48]. Several studies have demonstrated that overproduction or high serum concentration of TNF-α on day 7 post infection is closely associated with the fatality in severe HME [54;55]. This study provided evidence that E. chaffeensis Ank200 may contribute to the induction of TNF-α by binding directly with promoter and upregulating gene transcription.
It is known that E. chaffeensis enters host monocytes by clathrin-independent, receptor-mediated endocytosis [29;56]. However, the detailed mechanism of entry is still unclear, and the identity of the receptor remains undefined. In recent studies, the entry and establishment of infection of E. chaffeensis has been associated with caveolae and unidentified host GPI-anchored proteins [57]. CD48, a caveolae-associated GPI-anchored protein, recognized as a receptor for bacterial uptake is strongly upregulated during E. chaffeensis infection and is a Ank200 target gene. This situation suggests that E. chaffeensis may modulate gene transcription of cellular receptors associated with entry [48].
One of the primary mechanisms by which E. chaffeensis survives in the host cell appears to be the ability to block macrophage responsiveness to IFN-γ. E. chaffeensis blocks IFN-γ induced tyrosine phosphorylation of Jak and Stat by raising PKA activity in THP-1 cells [7]. Furthermore, Jak2 transcription appears to be silenced during E. chaffeensis infection and Jak/Stat genes are also Ank200 targets, suggesting that E. chaffeensis uses multiple strategies, including directly modulating genes associated with the Jak/Stat pathway [4]. It is not clear if Ank200 modulates the expression of these genes, but the fact that these genes are associated with ehrlichial pathobiology and pathogenesis and are Ank200 targets suggests that Ehrlichia modulates these innate immune response effectors by multiple mechanisms.
10. Conclusions
Ehrlichia TRPs and Ank proteins appear to be key effectors in novel host-pathogen interactions associated with important host cell processes. Recent studies have provided new insight into the complex host-pathogen interactions that require further exploration and definition of the molecular strategies and specific mechanisms involved in evading host defenses and modulation of host cell processes by Ehrlichia. New information on E. chaffeensis TRP47 interactions with PCGF5, FYN, PTPN2, CAP1, and IGLL1 demonstrates the multifunctional nature of ehrlichial effector proteins and the complexity of host interactions that may ultimately regulate gene transcription, vesicle trafficking, and cell signaling pathways. Modulation of host cell transcription by Ehrlichia and the mechanisms involved have been advanced significantly by the identification of a nuclear effector, Ank200, and provide insight into the mechanisms that the pathogen uses to directly modulate host gene transcription. Functional studies on Ehrlichia effectors will advance our understanding of the complex network of interactions between obligatory intracellular pathogens and their hosts. Such studies will likely reveal more complex interactions that are unique to these organisms as well as provide new insights into the cell biology and the complex relationship between the pathogen and the host. Future studies focusing on these bacterial effector mechanisms may not only increase our understanding E. chaffeensis pathobiology, but also provide us clues to rationally design preventive and therapeutic compounds that can control HME and other intracellular pathogens.
ACKNOWLEDGEMENTS
This work was supported by National Institute of Allergy and Infectious Diseases grant (AI 071145, AI 069270), and additional support was provided by the Clayton Foundation for Research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Paddock CD, Childs JE. Ehrlichia chaffeensis: a prototypical emerging pathogen. Clin.Microbiol.Rev. 2003;16:37–64. doi: 10.1128/CMR.16.1.37-64.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rikihisa Y. Ehrlichia subversion of host innate responses. Curr.Opin.Microbiol. 2006;9:95–101. doi: 10.1016/j.mib.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 3.Nandi B, Chatterjee M, Hogle K, McLaughlin M, MacNamara K, Racine R, Winslow GM. Antigen display, T-cell activation, and immune evasion during acute and chronic ehrlichiosis. Infect.Immun. 2009;77:4643–4653. doi: 10.1128/IAI.01433-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang JZ, Sinha M, Luxon BA, Yu XJ. Survival strategy of obligately intracellular Ehrlichia chaffeensis: novel modulation of immune response and host cell cycles. Infect.Immun. 2004;72:498–507. doi: 10.1128/IAI.72.1.498-507.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rikihisa Y. Molecular events involved in cellular invasion by Ehrlichia chaffeensis and Anaplasma phagocytophilum. Vet.Parasitol. 2009 doi: 10.1016/j.vetpar.2009.09.017. doi:10.1016/j.vetpar.2009.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin M, Rikihisa Y. Ehrlichia chaffeensis downregulates surface Toll-like receptors 2/4, CD14 and transcription factors PU.1 and inhibits lipopolysaccharide activation of NF-kappa B, ERK 1/2 and p38 MAPK in host monocytes. Cell Microbiol. 2004;6:175–186. doi: 10.1046/j.1462-5822.2003.00355.x. [DOI] [PubMed] [Google Scholar]
- 7.Lee EH, Rikihisa Y. Protein kinase A-mediated inhibition of gamma interferon-induced tyrosine phosphorylation of Janus kinases and latent cytoplasmic transcription factors in human monocytes by Ehrlichia chaffeensis. Infect.Immun. 1998;66:2514–2520. doi: 10.1128/iai.66.6.2514-2520.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mavromatis K, Doyle CK, Lykidis A, Ivanova N, Francino MP, Chain P, Shin M, Malfatti S, Larimer F, Copeland A, Detter JC, Land M, Richardson PM, Yu XJ, Walker DH, McBride JW, Kyrpides NC. The genome of the obligately intracellular bacterium Ehrlichia canis reveals themes of complex membrane structure and immune evasion strategies. J.Bacteriol. 2006;188:4015–4023. doi: 10.1128/JB.01837-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hotopp JC, Lin M, Madupu R, Crabtree J, Angiuoli SV, Eisen JA, Seshadri R, Ren Q, Wu M, Utterback TR, Smith S, Lewis M, Khouri H, Zhang C, Niu H, Lin Q, Ohashi N, Zhi N, Nelson W, Brinkac LM, Dodson RJ, Rosovitz MJ, Sundaram J, Daugherty SC, Davidsen T, Durkin AS, Gwinn M, Haft DH, Selengut JD, Sullivan SA, Zafar N, Zhou L, Benahmed F, Forberger H, Halpin R, Mulligan S, Robinson J, White O, Rikihisa Y, Tettelin H. Comparative genomics of emerging human ehrlichiosis agents. PLoS.Genet. 2006;2:e21. doi: 10.1371/journal.pgen.0020021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collins NE, Liebenberg J, de Villiers EP, Brayton KA, Louw E, Pretorius A, Faber FE, van HH, Josemans A, van KM, Steyn HC, van Strijp MF, Zweygarth E, Jongejan F, Maillard JC, Berthier D, Botha M, Joubert F, Corton CH, Thomson NR, Allsopp MT, Allsopp BA. The genome of the heartwater agent Ehrlichia ruminantium contains multiple tandem repeats of actively variable copy number. Proc.Natl.Acad.Sci.U.S.A. 2005;102:838–843. doi: 10.1073/pnas.0406633102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Frutos R, Viari A, Ferraz C, Morgat A, Eychenie S, Kandassamy Y, Chantal I, Bensaid A, Coissac E, Vachiery N, Demaille J, Martinez D. Comparative genomic analysis of three strains of Ehrlichia ruminantium reveals an active process of genome size plasticity. J.Bacteriol. 2006;188:2533–2542. doi: 10.1128/JB.188.7.2533-2542.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Frutos R, Viari A, Vachiery N, Boyer F, Martinez D. Ehrlichia ruminantium: genomic and evolutionary features. Trends Parasitol. 2007;23:414–419. doi: 10.1016/j.pt.2007.07.007. [DOI] [PubMed] [Google Scholar]
- 13.Chen SM, Dumler JS, Feng HM, Walker DH. Identification of the antigenic constituents of Ehrlichia chaffeensis. Am.J.Trop.Med.Hyg. 1994;50:52–58. [PubMed] [Google Scholar]
- 14.Doyle CK, Nethery KA, Popov VL, McBride JW. Differentially expressed and secreted major immunoreactive protein orthologs of Ehrlichia canis and E. chaffeensis elicit early antibody responses to epitopes on glycosylated tandem repeats. Infect.Immun. 2006;74:711–720. doi: 10.1128/IAI.74.1.711-720.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Luo T, Zhang X, Wakeel A, Popov VL, McBride JW. A variable-length PCR target protein of Ehrlichia chaffeensis contains major species-specific antibody epitopes in acidic serine-rich tandem repeats. Infect.Immun. 2008;76:1572–1580. doi: 10.1128/IAI.01466-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo T, Zhang X, McBride JW. Major species-specific antibody epitopes of the Ehrlichia chaffeensis p120 and E. canis p140 orthologs in surface-exposed tandem repeat regions. Clin.Vaccine Immunol. 2009;16:982–990. doi: 10.1128/CVI.00048-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luo T, Zhang X, Nicholson WL, Zhu B, McBride JW. Molecular characterization of antibody epitopes of Ehrlichia chaffeensis ankyrin protein 200 and tandem repeat protein 47 and evaluation of synthetic immunodeterminants for serodiagnosis of human monocytotropic ehrlichiosis. Clin.Vaccine Immunol. 2010;17:87–97. doi: 10.1128/CVI.00331-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McBride JW, Doyle CK, Zhang X, Cardenas AM, Popov VL, Nethery KA, Woods ME. Identification of a glycosylated Ehrlichia canis 19-kilodalton major immunoreactive protein with a species-specific serine-rich glycopeptide epitope. Infect.Immun. 2007;75:74–82. doi: 10.1128/IAI.01494-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nethery KA, Doyle CK, Zhang X, McBride JW. Ehrlichia canis gp200 contains dominant species-specific antibody epitopes in terminal acidic domains. Infect.Immun. 2007;75:4900–4908. doi: 10.1128/IAI.00041-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohashi N, Rikihisa Y, Unver A. Analysis of transcriptionally active gene clusters of major outer membrane protein multigene family in Ehrlichia canis and E. chaffeensis. Infect.Immun. 2001;69:2083–2091. doi: 10.1128/IAI.69.4.2083-2091.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Unver A, Rikihisa Y, Stich RW, Ohashi N, Felek S. The omp-1 major outer membrane multigene family of Ehrlichia chaffeensis is differentially expressed in canine and tick hosts. Infect.Immun. 2002;70:4701–4704. doi: 10.1128/IAI.70.8.4701-4704.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Unver A, Ohashi N, Tajima T, Stich RW, Grover D, Rikihisa Y. Transcriptional analysis of p30 major outer membrane multigene family of Ehrlichia canis in dogs, ticks, and cell culture at different temperatures. Infect.Immun. 2001;69:6172–6178. doi: 10.1128/IAI.69.10.6172-6178.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang JZ, Guo H, Winslow GM, Yu XJ. Expression of members of the 28-kilodalton major outer membrane protein family of Ehrlichia chaffeensis during persistent infection. Infect.Immun. 2004;72:4336–4343. doi: 10.1128/IAI.72.8.4336-4343.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kumagai Y, Huang H, Rikihisa Y. Expression and porin activity of P28 and OMP-1F during intracellular Ehrlichia chaffeensis development. J.Bacteriol. 2008;190:3597–3605. doi: 10.1128/JB.02017-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Popov VL, Chen SM, Feng HM, Walker DH. Ultrastructural variation of cultured Ehrlichia chaffeensis. J.Med.Microbiol. 1995;43:411–421. doi: 10.1099/00222615-43-6-411. [DOI] [PubMed] [Google Scholar]
- 26.Popov VL, Yu X, Walker DH. The 120 kDa outer membrane protein of Ehrlichia chaffeensis: preferential expression on dense-core cells and gene expression in Escherichia coli associated with attachment and entry. Microb.Pathog. 2000;28:71–80. doi: 10.1006/mpat.1999.0327. [DOI] [PubMed] [Google Scholar]
- 27.de la FJ, Garcia-Garcia JC, Barbet AF, Blouin EF, Kocan KM. Adhesion of outer membrane proteins containing tandem repeats of Anaplasma and Ehrlichia species (Rickettsiales: Anaplasmataceae) to tick cells. Vet.Microbiol. 2004;98:313–322. doi: 10.1016/j.vetmic.2003.11.001. [DOI] [PubMed] [Google Scholar]
- 28.Lin M, den Dulk-Ras A, Hooykaas PJ, Rikihisa Y. Anaplasma phagocytophilum AnkA secreted by type IV secretion system is tyrosine phosphorylated by Abl-1 to facilitate infection. Cell Microbiol. 2007;9:2644–2657. doi: 10.1111/j.1462-5822.2007.00985.x. [DOI] [PubMed] [Google Scholar]
- 29.Mott J, Barnewall RE, Rikihisa Y. Human granulocytic ehrlichiosis agent and Ehrlichia chaffeensis reside in different cytoplasmic compartments in HL-60 cells. Infect.Immun. 1999;67:1368–1378. doi: 10.1128/iai.67.3.1368-1378.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Alvarez-Martinez CE, Christie PJ. Biological diversity of prokaryotic type IV secretion systems. Microbiol.Mol.Biol.Rev. 2009;73:775–808. doi: 10.1128/MMBR.00023-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wakeel A, Kuriakose JA, McBride JW. An Ehrlichia chaffeensis tandem repeat protein interacts with multiple host targets involved in cell signaling, transcriptional regulation, and vesicle trafficking. Infect.Immun. 2009;77:1734–1745. doi: 10.1128/IAI.00027-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Faigle W, Colucci-Guyon E, Louvard D, Amigorena S, Galli T. Vimentin filaments in fibroblasts are a reservoir for SNAP23, a component of the membrane fusion machinery. Mol.Biol.Cell. 2000;11:3485–3494. doi: 10.1091/mbc.11.10.3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alvarez-Dominguez C, Barbieri AM, Beron W, Wandinger-Ness A, Stahl PD. Phagocytosed live Listeria monocytogenes influences Rab5-regulated in vitro phagosome-endosome fusion. J.Biol.Chem. 1996;271:13834–13843. doi: 10.1074/jbc.271.23.13834. [DOI] [PubMed] [Google Scholar]
- 34.Lin M, Zhu MX, Rikihisa Y. Rapid activation of protein tyrosine kinase and phospholipase C-gamma2 and increase in cytosolic free calcium are required by Ehrlichia chaffeensis for internalization and growth in THP-1 cells. Infect.Immun. 2002;70:889–898. doi: 10.1128/IAI.70.2.889-898.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Coyne CB, Bergelson JM. Virus-induced Abl and Fyn kinase signals permit coxsackievirus entry through epithelial tight junctions. Cell. 2006;124:119–131. doi: 10.1016/j.cell.2005.10.035. [DOI] [PubMed] [Google Scholar]
- 36.Clifton DR, Fields KA, Grieshaber SS, Dooley CA, Fischer ER, Mead DJ, Carabeo RA, Hackstadt T. A chlamydial type III translocated protein is tyrosine-phosphorylated at the site of entry and associated with recruitment of actin. Proc.Natl.Acad.Sci.U.S.A. 2004;101:10166–10171. doi: 10.1073/pnas.0402829101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Doody KM, Bourdeau A, Tremblay ML. T-cell protein tyrosine phosphatase is a key regulator in immune cell signaling: lessons from the knockout mouse model and implications in human disease. Immunol.Rev. 2009;228:325–341. doi: 10.1111/j.1600-065X.2008.00743.x. [DOI] [PubMed] [Google Scholar]
- 38.Stuible M, Doody KM, Tremblay ML. PTP1B and TC-PTP: regulators of transformation and tumorigenesis. Cancer Metastasis Rev. 2008;27:215–230. doi: 10.1007/s10555-008-9115-1. [DOI] [PubMed] [Google Scholar]
- 39.Hubberstey AV, Mottillo EP. Cyclase-associated proteins: CAPacity for linking signal transduction and actin polymerization. FASEB J. 2002;16:487–499. doi: 10.1096/fj.01-0659rev. [DOI] [PubMed] [Google Scholar]
- 40.Kessels MM, Engqvist-Goldstein AE, Drubin DG, Qualmann B. Mammalian Abp1, a signal-responsive F-actin-binding protein, links the actin cytoskeleton to endocytosis via the GTPase dynamin. J.Cell Biol. 2001;153:351–366. doi: 10.1083/jcb.153.2.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stevens JM, Galyov EE, Stevens MP. Actin-dependent movement of bacterial pathogens. Nat.Rev.Microbiol. 2006;4:91–101. doi: 10.1038/nrmicro1320. [DOI] [PubMed] [Google Scholar]
- 42.Verbeke P, Welter-Stahl L, Ying S, Hansen J, Hacker G, Darville T, Ojcius DM. Recruitment of BAD by the Chlamydia trachomatis vacuole correlates with host-cell survival. PLoS.Pathog. 2006;2:e45. doi: 10.1371/journal.ppat.0020045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xiong Q, Bao W, Ge Y, Rikihisa Y. Ehrlichia ewingii infection delays spontaneous neutrophil apoptosis through stabilization of mitochondria. J.Infect.Dis. 2008;197:1110–1118. doi: 10.1086/533457. [DOI] [PubMed] [Google Scholar]
- 44.Wang C, Zhou GL, Vedantam S, Li P, Field J. Mitochondrial shuttling of CAP1 promotes actin- and cofilin-dependent apoptosis. J.Cell Sci. 2008;121:2913–2920. doi: 10.1242/jcs.023911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Herzog S, Reth M, Jumaa H. Regulation of B-cell proliferation and differentiation by pre-B-cell receptor signalling. Nat.Rev.Immunol. 2009;9:195–205. doi: 10.1038/nri2491. [DOI] [PubMed] [Google Scholar]
- 46.Borden KL, Freemont PS. The RING finger domain: a recent example of a sequence-structure family. Curr.Opin.Struct.Biol. 1996;6:395–401. doi: 10.1016/s0959-440x(96)80060-1. [DOI] [PubMed] [Google Scholar]
- 47.Saurin AJ, Borden KL, Boddy MN, Freemont PS. Does this have a familiar RING? Trends Biochem.Sci. 1996;21:208–214. [PubMed] [Google Scholar]
- 48.Zhu B, Nethery KA, Kuriakose JA, Wakeel A, Zhang X, McBride JW. Nuclear translocated Ehrlichia chaffeensis ankyrin protein interacts with a specific adenine-rich motif of host promoter and intronic Alu elements. Infect.Immun. 2009;77:4243–4255. doi: 10.1128/IAI.00376-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tomilin NV, Bozhkov VM, Bradbury EM, Schmid CW. Differential binding of human nuclear proteins to Alu subfamilies. Nucleic Acids Res. 1992;20:2941–2945. doi: 10.1093/nar/20.12.2941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Matera AG, Hellmann U, Schmid CW. A transpositionally and transcriptionally competent Alu subfamily. Mol.Cell Biol. 1990;10:5424–5432. doi: 10.1128/mcb.10.10.5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Polak P, Domany E. Alu elements contain many binding sites for transcription factors and may play a role in regulation of developmental processes. BMC.Genomics. 2006;7:133. doi: 10.1186/1471-2164-7-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee EH, Rikihisa Y. Absence of tumor necrosis factor alpha, interleukin-6 (IL-6), and granulocyte-macrophage colony-stimulating factor expression but presence of IL-1beta, IL-8, and IL-10 expression in human monocytes exposed to viable or killed Ehrlichia chaffeensis. Infect.Immun. 1996;64:4211–4219. doi: 10.1128/iai.64.10.4211-4219.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee EH, Rikihisa Y. Anti-Ehrlichia chaffeensis antibody complexed with E. chaffeensis induces potent proinflammatory cytokine mRNA expression in human monocytes through sustained reduction of IkappaB-alpha and activation of NF-kappaB. Infect.Immun. 1997;65:2890–2897. doi: 10.1128/iai.65.7.2890-2897.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ismail N, Soong L, McBride JW, Valbuena G, Olano JP, Feng HM, Walker DH. Overproduction of TNF-alpha by CD8+ type 1 cells and down-regulation of IFN-gamma production by CD4+ Th1 cells contribute to toxic shock-like syndrome in an animal model of fatal monocytotropic ehrlichiosis. J.Immunol. 2004;172:1786–1800. doi: 10.4049/jimmunol.172.3.1786. [DOI] [PubMed] [Google Scholar]
- 55.Bitsaktsis C, Winslow G. Fatal recall responses mediated by CD8 T cells during intracellular bacterial challenge infection. J.Immunol. 2006;177:4644–4651. doi: 10.4049/jimmunol.177.7.4644. [DOI] [PubMed] [Google Scholar]
- 56.Barnewall RE, Rikihisa Y, Lee EH. Ehrlichia chaffeensis inclusions are early endosomes which selectively accumulate transferrin receptor. Infect.Immun. 1997;65:1455–1461. doi: 10.1128/iai.65.4.1455-1461.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lin M, Rikihisa Y. Obligatory intracellular parasitism by Ehrlichia chaffeensis and Anaplasma phagocytophilum involves caveolae and glycosylphosphatidylinositol-anchored proteins. Cell Microbiol. 2003;5:809–820. doi: 10.1046/j.1462-5822.2003.00322.x. [DOI] [PubMed] [Google Scholar]