Abstract
Genome sequencing projects have uncovered thousands of uncharacterized enzymes in eukaryotic and prokaryotic organisms. Deciphering the physiological functions of enzymes requires tools to profile and perturb their activities in native biological systems. Activity-based protein profiling has emerged as a powerful chemoproteomic strategy to achieve these objectives through the use of chemical probes that target large swaths of enzymes that share active-site features. Here, we review activity-based protein profiling and its implementation to annotate the enzymatic proteome, with particular attention given to probes that target serine hydrolases, a diverse superfamily of enzymes replete with many uncharacterized members.
Keywords: Enzyme Catalysis, Enzyme Inhibitors, Hydrolases, Peptidases, Proteomics
Introduction
Complete genome sequences have revolutionized our view of living systems. The number of genes possessed by organisms ranging from bacteria to yeast to humans is now more or less confidently assigned and has provided a framework for understanding complex cellular and physiological processes at a biochemical level. This framework is, however, plagued by huge knowledge gaps represented, perhaps most notably, by a daunting number of uncharacterized gene products. These include many predicted proteins that lack discernible sequence homology to other proteins of known function, as well as expansive protein families, of which only a modest subset of members have been assigned biochemical activities (1, 2).
Working under the Darwinian assumption that every protein has evolved to perform a unique function that ultimately benefits the host organism, it follows that significant gaps in our knowledge of the proteome imperil ongoing computational and experimental attempts to build molecular networks that explain higher order life processes. This problem is accentuated by the realization that among human protein families containing many poorly characterized members are the fundamental components of signal transduction (receptors, ion channels), gene regulation (transcription factors), and metabolic (enzymes, transporters) pathways. Ongoing and future “systems biology” endeavors would thus greatly benefit from new technologies that facilitate assignment of protein function on a global scale. These technologies can take the form of methods that map fundamental features of protein behavior, including intermolecular (e.g. gene-gene, protein-protein, protein-DNA, protein-metabolite) interactions (3, 4), cellular and subcellular localization (5), post-translational modification state (6), and biochemical activities (7–9). In this minireview, we will focus on the last category in our discussion of the chemoproteomic method activity-based protein profiling (ABPP),2 which aims to globally characterize the functional state of enzymes in native biological systems (8, 9). Our goal is to showcase how, in little more than a decade, ABPP has developed into a versatile platform for enzyme annotation in the genomic era.
Enzyme Analysis by ABPP: Serine Hydrolases as a Case Study
ABPP has been successfully applied to many enzyme classes (8). For the purposes of this minireview, however, we will focus our attention on its use to study serine hydrolases (SHs), which have served as a versatile testing group for ABPP and its various applications. SHs utilize a conserved serine nucleophile to hydrolyze amide, ester, and thioester bonds in both protein and small molecule (metabolite) substrates. They are one of the largest and most widely distributed enzyme classes in all three kingdoms of life, including humans, where SHs constitute ∼1% of the proteome (10). These include >100 serine proteases (e.g. trypsin, elastase, thrombin) that hydrolyze principally peptide bonds in proteins, as well as another 110+ SHs that act on metabolites and peptides and are hereafter referred to as mSHs. The mSHs include esterases (e.g. acetylcholinesterase), lipases (e.g. cytosolic phospholipase A2), peptidases (e.g. dipeptidyl peptidase IV), and amidases (e.g. FAAH) and can be organized into several distinct subfamilies based on three-dimensional structure and catalytic mechanism. The majority of mSHs (>60%) adopt an α,β-hydrolase fold and employ a Ser-His-Asp catalytic triad (11); however, multiple distinct and evolutionarily unrelated subclasses of mSHs also exist that utilize different folds and catalytic machinery, including the amidase signature enzymes (Ser-Ser-Lys triad) (12, 13) and patatin domain-containing lipases (Ser-Asp dyad) (14). Although many mSHs are well studied enzymes with established biochemical activities, it is striking to realize that nearly half of the human mSHs are completely uncharacterized with respect to physiological substrates and functions (Fig. 1A, blue enzymes). As will be described below, ABPP has proven especially well suited for investigating uncharacterized SHs.
FIGURE 1.
Human mSH superfamily. A, dendrogram showing all 116 members of the mSH family in humans. Branch length depicts sequence relatedness based on a sequence alignment anchored around the serine nucleophiles. The enzyme names are colored according to their degree of characterization, with characterized enzymes shown in black and uncharacterized enzymes shown in blue. B, sequence-derived evidence that most mSHs, regardless of their degree of annotation, perform unique and conserved functions in mammals. The scatter plot shows the 116 mSHs arranged according to the percent sequence identity for their nearest human mSH neighbor (homology, y axis) and mouse ortholog (x axis). Proteins appearing in the lower right-hand corner are both unique within the human proteome (as they display low sequence identity with their nearest human neighbor) and well conserved across mammalian species (as their mouse and human orthologs display strong sequence conservation).
Before proceeding further, however, it is first worth considering the potential evolutionary and functional significance of the large number of unannotated SHs found in the human proteome. Certain enzyme families, such as the cytochrome P450 class, contain many members that are subject to substantial evolutionary drift to the extent that they do not display reciprocal orthology between humans and other mammals (15). These non-orthologous enzymes also tend to exhibit promiscuous and overlapping substrate specificities, suggestive of some degree of functional redundancy. On the basis of a similar evolutionary analysis, we believe that mSHs present a different case, where most members, including those that are uncharacterized, possess conserved non-redundant biochemical functions. A visual depiction of this evolutionary argument is shown in Fig. 1B, where each human mSH is distributed across a two-dimensional plane that compares sequence similarity between 1) the most closely related mouse enzyme (orthology; x axis) and 2) the most closely related (non-identical) human enzyme (homology; y axis). The vast majority of mSHs cluster in the lower right-hand corner of the plot, reflecting both very low homology to their nearest neighbors in humans (indicating functional uniqueness) and high sequence identity with their respective mouse orthologs (indicating strong evolutionary conservation). One striking feature of this plot is that well characterized enzymes that perform specific biochemical functions in mammalian physiology, such as FAAH and acetylcholinesterase, appear alongside uncharacterized enzymes, such as ABHD13, LACTB, and BPHL. One can extrapolate from this analysis that the ∼50% of mSHs that remain unannotated are likely to play just as critical roles in human biology as the 50% of enzymes from this class that are characterized, thus underscoring the need for new functional profiling methods, like ABPP, that can be broadly applied to SHs regardless of their degree of annotation.
Fluorophosphonates as Prototypical Activity-based Probes for SHs
The principal currency of ABPP is chemistry and, more specifically, chemical probes (Fig. 2A) (8, 16). These activity-based probes contain at least two elements: 1) a reactive group to label mechanistically related enzymes in an active site-directed manner and 2) a reporter tag to visualize, enrich, and identify probe-labeled enzymes. Much of the success of ABPP has hinged on the adaptation of classical affinity labels to serve as reactive groups for probe design. For SHs, their catalytic mechanism involves formation of a covalent acyl-enzyme intermediate that has enabled development of activity-based probes that incorporate the fluorophosphonate (FP) affinity label (Fig. 2B). Early studies showed that biotinylated and fluorescently tagged FPs could be used in combination with gel-based readouts to profile dozens of SH activities in complex proteomes (17–20). Contemporaneous studies using epoxide-based probes for cysteine proteases were also described (21, 22). These efforts laid the foundation for future technical advances and biological applications of ABPP that have converged on achieving two major goals: enzyme discovery and inhibitor discovery.
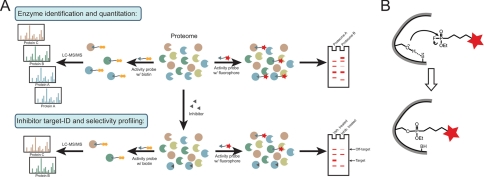
FIGURE 2.
Platforms and probes for ABPP of SHs. A, common ABPP platforms. In the upper panel, an activity-based probe conjugated to a fluorophore (right) or biotin (left) tag is shown labeling a proteome. Probe-labeled proteins can then be visualized directly via SDS-PAGE and fluorescent gel scanning (fluorophore probes) or enriched with avidin and identified by LC/LC-MS/MS (ABPP-MudPIT; biotin probes). The lower panel shows both of these techniques being applied to the characterization of inhibitors (competitive ABPP). B, covalent modification of the active-site serine nucleophile of SHs by a FP activity-based probe. The red stars indicate a tag, such as a fluorophore or biotin.
SH Discovery by ABPP
As with most proteomic methods (23), ABPP data are most straightforward to interpret when acquired in a comparative context, where differences in enzyme activities are measured across multiple biological samples. Comparative ABPP has proven particularly fruitful for discovering SH activities that are dysregulated in cancer. For instance, ABPP of a panel of human cancer cell lines that differ in pathogenicity has identified several serine proteases and mSHs that are selectively elevated in aggressive cancer cells (20, 24, 25). These enzymes include the urokinase- and tissue-type plasminogen activators, which are known to contribute to cancer malignancy, and MAGL and the uncharacterized mSH KIAA1363 (or AADACL1), for which no prior functional links to cancer had been made. Notably, some of these enzymes showed alterations in activity without corresponding changes in transcription (25), underscoring the value of ABPP for characterizing biochemical events that might evade detection by genomic methods. As will be described below, ABPP also proved critical for developing selective inhibitors of KIAA1363 and MAGL, which were then used to characterize the function of these enzymes in cancer.
The aforementioned SH activities were detected using fluorescent FP probes and one-dimensional SDS-PAGE readouts, which are highly useful methods because of their technical simplicity (Fig. 2A). However, the inherent resolution of one-dimensional SDS-PAGE limits the number of enzymes that can be comparatively analyzed in a single ABPP experiment. More recently, ABPP has been coupled with a shotgun LC-MS-based platform termed MudPIT (multidimensional protein identification technology) to greatly improve the depth of proteome coverage achievable in ABPP experiments (Fig. 2A) (26). This technique, known as ABPP-MudPIT, has been used to identify 50–100+ enzyme activities per proteomic sample and can provide semiquantitative information on the relative levels of enzyme activities across different samples. Additional LC-MS platforms for ABPP have also been described, including a method in which the probe-labeled peptides (instead of probe-labeled proteins) are enriched and identified (27, 28). These platforms facilitated the annotation of sialic acid 9-O-acetylesterase as an SH that lacks sequence homology to other enzymes from this class (29).
ABPP-MudPIT has seen diverse utility, including the characterization of SH activities in primary human breast tumors (26). In this study, more than one-third of the >50+ enzymes identified by ABPP represented uncharacterized SHs, demonstrating that FP probes are broadly applicable for the analysis of enzymes from this class regardless of their degree of annotation. Indeed, a survey of the literature (24, 26, 27, 30) and our own unpublished studies3 indicates that >80% of human/mouse mSHs have already been detected in one or more ABPP experiment using FP probes.
In a more recent application, ABPP-MudPIT was used to comprehensively profile enzymes in mouse brain and to identify novel mSHs that degrade 2-arachidonoylglycerol (2-AG) (30), an endogenous ligand for cannabinoid receptors (31, 32). This latter approach began with the observation that FP probes completely ablated 2-AG hydrolysis in brain lysates, suggesting that one or more SHs catalyzed this reaction. ABPP-MudPIT identified >30 brain SHs, which were then heterologously expressed and individually screened for 2-AG-hydrolyzing activity. Semiquantitative estimates of SH activity levels in brain (calculated by spectral counting of ABPP-MudPIT data) were then used to designate MAGL as the major brain 2-AG hydrolase, accounting for ∼85% of the total activity, with most of the remaining activity being assigned to two uncharacterized mSHs, ABHD6 and ABHD12. These findings and others (33) suggest that multiple enzymes can contribute to 2-AG hydrolysis in mammalian tissues. Other biological applications of ABPP include characterization of SH activities in adipose tissue (34), immune cell function (35), cancer metastasis (36), bacterial (37, 38) and fungal (39) pathogenesis, and natural product biosynthesis (40), as well as the analysis of other enzyme classes in cancer (41, 42) and infectious diseases (43).
SH Inhibitor Discovery by ABPP
As may be surmised from the aforementioned section, a major feature of ABPP is its capacity to acquire functional information on uncharacterized enzymes (e.g. a link between KIAA1363 and cancer, a potential role for ABHD6/ABHD12 in endocannabinoid metabolism). Still, the more complete annotation of these enzymes, including assignment of their endogenous substrates and products and (patho)physiological functions, requires selective inhibitors to perturb their activity in living systems. Indeed, one could make a similar argument for many other SHs that fall into the category of “characterized” enzymes because their functional analysis has often been restricted to in vitro (“test tube”) biochemistry experiments due to a lack of selective inhibitors for in vivo studies. Here, ABPP has found its second major application, viz. as a platform for the discovery and optimization of enzyme inhibitors (18, 22, 44).
Inhibitor discovery by ABPP is possible because small molecules can compete with activity-based probes for binding to enzyme active sites, thereby slowing the rate of probe labeling. These reductions in probe labeling can be detected by either gel-based (22, 44, 45) or LC-MS-based (45, 46) platforms (Fig. 2A). Competitive ABPP has several advantages for inhibitor discovery over classical substrate assays. First, enzymes can be screened in their native proteomes without requiring recombinant expression or purification. Second, the activity-based probe serves as a “universal” assay for its enzyme targets regardless of their degree of functional annotation. Thus, inhibitors can be developed for totally uncharacterized enzymes. Third, inhibitors are screened against many enzymes in parallel (all of the protein targets of an activity-based probe present in a test proteome), enabling concurrent optimization of inhibitor potency and selectivity.
Competitive ABPP has played a central role in the development of inhibitors for several mSHs, including the endocannabinoid-degrading enzymes FAAH (44, 47) and MAGL (46, 48), as well as the cancer-associated enzyme KIAA1363 (49). These inhibitors have been used, in combination with phenotypic and metabolic profiling, to confirm the role of FAAH and MAGL in endocannabinoid metabolism (44, 46–48) and to demonstrate that KIAA1363 and MAGL regulate ether lipid (49) and fatty acid (24) metabolism in aggressive cancer cells, respectively. More recently, competitive ABPP has been combined with library screening to identify lead inhibitors for uncharacterized mSHs. These screens can take on multiple forms, including lower throughput gel-based profiling (45) and high-throughput solution assays that use fluorescence polarization (fluopol) for readout of probe-enzyme labeling (50). The latter format, termed fluopol-ABPP, can be used in a 384-well format to screen 100,000s of compounds against an enzyme of interest but does require purified protein.
Conclusions and Future Challenges
Approximately a decade since its inception (17, 21), ABPP has gained considerable use as a technology that bridges the fields of chemistry, enzymology, and proteomics toward the goal of characterizing proteins and pathways in complex biological systems. Here, we have attempted to showcase some of the key attributes of ABPP using the SH superfamily as a case study.
The size and diversity of SHs, combined with the availability of broad-spectrum activity-based probes that target these enzymes, have made them a valuable prototype for ABPP. Using ABPP, investigators have discovered SH activities that are dysregulated in important diseases, such as cancer, as well as selective inhibitors for these enzymes to test their metabolic and (patho)physiological functions. In some of these cases, the enzymes under investigation were previously uncharacterized, underscoring an important feature of activity-based probes, which are generally agnostic to the degree of annotation of their enzyme targets.
Studies to date with SHs have also highlighted many of the future challenges that face the characterization of enzymes by ABPP. First, when applied in isolation, ABPP offers only limited insights into enzyme function. Only through integrating ABPP with other methods, such as emerging proteomic (51–53), peptidomic (54), and metabolomic (55) strategies to discover enzyme substrates in native biological systems, can the actual physiological activities of enzymes be defined. In cases in which this integrated approach has been applied to uncharacterized mSHs, specific metabolic functions have been discovered (e.g. KIAA1363 and its role in ether lipid metabolism in cancer cells (49)). These initial findings provide support that uncharacterized mSHs are likely to possess unique biochemical functions in cells and tissues. In this regard, it is also worth noting that even enzymes that are considered well characterized can offer surprises when studied through the lens of such large-scale profiling methods. For instance, ABPP, in combination with metabolomics, revealed that MAGL regulates fatty acids levels in aggressive cancer cells (24) even though these lipids are not under the control of MAGL in most normal tissues (56). Thus, enzymes can exhibit context-dependent differences in their metabolic functions.
A second challenge relates to the broader use of competitive ABPP to develop enzyme inhibitors. Efforts to date with SHs have yielded considerable fruit, including useful pharmacological tools for multiple characterized and uncharacterized enzymes. Still, we estimate that selective inhibitors currently exist for <20% of the SH superfamily. Expanding our pharmacological coverage of this enzyme class (and others) will likely benefit from emerging ABPP platforms (such as fluopol-ABPP) that enable much larger libraries of small molecules to be screened against enzymes in a high-throughput manner. We should also mention that competitive ABPP examines only the selectivity of inhibitors within the target enzyme class; these compounds could still have additional unrelated targets in the proteome. In cases in which lead inhibitors are covalent, a more comprehensive survey of their proteomic targets can be achieved by offshoots of ABPP in which clickable analogs are used to directly identify inhibitor-modified proteins in proteomes (57, 58).
Finally, we should briefly discuss the extent to which findings on SHs can be applied to other enzyme classes. Of the many lessons learned from ABPP studies of SHs, perhaps none is more important than recognizing the value of high-quality, broad-spectrum, activity-based probes. The FPs (and arylphosphonates) are, in many ways, ideal activity-based probes in that they show broad reactivity across the SH class but little or no cross-reactivity with other enzymes. Similar success stories can be found for a limited number of other enzyme classes (such as the cysteine proteases (59)), but most activity-based probes show either more restricted reactivity within a given enzyme class (41) or labeling of enzymes from different mechanistic classes (60). These features are not necessarily drawbacks, especially with the advent of higher resolution LC-MS platforms for identifying targets of activity-based probes (i.e. there is no a priori reason why an activity-based probe needs to selectively label enzymes from one class as long as all of its targets in the proteome can be resolved). Nonetheless, they do emphasize that chemistry will continue to play a prominent role in advancing and refining new reactive chemotypes for use in ABPP (60). From a biological perspective, SHs are certainly not special in terms of the large number of uncharacterized enzymes from this class that currently populate eukaryotic and prokaryotic proteomes. One could make a similar argument for virtually all major enzyme families. To the extent that one believes that the ∼50% of the proteome that remains uncharacterized is as interesting as (or more than) the ∼50% of the proteome that we know something about, ABPP should remain a forefront technology for ongoing and future research aimed at understanding the biochemical basis of life.
This work was supported, in whole or in part, by National Institutes of Health Grants CA087660, CA132630, and DA025285. This work was also supported by the ARCS Foundation and a Koshland Graduate Fellowship in Enzyme Biochemistry (to G. M. S.) and by the Skaggs Institute for Chemical Biology. This is the fourth of six articles in the “Chemical Biology Meets Biological Chemistry Minireview Series.” This minireview will be reprinted in the 2010 Minireview Compendium, which will be available in January, 2011.
G. M. Simon and B. F. Cravatt, unpublished data.
- ABPP
- activity-based protein profiling
- SH
- serine hydrolase
- mSH
- metabolic SH
- FAAH
- fatty acid amide hydrolase
- FP
- fluorophosphonate
- MAGL
- monoacylglycerol lipase
- LC-MS
- liquid chromatography-mass spectrometry
- 2-AG
- 2-arachidonoylglycerol
- fluopol
- fluorescence polarization.
REFERENCES
- 1.Galperin M. Y., Koonin E. V. (2004) Nucleic Acids Res. 32, 5452–5463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gerlt J. A., Babbitt P. C. (2001) Annu. Rev. Biochem. 70, 209–246 [DOI] [PubMed] [Google Scholar]
- 3.Köcher T., Superti-Furga G. (2007) Nat. Methods 4, 807–815 [DOI] [PubMed] [Google Scholar]
- 4.Collins S. R., Weissman J. S., Krogan N. J. (2009) Nat. Methods 6, 721–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., O'Shea E. K. (2003) Nature 425, 686–691 [DOI] [PubMed] [Google Scholar]
- 6.Zhao Y., Jensen O. N. (2009) Proteomics 9, 4632–4641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bertone P., Snyder M. (2005) FEBS J. 272, 5400–5411 [DOI] [PubMed] [Google Scholar]
- 8.Cravatt B. F., Wright A. T., Kozarich J. W. (2008) Annu. Rev. Biochem. 77, 383–414 [DOI] [PubMed] [Google Scholar]
- 9.Berger A. B., Vitorino P. M., Bogyo M. (2004) Am. J. Pharmaco Genomics 4, 371–381 [DOI] [PubMed] [Google Scholar]
- 10.Rawlings N. D., Barrett A. J., Bateman A. (2010) Nucleic Acids Res. 38, D227–D233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holmquist M. (2000) Curr. Protein Pept. Sci. 1, 209–235 [DOI] [PubMed] [Google Scholar]
- 12.Shin S., Lee T. H., Ha N. C., Koo H. M., Kim S. Y., Lee H. S., Kim Y. S., Oh B. H. (2002) EMBO J. 21, 2509–2516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bracey M. H., Hanson M. A., Masuda K. R., Stevens R. C., Cravatt B. F. (2002) Science 298, 1793–1796 [DOI] [PubMed] [Google Scholar]
- 14.Kienesberger P. C., Oberer M., Lass A., Zechner R. (2009) J. Lipid Res. 50, (suppl.) S63–S68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guengerich F. P., Wu Z. L., Bartleson C. J. (2005) Biochem. Biophys. Res. Commun. 338, 465–469 [DOI] [PubMed] [Google Scholar]
- 16.Sadaghiani A. M., Verhelst S. H., Bogyo M. (2007) Curr. Opin. Chem. Biol. 11, 20–28 [DOI] [PubMed] [Google Scholar]
- 17.Liu Y., Patricelli M. P., Cravatt B. F. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 14694–14699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kidd D., Liu Y., Cravatt B. F. (2001) Biochemistry 40, 4005–4015 [DOI] [PubMed] [Google Scholar]
- 19.Patricelli M. P., Giang D. K., Stamp L. M., Burbaum J. J. (2001) Proteomics 1, 1067–1071 [DOI] [PubMed] [Google Scholar]
- 20.Jessani N., Liu Y., Humphrey M., Cravatt B. F. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 10335–10340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Greenbaum D., Medzihradszky K. F., Burlingame A., Bogyo M. (2000) Chem. Biol. 7, 569–581 [DOI] [PubMed] [Google Scholar]
- 22.Greenbaum D., Baruch A., Hayrapetian L., Darula Z., Burlingame A., Medzihradszky K. F., Bogyo M. (2002) Mol. Cell. Proteomics 1, 60–68 [DOI] [PubMed] [Google Scholar]
- 23.Cravatt B. F., Simon G. M., Yates J. R., 3rd (2007) Nature 450, 991–1000 [DOI] [PubMed] [Google Scholar]
- 24.Nomura D. K., Long J. Z., Niessen S., Hoover H. S., Ng S. W., Cravatt B. F. (2010) Cell 140, 49–61 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jessani N., Humphrey M., McDonald W. H., Niessen S., Masuda K., Gangadharan B., Yates J. R., 3rd, Mueller B. M., Cravatt B. F. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 13756–13761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jessani N., Niessen S., Wei B. Q., Nicolau M., Humphrey M., Ji Y., Han W., Noh D. Y., Yates J. R., 3rd, Jeffrey S. S., Cravatt B. F. (2005) Nat. Methods 2, 691–697 [DOI] [PubMed] [Google Scholar]
- 27.Okerberg E. S., Wu J., Zhang B., Samii B., Blackford K., Winn D. T., Shreder K. R., Burbaum J. J., Patricelli M. P. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 4996–5001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Speers A. E., Cravatt B. F. (2005) J. Am. Chem. Soc. 127, 10018–10019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jessani N., Young J. A., Diaz S. L., Patricelli M. P., Varki A., Cravatt B. F. (2005) Angew. Chem. Int. Ed. Engl. 44, 2400–2403 [DOI] [PubMed] [Google Scholar]
- 30.Blankman J. L., Simon G. M., Cravatt B. F. (2007) Chem. Biol. 14, 1347–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mechoulam R., Ben-Shabat S., Hanus L., Ligumsky M., Kaminski N. E., Schatz A. R., Gopher A., Almog S., Martin B. R., Compton D. R., Pertwee R. G., Griffin G., Bayewitch M., Barg J., Vogel Z. (1995) Biochem. Pharmacol. 50, 83–90 [DOI] [PubMed] [Google Scholar]
- 32.Sugiura T., Kondo S., Sukagawa A., Nakane S., Shinoda A., Itoh K., Yamashita A., Waku K. (1995) Biochem. Biophys. Res. Commun. 215, 89–97 [DOI] [PubMed] [Google Scholar]
- 33.Muccioli G. G., Xu C., Odah E., Cudaback E., Cisneros J. A., Lambert D. M., López Rodríguez M. L., Bajjalieh S., Stella N. (2007) J. Neurosci. 27, 2883–2889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morak M., Schmidinger H., Krempl P., Rechberger G., Kollroser M., Birner-Gruenberger R., Hermetter A. (2009) J. Lipid Res. 50, 1281–1292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mahrus S., Craik C. S. (2005) Chem. Biol. 12, 567–577 [DOI] [PubMed] [Google Scholar]
- 36.Madsen M. A., Deryugina E. I., Niessen S., Cravatt B. F., Quigley J. P. (2006) J. Biol. Chem. 281, 15997–16005 [DOI] [PubMed] [Google Scholar]
- 37.Böttcher T., Sieber S. A. (2008) J. Am. Chem. Soc. 130, 14400–14401 [DOI] [PubMed] [Google Scholar]
- 38.Staub I., Sieber S. A. (2009) J. Am. Chem. Soc. 131, 6271–6276 [DOI] [PubMed] [Google Scholar]
- 39.Kaschani F., Gu C., Niessen S., Hoover H., Cravatt B. F., van der Hoorn R. A. (2009) Mol. Cell. Proteomics 8, 1082–1093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Meier J. L., Niessen S., Hoover H. S., Foley T. L., Cravatt B. F., Burkart M. D. (2009) ACS Chem. Biol. 4, 948–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sieber S. A., Niessen S., Hoover H. S., Cravatt B. F. (2006) Nat. Chem. Biol. 2, 274–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Joyce J. A., Baruch A., Chehade K., Meyer-Morse N., Giraudo E., Tsai F. Y., Greenbaum D. C., Hager J. H., Bogyo M., Hanahan D. (2004) Cancer Cell 5, 443–453 [DOI] [PubMed] [Google Scholar]
- 43.Greenbaum D. C., Baruch A., Grainger M., Bozdech Z., Medzihradszky K. F., Engel J., DeRisi J., Holder A. A., Bogyo M. (2002) Science 298, 2002–2006 [DOI] [PubMed] [Google Scholar]
- 44.Leung D., Hardouin C., Boger D. L., Cravatt B. F. (2003) Nat. Biotechnol. 21, 687–691 [DOI] [PubMed] [Google Scholar]
- 45.Li W., Blankman J. L., Cravatt B. F. (2007) J. Am. Chem. Soc. 129, 9594–9595 [DOI] [PubMed] [Google Scholar]
- 46.Long J. Z., Li W., Booker L., Burston J. J., Kinsey S. G., Schlosburg J. E., Pavón F. J., Serrano A. M., Selley D. E., Parsons L. H., Lichtman A. H., Cravatt B. F. (2009) Nat. Chem. Biol. 5, 37–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ahn K., Johnson D. S., Fitzgerald L. R., Liimatta M., Arendse A., Stevenson T., Lund E. T., Nugent R. A., Nomanbhoy T. K., Alexander J. P., Cravatt B. F. (2007) Biochemistry 46, 13019–13030 [DOI] [PubMed] [Google Scholar]
- 48.Long J. Z., Nomura D. K., Vann R. E., Walentiny D. M., Booker L., Jin X., Burston J. J., Sim-Selley L. J., Lichtman A. H., Wiley J. L., Cravatt B. F. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 20270–20275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chiang K. P., Niessen S., Saghatelian A., Cravatt B. F. (2006) Chem. Biol. 13, 1041–1050 [DOI] [PubMed] [Google Scholar]
- 50.Bachovchin D. A., Brown S. J., Rosen H., Cravatt B. F. (2009) Nat. Biotechnol. 27, 387–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dix M. M., Simon G. M., Cravatt B. F. (2008) Cell 134, 679–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mahrus S., Trinidad J. C., Barkan D. T., Sali A., Burlingame A. L., Wells J. A. (2008) Cell 134, 866–876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Doucet A., Butler G. S., Rodríguez D., Prudova A., Overall C. M. (2008) Mol. Cell. Proteomics 7, 1925–1951 [DOI] [PubMed] [Google Scholar]
- 54.Tagore D. M., Nolte W. M., Neveu J. M., Rangel R., Guzman-Rojas L., Pasqualini R., Arap W., Lane W. S., Saghatelian A. (2009) Nat. Chem. Biol. 5, 23–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saghatelian A., Trauger S. A., Want E. J., Hawkins E. G., Siuzdak G., Cravatt B. F. (2004) Biochemistry 43, 14332–14339 [DOI] [PubMed] [Google Scholar]
- 56.Long J. Z., Nomura D. K., Cravatt B. F. (2009) Chem. Biol. 16, 744–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Alexander J. P., Cravatt B. F. (2005) Chem. Biol. 12, 1179–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cohen M. S., Hadjivassiliou H., Taunton J. (2007) Nat. Chem. Biol. 3, 156–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kato D., Boatright K. M., Berger A. B., Nazif T., Blum G., Ryan C., Chehade K. A., Salvesen G. S., Bogyo M. (2005) Nat. Chem. Biol. 1, 33–38 [DOI] [PubMed] [Google Scholar]
- 60.Weerapana E., Simon G. M., Cravatt B. F. (2008) Nat. Chem. Biol. 4, 405–407 [DOI] [PMC free article] [PubMed] [Google Scholar]