Abstract
The functions of many cellular proteins have been elucidated by selective gene inactivation and subsequent phenotypic analysis. For example, genetic mutations, gene knock-out generation, and the use of RNA interference to target mRNA for degradation can all result in decreased production of a specific protein, yielding informative cellular phenotypes. However, these techniques each have certain inherent limitations. This minireview focuses on the recent development of new approaches to study protein function at the post-translational level, namely chemical induction of targeted protein degradation.
Keywords: Enzymes/Peptidases, Enzymes/Proteolytic, Proteases/Proteasomes, Proteases/Ubiquitin, Proteases/Ubiquitination, Protein/Targeting, Protein/Turnover, Chemical Biology
Introduction
Traditionally, protein function has been investigated by making changes in DNA sequences that encode a protein and monitoring the resulting phenotype. Chemical genetics represents an orthogonal approach to study protein function whereby cell-permeable small molecules are used to interfere with gene expression at the DNA, RNA, or post-translational levels. In this manner, the activity of a particular protein can be modulated and its role in cell biology studied through the resultant phenotypic changes. At the post-transcriptional level, RNAi2 has emerged as a useful tool for gene silencing due to its ability to knock down levels of any protein with a known sequence. Since its discovery, RNAi has been seen in plants (1), fungi (2), and a variety of other organisms, including mammals (3, 29). Significantly, the production of large-scale small interfering RNA and short hairpin RNA libraries has made genome-wide RNAi analysis possible.
Even though RNAi has generated an explosion of interest due to its possible therapeutic applications, it has certain limitations. First, RNAi causes no decrease in levels of protein already present within cells, leaving stable proteins with a long half-life unaffected. Second, because RNAi is a catalytic process, it lacks concentration dependence and offers limited temporal control over levels of protein expression. Moreover, its effect is gradual and irreversible compared with small molecules. This can lead to false negatives when it is used as a screening tool and also to complex phenotypes due to cells adapting to the slow protein depletion. Third, RNAi can lead to unintentional degradation of mRNA containing a partial sequence overlap with the target mRNA (4) and to saturation of the endogenous RNAi apparatus, resulting in impaired regulation of other cellular processes. Finally, even short small interfering RNAs can trigger the interferon response at high concentrations (5).
Despite the success of RNAi-based approaches, there clearly remains a need for general techniques to regulate protein levels directly. A variety of methods for post-translational protein knockdown have been developed. These techniques differ significantly from RNAi in that rather than preventing the synthesis of new proteins, they destroy proteins that have already been synthesized. A major advantage of post-translational inactivation lies in the fact that stable proteins with long half-lives that are inaccessible with RNAi can be effectively targeted.
Degradation by Localization to the Proteasome
The ATP-dependent ubiquitin-proteasome system is the major route for breakdown of intracellular proteins (6) and has been implicated in the regulation of diverse cellular processes such as cell cycle progression (7), transcription (8), and the inflammatory response (9). The pathway involves a cascade of enzymatic reactions, resulting in the covalent tagging of a protein for degradation through its lysine ϵ-amino groups with a highly conserved 76-amino acid protein called ubiquitin (10). Following this initial attachment, successive conjugation reactions occur to add additional copies of ubiquitin to the initial monomer, leading to the formation of a polyubiquitin chain (11), which is recognized by the 26 S proteasome. The target protein is subsequently deubiquitinated, unfolded, and threaded into the proteolytic core of the proteasome, where it is degraded into short polypeptide fragments. Substrate specificity of the ubiquitin-proteasome system is mediated by E3 ubiquitin ligases, which are part of the initial enzyme cascade responsible for tagging the substrate protein with ubiquitin. Each E3 ligase or recognition subunit of a multiprotein E3 ligase complex has its own cognate set of substrate proteins that it recognizes and helps tag for destruction.
One approach to targeting specific proteins for degradation based on localization of the target protein directly to the proteasome by utilizing the FKBP-rapamycin-FRB interaction in yeast was reported by Janse et al. (12). The authors fused the C termini of seven different non-catalytic proteasomal subunits ranging in distance from the 20 S core with FKBP12. The chromosomal copy of the auxotrophic marker HIS3 was deleted from the four strains that proved to be viable, making the expression of exogenous His3 necessary for a normal growth phenotype upon culture in histidine-dropout medium. Two reporter protein constructs were then designed for use in a screen for growth-deficient phenotypes. The ligand-binding domain of Tor1 (FRB) was fused to full-length His3 and used as the reporter in the FKBP-tagged strains, whereas a His3-FRB sequence containing a Tor1(S1972R) mutant with decreased affinity for the rapamycin-FKBP complex was used as the control. Each transformant was then spotted on experimental plates with histidine-dropout medium either containing or lacking rapamycin. Of the four strains, FKBP-tagged Rpn10 and Pre10 transformants showed a significant rapamycin-dependent growth-deficient phenotype compared with the low-affinity control mutant. Subsequent immunoblot analysis demonstrated that degradation of the His3-FRB reporter in these strains was brought about by FKBP-rapamycin-FRB heterotrimer formation at the proteasome.
Although this result indicates that polyubiquitination of proteins may not strictly be required for proteolysis, the efficacy of targeting specific proteins for degradation by direct localization to the proteasome has not yet been demonstrated in other systems. However, if shown to have wider applicability, this technique might represent a powerful general means of selective protein knockdown.
Destabilizing Domain Method
A complementary means to regulate cellular concentrations of specific proteins using a chemical genetic technique involves the fusion of the target protein with a degron, which is a small protein domain that is unstable when expressed in cells and confers this instability to its fusion partner. The target protein can be rescued from degradation by introduction of a small molecule that binds and stabilizes the degron, thus offering a rapid and tunable method of controlling the concentration of a protein within a cell.
Pioneering work in this direction was carried out by Szostak and co-workers (13), who showed that N-terminal fusion of a small unstable peptide sequence to a protein of interest was sufficient to cause degradation of the latter in yeast. Varshavsky and co-workers (14) added temporal control and reversibility to this strategy by engineering a mutant dihydrofolate reductase degron that was stabilized in the presence of the high-affinity ligand methotrexate. More recently, Crabtree and co-workers (15) used an 89-amino acid mutant FRB domain (FRB*) as a degron to transmit instability to the kinase glycogen synthase kinase-3β, which could be rescued by addition of the rapamycin analog MaRap, which binds to FRB* but not FRB in cultured cells from knock-in mice as well as in mouse embryos.
However, MaRap has limitations as a stabilizing ligand because of its poor pharmacokinetic properties and the need to bind to FKBP12 before it can recruit FRB*, necessitating two separate binding events. To circumvent these issues, Wandless and co-workers (16) selected FKBP12 itself as a potential degron and generated a library of FKBP12 mutants through error-prone PCR, which were then fused to yellow fluorescent protein and expressed in NIH3T3 cells. Several rounds of screening yielded a construct that exhibited fluorescence in the presence of a high-affinity FKBP12 ligand termed Shield-1 but not in its absence. The efficacy of this kind of mutant FKBP-Shield-1 system in controlling protein function was demonstrated with a variety of proteins in cultured cells (16) and in mice (17). This method has also been shown to work when the degron is spliced into the middle of a protein. Additional FKBP mutants that lend themselves better to C-terminal fusions have been characterized (18).
SURF Technology
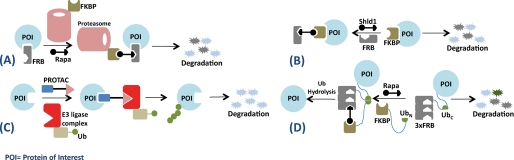
SURF technology was recently developed by Pratt et al. (19) and makes use of the previously described degron approach with an additional feature enabling release of the native protein from the degron sequence upon small molecule-induced rescue of the protein-degron chimera from degradation. The authors split ubiquitin into N-terminal UbN(I13A) (residues 1–37) and C-terminal UbC (residues 35–76) fragments. Two protein constructs were then prepared: one consisting of the protein of interest N-terminally fused to UbC, which was further linked to a FRB(W2101F) mutant degron, and the other containing FKBP fused to UbN(I13A). The I13A mutation in UbN was made to ensure that the ubiquitin fragments complemented and folded and subsequently cleaved from the fusion construct to release the native protein of interest only in the presence of the small molecule signal. The authors utilized the strategy described above to control the amounts of caspase-3, v-Src, and Smad3 in HeLa cells with rapamycin as the small molecule effector of stabilization through the formation of the FKBP-rapamycin-FRB heterotrimer. An important advantage of this approach over the destabilizing domain method described earlier is the release of the fully functional native protein upon rapamycin-induced rescue from degradation (Fig. 1).
FIGURE 1.
Chemical means of controlling protein function. A, direct localization to the proteasome; B, the destabilizing domain approach; C, PROTAC-mediated degradation; D, SURF technology. Ub, ubiquitin.
PROTACs
Several years ago, our group, in collaboration with the Deshaies laboratory at California Institute of Technology, developed the PROTAC approach, which utilizes the endogenous proteolytic pathway to target proteins for degradation (20). PROTACs are heterobifunctional molecules that contain a recognition element for the target protein attached to a recognition element for an E3 ligase. These molecules induce proximity between the E3 ligase and the targeted protein substrate and the resulting ubiquitination of the latter, tagging it for degradation.
This approach was first validated in an in vitro system targeting MetAP-2 (20). Ovalicin, a covalent binder of MetAP-2, was conjugated to a 10-amino acid phosphopeptide sequence derived from IκBα. This sequence mediates binding of IκBα to the mammalian F-box β-transducin repeat-containing protein (β-TRCP), which results in degradation of the former in the proteasome. In Xenopus egg extracts, the MetAP-2-PROTAC construct was degraded in a proteasome-dependent manner as evidenced by attenuated degradation in extracts supplemented with proteasome inhibitors but not in the presence of other protease inhibitors.
Given their role in prostate and breast cancer, the AR and ER were targeted by the next generation of PROTACs. An estradiol-IκBα PROTAC was shown to degrade the ER in a cell-free system, whereas microinjection of a dihydroxytestosterone-IκBα PROTAC into an HEK293 cell line stably expressing AR-GFP resulted in a decrease in the levels of GFP due to proteasome-dependent degradation (21). These experiments demonstrate that PROTACs can recruit target proteins via non-covalent interactions.
The first cell-permeable PROTAC contained the artificial ligand AP21998, known to bind the F36V mutant of FKBP12 (22), coupled to a 7-amino acid sequence that is the part of HIF1α responsible for its recognition by the Von Hippel-Lindau E3 ligase complex (23). A poly-d-Arg tag was incorporated into this PROTAC to enhance cellular uptake. This PROTAC effectively degraded a mutant FKBP-GFP hybrid protein upon addition to HeLa cells. In addition, a dihydroxytestosterone-HIF-Arg8 construct was shown to degrade the AR in HEK293 cells expressing AR-GFP. Cell-permeable PROTACs consisting of fumagillin, apigenin, and estrogen derivatives conjugated via a linker to the HIF1α peptide sequence, designed to target MetAP-2, the aryl hydrocarbon receptor, and the ER, respectively, have subsequently been reported by Kim and co-workers (24, 25). To address the cell permeability issue associated with the high molecular weight of earlier PROTACs, our group reported the first all-small molecule PROTAC targeting the AR in HeLa cells (26). This PROTAC consisted of a selective androgen receptor modulator (SARM) ligand attached via a soluble polyethylene glycol linker to an imidazoline derivative known to bind the E3 ligase MDM2. Substantial degradation of the AR was achieved upon incubation of HeLa cells with micromolar concentrations of the PROTAC.
Extensions of the PROTAC Methodology
Although the degron approach has been shown to be useful in cultured cells, the apicomplexan parasite Toxoplasma gondii (27), and mouse xenograft models, it has not yet been demonstrated in non-xenograft vertebrate models. This would require the creation of transgenic organisms, which is a significant undertaking. The need for purely small molecule routes to targeted protein degradation is thus further enhanced.
In addition to advantages an effective PROTAC methodology has over RNAi, such as dose-dependent knockdown, high permeability, rapid effect, and destruction even of existing copies of the protein of interest, it offers an edge by virtue of the additional layer of information offered by the three-dimensional structure of the protein that is unavailable with the use of RNAi. For example, specific conformational subpopulations of important oncogenic signaling proteins such as Ras are inaccessible through RNAi but could be targeted for degradation with a PROTAC containing the appropriate conformation-specific target-binding ligand.
PROTACs also offer a distinct advantage over conventional small molecule drugs that depend on functional inhibition of enzymatic activity to disrupt a specific pathway. PROTACs are not dependent on an enzyme active site and can function purely through binding to any accessible protein surface, and this lends them broader applications as a potential therapeutic tool in view of the fact that ∼80% of the eukaryotic proteome has no enzymatic activity (28).
Although PROTACs open up a wide spectrum of exciting applications, a number of issues remain outstanding and will be addressed by future research in this area. The length of the linker between the two affinity domains has to be optimized for each E3 ligase-target protein pair, and there is still no control over the subcellular distribution of PROTACs. In addition, the lack of high-affinity ligands for E3 ligases limits the potency of current PROTAC technology. However, it is not inconceivable that with the development of more expansive and more rapid screening technology, PROTACs could emerge as a highly specific, robust, and versatile tool to complement the traditional reverse genetic tools such as RNAi, ribozymes, and gene knock-outs in the study of protein function. The independence of this technology from any genetic modifications also gives it substantial potential value in therapeutics. The design of PROTAC libraries to illuminate new signaling pathways and novel proteins would go a long way in making it a useful investigative tool in disease research. Much more work needs to be done, however, before the true breadth of applications of targeted protein degradation through chemical means can be determined.
Supplementary Material
Acknowledgments
We thank Tom Sundberg and John Hines for critical evaluation of the manuscript.
This is the fifth of six articles in the “Chemical Biology Meets Biological Chemistry Minireview Series.” This minireview will be reprinted in the 2010 Minireview Compendium, which will be available in January, 2011.
- RNAi
- RNA interference
- FKBP
- FK506-binding protein
- SURF
- split ubiquitin rescue of protein function
- PROTAC
- proteolysis-targeting chimeric molecule
- MetAP-2
- methionine aminopeptidase-2
- AR
- androgen receptor
- ER
- estrogen receptor
- GFP
- green fluorescent protein
- HIF1α
- hypoxia-inducing factor 1α
- FRB
- FKBP12-rapamycin-binding domain of FRAP.
REFERENCES
- 1.Vaucheret H. (2006) Genes Dev. 20, 759–771 [DOI] [PubMed] [Google Scholar]
- 2.Nakayashiki H., Kadotani N., Mayama S. (2006) J. Mol. Evol. 63, 127–135 [DOI] [PubMed] [Google Scholar]
- 3.McCaffrey A. P., Meuse L., Pham T. T., Conklin D. S., Hannon G. J., Kay M. A. (2002) Nature 270, 38–39 [DOI] [PubMed] [Google Scholar]
- 4.Jackson A. L., Bartz S. R., Schelter J., Kobayashi S. V., Burchard J., Mao M., Li B., Cavet G., Linsley P. S. (2003) Nat. Biotechnol. 21, 635–637 [DOI] [PubMed] [Google Scholar]
- 5.Sledz C. A., Holko M., de Veer M. J., Silverman R. H., Williams B. R. (2003) Nat. Cell Biol. 5, 834–839 [DOI] [PubMed] [Google Scholar]
- 6.Rock K. L., Gramm C., Rothstein L., Clark K., Stein R., Dick L., Hwang D., Goldberg A. L. (1994) Cell 78, 761–771 [DOI] [PubMed] [Google Scholar]
- 7.Koepp D. M., Harper J. W., Elledge S. J. (1999) Cell 97, 431–434 [DOI] [PubMed] [Google Scholar]
- 8.Muratani M., Tansey W. P. (2003) Nat. Rev. Mol. Cell Biol. 4, 192–201 [DOI] [PubMed] [Google Scholar]
- 9.Ben-Neriah Y. (2002) Nat. Immunol. 3, 20–26 [DOI] [PubMed] [Google Scholar]
- 10.Vijay-Kumar S., Bugg C. E., Wilkinson K. D., Vierstra R. D., Hatfield P. M., Cook W. J. (1987) J. Biol. Chem. 262, 6396–6399 [PubMed] [Google Scholar]
- 11.Pickart C. M. (2001) Annu. Rev. Biochem. 3, 503–533 [DOI] [PubMed] [Google Scholar]
- 12.Janse D. M., Crosas B., Finley D., Church G. M. (2004) J. Biol. Chem. 279, 21415–21420 [DOI] [PubMed] [Google Scholar]
- 13.Park E. C., Finley D., Szostak J. W. (1992) Proc. Natl. Acad. Sci. U.S.A. 89, 1249–1252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnston J. A., Johnson E. S., Waller P. R., Varshavsky A. (1995) J. Biol. Chem. 270, 8172–8178 [DOI] [PubMed] [Google Scholar]
- 15.Stankunas K., Bayle J. H., Gestwicki J. E., Lin Y. M., Wandless T. J., Crabtree G. R. (2003) Mol. Cell 12, 1615–1624 [DOI] [PubMed] [Google Scholar]
- 16.Banaszynski L. A., Chen L. C., Maynard-Smith L. A., Ooi A. G., Wandless T. J. (2006) Cell 126, 995–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Banaszynski L. A., Sellmyer M. A., Contag C. H., Wandless T. J., Thorne S. H. (2008) Nat. Med. 14, 1123–1127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chu B. W., Banaszynski L. A., Chen L. C., Wandless T. J. (2008) Bioorg. Med. Chem. Lett. 18, 5941–5944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pratt M. R., Schwartz E. C., Muir T. W. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 11209–11214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sakamoto K. M., Kim K. B., Kumagai A., Mercurio F., Crews C. M., Deshaies R. J. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 8554–8559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sakamoto K. M., Kim K. B., Verma R., Ransick A., Stein B., Crews C. M., Deshaies R. J. (2003) Mol. Cell. Proteomics 2, 1350–1358 [DOI] [PubMed] [Google Scholar]
- 22.Rollins C. T., Rivera V. M., Woolfson D. N., Keenan T., Hatada M., Adams S. E., Andrade L. J., Yaeger D., van Schravendijk M. R., Holt D. A., Gilman M., Clackson T. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 7096–7101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hon W. C., Wilson M. I., Harlos K., Claridge T. D., Schofield C. J., Pugh C. W., Maxwell P. H., Ratcliffe P. J., Stuart D. I., Jones E. Y. (2002) Nature 417, 975–978 [DOI] [PubMed] [Google Scholar]
- 24.Zhang D., Baek S. H., Ho A., Kim K. B. (2004) Bioorg. Med. Chem. Lett. 14, 645–648 [DOI] [PubMed] [Google Scholar]
- 25.Lee H., Puppala D., Choi E. Y., Swanson H., Kim K. B. (2007) ChemBioChem 8, 2058–2062 [DOI] [PubMed] [Google Scholar]
- 26.Schneekloth A. R., Pucheault M., Tae H. S., Crews C. M. (2008) Bioorg. Med. Chem. Lett. 18, 5904–5908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Herm-Götz A., Agop-Nersesian C., Münter S., Grimley J. S., Wandless T. J., Frischknecht F., Meissner M. (2007) Nat. Methods 4, 1003–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arakaki A. K., Tian W., Skolnick J. (2006) BMC Genomics 7, 315–332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lewis D. L., Hagstrom J. E., Loomis A. G., Wolff J. A., Herweijer H. (2002) Nat. Genet. 32, 107–108 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.