Abstract
Meiotic recombination requires the formation of programmed Spo11-dependent DNA double strand breaks (DSBs). In Saccharomyces cerevisiae, the Sae2 protein and the Mre11-Rad50-Xrs2 complex are necessary to remove the covalently attached Spo11 protein from the DNA ends, which are then resected by so far unknown nucleases. Here, we demonstrate that phosphorylation of Sae2 Ser-267 by cyclin-dependent kinase 1 (Cdk1) is required to initiate meiotic DSB resection by allowing Spo11 removal from DSB ends. This finding suggests that Cdk1 activity is required for the processing of Spo11-induced DSBs, thus providing a mechanism for coordinating DSB resection with progression through meiotic prophase. Furthermore, the helicase Sgs1 and the nucleases Exo1 and Dna2 participate in lengthening the 5′-3′ resection tracts during meiosis by controlling a step subsequent to Spo11 removal.
Keywords: DNA Damage, DNA Enzymes, DNA Recombination, DNA Repair, Mutant, Yeast Genetics
Introduction
During the first meiotic division, homologous maternal and paternal chromosomes are segregated. In most organisms, homologs must be physically connected to ensure their proper segregation (1). By virtue of cohesion between sister chromatids, the exchange of chromosome arms through chiasmata formation provides the physical connections between homologous chromosomes. Chiasmata are generated by recombination events, which are initiated by the formation of self-inflicted DNA double strand breaks (DSBs).3
DSB formation requires meiosis-specific gene products, including the evolutionary conserved topoisomerase-like enzyme Spo11, as well as the three components of the MRX complex (Mre11-Rad50-Xrs2) (2). In particular, a Spo11 dimer coordinately breaks both DNA strands, creating a DSB with covalent linkages between the 5′ DNA ends and the catalytic tyrosine residue of each Spo11 monomer (3). Then, Spo11 must be removed by endonucleolytic cleavage to allow further DSB end processing by 5′-3′ resection that is required to initiate homologous recombination (4). This event is promoted by the Sae2 protein and the MRX complex, which are required to catalyze the endonucleolytic removal of Spo11-linked oligonucleotides (3, 5, 6). In fact, budding yeast sae2Δ cells and rad50s separation-of-function mutants allow DSB formation but are totally defective in Spo11 removal from DSB ends (3, 5, 7–9). Similarly, mre11 alleles impairing Mre11 nuclease activity allow Spo11-induced DSB formation, but not Spo11 removal (10–12), suggesting that the latter may take place by Mre11-catalyzed endonucleolytic cleavage and that Sae2 participates in this process. As recently shown, also Sae2 exhibits an endonuclease activity (13), suggesting that this protein, possibly in cooperation with MRX, may allow Spo11 removal by mediating an endonucleolytic cleavage close to the DNA end.
Because DSBs are highly hazardous for genome stability, commitment to DSB resection and meiotic progression must be tightly regulated to ensure proper DSB repair. In vegetative Saccharomyces cerevisiae cells, DSB resection is promoted by the activity of the cyclin-dependent protein kinase Cdk1 (Cdc28/Clb) during the S and G2 cell cycle phases (14, 15). This control relies on the phosphorylation of Sae2 Ser-267 by Cdk1 (16), a mechanism that is conserved in the vertebrate homologue of Sae2, CtIP (17, 18). Because Cdk1 activity is required to generate Spo11-induced DSBs (19, 20), its involvement in allowing their processing has not been assessed.
After Spo11 removal from the 5′ DSB ends, one or more so far unknown nucleases have to resect the break to generate 3′-ended single-stranded DNA (ssDNA) overhangs to initiate homologous recombination. Candidates for such activity are the nucleases Exo1 and Dna2 and the helicase Sgs1, which all contribute to resect DSB and chromosome ends in mitotic S. cerevisiae cells (21–24). Consistent with this hypothesis, EXO1 deletion has been shown to impair repair of meiotic DSBs and to reduce meiotic crossing over (25).
Here we show that phosphorylation by Cdk1 of the Ser-267 residue of S. cerevisiae Sae2 is required to initiate resection of meiotic DSBs. In fact, substitution of Sae2 Ser-267 with a non-phosphorylatable residue severely impairs both Spo11 removal and DNA-end processing, which instead take place efficiently when an aspartic residue mimicking constitutive phosphorylation replaces Sae2 Ser-267. Moreover, we demonstrate that further processing of Spo11-induced DSB ends depends on the nuclease Exo1 and the helicase Sgs1 that act in two different pathways.
EXPERIMENTAL PROCEDURES
Yeast Strains
Yeast strains used for this work are listed in supplemental Table S1. All the strains were SK1 derivatives that were isogenic with the NKY3000 strain (MATa/MATα, HO/HO, lys2/lys2, ura3::hisG/ura3::hisG, leu2::hisG/leu2::hisG), kindly provided by N. Kleckner (Harvard University, Cambridge, MA). Heterozygous diploid strains carrying deletions of SAE2, DMC1, EXO1, and DNA2 genes were obtained by one-step PCR disruption. The diploid strain carrying the cdc28-as allele was kindly provided by S. Keeney (New York, NY). The pif1-M2 mutation was introduced into an SK1 derivative strain as described (26). The SGS1 promoter was replaced with the CLB2 promoter using the pFA6a-KANMX6-pCLB2 cassette as described previously (27). The sae2-S267A, sae2-S267D, sae2-S134A, sae2-S134D, and sae2-S179A alleles were constructed by site-directed mutagenesis (Stratagene). ApaI digestion of the integrative plasmids pML469, pML674, pML673, pML692, pML703, pML691.3, pML691.5, and pML704 was used to direct the integration of these plasmids to the SAE2 promoter region of a SK1-derivative sae2Δ strain, giving rise to heterozygous diploid strains carrying single copies of the SAE2, sae2-S267A, sae2-S267D, sae2-S134A, sae2-S134D, sae2-S267A-S179A, sae2-S267A-S134A, and sae2-S267A-S134D alleles, respectively, at the SAE2 chromosomal locus. Diploid strains homozygous for the above deletions or mutations were obtained after tetrad dissection of the corresponding heterozygous strains and self-diploidization of the spore carrying the desired alleles.
PCR one-step tagging was used to obtain strains carrying myc-tagged SPO11 and HA-tagged SAE2, sae2-S267A, or MEK1 alleles. The SAE2-HA3, MEK1-HA3, and SPO11-MYC9 alleles were shown to be fully functional, since diploid strains homozygous for MEK1-HA3, SAE2-HA3, or SPO11-MYC9 alleles were undistinguishable from the isogenic untagged strains with respect to meiotic progression and meiotic DSB repair. The accuracy of all gene replacements and integrations was verified by Southern blot analysis or PCR.
Synchronous Meiotic Time Course
To obtain synchronous G1/G0 cell population, overnight liquid YEPD (yeast extract peptone dextrose) cell cultures were diluted to a final concentration of 1 × 107 cells/ml in 200 ml YPA (1% yeast extract, 2% Bacto-peptone, 1% potassium acetate) in a 2-liter flask, and grown with vigorous shaking for 13 h at 30 °C. Cells were then washed and transferred into the same volume of SPM (0.3% potassium acetate, 0.02% raffinose) to induce meiosis.
Meiotic DSB Formation and Processing
DSB formation and repair analysis were performed at the YCR048W locus as described (28). To detect DSB end resection at the YCR048W hotspot, genomic DNA was digested with DraIII and EcoRV and separated on alkaline agarose gels. The single-stranded Riboprobe used to detect DSB resection was complementary to part of the YCR048W locus on chromosome III (coordinates 212503 to 213199). Quantitative analysis of DSB processing was performed by calculating the ratio of band intensities for ssDNA and parental DNA.
ChIP Analysis
ChIP analysis was performed as described (29). After exposure to formaldehyde, chromatin samples were immunoprecipitated with anti-Myc antibody. Quantification of immunoprecipitated DNA was achieved by quantitative real-time PCR on a Bio-Rad MiniOpticon using primers located 162 bp (DSB) and 2319 bp (CON) distal to the DSB site of the YCR048W hotspot and normalized to input signal for each primer set; data are expressed as the fold enrichment of DSB over the amount of CON in the immunoprecipitates.
Other Techniques
Flow cytometric DNA analysis was determined on a BD Biosciences FACScan. Nuclear division was scored with a fluorescence microscope in propidium iodide stained cells. Immunoprecipitation was performed as described (28). Western blot analyses on protein extracts prepared by trichloroacetic acid precipitation and immunoprecipitation were performed by standard conditions. Secondary antibodies were purchased from Amersham Biosciences, and proteins were visualized by using an enhanced chemiluminescence system according to the manufacturer's instructions.
RESULTS
Sae2 Ser-267 Is Phosphorylated by Cdk1 in Meiosis
Effective DSB resection in vegetative S. cerevisiae cells is promoted by Cdk1 activity during the S and G2 phases of the cell cycle (14, 15). Because Cdk1 activity is required to generate meiosis-specific DSBs (19, 20), we could not assess directly its involvement in Spo11-induced DSB resection. To overcome this problem, we exploited the fact that Cdk1-mediated control of DSB resection during mitosis relies on the phosphorylation of Sae2 Ser-267 by Cdk1 (16). Thus, we asked whether Spo11-induced DSB resection requires Cdk1-mediated phosphorylation of Sae2 Ser-267.
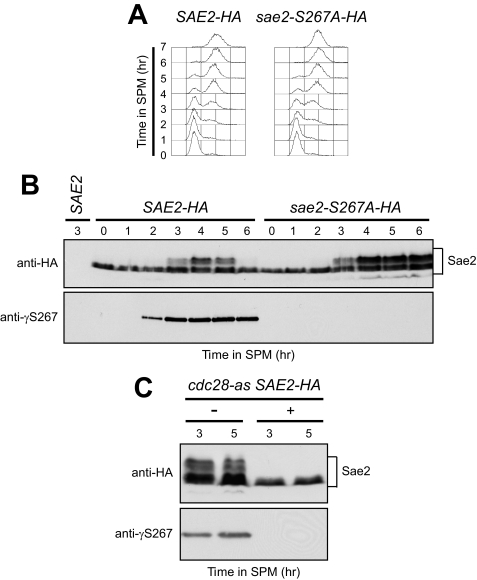
First, we examined if Sae2 is phosphorylated on Ser-267 during meiosis by using a phosphospecific antibody against this site (anti-γS267, kindly provided by S. Jackson, University of Cambridge, UK). Synchronous meiosis (Fig. 1A) was induced in diploid cells expressing either Sae2-HA or the Sae2-S267A-HA variant, where Ser-267 was substituted by a non phosphorylatable alanine residue. Western blot analysis of anti-HA immunoprecipitates revealed that the anti-γS267 antibody specifically detected wild-type Sae2-HA concomitantly with premeiotic S phase onset, but not Sae2-S267A-HA (Fig. 1B). By contrast, anti-HA antibodies detected both Sae2-HA and Sae2-S267A-HA (Fig. 1B). Thus, Sae2 Ser-267 is phosphorylated in a Cdk1-dependent manner after meiosis induction. Notably, both Sae2-HA and Sae2-S267A-HA underwent electrophoretic mobility shifts (Fig. 1B), known to be due to Mec1- and Tel1-dependent phosphorylation events that take place concomitantly with premeiotic DNA replication and increase with Spo11-induced DSB formation (28). Thus, Ser-267 phosphorylation does not influence Sae2 mobility under this electrophoretic condition. This finding is consistent with previous data showing that DSB- and S phase-induced Sae2 electrophoretic mobility shifts during both meiosis (28) and mitosis (30) are undetectable in both mec1Δ tel1Δ double mutants, and in cells carrying multiple changes to alanine of the five serine or threonine residues (Ser-73, Thr-90, Ser-249, Thr-279, and Ser-289) located in the (S/T)Q Sae2 motifs, which are favored for phosphorylation by Mec1/Tel1.
FIGURE 1.
Cdk1-dependent phosphorylation of Sae2 Ser-267 during meiosis. A and B, SAE2-HA and sae2-S267A-HA diploid cells were transferred to sporulation medium (SPM). A, at the indicated times after meiosis induction, DNA content was analyzed by FACS. B, protein extracts were immunoprecipitated with anti-HA antibody and subjected to Western blot analysis with anti-HA and anti-γS267 antibodies. Immunoprecipitation was also performed on untagged diploid cells (SAE2) 3 h after transfer to SPM. C, cdc28-as SAE2-HA diploid cells were transferred to SPM in the absence (−) or presence (+) of 5 μm 1-NM-PP1. At the indicated times after meiosis induction, protein extracts were immunoprecipitated with anti-HA antibody and subjected to Western blot analysis as in B.
We then evaluated Ser-267 phosphorylation dependence on Cdk1 by using the analogue-sensitive Cdk1 version Cdc28-as, which can be inactivated in vivo by the adenine analogue 1-NM-PP1 (31). As shown in Fig. 1C, cdc28-as meiotic cells allowed Sae2-HA Ser-267 phosphorylation in the absence of 1-NM-PP1, but not when the latter was added to the sporulation medium. In fact, anti-γS267 failed to detect wild-type Sae2-HA in immunoprecipitates from 5 μm 1-NM-PP1-treated cdc28-as cells (Fig. 1C). According to the knowledge that 5 μm 1-NM-PP1 prevents both DNA replication and DSB formation (19), the anti-HA antibody failed to detect Mec1- and Tel1-dependent Sae2-HA mobility shifts in 1-NM-PP1-treated cdc28-as immunoprecipitates (Fig. 1C). Altogether, these data indicate that Sae2 Ser-267 phosphorylation during meiosis is Cdk1-dependent, suggesting that Cdk1 might regulate processing/repair of Spo11-induced DSBs through Sae2 phosphorylation.
Meiotic DSB Repair Requires Sae2 Ser-267 Phosphorylation
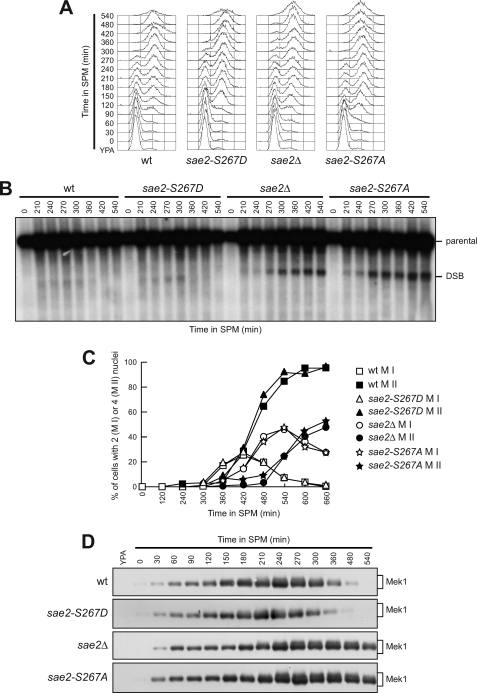
Because Sae2 is required to repair Spo11-induced DSBs by allowing Spo11 removal and generation of 3′-ended ssDNA (3), we asked whether Cdk1-dependent Sae2 Ser-267 phosphorylation is required for this meiotic function. Thus, we analyzed the kinetics of DSB repair at the natural YCR048W meiotic recombination hotspot in cells expressing Sae2 variants where Ser-267 was substituted by either a non-phosphorylatable alanine residue (Sae2-S267A) or an aspartic residue mimicking constitutive phosphorylation (Sae2-S267D). The DSB appeared in all cell cultures undergoing synchronous meiosis as soon as cells completed premeiotic DNA replication (Fig. 2, A and B). However, the DSB signal disappeared ∼360 min after transfer to sporulation medium in wild-type and sae2-S267D cells, whereas it persisted until the end of the experiment in both sae2Δ and sae2-S267A cells (Fig. 2B).
FIGURE 2.
Sae2 Ser-267 phosphorylation is required for DSB repair. Synchronous meiotic cultures of cells with the indicated genotypes and expressing Mek1-HA from the MEK1 promoter were analyzed at the indicated times for DNA content by FACS (A), for DSB formation/repair at the YCR048W hotspot by Southern blot (B), for the percentages of binucleate (completed meiosis I, MI) and tetranucleate (completed meiosis II, MII) cells (C), and for Mek1 amount/phosphorylation by Western blot analysis with anti-HA antibody (D). The Southern blot in B was probed with a DNA fragment complementary to the 5′ non-coding region of the YCR048W gene, which reveals an intact parental EcoRI fragment (parental) of 7.9-kb and a band of 5.7-kb corresponding to the prominent meiotic DSB site (DSB).
The inability of sae2Δ cells to repair meiotic DSBs is known to cause the hyperactivation of the Mek1-dependent recombination checkpoint that delays progression through meiosis (28). Strikingly, similarly to sae2Δ cells, sae2-S267A cells started to undergo meiosis I and II ∼60 and 120 min later, respectively, than wild-type and sae2-S267D cells (Fig. 2C). Moreover, this delay correlated with Mek1 phosphorylation, whose amount remained constant until the end of the experiment in both sae2Δ and sae2-S267A cells, while it decreased in both wild-type and sae2-S267D cells within 360 min after transfer to sporulation medium (Fig. 2D). Consistent with hyperactivation of the recombination checkpoint, Mec1- and Tel1-dependent phosphorylation of Sae2-S267A persisted longer after meiosis induction than that of wild-type Sae2 (Fig. 1B). Thus, phosphorylation of Sae2 Ser-267 by Cdk1 is required for repair of Spo11-induced DSBs, which in turn allows deactivation of the meiotic recombination checkpoint.
Meiotic DSB Resection Requires Sae2 Ser-267 Phosphorylation
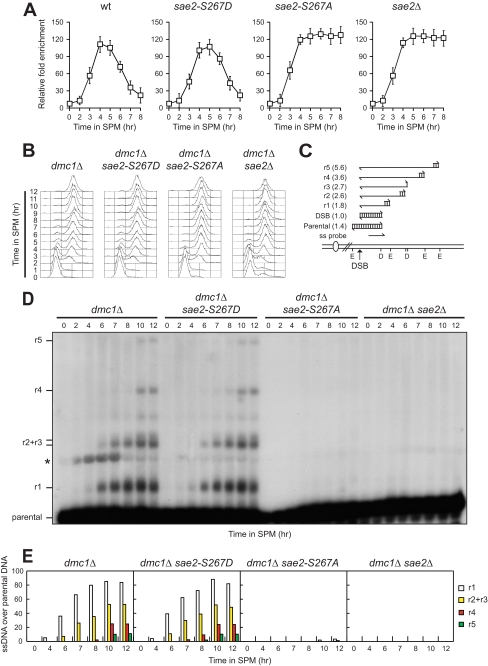
Because Sae2 is known to be required for Spo11 removal, we asked whether the Sae2-S267A variant might prevent Spo11 dissociation from the meiotic recombination YCR048W hotspot. To verify this hypothesis, we performed chromatin immunoprecipitation (ChIP) with anti-Myc antibody in strains expressing a fully functional Myc-tagged Spo11 variant, followed by quantitative real-time PCR to monitor coimmunoprecipitation of DNA fragments located either 162 bp (DSB) or 2319 bp (CON) distal to the natural YCR048W recombination hotspot. In all cell cultures, Spo11 associated with the DNA fragment closest to the YCR048W hotspot during the course of meiosis, as measured by an increase of the DSB/CON ratio (Fig. 3A). This Spo11-hotspot association decreased ∼6 h after meiosis induction in both wild-type and sae2-S267D, whereas it persisted in both sae2Δ and sae2-S267A cells (Fig. 3A). Thus, Spo11 removal requires phosphorylation of Sae2 Ser-267.
FIGURE 3.
Phosphorylation of Sae2 Ser-267 is essential for both Spo11 removal and DSB resection. A, Spo11-DNA association. Chromatin samples taken at different time points after meiosis induction were immunoprecipitated with anti-Myc antibody. Coimmunoprecipitated DNA was analyzed by quantitative real-time PCR using primer pairs located 162 bp (DSB) and 2319 bp (CON) distal to the DSB site of the YCR048W hotspot. Data were expressed as the -fold enrichment of DSB over CON signal after normalization to input signals for each primer set. The data presented are the mean of those obtained in three independent experiments. Error bars indicate ±S.D. B, synchronous meiotic cultures of cells with the indicated genotypes were analyzed at the indicated times for DNA content by FACS. C, scheme of the system used to detect DSB resection at the YCR048W hotspot. Genomic DNA was digested with both DraIII (D) and EcoRV (E), and DNA fragments were separated on alkaline agarose gel. Gel blots were hybridized with a single-stranded RNA probe, which reveals an uncut fragment of 1.4-kb (parental). DSB formation and subsequent 5′-to-3′ resection eliminate DraIII and EcoRV sites, thus producing larger DNA fragments (r1, r2, r3, r4, and r5) detected by the probe. D, genomic DNA prepared from samples taken at the indicated times during the experiment in B was analyzed for ssDNA formation as described in C. The asterisk points out an unspecific signal. E, densitometric analysis of the representative experiment shown in D. Values are expressed as arbitrary units. Three independent experiments were performed with very similar results.
Because Spo11-DNA association occurs independently of DSB formation (32), we followed more directly the kinetics of 3′-ended ssDNA formation by Southern blot analysis of genomic DNA that was run on an alkaline agarose gel, followed by hybridization with a single-stranded RNA probe specific for the YCR048W gene (Fig. 3C). To ensure the visualization of all the resection products, all the strains carried the deletion of the DMC1 gene, thus preventing the disappearance of the 3′-ended ssDNA regions due to homologous recombination between the homologous non-sister chromatids. Strikingly, after transfer to sporulation medium (Fig. 3B), 3′-ended ssDNA resection products were below the detection level in both sae2-S267A dmc1Δ and sae2Δ dmc1Δ cells, whereas they accumulated in both dmc1Δ and sae2-S267D dmc1Δ cells (Fig. 3, D and E). Altogether, these data indicate that Cdk1-dependent phosphorylation of Sae2 Ser-267 is required to resect Spo11-induced DSB ends.
It has been shown that the sae2-S267A allele causes a strong reduction in spore viability (6, 16) (Table 1). However, although both 3′-ended ssDNA and Spo11 removal were under the detection level in both sae2Δ and sae2-S267A cells, spore viability was reduced to a lesser extent in sae2-S267A cells compared with sae2Δ cells (Table 1). These findings suggest that full Sae2 activity might require Cdk1-dependent phosphorylation of additional residues. Besides Ser-267, Sae2 contains two other potential Cdk1 target sites, Ser-134 and Ser-179, with Ser-134 receiving a higher score for predicted phosphorylation site (16). When Ser-179 was substituted with a non-phosphorylatable alanine residue, sae2-S267A-S179A and sae2-S267A mutant cells showed similar spore viability (Table 1), suggesting that Ser-179 does not contribute to support Sae2 activity. By contrast, a slight reduction in spore viability was caused by the sae2-S134A mutation (Table 1). Furthermore, sae2-S267A-S134A spore viability was reduced compared with sae2-S267A, and it was similar to that caused by SAE2 deletion (Table 1). This loss of viability was likely due to the lack of Ser-134 phosphorylation and not to protein folding alterations, as sae2-S134D cells showed wild-type spore viability and sae2-S267A-S134D spore viability was similar to that of sae2-S267A cells (Table 1). Altogether, these data suggest that, in addition to Ser-267, Ser-134 phosphorylation might contribute to support Sae2 function in promoting meiotic DSB resection.
TABLE 1.
Spore viability in sae2 mutants
Diploid strains homozygous for the indicated SAE2 alleles were allowed to sporulate, and tetrads were dissected on YEPD plates. Spore viability was determined by scoring colony-forming spores after incubation at 28 °C for 3 days.
| Allele | Spore viability | No. viable spores/total |
|---|---|---|
| % | ||
| SAE2 | 96 | 146/152 |
| sae2Δ | 1 | 2/156 |
| sae2-S267A | 15 | 30/200 |
| sae2-S267D | 95 | 122/128 |
| sae2-S134A | 75 | 81/108 |
| sae2-S134D | 95 | 76/80 |
| sae2-S267A-S179A | 16 | 19/120 |
| sae2-S267A-S134A | 3 | 8/264 |
| sae2-S267A-S134D | 16 | 22/136 |
Exo1 and Sgs1 Are Involved in Meiotic DSB Resection
After Spo11 removal by Sae2, the 3′-ended DNA strands are rapidly processed through a still unknown mechanism. Possible candidates for such activity are Exo1, Dna2, and/or the helicase Sgs1, because they contribute to resect chromosome ends that are trimmed by Sae2 and MRX in vegetative cells (21–24).
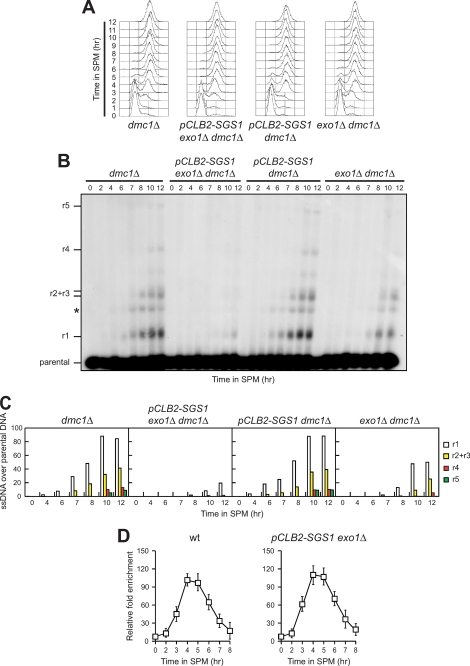
We then monitored the kinetics of 3′-ended ssDNA generation at the natural YCR048W meiotic recombination hotspot in cells lacking Exo1, Sgs1, and/or Dna2. Because sgs1Δ single mutant cells displayed vegetative growth defects and the simultaneous deletion of SGS1 and EXO1 caused cell lethality in the SK1 background (data not shown), we constructed a meiosis-specific pCLB2-SGS1 conditional allele by replacing the native SGS1 promoter with the CLB2 promoter, which is strongly repressed during meiosis (27). Normal vegetative growth phenotypes and efficient premeiotic synchronization were observed in both pCLB2-SGS1 dmc1Δ and pCLB2-SGS1 exo1Δ dmc1Δ cells (Fig. 4A), where DMC1 was deleted to ensure visualization of the resection products. Although DSB formation occurred with similar kinetics in all cell cultures after meiosis induction (data not shown), generation of 3′-ended ssDNA resection products was delayed in exo1Δ dmc1Δ compared with dmc1Δ cells, indicating that Exo1 contributes to meiotic DSB processing (Fig. 4, B and C). The residual resection of the Spo11-induced DSB in exo1Δ cells depends partially on Sgs1 activity. In fact, resection of this DSB was severely reduced in pCLB2-SGS1 exo1Δ dmc1Δ compared with exo1Δ dmc1Δ, although it was not defective in pCLB2-SGS1 dmc1Δ cells (Fig. 4, B and C). Thus, meiotic DSB end processing is controlled by at least two distinct mechanisms involving Sgs1 and Exo1, respectively, with Exo1 playing the major role.
FIGURE 4.
Meiotic DSB resection involves both Exo1 and Sgs1. A–C, synchronous meiotic cultures of cells with the indicated genotypes were analyzed at the indicated times for DNA content by FACS (A) and for DSB resection by Southern blot (B) as described in Fig. 3C. C, densitometric analysis of the representative experiment shown in B. Values are expressed as arbitrary units. Three independent experiments were performed with very similar results. D, Spo11-DNA association. Synchronous meiotic cultures of cells with the indicated genotypes were analyzed at the indicated times by ChIP and quantitative real-time PCR as described in Fig. 3A.
The inability to remove Spo11 from the DSB ends inhibits their processing, prompting us to ask whether the resection defects of cells crippled for both Sgs1 and Exo1 activities might be due to persistence of Spo11 binding to DSB ends. To test this hypothesis, we monitored Spo11 association to the meiotic YCR048W recombination hotspot by ChIP analysis with anti-Myc antibody from cells expressing Myc-tagged Spo11. A transient association of Spo11 to the DNA fragment located 162 bp distal to the natural YCR048W recombination hotspot was observed in both wild-type and pCLB2-SGS1 exo1Δ cells (Fig. 4D), indicating that Sgs1 and Exo1 are not involved in terminating Spo11-hotspot interaction. Thus, Sgs1 and Exo1 appear to participate in DSB processing by controlling a step subsequent to Spo11 removal.
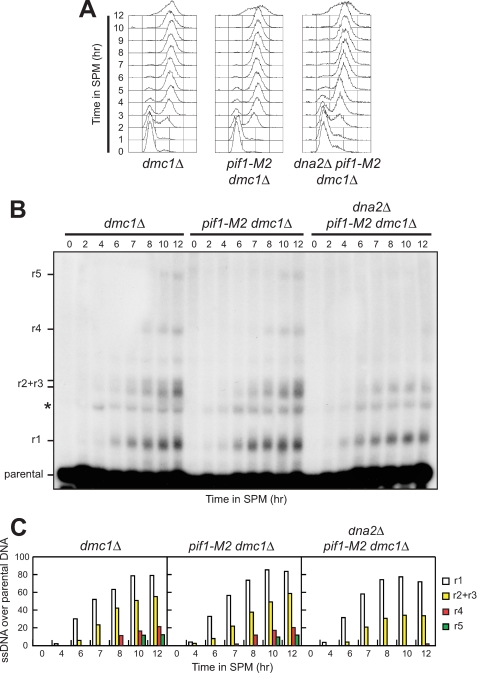
In addition to Exo1 and Sgs1, resection of chromosome ends in vegetative cells depends also of the nuclease/helicase Dna2 (23, 24). Thus, we analyzed whether Dna2 also promotes resection of meiotic DSBs by using a strain where its essential function in cell viability is bypassed by the pif1-M2 mutation, which specifically impairs Pif1 nuclear function (33). We found that initiation of YCR048W DSB resection seems to occur with similar kinetics in dmc1Δ, pif1-M2 dmc1Δ, and dna2Δ pif1-M2 dmc1Δ cells (Fig. 5). However, generation of the longest resection products (r4 and r5) was defective in dna2Δ pif1-M2 dmc1Δ cells compared with dmc1Δ and pif1-M2 dmc1Δ cells (Fig. 5, B and C). This finding suggests that Dna2 might contribute to the formation of long ssDNA tails, in agreement with the notion that Dna2 is involved in long range resection of DSB ends in vegetative Saccharomyces cerevisiae cells (23). Unfortunately, we were unable to assess whether Dna2 could resect Spo11-induced DSBs in the absence of Exo1 or Sgs1, because both dna2Δ pif1-M2 exo1Δ and dna2Δ pif1-M2 pCLB2-EXO1 cells were unviable, and dna2Δ pif1-M2 pCLB2-SGS1 cells did not enter meiosis synchronously (data not shown).
FIGURE 5.
Dna2 participates in meiotic DSB resection. Synchronous meiotic cultures of cells with the indicated genotypes were analyzed at the indicated times for DNA content by FACS (A) and for DSB resection by Southern blot (B) as described in Fig. 3C. C, densitometric analysis of the representative experiment shown in B. Values are expressed as arbitrary units. Three independent experiments were performed with very similar results.
DISCUSSION
DNA DSBs are highly hazardous for genome integrity, but meiotic cells deliberately introduce them into their genome to initiate homologous recombination. To minimize the risk of deleterious effects, meiotic DSB formation, processing, and repair must be tightly regulated to occur only at the right time and place. In this work, we have investigated the mechanism by which S. cerevisiae cells control Spo11 removal and resection of meiosis-specific DSBs. Overall, our data indicate that the requirements for resecting Spo11-induced DSBs, in terms of nucleases and Cdk1-dependent Sae2 phosphorylation, are similar to those of the processing events at an accidental DSB, indicating that cells have evolved the same mechanism to process both programmed and un-programmed DSBs.
Regulation of Spo11 Removal from Meiotic DSBs
Cdk1 activity accumulates during premeiotic S phase, increases through prophase, and peaks at about meiosis I (34). We show that Sae2 Ser-267 is phosphorylated in a Cdk1-dependent manner during meiosis. Moreover, substitution of Ser-267 with a non-phosphorylatable residue causes phenotypes comparable to those of sae2 null mutants, including severely impaired Spo11 removal and DNA-end processing. These defects are caused by the inability of Cdk1 to phosphorylate Sae2 Ser-267, because the same processes take place efficiently when an aspartic residue mimicking constitutive phosphorylation replaces Sae2 Ser-267. Thus, Cdk1-dependent phosphorylation of Ser-267 is required for Sae2 function in Spo11 removal from meiotic DSB ends and subsequent resection of the latter. This finding implies that Cdk1 activity is required not only for generation of meiotic DSBs, but also for their resection, thus providing a mechanism for coordinating DSB resection with progression through meiotic prophase.
Although formation of single-stranded DNA at Spo11-induced DSBs is undetectable in sae2-S267A cells, spore viability of the latter is still 15% compared with 1% of sae2Δ cells. One possibility is that additional Cdk1-dependent phosphorylation events on Sae2 might be needed for optimal resection. In agreement with this hypothesis, we found that sae2-S267A-S134A cells displayed a strong reduction in spore viability compared with sae2-S267A cells. This loss of viability was likely due to the lack of Ser-134 phosphorylation, because sae2-S267A-S134D spore viability was similar to that of sae2-S267A cells. Although we were unable to demonstrate that Sae2 Ser-134 is phosphorylated in vivo by using phosphospecific antibodies, these observations suggest that, in addition to Ser-267, phosphorylation of Ser-134 might be important for Sae2 meiotic functions.
How phosphorylation of Sae2 modulates Spo11 removal is still unknown. Sae2 has been shown to be an endonuclease that acts cooperatively with the MRX complex in vitro (13). These in vitro results were obtained in the absence of phosphorylation events, suggesting that phosphorylation of Sae2 is not absolutely required for its observed biochemical activity. Nevertheless, Sae2 function during meiosis and mitosis in vivo requires both Cdk1-dependent and checkpoint-dependent phosphorylation events (this work and Refs. 16, 28, 30). These apparent differences between the in vivo and in vitro data may suggest that unknown proteins inhibit Sae2 activity within the cell, such that its function is only exhibited upon phosphorylation events that relieve this inhibition. Alternatively, or in addition, Sae2 activity might be enhanced in vivo by positive regulators of DSB resection requiring Cdk1-dependent phosphorylations to exert their actions.
DSB Processing after Spo11 Removal
It has been recently shown that Sae2, in conjunction with the MRX complex, functions in the initial trimming of accidental DSBs to generate short 3′ overhangs (22, 23). Then, a secondary processing, redundantly executed by either the Sgs1 helicase and the Dna2 nuclease or the 5′-3′ exonuclease Exo1, exposes extensive 3′ single-stranded tails (22, 23). The lengthening of single-stranded DNA tracts at Spo11-induced meiotic DSBs appears to have similar requirements in terms of nucleases as the processing events at accidental DSBs. In fact, both Sgs1 and Exo1 turned out to be involved in 3′-ended ssDNA generation after the initial endonucleolytic removal of Spo11, likely controlling two distinct but partially complementary pathways. On the contrary, Exo1 and Sgs1 are not required for Spo11 removal, indicating that initiation and lengthening of meiotic DSB resection are controlled by different sets of nucleases.
Moreover, we demonstrate that generation of ssDNA at Spo11-induced DSBs depends also on the nuclease Dna2, which appears to contribute mainly to long range resection. Unfortunately, we could not establish the epistatic relationships between Dna2, Exo1, and Sgs1, because both dna2Δ pif1-M2 exo1Δ and dna2Δ pif1-M2 pCLB2-EXO1 cells were unviable, and dna2Δ pif1-M2 pCLB2-SGS1 cells did not enter meiosis synchronously (data not shown). Nonetheless, the finding that resection of Spo11-induced DSBs is reduced to a lesser extent in exo1Δ than in exo1Δ pCLB2-SGS1 cells indicates that Exo1 is not the only nuclease that can be targeted by Sgs1. Thus, we speculate that, although Exo1 can act independently of Sgs1, the helicase activity of the latter might unwind DSB ends to yield a fayed structure with both 5′ and 3′ single-stranded regions, thus facilitating nuclease access. Exo1 and/or other nuclease(s) such as Dna2 could then digest the 5′-terminal strand, resulting in a 3′ tail that can be engaged into homologous recombination. The combined use of enzymes with helicase and nuclease activities has been found also in bacteria, where the RecQ helicase and the RecJ 5′-3′ exonuclease function in DSB resection in the absence of the dominant RecBCD activity (35).
There are compelling evidences that premeiotic DNA replication and DSB formation are coupled by a still unknown mechanism. In fact, delaying replication of the left arm of chromosome III locally delays DSB formation by the same margin, without affecting timing on the right arm (36, 37). One possibility is that replication fork passage (or associated processes) promotes installation of chromosome features that constrain subsequent DSB formation. Because Dna2 is involved in the removal of the Okazaki fragments during DNA replication (38), it is tempting to speculate that its function in the processing of Spo11-induced DSBs might be exerted only after the passage of the replication fork, thus linking premeiotic DNA replication not only with formation of DSBs, but also with their subsequent processing.
Supplementary Material
Acknowledgments
We thank A. Amon for providing pFA6a-KANMX6 plasmid, S. Keeney for the cdc28-as allele, and N. Kleckner for the NKY3000 strain. The anti-γS267 antibody was kindly provided by S. Jackson. We also thank P. Huertas for technical advices and Michela Clerici for critical reading of the manuscript.
This work was supported by grants from Associazione Italiana Ricerca sul Cancro and Cofinanziamento 2008 Ministero dell'Università et della Ricerca/Università di Milano-Bicocca (to M. P. L.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1.
- DSB
- double strand break
- ssDNA
- single-stranded DNA
- Cdk1
- cyclin-dependent kinase 1
- FACS
- fluorescence-activated cell sorting
- HA
- hemagglutinin
- ChIP
- chromatin immunoprecipitation
- Mek1
- meiotic kinase 1
- 1-NM-PP1
- 4-amino-1-tert-butyl-3-(1-naphthyl)-pyrazolo-[3,4,d]pyrimidine.
REFERENCES
- 1.Petronczki M., Siomos M. F., Nasmyth K. (2003) Cell 112, 423–440 [DOI] [PubMed] [Google Scholar]
- 2.Longhese M. P., Bonetti D., Guerini I., Manfrini N., Clerici M. (2009) DNA Repair 8, 1127–1138 [DOI] [PubMed] [Google Scholar]
- 3.Keeney S., Kleckner N. (1995) Proc. Natl. Acad. Sci. U.S.A. 92, 11274–11278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Neale M. J., Pan J., Keeney S. (2005) Nature 436, 1053–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Usui T., Ohta T., Oshiumi H., Tomizawa J., Ogawa H., Ogawa T. (1998) Cell 95, 705–716 [DOI] [PubMed] [Google Scholar]
- 6.Uanschou C., Siwiec T., Pedrosa-Harand A., Kerzendorfer C., Sanchez-Moran E., Novatchkova M., Akimcheva S., Woglar A., Klein F., Schlögelhofer P. (2007) EMBO J. 26, 5061–5070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alani E., Padmore R., Kleckner N. (1990) Cell 61, 419–436 [DOI] [PubMed] [Google Scholar]
- 8.McKee A. H., Kleckner N. (1997) Genetics 146, 797–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Prinz S., Amon A., Klein F. (1997) Genetics 146, 781–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Furuse M., Nagase Y., Tsubouchi H., Murakami-Murofushi K., Shibata T., Ohta K. (1998) EMBO J. 17, 6412–6425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tsubouchi H., Ogawa H. (1998) Mol. Cell. Biol. 18, 260–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Moreau S., Ferguson J. R., Symington L. S. (1999) Mol. Cell. Biol. 19, 556–566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lengsfeld B. M., Rattray A. J., Bhaskara V., Ghirlando R., Paull T. T. (2007) Mol. Cell 28, 638–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aylon Y., Liefshitz B., Kupiec M. (2004) EMBO J. 23, 4868–4875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ira G., Pellicioli A., Balijja A., Wang X., Fiorani S., Carotenuto W., Liberi G., Bressan D., Wan L., Hollingsworth N. M., Haber J. E., Foiani M.(2004) Nature 431, 1011–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huertas P., Cortés-Ledesma F., Sartori A. A., Aguilera A., Jackson S. P. (2008) Nature 455, 689–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huertas P., Jackson S. P. (2009) J. Biol. Chem. 284, 9558–9565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yun M. H., Hiom K. (2009) Nature 459, 460–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henderson K. A., Kee K., Maleki S., Santini P. A., Keeney S. (2006) Cell 125, 1321–1332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wan L., Niu H., Futcher B., Zhang C., Shokat K. M., Boulton S. J., Hollingsworth N. M. (2008) Genes Dev. 22, 386–397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gravel S., Chapman J. R., Magill C., Jackson S. P. (2008) Genes Dev. 22, 2767–2772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mimitou E. P., Symington L. S. (2008) Nature 455, 770–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhu Z., Chung W. H., Shim E. Y., Lee S. E., Ira G. (2008) Cell 134, 981–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bonetti D., Martina M., Clerici M., Lucchini G., Longhese M. P. (2009) Mol. Cell 35, 70–81 [DOI] [PubMed] [Google Scholar]
- 25.Tsubouchi H., Ogawa H. (2000) Mol. Biol. Cell 11, 2221–2233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schulz V. P., Zakian V. A. (1994) Cell 76, 145–155 [DOI] [PubMed] [Google Scholar]
- 27.Lee B. H., Amon A. (2003) Science 300, 482–486 [DOI] [PubMed] [Google Scholar]
- 28.Cartagena-Lirola H., Guerini I., Viscardi V., Lucchini G., Longhese M. P. (2006) Cell Cycle 5, 1549–1559 [DOI] [PubMed] [Google Scholar]
- 29.Viscardi V., Bonetti D., Cartagena-Lirola H., Lucchini G., Longhese M. P. (2007) Mol. Biol. Cell 18, 3047–3058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Baroni E., Viscardi V., Cartagena-Lirola H., Lucchini G., Longhese M. P. (2004) Mol. Cell. Biol. 24, 4151–4165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bishop A. C., Ubersax J. A., Petsch D. T., Matheos D. P., Gray N. S., Blethrow J., Shimizu E., Tsien J. Z., Schultz P. G., Rose M. D., Wood J. L., Morgan D. O., Shokat K. M. (2000) Nature 407, 395–401 [DOI] [PubMed] [Google Scholar]
- 32.Prieler S., Penkner A., Borde V., Klein F. (2005) Genes Dev. 19, 255–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Budd M. E., Reis C. C., Smith S., Myung K., Campbell J. L. (2006) Mol. Cell. Biol. 26, 2490–2500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marston A. L., Amon A. (2004) Nat. Rev. Mol. Cell Biol. 5, 983–997 [DOI] [PubMed] [Google Scholar]
- 35.Amundsen S. K., Smith G. R. (2003) Cell 112, 741–744 [DOI] [PubMed] [Google Scholar]
- 36.Borde V., Goldman A. S., Lichten M. (2000) Science 290, 806–809 [DOI] [PubMed] [Google Scholar]
- 37.Murakami H., Borde V., Shibata T., Lichten M., Ohta K. (2003) Nucleic Acids Res. 31, 4085–4090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bae S. H., Bae K. H., Kim J. A., Seo Y. S. (2001) Nature 412, 456–461 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.