Abstract
Components of lymphotoxin beta receptor (LTBR)-associated signaling complexes, including TRAF2, TRAF3, NIK, IKK1, and IKK2 have been shown to participate in the coupling of LTBR to NFκB. Here, we report that TRAF3 functions as a negative regulator of LTBR signaling via both canonical and non-canonical NFκB pathways by two distinct mechanisms. Analysis of NFκB signaling in cell lines with functionally intact NFκB pathway but lacking LTBR-mediated induction of NFκB target genes revealed an inverse association of cellular TRAF3 levels with LTBR-specific defect in canonical NFκB activation. Increased expression of TRAF3 correlated with its increased recruitment to LTBR-induced signaling complexes, decreased recruitment of TRAF2, and attenuated phosphorylation of IκBα and RelA. In contrast, activation of NFκB by TNF did not depend on TRAF3 levels. siRNA-mediated depletion of TRAF3 promoted recruitment of TRAF2 and IKK1 to activated LTBR, enabling LTBR-inducible canonical NFκB signaling and NFκB target gene expression. TRAF3 knock-down also increased mRNA and protein expression of several non-canonical NFκB components, including NFκB2/p100, RelB, and NIK, accompanied by processing of NFκB2/p100 into p52. These effects of TRAF3 depletion did not require LTBR signaling and were consistent with autonomous activation of the non-canonical NFκB pathway. Our data illustrate the function of TRAF3 as a dual-mode repressor of LTBR signaling that controls activation of canonical NFκB, and de-repression of the intrinsic activity of non-canonical NFκB. Modulation of cellular TRAF3 levels may thus contribute to regulation of NFκB-dependent gene expression by LTBR by affecting the balance of LTBR-dependent activation of canonical and non-canonical NFκB pathways.
Keywords: Cytokines/Induction, Cytokines/Tumor Necrosis Factor, GENE/Transcription, Phosphorylation/Transcription Factors, Protein/Protein-Protein Interactions, Receptors, Signal Transduction/Adapter Proteins, Transcription/NF-κB
Introduction
Regulation of NFκB-mediated gene expression is central to the function of the lymphotoxin beta receptor (LTBR)3 in lymphoid organogenesis and immune response (reviewed in Ref. 1). Unlike the prototypical TNF receptors (TNFR) that activate exclusively the classical arm of NFκB, LTBR signals via both the canonical (NFκB1) and non-canonical (NFκB2) NFκB mechanisms and shares this property with several other members of the TNFR family, including CD40, BAFF-R, Fn14, and RANK (2, 3). TNFR-induced activation of NFκB1 is generally rapid and involves the Inhibitor of kappa-B kinase (IKK)-complex mediated phosphorylation of the inhibitor IκBα followed by its degradation to allow p50-mediated gene transcription (4). In contrast, activation of NFκB2 is more gradual and involves NFκB-inducing kinase (NIK)-dependent processing of NFκB2/p100 into its transcription-regulatory fragment p52 (4). Biological signals mediated by NFκB1 are central to inflammatory and innate immune responses (5) and self-limiting via NFκB1-dependent resynthesis of the inhibitor IκBα (6), whereas the NFκB2 signals are longer lasting and shown to regulate developmental processes, such as peripheral lymphoid organogenesis (7–9) and osteoclastogenesis (10).
LTBR-induced activation of NFκB1 and NFκB2 has been shown to produce distinct patterns of gene expression differentially controlled by the canonical and non-canonical NFκB pathways (11, 12). These findings raise the possibility that the balance of the pro-inflammatory and lymphoid histogenetic LTBR signaling may be regulated via differential utilization of NFκB1 and NFκB2. Interestingly, activation of NFκB by LTBR has been reported to occur by two apparently different scenarios, indicative of two alternative configurations of the LTBR signaling pathway. In one such configuration, LTBR initially induces rapid and transient activation of canonical NFκB, which is followed by gradual activation of the non-canonical pathway promoted by NFκB1-dependent synthesis of p100 (11). In the alternative configuration, the early activation of canonical NFκB is not observed (13). Instead the LTBR-induced signal is transmitted through the non-canonical NFκB pathway to eventually produce NFκB dimers containing both RelA and RelB (13). If recapitulated in vivo, the two different modes of NFκB activation could have significantly different immunological implications resulting in significantly different relative timing of LTBR-dependent inflammatory events and lymphoid histogenesis. Specific molecular mechanisms that define and coordinate the mode of LTBR coupling to the two arms of NFκB remain to be elucidated.
Ligand-induced activation of LTBR triggers the formation of receptor-associated cytoplasmic signaling complexes containing TNFR-associated factors (TRAFs) that regulate interactions of the receptor with downstream kinases (14–17). Analysis of the individual TRAF functions in LTBR signaling has demonstrated an essential role of TRAF2 as a mediator of NFκB activation (17, 18), whereas TRAF3 has been shown to mediate activation of JNK and induction of cell death by LTBR (19, 20). Studies of signal transduction mediated by other TNFRs coupled to both NFκB arms (CD40 and BAFF-R) have identified TRAF2 as a mediator and TRAF3 as an inhibitor of NFκB activation (21, 22) and suggested that TRAF2 and TRAF3 can reside in close proximity within CD40-associated complexes so that increased recruitment of TRAF3 to the receptor can inhibit TRAF2-mediated NFκB activation (23). Furthermore, overexpression of TRAF3 has been shown to inhibit NFκB2 activation (24), a finding consistent with TRAF3-mediated repression NIK (25), a kinase required for NFκB2 activation (26).
Herein, we report that TRAF3 controls LTBR-dependent activation of both the canonical and non-canonical NFκB pathways by two distinct mechanisms. We show that high cellular levels of TRAF3 can inhibit LTBR-mediated activation of NFκB1 and interfere with the recruitment of TRAF2 and IKK1 to LTBR-induced signaling complexes. We also show that TRAF3 inhibits the basal activity of NFκB2 via suppression of NIK mediated p100 processing and inhibition of a positive-autoregulatory NFκB2 loop that involves NFκB2-dependent transcription and resynthesis of RelB and p100. Our data suggest that modulation of TRAF3 levels can provide a mechanism that regulates preferential activation of the canonical or non-canonical NFκB by LTBR and modulates receptor-independent autonomous activity of the non-canonical NFκB.
MATERIALS AND METHODS
Cells and Antibodies
DLD-1 and WiDr colon carcinoma cell lines were obtained from ATCC (Manassas, VA), and cultured in MEM Earle's medium supplemented with 10% fetal bovine serum. Antibodies against LTBR (N-15), TRAF3 (H-20 and H-122), TRAF2 (H-249), and NFκB2 (C-5) were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Antibodies against phosphorylated IκBα (5A5), phosphorylated RelA, and NIK were purchased from Cell Signaling Technologies (Beverly, MA). Horseradish peroxidase (HRP)-conjugated secondary antibodies (anti-rabbit and anti-mouse) were from GE Healthcare/Amersham Biosciences (Piscataway, NJ), and HRP-conjugated anti-goat TrueBlot antibodies were from eBioscience (San Diego, CA). To activate LTBR or TNFR signaling, cells were stimulated with a humanized tetravalent LTBR agonist antibody BS-1 (100 ng/ml) developed at Biogen Idec, Inc. or TNFα (20 ng/ml), respectively.
Immunoprecipitation and Western Blotting
Cells were treated with the LTBR or TNFR agonists, washed twice in ice-cold phosphate-buffered saline with protease and phosphatase inhibitors, and harvested by scraping into ice-cold lysis buffer (50 mm PIPES pH 6.8, 100 mm KCl, 2 mm MgCl2, 1 mm EGTA, 0.2% Nonidet P-40, and 10% glycerol), supplemented with Complete EDTA-free protease inhibitor mixture (Roche, Indianapolis, IN), 10 mm sodium fluoride, and 100 μm sodium orthovanadate. The lysates were centrifuged at 14,000 rpm for 30 min at 4 °C, and supernatants were precleared by incubation with normal goat IgG-agarose beads (Sigma) for 1 h at 4 °C followed by centrifugation at 14,000 rpm for 15 min at 4 °C. For immunoprecipitation of LTBR signaling complexes, the precleared supernatants from BS-1-treated cells were incubated with goat anti-human IgG-agarose beads (Sigma) for 1 h at 4 °C on a rotator. The beads were collected into Handee mini spin-columns (Pierce), washed five times in the lysis buffer, and the bound material was eluted into Criterion XT loading buffer supplemented with XT-reducing agent (Bio-Rad) and protease and phosphatase inhibitors. The immunoprecipitated proteins were separated by denaturing SDS-PAGE in Criterion XT precast gels (Bio-Rad) and transferred onto nitrocellulose membranes. The membranes were incubated overnight at 4 °C and for 1 h at room temperature with primary and secondary antibodies, respectively, and washed by 5 × 5 min at room temperature in TBST after each antibody incubation. The blots were developed with SuperSignal chemiluminescent detection substrates (Pierce), and exposed on Biomax x-ray films (Kodak, Rochester, NY) or on the Kodak ImageStation 2000R luminescence detector. For quantitative densitometry, images of the films or directly exposed membranes acquired on the Kodak ImageStation were analyzed using Phoretix 1D gel analysis software (Nonlinear Dynamics, Durham, NC).
RNA Interference
Cells were transfected with siGENOME SMARTPool siRNA pools (Dharmacon, Lafayette, CO) against selected targets at concentrations of 100–200 nm using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). 48–72 h after transfection, the cells were stimulated as described for each figure and harvested for Western blotting, immunoprecipitation, or RNA isolation.
Microarray and Real-time PCR Procedures
RNA was isolated by extraction in QIAzol, purified on RNeasy columns (Qiagen) and analyzed for purity and integrity by capillary electrophoresis on Agilent Bioanalyzer 2100. Global transcript profiling was done using Affymetrix GeneChip U133 v.2 arrays. Hybridization probe synthesis, microarray hybridization, and scanning were performed according to the manufacturer's protocols. Probeset-level data were normalized using the robust microarray average (RMA) algorithm and analyzed with GeneSpring 7.3 data mining software (Agilent Technologies). For real-time RT-PCR, primer and fluorescent probe sets for selected genes, as well as the GAPDH housekeeping control probe and primers were obtained from Applied Biosystems (Foster City, CA). RT-PCR was performed using Quanti-tect Probe RT-PCR reagent kit (Qiagen) in duplex reactions combining probes for the selected gene and housekeeping control target. Thermal cycling was done on the Chromo4 thermal cycler equipped with fluorescence detector (Bio-Rad). Relative differences in target transcript abundance were calculated using the ΔΔCt method.
RESULTS
TRAF3 Inhibits Activation of Classical NFκB by LTBR
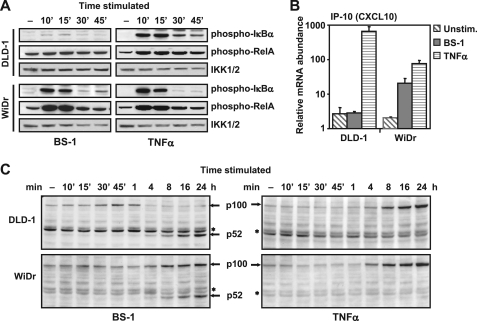
We had previously reported that LTBR ligation by agonist antibodies can cause death in a subset of colon carcinoma cell lines (27). Subsequent analysis of LTBR signaling in these cell lines has revealed two distinct cellular phenotypes, represented by the WiDr and DLD1 cell lines, which differed in their canonical NFκB responses to LTBR activation. Stimulation of LTBR with a tetravalent agonist mAb BS-1 induced rapid phosphorylation of IκBα and RelA in the WiDr but not in the DLD-1 cells (Fig. 1A). Consistent with the defective IκBα and RelA phosphorylation, LTBR-dependent induction of IP-10 that was identified in our preliminary experiments as a specific NFκB1 target (supplemental Fig. S1), as well as the production of another NFκB1 target gene product, NFκB2/p100,were readily detectable in the WiDr but virtually absent in the DLD-1 cells (Fig. 1, B and C). Contrary to the defective NFκB1 response, LTBR-induced processing of NFκB2/p100 to p52 in DLD1 cells was intact and comparable to that observed in the WiDr cells (Fig. 1C), indicating that LTBR-dependent NFκB2 signaling in DLD-1 cells was intact. Activation of NFκB1 by a TNFR agonist (TNFα) was identical in both cell lines (Fig. 1, A and C), showing that the defect in LTBR-mediated NFκB1 activation was selective for LTBR and unlikely to result from a general deficiency in the NFκB1 signaling machinery. Cell surface levels of LTBR were comparable in WiDr and DLD1, and full-length LTBR cDNA sequencing revealed no differences between the two cell lines (data not shown). Taken together, these results suggested the presence of a molecular switch or defect selectively blocking the coupling of LTBR to NFκB1 in DLD-1 cells.
FIGURE 1.
LTBR-specific activation of canonical NFκB is uncoupled in certain cells and correlates with differential cytokine gene activation. DLD-1 and WiDr cells were treated with agonist LTBR antibody (BS-1) at 100 ng/ml, or TNFα (20 ng/ml) for indicated times. Cell lysates were analyzed by Western blot for (A) phosphorylated IκBα (Ser-32/Ser-36), phosphorylated RelA (Ser-536), or IKK1/2 (to control for loading), and (C) NFκB2 (*, nonspecific bands). B, RNA isolated from untreated cells (▧), cells treated with BS-1 (■), or with TNFα (▤) for 4 h were analyzed by real-time qPCR for IP-10 transcripts. The data are shown as housekeeping gene (GAPDH)-normalized values from quadruplicate samples (average + S.D.).
Consistent with reports that NFκB1 activation is oscillatory with temporally decreasing amplitudes (28, 29), we found that activation of LTBR resulted in oscillatory IκBα phosphorylation in WiDr cells. A rapid and transient increase in IκBα phosphorylation occurred at 10 min and another, lower in magnitude wave of phosphorylation was detectable at 4 h (Fig. 2A). We found that while DLD-1 cells failed to display the first wave of IκBα phosphorylation at 10 min, the second wave of phosphorylation at 4 h was detectable and similar in magnitude to that observed in WiDr cells (Fig. 2A). These results indicated that the delayed NFκB1 signaling response observed in DLD1 cells could be enabled by specific molecular events promoted by persistent stimulation of LTBR.
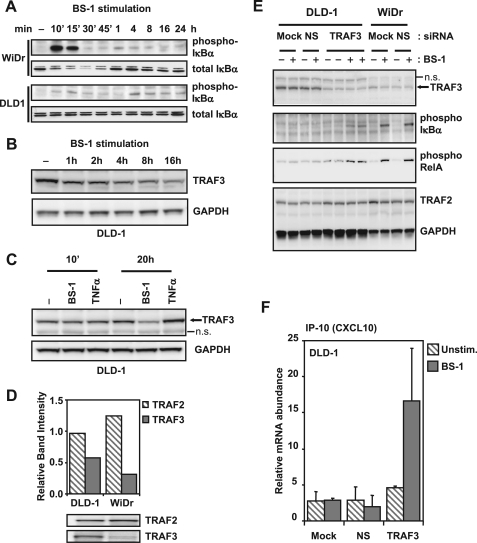
FIGURE 2.
TRAF3 inhibits LTBR-induced canonical NFκB activation. A, cells were treated with 100 ng/ml agonist LΤΒR antibody (BS-1) for indicated times, and the lysates were analyzed by Western blot for phosphorylated IκBα (Ser-32/Ser-36) and total IκBα. B, samples from BS-1 stimulated DLD-1 cells were also probed for TRAF3 or GAPDH (loading control). C, DLD-1 cells were left untreated, or treated for 10 min and 20 h with 100 ng/ml BS-1, or 20 ng/ml TNFα, and lysates analyzed for TRAF3 and GAPDH (loading control) by Western blot (n.s., nonspecific bands). D, total lysates from untreated DLD-1 and WiDr cells were analyzed by Western blots for TRAF2 and TRAF3, and band intensities were quantified by densitometry. E, cells were mock transfected (Mock), or transfected with either a nonspecific control siRNA (NS), or TRAF3 siRNA for 48 h. The cells were then left untreated (− lanes), or treated (+ lanes) with BS-1 for 10 min, and samples analyzed by Western blots. Blots were probed for different proteins as indicated (n.s., nonspecific band). F, DLD-1 cells were transfected as in C, and left untreated (unstim.) or stimulated with BS-1 for 4 h. RNA was collected and analyzed by real-time qPCR for IP-10 transcripts (unstimulated: ▧; BS-1 treated: ■). Results are normalized to GAPDH transcripts and shown as average + S.D. from quadruplicate samples.
We have analyzed effects of LTBR activation on the status of several known receptor-associated molecules involved in TNFR signaling and found that the late activation of IκBα phosphorylation in DLD1 correlated with a reduction in the cellular levels of TRAF3 (Fig. 2B). TRAF3 has been implicated as a negative regulator of CD40 signaling (21, 23). Therefore the down-regulation of TRAF3 following LTBR activation (Fig. 2C), as well as previous reports of TRAF3 down-regulation during CD40 and BAFF signaling (30, 31), suggested that the differences between early LTBR-induced NFκB1 activation in WiDr and DLD1 cells could be caused by different TRAF3 status of these cell lines. In accordance with this hypothesis, we found higher levels of TRAF3 in DLD-1 compared with WiDr cells, particularly apparent relative to the corresponding cellular levels of the NFκB activation mediator TRAF2 (Fig. 2D).
To determine whether the higher levels of TRAF3 in DLD-1 could account for the lack of NFκB1 response to LTBR stimulation, we transfected DLD-1 cells with siRNA targeting TRAF3 and stimulated the transfected cells with agonist LTBR antibodies for 10 min. The siRNA-mediated depletion of TRAF3 enabled clearly detectable induction of IκBα and RelA phosphorylation (Fig. 2E), as well IP-10 expression (Fig. 2F). Specificity of this effect was confirmed by overexpression of TRAF3 in the siRNA-transfected cells (supplemental Fig. S2).
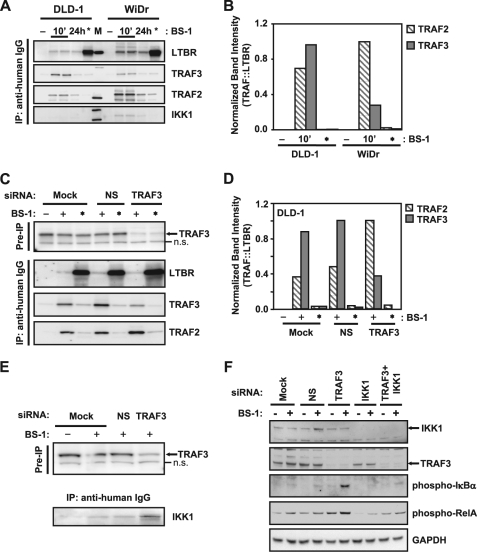
TRAF3 Regulates the Composition of Activation-induced LTBR Signaling Complexes
To determine effects of cellular TRAF3 levels on the formation of LTBR-associated signaling complexes, Western blot analysis of the activation-induced LTBR signaling complexes immunoprecipitated from WiDr and DLD-1 cells revealed a higher ratio of TRAF3 to LTBR in the complexes isolated from DLD1 cells compared with those from WiDr cells (Fig. 3A). We quantified amounts of LTBR-associated TRAF2 and TRAF3 by densitometry and normalized them by the corresponding levels of LTBR in the same immunoprecipitates. The resulting TRAF2/LTBR and TRAF3/LTBR ratios are shown in Fig. 3, B and D. Our use of these ratios instead of direct TRAF2 and TRAF3 levels ensures that differences in IP efficiency are internally controlled for variation in immunoprecipitation efficiency among samples. In addition to the higher levels of TRAF3, the LTBR-associated complexes isolated from DLD1 cells contained less TRAF2 and virtually no detectable IKK1, compared with the complexes immunoprecipitated from WiDr cells (Fig. 3, A and B). No significant association of TRAF2, TRAF3, or IKK1 with LTBR was observed when LTBR was immunoprecipitated from unstimulated cells (Fig. 3, A and B, * lanes). siRNA-mediated depletion of TRAF3 in DLD1 cells caused decreased TRAF3 association with LTBR and enhanced the recruitment of TRAF2 and IKK1 to LTBR-induced signaling complexes (Fig. 3, C–E). These results suggested that cytoplasmic levels of TRAF3 can modulate the formation of LTBR-induced signaling complexes and inhibit interactions of LTBR with mediators of NFκB activation. IKK1 and IKK2 are essential to the LTBR-mediated phosphorylation of IκBα and RelA in TRAF3-depleted DLD1 cells, as double knock-down of TRAF3 and IKK1 (or IKK2) markedly reduces the phosphorylation of these proteins (Fig. 3F and supplemental Fig. S3).
FIGURE 3.
Cellular TRAF3 level controls the composition of LTBR signaling complexes. A, cells were left untreated (− lanes), or treated for 10 min (10′ lanes) and 24 h (24 h lanes) with 100 ng/ml agonist LTBR antibody (BS-1). For positive controls, cells were lysed first and BS-1 added to the unstimulated lysates (*, lanes). BS-1-bound complexes were immunoprecipitated using anti-human IgG conjugated agarose beads, and analyzed by Western blots for LTBR, TRAF3, TRAF2, and IKK1, as indicated. B, band intensities were analyzed by densitometry, and data shown as ratio of either TRAF2 (▧), or TRAF3 (■), to LTBR. C, DLD-1 cells were mock transfected or transfected with non-silencing control siRNA (NS), or siRNA directed against TRAF3, and treated as in A. Pre-IP samples were probed for TRAF3 (n.s., nonspecific band), and immunoprecipitates were analyzed as in A for LTBR, TRAF2, and TRAF3. D, bands from C were quantified by densitometry as described in B. E, DLD-1 cells were treated as in C, lysates were probed for TRAF3 levels before immunoprecipitation to ensure knock-down, and immunoprecipitates were analyzed for IKK1 by Western blot. F, DLD-1 cells were transfected with siRNA against TRAF3 and IKK1, alone or in combination. 48 h after transfection, cells were left unstimulated or stimulated with BS-1 for 10 min and lysates analyzed by Western blots for IKK1, TRAF3, phospho-IκBα (Ser-32/Ser-36), phospho-RelA (Ser-536), and GAPDH (to control for loading).
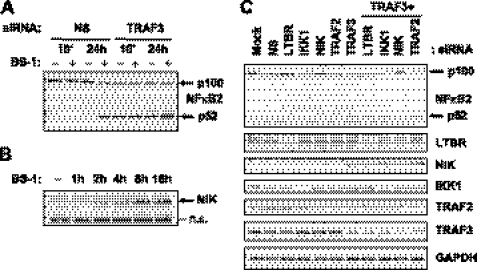
TRAF3 Modulates Intrinsic Activity of the Alternative NFκB and Expression of a Specific Set of NFκB Regulators
TRAF3 has previously been shown to repress NIK-mediated processing of NFκB2 p100 into p52 by promoting proteasomal degradation of NIK (25). During the preparation of this manuscript, He et al. (32) reported that genetic ablation of TRAF3 causes accumulation of NIK and sustained NFκB2 activity. In our experiments, we assessed impact of siRNA-mediated TRAF3 depletion on NFκB2 function and its regulation by LTBR. We found that depletion of TRAF3 in DLD-1 cells caused stimulus-independent processing of NFκB2/p100 to p52 and increased levels of NIK (Fig. 4, A and B). Consistent with previous data from TRAF3 overexpression studies (25), the processing of NFκB2/p100 in TRAF3 siRNA-transfected cells required NIK and was not observed in cells with combined knock-down of TRAF3 and NIK (Fig. 4C). Importantly, siRNA-mediated depletion of LTBR did not block TRAF3 siRNA-mediated autonomous activation of NFκB2 (Fig. 4C), thus confirming that NFκB2 activity caused by down-regulation of TRAF3 did not depend on signaling events upstream of NIK.
FIGURE 4.
TRAF3 knock-down leads to constitutive, NIK-dependent processing of NFκB2/p100. A, DLD-1 cells transfected with a non-silencing control siRNA (NS) or TRAF3 siRNA for 48 h were left untreated (− lanes) or treated (+ lanes) with 100 ng/ml anti-LTBR antibody (BS-1) for 10 min and 24 h. Lysates were analyzed by Western blot for NFκB2 (p100, and processed p52 are indicated by arrows). B, DLD-1 cells treated for indicated times with BS-1 were analyzed by Western blot for NIK (indicated by an arrow; n.s., nonspecific band). C, DLD-1 cells were mock transfected (Mock), transfected with non-silencing control siRNA (NS), or with siRNA against indicated targets either alone or in combination with TRAF3 siRNA. Cells were cultured for 48 h after transfection in the absence of any stimulus, and samples were probed for the indicated proteins by Western blots.
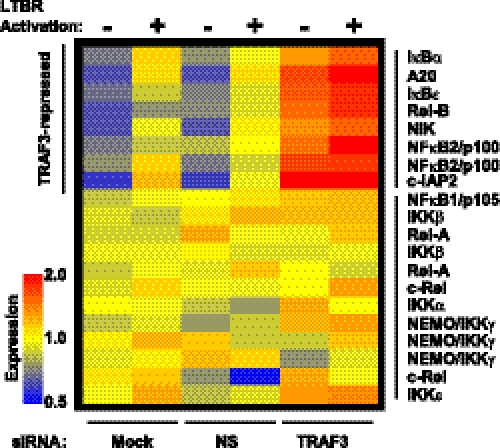
To characterize the gene regulatory effect of NFκB activation caused by the depletion of TRAF3, we have performed transcript profiling of RNA isolated from the TRAF3 knock-down cells. Depletion of TRAF3 caused specific accumulation of transcripts encoding a distinct subset of NFκB regulators, which included the key components of NFκB2 (p100, RelB, and NIK), and a group of NFκB1 inhibitors including IκBα, IκBϵ, A20, and c-IAP2 (Fig. 5). This pattern of NFκB-related gene regulation suggests that cellular levels of TRAF3 can regulate availability of the key components required for NFκB2 activity, as well as the activity of the negative feedback mechanisms that have been shown to suppress both NFκB1 and NFκB2 function at several different points.
FIGURE 5.
TRAF3 represses a group of NFκB-related genes. DLD-1 cells were mock transfected (Mock), or transfected with a non-silencing siRNA (NS) or TRAF3 siRNA. 48 h post-transfection, cells were left untreated (−), or treated (+) with agonist anti-LTBR antibody (BS-1) for 4 h. RNA was collected and profiled for NFκB-related transcripts by microarray.
FIGURE 6.
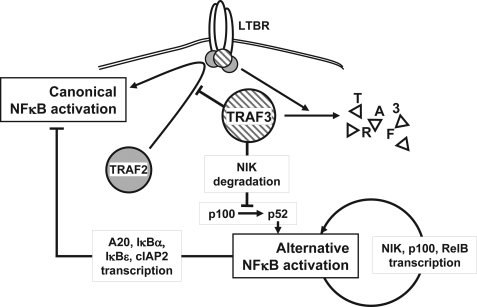
Proposed model for NFκB repression by TRAF3 in LTBR activation. TRAF3 inhibits activation of the classical NFκB pathway by interfering with the formation of productive signaling complexes at LTBR. A high level of TRAF3 interferes with the recruitment of TRAF2 to the receptor. In parallel, TRAF3 induces NIK degradation and consequently inhibits the activation of the alternative arm of NFκB. During LTBR signaling, the alternative arm is activated as a result of TRAF3 degradation and NIK derepression. The sustained activation of the alternative arm in the absence of TRAF3 is maintained by an autoactivation loop. Alternative arm activation up-regulates NIK, NFκB2/p100, and RelB transcription by p52 and RelB, and the continued processing of NFκB2/p100 into p52 is induced by derepressed NIK. Additionally, persistent alternative arm activation up-regulates NFκB1 inhibitors such as IκBα and A20, keeping the classical NFκB pathway inactive in the absence of activation signals.
DISCUSSION
For the subset of TNF receptors that activate both NFκB pathways, the physiological significance of the overlap with the other TNFR members that exclusively activate the canonical NFκB pathway is not fully understood. Additionally, and in the specific case of LTBR, receptor-induced activation of NFκB has been seen to assume more than a single configuration. In one such configuration, LTBR sequentially activates the canonical NFκB pathway followed by the alternative pathway (11). In a different configuration, the early activation of the canonical NFκB pathway by LTBR was not seen (13). Instead, LTBR activation has been reported to proceed exclusively through the non-canonical NFκB pathway to finally induce both RelA and RelB containing NFκB dimers (13). Why and how these different modes of NFκB activation are controlled downstream of a single receptor, LTBR, is not known.
We have found that TRAF3 is a molecular switch that inhibits the LTBR-dependent activation of NFκB1. This is consistent with the reported role of TRAF3 as an inhibitor in CD40 and BAFF-R signaling (21, 22), both members of the LTBR-subset of TNFRs. The mechanism of TRAF3-mediated inhibition of NFκB1 signaling is not known, but a reasonable possibility, which is in keeping with our results, and of others (23, 33), is that excess TRAF3 prevents recruitment of components in receptor complexes that are necessary for NFκB1 activation, such as TRAF2 and IKK1. The role for TRAF2 in this activation is more apparent, as TRAF2 is a required component of CD40 and LTBR-dependent NFκB1 signaling (18, 21, 34), and regulates the degradation of itself and TRAF3 (30, 35). The role for IKK1 in NFκB1 activation, however, is a little less clear. Although IKK1 is a part of the IKK-complex that is involved in IκBα and RelA phosphorylation, IKK1 involvement has been reported to be more important for NFκB2 activation (3), and IKK2 and NEMO/IKKγ instead have been shown to be more crucial mediators of NFκB1 activation, because IKK1 knock-out mice retain the ability to phosphorylate IκBα (36, 37). Despite these results, other studies of NFκB1 signaling using IKK1 and IKK2 knock-out and knock-down cells reveal a more active role for IKK1 in RelA phosphorylation (38, 39). Moreover, LTBR activated nuclear lysates from IKK1 knock-out cells fail to form any RelA/p50 DNA binding activity, while IKK2 knock-out cells display diminished, but clearly detectable, DNA binding activity (40). Our results suggest an involvement of IKK1 in LTBR signaling complexes for the activation of the classical arm, because IKK1 is a part of classical NFκB signal-competent receptor complexes (Fig. 3, A and E), but not of non-signaling complexes. Moreover, our results also suggest that IKK1 is not necessary for basal activation of the alternative arm induced by the loss of TRAF3 (Fig. 4C). This is a surprising finding given the conventional role of IKK1 in p100 processing; however, it is consistent with previous results that while the elimination of IKK1 allows residual p100 to p52 processing during LTBR stimulation, the elimination of both IKK1 and IKK2 eliminates this processing (11). More studies will need to be done to further define the role of IKK1 in p100 processing.
Unlike the role of TRAF3 in inhibiting stimulus-dependent NFκB1 activation, its mode of NFκB2 inhibition is stimulus-independent (Fig. 4A). TRAF3 has been shown to be a negative regulator of NFκB2 activation (24). This occurs most likely by the ability of TRAF3 to destabilize NIK, a kinase that activates IKK1-dependent NFκB2 p100 processing into p52 (25). Co-overexpression of NIK with TRAF3 in 293 cells results in NIK degradation (25), and this is consistent with another report that infers, albeit in an overexpression system, the inhibitory role of TRAF3 in NFκB2 activation (24). Also consistent with the role of TRAF3 as an inhibitor of NIK is the observation (ours and Refs. 25, 30) that signaling through receptors that activate NFκB2, such as LTBR, but not by TNFα (which actually increases the levels of TRAF3, see Fig. 2C), results in the degradation of TRAF3. This explains the ability of these receptors to selectively activate NFκB2, and suggests that the activation of NFκB2 is minimally dependent on reduction of TRAF3 levels to relieve its destabilization of NIK. This is consistent with our results of TRAF3 knock-down in DLD-1 cells, and demonstrates that NFκB2 p100 processing is not dependent on a stimulus to activate LTBR. Whereas NFκB2 activating receptors still must depend on a stimulus to initiate TRAF3 degradation, we show here that NFκB2 p100 processing is directly dependent on this degradation and not on the stimulus per se. TRAF2 has also been shown to be a negative regulator of NFκB2 p100 processing (34), and unstimulated lymph node B cells from TRAF2-deleted mice have constitutively high levels of p52. Likewise, two recent studies using TRAF2-null MEFs also show constitutive p100 processing (41, 42). However, evidence of the inhibitory role of TRAF2 in p100 processing is somewhat controversial as we (Fig. 4C) and others (18, 43) do not observe enhanced p100 processing as a result of TRAF2 depletion. One possibility is that TRAF2 and TRAF3 together, in a non-exclusive process, control NIK destabilization, and that the absence of either one of the components is sufficient to derepress NIK. The studies by Vallabhapurapu et al. (41) and Zarnegar et al. (42) suggest that p100 processing occurs via combined action of TRAF2 recruiting cIAPs and TRAF3 recruiting NIK to a molecular complex to allow NIK degradation. However, they do not explain how cIAPs would be recruited to this molecular complex in the absence of TRAF2 to degrade NIK. Thus, the role of TRAF2 in p100 processing still needs verification. Furthermore, as these studies show, TRAF2 KO mice, unlike TRAF3 KO mice, can be rescued with a heterozygous deletion of NIK, and show only a modest increase in inflammatory cytokines when compared with those in TRAF3 KO mice. This suggests that TRAF2 has a lesser inhibitory role in NFκB activation and cytokine production than TRAF3, and its ablation in certain cell types, such as ours, might not be sufficient to activate non-canonical NFκB.
Both DLD-1 and WiDr cells are intestinal epithelial origin cells which, as shown here, have functionally different levels of TRAF3, suggesting that epithelial cells might have the means to modulate TRAF3 levels. It is possible that these cells represent different stages of epithelial cells during the course of an intestinal inflammation. Epithelial cells in the gut mucosa continuously sample microbial pathogens as well as commensal bacteria, but only initiate inflammatory responses against pathogens (44) to recruit lymphocytes. A high level of TRAF3, which can selectively decouple the classical NFκB pathway and the associated production of inflammatory cytokines from a subset of TNFRs, could counteract the inflammation that would otherwise ensue during lymphocyte infiltration to the gut mucosa. This is consistent with the high levels of TRAF3 that we have observed in DLD-1 cells. On the other hand, WiDr cells could represent epithelial cells that have already encountered CD40L or lymphotoxin beta from infiltrating lymphocytes, and as a result, downregulated their levels of TRAF3. Modulation of TRAF3 levels in the epithelial cells of the intestine might thus have a physiological role in inflammation and associated disorders.
Likewise, normal function of dendritic cells depends on their ability to pass through the steady-state inflammatory milieu without T- or B-cell immunogenic activity unless directly contacted by pathogens or pathogen-activated lymphocytes (45). The immunogenic capacity of DCs is controlled by their maturation stage: immature or semi-mature DCs induce anergy or tolerance, while only fully mature DCs are able to prime T cells (46, 47). Maturation and immunogenic activity of dendritic cells require either TLR stimulation by pathogens or the activation of alternative NFκB pathway by antigen-stimulated lymphocytes (48, 49). The ability of either LTBR or CD40 to efficiently induce DC maturation and immunogenic activity (49, 50) suggests the involvement of a shared component, such as TRAF3, in the activation of the alternative NFκB pathway by these receptors. Indeed, both CD40 and LTBR similarly induce TRAF3 degradation to activate the alternative NFκB pathway (our data and Ref. 30). On the other hand, the repression of classical NFκB pathway in dendritic cells, while interfering with their innate immune functions and their activation by various TLR ligands, is unable to inhibit T-cell induced maturation through the alternative NFκB pathway (48). The requirement for either TLR or lymphocyte induced signals for the full maturation of DCs is consistent with the ability of tissue-resident as well as semi-mature migratory DCs to generally ignore self-antigens or commensal flora and prevent priming of bystander lymphocytes (46). Because the activation of several TLRs has been shown to be TRAF3-dependent (51, 52), having high levels of TRAF3 could ensure efficient TLR-mediated maturation of tissue resident DCs by infiltrating pathogens. At the same time, high TRAF3 would also inhibit the inflammatory activity generated via the classical NFκB pathway when the DCs come into contact with lymphocyte-derived CD40L or lymphotoxin beta. Similarly, high initial levels of TRAF3 would also limit inflammatory cytokine production by DCs receiving maturation signals through CD40 or LTΒR by selectively decoupling the classical NFκB pathway from these receptors.
Supplementary Material

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
- LTBR
- lymphotoxin beta receptor
- TRAF
- TNF receptor-associated factor
- IKK
- inhibitor of κB kinase
- NIK
- NF-κB-inducing kinase
- siRNA
- small interfering RNA
- PIPES
- 1,4-piperazinediethanesulfonic acid
- KO
- knock-out
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase.
REFERENCES
- 1.Schneider K., Potter K. G., Ware C. F. (2004) Immunol. Rev. 202, 49–66 [DOI] [PubMed] [Google Scholar]
- 2.Bonizzi G., Karin M. (2004) Trends Immunol. 25, 280–288 [DOI] [PubMed] [Google Scholar]
- 3.Hayden M. S., Ghosh S. (2004) Genes Dev. 18, 2195–2224 [DOI] [PubMed] [Google Scholar]
- 4.Beinke S., Ley S. C. (2004) Biochem. J. 382, 393–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Locksley R. M., Killeen N., Lenardo M. J. (2001) Cell 104, 487–501 [DOI] [PubMed] [Google Scholar]
- 6.Sun S. C., Ganchi P. A., Ballard D. W., Greene W. C. (1993) Science 259, 1912–1915 [DOI] [PubMed] [Google Scholar]
- 7.Franzoso G., Carlson L., Poljak L., Shores E. W., Epstein S., Leonardi A., Grinberg A., Tran T., Scharton-Kersten T., Anver M., Love P., Brown K., Siebenlist U. (1998) J. Exp. Med. 187, 147–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gommerman J. L., Browning J. L. (2003) Nat. Rev. Immunol. 3, 642–655 [DOI] [PubMed] [Google Scholar]
- 9.Senftleben U., Cao Y., Xiao G., Greten F. R., Krähn G., Bonizzi G., Chen Y., Hu Y., Fong A., Sun S. C., Karin M. (2001) Science 293, 1495–1499 [DOI] [PubMed] [Google Scholar]
- 10.Franzoso G., Carlson L., Xing L., Poljak L., Shores E. W., Brown K. D., Leonardi A., Tran T., Boyce B. F., Siebenlist U. (1997) Genes Dev. 11, 3482–3496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dejardin E., Droin N. M., Delhase M., Haas E., Cao Y., Makris C., Li Z. W., Karin M., Ware C. F., Green D. R. (2002) Immunity 17, 525–535 [DOI] [PubMed] [Google Scholar]
- 12.Müller J. R., Siebenlist U. (2003) J. Biol. Chem. 278, 12006–12012 [DOI] [PubMed] [Google Scholar]
- 13.Basak S., Kim H., Kearns J. D., Tergaonkar V., O'Dea E., Werner S. L., Benedict C. A., Ware C. F., Ghosh G., Verma I. M., Hoffmann A. (2007) Cell 128, 369–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bradley J. R., Pober J. S. (2001) Oncogene 20, 6482–6491 [DOI] [PubMed] [Google Scholar]
- 15.Chung J. Y., Park Y. C., Ye H., Wu H. (2002) J. Cell Sci. 115, 679–688 [DOI] [PubMed] [Google Scholar]
- 16.Force W. R., Glass A. A., Benedict C. A., Cheung T. C., Lama J., Ware C. F. (2000) J. Biol. Chem. 275, 11121–11129 [DOI] [PubMed] [Google Scholar]
- 17.Kuai J., Nickbarg E., Wooters J., Qiu Y., Wang J., Lin L. L. (2003) J. Biol. Chem. 278, 14363–14369 [DOI] [PubMed] [Google Scholar]
- 18.Kim Y. S., Nedospasov S. A., Liu Z. G. (2005) Mol. Cell. Biol. 25, 2130–2137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Force W. R., Cheung T. C., Ware C. F. (1997) J. Biol. Chem. 272, 30835–30840 [DOI] [PubMed] [Google Scholar]
- 20.VanArsdale T. L., VanArsdale S. L., Force W. R., Walter B. N., Mosialos G., Kieff E., Reed J. C., Ware C. F. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 2460–2465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hostager B. S., Bishop G. A. (1999) J. Immunol. 162, 6307–6311 [PubMed] [Google Scholar]
- 22.Xu L. G., Shu H. B. (2002) J. Immunol. 169, 6883–6889 [DOI] [PubMed] [Google Scholar]
- 23.He L., Grammer A. C., Wu X., Lipsky P. E. (2004) J. Biol. Chem. 279, 55855–55865 [DOI] [PubMed] [Google Scholar]
- 24.Hauer J., Püschner S., Ramakrishnan P., Simon U., Bongers M., Federle C., Engelmann H. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 2874–2879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liao G., Zhang M., Harhaj E. W., Sun S. C. (2004) J. Biol. Chem. 279, 26243–26250 [DOI] [PubMed] [Google Scholar]
- 26.Xiao G., Harhaj E. W., Sun S. C. (2001) Mol. Cell 7, 401–409 [DOI] [PubMed] [Google Scholar]
- 27.Browning J. L., Miatkowski K., Sizing I., Griffiths D., Zafari M., Benjamin C. D., Meier W., Mackay F. (1996) J. Exp. Med. 183, 867–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nelson D. E., Ihekwaba A. E., Elliott M., Johnson J. R., Gibney C. A., Foreman B. E., Nelson G., See V., Horton C. A., Spiller D. G., Edwards S. W., McDowell H. P., Unitt J. F., Sullivan E., Grimley R., Benson N., Broomhead D., Kell D. B., White M. R. (2004) Science 306, 704–708 [DOI] [PubMed] [Google Scholar]
- 29.Hoffmann A., Levchenko A., Scott M. L., Baltimore D. (2002) Science 298, 1241–1245 [DOI] [PubMed] [Google Scholar]
- 30.Moore C. R., Bishop G. A. (2005) J. Immunol. 175, 3780–3789 [DOI] [PubMed] [Google Scholar]
- 31.Morrison M. D., Reiley W., Zhang M., Sun S. C. (2005) J. Biol. Chem. 280, 10018–10024 [DOI] [PubMed] [Google Scholar]
- 32.He J. Q., Zarnegar B., Oganesyan G., Saha S. K., Yamazaki S., Doyle S. E., Dempsey P. W., Cheng G. (2006) J. Exp. Med. 203, 2413–2418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xie P., Hostager B. S., Bishop G. A. (2004) J. Exp. Med. 199, 661–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grech A. P., Amesbury M., Chan T., Gardam S., Basten A., Brink R. (2004) Immunity 21, 629–642 [DOI] [PubMed] [Google Scholar]
- 35.Brown K. D., Hostager B. S., Bishop G. A. (2002) J. Biol. Chem. 277, 19433–19438 [DOI] [PubMed] [Google Scholar]
- 36.Li Q., Estepa G., Memet S., Israel A., Verma I. M. (2000) Genes Dev. 14, 1729–1733 [PMC free article] [PubMed] [Google Scholar]
- 37.Hu Y., Baud V., Delhase M., Zhang P., Deerinck T., Ellisman M., Johnson R., Karin M. (1999) Science 284, 316–320 [DOI] [PubMed] [Google Scholar]
- 38.Sakurai H., Suzuki S., Kawasaki N., Nakano H., Okazaki T., Chino A., Doi T., Saiki I. (2003) J. Biol. Chem. 278, 36916–36923 [DOI] [PubMed] [Google Scholar]
- 39.Sizemore N., Lerner N., Dombrowski N., Sakurai H., Stark G. R. (2002) J. Biol. Chem. 277, 3863–3869 [DOI] [PubMed] [Google Scholar]
- 40.Derudder E., Dejardin E., Pritchard L. L., Green D. R., Korner M., Baud V. (2003) J. Biol. Chem. 278, 23278–23284 [DOI] [PubMed] [Google Scholar]
- 41.Vallabhapurapu S., Matsuzawa A., Zhang W., Tseng P. H., Keats J. J., Wang H., Vignali D. A., Bergsagel P. L., Karin M. (2008) Nat. Immunol. 9, 1364–1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zarnegar B. J., Wang Y., Mahoney D. J., Dempsey P. W., Cheung H. H., He J., Shiba T., Yang X., Yeh W. C., Mak T. W., Korneluk R. G., Cheng G. (2008) Nat. Immunol. 9, 1371–1378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saitoh T., Nakayama M., Nakano H., Yagita H., Yamamoto N., Yamaoka S. (2003) J. Biol. Chem. 278, 36005–36012 [DOI] [PubMed] [Google Scholar]
- 44.Macdonald T. T., Monteleone G. (2005) Science 307, 1920–1925 [DOI] [PubMed] [Google Scholar]
- 45.Nolte M. A., Leibundgut-Landmann S., Joffre O., Reis E., Sousa C. (2007) J. Exp. Med. 204, 1487–1501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lutz M. B., Schuler G. (2002) Trends Immunol. 23, 445–449 [DOI] [PubMed] [Google Scholar]
- 47.Reis e Sousa C. (2006) Nat. Rev. Immunol. 6, 476–483 [DOI] [PubMed] [Google Scholar]
- 48.Moore F., Buonocore S., Aksoy E., Ouled-Haddou N., Goriely S., Lazarova E., Paulart F., Heirman C., Vaeremans E., Thielemans K., Goldman M., Flamand V. (2007) J. Immunol. 178, 1301–1311 [DOI] [PubMed] [Google Scholar]
- 49.Summers-DeLuca L. E., McCarthy D. D., Cosovic B., Ward L. A., Lo C. C., Scheu S., Pfeffer K., Gommerman J. L. (2007) J. Exp. Med. 204, 1071–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fujii S., Liu K., Smith C., Bonito A. J., Steinman R. M. (2004) J. Exp. Med. 199, 1607–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oganesyan G., Saha S. K., Guo B., He J. Q., Shahangian A., Zarnegar B., Perry A., Cheng G. (2006) Nature 439, 208–211 [DOI] [PubMed] [Google Scholar]
- 52.Häcker H., Redecke V., Blagoev B., Kratchmarova I., Hsu L. C., Wang G. G., Kamps M. P., Raz E., Wagner H., Häcker G., Mann M., Karin M. (2006) Nature 439, 204–207 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.