Abstract
c-Myc (Myc) oncoprotein induction of genomic instability (GI) contributes to its initial transforming function and subsequent tumor cell evolution. We describe here a pathway by which Myc, via its target protein glycoprotein Ibα (GpIbα), mediates GI. Proteomic profiling revealed that the serine/threonine kinase Aurora B is down-regulated by GpIbα in p53-deficient primary human fibroblasts. The phenotypes of Aurora B deficiency are strikingly reminiscent of Myc or GpIbα overexpression and include double-stranded DNA breaks, altered nuclear size and morphology, chromatin bridges, cleavage furrow regression, and tetraploidy. During mitosis, GpIbα and Aurora B redistribute to the cleavage furrow along with other cleavage furrow proteins. GpIbα overexpression at levels comparable with those seen in some tumor cells causes the dispersal of these proteins but not Aurora B, resulting in furrow regression and cytokinesis failure. Aurora B normalization redirects the mislocalized furrow proteins to their proper location, corrects the cleavage furrow abnormalities, and restores genomic stability. Aurora B thus appears necessary for a previously unrecognized function in guiding and positioning a number of key proteins, including GpIbα to the cleavage furrow. These findings underscore the importance of maintaining a delicate balance among cleavage furrow-associated proteins during mitosis. Suppression of Aurora B via GpIbα provides a unifying and mechanistic explanation for several types of Myc-mediated GI.
Keywords: DNA Damage, Mitosis, Myc, Oncogene, p53, Aneuploidy, Cleavage Furrow Regression, Double-stranded Breaks, Endoreduplication, Tetraploidy
Introduction
As an oncogenic transcription factor, c-Myc (hereafter Myc) regulates as much as 10–20% of the protein-encoding genome and influences numerous additional cellular phenotypes pertaining to proliferation, cell cycle regulation, apoptosis, differentiation, genomic stability, and metabolism (1–7). Although in some cases these phenotypes are genetically separable, collectively they contribute to the fully transformed phenotype (8).
Apart from its function as an oncoprotein, Myc promotion of genomic instability (GI)2 is critical for cancer pathogenesis by driving other key mutational events needed for tumor initiation (5, 6, 9). Ongoing mutations may further contribute to invasive and metastatic behaviors, angiogenesis, and development of resistance to chemotherapeutic drugs thus driving tumor evolution (6).
Myc-mediated GI can involve the amplification of specific genomic loci, the induction of double-stranded DNA breaks (DSBs), palindrome formation, oxidative damage to DNA bases, the promotion of cryptic replication origin firings, and the promotion of tetraploidy (5, 6, 10–15). The promotion of tetraploidy is particularly insidious because it can lead to centrosome duplication and chromosomal mis-segregation. The resultant aneuploidy is common to most cancers and may be critical for their initial generation (16–23).
Numerous direct, downstream targets of Myc have been identified that recapitulate one or more of its phenotypes (24–31). Among these targets is MT-MC1, which is unique among Myc targets in that its enforced expression can recapitulate multiple Myc-like phenotypes including transformation, the promotion of proliferation and apoptosis, the inhibition of differentiation, and the induction of several forms of GI (32–34).
To understand the basis for the multiple Myc-like phenotypes of MT-MC1, we previously conducted genome-wide transcriptional profiling of MT-MC1-overexpressing cells (32). Surprisingly, only 47 genes were deregulated, and all of those examined were also Myc targets. Shared promoter features suggested a common means of regulation. We proposed that these genes might represent a particularly important subset of Myc targets with critical roles in the promotion of Myc-like functions.
Among the more intriguing of the above Myc and MT-MC1 target genes is GPIBA, which encodes GpIbα, a subunit of the von Willebrand factor receptor that typically resides on the surface of platelets and megakaryocytes and that plays an additional role in regulating megakaryocyte ploidy (35, 36). GpIbα is also expressed in nonhematopoietic normal and tumor cells, particularly those with high endogenous levels of Myc; other von Willebrand factor receptor subunits, however, are generally not co-expressed (37). In their absence and unable to form a functional von Willebrand factor receptor, GpIbα localizes to the endoplasmic reticulum (ER), where it drives numerous Myc-like phenotypes, including GI, altered proliferative and apoptotic properties, and frank transformation (38).
The ability of both Myc and MT-MC1 to induce tetraploidy in several immortalized, untransformed cell lines requires the induction of GpIbα and its trafficking to the ER (37, 39). The importance of GpIbα to this pathway is further underscored by the finding that its solitary enforced overexpression is sufficient to promote not only tetraploidy but several other forms of GI, including DSBs, chromosomal bridges, and numerical and structural nuclear abnormalities (37, 39).
Unlike the case in established cells, GpIbα overexpression in primary cells, at levels comparable with those found in some tumor cells, only promotes GI (37). This leads to a marked up-regulation of the p53 tumor suppressor and p53 target genes and causes a form of proliferative arrest indistinguishable from oncogene-induced senescence (OIS) (40, 41). Disabling p53 allows the cells to escape OIS and continue proliferating in the face of ongoing DNA damage (37).
Aurora B is one of three mammalian Aurora serine/threonine kinases that play important and distinct roles in mitosis and that are overexpressed in various human cancers (42–46). Aurora B is particularly critical during the latter stages of mitosis, when it supervises mitotic spindle assembly, chromosome segregation, and cleavage furrow formation and abscission, all of which are highly coordinated and interdependent processes (47–55). Abnormalities in any of these can activate Aurora B and delay abscission until all of the irregularities are resolved (53–55). Conversely, inhibition of Aurora B can lead to cleavage furrow regression in cells with lagging or bridged chromosomes (53). The resultant failure of cytokinesis can lead to tetraploidy and aneuploidy (16, 17, 21, 51).
Given that GI is the only recognizable GpIbα-mediated phenotype in p53-compromised primary cells, they should be ideally suited for identifying GI-associated pathways. As reported here, we subjected genomically unstable GpIbα-overexpressing immortalized human foreskin fibroblasts (HFFs) to proteomic profiling and observed Aurora B to be markedly down-regulated. All of the previously reported forms of GpIbα-mediated GI could be reversed to varying degrees by normalizing Aurora B expression. Taken together, these findings define a pathway by which Myc and its downstream targets, MT-MC1 and GpIbα, regulate GI by perturbing Aurora B. Aurora B is therefore a critical but indirect Myc target.
EXPERIMENTAL PROCEDURES
Cell Lines and Vectors
HFFs were obtained from The American Type Culture Collection (Manassas, VA) and cultured as described previously (37). Low passage number cells were immortalized by transduction with the pBABE retroviral vector encoding the catalytic subunit of telomerase (human telomerase reverse transcriptase) (Addgene, Inc., Cambridge, MA) and selected in hygromycin. Subsequent sequential transductions were performed with a pLXSN-EYFP retroviral vector encoding a full-length Myc epitope-tagged human GpIbα (37–38) and a pRetroSUPER-puro vector encoding a small hairpin RNA directed against human p53 (37). Selections were performed by cell sorting for EYFP-positive cells and by selection in puromycin (33, 37), respectively. Control transductions were performed with the empty pLXSN vector or the pRetroSUPER vector encoding a scrambled small hairpin sequence (39). All of the retroviral packaging was performed in the Phoenix A amphotropic cell line as described previously (33). For the sake of simplicity, the four HFF cell lines, all green fluorescent protein-positive and puromycin-resistant, are denoted hereafter as vector, shp53, GpIbα, and GpIbα+shp53.
Human Aurora B was expressed as a full-length, V5 epitope-tagged protein in the pLenti6-TOPO-V5 vector (Invitrogen) after amplifying an Aurora B cDNA by PCR. Automated DNA sequencing was performed to confirm the identity and sequence of the Aurora B insert. Packaging of this vector, or the empty parental control vector, together with the necessary packaging plasmids was performed in ∼80% confluent 293-FT cells according to the directions of the supplier (Invitrogen) using Superfect reagent (Qiagen). Following a 48–72-h incubation period, the supernatants were filtered through 0.45-μm nitrocellulose filters (Millipore, Inc., Bedford, NY) and applied to HFF cells plated the previous day. Following a 48-h incubation in the presence of 8 μg/ml Sequebrene (Sigma-Aldrich), fresh medium containing 2 μg/ml blasticidin (Invitrogen) was added. Surviving blasticidin-resistant clones were then pooled and used for all further studies.
Antibody Microarrays
shp53 cells and shp53+GpIbα cells were grown to ∼80–90% confluency in T175 tissue culture flasks. The monolayers were washed three times with phosphate-buffered saline (PBS), trypsinized, and pelleted by low speed centrifugation followed by three additional washes in PBS. The cell pellets were then lysed and fluorescently labeled according to protocols supplied by the Kinexus Bioinformatics Corp. Antibody microarray profiling and bioinformatics processing was performed with over 650 antibodies against a variety of human proteins and phosphoproteins as described on the company website. From among the differentially expressed proteins, we selected 18 and confirmed differences in expression of ≥1.5-fold in 15 by quantitative Western blotting (supplemental Fig. S2).
Immunoblotting and Immunostaining
Total cell lysates were prepared and processed by SDS-PAGE and immunoblotting using described previously methods and antibodies (33). Additional antibodies consisted of an anti-Aurora B murine monoclonal antibody (mAb) (BD Transduction Laboratories, San Jose, CA) that was used at a dilution of 1:500 for immunoblotting and at 1:200 for immunostaining of coverslips and a Ser(p)1981-ATM mAb (Rockland Immunochemicals, Gilbertsville, PA) that was used at a 1:200 dilution for immunostaining.
For immunostaining, the cells were grown on glass coverslips and then fixed in 4% paraformaldehyde at room temperature and washed in PBS. 0.1% Triton X-100 in PBS was used to permeabilize the cells, and 1.5% bovine serum albumin/PBS was used as a blocking solution. Primary antibodies included a rabbit mAb against myosin heavy chain (Sigma-Aldrich) (1:500), a rat mAb against GpIbα (Emfret Analytics, Eibelstadt, Germany) (1:100), and murine mAbs against actin (Cytoskeleton, Inc., Denver, CO) (1:100), filamin (a gift from Dr. F. Nakamura, Brigham and Women's Hospital, Boston, MA) (1:500), RhoA (Santa Cruz Biotechnology, Santa Cruz, CA) (1: 100), and CD44 (BD Pharmingen, Franklin Lakes, NJ) (1:1000). All of the primary antibodies were diluted in the blocking solution and incubated with cells for 30 min at room temperature. The secondary antibodies, consisting of fluorescently labeled goat anti-rabbit, mouse, and rat IgG (Invitrogen), were also diluted in the blocking solution (1:500) and applied for 30 min. After additional PBS washing, the cells were finally stained with 4,6-diamidino-2-phenylindole (DAPI) at 1 μg/ml (Sigma) for 5 min.
Microscopy
For fixed cell imaging, stained coverslips were mounted and examined under an Olympus BX60 epifluorescence microscope with 100× oil immersion objectives. A Hamamatsu Argus-20 CCD camera was used to capture the images. Confocal microscopy was performed using either an Olympus Fluoview 1000 microscope or a Nikon Eclipse E800 microscope equipped with a Bio-Rad Radiance 2000 system. For live cell microscopy, 2 × 105 cells were seeded on 35-mm glass-bottomed Petri dishes (MatTek Corporation, Ashland, MA) and subjected to live cell imaging with or without transient transfection with the indicated plasmids. The cells were video recorded while being maintained at 37 °C with a moisturized warm air microscope chamber (Life Imaging Services, Reinach, Switzerland). Differential interference contrast microscopy and epifluorescence microscopy were performed on Nikon Eclipse TE2000-U inverted microscope with Coolsnap HQ digital camera (Roper Scientific Photometrics, Tuczon, AZ). The images were taken and analyzed using MetaMorph software (Molecular Devices, Sunnyvale, CA). Cleavage furrow regression was determined visually from differential interference contrast microscopy images recorded at sequential time points.
Flow Cytometry
HFFs were analyzed for DNA content by flow cytometry of propidium iodide-stained nuclei as described previously (33, 39). Nonidet P-40 detergent-treated nuclei were isolated from either log phase cells or following a 16-h incubation with the 20 ng/ml colcemid. The data were analyzed using single histogram statistics as described previously (33).
RESULTS
Characterization of HFF Cell Lines and Suppression of Aurora B by GpIbα
In preliminary experiments, we found that human telomerase reverse transcriptase-immortalized HFFs possessed properties similar to those of previously described cell lines (37). These properties included the up-regulation of p53 and the p53 target p21WAF1/CIP1 in GpIbα cells (supplemental Fig. S1A) and a loss of this capacity in shp53+GpIbα cells (supplemental Fig. S1B). The levels of ectopic GpIbα expression in these latter two cell lines were also comparable with or less than those previously seen in some tumor cell lines (37 and data not shown). In contrast to nonimmortalized cells, however, GpIbα cells proliferated continuously and did not assume a senescent-like phenotype (supplemental Fig. S1C and data not shown).
The high levels of GI in sh53+GpIbα cells might be due to qualitative and/or qualitative differences in proteins normally involved in maintaining genomic integrity. To test this, we employed an antibody microarray to compare the proteomic profiles of shp53 and shp53+GpIbα cells. This approach (Kinexus, Inc.) involved the use of fluorescently tagged total cell lysates, which were used to probe duplicate arrays containing antibodies to >650 proteins. From the group of differentially expressed proteins (≥ or ≤1.5-fold differences), we confirmed 15 of these by quantitative immunoblotting (supplemental Fig. S2). The range of expression differences varied from 3.1-fold down-regulation by GpIbα to 5.5-fold up-regulation. Among the proteins in the former group was Aurora B (2.6-fold down-regulation) (supplemental Fig. S2). Immunoblotting of the four HFF cell lines reconfirmed this differential expression (Fig. 1A).
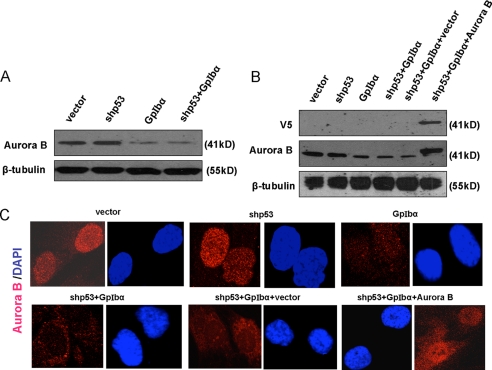
FIGURE 1.
Detection of Aurora B. A, endogenous Aurora B levels. Lysates from the four indicated human telomerase reverse transcriptase-immortalized HFF cell lines (see supplemental Fig. 1) were immunoblotted for Aurora B expression. Note the down-regulation of Aurora B in both GpIbα overexpressing cell lines, thus confirming the results depicted in supplemental Fig. 2. B, shp53+GpIbα cells were transduced with a lentiviral vector encoding V5 epitope-tagged Aurora B or the control empty vector. The resultant blasticidin-resistant clones were pooled and used for immunoblotting. The top panel shows the results obtained with an anti V5 mAb, and the middle panel shows repeat immunoblotting with the anti-Aurora B mAb used in A. Note that transduction with the Aurora B-encoding lentiviral vector restored Aurora B to a level equal to or only modestly exceeding that of the endogenous protein in vector control cells (compare first and last lanes). C, immunolocalization of Aurora B. The indicated cell lines were grown on glass coverslips and then fixed and immunostained for Aurora B using the antibodies described under “Experimental Procedures.” Note the common and generally nuclear localization of Aurora B in all cell lines and the weaker signal in those lines expressing GpIbα.
Aurora B depletion can produce a variety of mitotic defects, culminating in the development of chromosomal instability and aneuploidy (49, 53, 55, 56). We therefore asked whether the GpIbα-associated reduction of Aurora B might account for the high levels of GI. Using lentiviral-mediated transduction, we expressed V5 epitope-tagged Aurora B in shp53+GpIbα cells and detected the ectopically expressed protein with antibodies against either the V5 epitope or Aurora B itself (Fig. 1B). In the latter case, this showed that the restored Aurora B levels were generally equal to or only modestly elevated relative to endogenous GpIbα in vector cells. Finally, we employed immunostaining to demonstrate that, irrespective of the levels of Aurora B in the different cell lines, its subcellular localization was consistent, with the majority of the protein localizing to the nuclei of nonmitotic cells (Fig. 1C).
Restoration of Aurora B Suppresses DSBs Induced by GpIbα
DSBs are frequently used as a surrogate marker of replicative senescence or OIS (57–60). Indeed, the DSBs seen in nonimmortalized GpIbα cells are associated with a proliferative arrest and other senescence phenotypes such as a flattened morphology and the activation of p16INK4a and senescence-associated β-galactosidase (37). However, upon abrogating p53 and reversing OIS, DSBs persist (37). We proposed that DSBs were an OIS-associated but otherwise independent property of GpIbα overexpression and hypothesized that this damage leads to other forms of GI in the face of OIS suppression and a resumption of proliferation (37). Similarly, the proliferating immortalized GpIbα cells described here also demonstrated high levels of DSBs, as indicated by intense γ-H2AX staining (Fig. 2, A and B, and data not shown). As confirmation that γ-H2AX staining was identifying actual DSBs, the above studies were repeated using a mAb directed against phosphorylated ATM (ataxia telangiectasia mutated) protein with similar overall results (Fig. 2, C and D). From these studies, we conclude that the restoration of Aurora B almost completely reverses the high incidence of DSBs induced by GpIbα overexpression.
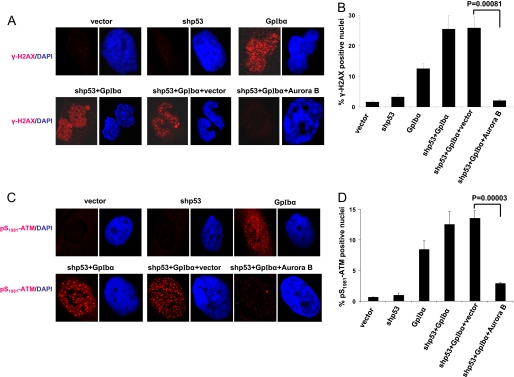
FIGURE 2.
Re-expression of Aurora B is associated with a suppression of DSBs. A, DSBs assessed by immunostaining for γ-H2AX. Coverslips containing each of the indicated cell types were fixed and stained with a mAb directed against γ-H2AX (red), a surrogate indicator of DSBs (31, 60). Nuclear counterstaining (blue) was performed with DAPI as described previously (37). Typical examples of positive nuclei are shown here. B, quantification of DSBs as determined by immunostaining for γ-H2AX. At least 100 cells from each of three different fields were observed, and the fraction of DSBs was quantified (37). The average percentage of positive cells ± S.E. is depicted. The p values were determined by Student's unpaired two-tailed t test. Note that the restoration of Aurora B in shp53+GpIbα cells significantly reduced the fraction of cells with DSBs. C, the above findings were corroborated by staining of separate coverslips for an independent marker of DSBs, Ser(P)1981-ATM (30). D, similar quantification performed with Ser(P)1981-ATM immunostaining.
Restoration of Aurora B Corrects the Nuclear and Ploidy Abnormalities Associated with GpIbα Overexpression
shp53+GpIbα cells often contain micronuclei and multiple nuclei, many of which are dysmorphic and/or of unequal size (37, 38 and supplemental Fig. S3). Moreover, although these cells are referred to as being diploid, they are in fact pseudo-diploid and harbor numerous and complex nonclonal chromosomal gains, losses, and translocations (supplemental Fig. S3).
We compared the frequency of various nuclear aberrations prior to and after the restoration of Aurora B. As shown in Fig. 3 and supplemental Fig. S3, shp53+GpIbα cells contained a high proportion of these abnormalities, all of which were reduced to nearly background levels following Aurora B re-expression.
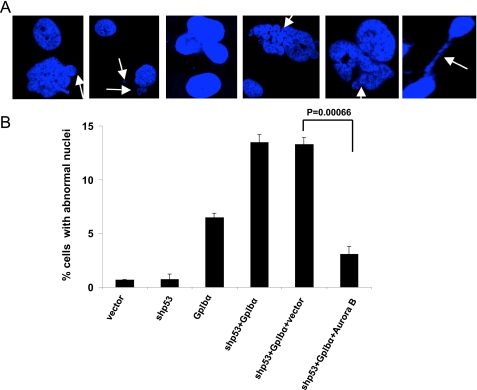
FIGURE 3.
Reduced incidence of abnormal nuclei following restoration of Aurora B. A, representative microscopic fields of HFF cell lines. Note examples of cells with micronuclei (arrows, first and second panels), ≥2 nuclei (third panel), and dysmorphic and/or abnormally sized nuclei (fourth and fifth panels). Also note the presence of chromatin bridges (sixth panel) (see supplemental Fig. S3 and Fig. 2a for additional examples. B, quantification of nuclear defects from A. The nuclei from at least 200 cells from three different coverslips were examined. The bars indicated S.E.
shp53+GpIbα cells are also highly prone to the development of tetraploidy—or, more correct pseudo-tetraploidy—when exposed to mitotic spindle poisons (37, 39). We therefore examined the frequency with which the shp53+GpIbα cells described here developed pseudo-tetraploidy under similar circumstances prior to or following the re-expression of Aurora B. As shown in Fig. 4 and as previously reported for nonimmortalized HFFs (37), the individual overexpression of GpIbα or the knockdown of p53 led to only a minimal accumulation of pseudo-tetraploid cells following microtubule inhibition with colcemid, whereas shp53+GpIbα cells repeatedly accumulated a significant pseudo-tetraploid population. In contrast, shp53+ GpIbα cells in which Aurora B levels had been normalized showed a nearly total reversal of this tendency. From these studies, we conclude that Aurora B also blocks the induction of tetraploidy by GpIbα in the appropriately permissive environment. These findings are also consistent with our previous reports that GpIbα is a key downstream mediator of Myc and MT-MC1 promotion of tetraploidy (39).
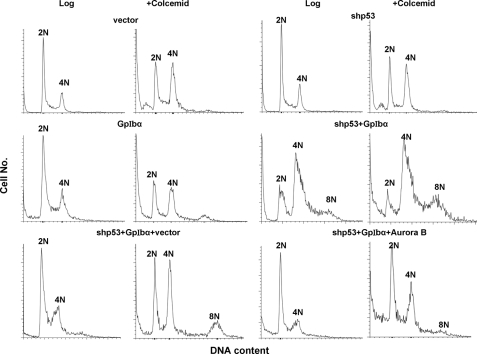
FIGURE 4.
Inhibition of pseudo-tetraploidy/aneuploidy induction by restoration of Aurora B. Flow cytometric profiles of propidium iodide-stained nuclei from each of the indicated cell lines were obtained on either log phase populations or following a 16-h exposure to colcemid as described previously (39). Propidium iodide staining was performed on isolated nuclei to eliminate the contributions of cells with multiple nuclei. Typical results are shown from a total of three to five individual experiments performed with each cell line.
Cleavage Furrow Defects and Cytokinesis Failure Are Corrected by Restoration of Aurora B
In interphase HFFs, endogenous GpIbα diffusely distributes throughout the cytoplasm in a pattern that is consistent with its previously demonstrated association with the ER (37). During the latter stages of mitosis, however, endogenous GpIbα also transiently accumulates at the cleavage furrow along with other furrow-associated proteins such as actin, filamin A, and RhoA.3 Aurora B also localizes to the cleavage furrow and is essential for abscission, the late stage assembly of the plasma membrane barrier separating daughter cells (61). More recently, Aurora B has been shown to be required for the “no cut pathway,” which blocks cytokinesis in response to lagging chromosomes and chromatin bridges (53, 62). We therefore examined the localization of the above cleavage furrow proteins in shp53+GpIbα cells following the restoration of Aurora B. As shown in Fig. 5(A–D) and supplemental Fig. S5 and as we have previously shown,3 endogenous GpIbα, actin, filamin A, and RhoA were absent from or asymmetrically localized to the cleavage furrow in the majority of shp53+GpIbα cells. In contrast, the remaining Aurora B present in these cells localized normally to the cleavage furrow (Fig. 5C). When Aurora B levels were normalized by exogenous expression, actin, filamin A, and RhoA localization was mostly restored to the cleavage furrow, and Aurora B remained confined to the cleavage furrow (Fig. 5, C and D). Therefore, Aurora B was necessary to maintain the proper organization of the contractile structures at the cleavage furrow.
FIGURE 5.
Normalization of mislocalized cleavage furrow proteins and cytokinesis failure following restoration of Aurora B. A, localization of endogenous GpIbα, actin, filamin, RhoA, and Aurora B at the cleavage furrow in mitotic shp53 cells. Typical examples are depicted. Each of the indicated endogenous proteins was detected by immunostaining with appropriate monoclonal antibodies as described previously.3 The patterns of staining are indistinguishable from those seen in vector cells (not shown). B, mislocalization of GpIbα, actin, filamin, and RhoA in shp53+GpIbα cells but retention of Aurora B at the cleavage furrow. C, normalization of all proteins at the cleavage furrow of shp53+GpIbα+Aurora B cells. D, quantification of mitotic cells with mislocalization of the above cleavage furrow-associated proteins. At least 100 mitotic cells from each group were evaluated following immunostaining. The percentage of cells with mislocalized proteins was determined on mitotic cells with obvious cleavage furrows. The error bars represent S.D. The p values are based on a two-tailed Student's t test in a comparison with shp53+GpIbα+vector cells. Note that the localization of endogenous Aurora B did not appreciably change among the different cell lines. E, cytokinesis failure is corrected by Aurora B restoration. The percentage of cells undergoing cytokinesis failure in each of the indicated cell lines was determined by live cell differential interference contrast microscopy as described previously.3 A total of 91 shp53 cells, 53 shp53+Gp1bα+vector cells, and 76 shp53+Gp1bα+Aurora B cells were scored.
Coincident with the correction of cleavage furrow-associated protein localization by Aurora B repletion, we observed a reversal of the prominent cytokinesis failure that accompanies GpIbα deregulation and mislocalization (Fig. 5E). These findings are consistent with our previous results obtained in HeLa cells, where high levels of endogenous GpIbα are associated with the cytoplasmic dispersion of cleavage furrow proteins and a high incidence of cleavage furrow regression, all of which are normalized by reducing GpIbα levels.3 The current studies show that GpIbα deregulation, and by extension Myc activation, induced failure of cell division by reducing Aurora B expression.
Aurora B Restoration Corrects Abnormal Chromatin Bridges
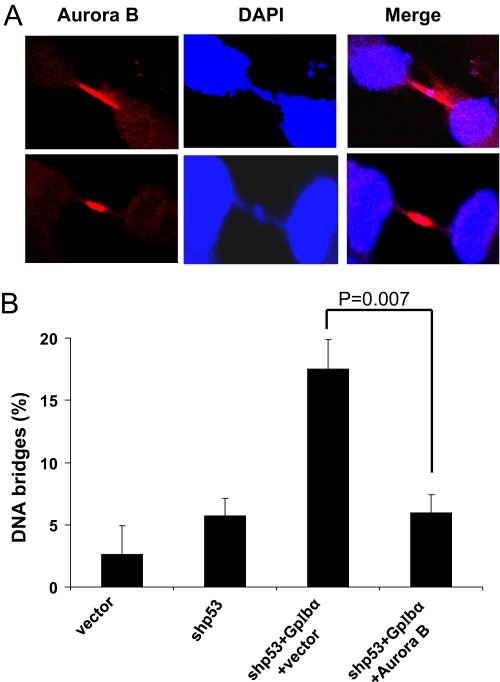
GpIbα overexpression causes chromatin bridge formation in HFFs and other cell types (37). Aurora B can prevent tetraploidy or the inheritance of broken chromosomes by delaying abscission in cells with chromatin in the cleavage furrow (53). A close inspection of DAPI-stained nuclei from shp53+GpIbα cells that were concurrently immunostained for Aurora B showed the frequent presence of highly concentrated bands of Aurora B located along chromatin bridges (Fig. 6A). The re-establishment of normal levels of Aurora B was associated with a greatly reduced incidence of chromatin bridges (Fig. 6B). Taken together, our observations are consistent with the idea that Aurora B not only localizes to the cleavage furrow in association with other furrow proteins to prevent premature abscission but also serves an additional role in chromatin bridge recognition and/or repair (53).
FIGURE 6.
Chromatin bridges in shp53+GpIbα cells are reduced by restoration of Aurora B. A, representative images of chromatin bridges associated with co-localization of Aurora B. shp53+GpIbα cells were immunostained to localize endogenous Aurora B and then counterstained with DAPI. B, quantification of mitotic nuclei with chromatin bridges. Each of the indicated cell lines was stained with DAPI, and the percentage of cells with chromatin bridges as a percentage of all mitotic nuclei was determined. Note the normalization of GpIbα-induced bridging defects following the restoration of Aurora B.
DISCUSSION
The studies described here ultimately address the means by which Myc and its downstream targets promote widespread and seemingly disparate forms of GI ranging from the damage of individual DNA bases to whole chromosome gains/losses and tetraploidy (5, 6, 62). Among the various Myc targets that we previously identified are MT-MC1 and GpIbα, both of which can promote several forms of GI and other Myc phenotypes (32, 34, 37, 39). Indeed, our previous finding that Myc and MT-MC1 can each independently up-regulate GpIbα (32), together with the fact that GpIbα is necessary for both Myc and MT-MC1-mediated GI (39), suggest that the proper regulation of GpIbα is central to maintaining genomic integrity. With this in mind, we used a proteomic-based approach with human telomerase reverse transcriptase-immortalized HFFs, and, as described here, identified Aurora B as one of the most strongly down-regulated proteins in response to GpIbα deregulation.
Aurora B is a serine/threonine kinase that promotes chromosome condensation and adhesion and mitotic spindle assembly (55). The single Aurora kinase of yeast, which is most closely related to mammalian Aurora B, is involved in the so-called no cut checkpoint that senses and regulates abscission timing in response to midspindle chromosome defects or retention of chromatin in the cleavage plane (62–64). In HeLa cells, chromatin bridges increase Aurora B activity and delay abscission, whereas the inactivation of Aurora B promotes cleavage furrow regression (53). Aurora B was proposed to be a component of a pathway that recognizes DNA damage in the form of unsegregated chromatin or chromatin bridges, to delay abscission until the resolution of these abnormalities, and to prevent the generation of tetraploidy as a result of failed cytokinesis stemming from cleavage furrow regression (48, 65–67). Defects in this mechanism are believed to underlie the chromosomal segregation abnormalities of many transformed cells (17, 22). Indeed, the roles for Aurora B thus far described permit us to understand in general terms how either its over- or under-expression could lead to GI (68). In the former case, prolonged abscission delay or overt abscission failure would be expected to greatly increase the likelihood of developing tetraploidy or bi-nucleation. In the latter case, cells with misaligned chromosomes or chromatin bridges would divide prematurely, leading to asymmetric chromosome partitioning and/or broken or translocated chromosomes.
The forms of GI arising as a consequence of GpIbα overexpression are quite varied and in some cases indistinguishable from those detected in cells with misregulated Myc or MT-MC1. These include DSBs, chromatin bridges, failed cytokinesis, tetraploidy, multinucleation, and aberrantly sized and shaped nuclei. Such a causal relationship between Myc, MT-MC1, and GpIbα and the promotion of tetraploidy and other forms of GI in a variety of cell types has been previously shown (39). The direct karyotypic comparisons of shp53 and shp53+GpIbα cells have provided further evidence for complex forms of chromosomal instability in the latter (supplemental Fig. S4). It is likely that the chromosomal profiles of these interphase cells represent the cumulative consequences of the widespread and ongoing GpIbα-mediated GI and cytokinesis failure described here and elsewhere (37, 39).
Based upon our current results and those of others, we can propose a model by which GpIbα and, by extension, Myc and MT-MC1, promote most, if not all, of the described previously forms of GI (Fig. 7). In this model, both Myc and MT-MC1 up-regulate GpIbα as previously reported (32), leading to the accumulation of DSBs. This occurs through the GpIbα-mediated suppression of Aurora B, which normally inhibits DSB formation. However, GpIbα-independent mechanisms may also contribute. The resultant DNA damage leads to p53 up-regulation and eventual p53-dependent OIS as previously demonstrated (37). Absent p53, however, OIS is circumvented, and damaged cells continue to cycle and further accumulate DSBs. These DSBs in turn lead to an increase in nonhomologous end joining and chromatin bridging (69, 70). Acting as a sensor of this type of damage, Aurora B normally activates the no cut pathway so as to immobilize the advancing cleavage furrow until the chromatin bridges are repaired (53). However, because Aurora B is down-regulated, the cleavage furrow regresses, leading to tetraploidy and centrosomal amplification.
FIGURE 7.
Model for GI mediated by the Myc → MT-MC1 → GpIbα pathway. MT-MC1 is a direct target of Myc and GpIbα is up-regulated both by Myc and MT-MC1 (32, 34). The induction of DSBs by Myc occurs through both reactive oxygen species-dependent (dotted arrow) and -independent pathways, which, in the latter case, involve GpIbα (6, 11, 65 and Fig. 2). Concurrently, GpIbα down-regulates Aurora B, which, as described in the current work, also suppresses DSBs (Fig. 2). Aberrant repair of DSBs via nonhomologous end joining can lead to chromatin bridges (13, 69, 70, 74, 75), resulting in delayed cleavage furor formation and abscission. The delay would normally persist until chromatin bridges had been resolved or until lagging chromosomes had further migrated away from the cleavage plane. Aurora B deficiency, possibly in combination with an overwhelming number of chromatin bridges, would prevent a timely resolution of these defects, leading to eventual cleavage furrow regression, cytokinesis failure, and the development of either true tetraploidy or multinucleation (Figs. 4 and 5).
The dispersal of certain cleavage furrow proteins such as actin, filamin, and RhoA in p53+GpIbα cells appears to be less a direct consequence of GpIbα deregulation than of a relative paucity of Aurora B activity at the cleavage furrow. This provides the best explanation as to why restoring Aurora B corrects the localization defect even in the face of persistently high levels of GpIbα (Fig. 5). That Aurora B remains at the cortex and is not itself disrupted by GpIbα overexpression further argues that these two proteins, although communicating with one another, may do so indirectly and are components of functionally distinct cortical complexes.
In addition to serving as a subunit of the von Willebrand factor receptor, GpIbα also plays an apparently unrelated and little explored role in regulating megakaryocyte endoreduplication, a process by which these cells acquire highly polyploidy nuclei via multiple rounds of DNA replication without cell division (35). The current work suggests that the prevention of abscission by promoting cleavage furrow regression could represent a mechanism by which GpIbα allowed megakaryocytes to attain their high ploidy state. Consistent with this idea, recent work has demonstrated that megakaryocyte endoreduplication is in fact associated with a high incidence of cleavage furrow regression (71). Our current findings suggest that cleavage furrow regression in megakaryocytes may be a consequence of changes in GpIbα activity and/or localization, although such a direct link has yet to be made.
The link between DNA breaks, Aurora B inhibition, and cytokinesis failure suggested that chromatin bridges could be an underlying consequence of GpIbα deregulation. However, careful analysis revealed no correlation between bridges and cytokinesis protein mislocalization in shp53+GpIbα cells (data not shown). The Aurora B-mediated no cut pathway identifies misplaced chromatin in the cleavage furrow as the contractile furrow condenses. Therefore, there is an early period of persistent cleavage furrow structure even in cells with lagging chromatin. Furthermore, when Aurora B is inhibited, cleavage furrow regression and protein mislocalization may perhaps continue even after a bridge has broken. These changing relationships may obscure a correlation between cleavage furrow disruption and chromatin bridges until the appropriate green fluorescent protein-tagged probes for live cell imaging are developed to test this model.
The means by which GpIbα communicates with Aurora B is currently unknown and is beyond the scope of the current work. However, it presumably involves signaling from the ER, where virtually all GpIbα is sequestered (38), to the nucleus, which is the primary domain of Aurora B. The critical nature of the ER association for GpIbα is underscored by the fact that a mutant of GpIbα, lacking its signal peptide and unable to localize to the ER, is defective at promoting GI as well as all other previously identified functions (38). ER-to-nuclear signaling has been well documented by such examples as the unfolded protein response, which involves transcriptional reprogramming in response to the accumulation of misfolded or unfolded proteins (54, 72, 73). The identification of other proteins that are clearly deregulated as a result of GpIbα overexpression (supplemental Fig. S2) provides a number of potential candidates for this task that will require further investigation.
Supplementary Material
Acknowledgment
We thank Laura Niedernhofer for valuable discussions and suggestions.
This work was supported, in whole or in part, by National Institutes of Health Grants RO1 CA105033 (to E. V. P.) and ROI DE 016086 (to W. S. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S5.
Q. Wu, F. L. Xu, Y. Li, E. V. Prochownik, and W. S. Saunders, submitted for publication.
- GI
- genomic instability
- DAPI
- 4,6-diamidino-2-phenylindole
- DSB
- double-stranded DNA break
- ER
- endoplasmic reticulum
- γ-H2AX
- serine 139-phosphorylated histone A2X
- GpIbα
- glycoprotein Ibα
- HFF
- human foreskin fibroblast
- mAb
- monoclonal antibody
- OIS
- oncogene-induced senescence
- PBS
- phosphate-buffered saline.
REFERENCES
- 1.Eilers M. (1999) Mol. Cells 9, 1–6 [PubMed] [Google Scholar]
- 2.Hoffman B., Liebermann D. A. (2008) Oncogene 27, 6462–6472 [DOI] [PubMed] [Google Scholar]
- 3.Kim J. W., Dang C. V. (2006) Cancer Res. 66, 8927–8930 [DOI] [PubMed] [Google Scholar]
- 4.Prendergast G. C. (1999) Oncogene 18, 2967–2987 [DOI] [PubMed] [Google Scholar]
- 5.Prochownik E. V., Li Y. (2007) Cell Cycle 6, 1024–1029 [DOI] [PubMed] [Google Scholar]
- 6.Prochownik E. V. (2008) Curr. Mol. Med. 8, 446–458 [DOI] [PubMed] [Google Scholar]
- 7.Zhou Z. Q., Hurlin P. J. (2001) Trends Cell Biol. 11, S10–14 [DOI] [PubMed] [Google Scholar]
- 8.Oster S. K., Mao D. Y., Kennedy J., Penn L. Z. (2003) Oncogene 22, 1998–2010 [DOI] [PubMed] [Google Scholar]
- 9.Kuttler F., Mai S. (2006) Genome Dyn. 1, 171–190 [DOI] [PubMed] [Google Scholar]
- 10.Neiman P. E., Kimmel R., Icreverzi A., Elsaesser K., Bowers S. J., Burnside J., Delrow J. (2006) Oncogene 25, 6325–6335 [DOI] [PubMed] [Google Scholar]
- 11.Vafa O., Wade M., Kern S., Beeche M., Pandita T. K., Hampton G. M., Wahl G. M. (2002) Mol. Cell 9, 1031–1044 [DOI] [PubMed] [Google Scholar]
- 12.Ruchaud S., Carmena M., Earnshaw W. C. (2007) Nat. Rev. Mol. Cell Biol. 8, 798–812 [DOI] [PubMed] [Google Scholar]
- 13.Sallmyr A., Fan J., Rassool F. V. (2008) Cancer Lett. 270, 1–9 [DOI] [PubMed] [Google Scholar]
- 14.Yin X. Y., Grove L., Datta N. S., Long M. W., Prochownik E. V. (1999) Oncogene 18, 1177–1184 [DOI] [PubMed] [Google Scholar]
- 15.Yin X. Y., Grove L., Datta N. S., Katula K., Long M. W., Prochownik E. V. (2001) Cancer Res. 61, 6487–6493 [PubMed] [Google Scholar]
- 16.Caldwell C. M., Green R. A., Kaplan K. B. (2007) J. Cell Biol. 178, 1109–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cimini D., Mattiuzzo M., Torosantucci L., Degrassi F. (2003) Mol. Biol. Cell 14, 3821–3833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dominguez-Sola D., Ying C. Y., Grandori C., Ruggiero L., Chen B., Li M., Galloway D. A., Gu W., Gautier J., Dalla-Favera R. (2007) Nature 448, 445–451 [DOI] [PubMed] [Google Scholar]
- 19.Fujiwara T., Bandi M., Nitta M., Ivanova E. V., Bronson R. T., Pellman D. (2005) Nature 437, 1043–1047 [DOI] [PubMed] [Google Scholar]
- 20.Ganem N. J., Storchova Z., Pellman D. (2007) Curr. Opin. Gene. Dev. 17, 157–162 [DOI] [PubMed] [Google Scholar]
- 21.Ganem N. J., Godinho S. A., Pellman D. (2009) Nature 460, 278–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gisselsson D., Pettersson L., Höglund M., Heidenblad M., Gorunova L., Wiegant J., Mertens F., Dal Cin P., Mitelman F., Mandahl N. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 5357–5362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pellman D. (2007) Nature 446, 38–39 [DOI] [PubMed] [Google Scholar]
- 24.Ghiorghi Y. K., Zeller K. I., Dang C. V., Kaminski P. A. (2007) J. Biol. Chem. 282, 8150–8156 [DOI] [PubMed] [Google Scholar]
- 25.Hermeking H., Rago C., Schuhmacher M., Li Q., Barrett J. F., Obaya A. J., O'Connell B. C., Mateyak M. K., Tam W., Kohlhuber F., Dang C. V., Sedivy J. M., Eick D., Vogelstein B., Kinzler K. W. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 2229–2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lewis B. C., Shim H., Li Q., Wu C. S., Lee L. A., Maity A., Dang C. V. (1997) Mol. Cell. Biol. 17, 4967–4978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nikiforov M. A, Chandriani S., O'Connell B., Petrenko O., Kotenko I., Beavis A., Sedivy J. M., Cole M. D. (2002) Mol. Cell. Biol. 22, 5793–5800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Hagan R. C., Ohh M., David G., de Alboran I. M., Alt F. W., Kaelin W. G., Jr., DePinho R. A. (2000) Genes Dev. 14, 2185–2191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wood L. J., Mukherjee M., Dolde C. E., Xu Y., Maher J. F., Bunton T. E., Williams J. B., Resar L. M. (2000) Mol. Cell. Biol. 20, 5490–5502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lavin M. F. (2007) Oncogene 26, 7749–7758 [DOI] [PubMed] [Google Scholar]
- 31.Kuo L. J., Yang L. X. (2008) In Vivo 22, 305–309 [PubMed] [Google Scholar]
- 32.Rogulski K. R., Cohen D. E., Corcoran D. L., Benos P. V., Prochownik E. V. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 18968–18973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rothermund K., Rogulski K., Fernandes E., Whiting A., Sedivy J., Pu L., Prochownik E. V. (2005) Cancer Res. 65, 2097–2107 [DOI] [PubMed] [Google Scholar]
- 34.Yin X., Grove L., Rogulski K., Prochownik E. V. (2002) J. Biol. Chem. 277, 19998–20010 [DOI] [PubMed] [Google Scholar]
- 35.Kanaji T., Russell S., Cunningham J., Izuhara K., Fox J. E., Ware J. (2004) Blood 104, 3161–3168 [DOI] [PubMed] [Google Scholar]
- 36.López J. A., Dong J. F. (1997) Curr. Opin. Hematol. 4, 323–329 [DOI] [PubMed] [Google Scholar]
- 37.Li Y., Lu J., Cohen D., Prochownik E. V. (2008) Oncogene 27, 1599–1609 [DOI] [PubMed] [Google Scholar]
- 38.Li Y., Lu J., Prochownik E. V. (2009) J. Biol. Chem. 284, 1410–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li Y., Lu J., Prochownik E. V. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 3490–3495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bartek J., Bartkova J., Lukas J. (2007) Oncogene 26, 7773–7779 [DOI] [PubMed] [Google Scholar]
- 41.Courtois-Cox S., Jones S. L., Cichowski K. (2008) Oncogene 27, 2801–2809 [DOI] [PubMed] [Google Scholar]
- 42.Carmena M., Earnshaw W. C. (2003) Nat. Rev. Mol. Cell Biol. 4, 842–854 [DOI] [PubMed] [Google Scholar]
- 43.Nair J. S, Ho A. L., Tse A. N., Coward J., Cheema H., Ambrosini G., Keen N., Schwartz G. K. (2009) Mol. Biol. Cell 20, 2218–2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Natarajan A. T., Palitti F. (2008) Mutat. Res. 657, 3–7 [DOI] [PubMed] [Google Scholar]
- 45.Neef R., Klein U. R., Kopajtich R., Barr F. A. (2006) Curr. Biol. 16, 301–307 [DOI] [PubMed] [Google Scholar]
- 46.Meraldi P., Honda R., Nigg E. A. (2002) EMBO J. 21, 483–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Andrews P. D., Knatko E., Moore W. J., Swedlow J. R. (2003) Curr. Opin. Cell Biol. 15, 672–683 [DOI] [PubMed] [Google Scholar]
- 48.Eggert U. S., Mitchison T. J., Field C. M. (2006) Annu. Rev. Biochem. 75, 543–566 [DOI] [PubMed] [Google Scholar]
- 49.Glotzer M. (2005) Science 307, 1735–1739 [DOI] [PubMed] [Google Scholar]
- 50.Guse A., Mishima M., Glotzer M. (2005) Curr. Biol. 15, 778–786 [DOI] [PubMed] [Google Scholar]
- 51.May K. M., Hardwick K. G. (2006) J. Cell Sci. 119, 4139–4142 [DOI] [PubMed] [Google Scholar]
- 52.Musacchio A., Salmon E. D. (2007) Nat. Rev. Mol. Cell Biol. 8, 379–393 [DOI] [PubMed] [Google Scholar]
- 53.Steigemann P., Wurzenberger C., Schmitz M. H., Held M., Guizetti J., Maar S., Gerlich D. W. (2009) Cell 136, 473–484 [DOI] [PubMed] [Google Scholar]
- 54.Tsuno T., Natsume A., Katsumata S., Mizuno M., Fujita M., Osawa H., Nakahara N., Wakabayashi T., Satoh Y., Inagaki M., Yoshida J. (2007) J. Neurooncol. 83, 249–258 [DOI] [PubMed] [Google Scholar]
- 55.Vader G., Lens S. M. (2008) Biochim. Biophys. Acta 1786, 60–72 [DOI] [PubMed] [Google Scholar]
- 56.Yasui Y., Urano T., Kawajiri A., Nagata K., Tatsuka M., Saya H., Furukawa K., Takahashi T., Izawa I., Inagaki M. (2004) J. Biol. Chem. 279, 12997–13003 [DOI] [PubMed] [Google Scholar]
- 57.Engels W. R., Johnson-Schlitz D., Flores C., White L., Preston C. R. (2007) Cell Cycle 6, 131–135 [DOI] [PubMed] [Google Scholar]
- 58.Halazonetis T. D., Gorgoulis V. G., Bartek J. (2008) Science 319, 1352–1355 [DOI] [PubMed] [Google Scholar]
- 59.Nijnik A., Woodbine L., Marchetti C., Dawson S., Lambe T., Liu C., Rodrigues N. P., Crockford T. L., Cabuy E., Vindigni A., Enver T., Bell J. I., Slijepcevic P., Goodnow C. C., Jeggo P. A., Cornall R. J. (2007) Nature 447, 686–690 [DOI] [PubMed] [Google Scholar]
- 60.Sedelnikova O. A., Horikawa I., Bonner W. M., Barrett J. C. (2004) Nat. Cell Biol. 6, 168–170 [DOI] [PubMed] [Google Scholar]
- 61.Doxsey S. J. (2005) Mol. Cell 20, 170–172 [DOI] [PubMed] [Google Scholar]
- 62.Mendoza M., Barral Y. (2008) Biochem. Soc. Trans. 36, 387–390 [DOI] [PubMed] [Google Scholar]
- 63.Ray S., Atkuri K. R., Deb-Basu D., Adler A. S., Chang H. Y., Herzenberg L. A., Felsher D. W. (2006) Cancer Res. 66, 6598–6605 [DOI] [PubMed] [Google Scholar]
- 64.Norden C., Mendoza M., Dobbelaere J., Kotwaliwale C. V., Biggins S., Barral Y. (2006) Cell 125, 85–98 [DOI] [PubMed] [Google Scholar]
- 65.Barr F. A., Gruneberg U. (2007) Cell 131, 847–860 [DOI] [PubMed] [Google Scholar]
- 66.Giet R., Glover D. M. (2001) J. Cell Biol. 152, 669–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Weaver B. A., Silk A. D., Cleveland D. W. (2006) Nature 442, E9–10 [DOI] [PubMed] [Google Scholar]
- 68.Nguyen H. G., Makitalo M., Hilaire C., Ravid K. (2009) FASEB J. 23, 2741–2748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Acilan C., Potter D. M., Saunders W. S. (2007) Genes Chromosomes Cancer 46, 522–531 [DOI] [PubMed] [Google Scholar]
- 70.van Gent D. C., van der Burg M. (2007) Oncogene 26, 7731–7740 [DOI] [PubMed] [Google Scholar]
- 71.Geddis A. E., Fox N. E., Tkachenko E., Kaushansky K. (2007) Cell Cycle 6, 455–460 [DOI] [PubMed] [Google Scholar]
- 72.Malhotra J. D., Kaufman R. J. (2007) Semin. Cell Dev. Biol. 18, 716–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Szegezdi E., Logue S. E., Gorman A. M., Samali A. (2006) EMBO Rep. 7, 880–885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shields B. J., Hauser C., Bukczynska P. E., Court N. W., Tiganis T. (2008) Cancer Cell 14, 166–179 [DOI] [PubMed] [Google Scholar]
- 75.Shrivastav M., De, Haro L. P., Nickoloff J. A. (2008) Cell Res. 18, 134–147 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.