Abstract
RNAs often exhibit a high degree of conformational dynamics and heterogeneity, leading to a rugged energy landscape. However, the roles of conformational heterogeneity and rapid dynamics in molecular recognition or RNA function have not been extensively elucidated. Ultrafast time-resolved fluorescence spectroscopic experiments were used here to probe picosecond dynamics of the theophylline-binding RNA aptamer. These studies showed that multiple conformations are populated in the free RNA indicating that this aptamer employs a conformational capture mechanism for ligand binding. The base on residue 27 in an internal loop exists in at least three conformational states in the free RNA, including binding competent and incompetent states that have distinct fluorescence decay signatures indicating different base stacking interactions. Picosecond dynamics were also detected by anisotropy experiments, where these motions indicate additional dynamics for base 27. The picosecond data show that theophylline binding shifts the equilibrium for conformations of base 27 from primarily stacked in the free RNA to mostly unstacked in the RNA-theophylline complex, as observed in the previous NMR structure. In contrast, base 10 in a second internal loop is mostly pre-organized in the free RNA, consistent with it being stacked between G11 and G25, as is observed in the bound state. Picosecond dynamics were also measured on a modified aptamer that binds with higher affinity to 3-methylxanthine than theophylline. The modified aptamer shows less heterogeneity in the aptamer-3-methylxanthine complex than what is observed in the theophylline aptamer-theophylline complex.
Keywords: RNA aptamer, conformational heterogeneity, conformational dynamics, conformational capture, ligand recognition, theophylline, 3-methylxanthine
RNAs often exist as an ensemble of conformational states (1) that interconvert on various timescales (2–5). Consequently, the folding landscapes can be very complicated due to the dynamic nature of RNAs. Ligand binding can dramatically modify this energy landscape, where one conformation or a small subset of conformations is preferentially stabilized via additional interactions, by employing a so-called conformational capture or conformational selection mechanism (3, 4). Thus, conformational dynamics are expected to play an important role in the functional diversity of RNAs (6, 7). To better understand the role of conformational capture in RNA recognition, it is crucial to develop tools that quantitatively define the conformational ensemble for RNAs. NMR is one method for probing RNA conformational dynamics (6, 8), and improvements in the potentials for nucleic acids are leading to more robust molecular dynamic simulations of fast motions in RNAs (9). More recently, single molecule methods have been used to directly observe transitions between conformation states in RNAs (10, 11). However, even with these recent advances we still have only a crude understanding of the roles that conformational dynamics and conformational heterogeneity play in the biological function of RNAs.
We recently developed a femtosecond fluorescence dynamics-based approach that quantitatively probes conformational heterogeneity in RNAs (12). This method is sensitive to changes in base stacking patterns, which is a prominent feature in RNA structure. When 2-amino purine (2AP) is incorporated into nucleic acids, the quenching dynamics of this fluorescent base are sensitive to base-specific stacking interactions that give rise to fluorescence decay profiles on the ps-ns timescale (13, 14). A multi-exponential fluorescence decay profile for the 2AP base in an RNA is therefore indicative of conformational heterogeneity of the stacking interactions for the 2AP. The differential rates for fluorescence quenching can arise from charge transfer between the 2AP excited state and other surrounding bases, and analysis of these rates can be used to identify specific base stacking interactions as well as the heterogeneity of these interactions (12, 15–17). By resolving the timescales and the amplitudes of each component in a fluorescence decay profile, it is possible to obtain direct information on the number of distinct stacking conformations and their equilibrium distribution, respectively (12). This technique is used here to study conformational heterogeneity in two closely related RNA aptamers (Figure 1A), evolved via SELEX that either specifically recognizes theophylline (18), a naturally occurring alkaloid widely used as a bronchodilator in the treatment of asthma and bronchitis (19), or with a single base change evolved as part of an allosteric ribozyme (20) that specifically recognizes the structurally similar 3-methylxanthine (3MX, Figure 1B).
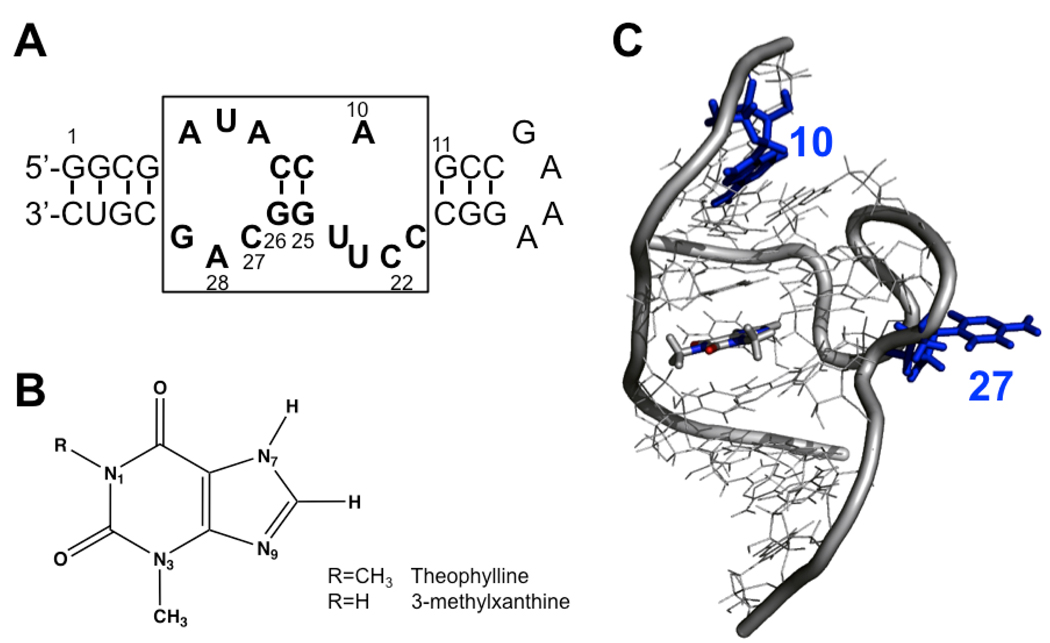
Figure 1.
(A) Sequence and secondary structure of the theophylline-binding aptamer. The conserved region is boxed with bases in bold. The 3MX-binding aptamer used here has C22 changed to A22 and C27 changed to A27. (B) Chemical structures of theophylline and 3-methylxanthine. (C) NMR structure of the theophylline binding aptamer-theophylline complex (1EHT) showing only the conserved region, with bases 10 and 27 labeled in blue. The theophylline ligand is shown in sticks.
The theophylline-binding aptamer has a central core composed of 15 conserved residues forming two internal loops separated by a short helical region (Figure 1A) (18). The aptamer discriminates against a number of similarly structured small molecules and shows 10,000-fold lower binding affinity to caffeine, which differs from theophylline by addition of a single methyl at N7 (Figure 1B). An S-turn in the backbone allows residues in the two internal loops to interact and recognize the functional groups on the theophylline ligand (Figure 1C). Extensive hydrogen bonding and stacking interactions likely provide the thermodynamic stabilization energy for ligand binding by the aptamer (18, 21). Interestingly, a single base change at position 22 from C to A, which is directly involved in hydrogen bonding with the ligand, can switch the aptamer’s specificity to 3-methylxanthine (20). Such specificity switches with only a small number of mutations have also been observed in amino acid binding aptamers (22, 23), and that these mutations would yield the observed changes in specificity were not readily predicted from the structure. Therefore, studies of the mechanism by which this aptamer recognizes theophylline with such high affinity and specificity will lead to an improved understanding of the general mechanism that functional RNAs, such as riboswitches (24–26), use to recognize their small molecule ligands.
The theophylline-binding aptamer shows a very high level of conserved nucleotides around the theophylline binding site (Figure 1A), but residue 27 is only semi-conserved and can be A, C or even abasic without significantly affecting the Kd for theophylline binding (27). In the NMR structure of the RNA-theophylline complex, the base of C27 is flipped out of the loop (Figure 1C) and not interacting with any groups (21). RNAs are notorious for forming alternate, often inactive, structures and this is exemplified by residue 27 in this aptamer. This position cannot be U or G, because that leads to stable interaction in free RNA (such as an A7-U27 base pair) (27), which prevents A7 from forming an AC-platform with C8 as observed in the complex (21). In addition, C27 can form a protonated A7H+-C27 pair at lower pH, again preventing ligand binding (27). These results demonstrate that certain interactions in the free RNA state can have profound effects on ligand binding. Thus, to fully understand ligand binding by RNAs requires structural and dynamic information on both the free RNA and the RNA-ligand complex. NMR studies suggested that the free RNA aptamer is conformationally heterogeneous (18, 21, 27, 28) and fluorescence-detected stopped flow kinetics measurements showed that theophylline binding is over 1000-fold slower than diffusion-controlled process, where the high affinity mostly comes from a slow off-rate (28, 29). The stopped flow data led to a model where the free RNA exists as a population of two inter-converting non-fluorescent conformations (a binding competent and incompetent species) consistent with this aptamer employing a conformational capture mechanism for binding of theophylline (28, 29).
When an RNA employs conformational selection, there must be a ligand-binding competent state, often in low abundance, that is selectively stabilized by additional interactions with the ligand. To characterize such a mechanism, it is necessary to quantitatively resolve the pre-existing equilibrium of multiple states in the free RNA and identify which states are (in)competent for ligand binding. Ultrafast time-resolved fluorescence spectroscopy was used here to show that the free theophylline aptamer has extensive conformational heterogeneity and that this heterogeneity and conformational dynamics play a crucial role in ligand recognition. It was shown that in the free RNA, the internal loop containing residue 27 has a high degree of conformational flexibility. There is high population that has extensive stacking interactions with base 26 and 28 and therefore is “inactive” for ligand binding, and a low population of the “active” (ligand binding competent) conformation where base 27 is flipped out of the helix. In the theophylline-complex, base 27 mostly samples the flipped-out conformation. In contrast, residue 10 in a second internal loop is largely pre-organized in the free RNA and shows no evidence for changes in stacking interactions upon theophylline binding. Ultrafast dynamics were also measured on a modified aptamer that binds with higher affinity to 3-methylxanthine. These data show that there is much less conformational heterogeneity for the RNA in this modified aptamer-3MX complex than in the wild-type aptamer-theophylline complex.
MATERIALS AND METHODS
Materials and Steady-State Fluorescence Measurements
All RNAs (Figure 2) were purchased from Dharmacon (Lafayette, CO) and purified by PAGE. RNA concentrations were calculated from UV absorbance at 260 nm using extinction coefficients provided by Dharmacon. Unless noted, measurements were performed in a buffer of 100 mM HEPES, 100 mM NaCl, 10 mM MgCl2, 0.1 mM EDTA, pH 7.5. Steady state fluorescence titration experiments were conducted by monitoring the change in fluorescence of a 2AP probe incorporated in the RNA (100 nM), upon titration of ligands. Titrations were performed at 25 °C on a Shimadzu spectrofluorophotometer (RF-5301PC) equipped with a temperature control module where the sample was constantly stirred during data collection. The excitation and emission wavelengths were 310 nm and 370 nm, respectively. Kd values were determined by fitting to a binding isotherm using the DynaFit program (30).
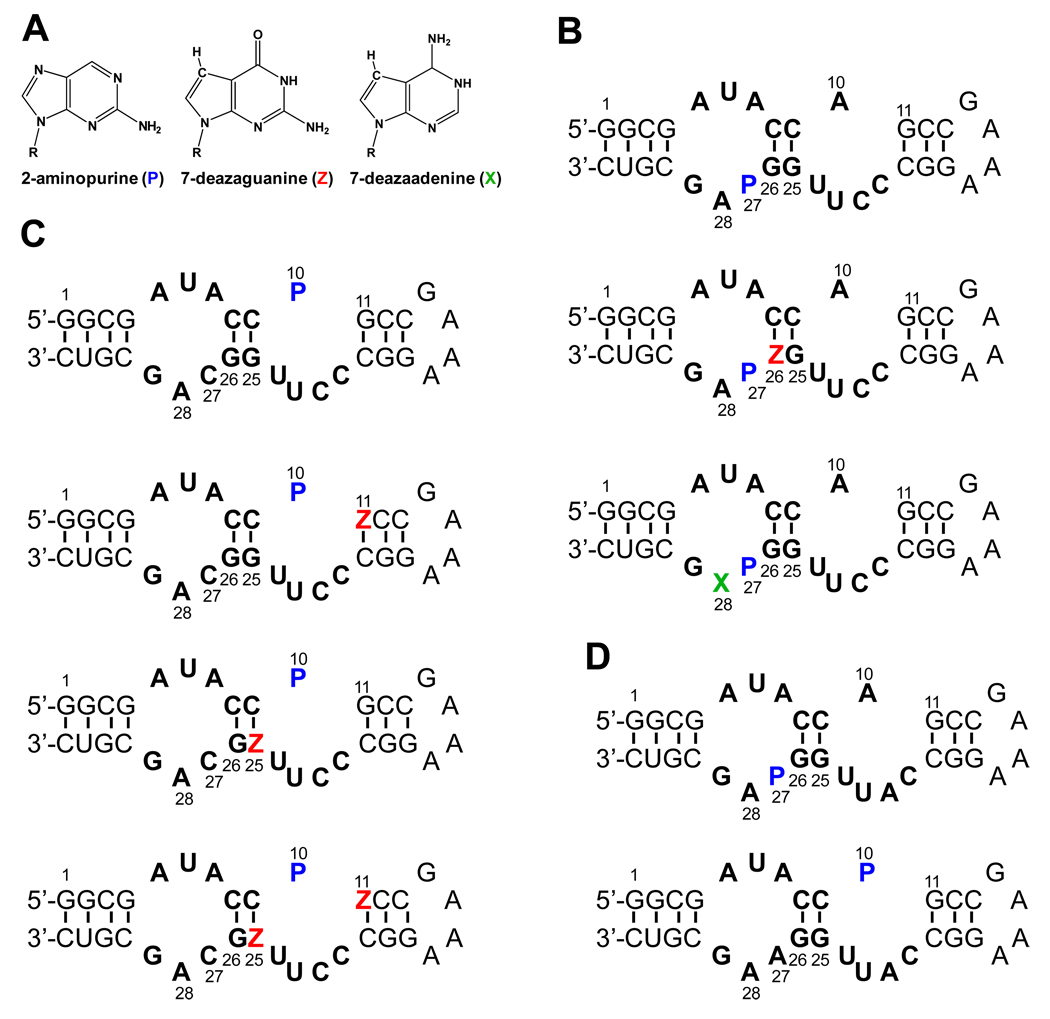
Figure 2.
(A) Structures of 2-aminopurine (P), 7-deazaguanine (Z), and 7-deazaadenine (X). Sequences and secondary structures of (B) the theophylline-binding aptamer RNA constructs with 2AP (P) labeled at residue 27; (C) the theophylline-binding aptamer RNA constructs with 2AP labeled at residue 10; and (D) 3MX-binding aptamer RNA constructs with 2AP at residues 27 and 10.
Isothermal Titration Calorimetry
ITC measurements were performed on a MicroCal iTC200 microcalorimeter (Microcal, Inc.) at 25 °C using RNA concentrations of 10 µM (WT), 19 µM (P27), and 26.4 µM (P10) for theophylline aptamer constructs, and 10 or 20 µM for 3MX aptamer constructs. Theophylline (є270 = 10,470 M−1cm−1) or 3MX (є275 = 11,500 M−1cm−1) were dissolved in identical buffer at a concentration 20-fold higher than the RNA. The theophylline or 3MX ligand was titrated into the sample cell containing RNA in twenty-five 1.5 µL injections with reference power 3.5–11 µcal·s−1 using an initial delay of 60 s with 120 s between injections and a 1000 rpm stirring speed. Data collected from each experiment were fit to a single-site binding model using Origin 7.0 ITC software (Microcal Software Inc.).
Femtosecond Fluorescence Upconversion and Data Analyses
Femtosecond pulses (120 fs, 800 nm, 2.3 mJ) were generated using a Ti:sapphire laser system (Spectra Physics). The pulse was split equally into two beams, where one beam was directed to pump an optical parametric amplifier (OPA) and the signal output from the OPA was then quadrupled to generate the excitation pump pulse at 322 nm for 2AP excitation. The remainder of the fundamental 800 nm was used as the probe pulse. The emission was collected by a pair of parabolic focus mirrors and mixed with the fundamental in the BBO crystal. The upconverted signal (257 nm, corresponding to 380 nm fluorescence) was detected by a photomultiplier after passing through a double-grating monochromator. RNA concentrations were 100–200 µM and theophylline or 3MX were 600–2000 µM at 5 mM MgCl2 in the same buffer used for the Kd measurements. All measurements were carried out at room temperature (22 °C) with the sample being constantly stirred during data collection. For these magic angle fluorescence experiments, the pump beam polarization was set to 54.7° with respect to the fluorescence emission polarization of the BBO crystal, to avoid complications from orientational motions. The femtosecond transients were collected up to 400 ps and were analyzed using a least-square fitting procedure, in which the multi-exponential decays (three or four terms) were convoluted with a Gaussian response function by floating the value of the full width at half maximum (FWHM) (13):
| (1) |
where τi’s and Ai’s are the decay lifetimes and the pre-exponential amplitudes, respectively, for the ith decay component; t0 is time-zero, Δ is the width of the instrument response function (cross correlation, typically 500–600 fs determined by recording Raman emission profile for solvent water), and erf is the error function.
Mathematical software Scientis was used to analyze the ultrafast dynamics data at magic angle. Statistical tests including χ2- and F-tests were used to analyze whether models with more parameters significantly improved the fits (31). The F-test compares the reduced χ2 for the two models, with the F-value (the ratio of the two reduced χ2’s) distributed according to the F-distribution (31). A small F probability (<0.5) indicates high significance of the difference between the two model fits. Except for 3MX (P27) cognate complex which only needs one parameter to fit, none of the decay profiles fit well to a two-exponential function (Figure S3 of Supplementary Information). A three-exponential function significantly improved the fit for all profiles (F values range from 10−44 to 0.11); however, the residuals still showed systematic deviation for most experiments. For those profiles where a three-exponential fit was sufficient, the F-values range from 0.88 to 0.99; but for others, a four-exponential function produced the best fit (F values range from 10−5 to 0.29), with the residuals more randomly distributed. The use of a five-exponential function was not tested. The time window of the femtosecond experiment is too short to reliably determine slow decay components (> 5 ns). Thus, the parameter corresponding to the longest lifetime, τ4, was fixed at 11.3 ns, which is the average observed lifetime for the free 2-aminopurine base (9-methyl-2-aminopurine) and the 2-aminopurine ribonucleoside (32). The specific value of this component (for a range of 10–12 ns) does not affect the fit of the three faster components; however, the fit does not converge if τ4 was not fixed. The third component, which is typically on the order of hundreds of picoseconds, could be accurately determined. Values of τ1, τ2, τ3, and τ4 typically differ by one order of magnitude, further indicating the necessity of using four-parameter fits for most of decay profiles.
Femtosecond Time-Resolved Fluorescence Anisotropy
Separate fluorescence upconversion transients were also collected with the emission polarization parallel or perpendicular to that of the excitation. To account for possible excitation power fluctuation between the two measurements, the fluorescence intensities for a solution of free 2AP base were measured at two time delay points of −10 and 400 ps following each transient acquisition on an RNA sample. The fluorescence intensity of 2-aminopurine at −10 ps time delay was used as background. For a time delay of 400 ps, the anisotropy decay of free 2-aminopurine base is complete, and identical intensity should be observed between parallel and perpendicular polarization. The differences in intensity between these two time delays (−10 and 400 ps) for either parallel polarization or perpendicular polarization were used to calibrate the two polarization transients of RNA samples. The femtosecond time-resolved anisotropy, r(t), was constructed using the equation (33):
| (2) |
where I∥(t) and I⊥(t) are the calibrated fluorescence transients with emission polarization parallel and perpendicular to the excitation polarization, respectively. The time-resolved anisotropy r(t) was then fit with a function containing 2 or 3 exponential terms.
RESULTS AND DISCUSSION
Design and biophysical characterization of the RNA aptamers
Ultrafast fluorescence spectroscopic data were collected on various RNA constructs to help assign decay components that arise from specific stacking interactions in both the free aptamers and the RNA aptamer-ligand complexes (34). Two sets of modified RNAs were prepared for both the theophylline-binding (Figure 2B & 2C) or 3MX-binding aptamers (Figure 2D). One set of RNAs had C27 replaced by 2AP (denoted P thereafter, Figure 2B, D), because this modification was previously used to monitor ligand binding in a fluorescence-detected stopped-flow investigation (29). A second modified base, either 7-deazaguanine (Z) or a 7-deazaadenine (X) base (Figure 2A), was also incorporated at either position 26 or 28, respectively, in the theophylline-binding aptamer. For the second set of RNAs, A10 is replaced by 2AP (Figure 2C and 2D) and the other constructs had either G11 or G25 or both modified by Z in the theophylline-binding aptamer. As demonstrated earlier, these types of modifications facilitate base-specific assignment of the ultrafast dynamic decay components of the RNA (15–17). For example, if a purine base forms a stacking interaction with the probe base, then an N7 to CH modification of the purine leads to faster quenching of the 2AP (14); therefore changing the observed fluorescence decay profiles in a predictable way (15–17, 34).
The P27 construct of theophylline aptamer binds theophylline with a Kd of 0.72 and 1.4 µM at 10 mM [Mg2+] based on steady state fluorescence titrations and ITC measurements (Table 1 and Figures S1 and S2), compared to 0.86 µM for the unlabeled WT RNA, similar to previous studies (27, 29). The large enhancement of steady state fluorescence intensity for P27 upon ligand binding (7-fold) is consistent with flipping out of this base in the RNA-ligand complex (Figure 1C). The second modification, Z at 26 or X at 28, dramatically reduced the binding affinity, likely due to disruption of specific hydrogen bonds (e.g., N7 of A28 is involved in U6-A28-U23 base triple), consistent with previous studies (35). However, these constructs are still useful for identifying the source of ultrafast charge transfer in the free RNA (see below). There was very little fluorescence change for constructs P10 and P10Z11 in theophylline aptamer upon theophylline binding (data not shown), making it hard to accurately determine the Kd values from the fluorescence titrations, but ITC measurements provided estimates of the Kd values of ~10 µM for both constructs (Table 1 and Figure S2). The P10Z25 and P10Z11Z25 constructs showed much lower affinities, with Kd values > 100 µM. These data suggest that N7 of G25 may form critical interaction in the bound state, but again the constructs were only used here to help identify the source of ultrafast charge transfer in the free RNA.
Table 1.
Dissociation constant (Kd in µM) of RNA aptamers with theophylline or 3MX ligands a
| Aptamer Constructs |
Ligand | Steady-State Fluorescence | ITC |
|---|---|---|---|
| Theophylline-binding Aptamer | |||
| WT | theophylline | — | 0.86 ± 0.12 |
| 3MX | — | 1.6 ± 0.5 | |
| P27 | theophylline | 0.72 ± 0.01 | 1.4 ± 0.12 |
| 3MX | 1.9 ± 0.7 | 2.6 ± 0.6 | |
| P27Z26 | theophylline | 220 ± 120 | 281 ± 38 |
| P27X28 | theophylline | ND | 1050 ± 280 |
| P10 | theophylline | ~ 4 b | 9.4 ± 0.8 |
| P10Z11 | theophylline | ~ 12 b | 9.4 ± 0.7 |
| P10Z25 | theophylline | > 100 b | ND |
| P10Z11Z25 | theophylline | > 100 b | ND |
| 3MX-binding Aptamer | |||
| WT | 3MX | — | 2.7 ± 2.0 |
| P27 | 3MX | 3.6 ± 0.3 | 3 ± 0.5 |
| theophylline | >1000 | ND | |
| P10 | 3MX | ND | 24 ± 5 |
Uncertainties in parameters were estimated from multiple measurements. All measurements were performed in a buffer of 100 mM HEPES, 100 mM NaCl, 10 mM MgCl2, 0.1 mM EDTA, pH 7.5, and at 25 °C.
Kd was only estimated due to extremely small change of the fluorescence intensity upon ligand binding. ND = not determined due to weak binding or lack of fluorescence change upon binding.
Breaker and coworkers developed a variant of the theophylline-binding aptamer that binds 3MX where a single C22A mutation changes the specificity of the aptamer for these two ligands (20). Binding studies were performed here on the 3MX aptamer and showed that this variant discriminates against theophylline because of its reduced affinity toward theophylline (> mM) compared to that for 3MX, as opposed to increased affinity toward 3MX (Table 1). 3MX binds both aptamers with similar affinities (Kd = ~1.6 – 3 µM), which is somewhat weaker than the cognate theophylline aptamer/theophylline affinity (~1 µM). Binding of the 3MX ligand induces significant fluorescence enhancement at P27 in both the theophylline- and 3MX-binding aptamers, similar to that observed for the cognate theophylline complex (Figure S1), suggesting that P27 undergoes similar conformational changes when forming both these complexes. No significant change of fluorescence was observed upon binding of 3MX for the P10 RNA.
Ultrafast fluorescence dynamics at residue 27 indicate significant ligand binding-dependent conformational heterogeneity
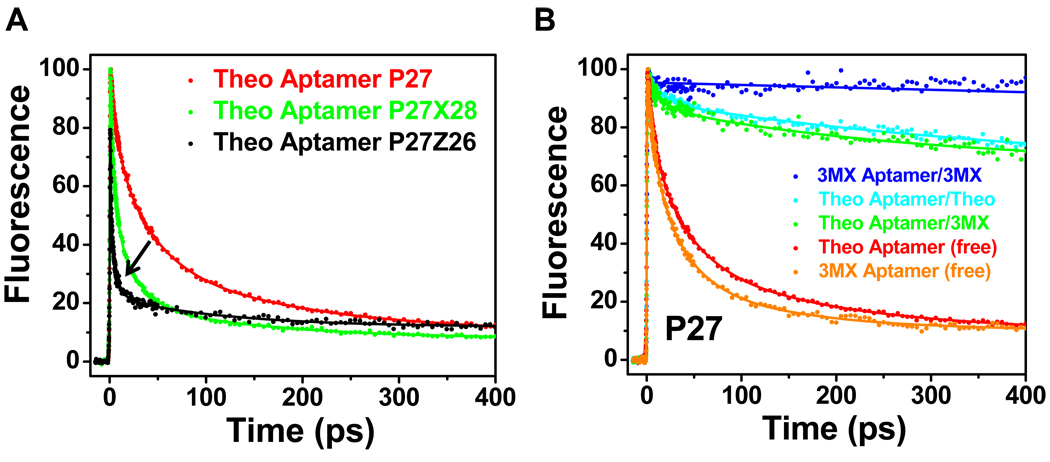
We have previously shown that measurement of the ultrafast fluorescence decay dynamics of 2AP-labeled RNAs can provide unique structural information, particularly in identification of conformational heterogeneity (12, 15–17). Figure 3A shows the ultrafast fluorescence decay profiles for various modified theophylline aptamer constructs (P27, P27Z26, and P27×28) in the ligand-free state. As is typically observed for RNAs where 2AP has been incorporated as a probe in a helical region, all three profiles display multi-phasic decays in the ps-ns regime. Analysis of the fluorescence profiles and comparison with previous RNAs (15–17) indicate that the two fastest components for P27 represent quenching arising from stacking interactions with neighboring bases (G26 and/or A28) either via direct static quenching (6.8 ps), or charge transfer modulated by rapid changes in base stacking (37 ps) (16). The latter may result from rapid small angle rotations of bases about their glycosidic bond (7, 36). The amplitudes of the 6.8 and 37 ps components (Table 2) show that P27 is stacked with G26 and/or A28 more than half the time (~ components 1 and 2 correspond to 63% of the population). Analysis of the decay profiles for P27Z26 and P27X28 support these interpretations (Table 2). As previously described (15–17), the double modifications facilitate base-specific assignments of the ultrafast dynamic decay components, where these N7 to CH modifications lead to more rapid fluorescence quenching for the stacked base. The presence of the 1.1 ps component for P27Z26 is consistent with the strong base stacking interaction between P27 and Z26 (15, 16). The amplitudes of the two fastest decays indicate that these two bases are stacked ~78% of the time in the P27Z26 construct. Also note that the fastest component for P27X28 (3.7 ps) is somewhat faster than that for P27 alone (6.8 ps), indicating that this component is due to stacking of 27 by both 26 and 28 in a “sandwiched” state. Thus the data are consistent with the two fastest components in the decay profile arising from populations where P27 is stacked with both G26 and A28.
Figure 3.
Ultrafast decay profiles measured at the magic angle for the P27 constructs. (A) Decay profiles for the free theophylline-binding aptamer constructs P27 (red), P27Z26 (black), and P27X28 (green). The arrow indicates the change of dynamic timescale from slower in P27 to faster in P27Z26 and P27ZX28 (see text). (B) Comparison of decay profiles between free aptamer P27 constructs (theophylline, red; 3MX, orange) and complexes with theophylline (cyan) or 3MX (green and blue).
Table 2.
Parameters of femtosecond dynamics decay for RNA aptamer constructs
| Aptamer Constructs |
Ligand | τ1 (ps) |
A1 (%) |
τ2 (ps) |
A2 (%) |
τ3 (ps) |
A3 (%) |
τ4 (ns) |
A4 (%) |
|---|---|---|---|---|---|---|---|---|---|
| Theophylline-binding Aptamer | |||||||||
| none (and no Mg2+) |
6.5 | 24 | 43 | 45 | 261 | 17 | 11.3 | 14 | |
| P27 | none | 6.8 | 22 | 37 | 41 | 169 | 27 | 11.3 | 10 |
| theophylline | 18 | 9 | 471 | 21 | 11.3 | 70 | |||
| 3MX | 16 | 10 | 235 | 16 | 11.3 | 74 | |||
| P27Z26 | none | 1.1 | 55 | 6 | 23 | 108 | 10 | 11.3 | 12 |
| P27X28 | none | 3.7 | 47 | 21 | 33 | 145 | 12 | 11.3 | 8 |
| P10 | none | 6 | 63 | 73 | 25 | 11.3 | 12 | ||
| theophylline | 6 | 62 | 75 | 25 | 11.3 | 13 | |||
| P10Z11 | none | 1.5 | 43 | 10 | 33 | 120 | 16 | 11.3 | 8 |
| theophylline | 1.7 | 46 | 11 | 28 | 172 | 17 | 11.3 | 9 | |
| P10Z25 | none | 1.4 | 45 | 11 | 28 | 108 | 17 | 11.3 | 10 |
| P10Z11Z25 | none | 1.3 | 54 | 11 | 19 | 137 | 17 | 11.3 | 10 |
| 3MX-binding Aptamer | |||||||||
| P27 | none | 8 | 27 | 34 | 44 | 131 | 19 | 11.3 | 10 |
| 3MX | 11.3 | 100 | |||||||
| P10 | none | 4.3 | 56 | 32 | 36 | 11.3 | 8 | ||
| 3MX | 4.3 | 60 | 36 | 27 | 11.3 | 13 | |||
Decay life times of amplitudes are represented by τ (ps) and A (%), respectively. Estimated errors are ~5%. All measurements were performed in a buffer of 100 mM HEPES, 100 mM NaCl, 5 mM MgCl, 0.1 mM EDTA, pH 7.5, and at 22 °C.
The third component in P27 (169 ps) could arise from a population where the decay arises from static quenching of P27 from only one base, G26 or A28, or a dynamically-gated charge transfer process, where some conformational dynamic process is occurring on this timescale, or combination of both these processes (16). The decay profiles for P27Z26 and P27X28 help resolve this third component, where the amplitudes of τ3 are reduced compared to P27, consistent with the model that at least part of this component is due to quenching of P27 by the purine base on residue 26 or 28. The remaining amplitude may represent dynamics-gated charge transfer (see discussion below on anisotropy). The 4th component with ns timescale and small amplitude represents the population of P27 base that is totally unstacked, and therefore not quenched. These observations indicate that P27 exists in multiple conformations in the free RNA where its base forms various stacking interactions with the surrounding bases.
All three RNAs (P27, P27Z26 and P27X28) have similar total populations for the sub-ns components indicting that they have equivalent distribution between stacked and unstacked states. The ultrafast dynamics data show that there are at least three distinct populations for the base P27 in the free RNA; one population (in the range of 60 – 70%) features a triple stack of G26/P27/A28, a second population (20 – 30%) features double stack of P27/A28 or G26/P27, and a third population (~10%) where P27 is not stacked with any base. Apparently, the populations where P27 is stacked cannot be the active conformation for theophylline binding (29), because base 27 is flipped out in the complex (21). We propose that both stacked states are inactive (binding incompetent) since P27 has to unstack from G26 and A28 to allow formation of the base triples in the complex (21, 29). It was previously proposed that both active and inactive states are non-fluorescent for the P27 RNA (29). It is apparent from the decay profile data here, that the presence of significant quenching dynamics on fast ps timescale renders these species fluorescently “dark” in steady state measurements. Thus, since the free RNA has a low population of active state with P27-flipped out, the total fluorescence of free RNA is low.
When theophylline is added to P27, the decay profile is dramatically changed to one that shows much reduced populations of rapid decays (Figure 3B), and the long ns component becomes dominant (70%). Thus, the population of aptamer with P27 unstacked increases ~7-fold upon addition of theophylline consistent with the steady-state measurements (29) and the high-resolution structure of the complex (21). The 30% population with rapid decays for the P27-theophylline sample is not due to free RNA, since the theophylline is in large excess and well above the Kd. Thus the ultrafast dynamics experiments are consistent with the base of P27 dynamically interacting with other parts of the RNA, even in the complexed state. Comparisons of the decay profiles between the free P27 and P27-theophylline complex clearly demonstrate that the RNA transitions from a heterogeneous population distribution of largely inactive states, where there is a pre-existing low population of binding competent active state, to a complex that has a more homogenous population but one that still shows considerable dynamics and is not in a single state.
Ultrafast dynamics experiments were also performed on the theophylline-binding aptamer in complex with 3MX, the free 3MX-binding aptamer and the 3MX-binding aptamer complexed with 3MX (Figure 3B). The decay profile for the free 3MX-binding aptamer is very similar to the free theophylline-binding aptamer and only small differences were observed for the decay profiles for the theophylline-binding aptamer bound to theophylline or 3MX (Table 2). Interestingly, the decay profile of the 3MX-binding aptamer in complex with 3MX shows no evidence for any fast dynamics. This is consistent with this complex being in a single state, where P27 is flipped out and does not interact with other bases on this timescale. Comparison of the 3MX-binding aptamer-3MX complex with the theophylline binding aptamer-3MX complex indicates that it is the RNA, and not the ligand, that dictates the degree of conformational heterogeneity in the RNA-ligand complex.
For the free theophylline-binding aptamer, the decay components (1+2), 3, and 4, have 60–70%, 20–30%, and 10% relative population distributions, respectively. These populations give ΔΔG°22°C values for the first two inactive states of −1.0 to −1.1 and −0.4 to −0.6 kcal/mol, respectively, relative to the active state (component 4). These data indicate that there are small free energy differences between these states.
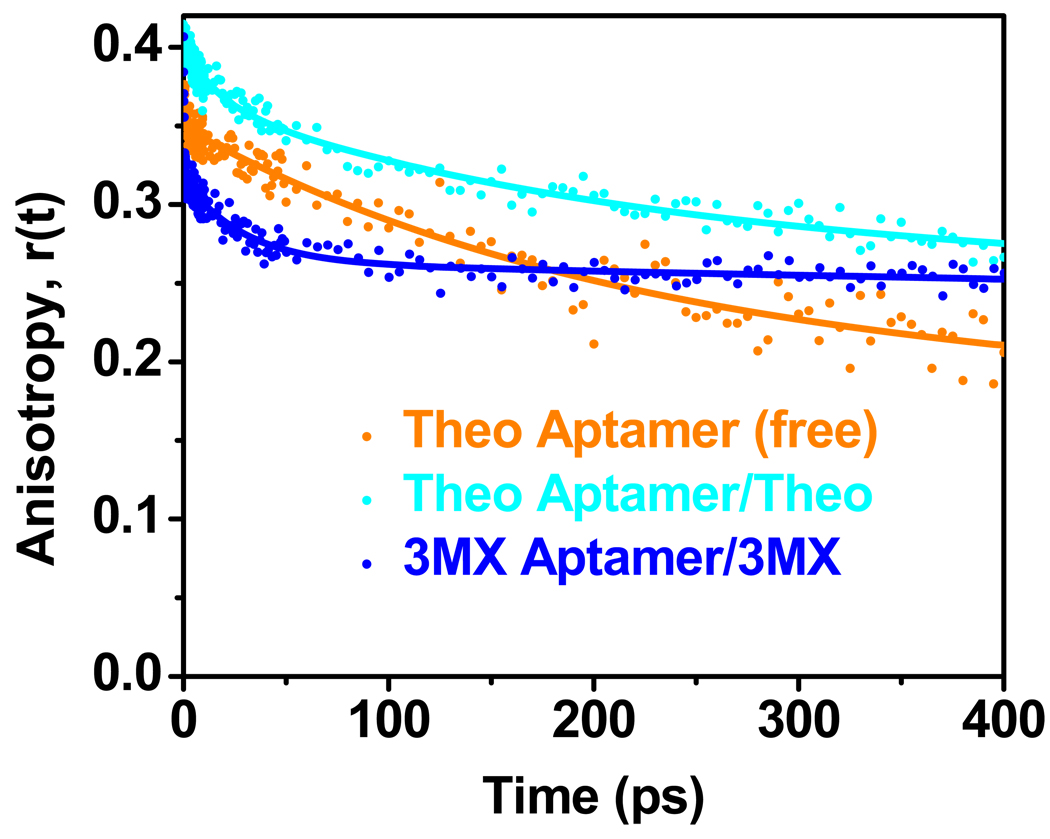
We also measured the femtosecond time-resolved anisotropy decay of theophylline P27 with and without theophylline, and 3MX aptamer/3MX complex (Figure 4 and Table 3). These measurements can directly reveal the timescales of the base motions. For both free and bound profiles, there is a ~10 ns decay component consistent with the tumbling of the entire molecule. The free theophylline P27 RNA has a dynamic component of ~220 ps, indicating internal base dynamics on this timescale, that can be attributed to hindered base stacking/unstacking motions as typically observed for base paired regions (16). This motion is likely the rate-limiting step in the charge transfer reaction with a base quencher (e.g. G26), partly contributing to a decay component on this timescale (τ3 in Table 2). When complexed with theophylline, there is a 16 ps minor component in addition to the major 200 ps component. This indicates that base 27 is more dynamic in the complex, consistent with previous NMR measurements that revealed sub-ns motions for this base and its ribose moiety (27). Anisotropy decays were also measured for the 3MX-binding aptamer-3MX complex and as seen in Figure 4 and Table 3 this complex does not have a 200 ps component and only has the faster 29 ps component which likely arises from dynamics of the P27 base, such as rotation around the glycosidic bond. This is also consistent with the absence of any fast decay components on hundreds of ps in the magic angle experiment for this complex (Figure 3 and Table 2).
Figure 4.
Ultrafast anisotropy decay profiles for theophylline aptamer construct P27 as free RNA (orange) and complexed with theophylline (cyan), and 3MX-binding aptamer complexed with 3MX (blue).
Table 3.
Parameters of femtosecond time-resolved anisotropy decay for P27 constructs a
| Constructs | τ1 (ps) | r1 | τ2 (ps) | r2 | τ3 (ns) | r3 | r0b |
|---|---|---|---|---|---|---|---|
| Theophylline aptamer (free) |
220 | 0.155 | ~10 | 0.193 | 0.348 | ||
| Theophylline complex |
16 | 0.03 | 200 | 0.094 | ~10 | 0.273 | 0.397 |
| 3MX complex | 29 | 0.054 | ~10 | 0.263 | 0.317 |
Anisotropy decay profiles are fit to two or three exponential terms, each with a decay lifetime (τ1 to τ3). τ3 should be considered as an estimate of decay on the nanosecond (ns) time scale, due to the limited exponential time window. Estimated errors are ~10%. All measurements were performed in a buffer of 100 mM HEPES, 100 mM NaCl, 5 mM MgCl, 0.1 mM EDTA, pH 7.5, and at 22 °C.
The fundamental anisotropy at time zero r0 is calculated as r0 = r1 +r2 + r3.
The presence of the ps base dynamics from the anisotropy decays indicates that the energy barrier between these conformational transitions is relatively low. It is possible that the ps base dynamics represent only the fast local stacking/unstacking of P27 from G26 or A28, where formation of other features, such as the base triples, occurs on longer timescales (µs-ms) (29). Previous stopped-flow kinetics measurements revealed that 30–60% of RNA is in active conformation amenable to binding, but these measurements had a minimum resolution of 2 ms (29). The higher population of binding incompetent species observed here is a result of the ability of the femtosecond dynamics experiments to probe much faster timescales than can be obtained with steady-state rapid mixing techniques.
Base 10 is largely pre-organized in the free RNA
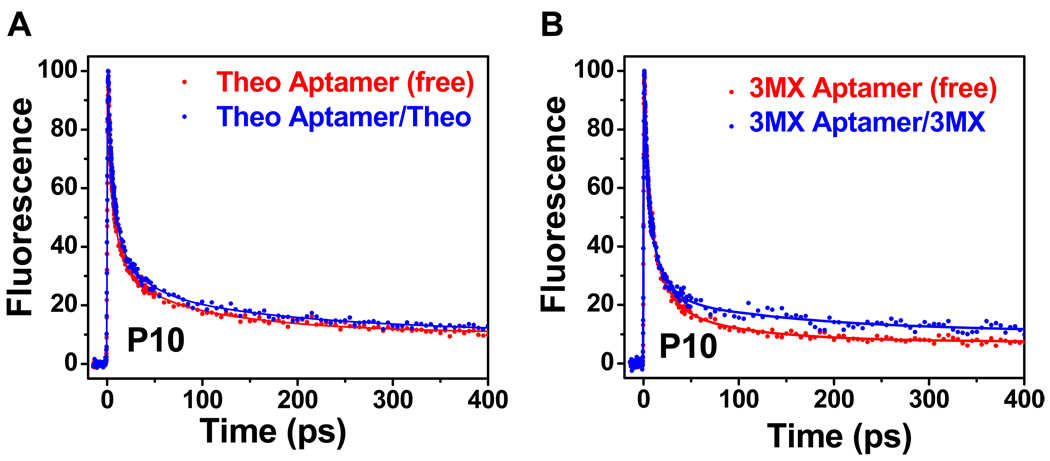
In the structure of the RNA-theophylline complex, the base of A10 is sandwiched between the bases of G11 and G25, and therefore is involved in both intra- and inter-strand stacking (21). To test whether such favorable stacking interactions are pre-organized or are induced at this site, ultrafast fluorescence data were collected on a set of RNAs containing a 2AP base on residue 10 (Figure 1C, D). Figure 5A shows the decay dynamics for the P10 RNAs for the theophylline aptamer. The free RNA exhibits several large amplitude components with fast decays (6 ps, 63%; 73 ps, 25%), and only ~12% for the ns component (Table 2). This indicates that this purine base is largely stacked in the free RNAs, and the decay timescales are consistent with quenching by G bases, presumably by G11 and G25. This decay profile is similar to that observed for a purine as a 3’-dangling end on a CG pair (C9-G25) (16). The presence of an ultrafast ~1 ps component in the decay profiles for constructs P10Z11, P10Z25, and P10Z11Z25 in the free RNAs (Figure S4) confirm the direct base stacking of base 10 with both bases on residues 11 and 25. In contrast to P27, addition of ligand does not produce significant changes in either the timescales or amplitudes of the decay components (Table 2), suggesting there are no large conformational rearrangements at this position upon ligand binding. Therefore, unlike base 27, the purine base 10 in the free state is mostly pre-organized in a conformation similar to the bound RNA. The decay profiles for both the free and bound 3MX-binding P10 aptamers (Figure 5B) are similar to the corresponding profiles for the theophylline-binding P10 aptamer, where again there are only minor changes in decay lifetimes or amplitudes upon ligand binding. It was previously shown that flipping G11-C20 to CG reduces the binding (35) which may arise from altered stacking interactions of these bases with A10 in free and/or bound state. This shows that the orientation of base pairs in the stem regions can also play a direct role in ligand binding.
Figure 5.
Ultrafast decay profiles measured at magic angle for the P10 construct as free RNA (red) and complex (blue) for (A) the theophylline-binding aptamer and (B) the 3MX-binding aptamer.
Conclusions
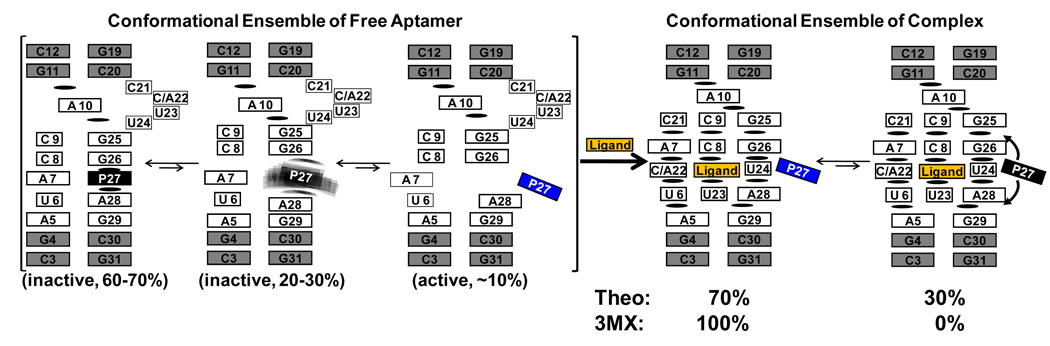
The results here are consistent with both the theophylline-binding and 3MX-binding aptamers employing a conformational capture mechanism (3, 4) for binding of ligands, where parts of the free RNA are pre-organized and other regions are highly dynamic. Based on the ultrafast dynamics decay experiments, we propose a revised model for these two aptamer RNAs (Figure 6), consisting of three states with distinct stacking patterns for base 27 in the free RNAs. These states undergo inter-conversion on a range of timescales that have previously been probed by NMR (27, 28), stopped flow (29), and now here by ultrafast fluorescence decay spectroscopy. The populations of the various states in the free RNAs were determined from the amplitudes of the decay profiles in the magic angle fluorescence decay experiments, and show that only a 10% population has base 27 in a binding competent state. It should be noted that the time-resolved spectroscopic approach only provides local structural information at the 2AP site. Although an extruded base at 27 is consistent with its conformation in the ligand-aptamer complex, the dynamics data alone do not provide any direct information on the rest of the RNA. Therefore the 10% population should be regarded as the upper limit of the active conformation. Given the secondary structure of the core, however, most of this population likely resembles that of the bound form. This low population state can be captured by a binding event, which induces additional interactions that stabilize the bound form. Upon ligand binding, the conformational landscape of the theophylline-binding aptamer is altered, where the dominant population (70%) has P27 flipped out but there is also a sub-population (30%) where this base is interacting with part of the aptamer. The 3MX ligand induces a similar set of populations when binding to the theophylline aptamer. However, the 3MX-binding aptamer-3MX complex shows no fast decays and is in a conformation where P27 is completely flipped out (Figure 6). Thus, for these aptamers it is the RNA and not the ligand that determines the conformational distribution and heterogeneity in the complex. In contrast to base 27, the fluorescence decay data here indicate that base 10 in the free RNA is mostly pre-organized with similar stacking interactions as observed in the complex. Although P10 substitution caused a ~10-fold reduction in binding affinity, the lack of any significant changes in the decay profiles between the free and bound states indicated that the conformational distributions were not affected by the substitution, and ligand binding does not alter the local dynamics, in contrast to position 27. Together, these results show that some regions of the aptamers are conformationally heterogeneous in the free state and others regions are pre-organized in their bound-state conformation. The observed conformational heterogeneity is necessarily the consequence of conformational dynamics. Conformational dynamics on ps-ns timescales were captured by time-resolved anisotropy measurements. These motions are likely base stacking/unstacking dynamics, which are necessary steps that lead to transitions from the inactive state to the active state and open up the binding pocket to accommodate the ligand. Therefore, the data here demonstrate that conformational heterogeneity and dynamics play a crucial role in ligand recognition for these aptamers. The ultrafast dynamics methods used here should prove valuable for understanding the mechanisms and timescales involved in other RNA-ligand interactions, such as the riboswitch regulatory elements (37–39).
Figure 6.
Dynamic model for theophylline- and 3MX-binding aptamers that illustrates the transition from an ensemble of conformations in free RNA to complexes. The black and blue colors of base P27 indicate dark and bright fluorescent states, respectively. The percentages indicate the relative populations of each different states for base 27 in the free RNA, two are identified as inactive, one as active. The dynamic nature of the base 27 in one of the inactive state is represented by the blurred black boxes. The differences in the conformational distribution of the complex state between the theophylline- and 3MX-binding aptamers are indicated in the conformational ensemble of complex. Curved arrows indicate some undefined interactions between P27 and the aptamers in the complex state.
Supplementary Material
Acknowledgments
This work was supported in part by grants from Robert A. Welch Foundation (AT-1645) and THECB Norman Hackerman Advanced Research Program (009741-0004-2006 and 009741-0015-2007) to T.X. and NIH GM086862 to A.P.
Abbreviations
- 2AP or P
2-aminopurine
- X
7-deaza adenine
- Z
7-deaza guanine
- 3MX
3-methylxanthine
- ITC
isothermal calorimetry
- FWHM
full width at half-maximum
- OPA
optical parametric amplifier
- fs
femtosecond
- ps
picoseconds
- ns
nanosecond
- SELEX
systematic evolution of ligands by exponential enrichment
- PAGE
polyacrylamide gel electrophoresis
Footnotes
Supporting Information Available: Steady state fluorescence titration curves, ITC data, femtosecond fluorescence decay profiles and residual plot of fittings. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Uhlenbeck OC. Keeping RNA Happy. RNA. 1995;1:4–6. [PMC free article] [PubMed] [Google Scholar]
- 2.Crothers DM. RNA Conformational Dynamics. In: Soll D, Nishimura S, Moore PB, editors. RNA. Oxford: Elsevier Science Ltd; 2001. pp. 61–70. [Google Scholar]
- 3.Williamson JR. Induced fit in RNA-protein recognition. Nat Struct Biol. 2000;7:834–837. doi: 10.1038/79575. [DOI] [PubMed] [Google Scholar]
- 4.Leulliot N, Varani G. Current topics in RNA-protein recognition: control of specificity and biological function through induced fit and conformational capture. Biochemistry. 2001;40:7947–7956. doi: 10.1021/bi010680y. [DOI] [PubMed] [Google Scholar]
- 5.Al-Hashimi HM, Walter NG. RNA Dynamics: it is about time. Curr Opin Struct Biol. 2008;18:321–329. doi: 10.1016/j.sbi.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Al-Hashimi HM. Dynamics-based amplification of RNA function and its characterization by using NMR spectroscopy. ChemBioChem. 2005;6:1506–1519. doi: 10.1002/cbic.200500002. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Q, Sun XY, Watt ED, Al-Hashimi HM. Resolving the motional modes that code for RNA adaptation. Science. 2006;311:653–656. doi: 10.1126/science.1119488. [DOI] [PubMed] [Google Scholar]
- 8.Palmer AG. NMR characterization of the dynamics of biomacromolecules. Chem Rev. 2004;104:3623–3640. doi: 10.1021/cr030413t. [DOI] [PubMed] [Google Scholar]
- 9.Hall KB. RNA in motion. Curr Opin Chem Biol. 2008;12:612–618. doi: 10.1016/j.cbpa.2008.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bokinsky G, Zhuang XW. Single-molecule RNA folding. Acc Chem Res. 2005;38:566–573. doi: 10.1021/ar040142o. [DOI] [PubMed] [Google Scholar]
- 11.Greenleaf WJ, Frieda KL, Foster DAN, Woodside MT, Block SM. Direct observation of hierarchical folding in single riboswitch aptamers. Science. 2008;319:630–633. doi: 10.1126/science.1151298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xia T. Taking femtosecond snapshots of RNA conformational dynamics and complexity. Curr Opin Chem Biol. 2008;12:604–611. doi: 10.1016/j.cbpa.2008.08.033. [DOI] [PubMed] [Google Scholar]
- 13.Fiebig T, Wan C, Zewail AH. Femtosecond Charge Transfer Dynamics of a Modified DNA Base: 2-Aminopurine in Complexes with Nucleotides. ChemPhysChem. 2002;3:781–788. doi: 10.1002/1439-7641(20020916)3:9<781::AID-CPHC781>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 14.Wan CZ, Xia TB, Becker HC, Zewail AH. Ultrafast unequilibrated charge transfer: A new channel in the quenching of fluorescent biological probes. Chem Phys Lett. 2005;412:158–163. [Google Scholar]
- 15.Zhao L, Xia T. Direct Revelation of Multiple Conformations in RNA by Femtosecond Dynamics. J Am Chem Soc. 2007;129:4118–4119. doi: 10.1021/ja068391q. [DOI] [PubMed] [Google Scholar]
- 16.Liu JD, Zhao L, Xia T. The Dynamic Structural Basis of Differential Enhancement of Conformational Stability by 5'- and 3'-Dangling Ends in RNA. Biochemistry. 2008;47:5962–5975. doi: 10.1021/bi800210t. [DOI] [PubMed] [Google Scholar]
- 17.Kadakkuzha BM, Zhao L, Xia T. Conformational Distribution and Ultrafast Base Dynamics of Leadzyme. Biochemistry. 2009;48:3807–3809. doi: 10.1021/bi900256q. [DOI] [PubMed] [Google Scholar]
- 18.Jenison RD, Gill SC, Pardi A, Polisky B. High-Resolution Molecular Discrimination by RNA. Science. 1994;263:1425–1429. doi: 10.1126/science.7510417. [DOI] [PubMed] [Google Scholar]
- 19.Hendeles L, Weinberger M. Theophylline - a State of the Art Review. Pharmacotherapy. 1983;3:2–44. doi: 10.1002/j.1875-9114.1983.tb04531.x. [DOI] [PubMed] [Google Scholar]
- 20.Soukup GA, Emilsson GAM, Breaker RR. Altering molecular recognition of RNA aptamers by allosteric selection. J Mol Biol. 2000;298:623–632. doi: 10.1006/jmbi.2000.3704. [DOI] [PubMed] [Google Scholar]
- 21.Zimmermann GR, Jenison RD, Wick CL, Simorre JP, Pardi A. Interlocking structural motifs mediate molecular discrimination by a theophylline-binding RNA. Nat Struct Biol. 1997;4:644–649. doi: 10.1038/nsb0897-644. [DOI] [PubMed] [Google Scholar]
- 22.Famulok M. Molecular Recognition of Amino-Acids by RNA-Aptamers - an L-Citrulline Binding RNA Motif and Its Evolution into an L-Arginine Binder. J Am Chem Soc. 1994;116:1698–1706. [Google Scholar]
- 23.Yang YS, Kochoyan M, Burgstaller P, Westhof E, Famulok M. Structural basis of ligand discrimination by two related RNA aptamers resolved by NMR spectroscopy. Science. 1996;272:1343–1347. doi: 10.1126/science.272.5266.1343. [DOI] [PubMed] [Google Scholar]
- 24.Wakeman CA, Winkler WC, Dann CE. Structural features of metabolite-sensing riboswitches. Trends Biochem Sci. 2007;32:415–424. doi: 10.1016/j.tibs.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kim JN, Breaker RR. Purine sensing by riboswitches. Biology of the Cell. 2008;100:1–11. doi: 10.1042/BC20070088. [DOI] [PubMed] [Google Scholar]
- 26.Montange RK, Batey RT. Riboswitches: Emerging themes in RNA structure and function. Ann Rev Biophys. 2008;37:117–133. doi: 10.1146/annurev.biophys.37.032807.130000. [DOI] [PubMed] [Google Scholar]
- 27.Zimmermann GR, Shields TP, Jenison RD, Wick CL, Pardi A. A semiconserved residue inhibits complex formation by stabilizing interactions in the free state of a theophylline-binding RNA. Biochemistry. 1998;37:9186–9192. doi: 10.1021/bi980082s. [DOI] [PubMed] [Google Scholar]
- 28.Latham MP, Zimmermann GR, Pardi A. NMR Chemical Exchange as a Probe for Ligand-Binding Kinetics in a Theophylline-Binding RNA Aptamer. J Am Chem Soc. 2009;131:5052–5053. doi: 10.1021/ja900695m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jucker FM, Phillips RM, McCallum SA, Pardi A. Role of a heterogeneous free state in the formation of a specific RNA-theophylline complex. Biochemistry. 2003;42:2560–2567. doi: 10.1021/bi027103+. [DOI] [PubMed] [Google Scholar]
- 30.Kuzmic P. Program DYNAFIT for the analysis of enzyme kinetic data: application to HIV proteinase. Anal Biochem. 1996;237:260–273. doi: 10.1006/abio.1996.0238. [DOI] [PubMed] [Google Scholar]
- 31.Meyer SL. Data Analysis For Scientists and Engineers. 2nd ed. Evanston: Peer Management Consultants, Ltd; 1992. [Google Scholar]
- 32.Holmen A, Norden B, Albinsson B. Electronic Transition Moments of 2-Aminopurine. J Am Chem Soc. 1997;119:3114–3121. [Google Scholar]
- 33.Lakowicz JR. Principles of Fluorescence Spectroscopy. 2nd ed. New York: Kluwer Academic/Plenum Plublishers; 1999. [Google Scholar]
- 34.Zhao L, Xia T. Probing RNA Conformational Dynamics and Heterogeneity Using Femtosecond Time-resolved Fluorescence Spectroscopy. Methods. 2009;49:128–135. doi: 10.1016/j.ymeth.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 35.Zimmermann GR, Wick CL, Shields TP, Jenison RD, Pardi A. Molecular interactions and metal binding in the theophylline-binding core of an RNA aptamer. RNA. 2000;6:659–667. doi: 10.1017/s1355838200000169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Akke M, Fiala R, Jiang F, Patel D, Palmer AG. Base dynamics in a UUCG tetraloop RNA hairpin characterized by N-15 spin relaxation: Correlations with structure and stability. RNA. 1997;3:702–709. [PMC free article] [PubMed] [Google Scholar]
- 37.Winkler WC, Breaker RR. Genetic control by metabolite-binding riboswitches. ChemBiochem. 2003;4:1024–1032. doi: 10.1002/cbic.200300685. [DOI] [PubMed] [Google Scholar]
- 38.Nudler E, Mironov AS. The riboswitch control of bacterial metabolism. Trends Biochem Sci. 2004;29:11–17. doi: 10.1016/j.tibs.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 39.Tucker BJ, Breaker RR. Riboswitches as versatile gene control elements. Curr Opin Struc Biol. 2005;15:342–348. doi: 10.1016/j.sbi.2005.05.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.