Abstract
Spinal muscular atrophy (SMA) is a neurodegenerative disease caused by loss of motor neurons in patients with null mutations in the SMN1 gene. An almost identical SMN2 gene is unable to compensate for this deficiency because a single C-to-T transition at position +6 in exon-7 causes skipping of the exon by a mechanism not yet fully elucidated. We observed that the C-to-T transition in SMN2 creates a putative binding site for the RNA-binding protein Sam68. RNA pull-down assays and UV-crosslink experiments showed that Sam68 binds to this sequence. In vivo splicing assays showed that Sam68 triggers SMN2 exon-7 skipping. Moreover, mutations in the Sam68-binding site of SMN2 or in the RNA-binding domain of Sam68 completely abrogated its effect on exon-7 skipping. Retroviral infection of dominant-negative mutants of Sam68 that interfere with its RNA-binding activity, or with its binding to the splicing repressor hnRNP A1, enhanced exon-7 inclusion in endogenous SMN2 and rescued SMN protein expression in fibroblasts of SMA patients. Our results thus indicate that Sam68 is a novel crucial regulator of SMN2 splicing.
Keywords: alternative splicing, hnRNP A1, Sam68, SMA, SMN2
Introduction
Spinal muscular atrophy (SMA) is an autosomal recessive neuromuscular disorder with an incidence of 1 in 6000 in the human population, representing the primary genetic cause of infant mortality (Monani, 2005; Burghes and Beattie, 2009). SMA is generally classified as three types on the basis of disease severity, with type-I being the most severe and type-III the mildest form (Pearn, 1980). The disease is characterized by degeneration of α-motor neurons in the anterior horn of the spinal cord and by consequent skeletal muscle atrophy (Monani, 2005; Burghes and Beattie, 2009). The genetic cause of SMA is a homozygous loss of SMN1, a gene located in the telomeric region of chromosome-5 that encodes the ‘survival motor neuron' protein SMN, an essential regulator of the biogenesis of the small nuclear ribonucleoproteins (snRNPs) (Battle et al, 2006).
Although SMN's deficiency leads to a wide range of defects in splicing regulation in many cell types (Zhang et al, 2008), motor neurons are the most affected in terms of function and survival (Burghes and Beattie, 2009). Notably, all SMA patients retain at least one copy of the centromeric and almost identical SMN2 gene. However, although this gene encodes a virtually identical protein, the expression levels of SMN2 are not sufficient to restore the activity of SMN (Monani, 2005; Battle et al, 2006; Burghes and Beattie, 2009). The instability of SMN2 protein derives principally from a single substitution, C-to-T at position +6 in exon-7, which is translationally silent but causes skipping of this exon in most SMN2 transcripts (Lorson et al, 1999; Monani et al, 1999). The resulting truncated protein is highly unstable and does not support the survival and function of spinal α-motor neurons, thereby causing the disease. Thus, recovery of full-length SMN2 protein is considered a valuable therapeutic approach for treatment of SMA and regulation of exon-7 inclusion in the SMN2 mRNA represents a clinically important model to investigate the impact of splicing regulation in human pathologies (Cartegni et al, 2002; Pellizzoni, 2007; Wang and Cooper, 2007).
The C-to-T substitution in SMN2 exon-7 has been suggested to induce exon skipping by two different mechanisms. The first model suggested that this substitution disrupts an exonic splicing enhancer (ESE) and impairs the binding of the splicing factor ASF/SF2, thereby affecting exon recognition (Cartegni and Krainer, 2002; Cartegni et al, 2006). An alternative model subsequently proposed that this single-nucleotide change instead creates an exonic splicing silencer (ESS) to which the splicing repressor hnRNP A1 binds, thereby favouring exon-7 exclusion from the SMN2 pre-mRNA (Kashima and Manley, 2003; Kashima et al, 2007a). SMN2 exon-7 skipping can be counteracted by overexpression of TRA2β (Hofmann et al, 2000), a positive regulator of exon-7 inclusion, indicating that the relative expression levels of specific splicing factors can strongly affect SMN2 splicing. As disease severity is inversely correlated with the levels of SMN2 protein (Lefebvre et al, 1997; Monani et al, 2000) and restoration of exon-7 inclusion is required to improve SMN2 protein stability (Monani, 2005; Battle et al, 2006; Burghes and Beattie, 2009), regulation of SMN2 exon-7 splicing represents an attractive therapeutic strategy for SMA.
Alternative splicing (AS) is normally orchestrated by a large number of RNA-binding proteins (RBPs) that are transiently recruited nearby splice sites and influence each other through specific protein–protein and protein–RNA interactions (Black, 2003; Matlin et al, 2005; Singh and Valcárcel, 2005; Chen and Manley, 2009). Analysis of the SMN2 exon-7 sequence showed the presence of a potential binding site for Sam68 just upstream from the consensus sequence for hnRNP A1. Sam68 (gene name: KHDRBS1) is a member of the Signal Transduction and Activation of RNA (STAR) family of RBPs (Lukong and Richard, 2003) and it regulates AS of CD44, BCL2L1, CCND1 and of genes involved in neurogenesis (Matter et al, 2002; Paronetto et al, 2007, 2010; Chawla et al, 2009). Furthermore, we previously showed that Sam68 and hnRNP A1 physically associate and cooperate in the regulation of BCL2L1 AS (Paronetto et al, 2007). Here, we have investigated whether Sam68 has a role in the regulation of SMN2 AS. Our results identify a novel ESS in SMN2 exon-7 that is bound by Sam68. Upregulation of Sam68 expression strongly triggers SMN2 exon-7 skipping, whereas dominant-negative versions of this splicing regulator revert this effect. Remarkably, interference with Sam68 RNA-binding activity or with its association with hnRNP A1 restores exon-7 inclusion in the endogenous SMN2 mRNA and promotes accumulation of SMN protein in cells obtained from SMA patients. Thus, Sam68 is a previously unidentified regulator of SMN2 splicing that might affect disease severity, suggesting that interference with its function offers a potential therapeutic approach for SMA.
Results
Sam68 affects SMN2 exon-7 splicing
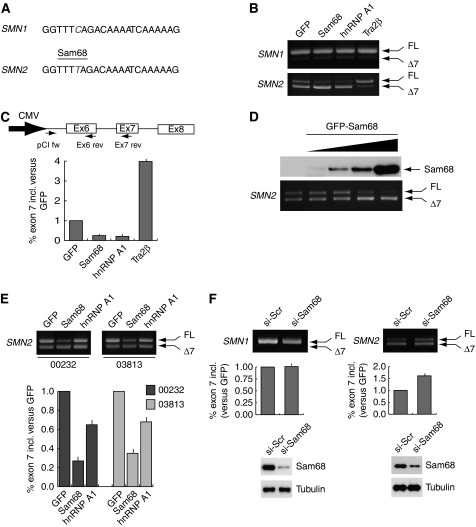
Sam68 binds to U-rich and A-rich RNA sequences (Lin et al, 1997). We observed that the C-to-T transition at position +6 in exon-7 (underlined below) creates a potential binding site for Sam68 (UUUUA) (Figure 1A). To determine whether Sam68 affects SMN2 exon-7 splicing, we performed in vivo splicing assays using a minigene that spans the whole alternatively spliced region from exon-6 to exon-8 of human SMN2 (Lorson et al, 1999). Co-transfection of Sam68 with the SMN2 minigene caused exon-7 skipping (Figure 1B). The effect of Sam68 was similar to that exerted by hnRNP A1 and opposite to that elicited by TRA2β (Figure 1B). By contrast, Sam68 was unable to stimulate exon-7 skipping in SMN1 (Figure 1B), in which its potential binding site is disrupted by the presence of the C at position +6. Dose-dependent experiments showed that even small changes in Sam68 levels affected SMN2 AS (Figure 1D), within the same range of concentrations in which hnRNP A1 and TRA2β were active (Supplementary Figure S1D), whereas upregulation of ASF/SF2 was ineffective (Supplementary Figure S1D). The effect of Sam68 was not cell-specific, because its overexpression also promoted exon-7 skipping in SH-SY5Y neuroblastoma cells and in SW480 colon cancer cells (Supplementary Figure S1E). hnRNP A1 was much less effective in these cells, suggesting that its activity might depend on the cell context. All reactions were performed within the linear range of PCR amplification, as shown by analysis of cells transfected with suboptimal doses of Sam68 (Supplementary Figure S1A and B). Quantitative real-time PCR analysis indicated that Sam68 caused an ∼4-fold increase in exon-7 skipping (Figure 1C and Supplementary Figure S1C).
Figure 1.
Sam68 induces exon-7 skipping in the SMN2 pre-mRNA. (A) The SMN1 and SMN2 exon-7 sequences are schematically represented and the C-to-T transition is highlighted in grey (italics font). The binding site for Sam68 in SMN2 exon-7 is indicated. (B) RT–PCR of splicing assays performed using HEK293T cells co-transfected with the pCI-SMN1 (upper panel) or the pCI-SMN2 (bottom panel) minigenes and GFP, or GFP-Sam68, or GFP-hnRNP A1 or TRA2β, as indicated above the panels. (C) Quantitative real-time analysis of exon-7 inclusion in the splicing assays shown in panel B. A scheme of the SMN2 minigene sequence amplified and of the oligonucleotides used is shown. Data were expressed as the ratio of full-length (pCIfw-Ex7rev)/total (pCIfw-Ex6rev) SMN2 transcripts. The ΔΔCt for each sample was calculated versus the GFP sample used as standard control and expressed as fold increase with respect to GFP (see Materials and methods). (D) RT–PCR analysis of the splicing assays performed by co-transfection of the pCI-SMN2 minigene (0.5 μg) and increasing amount of GFP-Sam68 (0.1–0.6 μg) in HEK293T cells. Western blot analysis of GFP-Sam68 expression is shown above the PCR panel. (E) RT–PCR analysis of SMN2 splicing assays performed by co-transfection of the minigene (0.5 μg) with constructs encoding GFP, GFP-Sam68 or GFP-hnRNP A1 (0.6 μg each) in fibroblasts derived from two SMA patients affected by a milder (03813) or more dramatic (00232) form of the disease. Real-time analysis of SMN2 exon-7 inclusion was performed as in panel C. (F) RT–PCR analysis of splicing assays performed by transfection of the SMN1 or SMN2 minigenes in HEK293T cells that had been previously transfected with either scrambled or Sam68 siRNAs as indicated. Real-time analysis of SMN2 exon-7 inclusion was performed as in panel C. Western blot analysis of Sam68 depletion by RNAi is reported below the graphs. Real-time analyses of all the splicing assays represent the mean±s.d. of three independent experiments.
We next tested whether Sam68 could affect SMN2 AS in a more physiologically relevant context. Fibroblast cell lines obtained from SMA patients with milder (03813) or more dramatic (00232) phenotype were co-transfected with the SMN2 minigene and GFP-, GFP-Sam68- or GFP-hnRNP A1-encoding plasmids. Sam68 was in fact stronger than hnRNP A1 in promoting exon-7 skipping also in SMA fibroblasts (Figure 1D). To rule out that forced upregulation redirected Sam68 towards a non-physiological substrate, we depleted the endogenous Sam68 from HEK293T cells by RNAi and tested the effect on SMN2 or SMN1 AS. As shown in Figure 1E, depletion of Sam68 caused an increase in exon-7 inclusion in SMN2, whereas it did not affect SMN1 splicing, showing that the endogenous protein participates in this AS event. These results indicate that Sam68 can specifically affect SMN2 exon-7 splicing.
The RNA-binding activity of Sam68 is required for SMN2 exon-7 skipping
Sam68 is a multifunctional RBP implicated in several cellular processes. In addition to its RNA-binding-dependent regulation of AS (Matter et al, 2002; Paronetto et al, 2007, 2010; Chawla et al, 2009) and mRNA translation (Paronetto et al, 2009), Sam68 can regulate transcription (Taylor et al, 2004; Rajan et al, 2008) and participate in signal transduction cascades in a RNA-binding-independent manner (Paronetto et al, 2003; Huot et al, 2009). To test whether the RNA-binding activity of Sam68 was required for SMN2 exon-7 skipping, we used two site-specific mutants (Figure 2A). Sam68V229F carries a point mutation in the RNA-binding domain (GSG) that strongly impairs its affinity for RNA (Paronetto et al, 2007) but retains transcriptional activity (Rajan et al, 2008); Sam68NLS−KO contains mutations in the nuclear localization signal (NLS), which interfere with the ability of the protein to affect splicing in the nucleus (Paronetto et al, 2007). In vivo splicing assays showed that both mutations completely suppressed the ability of Sam68 to inhibit SMN2 exon-7 inclusion (Figure 2B), whereas they did not affect SMN1 splicing (Figure 2C). Furthermore, as affinity of Sam68 for RNA (Wang et al, 1995) and its splicing activity towards BCL2L1 (Paronetto et al, 2007) are strongly decreased by tyrosine phosphorylation at the C-terminus of the protein, we tested whether coexpression of the tyrosine kinase Fyn affected Sam68-mediated SMN2 AS. In line with the requirement of RNA binding, we observed that coexpression of Fyn suppressed the ability of Sam68 to inhibit SMN2 exon-7 inclusion (Figure 2D). These results indicate that the RNA-binding activity and nuclear localization of Sam68 are required for SMN2 exon-7 skipping.
Figure 2.
Binding of Sam68 to the SMN2 mRNA is required for exon-7 skipping. (A) A schematic representation of Sam68 and the mutations introduced in the RNA-binding domain (V229F) and NLS (R436/442A). (B, C) RT–PCR (upper panel) and anti-GFP and anti-tubulin Western blot (bottom panels) analyses of splicing assays performed using HEK293T cells co-transfected with the SMN2 (B) or SMN1 (C) minigene and the indicated constructs. Quantitative real-time analysis of exon-7 inclusion is shown in the bar graph below. (D) Splicing assay of the SMN2 minigene using HEK293T cells co-transfected with either GFP or GFP-Sam68 alone (0.6 μg each) or in combination with Fyn (0.25 μg). Cells extracts were analysed by RT–PCR of SMN2 AS, quantified by real-time PCR (left panels), and by Western blotting for Sam68, tyrosine phosphorylation of Sam68 with the PY20 antibody and Fyn (right panels). Real-time analysis of SMN2 exon-7 inclusion in all the splicing assays was performed as described in Figure 1 and represent the mean±s.d. of three independent experiments.
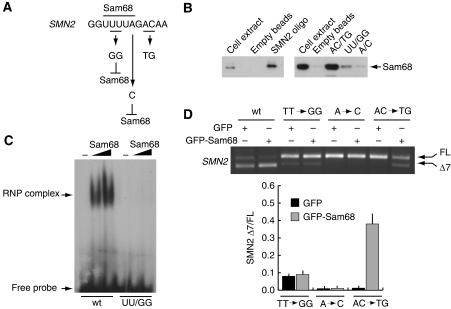
Sam68 binds to the UUUUA consensus site in SMN2 exon-7
Next, we sought to determine whether Sam68 binds to SMN2 exon-7 RNA. First, we performed a streptavidin pull-down assay using either SMN2 or SMN1 exon-7 biotin-labelled RNAs and nuclear extracts from HEK293T cells. As shown in Figure 3A, Sam68 associated preferentially with SMN2 exon-7 in vitro. To determine whether this binding was direct, we performed UV cross-linking experiments. First, splicing-competent HeLa nuclear extracts were irradiated with UV light in the presence of radiolabelled SMN2 exon-7, treated with RNAse and immunoprecipitated with an anti-Sam68 antibody or control IgGs. Following SDS–PAGE, autoradiographic analysis showed several strong radioactive bands, including one of ∼68 kDa that was specifically immunoprecipitated with the anti-Sam68 antibody (Figure 3B), suggesting that endogenous Sam68 directly bound to SMN2 exon-7. To confirm this result, we performed a similar UV cross-linking experiment using nuclear extracts from HEK293T cells transfected with an either empty vector or with myc-Sam68. Immunoprecipitation of extracts from both the experiments with an anti-myc antibody revealed a strong radioactive band of 70 kDa, the expected size of myc-Sam68, only in nuclear extracts expressing the protein (Figure 3C), confirming that Sam68 binds to SMN2 exon-7. Furthermore, purified GST-Sam68 and GST-Sam681−277, containing only the N-terminus and the GSG RNA-binding domain of the protein, were efficiently cross-linked to SMN2 exon-7, as was GST-hnRNP A1, used as positive control (Figure 3D and E).
Figure 3.
Sam68 directly binds to SMN2 exon-7 mRNA. (A) Streptavidin pull-down experiment. Western blot analysis of the endogenous Sam68 present in HEK293T nuclear extracts pulled down with streptavidin-bound biotinylated SMN2 (upper panel) or SMN1 (bottom panel) exon-7 RNA synthesized in vitro or control streptavidin beads. (B–D) UV cross-linking experiments. Nuclear extract from splicing-competent HeLa cells (B) or from HEK293T transfected with either empty vector or myc-Sam68, were incubated for 30 min with 32P-labelled SMN2 exon-7 RNA, cross-linked by UV radiation and immunoprecipitated with anti-Sam68 (B) or anti-myc (C) antibodies. The cross-linked proteins were then analysed by SDS–PAGE and autoradiography. (D) 32P-labelled SMN2 exon 7 RNA was incubated with GST, GST-Sam68, GST-Sam68(1–277) or GST-hnRNP A1. The mixture reactions were UV-irradiated and the cross-linked proteins were analysed by SDS–PAGE and autoradiography. (E) Coomassie blue staining of the purified proteins used for the experiment in panel D.
To test whether Sam68 binds to the UUUUA consensus, we used biotinylated RNA oligonucleotides containing the first 12 bases of exon-7 (Figure 4A). Sam68 was specifically associated with the wild-type RNA oligonucleotide in pull-down experiments with streptavidin-conjugated beads (Figure 4B). By contrast, substitution of the U at positions +4 and +5 with G (UU/GG oligo), or of the A at position +7 with C (A/C oligo), to disrupt the Sam68-binding site (Figure 4A and B), strongly reduced or completely abolished binding of Sam68 to the SMN2 oligonucleotide, respectively. Conversely, substitution of the A and C at position +9 and +10 with U and G (AC/UG oligo), outside of the Sam68-binding site, did not exert any effect (Figure 4A and B). To confirm that the integrity of UUUUA sequence is required for binding of Sam68 to the SMN2 ESS, we performed gel-shift assays. Sam68 specifically bound to the SMN2 probe (Figure 4C and Supplementary Figure S2A), whereas mutation of the consensus to UGGUA completely suppressed the formation of this complex.
Figure 4.
Sam68 binds to the UUUUA consensus site in SMN2 exon-7. (A) A schematic diagram indicating the mutations introduced in the Sam68-binding site. (B) Western blot analysis of pull-down experiments of the endogenous Sam68 present in HEK293T nuclear extracts using wild-type or mutated biotinylated SMN2 RNA oligonucleotides or control streptavidin beads. (C) Electrophoretic mobility-shift analysis of binding of Sam68 to 32P-labelled SMN2 RNA sequences. Wild-type or mutated SMN2 probes were incubated in the presence or absence of purified GST-Sam68 (50 and 100 ng) and reactions were separated on 5% acrylamide gels under native conditions. (D) HEK293T cells were co-transfected with the indicated wild-type or mutated SMN2 minigene plasmid and GFP-Sam68. After 24 h, cells were harvested and analysed by RT–PCR for SMN2 AS. Densitometric analyses of the ratio between SMN2 Δexon-7/full-length for the mutated minigenes are reported in the bar graph to the right of the PCR panel (mean±s.d. of three experiments).
The mutations in exon-7 were next introduced into the SMN2 minigene and tested in splicing assays in vivo using HEK293T cells. Mutation of the Sam68-binding site by the TT/GG substitution strongly impaired exon-7 skipping showing that this sequence has ESS activity (Figure 4D). Recovery of exon-7 inclusion was not cell-specific, as it was also observed in SH-SY5Y and SW480 cells (Supplementary Figure S2B). Furthermore, this mutation also abolished the effect of Sam68 overexpression on SMN2 AS (Figure 4D). An even stronger suppression of exon-7 skipping was obtained by mutating the A at position +7 (Figure 4C), and upregulation of Sam68 was unable to affect AS of the exon also in this case. Conversely, when mutations were introduced outside its consensus site (Figure 4A), Sam68 still triggered exon-7 skipping (Figure 4D). As both the TT/GG and the A/C mutations rescued exon-7 inclusion and prevented the effect of overexpressed Sam68, our results suggest that binding of Sam68 to the UUUUA consensus in the SMN2 exon-7 is required for efficient skipping.
Sam68 and hnRNP A1 cooperate in SMN2 exon-7 skipping
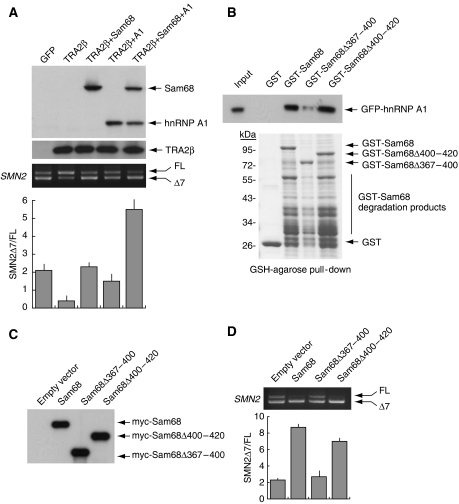
Sam68 and hnRNP A1 physically interact and cooperate in promoting BCL2L1 AS (Paronetto et al, 2007). To investigate whether the concerted action of Sam68 and hnRNP A1 enhances SMN2 exon-7 skipping, we analysed their ability to counteract TRA2β, a positive regulator of SMN2 exon-7 inclusion (Hofmann et al, 2000). We observed that coexpression of either Sam68 or hnRNP A1 inhibited TRA2β-induced exon-7 inclusion (Figure 5A). Furthermore, coexpression of Sam68 and hnRNP A1 together elicited an almost complete suppression of exon-7 inclusion even in the presence of excess TRA2β (Figure 5A), suggesting that the two proteins cooperate in the regulation of this AS event.
Figure 5.
The interaction between Sam68 and hnRNP A1 enhances SMN2 exon-7 skipping. (A) HEK293T cells were transfected with pCI-SMN2 (0.5 μg) and pCDNA3.1-TRA2β (0.5 μg) with or without Sam68 or hnRNP A1 either alone or in combination (0.2 μg each). After 24 h, cells were analysed by RT–PCR for alternative splicing of exon-7. Densitometric analyses of the ratio between SMN2 Δexon-7/full-length (mean±s.d. of three experiments) are reported. Western blot analysis of GFP-Sam68, Flag-TRA2β and GFP-hnRNP A1 is shown above the PCR panel. (B) Pull-down assay of GFP-hnRNP A1 with GST-fusion proteins of Sam68-deletion mutants. The upper panel shows anti-hnRNP A1 Western blot analysis; the bottom panel shows Coomassie blue staining of purified GST-fusion proteins used in the experiment. Position of the GST-fusion proteins is indicated by arrows. The lower bands present in the Sam68 lanes are due to degradation products of the GST-fusion proteins. (C, D) SMN2 splicing assays using HEK293T cells transfected with the indicated constructs encoding wild-type Sam68 or its deletion mutants. (C) Western blot analysis of the myc-Sam68 proteins used in the assay. (D) RT–PCR analysis of the in vivo splicing assay. The lower graph shows the results of densitometric analyses of three experiments performed as described in panel A.
As interaction of Sam68 with hnRNP A1 requires the Sam68 C-terminal region (aa 351–443) (Paronetto et al, 2007), we produced two internal deletion mutants (Sam68Δ367−400 and Sam68Δ400−420) that maintained the NLS (Ishidate et al, 1997) and tested their ability to interact with hnRNP A1 in pull-down assays. Deletion of residues 367–400 reduced hnRNP A1 binding, whereas residues 400–420 were not required for this interaction (Figure 5B). Significantly, the ability of Sam68Δ367−400 to induce SMN2 exon-7 skipping was strongly impaired, whereas Sam68Δ400−420 behaved like the wild-type protein (Figure 5D). The lack of effect of Sam68Δ367−400 was not due to production of a non-functional protein, as this mutant was expressed at high levels (Figure 5C) and was as effective as Sam68Δ400−420 in inducing CD44 exon-v5 inclusion (Supplementary Figure S3), a splicing event in which Sam68 does not require hnRNP A1 binding (Matter et al, 2000, 2002). These results suggest that the interaction of Sam68 with hnRNP A1 favours SMN2 exon-7 skipping.
Mutations that interfere with Sam68 activity restore exon-7 inclusion in the SMN2 pre-mRNA
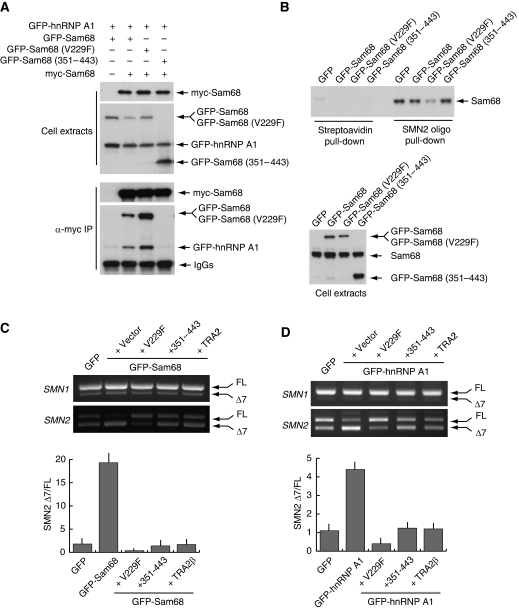
Sam68 binds to RNA as a dimer in vivo (Chen et al, 1997) and it interacts with hnRNP A1 through its C-terminal region (Paronetto et al, 2007). We reasoned that if an interaction between Sam68 and hnRNP A1 promotes exon-7 skipping, interference with either of these functions of Sam68 might limit or revert its effect on SMN2 AS. In line with our hypothesis, we observed that Sam68V229F, which is defective in RNA-binding activity (Paronetto et al, 2007) but homodimerizes with wild-type Sam68 (Figure 6A), strongly enhanced exon-7 inclusion when overexpressed in HEK293T cells (Figure 2B), suggesting that it exerted a dominant-negative effect on Sam68's function. In line with this hypothesis, RNA pull-down assays showed that expression of GFP-Sam68V229F interfered with binding of endogenous Sam68 to SMN2 exon-7 (Figure 6B). A similar recovery of exon-7 inclusion was obtained by overexpression of Sam68351−443 (Supplementary Figure S4). This truncated nuclear form of Sam68 contains the hnRNP A1 binding site but lacks the RNA-binding and the homodimerization domain (GSG in Figure 2A), and co-immunoprecipitation experiments showed that it interfered with the association between wild-type Sam68 and hnRNP A1 (Figure 6A) without affecting Sam68 binding to SMN2 exon-7 RNA (Figure 6B). To determine whether these Sam68 mutants could act as dominant-negative alleles and attenuate or inhibit SMN2 exon-7 skipping in live cells, we coexpressed them in HEK293T cells together with either GFP-Sam68 or GFP-hnRNP A1. We found that both Sam68V229F and Sam68351−443 functioned as dominant-negative alleles and suppressed exon-7 skipping induced by overexpression of Sam68 (Figure 6C) and hnRNP A1 (Figure 6D). Furthermore, the effect of GFP-Sam68V229F was even stronger than that exerted by TRA2β. The Sam68 dominant-negative derivatives caused an almost complete reversion of the AS event and accumulation of the full-length form of SMN2 above basal levels, even in the presence of excess Sam68 or hnRNP A1. The effect of the Sam68 dominant-negative alleles was specific because it did not affect AS of SMN1 (Figure 6C and D) or of the Sam68-independent AS of RON exon-11 (Supplementary Figure S5). These experiments suggest that GFP-Sam68V229F and GFP-Sam68351−443 are efficient competitors of SMN2 exon-7 skipping in vivo.
Figure 6.
Dominant-negative mutants of Sam68 rescue exon-7 inclusion in SMN2. (A) Nuclear extracts from HEK293T transfected with the indicated constructs were immunoprecipitated with an anti-myc antibody. The extracts and immunoprecipitated proteins were analysed by Western blotting. (B) RNA pull down of endogenous Sam68 present in HEK293T nuclear extracts using biotinylated SMN2 exon-7 RNA or control empty streptavidin beads. Extracts were prepared from cells transfected with GFP, GFP-Sam68 or the indicated Sam68 mutants. The upper panel shows the Western blot analysis of endogenous Sam68 pulled down from the corresponding extracts. The lower panel show Western blot analysis of recombinant GFP-Sam68 and endogenous Sam68 proteins from the extracts used for the pull-down experiment. (C, D) HEK293T cells were transfected with pCI-SMN1 or pCI-SMN2 and a plasmid encoding GFP-Sam68 (C) or GFP-hnRNP A1 (D) either alone or together with pGFP-Sam68V229F, or pGFP-Sam68351−443 or pCDNA3.1-TRA2β. After 24 h, cells were analysed by RT–PCR for SMN2 AS as described in Figure 5. Densitometric analyses of the ratio between SMN2 Δexon-7/full-length is reported in the bar graph below the PCR panel (mean±s.d. of three experiments).
GFP-Sam68V229F and GFP-Sam68351−443 restore exon-7 inclusion and allow SMN2 protein accumulation in SMA cells
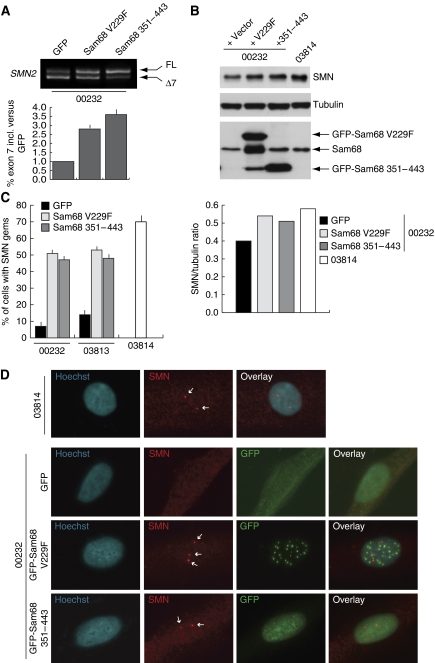
To determine whether Sam68 dominant-negative alleles could affect endogenous SMN2 AS in a physiological setting, we infected SMA fibroblasts with retroviral constructs encoding these GFP-Sam68 mutants or GFP as control. Infected cells were sorted for GFP fluorescence and RNA and proteins were extracted. Expression of GFP-Sam68V229F and GFP-Sam68351−443 enhanced exon-7 inclusion in the endogenous SMN2 pre-mRNA in patient cells as compared with that in cells infected with GFP alone (Figure 7A). This effect on AS resulted in increased SMN protein production (Figure 7B). Remarkably, the amount of SMN protein produced after expression of GFP-Sam68V229F and GFP-Sam68351−443 was comparable to that observed in fibroblasts from a heterozygote donor (03814) not affected by the disease. In normal cells, SMN protein accumulates in discrete nuclear foci known as gems (Monani, 2005; Battle et al, 2006; Burghes and Beattie, 2009). These structures are strongly decreased or absent in SMA cells. Immunofluorescence analysis showed that GFP-Sam68V229F and GFP-Sam68351−443 also restored the typical localization of functional SMN in infected cells, as shown by the formation of gems in the nucleus as in the healthy donor fibroblasts (Figure 7C and D). The effect of the Sam68 dominant-negative alleles was similar in fibroblasts obtained from SMA patients with milder (03813) or more dramatic (00232) phenotype (Figure 7C). These experiments show that interference with the splicing activity of Sam68 in vivo by expressing dominant-negative Sam68 mutant proteins restores SMN2 exon-7 splicing and SMN protein levels in SMA cells.
Figure 7.
Dominant-negative mutants of Sam68 rescue exon-7 inclusion in SMN2. (A) Type-I SMA fibroblasts (00232) were infected with retroviruses encoding GFP, GFP-Sam68V229F or GFP-Sam68351−443. After selection by sorting for the GFP fluorescence, cells were analysed by RT–PCR for endogenous SMN2 transcripts. Real-time analysis of SMN2 exon-7 inclusion in the splicing assays was performed as described in Figure 1 (using the SMN_exon6-F instead of pCIfw as forward primer) and represent the mean + s.d. of three independent experiments. (B) Western blot analysis of SMN protein, GFP-fusion proteins and tubulin of the samples analysed in (A). Fibroblasts from a healthy donor (03814) are shown as control. Densitometric analysis of the ratio between SMN and tubulin signals is shown in the bar graph. (C) A bar graph of the nuclear gems observed in fibroblasts obtained from two SMA patients (03813 and 00232) and infected with retroviruses encoding GFP, GFP-Sam68V229F or GFP-Sam68351−443. For comparison, nuclear gems were also counted in fibroblasts from a healthy donor (03814). At least 100 nuclei for each sample from three separate immunofluorescence experiments were analysed. The data are represented as mean±s.d. Representative examples of the detection of the nuclear gems (indicated by white arrows) in the different cell lines are illustrated in panel D. The immunofluorescence analyses show SMN protein (red signal), GFP-fusion proteins (green) and DNA (blue) in the indicated fibroblast cell lines.
Discussion
Our work identifies a novel ESS element in SMN2 exon-7, shows that Sam68 binds this ESS and acts as a crucial regulator of SMN2 AS, provides tools to interfere with Sam68 activity in vivo, and highlights a novel approach to recover SMN protein expression in SMA cells. We provide evidence that Sam68 promotes exon-7 skipping and that this effect relies on its ability to bind to RNA and to associate physically and functionally with hnRNP A1, a known regulator of SMN2 AS (Kashima and Manley, 2003; Kashima et al, 2007a). These findings identify Sam68 as a novel regulator of SMN2 splicing that offers a new and potentially valuable therapeutic target for SMA (Figure 8).
Figure 8.
A hypothetical model of the role of Sam68 in alternative splicing of the SMN2 pre-mRNA. Position of the Sam68-binding site, as well as the region where hnRNP A1 and TRA2β bind, in SMN2 exon-7 is indicated. Binding of Sam68 to exon-7, possibly through the recruitment of hnRNP A1, favours skipping of the exon by blocking the activity of TRA2β to enhance interactions at the upstream 3′ splice site, resulting in the production of SMN2 Δ7 mRNA; inhibition of Sam68 function rescues exon-7 inclusion and production of full-length SMN2 mRNA.
SMA is caused by the inability of the SMN2 gene to compensate for the functional loss of SMN1 in patients. Since exon-7 skipping is the major cause of the decreased function of SMN2, characterization of the splicing factors causing this AS event is of crucial relevance for the design of therapeutic approaches to the disease (Monani, 2005; Burghes and Beattie, 2009). An advance in this direction was provided by the identification of an ESS (UAGACA) created by the C-to-T transition in SMN2 exon-7 (Kashima and Manley, 2003). This ESS is a binding site for hnRNP A1, an abundant RBP that usually acts as a negative regulator of AS (Matlin et al, 2005). We have now identified an additional ESS created by the C-to-T transition in exon 7 (UUUUA) and shown that its mutation rescues exon-7 inclusion. The new ESS is located just upstream from, and indeed it overlaps with (see below), the hnRNP A1 site, and we have observed that it is a binding site for Sam68. Our results, showing that Sam68 directly binds to SMN2 exon-7 and that fluctuations in its expression levels modulate SMN2 splicing, strongly support a direct role for Sam68 in exon-7 skipping.
We found that Sam68-mediated exon-7 skipping required the integrity of its UUUUA-binding site. First, Sam68 was unable to promote exon skipping in SMN1 in which the C at position +6 (UUUCA) disrupts the Sam68 consensus. Moreover, loss of the Sam68-elicited effect was also observed when flanking positions in the SMN2-binding site were modified (UGGUA and UUUUC). Notably, these mutations caused an almost complete inclusion of SMN2 exon-7, as in SMN1, suggesting that destroying the Sam68-binding site rescues the splicing defect. The UGGUA mutation was previously shown to affect exon-7 inclusion at most modestly (Kashima and Manley, 2003). However, we now show a strong and statistically significant recovery of exon-7 inclusion in three different cell lines tested, suggesting that this mutation also destroys an ESS in SMN2. The apparent discrepancy might reflect the different experimental conditions used (e.g., the amount of DNA transfected, 4 versus 0.5 μg in our experiments, might have produced sufficient SMN2 pre-mRNA to saturate the ability of endogenous Sam68 to affect SMN2 AS without compromising that of the highly expressed hnRNP A1). Furthermore, the possibility that Sam68 acts as a non-specific negative regulator of SMN2 exon-7 splicing is unlikely, because its overexpression does not affect SMN1 AS or mutated SMN2 minigenes in which its specific binding site is disrupted. These results indicate that Sam68 is a specific regulator of SMN2 exon-7 splicing in live cells.
Interestingly, we found that overexpression of Sam68V229F exerted the opposite effect of wild-type Sam68, strongly enhancing exon-7 inclusion. As this mutant protein maintains the ability to homodimerize, we interpret this result as due to a dominant-negative effect caused by its interaction with the endogenous Sam68 and its sequestration in non-functional complexes. Indeed, we found that Sam68V229F reduced exon-7 binding by endogenous Sam68 and competed with the overexpressed wild-type Sam68 even more than TRA2β. A similar dominant-negative effect was also observed with two other RNA-binding-defective alleles of Sam68 (G178E and I184N) in the regulation of Sgce AS (Chawla et al, 2009). As all these Sam68 mutants homodimerize with the wild-type protein, they might either impair recognition of targets by the dimeric Sam68 complex or compete with the recruitment of additional factors involved in splicing regulation. In support of this hypothesis, we found that Sam68 dominant-negative alleles could also repress hnRNP A1-induced exon-7 skipping, whereas a Sam68-deletion mutant lacking the binding site for hnRNP A1 was strongly defective in SMN2 AS modulation.
The binding sites for Sam68 and hnRNP A1 in SMN2 exon-7 partially overlap. Thus, we consider concomitant binding to be unlikely. However, Sam68 binds to hnRNP A1 and this feature of the protein is important for regulation of SMN2 AS. Indeed, Sam68351−443, which is unable to bind to RNA but competes with the interaction between Sam68 and hnRNP A1, acts as dominant negative towards both Sam68- and hnRNP A1-mediated SMN2 exon-7 skipping. A possible interpretation is that under some conditions Sam68 recruits hnRNP A1 to the SMN2 pre-mRNA, and this might facilitate spreading of hnRNP A1 by low-affinity interactions with RNA sequences located downstream or upstream from its binding site (Okunola and Krainer, 2009). Additionally, an SMN2-specific hnRNP A1 site in exon-7 has been shown to be important for maximal exon-7 exclusion, perhaps by facilitating looping between hnRNP A1 molecules (Kashima et al, 2007b). In any case, depending on the relative expression levels of endogenous Sam68 and hnRNP A1 in different cell types, direct binding of one or the other would be favoured, which could then recruit the other. Thus, our results suggest that binding of a functional Sam68/hnRNP A1 complex to the SMN2 pre-mRNA promotes exon-7 skipping.
Various experimental approaches have been proposed to rescue SMN protein production in SMA cells. Bifunctional RNAs can efficiently rescue exon-7 inclusion and SMN2 protein expression in cell and animal models. These RNAs were engineered to contain antisense sequences that target either exonic (Baughan et al, 2006) or intronic (Baughan et al, 2009) SMN2 regions fused to binding sites for SR proteins to promote exon recognition. Similar results were obtained with injection of chemically modified antisense oligonucleotides (ASOs), targeting either exon-7 or the downstream intron (Hua et al, 2007, 2008; Singh et al, 2009). In both cases, recovery of exon-7 inclusion and/or SMN protein expression and localization in nuclear gems was reported. Here, we have shown that a similar recovery can be achieved by targeting Sam68 function through expression of dominant-negative mutants of the protein. Sam68V229F and Sam68351−443 inhibited SMN2 exon-7 skipping even under conditions in which Sam68 or hnRNP A1 were upregulated, suggesting that these mutants might rescue SMN activity in SMA cells. Indeed, both Sam68 alleles enhanced exon-7 inclusion and promoted accumulation of endogenous SMN protein in stably infected SMA fibroblasts. The levels of SMN protein obtained and the number of SMN nuclear gems, which is indicative of functional assembly of the protein, almost reached those observed in control fibroblasts from a healthy donor. Although other splicing factors, such as TRA2β (Hofmann et al, 2000), hnRNP Q (Chen et al, 2008) and TDP-43 (Bose et al, 2008), have been shown to enhance SMN2 exon-7 inclusion, in none of these cases was an effect on SMN recovery in SMA cells observed. Thus, this is the first report showing that direct interference with a splicing regulator rescues endogenous SMN2 expression in patient cells.
Our results show that inhibition of Sam68's activity, through competition with dominant-negative mutants, provides a valuable approach for the rescue of SMN activity. As these dominant-negative mutants affect two specific functions of Sam68 (i.e., its RNA-binding affinity and its ability to interact with hnRNP A1), it is conceivably possible to obtain similar effects through development of small molecules that interfere either with the Sam68 RNA-binding domain or with its surface of interaction with hnRNP A1. With this in mind, our results that limit the binding site for this RBP to only 34 residues might open the way to develop short interfering peptides that mimic this region, as this approach has been proven effective in interfering with protein-protein interactions in other systems (Loiarro et al, 2005, 2007; Toshchakov et al, 2007). Nevertheless, indiscriminate inhibition of Sam68 activity might result in changes in a large spectrum of genes, thereby eliciting unwanted negative effects, and direct studies using animal models are required to support this approach. In this regard, the observations that Sam68-knockout mice survive to adulthood and show only minor motor coordination defects is promising (Lukong and Richard, 2008). Moreover, inhibition of Sam68 splicing activity and/or interaction with hnRNP A1 also affects AS of the BCL2L1 pre-mRNA, towards the form encoding the pro-survival long isoform (Paronetto et al, 2007), and upregulation of this isoform improved viability of SMA mice independently of SMN protein expression (Tsai et al, 2008). Thus, our results suggest that interfering with Sam68's function in the α-motor neurons of SMA animal models might exert both direct effects on SMN expression and indirect effects on cell survival through modulation of two of its splicing targets.
Materials and methods
Plasmid constructs
pCI-SMN2 and pCI-SMN1 minigenes (Lorson et al, 1999), wild-type and mutant pCDNA3-SMN2 minigenes, pCDNA3-Ron (Ghigna et al, 2005) and pCDNA3-CD44 (Matter et al, 2002) minigenes, pCDNA3-TRA2β (Kashima and Manley, 2003), wild-type and mutant pEGFPC1-Sam68, pEGFPC3-hnRNP A1 and p3xFlag–ASF/SF2 (Paronetto et al, 2007) have been previously described. pGEX4T1-Sam68Δ367−400 was generated using the NdeI restriction site in the Sam68 sequence at position +1095 bp. An NdeI–SalI fragment from aa 400 to aa 443 of Sam68 was amplified by PCR using Pwo Super Yield DNA Polymerase (Roche) using the F1 and R1 primers (Supplementary Table S1) and human Sam68 cDNA as template (Paronetto et al, 2003). The NdeI–SalI fragment was then cloned into the NdeI–SalI restriction site of pGEX4T1-Sam68 (Paronetto et al, 2007). pGEX4T1-Sam68Δ400−420 was generated by the overlap extension PCR strategy (Aiyar et al, 1996) using the F2 and R2 primers (Supplementary Table S1) in the first-step PCR and F3 (Supplementary Table S1) and the product of the first-step PCR as reverse primer in the second step. The EcoRI–SalI fragment was then cloned into EcoRI–SalI restriction sites of pGEX4-T1. The myc-tagged derivatives were subcloned from the pGEX4T1 constructs into the EcoRI–XhoI restriction sites of the pCDNA-myc vector (Paronetto et al, 2007). The retroviral constructs were generated by subcloning the full-length Sam68 wild-type, Sam68V229F Sam68351−443 from pGEX4-T (Paronetto et al, 2007) into the EcoRI–SalI restriction sites of pCLPCX-GFP. All constructs were sequenced (BMR Genomics, Italy) before their use.
Cell cultures, transfections and in vivo splicing assays
HEK293T (purchased from ATCC) and human SMA cell lines GM03814, GM03813 and GM00232 (purchased from Coriell Repositories) were maintained in Dulbecco's modified Eagles medium (DMEM; Gibco BRL) supplemented with 10% foetal bovine serum (BioWhittaker Cambrex Bioscience), penicillin and streptomycin. Transfections were performed with 0.1–1 μg of appropriate constructs in the presence or absence of 0.5 μg of pCI–SMN2 minigene using Lipofectamine 2000 (Invitrogen). For RNAi, HEK293T cells were plated the day before transfection at 2.5 × 105 per well in six-well plates and transfected with 20 μM scramble or Sam68 siRNA oligonucleotides (Busà et al, 2007) using Lipofectamine RNAiMAX (Invitrogen). Forty-eight hours after transfections, cells were collected for RNA or biochemical analyses were performed according to published procedures (Paronetto et al, 2007). Briefly, total RNA was isolated using cold TRIzol reagent (Invitrogen) and resuspended in RNAse-free water (Sigma-Aldrich). For protein extraction, cells were rinsed in phosphate-buffered saline and resuspended in a lysis buffer (100 mM NaCl, 10 mM MgCl2, 30 mM Tris–HCl (pH 7.5), 1 mM dithiothreitol (DTT), 10 mM β-glycerophosphate, 0.5 mM NaVO4 and protease inhibitor cocktail (Sigma-Aldrich)) supplemented with 0.5% Triton X-100. For analysis of SMN protein, cell extracts were also sonicated for 30 s using a small size probe. Extracts were centrifuged for 10 min at 12 000 × g at 4°C and the supernatant fraction was collected and used for Western blotting or immunoprecipitation experiments.
RT–PCR analysis
Total RNA (0.2–1 μg) from transfected cells was used for RT–PCR using M–MLV reverse transcriptase (Invitrogen). Five percent of the RT reaction was used as template together with the following primers (Supplementary Table S1): pCI fw (for the pCI minigenes), T7 fw (for the pCDNA3 minigenes), SMN2 Ex6 fw (for endogenous SMN1 and SMN2) and SMN2 Ex8 rev. For all reactions, 25 cycles of amplification, within the linear range of PCR amplification (Supplementary Figure S1A and B), were performed. The reactions were analysed by ethidium bromide agarose gel and intensity of the bands was quantified by densitometric analysis using the ImageQuant 5.0 software. Each experiment was performed in triplicate. Densitometric data in Figures 4, 5 and 6 are represented as the mean±s.d. of the ratio between SMN2 Δexon-7 and full length.
Quantitative real-time PCR reactions were performed with Sybr Green mix (for Light-Cycler480; Roche) according to the manufacturer's instructions. The ΔΔCt method was used to calculate the fold change in the exon-7-containing SMN2 mRNA. ΔCt values were obtained by subtracting the Ct value of total (exon-6-containing) SMN2 mRNA (amplified with primer pCIfw-SMN_Ex6-R or SMN_Ex6-F-SMN_Ex6-R for the endogenous SMN2) from the Ct value for the exon-7-containing mRNA (amplified with primers pCIfw and SMN_Ex7-R or SMN_Ex6-F and SMN_Ex7-R for the endogenous SMN2) for each sample. Each ΔCt value represents the mean±s.d. of three independent experiments. ΔΔCt was calculated as the difference in ΔCt between different samples (with respect to GFP sample). The data are represented as fold increase versus GFP sample calculated by the 2−(ΔΔC)t formula.
Electrophoretic mobility-shift assay
The radiolabelled wild-type and mutated probes used for the gel-shift experiment were generated by in vitro annealing of the oligonucleotides fw T7 SMN2 and rev T7 SMN2; fw T7 SMN2-GG and rev T7 SMN2-GG and T7 through in vitro transcription using the MAXIScript RNA synthesis kit (Ambion) in the presence or absence of a labelled nucleotide (32P α-UTP and 32P α-GTP). The RNA probes (20 000 c.p.m.) were added to the reaction mixture in the presence or absence of GST-Sam68 (50 and 100 nM), pre-equilibrated in binding buffer (10 mM HEPES (pH 7.4); 10 mM KCl; 2 mM MgCl2; 100 mM NaCl; 200 ng/μl yeast tRNA; 1 mM DTT; protease inhibitor cocktail; RNAse OUT 30 U/μl) for 5 min at 30 °C, and incubated for 30 min at 30 °C. In some experiments, 5, 25 and 50-fold excess of cold probe was added to the reaction. The samples were diluted in a loading buffer, loaded onto pre-run native 5% polyacrylamide gel in 0.5% TBE and resolved at 100 V for 3 h. The gel was then dried and analysed by autoradiography.
Protein-protein co-immunoprecipitation experiments
HEK293T nuclear extracts were prepared by resuspending cells in buffer-A (10 mM Tris–HCl (pH 7.4), 10 mM NaCl, 2.5 mM MgCl2, 1 mM DTT, protease inhibitor cocktail, 15 mM β-glycerophosphate, 0.5 mM NaVO4). After incubation on ice for 15 min, the samples were centrifuged at 700 g for 7 min. The pellet containing the nuclei was resuspended in buffer-A supplemented with 100 mM NaCl and 0.5% Triton X-100, sonicated and stratified on 30% sucrose. After centrifugation at 7000 g for 15 min, nuclear extracts were precleared for 1 h on protein-A–Sepharose beads and for 1 h on protein-A-Sepharose beads in the presence of 0.01% bovine serum albumin (BSA) and 2 μg of rabbit IgGs. After centrifugation for 1 min at 1000 g, the supernatants were incubated with 2 μg of mouse anti-myc IgGs (Santa Cruz Biotechnology Inc.) or control mouse IgGs for 3 h at 4°C under constant rotation. Beads were washed three times with lysis buffer and were eluted in SDS sample buffer for Western blot analysis.
Retrovirus generation and infection of SMA cells lines
For retroviral expression, 15 μg of the retroviral vectors pCLPCX-GFP or GFP-Sam68(V229F) or GFP-Sam68(351-443) were co-transfected with 5 μg of an expression plasmid for the vesicular stomatitis virus-G protein into a cell line HEK293T gp/bsr by using the calcium phosphate method. Forty-eight hours later, the supernatant containing the retroviral particles were recovered and supplemented with polybrene (4 μg/ml). The human SMA cell lines GM03813 and GM00232 (5 × 104) were infected by incubation with the retroviruses. Briefly, infection were performed in three steps: (1) Cells were incubated with the retroviruses for 4 h; (2) the supernatant was removed and infection was repeated with fresh viruses for further 4 h; and (3) the supernatant was removed and fresh viral preparation was added and infection proceeded overnight. At the end, cells were rinsed and 24–48 h later selected for GFP expression by cell sorting.
In vitro synthesis of RNAs
The templates for RNA synthesis were generated using PCR products amplified with forward oligonucleotides containing at the 5′-end the T7 promoter sequence upstream from sequences in intron-6 or exon-7 of SMN2. Reverse primers were complementary to the 3′-end of exon-7. Amplified bands were gel-purified and used as templates (1 μg) for in vitro RNA transcription using the MAXIScript RNA synthesis kit (Ambion) in the presence or absence of labelled nucleotides (biotin or 32P CTP) depending on the experiment. RNA was purified using ProbeQuant G-50 Micro Columns (GE Healthcare) and diluted in RNAse-free water (Sigma-Aldrich) for subsequent experiments.
Biotin–RNA pull-down experiments
HEK293T-cell nuclear extracts prepared as described above were precleared for 60 min on protein-A–Sepharose beads and for 60 min on streptavidin–Sepharose beads in the presence of 0.25 mg/ml yeast tRNA to diminish non-specific interactions of proteins with RNAs. Precleared extracts were then incubated with streptavidin–Sepharose beads preincubated for 1 h with 0.05% BSA and the specific biotinylated RNA oligonucleotide to be tested. Incubation was performed for 60 min at 4°C under rotation. Bound complexes were rinsed three times with buffer-B used for preparation of the nuclear extracts and eluted in an SDS–PAGE sample buffer. Samples were analysed by Western blotting according to standard procedures (Paronetto et al, 2003, 2007, 2009).
UV-crosslink immunoprecipitation
In vitro synthesized, labelled exon-7 RNAs (50–60 fmol; 1 × 105 c.p.m.) were incubated for 30 min at 30°C with splicing-competent HeLa nuclear extracts (Cell Biotech, Belgium) in 50 μl reaction volume. Samples were irradiated with UV light (254 nm, 0.6 J, 2 cm from the light source) using a Stratalinker (Stratagene), digested with 1 μg/μl RNAse-A and immunoprecipitated with anti-Sam68 or control antibodies (1 μg) as described above. The experiment was also performed by incubating the same labelled RNAs with nuclear extracts from HEK293T transfected with empty vector or myc-Sam68 plasmid and immunoprecipitated with an anti-myc antibody (1 μg). Alternatively, purified GST, GST-Sam68, GST-Sam68(1–277) or GST-hnRNP A1 (0.1 μg) was incubated and irradiated as detailed above and directly diluted in SDS sample buffer after RNAse-A treatment. Samples were then resolved by 10% SDS–PAGE and analysed by autoradiography.
Supplementary Material
Acknowledgments
We wish to thank Drs Adamo Diamantini and Luca Battistini for cell sorting, Dr Claudia Ghigna for the pCDNA3-Ron plasmid, Dr Roberta Busà for assistance with real time PCR analyses, Prof Pellegrino Rossi, Raffaele Geremia, Daniela Barilà and Antonio Musarò for the critical reading of the paper, all members of our laboratories for discussion and suggestions throughout the study. This work was supported by Grants from Telethon (GGP09154), The Associazione Italiana Ricerca sul Cancro (AIRC), the Istituto Superiore della Sanità (ISS Project n.527/B/3A/5) and the Association for International Cancer Research (AICR). JLM acknowledges support from the NIH. SS acknowledges support from NIH and the Muscular Dystrophy Association.
Footnotes
The authors declare that they have no conflict of interest.
References
- Aiyar A, Xiang Y, Leis J (1996) Site-directed mutagenesis using overlap extension PCR. Methods Mol Biol 57: 177–191 [DOI] [PubMed] [Google Scholar]
- Battle DJ, Kasim M, Yong J, Lotti F, Lau CK, Mouaikel J, Zhang Z, Han K, Wan L, Dreyfuss G (2006) The SMN complex: an assembly machine for RNPs. Cold Spring Harb Symp Quant Biol 71: 313–320 [DOI] [PubMed] [Google Scholar]
- Baughan T, Shababi M, Coady TH, Dickson AM, Tullis GE, Lorson CL (2006) Stimulating full-length SMN2 expression by delivering bifunctional RNAs via a viral vector. Mol Ther 14: 54–62 [DOI] [PubMed] [Google Scholar]
- Baughan TD, Dickson A, Osman EY, Lorson CL (2009) Delivery of bifunctional RNAs that target an intronic repressor and increase SMN levels in an animal model of spinal muscular atrophy. Hum Mol Genet 18: 1600–1611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black DL (2003) Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem 72: 291–336 [DOI] [PubMed] [Google Scholar]
- Bose JK, Wang IF, Hung L, Tarn WY, Shen CK (2008) TDP-43 overexpression enhances exon 7 inclusion during the survival of motor neuron pre-mRNA splicing. J Biol Chem 283: 28852–28859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burghes AH, Beattie CE (2009) Spinal muscular atrophy: why do low levels of survival motor neuron protein make motor neurons sick? Nat Rev Neurosci 10: 597–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busà R, Paronetto MP, Farini D, Pierantozzi E, Botti F, Angelini DF, Attisani F, Vespasiani G, Sette C (2007) The RNA-binding protein Sam68 contributes to proliferation and survival of human prostate cancer cells. Oncogene 26: 4372–4382 [DOI] [PubMed] [Google Scholar]
- Cartegni L, Chew SL, Krainer AR (2002) Listening to silence and understanding nonsense: exonic mutations that affect splicing. Nat Rev Genet 3: 285–298 [DOI] [PubMed] [Google Scholar]
- Cartegni L, Hastings ML, Calarco JA, de Stanchina E, Krainer AR (2006) Determinants of exon 7 splicing in the spinal muscular atrophy genes, SMN1 and SMN2. Am J Hum Genet 78: 63–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cartegni L, Krainer AR (2002) Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nat Genet 30: 377–384 [DOI] [PubMed] [Google Scholar]
- Chawla G, Lin CH, Han A, Shiue L, Ares M Jr, Black DL (2009) Sam68 regulates a set of alternatively spliced exons during neurogenesis. Mol Cell Biol 29: 201–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HH, Chang JG, Lu RM, Peng TY, Tarn WY (2008) The RNA binding protein hnRNP Q modulates the utilization of exon 7 in the survival motor neuron 2 (SMN2) gene. Mol Cell Biol 28: 6929–6938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M, Manley JL (2009) Diverse mechanisms of alternative splicing regulation: Insights from molecular and genomic analyses. Nat Rev Mol Cell Biol 10: 741–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen T, Damaj BB, Herrera C, Lasko P, Richard S (1997) Self-association of the single-KH-domain family members Sam68, GRP33, GLD-1, and Qk1: role of the KH domain. Mol Cell Biol 17: 5707–5718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghigna C, Giordano S, Shen H, Benvenuto F, Castiglioni F, Comoglio PM, Green MR, Riva S, Biamonti G (2005) Cell motility is controlled by SF2/ASF through alternative splicing of the Ron proto oncogene. Mol Cell 20: 881–890 [DOI] [PubMed] [Google Scholar]
- Hofmann Y, Lorson CL, Stamm S, Androphy EJ, Wirth B (2000) Htra2-beta 1 stimulates an exonic splicing enhancer and can restore full-length SMN expression to survival motor neuron 2 (SMN2). Proc Natl Acad Sci USA 97: 9618–9623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Y, Vickers TA, Baker BF, Bennett CF, Krainer AR (2007) Enhancement of SMN2 exon 7 inclusion by antisense oligonucleotides targeting the exon. PLoS Biol 5: e73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Y, Vickers TA, Okunola HL, Bennett CF, Krainer AR (2008) Antisense masking of an hnRNP A1/A2 intronic splicing silencer corrects SMN2 splicing in transgenic mice. Am J Hum Genet 82: 834–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huot ME, Brown CM, Lamarche-Vane N, Richard S (2009) An adaptor role for cytoplasmic Sam68 in modulating Src activity during cell polarization. Mol Cell Biol 29: 1933–1943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishidate T, Yoshihara S, Kawasaki Y, Roy BC, Toyoshima K, Akiyama T (1997) Identification of a novel nuclear localization signal in Sam68. FEBS Lett 409: 237–241 [DOI] [PubMed] [Google Scholar]
- Kashima T, Manley JL (2003) A negative element in SMN2 exon 7 inhibits splicing in spinal muscular atrophy. Nat Genet 34: 460–463 [DOI] [PubMed] [Google Scholar]
- Kashima T, Rao N, David CJ, Manley JL (2007a) hnRNP A1 functions with specificity in repression of SMN2 exon 7 splicing. Hum Mol Genet 16: 3149–3159 [DOI] [PubMed] [Google Scholar]
- Kashima T, Rao N, Manley JL (2007b) An intronic element contributes to splicing repression in spinal muscular atrophy. Proc Natl Acad Sci USA 104: 3426–3431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre S, Burlet P, Liu Q, Bertrandy S, Clermont O, Munnich A, Dreyfuss G, Melki J (1997) Correlation between severity and SMN protein level in spinal muscular atrophy. Nat Genet 16: 265–269 [DOI] [PubMed] [Google Scholar]
- Lin Q, Taylor SJ, Shalloway D (1997) Specificity and determinants of Sam68 RNA binding. Implications for the biological function of K homology domains. J Biol Chem 272: 27274–27280 [DOI] [PubMed] [Google Scholar]
- Loiarro M, Capolunghi F, Fantò N, Gallo G, Campo S, Arseni B, Carsetti R, Carminati P, De Santis R, Ruggiero V, Sette C (2007) Pivotal Advance: Inhibition of MyD88 dimerization and recruitment of IRAK1 and IRAK4 by a novel peptidomimetic compound. J Leukoc Biol 82: 801–810 [DOI] [PubMed] [Google Scholar]
- Loiarro M, Sette C, Gallo G, Ciacci A, Fantò N, Mastroianni D, Carminati P, Ruggiero V (2005) Peptide-mediated interference of TIR domain dimerization in MyD88 inhibits interleukin-1-dependent activation of NF-{kappa}B. J Biol Chem 280: 15809–15814 [DOI] [PubMed] [Google Scholar]
- Lorson CL, Hahnen E, Androphy EJ, Wirth B (1999) A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc Natl Acad Sci USA 96: 6307–6311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukong KE, Richard S (2003) Sam68, the KH domain-containing superSTAR. Biochim Biophys Acta 1653: 73–86 [DOI] [PubMed] [Google Scholar]
- Lukong KE, Richard S (2008) Motor coordination defects in mice deficient for the Sam68 RNA-binding protein. Behav Brain Res 189: 357–363 [DOI] [PubMed] [Google Scholar]
- Matlin AJ, Clark F, Smith CW (2005) Understanding alternative splicing: towards a cellular code. Nat Rev Mol Cell Biol 6: 386–398 [DOI] [PubMed] [Google Scholar]
- Matter N, Herrlich P, Konig H (2002) Signal-dependent regulation of splicing via phosphorylation of Sam68. Nature 420: 691–695 [DOI] [PubMed] [Google Scholar]
- Matter N, Marx M, Weg-Remers S, Ponta H, Herrlich P, König H (2000) Heterogeneous ribonucleoprotein A1 is part of an exon-specific splice-silencing complex controlled by oncogenic signaling pathways. J Biol Chem 275: 35353–35360 [DOI] [PubMed] [Google Scholar]
- Monani UR (2005) Spinal muscular atrophy: a deficiency in a ubiquitous protein; a motor neuron-specific disease. Neuron 48: 885–896 [DOI] [PubMed] [Google Scholar]
- Monani UR, Lorson CL, Parsons DW, Prior TW, Androphy EJ, Burghes AH, McPherson JD (1999) A single nucleotide difference that alters splicing patterns distinguishes the SMA gene SMN1 from the copy gene SMN2. Hum Mol Genet 8: 1177–1183 [DOI] [PubMed] [Google Scholar]
- Monani UR, Sendtner M, Coovert DD, Parsons DW, Andreassi C, Le TT, Jablonka S, Schrank B, Rossoll W, Prior TW, Morris GE, Burghes AH (2000) The human centromeric survival motor neuron gene (SMN2) rescues embryonic lethality in Smn(−/−) mice and results in a mouse with spinal muscular atrophy. Hum Mol Genet 9: 333–339 [DOI] [PubMed] [Google Scholar]
- Okunola HL, Krainer AR (2009) Cooperative-binding and splicing-repressive properties of hnRNP A1. Mol Cell Biol 29: 5620–5631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paronetto MP, Achsel T, Massiello A, Chalfant CE, Sette C (2007) The RNA-binding protein Sam68 modulates the alternative splicing of Bcl-x. J Cell Biol 176: 929–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paronetto MP, Cappellari M, Busà R, Pedrotti S, Vitali R, Comstock C, Hyslop T, Knudsen KE, Sette C (2010) Alternative splicing of the cyclin D1 proto-oncogene is regulated by the RNA-binding protein Sam68. Cancer Res 70: 229–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paronetto MP, Messina V, Bianchi E, Barchi M, Vogel G, Moretti C, Palombi F, Stefanini M, Geremia R, Richard S, Sette C (2009) Sam68 regulates translation of target mRNAs in male germ cells, necessary for mouse spermatogenesis. J Cell Biol 185: 235–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paronetto MP, Venables JP, Elliott DJ, Geremia R, Rossi P, Sette C (2003) Tr-kit promotes the formation of a multimolecular complex composed by Fyn, PLCgamma1 and Sam68. Oncogene 22: 8707–8715 [DOI] [PubMed] [Google Scholar]
- Pearn J (1980) Classification of spinal muscular atrophies. Lancet 1: 919–922 [DOI] [PubMed] [Google Scholar]
- Pellizzoni L (2007) Chaperoning ribonucleoprotein biogenesis in health and disease. EMBO Rep 8: 340–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajan P, Gaughan L, Dalgliesh C, El-Sherif A, Robson CN, Leung HY, Elliott DJ (2008) The RNA-binding and adaptor protein Sam68 modulates signal-dependent splicing and transcriptional activity of the androgen receptor. J Pathol 215: 67–77 [DOI] [PubMed] [Google Scholar]
- Singh NN, Shishimorova M, Cao LC, Gangwani L, Singh RN (2009) A short antisense oligonucleotide masking a unique intronic motif prevents skipping of a critical exon in spinal muscular atrophy. RNA Biol 6: 341–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Valcárcel J (2005) Building specificity with nonspecific RNA-binding proteins. Nat Struct Mol Biol 12: 645–653 [DOI] [PubMed] [Google Scholar]
- Taylor SJ, Resnick RJ, Shalloway D (2004) Sam68 exerts separable effects on cell cycle progression and apoptosis. B M C Cell Biol 22: 5:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toshchakov VY, Fenton MJ, Vogel SN (2007) Cutting Edge: Differential inhibition of TLR signaling pathways by cell-permeable peptides representing BB loops of TLRs. J Immunol 178: 2655–2660 [DOI] [PubMed] [Google Scholar]
- Tsai LK, Tsai MS, Ting CH, Wang SH, Li H (2008) Restoring Bcl-x(L) levels benefits a mouse model of spinal muscular atrophy. Neurobiol Dis 31: 361–367 [DOI] [PubMed] [Google Scholar]
- Wang GS, Cooper TA (2007) Splicing in disease: disruption of the splicing code and the decoding machinery. Nat Rev Genet 8: 749–761 [DOI] [PubMed] [Google Scholar]
- Wang LL, Richard S, Shaw AS (1995) P62 association with RNA is regulated by tyrosine phosphorylation. J Biol Chem 270: 2010–2013 [DOI] [PubMed] [Google Scholar]
- Zhang Z, Lotti F, Dittmar K, Younis I, Wan L, Kasim M, Dreyfuss G (2008) SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell 133: 585–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.