Abstract
Recent research efforts in oral biology have resulted in elucidation of the proteomes of major human salivary secretions and whole saliva. One might hypothesize that the proteome of minor gland secretions may show significantly different characteristics when compared with the proteomes of parotid or submandibular/sublingual secretions. To test this hypothesis, we conducted the first exploration into the proteome of minor salivary gland secretion. Minor gland secretion was obtained from healthy volunteers, and its components were subjected to liquid-chromatography-electrospray-ionization-tandemmass-spectrometry. This led to the identification of 56 proteins, 12 of which had never been identified in any salivary secretion. The unique characteristics of the minor salivary gland secretion proteome are related to the types as well as the numbers of components present. The differences between salivary proteomes may be important with respect to specific oral functions.
Keywords: minor salivary gland secretion, proteomics, oral, protein, saliva
INTRODUCTION
Major efforts in salivary research have been made to characterize the salivary proteins and peptides present in parotid, submandibular, and sublingual glands and whole saliva, both by classic biochemical methods (Keller et al., 1971; Oppenheim et al., 1971, 2007; Dawes, 1972; Baum et al., 1976) and by modern state-of-the-art proteomics approaches (Yao et al., 2003; Wilmarth et al., 2004; Hardt et al., 2005; Hu et al., 2005). Minor salivary gland secretion has not been subjected to the same biochemical characterization scrutiny as those of major glands, despite the fact that minor salivary gland secretion may play important roles in host tissue protection in the oral cavity (Dawes and Wood, 1973; Ferguson, 1999). This led to the hypothesis that the minor gland proteome may also be composed of unique proteins, some of which may not be common to major gland secretions.
High-sensitivity tools for the identification of proteins/peptides required for the characterization of the minor gland secretion proteome have become available with the advent of the current arsenal of proteomics. This study describes the first proteome determination of labial minor salivary gland secretion by mass spectrometry.
MATERIALS & METHODS
Human Participants
Ten healthy individuals, four males and six females, with an overall mean age of 32.3 yrs, participated in this study. The collection protocol for obtaining minor salivary gland secretion was approved by the Institutional Review Board of Boston University Medical Center, and prior to collection, all participants signed an informed consent document.
Collection of Labial Minor Salivary Gland Secretion
In preparation for the collection of labial minor salivary gland secretion, the lower lip mucosa was made accessible by means of cheek retractors. The mucosa of the lower lip was isolated with cotton rolls, washed 3 times with water spray, air-dried, and kept isolated with a new set of cotton rolls. Salivary flow was stimulated by the application of 5% citric acid solution to the lateral border of the tongue by means of a cotton applicator. Once secretion droplets formed over the glandular ducts, the secretion was collected by means of a microliter plastic pipette tip connected to a P10 Gilson pipette, and the fluid was expelled into a 1.5-mL polypropylene Eppendorf tube. Volumes of 10 to 15 µL of labial minor salivary secretion were collected per person. The samples were then centrifuged at 4000 g over a five-minute interval and kept at −20°C until used. Analyses were performed with a 140-µL pool of secretion obtained from the ten participants. The total protein concentration was measured by the bicinchoninic acid (BCA) assay (Pierce Chemical, Co., Rockford, IL, USA) and bovine serum albumin protein standard. The mean protein concentration of the labial minor salivary gland secretion pool was 1.2 mg/mL.
Sample Preparation
Method A
A 47-µL aliquot of pooled labial minor salivary gland secretion was diluted 1:1 with Bis-Tris sample buffer, boiled for 5 min, and divided into 3 equal aliquots, each of which was subjected to precast 12% Bis-Tris polyacrylamide gel electrophoresis (Bis-Tris PAGE, Invitrogen, Carlsbad, CA, USA). Proteins were simply forced to enter the separating gel by means of a constant voltage of 120 V for 15 min. After 30 minutes' staining in 0.1% Coomassie Brilliant Blue R-250 in methanol/acetic acid/water (40%/10%/50% v/v), and de-staining for 2 hrs in the same solution lacking dye, the stained gel portions of each lane were excised with a razor blade, and cut lengthwise into 2 equal parts. Each gel sample was cut into small fragments of approximately 1 mm3, and subjected to in-gel trypsinization. The enzymatic cleavage was carried out in 50 mM NH4HCO3, pH 8.0, solution containing 2% (w/w) sequencing-grade trypsin (Promega, Madison, WI, USA). Peptides were extracted by multiple wash and dehydration/hydration steps as described previously (Shevchenko et al., 1996).
Method B
A 93-µL aliquot of pooled labial minor salivary gland secretion was dried by Speedvac, denatured, and reduced for 1 hr by the addition of 200 µL of 4 M urea, 10 mM dithiothreitol (DTT), and 50 mM NH4HCO3, pH 8.0. After four-fold dilution with 50 mM NH4HCO3, pH 8.0, tryptic digestion was carried out for 16 hrs at 37°C, after the addition of 2% (w/w) sequencing-grade trypsin (Promega, Madison, WI, USA). After trypsinization, the 93-µL aliquot was divided into 2 portions, one of which was subjected directly to mass spectrometric analysis and the other to further fractionation (method C).
Method C
Following the treatment described in Method B, a 47-µL aliquot was dried, dissolved in 100 µL of 20 mM maleic acid, pH 2.0, and subjected to fractionation by cation exchange chromatography. For this purpose, the sample was applied to a 1.6 mm × 50 mm sulphonate cation exchange column (Mono S PC 1.6/5, Smart System, GE Healthcare, Piscataway, NJ, USA), which had been equilibrated with the same buffer (buffer A) at a flow rate of 50 µL/min. Elution was carried out with a linear gradient ranging from 0 to 100% buffer B, 20 mM maleic acid, and 1 mM NaCl, pH 2.0, over 60 min, and the eluent was monitored by absorbance at 219 nm. Twelve consecutive five-minute fractions (250 µL) were collected and dried.
Liquid Chromatography Electrospray Ionization Tandem Mass Spectrometry (LC-ESI-MS/MS)
The dried trypsinized aliquots were re-suspended in 100 µL of 0.1% trifluoroacetic acid and de-salted by means of a Zip TIPc18 (Millipore, Bedford, MA, USA). The eluted portion was dried, re-suspended in 25 µL of 97.5% H2O/24% acetonitrile/0.1% formic acid, and then subjected to reverse-phase LC-ESI-MS/MS. For mass spectrometric analyses, samples were applied to a nano-flow reverse-phase HPLC capillary column, 50 µm × 10 cm (Pico Tip™ EMITTER, New Objective, Woburn, MA, USA), packed in-house with Magic C18 resin (diameter, 5 µm; pore size, 20 nm) (Michrom BioResources, Auburn, CA, USA), connected to a LTQ-linear-ion-trap (Thermo-Finnigan, San Jose, CA, USA). A linear 40-minute gradient was used, ranging from 5% to 50% of solvent B (97.5% acetonitrile, 0.1% formic acid) at a flow rate of 110 nL/min. Electrospray voltage and the temperature of the ion transfer capillary were 1.8 kV and 230°C, respectively. The normalized collision energy was set to 35.0%, and the mass spectrometer was operated in the positive-ion mode according to a data-dependent acquisition method initiated by a survey scan in the range of m/z values 400–2000, followed by MS/MS analysis of the selected peptide ions.
Data Analysis
MS/MS spectral data were searched against human protein databases (Swiss Prot and TrEMBL, Swiss Institute of Bioinformatics, Geneva, Switzerland, http://expasy.org/sprot) by SEQUEST (Bioworks Browser 3.2, Thermo-Finnigan, San Jose, CA, USA). Search parameters and filter criteria were used as described previously (Siqueira et al., 2007b). For protein identification, at least 2 or more peptides had to be characterized, and protein annotations were obtained from the Web-based databases Babelomics (http://babelomics.bioinfo.cipf.es/index.html), AmiGO (http://amigo.geneontology.org), and Swiss protein (http://ca.expasy.org/).
RESULTS
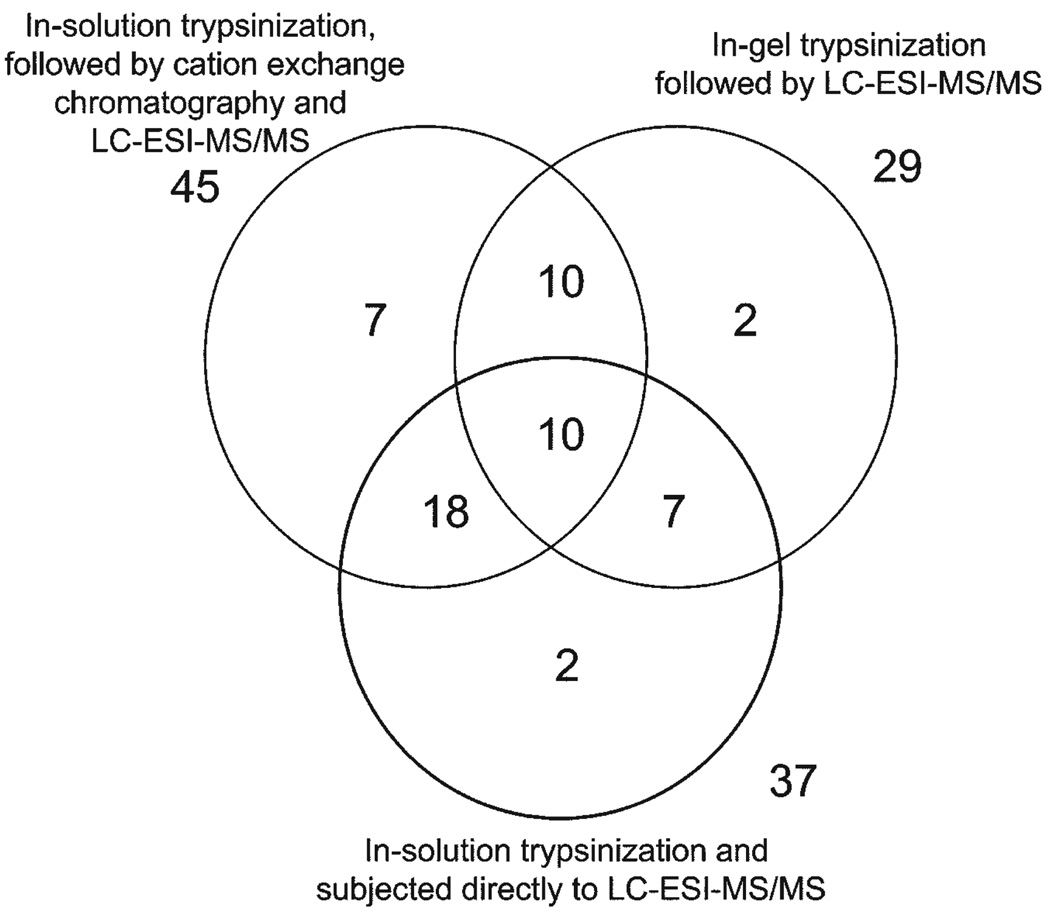
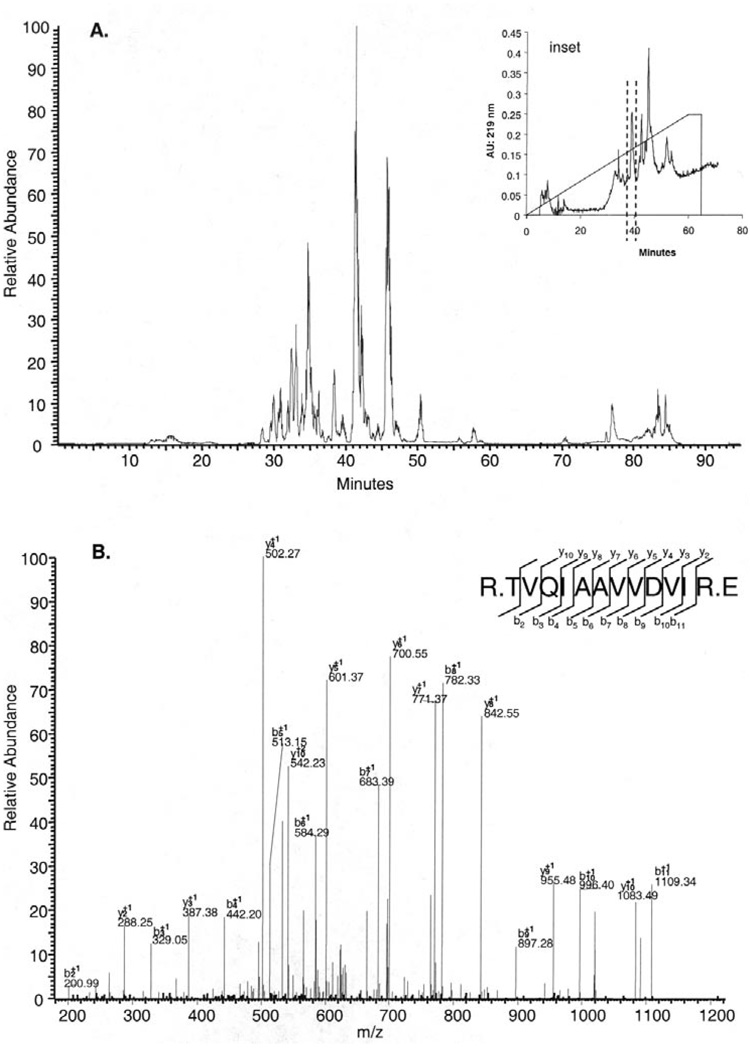
For the characterization of the human labial minor salivary gland secretion proteome, 3 different sample preparation methods were used. PAGE and in-gel trypsinization (method A) allowed for the identification of 29 proteins, in-solution trypsinization (method B) made it possible for 37 proteins to be identified, and in-solution trypsinization followed by cation exchange chromatography (method C) led to the identification of 45 proteins (Fig. 1). The combination of the data from all sample preparation methods resulted in the identification of a total of 56 different proteins in labial minor salivary gland secretion. Among these proteins, 45 members were found in at least 2 of the 3 sample preparations, and 10 of those were actually found in all 3 sample preparations (Fig. 1). The largest number of proteins identified was with in-solution trypsinization in combination with cation exchange chromatography, amounting to 79% of all proteins. The superiority of this approach seems to be related to the high resolution of labial minor salivary gland secretion protein separation with cation exchange chromatography (Fig. 2A, inset). Each of the 12 fractions yielded base peak chromatograms similar in complexity to those of the 40- to 45-minute fraction (Fig. 2A). Final identification of proteins was based on peptide sequence determination as shown for the prolactin-inducible protein (Fig. 2B). The lowest number of proteins identified was associated with PAGE and in-gel trypsinization, providing 52% of all proteins identified. The three-pronged approach of sample preparation for MS analysis not only increased the number of proteins identified, but also allowed for different proteins to be captured. (Some salient characteristics of all 56 identified proteins are listed in the APPENDIX Table.) Arranging the labial minor salivary gland secretion components based on the number of unique peptides found for each protein showed that 14 of the 56 proteins were identified on the basis of 5 or more peptides, whereas 15 labial minor salivary gland secretion macromolecules were represented by only 2 peptides found by MS.
Figure 1.
Summary of the distribution of 56 proteins identified from labial minor salivary gland secretion with 3 different sample preparations.
Figure 2.
Base-peak chromatogram of one of the 12 fractions separated by cation exchange chromatography and analyzed by LC-ESI-MS/MS with a nano-flow reverse-phase HPLC column, with gradient elution ranging from 5 to 50% solvent B in 40 min (A). Cation exchange chromatogram of the tryptic peptides derived from proteins associated with labial minor salivary gland secretion (Inset). MS/MS spectrum and sequence analysis of a tryptic peptide. The sequence of R.TVQIAAVVDVIR.E led to the identification of "prolactin inducible protein" (P12273). Matching b- and y-ion series are indicated in the upper right (B).
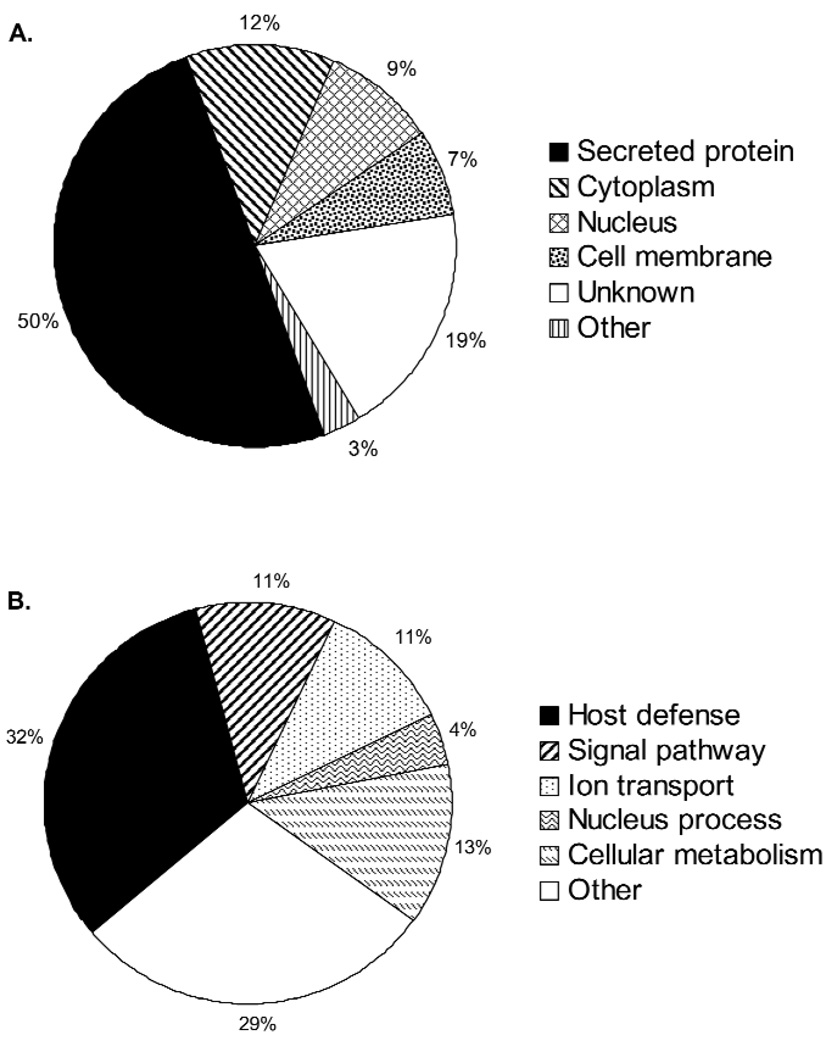
With the information provided via the accession number, the labial minor salivary gland secretion components could be grouped into categories of cellular localization and biological function (Fig. 3). It is not surprising that at least 50% of the proteins identified are known to be secretory proteins, while 28% of the proteins could be associated with the cytoplasm, nucleus, or the cell membrane. The distribution of the labial minor salivary gland secretion proteins with respect to biological function (Fig. 3B) demonstrated that one-third of all components are related to host defense mechanisms.
Figure 3.
Classification of labial minor salivary gland secretion proteins according to their origin (A) and their biological functions (B). Information on origin and biological function was derived from protein annotations. Proteins with more than one origin and more than one biological function were counted multiple times.
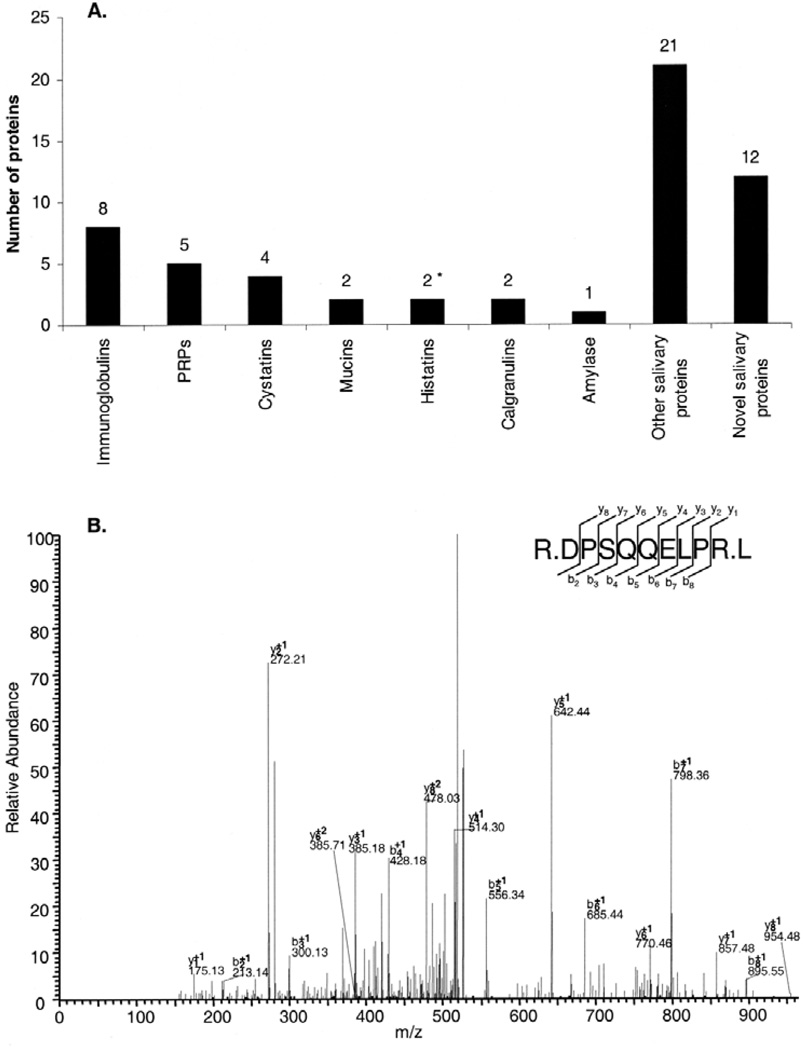
Among the secretory group of labial minor salivary gland secretion proteins, typical members of salivary polymorphic families of immunoglobulins, PRPs, cystatins, mucins, histatins, calgranulins, and amylase were identified (Fig. 4A). Analysis of the data indicated that these salivary constituents occur in labial minor salivary gland secretion in relative proportions similar to those found in major gland secretions. Labial minor salivary gland secretion contains a significant number of proteins never found in any other salivary secretions (Fig. 4A). These novel salivary proteins comprised a group of 12 components amounting to 21% of all proteins identified. An example of the identification of one of those newly discovered proteins is shown (Fig. 4B), providing the amino acid sequence evidence leading to the identification of fatty acid synthase.
Figure 4.
Relative proportions of major, minor, and novel salivary proteins in labial minor salivary gland secretion. *Histatin 1 was included based on the presence of the C-terminal 16-residue peptide found in all 3 sample preparations (A). MS/MS spectrum and sequence analysis of a tryptic peptide. The sequence of R.DPSQQELPR.L is indicative of the presence of "fatty acid synthase" (P49327). Matching b-and y-ion series are indicated in the upper right (B).
DISCUSSION
Minor salivary glands contribute only about 10% to the total volume of human saliva released into the oral cavity (Dawes and Wood, 1973). Despite this small volume, minor glands have long been considered to be of significant importance for oral physiology and the maintenance of oral health, due to their unique anatomical distribution, their proximity to mucosal surfaces, and their relative enrichment with respect to mucins and immunoglobulins when compared with major salivary glands (Smith et al., 1991; Ferguson, 1999; Hand et al., 1999). Indeed, the current study identified MUC5B and MUC7, the major members of the salivary mucin family. These macromolecules are well-known to protect the oral mucosa from desiccation, mechanical injury, and microbial attacks (Tabak et al., 1982; Loomis et al., 1987). In addition, mucins have been shown to form complexes with other salivary proteins, such as sIgA, lysozyme, histatins, and PRPs (Bruno et al., 2005), all of which were identified in this study as well. Such complexes may protect complexed proteins from enzymatic degradation, or may enhance their function by acting as 'chaperones', targeting mucosal and enamel surfaces.
Another interesting attribute of labial minor salivary gland secretion is its enrichment with respect to immunoglobulins. The identification of 8 different immunoglobulins supports the results of previous studies indicating that this specific oral fluid represents a major source of orally secreted immunoglobulins (Crawford et al., 1975; Smith et al., 1987; Smith et al., 1991). The relative abundance of proteins belonging to the acquired immune system released locally throughout the major mucosal surfaces may play a crucial role in host defense mechanisms. Several of the identified salivary proteins belong to the cystatin family (cystatins A, B, D, SN). Traditionally, cystatins are considered to be cysteine proteinase inhibitors that mitigate the deleterious effects of inflammatory processes by minimizing proteolysis-associated tissue damage (Dickinson et al., 2002). A surprising finding was the presence of 5 members of the large salivary-proline-rich protein family, which are dominant constituents of the major gland secretions. These proteins are functionally related to mineral homeostasis and the maintenance of tooth enamel integrity (Oppenheim et al., 2007). The existence of this type of protein in labial minor salivary gland secretion suggests that this secretion could act directly in the protection of hard tissue and help in the formation of enamel pellicle. Enamel pellicle is an important protein film that acts as a selective permeability barrier that regulates demineralization/remineralization processes and dictates the initial colonization of tooth surfaces (Siqueira et al., 2007a). The labial minor salivary gland secretion proteome also contained histatin 3 and one peptide of histatin 1. The peptide found for histatin 1 exhibited the sequence R.EFPFYGDYGSNYLYDN, representing its 16-carboxyl-terminal fragment, and it is noteworthy that this peptide was detected in all 3 sample preparations. Histatins are considered to be important components of the innate oral host defense system, since they kill the pathogenic yeast Candida albicans (Helmerhorst et al., 2006) and exhibit growth-inhibitory activity against Streptococcus mutans (MacKay et al., 1984).
A very important finding in determination of the proteome of labial minor salivary gland secretion was the discovery of 12 new salivary proteins, demonstrating that labial minor salivary gland secretion is a body fluid with a unique protein composition. Although some of these proteins have already been assigned distinct biological functions elsewhere in the human body, their functions in the oral cavity have not yet been clarified. For example, the protein "fatty acid synthase" is also expressed at high levels in several human cancer tissues (Pizer et al., 1998), and its biological role in the oral cavity needs to be addressed. The present study is the first exploration into the proteome of labial minor salivary gland secretion, and analysis of the data clearly shows that this proteome differs in major ways from the proteomes of major salivary secretions. First, proteomic analyses yielded only 56 proteins in labial minor salivary gland secretion, in contrast to parotid and submandibular/sublingual secretions, where over a thousand proteins could be identified by almost identical proteomic approaches (Siqueira et al., unpublished observations; Wong, 2007, personal communication). Second, one-quarter of the labial minor salivary gland secretion proteins are novel salivary components not detected in major salivary secretions. The methods used in this study allowed for an exploration into the proteome of labial minor salivary gland secretion and thus provide major insights into the hitherto largely unknown composition of this exocrine body fluid. The functional classification of labial minor salivary gland secretion constituents suggests the presence of a wide variety of host-protective proteins that not only play a pivotal role in oral tissue homeostasis, but also could be critically involved in processes influencing infectious diseases. This may be particularly relevant for dental caries and periodontal disease, considering the proximity of minor salivary glands to the affected oral targets.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by NIH/NIDCR Grants DE05672, DE07652, DE17788, and DE 18132.
REFERENCES
- Baum BJ, Bird JL, Millar DB, Longton RW. Studies on histidine-rich polypeptides from human parotid saliva. Arch Biochem Biophys. 1976;177:427–436. doi: 10.1016/0003-9861(76)90455-0. [DOI] [PubMed] [Google Scholar]
- Bruno LS, Li X, Wang L, Soares RV, Siqueira CC, Oppenheim FG, et al. Two-hybrid analysis of human salivary mucin MUC7 interactions. Biochim Biophys Acta. 2005;1746:65–72. doi: 10.1016/j.bbamcr.2005.08.007. [DOI] [PubMed] [Google Scholar]
- Crawford JM, Taubman MA, Smith DJ. Minor salivary glands as a major source of secretory immunoglobin A in the human oral cavity. Science. 1975;190:1206–1209. doi: 10.1126/science.1198107. [DOI] [PubMed] [Google Scholar]
- Dawes C. Circadian rhythms in human salivary flow rate and composition. J Physiol. 1972;220:529–545. doi: 10.1113/jphysiol.1972.sp009721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawes C, Wood CM. The contribution of oral minor mucous gland secretions to the volume of whole saliva in man. Arch Oral Biol. 1973;18:337–342. doi: 10.1016/0003-9969(73)90156-8. [DOI] [PubMed] [Google Scholar]
- Dickinson DP, Thiesse M, Hicks MJ. Expression of type 2 cystatin genes CST1-CST5 in adult human tissues and the developing submandibular gland. DNA Cell Biol. 2002;21:47–65. doi: 10.1089/10445490252810311. [DOI] [PubMed] [Google Scholar]
- Ferguson DB. The flow rate and composition of human labial gland saliva. Arch Oral Biol. 1999;44 Suppl 1:11–14. doi: 10.1016/s0003-9969(99)90004-3. [DOI] [PubMed] [Google Scholar]
- Hand AR, Pathmanathan D, Field RB. Morphological features of the minor salivary glands. Arch Oral Biol. 1999;44 Suppl 1:3–10. doi: 10.1016/s0003-9969(99)90002-x. [DOI] [PubMed] [Google Scholar]
- Hardt M, Thomas LR, Dixon SE, Newport G, Agabian N, Prakobphol A, et al. Toward defining the human parotid gland salivary proteome and peptidome: identification and characterization using 2D SDS-PAGE, ultrafiltration, HPLC, and mass spectrometry. Biochemistry. 2005;44:2885–2899. doi: 10.1021/bi048176r. [DOI] [PubMed] [Google Scholar]
- Helmerhorst EJ, Alagl AS, Siqueira WL, Oppenheim FG. Oral fluid proteolytic effects on histatin 5 structure and function. Arch Oral Biol. 2006;51:1061–1070. doi: 10.1016/j.archoralbio.2006.06.005. [DOI] [PubMed] [Google Scholar]
- Hu S, Xie Y, Ramachandran P, Ogorzalek Loo RR, Li Y, Loo JA, et al. Large-scale identification of proteins in human salivary proteome by liquid chromatography/mass spectrometry and two-dimensional gel electrophoresis-mass spectrometry. Proteomics. 2005;5:1714–1728. doi: 10.1002/pmic.200401037. [DOI] [PubMed] [Google Scholar]
- Keller PJ, Kauffman DL, Allan BJ, Williams BL. Further studies on the structural differences between the isoenzymes of human parotid-amylase. Biochemistry. 1971;10:4867–4874. doi: 10.1021/bi00802a006. [DOI] [PubMed] [Google Scholar]
- Loomis RE, Prakobphol A, Levine MJ, Reddy MS, Jones PC. Biochemical and biophysical comparison of two mucins from human submandibular-sublingual saliva. Arch Biochem Biophys. 1987;258:452–464. doi: 10.1016/0003-9861(87)90366-3. [DOI] [PubMed] [Google Scholar]
- MacKay BJ, Denepitiya L, Iacono VJ, Krost SB, Pollock JJ. Growth-inhibitory and bactericidal effects of human parotid salivary histidine-rich polypeptides on Streptococcus mutans. Infect Immun. 1984;44:695–701. doi: 10.1128/iai.44.3.695-701.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppenheim FG, Hay DI, Franzblau C. Proline-rich proteins from human parotid saliva. I. Isolation and partial characterization. Biochemistry. 1971;10:4233–4238. doi: 10.1021/bi00799a013. [DOI] [PubMed] [Google Scholar]
- Oppenheim FG, Salih E, Siqueira WL, Zhang W, Helmerhorst EJ. Salivary proteome and its genetic polymorphisms. Ann NY Acad Sci. 2007;1098:22–50. doi: 10.1196/annals.1384.030. [DOI] [PubMed] [Google Scholar]
- Pizer ES, Lax SF, Kuhajda FP, Pasternack GR, Kurman RJ. Fatty acid synthase expression in endometrial carcinoma: correlation with cell proliferation and hormone receptors. Cancer. 1998;83:528–537. [PubMed] [Google Scholar]
- Shevchenko A, Jensen ON, Podtelejnikov AV, Sagliocco F, Wilm M, Vorm O, et al. Linking genome and proteome by mass spectrometry: large-scale identification of yeast proteins from two dimensional gels. Proc Natl Acad Sci USA. 1996;93:14440–14445. doi: 10.1073/pnas.93.25.14440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siqueira WL, Helmerhorst EJ, Zhang W, Salih E, Oppenheim FG. Acquired enamel pellicle and its potential role in oral diagnostics. Ann NY Acad Sci. 2007a;1098:504–509. doi: 10.1196/annals.1384.023. [DOI] [PubMed] [Google Scholar]
- Siqueira WL, Zhang W, Helmerhorst EJ, Gygi SP, Oppenheim FG. Identification of protein components in in vivo human acquired enamel pellicle using LC-ESI-MS/MS. J Proteome Res. 2007b;6:2152–2160. doi: 10.1021/pr060580k. [DOI] [PubMed] [Google Scholar]
- Smith DJ, Taubman MA, King WF. Immunological features of minor salivary gland saliva. J Clin Immunol. 1987;7:449–455. doi: 10.1007/BF00915054. [DOI] [PubMed] [Google Scholar]
- Smith DJ, Taubman MA, Ali-Salaam P. Immunoglobulin isotypes in human minor gland saliva. J Dent Res. 1991;70:167–170. doi: 10.1177/00220345910700030201. [DOI] [PubMed] [Google Scholar]
- Tabak LA, Levine MJ, Mandel ID, Ellison SA. Role of salivary mucins in the protection of the oral cavity. J Oral Pathol. 1982;11:1–17. doi: 10.1111/j.1600-0714.1982.tb00138.x. [DOI] [PubMed] [Google Scholar]
- Wilmarth PA, Riviere MA, Rustvold DL, Lauten JD, Madden TE, David LL. Two-dimensional liquid chromatography study of the human whole saliva proteome. J Proteome Res. 2004;3:1017–1023. doi: 10.1021/pr049911o. [DOI] [PubMed] [Google Scholar]
- Yao Y, Berg EA, Costello CE, Troxler RF, Oppenheim FG. Identification of protein components in human acquired enamel pellicle and whole saliva using novel proteomics approaches. J Biol Chem. 2003;278:5300–5308. doi: 10.1074/jbc.M206333200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.