Summary
DNA cytosine methylation is a central epigenetic modification that plays essential roles in cellular processes including genome regulation, development and disease. Here we present the first genome-wide, single-base resolution maps of methylated cytosines in a mammalian genome, from both human embryonic stem cells and fetal fibroblasts, along with comparative analysis of mRNA and small RNA components of the transcriptome, several histone modifications, and sites of DNA-protein interaction for several key regulatory factors. Widespread differences were identified in the composition and patterning of cytosine methylation between the two genomes. Nearly one-quarter of all methylation identified in embryonic stem cells was in a non-CG context, suggesting that they may utilize different methylation mechanisms to affect gene regulation. Methylation in non-CG contexts showed enrichment in gene bodies and depletion in protein binding sites and enhancers. Non-CG methylation disappeared upon induced differentiation of the embryonic stem cells, and was restored in induced pluripotent stem cells. We identified hundreds of differentially methylated regions proximal to genes involved in pluripotency and differentiation, and widespread reduced methylation levels in fibroblasts associated with lower transcriptional activity. These reference epigenomes provide a foundation for future studies exploring this key epigenetic modification in human disease and development.
Thirty-four years have passed since Riggs, Holliday and Pugh proposed that cytosine DNA methylation in eukaryotes could act as a stably inherited modification affecting gene regulation and cellular differentiation1,2. Since then, intense research effort has expanded our understanding of diverse aspects of DNA methylation in higher eukaryotic organisms. These include elucidation of molecular pathways required for establishing and maintaining DNA methylation, cell-type specific variation in methylation patterns, and the involvement of methylation in multifarious cellular processes such as gene regulation, DNA-protein interactions, cellular differentiation, suppression of transposable elements, embryogenesis, X-inactivation, genomic imprinting and tumorigenesis3–9. DNA methylation, together with covalent modification of histones, is thought to alter chromatin density and accessibility of the DNA to cellular machinery, thereby modulating the transcriptional potential of the underlying DNA sequence10.
Genome-wide studies of mammalian DNA methylation have previously been conducted, however they have been limited by low resolution11, sequence-specific bias, or complexity reduction approaches that analyze only a very small fraction of the genome12–14. In order to improve our understanding of the genome-wide patterns of DNA methylation we have generated single-base resolution DNA methylation maps throughout the majority of the human genome in both embryonic stem cells and fibroblasts. Furthermore, we have profiled several important histone modifications, protein-DNA interaction sites of regulatory factors, and the mRNA and small RNA components of the transcriptome to better understand how changes in DNA methylation patterns and histone modifications may affect readout of the proximal genetic information.
Single-base resolution maps of DNA methylation for two human cell lines
Single-base DNA methylomes of the flowering plant Arabidopsis thaliana were previously achieved using MethylC-Seq15 or BS-Seq16. In this method, genomic DNA is treated with sodium bisulfite (BS) to convert cytosine, but not methylcytosine, to uracil, and subsequent high-throughput sequencing. We performed MethylC-Seq for two human cell lines, H1 human embryonic stem cells17 and IMR90 fetal lung fibroblasts18, generating 1.16 and 1.18 billion reads, respectively, that aligned uniquely to the human reference sequence (NCBI build 36/HG18). The total sequence yield was 87.5 and 91.0 gigabases (Gb), with an average read depth of 14.2× and 14.8× per strand for H1 and IMR90, respectively (Supplementary Fig. 1a). In each cell type, over 86% of both strands of the 3.08 Gb human reference sequence are covered by at least one sequence read (Supplementary Fig. 1b), accounting for 94% of the cytosines in the genome.
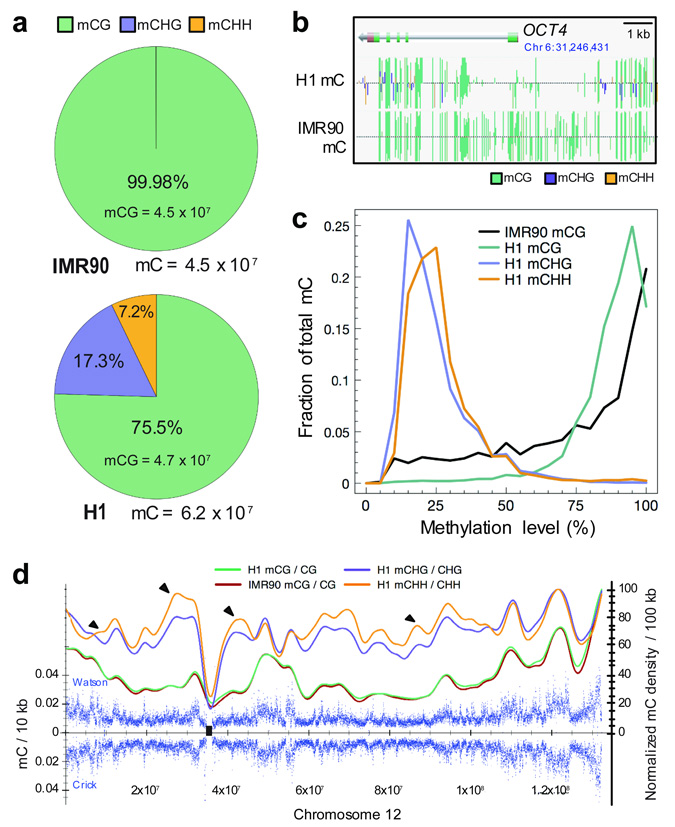
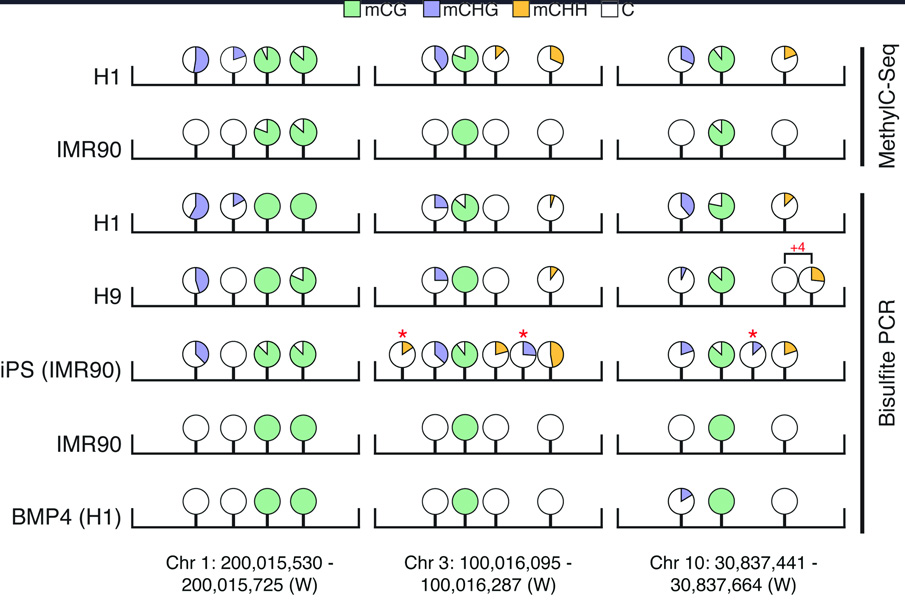
We detected approximately 62 million and 45 million methylcytosines in H1 and IMR90 cells, respectively (1% FDR, see Supplementary Materials, Fig. 1a), comprising 5.83% and 4.25% of the cytosines with sequence coverage. Full browsing of the entire dataset at single base resolution can be performed at http://neomorph.salk.edu/human_methylome using the AnnoJ browser (www.annoj.org). Of the methylcytosines detected in the IMR90 genome, 99.98% were in the CG context, and the total number of mCG sites was very similar in both cell types. In the H1 stem cells we detected abundant DNA methylation in non-CG contexts (mCHG and mCHH, where H = A, C or T), comprising almost 25% of all cytosines at which DNA methylation is identified, and accounting for most of the difference in total methylcytosine number between the cell types (Fig. 1a). The prevailing assumption is that mammalian DNA methylation is located almost exclusively in the CG context, however a handful of studies have previously detected non-CG methylation in human cells, and in particular in embryonic stem cells19,20. Bisulfite-PCR, cloning and sequencing of selected loci displaying H1 non-CG methylation in several human cell lines revealed that a second embryonic stem cell line, H917, displayed mCHG and mCHH at conserved positions, confirming that non-CG methylation is likely a general feature of human ES cells (Fig. 2, Supplementary Table 2). In addition, like IMR90 cells, BMP4-induced H1 cells lost non-CG methylation at several loci examined while methylation in the CG context was maintained, indicating that the pervasive non-CG methylation is lost upon differentiation. Furthermore, analysis of these loci in IMR90 induced pluripotent stem (iPS) cells revealed restored non-CG methylation (Fig. 2). Overall this demonstrates that the CHG and CHH methylation identified in H1 and absent in IMR90 are not simply due to genetic differences between the two cell types, but rather the presence of non-CG methylation is characteristic of an embryonic stem cell state. For each cell type, two biological replicates were performed with cells of different passage number (see Supplementary Materials), and comparison of the methylcytosines identified independently in each replicate revealed a high concordance of cytosine methylation status between replicates (Supplementary Fig. 2). For each cell type, the final DNA methylation map presented in this study represents the composite of the two biological replicates. The OCT4 gene exemplifies both cell-specific differential methylation and the presence of non-CG methylation (Fig. 1b), and in addition displayed a ~50-fold reduction in OCT4 transcript in IMR90 (data not shown). The absence of mCHG and mCHH methylation in IMR90 coincided with significantly lower transcript abundance of the de novo DNA methyltransferases (DNMTs) DNMT3A and DNMT3B and the associated DNMT3L in IMR90 cells (Supplementary Fig. 3), which is supported by a previous study of DNA methylation in ES cells and somatic cells19 and by the determined target sequence specificity of these DNMTs21,22.
Figure 1. Global trends of Human DNA methylomes.
a, The percent of methylcytosines identified for H1 and IMR90 cells in each sequence context. b, AnnoJ browser representation of OCT4. c, Distribution of the methylation level in each sequence context. The y-axis indicates the fraction of all methylcytosines that display each methylation level (x-axis), where methylation level is the mC/C ratio at each reference cytosine (at least 10 reads required) d, Blue dots indicate methylcytosine density in H1 cells in 10 kb windows throughout chromosome 12 (black rectangle, centromere). Smoothed lines represent the methylcytosine density in each context in H1 and IMR90. Black triangles indicate various regions of contrasting trends in CG and non-CG methylation. Abbreviations: mC, methylcytosine.
Figure 2. Bisulfite-PCR validation of non-CG DNA methylation in differentiated and stem cells.
DNA methylation sequence context is displayed according to the key and the percent methylation at each position is represented by the fill of each circle (see Supplementary Table 2 for values). Non-CG methylated positions indicated by an asterisk are unique to that cell type and “+4” indicates a mCHH that is shifted 4 bases downstream in H9 cells. Abbreviations: iPS, induced pluripotent stem cell.
Multiple reads covering each methylcytosine can be used as a readout of the fraction of the sequences within the sample that are methylated at that site16, here referred to as the methylation level of a specific cytosine. Similar to the Arabidopsis genome15, in the H1 genome we observed that 77% of mCG sites were 80–100% methylated, whereas 85% of mCHG and mCHH sites were 10–40% methylated (Fig. 1c), indicating that at sites of non-CG methylation only a fraction of the surveyed genomes in the sample were methylated. Notably, 56% of mCG sites in IMR90 were highly methylated (80–100%, Fig. 1c), indicating that although the total number of mCG sites in H1 and IMR90 is similar, in general the IMR90 mCG sites were typically less frequently methylated. In support of this, considering all CG site sequencing events, 82.7% and 67.7% were methylated in H1 and IMR90, respectively. A global-scale view of DNA methylation levels revealed that the density of DNA methylation showed large variations throughout each chromosome (Fig. 1d). Sub-telomeric regions of the chromosomes frequently showed higher DNA methylation density (Fig. 1d, Supplementary Fig. 4), which was previously reported as important for control of telomere length and recombination23,24. The smoothed profile of DNA methylation density in 100 kb windows indicated that on the chromosomal level the density profile of mCG in H1 and IMR90 was similar. The density profiles of mCHG and mCHH revealed that non-CG methylation was present throughout the entire chromosome. These two non-CG methylation marks showed a moderate correlation and did not always occur together (Pearson correlation 0.5 in 1 kb windows; Supplementary Fig. 2d). Notably, changes in density of the non-CG methylation were distinct from that of mCG in a number of regions.
Pervasive non-CG DNA methylation in embryonic stem cells
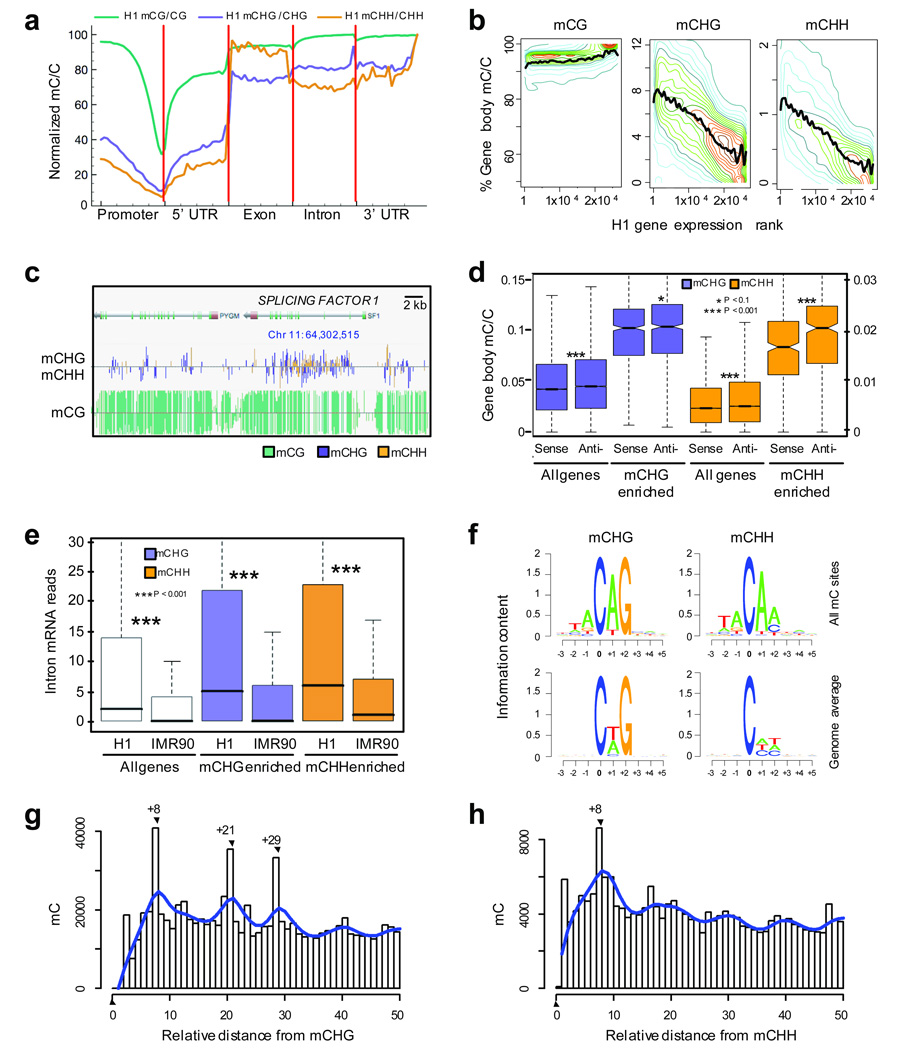
To characterize the abundant non-CG methylation in the H1 genome, we compared the average density of methylation relative to the underlying density of all potential sites of methylation in each context (henceforth referred to as the relative methylation density), throughout various genomic features (Fig. 3a, Supplementary Fig. 5). We observed a correlation in the density of mCG and the distance from the transcriptional start site (TSS), with mCG density increasing in the 5’ UTR to a similar level in exons, introns and the 3’ UTR as to 2 kb upstream of the TSS (Fig. 3a). We generally observed lower relative densities of methylation at CG islands and TSS, however a subset of these regions did not display this depletion (Supplementary Fig. 6)13,14,25. mCHG and mCHH methylation densities also decreased significantly toward the TSS and returned to the same level as 2 kb upstream at the end of the 5’ UTR, however within exons, introns and 3’ UTRs the non-CG methylation densities were twice as high. Intriguingly, the mCHH density was approximately 15–20% higher in exons than within introns and the 3’ UTR. To identify links between gene activity and non-CG methylation level within the gene body we performed strand-specific RNA-Seq15 and observed a positive correlation between gene expression and mCHG (R = 0.60) or mCHH (R = 0.58) density (Fig. 3b), with highly expressed genes containing 3-fold higher non-CG methylation density than non-expressed genes (Supplementary Fig. 7a). However, no correlation was observed between CG methylation density and gene expression in the H1 cells (Fig. 3b).
Figure 3. Non-CG DNA methylation in H1 embryonic stem cells.
a, Relative methylation density (the ratio of methylcytosines to reference cytosines) in H1 throughout different gene-associated regions (promoters encompass 2 kb upstream of the transcriptional start site).The mean mC/C profile was normalized to the maximum value. b, Relative methylation density within gene bodies (y-axis) as a function of gene expression (x-axis), with transcript abundance increasing from right to left. Colored lines represent data point density and smoothing with cubic splines is displayed in black. c, Graphical representation of methylation at a non-CG methylation enriched gene, SPLICING FACTOR 1. d, Average relative methylation densities in each sequence context within gene bodies on the sense or anti-sense strand relative to gene directionality. P-values for differences between sense and antisense densities are indicated. Boxes in panels d and e represent the quartiles and whiskers mark the minimum and maximum values. e, Number of mRNA intronic reads in all genes or genes associated with non-CG enriched regions, in H1 and IMR90. P-values for differences between H1 and IMR90 reads are indicated. f, Logo plots of the sequences proximal to sites of non-CG DNA methylation in each sequence context in H1 cells. g, h, Prevalence of mCHG/mCHH sites (y-axis) as a function of the number of bases between adjacent mCHG/mCHH sites (x-axis) based on all non-redundant pair-wise distances up to 50 nt in all introns. Blue line represents smoothing with cubic splines.
We identified 447 and 226 genes that were proximal to genomic regions highly enriched for mCHG and mCHH, respectively, with 180 genes in common. An example of non-CG methylation enrichment in such a gene, Splicing Factor 1, is shown in Fig. 3c. Analysis of gene ontology terms for each set revealed significant enrichment for genes involved in RNA processing, RNA splicing, and RNA metabolic processes (P 2 × 10−11, Supplementary Fig. 7b). Unexpectedly, we found a significant enrichment of non-CG methylation on the anti-sense strand of gene bodies, for both mCHG and mCHH enriched sets of genes (P < 0.1 and P < 0.001, respectively, Fig. 3d). The anti-sense strand serves as the template for RNA polymerization, and further investigation will be required to determine if there are functional repercussions of this non-CG methylation strand bias. We also observed that genes in H1 had significantly more RNA originating from introns than in IMR90, relative to the total number of sequenced reads in each sample, and this discrepancy in intronic read abundance was significantly enhanced in the mCHG and mCHH enriched genes (P < 0.001, Fig. 3e). The higher abundance of intronic reads was associated with higher non-CG methylation within gene bodies, rather than differential non-CG methylation of exons versus introns.
In the Arabidopsis genome, the methylation state of a cytosine in the CG and CHG contexts is highly correlated with the methylation of the cytosine on the opposite strand within the symmetrical site15,16. While we observed that 99% of mCG sites from the human cell lines were methylated on both strands, surprisingly mCHG was highly asymmetrical, with 98% of mCHG sites being methylated on only one strand. This raises an interesting question as to how these sites of DNA methylation are consistently methylated in a considerable fraction of the genomes without two hemi-methylated CHG sites as templates for faithful propagation of the methylation state (Fig. 1c). It is not yet known whether continual, but indiscriminate, de novo methyltransferase activity preferentially methylates particular CHG sites after replication, or if a persistent targeting signal is present that drives CHG methylation.
We analyzed the genome sequence proximal to sites of non-CG methylation to determine whether enrichment of particular local sequences were evident, as previously reported in the Arabidopsis DNA methylomes15,16. Whereas no local sequence enrichment was observed for mCG sites, a preference for the TA dinucleotide upstream of non-CG methylation was observed (Fig. 3f and Supplementary Fig. 8). Furthermore, the base following a non-CG methylcytosine was most commonly an A, with a T also observed relatively frequently, a sequence preference observed in previous in vitro studies of the mammalian DNMT3 methyltransferases21,22.
To determine whether there was any preference for the distance between adjacent sites of DNA methylation in the human genome, we analyzed the relative distance between methylcytosines in each context within 50 nucleotides in introns. We focused on introns because these are genomic regions enriched in non-CG methylation, but unlike exons, are not constrained by protein coding selective pressures (Fig. 3g,h). Analyses for random genomic sequences and exons are presented in Supplementary Fig. 9, together with mCG spacing patterns. For methylcytosines in all contexts, a periodicity of 8–10 bases was evident (Fig. 3g, h and Supplementary Fig. 9), but interestingly a strong tendency was observed for two pairs of 8-base separated mCHG sites spaced with 13 bases between them. An 8–10 base periodicity was also evident for mCHH sites, corresponding to a single turn of the DNA helix, as previously observed in the Arabidopsis genome16, indicating that the molecular mechanisms governing de novo methylation at CHH sites may be common between the plant and animal kingdoms. A structural study of the mammalian de novo methyltransferase DNMT3A and its partner protein DNMT3L found that 2 copies of each form a heterotetramer that contains two active sites separated by the length of 8–10 nucleotides in a DNA helix26,27. The consistent 8–10 nucleotide spacing we observed in the human genome suggests that DNMT3A may be responsible for catalysing the methylation at non-CG sites. Notably, the mCHG and mCHH relative spacing patterns were distinct, suggesting that this sub-categorization of the non-CG methylation is appropriate, and that distinct pathways may be responsible for the deposition of mCHG and mCHH in the human genome.
DNA methylation is depleted at DNA-protein interaction sites
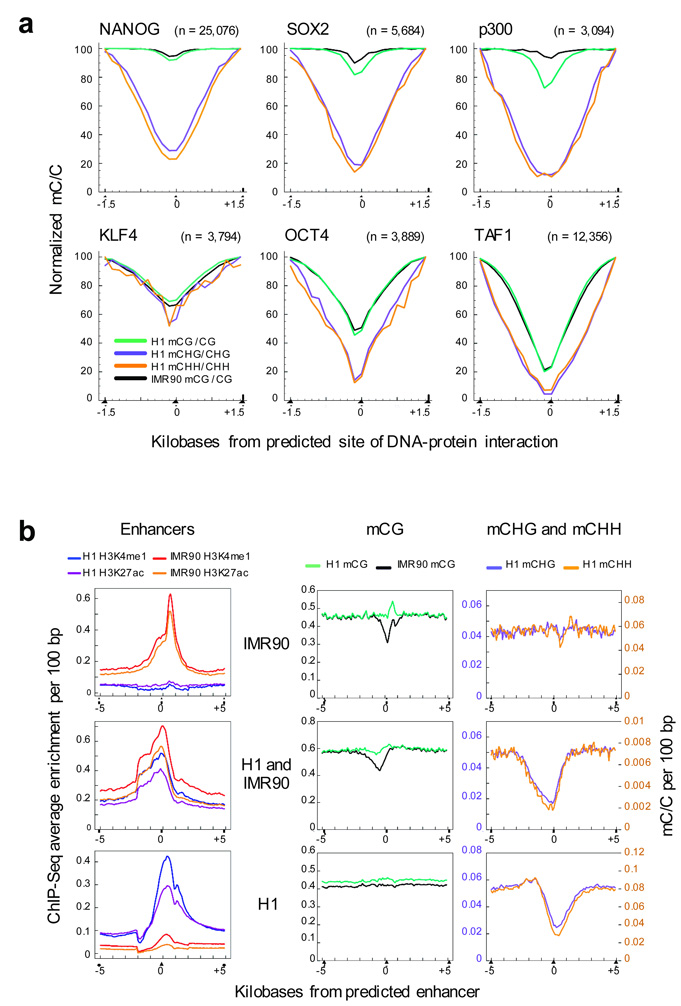
Numerous past studies have documented that DNA methylation can alter the ability of some DNA binding proteins to interact with their target sequences28–32. In order to further investigate this relationship we used ChIP-Seq33 to identify sites of protein-DNA interaction in H1 cells for a set of proteins important for gene expression in the pluripotent state, namely NANOG, SOX2, KLF4, and OCT4, as well as proteins involved in the transcription initiation complex and in enhancers (TAF1 and p300, respectively) (Supplementary Tables 3–8). In general we observed a decrease in the profile of relative methylation density toward the site of interaction, particularly in the non-CG context, independently from proximity to the TSS (Fig. 4a and Supplementary Fig. 10). The IMR90 genome showed higher average density of methylation at H1 SOX2 and p300 interaction sites, but had similar CG methylation densities for the H1 NANOG and OCT4 interaction sites, even though the genes encoding these proteins are transcribed at a very low level in IMR90 relative to H1 (47 – 50 fold less mRNA), and are not considered to be functional in fibroblasts. This suggests that these genomic regions are generally maintained in a less methylated state in multiple cell types regardless of the occupancy of these specific DNA binding proteins.
Figure 4. Density of DNA methylation at sites of DNA-protein interaction.
a, Average relative DNA methylation densities 1.5 kb up- and down-stream of predicted sites of DNA-protein interaction. b, Co-localization of H3K4me1 and H3K27ac ChIP-seq tag enrichment indicative of enhancer sites that have been grouped into three sets: specific to IMR90 cells (top), H1 cells (bottom), or common to both H1 and IMR90 cells (middle). Average relative DNA methylation densities in each sequence context in 100 bp windows are displayed throughout 5 kb up- and down-stream of the enhancers in each of the sets.
We next analyzed the patterns of DNA methylation in sets of enhancers either unique to each cell type or shared. ChIP-Seq was utilized to detect the location of enhancers throughout the H1 and IMR90 genomes, defined as regions of simultaneous enrichment of the histone modifications H3K4me1 and H3K27ac34 (Fig. 4b). We examined the average relative DNA methylation density at enhancer sites, as well as the flanking genomic regions, and found a depletion of CG methylation at IMR90-specific enhancers, yet enrichment in mCG density in H1 at the same genomic locations (Fig. 4b). In contrast, at H1-specific enhancers there was no change in mCG density in either the H1 or IMR90 genome, but non-CG methylation density decreased approximately 3-fold at the enhancer sites, relative to the density 5 kb up- and downstream. This is in agreement with the depletion of non-CG methylation in the H1 genome at predicted sites of p300 interaction (Fig. 4a), a strong indicator of enhancer activity34. The set of enhancer sites present in both H1 and IMR90 cells showed both of these cell-specific patterns: lower mCG density in IMR90 and lower non-CG methylation density in H1. The specific depletion of DNA methylation at active enhancers in each cell type (also recently reported on a limited basis35) indicates maintenance of these elements in an unmethylated state, potentially preventing interference in the process of protein-DNA interaction at these sites. Notably, H1 cells had depleted non-CG methylation but not mCG, in contrast to the mCG depletion at IMR90 enhancers. These data could indicate cell-type specific utilization of different categories of DNA methylation, possibly coupled with novel stem-cell specific factors that are able to recognize non-CG methylation, akin to the specific binding of the H3K9 histone methyltransferase KRYPTONITE to non-CG methylation sites in Arabidopsis 36.
Widespread cell-specific patterns of DNA methylation
The paradigm of DNA methylation controlling aspects of cellular differentiation necessitates that patterns of methylation vary in different cell types. Numerous studies have previously documented differences in DNA methylation between cell types and disease states7,8,10,37. With comprehensive maps of DNA methylation throughout the genomes of the two distinct cell types, we next characterized changes in DNA methylation evident between the H1 and IMR90 DNA methylomes, and explored how these changes may relate to the distinctiveness of these cells.
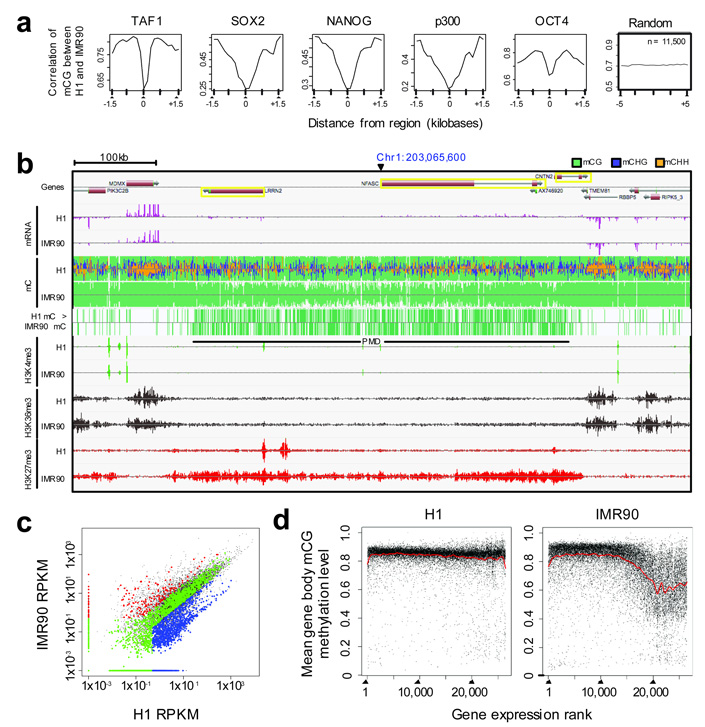
Pairwise comparison of methylation at the same genomic coordinates between H1 and IMR90 is required to reveal cell-specific methylation patterns potentially masked by average profiles. The Pearson correlation coefficient of the mCG methylation state between H1 and IMR90 was calculated for 20 equally sized windows flanking or within various genomic features (Fig. 5a and Supplementary Fig 11), providing a measure of methylation state conservation at these genomic features between the two cell types, and distinct from the average methylation density profiles presented above (Fig. 4). At the sites of protein-DNA interaction surveyed in Fig. 4a, we observed a decrease in the correlation of methylation compared to the flanking 1.5 kb of the genome (Fig. 5a), except for KLF4 (data not shown). This decrease was most pronounced at the predicted site of protein-DNA interaction, indicating that even though the mCG depletion was a general feature of the surveyed protein binding sites (Fig. 4a), when a pairwise comparison of the methylation status at each cytosine associated with the protein binding site between H1 and IMR90 was performed a significant decrease in the conservation of methylation was observed (Fig. 5a).
Figure 5. Cell-type variation in DNA methylation.
a, Pearson correlation coefficient of mCG methylation density (y-axis) between H1 and IMR90 at various genomic features. Regions were divided in 20 equally sized bins from 5' to 3'. Pearson correlation was determined in each bin considering all the H1 and IMR90 occurrences of the given genomic region. b, DNA methylation, mRNA, and histone modifications in H1 and IMR90 associated with a PMD. Vertical lines above and below the dotted central line in DNA methylation tracks indicate methylcytosines on the Watson and Crick strands, respectively. Line vertical height indicates the methylation level. The H1 > IMR90 mC track indicates methylcytosines significantly more methylated in H1 than IMR90 at a 5% FDR (FET). Vertical bars in the mRNA and histone modification tracks represent sequence tag enrichment. A yellow box indicates any gene with ≥30 fold higher mRNA abundance in H1 than IMR90. c, Comparison of transcript abundance between H1 and IMR90 of genes with a transcriptional start site located in or within 10 kb of a PMD. Black dots indicate all genes in the genome; blue, red and green indicate PMD genes expressed ≥3-fold higher in H1, IMR90 or not differentially expressed, respectively. d Mean gene body mCG methylation (at least 10 reads required) as a function of the gene expression rank, 1 being the most expressed. Abbreviations: mC, methylcytosine. PMD, partially methylated domain.
Surprisingly, we found that a large proportion of the IMR90 genome displayed lower levels of CG methylation than H1 (Fig. 1c). Contiguous regions with an average methylation level less than 70% were identified (mean length = 153 kb), which we termed partially methylated domains (PMDs) (Fig. 5b, Supplementary Fig. 12 and Supplementary Table 9). The PMDs comprised a large proportion of every autosome (average = 38.4%), and 80% of the IMR90 × chromosome (Supplementary Fig. 12), consistent with the lower levels of DNA methylation reported in the inactive × chromosome38. As IMR90 cells are derived from a female (XX), it is anticipated that simultaneous sequencing of BS-converted genomic DNA from both the inactive and the active × chromosomes will manifest as PMDs throughout the majority of the × chromosome. However, the widespread prevalence of PMDs on the autosomes was unexpected. We analyzed the ratio of methylated to unmethylated CG sites within individual MethylC-Seq reads. The IMR90 reads located within PMDs were more frequently partially- or un-methylated compared to all IMR90 reads aligned to the autosomes (Supplementary Fig. 12b). The decrease in PMD methylation manifested similarly in IMR90 autosomes and chromosome ×, however currently we cannot determine whether common pathways are responsible for altering methylation patterns in all chromosomes.
Upon inspection of 5,644 genes with a TSS located in or within 10 kb of a PMD, we found a strong enrichment for these genes to be less expressed in IMR90 (P = 2 × 10−47, Fisher’s Exact Test, FET). Specifically, of all of the genes that were more highly expressed in H1 (H1 ≥ 3 × IMR90 transcript abundance), 42% were located within PMDs, compared to only 13% of all more highly expressed genes in IMR90 cells being located in PMDs (Fig. 5b,c, Supplementary Tables 10,11). Many of the partially methylated and down-regulated genes in IMR90 displayed lower proximal H3K4me3 and H3K36me3 modifications, and higher proximal H3K27me3 levels (Fig. 5b, Supplementary Fig. 13, and Hawkins et al., submitted). While in IMR90 cells we observed a positive correlation between the mean gene body mCG methylation level and gene expression, no such relationship was discernible in H1 cells (Fig. 5d). Consequently, the positive correlation between gene expression and gene body methylation recently reported12 could be re-interpreted as a depletion of methylation in repressed genes in differentiated cells.
Epigenetic and transcriptional changes associated with stem cell hypomethylation
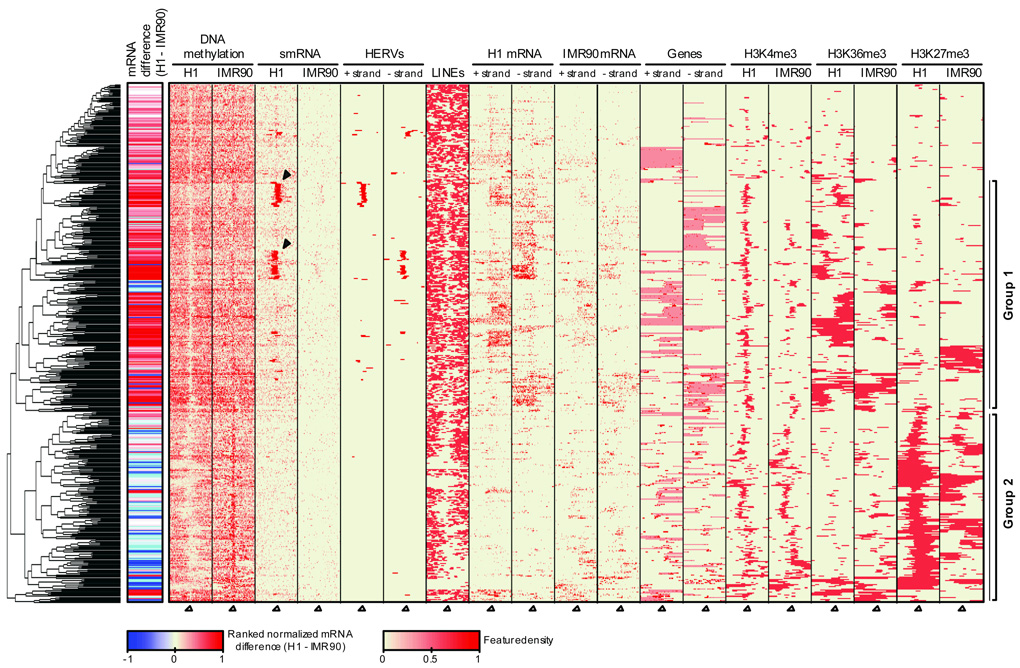
A sliding window approach was used to identify differentially methylated regions (DMRs) enriched for cytosines where IMR90 was more highly methylated than H1 (5% FDR, FET, Supplementary Fig. 14). We identified 491 DMRs, and in a window spanning 20 kb up- and down-stream of each DMR we surveyed mCG density, mRNAs, smRNAs, H3K4me3, H3K36me3, H3K27me3, genes, and repetitive elements (Fig. 6, Supplementary Table 12, and Hawkins et al., submitted). The DMRs were associated with 139 and 113 genes more highly expressed in H1 and IMR90, respectively. More than half of these genes were associated with DMRs located within 2 kb upstream of the TSS or the 5’ UTR, which include factors previously defined as playing a role in embryonic stem cell function39 (Supplementary Fig. 15 and Supplementary Tables 13,14).
Figure 6. Clustering of genomic, epigenetic and transcriptional features at differentially methylated regions.
The density of DNA methylation, smRNA reads, strand-specific mRNA reads, and the presence of domains of H3K4me3, H3k36me3 and H3K27me3 in H1 and IMR90 was profiled through 20 kb up- and down-stream of each of the 491 DMRs where DNA methylation was more prevalent in IMR90 than H1. Open triangles indicate the central point in each window. The side colorbar indicates the difference between H1 and IMR90 mRNA levels. The location of HERVs, LINEs, and genes are displayed on each strand, where pink coloring indicates the gene body and dark red boxes represent exons. Black triangles indicate regions enriched for smRNAs that are coincident with HERVs. Group 1 and 2 are discussed in the text. Abbreviations: DMRs, Differentially Methylated Regions. HERVs, Human Endogenous Retroviruses.
Complete linkage hierarchical clustering of these data revealed two broad categories of transcriptional activity, histone modifications and DNA methylation proximal to the DMRs (Fig. 6). Group 1 DMRs are associated with high proximal H3K4me3, H3K36me3, and transcriptional activity relative to IMR90, and are unmarked by H3K27me3 in both cell types. Although we did not observe widespread association of small RNA molecules with enrichment of DNA methylation, we found that a subset of Group 1 DMRs co-localize with dense clusters of small RNAs that map to annotated Human Endogenous Retroviruses (HERVs)40. Notably, the HERVs were less densely methylated in H1 and frequently associated with high downstream transcriptional activity, in contrast to the more methylated state in IMR90 that was not associated with abundant small RNAs and showed little proximal transcription (Fig. 6 and Supplementary Fig. 16). Accurate targeting of DNA methylation by small RNAs is a well-established process in plants41. While our data did not provide evidence for the existence of an analogous process in the human cells, further experiments may be required to investigate this relationship in greater detail, such as DNA methylation profiling following silencing of components of the RNAi machinery.
Group 2 DMRs were associated with gene-rich sequences that were more highly expressed in IMR90 cells and generally displaed a depletion of LINEs in the flanking sequence, with concomitant H3K27me3 modification and less DNA methylation, as observed in many IMR90 PMDs. Furthermore, Group 2 regions in H1 frequently displayed both H3K4me3 and H3K27me3 modifications, characteristic of the bivalent state that is thought to instil a suppressed but poised transcriptional status42,43. Many of these regions showed markedly less H3K27me3 in IMR90 in addition to more DNA methylation, suggesting that prior repression may have been relieved, and defining a set of genes potentially regulated by DNA methylation and involved in the developmental transition from a pluripotent to differentiated state.
Concluding Remarks
We found extensive differences between the DNA methylomes of two human cell types, revealing the highly dynamic nature of this epigenetic modification. The genomic context of the DNA methylation is resolved, here revealing abundant methylation in the non-CG context, which is typically overlooked in alternative methodologies. Profiling of enhancers and different patterning of CG and non-CG methylation in gene bodies and their different correlation with gene expression suggest possible alternative roles for DNA methylation in these two contexts. The exclusivity of non-CG methylation in stem cells, likely maintained by continual de novo methyltransferase activity and not observed in differentiated cells, suggests that it may play a key role in the origin and maintenance of this pluripotent state. Essential future studies will need to explore the prevalence of non-CG methylation in diverse cell types, including variation throughout differentiation and its potential reestablishment in induced pluripotent states.
Methods Summary
Biological materials and sequencing libraries
Human H1, H9, BMP4-induced H1, and IMR90 cells were cultured as described previously34,44,45. smRNA-seq libraries were generated from 10 – 50 nt small RNAs using the Small RNA Sample Prep v1.5 kit (Illumina, San Diego, CA), as per manufacturer’s instructions. Strand-specific mRNA-seq libraries were produced using a modification of a protocol described previously15. Unique 5’ and 3’ RNA oligonucleotides were sequentially ligated to the ends of fragments of RNA isolated by depletion of rRNA from total RNA samples. MethylC-seq libraries were generated by ligation of methylated sequencing adapters to fragmented genomic DNA followed by gel purification, sodium bisulfite conversion and 4 cycles of PCR amplification. ChIP-Seq libraries were prepared following Illumina protocols with minor modifications (See Supplementary Materials). Sequencing was performed using the Illumina Genome Analyzer II as per the manufacturer’s instructions.
Read processing and alignment
MethylC-seq sequencing data was processed using the Illumina analysis pipeline and FastQ format reads were aligned to the human reference genome (hg18) using the Bowtie alignment algorithm46. The base calls per reference position on each strand were used to identify methylated cytosines at 1% FDR. mRNA-seq reads were aligned to the human reference and splice junctions of UCSC Known Genes using the ELAND algorithm (Illumina, San Diego, CA). smRNA-seq reads that contained a subset of the 3’ adapter sequence were selected and this adapter sequence removed, retaining trimmed reads that were from 16 to 37 nt in length. These processed reads were aligned to the human reference genome (NCBI build 36/HG18) using the Bowtie alignment algorithm, and any read that aligned with no mismatches and to no more than 1000 locations in the reference genome was retained. Base calling, and mapping of Chip-Seq reads was performed using the Illumina pipeline.
Supplementary Material
Acknowledgements
We thank A. Elwell and A. Hernandez for assistance with sequence library preparation and Illumina sequencing. R.L. is supported by a Human Frontier Science Program Long-term Fellowship. R.D.H. is supported by an American Cancer Society Postdoctoral Fellowship. This work was supported by grants from the following: Mary K. Chapman Foundation, The National Institutes of Health (U01 ES017166 and U01 1U01ES017166-01), the California Institute for Regenerative Medicine (RS1-00292-1), the Australian Research Council Centre of Excellence Program (CE0561495, DP0771156) and Morgridge Institute for Research, Madison, Wisconsin. We thank the NIH Roadmap Reference Epigenome Consortium (http://nihroadmap.nih.gov/epigenomics/referenceepigenomeconsortium.asp) and C. Gunter (Hudson-Alpha Institute) for assistance.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions Experiments were designed by J.R.E., B.R., R.L., J.A.T. and R.D.H. Cells were grown by J.A.-B. and Q.-M.N. MethylC-Seq, RNA-Seq and smRNA-Seq experiments were conducted by R.L. and J.R.N. ChIP-Seq experiments were conducted by R.D.H., L.L. and Z.Y. ChIP-seq data analysis was performed by G.H., R.D.H. and L.E. BS-PCR validation was performed by R.H.D. Sequencing data processing was performed by R.L., J.T.-F., L.E., V.R. and G.H. Bioinformatic and statistical analyses were conducted by M.P., R.L., G.H., J.T.-F., R.H.D., R.S. and A.H.M. AnnoJ development was performed by J.T.F and A.H.M. The manuscript was prepared by R.L., M.P., R.H.D., A.H.M. and J.R.E.
Author Information Sequence data is available under the GEO accessions GSM429321-23, GSM432685-92, GSM438361-64, GSE17917 and GSE16256, and the SRA accessions SRX006782-89, SRX006239-41, SRX007165.1-68.1 and SRP000941. Analysed data sets can be obtained from http://neomorph.salk.edu/human_methylome. Reprints and permissions information is available at www.nature.com/reprints.
References
- 1.Holliday R, Pugh JE. DNA modification mechanisms and gene activity during development. Science. 1975;187:226–232. [PubMed] [Google Scholar]
- 2.Riggs AD. X inactivation, differentiation, and DNA methylation. Cytogenet Cell Genet. 1975;14:9–25. doi: 10.1159/000130315. [DOI] [PubMed] [Google Scholar]
- 3.Bestor TH. The DNA methyltransferases of mammals. Human Molecular Genetics. 2000;9:2395–2402. doi: 10.1093/hmg/9.16.2395. [DOI] [PubMed] [Google Scholar]
- 4.Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- 5.Lippman Z, et al. Role of transposable elements in heterochromatin and epigenetic control. Nature. 2004;430:471–476. doi: 10.1038/nature02651. [DOI] [PubMed] [Google Scholar]
- 6.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247–257. doi: 10.1016/s0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 7.Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007;447:425–432. doi: 10.1038/nature05918. [DOI] [PubMed] [Google Scholar]
- 8.Straussman R, et al. Developmental programming of CpG island methylation profiles in the human genome. Nat Struct Mol Biol. 2009;16:564–571. doi: 10.1038/nsmb.1594. [DOI] [PubMed] [Google Scholar]
- 9.Weber M, Schübeler D. Genomic patterns of DNA methylation: targets and function of an epigenetic mark. Curr Opin Cell Biol. 2007;19:273–280. doi: 10.1016/j.ceb.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 10.Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet. 2009;10:295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 11.Rauch TA, Wu X, Zhong X, Riggs AD, Pfeifer GP. A human B cell methylome at 100-base pair resolution. Proc. Natl. Acad. Sci. U.S.A. 2009;106:671–678. doi: 10.1073/pnas.0812399106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ball M, et al. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat Biotechnol. 2009;27:361–368. doi: 10.1038/nbt.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deng J, et al. Targeted bisulfite sequencing reveals changes in DNA methylation associated with nuclear reprogramming. Nat Biotechnol. 2009;27:353–360. doi: 10.1038/nbt.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Meissner A, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lister R, et al. Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell. 2008;133:523–536. doi: 10.1016/j.cell.2008.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cokus SJ, et al. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452:215–219. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thomson JA, et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 18.Nichols WW, et al. Characterization of a new human diploid cell strain, IMR-90. Science. 1977;196:60–63. doi: 10.1126/science.841339. [DOI] [PubMed] [Google Scholar]
- 19.Ramsahoye BH, et al. Non-CpG methylation is prevalent in embryonic stem cells and may be mediated by DNA methyltransferase 3a. Proc. Natl. Acad. Sci. U.S.A. 2000;97:5237–5242. doi: 10.1073/pnas.97.10.5237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Woodcock DM, Crowther PJ, Diver WP. The majority of methylated deoxycytidines in human DNA are not in the CpG dinucleotide. Biochem Biophys Res Commun. 1987;145:888–894. doi: 10.1016/0006-291x(87)91048-5. [DOI] [PubMed] [Google Scholar]
- 21.Aoki A, et al. Enzymatic properties of de novo-type mouse DNA (cytosine-5) methyltransferases. Nucleic Acids Research. 2001;29:3506–3512. doi: 10.1093/nar/29.17.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gowher H, Jeltsch A. Enzymatic properties of recombinant Dnmt3a DNA methyltransferase from mouse: the enzyme modifies DNA in a non-processive manner and also methylates non-CpG [correction of non-CpA] sites. Journal of Molecular Biology. 2001;309:1201–1208. doi: 10.1006/jmbi.2001.4710. [DOI] [PubMed] [Google Scholar]
- 23.Gonzalo S, et al. DNA methyltransferases control telomere length and telomere recombination in mammalian cells. Nat Cell Biol. 2006;8:416–424. doi: 10.1038/ncb1386. [DOI] [PubMed] [Google Scholar]
- 24.Steinert S, Shay JW, Wright WE. Modification of subtelomeric DNA. Molecular and Cellular Biology. 2004;24:4571–4580. doi: 10.1128/MCB.24.10.4571-4580.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brunner AL, et al. Distinct DNA methylation patterns characterize differentiated human embryonic stem cells and developing human fetal liver. Genome Res. 2009;19:1044–1056. doi: 10.1101/gr.088773.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ferguson-Smith AC, Greally J. Epigenetics: perceptive enzymes. Nature. 2007;449:148–149. doi: 10.1038/449148a. [DOI] [PubMed] [Google Scholar]
- 27.Jia D, Jurkowska RZ, Zhang X, Jeltsch A, Cheng X. Structure of Dnmt3a bound to Dnmt3L suggests a model for de novo DNA methylation. Nature. 2007;449:248–251. doi: 10.1038/nature06146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bell AC, Felsenfeld G. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000;405:482–485. doi: 10.1038/35013100. [DOI] [PubMed] [Google Scholar]
- 29.Clark SJ, Harrison J, Molloy PL. Sp1 binding is inhibited by (m)Cp(m)CpG methylation. Gene. 1997;195:67–71. doi: 10.1016/s0378-1119(97)00164-9. [DOI] [PubMed] [Google Scholar]
- 30.Hark AT, et al. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000;405:486–489. doi: 10.1038/35013106. [DOI] [PubMed] [Google Scholar]
- 31.Kitazawa S, Kitazawa R, Maeda S. Transcriptional regulation of rat cyclin D1 gene by CpG methylation status in promoter region. J. Biol. Chem. 1999;274:28787–28793. doi: 10.1074/jbc.274.40.28787. [DOI] [PubMed] [Google Scholar]
- 32.Mancini DN, Singh SM, Archer TK, Rodenhiser DI. Site-specific DNA methylation in the neurofibromatosis (NF1) promoter interferes with binding of CREB and SP1 transcription factors. Oncogene. 1999;18:4108–4119. doi: 10.1038/sj.onc.1202764. [DOI] [PubMed] [Google Scholar]
- 33.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 34.Heintzman N, et al. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature. 2009;459:108–112. doi: 10.1038/nature07829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schmidl C, et al. Lineage-specific DNA methylation in T cells correlates with histone methylation and enhancer activity. Genome Res. 2009:1–11. doi: 10.1101/gr.091470.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson LM, et al. The SRA Methyl-Cytosine-Binding Domain Links DNA and Histone Methylation. Curr. Biol. 2007;17:379–384. doi: 10.1016/j.cub.2007.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128:683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hellman A, Chess A. Gene body-specific methylation on the active × chromosome. Science. 2007;315:1141–1143. doi: 10.1126/science.1136352. [DOI] [PubMed] [Google Scholar]
- 39.Initiative ISC, et al. Characterization of human embryonic stem cell lines by the International Stem Cell Initiative. Nat Biotechnol. 2007;25:803–816. doi: 10.1038/nbt1318. [DOI] [PubMed] [Google Scholar]
- 40.Villesen P, Aagaard L, Wiuf C, Pedersen FS. Identification of endogenous retroviral reading frames in the human genome. Retrovirology. 2004;1:32. doi: 10.1186/1742-4690-1-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chan SW. Inputs and outputs for chromatin-targeted RNAi. Trends in Plant Science. 2008;13:383–389. doi: 10.1016/j.tplants.2008.05.001. [DOI] [PubMed] [Google Scholar]
- 42.Azuara V, et al. Chromatin signatures of pluripotent cell lines. Nat Cell Biol. 2006;8:532–538. doi: 10.1038/ncb1403. [DOI] [PubMed] [Google Scholar]
- 43.Bernstein BE, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006;125:315–326. doi: 10.1016/j.cell.2006.02.041. [DOI] [PubMed] [Google Scholar]
- 44.Ludwig T, et al. Feeder-independent culture of human embryonic stem cells. Nature Methods. 2006;3:637–646. doi: 10.1038/nmeth902. [DOI] [PubMed] [Google Scholar]
- 45.Ludwig T, et al. Derivation of human embryonic stem cells in defined conditions. Nat Biotechnol. 2006;24:185–187. doi: 10.1038/nbt1177. [DOI] [PubMed] [Google Scholar]
- 46.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.