Conspectus
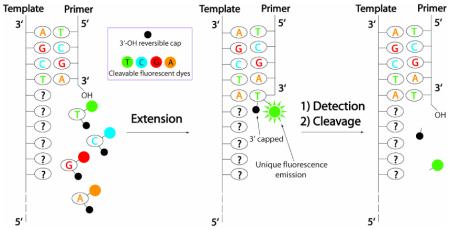 The Human Genome Project has concluded, but its successful completion has increased, rather than decreased, the need for high-throughput DNA sequencing technologies. The possibility of clinically screening a full genome for an individual's mutations offers tremendous benefits, both for pursuing personalized medicine as well as uncovering the genomic contributions to diseases. The Sanger sequencing method—although enormously productive for more than 30 years—requires an electrophoretic separation step that, unfortunately, remains a key technical obstacle for achieving economically acceptable full-genome results. Alternative sequencing approaches thus focus on innovations that can reduce costs.
The Human Genome Project has concluded, but its successful completion has increased, rather than decreased, the need for high-throughput DNA sequencing technologies. The possibility of clinically screening a full genome for an individual's mutations offers tremendous benefits, both for pursuing personalized medicine as well as uncovering the genomic contributions to diseases. The Sanger sequencing method—although enormously productive for more than 30 years—requires an electrophoretic separation step that, unfortunately, remains a key technical obstacle for achieving economically acceptable full-genome results. Alternative sequencing approaches thus focus on innovations that can reduce costs.
The DNA sequencing by synthesis (SBS) approach has shown great promise as a new sequencing platform, with particular progress reported recently. The general fluorescent SBS approach involves (i) incorporation of nucleotide analogs bearing fluorescent reporters, (ii) identification of the incorporated nucleotide by its fluorescent emissions, and (iii) cleavage of the fluorophore, along with the reinitiation of the polymerase reaction for continuing sequence determination. In this Account, we review the construction of a DNA-immobilized chip and the development of novel nucleotide reporters for the SBS sequencing platform.
Click chemistry, with its high selectivity and coupling efficiency, was explored for surface immobilization of DNA. The first generation (G-1) modified nucleotides for SBS feature a small chemical moiety capping the 3′-OH and a fluorophore tethered to the base through a chemically cleavable linker; the design ensures that the nucleotide reporters are good substrates for the polymerase. The 3′-capping moiety and the fluorophore on the DNA extension products, generated by the incorporation of the G-1 modified nucleotides, are cleaved simultaneously to reinitiate the polymerase reaction. The sequence of a DNA template immobilized on a surface via click chemistry is unambiguously identified with this chip-SBS system.
The second generation (G-2) SBS system was developed based on the concept that the closer the structures of the added nucleotide and the primer are to their natural counterparts, the more faithfully the polymerase would incorporate the nucleotide. In this approach, the polymerase reaction is performed with the combination of 3′-capped nucleotide reversible terminators (NRTs) and cleavable fluorescent dideoxynucleotides (ddNTPs). By sacrificing a small amount of the primers permanently terminated by ddNTPs, the majority of the primers extended by the reversible terminators are reverted to the natural ones after each sequencing cycle. We have also developed the 3′-capped nucleotide reversible terminators to solve the problem of deciphering the homopolymeric regions of the template in conventional pyrosequencing. The 3′-capping moiety on the DNA extension product temporarily terminates the polymerase reaction, which allows only one nucleotide to be incorporated during each sequencing cycle. Thus, the number of nucleotides in the homopolymeric regions are unambiguously determined using the 3′-capped NRTs.
It has been established that millions of DNA templates can be immobilized on a chip surface through a variety of approaches. Therefore, the integration of these high-density DNA chips with the molecular-level SBS approaches described in this Account is expected to generate a high-throughput and accurate DNA sequencing system with wide applications in biological research and health care.
Introduction
The completion of the Human Genome Project set the stage for a drive to routinely and economically screen genetic mutations to identify disease genes on a genome-wide scale.1 The decreased cost of sequencing, with the “$1000 genome analysis” as a holy grail, is critical to the ultimate goals of personalized medicine based on genetic and genomic information. Accuracy, speed, and size of the instrument are among the vital considerations for the cost effective development of new DNA analysis methods that can be implemented directly in the hospital and clinical settings.
The Sanger dideoxy chain-termination method2 has been the gold standard in genome research for over three decades. Despite the success of Sanger sequencing, its requirement for electrophoresis to separate the DNA products has some limitations due to the difficulty in achieving high throughput and to the complexity involved in the automation. In order to overcome the limitations of the Sanger sequencing technology, a variety of new methods have been investigated. Such approaches include sequencing by hybridization,3 mass spectrometry sequencing,4-6 nanopores sequencing of single-stranded DNA,7 sequencing by ligation,8 and single molecule DNA sequencing.9,10
The massive parallel DNA sequencing technologies have advanced biomedical research in many aspects, including elucidation of DNA-protein interactions on a genome-wide scale,11-13 study of gene expression with single copy transcript sensitivity,14 and discovery of noncoding RNAs.15 Among these novel approaches for DNA sequencing, the sequencing by synthesis (SBS) approach, which will be the centerpiece of this account, has emerged as a viable candidate for massive parallel high throughput sequencing platform. SBS is a sequencing platform taking advantage of DNA polymerase reaction, a key process for DNA replication inside cells. The basic concept of SBS is to use DNA polymerase to extend a primer that is hybridized to a template by a single nucleotide, determine the identity of the nucleotide, and then proceed to incorporate, through the polymerase reaction, the next nucleotide, eventually reading out the entire DNA sequence serially. In contrast to Sanger sequencing, in which fluorescently labeled DNA fragments of different sizes are all generated in a single reaction and then separated and detected, SBS approaches have an advantage in that individual bases on a high density array are detected simultaneously without the need for separations. Furthermore, SBS methods can easily be scaled up over Sanger's dideoxy-sequencing techniques by using massive parallel microarray chip method. Currently, over tens of millions of DNA sample spots can be arrayed on a glass chip surface for sequencing.16
Consideration of the stringent requirements for employing SBS on a chip leads in turn to stringent design features of the chip and the SBS process. In this account, we will first describe the design and construction of DNA-immobilized chips for SBS and then detail the design and implementation of several generations of SBS technologies that have culminated in the current chip-SBS system. Because of its mild reaction conditions and high selectivity and efficiency, click chemistry17 has been explored for covalent immobilization of DNA fragments on a surface. By taking the surface immobilized DNA strands as primers, it has been established that millions of different DNA templates immobilized on a sequencing platform can be generated through emulsion PCR or clonal amplification.13,14 Therefore, further implementation of the SBS approaches discussed in this account should lead to a high-throughput DNA sequencing system with wide applications in genome biology and health care. This account mainly reviews the authors' contribution to sequencing by synthesis. Readers are referred to other excellent reports concerning the solid phase DNA immobilization and amplification18,19 and the development of the sequencing chemistry.20-24 We hope to demonstrate that the challenges of design and development of a SBS method on a chip have led to a fruitful marriage integrating the basic science of chemical biology and the current and developing technologies involving microanalysis and informatics.
Design and Exemplar of a Chip for SBS
“Chip” for SBS is a multiplex microarray built on a flat surface of glass or some other suitable material. Each spot will contain a specific DNA target sequence that has been obtained from the fragmented genome whose overall sequence is to be determined. The surface properties of chips, which may possess massive dimensionalities of data, are designed and engineered to obtain high selectivity in the faithfulness of identification of the correctly added probe and high sensitivity with respect to signal to noise features.
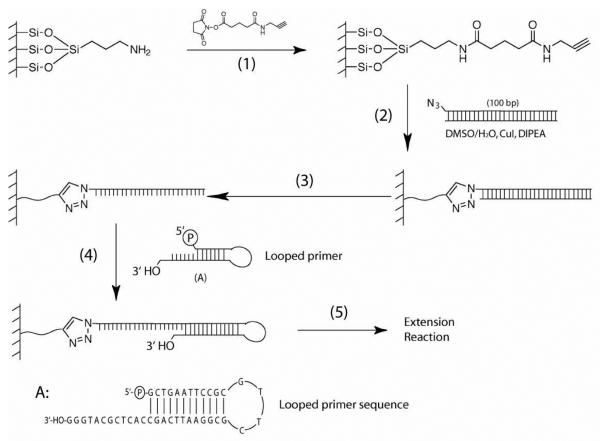
Figure 1 shows a general procedure for DNA chip construction. The target sequences are covalently bonded to the chip to insure that they survive and remain attached to the chip through multiple chemical procedures involved in the SBS process. We selected click chemistry involving the [2 + 3] cycloaddition of a terminal acetylene to an azide as a means of covalently attaching target fragments to the glass surface.25,26 The process involves several steps, the first being the modification of an available chip surface and then followed by attachment of the target sequence via click chemistry.
Figure 1.
DNA immobilization on a surface with click chemistry. An amino modified glass surface isreacted with a linker containing an NHS-ester on one end and an alkyne on the other to functionalizethe surface (step 1). Azide labeled PCR product is then attached to the surface using click chemistry(step 2). The unattached DNA strand is removed under alkaline condition (step 3). A loop DNA primeris ligated to the single-stranded DNA template (step 4). DNA extension reaction is carried out toidentify the sequence of the template (step 5). The sequence of the loop primer is shown in (A).
Construction of DNA chips starts with commercially available glass slides with amino-functionalized surfaces (Figure 1). In step (1), the acetylene partner of the click reaction is first attached to the surface. This effectively converts the surface to one that is rich in the acetylene functionality and ready for reactions with the azide functionality upon contact. In step (2), the PCR products labeled with the azide functionality are then covalently attached to the surface with click chemistry. In step (3), the surface-bound double stranded PCR products were denatured under alkaline conditions to remove the complementary strand without the azide group, generating a single-stranded DNA on the surface. In step (4), in order to prevent the dissociation of primers from templates during SBS, a looped primer is covalently ligated to the 3′ end of the DNA template. In step (5), the sequence of the PCR products is deciphered by performing SBS on the looped DNA strand.
SBS on a Chip: Design and Synthesis of Novel Reporter Nucleotides
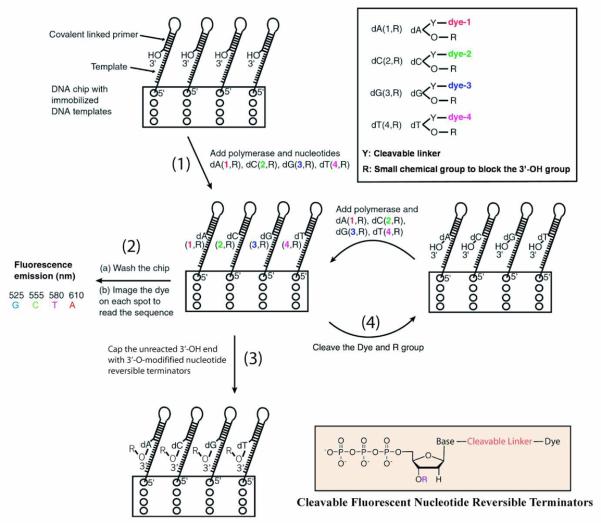
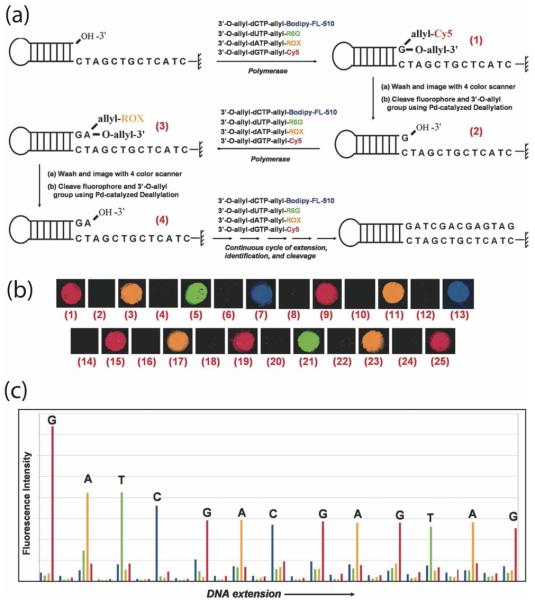
Figure 2 summarizes the general strategy for the design of a SBS approach on a chip.27 As shown in Figure 1, the chip is constructed by immobilizing DNA template fragments that are capable of self-priming to initiate the polymerase incorporation of the first complementary nucleotide after the last base on the primer. The first generation (G-1) SBS chip system (Figure 2) developed in our laboratory possesses the following general features. In step (1), the functionalized chip prepared as shown in Figure 1 is treated with the four modified nucleotide analogues of A, C, G and T (actual examples of structures of the modified nucleotides are shown in Chart 2). Each of the nucleotide analogues possesses a fluorescence label that is characteristic of the base and a small 3′-OH capping group. Upon adding DNA polymerase, the modified nucleotide that is complementary to the next nucleotide on the template is incorporated on each spot of the chip. In step (2), the excess reagents are then removed along with any unincorporated nucleotide analogues and the chip is analyzed by a 4-color imager to determine the unique fluorescence emission of each base, revealing the identity of the incorporated nucleotide for each template on the chip. In order to minimize any lagging fluorescence signal caused by previously un-extended strands, step (3) is carried out to synchronize the incorporation by using excessive amount of 3′-OH capped nucleotide analogues along with the DNA polymerase to extend any remaining priming strands that retain a free 3′-OH group. In step (4), the fluorophore and the 3′-OH capping moiety are removed simultaneously for each of the templates and the system is ready for a repetition of steps (1), (2), (3), and (4) to continue the sequencing.
Figure 2.
In the SBS approach, a chip is constructed with immobilized DNA templates that are able to self-primefor initiating the polymerase reaction. Four nucleotide analogues are designed such that each is labeled with aunique fluorescent dye on the specific location of the base, and a small chemical group (R) to cap the 3′-OH group. Upon adding the four nucleotide analogues and DNA polymerase, only the nucleotide analogue complementary tothe next nucleotide on the template is incorporated by polymerase on each spot of the chip (step 1). After removingthe excess reagents and washing away any unincorporated nucleotide analogues, a 4 color fluorescence imager isused to image the surface of the chip, and the unique fluorescence emission from the specific dye on the nucleotideanalogues on each spot of the chip will yield the identity of the nucleotide (step 2). After imaging, the small amountof unreacted 3′-OH group on the self-primed template moiety will be capped by excess 3′-O-modified nucleotidereversible terminators and DNA polymerase to avoid interference with the next round of synthesis forsynchronization (step 3). The dye moiety and the R protecting group will be removed to generate a free 3′-OH groupwith high yield (step 4). The self-primed DNA moiety on the chip at this stage is ready for the next cycle of thereaction by repetition of steps 1, 2, 3, and 4 to identify the next nucleotide sequence of the template DNA.
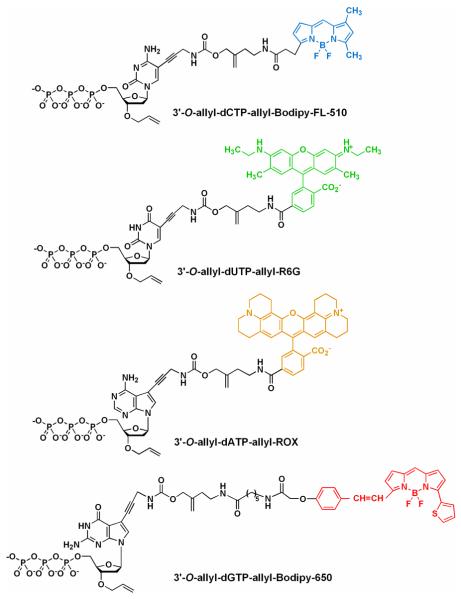
Chart 2.
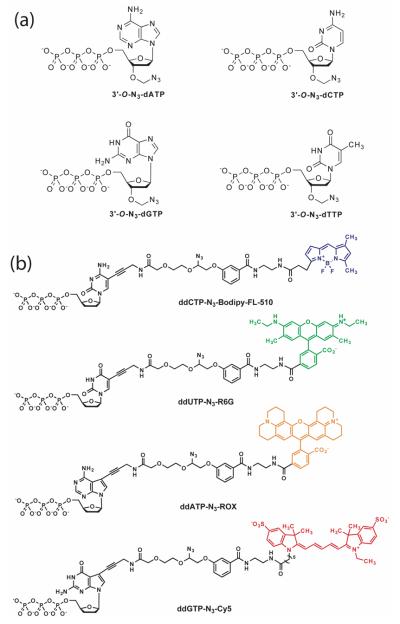
Structures of an example of G-1 modified nucleotides (3′-O-ally-dNTP-allyl-fluorophores), with the 4 fluorophores having distinct fluorescent emissions: 3′-O-allyl-dCTPallyl-bodipy-FL-510 [λabs(max) = 502 nm; λem(max) = 510 nm], 3′-O-allyl-dUTP-allyl-R6G [λabs(max) = 525 nm; λem(max) = 550 nm], 3′-O-allyl-dATP-allyl-ROX [λabs(max) = 585 nm; λem(max)= 602 nm], and 3′-O-allyl-dGTP-allyl-bodipy-650 [λabs(max) = 649 nm; λem(max) = 670 nm].
Design and optimization of the structural features of the modified nucleotide through examination of the 3-D structure of the active site of a DNA polymerase complex
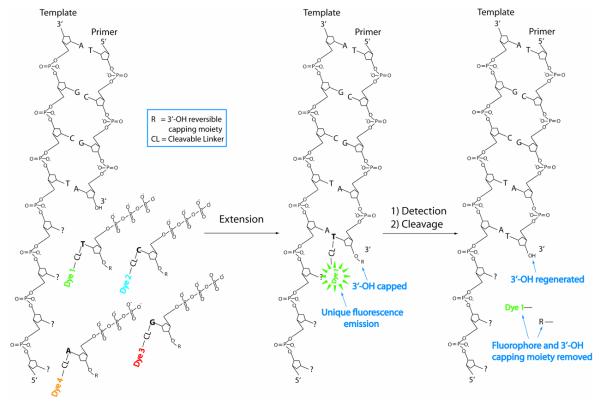
One of the most challenging tasks for developing a successful SBS platform is the design of modified nucleotides that can be faithfully incorporated by polymerase. In order to design the first generation cleavable fluorescent nucleotide reversible terminators (CF-NRTs) used in the SBS extension reaction, we examined the 3-D structure of the polymerase complex with a DNA template, a primer and an incoming nucleotide (ddCTP) in polymerase reaction.28 Inspection of the structure indicated that the 5-position of the cytosine points away from the catalytic pocket of the enzyme into a region with considerable open space; on the other hand, the 3′-position of the ribose ring in ddCTP occupied a very crowded space near the active amino acid residues of the polymerase. With this model as a guide, we concluded that any group that is attached at the 3′ position of the sugar must be as small as possible or there would be a large steric hindrance to the polymerase activity. It is also known that some modified DNA polymerase are highly tolerable for nucleotides with extensive modifications with bulky groups at the 5-position of the pyrimidines (T and C) and 7-position of purines (G and A).29, 30 Thus we decided to use these features to design modified nucleotide for which a unique fluorophore is attached to the 5-position of the pyrimidines (T and C) and the 7-position of purines (G and A) through a cleavable linker, and a small chemical moiety is used to cap the 3′-OH group. The general SBS approach using modified nucleotides with the described features is shown in Figure 3. After the incorporation of a fluorescently labeled nucleotide, which is complementary to the next available base on the DNA template, the fluorescent signal is detected to determine the identity of the incorporated nucleotide. Afterwards, the fluorophore and the 3′ capping moiety are efficiently removed in a manner compatible with DNA stability and function for subsequent sequence determination.
Figure 3.
DNA sequencing by synthesis with cleavable fluorescent nucleotide reversible terminators(CF-NRTs). DNA polymerase catalyzes the incorporation of a CF-NRT, which is complementary tothe next base on the unknown DNA template. After removing unincorporated nucleotide analoguesand other excess reagents, a fluorescence imager is used to identify the base just incorporated. Thenthe fluorophore and the 3′ capping moiety are chemically cleaved to reinitiate the DNA polymerasereaction for subsequent sequence determination.
In summary, the key structural features of the modified nucleotides are: (1) 4 distinct fluorophores (FA, FT, FG, FC) attached to the 5-position of pyrimidines and the 7-position of purines that can be removed after sequence identification; and (2) a small reversible capping moiety for the 3′-OH group that can be removed after incorporation and identification of sequence. These structural design features have proven successful for the incorporation of modified nucleotides in the growing DNA strand in the SBS processes to be described below. The reversible termination group and the fluorophore can be removed by chemical reagents or photons.31, 32, 33
First generation design of modified nucleotides for SBS
Chart 1 shows a cartoon of the first generation (G-1) modified nucleotides (CF-NRTs) for SBS based on the above analysis. A specific example of G-1 modified nucleotides containing a 3′-O-allyl group and a unique fluorophore tethered by a cleavable allyl linker for SBS is shown in Chart 2.31 To establish proof of principle, we first demonstrated that these modified nucleotides were successful substrates for DNA polymerase in a solution phase DNA extension reaction and that the fluorophore and the 3′ reversible termination groups could be removed with high speed and efficiency. We then performed SBS using four CF-NRTs to identify the sequence of a DNA template immobilized on a chip via click chemistry.
Chart 1.
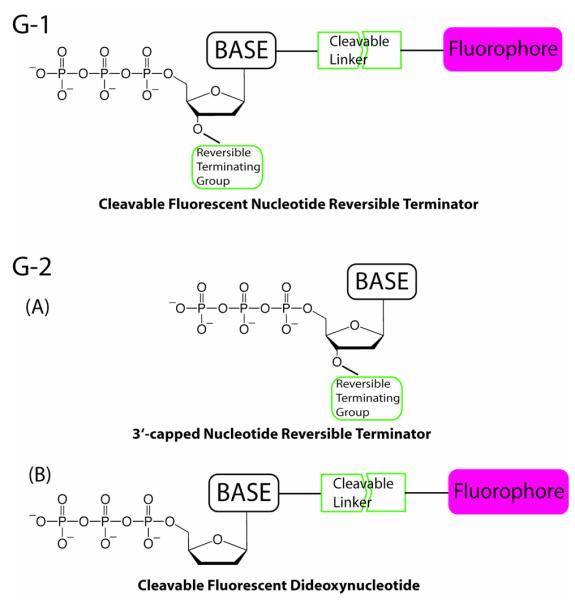
Cartoon structures of G-1 and G-2 modified nucleotides for SBS.
A schematic and example of a first generation SBS system on a chip is shown in Figure 4. Figure 4a shows a schematic of the operation of the SBS through several cycles with the first step incorporating a G nucleotide, then washing and removal of all fluorescence materials and the 3′-capped group via a Pd-catalyzed deallylation reaction,31 followed by the second step incorporating an A nucleotide, etc. In Figure 4b, the observed experimental fluorescence color changes are shown as a function of incorporation, sequential cleavages and incorporations. Finally, the data is digitized for sequence display and is available for informatics analysis (Figure 4c).31 We note that there is a degradation of the signal to noise with respect to the distinction of the added base. This degradation could result from a number of factors such as less than 100% incorporation of the fluorescently labeled nucleotides by the polymerase, the non-natural chemical groups left on the synthesized strand, and incomplete removal of the residual modified nucleotides, etc.
Figure 4.
(a) Reaction scheme of G-1 SBS on a chip using 3′-O-ally-dNTP-allyl-fluorophores. (b)The scanned four-color fluorescence images for each step of SBS on a chip: (1) incorporation of 3′-O-allyl-dGTP-allyl-Cy5; (2) cleavage of allyl-Cy5 and 3′-allyl group; (3) incorporation of 3′-O-allyldATP-allyl-ROX; (4) cleavage of allyl-ROX and 3′-allyl group; (5) incorporation of 3′-O-allyldUTP-allyl-R6G; (6) cleavage of allyl-R6G and 3′-allyl group; (7) incorporation of 3′-O-allyl-dCTPallyl-bodipy-FL-510; (8) cleavage of allyl-bodipy-FL-510 and 3′-allyl group; images 9-25 aresimilarly produced. (c) A plot (four-color sequencing data) of raw fluorescence emission intensity atthe four designated emission wavelength of the four chemically cleavable fluorescent nucleotides vs. the progress of sequencing extension.
Second generation design of modified nucleotides for SBS
In order to increase the number of nucleotides that could be sequenced, a new strategy for SBS was designed.32 It was apparent that higher incorporation efficiency of the modified nucleotides and lower polymerase complex hindrance could drastically improve the read length of SBS. The incorporation of the G-1 modified nucleotides extends the DNA strand with a propargyl amino group modification on the base during SBS,31 which might interfere with the activity of the polymerase for the DNA chain elongation. To eliminate this chemical modification on the base, we have developed the second generation (G-2) modified nucleotides for the SBS system.32 In this method, the nucleotide reversible terminators with a chemical moiety capping the 3′-OH, are combined with cleavable fluorescent dideoxynucleotides to extend the DNA strand during SBS (Chart 1, G-2). DNA sequences are determined by the unique fluorescence emission of each fluorophore on the DNA products terminated by ddNTPs. Upon removing the 3′-OH capping group from the incorporated nucleotide reversible terminators and the fluorophore from the DNA products terminated by ddNTPs, the polymerase reaction reinitiates to continue the sequence determination. The DNA products extended by cleavable fluorescent dideoxynucleotides, after fluorescence detection and cleavage, are no longer involved in the subsequent polymerase reaction cycles because they are permanently terminated. The further polymerase reaction only occurs on DNA strands that incorporate the nucleotide reversible terminators, which subsequently are reverted to natural nucleotides upon cleavage of the 3′-OH capping group. Therefore, during the whole SBS process, the polymerase reaction is only carried out on the priming strands consisting of natural nucleotides, which should have no deleterious effect on the polymerase binding to faithfully incorporate the next modified nucleotides.
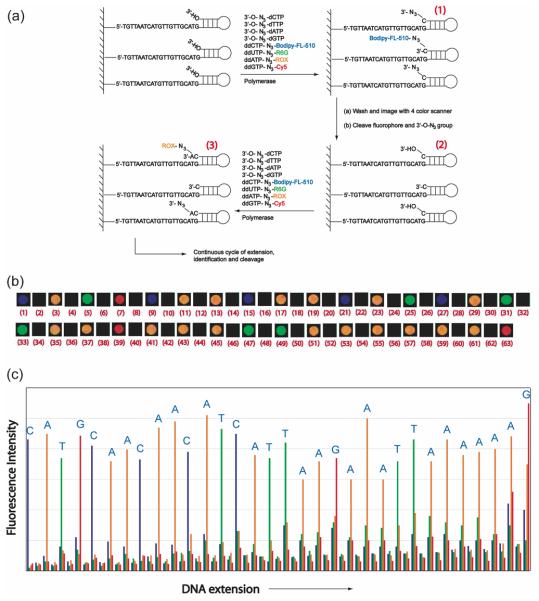
A specific example of the G-2 modified nucleotides is shown in Chart 3. Four 3′-O-azidomethyl-modified nucleotide analogues (3′-O-N3-dNTPs) and four cleavable fluorescent dideoxynucleotides (ddNTP-N3-fluorophores) have been designed and synthesized to demonstrate the proof of concept. In this approach, the identity of the incorporated nucleotide is determined by the unique fluorescence emission from the four cleavable fluorescent ddNTPs, whereas the role of the 3′-O-modified nucleotide reversible terminators is to further extend the DNA strand. With a finite amount of immobilized DNA template on a solid surface, initially the majority of the priming strands should be extended with 3′-O-N3-dNTPs, while a relatively smaller amount should be extended with ddNTP-N3-fluorophores. As the sequencing cycle continues, the amount of the cleavable fluorescent ddNTPs needs to be gradually increased to maintain sufficient fluorescent signals that are above the fluorescence detection system's threshold for sequence determination. Following these guidelines, we performed SBS on a chip-immobilized DNA template (Figure 5). The scheme of this sequencing methodology is shown in Figure 5a. The de novo sequencing reactions were initiated by extending the self-priming DNA template using a polymerase reaction mixture containing four 3′-O-N3-dNTPs and four ddNTP-N3-fluorophores. To negate any lagging fluorescence signal caused by a previously unextended priming strand, a synchronization reaction mixture consisting of just the four 3′-O-N3-dNTPs and the DNA polymerase was used to extend any priming strands that retain a free 3′-OH group. After the removal of the fluorophores and the 3′-OH capping moieties via the Staudinger reaction by using aqueous Tris(2-carboxyethyl) phosphine (TCEP) solution,32 the DNA extension reactions were reinitiated to identify the next base on the template. Figure 5b shows the experimental fluorescence color changes during the sequencing process and Figure 5c shows the digitized sequencing data for further informatics analysis. The entire process of incorporation, synchronization, detection and cleavage was performed multiple times to identify 32 successive bases in the DNA template.
Chart 3.
Structures of an example of G-2 modified nucleotides. (a) Structures of the nucleotide reversibleterminators, 3′-O-N3-dATP, 3′-O-N3-dCTP, 3′-O-N3-dGTP, 3′-O-N3-dTTP. (b) Structures of cleavablefluorescent dideoxynucleotide terminators ddNTP-N3-fluorophores, with the 4 fluorophores having distinctfluorescent emissions: ddCTP-N3-Bodipy-FL-510 (λabs (max) = 502 nm; λem (max) = 510 nm), ddUTP-N3-R6G (λabs (max) = 525 nm; λem (max) = 550 nm), ddATP-N3-ROX (λabs (max) = 585 nm; λem (max) = 602 nm), and ddGTP-N3-Cy5 (λabs (max) = 649 nm; λem (max) = 670 nm).
Figure 5.
(a) G-2 SBS scheme for 4-color sequencing on a chip using the nucleotide reversible terminators(3′-O-N3-dNTPs) and cleavable fluorescent dideoxynucleotide terminators (ddNTP-N3-fluorophores). (b) The4-color fluorescence images for each step of SBS: (1) incorporation of 3′-O-N3-dCTP and ddCTP-N3-BodipyFL-510; (2) cleavage of N3-Bodipy-FL-510 and 3′-CH2N3 group; (3) incorporation of 3′-O-N3-dATP andddATP-N3-Rox; (4) cleavage of N3-ROX and 3′-CH2N3 group; (5) incorporation of 3′-O-N3-dTTP andddTTP-N3-R6G; (6) cleavage of N3-R6G and 3′-CH2N3 group; (7) incorporation of 3′-O-N3-dGTP andddGTP-N3-Cy5; (8) cleavage of N3-Cy5 and 3′-CH2N3 group; images 9-63 are similarly produced. (c) A plot(4-color sequencing data) of raw fluorescence emission intensity obtained using 3′-O-N3-dNTPs and ddNTPN3-fluorophores at the four designated emission wavelengths of the four cleavable fluorescentdideoxynucleotides.
3′-capped nucleotide reversible terminators for pyrosequencing
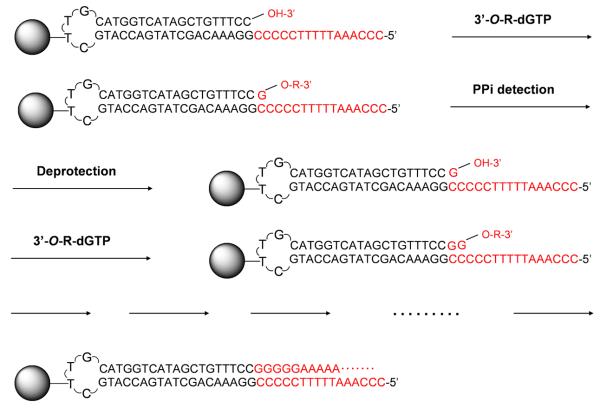
Instead of using fluorescently labeled nucleotide analogues, pyrosequencing, an alternative DNA sequencing by synthesis method, deciphers the sequence of DNA by performing the polymerase reaction with natural nucleotides.34 In this approach, each of the four natural nucleotides (A, C, G and T) is added sequentially during the DNA polymerase reaction. If the added nucleotide is complementary with the first available base on the template, the nucleotide will be incorporated, leading to the release of a pyrophosphate (PPi). The pyrophosphate is then used by ATP sulfurylase to convert adenosine 5′-phosphosulfate to ATP. The ATP acts as a cofactor for the luciferase-catalized conversion of luciferin to oxyluciferin, generating visible light that can be detected. However, conventional pyrosequencing has inherent difficulties to decipher homopolymeric regions of the DNA templates.35 The reason is that the light signal intensity is not exactly proportional to the amount of PPi released, especially when the homopolymeric region has more than five bases. We have solved this problem of determining the number of nucleotides in the homopolymeric regions by using the 3′-capped NRTs (G-2A, Chart 1).33 As shown in Figure 6, these nucleotide analogues are modified with a reversible chemical moiety capping the 3′-OH group to temporarily terminate the polymerase reaction. In this way, only one nucleotide is incorporated into the growing DNA strand even in homopolymeric regions. After sequence determination, the reversible capping moiety is removed to regenerate the 3′-OH of the extended primer, which is then ready for the next sequencing cycle. Figure 7 shows a scheme comparing sequencing data from our NRT pyrosequencing and conventional pyrosequencing. All bases in the homopolymeric regions are clearly identified with the NRTs, whereas the pyrosequencing data obtained by using natural nucleotides show large peaks corresponding to stretches of G and A bases, causing ambiguity for sequence identification.
Figure 6.
Reaction scheme of pyrosequencing using 3′-capped NRTs. The incorporation of G-2Amodified nucleotides (3′-O-R-dNTPs) releases a pyrophosphate (PPi), which triggers enzymaticreactions to produce the light signal for sequence detection. Upon the removal of the reversiblecapping moiety (deprotection), the 3′-OH of the extended primer is regenerated to reinitiate thepolymerase reaction. The pyrosequencing reaction is repeated continuously to identify the sequenceof the target DNA template.
Figure 7.
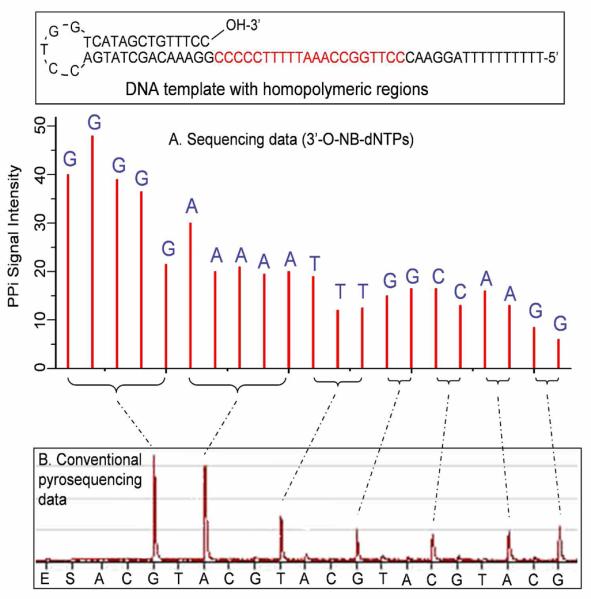
Comparison of reversible terminator pyrosequencing using 3′-O-(2-nitrobenzyl)-dNTPswith conventional pyrosequencing using natural nucleotides. (A) The self-priming DNA templatewith stretches of homopolymeric regions was sequenced by using 3′ capped NRTs [3′-O-(2-nitrobenzyl)-dNTPs]. The homopolymeric regions are clearly identified, with each peakcorresponding to the identity of each base in the DNA template. (B) Pyrosequencing data usingnatural nucleotides. The homopolymeric regions produced two large peaks corresponding to thestretches of G and A bases and five smaller peaks corresponding to stretches of T, G, C, A, and Gbases. However, it is very difficult to decipher the exact sequence from the data.
Summary
In this account, we have described some of the advances in making DNA sequencing by synthesis as a viable technology for genomic research. This includes the development of new surface attachment chemistries for DNA templates. Due to its high efficiency, selectivity and great biocompatibility, click chemistry has been explored to covalently immobilize DNA templates on surface for SBS. Several generations of modified nucleotide reporters have been designed and evaluated for SBS based on the structural-directed design rationale of nucleotide analogues that will cause minimum interference for polymerase recognition. The G-1 modified nucleotides feature a 3′-OH capping moiety and unique fluorophores tethered to the 5-position of pyrimidines and the 7-position of purines through a cleavable linker. These nucleotides have been demonstrated to be good substrates for the polymerase. The fluorophore and the 3′ capping moiety on the DNA extension products, generated by the incorporation of the G-1 modified nucleotides, are removed simultaneously after sequence determination. The regenerated 3′-OH allows the reinitiation of the polymerase reaction for subsequent sequence determination. This G-1 SBS system has successfully demonstrated its potential to sequence DNA templates immobilized on a surface on a massively parallel manner. Nonetheless, the chemical group leftover on the base during each SBS cycle may interfere with the efficiency and accuracy of the polymerase in the subsequent sequencing cycles. To address this issue, the G-2 SBS system has been developed by using 3′-OH capped NRTs combined with cleavable fluorescent dideoxynucleotides. Once the fluorescent dideoxynucleotides are incorporated into the growing DNA strand, the polymerase reaction is permanently terminated. Thus the continuous extension reactions occur only on DNA strands that incorporate the nucleotide reversible terminators, which subsequently are reverted to natural nucleotides upon the cleavage of the 3′-OH capping moiety.
The major advantage of fluorescent SBS approach described in this account compared to the Sanger sequencing method is the high throughput and simplicity of the SBS platform in which all sequencings are conducted on a chip without any separation steps. The ultimate read length of fluorescent SBS depends on three factors: the number of starting DNA molecules on each spot of a DNA chip, the nucleotide incorporation and cleavage efficiency, and the sensitivity of the detection system. The read length with the Sanger sequencing method commonly reaches more than 700 base pairs. The fluorescent SBS system has the potential to approach this read length, if stepwise yield can reach over 99% and the sensitivity of the fluorescent detection system can be further improved.
Conventional pyrosequencing technology carries out the polymerase reaction by using natural nucleotides. However, it has inherent difficulties to decipher the homopolymeric regions of the DNA template. We have used 3′ capped NRTs to address this problem. The 3′ capping moiety on the DNA extension product, generated by the incorporation of each individual NRT, temporarily terminates the polymerase reaction. With only one nucleotide incorporated during each SBS cycle, the numbers of nucleotides in homopolymeric regions of the template are unambiguously determined.
At present, three platforms are in widespread use for massive parallel DNA sequencing: the Roche/454 FLX Pyrosequencer,36 the Illumina Genome Analyzer,37 and the Applied Biosystems SOLiD™ System.38 The Roche/454 Pyrosequencer and the Applied Biosystems SOLiD sequencer use emulsion PCR for DNA template preparation,39 while the bridge PCR amplification is used by Illumina Genome Analyzer.37, 40 Recently, two platforms for single molecule DNA sequencing by synthesis have been reported. For Helicos Heliscope sequencer,9 single-molecule DNA template is directly immobilized on a solid surface to allow SBS to take place. Pacific Biosciences SMRT single molecule sequencing system10 captures the polymerase rather than the DNA library on surface to carry out real-time sequencing determination. With these next generation DNA sequencing systems, massive parallel digital gene expression analogous to a high-throughput SAGE approach has been reported reaching single copy transcript sensitivity,14 and CHIP-Seq11-13 based on sequencing tags of around 25 bases has led to many new discoveries in genome function and regulation. It is well established that millions of different DNA templates can be generated on a solid surface to construct the massive parallel DNA sequencing platform.13,14 Therefore, future implementation of the molecular level SBS approaches described in this account on a high-density DNA array will provide a high-throughput and accurate DNA sequencing system with wide applications in genome biology and biomedical research.
Acknowledgements
We thank Dr. Steffen Jockusch for generous discussions and technical support. This work was supported by National Institutes of Health Grants R01HG003582, R21HG004404, R01HG005109, and R01NS060762. The authors also thank the NSF for generous support of this research through grant CHE 07-17518.
Biographies
Jia Guo received his B.S. degree in chemical physics from the University of Science and Technology of China in 2005. He received a Ph.D. degree in chemistry in 2009 under the direction of Professor Jingyue Ju and Professor Nicholas J. Turro at Columbia University, where he has worked on the design and synthesis of novel nucleotide analogues for DNA sequencing and analysis.
Lin Yu received his B. S. degree in chemical engineering from the Cooper Union for Advancement of Science and Art, in New York, in 2005. He joined Professor Jingyue Ju's group at Columbia University as a Ph.D. graduate student the same year. He has been working in the area of DNA sequencing technology with a focus on the design and synthesis of novel fluorescently labeled nucleotide analogues for DNA sequencing by synthesis.
Nicholas J. Turro has been teaching at Columbia University since 1964 and is currently the Wm. P. Schweitzer Professor of Chemistry and Professor of Chemical Engineering, as well as Professor of Earth and Environmental Engineering. He has written over 800 scientific publications and 2 textbooks on molecular photochemistry. Recent awards include the 2005 Theodor Förster Award from the German Chemical Society and the 2007 Nichols Medal by the New York Section of the American Chemical Society. He is a member of the National Academy of Sciences and the American Academy of Arts and Sciences.
Jingyue Ju received his Ph.D. degree in bio-organic chemistry from the University of Southern California in 1994. He was a U.S. Department of Energy Human Genome Distinguished Postdoctoral Fellow at the University of California at Berkeley from 1994 to 1995, during which time he co-invented the fluorescent energy transfer labeling technology for DNA sequencing and analysis. He joined the Columbia University faculty in 1999 and is currently Professor of Chemical Engineering, Director of Center for Genome Technology & Biomolecular Engineering, and Head of DNA Sequencing and Chemical Biology at the Columbia Genome Center. His research interests include the design and synthesis of novel molecular tags for biological labeling and imaging and developing new technologies for genomic research.
REFERENCES
- 1.Collins FS, Green ED, Guttmacher AE, Guyer MS. A vision for the future of genomics research. Nature. 2003;422:835–847. doi: 10.1038/nature01626. [DOI] [PubMed] [Google Scholar]
- 2.Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Drmanac S, Kita D, Labat I, Hauser B, Schmidt C, Burczak JD, Drmanac R. Accurate sequencing by hybridization for DNA diagnostics and individual genomics. Nat Biotechnol. 1998;16:54–58. doi: 10.1038/nbt0198-54. [DOI] [PubMed] [Google Scholar]
- 4.Fu DJ, Tang K, Braun A, Reuter D, Iverson BL, Darnhofer-Demar B, Little DP, O'Donnell MJ, Cantor CR, Koster H. Sequencing exons 5 to 8 of the p53 gene by MALDI-TOF mass spectrometry. Nat Biotechnol. 1998;16:381–384. doi: 10.1038/nbt0498-381. [DOI] [PubMed] [Google Scholar]
- 5.Roskey MT, Juhasz P, Smirnov IP, Takach EJ, Martin SA, Haff LA. DNA sequencing by delayed extraction-matrix-assisted laser desorption/ionizaton time of flight mass spectrometry. Proc Natl Acad Sci U S A. 1996;93:4724–4729. doi: 10.1073/pnas.93.10.4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Edwards JR, Itagaki Y, Ju J. DNA sequencing using biotinylated dideoxynucleotides and mass spectrometry. Nucleic Acids Res. 2001;29:E104. doi: 10.1093/nar/29.21.e104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kasianowicz JJ, Brandin E, Branton D, Deamer DW. Characterization of individual polynucleotide molecules using a membrane channel. Proc Natl Acad Sci U S A. 1996;93:13770–13773. doi: 10.1073/pnas.93.24.13770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shendure J, Porreca GJ, Reppas NB, Lin X, McCutcheon JP, Rosenbaum AM, Wang MD, Zhang K, Mitra RD, Church GM. Accurate Multiplex Polony Sequencing of an Evolved Bacterial Genome. Science. 2005;309:1728–1732. doi: 10.1126/science.1117389. [DOI] [PubMed] [Google Scholar]
- 9.Harris TD, Buzby PR, Babcok H, Beer E, Bowers J, Braslavsky I, Causey M, Colonell J, DiMeo J, Efcavitch JW, Giladi E, Gill J, Healy J, Jarosz M, Lapen D, Moulton K, Quake SR, Steinmann K, Thayer E, Tyurina A, Ward R, Weiss H, Xie Z. Single-molecule DNA sequencing of a viral genome. Science. 2008;320:106–109. doi: 10.1126/science.1150427. [DOI] [PubMed] [Google Scholar]
- 10.Eid J, Fehr A, Gray J, Luong K, Lyle J, Otto G, Peluso P, Rank D, Baybayan P, Bettman B, Bibillo A, Bjornson K, Chaudhuri B, Christians F, Cicero R, Clark S, Dalal R, deWinter A, Dixon J, Foquet M, Gaertner A, Hardenbol P, Heiner C, Hester K, Holden D, Kearns G, Kong X, Kuse R, Lacroix Y, Lin S, Lundquist P, Ma C, Marks P, Maxham M, Murphy D, Park I, Pham T, Phillips M, Roy J, Sebra R, Shen G, Sorenson J, Tomaney A, Travers K, Trulson M, Vieceli J, Wegener J, Wu D, Yang A, Zaccarin D, Zhao P, Zhong F, Korlach J, Turner S. Real-time DNA sequencing from single polymerase molecule. Science. 2008;323:133–138. doi: 10.1126/science.1162986. [DOI] [PubMed] [Google Scholar]
- 11.Mikkelson TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim T, Koche RP, Lee W, Mendenhall E, O'Donovan A, Presser A, Russ C, Xie X, Meissner A, Wernig M, Jaenisch R, Nusbaum C, Lander ES, Bernstein BE. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-wide mapping of in vivo protein-DNA interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 13.Barski A, Cuddapah S, Cui K, Roh T, Schones D, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 14.Kim JB, Porreca GJ, Song L, Greenway SC, Gorham JM, Church GM, Seidman CE, Seidman JG. Polony multiplex analysis of gene expression (PMAGE) in mouse hypertrophic cardiomyopathy. Science. 2007;316:1481–1484. doi: 10.1126/science.1137325. [DOI] [PubMed] [Google Scholar]
- 15.Czech B, Malone CD, Zhou R, Stark A, Schingeheyde C, Dus M, Perrimon N, Kellis M, Wohlschlegel JA, Sachidanandam R, Hannon GJ, Brennecke J. An endogenous small interfering RNA pathway in Drosophila. Nature. 2008;453:798–802. doi: 10.1038/nature07007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bubnoff A. Next-Generation Sequencing: The Race is On. Cell. 2008;132:721–723. doi: 10.1016/j.cell.2008.02.028. [DOI] [PubMed] [Google Scholar]
- 17.Kolb HC, Finn MG, Sharpless KB. Click chemistry: diverse chemical function from a few good reactions. Angew. Chem. Int. Ed. 2001;40:2004–2021. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 18.Adessi C, Matton G, Ayala G, Turcatti G, Mermod J, Mayer P, Kawashima E. Solid phase DNA amplification: characterisation of primer attachment and amplification mechanisms. Nucleic Acids Res. 2000;28:E87. doi: 10.1093/nar/28.20.e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fedurco M, Romieu A, Williams S, Lawrence I, Turcatti G. BTA, a novel reagent for DNA attachment on glass and efficient generation of solid-phase amplified DNA colonies. Nucleic Acids Res. 2006;34:E22. doi: 10.1093/nar/gnj023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bowers J, Mitchell J, Beer E, Buzby PR, Causey M, Efcavitch JW, Jarosz M, Krzymanska-Olejnik E, Kung L, Lipson D, Lowman GM, Marappan S, McInerney P, Platt A, Roy A, Siddiqi SM, Steinmann K, Thompson JF. Virtual terminator nucleotides for next-generation DNA sequencing. Nature Methods. 2009;6:593–595. doi: 10.1038/nmeth.1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Braslavsky I, Hebert B, Kartalov E, Quake SR. Sequence information can be obtained from single DNA molecules. Proc Natl Acad Sci U S A. 2003;100:3960–3964. doi: 10.1073/pnas.0230489100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mitra RD, Shendure J, Olejnik J, Edyta-Krzymanska-Olejnik. Church GM. Fluorescent in situ sequencing on polymerase colonies. Anal Biochem. 2003;320:55–65. doi: 10.1016/s0003-2697(03)00291-4. [DOI] [PubMed] [Google Scholar]
- 23.Smith T. Whole genome variation analysis using single molecule sequencing. Drug Discovery Today: TARGETS. 2004;3:112–116. [Google Scholar]
- 24.Turcatti G, Romieu A, Fedurco M, Tairi A. A new class of cleavable fluorescent nucleotides: synthesis and optimization as reversible terminators for DNA sequencing by synthesis. Nucleic Acids Res. 2008;36:E25. doi: 10.1093/nar/gkn021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Seo TS, Bai X, Ruparel H, Li Z, Turro NJ, Ju J. Photocleavable fluorescent nucleotides for DNA sequencing on a chip constructed by site-specific coupling chemistry. Proc Natl Acad Sci U S A. 2004;101:5488–5493. doi: 10.1073/pnas.0401138101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seo TS, Bai X, Kim DH, Meng Q, Shi S, Ruparel H, Li Z, Turro NJ, Ju J. Four-color DNA sequencing by synthesis on a chip using photocleavable fluorescent nucleotides. Proc Natl Acad Sci U S A. 2005;102:5926–5931. doi: 10.1073/pnas.0501965102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ju J, Li Z, Edwards JR, Itagaki Y. Massive parallel method for decoding DNA and RNA. US Patent 6,664,079. 2003 [Google Scholar]
- 28.Pelletier H, Sawaya MR, Kumar A, Wilson SH, Kraut J. Structures of ternary complexes of rat DNA polymerase beta, a DNA template-primer, and ddCTP. Science. 1994;264:1891–1903. [PubMed] [Google Scholar]
- 29.Rosenblum BB, Lee LG, Spurgeon SL, Khan SH, Menchen SM, Heiner CR, Chen SM. New dye-labeled terminators for improved DNA sequencing patterns. Nucleic Acids Res. 1997;25:4500–4504. doi: 10.1093/nar/25.22.4500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu Z, Chao J, Yu H, Waggoner AS. Directly labeled DNA probes using fluorescent nucleotides with different length linkers. Nucleic Acids Res. 1994;22:3418–3422. doi: 10.1093/nar/22.16.3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ju J, Kim DH, Bi L, Meng Q, Bai X, Li Z, Li X, Marma MS, Shi S, Wu J, Edwards JR, Romu A, Turro NJ. Four-color DNA sequencing by synthesis using cleavable fluorescent nucleotide reversible terminators. Proc Natl Acad Sci U S A. 2006;103:19635–19640. doi: 10.1073/pnas.0609513103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo J, Xu N, Li Z, Zhang S, Wu J, Kim DH, Marma MS, Meng Q, Cao H, Li X, Shi S, Yu L, Kalachikov S, Russo JJ, Turro NJ, Ju J. Four-color DNA sequencing with 3′-O-modified nucleotide reversible terminators and chemically cleavable fluorescent dideoxynucleotides. Proc Natl Acad Sci U S A. 2008;105:9145–4150. doi: 10.1073/pnas.0804023105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu J, Zhang S, Meng Q, Cao H, Li Z, Li X, Shi S, Kim DH, Bi L, Turro NJ, Ju J. 3′-O-modified nucleotides as reversible terminators for pyrosequencing. Proc Natl Acad Sci U S A. 2007;104:16462–16467. doi: 10.1073/pnas.0707495104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ronaghi M, Karamohamed S, Pettersson B, Uhlen M, Nyren P. Real-time DNA sequencing using detection of pyrophosphate release. Anal Biochem. 1996;242:84–89. doi: 10.1006/abio.1996.0432. [DOI] [PubMed] [Google Scholar]
- 35.Ronaghi M, Uhlen M, Nyren P. A sequencing method based on real-time pyrophosphate. Science. 1998;281:363–365. doi: 10.1126/science.281.5375.363. [DOI] [PubMed] [Google Scholar]
- 36.Wheeler DA, Srinivasan M, Egholm M, Shen Y, Chen L, McGuire A, He W, Chen Y, Makhijani V, Roth GT, Gomes X, Tartaro K, Niazi F, Turcotte CL, Irzyk GP, Lupski JR, Chinault C, Song X, Liu Y, Yuan Y, Nazareth L, Qin X, Muzny DM, Margulies M, Weinstock GM, Gibbs RA, Rothberg JM. The complete genome of an individual by massively parallel DNA sequencing. Nature. 2008;452:872–877. doi: 10.1038/nature06884. [DOI] [PubMed] [Google Scholar]
- 37.Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, Boutell JM, Bryant J, Carter RJ, Cheetham RK, Cox AJ, Ellis DJ, Flatbush MR, Gormley NA, Humphray SJ, Irving LJ, Karbelashvili MS, Kirk SM, Li H, Liu X, Maisinger KS, Murray LJ, Obradovic B, Ost T, Parkinson ML, Pratt MR, Rasolonjatovo IMJ, Reed MT, Rigatti R, Rodighiero C, Ross MT, Sabot A, Sankar SV, Scally A, Schroth GP, Smith ME, Smith JP, Spiridou A, Torrance PE, Tzonev SS, Vermaas EH, Walter K, Wu X, Zhang L, Alam MD, Anastasi C, Aniebo IC, Bailey DMD, Bancarz IR, Banerjee S, Barbour SG, Baybayan PA, Benoit VA, Benson KF, Bevis C, Black PJ, Boodhun A, Brennan JA, Bridgham JA, Brown RC, Brown AA, Buermann DH, Bundu AA, Burrows JC, Carter NP, Castillo N, Catenazzi MCE, Chang S, Cooley RN, Crake NR, Dada OO, Diakoumakos K,D, Dominguez-Fernandez B, Earnshaw DJ, Egbujor UC, Elmore DW, Etchin SS, Ewan MR, Fedurco M, Fraser LJ, Fajardo KVF, Furey WS, George D, Gietzen KJ, Goddard CP, Golda GS, Granieri PA, Green DE, Gustafson DL, Hansen NF, Harnish K, Haudenschild CD, Heyer NI, Hims MM, Ho JT, Horgan AM, Hoschler K, Hurwitz S, Ivanov DV, Johnson MQ, James T, Jones TAH, Kang G, Kerelska TH, Kersey AD, Khrebtukova I, Kindwall AP, Kingsbury Z, Kokko-Gonzales PI, Kumar A, Laurent MA, Lawley CT, Lee SE, Lee X, Liao AK, Loch JA, Lok M, Luo S, Mammen RM, Martin JW, McCauley PG, McNitt P, Mehta P, Moon KW, Mullens JW, Newington T, Ning Z, Ng BL, Novo SM, O'Neill MJ, Osborne MA, Osnowski A, Ostadan O, Paraschos LL, Pickering L, Pike AC, Pinkard DC, Pliskin DP, Podhasky J, Quijano VJ, Raczy C, Rae VH, Rawlings SR, Rodriguez AC, Roe PM, Rogers J, Bacigalupo MCR, Romanov N, Romieu A, Roth RK, Rourke NJ, Ruediger ST, Rusman E, Sanches-Kuiper RM, Schenker MR, Seoane JM, Shaw RJ, Shiver MK, Short SW, Sizto NL, Sluis JP, Smith MA, Sohna JE, Spence EJ, Stevens K, Sutton J, Szajkowski L, Tregidgo CL, Turcatti G, vandeVondele S, Verhovsky Y, Virk SM, Wakelin S, Walcott GC, Wang J, Worsley GJ, Yan J, Yau L, Zuerlein M, Rogers J, Mullikin JC, Hurles ME, McCooke NJ, West JS, Oaks FL, Lundberg PL, Klenerman D, Durbin R, Smith AJ. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McKernan KJ, Peckham HE, Costa GL, McLaughlin SF, Fu Y, Tsung EF, Clouser CR, Duncan C, Ichikawa JK, Lee CC, Zhang Z, Ranade SS, Dimalanta ET, Hyland FC, Sokolsky TD, Zhang L, Sheridan JA, Fu H, Hendrickson CL, Li B, Kotler L, Stuart JR, Malek JA, Manning JM, Antipova AA, Perez DS, Moore MP, Hayashibara KC, Lyons MR, Beaudoin RE, Coleman BE, Laptewicz MW, Sannicandro AE, Rhodes MD, Gottimukkala RK, Yang S, Bafna V, Bashir A, MacBride A, Alkan C, Kidd JM, Eichler EE, Reese MG, De La Vega FM, Blanchard AP. Sequence and structural variation in a human genome uncovered by short-read, massively parallel ligation sequencing using two base encoding. Genome Research. 2009;19:1527–1541. doi: 10.1101/gr.091868.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Margulies M, Egholm M, Altman WE, Attiya S, Bader JS, Bemben LA, Berka J, Braverman MS, Chen Y, Chen Z, Dewell SB, Du L, Fierro JM, Gomes XV, Goodwin BC, He W, Helgesen S, Ho CH, Irzyk GP, Jando SC, Alenquer MLI, Jarvie TP, Jirage KB, Kim J, Knight JR, Lanza JR, Leamon JH, Lefkowitz SM, Lei M, Li M, Lohman KL, Lu H, Makhijani VB, McDade KE, McKenna MP, Myers EW, Nickerson E, Nobile JR, Plant R, Puc BP, Ronan MT, Roth GT, Sarkis GJ, Simons JF, Simpson JW, Srinivasan M, Tartaro KR, Tomasz A, Vogt KA, Volkmer GA, Wang SH, Wang Y, Weiner MP, Yu P, Begley RF, Rothberg JM. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mardis ER. Next-generation DNA sequencing methods. Annu. Rev. Genomics Hum.Genet. 2008;9:387–402. doi: 10.1146/annurev.genom.9.081307.164359. [DOI] [PubMed] [Google Scholar]