Abstract
Selenocysteine is the only genetically encoded amino acid in humans whose biosynthesis occurs on its cognate transfer RNA (tRNA). O-Phosphoseryl-tRNA:selenocysteinyl-tRNA synthase (SepSecS) catalyzes the final step of selenocysteine formation by a poorly understood tRNA-dependent mechanism. The crystal structure of human tRNASec in complex with SepSecS, phosphoserine, and thiophosphate, together with in vivo and in vitro enzyme assays, supports a pyridoxal phosphate–dependent mechanism of Sec-tRNASec formation. Two tRNASec molecules, with a fold distinct from other canonical tRNAs, bind to each SepSecS tetramer through their 13–base pair acceptor-TΨC arm (where Ψ indicates pseudouridine). The tRNA binding is likely to induce a conformational change in the enzyme’s active site that allows a phosphoserine covalently attached to tRNASec, but not free phosphoserine, to be oriented properly for the reaction to occur.
The 21st amino acid, selenocysteine (Sec), is distinct from other amino acids not only because it lacks its own tRNA synthetase, but also because it is the only one that is synthesized on the cognate tRNA in all domains of life [reviewed in (1–4)] in a process that is reminiscent of the tRNA-dependent synthesis of glutamine, asparagine, and cysteine in prokaryotes (4). The importance of Sec is illustrated by the embryonic lethal phenotype of the tRNASec knockout mouse (5) and by the presence of Sec in the active sites of enzymes involved in removing reactive oxidative species and in thyroid hormone activation (6, 7). It is intriguing that the codon for Sec is UGA, which is normally a translational stop signal (1). During translation of selenoprotein mRNAs, UGA is recoded by the interaction of a specialized elongation factor, SelB in bacteria and EFsec in humans, with a downstream Sec-insertion sequence element that forms a stem loop (1, 3).
The first step in Sec formation involves the misacylation of tRNASec by seryl-tRNA synthetase (SerRS) to give Ser-tRNASec [reviewed in (3, 8)]. Although the tertiary structure of tRNASec was unknown, it was proposed that the mischarging reaction is possible because of similarities between the tRNASec and tRNASer structures (3, 8). In archaea and eukaryotes, the γ-hydroxyl group of Ser-tRNASec is subsequently phosphorylated by O-phosphoseryl–tRNA kinase (PSTK) (9) to give O-phosphoseryl–tRNASec (Sep-tRNASec), which is then used as a substrate for the last synthetic enzyme, SepSecS (8, 10). SepSecS catalyzes the conversion of the phosphoseryl moiety into the selenocysteinyl group by using selenophosphate as the selenium donor. An early observation that autoantibodies isolated from patients with type I autoimmune hepatitis targeted a ribonucleoprotein complex containing tRNASec led to the identification and characterization of the archaeal and the human SepSecS (2, 11). The crystal structures of the archaeal and murine SepSecS apo-enzymes and phylogenetic analysis suggested that SepSecS forms its own branch in the family of fold-type I pyridoxal phosphate (PLP) enzymes that goes back to the last universal common ancestor (12, 13).
In contrast to its closest homologue, SepCysS, which functions as a dimer to convert Sep-tRNACys to Cys-tRNACys in archaea (14, 15), SepSecS forms a stable tetramer (12, 13). SepSecS acts on phosphoserine that is linked to tRNASec and not on free phosphoserine or Ser-tRNASec (8, 13). However, the molecular basis for substrate discrimination and the roles of PLP and tRNASec in the mechanism of Sep to Sec conversion are not clear. To explore these questions, we have determined the crystal structure of the quaternary complex between human SepSecS, unacylated tRNASec, and a mixture of O-phosphoserine (Sep) and thiophosphate (Thiop) to 2.8 Å resolution. The observed intensity divided by its standard deviation [I/σ(I)] of the x-ray diffraction data are 2 at 3.0 Å resolution, but the data out to 2.8 Å resolution, where I/σ (I) drops to 1, were included in the structure refinement.
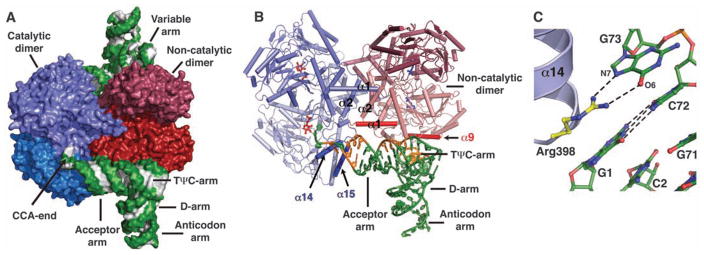
Human SepSecS forms a tetramer that is bound to two tRNASec molecules in the crystal (Fig. 1, A and B, and fig. S1). Computational modeling suggests that the tetrameric enzyme could potentially bind up to four tRNASec molecules (fig. S2), but these additional tRNA-binding sites are blocked by crystal packing in our crystal (16). Electron density for the 23 N-terminal and 15 C-terminal residues of SepSecS, as well as for the anticodon loop (nucleotides 31 to 38) and A76 of tRNASec, was of poor quality, and these residues were not included in the final model. Each SepSecS monomer has a PLP cofactor covalently linked to the Nε-amino group of the conserved Lys284 by means of formation of a Schiff base (internal aldimine). Two SepSecS monomers form a homodimer, and two active sites are formed at the dimer interface. The two homodimers associate into a tetramer through interactions between the N-terminal α1-loop-α2 motifs (Fig. 1B). Given that SepCysS is a dimer and that the active sites of one homodimer do not communicate with the active sites of the other homodimer in the apo-SepSecS tetramer (12, 13), the reason for the tetrameric organization of SepSecS was not known. The mode of tRNASec binding to SepSecS provides an answer to this question.
Fig. 1.
Structure of human SepSecS in complex with unacylated tRNASec. (A) Surface representation of the physiological complex of SepSecS with tRNASec. The subunits of the catalytic dimer are dark and light blue, those of the noncatalytic dimer are dark and light red; the backbone and the bases of tRNASec are green and gray, respectively. (B) The catalytic dimer interacts with the acceptor arm of tRNASec through helices α14 and α15 (blue). The α1 helix (red) of the noncatalytic dimer interacts with the rest of the acceptor-TΨC arm. The regions of tRNASec that interact with SepSecS are shown in orange; the rest is green. One tRNASec molecule is shown for clarity. (C) Interactions between the discriminator base G73 and the conserved Arg398 in the α14-β11 loop. The protein side chains are gold, and tRNASec is green.
The CCA ends of both tRNASec molecules point to the active sites of the same homodimer, which we shall refer to as the catalytic dimer (Fig. 1A). The other homodimer, which we shall refer to as the noncatalytic dimer, serves as a binding platform that orients tRNASec for catalysis (Fig. 1A). The structure reveals that SepSecS binds only to the acceptor-, TΨC-, and variable arms of tRNASec (Fig. 1B and fig. S3A). (Ψ, pseudo-uridine.) The tip of the acceptor arm interacts with the C terminus of the catalytic dimer, whereas the rest of the acceptor-, TΨC-, and variable arms wrap around a monomer from the noncatalytic dimer (Fig. 1, A and B, and fig. S3A). The most important binding element is an interaction between the discriminator base G73 of tRNASec and the conserved Arg398 of the catalytic dimer (Fig. 1C). The guanidinium group of Arg398 forms hydrogen bonds with the Hoogsteen face of G73. The discriminator base G73 of tRNASec is universally conserved in archaea and eukaryotes. Neither adenine nor cytidine in position 73 could form hydrogen bonds with Arg398 because they have amino groups instead of the keto group. Also, if a cytidine or uridine were in position 73, the C5 and C6 atoms of the pyrimidine ring would clash with the side chain of Thr397 and thus prevent the interaction of these bases with Arg398.
Moreover, the side chain of Lys463 from the C-terminal helix α15 forms a hydrogen bond with the backbone oxygen of G69 from the acceptor arm. We propose that autoantibodies bind to an interface that lies between the α15 helix of SepSecS and the tip of the acceptor arm of tRNASec (Fig. 1B), which enables them to precipitate the entire ribonucleoprotein complex (11). The interaction of the antibody with a region that lies close to the acceptor stem–active site interface may inhibit the function of SepSecS. This would be similar to the mechanism of autoimmune hepatitis type 2, where LKM-1 autoantibodies (anti–liver-kidney microsomal antibodies) inhibit cytochrome P450 isoenzyme 2D6 (CYP2D6) and contribute to the pathogenesis of the disease (17).
The tRNASec molecule is anchored to the SepSecS tetramer, on one end by interactions between the tip of its acceptor arm (G73) and the catalytic dimer (Arg398) (Fig. 1C) and on the other end by interactions between its variable arm (C46L) and the noncatalytic dimer (Arg271) (fig. S3B). The rest of the interactions between the α1 helix of the noncatalytic dimer of SepSecS (Arg26, Lys38, and Lys40) and the acceptor-TΨC arm (C2, G50, and C64) further stabilize the complex (fig. S3C). For instance, the side chain of Arg26 forms hydrogen bonds with the phosphate backbone of C2–C3 of the acceptor arm and the Nε-amino group of Lys38 is within the hydrogen bonding distance from both the O2′-hydroxyl group and the O2 atom of C64 (fig. S3C), whereas the side chain of Lys40 interacts with the phosphate oxygen of G50 from the TΨC arm (fig. S3C). The interactions between SepSecS and tRNASec seen in the crystal are consistent with in vivo activity assays of SepSecS mutants (fig. S4A) (16). For example, replacing Arg398 with either alanine or glutamate renders the enzyme completely inactive, which suggests that the interaction between the discriminator base and the highly conserved Arg398 of the catalytic dimer is critical for tRNASec recognition (fig. S4A).
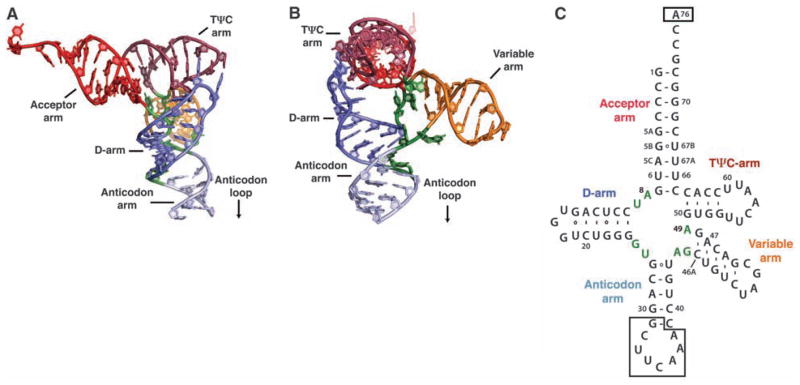
Human tRNASec contains 90 nucleotides rather than the conventional 75 nucleotides of canonical tRNA molecules. The structure shows that human tRNASec adopts a unique 9/4 fold with a 13–base pair (bp) acceptor-TΨC arm (where 9 and 4 reflect the number of base pairs in the acceptor and TΨC arms, respectively) and a long variable arm (Fig. 2) (16). This resolves a controversy between conflicting models in which the 7/5 model suggested 12 bp in the acceptor-TΨC arm, as found in all known tRNA structures (18), whereas the 9/4 model suggested a unique 13-bp acceptor-TΨC arm (19). The 13-bp acceptor-TΨC arm and a long variable arm are distinct structural features that serve as major recognition motifs for binding to SepSecS (Fig. 1, A and B, and fig. S3). Indeed, based on our modeling analysis with tRNAAsp, we suggest that tRNASer, which contains both the G73 discriminator base and a long variable arm, is not able to bind to SepSecS because of its shorter acceptor-TΨC arm (fig. S5, A, B, and C). This is reminiscent of the rejection of tRNASec by bacterial EF-Tu (20). However, modeling of the SerRS-tRNASec complex based on the crystal structure of the bacterial SerRS in complex with tRNASer (21) suggests that SerRS can recognize the variable arm of tRNASec similarly to tRNASer, which provides an explanation for the inability of SerRS to discriminate between tRNASer and tRNASec (fig. S5D). Finally, the observations that the archaeal PSTK and SepSecS can act on E. coli tRNASec in vivo; that human SepSecS can use E. coli tRNASec in vivo, as well as the archaeal tRNASec in vitro (8); and that the bacterial selenocysteine synthase can use both archaeal and murine tRNASec in vitro (10) suggest that the length of the acceptor-TΨC arm of tRNASec, the position of the variable arm, and the mode of tRNASec recognition are likely to be conserved in all domains of life.
Fig. 2.
Structure of human tRNASec. (A) Ribbon diagram of the human tRNASec molecule observed in complex with SepSecS. The major structural elements are colored as follows: the acceptor arm is red, the D-arm is blue, the anticodon arm is light blue, the variable arm is orange, and the TΨC arm is dark red. (B) The view is rotated by ~90° clockwise around the vertical axis. (C) Secondary structure diagram of human tRNASec derived from the crystal structure.
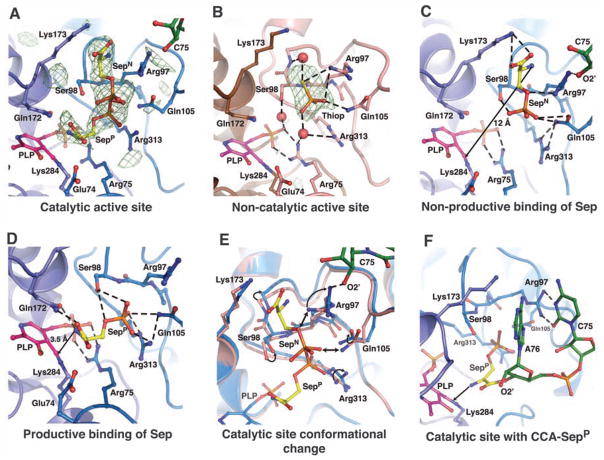
To explore the mechanism by which the phosphoryl group is converted to the selenocysteinyl moiety, we soaked crystals of the binary SepSecS-tRNASec complex in a solution containing a mixture of O-phosphoserine and thiophosphate. Sep and Thiop were used as mimics of the phosphoseryl group attached to tRNASec and selenophosphate, respectively. Although both ligands were used in the soaking experiments, Sep bound only to the active sites of the catalytic dimer (Fig. 3A), whereas Thiop bound only to the active sites of the noncatalytic dimer (Fig. 3B), i.e., Sep can bind to an active site only in the presence of tRNASec. Both Thiop and the phosphoryl group of Sep bind to the same binding pocket (Fig. 3, A and B), which suggests that a specific active site can accommodate only one ligand at a time. Sep binds to the catalytic active site in either of two different orientations (Fig. 3A). The phosphoryl group occupies a similar location in the two orientations, whereas the seryl moieties are rotated by ~90° around the phosphate group. In the non-productive orientation of Sep (SepN), its seryl group is sandwiched between the side chains of Arg97 and Lys173 at the catalytic dimer interface, whereas its amino group lies ~12 Å away from the PLP Schiff base (Fig. 3C). Thus, it is unlikely that SepSecS would act on SepN, unless a PLP-independent mechanism is utilized. In the productive-like orientation of Sep (SepP), the amino group of Sep is ~3.5 Å away from the Schiff base, yet it is not positioned for attack because of a hydrogen bond with Gln172 (Fig. 3D). The carboxyl group of SepP also forms a hydrogen bond with the side chain of Gln172, whereas its phosphoryl group anchors the ligand into the active site through its interactions with the side chains of Ser98, Gln105, and Arg313. That the phosphoryl group is required to properly position the ligand for catalysis explains why the obligate substrate for SepSecS is Sep-tRNASec and not Ser-tRNASec, which is the physiological substrate for the bacterial selenocysteine synthase.
Fig. 3.
Ligand binding to the active sites in the SepSecS-tRNASec complex. In (A), (C), (D), (E), and (F), the catalytic PLP-monomer is purple, the P-loop monomer is light blue, Sep is gold, tRNA is green, and PLP is magenta. In (A) and (B), the unbiased omit electron density map (green mesh) is contoured at 3.5 σ. (A) Phosphoserine binds to the catalytic sites in two orientations (SepP and SepN). (B) Thiop (orange) binds only to the non-catalytic site. The PLP-monomer is brown, and the P-loop monomer is pink. (C) The SepN amino group is ~12 Å away from the Schiff base. Arg97, Gln105, and Arg313 coordinate phosphate, and the carboxyl group interacts with Lys173. (D) The amino group of SepP interacts with Gln172 and is 3.5 Å away from the Schiff base. The carboxyl group interacts with Gln172, whereas Ser98, Gln105, and Arg313 coordinate phosphate. (E) The P-loop adopts different conformation after tRNASec binding. The noncatalytic dimer is pink, the catalytic dimer is light blue. Steric clashes between the noncatalytic P-loop and SepN are shown (double arrow). (F) A model of CCA-SepP (gold) in the catalytic active site. A76 binds between the side chains of Arg97 and Lys173, whereas C75 interacts with Arg97.
A comparison of the catalytic and noncatalytic sites reveals that Sep binds to the active site only in the presence of tRNASec because the conformation of the P-loop (residues Gly96 to Lys107) differs between the two active sites (Fig. 3E). In the noncatalytic dimer, the guanidinium group of Arg97 and the side chain of Gln105 are rotated toward the phosphate-binding groove, where they coordinate Thiop (Fig. 3, B and E). In the catalytic dimer, the side chain of Arg97 rotates away from the phosphate-binding groove and forms a hydrogen bond with the 2′-OH group of C75. Gln105 also rotates away from the phosphate, and this concerted movement of Arg97 and Gln105 in the P-loop on tRNASec binding allows Sep to bind to the active site (Fig. 3E). Free Sep enters the active site through the gate formed by the side chains of Arg97, Gln105, and Lys173 and gets trapped in the SepN orientation. Electron density for both the amino and the carboxyl groups of SepP is weak, which suggests higher mobility in this part of SepP, presumably because of free rotation around the Cα-Cβ bond. This explains why the free amino group of SepP cannot be positioned appropriately for attack onto the Schiff base and why the reaction does not occur in the crystal. Thus, the covalent attachment of Sep to tRNASec is necessary for the proper placement of the Sep moiety into the active site and for orienting the amino group of Sep for attack onto the Schiff base of PLP (Fig. 3F).
We used both in vivo and in vitro activity assays to investigate the mechanism of Sep-tRNA to Sec-tRNA conversion by human SepSecS. First, the reduction of the Schiff base by sodium borohydride to form a chemically stable secondary amine and thus to cross-link PLP to Lys284 renders SepSecS completely inactive in vitro (fig. S4B). The catalytic activity of SepSecS is also quenched on removal of PLP by treatment with hydroxylamine (fig. S4B). Some residual activity that is observed after hydroxylamine treatment is probably because of incomplete removal of PLP (fig. S4B). Second, we show that the Arg75Ala, Gln105Ala, and Arg313Ala mutants are inactive in vivo (fig. S4A). These residues are involved in coordinating either the phosphate group of PLP or that of Sep. Finally, the in vivo activities of the Arg97Ala, Arg97Gln, Lys173Ala, and Lys173Met mutants are indistinguishable from that of the wild-type enzyme, which confirms that Arg97 and Lys173 are involved only in the nonproductive binding of free Sep (fig. S4A).
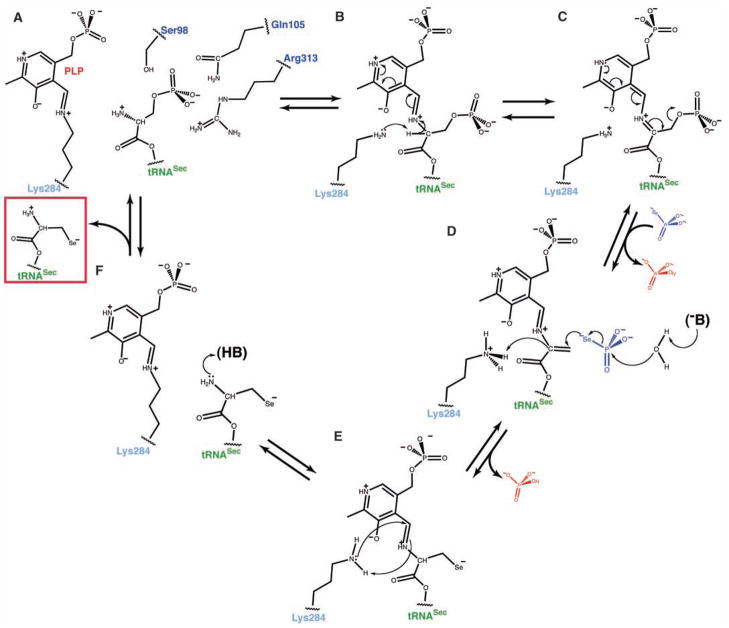
On the basis of our findings, we propose the following PLP-based mechanism of Sep-tRNA to Sec-tRNA conversion. The reaction begins by the covalently attached Sep being brought into the proximity of the Schiff base when Sep-tRNASec binds to SepSecS. The amino group of Sep can then attack the Schiff base formed between Lys284 and PLP, which yields an external aldimine (Fig. 4, A and B). The reoriented side chain of Lys284 abstracts the Cα proton from Sep (Fig. 4C), and the electron delocalization by the pyridine ring assists in rapid β-elimination of the phosphate group, which produces an intermediate dehydroalanyl-tRNASec (Fig. 4, C and D). After phosphate dissociation and binding of selenophosphate, the concomitant attack of water on the selenophosphate group and of the nucleophilic selenium onto the highly reactive dehydroalanyl moiety yield an oxidized form of Sec-tRNASec (Fig. 4D). The protonated Lys284, returns the proton to the Cα carbon and then attacks PLP to form an internal aldimine (Fig. 4E). Finally, Sec-tRNASec is released from the active site (Fig. 4F).
Fig. 4.
The PLP-dependent mechanism of Sep to Sec conversion. (A) The phosphoseryl moiety of Sep-tRNASec is bound to the active site similar to SepP. The amino group is oriented for attack on the Schiff base, whereas the phosphoryl group is stabilized by the side chains of Ser98, Gln105, and Arg313. Hydrogen bonds are shown in dashed lines. (B) After the formation of the external aldimine, the side chain of Lys284 rearranges and abstracts the Cα proton from Sep. The protonated pyridine ring of PLP stabilizes the carbanion. (C) Electron delocalization leads to a rapid β-elimination of phosphate and to the formation of dehydroalanyl-tRNASec. Free phosphate dissociates, and selenophosphate binds to the active site. (D) An unidentified base (–B) activates water that hydrolyzes selenophosphate. Free phosphate dissociates again, and selenium attacks the dehydroalanyl-tRNASec. Lys284 returns the proton to the Cα carbon, and the selenocysteinyl moiety is formed. (E) The reaction of reverse transaldimination is shown. Lys284 forms the Schiff base, with PLP leading to a release of the oxidized form of Sec-tRNASec (red box). (F) The free amino group of Sec-tRNASec is protonated, and the active site of SepSecS is regenerated.
This mechanism is clearly distinct from the persulfide-intermediate mechanism in the Sep-tRNACys to Cys-tRNACys reaction (22) and explains why SepSecS does not group together with its closest homolog, SepCysS, in the family tree of fold-type I PLP enzymes (12). Moreover, the proposed mechanism for SepSecS is similar to the one used by the bacterial SelA that also proceeds through a dehydroalanyl-tRNASec intermediate (23). SepSecS therefore uses a primordial tRNA-dependent catalytic mechanism in which the PLP cofactor is directly involved, while using a tetrameric fold-type I architecture as the scaffold for binding the distinct structure of tRNASec.
Supplementary Material
Acknowledgments
We thank C. Axel Innis, Gregor Blaha, Robin Evans, Michael Strickler, Michael E. Johnson, Bernard D. Santarsiero, Jimin Wang, and the NE-CAT beamline staff (APS, ANL, Chicago) for their help during data collection and structure determination. We thank Dan Su, Theodoros Rampias, and Kelly Sheppard for helpful discussions. Atomic coordinates and structure factors have been deposited in the Protein Data Bank (code 3HL2). Supported by grants from DOE (to D.S.) and NIGMS (to T.A.S. and D.S.). In the initial phase of this study M.S. was supported by HHMI at Yale University. S.P. holds a fellowship of the Yale University School of Medicine MD/PhD Program. R.L.S. was supported by a Ruth L. Kirschstein National Research Service Award.
Footnotes
Authors’ contributions: S.P. designed the research, collected and analyzed the data, and wrote the manuscript; R.L.S. did research and read the manuscript; T.A.S. designed the research and wrote the manuscript; D.S. designed the research and wrote the manuscript; M.S. designed the research, collected and analyzed the data, and wrote the manuscript.
Supporting Online Material
www.sciencemag.org/cgi/content/full/325/5938/321/DC1
Materials and Methods
SOM Text
References
References and Notes
- 1.Böck A, Thanbichler M, Rother M, Resch A. In: Aminoacyl-tRNA Synthetases. Ibba M, Francklyn C, Cusack S, editors. Landes Bioscience; Georgetown, TX: 2005. pp. 320–327. [Google Scholar]
- 2.Su D, et al. IUBMB Life. 2009;61:35. doi: 10.1002/iub.136. [DOI] [PubMed] [Google Scholar]
- 3.Ambrogelly A, Palioura S, Söll D. Nat Chem Biol. 2007;3:29. doi: 10.1038/nchembio847. [DOI] [PubMed] [Google Scholar]
- 4.Sheppard K, et al. Nucleic Acids Res. 2008;36:1813. doi: 10.1093/nar/gkn015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bösl MR, Takaku K, Oshima M, Nishimura S, Taketo MM. Proc Natl Acad Sci USA. 1997;94:5531. doi: 10.1073/pnas.94.11.5531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rayman MP. Lancet. 2000;356:233. doi: 10.1016/S0140-6736(00)02490-9. [DOI] [PubMed] [Google Scholar]
- 7.Kryukov GV, et al. Science. 2003;300:1439. doi: 10.1126/science.1083516. [DOI] [PubMed] [Google Scholar]
- 8.Yuan J, et al. Proc Natl Acad Sci USA. 2006;103:18923. doi: 10.1073/pnas.0609703104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carlson BA, et al. Proc Natl Acad Sci USA. 2004;101:12848. doi: 10.1073/pnas.0402636101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu XM, et al. PLoS Biol. 2007;5:e4. doi: 10.1371/journal.pbio.0050004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gelpi C, Sontheimer EJ, Rodriguez-Sanchez JL. Proc Natl Acad Sci USA. 1992;89:9739. doi: 10.1073/pnas.89.20.9739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Araiso Y, et al. Nucleic Acids Res. 2008;36:1187. doi: 10.1093/nar/gkm1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ganichkin OM, et al. J Biol Chem. 2008;283:5849. doi: 10.1074/jbc.M709342200. [DOI] [PubMed] [Google Scholar]
- 14.Sauerwald A, et al. Science. 2005;307:1969. doi: 10.1126/science.1108329. [DOI] [PubMed] [Google Scholar]
- 15.Fukunaga R, Yokoyama S. J Mol Biol. 2007;370:128. doi: 10.1016/j.jmb.2007.04.050. [DOI] [PubMed] [Google Scholar]
- 16.Materials and methods are available as supporting material on Science Online.
- 17.Herkel J, Manns MP, Lohse AW. Hepatology. 2007;46:275. doi: 10.1002/hep.21807. [DOI] [PubMed] [Google Scholar]
- 18.Ioudovitch A, Steinberg SV. RNA. 1998;4:365. [PMC free article] [PubMed] [Google Scholar]
- 19.Sturchler C, Westhof E, Carbon P, Krol A. Nucleic Acids Res. 1993;21:1073. doi: 10.1093/nar/21.5.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rudinger J, Hillenbrandt R, Sprinzl M, Giege R. EMBO J. 1996;15:650. [PMC free article] [PubMed] [Google Scholar]
- 21.Biou V, Yaremchuk A, Tukalo M, Cusack S. Science. 1994;263:1404. doi: 10.1126/science.8128220. [DOI] [PubMed] [Google Scholar]
- 22.Hauenstein SI, Perona JJ. J Biol Chem. 2008;283:22007. doi: 10.1074/jbc.M801839200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Forchhammer K, Böck A. J Biol Chem. 1991;266:6324. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.