Abstract
In approximately one billion people, a point mutation inactivates a key detoxifying enzyme, aldehyde dehydrogenase (ALDH2). This mitochondrial enzyme metabolizes toxic biogenic and environmental aldehydes, including the endogenously produced 4-hydroxynonenal (4HNE) and the environmental pollutant, acrolein. ALDH2 also bioactivates nitroglycerin, but it is best known for its role in ethanol metabolism. The accumulation of acetaldehyde following the consumption of even a single alcoholic beverage leads to the Asian Alcohol-induced Flushing Syndrome in ALDH2*2 homozygotes. The ALDH2*2 allele is semi-dominant and heterozygotic individuals exhibit a similar, but not as severe phenotype. We recently identified a small molecule, Alda-1, which activates wild-type ALDH2 and restores near wild-type activity to ALDH2*2. The structures of Alda-1 bound to ALDH2 and ALDH2*2 reveal how Alda-1 activates the wild-type enzyme and how it restores the activity of ALDH2*2 by acting as a structural chaperone.
Introduction
Mitochondrial aldehyde dehydrogenase (ALDH2) is most commonly associated with its role in ethanol metabolism, catalyzing the oxidation of ethanol-derived acetaldehyde to acetate1. However, recent data suggest ALDH2 plays a role in cardiovascular disease including its ability to bioactivate nitroglycerin as an anti-anginal and in the cardio-protective effects of ethanol prior to myocardial infarction2,3. Approximately 40% of the East Asian population has a semi-dominant polymorphism of ALDH2, ALDH2*2, that is essentially inactive in vivo1. Those individuals who carry the gene for ALDH2*2 experience a disulfiram-like reaction to ethanol consumption, with a more severe reaction observed in individuals who are homozygous for ALDH2*2(ref. 1). This reaction, also referred to as the Asian Ethanol-induced flushing syndrome, is associated with a lower risk for alcoholism4 and an approximate 100-fold increased risk for esophageal cancer5. Additional clinical outcomes associated with the ALDH2*2 allele include a reduced efficacy of nitroglycerin for the treatment of angina, increased morbidity following myocardial infarction and increased risk for Alzheimer’s disease and other neurodegenerative diseases associated with aging6–8.
From a biochemical point of view, the low activity of ALDH2*2 is the result of a single substitution of lysine for glutamate at position 487 of the 500-amino-acid mature enzyme or position 504 of the 517 amino acid precursor protein. This substitution, at the subunit interface of this homotetrameric enzyme9, increases the KM for NAD+ by 200 fold and reduces the kcat by 10 fold compared to the wild-type enzyme10. The KMNAD+ exceeds the in vivo concentration of the coenzyme by 15-fold, which, combined with the 10-fold lower catalytic rate, is responsible for its severely reduced activity in vivo. In addition to these kinetic changes, the ALDH2*2 enzyme also does not exhibit the same pre-steady-state burst of NADH production observed for the wild-type enzyme (9). The lack of a burst is evidence for a change in rate-limiting step from acyl-enzyme hydrolysis to some step preceding hydride transfer in ALDH2*2 (ref. 9). Structural analysis of the apo- and holoenzyme forms of ALDH2*2 reveals structural deficits in both the coenzyme-binding and active sites that are consistent with the observed kinetic properties9,11. NAD(H) binding ameliorates much of the structural disorder in the coenzyme-binding site, but does not improve the positioning of critical residues involved in catalysis11.
The recent discovery of a novel small molecule, Alda-1, that activates catalysis for both wild-type ALDH2 (ALDH2*1) and ALDH2*2 led us to characterize further the mechanism behind this novel pharmacological chaperone3. Small molecule chemical chaperones are still a unique occurrence in drug development12 and a greater understanding of their structure-activity relationship may permit the design of more efficacious compounds for both basic research of the role in which ALDH2 plays in the basic aspects of alcohol and lipid peroxidation product metabolism3, as well as for possible treatments associated with the lower activity of the ALDH2*2 gene product.
We sought to determine the molecular mechanism underlying the activation of human ALDH2*1 and ALDH2*2 by Alda-1. We determined the structure of Alda-1 bound to ALDH2*1 and to ALDH2*2 and performed a series of biochemical assays designed to characterize these novel functions. Rather than binding near the site of mutation in ALDH2*2, Alda-1 binds near the exit of the substrate-binding tunnel, at a position that overlaps the inhibitory site occupied by the isoflavone daidzin, but leaves the critical active site residues Cys302 and Glu268 free to function. The ability of Alda-1 to activate the wild-type enzyme, ALDH2*1, is dependent on substrate size and Alda-1 can also activate the esterase activities of ALDH2*1 and ALDH2*2. Both the enzymatic assays and structural data demonstrate that Alda-1 functions to increase the efficiency of acyl-enzyme hydrolysis for smaller aliphatic substrates in ALDH2*1 and provides structural stability to the ALDH2*2 enzyme, which rescues a large fraction of the functional deficits associated with the substitution of the critical Glu487 by a lysine.
Results
Structure of Alda-1 bound to ALDH2*1 and 2*2
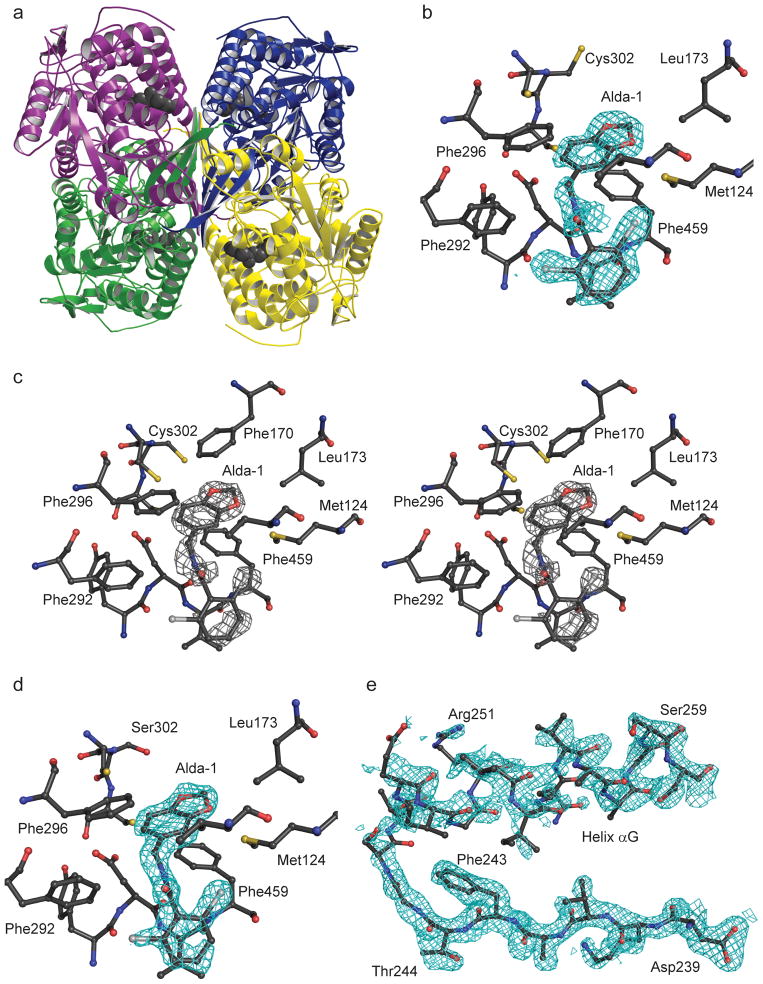
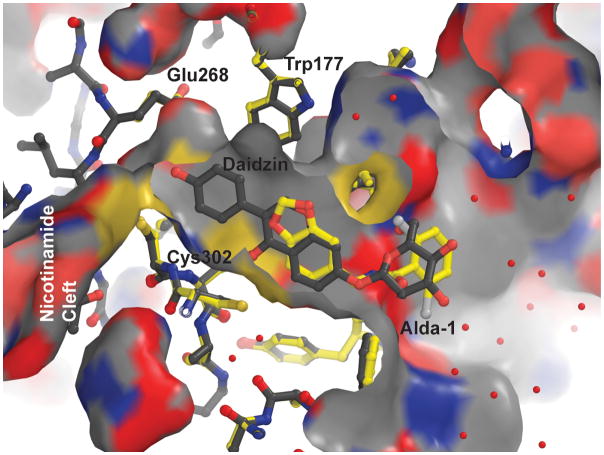
In order to better define the manner in which Alda-1 influences the activity of ALDH2, we determined the structures of wild-type ALDH2*1 and a C302S mutant of ALDH2*2 in binary complexes with Alda-1 to 1.69 Å and 1.86 Å, respectively (Table 1). The C302S mutant of ALDH2*2 was chosen for the structural work to eliminate the covalent modification of Cys302 that occurs only in the crystallization solution for the ALDH2*2 enzyme9. The decreased nucleophilicity of Ser302 prevents this modification from competing for Alda-1 binding. Alda-1 binds to the apo-form of both enzymes at the exit of the substrate tunnel and extends in toward the active site (Fig. 1). This mode of binding leaves the catalytic Cys302 unimpeded, although its side chain adopts two distinct rotamer positions. The benzodioxol group of Alda-1 is bound within an aromatic and hydrophobic collar comprised of amino acids Val120, Met124, Phe170, Leu173, Phe292, Phe296, and Phe459 solely through hydrophobic interactions. A single hydrogen bond is formed between the amide nitrogen that links the two ring structures in Alda-1 and the mainchain carbonyl oxygen atom of Asp457. The dichlorobenzamide ring is bound also primarily through hydrophobic interactions between the benzamide ring and Val458, Phe292 and Met124. It is interesting to note that diadzin, an ALDH2 inhibitor, occupies a site that overlaps with Alda-1 (ref. 13, PDB code 2VLE). However, the additional phenolic arm of daidzin reaches further into the catalytic site and contacts Cys302 and Glu268, thus blocking catalytic function (Fig. 2).
Table 1.
Data collection and refinement statistics
| ALDH2 | C302S ALDH2*2 | |
|---|---|---|
| Data collection | ||
| Space group | P21 | P21 |
| Cell dimensions | ||
| a, b, c (Å) | 101.9,176.8, 102.5 | 102.1,176.9,102.3 |
| α, β, γ (°) | 90.0, 94.5, 90.0 | 90.0, 94.6, 90.0 |
| Resolution (Å) | 1.69 (1.74–1.69)a | 1.86 (1.89–1.86) |
| Rmerge | 0.077 (0.27) | 0.090 (0.46) |
| I/σI | 11.4 (2.7) | 14.0 (2.2) |
| Completeness (%) | 93.1 (90.9) | 98.7 (75) |
| Redundancy | 2.7 (2.4) | 3.7 (2.9) |
| Refinement | ||
| Resolution (Å) | 46.0–1.69 | 50.0–1.86 |
| No. reflections | 363,986 (32,124) | 293,169 (25,560) |
| Rwork/Rfree | 0.18/0.20 (0.28/0.34) | 0.14/0.17 (0.21/0.22) |
| No. atoms | ||
| Protein | 30,853 | 30,736 |
| Ligand/ion | 276 | 300 |
| Water | 4,009 | 3,408 |
| B-factors | ||
| Protein | 19.2 | 32.2 |
| Ligand/ion | 21.6 | 33.6 |
| Water | 24.5 | 36.9 |
| r.m.s. deviations | ||
| Bond lengths (Å) | 0.008 | 0.005 |
| Bond angles (°) | 1.02 | 0.91 |
Each structure was determined from a single crystal. Twin fractions are 0.28 for ALDH2 and 0.38 for C302S ALDH2*2.
Values in parentheses are for highest-resolution shell.
Figure 1. Structure of ALDH2 with Alda-1 bound.
(A) Ribbon diagram of the ALDH2 tetramer with different color denoting the individual subunits and the four bound Alda-1 molecules indicated using the gray space-filling atom representation. (B) Simulated annealing omit 2Fo-Fc electron density (contoured at 1.0 standard deviation of the map) for Alda-1 bound to ALDH2*1. (C) Stereoview of the original Fo-Fc electron density map prior to inclusion and refinement of the Alda1-1 ligand; contoured at 3 standard deviations of the map. (D) Simulated annealing omit 2Fo-Fc electron density for Alda-1 bound to the Ser302 mutant of ALDH2*2 (contoured at one standard deviation of the map). (E) Simulated annealed omit 2Fo-Fc electron density map for residues 239–260 in the ALDH2*2/Alda-1 binary complex structure (contoured at one standard deviation of the map). Produced using PyMol for Windows25.
Figure 2. Overlay of the aligned structures of ALDH2 with bound Alda-1 (yellow) and with bound daidzin (white, pdb entry 1vle).
The alignment was generated using PyMol and atom type coloring is utilized for both structures (white for carbon atoms in the daidzin structure and yellow for carbon atoms in the Alda-1 structure). The available molecular surface in this region for ALDH2 is displayed using the daidzin structure and the molecule is sliced above the plane of the bound ligands. Critical active site residues are labeled. The cleft through which the nicotinamide moiety accesses the active site lies to the left of Glu268 and Cys302 in this view and is labeled. Produced using PyMol for Windows25.
An intriguing feature of the ALDH2*2 binary complex with Alda-1 is the observation of electron density for residues 245–262 (helix αG, Fig. 1e) and residues 466–478 (active site loop) of the ALDH2*2 structure even though Alda-1 makes no direct contact with these residues. Prior studies from our laboratory has demonstrated an almost total lack of electron density for these regions of the coenzyme-binding and active sites in the absence of coenzyme, whereas the binding of coenzyme restored the main chain structure to near native appearance9,11. In the ALDH2*2•Alda-1 binary complex, we observe a similar state of structural restoration within the coenzyme-binding site without direct contact between Alda-1 and these residues.
Kinetics of Alda-1 activation on ALDH2 dehydrogenase activity
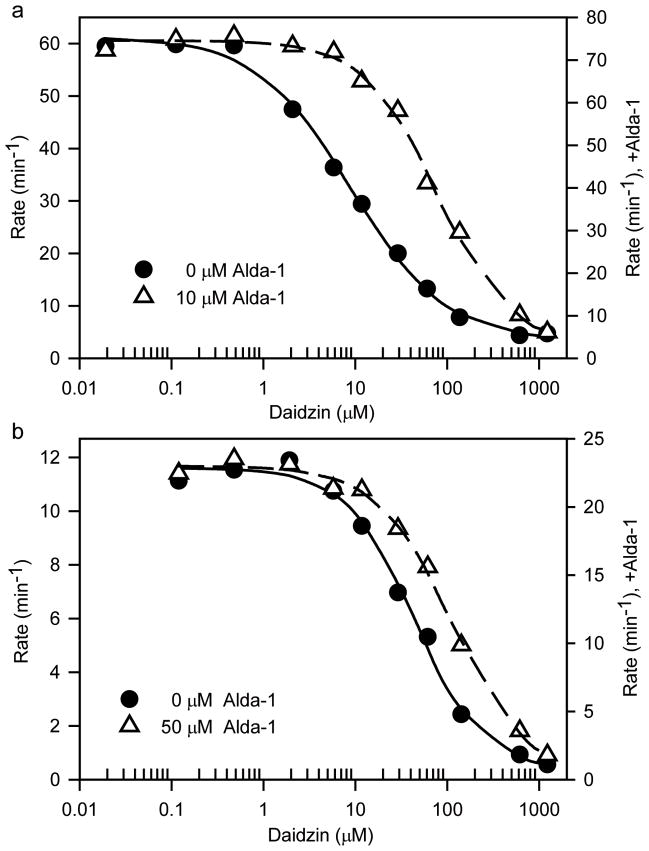
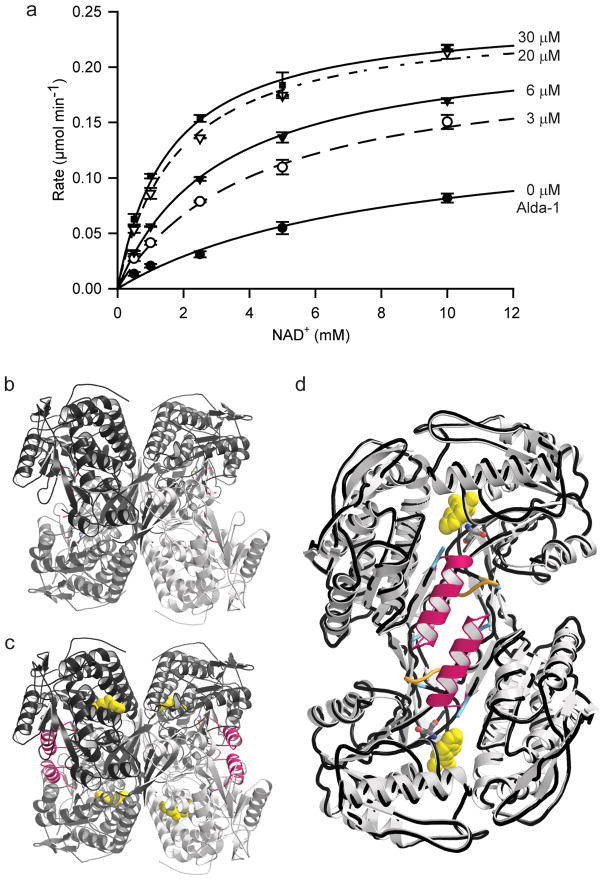
The location of Alda-1 within the substrate-binding tunnel of ALDH2 led us to investigate whether the activation effects of Alda-1 were dependent on the substrate length and whether Alda-1 activation and daidzin inhibition were mutually exclusive. At pH 7.5, we find a strong dependence of activation on the length and nature of the substrate aldehydes with linear aliphatic aldehydes up to butyraldehyde activated by Alda-1 with μM KAct values (Table 2) and aromatic aldehydes, such as benzaldehyde, phenylacetaldehyde and 4-trans-(N,N-dimethylamino)-cinnamaldehyde (DACA), exhibiting minimal effects at 20 μM Alda-1 and saturating concentrations of substrate. Alda-1 antagonized daidzin inhibition of both ALDH2 and ALDH2*2 in a manner consistent with their overlapping binding sites in ALDH2 (Table 3 and Fig. 3). We analyzed, in detail, the nature of the activation of ALDH2*2 through a covariation experiment between NAD+ and Alda-1 and fit the data to the non-essential activator equation. This analysis showed that Alda-1 increases the Vmax of ALDH2*2 by 2-fold and decreases the apparent Km for NAD+ by 6.7-fold (Fig. 4).
Table 2.
Substrate Dependence for Alda-1 Activationa
| Substrate | Vmax(app) (min−1) | KAct(app) (μM) | Vmax/Vo |
|---|---|---|---|
| 100 μM Acetaldehyde | 107 +/− 12 | 0.98 +/− 0.20 | 1.8 +/− 0.1 |
| 100 μM Propionaldehyde | 78.5 +/− 9.6 | 5.1 +/− 1.2 | 1.7 +/− 0.1 |
| 100 μM Butyraldehyde | 86.6 +/− 3.1 | 1.8 +/− 0.5 | 1.3 +/− 0.1 |
Data were fit to the non-essential activator expression (see Methods), where Vo is the initial velocity for the reaction in the absence of activator, Vmax is the maximal velocity and KAct is the concentration of activator required for half-maximal activation
Table 3.
Alda-1 induced antagonism of Daidzin Inhibitiona
| ALDH2 (no Alda-1) | ALDH2 (10 μM Alda-1) | ALDH2*2 (no Alda-1) | ALDH2*2 (50 μM Alda-1) | |
|---|---|---|---|---|
| Vmax(app) (min−1) | 61.5 +/− 6.1 | 74.9 +/− 12.1 | 11.6 +/− 1.3 | 23.0 +/− 1.7 |
| Vmin(app) (min−1) | 5.1 +/− 4.7 | 2.5 +/− 5.2 | 0.2 +/− 0.6 | 0.2 +/− 0.2 |
| Daidzin IC50 (μM) | 8.0 +/− 0.6 | 72.0 +/− 16.9 | 44.8 +/− 10.4 | 113.0 +/− 15.6 |
| Hill Slope | 0.9 +/− 0.3 | 1.0 +/− 0.1 | 1.1 +/− 0.1 | 1.1 +/− 0.1 |
The daidzin inhibition data were fit to the four parameter EC50 equation (see Methods) where Vmax is the velocity in the absence of inhibitor, Vmin is the minimum velocity from the fit EC50 is the concentration required for 50% inhibition, and Hill Slope is the Hill coefficient as an indicator of potential cooperativity.
Figure 3. Alda-1 competition with daidzin inhibition.
Dehydrogenase activity was measured at 0.1 mM propionaldehyde, varying concentrations of daidzin for (A) Wild-type ALDH2 at 0 (solid line and circles) or 10 μM Alda-1 (dashed line and open triangles) and (B) ALDH2*2 at 0 (solid line and circles) or 50 μM Alda-1 (dashed line and open triangles). NAD+ concentrations were 0.5 mM for wild-type ALDH2 and 10 mM for ALDH2*2. The lines demonstrate the fits to the 4-parameter logistic curve.
Figure 4. Restoration of Coenzyme-binding properties in ALDH2*2 upon Alda-1 Binding.
(A) Michaelis-Menton plot for the effects of Alda-1 on the dehydrogenase activity of ALDH2*2 against varied NAD+. The plot shows the average values from 3 experiments: Kact = 16 +/− 3 μM; KMNAD = 7.4 +/− 0.7 mM; α-factor = 0.15 +/− 0.03; β-factor = 2.0 +/− 0.2. The concentrations of Alda-1 used in each trace are labeled (0, 3, 6, 20 and 30 μM). The α- and β-factors describe the manner in which Alda-1 impacts the observed KMNAD and Vmax, respectively (See Methods). Thus, Alda-1 restores the KM for NAD+ from 7.4 mM to 1.1 mM and increases the Vmax 2-fold. (B) Ribbon diagram of human ALDH2*2 in the absence Alda-1 (C) Ribbon diagram of human ALDH2*2 in the presence Alda-1. (D) An overlay of a dimer from the ALDH2*2 structures with (light grey ribbons) and without Alda-1 (black worm tracing). For panels (C) and (D), the positions of the Alda-1 molecules are indicated using yellow space-filling representations and the positions of the now ordered αG helices in each subunit are highlighted with pink coloring. Panels B, C and D were produced using PyMol for Windows25.
Kinetics of Alda-1 activation on ALDH2 esterase activity
In addition to the dehydrogenation reaction, members of the aldehyde dehydrogenase family also exhibit the ability to hydrolyze esters, such as p-nitrophenylacetate, and the coenzyme, NAD+, is known to stimulate the hydrolytic activity of ALDH2 (refs. 12, 13). We examined the ability of Alda-1 to stimulate the hydrolysis of p-nitrophenylacetate both in the presence and absence of coenzyme. Alda-1 alone activates the esterase activity of both ALDH2 and ALDH2*2 between 6- and 7-fold and the combined activatory effects of both Alda-1 and NAD+ increase ester hydrolysis 10-fold for ALDH2*1 and over 100-fold for ALDH2*2 (Table 4). At higher concentrations Alda-1 and NAD+ become antagonistic for ALDH2, a behavior not noted for ALDH2*2 at the concentrations examined in our assays. Similar to that observed for coenzyme-binding kinetics in the dehydrogenation reaction catalyzed by ALDH2*2, Alda-1 lowered the half-maximal activating concentration of NAD+ for the esterase reaction from 7.5 mM10 to 2.8 mM for ALDH2*2 (Table 4). We sought to determine whether the binding of Alda-1 to wild-type ALDH2 is associated with a lowering of the pKa for Cys302. Prior work has shown that esterase activity in ALDH2 is limited in rate by the formation of the acyl-enzyme intermediate12,13. Consequently, we examined the pH dependence of the esterase reaction in the absence and presence of Alda-1 and found well-defined pKa values of 7.6 ± 0.1 and 7.8 ± 0.1 for ALDH2*1 in the presence of Alda-1 and for ALDH2*1 alone, respectively (Supplementary Fig. 1).
Table 4.
Esterase Activity Activation Constants for ALDH2 and ALDH2*2a
| Constant | ALDH2 | ALDH2 (0.5 mM NAD+) | ALDH2*2 | ALDH2*2 (1. 0 mM NAD+) | ALDH2*2 (50 μM Alda-1) |
|---|---|---|---|---|---|
| Vo (min−1) | 24.9 +/− 2.0 | 96.3 +/− 2.2 | 0.40 +/− 0.03 | 1.36 +/− 0.28 | 0.64 +/− 0.19 |
| Vmax (min−1) | 181 +/− 6.8 | 248 +/− 22 | 2.3 +/− 0.2 | 14.7 +/− 1.3 | 49.5 +/− 4.3 |
| KAct(app) (μM) | 3.4 +/− 0.5 | 2.6 +/− 0.1 | 16.1 +/− 5.8 | 11.2 +/− 1.3 | 2,820 +/− 330 |
| Vmax (min−1) | -- | 242 +/− 14 | -- | -- | -- |
| Ki(app) (μM) | -- | 328 +/− 24 | -- | -- | -- |
Data were fit to the non-essential activator expression (see Methods), where Vo is the initial velocity for the reaction in the absence of activator, Vmax is the maximal velocity and KAct is the concentration of activator required for half-maximal activation and Ki is the concentration required for half-maximal inhibition.
Discussion
The discovery of a novel activator of ALDH2*1 and ALDH2*2 (ref. 3), and the desire to improve the properties of this interesting lead compound led us to characterize the mechanism of activation of Alda-1. First, we sought structural insight into the interaction of Alda-1 with ALDH2. The structure of the binary complexes between Alda-1 and ALDH2*1 and ALDH2*2 were solved by X-ray crystallography. The site to which Alda-1 binds was surprising and generated several working hypotheses for how ALDH2*1 and ALDH2*2 are activated. With this new information in mind, we designed a series of kinetics experiments to characterize the substrate and reaction specificity of activation.
The location of Alda-1 binding within the substrate entrance tunnel of ALDH2 is reminiscent of the binding of daidzin, a known potent inhibitor of ALDH2 (ref. 13). If the positions of Alda-1 and daidzin in their respective crystal structures are correct, we reasoned that Alda-1 should antagonize daidzin inhibition. As shown in Table 3 and Figure 3 (see also ref. 3), we found this to be true for both the wild-type, ALDH2*1, and for the mutant, ALDH2*2, confirming that Alda-1 and daidzin share overlapping binding sites. The very different effects of daidzin and Alda-1 on ALDH2 activity can be explained, in part, from their crystal structures. In the daidzin bound structure13, the phenolic moiety interacts directly with two essential active site residues, Cys302 and Glu268, inhibiting the enzyme by restricting substrate binding and catalysis. In contrast, our structure shows that Alda-1 binds at the entrance to the active site, but does not sterically interfere with the catalytic residues.
Because Alda-1 does block part of the substrate site, we predicted that ALDH2 activation would depend on substrate size. Modeling of the complex suggests that the space between Cys302 and the benzodioxol ring of Alda-1 could accommodate acyl-enzyme intermediates up to 4 carbons in length (Fig. 5). Hence, we examined the concentration dependence of Alda-1 activation at saturating concentrations of acetaldehyde, propionaldehyde, butyraldehyde, benzaldehyde, phenylacetaldehyde, and 4-trans-(N,N-dimethylamino)-cinnamaldehyde (DACA). We found that only the smaller linear aliphatic aldehydes were activated by Alda-1 and the extent of activation decreased with length (Table 2). Alda-1 had little effect on activity with benzaldehyde, even at the maximum concentration used; 200 μM. Alda-1 also had little effect on ALDH2 activity with phenylaceteldehyde or DACA, although high concentrations of Alda-1 (>100 μM) were weakly inhibitory. Of course, these data do not explain how substrate is able to gain access to the active site when Alda-1 is bound in the substrate entrance tunnel, nor do they shed light on why Alda-1 is an activator rather than an inhibitor of ALDH2.
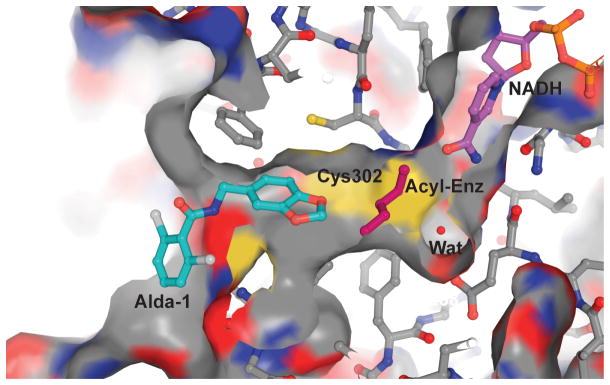
Figure 5. Model of the potential mechanism for activation of ALDH2 by Alda-1.
The structure of the Alda-1 complex was used to model a quaternary complex between ALDH2, coenzyme, acyl-enzyme intermediate and Alda-1. For this figure, the position of reduced coenzyme is shown in the ‘hydrolysis position’, whereas the hydride transfer position puts the nicotinamide ring in the space between Cys302 and Glu268. A butyryl group (magenta) is shown as the acyl-enzyme intermediate and a crystallographically observed water molecule (magenta sphere) is shown between Cys302 and Glu 268. The figure was generated using PyMol for Windows25.
The answers to these questions reside in the unique nature of coenzyme binding to ALDHs. In all the ALDH family members, the active site and the NAD+ coenzyme binding site are connected, forming a tunnel through the enzyme. One rationale for the ordered binding of coenzyme prior to that of aldehyde substrate is that the binding of coenzyme helps to retain the substrate within the active site by lowering the frequency of its exit from the coenzyme-binding site side of this tunnel. Figure 5 shows a hypothetical quaternary complex of ALDH2, an acyl-enzyme intermediate of butyraldehyde, NAD+, and Alda-1. Although Alda-1 blocks the substrate entrance tunnel, we propose that substrate access and product release can still occur - through the nicotinamide cleft. This is possible because of the well-characterized flexibility of the nicotinamide mononucleotide moiety in ALDH2 (refs. 14,15). We and others have demonstrated that the nicotinamide half of the NAD+ coenzyme retains substantial flexibility when bound to ALDH isozymes. In ALDH2, there appear to be two relatively stable conformations relevant to catalysis; with the nicotinamide ring adjacent to or rotated away from Cys302 (ref. 15). We also know from NMR studies that both NAD+ and NADH are rapidly sampling a number of conformations when bound to ALDH2 (ref. 14). In addition, several alternate cofactor conformations have been observed in crystal structures of other ALDH family members16–18. Hence, the coenzyme can - and probably does - sample multiple conformations during catalysis which permit both substrate binding and product release even when Alda-1 is bound.
Activation of dehydrogenase activity by Alda-1 is most likely achieved through acceleration of acyl-enzyme hydrolysis, the rate-limiting step in ALDH2*1. Clearly, limiting the substrate access to the nicotinamide cleft could slow the rate of substrate binding or product release. However, the Vmax/KM values for aldehyde oxidation by ALDH2*1 are close to diffusion-limited conditions, so even a substantial decrease in aldehyde association rates will not be observed in steady state measurements, where the rates of catalysis are approximately five orders of magnitude lower. On the other hand, we know from the effects of Alda-1 and NAD+ on the esterase reaction that the simultaneous presence of both molecules accelerates ester hydrolysis, even though the rate-limiting step, in the absence of activators, is known to be acylation19,20. Consequently, access through the nicotinamide cleft may not appreciably slow substrate access. The fact that Alda-1 activates ALDH2*1 approximately 2-fold for the dehydrogenase reaction suggests that the activated water molecule, on average, has half as many non-productive encounters within the active site. Thus, we propose that by binding at and blocking one of the exits from the active site, Alda-1 increases the likelihood of a productive encounter between the activated water molecule and the thioacyl intermediate (Fig. 5), moderately accelerating acyl-enzyme hydrolysis in ALDH2*1.
Another important feature of Alda-1 is its ability to protect ALDH2 from inactivation due to adduct formation between the substrate and key Cys residues in the enzyme. Reduction in ALDH activity occurs in a number of disease states associated with oxidative stress21,22. We previously reported that in the presence of 100 μM of 4-hydroxynonenal, ALDH2 activity is abolished within a couple of minutes3. However, 20 μM Alda-1 prevented 4HNE-induced ALDH2 inactivation. It appears likely that Alda-1 reduces accessibility of Cys302, as well as the adjacent Cys301 and Cys303 residues, in the catalytic channel to 4-HNE adduct formation and the resulting enzyme inactivation3.
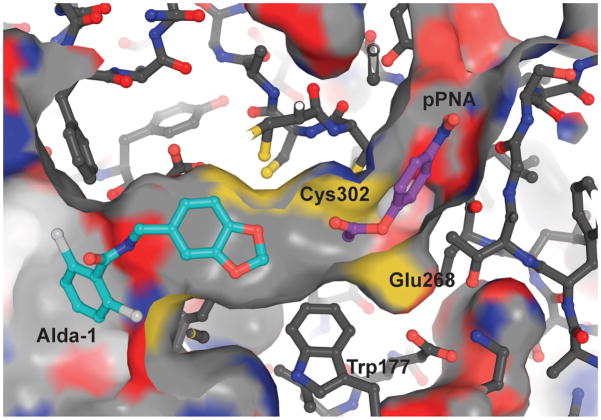
It is interesting that both coenzyme and Alda-1 are nonessential activators for the esterase reaction in ALDH2. Neither coenzyme nor Alda-1 participates in the chemical events, but the presence of either activates catalysis in a saturable manner. The ability of NAD+ and NADH to increase the rate of ester hydrolysis in ALDH2 isozymes is well-established and it is generally thought that this occurs through an increase in the nucleophilicity of Cys30217. Indeed, there is evidence that coenzyme binding is associated with a decrease in the pKa of the active site nucleophile in ALDH family members and provides a chemical and kinetic rationale for the ordered binding of coenzyme prior to the substrate aldehyde in the dehydrogenation reaction23. We show here that Alda-1 induces a similar increase in reactivity towards esters (Table 4), but this does not appear to be associated with an appreciable shift in the pKa of Cys302 upon Alda-1 binding (apparent pKa values of 7.8 and 7.6, respectively). Thus, it is likely that esterase activation occurs primarily by preventing the ester from non-productively transiting the active site through the substrate binding tunnel (Fig. 6). This is consistent with our proposed mechanism of dehydrogenase activation, where blocking one exit would effectively increase the number of productive encounters with Cys302. This mechanism could also be a component of esterase activation by coenzyme; whether coenzyme is bound in the hydride transfer or hydrolysis positions15, it would reduce the chances of non-productive escape through the coenzyme binding cleft.
Figure 6. Modeled complex between ALDH2, Alda-1 and para-nitrophenylacetate (pNPA).
The position of pPNA was modeled using local geometric constraints with the acetyl moiety in position for attack by Cys302. Cys302 is 3.1 Å from the carbonyl carbon and no other contacts between pPNA and ALDH2 or Alda-1 are closer than 2.9 Å. The figure was generated using PyMol for Windows25.
The fact that the actions of coenzyme and Alda-1 are additive suggests that the presence of both activators does not restrict access to Cys302 enough to slow the rate limiting acylation step for ester hydrolysis19,20. Thus, we propose that nonessential activator binding at either end of the substrate-to-coenzyme tunnel in ALDH would limit diffusion of the substrate straight through the enzyme and thereby increase the effective concentration of the reacting groups within the active site. Like the dehydrogenase reaction, we predict that activation of the esterase reaction by Alda-1 will depend on the size of the ester; the leaving group will need to fit within the nicotinamide cleft and the acylating portion will likely be limited to four or fewer carbons (Fig. 6). A dependence of the rate of ester hydrolysis on the size of the acylating portion has been reported and is in agreement with this prediction24. Here again, the conformational flexibility of the nicotinamide moiety is essential to facilitate both the binding of the ester and release of the products through the nicotinamide cleft.
Consistent with the structural information gleaned from the complex between Alda-1 and ALDH2*2, Alda-1 partially restores both the kcat and KM values for NAD+ in ALDH2*2 (Fig. 4). We also show that, in the presence of both coenzyme and Alda-1, esterase activity is increased to a greater extent in ALDH2*2 than in wild-type ALDH2 (100-fold vs 10-fold, respectively; Table 3). Together, these results suggest that Alda-1 binding to ALDH2*2 does more than increase the effective concentration of substrates within the active site; namely that Alda-1 directly promotes the structural and functional rescue of ALDH2*2 (Fig. 4). Our lab has previously shown that the exchange of Lys for Glu487 in ALDH2*2 results in the loss of structural integrity to the coenzyme-binding and active sites9,11. This disordering gives rise to the 200-fold elevation in KM for NAD+ and the 10-fold reduction in maximal velocity10. In general, the disorder of the coenzyme binding site begins at residue 246 and continues through Glu268 and the catalytic site is disordered in the immediate vicinity of Glu268, beginning at Phe465 and continuing on through Glu478 (ref. 9). Even when the structure of the coenzyme-binding site is restored upon coenzyme binding, the regions surrounding the coenzyme and active sites remain less ordered than in the wild-type enzyme11. Two of the active site residues that remain disordered are Glu268, the general base for catalysis, and Glu399, which stabilizes the position of the nicotinamide ring. Alda-1 does not directly contact any of the aforementioned residues, but does form close interactions with Phe459 and Trp177, both of which are in immediate proximity to Phe465 and Glu268. Thus, we hypothesize that through these interactions, binding of Alda-1 could redirect the aberrant dynamics in both regions of the enzyme to improve catalytic efficiency. Similarly, Alda-1 increases the maximal velocity of ALDH2*1 by reducing the number of non-productive encounters between the reacting groups in the active site and thereby accelerates catalysis. Thus, Alda-1 represents a new pharmacological agonist: it increases productive substrate-enzyme interaction and protects the enzyme from substrate-induced inactivation. Further, Alda-1 acts as a chemical chaperone by stabilizing the structurally-impaired enzyme at the tetrameric interface as well as within the catalytic tunnel, leading to catalytic recovery. This is a variation on the idea for chemical chaperone design discussed for lysosomal storage diseases12, where inhibitors are targeted to improve the delivery of structurally impaired enzymes to the lysosomes. Once delivered to the lysosome, the low pH promotes dissociation of the inhibitor, thereby alleviating any unwanted inhibition while increasing the yield of properly folded enzyme to the lysosomal compartment12. This work suggests that a rational design for similar molecular chaperones for other mutant enzymes may be possible by exploiting binding of compounds to sites adjacent to the structurally disrupted regions, thus avoiding the possibility of enzymatic inhibition entirely independent of the conditions in which the enzyme operates.
Methods
ALDH Expression, Purification and Kinetic studies
We produced and purified ALDH2*1 and ALDH2*2 using an E. coli expression system as previously described9,11,26. We assayed for ALDH2 activity in a standard reaction mixture comprised of 25 mM BES, pH 7.5, with 100 μM propionaldehyde and either 0.5 mM (ALDH2*1) or 10 mM NAD+ (ALDH2*2). All assays for Alda-1 characterization included a final concentration of 2% (v/v) DMSO as a cosolvent whether or not Alda-1 was present. Enzyme concentrations in the dehydrogenase assays were set between 0.03 and 0.06 μM for ALDH2*1 and between 0.3 and 0.5 μM for ALDH2*2. Esterase assays utilized 0.97 mM para-nitrophenylacetate (pNPA) as a standard substrate concentration and enzyme concentrations of 0.06 μM for ALDH2 and 0.7 μM for ALDH2*2. We analyzed all kinetic data using the program package SigmaPlot (v10.0, StatSys). We fit all activation data for the dehydrogenase and esterase reactions to the expression v = Vo+{(Vmax[S])/(KAct+[S])}, where Vo is the initial velocity for the reaction in the absence of activator and KAct is the concentration of activator required for half-maximal activation. All single-vary experiments for Alda-1 activation utilized a minimum of 10 different concentrations across a range from 0–200 μM. We performed the covariation data between NAD+ and Alda-1 for ALDH2*2 using Alda-1 concentrations between 0–30 μM and NAD+ concentrations between 0.5–10 mM and fit the resulting data to the nonessential activator equation:
where [S] is the varied concentration of coenzyme, [A] the varied concentration of activator, α is the modifier on KM, β is the modifier on Vmax and KA is the half-maximal concentration of activator. We fit the daidzin inhibition data to the four parameter EC50 equation, v = Vmin+{(Vmax−Vmin)/(1+[S]/EC50)Hillslope}. All tabulated data represent the average of a minimum of three independent experiments from at least two different enzyme preparations. We determined of the pKa values associated with esterase catalysis in ALDH2 with and without Alda-1 across a pH range of 5.0 to 9.5 using the following buffer system; 15 mM acetic acid, 15 mM 2-(N-morpholino)ethanesulfonic acid (MES) and 60 mM Tris, where the pH was adjusted using either HCl or NaOH to achieve the desired pH value. We chose this buffer system to minimize buffer-assisted ester hydrolysis and variations in ionic strength across the pH range. DMSO was present at a final concentration of 2% (v/v) in both control and Alda-1 containing reactions. The apparent pKa values were obtained through fits of the pH versus velocity plots using the single ascending pKa function available in SigmaPlot (v 10.0, StatSys).
Crystallization and Structure determination
We grew crystals of the wild-type ALDH2 and the C302S mutant of ALDH2*2 under conditions similar to that previously reported15,26. The apo-enzymes were concentrated to 8 mg ml-1 and equilibrated against a crystallization solution that contained 100 mM ACES, pH 6.4, 100 mM guanidine-HCl, 10 mM MgCl2 and 16–17% (w/v) PEG 6000. We prepared the complexes with Alda-1 through a direct soaking experiment in which the apo-enzyme crystals were first equilibrated against 2% DMSO in a crystal stabilization solution (100 mM ACES, pH 6.4, 100 mM guanidine-HCl, 10 mM MgCl2 and 19% (w/v) PEG 6000). Following an overnight incubation, the stabilization solution was replaced with an identical solution to which 250 μM Alda-1 was added. We allowed the crystals to incubate overnight and prepared them for cryogenic freezing using a two-step protocol to introduce 18% (v/v) ethylene glycol into the soaking solution. We collected the diffraction data at beamline 19-ID which is operated by the Structural Biology Consortium located within the Advanced Photon Source at Argonne National Laboratory. All diffraction data were indexed, integrated and scaled using the HKL3000 program suite27. Crystals of ALDH2 tend to form two different lattice groups primitive orthorhombic P212121 and a pseudo-centered monoclinic lattice that indexes in the C2221 space group with the same cell dimensions as the primitive orthorhombic lattice. However, the intensities of the diffraction pattern lack the strict centered orthorhombic symmetry and generally require integration in a primitive monoclinic lattice. The monoclinic crystal form generally exhibits pseudomerohedral twinning and both datasets used in this study suffered from this effect, though to varying degrees. We obtained initial estimates for the twin-fraction and the twinning operator using the Phenix-xtriage program28. As the structure is essentially isomorphous with the wild-type ALDH2 monoclinic data set, the structures were solved by direct refinement using the wild-type human ALDH2 structure (with ligands and solvent removed) as the starting model (PDB code 1cw3, ref. 26). Confirmation of the binding of Alda-1 was evaluated through inspection of the initial Fo-Fc electron density maps. Refinement of the structure utilized the program Phenix28 (version 1.4–3) with the twin refinement option and a selection of Rfree reflections that account for the twin operations. The resulting structures were visually inspected and adjusted using the visualization program Coot29.
Supplementary Material
Acknowledgments
TDH wishes to thank the staff at 19-ID, especially Marianne Cuff, Norma Duke and Steve Ginell. Results shown in this report are derived from work performed at Argonne National Laboratory, Structural Biology Center at the Advanced Photon Source. Argonne is operated by UChicago Argonne, LLC, for the U.S. Department of Energy, Office of Biological and Environmental Research under contract DE-AC02-06CH11357. This work was supported in part by NIH grants AA011982 and AA018123 to TDH and AA11147 to DM-R; SP-M was supported by T32-DK064466.
Footnotes
Accession Codes
The structure factors and derived atomic coordinates are deposited in the RCSB under ascension codes 1INJ and 1INL.
Author contributions statement:
SP-M performed experiments, data analysis and manuscript preparation
HY performed experiments and helped with data analysis
RV performed experiments
C-HC helped with data analysis and manuscript preparation
DM-R initiated the study and helped with study design, data analysis and manuscript preparation
TDH study design, data analysis, performed experiments, and manuscript preparation
References
- 1.Hurley TD, Edenberg HJ, Li TK. Pharmacogenomics of Alcholism. In: Licinio J, Wong M, editors. Pharmacogenomics: The Search for Individualized Therapies. 2002. pp. 417–441. [Google Scholar]
- 2.Chen Z, Foster MW, Zhang J, Mao L, Rockman HA, Kawamoto T, Kitagawa K, Nakayama KI, Hess DT, Stamler JS. An essential role for mitochondrial aldehyde dehydrogenase in nitroglycerin bioactivation. Proc Nat Acad Sci (USA) 2005;102(34):12159–12164. doi: 10.1073/pnas.0503723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen CH, Budas GR, Churchill EN, Disatnik MH, Hurley TD, Mochly-Rosen D. Activation of aldehyde dehydrogenase-2 reduces ischemic damage to the heart. Science. 2008;321:1493–1495. doi: 10.1126/science.1158554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Higuchi S, Matsushita S, Imazeki H, Kinoshita T, Takagi S, Kono H. Aldehyde dehydrogenase genotypes in Japanese alcoholics. Lancet. 1994;343:741–742. doi: 10.1016/s0140-6736(94)91629-2. [DOI] [PubMed] [Google Scholar]
- 5.Brooks PJ, Enoch MA, Goldman D, Li TK, Yokoyama A. The alcohol flushing response: an unrecognized risk factor for esophageal cancer from alcohol consumption. PLoS Medicine. 2009;6:258–263. doi: 10.1371/journal.pmed.1000050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Y, Zhang D, Jin W, Shao C, Yan P, Xu C, Sheng H, Liu Y, Yu J, Xie Y, Zhao Y, Lu D, Nebert DW, Harrison DC, Huang W, Jin L. Mitochondrial aldehyde dehydrogenase-2 (ALDH2) Glu504Lys polymorphism contributes to the variation in efficacy of sublingual nitroglycerin. J Clin Invest. 2006;116:506–511. doi: 10.1172/JCI26564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jo SA, Kim EK, Park MH, Han C, Park HY, Jang Y, Song BJ, Jo I. A Glu487Lys polymorphism in the gene for mitochondrial aldehyde dehydrogenase 2 is associated with myocardial infarction in elderly Korean men. Clin Chim Acta. 2007;382:43–47. doi: 10.1016/j.cca.2007.03.016. [DOI] [PubMed] [Google Scholar]
- 8.Marchitti SA, Deitrich RA, Vasiliou V. Neurotoxicity and metabolism of the catecholamine-derived 3,4-dihydroxyphenylacetaldehyde and 3,4-dihydroxyphenylglycolaldehyde: the role of aldehyde dehydrogenase. Pharmacol Rev. 2007;59:125–150. doi: 10.1124/pr.59.2.1. [DOI] [PubMed] [Google Scholar]
- 9.Larson HN, Weiner H, Hurley TD. Disruption of the coenzyme binding site and dimer interface revealed in the crystal structure of mitochondrial aldehyde dehydrogenase “asian” variant. J Biol Chem. 2005;280:30550–30556. doi: 10.1074/jbc.M502345200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Farres J, Wang X, Takahashi K, Cunningham SJ, Wang TT, Weiner H. Effect of changing glutamate to lysine in rat and human liver mitochondrial aldehyde dehydrogenase: a model to study human (Oriental type) class 2 aldehyde dehydrogenase. J Biol Chem. 1994;269:13854–13860. [PubMed] [Google Scholar]
- 11.Larson H, Zhou J, Chen Z, Stamler JS, Weiner H, Hurley TD. Structural and functional consequences of coenzyme binding to the inactive asian variant of mitochondrial aldehyde dehydrogenase: Roles of residues 475 and 487. J Biol Chem. 2007;282:12940–12950. doi: 10.1074/jbc.M607959200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Suzuki Y, Ogawa S, Sakakibara Y. Chaperone therapy for neuronopathic lysosomal diseases: Competitive inhibitors as chemical chaperones for enchancement of mutant enzyme activities. Perspect Med Chem. 2009;3:7–19. doi: 10.4137/pmc.s2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lowe ED, Gao GY, Johnson LN, Keung WM. Structure of Daidzin, a Naturally Occurring Anti-Alcohol-Addiction Agent, in Complex with Human Mitochondrial Aldehyde Dehydrogenase. J Med Chem. 2008;51:4482–4487. doi: 10.1021/jm800488j. [DOI] [PubMed] [Google Scholar]
- 14.Hammen PK, Allali-Hassani A, Hallenga K, Hurley TD, Weiner H. Multiple Conformations of NAD and NADH When Bound to Human Cytosolic and Mitochondrial Aldehyde Dehydrogenase. Biochemistry. 2002;41:7156–7168. doi: 10.1021/bi012197t. [DOI] [PubMed] [Google Scholar]
- 15.Perez-Miller S, Hurley TD. Coenzyme isomerization is integral to catalysis in human aldehyde dehydrogenase. Biochemistry. 2003;42:7100–7109. doi: 10.1021/bi034182w. [DOI] [PubMed] [Google Scholar]
- 16.Liu ZJ, Sun YJ, Rose J, Chung YJ, Hsiao CD, Chang WR, Kuo I, Perozich J, Lindahl R, Hempel J, Wang BC. The first structure of an aldehyde dehydrogenase reveals novel interactions between NAD and the Rossmann fold. Nat Struct Biol. 1997;4:317–326. doi: 10.1038/nsb0497-317. [DOI] [PubMed] [Google Scholar]
- 17.Lamb AL, Newcomer ME. The structure of retinal dehydrogenase type II at 2.7 Å resolution: Implications for retinal specificity. Biochemistry. 1999;38:6003–6011. doi: 10.1021/bi9900471. [DOI] [PubMed] [Google Scholar]
- 18.D’Ambrosio K, Pailot A, Talfournier F, Didierjean C, Benedetti E, Aubry A, Branlant G, Corbier C. The first crystal structure of a thioacylenzyme intermediate in the ALDH family: new coenzyme conformation and relevance to catalysis. Biochemistry. 2006;45:2978–2986. doi: 10.1021/bi0515117. [DOI] [PubMed] [Google Scholar]
- 19.Feldman RI, Weiner H. Horse liver aldehyde dehydrogenase. II. Kinetics and mechanistic implications of the dehydrogenase and esterase activity. J Biol Chem. 1972;247:267–272. [PubMed] [Google Scholar]
- 20.Takahashi K, Weiner W. Nicotinamide adenine dinucleotide activation of the esterase reaction of horse liver aldehyde dehydrogenase. Biochemistry. 1981;20:2720–2726. doi: 10.1021/bi00513a003. [DOI] [PubMed] [Google Scholar]
- 21.Wenzel P, Muller J, Zurmeyer S, Schuhmacher S, Schulz E, Oelze M, Pautz A, Kawamoto T, Wojnowski L, Kleinert H, Munzel T, Daiber A. ALDH-2 deficiency increases cardiovascular oxidative stress--evidence for indirect antioxidative properties. Biochem Biophys Res Commun. 2008;367:137–43. doi: 10.1016/j.bbrc.2007.12.089. [DOI] [PubMed] [Google Scholar]
- 22.Jinsmaa Y, Florang VR, Rees JN, Anderson DG, Strack S, Doorn JA. Products of oxidative stress inhibit aldehyde oxidation and reduction pathways in dopamine catabolism yielding elevated levels of a reactive intermediate. Chem Res Toxicol. 2009;22:835–41. doi: 10.1021/tx800405v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marchal S, Branlant G. Evidence for the Chemical Activation of Essential Cys-302 upon Cofactor Binding to Nonphosphorylating Glyceraldehyde 3-Phosphate Dehydrogenase from Streptococcus mutans. Biochemistry. 1999;38:12950–12958. doi: 10.1021/bi990453k. [DOI] [PubMed] [Google Scholar]
- 24.Mukerjee N, Pietruszko R. Human mitochondrial aldehyde dehydrogenase substrate specificity: comparison of esterase with dehydrogenase reaction. Arch Biochem Biophys. 1992;299:23–29. doi: 10.1016/0003-9861(92)90239-s. [DOI] [PubMed] [Google Scholar]
- 25.DeLano WL. The PyMOL Molecular Graphics System. DeLano Scientific; San Carlos, CA: 2002. [Google Scholar]
- 26.Ni L, Zhou J, Hurley TD, Weiner H. Human liver mitochondrial aldehyde dehydrogenase: Three-dimensional structure and the restoration of solubility and activity of chimeric forms. Protein Science. 1999;8:2784–2790. doi: 10.1110/ps.8.12.2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Meth Enz. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 28.Adams PD, Grosse-Kunstleve RW, Hung L-W, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. Acta Cryst. D58. 2002. PHENIX: building new software for automated crystallographic structure determination; pp. 1948–1954. [DOI] [PubMed] [Google Scholar]
- 29.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.