Abstract
Hausp is a deubiquitinase that has been shown to regulate the p53–Mdm2 pathway. Cotransfection of p53 and Hausp stabilizes p53 through the removal of ubiquitin moieties from polyubiquitinated p53. Interestingly, knockout or RNA interference-mediated knockdown of Hausp in human cells also resulted in the stabilization of p53 due to the destabilization of Mdm2, suggesting a dynamic role of Hausp in p53 activation. To understand the physiological functions of Hausp, we generated hausp knockout mice. Hausp knockout mice die during early embryonic development between embryonic days E6.5 and E7.5. The hausp knockout embryos showed p53 activation, but no apparent increase in apoptosis. Embryonic lethality was caused by a dramatic reduction in proliferation and termination in development, in part due to p53 activation and/or abrogation of p53-independent functions. Although deletion of p53 did not completely rescue the embryonic lethality of the hausp knockout, embryonic development was extended in both hausp and p53 double knockout embryos. These data show that Hausp has a critical role in regulating the p53–Mdm2 pathway.
Keywords: hausp, p53, mdm2, deubiquitination, gene knockout, mouse
Introduction
Posttranslational modification of proteins by ubiquitination has been shown to be an important regulatory mechanism, not only for controlling protein stability but also for the regulation of important cellular functions. The balance between ubiquitinated and non-ubiquitinated forms of a protein is delicately maintained to achieve the spatial and temporal regulations of the protein, as well as its physiological functions. These regulations are especially evident for p53, a tumor suppressor, which is well studied because of its critical functions in maintaining cellular homeostasis. Under unstressed conditions, p53 is maintained at a low level in normal cells, through ubiquitination catalyzed by Mdm2 and degradation mediated by the 26S protea-some. The importance of this regulation is underscored by the embryonic lethality of mdm2 knockout mice due to apoptosis resulting from p53 activation. However, combining a p53-null background with the mdm2 knockout rescues this phenotype, suggesting that Mdm2 is a major negative regulator of p53 (Jones et al., 1995; Montes de Oca Luna et al., 1995). Recent studies have also shown that other ubiquitin ligases, such as Pirh2, Cop1 and Arf-bp1/Mule1, ubiquitinate p53 and potentially contribute to the regulation of the stability and functions of p53 (Leng et al., 2003; Dornan et al., 2004; Chen et al., 2005). Under stress and DNA damage conditions, p53 is stabilized and activates downstream targets to initiate cell-cycle control, DNA repair mechanisms and apoptosis. Stabilization of p53 can be achieved by inhibiting Mdm2-mediated ubiquitination, through mechanisms such as the activation of p14Arf (Sherr and Webber, 2000). Alternatively, stabilization of p53 can also be achieved by deubiquitination, catalyzed by deubiquitinases (DUBs), to reduce ubiquitination of p53. Through purification of p53-binding proteins, the ubiquitin-specific protease Hausp, also known as USP7, was identified as a specific p53-binding protein. Subsequently, Hausp was shown to stabilize p53 through deubiquitination and to upregulate p53 functions (Li et al., 2002).
Further studies have shown that Hausp can bind to multiple substrates, including Mdm2, MdmX and FoxO4a, as well as regulate their stability and functions through deubiquitination. The dual control of p53 and Mdm2 by Hausp indicates a complicated p53–Mdm2– Hausp regulatory pathway. Evidence of this interplay came from the somatic knockout of Hausp in HCT116 cells. These cells exhibited destabilization of Mdm2, and surprisingly, a dramatic stabilization of p53 due to the absence of Mdm2 (Cummins et al., 2004). Interestingly, moderate reduction of Hausp protein levels by RNA interference in U2OS cells resulted in the destabilization of p53. However, complete ablation of the Hausp protein resulted in the stabilization of p53 due to the loss of Mdm2 (Li et al., 2004). These results further highlight the importance of Mdm2 in the regulation of p53 stability, and implicate a critical regulation of p53 stability by Hausp through deubiquitination of both Mdm2 and p53. In addition, MdmX was also found to interact with Hausp and its stability was increased after overexpression of Hausp, adding more complexity to the regulation of the p53–Mdm2 pathway by Hausp (Meulmeester et al., 2005).
These complex findings can be supported at the molecular level as well. Through the determination of crystal structures of Hausp and the complexes formed between Hausp/Mdm2 and Hausp/p53, it has been shown that both Mdm2 and p53 bind to the same domain on Hausp. In addition, the binding affinity for Mdm2 is higher than that for p53, suggesting that Mdm2 is preferentially stabilized under normal cellular conditions (Hu et al., 2002, Holowaty et al., 2003; Hu et al., 2006). In fact, experiments have shown that the half-life of Mdm2 shortens drastically with a concomitant increase of p53 levels in hausp knockout HCT116 cells and in cells with complete knockdown of hausp by small interfering RNA (Cummins et al., 2004; Li et al., 2004). Under genotoxic stress and DNA damage conditions, the affinity of Mdm2 for Hausp changes because of protein modifications. This may lead to an affinity switch of Hausp for p53, resulting in the destabilization of Mdm2 and stabilization of p53.
Deubiquitinases have been shown to function in a wide variety of cellular processes (Hershko and Ciechan-over, 1998; Laney and Hochstrasser, 1999; Pickart and Eddins, 2004). Besides the regulation of protein stability, DUBs have been shown to regulate chromatin structure and to facilitate both gene activation and gene silencing. Studies of Hausp have provided a glimpse of its potential physiological significance. Besides having a dynamic role in the p53–Mdm2 pathway, Hausp was initially discovered as a protein associated with Herpes virus protein vmw110 that facilitated the activation of type 1 herpes simplex virus expression and viral replication (Everett et al., 1999). Hausp has also been shown to interact with Guanosine 5′-monophosphate synthetase in Drosophila, and together mediate the deubiquitination of histone H2B, which is important for the epigenetic silencing of homeotic genes mediated by the Polycomb protein (van der Knaap et al., 2005). Both of these findings implicate specific regulation of chromatin structure through histone deubiquitination and the wide range of effects on transcriptional activation. Other mammalian proteins have been shown to interact with Hausp as well, including Ataxin-1, Daxx and FoxO4a (Hong et al., 2002; Tang et al., 2006; van der Horst et al., 2006). Interestingly, Hausp modulates the balance of mammalian FoxO4a and its mono-ubiquitinated form, which determines FoxO4a cellular localization and its transcriptional activity in response to oxidative stress (van der Horst et al., 2006).
To validate the diverse functions of Hausp, it is critical to investigate its physiological functions in vivo. To accomplish this, a mouse model carrying a null allele of hausp was generated. Knockout of the hausp gene in mice caused early embryonic lethality between embryonic days 6.5 (E6.5) and E7.5. The hausp knockout embryos showed p53 stabilization and cell growth arrest. Although the lethality in hausp knockout mice cannot be fully rescued by deletion of p53, the development of the embryos of the hausp and p53 double knockout mice was extended, suggesting a partial rescue of the hausp knockout phenotype by deletion of p53. Furthermore, mouse embryonic fibroblast (MEF) cells from hausp heterozygote mice showed modestly reduced protein half-lives of Mdm2 and p53, as well as decreased p53 activation after DNA damage, potentially due to the reduced Hausp protein level. These results suggest that Hausp has a critical role in regulating p53 stability. The results also imply that Hausp may have roles in p53-independent functions that are essential in both proliferation and differentiation.
Results
Genetargeting of hausp
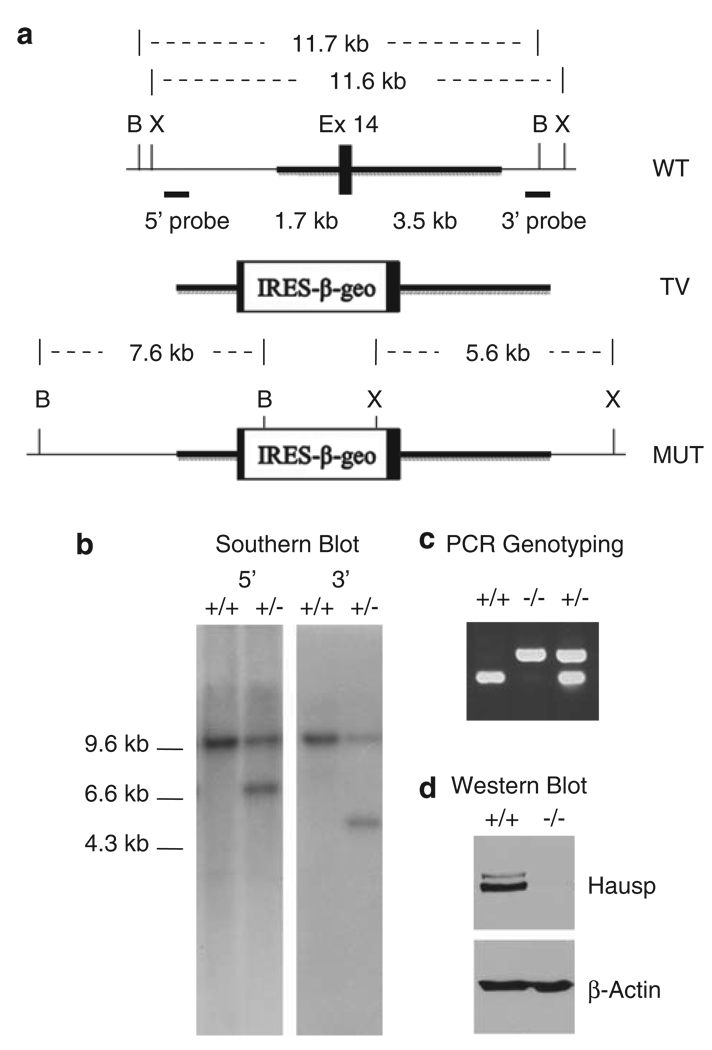
To investigate the physiological functions of Hausp, a hausp knockout mouse was generated using homologous recombination. A conventional targeting vector was created to insert an IRES LacZ-neo cassette into exon 14 of hausp and to delete 29 nucleotides within exon 14 (Figure 1a). The resulting mutation causes C-terminal truncation and partial deletion of DUB active site domain due to a reading frame shift in protein translation. The targeting event was confirmed by Southern blotting using external probes (Figure 1b). Mutated mouse embryonic stem cells were microinjected into blastocysts to derive chimeras, from which germline transmission of the mutant allele of the hausp gene was achieved. Hausp heterozygote mice were maintained in a 129 and C57BL/6J mix background. Initially, mice were genotyped by Southern blot. Subsequently, offspring were genotyped by PCR (Figure 1c).
Figure 1.
Targeted disruption of the mouse hausp gene. (a) Diagram of the hausp genomic region containing exon 14 is shown with restriction fragments of BamHI and XbaI indicated. Targeting vector contains IRES-lac Z and a neo cassette was inserted into exon 14, flanking with 1.7 kb 5′-homology and 3.5kb 3′-homology (thicker lines). The diagram of the targeted mutant allele is shown, with altered restriction fragments of BamHI and XbaI after correct gene targeting. (b) Genotyping by Southern blot using genomic DNA digested by BamHI and XbaI, respectively. Hybridization with a 5′-external probe detected an 11.7kb wild-type band and an additional 7.6-kb mutant band in heterozygote mouse. Hybridization using 3′-external probe detected an 11.6 wild-type band and a 5.6-kb mutant band. (c) Genotyping by PCR showed a 202-bp band for wild-type allele and a 433-bp band for mutant allele, simultaneously. (d) The Hausp protein is absent in hausp knockout embryo. Protein extracts prepared from wild-type embryo and hausp knockout embryo at day E8.5, were analyzed by western blot using an anti-Hausp polyclonal antibody and an anti-β-actin monoclonal antibody. The Hausp protein was shown absent in hausp knockout embryo, whereas the total proteins, indicated by β-actin level, were comparable. B, BamHI; X, XbaI.
Targeted disruption of hausp resulted in a null allele
To prove that the targeted disruption of the hausp locus created a null mutation, two experiments were conducted. First, the protein containing the N terminus of Hausp from exons 1 to 13 was expressed in cultured cells. The truncated protein failed to stabilize p53 through deubiquitination, suggesting loss of DUB activity for the C-terminal truncated Hausp protein (Li M and Gu W, unpublished results). Second, protein extracts were prepared from wild-type and hausp knockout embryos of day E8.5 from mating of hausp heterozygote mice and analyzed by western blot. The Hausp protein was readily detected in protein extracts collected from the wild-type embryo, whereas no Hausp protein was detected in protein extracts from hausp homozygote knockout embryos (Figure 1d). We also did not detect the C-terminal truncated Hausp protein using polyclonal antibody against the N-terminus of Hausp, suggesting that the truncated protein was not stable. Therefore, we concluded that the disruption of the hausp locus created a null mutation.
Expression pattern of Hausp during early embryogenesis
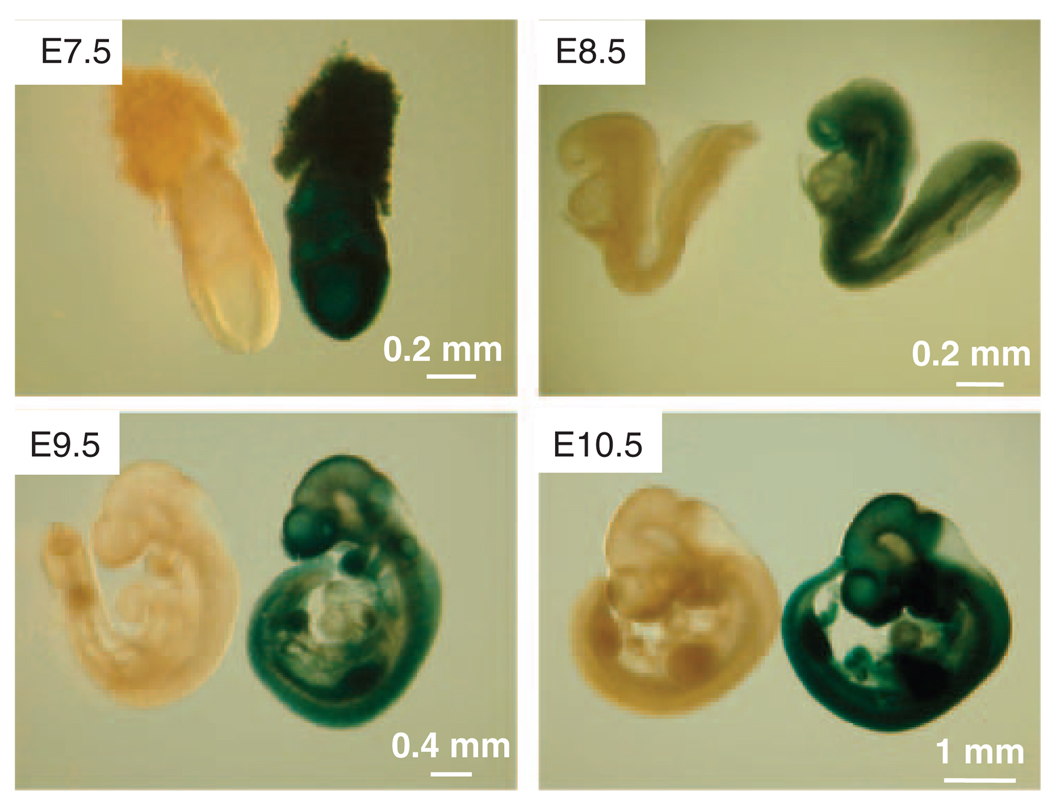
The inserted IRES-lacZ into the hausp locus allows bicistronic expression of β-galactosidase controlled by the hausp promoter. Therefore, the expression pattern of hausp can be identified by the presence of β-galactosidase activity, which was visualized by X-gal staining. As shown by the blue color resulting from the X-gal staining, hausp expression was detected as early as the blastocyst stage (day E3.5) (data not shown), and the expression pattern was ubiquitous during early embryonic development from day E7.5 to E10.5 (Figure 2). Consistent with X-gal staining, immunostaining using a polyclonal antibody against Hausp showed that Hausp was a nuclear protein and was expressed ubiquitously in all cells derived from the embryo both in embryonic and in trophoblastic cells (Figure 3e).
Figure 2.
Expression pattern of Hausp during mouse early embryonic development. The inserted IRES-lacZ allowed the bicistronic expression of β-galactosidase under the control of hausp promoter, which can be detected by staining with X-gal. Embryos from embryonic days E7.5, E8.5, E9.5 and E10.5 were collected and stained with X-gal. As indicated by blue color, Hausp was shown expressed ubiquitously during early embryonic development. Wild-type embryos were used as negative controls for staining.
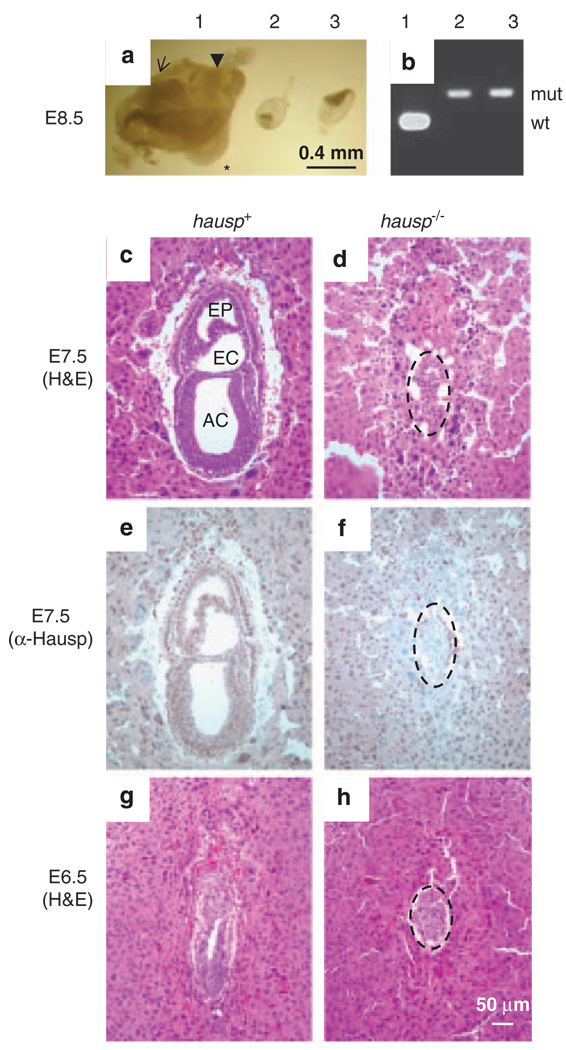
Figure 3.
Phenotypes of hausp knockout embryos. (a) Embryos at day E8.5 from the hausp heterozygote intercross are shown. Embryo no. 1 shows head fold (arrow), somites (*) and tail (arrow head), which are typically present in wild-type embryos at this stage; embryo nos 2 and 3 were abnormal, showing severely reduced cell mass and no advanced structures. (b) Genotyping results for the embryos showed in panel a, embryo no. 1 was wild-type and embryo nos 2 and 3 were hausp knockout embryos. (c) Deciduae from earlier developmental stages were analyzed by histology and immunostaining. Normal embryos at day E7.5 showed development of cavities, such as amniotic cavity (AC), exocoelom (EC) and ectoplacental cavity (EP). (d) Abnormal embryo at day E7.5 failed to develop any noticeable structures. Embryonic cell numbers were greatly reduced (dotted circle). (e) Genotyping of embryos from early embryonic development was performed by immunostaining using a polyclonal antibody anti-Hausp C-terminus. Embryonic cells from the embryo shown in panel c showed brown nuclear staining, suggesting it was either a wild-type or a hausp heterozygote embryo. (f) Embryonic cells (dotted circle) from panel d showed no Hausp staining, suggesting it was a hausp knockout embryo. The surrounding cells from the deciduae were stained by the Hausp antibody, as they were derived from a hasup heterozygote mother. (g) Wild-type embryos at E6.5 show formation of proamniotic cavity, and (h) hausp knockout embryo at E6.5 is smaller and delayed in development. Dotted circles in panels d, f and h, outline the hausp knockout embryos.
Hausp knockout led to early embryonic lethality
Among more than 200 weaning age offspring from mating of hausp heterozygote mice, no homozygote mice were identified through genotypic analysis. The ratio between the resulting wild-type mice to heterozygote mice was approximately 1:2, suggesting that the hausp homozygote mice were embryonic lethal (Table 1). To determine the timing of the embryonic lethality in hausp knockout mice, embryos from timed pregnancies were analyzed. At day E8.5, although most embryos showed normal structures of head fold and development of somites in the body trunk (Figure 3a, embryo no. 1), approximately one-quarter of the embryos showed severe abnormality in development. Most of these embryos had dramatic reduction in size and contained mainly trophoblastic giant cells, two of which were dissected and appeared to have only the amniotic membrane and limited cells from the embryonic proper (Figure 3a, embryo nos 2 and 3). The PCR genotyping confirmed that these abnormal embryos were hausp homozygote knockouts (Figure 3b). We went on to analyze earlier stage embryos from days E7.5 and E6.5, using deciduae that resulted from mating of hausp heterozygote mice. The deciduae were embedded, sectioned and analyzed together. To genotype the embryos, sections were stained with an anti-Hausp C-terminus polyclonal antibody. Cells from wild-type or hausp heterozygote embryos showed positive staining in the nuclei (Figure 3e). In contrast, cells from hausp homozygote knockout embryos could not be stained because of the C-terminal truncation in Hausp (Figure 3f).
Table 1.
Analysis of offspring and embryos from hausp heterozygote intercross
| Stage |
Genotype |
||
|---|---|---|---|
| +/+ | +/− | −/− | |
| 3.5 dpc | 15 | 21 | 15 |
| 6.5 dpc | 17a | 12b | |
| 7.5 dpc | 18a | 6b | |
| 8.5 dpc | 14 | 20 | 8b |
| Offspring | 88 | 121 | 0 |
Abbreviation: dpc, days post coitum.
Hausp staining positive embryos.
Abnormal structures.
At day E6.5, Hausp-positive embryos showed normal development with the appearance of organized germinal layers and a prominent proamniotic cavity (Figure 3g), whereas hausp homozygote mutant embryos were smaller and had disorganized cells without the formation of a proamniotic cavity (Figure 3h). At day E7.5, the abnormalities for hausp knockout embryo were much more severe. Hausp-positive embryos showed structures of amniotic cavity, exocoelom and ectoplacental cavity, as well as formation of germinal layers of the ectoderm, endoderm and mesoderm (Figure 3c). In contrast, hausp knockout embryos showed no recognizable structures; embryonic cells were disorganized and greatly reduced in numbers. Many of the surviving cells were trophoblastic giant cells with large nuclei (Figure 3d). These results suggested that hausp knockout embryos died between days E6.5 and E7.5 and that Hausp was essential for early embryonic development in mouse.
Hausp knockout embryos showed p53 activation
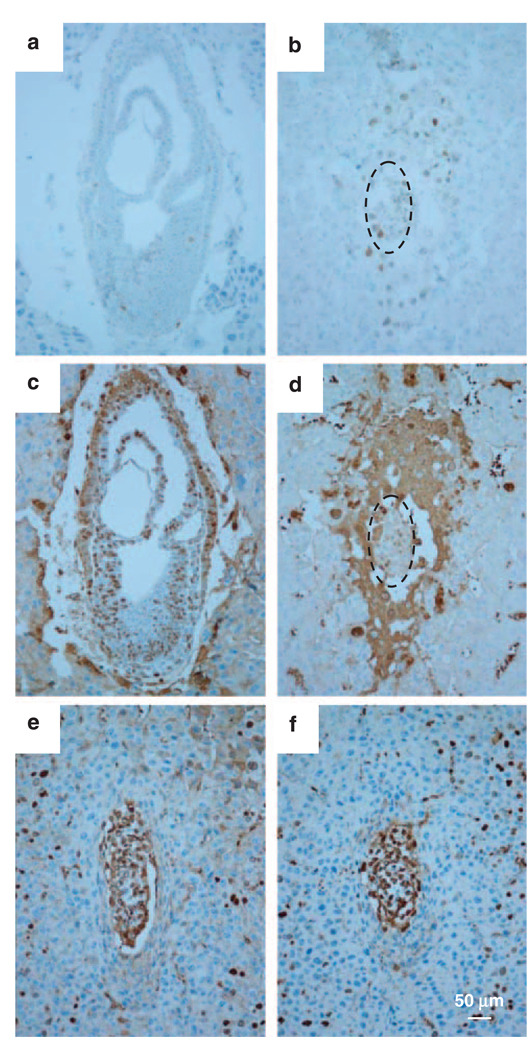
Owing to the potential regulation of the p53–Mdm2 pathway by Hausp, we first investigated the p53 level in hausp knockout embryos by immunostaining using an anti-p53 polyclonal antibody (CM5). The staining conditions were optimized using spleen tissue obtained from irradiated mice of wild-type and p53 knockout mice. Although p53 protein was readily detected in the spleen from wild-type mice after irradiation, no p53 staining was present in the spleen from p53 knockout mice (data not shown). Using the same conditions, a few cells stained positive for p53 in hausp-positive E7.5 embryos (Figure 4a), whereas most of the cells in the hausp knockout E7.5 embryos showed positive p53 staining, suggesting that p53 was stabilized in hausp knockout embryos (Figure 4b).
Figure 4.
Activation of p53 and assay of proliferative activity by BrdU incorporation. Activation of p53 was detected by immunostaining using anti-p53 polyclonal antibody. (a) The wild-type embryos at E7.5 from showed a few p53-positive cells, in contrast, (b) almost all embryonic cells were positive for p53 in hausp knockout embryo. Proliferative activity in embryos was determined by BrdU staining after BrdU incorporation. (c) More than 70% of cells in wild-type embryos at day E7.5 showed BrdU-positive staining, whereas (d) hausp knockout embryos had markedly reduced number of BrdU-positive cells, which was ~35%. However, BrdU-positive cells in wild-type embryo (e) and hausp knockout embryos (f) from day E6.5 were comparable. Dotted circles in panels b and d outline the hausp knockout embryos. BrdU, 5-bromo-2-deoxyuridine.
Hausp knockout resulted in growth arrest
To explore the mechanisms that cause early embryonic lethality in hausp knockout mice, we determined the proliferative activity of the embryos by labeling proliferative cells with 5-bromo-2-deoxyuridine (BrdU). Although the hausp homozygote mutant embryos were smaller than wild-type embryos at day E6.5, the percentage of BrdU-labeled cells was similar for both genotypes (Figures 4e and f), that is, 81.4% for wild-type embryos and 75.4% for hausp knockout embryos, averaged from three individual embryos. In contrast, far fewer cells in the hausp knockout embryos were labeled by BrdU at day E7.5, with 34.9% labeled in the hausp knockout embryos (Figure 4d) compared with 72.9% labeled in the wild-type embryos (Figure 4c), suggesting a dramatic reduction of proliferation in the hausp knockout embryonic cells at E7.5. We concluded that hausp embryonic lethality was due at least in part to deficiency in cell proliferation.
No increase of apoptosis in hausp knockout mice
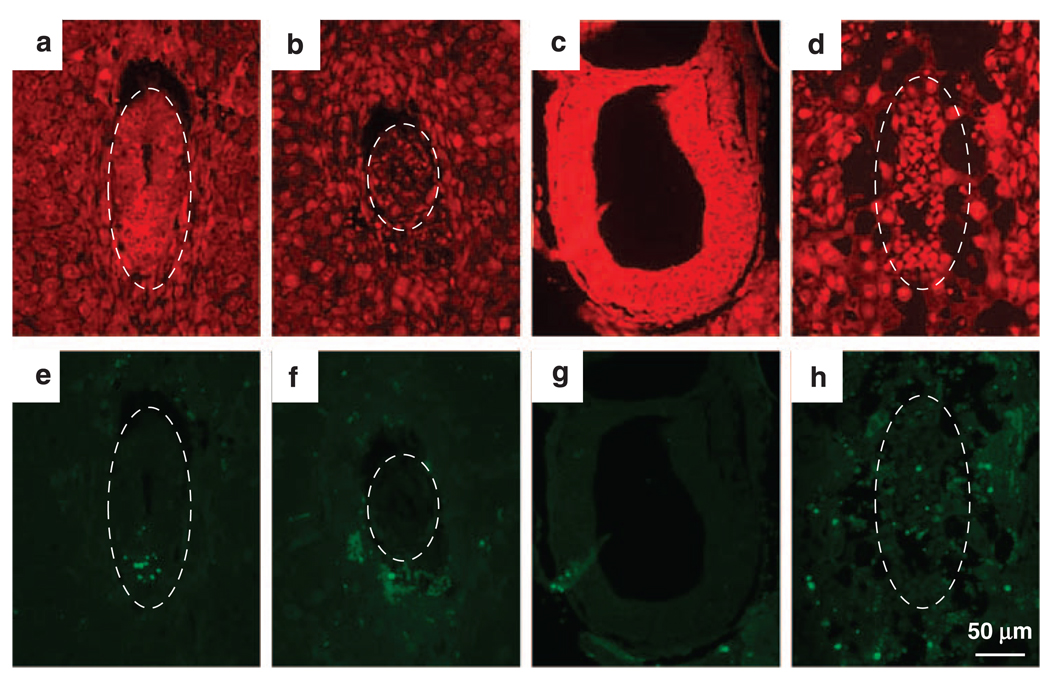
To determine whether there was increased apoptosis due to p53 activation in hausp knockout embryos, a TUNEL (TdT-mediated dUTP Nick-End Labeling) assay was performed on wild-type and hausp knockout embryos of both days E6.5 (Figures 5a – b and e–f) and E7.5 (Figures 5c – d and g–h). As expected, some cells were apoptotic in wild-type embryos, indicated by the presence of green fluorescent nuclei (Figures 5e and g). However, there was no significant increase of apoptotic cells in mutant embryos and most apoptotic cells were trophoblastic giant cells in days E6.5 and E7.5 mutant embryos (Figures 5f and h). Moreover, no increase in apoptosis was detected in these mutant embryos determined by staining for cleaved Caspase3 (data not shown). These data, combined with the results obtained from BrdU-labeling experiments, suggested that growth arrest, but not apoptosis, likely had an important role in causing embryonic lethality in hausp knockout embryos.
Figure 5.
Detection of apoptotic cells by TdT-mediated dUTP Nick-End Labeling (TUNEL) assay. Embryos from hausp heterozygote intercross were fixed and sectioned. The apoptotic cells were detected by TUNEL, followed by fluorescence microscopy (e–h). Nuclei were counter stained red by propidine iodide (a–d). Wild-type embryos are shown in panels a and e from day E6.5 and in panels c and g from day E7.5. Hausp knockout embryos are shown in panels b and f from day E6.5 and in panels d and h from day E7.5. There were minimum numbers of apoptotic cells detected in both wild-type embryos and hausp knockout embryos. Only wild-type embryo at day E6.5 (e) showed some apoptotic cells, which was likely normal during embryonic development. Dotted circles outline the embryos.
Blastocyst of hausp knockout failed to proliferate
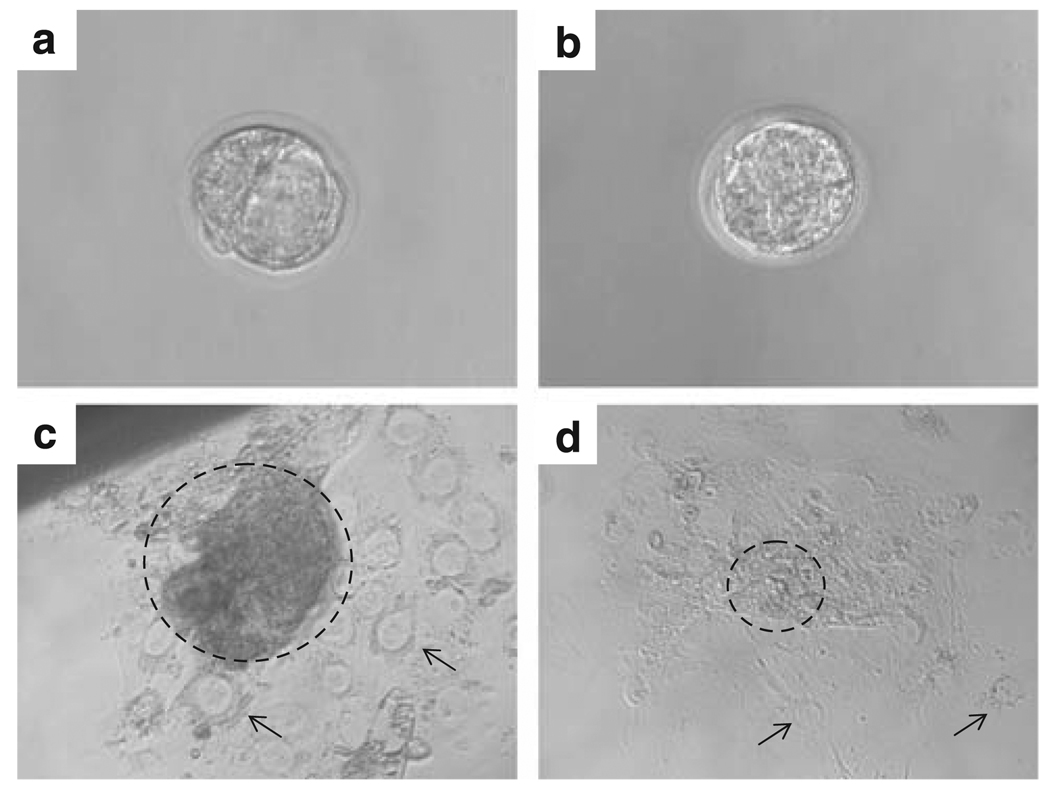
To further show the deficiency of proliferative activity in hausp knockout embryos, blastocysts (day E3.5) were collected and cultured individually in vitro. As shown in Figure 6a, wild-type blastocysts had a well-organized inner cell mass. These embryos showed extensive proliferation in culture, indicated by the expansion of cells from the inner cell mass after being cultured for 5 days (Figure 6c). In contrast, embryonic cells from hausp knockout blastocysts failed to proliferate extensively and the resulting culture showed mostly trophoblastic cells from the mutant embryos (Figure 6d). These data further showed that hausp knockout cells had decreased proliferative activities.
Figure 6.
Proliferative activity assay by blastocyst outgrowth in vitro. Blastocysts, (a) wild-type and (b) hausp knockout embryos, were collected from hausp heterozygote intercross at day E3.5 and cultured individually in vitro for 5 days. (c) Wild-type blastocysts showed prominent outgrowth of the embryonic cells (dotted circle) from the inner cell mass of the blastocyst on day 5. Trophoblastic giant cells are indicated by arrows. (d) Hausp knockout embryo showed virtually no outgrowth of the embryonic cells (dotted circle), the majority of the surviving cells were trophoblastic giant cells (arrows).
Hausp knockout was partially rescued by p53 knockout
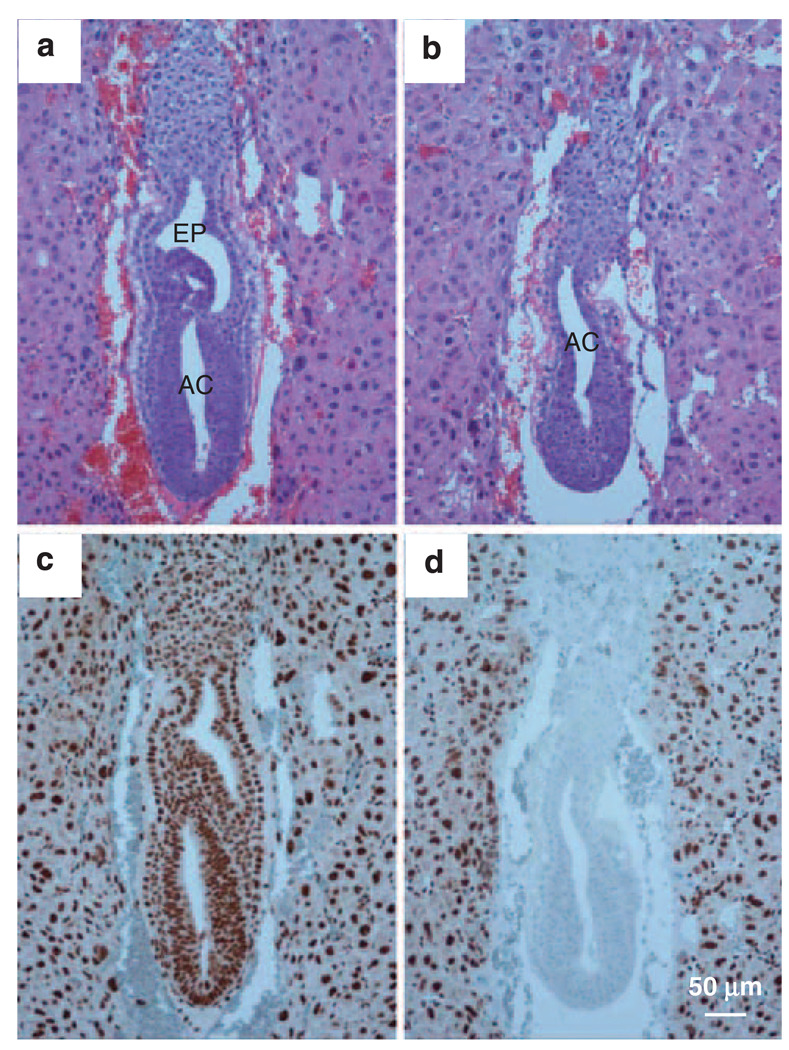
To determine whether the lethality of hausp knockout embryos could be rescued by the deletion of p53, heterozygote mice for both hausp and p53 were intercrossed. Among more than 100 surviving mice, no hausp and p53 double knockout mice were obtained, whereas the ratio of hausp wild-type mice to hausp heterozygote mice was close to 1:2, regardless of the genotype for p53. The results showed that knockout of p53 did not rescue the lethality of the hausp knockout. To determine whether there were any phenotypic changes at the embryonic level due to deletion of p53 in hausp knockout embryos, we analyzed day E7.5 embryos obtained from mating of hausp heterozygote mice, which were also knockouts for p53. The resulting embryos, all knockouts for p53, were stained using anti-Hausp antibody to identify hausp knockout embryos (Figures 7c and d). The hausp and p53 double null embryo (Figure 7b) showed formation of a proamniotic cavity and organized germinal layers at day E7.5, all of which were absent in the hausp-null, p53 wild-type embryos at the same stage (Figure 3f). However, the hausp and p53 double null embryos (Figure 7b) were closer in resemblance to the hausp-positive embryos at day E6.5 (Figure 3g) than those at day E7.5 (Figure 3c and Figure 7a), suggesting that development was delayed because of the loss of p53-independent functions in the double knockout embryos. Nevertheless, developmental improvements of the hausp and p53 double knockout embryos suggested that deletion of p53 partially rescued the phenotype in hausp knockout embryos.
Figure 7.
Deletion of p53 extended the embryonic development in hausp knockout embryo. Embryos from the intercross of hausp heterozygote, p53 homozygote mice were collected at day E7.5. The genotype of embryos shown in (a) and (b) was determined by immunostaining using anti-Hausp C-terminus antibody shown in (c) and (d), respectively. The hausp-positive, p53 knockout embryos (panel a) showed normal development, such as formation of amniotic cavity (AC) and ectoplacental cavity (EP). Similarly, the hausp and p53 double knockout embryo (b) also showed formation of amniotic cavity. However, the development of ectoplacental cavity was missing or delayed.
Mouse embryonic fibroblast cells from hausp heterozygote mice showed reduced half-lives of Mdm2 and p53, and p53 activation was reduced after DNA damage
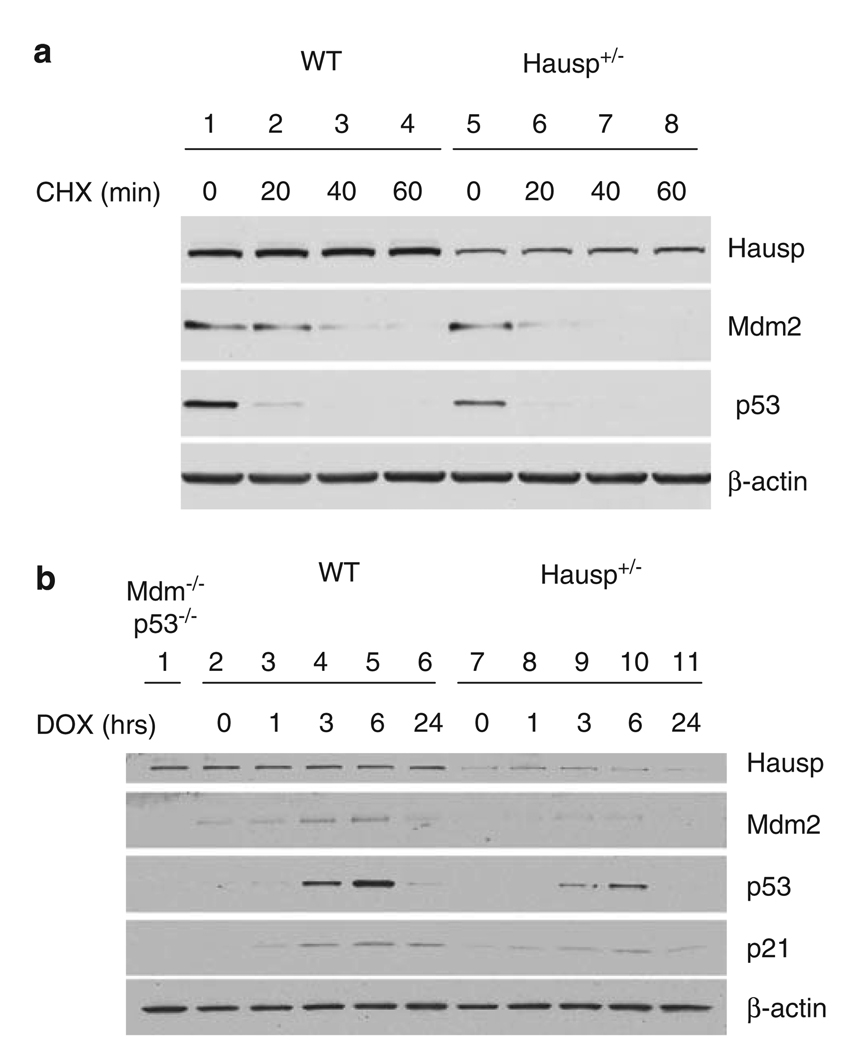
To provide a detailed analysis of regulation of the p53–Mdm2 pathway by Hausp, hausp heterozygote MEF cells were generated. As previously shown, a modest reduction in Hausp by RNA interference decreased p53 stability (Li et al., 2004). Therefore, we expected that hausp heterozygote MEF cells may replicate some of that effect, particularly when p53 stabilization is required after DNA damage. First, the protein half-lives of Mdm2 and p53 were determined. Protein extracts were prepared after incubation in cycloheximide to inhibit translation and analyzed by western blot. As shown in Figure 8a, both p53 and Mdm2 had decreased half-lives in hausp heterozygote MEF cells (lanes 5–8) compared with that in wild-type MEF cells (lanes 1–4). Hausp appeared relatively stable during the time course. Second, the effect of a reduction in Hausp protein levels on p53 stabilization and Mdm2 protein levels was studied after DNA damage by treating MEF cells with doxorubicin. Before addition of doxorubicin, p53 was barely detectable in both wild-type MEF cells (Figure 8b, lane 2) and hausp heterozygote MEF cells (Figure 8b, lane 6). Upon addition of doxorubicin, p53 was stabilized as early as 1 h (Figure 8b, lane 3) and continued to accumulate with a maximum level reached at ~6h (Figure 8b, lane 5) in wild-type MEF cells. In contrast, although p53 in hausp heterozygote MEF cells was stabilized and accumulated following a similar time course in the presence of doxorubicin, the extent of p53 stabilization was significantly less at the same time point when comparing lanes 8–10 with lanes 3–5 (Figure 8b). Moreover, mdm2 and p21, the transcriptional targets of p53, were upregulated to a lesser extent in hausp heterozygote MEF cells than in hausp wild-type MEF cells, because of decreased stabilization of p53. No significant change was observed for the Hausp protein. These data suggested that the reduction of Hausp by 50% resulted in a decrease in p53 stability at steady-state condition and after DNA damage.
Figure 8.
Stability of Mdm2 and p53 in hausp heterozygote MEF cells. (a) Comparison of the protein half-lives of p53 and mdm2 in MEF cells. Total cell extracts were made from wild-type MEF cells (lanes 1–4) and hausp heterozygote MEF cells (lanes 5–8) after cells were incubated for 0 (lanes 1 and 5), 20 (lanes 2 and 6), 40 (lanes 3 and 7) and 60 min (lanes 4 and 8) in the presence of 10 µg/ml cycloheximide. The protein levels of Hausp, p53 and Mdm2, were determined by western blot and sensitive enhanced chemiluminesence (ECL). The level of β-actin was used as internal control for total protein. (b) Reduced activation of p53 in hausp heterozygote MEF cells after DNA damage. Total cell extracts were made from wild-type MEF cells (lanes 2–6) and hausp heterozygote MEF cells (lanes 7–11) after cells were incubated for 1 h (lanes 3 and 8), 3 h (lanes 4 and 9), 6 h (lanes 6 and 10) and 24 h (lanes 7 and 11) in the presence of 0.2 µg/ml Doxorubicin, respectively. Cell extracts from wild-type and hausp heterozygote MEF cells without doxorubicin treatment were loaded in lanes 2 and 7, respectively. Cell extracts from p53 and mdm2 double knockout MEF cells were used to facilitate identification of p53 and Mdm2 (lane 1). The protein levels of Hausp, p53, Mdm2 and p53 downstream target p21 were determined by western blot and regular ECL. The level of β-actin was used as internal control for total proteins. MEF, mouse embryonic fibroblast.
Discussion
There are less than 100 DUBs identified on the basis of sequence homology in the human genome (Nijman et al., 2005), and their unique functions have gradually come to light as more research focuses on regulations involving ubiquitination. Although studies in yeast have suggested that many of the DUBs are redundant (Amerik et al., 2000), deletion of DUBs has proven their necessity in higher eukaryotic organisms. Recently, knockout of usp8 in mice resulted in embryonic lethality because of its essential role in receptor tyrosine kinase stability and endocytic trafficking (Niendorf et al., 2007). Again, in this study, we showed that deletion of hausp (usp7) in mice resulted in early embryonic lethality. Hausp-null embryos died between days E6.5 and E7.5 post coitum, suggesting that fundamental processes were affected during early embryonic development in hausp knockout mice. These results were not surprising, as Hausp has been implicated in regulating many proteins with important cellular functions.
On the basis of our previous findings, we focused our analysis of hausp knockout mice on the regulation of p53 and on the subsequent effects on cell growth and apoptosis. Consistently, p53 was found to be stabilized in hausp knockout embryos, suggesting that activation of p53 was a potential cause of embryonic lethality in hausp knockout embryos. Our data showed that the hausp-null embryos had a drastic decrease in cell-proliferative activity, as indicated by reduced BrdU incorporation. Decrease in proliferation likely contributed to the reduction in embryo size and the delay in differentiation in hausp knockout mice. As activation of p53 is known to induce apoptosis, TUNEL assay was performed and showed no increase of apoptosis in hausp knockout embryos. These results suggested that cell growth arrest, but not apoptosis, likely caused embryonic lethality. As previously reported, the level of p53 protein dictates the extent of cellular response; low levels of p53 lead to cell growth arrest, whereas higher levels of p53 lead to apoptosis (Chen et al., 1996). It is possible that the stabilization of p53 in hausp knockout mice may not be high enough to trigger an apoptotic response but is high enough to cause growth arrest. Owing to the observation of p53 activation in hausp knockout embryos, we attempted to rescue the lethality of hausp knockout mice by generating hausp and p53 double knockout mice. Although no live hausp and p53 double knockout mice were observed, the development of hausp and p53 double knockout embryos was extended when compared with hausp knockout embryos in a p53 wild-type background. These data suggest that Hausp has p53-independent functions that are indispensable for proliferation and differentiation. One of those functions may be related to the regulation of histone ubiquitination and to that of gene expression, which potentially affects a wide range of functions in proliferation and differentiation. More recently, Hausp was found to associate with important regulatory proteins through a systematic proteomic analysis (Sowa et al., 2009). Among those identified were DNMT1, RAE1, Bub3 and Nup98, the stability and functions of which are potentially regulated by Hausp. In particular, loss of DNMT1 resulted in a gross reduction of DNA methylation and led to early embryonic lethality at day E9.5 (Li et al., 1992). Similarly, RAE1 and Bub3 cooperatively function to ensure faithful chromosomal segregation during mitosis. Rae1 knockout embryos failed to develop during early embryogenesis, and Rae1/ Bubs double heterozygote MEF cells had accelerated genomic instability (Babu et al., 2003). Knockout of nup98 in mice resulted in disorganized embryonic development at day E8.5 and failure of gastrulation (Wu et al., 2001). Crippling of these critical functions in hausp knockout mice may cause embryonic lethality after deletion of p53.
To further evaluate the significance of p53 regulation by Hausp, hausp heterozygote MEF cells were used to test the effect of reducing Hausp on p53 stabilization and Mdm2 levels, at steady-state condition and after DNA damage. Consistent with a previous study (Li et al., 2004), the protein half-lives of Mdm2 and p53 were reduced in hausp heterozygote MEF cells. Moreover, p53 activation in response to DNA damage was also reduced, which was potentially resulted from reduced Hausp protein in hausp heterozygote MEF cells. Mdm2 was also reduced after DNA damage, which could be due to a more complicated mechanism, including regulations in transcription and protein stabilization. In summary, these results again support the critical role of Hausp in regulation of the p53–Mdm2 pathway. Conversely, activation of p53 in hausp knockout cells potentially leads to reduction in tumor formation. This is supported by the observation that no hausp mutations were identified when surveying mutations in non-small-cell lung carcinomas, suggesting that complete loss of Hausp function is unfavorable for tumor growth (Mori et al., 2004). Thus, it is perceivable that Hausp inhibitors can be effective in eliminating cancer cells carrying wild-type p53. Finally, it has been shown that both upregulation and downregulation of Hausp could increase radiation sensitivity of xenografted tumor cells in mouse with concomitant p53 stabilization under both conditions (Becker et al., 2008). These findings again point to the critical role of Hausp in the regulation of the p53–Mdm2 pathway and implicated novel strategies in cancer therapy through modulating Hausp activity.
Materials and methods
Gene targeting and generation of Hausp mutant mice
Targeted disruption of the hausp locus was accomplished by the insertion of an IRES-β-galactosidase (IRES-lac Z) and neomycin resistance gene cassette into exon 14, as well as by the deletion of 29 nucleotides within exon 14, using a targeting vector containing 1.7 kb of 5′-homology and 3.5kb of 3′-homology (Figure 1a). The mutated mouse embryonic stem cells were injected into blastocysts to generate chimeras, from which germline transmission of the mutated allele of the hausp gene was accomplished. The targeting event was confirmed by Southern blot using external probes. PCR assay was designed to genotype hausp knockout mice, using hausp-specific primers 5′-ATGATGACGTGGTATCCAGGTGTAC-3′ and 5′-CCA AGGCAGTGAAGATTCTTTGCAG-3′ in combination with a neo-specific primer 5′-GGGCCAGCTCATTCCTCCCACT CAT-3′ to detect wild-type allele (202 bp) and mutant allele (433 bp) simultaneously. The hausp knockout mice were maintained on a mixed background of 129Sv and C57BL/6J through breeding between littermates. Maintenance and handling protocols were approved by the IACUC (Institutional Animal Care and Use Committee) of Columbia University.
Embryo collection, X-Gal staining and histology
Embryos were collected from timed pregnancy at various stages of gestation for X-gal staining and histological analysis. For X-gal staining, the embryos were fixed for 15min in 0.2% glutaraldehyde, 2% formaldehyde, 5mM EGTA (ethylene glycol tetraacetic acid) and 2mM MgCl2 in 1 × phosphate-buffered saline, and stained for X-Gal (5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside) at 37 °C for 4h. For histological study, deciduae were dissected out and fixed in 4% formaldehyde overnight at 4°C, dehydrated and embedded in paraffin. Serial 5-µm sections were collected and stained by hematoxylin and eosin according to standard procedures.
Immunohistochemistry
For BrdU staining, BrdU, at 100 µg BrdU per gram of body weight, was injected into the peritoneal cavity of pregnant female mice. The deciduae were collected 1h later and sections were prepared according to standard procedures. The sections were immunostained using antibodies of anti-Hausp C terminus at 1.1µg/ml (Li et al., 2002), anti-BrdU (BD Biosciences, San Jose, CA, USA) (1:20 dilution) and anti-p53 CM5 (Leica Microsystems Inc., Bannockburn, IL, USA) (1:5000 dilution), and counter stained with hematoxylin. The TUNEL assay was performed using the DEAD End Fluorometric TUNEL system (Promega, Madison, WI, USA).
Blastocyst outgrowth assay
Blastocysts were collected at day E3.5 and cultured individually in 200 µl of Dulbecco’s modified Eagle’s medium containing 20% fetal bovine serum without leukemia inhibitory factor. The cultures were photographed daily. The genomic DNA was prepared by lysing the cells in 50 µl of buffer containing proteinase K on day 5. After inactivation of proteinase K at 95°C for 10 min, PCR genotyping was performed using 5 µl of DNA samples.
Analysis of changes in the p53–Mdm2 pathway using hausp heterozygote MEF cells
The half-life of p53 and Mdm2 was determined by treating MEF cells with cycloheximide (10 µg/ml) for 0, 20, 40 and 60 min. Total cell extracts were prepared with the radio-immunoprecipitation assay buffer in the presence of dithiothreitol (1 mM) and proteinase inhibitors and analyzed by western blot. To induce DNA damage in cells, MEF cells were treated with doxorubicin (0.2 µg/ml) for 1, 3, 6 and 24 h. Cell extracts were prepared with the radioimmunoprecipitation assay buffer and analyzed by western blot.
Acknowledgements
We thank Qiong Li and Xi Sun for their expert assistance with histology. We also thank Dr Chi-Ming Li for helpful suggestions. This study was supported in part by a postdoctoral fellowship from NIH to NK and grants from NIH/ NCI to TL and WG.
Footnotes
Conflict of interest
The authors declare no conflict of interest.
References
- Amerik AY, Li S, Hochstrasser M. Analysis of the deubiquitinating enzymes of the yeast Saccharomyces cerevisiae. Biol Chem. 2000;381:981–992. doi: 10.1515/BC.2000.121. [DOI] [PubMed] [Google Scholar]
- Babu JR, Jeganathan KB, Baker DJ, Wu X, Kang-Decker N, van Deursen JM. Rae1 is an essential mitotic checkpoint regulator that cooperates with Bub3 to prevent chromosome missegregation. J Cell Biol. 2003;3:341–353. doi: 10.1083/jcb.200211048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker K, Marchenko ND, Palacios G, Moll UM. A role of HAUSP in tumor suppression in a human colon carcinoma xenograft model. Cell Cycle. 2008;7:1205–1213. doi: 10.4161/cc.7.9.5756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Kon N, Li M, Zhang W, Qin J, Gu W. ARF-BP1/ Mule is a critical mediator of the ARF tumor suppressor. Cell. 2005;121:1071–1083. doi: 10.1016/j.cell.2005.03.037. [DOI] [PubMed] [Google Scholar]
- Chen X, Ko LJ, Jayaraman L, Prives C. p53 levels, functional domains, and DNA damage determine the extent of the apoptotic response of tumor cells. Genes Dev. 1996;1:2438–2451. doi: 10.1101/gad.10.19.2438. [DOI] [PubMed] [Google Scholar]
- Cummins JM, Rago C, Kohli M, Kinzler KW, Lengauer C, Vogelstein B. Tumour suppression: disruption of HAUSP gene stabilizes p53. Nature. 2004;428:1. doi: 10.1038/nature02501. [DOI] [PubMed] [Google Scholar]
- Dornan D, Wertz I, Shimizu H, Arnott D, Frantz GD, Dowd P, et al. The ubiquitin ligase COP1 is a critical negative regulator of p53. Nature. 2004;429:86–92. doi: 10.1038/nature02514. [DOI] [PubMed] [Google Scholar]
- Everett RD, Meredith M, Orr A. The ability of herpes simplex virus type 1 immediate-early protein Vmw110 to bind to a ubiquitin-specific protease contributes to its roles in the activation of gene expression and stimulation of virus replication. J Virol. 1999;73:417–426. doi: 10.1128/jvi.73.1.417-426.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- Holowaty MN, Sheng Y, Nguyen T, Arrowsmith C, Frappier L. Protein interaction domains of the ubiquitin-specific protease, USP7/HAUSP. J Biol Chem. 2003;278:47753–47761. doi: 10.1074/jbc.M307200200. [DOI] [PubMed] [Google Scholar]
- Hong S, Kim SJ, Ka S, Choi I, Kang S. USP7, a ubiquitin-specific protease, interacts with ataxin-1, the SCA1 gene product. Mol Cell Neurosci. 2002;20:298–306. doi: 10.1006/mcne.2002.1103. [DOI] [PubMed] [Google Scholar]
- Hu M, Li P, Li M, Li W, Yao T, Wu JW, et al. Crystal structure of a UBP-family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde. Cell. 2002;111:1041–1054. doi: 10.1016/s0092-8674(02)01199-6. [DOI] [PubMed] [Google Scholar]
- Hu M, Gu L, Li M, Jeffrey PD, Gu W, Shi Y. Structural basis of competitive recognition of p53 and MDM2 by HAUSP/USP7: implications for the regulation of the p53-MDM2 pathway. PLoS Biol. 2006;4:e27. doi: 10.1371/journal.pbio.0040027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones SN, Roe AE, Donehower LA, Bradley A. Rescue of embryonic lethality in Mdm2-deficient mice by absence of p53. Nature. 1995;378:206–208. doi: 10.1038/378206a0. [DOI] [PubMed] [Google Scholar]
- Laney JD, Hochstrasser M. Substrate targeting in the ubiquitin system. Cell. 1999;14:427–430. doi: 10.1016/s0092-8674(00)80752-7. [DOI] [PubMed] [Google Scholar]
- Leng RP, Lin Y, Ma W, Wu H, Lemmers B, Chung S, et al. Pirh2, a p53-induced ubiquitin-protein ligase, promotes p53 degradation. Cell. 2003;112:779–791. doi: 10.1016/s0092-8674(03)00193-4. [DOI] [PubMed] [Google Scholar]
- Li E, Bestor TH, Jaenisch R. Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell. 1992;69:915–926. doi: 10.1016/0092-8674(92)90611-f. [DOI] [PubMed] [Google Scholar]
- Li M, Chen D, Shiloh A, Luo J, Nikolaev AY, Qin J, et al. Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature. 2002;416:648–653. doi: 10.1038/nature737. [DOI] [PubMed] [Google Scholar]
- Li M, Brooks CL, Kon N, Gu W. A dynamic role of HAUSP in the p53-Mdm2 pathway. Mol Cell. 2004;13:879–886. doi: 10.1016/s1097-2765(04)00157-1. [DOI] [PubMed] [Google Scholar]
- Meulmeester E, Maurice MM, Boutell C, Teunisse AF, Ovaa H, Abraham TE, et al. Loss of HAUSP-mediated deubiquitination contributes to DNA damage-induced destabilization of Hdmx and Hdm2. Mol Cell. 2005;18:565–576. doi: 10.1016/j.molcel.2005.04.024. [DOI] [PubMed] [Google Scholar]
- Montes de Oca Luna R, Wagner DS, Lozano G. Rescue of early embryonic lethality in mdm2-deficient mice by deletion of p53. Nature. 1995;378:203–206. doi: 10.1038/378203a0. [DOI] [PubMed] [Google Scholar]
- Mori S, Ito G, Usami N, Yoshioka H, Ueda Y, Kodama Y, et al. p53 apoptotic pathway molecules are frequently and simultaneously altered in nonsmall cell lung carcinoma. Cancer. 2004;100:1673–1682. doi: 10.1002/cncr.20164. [DOI] [PubMed] [Google Scholar]
- Niendorf S, Oksche A, Kisser A, Löhler J, Prinz M, Schorle H, et al. Essential role of ubiquitin-specific protease 8 for receptor tyrosine kinase stability and endocytic trafficking in vivo. Mol Cell Biol. 2007;27:5029–5039. doi: 10.1128/MCB.01566-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nijman SM, Luna-Vargas MP, Velds A, Brummelkamp TR, Dirac AM, Sixma TK, et al. A genomic and functional inventory of deubiquitinating enzymes. Cell. 2005;123:773–786. doi: 10.1016/j.cell.2005.11.007. [DOI] [PubMed] [Google Scholar]
- Pickart CM, Eddins MJ. Ubiquitin: structures, functions, mechanisms. Biochim Biophys Acta. 2004;29:55–72. doi: 10.1016/j.bbamcr.2004.09.019. [DOI] [PubMed] [Google Scholar]
- Sherr CJ, Webber JD. The ARF/p53 pathway. Curr Opin Genet Dev. 2000;10:94–99. doi: 10.1016/s0959-437x(99)00038-6. [DOI] [PubMed] [Google Scholar]
- Sowa ME, Bennett EJ, Gygi SP, Harper JW. Defining the human deubiquitinating enzyme interaction landscape. Cell. 2009;23:389–403. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Qu LK, Zhang J, Wang W, Michaelson JS, Degenhardt YY, et al. Critical role for Daxx in regulating Mdm2. Nat Cell Biol. 2006;8:855–862. doi: 10.1038/ncb1442. [DOI] [PubMed] [Google Scholar]
- van der Horst A, de Vries-Smits AM, Brenkman AB, van Triest MH, van den Broek N, Colland F, et al. FOXO4 transcriptional activity is regulated by monoubiquitination and USP7/HAUSP. Nat Cell Biol. 2006;8:1064–1073. doi: 10.1038/ncb1469. [DOI] [PubMed] [Google Scholar]
- van der Knaap JA, Kumar BR, Moshkin YM, Langenberg K, Krijgsveld J, Heck AJ, et al. GMP synthetase stimulates histone H2B deubiquitylation by the epigenetic silencer USP7. Mol Cell. 2005;17:695–707. doi: 10.1016/j.molcel.2005.02.013. [DOI] [PubMed] [Google Scholar]
- Wu X, Kasper LH, Mantcheva RT, Mantchev GT, Springett MJ, van Deursen JM. Disruption of the FG nucleoporin NUP98 causes selective changes in nuclear pore complex stoichiometry and function. Proc Natl Acad Sci USA. 2001;13:3191–3196. doi: 10.1073/pnas.051631598. [DOI] [PMC free article] [PubMed] [Google Scholar]